Abstract
The development of nanoparticle probes has opened up new possibilities for molecular imaging in the era of precision medicine. There are a wide range of nanoprobes that are being used for various modalities that have demonstrated promising potential in early detection, disease monitoring and theranostics. However, the rate of clinical translation of the nanoprobes is very low and is affected by the lack of our understanding about nanoparticle interaction with biological fluids after systemic administration thus representing an unmet clinical need. One of the poorly understood issues relates to the formation of biomolecular corona, a layer of biomolecules formed on the surface of nanoscale materials during their interactions with biological fluids. The biomolecular corona has several significant effects on the biodistribution of nanoprobes and their imaging ability by i) reducing their targeting efficacy and ii) affecting the intrinsic imaging properties (e.g., contrast capacity of magnetic nanoprobes). This review provides insights on the importance of considering biomolecular corona in the development of nanoprobes, which may enable their more efficient utilization for molecular imaging applications.
Keywords: molecular imaging, nanoparticle, biomolecular corona, targeting
Introduction
Development of non-invasive strategies, capable of ultra-sensitive imaging of desired biosystems is a major unmet clinical need [1–2]. Successful development of such strategies enables clinicians to precisely identify diseases at their early stages which, in turn, can save many lives and significantly reduce the economic and social burden of catastrophic diseases such as cancer, cardiovascular and neurodegenerative disorders. In addition, various events can be monitored during the course of the treatment such as the efficacy of drug/molecular therapies, apoptosis, activation of immune system, appearance/disappearance of certain drug metabolites and others. Moreover, monitoring of disease relapse that may have different biomarkers compared to the initial diagnosis is also required. Finally, a combination of imaging and therapeutic probes (“theranostics”) is gaining more momentum as it allows to deliver drugs and monitor this delivery simultaneously in in vivo setting.
In the past few decades, a wide span of nanoprobes including theragnostics have been developed for a variety of molecular imaging applications. These nanoprobes have been used as contrast agents across multiple imaging modalities including fluorescence [3–6], computed tomography (CT) [3, 7–9], single-photon emission computed tomography (SPECT) [10–11], positron emission tomography (PET) [12–14], magnetic resonance imaging (MRI) and magnetic particle imaging (MPI) [3, 15–19]. Nanoparticles offer significant advantages over contrast agents that represent small molecules in that they have longer blood half-life, allow for accumulation in tumors or areas of inflammation via enhanced permeability and retention effect (EPR), allow for delivery and controlled release of drugs (encapsulated or conjugated). In certain situations, long half-life of the nanoparticles leading to the EPR effect could create the necessity to wait for their clearance from the circulation. Most importantly, however, is the ability to perform surface modification of nanoparticles with various moieties (targeting, pH-sensitive, therapeutic), which provides for exceptional versatility in various applications [20–24]. Although the developed nanoprobes in some cases significantly improved the sensitivity of the molecular imaging modalities, our knowledge of their interaction with biological systems is not adequate for direct clinical translation.
One issue that has been largely overlooked deals with the formation of biomolecular protein corona on the surface of nanoprobes. In vivo targeting and delivery of nanoparticles to the tissue of interest could be dramatically altered by biomolecular corona and prevent researchers from attaining the required optimal nanoprobes for molecular imaging applications. Here, we discuss the adverse effects of biomolecular corona on molecular imaging, together with insights on minimizing these effects and the ways to use it to our advantage.
Biomolecular corona and its interaction with biological systems
Nanoscale materials tend to absorb biomolecules at their surfaces upon their interactions with biological fluids [25]. These biomolecules interact with nanoparticles, creating a shell on their surfaces called “biomolecular corona” or, sometimes, “protein corona”. The biomolecular corona provides a new biological identity to the nanoparticles which could be completely different from their intended synthetic identity [25]. In other words, what the cells actually interact with and respond to, is the nanoparticle coated with biomolecular corona and not the synthesized nanoparticle coating (Figure 1a). In principal, the initial formation of biomolecular corona is called soft corona which consists of reversible and loosely attached biomolecules. Over time soft corona evolves to hard corona consisting of irreversible tightly attached biomolecules (Figure 1b).
Figure 1:
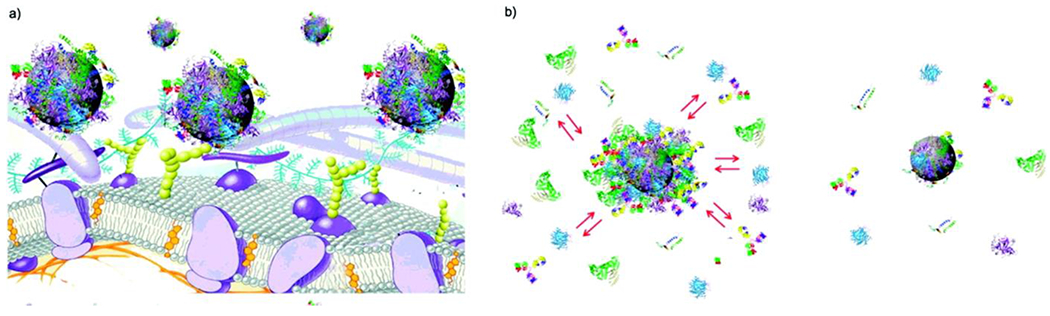
Schematics showing a) the biomolecular corona as the interaction site between nanoparticles and cells and (b) the formation of “soft” and “hard” biomolecular corona. Copyright American Chemical Society 2011 [28].
Biomolecular corona consists of proteins, lipids, metabolomes, nucleic acids, and other biologically active materials [26]. However, the lion share of the biomolecules in the corona layer is occupied by proteins [26]. The composition and decoration of the corona layer at the surface of nanoparticles strongly depend on several variables including physicochemical properties of the nanoparticles [27], incubation time [28] and temperature [29], and the protein source (e.g., fetal bovine serum, animal serum/plasma, and human serum/plasma) [30–31]. The biomolecular corona evolves from a dynamic state (i.e., soft corona) to semi-static state (i.e., hard corona) over the interaction time between nanoparticles and biological fluids [25]. However, various conditions including changing the type and concentration of biological fluids, have a capacity to change the composition of hard corona as well [32].
There are several techniques for probing interactions between nanoparticles and biomolecules, each of which has its own advantages and disadvantages. Table 1 provides a comprehensive information regarding the available analytical techniques for analysis of the nano-bio interfaces with their pros and cons. Very recently, we introduced another analytical technique, based on magnetic levitation (MagLev) approach, for the analysis of the homogeneity of biomolecular corona [33]. The MagLev approach is a fast, portable, and robust way to provide reliable and highly reproducible information on homogeneity of biomolecular corona; the major limitation of the MagLev system for corona analysis is its inability to analyze non-diamagnetic nanoparticles [34].
