Abstract

Provided herein are novel compounds as SMARCA degraders, pharmaceutical compositions, use of such compounds in treating cancer, and processes for preparing such compounds.
Important Compound Classes

Title
SMARCA Degraders and Uses Thereof
Patent Publication Number
WO 2022/125804 A1
Publication Date
June 16, 2022
Priority Application
US 63/123,427
Priority Date
December 9, 2020
Inventors
Zhang, Y.; Zheng, X.; Zhu, X.
Assignee Company
Kymera Therapeutics Inc., USA
Disease Area
Cancer
Biological Target
SMARCA
Summary
The field of targeted protein degradation promoted by small molecules has been intensively studied. The ubiquitin-proteasome pathway (UPP) is a critical pathway that regulates key regulator proteins and degrades misfolded or abnormal proteins. UPP is central to multiple cellular processes and if defective or imbalanced, it leads to pathogenesis of a variety of diseases. The covalent attachment of ubiquitin to specific protein substrates is achieved through the action of E3 ubiquitin ligases.
UPP plays a key role in the degradation of short-lived and regulatory proteins important in a variety of basic cellular processes, including regulation of the cell cycle, modulation of cell cycle receptors and ion channels, and antigen presentation. The UPP is used to induce selective protein degradation, including use of fusion proteins to artificially ubiquitinate target proteins and small molecule probes to induce proteasome-dependent degradation.
The present application describes a series of novel compounds as SMARCA (Switch/Sucrose Non-Fermentable (SWI/SNF)-Related, Matrix-Associated, Actin-Dependent Regulator of Chromatin, Subfamily A) degraders for the treatment of cancer. Further, the application discloses compounds, their preparation, use, and pharmaceutical composition, and treatment.
Definitions
SMARCA is a protein binding moiety capable of binding to one or more of SMARCA2, SMARCA4, and PB1;
L is a bivalent moiety that connects SMARCA to DIM; and
DIM is a degradation inducing moiety.
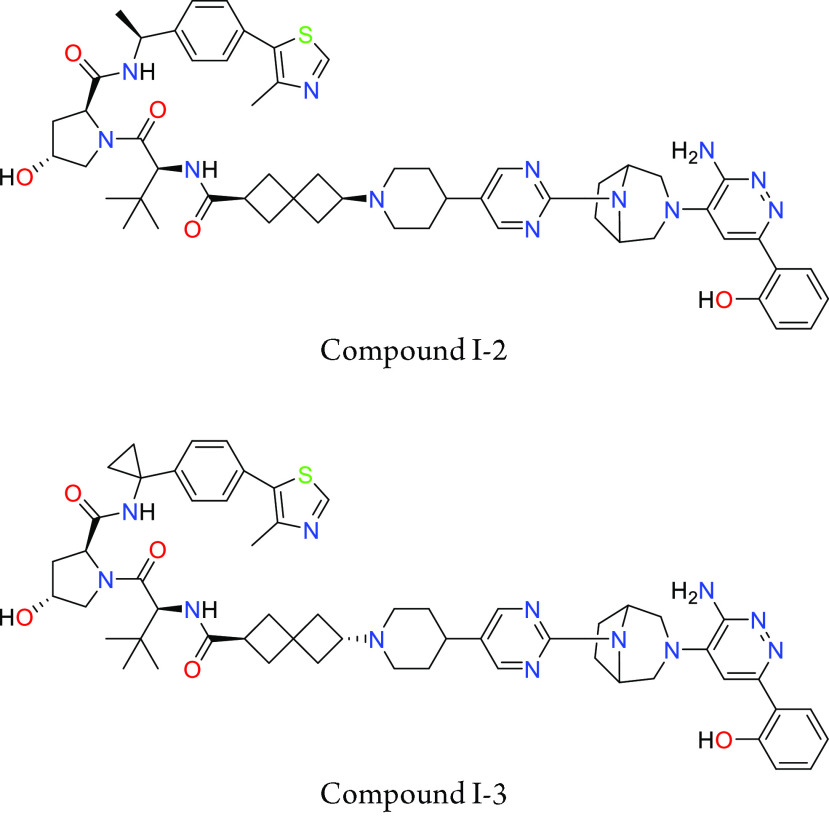
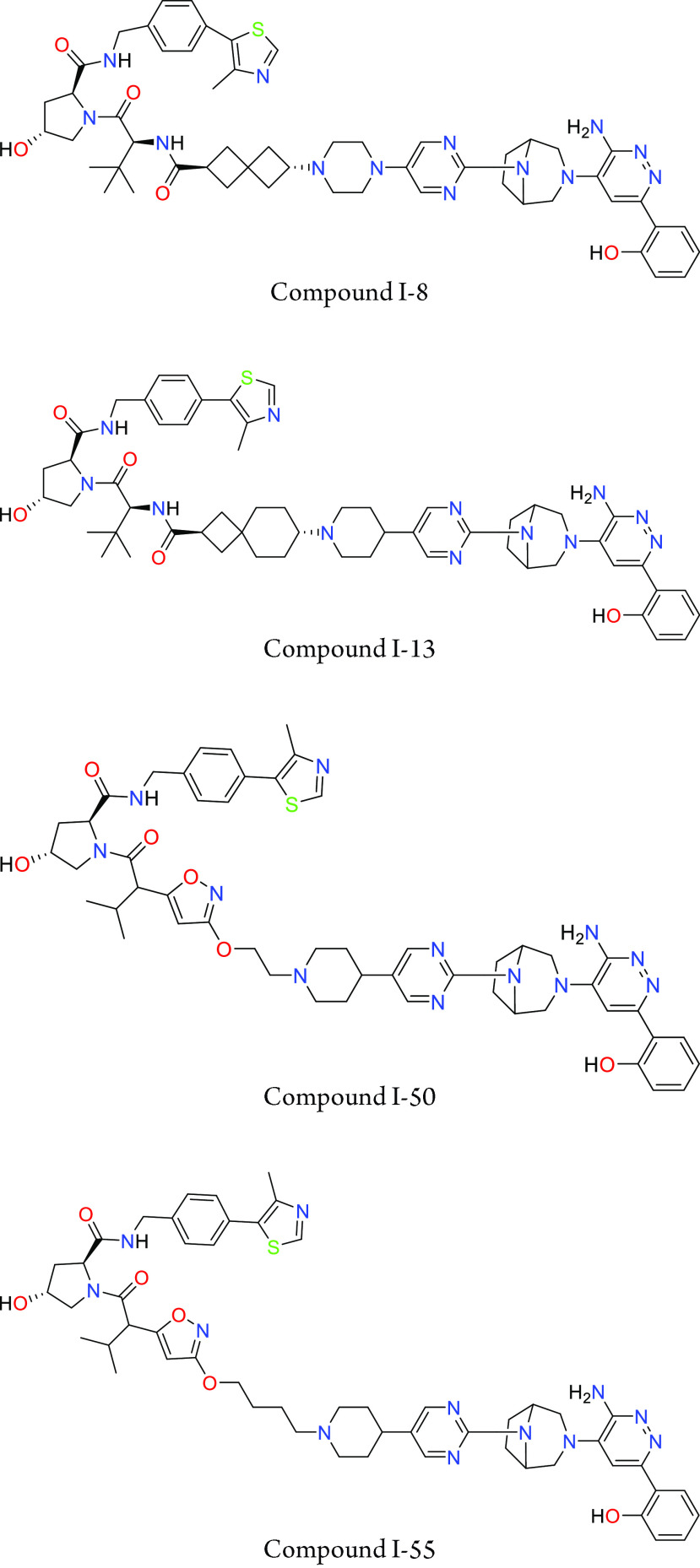
Key Structures
Biological Assay
