This review provides an overview of the advances made with the Arabidopsis thaliana–Fusarium oxysporum5176 pathosystem in understanding the cause and defense against wilt disease in plants.
Keywords: Arabidopsis thaliana, Fo5176, fungal pathogen, Fusarium oxysporum, immunity, plant–microbe interactions, vascular wilt
Abstract
Fusarium oxysporum is a soil-borne fungal pathogen of several major food crops. Research on understanding the molecular details of fungal infection and the plant’s defense mechanisms against this pathogen has long focused mainly on the tomato-infecting F. oxysporum strains and their specific host plant. However, in recent years, the Arabidopsis thaliana–Fusarium oxysporum strain 5176 (Fo5176) pathosystem has additionally been established to study this plant–pathogen interaction with all the molecular biology, genetic, and genomic tools available for the A. thaliana model system. Work on this system has since produced several new insights, especially with regards to the role of phytohormones involved in the plant’s defense response, and the receptor proteins and peptide ligands involved in pathogen detection. Furthermore, work with the pathogenic strain Fo5176 and the related endophytic strain Fo47 has demonstrated the suitability of this system for comparative studies of the plant’s specific responses to general microbe- or pathogen-associated molecular patterns. In this review, we highlight the advantages of this specific pathosystem, summarize the advances made in studying the molecular details of this plant–fungus interaction, and point out open questions that remain to be answered.
Introduction
Fusarium oxysporum is a soil-inhabiting species of ascomycete fungi. While most strains within the species are harmless, and typically isolated from asymptomatic crops, several are plant pathogens (Gordon and Martyn, 1997). Fusarium oxysporum reproduces asexually, and thus the different strains represent individual clonal lineages (Summerell, 2019). These strains are collectively referred to as the F. oxysporum species complex. The genetic differences between strains within the complex reflect the individually evolved host speciation, and each strain is classified as forma specialis (f. sp.) based on its host range. Over 120 such formae speciales (ff. spp.) have been described so far (Summerell, 2019). Pathogenic ff. spp. target several important crop plants on which they cause the Fusarium wilt disease (Gordon, 2017). Typical disease symptoms are leaf and vein clearing and necrosis, wilting, and eventual plant death (Thatcher et al., 2016b). Among the economically most important crops targeted are banana, cotton, and tomato (Gordon, 2017; Dita et al., 2018; Cox et al., 2019; Srinivas et al., 2019). With tomato also being an established research model for plant genetics and development, the Solanum lycopersicum–F. oxysporum f. sp. lycopersici pathosystem became the predominant system to study plant–Fusarium interactions (di Pietro et al., 2003; Kimura and Sinha, 2008; Takken and Rep, 2010). Several comprehensive reviews have been published on this pathosystem over the past years, so we will here focus on the Arabidopsis thaliana–F. oxysporum pathosystem (Takken and Rep, 2010; de Sain and Rep, 2015; Srinivas et al., 2019).
Most pathogenic F. oxysporum ff. spp. can be considered hemibiotrophs, as they begin their infection cycle as a biotroph (roughly at days 1–5), before becoming necrotrophic at the later stages of infection (roughly day 6 onwards) (Thaler et al., 2004; Gordon, 2017). After the spores germinate in the soil, the fungal hyphae grow toward the plant and attach to its root. Growing along the root, it is generally assumed that they enter the root via natural openings, such as wounds or the sites of lateral root emergence (de Sain and Rep, 2015; Thatcher et al., 2016b). Specifically for A. thaliana, however, it appears that the hyphae preferentially enter the root at the meristematic zone, before the Casparian strips are formed to protect the vasculature from colonization (Czymmek et al., 2007). In the root, the fungus grows in the apoplast until it reaches the vasculature of the plant, where it then colonizes xylem cells and drains water and nutrients from the plant. During these early stages, the fungus lives biotrophically (de Sain and Rep, 2015; Thatcher et al., 2016b). Subsequently, mycelia growth in the vasculature, and the production of new spores will result in blockage of the xylem, at which stage the above-ground parts of the plant will start wilting due to an undersupply of water and nutrients. This starts the necrotrophic phase of the infection cycle, which eventually results in the death of the host plant and the release of new fungal spores (de Sain and Rep, 2015; Thatcher et al., 2016b).
Three ff. spp. pathogenic to A. thaliana were described in 1987 as f. sp. conglutinans [isolated from cabbage (Brassica species)], f. sp. matthioli [from garden stock (Matthiola incana)], and f. sp. raphani [from radish (Raphanus sativus)] (Bosland and Williams, 1987). Interestingly, these ff. spp. do not infect all, or the same, A. thaliana natural accessions. While plants from most accessions, including Columbia (Col), were susceptible to infection by F. oxysporum ff. spp. conglutinans and raphani, several, including Col, exhibited full resistance against F. oxysporum f. sp. matthioli (Diener and Ausubel, 2005). The accession Taynuilt-0 (Ty), on the other hand, is susceptible to all three ff. spp. (Diener and Ausubel, 2005). This observation, together with the availability of beneficial and completely incompatible strains, makes the F. oxysporum species complex an interesting subject to study specialization and pathogenesis in the context of natural variation, especially in conjunction with the well-described collection of A. thaliana natural accessions (Alonso-Blanco et al., 2016). Moreover, it also highlights the importance of establishing a specific fungal strain–plant accession pair as a reference pathosystem, to correctly interpret results without additional convolution coming from such fungal strain- and/or plant accession-specific effects.
The specific F. oxysporum f. sp. conglutinans strain 5176 is maintained by the Brisbane Pathology (BRIP) Plant Pathology Herbarium in Queensland, Australia (accession number BRIP 5176 a). It was collected in 1971 (collection number 19142) from white cabbage [Brassica oleracea var. capitata (L.)] in a glasshouse in Indooroopilly, Australia. It had been classified as f. sp. conglutinans based on it being isolated from Brassica oleracea, and this classification was confirmed when an updated genome assembly placed it in the same phylogenetic group as the other four ff. spp. conglutinans strains included in the analysis (Fokkens et al., 2020). The availability of this genome facilitates genetic work, directed mutagenesis, or cloning of fungal genes, and Agrobacterium tumefaciens-mediated transformation protocols to create transgenic lines have been established as well (Mullins et al., 2001; Kidd et al., 2011). Due to the asexual reproduction of the fungus, individual transgenic and mutant lines can be readily maintained either dried on filter paper or as spores in glycerol stocks at –80 °C. Its use as a model pathogen for A. thaliana research started in the early 2000s at the University of Queensland, Australia (Campbell et al., 2003). In the following, we will refer to Fusarium oxysporum f. sp. conglutinans strain 5176 as ‘Fo5176’, and to other Fusarium oxysporum f. sp. conglutinans strains, for which the authors of the original work did not explicitly state that they used Fo5176, as ‘FoCon’.
The role of phytohormones
Salicylic acid
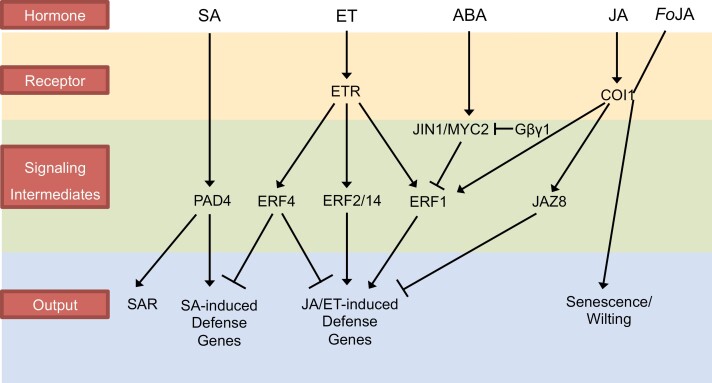
Salicylic acid (SA) and jasmonic acid (JA) are considered to be the two main hormone signals coordinating a plant’s response to pathogens (Hou et al., 2022). While the two pathways are interconnected and embedded within a more comprehensive phytohormone network, it is assumed that SA is specifically involved in conferring resistance to biotrophs, while JA signaling is activated in response to necrotrophs (Beckers and Spoel, 2006; Hou et al., 2022). SA is furthermore involved in providing long-lasting systemic acquired resistance (SAR). Regarding the latter, it was shown that treating the leaves of A. thaliana plants with SA indeed also conferred SAR toward Fo5176 infection, since less severe disease symptoms were observed in the leaves of SA-treated plants (Edgar et al., 2006). It is noteworthy however, that up-regulation of the typical SA-responsive defense gene PATHOGENESIS-RELATED 1 (PR1) was only observed in the treated leaves, and not in the root, where actual infection occurs. Hence, the role of SAR could be to prevent spread of the disease, rather than infection. A potential link between SA and an effector-triggered immunity (ETI) response to FoCon infection may be inferred from the observation that phytoalexin deficient 4 (pad4) mutants are more sensitive to FoCon infection in an SA-dependent manner, but this has not been further substantiated (Diener and Ausubel, 2005). Transcriptomic data regarding the role of SA in protection against Fo5176 are inconclusive. Inoculation of plants with Fo5176 without prior SA treatment did not affect PR1 expression in the shoot at early stages of infection based on quantitative real-time PCR (qRT-PCR) data, while PR1 expression was slightly suppressed in the root, an observation corroborated by microarray data (Edgar et al., 2006; Kidd et al., 2011). In an RNA-Seq experiment, SA-dependent genes were mostly unresponsive to infection by Fo5176, though PR1 was also up-regulated in leaves during the early stages of infection [1 day post-inoculation (dpi)], when the fungus is supposedly in the biotrophic stage of infection, and was later down-regulated (6 dpi), possibly by an antagonistic effect of JA signaling, induced during the transition of the fungus to its necrotrophic stage of infection (Lyons et al., 2015). Curiously, silencing SA signaling by insertion of the SA draining NahG transgene from Pseudomonas putida or in the SA biosynthesis mutant sa induction-deficient 2 (sid2) reduced the severity of disease symptoms in response to infection by Fo5176 or FoCon (Delaney et al., 1994; Diener and Ausubel, 2005; Trusov et al., 2009). Hence, it is more likely that SAR and SA signaling aid in preventing progression of the infection/disease in the plant, rather than limiting the infection itself (Fig. 1).
Fig. 1.
The phytohormone network involved in the defense against Fo5176. The different hormones (SA, ET, ABA, JA, and FoJA) are shown in the top row. Identified receptors and signaling intermediates are indicated downstream of the specific hormones. The distinct transcriptional outputs observed in this network are indicated below. Black arrows and lines with bars indicate positive and negative effects, respectively.
