ABSTRACT
Cells in microbial communities on surfaces live and divide in close proximity, which greatly enhances the potential for social interactions. Spatiogenetic structures are manifested through competitive and cooperative interactions among the same and different genotypes within a shared space, and extracellular secretions appear to function dynamically at the forefront. A previous experimental evolution study utilizing Pseudomonas fluorescens Pf0-1 colonies demonstrated that diverse mutations in the rsmE gene were repeatedly and exclusively selected through the formation of a dominant spatial structure. RsmE’s primary molecular function is translation repression, and its homologs regulate various social and virulence phenotypes. Pseudomonas spp. possess multiple paralogs of Rsm proteins, and RsmA, RsmE, and RsmI are the most prevalent. Here, we demonstrate that the production of a mucoid polymer and a biosurfactant are exclusively regulated through RsmE, contradicting the generalized notion of functional redundancy among the Rsm paralogs. Furthermore, we identified the biosurfactant as the cyclic lipopeptide gacamide A. Competition and microscopy analyses showed that the mucoid polymer is solely responsible for creating a space of low cellular density, which is shared exclusively by the same genotype. Gacamide A and other RsmE-regulated products appear to establish a physical boundary that prevents the encroachment of the competing genotype into the newly created space. Although cyclic lipopeptides and other biosurfactants are best known for their antimicrobial properties and reducing surface tension to promote the spreading of cells on various surfaces, they also appear to help define spatial structure formation within a dense community.
IMPORTANCE In densely populated colonies of the bacterium Pseudomonas fluorescens Pf0-1, diverse mutations in the rsmE gene are naturally selected by solving the problem of overcrowding. Here, we show that RsmE-regulated secretions function together to create and protect space of low cell density. A biosurfactant generally promotes the spreading of bacterial cells on abiotic surfaces; however, it appears to function atypically within a crowded population by physically defining genotypic boundaries. Another significant finding is that these secretions are not regulated by RsmE’s paralogs that share high sequence similarity. The experimental pipeline described in this study is highly tractable and should facilitate future studies to explore additional RsmE-regulated products and address why RsmE is functionally unique from its paralogs.
KEYWORDS: biosurfactants, cell-cell interaction, extracellular matrix, mutational studies, variable phenotypes
INTRODUCTION
Central to the architecture of microbial communities is the extracellular matrix (1–8), a dynamic cumulus of compounds produced by individual cells that physically define both the spatial arrangements within the community and the three-dimensional boundaries. Micrometer-scale spatiogenetic structures readily emerge within surface-grown communities as individual cells produce identical copies of themselves in a given area (9–11). Competition between different genotypes leads to the spatial enrichment of a particular genotype, producing macroscopic regions that stem from a recent common ancestor (10, 12–15). Individual phenotypes can positively or negatively impact the fitness of neighboring cells, including the consumption of limiting nutrients and the secretion of enzymes and toxins that promote or discourage the growth of neighboring cells (8, 16–18). Mechanistic understanding of how individual phenotypes antagonize or synergize with another clearly carries both fundamental and clinical significance.
Researchers employ various experimental approaches to study the interactive dynamics of microbial cells within a community, whether the approaches be computational (9, 10, 19) or empirical on a variety of abiotic surfaces (5, 20–22). We previously described a model system based on Pseudomonas fluorescens colonies which showed how spatial structures rapidly evolved within clonal aggregates (23). Mucoid patches repeatedly emerge on the surface of aging colonies due to the activities of specific mutants, where they expand space and decrease local density. Remarkably, a mutation in a single gene, rsmE, was responsible for each and every case of over 500 independently derived mucoid patches. Importantly, rsmE mutants shared the same growth rate in isolation as the parent cells, and the evolutionary advantage specifically required the proximal presence of the parent cells. These observations collectively suggest that RsmE-regulated phenotypes physically act to create dominant spatial structures in a densely populated bacterial colony.