The MSD SMARCA2 degradation in A549 cell line assay was performed. The compounds described in this application were tested for their ability to degrade SMARCA2. The SMARCA2 DC50 (nM) are shown in the table below.
Biological Data
The following table shows representative compounds tested for SMARCA2 degradation and the biological data obtained from testing representative examples.
For DC50, “A” means <100 nM,
“B” means 100–500 nM, “C” means
501–1000 nM, and “D” means >1000
nM.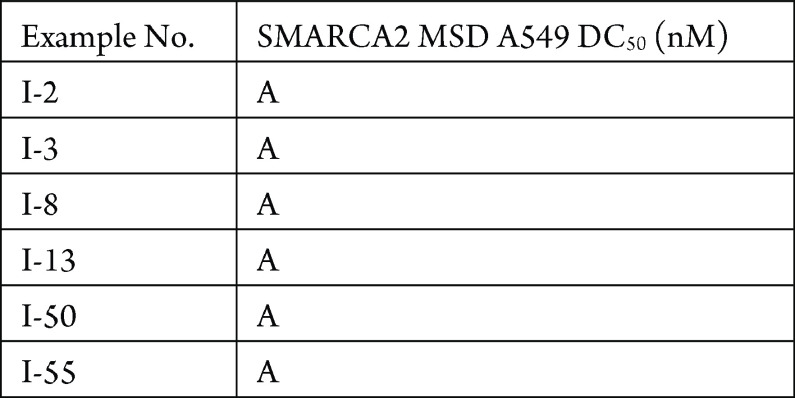
Claims
Total claims: 30
Compound claims: 23
Pharmaceutical composition claims: 2
Method of treatment claims: 4
Method of degradation claims: 1
The author declares no competing financial interest.
Special Issue
Published as part of the ACS Medicinal Chemistry Letters virtual special issue “New Drug Modalities in Medicinal Chemistry, Pharmacology, and Translational Science”.
Recent Review Articles. References
- Webb T.; Craigon C.; Ciulli A. Targeting epigenetic modulators using PROTAC degraders: Current status and future perspective. Bioorg. Med. Chem. Lett. 2022, 63, 128653. 10.1016/j.bmcl.2022.128653. [DOI] [PubMed] [Google Scholar]
- Lv M.; Hu W.; Zhang S.; He L.; Hu C.; Yang S. Proteolysis-targeting chimeras: A promising technique in cancer therapy for gaining insights into tumor development. Cancer Lett. 2022, 539, 215716. 10.1016/j.canlet.2022.215716. [DOI] [PubMed] [Google Scholar]
- Xi J.; Zhang R.; Chen K.; Yao L.; Li M.; Jiang R.; Li X.; Fan L. Advances and perspectives of proteolysis targeting chimeras (PROTACs) in drug discovery. Bioorg. Chem. 2022, 125, 105848. 10.1016/j.bioorg.2022.105848. [DOI] [PubMed] [Google Scholar]
- Cresser-Brown J. O.; Marsh G. P.; Maple H. J. Reviewing the toolbox for degrader development in oncology. Curr. Opin. Pharmacol. 2021, 59, 43. 10.1016/j.coph.2021.04.009. [DOI] [PubMed] [Google Scholar]
- Coughlan A. Y.; Testa G. Exploiting epigenetic dependencies in ovarian cancer therapy. Int. J. Cancer 2021, 149, 1732. 10.1002/ijc.33727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnaby W.; Koegl M.; McConnell D. B.; Ciulli A. Transforming targeted cancer therapy with PROTACs: A forward-looking perspective. Curr. Opin. Pharmacol. 2021, 57, 175. 10.1016/j.coph.2021.02.009. [DOI] [PubMed] [Google Scholar]