Jasmonic acid
In contrast to SA, pre-treating the leaves of A. thaliana plants with JA did not result in acquired resistance to F. oxysporum, even though the JA-responsive gene PLANT DEFENSIN 1.2 (PDF1.2) was induced in such leaves. Again, no response to this leaf treatment was detected in the root (Edgar et al., 2006). Similarly, RNA-Seq data for 1 dpi and microarray data for 2 dpi showed a strong up-regulation of JA biosynthesis, signaling, and response gene expression in the shoot following infection by Fo5176, but only a slight induction in the root, with some defense genes even showing slight down-regulation in this tissue (Kidd et al., 2011; Chen et al., 2014; Lyons et al., 2015). Poncini et al. (2017) reported a minor up-regulation of JA biosynthesis [ALLENE OXIDE SYNTHASE (AOS)] and signaling (PR4) marker genes in roots at 2 dpi via fluorescent reporter lines. Regarding the observed down-regulation of JA-related genes in roots at early time points, Chen et al. (2014) showed that JASMONATE ZIM DOMAIN 8 (JAZ8), a repressor of JA-responsive gene expression, was among the most strongly up-regulated genes in the root following Fo5176 inoculation, indicating that this could be one reason for the weak response in this tissue compared with the leaf. The strongest up-regulation of JA-related genes in the roots was observed at 6 dpi, again at the switch from biotrophic to necrotrophic behavior by Fo5176, with several JA biosynthesis genes, signaling regulators [e.g. JASMONATE ZIM-DOMAIN (JAZ) family members], and JA-induced defense genes [e.g. PDF1.2 and DEFENSIN-LIKE (DEFL) family members] being up-regulated (Lyons et al., 2015). Thatcher et al. (2016a) also found that JAZ family genes (JAZ5, JAZ7, JAZ8, JAZ9, and JAZ10) were quickly (peaking at 3 h and 48 h) up-regulated in the root in response to Fo5176, and added that induction in leaves (JAZ6, JAZ7, JAZ8, JAZ9, and JAZ10) can only be observed at later time points, when symptoms become apparent. Interestingly, none of the mutants for these JAZ proteins showed any changes in Fo5176 susceptibility, except for an activation-tagged allele of JAZ7, jaz7-1D, which exhibited stronger wilt symptoms and reduced survival rate. However, a JAZ7 overexpression line did not reproduce these phenotypes, so the role of JAZ7 remains unclear (Thatcher et al., 2016a). JAZ7 interacts with the co-repressor TOPLESS, thus JAZ7 may function as a repressor in the JA signaling pathway in response to Fo5176 infection; however, due to the conflicting results reported by Thatcher et al. (2016a), this requires further examination.
Curiously, JA biosynthesis mutants such as aos do not exhibit an altered susceptibility to Fo5176, while the JA receptor mutant coronatine insensitive 1 (coi1) does not show any disease phenotypes in response to Fo5176 infection, despite still being colonized by the fungus, albeit at a reduced efficiency (Thatcher et al., 2009; Cole et al., 2014). COI1 is a co-receptor for the bioactive form of JA, (3R,7S)-jasmonoyl-l-isoleucine (JA-Ile), and JA-induced defense gene expression is indeed abolished in coi1 mutants (Thomma et al., 1998; Thatcher et al., 2009; Sheard et al., 2010). However, while no disease symptoms can be observed in the coi1 mutant in response to Fo5176 infection, the mutants are still susceptible to F. oxysporum f. sp. raphani (Thatcher et al., 2009; Cole et al., 2014). These observations indicate that the disease symptoms are not caused by endogenous JA signaling, but may be the result of a fungus-derived signal that hijacks the COI1-dependent JA signaling pathway in the plant. Indeed, the ff. spp. conglutinans and matthioli appear to secrete JA, JA-Ile, and jasmonoyl-l-leucine (JA-Leu), while these compounds were not detectable in filtrates from f. sp. raphani (Cole et al., 2014). In accordance with the absence of the typical leaf senescence and necrosis phenotypes normally caused by the wilt disease, senescence marker genes are down-regulated in the coi1 mutant. Accordingly, COI1 appears to play a dual role in response to pathogenic infection, one being to induce defense gene expression in response to endogenous JA signaling, and another being the induction of senescence and thus disease symptoms, potentially via the senescence-associated protein SEN1, in response to F. oxysporum-derived JA (Fig. 1) (Schenk et al., 2005; Thatcher et al., 2009; Cole et al., 2014). Regarding the former, it is somewhat surprising then that coi1 mutants are not more susceptible to colonization, as their defense genes should at least be partially suppressed (Thatcher et al., 2009; Cole et al., 2014). The observation that coi1 mutants do not exhibit the typical wilt symptoms prior to death also indicated that wilt symptoms are an insufficient proxy for resistance or susceptibility of plants to infection by Fo5176. Colonization efficiency, disease symptoms, and survival rates need to be scored independently to properly assess a plant’s susceptibility to fungal infection.
Ethylene
Ethylene (ET) and JA often act synergistically in the plant’s defense network, and several ETHYLENE RESPONSE FACTORs (ERFs) appear to act downstream of the JA receptor COI1 and the ETHYLENE RECEPTOR (ETR) to integrate the two pathways (Fig. 1) (Li et al., 2019). ERF1 is one such integrator, as ERF1 expression is induced in response to infection by FoCon, and this induction is abolished in ET or JA receptor mutants (ein2-5 or coi1-1) (Berrocal-Lobo and Molina, 2004). ERF1 is a positive regulator of defense gene activation, such as of PDF1.2, and overexpression enhances both PDF1.2 expression and resistance to FoCon (based on plant fresh weight after inoculation with the fungus) (Berrocal-Lobo and Molina, 2004). ERF2 and 4 are also induced by Fo5176, but while erf4-1 mutants are more resistant to Fo5176 than the wild type, and ERF4 overexpression results in susceptibility, ERF2 overexpression increases the plant’s resistance to Fo5176 (McGrath et al., 2005). Hence, ERF4 is a negative regulator of Fo5176 resistance whereas ERF2 acts as a positive regulator. This opposite effect is also reflected in PDF1.2 defense gene expression, which is positively regulated by ERF2 and negatively by ERF4 (McGrath et al., 2005). In both cases, susceptibility was scored via wilting symptoms, and thus may also be influenced by altered JA signaling. ERF4 may furthermore integrate SA signaling, as PR1 expression was also elevated in erf4-1 mutants (Edgar et al., 2006). The increased resistance of the erf4-1 mutant to Fo5176 may therefore be the result of elevated SA- and JA-dependent defense gene activation (McGrath et al., 2005; Edgar et al., 2006). Another ERF involved in defense against Fo5176 is ERF14, which may act upstream of ERF1 and 2. ERF14 induces the expression of those two ERF genes, as well as JA- (PDF1.2), SA- (PR1), or ET-inducible (CHITINASE B) defense genes (Oñate-Sánchez et al., 2007). ERF14 itself is induced by ET and is required for resistance to Fo5176, as erf14 mutants are more susceptible to infection by Fo5176, based on a reduced survival rate following inoculation with the fungus (Oñate-Sánchez et al., 2007). These ERF-mediated responses probably capture a very early response, as the induction occurred within the first 24 h post-inoculation. However, it was not clear if the induction was measured in roots or shoots. Nevertheless, based on the complementary data available on PDF1.2 and PR1 expression, this induction most probably occurred primarily in leaves. While ERF14 is likely to be induced through an ET-dependent pathway, ERF1 and PDF1.2 expression may be the result of ET/JA crosstalk, while induction of PR1 could result from ET/SA crosstalk (Fig. 1).
Abscisic acid
Abscisic acid (ABA) appears to antagonize JA/ET signaling with respect to the plant’s defense pathways (Anderson et al., 2004). Exogenous application of ABA suppresses JA/ET-responsive gene expression (e.g. PDF1.2and PR4) even in the presence of applied JA or ET, and in an ABA biosynthesis mutant (aba2-1) these genes are constitutively up-regulated (Anderson et al., 2004). Inoculation with Fo5176 rapidly induces the expression of JASMONATE INSENSITIVE 1 (JIN1; aka MYC2), a positive regulator of ABA, and negative regulator of JA/ET signaling. Expression peaks between the first 6 h and 12 h after inoculation with the fungus, and subsequently declines (Anderson et al., 2004). jin1-9 and aba2-1 mutants both appear to be more resistant to Fo5176 when scoring wilt symptoms, and JA/ET-responsive defense genes (PDF1.2 and PR4) are constitutively up-regulated in these mutants. In JIN1-overexpressing lines, on the other hand, PDF1.2 expression is suppressed in an ERF1-dependent manner (Anderson et al., 2004). Addition of exogenous ABA to the JIN1-overexpressing line reduces PDF1.2 expression even further, indicating that ABA affects expression not just via JIN1. Also, fittingly, ABA could still suppress PDF1.2 in the jin1-9 mutant (Anderson et al., 2004). Thus, ABA negatively regulates JA/ET-dependent defense gene expression in response to infection by Fo5176, possibly via positive regulation of ERF4 (Anderson et al., 2004; Yang et al., 2005). Conversely, ABA-responsive genes are de-repressed in ET signaling mutants, which are furthermore hypersensitive to exogenous ABA application. This implies that ET in turn negatively affects ABA-dependent gene expression (Fig. 1) (Anderson et al., 2004). Hence, the increased resistance of jin1-9 and aba2-1 mutants to Fo5176 may be the result of increased JA/ET-related defense gene expression. However, this raises the question of why Fo5176 colonization of the root results in ABA-dependent JIN1 expression, if this decreases the plant’s fitness and resistance. In this respect, one should note that Fo5176 may synthesize its own ABA compound as an effector to suppress the host’s immune response during the early stages of infection. This has been demonstrated for F. oxysporum f. sp. lycopersici and other pathogenic fungi (Dörffling et al., 1984). Similar assessments are so far lacking for Fo5176 and FoCon, but it is conceivable that the observed JIN1-dependent suppression of JA/ET defense gene expression may at least in part stem from Fo5176 hijacking this pathway to promote pathogenesis of the fungus.
Auxin
So far, no direct role for auxin has been established in the defense against Fo5176, but in a microarray analysis of the leaf transcriptome 48 h after inoculation of the plant with the pathogen, a whole set of tryptophan, indole-3-acetic acid (IAA), and indole-3-methyl-glucosinolate biosynthesis and metabolism genes were up-regulated (Kidd et al., 2011). The auxin signaling mutants auxin resistant (axr) 1, 2, and 3 showed slightly increased resistance to Fo5176, scored by a delay in wilt symptom appearance, as did two auxin transport mutants (auxin resistant 1 and transport inhibitor response 3) (Kidd et al., 2011). However, neither the analysis of auxin biosynthesis mutants, nor overexpressing lines or exogenous auxin application resulted in any observable differences in resistance or susceptibility to the fungus (Kidd et al., 2011). Thus, it is more likely that the observed expression changes in tryptophan and auxin biosynthesis pathway genes reflect a general shift of the plant’s metabolism from development to defense, rather than an indication that auxin is directly involved in the defense against Fo5176.
