Abstract
Water suppression is of paramount importance for many biological and analytical NMR spectroscopy applications. Here, we report the design of a new set of binomial-like radio frequency (RF) pulses that elude water irradiation while exciting or refocusing the remainder of the 1H spectrum. These pulses were generated using a combination of an evolutionary algorithm and artificial intelligence. They display higher sensitivity relative to classical water suppression schemes and tunable water selectivity to avoid suppressing 1H resonances near the water signal. The broad bandwidth excitation obtained with these RF pulses makes them suitable for several NMR applications at high and ultra-high-field magnetic fields.
Solvent suppression is indispensable for 1H-detected biomolecular NMR experiments as the H2O signal is beyond the dynamic range of NMR receivers.1 At high and ultra-high magnetic fields, the intense water signal generates an additional field (radiation damping), affecting the apparent nuclear spin relaxation of the solute resonances.2 The first water suppression scheme was developed by Bax and co-workers, consisting of selective irradiation (or presaturation) of the water signal using a continuous radiofrequency (RF) field.3 Though presaturation is highly selective and easy to implement into pulse schemes, it attenuates the signals of rapidly exchanging protons and often creates unwanted spin diffusion effects and/or nuclear Overhauser effects.4 More elaborate pulse schemes have been implemented in the past decades, thanks to the parallel development of magnetic field gradients and shaped RF pulses.4,5–11 These advanced techniques mitigate signal losses by aligning the water polarization along the z-axis and performing water suppression before signal acquisition. Among the most effective methods is the Water-suppression by GrAdient-Tailored Excitation or WATERGATE12 WATERGATE utilizes two selective π/2 pulses and a broadband π pulse resulting in an effective identity operation performed on the water signal, which is dephased by two flanking field gradients applied along the z-axis. The efficiency of water irradiation depends on the selectivity of the two pulses, whose excitation profiles often cause signal reductions in the range of 5–20%. WATERGATE sequences of short duration were also implemented using binomial-like sequences such as 3-9-19 and W5.13, 14 With the advent of high and ultra-high magnetic fields; however, the selective irradiation of the water is becoming a challenge, even for binomial sequences due to the increased bandwidths. An ideal solution is to avoid water excitation throughout an NMR pulse sequence, leaving the water magnetization along the z-axis. A remarkable example is a family of Band-selective Excitation Short-Transient (BEST) pulse sequences,15, 16 which offers excellent water suppression and the benefit of fast repetition rates speeding up multidimensional NMR spectra acquisition. The BEST sequences utilize selective pulses for amide excitation and refocusing, and have been very successful for intrinsically disordered proteins that display narrow 1H chemical shift breadth.17 However, due to the limited 1H bandwidth of the selective pulses, these experiments have had sparse applications for folded biomolecules with broader 1H chemical shift dispersion.
Here, we used a new software, GENErator of TrIply Compensated pulSes via Artificial Intelligence or GENETICS-AI,18 to design high-fidelity RF pulses that completely elude the excitation of the water signal. These Water irrAdiation DEvoid (WADE) pulses create an identity operation for the on-resonance signals and refocus both sides of the 1H spectrum. As with all binomial-like sequences, including jump-return,1, 19 3-9-19,20 W5,14 the WADE pulses are symmetric and have a constant amplitude with a 180° phase shift (π -shifted) symmetry in the phase shape profile. For the on-resonance spins, the π-shifted symmetry on the right half of the phase shape cancels the effects of the RF pulse on the left half, resulting in an identity operation (I). To illustrate this concept, let’s consider a 10-point RF shape with π-shifted symmetry (Fig. S1A, ESI†). The evolution operators for the pulse segments in the center of the phase shape (U5 and U6) are:
which can be rewritten as
Multiplying the two operators, we obtain an identity operation:
The latter relationship is also valid for the following pairs: {U4, U7}, {U3, U8}, {U2, U9}, and {U1, U10}. However, this relationship does not hold for the off-resonance spins since they are under chemical shift evolution. Therefore, we created a universal π-pulse using numerical optimization for an effective off-resonance refocusing operation. Since spatial RF inhomogeneity does not affect the symmetry of the RF shape, the relation for on-resonance spins holds for any RF inhomogeneity. A prototype of the WADE-π pulse is shown in Fig. S1B (ESI†). This specific pulse performs an identity operation on-resonance and refocuses the spins on both sides of the bandwidth. We compared the RF offset dependence of the WADE-π pulse with the 3-9-19 sequence, which is typically used to suppress the water signal for triple-resonance NMR experiments. (Fig. S1C, ESI†). Specifically, we assess the pulse responses to the RF offset. From the simulated trajectories, it is apparent that the WADE-π pulse does not affect the Mx component of the magnetization, while the 3-9-19 sequence shows periodic excitation bands throughout the bandwidth (~ 30 kHz). The My component is fully inverted by the WADE-π pulse, with no off-resonances effects over the amide and aliphatic regions of the spectrum. In contrast, the 3-9-19 sequence displays a periodic modulation of My within the bandwidth considered. This analysis reveals that the WADE-π pulse suppresses the water signal with high efficiency and outperforms the 3-9-19 sequence for broader bandwidths. which occur at high- and ultra-high magnetic fields. An example of WADE- π pulse for the commercially available 1.2 GHz magnets is reported in Fig. S2, ESI†. Importantly, the WADE-π pulse amplitude is tunable, making it possible to match the desired selectivity for on-resonance solvent signals (Fig. S4, ESI†). Suppose the water signal is particularly broad or narrow. In that case, the user can modulate the WADE-π amplitude to achieve the desired water suppression, minimizing the effects of resonances near the water signal.
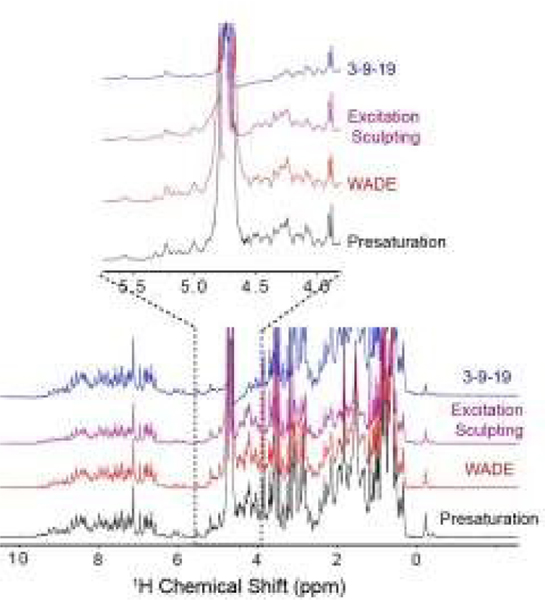
To demonstrate the performance of the WADE-π pulse, we acquired a 1D 1H spectrum of U-15N labeled K48C mutant of ubiquitin in H2O/D2O (90/10%) and compared it with the most commonly used water suppression schemes, i.e., presaturation, 3-9-19, and excitation sculpting9 (Fig. 1). While all the sequences suppress the water signal with similar efficiency, the WADE pulse has minimal effects on the 1H resonances near the water signal (~0.3 ppm, Fig. S3, ESI†). The suppression of the resonances near the water can be further minimized by adjusting the WADE-π pulse amplitude (Fig. S4, S2D, ESI†). Although it is possible to change the selectivity of the water suppression in the excitation sculpting sequence by tuning the pulse amplitude, the length of this sequence (two selective pulses and four gradients) may cause a reduction of the sensitivity due to transverse relaxation, particularly for large biomacromolecules. A direct comparison of the WADE-π pulse with other standard binomial-like pulses such as 3-9-19,20 W5,14 jump-and-return sandwich pulse,21 PM1, and PM222 is shown in Fig. S5 and S6 (ESI†). Increasing the pulse width or lowering the RF amplitude reduces the operational fidelity and bandwidth of the commonly used binomial-like sequences (Fig. S5, ESI†). On the contrary, the operational fidelity of the WADE pulses, which are designed considering the effects of the finite pulse duration in the optimization procedure, is not compromised (Fig. S6, ESI†). A comparison of the water suppression pulses expressed as bandwidths vs. duration and fidelity level is reported in Fig. S8 (ESI†). The 3-9-19 and the W5 sequences have the shortest duration, though. W5 excites a broader bandwidth with a higher fidelity level. PM1 and PM2 sequences, on the other hand, have an excitation bandwidth comparable to W5 but with a significantly longer duration. A similar duration is observed for the jump-and-return sandwich pulse, which excites approximately 10 ppm with a higher fidelity level. However, the duration of these sequences is significantly longer than the 3-9-19 or W5. We also generated three WADE-π pulses with the same duration but different RF amplitudes, i.e., WR1, WR2, and WR3 (Fig. S6, ESI†). The WR1 pulse (B1=6.25 kHz and duration 3 ms) has a slightly broader excitation profile with a significantly lower pulse duration relative to the W5 and 3-9-19 sequences. Similar behavior is observed for WR2 (B1=8.33 kHz) and WR3 (B1=11.31 kHz), with the latter able to excite up to 50 ppm. Notably, the evolution of J coupling during WADE-π pulse is negligible for both amide and aliphatic regions. To demonstrate this, we simulated the J coupling response of the WR1 pulse for J values up to 200 Hz and is shown in Fig. S7, ESI†. Compared to J evolution for equivalent delay, the WADE-π pulse on single spin or both spins suppress the coupling evolution. Taken altogether, the WADE pulses are more suitable for ultra-highfield NMR spectroscopy than the commonly used solvent suppression sequences.