Table 1:
A list of the most common analytical approaches, with their pros and cons, to probe the interactions between nanoparticles and biomolecules.
| Analytical method | Advantages | Disadvantages |
|---|---|---|
| UV/vis | cheap, fast, flexible, and simple; little sample preparation | nature of the solvent, pH of the solution, temperature, high electrolyte concentrations, and presence of interfering substances can influence the absorption spectrum; experimental variations such as the slit width (effective bandwidth) of the spectrophotometer will also alter the spectrum; to apply UV/vis spectroscopy to analysis, these variables must be controlled or accounted for to identify the substances present |
| fluorescence spectroscopy | sensitive | unstable |
| FTIR | quite cheap, versatile, easy to identify functional groups; sensitive to protein conformation; not constrained by substrate size or material | sample characterization is not possible in complex media; cannot get fine structural detail; time-consuming sample preparation; sample preparation destroys the sample |
| Raman spectroscopy | can be used with solids and liquids; no sample preparation needed; no interference from water; nondestructive; highly specific—like a chemical fingerprint of a material; Raman spectra are acquired quickly (within seconds); samples can be analyzed through glass or a polymer packaging; laser light and Raman scattered light can be transmitted by optical fibers over long distances for remote analysis; Raman spectra can be collected from a very small volume (<1 μm in diameter); inorganic materials are normally easily analyzed by Raman compard to infrared spectroscopy | cannot be used for metals or alloys; Raman effect is very weak; detection needs a sensitive and highly optimized instrumentation; fluorescence of impurities or of the sample itself can hide the Raman spectrum; sample heating through the intense laser radiation can destroy the sample or cover the Raman spectrum |
| mass spectrometry | high-resolution method for characterization of NP-bound proteins; unique technique to obtain protein identities | expensive; requires dedicated facility and trained user |
| NMR spectroscopy | can detect very fine structural components; works for organic and inorganic materials; qualitative and quantitative, versatile; it can be applied to a wide variety of samples for direct structural study and molecular dynamics studies, both in solution and in the solid state | expensive, time-consuming; spectra take a long time to interpret |
| DLS | nonperturbative, fast, and accurate, giving a measure of the vesicle hydrodynamic diameter as this dimension changes in solution | hydrodynamic diameters are influenced by the formation of hydration shells, the shape of the particles, and counterion binding; requires a monodisperse population |
| CD | monitoring conformational changes induced by protein-NP interaction | inherent inconsistency problems in absolute secondary structure determination; CD signal reflects an average of the entire molecular population; CD measurements cannot provide information regarding local structural alterations at the level of individual amino acids |
| ITC | can directly and quantitatively measure the binding affinity constant, enthalpy changes, and binding stoichiometry between NP and proteins in solution; no labeling or immobilization is required; not limited by the ligand or protein size; relatively artifact-free and not affected by the optical properties of the samples | requires relatively high concentrations of samples |
| ζ potential | straightforward method to measure surface charge and changes in surface charge; indicator of stability of NP dispersions | requires a minimum ionic strength and that the NPs be monodisperse as calculates a charge/size ratio |
| chromatography | very sensitive and reliable (provided that the method is carried out carefully without any contamination); complex mixtures can be separated accurately using only a few micrograms of sample; separation takes less time as compared to other techniques; the equipment setups are simple and easy | since the method is very sensitive, improper setup or contamination, even in nanograms, will give different results; sample is generally very diluted afterward and requires reconcentration; time consuming |
| electrophoresis | suitable for separation of complicated protein mixtures; suitable for qualitative and quantitative analysis | proteins are easily adsorbed onto the inner surface of the capillary, and the detection sensitivity is not high |
| SPR | sensitive to changes in the refractive index of the medium surrounding the sensor and to the thickness of the sensor layer; as any change in protein conformation will bring a modification in this parameter, SPR has also been extensively used to study the conformation of immobilized proteins in various environments | sensitivity of the system with a detection limit restricted to 1–10 nM of a 20 kDa protein and even higher for smaller molecules, particularly when the receptor displays a weak affinity |
| QCM | simple, cost-effective, high-resolution mass sensing technique; ease of setup and operation and low cost; QCMs are capable of measuring mass changes as small as a fraction of a monolayer or single layer of atoms; allows a label-free detection of molecules | variations in interfacial parameters, such as surface roughness, surface free energy, surface charge, and viscoelasticity, hamper interpretation of QCM results |
Copyright American Chemical Society 2011 [35]. Abbreviations: Fourier transform infrared (FTIR); nuclear magnetic resonance (NMR); dynamic light scattering (DLS); circular dichroism (CD), isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), and quartz crystal microbalance (QCM).
It is now well-documented that the type, conformation and density of the biomolecular corona have the capacity to dictate its interactions with biosystems and, therefore, alter the pharmacokinetics of nanoparticles [28, 36–37]. It could also lead to untoward systemic effects and induction of immune response. For example, it has been shown that the conformational changes of fibrinogen on the surface of gold nanoparticles can activate secretion of inflammatory cytokines [37]. As another example, liposomes with various surface properties create different patterns of biomolecular corona which affect their interactions with leukocytes and, therefore, significantly alter their blood circulation time [38].
The formation of biomolecular corona presents several issues related to the central goals of nanomedicine including development of efficient targeting image-guided nanoptherapeutics. Failure to comprehensively consider biomolecular corona in the interaction of nanoparticles with biosystems may lead to misinterpretation and misprediction of the safety and therapeutic efficacy of nanoparticles/nanoprobes [39]. For example, biomolecular corona can add another layer to the drug carriers and, thus, change drug release profile in both in vitro [40] and in vivo [41]. In another example, the formation of corona can change the nutrition balance of the cell culture media and induce false errors in the toxicological outcomes [42].
The formation of biomolecular corona at the surface of nanoparticles can significantly reduce their cellular uptake, compared to the uncoated nanoparticles [43]. This is mainly due to the stronger adhesion capacity of bare nanoparticles to cell membranes which leads to higher internalization efficiency [43–44]. Even smaller coverage of nanoparticles with proteins reduced this uptake.
Biomolecules that occupy the outer layer of biomolecular corona interacts with cell surfaces. Functionality of the exposed proteins to cell receptors together with protein conformational changes defines how cells interact and respond to the existence of nanoparticles. The cellular responses to the corona coated nanoparticles include macrophage uptake/activation [45–46], blood coagulation [47–48], and complement activation [49]. For example, gold nanoparticles with poly(acrylic acid) coating could induce unfolding of fibrinogen and expose its c-terminus of the γ chain to integrin receptor (Mac-1) of THP-1 (a human monocytic cell) and, therefore, increase the NF-κB signaling pathway leading to the release in inflammatory cytokines [37]. Besides the physicochemical properties of nanoparticles, the type of disease that plasma donor may have could affect the composition of corona and, therefore, activate inflammatory cytokine releases. For example, it was shown that silica and polystyrene nanoparticles, after interaction with plasma of hypofibrinogenemia patients, could not induce inflammatory cytokine release while identical nanoparticles promoted the release of inflammatory cytokines after interaction with plasma of healthy individuals [50–51].