The plant’s Fo5176 detection system
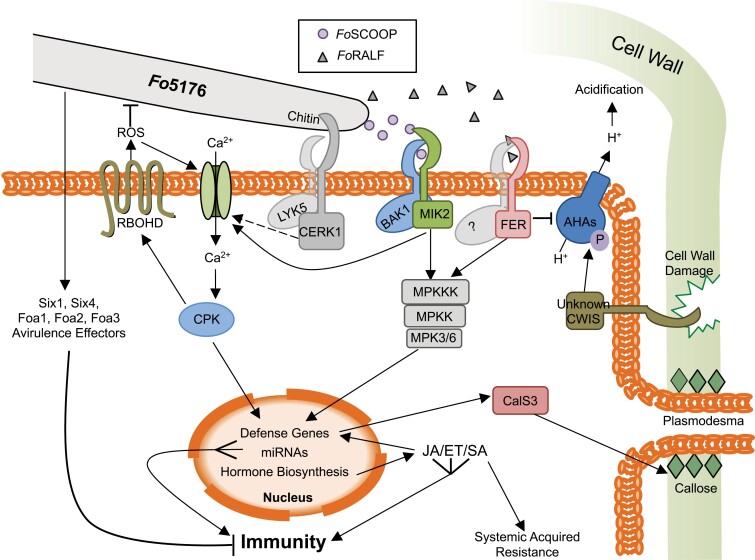
Plants can sense the presence of pathogens by a wide range of receptors (Fig. 2) (Ngou et al., 2022). Pattern recognition receptors recognize certain molecular patterns on the surface of a pathogen, such as the fungal cell wall polysaccharide chitin, which is detected by the plant’s CHITIN ELICITOR RECEPTOR KINASE 1 (CERK1) and its co-receptor LYSIN MOTIF RECEPTOR KINASE 5 (LYK5) (Cao et al., 2014; Yang et al., 2022). Since chitin is part of both pathogenic and harmless fungi, it is generally considered a microbe-associated molecular pattern (MAMP), rather than a pathogen-associated molecular pattern (PAMP), and, fittingly, CERK1 expression is suppressed by both the pathogenic Fo5176 and endophytic Fo47 to allow for colonization of the plant by these strains (Fig. 2) (Guo et al., 2021). Other receptors sense a pathogen indirectly by the damage they inflict, for example by sensing plant cell wall debris, produced when a pathogen breaches the cell wall [those elicitors are referred to as damage-associated molecular patterns (DAMPs)]. Additionally, cell wall integrity sensors typically are connected to the cell wall with their receptor domain, and can perceive instability induced in the wall by the pathogen.
Fig. 2.
Intracellular pathways involved in the plant’s defense response to Fo5176 infection. The Fo5176 hyphae are sensed in the apoplast by plasma membrane-localized receptors. CERK1/LYK5 bind to the fungal cell wall polysaccharide chitin, a MIK2/BAK1 receptor complex binds Fo5176-derived FoSCOOP12 peptides, while FER (with an unknown co-receptor) binds FoRALF peptides. An unknown cell wall integrity sensor (CWIS) senses the damage caused by Fo5176. Downstream, CERK1/LYK5, MIK2/BAK1, and FER may activate calcium channels and the MAP kinase cascade via phosphorelays from their kinase domains. MAP kinase cascade activation leads to defense gene activation in the nucleus. Calcium (Ca2+) influx via the activated calcium channels activates CPKs, which in turn activate RBOHD and defense gene expression. RBOHD releases ROS into the apoplast, which amplifies the Ca2+ influx and is toxic to Fo5176. The CWISs activate AHA proton (H+) pumps via phosphorylation (P) to acidify the apoplast in defense against Fo5176. FoRALF signaling via FER could antagonize this pathway by deactivating the AHAs, leading to alkalization of the apoplast. Defense genes activated in the nucleus include hormone (JA/ET/SA) biosynthesis genes, which activate these hormone pathways, in turn activating more defense genes, such as PDF1.2 or PR1, as well as systemic acquired resistance via SA. Expression of CalS3 leads to callose deposition at the plasmodesmata and blockage of cell to cell movement of the pathogen. Fo5176 releases Avr effectors (Six, Foa) into the cell, which counteract these defense responses and dampen the plant’s immune response.
WAKs and RFO1
A group of potential cell wall integrity sensors are the WALL-ASSOCIATED KINASEs (WAKs), and indeed several WAK family genes were identified as up-regulated in response to Fo5176, among them WAK1, 3, and 10 (Zhu et al., 2013). However, no further functional analysis has been done for these family members. The first WAK family member identified as a receptor for Fo5176 was RESISTANCE TO FUSARIUM OXYSPORUM 1 [RFO1; aka WALL-ASSOCIATED KINASE-LIKE 22 (WAKL22)]. The A. thaliana natural accession Ty is susceptible to infection by F. oxysporum f. sp. matthioli, while Col is not (Diener and Ausubel, 2005). Six loci (RFO1–ROF6) were identified to contribute to this resistance of Col, with RFO1 being the main locus responsible, since introduction of the Col RFO1 allele into Ty is sufficient to confer resistance to this accession (Diener and Ausubel, 2005). In contrast, the RECEPTOR-LIKE PROTEIN (RLP) RFO2 does not seem to contribute to resistance in Col, and resistance conferred to Ty by RFO2 is dependent on the RFO1 locus (Diener and Ausubel, 2005; Shen and Diener, 2013). Col rfo1 mutants are still resistant to f. sp. matthioli, however, indicating that resistance to f. sp. matthioli in Col is conferred by more than one locus (Diener and Ausubel, 2005). RFO1 expression is suppressed during colonization by both Fo5176 and the endophyte Fo47, indicating that it acts to generally suppress colonization (Guo et al., 2021). RFO3 is a receptor-like kinase that is expressed in the root vasculature of Col plants and confers resistance to F. oxysporum f. sp. matthioli, but does not seem to play a role in resistance to FoCon (Cole and Diener, 2013). As a WAK family protein, RFO1 may be involved in sensing pathogen-inflicted damage to the cell wall as a cell wall integrity sensor, but to date no ligand has been identified. It is somewhat curious that the role of RFO1 in resistance to F. oxysporum still remains unclear. However, the evidence gathered so far, especially the observation that rfo1 mutants in accessions other than Ty are still resistant to F. oxysporum f. sp. matthioli, points to a more indirect role for RFO1, rather then it being a direct receptor for a Fo5176-derived signal.
THE1, MIK2, and FER
Further potential cell wall integrity sensors are the malectin-like Catharanthus roseus receptor-like kinase 1-like (CrRLK1L) family members THESEUS1 (THE1), HERCULES1 (HERK1), and FERONIA (FER), as well as the LRR-RK MALE DISCOVERER 1-INTERACTING RECEPTOR LIKE KINASE 2 (MIK2). Among those, HERK1 has been found to be up-regulated in response to sensing both Fo5176 and the non-pathogenic endophyte Fo47, but no follow-up work has been reported so far (Guo et al., 2021). THE1 and MIK2 were initially shown to link cell wall integrity sensing to a transcriptional defense output (van der Does et al., 2017): treating plants with isoxaben results in reduced cell wall integrity via the inhibition of cellulose biosynthesis (Heim et al., 1991). This chemically induced cell wall weakening leads to JA/SA accumulation, lignin deposition, and an up-regulation of several immunity marker genes, such as FLG22-INDUCED RECEPTOR-LIKE KINASE 1 (FRK1) or At1g51890 (van der Does et al., 2017). mik2-1 and the1-1 mutants are impaired in this defense response, indicating that these receptors may act as cell wall integrity sensors. Additionally, both mutants exhibit increased susceptibility to infection by Fo5176, with more severe wilt symptoms and a reduced survival rate observed, though the results obtained for the1 are not as clear as those for mik2-1 (van der Does et al., 2017). This may be explained by THE1 being a more general cell wall integrity sensor, while MIK2 could play a role more specific in the defense against Fo5176. Indeed, recent work indicates a direct role for MIK2 in sensing Fo5176 (Hou et al., 2021; Rhodes et al., 2021). Plant SERINE RICH ENDOGENOUS PEPTIDEs (SCOOPs) are phytosulfokines involved in signaling pathways combating biotic and oxidative stress (Gully et al., 2019). Overexpression of A. thaliana SCOOP27 [named ENHANCER OF VASCULAR WILT RESISTANCE 1 (EWR1)] enhances the plant’s resistance to colonization by Fo5176 and F. oxysporum f. sp. raphani (Yadeta et al., 2014; Zhang et al., 2022). Furthermore, exogenous addition of synthetic SCOOP27 or SCOOP12 to A. thaliana seedlings induces immune responses such as defense gene expression (FRK1), reactive oxygen species (ROS) burst, callose deposition, and a reduction in root growth in a BAK1-dependent manner (Gully et al., 2019; Zhang et al., 2022). Eventually, a receptor complex consisting of MIK2 and BAK1 was identified as the direct receptor for SCOOP12 and 27 in A. thaliana (Hou et al., 2021; Rhodes et al., 2021; Zhang et al., 2022). Moreover, Rhodes et al. (2021) and Hou et al. (2021) show that the F. oxysporum genome encodes cytosolic proteins with SCOOP peptide-like sequences on their surface, that are transcriptionally up-regulated in response to inoculation with A. thaliana roots (Fig. 2) (Hou et al., 2021; Rhodes et al., 2021). When testing synthetic versions of these Fusarium SCOOP (FoSCOOP) peptides, the authors find that the SCOOP10 homolog of F. langsethiae, as well as FoSCOOP12 from Fo5176 and F. oxysporum ff. spp. lycopersici and cubense are perceived by MIK2 to trigger defense responses, while foscoop mutant strains of Fo5176 exhibit elevated virulence in A. thaliana (Hou et al., 2021; Rhodes et al., 2021). Thus, it is a possibility that A. thaliana senses Fo5176 via these SCOOP-like proteins.