Figure 1.
1D 1H spectra of U-15N K48C ubiquitin recorded on a 850 MHz spectrometer at 300 K. The 1D spectrum with 3-9-19 water suppression (blue) was recorded using the p3919gp pulse program with amplitude of 16.67 kHz and inter-pulse delay 200 μs for a total length of 3.2 ms. The 1D spectrum with the excitation sculpting (purple) was recorded using the zgesgp pulse program with 5 ms selective pulses for a total length of 15 ms. The 1D spectrum with the WADE-π pulse (red) was acquired with an RF amplitude of 1.25 kHz. A recycle delay of 1 s was used for all the experiments. All the spectra are plotted with identical noise level. The pulse sequences for all the 1D experiments are shown in Figs. S2B and S11, (ESI†).
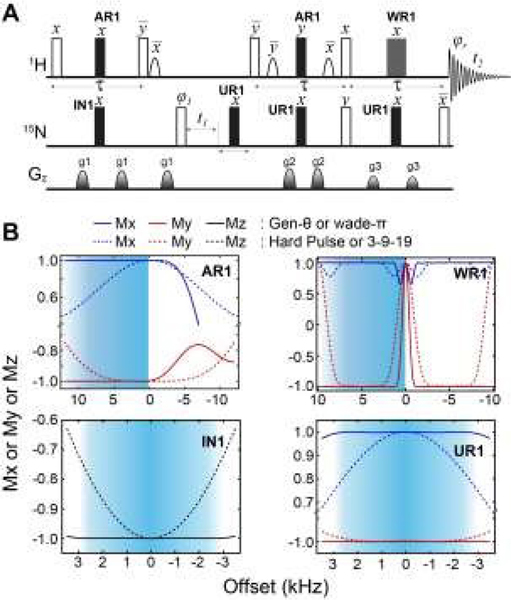
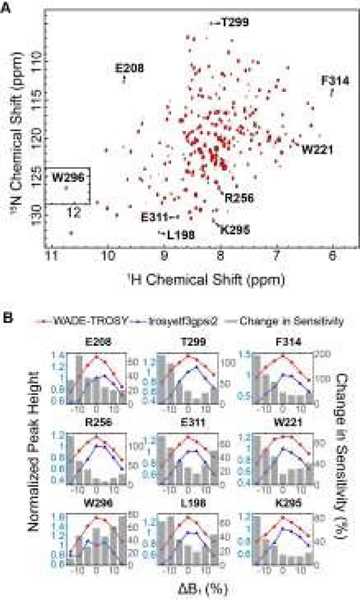
We then implemented the WADE pulses in the 2D [1H,15N]-TROSY (Transverse relaxation optimized spectroscopy) HSQC (heteronuclear single quantum coherence) pulse sequence (Fig. 2).23 This experiment images the amide protein fingerprints and constitutes the building block for multidimensional NMR spectroscopy of large biomacromolecules. Specifically, we replaced the rectangular π pulses, AR1, UR1, IN1, and WR1, with high-fidelity inversion and refocusing WADE-π pulses for both 1H and 15N channels in the basic TROSY-HSQC pulse scheme (Fig. 2A). The AR1 pulse in the middle of the INEPT sequence refocuses the 1JHN evolution of the amide proton resonances. The offset response of AR1 for the transverse components of the magnetization (Mx and My) is shown in Fig. 2B. The dashed lines represent the response of a rectangular π pulse of the same amplitude. AR1 is applied on-resonance, and the excitation of the off-resonance amide 1H resonances is achieved with a numerically designed phase ramp. The new amide pulses are shorter than our previously designed triply-compensated G5 pulses,24, 25 minimizing signal losses for fast relaxing systems. The WR1 pulse was substituted with the WADE-π pulse for water suppression before the signal acquisition (Fig. 2B and 1S, ESI†). The pulse length of WR1 is 1.29 ms with an RF amplitude of 5 kHz. A comparison of the offset responses of WR1 and the 3-9-19 sequence shows that the WADE-π pulse has a superior response in terms of bandwidth covered (Fig. 2B). The IN1 and UR1 pulses on the 15N channel are used to achieve inversion and refocusing operations, respectively, and were replaced by new π pulses with higher fidelity and broader bandwidth also designed with GENETICS-AI (Fig. 2B). We tested the performance of the TROSY-HSQC sequence with all WADE-π pulses (WADE-TROSY-HSQC) on a uniformly