Studies revealed that there are considerable differences between in vitro and in vivo biological identity of nanoparticles, and the degree of such differences is strongly dependent on the physicochemical properties of nanoparticles and the type of the animal model employed [52–53]. It is noteworthy that the majority of the current literature in the biomolecular corona field describes the studies performed in vitro. The main reason for the paucity of the in vivo studies is the difficulties in separation of the nanoparticle from the in vivo environment [52]. The major differences between in vitro and in vivo characteristics of biomolecular corona are presented in Figure 2 and described in more details in our recent review [54].
Figure 2:
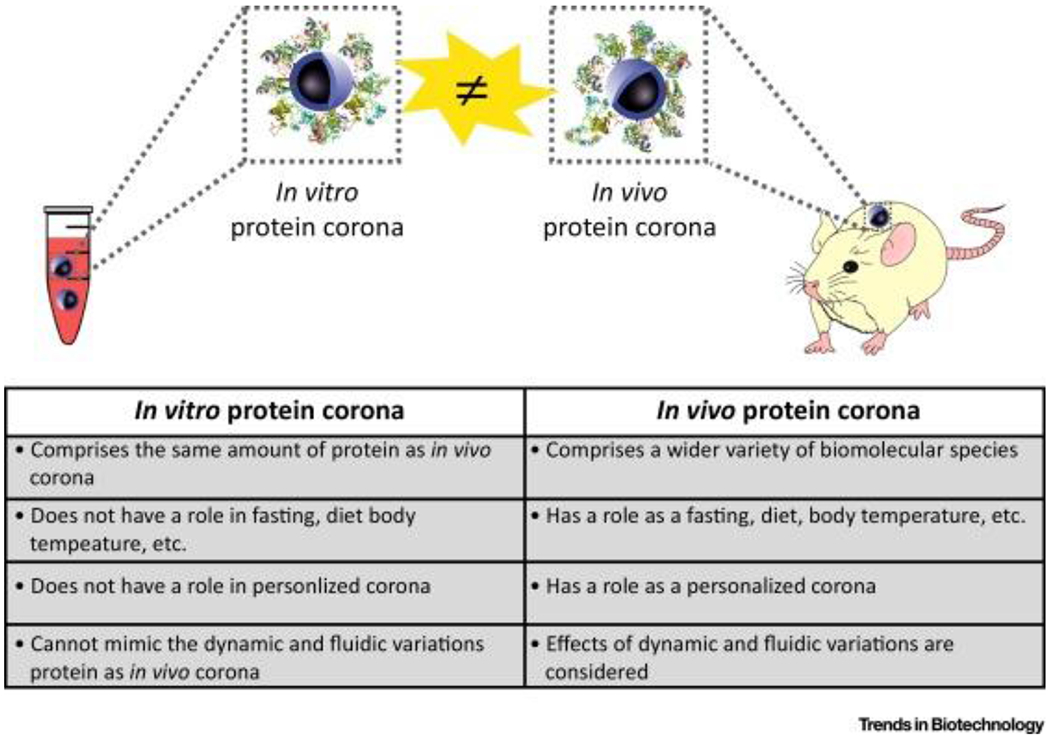
Major differences between in vitro and in vivo characteristics of biomolecular corona at the surface of nanoparticles. Copyright Cell Press 2017 [54].
Biomolecular corona and targeting efficacy of nanoprobes
One of the main approaches to create targeted nanoparticles is to attach/conjugate targeting moieties to the surface of nanoparticles so they can interact with specific receptors on the cells of interest [55]. Although the in vitro studies with targeted nanoparticles usually demonstrate excellent recognition and binding/uptake by these cells, significant number of in vivo investigations show much lower targeting efficacy and unfavorable distribution, which is one of the main reasons for failure of such nanoparticles in clinic. We and others proposed that the discrepancy between the in vitro and in vivo results is related, at least in large part, to the shielding effect of the protein corona [56–58]. It is noteworthy that although in vitro cell culture media contains proteins, the concentration of the protein source (e.g., fetal bovine serum) is as low as 10% with no contribution of plasma specific proteins (e.g., coagulation factors). In addition, in vivo condition is a dynamic environment while most of in vitro conditions are static which significantly affects the type of nanoparticles interactions with biomolecules and cells.
The shielding effect of biomolecular corona could be a great obstacle for the nanoprobes, as their delivery to the site of interest is the prerequisite for successful imaging outcome. Recent meta-analysis of the literature on the use of targeted nanoparticles revealed that only 0.7% of the administered nanoparticle dose was delivered to solid tumors [59]. There are many reasons for the observed low targeting efficacy of nanoparticles which include, but are not limited to, the shielding role of protein corona, physicochemical properties of nanoparticles, tumor models, cancer types, and sex of the recipients [59–61]. Therefore, the issue of the shielding effect produced by biomolecular corona needs to be carefully considered in development of targeted nanoprobes for a wide range of precise imaging applications. For example, in the case when formation of protein corona could significantly cover targeting moieties at the surface of nanoparticles (which can be examined through in vitro tests [57]), researchers may need to use specific types of coatings, including zwitterionic compounds [62], and/or alter the process under which targeting species are attached to the surface of nanoparticles (chemical vs. physical) to minimize the shielding role of biomolecular corona [63].
Biomolecular corona affects contrast agent properties
The formation of biomolecular corona may have the capacity to affect the imaging ability of the nanoprobes. For example, it was demonstrated that existence of biomolecular corona can affect the contrast agent capacity of magnetic nanoparticles [64]. In this study, 1H relaxometry was used to obtain the longitudinal, r1, and transverse, r2, the relaxivities of superparamagnetic iron oxide nanoparticles (SPIONs) in the presence or absence of biomolecular corona, as a function of the Larmor frequency. It was found that the transverse relaxivity that determines the efficiency of negative contrast agents was strongly dependent on the surface properties of SPIONs such as the presence of functional groups and surface charge of the coating. Specifically, the relaxivity of plain SPIONs was not changed by the formation of biomolecular corona; however, subtle increase and dramatic decrease were observed for the relaxivity of the negatively and positively charged nanoparticles (respectively) [64]. One of the reasons, among others, for significant reduction of relaxivity in positive nanoparticles could be due to particle agglomeration in which biomolecular corona plays significant role.
The underlying mechanisms of changing the contrast agent capacity of SPIONs is due to the fact that the formation of protein corona may affect the signal between water molecules interacting with magnetic nanoparticles. As the interactions between water and SPIONs occur primarily at their interfaces, the formation of biomolecular corona, together with the role of proteins in changing water molecule patterns [65–66] may play an important role in altering the magnetic properties of SPIONs and hence their imaging efficacy.