MIK2, together with FER, was also identified as a receptor involved in triggering immune responses to a crude elicitor mix, produced from lyophilized, ground up mycelia of F. oxysporum isolate 62292 (Coleman et al., 2021). Treating A. thaliana plants with this mix induced defense gene expression (FRK1), ROS burst, MAP KINASE (MPK) 3/6 phosphorylation, and calcium influx (Coleman et al., 2021). However, ROS burst and calcium influx were reduced in bak1, fer-4, and mik2 mutants. Transient expression of A. thaliana MIK2 in Nicotiana benthamiana plants furthermore conferred sensitivity of this plant to the crude elicitor mix, confirming that MIK2 is a receptor to a component of the mix (Coleman et al., 2021). In this case, MIK2 and FER most probably recognize a Fusarium-derived molecule present in the elicitor mix, but the exact compound detected by these receptors remains to be determined.
Expression of FER has furthermore been shown to be suppressed in response to colonization by the endophytic Fo47 but not Fo5176, which could allow for colonization of the plant by the endophyte, while restricting colonization by the pathogen (Guo et al., 2021). Masachis et al. (2016) previously observed that growing tomato plants progressively acidify the medium surrounding their roots. In the presence of F. oxysporum f. sp. lycopersici, this acidification was not observed, but instead the pH increased. This shift to a more alkaline pH correlated with an increased pathogenicity of the fungus, indicating that F. oxysporum may actively alkalize the soil around roots to facilitate efficient colonization (Masachis et al., 2016). In plants, cysteine-rich RAPID ALKALINIZATION FACTOR (RALF) peptides have a wide range of physiological functions, sometimes achieved by increasing the apoplastic pH (Abarca et al., 2021). Analyses of the F. oxysporum genome revealed that several strains indeed carry potential homologs to such RALF peptides (Masachis et al., 2016; Thynne et al., 2017). Adding a synthetic FoRALF to the growth medium led to alkalinization and growth inhibition of both tomato and A. thaliana seedlings (Masachis et al., 2016). In contrast, foralf mutants were no longer able to alkalinize the growth medium. Furthermore, A. thaliana immune marker genes (e.g. WRKY53 and PDF1.2) were expressed at higher levels during colonization of the plant by this mutant fungus, indicating that FoRALF normally suppressed the immune response in the plant to facilitate efficient colonization (Masachis et al., 2016). CrRLK1L receptor kinases are typical receptors for RALF peptides in A. thaliana, and indeed fer-4 mutants are insensitive to FoRALF peptide application with regards to growth inhibition (Masachis et al., 2016; Abarca et al., 2021). Exogenous alkalinization of the growth medium still inhibits root growth of the fer-4 mutant, however, indicating that FER acts upstream of this alkalinization response. In line with these observations, fer-4 plants show increased resistance to infection by FoCon, and hyphae growing in the mutant were regularly observed to undergo cell death. Contributing to this increased resistance is a constitutive expression of defense genes, such as FRK1, WRKY53, and PDF1.2 in the fer-4 mutant (Masachis et al., 2016). Thus, it appears that FoRALF targets A. thaliana FER to increase the apoplastic pH, thereby allowing for optimal colonization of the plant by F. oxysporum (Fig. 2) (Masachis et al., 2016).
Cell wall integrity sensing
While alkalinization of the apoplast may be the result of F. oxysporum hijacking a plant signaling pathway to facilitate colonization, manipulation of the apoplastic pH could also be a defense strategy employed by the plant. Infection of A. thaliana by Fo5176 was shown to induce acidification of the apoplast via the activation of Arabidopsis H+-ATPase (AHA) proton pumps (Kesten et al., 2019). The cellulose biosynthesis mutant companion of cellulose synthase 1, 2 has a constitutively acidic apoplast, and this mutant is more resistant to Fo5176. Similarly, several other cellulose synthesis mutants (e.g. cellulose synthase 3 and 6, korrigan, cobra, and procuste1) are more resistant to infection by Fo5176 (Menna et al., 2021). This effect seems to be independent of JA signaling, as double mutants of these cellulose synthesis mutants with jasmonate biosynthesis (aos) or signaling (coi1) mutants showed no altered response. However, combination with an ein2-5 mutant restored the Fo5176 sensitivity, indicating that this response is ethylene signaling dependent (Menna et al., 2021). How such cell wall biosynthesis mutants signal to the AHAs to modulate the plant’s apoplastic pH remains unclear, but most probably the signal originates from certain cell wall integrity sensors, such as the WAKs (Fig. 2).
Downstream of the detection system
Oxidases and peroxidases
ROS are highly reactive forms of oxygen that are produced by plants in response to a wide range of stresses. The production of a ROS burst in the apoplast is a typical marker for an activated immune response, and the produced ROS serve to induce cell wall fortifications, can act as signaling molecules to warn neighboring cells, and are also directly toxic to the pathogen (Waszczak et al., 2018). Apoplastic ROS are produced by NADPH oxidases and apoplastic peroxidases, which are activated in a calcium-dependent manner (Davies et al., 2006; Kadota et al., 2015). Early work with A. thaliana and a crude elicitor mix from F. oxysporum f. sp. matthioli suggested that F. oxysporum is also able to trigger this response, and subsequent work showed that inoculation of A. thaliana roots with Fo5176 induces the expression of the NADPH oxidase genes RBOHD and RBOHF, as well as the peroxidase gene PRX33 (Davies et al., 2006; Zhu et al., 2013; Lyons et al., 2015). Furthermore, Guo et al. (2021) found that the endophytic strain Fo47 suppressed RBOHD expression to successfully colonize the root, indicating that RBOHD normally acts to restrict colonization. Interestingly, RBOHD and PRX33 seem to act antagonistically to RBOHF, as rbohd and prx33-1 mutants are more resistant to infection by Fo5176, while rbohf mutants are more susceptible (Zhu et al., 2013; Lyons et al., 2015). PRX33 is specifically expressed in guard cells, hence the role of PRX33 may be to facilitate the closure of stomata in response to fungal infection, while RBOHD plays a more general role (Kadota et al., 2015; Arnaud et al., 2017). Detection of Fo5176 could hence activate RBOHD to generate a ROS burst in the apoplast (Fig. 2). However, rbohd mutants show less severe disease symptoms and have a higher survival rate after inoculation with Fo5176 (Zhu et al., 2013). Since RBOHD is assumed to function in defense against pathogens, and is up-regulated in response to Fo5176 infection, it is somewhat surprising that the mutants are more resistant to the pathogen. A possible explanation for this comes from the observation that rbohd mutants appear to accumulate excess SA, and have up-regulated ET signaling, which could partly negate the effects of the mutation (Kadota et al., 2015).
Heterotrimeric G proteins and MLO proteins
Heterotrimeric G proteins serve as intracellular signal transducers for membrane-localized G protein-coupled receptors, and consist of three subunits, Gα (encoded by GPA1), β (encoded by AGB1), and γ (encoded by AGG1 and AGG2) (Zhong et al., 2019). Such Gβγ complexes often function to integrate several different signaling pathways, and in regards to combating infection by Fo5176 they may be involved in integrating hormone pathways. The β-subunit mutants agb1-1 and agb1-2, as well as the γ-subunit mutant agg1-1c are more sensitive to Fo5176 or FoCon compared with the wild type, while the α-subunit mutant gpa1-4 and the γ-subunit mutant agg2-1 were unaffected (Llorente et al., 2005; Trusov et al., 2006, 2007). Interestingly, infection with Fo5176 leads to increased AGB1 and AGG1 expression in leaves, but not in the root, while AGG2 expression was slightly reduced in leaves (Trusov et al., 2007). The agb1-2 and agg1-1c mutants furthermore showed impaired JA-related responses, such as a lack of PDF1.2 up-regulation in response to exogenous JA treatment (Trusov et al., 2006, 2007). This suppression of JA responses in agb1-2 may be due to an up-regulation of JIN1 in the mutant (Trusov et al., 2009). From these studies, it can be concluded that Gβγ2 trimers are not involved in this immune pathway, while Gβγ1 trimers could signal to suppress JIN1 expression, thereby indirectly activating JA/ET-dependent defense gene expression (Fig. 1) (Trusov et al., 2007, 2009).
MILDEW RESISTANCE LOCUS O (MLO) proteins are calcium-activated seven transmembrane proteins that are generally considered to be susceptibility factors (Jacott et al., 2021). Initially identified as a mutant in barley that conferred resistance to powdery mildew fungi, it has since been established that MLO genes, which are conserved across the plant kingdom, act in a wide range of biological processes (Freisleben and Lein, 1942; Jacott et al., 2021). The overbearing principle uniting these processes is that some form of physical stimulus is involved (Jacott et al., 2021). The A. thaliana mlo2 mlo6 mlo12 triple mutant is resistant to powdery mildew, but generally shows different responses to a variety of plant-colonizing microbes, ranging from resistance to greater susceptibility (Consonni et al., 2006; Acevedo-Garcia et al., 2017). With regards to Fo5176, mlo2 mlo6 mlo12 is more susceptible to infection, with increased chlorosis and wilt symptoms. These observed differences in infection outcome by different microbes may depend on the mode of entry, with plants being more resistant toward pathogens that enter via direct penetration, and more susceptible to pathogens that enter via openings such as stomata or wounds (Acevedo-Garcia et al., 2017). As MLO also functions in the regulation of cell death, the enhanced disease symptoms observed in response to Fo5176 may be the result of a deregulation of this process in the triple mutant (Piffanelli et al., 2002; Acevedo-Garcia et al., 2017). In a recent study it was furthermore found that several MLO proteins, including MLO2 and 12, act as transmembrane calcium channels in a FER and RALF-dependent manner (Gao et al., 2022). Thus, the change in susceptibility to Fo5176 may be the result of impaired calcium signaling in the triple mutant.