U-13C,15N labeled maltose binding protein (MBP)26 and 2H, 15N, 13CH3 ILV- labeled catalytic subunit of protein kinase A (PKA-C)27 using a 900 and an 850 MHz Bruker spectrometers, respectively (Fig. S9, ESI†, and Fig. 3). For the MBP, we recorded four 2D spectra with commonly used TROSY-HSQC sequences (Fig. S9, ESI†), included in the TopSpin® Bruker library: trosyetf3gpsi: a gradient pulse sensitivity improved TROSY-HSQC; 23,28 trosyetf3gpsi.2: a clean TROSY-HSQC version that minimizes semi-TROSY artifacts;29 trosyetf3gpsi2: a sensitivity enhanced gradient selection TROSY-HSQC with suppression of anti-TROSY peaks;30 and trosyf3gpphsi19.2,31, 32 a sensitivity improved TROSY with 3-9-19 water suppression.23, 28–30, 32–34 We then compared the spectral sensitivity of all these experiments with our WADE-TROSY-HSQC sequence (Fig. S9, ESI†). Note that the trosyetf3gpsi and trosyf3gpphsi19.2 experiments were implemented with water flip-back pulses (Fig. S12, ESI†),12 which in our hand, display higher sensitivity (~30%) than their standard Bruker versions. We analyzed the signal-to-noise (S/N) ratio for the least sensitive peaks in the spectra to quantify the sensitivity gain. Specifically, we integrated the volumes of all the peaks and arranged them in ascending order, and calculated the average sensitivity gain for the 50 least sensitive peaks in all four TROSY-HSQC experiments (Fig. S10, ESI†). The average gain in sensitivity of the least sensitive peaks in the WADE-TROSY-HSQC is 70.5% compared to the trosyetf3gpsi.2 experiment, whereas the highest sensitive TROSY-HSQC sequence in Bruker standard library, trosyetf3gpsi2, is 31.7%. As further verification, we tested the WADE-TROSY-HSQC experiment on the U-2H, 15N, 13CH3 ILV labeled catalytic subunit of protein kinase A (PKA-C) [24]. The average gain in sensitivity relative to the trosyetf3gpsi2 sequence is up to 60% (Fig. 3). In this case, protein perdeuteration contributes to the overall signal enhancement. Finally, we assessed the tolerance of the pulse sequences to miscalibration of the 1H RF amplitude (or RF inhomogeneity) by scaling the pulse amplitude. Reducing the amplitude by 15%, the signal loss in the WADE-TROSY-HSQC experiment is approximately 20–25%, whereas for the trosyetf3gpsi2 experiment is about 60%. The sensitivity gain and higher tolerance to RF inhomogeneity of the WADE-TROSY experiment is due to the higher fidelity of AR1 and WR1 pulses and the efficiency of the WADE water suppression. We anticipate that these effects will be more pronounced at ultra-high fields.
Figure 2. Implementation of the WADE-π pulses in the 2D TROSY sequence.