RsmE belongs to the CsrA/Rsm family, and its homologs are regulators of social and virulence phenotypes in Gammaproteobacteria (24, 25). CsrA was the first member of the family to be discovered 3 decades ago in Escherichia coli (26), and its homologs are now known to be present in over 2,900 species (27). CsrA/Rsm proteins interact with diverse mRNA (25, 28–30) and primarily function as a translation repressor by either directly or indirectly blocking their respective Shine-Dalgarno sequence (31–34). CsrA also possesses additional regulatory functions that impact Rho-dependent transcription attenuation, mRNA stabilization and destabilization, and even activation of translation (32). In contrast to CsrA in Enterobacteriaceae, Pseudomonas spp. possess varied numbers of Rsm paralogs (27). Rsm paralogs were initially characterized to repress the production of secondary metabolites and are generally described to overlap or cumulate in function (35–41).
Although the three paralogs in P. fluorescens (RsmE, RsmA, and RsmI) share high sequence similarity, the exclusive selection of mutations in the rsmE locus (23) suggests functional specificity of RsmE from its paralogs. Here, we show that all three Rsm paralogs are expressed, but RsmE uniquely governs the production of both a mucoid polymer and a biosurfactant. The biosynthetic genes of the mucoid polymer were previously described (42), and we identified the biosynthetic genes of the biosurfactant in this study. Competition and microscopy analyses of the extracellular polysaccharide and biosurfactant mutants revealed that these extracellular secretions function collectively to confer a fitness benefit as a direct result of the spatial structures they form.
RESULTS
RsmE, RsmA, and RsmI in P. fluorescens Pf0-1 are highly conserved in sequence and all three respective genes are expressed.
P. fluorescens Pf0-1 possesses three Rsm paralogs, RsmA, RsmE, and RsmI, which share high sequence similarity (Fig. 1A). We sought to first determine whether or not the three corresponding genes are expressed. Quantitative PCR confirmed that all three genes are indeed expressed at the time of sampling (Fig. 1B). We extracted mRNA from wild-type (WT) cells growing in colonies after 3 days of incubation, which coincided with the timing of the natural emergence of rsmE mutants as visible mucoid patches (23). These results showed that the exclusive selection of rsmE mutations in our previous experimental evolution study (23) was not simply due to the absence of rsmA and rsmI expression under the same experimental conditions.
FIG 1.
Rsm paralogs in P. fluorescens Pf0-1 share a highly conserved sequence, and their respective genes are simultaneously expressed. (A) Sequence alignments of Rsm paralogs in Pseudomonas fluorescens Pf0-1 with ClustalX show similarities of amino acid sequences and chemical properties (65). (B) Expression of rsmE, rsmA, and rsmI genes assessed in WT by qPCR. Transcripts of all three genes were detected; shown here is the relative abundance of each transcript using the 2−ΔΔCT method, with comparison to that of the least abundantly expressed, rsmI. Plotted are the means of three biological replicates with three technical replicates for each biological replicate, and the error bars represent the standard deviations of the means.
RsmE specifically regulates the production of a mucoid polymer and biosurfactant.
Experimentally selected rsmE mutants visibly produce a mucoid polymer and/or a biosurfactant (23), which suggests that specific mutations differentially impact RsmE’s functions. To determine if these extracellular secretions are commonly regulated by the three Rsm homologs, we constructed deletion mutants for the three genes. Comparison of colony morphologies showed that only the rsmE mutant exhibited mucoidy (Fig. 2A). In addition, mucoid patches consistently emerged in colonies of the WT, rsmA mutant, and rsmI mutant (Fig. 2A); these colonies were characteristic of naturally mutated rsmE (23). These results confirmed that the production of the mucoid polymer is specifically regulated by RsmE. We next compared biosurfactant production on a polycarbonate membrane overlaid on the agar surface. Production of the biosurfactant on the shiny side of the membrane allows the colony to spread out radially, but the cells remained trapped on the dull side of the membrane while the biosurfactant spreads out unhindered (23). Only the rsmE mutant produced a visible ring on the dull side of the membrane and also spread out on the shiny side of the membrane (Fig. 2B). Furthermore, genetic complementation of the rsmE knockout strain with the native rsmE locus restored the WT phenotype (see Fig. S1 in the supplemental material). These results confirmed that the production of both the mucoid polymer and the biosurfactant is uniquely governed by RsmE from its paralogs.