Transcriptional regulators
The MEDIATOR complex is a cofactor for the transcriptional machinery that integrates signaling information from transcription factors to direct the activity of RNA polymerase II (Dolan and Chapple, 2016). The role of the MEDIATOR complex downstream of Fo5176 perception has been analyzed because plants with mutations in MEDIATOR18, 20, and 25 [aka PHYTOCHROME AND FLOWERING TIME1 (PFT1)] show increased resistance to Fo5176, based on less severe wilt symptoms and reduced colonization by the fungus (Kidd et al., 2009; Fallath et al., 2017). RNA-Seq analysis revealed an enrichment of immunity-related genes that were no longer induced in the med18 and 20 mutants, with candidates such as FLAGELLIN-SENSING 2 (FLS2), MPK3, PROPEP1, and several jasmonate-related genes (JIN1, JAZ1, 5, 7, 8, and 10, and AOS) being expressed at lower levels than in the wild type. Conversely, SA-dependent genes such as PR1 and 5 were up-regulated in the mutants (Fallath et al., 2017). Similar results were obtained for med25 by qRT-PCR, even though in this case JA- and SA-related genes were both down-regulated in the mutant (Kidd et al., 2009). The effects of the med25 mutant were furthermore enhanced by an additional mutation in med8 (Kidd et al., 2009). ROS-related genes, such as RBOHD, were only up-regulated in med20, but not in med18. MED18 acts, at least in part, by activating WRKY33 expression (Liao et al., 2016). WRKY33 is an activator of SA signaling and a suppressor of JA signaling, and hence could be responsible for the effects observed here (Birkenbihl et al., 2012). However, in the RNA-Seq data for med18, WRKY33 was not differentially expressed compared with the wild type (Fallath et al., 2017). From these observations, it appears that the MEDIATOR complex may be involved in integrating the signals from the different hormone pathways to produce the necessary transcriptional output to activate the plant’s defense pathways. Defense genes such as FLS2, MPK3, or PROPEP1 may well be among the targets, just like the different JAZ genes, which typically act as repressors of JA-induced defense genes and serve to fine-tune the output of the JA signaling pathway (Pauwels and Goossens, 2011; Fallath et al., 2017). However, the exact mode of action, as well as a possible role for WRKY33 in this pathway, still requires rigorous assessment.
A specific transcription factor identified to act in the defense pathways against Fo5167 is LATERAL ORGAN BOUNDARY DOMAIN 20 (LBD20). lbd20 mutants exhibit reduced wilt symptoms and a greater survival rate when challenged with Fo5176 (Thatcher et al., 2012b). The LBD20 gene is expressed specifically in the root, and is rapidly (3 h) induced in response to inoculation with Fo5176. This induction is JA and COI1 dependent, as both Fo5176- and exogenous JA-induced expression were suppressed in coi1 and jin1 mutants. Conversely, JIN1 overexpression also induces LBD20 (Thatcher et al., 2012b). In lbd20 mutants, some JA-dependent defense genes were more strongly induced than in the wild type [e.g. THIONIN2.1 (THI2.1) and VEGETATIVE STORAGE PROTEIN2 (VPS2)], while others were unaffected (PDF1.2), and overexpression of LBD20 resulted in suppression of JA-induced expression of THI2.1 and VPS2, but not PDF2.1. These observations suggest that LBD20 is a negative regulator for a subset of JA-dependent defense genes (Thatcher et al., 2012b).
MicroRNAs
miRNAs are also involved in regulating the plant’s response to pathogens and, when analyzing the plant’s transcriptome via RNA-Seq at 6 dpi with FoCon, several miRNAs were among the differentially regulated genes (Zhu et al., 2013). A total of 56 miRNA families, representing 25% of the total miRNA genes, showed up in the expression dataset. Out of all of those miRNAs, MIR398b and MIR398c, as well as MIR159b, stood out because their target genes were also detected as differentially regulated, indicating a functional significance of their altered expression (Zhu et al., 2013). MIR398b and MIR398c were both up-regulated, and their predicted targets were down-regulated, while MIR159b and its target were both induced (Zhu et al., 2013). However, no further function has been assigned to these miRNAs and their respective targets in contributing to resistance against Fo5176.
miR396, miR773, and miR858 have also been identified as potentially involved in the defense against Fo5176, because knocking out these three miRNAs increases the plant’s resistance to FoCon (Soto-Suárez et al., 2017; Camargo-Ramírez et al., 2018; Salvador-Guirao et al., 2018). Fittingly, infection with FoCon results in a down-regulation of MIR773 in shoots and roots, potentially to increase plant resistance (Salvador-Guirao et al., 2018). In the case of the miR858 knockout, the enhanced resistance may be the result of an increase in production of flavonoids and phenylpropanoid compounds with anti-fungal activity (naringenin, kaempferol, and p-coumaric acid), since genes for these biosynthetic products are normally among the targets of miR858 (Camargo-Ramírez et al., 2018).
The role of fungal avirulence effectors
Avirulence effector proteins (Avr) are utilized by most pathogenic microbes during the plant infection process. Fusarium oxysporum expresses and releases effector proteins once it has reached the xylem cells (de Sain and Rep, 2015; Redkar et al., 2022). The release is strictly dependent on the plant cell still being alive, indicating that a plant-derived molecule is a trigger for Avr expression (van der Does et al., 2008). Such effectors typically target the plant’s immune system to suppress the host’s defense response and facilitate colonization. At the same time, plants have evolved receptor proteins to detect such effectors and induce the appropriate defense response to combat the pathogen, in a prime example of an evolutionary arms race (de Sain and Rep, 2015). The best studied F. oxysporum-derived effectors are the small cysteine-rich SECRETED IN XYLEM (Six) family proteins, which were initially identified in the xylem sap of tomato plants infected by F. oxysporum f. sp. lycopersici (Rep et al., 2002; Takken and Rep, 2010). Six effectors are detected by the tomato plant’s IMMUNITY (I) gene products (R gene equivalents) (de Sain and Rep, 2015). The first tomato Six gene identified was Six1, and since the corresponding avirulence effector is detected by the I-3 gene product, it is also called Avr3 (Rep et al., 2004). Comparing the genome sequences of Fo5176 with that of F. oxysporum f. sp. lycopersici, Thatcher et al. (2012a) identified putative homologs for Six1, Six4, Six8, and Six9. Since the Fo5176 and f. sp. lycopersici Six4 homologs are 99.2% identical, and six4 mutants of F. oxysporum f. sp. lycopersici are no longer able to suppress the plant’s immune response, the authors focused on this gene for further analysis. Six4 is highly expressed during A. thaliana infection by Fo5176, and Fo5176 six4 mutants exhibit impaired virulence, pointing to a role for Fo5176 Six4 in suppressing the plant immune system and facilitating colonization (Thatcher et al., 2012a).
In order to identify and characterize additional Avr effectors of Fo5176, Tintor et al. (2020) further analyzed the Fo5176 genome sequence and, in addition to the four previously described Six genes, identified four novel effector candidates designated FoaEffector1–4 (Foa1–Foa4). When expressed transiently in N. benthamiana leaves, Foa2 and Foa3 suppressed the flg22- and chitin-induced ROS burst, while Six1 and Foa1 suppressed the flg22-induced burst, but not the chitin-induced burst. Additionally, Six1 and Foa1 had to be targeted to the apoplast to suppress the immune response, while Foa2 and 3 acted both in the apoplast and intracellularly (Tintor et al., 2020). This mode of action was confirmed in A. thaliana for Foa2, where flg22- and chitin-induced ROS burst and MPK3/6 phosphorylation were dampened in the presence of the Foa2 transgene. Using an elegant in vivo effector labeling approach, the authors furthermore confirmed that Foa2 and 3 are injected into the cell by Fo5176, while Six1 and Foa1 could only be detected in traces intracellularly, thereby confirming their action in the apoplast (Tintor et al., 2020). The function of Six4 could not be clearly determined in this study, but the authors could not confirm its earlier described pattern-triggered immunity (PTI) suppression function (Thatcher et al., 2012a). Thus, the exact mode of action of Six4 still requires further elucidation, while Six1, Foa1, Foa2, and Foa3 are all immune system-suppressing effectors (Fig. 2).
Transcriptomic datasets
Several large-scale transcriptomic datasets are available as valuable resources to study the response of A. thaliana to infection by Fo5176 or FoCon, and data from these sets have been used and referenced throughout this article (Table 1). Kidd et al. (2011) employed leaf tissue from Fo5176-infected A. thaliana plants (2 dpi) in an Affymetrix microarray experiment. The authors found that genes involved in JA-related processes, as well as tryptophan- and IAA-related biosynthesis are enriched among the up-regulated genes. Chen et al. (2014) later complemented this study with a microarray experiment analyzing root tissue at 2 dpi. They discovered that the root transcriptome is characterized by a down-regulation of defense genes, rather than an up-regulation as observed in shoot tissue. They also found that the genes suppressed upon infection with Fo5176 are typically also down-regulated in response to exogenous flg22 addition or Pseudomonas syringae infection. Among the down-regulated genes, the authors identified ERF72, and demonstrated that erf72 mutants were more resistant to Fo5176 infection. ERF72 was previously shown to act downstream of MPK3/6 to activate defense gene expression in response to the necrotropic fungus Botrytis cinerea, so a similar role is conceivable during Fo5176 infection, as these two MPKs are also activated in response to Fo5176 (Chen et al., 2014; Rhodes et al., 2021; Li et al., 2022). Zhu et al. (2013) performed RNA-Seq on whole-plant tissue, and thus could not differentiate between root- and shoot-specific responses. They did compare early and late time points, however (1 and 6 dpi). In addition to general immunity genes, they found several groups of genes to be enriched among the differentially expressed genes, including WAK1, 3, 10, WRKY51, 45, 63, 75, Lectin receptor kinases, TIR-NBS-LRR class genes, cytochrome P450 genes CYP70, 71, 82, and MYB15, 112, and 113 transcription factor genes. Furthermore, they studied different members of the NADPH oxidase family, such as RBOHD. Finally, Lyons et al. (2015) performed comparative RNA-Seq for root and leaf tissue of A. thaliana seedlings 1 and 6 dpi with Fo5176. The authors found that there is a tissue-specific response to the pathogen, with ~30% of the differentially expressed genes common to both root and leaf samples across the time points analyzed. Conversely, >50% of differentially expressed genes are tissue specific. Among the induced genes were JA-, ROS-, and fungal defense-related genes, while the repressed genes are primarily related to general biosynthesis- and metabolomic-related processes and photosynthesis (Lyons et al., 2015). They also identified CALLOSE SYNTHASE 3 as strongly inhibited at 6 dpi. As this enzyme is required for callose depositions at lateral root emergence sites, it may be important for closing the plasmodesmata around this typical area of Fo5176 entry (Fig. 2) (Vatén et al., 2011; Lyons et al., 2015). Thus, it seems counterintuitive that expression of this gene is suppressed, rather than induced. However, since 6 dpi is a time point long after infection/colonization has taken place, the down-regulation at this stage may be a response to successful callose deposition and plasmodesmata closure. In leaves, the ethylene response factor RAP2.6 is strongly up-regulated, as are JA-induced defense genes. Interestingly, plant senescence markers (SAG29 and SAG12) are initially induced in leaves at 1 dpi, but then repressed at day 6. This could indicate an active suppression of senescence by the fungus during the switch from the biotrophic to the necrotrophic stage of infection. Finally, the authors noted that the auxin-related genes ARF1 and 2 stand out as strongly repressed in the roots at 6 dpi. arf1-3 mutants have a slightly increased resistance to Fo5176, while arf2-6 and double mutants show strong resistance. Since ARF2 is involved in facilitating lateral root emergence, the resistance in this mutant may be indirectly caused by restricting entry of Fo5176 through lateral root emergence sites. Furthermore, the arf2 mutants also display delayed senescence, so down-regulation of this gene at 6 dpi may partly explain the repression of senescence markers at this later time point (Ellis et al., 2005; Lyons et al., 2015).