Similar to the magnetic nanoparticles, the formation of biomolecular corona may affect the efficacy of nanoprobes from other materials. This is mainly because the formation of biomolecular corona on many types of nanoparticles (e.g., silica [43], gold [67], zeolite [68], graphene oxide [69], and polystyrene [70]) is inevitable and well-documented. Such adverse effects of biomolecular corona on the imaging capacity of the nanoprobes need to be carefully considered in data analysis/interpretation of molecular imaging data.
Strategies to minimize the effect of biomolecular corona
There are several strategies to minimize the adverse effects created by biomolecular corona depicted in Figure 3 (details are provided in our recent review [71]). The conventional approach is to use PEG [poly(ethylene glycol)] polymer to minimize protein adsorption at the surface of nanoparticles [72–73]. However, PEGylation of nanoparticles cannot completely avoid the formation of protein corona. One strategy to preserve targeting capacity of nanoparticles is to use longer PEG chains at the surface of nanoparticles to keep the targeting ligands out of the corona shell. For example, it was shown that the use of longer PEG chains at the surface of gold nanoparticles could better preserve targeting molecules of Herceptin compared to the shorter PEG chains [74].
Figure 3:
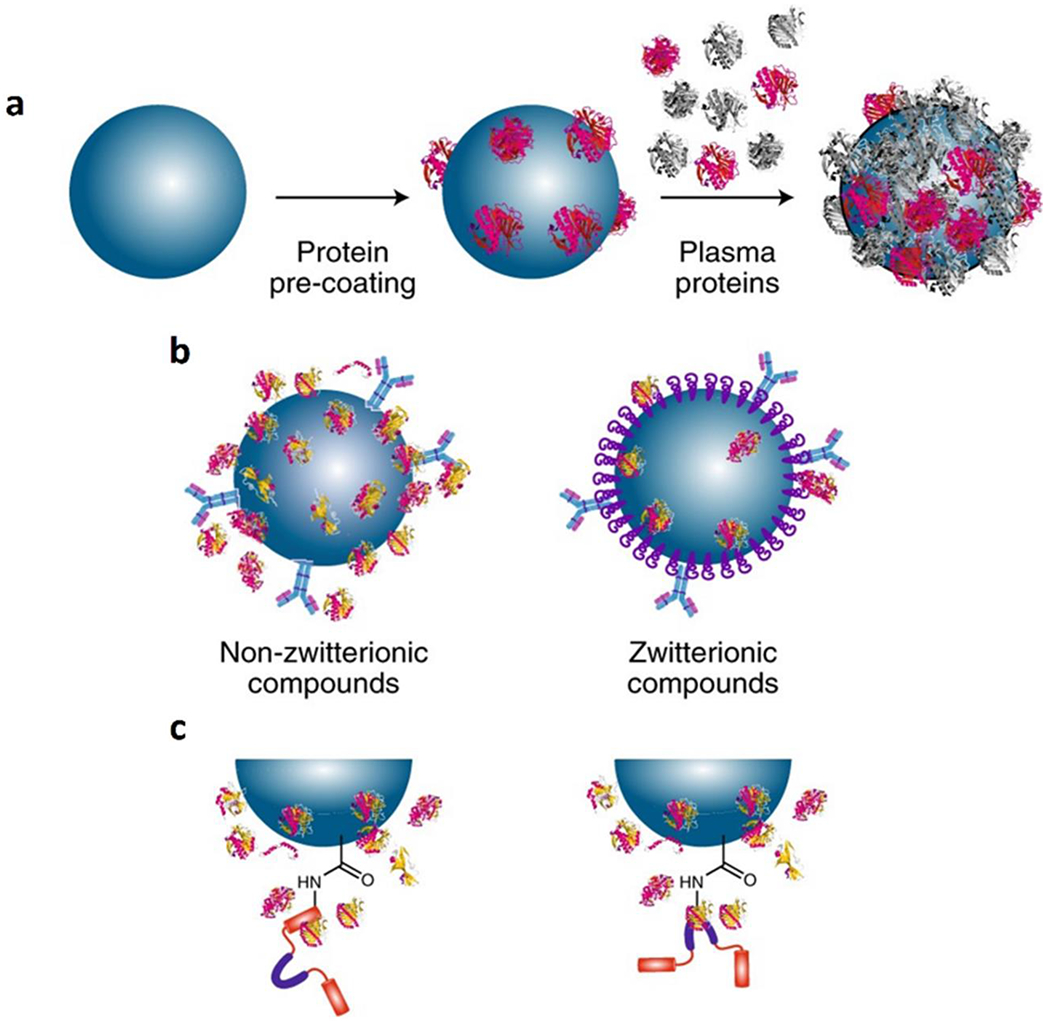
Schematics showing the newly introduced methods to minimize the formation of biomolecular corona: (a) recruitment of proteins with natural targeting capacity through pre-coating of nanoparticles with specific protein(s); (b) minimizing the affinity of biomolecules to the surface of nanoparticles through specific coatings including zwitterionic compounds; and (c) using pre-adsorption, rather than chemical conjugation, of targeting species to the surface of nanoparticles. Copyright Nature Publishing Group 2018 [39].
Another approach to mitigate the effect of protein corona is to pre-coat nanoparticles with specific proteins to improve recruitment of specific plasma proteins with intrinsic targeting capabilities [75]. For example, gamma globulin used as a pre-coating protein can recruit more immunoglobulins and activated complement factors in the corona composition which, in turn, can accelerate their cellular uptake through Fc receptor (e.g., on the surface of macrophages) [75]. In this case, one should also analyze the formed corona in terms of the targeting efficacy of the recruited proteins of interest, as the orientation of the targeting proteins is critical to achieve correct targeting. The orientations of targeting proteins are of crucial importance because their functional site need to be accessible to cell receptors.
Another alternative method is to use specific coating that have the capacity to minimize the formation of biomolecular corona. Zwitterionic coatings have shown such activity [62] mainly due to their dual charge and hydrophobicity. To this end, we have designed biotin-cysteine conjugated silica nanoparticles, where biotin was employed as a targeting molecule and cysteine was used as a zwitterionic ligand [76]. Using gel electrophoresis, we revealed that, as expected, the cysteine coating inhibited the formation of biomolecular corona. The in vitro cellular targeting analysis confirmed significant improvement in the targeting capacity of the nanoparticles with zwitterionic coatings compared to the zwitterionic-free nanoparticles.