A. [1H-15N] WADE-TROSY pulse sequence. The open rectangles are π/2 pulses and filled rectangles are π pulses for inversion and refocusing operations. The shaped pulses on the 1H channel are water flip-back pulses of 1 ms. The phase cycles for φ1 and receiver phase (φr) are y, -y, -x, x and x, -x, -y, y, respectively. The duration of all gradient pulses was 1 ms and their ratio were 30:45:50 for g1:g2:g3. B. Offset responses of AR1, WR1, IN1 and UR1 pulses. The shaded areas correspond to the amide region detected by the 1H-15N correlation experiment. The AR1 pulse length was 120 μs (B1 = 16.67 kHz). The offset response was evaluated for the initial magnetization Mx (blue) and My (red). The comparison with hard pulses of identical power is shown with dashed lines. WR1 is a binomial WADE-π pulse for water suppression, with an amplitude of 5 kHz and a pulse length of 1.29 ms. The responses of the 3-9-19 pulse (dotted lines) with an inter-pulse delay of 100 μs on both components of the magnetization, Mx and My, are reported in blue and red, respectively. The responses of IN1 and UR1 for an initial Mz magnetization are shown as black lines and the corresponding. responses for a hard pulse are shown as black dashed lines. The pulse lengths for inversion and refocusing pulses were 147.5 and 227 μs, respectively with RF power of 7.8 kHz. Details for all the RF shapes are summarized in Table S1.
Figure 3. Increase in sensitivity obtained with the 2D WADE-TROSY-HSQC sequence.
A. 2D WADE-TROSY-HSQC spectrum of U-2H, 15N, 13CH3 ILV PKA-C recorded on a 850 MHz Bruker spectrometer at 300 K. B. Responses of selected peak intensities upon changing the RF pulse amplitude. The blue and red lines (left y axis) are the peak intensities obtained with trosyetf3gpsi (IT) and WADE-TROSY-HSQC sequence (IW), respectively. All peaks were normalized relative to the trosyetf3gpsi spectrum (ΔB1 = 0). The grey bars (right y axis) are the changes in sensitivity of the WADE-TROSY-HSQC spectrum relative to the trosyetf3gpsi spectrum calculated using (IW – IT)/IT.
In conclusion, we present the design of novel WADE pulses and pulse sequences for applications in biological NMR spectroscopy. The WADE pulses were designed using an evolutionary algorithm and artificial intelligence to avoid the excitation of the water signal while performing high-fidelity inversion and refocusing operations in the remainder of the 1H spectrum. When implemented in the 2D TROSY-HSQC experiments, these pulses increase the sensitivity of large proteins spectra up to 60%. We anticipate that replacing the rectangular (hard) pulses for homo- and heteronuclear NMR experiments with the WADE pulses will significantly improve the sensitivity of the current experiments. Given their broadband nature, the WADE pulses will be highly efficient at ultra-high magnetic fields, propelling the structural biology of large macromolecules and the analytical applications of NMR spectroscopy. Another future development would be to create pulses with multiple solvent suppression bands for applications to NMR spectroscopy of organic compounds.
Supplementary Material
Acknowledgements
This work was financially supported by the National Institute of Health (GM 64742, HL144130, 1S10OD021536 to GV). The experiments were carried out at the Minnesota NMR Center, University of Minnesota.
Footnotes
Conflicts of interest
There are no conflicts of interest to declare.
Electronic Supplementary Information (ESI) available: Experimental details, Figures S1–S12, Table S1, pulse sequence and RF shapes.
Publisher's Disclaimer: Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available.
Data availability
RF shapes and pulse sequences are publicly available through the University of Minnesota repository site: https://hdl.handle.net/11299/228028 and GitHub https://github.com/manuvsub/WADE_TROSY_v1.0.