Table 1.
Available transcriptomic datasets
| Dataset | Focus of analysis | Tissue | Time points | Technique |
|---|---|---|---|---|
| Kidd et al. (2011) | Phytohormones, auxin | Leaves | 2 dpi | Microarray |
| Zhu et al. (2013) | NADPH oxidases | Whole seedling | 1 dpi, 6 dpi | RNA-Seq |
| Chen et al. (2014) | Phytohormones | Roots | 2 dpi | Microarray |
| Lyons et al. (2015) | Comparative analysis of leaf and root responses, as well as early and late time points. Phytohormones | Leaves and roots | 1 dpi, 6 dpi | RNA-Seq |
| Guo et al. (2021) | Comparative analysis of plants inoculated with Fo5176 or Fo47, as well as across several time points. Transcriptomic responses of the fungus. | Root and fungal | Several points from 12 to 96 hpi | RNA-Seq |
MAMP versus PAMP
How plants can engage with beneficial microbes while at the same time defending themselves against pathogens is one of the major unanswered questions in the field of molecular plant–microbe interactions (Harris et al., 2020; Thoms et al., 2021). Beneficial, pathogenic, and incompatible microbes all share some common features that are recognized by the plant. These are referred to as microbe-associated molecular patterns (MAMPs) and represent the first layer of the plant’s defense system. Once recognized by the MAMP perception system, endophytes will be allowed to interact with the plant, while pathogens are further recognized by their pathogen-associated molecular patterns (PAMPs), and recognition of these will result in a PTI response (Thoms et al., 2021). Pathogens that evade PAMP detection and the PTI response can release avirulence effectors into the host cell, thereby causing an ETI response by the plant, a heightened response that can eventually lead to targeted cell death to protect the surrounding cells (Ngou et al., 2022). How plants manage to maintain this delicate balance between promoting interaction with endophytes, while at the same time strictly excluding pathogens, is a highly interesting and important area of research. The interaction between A. thaliana and F. oxysporum provides a suitable model system to study these differences in plant–microbe interactions. The F. oxysporum species complex contains several ff. spp. that are either beneficial, pathogenic, or incompatible for A. thaliana, while at the same time being highly similar in biology and genetics. Hence, comparing the response of A. thaliana plants that are challenged with either kind of F. oxysporum f. sp. or strain can help to uncover differences between the plant’s MAMP and PAMP response.
In one early study comparing the response of A. thaliana with the pathogenic strain Fo5176 and the endophytic strain Fo47, it was shown that treatment of A. thaliana seedlings with spores of Fo47 triggers MPK3/6 activation and FRK1 expression, as was previously shown for Fo5176 (Babilonia et al., 2021). Similarly, crude cell wall extracts of both Fo47 and Fo5176 triggered MPK3/6 activation in A. thaliana in a CERK1/LYK5/BAK1- and SOBIR1-independent manner, indicating the presence of a cell wall-derived elicitor common to both strains that is not chitin, peptidoglycan, or unbranched β-1,3-1,4-glucan (Babilonia et al., 2021). However, this elicitor still needs to be identified.
Subsequently, Guo et al. (2021) conducted a large-scale study attempting to resolve differences in closer detail. The authors made use of the availability of high-quality genome data for Fo5176 and Fo47 to compare the transcriptional response of a plant and fungus in a metatranscriptomic analysis (Wang et al., 2020; Fokkens et al., 2020; Guo et al., 2021). The authors performed RNA-Seq on root and fungal tissue following inoculation of plants with either Fo5176 or Fo47 for 12, 24, 48, or 96 h, and compared the individual transcriptomes with a water-treated [12 hours post-inoculation (hpi)] control and with each other. Fusarium species carry lineage-specific accessory chromosomes as part of their genomes that contain all virulence factors and account for the host specificity of the different species and f. sp. (Ma et al., 2010). Introduction of such accessory chromosomes is sufficient to convert a non-pathogenic into a pathogenic strain. The strain Fo5176 carries four accessory chromosomes (chromosomes 2, 14, 15, and 18 with a combined 21.63 Mb), in addition to the shared 11 core chromosomes, while Fo47 carries only one (chromosome 7; 4.25 Mb) (Wang et al., 2020; Fokkens et al., 2020; Redkar et al., 2022). This difference again highlights the importance of the accessory chromosomes for pathogenicity, and accordingly these accessory chromosomes showed markedly different transcriptional responses during the plant colonization process (Guo et al., 2021). While mostly signaling-related genes, such as transcription factors and G protein regulators, were up-regulated in Fo47, the expression profile of Fo5176 showed up-regulation of avirulence effectors, proteases, or peptidases. Thus, this dataset provides valuable information on fungal genes required for successful colonization of the plant.
On the plant side, colonization with either strain resulted in a highly similar transcriptomic response (~60% overlap of differentially expressed genes compared with the control), with a relatively small subset of genes (~20%) apparently accounting for the difference between pathogenicity and endophytism (Guo et al., 2021). As expected, the transcriptome of Fo5176-infected plants showed up-regulation of genes involved in defense, toxin metabolism, small molecule biosynthesis, and drug response, while development- and growth-related genes were suppressed. Interestingly, in plants colonized by the endophyte, general immunity markers, JA response, and defense genes were strongly suppressed, while genes involved with nutrient metabolism and growth promotion were up-regulated (Guo et al., 2021). Clustering of genes differentially expressed in either Fo5176- or Fo47-treated plants compared with the wild type resulted in 24 co-expression clusters, four of which (C7, C15, C16, and C21) were enriched specifically with immunity/defense genes. The largest of these immunity clusters (C15), comprising ~1200 genes, showed nearly identical transcriptional patterns for both F. oxysporum strains, hence most probably reflecting general MAMP responses independent of pathogenicity or endophytism. Among the up-regulated genes in this cluster was the known MAMP receptor CERK1, which recognizes fungal chitin of either endophytic or pathogenic fungi, but also RFO1. The general defense marker FRK1 interestingly was suppressed by both strains. Fittingly, the genes in this cluster showed a strong up-regulation at the earliest time point, and then reverted to control levels as infection or colonization progressed (Guo et al., 2021). The other three of the four immunity clusters showed a stronger induction of immunity genes by the pathogenic strain Fo5176, thereby probably representing PAMP response genes, with predominantly PTI genes being induced. Conversely, in two of the clusters (C16 and C21), colonization by the endophyte Fo47 resulted in marked suppression of general immunity and JA response genes, such as BAK1, PEPR1 and 2, FER, RBOHD, RPM1, PAD4, and ZAR1, probably indicating that this suppression is part of the mechanism that allows the endophyte to colonize the plant without activation of detrimental defense responses. Next to this down-regulation of defense genes, colonization of A. thaliana with Fo47 also induced genes involved in nitrogen assimilation, demonstrating how colonization by this endophyte could be beneficial to the plant (Guo et al., 2021). Known ETI-related nucleotide-binding site-leucine-rich repeat (NLR) receptors were found in cluster C15, which also contained most of the known PTI-related genes, while uncharacterized NLR proteins were uniquely enriched in the two clusters C16 and C21 that featured the genes suppressed by the endophyte (Guo et al., 2021). This distribution of NLRs shows the tight interconnectedness of PTI and ETI, and indicates a unique role for uncharacterized NLRs in permitting colonization. The dataset created in this work is a valuable resource to the community and provides several starting points for follow-up work to disentangle the plant’s response to MAMPs and PAMPs (Guo et al., 2021). It also nicely demonstrated the suitability of the A. thaliana–Fo5176 pathosystem for such studies.
Future directions
For the immediate future, one main focus needs to be the move from large-scale, long-term analyses toward the cellular level. The available transcriptomic data provide several interesting starting points and candidate genes for such approaches (Table 1). The individual function of genes and their pathways in determining the outcome of the infection process needs to be investigated with high spatial and temporal resolution at the tissue and cellular level in planta. For this, cell biological and advanced microscopy approaches will be essential, for example by performing time-resolved live imaging of fluorescent-tagged proteins during certain infection stages at the cellular level. Studying the defense response of A. thaliana with such a microscopy-based approach on an individual cell level could also help to resolve some of the inconclusive observations described above. For example, the conflicting observations that JA and SA biosynthesis and signaling are only activated in the shoot, while the root shows either no response, just a very weak response, or even a slight down-regulation when challenged with this root pathogen may be the result of the data being based on an average of the whole-root tissue. Responses of small cell populations around the infection site, or only at a certain time point, could easily be lost in tissue-wide studies but resolved using a microscope.
The more recent identification of different plant receptor complexes involved in sensing the pathogen, such as MIK2/BAK1 or FER, as well as the SCOOP and RALF peptide ligands, opens the door to conducting similar detailed analyses as have been done for the flagellin pathway (Couto and Zipfel, 2016). Protein–protein interaction tools, such as traditional co-immunoprecipitation or the novel proximity labeling techniques, could be used to identify signaling components downstream of these receptor complexes, following artificial activation of these downstream signaling pathways by treatment with synthetic versions of the peptides (Lampugnani et al., 2018). Similarly, treatment with the different peptides could uncover different downstream pathways for these individual peptide ligand–receptor pairs. Receptor–protein interactions and turnover/internalization could also be analyzed to inform about pathways activated and crosstalk between them.
The roles of calcium signaling and ROS should also be investigated further (Kadota et al., 2015; Kim et al., 2022). Both play a conserved role in ROS release upon pathogen recognition, and signaling components for both have been identified in the transcriptomic work described earlier. Based on these results, the role of NADPH oxidases, such as RBOHD and RBOHF, should be investigated in closer detail, especially to investigate the biological significance of the differential regulation between these two. A closer investigation of calcium signaling pathways involved in the plant’s defense response against Fo5176 could furthermore provide new leads to study the role of NLRs, and ETI in general, in response to the fungus and its avirulence effectors (Kim et al., 2022). Calcium signaling is a convergence point between PTI and ETI, and recent work has shown that several NLRs function in so-called resistosomes as calcium-permeable cation channels (Bi et al., 2021; Jacob et al., 2021). Since the metatranscriptomic analysis performed by Guo et al. (2021) has uncovered several NLR proteins, of which several so far are functionally uncharacterized, this path may lead to a better understanding of the role of ETI in defending the plant from the pathogen (Guo et al., 2021).