Another way to reduce protein adsorption on the nanoparticle surface is to use pre-adsorption of targeting species, rather than their chemical conjugation. Studies revealed that pre-absorption of targeting species on the surface of nanoparticles compared with chemical conjugation, can significantly improve their targeting efficacies due to the i) reduced shielding effect of biomolecular corona and ii) increased availability of the antibody targeting sites [63]. In this study targeted polystyrene nanoparticles with carboxyl surface groups (PS-COOH) were made by attaching anti-CD63 antibodies by pre-adsorption and chemical conjugation [via EDC-NHS chemistry; EDC: (1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide); NHS: N-hydroxysuccinimide)]. Monocyte-derived dendritic cells (which have CD63 receptors on their surfaces), key players in orchestrating immunological responses, were selected as target cells. The targeted nanoparticles showed similar surface properties (e.g., surface charge) regardless of the mode of antibody attachment. Using secondary monoclonal anti-IgG1 antibody (against F(c) region), it was shown that similar amounts of anti-CD63 antibodies were attached to the surface of nanoparticles in both pre-adsorption and chemical conjugation strategies. In addition, in the absence of human serum or plasma, both attached antibodies were functional, as they could bind to the CD63 antigen on the surface of monocyte-derived dendritic cells and substantially increase uptake compared to control PS-COOH nanoparticles. However, in the presence of 10% serum or plasma and by increasing their concentrations to 100%, the cellular targeting efficacy of nanoparticles was substantially decreased, but the degree of reduction was strongly related to the mode of antibody attachment. More specifically, by increasing both serum or plasma to 100%, chemically attached targeted nanoparticles completely lost their targeting efficacy, while the pre-absorbed antibody-coated nanoparticles could still target the cells even after exposure to 100% serum or plasma [63].
In an effort to identify the mechanisms underlying the differences in targeting efficacy between pre-adsorption and chemical conjugation strategies, the authors found that antibody attachment can affect orientation and consequently accessibility to the active sites of the antibody, altering the targeting efficacy of nanoparticles [63]. Thus, instead of using a secondary antibody against the F(c) region, the authors investigated the accessibility of the F(ab) region, which contains an antigen binding site for the CD63 antibody. This study revealed significant differences in accessibility of this region between covalently bonded and pre-absorbed antibody, i.e., half of the region was inaccessible in the covalently bonded targeted nanoparticles while the entire region was active using the pre-adsorption strategy. One possible reason for the inaccessibility of the F(ab) region is the interaction between the activated carboxylated group of PS nanoparticles (due to the EDC-NHS chemistry) and the amine groups of the antibody. The N-terminal amino groups are located in the F(ab) region, much closer to the antigen-binding sites than the F(c) region, and thus the active sites of the antibodies may be affected/immobilized by chemical coupling to the nanoparticle surface. At this point, additional studies have to be conducted to evaluate this approach in vivo and to analyze the stability of protein pre-coating. We expect that based on the stability of hard corona itself, the pre-coating should be stable on the surface of the nanoparticles for at least 24 hours. This, however, needs to be demonstrated empirically.
Biomolecular corona has disease detection capacity
It is becoming increasingly clear that imaging and therapeutic applications of nanoparticle formulations could be altered by the presence of biomolecular corona on their surface. However, we can and we must in certain cases use this presence to our advantage. As such, in 2014, our group revealed the formation “personalized protein corona” meaning that corona profiles of identical nanoparticles are different after interaction with plasma of various donors [77]. Later, we discovered that this corona is disease-specific and can induce significant effect in cellular responses to identical nanoparticles dependent on the patient [78]. These findings opened up a new paradigm in the field of biomolecular corona indicating its diagnostic capacity. The significance of the concept of disease-specific protein corona was used and reproduced by several groups [79–84]. Very recently, we combined the concept of disease-specific protein corona with sensor array, and developed a sensor array protein corona for cancer and neurodegenerative diseases (Figure 4) that allows one to obtain comprehensive proteomics information on the “fingerprint” patterns specific to the disease type [85–87].
Figure 4.
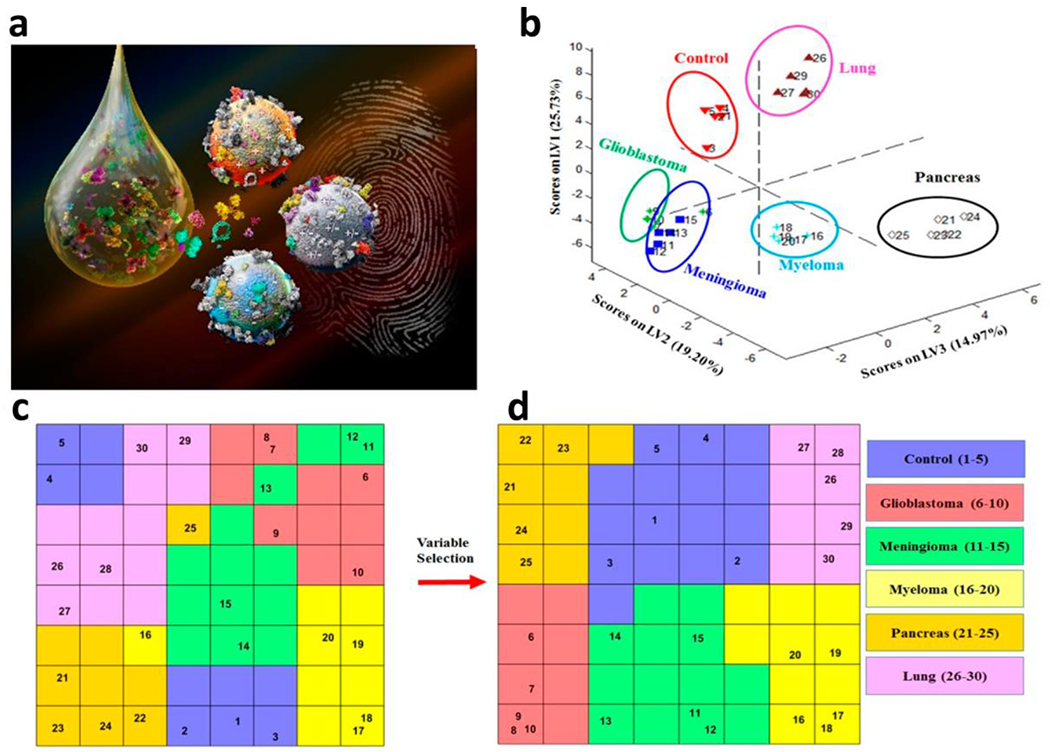
Development of protein corona sensor array for diagnostic purposes, (a) Scheme showing the formation of protein corona on the surface of multi-nanoparticles after interactions with patient plasmas to define “fingerprint” patterns specific to the diseases type, using advanced classifiers, (b-d) Use of two distinct supervised classifiers consisting of (b) partial least-squares discriminant analysis (PLS-DA) and (c,d) counter-propagation artificial neural network algorithm (CPANN) to analyze protein corona profiles for identification and separation of 5 distinct cancer types from each other and from healthy controls. Copyright 2019, Royal Society of Chemistry [85].
Conclusions and future perspectives
Consideration of the possible adverse effects of biomolecular corona is critical for the future design and development of efficient and safe nanoprobes which, in turn, can significantly enhance the sensitivity and specificity of molecular imaging techniques for early detection of diseases. Recent findings revealed that sex [88–89], age [90], and health status [77, 85] of people can significantly alter the composition of biomolecular corona and the interaction of the nanoparticles with biosystems, which adds more complications to the development of optimal nanoprobes for molecular imaging applications. Therefore, more information regarding sex, age, and type of diseases, together with comorbidities of patients or animal models should be fully documented in future reports. The existence of such a comprehensive dataset would enable the scientific community to develop safe and efficient nanoprobes suitable for personalized care.