Notes and references
- 1.Hore PJ, Journal of Magnetic Resonance (1969), 1983, 55, 283–300. [Google Scholar]
- 2.Bloembergen N. and Pound RV, Physical Review, 1954, 95, 8–12. [Google Scholar]
- 3.Hoult DI, Journal of Magnetic Resonance (1969), 1976, 21, 337–347. [Google Scholar]
- 4.Guéron M, Plateau P. and Decorps M, Progress in Nuclear Magnetic Resonance Spectroscopy, 1991, 23, 135–209. [Google Scholar]
- 5.McKay RT, in Annual Reports on NMR Spectroscopy, Academic Press, 2009, vol. 66, pp. 33–76. [Google Scholar]
- 6.Zheng G. and Price WS, Progress in Nuclear Magnetic Resonance Spectroscopy, 2010, 56, 267–288. [DOI] [PubMed] [Google Scholar]
- 7.Günther UL, Ludwig C. and Rüterjans H, Journal of Magnetic Resonance, 2002, 156, 19–25. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen BD, Meng X, Donovan KJ and Shaka AJ, J Magn Reson, 2007, 184, 263–274. [DOI] [PubMed] [Google Scholar]
- 9.Hwang TL and Shaka AJ, Journal of Magnetic Resonance, Series A, 1995, 112, 275–279. [Google Scholar]
- 10.Adams RW, Holroyd CM, Aguilar JA, Nilsson M. and Morris GA, Chemical Communications, 2013, 49, 358360. [DOI] [PubMed] [Google Scholar]
- 11.Ishima R, Concepts Magn Reson Part A Bridg Educ Res, 2015, 44A, 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grzesiek S. and Bax A, Journal of the American Chemical Society, 1993, 115, 12593–12594. [Google Scholar]
- 13.Piotto M, Saudek V. and Sklenář V, Journal of Biomolecular NMR, 1992, 2, 661–665. [DOI] [PubMed] [Google Scholar]
- 14.Liu M, Mao X.-a., Ye C, Huang H, Nicholson JK and Lindon JC, Journal of Magnetic Resonance, 1998, 132, 125–129. [DOI] [PubMed] [Google Scholar]
- 15.Schanda P, Progress in Nuclear Magnetic Resonance Spectroscopy, 2009, 55, 238–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schanda P, Van Melckebeke H. and Brutscher B, J Am Chem Soc, 2006, 128, 9042–9043. [DOI] [PubMed] [Google Scholar]
- 17.Solyom Z, Schwarten M, Geist L, Konrat R, Willbold D. and Brutscher B, J Biomol NMR, 2013, 55, 311–321. [DOI] [PubMed] [Google Scholar]
- 18.United States of America Pat., Patent No. US11221384B2, 2022.
- 19.Hore PJ, Journal of Magnetic Resonance (1969), 1983, 54, 539–542. [Google Scholar]
- 20.Sklenar V, Piotto M, Leppik R. and Saudek V, Journal of Magnetic Resonance, Series A, 1993, 102, 241–245. [Google Scholar]
- 21.Brenner T, Chen J, Stait-Gardner T, Zheng G, Matsukawa S. and Price WS, Journal of Magnetic Resonance, 2018, 288, 100–108. [DOI] [PubMed] [Google Scholar]
- 22.Zheng G, Torres AM and Price WS, Journal of Magnetic Resonance, 2008, 194, 108–114. [DOI] [PubMed] [Google Scholar]
- 23.Pervushin K, Riek R, Wider G. and Wüthrich K, Proceedings of the National Academy of Sciences, 1997, 94, 12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manu VS and Veglia G, Journal of Magnetic Resonance, 2015, 260, 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia Y, Rossi P, Subrahmanian MV, Huang C, Saleh T, Olivieri C, Kalodimos CG and Veglia G, J Biomol NMR, 2017, 69, 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spurlino JC, Lu GY and Quiocho FA, J Biol Chem, 1991, 266, 5202–5219. [DOI] [PubMed] [Google Scholar]
- 27.Turnham RE and Scott JD, Gene, 2016, 577, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weigelt J, Journal of the American Chemical Society, 1998, 120, 12706–12706. [Google Scholar]
- 29.Schulte-Herbrüggen T. and Sørensen OW, J Magn Reson, 2000, 144, 123–128. [DOI] [PubMed] [Google Scholar]
- 30.Nietlispach D, J Biomol NMR, 2005, 31, 161–166. [DOI] [PubMed] [Google Scholar]
- 31.Rance M, Loria JP and Palmer AGR, J Magn Reson, 1999, 136, 92–101. [DOI] [PubMed] [Google Scholar]
- 32.Czisch M. and Boelens R, J Magn Reson, 1998, 134, 158160. [DOI] [PubMed] [Google Scholar]
- 33.Meissner A, Schulte-Herbrüggen T, Briand J. and Sørensen OW, Molecular Physics, 1998, 95, 1137–1142. [Google Scholar]
- 34.Zhu G, Kong XM and Sze KH, J Biomol NMR, 1999, 13, 77–81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RF shapes and pulse sequences are publicly available through the University of Minnesota repository site: https://hdl.handle.net/11299/228028 and GitHub https://github.com/manuvsub/WADE_TROSY_v1.0.