Finally, the comparative work with the pathogenic strain Fo5176 and the endophytic strain Fo47 provides ample opportunity to study the differences in the plant’s PAMP and MAMP response. The metatranscriptomic analysis discussed above has provided the basis for several follow-up experiments with different candidate genes and pathways activated in response to both or only one of the fungal strains. Investigating these differential responses, ideally, again, with cellular resolution, will most probably provide the most valuable insight into which pathways are activated by both or only one of these strains.
Studies of this kind in combination with modern techniques such as gene editing, and new in silico tools for function and structure predictions will certainly advance our knowledge, not only regarding this pathosystem, but also by providing valuable clues on certain principles that may be valid in other related fungal–pathogen interactions.
Conclusions
Since the establishment of the A. thaliana–Fo5176 pathosystem in the early 2000s, significant progress has been made. Several large-scale transcriptomic datasets and a high-quality reference genome assembly of the fungus have been created (Table 1). Early work on establishing the role of phytohormones in the plant’s defense against Fo5176 mainly focused on exhaustive genetic analyses of single, double, and multiple mutants of the different hormone biosynthesis and signaling pathways. These efforts have created a solid basis for future projects to be developed. From the transcriptomic work, numerous candidate genes, both from the plant and the pathogen, have been discovered that can now be tested for their potential roles in determining the outcome of this host–pathogen interaction. The role of the different hormones is broadly characterized, and numerous proteins with distinct functions, from potential receptors and kinases, signal integrators, transcriptional and translational regulators, to signaling peptides and fungal effectors have been identified (Figs 1, 2). It is now time to move forward from the large-scale to the cellular description of how plants can effectively defend themselves against fungal infection.
Acknowledgements
The authors would like to thank Alexander Idnurm and Imre E. Somssich for critical reading and comments on the manuscript.
Contributor Information
Liu Wang, School of BioSciences, University of Melbourne, Parkville, VIC, 3010, Australia.
Jacob Calabria, School of BioSciences, University of Melbourne, Parkville, VIC, 3010, Australia.
Hsiang-Wen Chen, School of BioSciences, University of Melbourne, Parkville, VIC, 3010, Australia.
Marc Somssich, School of BioSciences, University of Melbourne, Parkville, VIC, 3010, Australia.
Monica Höfte, University of Ghent, Belgium.
Author contributions
MS: conceptualization, and writing the article with input from all authors. All authors discussed the data and manuscript. LW and MS: figure creation.
Conflict of interest
The authors declare no conflict of interest.
Funding
LW is supported by the China Scholarship Council. JC and MS are supported by the Australian Research Council (grant no. DE200101560 to MS). H-WC is supported by a Graduate Research Scholarship from the University of Melbourne.
References
- Abarca A, Franck CM, Zipfel C.. 2021. Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiology 187, 996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acevedo-Garcia J, Gruner K, Reinstädler A, et al. 2017. The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Scientific Reports 7, 9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Andrade J, Becker C, et al. 2016. 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166, 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K.. 2004. Antagonistic interaction between abscisic acid and jasmonate–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. The Plant Cell 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud D, Lee S, Takebayashi Y, Choi D, Choi J, Sakakibara H, Hwang I.. 2017. Cytokinin-mediated regulation of reactive oxygen species homeostasis modulates stomatal immunity in Arabidopsis. The Plant Cell 29, 543–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babilonia K, Wang P, Liu Z, et al. 2021. A nonproteinaceous Fusarium cell wall extract triggers receptor-like protein-dependent immune responses in Arabidopsis and cotton. New Phytologist 230, 275–289. [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Spoel SH.. 2006. Fine-tuning plant defence signalling: salicylate versus jasmonate. Plant Biology 8, 1–10. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A.. 2004. Ethylene Response Factor 1 mediates arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Molecular Plant-Microbe Interactions 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Bi G, Su M, Li N, et al. 2021. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE.. 2012. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiology 159, 266–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosland PW, Williams PH.. 1987. An evaluation of Fusarium oxysporum from crucifers based on pathogenicity, isozyme polymorphism, vegetative compatibility, and geographic origin. Canadian Journal of Botany 65, 2067–2073. [Google Scholar]
- Camargo-Ramírez R, Val-Torregrosa B, San Segundo B.. 2018. MiR858-mediated regulation of flavonoid-specific MYB transcription factor genes controls resistance to pathogen infection in Arabidopsis. Plant & Cell Physiology 59, 190–204. [DOI] [PubMed] [Google Scholar]
- Campbell EJ, Schenk PM, Kazan K, Penninckx IAMA, Anderson JP, Maclean DJ, Cammue BPA, Ebert PR, Manners JM.. 2003. Pathogen-responsive expression of a putative ATP-binding cassette transporter gene conferring resistance to the diterpenoid sclareol is regulated by multiple defense signaling pathways in Arabidopsis. Plant Physiology 133, 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A, Stacey G.. 2014. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Wong CL, Muzzi F, Vlaardingerbroek I, Kidd BN, Schenk PM.. 2014. Root defense analysis against Fusarium oxysporum reveals new regulators to confer resistance. Scientific Reports 4, 5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SJ, Diener AC.. 2013. Diversity in receptor-like kinase genes is a major determinant of quantitative resistance to Fusarium oxysporum f.sp. matthioli. New Phytologist 200, 172–184. [DOI] [PubMed] [Google Scholar]
- Cole SJ, Yoon AJ, Faull KF, Diener AC.. 2014. Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine- and leucine-conjugated jasmonates. Molecular Plant Pathology 15, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AD, Maroschek J, Raasch L, Takken FLW, Ranf S, Hückelhoven R.. 2021. The Arabidopsis leucine-rich repeat receptor-like kinase MIK2 is a crucial component of early immune responses to a fungal-derived elicitor. New Phytologist 229, 3453–3466. [DOI] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, et al. 2006. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nature Genetics 38, 716–720. [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C.. 2016. Regulation of pattern recognition receptor signalling in plants. Nature Reviews. Immunology 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Cox KL, Babilonia K, Wheeler T, He P, Shan L.. 2019. Return of old foes—recurrence of bacterial blight and Fusarium wilt of cotton. Current Opinion in Plant Biology 50, 95–103. [DOI] [PubMed] [Google Scholar]
- Czymmek KJ, Fogg M, Powell DH, Sweigard J, Park S-Y, Kang S.. 2007. In vivo time-lapse documentation using confocal and multi-photon microscopy reveals the mechanisms of invasion into the Arabidopsis root vascular system by Fusarium oxysporum. Fungal Genetics and Biology 44, 1011–1023. [DOI] [PubMed] [Google Scholar]
- Davies DR, Bindschedler LV, Strickland TS, Bolwell GP.. 2006. Production of reactive oxygen species in Arabidopsis thaliana cell suspension cultures in response to an elicitor from Fusarium oxysporum: implications for basal resistance. Journal of Experimental Botany 57, 1817–1827. [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, et al. 1994. A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- de Sain M, Rep M.. 2015. The role of pathogen-secreted proteins in fungal vascular wilt diseases. International Journal of Molecular Sciences 16, 23970–23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Ausubel FM.. 2005. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Pietro A, Madrid MP, Caracuel Z, Delgado-Jarana J, Roncero MIG.. 2003. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Molecular Plant Pathology 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Dita M, Barquero M, Heck D, Mizubuti ESG, Staver CP.. 2018. Fusarium wilt of banana: current knowledge on epidemiology and research needs toward sustainable disease management. Frontiers in Plant Science 9, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan WL, Chapple C.. 2016. Conservation and divergence of Mediator structure and function: insights from plants. Plant & Cell Physiology 58, pcw176. [DOI] [PubMed] [Google Scholar]
- Dörffling K, Petersen W, Sprecher E, Urbasch I, Hanssen H-P.. 1984. Abscisic acid in phytopathogenic fungi of the genera Botrytis, Ceratocystis, Fusarium, and Rhizoctonia. Zeitschrift für Naturforschung C 39, 683–684. [Google Scholar]
- Edgar CI, McGrath KC, Dombrecht B, Manners JM, Maclean DC, Schenk PM, Kazan K.. 2006. Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana. Australasian Plant Pathology 35, 581. [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW.. 2005. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563–4574. [DOI] [PubMed] [Google Scholar]
- Fallath T, Kidd BN, Stiller J, Davoine C, Björklund S, Manners JM, Kazan K, Schenk PM.. 2017. MEDIATOR18 and MEDIATOR20 confer susceptibility to Fusarium oxysporum in Arabidopsis thaliana. PLoS One 12, e0176022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkens L, Guo L, Dora S, Wang B, Ye K, Sánchez-Rodríguez C, Croll D.. 2020. A chromosome-scale genome assembly for the Fusarium oxysporum strain Fo5176 to establish a model Arabidopsis–fungal pathosystem. G3 10, 3549–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisleben R, Lein A.. 1942. Über die Auffindung einer mehltauresistenten Mutante nach Röntgenbestrahlung einer anfälligen reinen Linie von Sommergerste. Die Naturwissenschaften 30, 608–608. [Google Scholar]
- Gao Q, Wang C, Xi Y, Shao Q, Li L, Luan S.. 2022. A receptor–channel trio conducts Ca2+ signalling for pollen tube reception. Nature 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon TR. 2017. Fusarium oxysporum and the Fusarium Wilt Syndrome. Annual Review of Phytopathology 55, 23–39. [DOI] [PubMed] [Google Scholar]
- Gordon TR, Martyn RD.. 1997. The evolutionary biology of Fusarium oxysporum. Annual Review of Phytopathology 35, 111–128. [DOI] [PubMed] [Google Scholar]
- Gully K, Pelletier S, Guillou M-C, et al. 2019. The SCOOP12 peptide regulates defense response and root elongation in Arabidopsis thaliana. Journal of Experimental Botany 70, 1349–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Yu H, Wang B, et al. 2021. Metatranscriptomic comparison of endophytic and pathogenic Fusarium–Arabidopsis interactions reveals plant transcriptional plasticity. Molecular Plant-Microbe Interactions 34, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JM, Balint-Kurti P, Bede JC, et al. 2020. What are the Top 10 unanswered questions in molecular plant–microbe interactions? Molecular Plant-Microbe Interactions 33, 1354–1365. [DOI] [PubMed] [Google Scholar]
- Heim DR, Skomp JR, Waldron C, Larrinua IM.. 1991. Differential response to isoxaben of cellulose biosynthesis by wild-type and resistant strains of Arabidopsis thaliana. Pesticide Biochemistry and Physiology 39, 93–99. [Google Scholar]