Acknowledgments
Funding
This work was supported in part by R01CA135650 to A.M.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest Statement
The authors have declared that no competing interest exists.
References
- 1.Lee J-H, Huh Y-M, Jun Y-w, et al. (2007) Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat Med 13:95–99. [DOI] [PubMed] [Google Scholar]
- 2.Caracciolo G, Vali H, Moore A, Mahmoudi M (2019) Challenges in molecular diagnostic research in cancer nanotechnology. Nano Today 27:6–10. [Google Scholar]
- 3.Xing H, Bu W, Zhang S, et al. (2012) Multifunctional nanoprobes for upconversion fluorescence, MR and CT trimodal imaging. Biomaterials 33:1079–1089. [DOI] [PubMed] [Google Scholar]
- 4.Whiting GT, Nikolopoulos N, Nikolopoulos I, Chowdhury AD, Weckhuysen BM (2019) Visualizing pore architecture and molecular transport boundaries in catalyst bodies with fluorescent nanoprobes. Nat Chem 11:23–31. [DOI] [PubMed] [Google Scholar]
- 5.Pal S, Ray A, Andreou C, et al. (2019) DNA-enabled rational design of fluorescence-Raman bimodal nanoprobes for cancer imaging and therapy. Nature communications 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S, Liu L, Fan Y, et al. (2019) In vivo high-resolution ratiometric fluorescence imaging of inflammation using NIR-II nanoprobes with 1550 nm emission. Nano Let 19:2418–2427. [DOI] [PubMed] [Google Scholar]
- 7.Xu G, Qian Y, Zheng H, et al. (2019) Long-distance tracing of the lymphatic system with a computed tomography/fluorescence dual-modality nanoprobe for surveying tumor lymphatic metastasis. Bioconj Chem 30:1199–1209. [DOI] [PubMed] [Google Scholar]
- 8.Mancebo DG, Becerro Al, Corral A, et al. (2020) Design of a nanoprobe for high field magnetic resonance imaging, dual energy X-ray computed tomography and luminescent imaging. J Colloid Interface Sci 573:278–286. [DOI] [PubMed] [Google Scholar]
- 9.Zeng S, Tsang M-K, Chan C-F, Wong K-L, Hao J (2012) PEG modified BaGdF5: Yb/Er nanoprobes for multi-modal upconversion fluorescent, in vivo X-ray computed tomography and biomagnetic imaging. Biomaterials 33:9232–9238. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Sun Z, Chen Z, et al. (2019) In vivo photoacoustic/single-photon emission computed tomography imaging for dynamic monitoring of aggregation-enhanced photothermal nanoagents. Anal Chem 91:2128–2134. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Zhang L, Cai J, et al. (2016) Tumor angiogenesis targeted radiosensitization therapy using gold nanoprobes guided by MRI/SPECT imaging. ACS Appl Mater Interfaces 8:1718–1732. [DOI] [PubMed] [Google Scholar]
- 12.Jing B, Qian R, Gai Y, Lan X, An R (2019) Multimodality PET/CT and NIRF imaging for image-guided surgery of colon cancer with exosomes based nanoprobe. J Nucl Med 60:662–662. [Google Scholar]
- 13.Lahooti A, Shanehsazzadeh S, Laurent S (2019) Preliminary studies of 68Ga-NODA-USPION-BBN as a dual-modality contrast agent for use in positron emission tomography/magnetic resonance imaging. Nanotechnology 31:015102. [DOI] [PubMed] [Google Scholar]
- 14.Zhan Y, Ai F, Chen F, et al. (2016) Intrinsically zirconium-89 labeled Gd202S: Eu nanoprobes for in vivo positron emission tomography and gamma-ray-induced radioluminescence imaging. Small 12:2872–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park YI, Kim JH, Lee KT, et al. (2009) Nonblinking and nonbleaching upconverting nanoparticles as an optical imaging nanoprobe and T1 magnetic resonance imaging contrast agent. Advanced Mater 21:4467–4471. [Google Scholar]
- 16.Sharifi S, Seyednejad H, Laurent S, Atyabi F, Saei AA, Mahmoudi M (2015) Superparamagnetic iron oxide nanoparticles for in vivo molecular and cellular imaging. Contrast Media Mol Imaging 10:329–355. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Song M, Tang J, et al. (2016) Ultrahigh 19F loaded Cul. 75S nanoprobes for simultaneous 19F magnetic resonance imaging and photothermal therapy. ACS nano 10:1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yigit MV, Zhu L, Ifediba MA, et al. (2011) Noninvasive MRI-SERS imaging in living mice using an innately bimodal nanomaterial. ACS nano 5:1056–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Yoo B, Yang J, et al. (2014) GLP-1R–targeting magnetic nanoparticles for pancreatic islet imaging. Diabetes 63:1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Song J, Yung BC, Huang X, Xiong Y, Chen X (2018) Ratiometric optical nanoprobes enable accurate molecular detection and imaging. Chem Soc Rev 47:2873–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Won J, Kim M, Yi Y-W, Kim YH, Jung N, Kim TK (2005) A magnetic nanoprobe technology for detecting molecular interactions in live cells. Science 309:121–125. [DOI] [PubMed] [Google Scholar]
- 22.Zhu C, Zeng Z, Li H, Li F, Fan C, Zhang H (2013) Single-layer MoS2-based nanoprobes for homogeneous detection of biomolecules. J Am Chem Soc 135:5998–6001. [DOI] [PubMed] [Google Scholar]
- 23.Wabuyele MB, Vo-Dinh T (2005) Detection of human immunodeficiency virus type 1 DNA sequence using plasmonics nanoprobes. Anal Chem 77:7810–7815. [DOI] [PubMed] [Google Scholar]
- 24.Song S, Qin Y, He Y, Huang Q, Fan C, Chen H-Y (2010) Functional nanoprobes for ultrasensitive detection of biomolecules. Chem Soc Rev 39:4234–4243. [DOI] [PubMed] [Google Scholar]
- 25.Monopoli MP, Aberg C, Salvati A, Dawson KA (2012) Biomolecular coronas provide the biological identity of nanosized materials. Nat Nanotech 7:779. [DOI] [PubMed] [Google Scholar]
- 26.Kelly PM, Aberg C, Polo E, et al. (2015) Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat Nanotech 10:472. [DOI] [PubMed] [Google Scholar]
- 27.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA (2008) Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Nat Acad Sci USA 105:14265–14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA (2010) What the cell “sees” in bionanoscience. J Am Chem Soc 132:5761–5768. [DOI] [PubMed] [Google Scholar]
- 29.Mahmoudi M, Abdelmonem AM, Behzadi S, et al. (2013) Temperature: the “ignored” factor at the nanobio interface. ACS nano 7:6555–6562. [DOI] [PubMed] [Google Scholar]
- 30.Mirshafiee V, Kim R, Mahmoudi M, Kraft ML (2016) The importance of selecting a proper biological milieu for protein corona analysis in vitro: Human plasma versus human serum. Int J Biochem Cell Biol 75:188–195. [DOI] [PubMed] [Google Scholar]
- 31.Müller LK, Simon J, Rosenauer C, Mailänder V, Morsbach S, Landfester K (2018) The transferability from animal models to humans: challenges regarding aggregation and protein corona formation of nanoparticles. Biomacromolecules 19:374–385. [DOI] [PubMed] [Google Scholar]
- 32.Ghavami M, Saffar S, Abd Emamy B, et al. (2013) Plasma concentration gradient influences the protein corona decoration on nanoparticles. Rsc Advances 3:1119–1126. [Google Scholar]
- 33.Ashkarran AA, Dararatana N, Crespy D, Caracciolo G, Mahmoudi M (2020) Mapping the heterogeneity of protein corona by ex vivo magnetic levitation. Nanoscale 12:2374–2383. [DOI] [PubMed] [Google Scholar]
- 34.Ashkarran AA, Mahmoudi M (2020) Magnetic levitation systems for disease diagnostics. Trends Biotechnol, in press. [DOI] [PubMed] [Google Scholar]
- 35.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S (2011) Protein– nanoparticle interactions: opportunities and challenges. Chem Rev 111:5610–5637. [DOI] [PubMed] [Google Scholar]
- 36.Monopoli MP, Walczyk D, Campbell A, et al. (2011) Physical– chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc 133:2525–2534. [DOI] [PubMed] [Google Scholar]
- 37.Deng ZJ, Liang M, Monteiro M, Toth I, Minchin RF (2011) Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat Nanotechnol 6:39–44. [DOI] [PubMed] [Google Scholar]
- 38.Giulimondi F, Digiacomo L, Pozzi D, et al. (2019) Interplay of protein corona and immune cells controls blood residency of liposomes. Nat Commun 10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudi M (2018) Antibody orientation determines corona mistargeting capability. Nat Nanotechnol 13:775–776. [DOI] [PubMed] [Google Scholar]
- 40.Behzadi S, Serpooshan V, Sakhtianchi R, et al. (2014) Protein corona change the drug release profile of nanocarriers: the “overlooked” factor at the nanobio interface. Colloids Surf B Biointerfaces 123:143–149. [DOI] [PubMed] [Google Scholar]
- 41.Sharifi S, Caracciolo G, Mahmoudi M (2020) Biomolecular corona affects controlled release of drug payloads from nanocarriers. Trends Pharmacol Sci 41:641–652. [DOI] [PubMed] [Google Scholar]
- 42.Mahmoudi M, Simchi A, Imani M, et al. (2010) A new approach for the in vitro identification of the cytotoxicity of superparamagnetic iron oxide nanoparticles. Colloids Surf B Biointerfaces 75:300–309. [DOI] [PubMed] [Google Scholar]
- 43.Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A (2012) Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS nano 6:5845–5857. [DOI] [PubMed] [Google Scholar]
- 44.Lesniak A, Salvati A, Santos-Martinez MJ, Radomski MW, Dawson KA, Åberg C (2013) Nanoparticle adhesion to the cell membrane and its effect on nanoparticle uptake efficiency. J Am Chem Soc 135:1438–1444. [DOI] [PubMed] [Google Scholar]
- 45.Dutta D, Sundaram SK, Teeguarden JG, et al. (2007) Adsorbed proteins influence the biological activity and molecular targeting of nanomaterials. Toxicol Sci 100:303–315. [DOI] [PubMed] [Google Scholar]
- 46.Zanganeh S, Hutter G, Spitler R, et al. (2016) Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat Nanotechnol 11:986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE (2008) Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol Pharm 5:487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derakhshankhah H, Hajipour MJ, Barzegari E, et al. (2016) Zeolite nanoparticles inhibit Aβ–fibrinogen interaction and formation of a consequent abnormal structural clot. ACS Appl Mater Interfaces 8:30768–30779. [DOI] [PubMed] [Google Scholar]
- 49.Reddy ST, Van Der Vlies AJ, Simeoni E, et al. (2007) Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat Biotechnol 25:1159–1164. [DOI] [PubMed] [Google Scholar]
- 50.Rahman M, Mahmoudi M (2015) Disease specific protein corona [abstract], 9338: 93380VP. [Google Scholar]
- 51.Tirtaatmadja N, Mortimer G, Ng E-P, et al. (2015) Nanoparticles-induced inflammatory cytokines in human plasma concentration manner: an ignored factor at the nanobio-interface. J Iranian Chem Soc 12:317–323. [Google Scholar]
- 52.Sakulkhu U, Maurizi L, Mahmoudi M, et al. (2014) Ex situ evaluation of the composition of protein corona of intravenously injected superparamagnetic nanoparticles in rats. Nanoscale 6:11439–11450. [DOI] [PubMed] [Google Scholar]
- 53.Hadjidemetriou M, Al-Ahmady Z, Mazza M, Collins RF, Dawson K, Kostarelos K (2015) In vivo biomolecule corona around blood-circulating, clinically used and antibody-targeted lipid bilayer nanoscale vesicles. ACS nano 9:8142–8156. [DOI] [PubMed] [Google Scholar]
- 54.Caracciolo G, Farokhzad OC, Mahmoudi M (2017) Biological identity of nanoparticles in vivo: clinical implications of the protein corona. Trends Biotechnol 35:257–264. [DOI] [PubMed] [Google Scholar]
- 55.Behzadi S, Serpooshan V, Tao W, et al. (2017) Cellular uptake of nanoparticles: journey inside the cell. Chem Soc Rev 46:4218–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laurent S, Mahmoudi M (2011) Superparamagnetic iron oxide nanoparticles: promises for diagnosis and treatment of cancer. Int J Mol Epidemiol Genet 2:367–390. [PMC free article] [PubMed] [Google Scholar]
- 57.Mirshafiee V, Mahmoudi M, Lou K, Cheng J, Kraft ML (2013) Protein corona significantly reduces active targeting yield. Chem Comm 49:2557–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salvati A, Pitek AS, Monopoli MP, et al. (2013) Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat Nanotechnol 8:137–143. [DOI] [PubMed] [Google Scholar]
- 59.Wilhelm S, Tavares AJ, Dai Q, et al. (2016) Analysis of nanoparticle delivery to tumours. Nat Rev Mat 1:1–12. [Google Scholar]