- Hou S, Liu D, Huang S, et al. 2021. The Arabidopsis MIK2 receptor elicits immunity by sensing a conserved signature from phytocytokines and microbes. Nature Communications 12, 5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Tsouda K.. 2022. Salicylic acid and jasmonic acid crosstalk in plant immunity. Essays in Biochemistry doi: 10.1042/EBC20210090. [DOI] [PubMed] [Google Scholar]
- Jacob P, Kim NH, Wu F, et al. 2021. Plant ‘helper’ immune receptors are Ca2+-permeable nonselective cation channels. Science 373, 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacott CN, Ridout CJ, Murray JD.. 2021. Unmasking mildew resistance locus O. Trends in Plant Science 26, 1006–1013. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C.. 2015. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant & Cell Physiology 56, 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kesten C, Gámez-Arjona FM, Menna A, et al. 2019. Pathogen-induced pH changes regulate the growth–defense balance in plants. The EMBO Journal 38, 1–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K.. 2009. The Mediator complex subunit pft1 is a key regulator of jasmonate-dependent defense in Arabidopsis. The Plant Cell 21, 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Kadoo NY, Dombrecht B, Tekeoglu M, Gardiner DM, Thatcher LF, Aitken EAB, Schenk PM, Manners JM, Kazan K.. 2011. Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Molecular Plant-Microbe Interactions 24, 733–748. [DOI] [PubMed] [Google Scholar]
- Kim NH, Jacob P, Dangl JL.. 2022. Con-Ca2+-tenating plant immune responses via calcium-permeable cation channels. New Phytologist 234, 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Sinha NR.. 2008. Tomato (Solanum lycopersicum): a model fruit-bearing crop. Cold Spring Harbor Protocols 2008, pdb.emo105. [DOI] [PubMed] [Google Scholar]
- Lampugnani ER, Wink RH, Persson S, Somssich M.. 2018. The toolbox to study protein–protein interactions in plants. Critical Reviews in Plant Sciences 0, 1–27. [Google Scholar]
- Li N, Han X, Feng D, Yuan D, Huang L-J.. 2019. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? International Journal of Molecular Sciences 20, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu K, Tong G, Xi C, Liu J, Zhao H, Wang Y, Ren D, Han S.. 2022. MPK3/MPK6-mediated phosphorylation of ERF72 positively regulates resistance to Botrytis cinerea through directly and indirectly activating the transcription of camalexin biosynthesis enzymes. Journal of Experimental Botany 73, 413–428. [DOI] [PubMed] [Google Scholar]
- Liao C-J, Lai Z, Lee S, Yun DJ, Mengiste T.. 2016. Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. The Plant Cell 28, tpc.00105.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sánchez-Rodríguez C, Jordá L, Molina A.. 2005. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. The Plant Journal 43, 165–180. [DOI] [PubMed] [Google Scholar]
- Lyons R, Stiller J, Powell J, Rusu AG, Manners JM, Kazan K.. 2015. Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS One 10, e0121902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L-J, van der Does HC, Borkovich KA, et al. 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masachis S, Segorbe D, Turrà D, et al. 2016. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nature Microbiology 1, 16043. [DOI] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible W-R, Udvardi MK, Kazan K.. 2005. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiology 139, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna A, Dora S, Sancho-Andrés G, Kashyap A, Meena MK, Sklodowski K, Gasperini D, Coll NS, Sánchez-Rodríguez C.. 2021. A primary cell wall cellulose-dependent defense mechanism against vascular pathogens revealed by time-resolved dual transcriptomics. BMC Biology 19, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins ED, Chen X, Romaine P, Raina R, Geiser DM, Kang S.. 2001. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91, 173–180. [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Jones JDG, Ding P.. 2022. Plant immune networks. Trends in Plant Science 27, 255–273. [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Anderson JP, Young J, Singh KB.. 2007. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiology 143, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Goossens A.. 2011. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. The Plant Cell 23, 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P.. 2002. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiology 129, 1076–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncini L, Wyrsch I, Dénervaud Tendon V, Vorley T, Boller T, Geldner N, Métraux J-P, Lehmann S.. 2017. In roots of Arabidopsis thaliana, the damage-associated molecular pattern AtPep1 is a stronger elicitor of immune signalling than flg22 or the chitin heptamer. PLoS One 12, e0185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A, Sabale M, Schudoma C, et al. 2022. Conserved secreted effectors contribute to endophytic growth and multi-host plant compatibility in a vascular wilt fungus. The Plant Cell. doi: 10.1093/plcell/koac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Dekker HL, Vossen JH, de Boer AD, Houterman PM, Speijer D, Back JW, de Koster CG, Cornelissen BJC.. 2002. Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiology 130, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, van der Does HC, Meijer M, van Wijk R, Houterman PM, Dekker HL, de Koster CG, Cornelissen BJC.. 2004. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Molecular Microbiology 53, 1373–1383. [DOI] [PubMed] [Google Scholar]
- Rhodes J, Yang H, Moussu S, Boutrot F, Santiago J, Zipfel C.. 2021. Perception of a divergent family of phytocytokines by the Arabidopsis receptor kinase MIK2. Nature Communications 12, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador-Guirao R, Baldrich P, Weigel D, Rubio-Somoza I, San Segundo B.. 2018. The microRNA miR773 is involved in the Arabidopsis immune response to fungal pathogens. Molecular Plant-Microbe Interactions 31, 249–259. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Rusu AG, Manners JM, Maclean DJ.. 2005. The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiology and Biochemistry 43, 997–1005. [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. 2010. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Diener AC.. 2013. Arabidopsis thaliana RESISTANCE TO FUSARIUM OXYSPORUM 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genetics 9, e1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Suárez M, Baldrich P, Weigel D, Rubio-Somoza I, San Segundo B.. 2017. The Arabidopsis miR396 mediates pathogen-associated molecular pattern-triggered immune responses against fungal pathogens. Scientific Reports 7, 44898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas C, Nirmala Devi D, Narasimha Murthy K, et al. 2019. Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity—a review. Saudi Journal of Biological Sciences 26, 1315–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerell BA. 2019. Resolving Fusarium: current status of the genus. Annual Review of Phytopathology 57, 323–339. [DOI] [PubMed] [Google Scholar]
- Takken FLW, Rep M.. 2010. The arms race between tomato and Fusarium oxysporum. Molecular Plant Pathology 11, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Owen B, Higgins VJ.. 2004. The role of the jasmonate response in plant susceptibility to diverse pathogens with a range of lifestyles. Plant Physiology 135, 530–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Cevik V, Grant M, Zhai B, Jones JDG, Manners JM, Kazan K.. 2016a. Characterization of a JAZ7 activation-tagged Arabidopsis mutant with increased susceptibility to the fungal pathogen Fusarium oxysporum. Journal of Experimental Botany 67, 2367–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Gardiner DM, Kazan K, Manners JM.. 2012a. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Molecular Plant-Microbe Interactions 25, 180–190. [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Kidd BN, Kazan K.. 2016b. Belowground defence strategies against Fusarium oxysporum. In: Vos C, Kazan K, eds. Belowground defence strategies in plants. Cham: Springer, 71–98. [Google Scholar]
- Thatcher LF, Manners JM, Kazan K.. 2009. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. The Plant Journal 58, 927–939. [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Powell JJ, Aitken EAB, Kazan K, Manners JM.. 2012b. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiology 160, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF.. 1998. Separate jasmonate-dependent and salicylate-dependent defense–response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences, USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms D, Liang Y, Haney CH.. 2021. Maintaining symbiotic homeostasis: how do plants engage with beneficial microorganisms while at the same time restricting pathogens? Molecular Plant-Microbe Interactions 34, 462–469. [DOI] [PubMed] [Google Scholar]
- Thynne E, Saur IML, Simbaqueba J, et al. 2017. Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Molecular Plant Patholog y 18, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor N, Paauw M, Rep M, Takken FLW.. 2020. The root-invading pathogen Fusarium oxysporum targets pattern-triggered immunity using both cytoplasmic and apoplastic effectors. New Phytologist 227, 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR.. 2006. Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiology 140, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen J-G, Jones AM, Botella JR.. 2007. Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. The Plant Cell 19, 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Sewelam N, Rookes JE, Kunkel M, Nowak E, Schenk PM, Botella JR.. 2009. Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. The Plant Journal 58, 69–81. [DOI] [PubMed] [Google Scholar]
- van der Does D, Boutrot F, Engelsdorf T, et al. 2017. The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genetics 13, e1006832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Does HC, Duyvesteijn RGE, Goltstein PM, van Schie CCN, Manders EMM, Cornelissen BJC, Rep M.. 2008. Expression of effector gene SIX1 of Fusarium oxysporum requires living plant cells. Fungal Genetics and Biology 45, 1257–1264. [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, et al. 2011. Callose biosynthesis regulates symplastic trafficking during root development. Developmental Cell 21, 1144–1155. [DOI] [PubMed] [Google Scholar]
- Wang B, Yu H, Jia Y, et al. 2020. Chromosome-scale genome assembly of Fusarium oxysporum strain Fo47, a fungal endophyte and biocontrol agent. Molecular Plant-Microbe Interactions 33, 1108–1111. [DOI] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjärvi J.. 2018. Reactive oxygen species in plant signaling. Annual Review of Plant Biology 69, 209–236. [DOI] [PubMed] [Google Scholar]
- Yadeta KA, Valkenburg D-J, Hanemian M, Marco Y, Thomma BPHJ.. 2014. The Brassicaceae-specific EWR1 gene provides resistance to vascular wilt pathogens. PLoS One 9, e88230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang E, Liu J.. 2022. CERK1, more than a co-receptor in plant–microbe interactions. New Phytologist 234, 1606–1613. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K.. 2005. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology 58, 585–596. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao J, Yang Y, Bao Q, Li Y, Wang H, Hou S.. 2022. EWR1 as a SCOOP peptide activates MIK2-dependent immunity in Arabidopsis. Journal of Plant Interactions 17, 562–568. [Google Scholar]
- Zhong C-L, Zhang C, Liu J-Z.. 2019. Heterotrimeric G protein signaling in plant immunity. Journal of Experimental Botany 70, 1109–1118. [DOI] [PubMed] [Google Scholar]
- Zhu Q-H, Stephen S, Kazan K, Jin G, Fan L, Taylor J, Dennis ES, Helliwell CA, Wang M-B.. 2013. Characterization of the defense transcriptome responsive to Fusarium oxysporum-infection in Arabidopsis using RNA-seq. Gene 512, 259–266. [DOI] [PubMed] [Google Scholar]