- 60.Mahmoudi M (2018) Debugging nano–bio interfaces: systematic strategies to accelerate clinical translation of nanotechnologies. Trends Biotechnol 36:755–769. [DOI] [PubMed] [Google Scholar]
- 61.Serpooshan V, Sheibani S, Pushparaj P, et al. (2018) Effect of cell sex on uptake of nanoparticles: The overlooked factor at the nanobio interface. ACS nano 12:2253–2266. [DOI] [PubMed] [Google Scholar]
- 62.Moyano DF, Saha K, Prakash G, et al. (2014) Fabrication of corona-free nanoparticles with tunable hydrophobicity. ACS nano 8:6748–6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tonigold M, Simon J, Estupiñán D, et al. (2018) Pre-adsorption of antibodies enables targeting of nanocarriers despite a biomolecular corona. Nat Nanotechnol 13:862–869. [DOI] [PubMed] [Google Scholar]
- 64.Amiri H, Bordonali L, Lascialfari A, et al. (2013) Protein corona affects the relaxivity and MRI contrast efficiency of magnetic nanoparticles. Nanoscale 5:8656–8665. [DOI] [PubMed] [Google Scholar]
- 65.Guckeisen T, Hosseinpour S, Peukert W (2019) Isoelectric points of proteins at the air/liquid interface and in solution. Langmuir 35:5004–5012. [DOI] [PubMed] [Google Scholar]
- 66.Hosseinpour S, Roeters SJ, Bonn M, Peukert W, Woutersen S, Weidner T (2020) Structure and dynamics of interfacial peptides and proteins from vibrational sum-frequency generation spectroscopy. Chem Rev 120:3420–3465. [DOI] [PubMed] [Google Scholar]
- 67.Charbgoo F, Nejabat M, Abnous K, et al. (2018) Gold nanoparticle should understand protein corona for being a clinical nanomaterial. J Control Release 272:39–53. [DOI] [PubMed] [Google Scholar]
- 68.Rahimi M, Ng E-P, Bakhtiari K, et al. (2015) Zeolite nanoparticles for selective sorption of plasma proteins. Sci Reports 5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu W, Peng C, Lv M, et al. (2011) Protein corona-mediated mitigation of cytotoxicity of graphene oxide. ACS nano 5:3693–3700. [DOI] [PubMed] [Google Scholar]
- 70.Tenzer S, Docter D, Kuharev J, et al. (2013) Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat Nanotechnol 8:772–781. [DOI] [PubMed] [Google Scholar]
- 71.Mahmoudi M, Bertrand N, Zope H, Farokhzad OC (2016) Emerging understanding of the protein corona at the nano-bio interfaces. Nano Today 11:817–832. [Google Scholar]
- 72.Karakoti AS, Das S, Thevuthasan S, Seal S (2011) PEGylated inorganic nanoparticles. Angew Chem Int Ed Engl 50:1980–1994. [DOI] [PubMed] [Google Scholar]
- 73.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS (2011) Nanoparticle PEGylation for imaging and therapy. Nanomedicine 6:715–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dai Q, Walkey C, Chan WC (2014) Polyethylene glycol backfilling mitigates the negative impact of the protein corona on nanoparticle cell targeting. Angew Chem Int Ed Engl 53:5093–5096. [DOI] [PubMed] [Google Scholar]
- 75.Mirshafiee V, Kim R, Park S, Mahmoudi M, Kraft ML (2016) Impact of protein pre-coating on the protein corona composition and nanoparticle cellular uptake. Biomaterials 75:295–304. [DOI] [PubMed] [Google Scholar]
- 76.Safavi-Sohi R, Maghari S, Raoufi M, et al. (2016) Bypassing protein corona issue on active targeting: zwitterionic coatings dictate specific interactions of targeting moieties and cell receptors. ACS Appl Mat Interfaces 8:22808–22818. [DOI] [PubMed] [Google Scholar]
- 77.Hajipour MJ, Laurent S, Aghaie A, Rezaee F, Mahmoudi M (2014) Personalized protein coronas: a “key” factor at the nanobiointerface. Biomater Sci 2:1210–1221. [DOI] [PubMed] [Google Scholar]
- 78.Hajipour MJ, Raheb J, Akhavan O, et al. (2015) Personalized disease-specific protein corona influences the therapeutic impact of graphene oxide. Nanoscale 7:8978–8994. [DOI] [PubMed] [Google Scholar]
- 79.Caputo D, Caracciolo G (2020) Nanoparticle-enabled blood tests for early detection of pancreatic ductal adenocarcinoma. Cancer Let 470:191–196. [DOI] [PubMed] [Google Scholar]
- 80.Barui AK, Oh JY, Jana B, Kim C, Ryu JH (2020) Cancer-targeted nanomedicine: overcoming the barrier of the protein corona. Adv Therapeutics 3:1900124. [Google Scholar]
- 81.Quagliarini E, Di Santo R, Pozzi D, Caracciolo G (2020) Protein corona-enabled serological tests for early stage cancer detection. Sensors Int 1:100025. [Google Scholar]
- 82.Colapicchioni V, Tilio M, Digiacomo L, et al. (2016) Personalized liposome-protein corona in the blood of breast, gastric and pancreatic cancer patients. Int J biochem Cell Biol 75:180–187. [DOI] [PubMed] [Google Scholar]
- 83.Lazarovits J, Chen YY, Song F, et al. (2018) Synthesis of patient-specific nanomaterials. Nano Let 19:116–123. [DOI] [PubMed] [Google Scholar]
- 84.Hadjidemetriou M, Al-Ahmady Z, Buggio M, Swift J, Kostarelos K (2019) A novel scavenging tool for cancer biomarker discovery based on the blood-circulating nanoparticle protein corona. Biomaterials 188:118–129. [DOI] [PubMed] [Google Scholar]
- 85.Caracciolo G, Safavi-Sohi R, Malekzadeh R, et al. (2019) Disease-specific protein corona sensor arrays may have disease detection capacity. Nanoscale Horizons 4:1063–1076. [Google Scholar]
- 86.Digiacomo L, Jafari-Khouzani K, Palchetti S, et al. (2020) A protein corona sensor array detects breast and prostate cancers. Nanoscale 12:16697–16704. [DOI] [PubMed] [Google Scholar]
- 87.Hajipour MJ, Ghasemi F, Aghaverdi H, et al. (2017) Sensing of Alzheimer’s disease and multiple sclerosis using nano-bio interfaces. J Alzheimer’s Dis 59:1187–1202. [DOI] [PubMed] [Google Scholar]
- 88.Gao J, Lin L, Wei A, Sepúlveda MS (2017) Protein Corona Analysis of Silver Nanoparticles Exposed to Fish Plasma. Environ Sci Technol Lett 4:174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hayashi Y, Miclaus T, Murugadoss S, et al. (2017) Female versus male biological identities of nanoparticles determine the interaction with immune cells in fish. Environ Sci: Nano 4:895–906. [Google Scholar]
- 90.Foroozandeh P, Aziz AA, Mahmoudi M (2019) Effect of Cell Age on Uptake and Toxicity of Nanoparticles: The Overlooked Factor at the Nanobio Interface. ACS Appl Mat Interfaces 11:39672–39687. [DOI] [PubMed] [Google Scholar]