FIG 2.
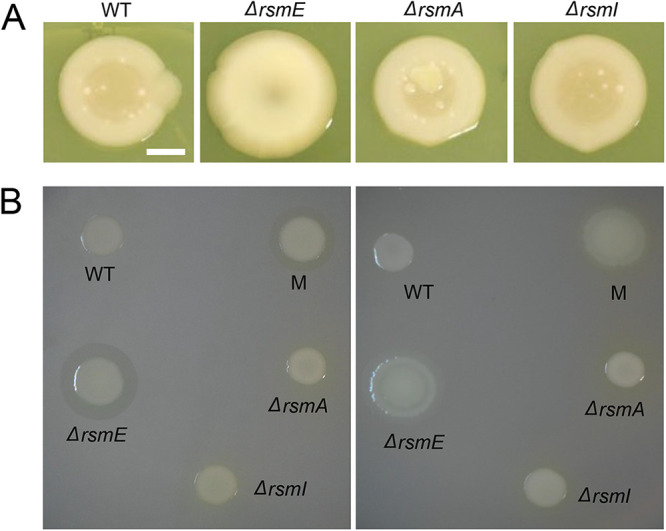
Both the mucoid polymer and the biosurfactant are regulated by RsmE, but not by RsmA or RsmI. (A) Colony morphology comparisons of WT and deletion mutants of rsmE, rsmA, and rsmI. Liquid cultures were spotted on PAF plates 7 days prior to capturing the images. Only the ΔrsmE strain was mucoid in appearance, and new mucoid patches naturally emerged in WT, ΔrsmA, and ΔrsmI colonies that characteristically represented de novo rsmE mutations. Scale bar, 10 mm. (B) Comparison of biosurfactant production on the dull (left) and shiny (right) sides of the polycarbonate membrane overlaid on PAF. The M strain is a naturally selected mutant from a WT colony and harbors a frameshift mutation in rsmE. Only the ΔrsmE and M strains produced the biosurfactant ring on the dull side, which promoted spreading of cells on the shiny side of the membrane.
Identification of the biosurfactant gacamide A.
The biosynthetic genes of the mucoid polymer were previously characterized as encoding a glucose-rich extracellular polysaccharide, and a corresponding gene was deleted in a mucoid (M) strain with a frameshift mutation in rsmE (23) to produce the nonmucoid M* strain (42). To identify the biosynthetic genes of the biosurfactant, we carried out random transposon mutagenesis in the M* strain background. Seven mutants were independently isolated that no longer produced the secretion on the dull side of the polycarbonate membrane and failed to spread out on the shiny side of the membrane. All transposon insertion sites were mapped to three contiguous loci (annotated as Pfl01_2211, Pfl01_2212, and Pfl-1_2213), which were recently demonstrated to encode nonribosomal peptide synthetases (43) that produce the cyclic lipopeptide gacamide A (44). Cyclic lipopeptides are indeed classified as surfactants, and they contribute to surface motility and biofilm formation in many Pseudomonas spp. (45, 46). Given that four independent transposon insertions occurred in the Pfl01_2211 locus, we constructed a corresponding in-frame deletion mutant in the M strain to produce the MS strain, and the same mutation was also introduced in the nonmucoid M* strain to produce the MS* strain. Neither MS nor MS* produced the biosurfactant ring on the dull side of the membrane and the spreading phenotype on the shiny side of the membrane (Fig. 3), confirming that the Pfl01_2211-Pfl-1_2213 cluster encodes the production of the biosurfactant. Importantly, M* maintained the production of the biosurfactant and MS maintained the production of the mucoid polymer (Fig. 3), which showed that the biosynthesis of these two secreted products is not genetically linked but that both are regulated by RsmE.
FIG 3.
Deletion of the Pfl01_2211 locus abolished biosurfactant production. Shown are the results from the dull side (A) and the shiny side (B) of the polycarbonate membrane. M (rsmE mutant) and M* (M with the mucoid polymer biosynthesis gene [Pfl01_3834] deleted) produced the biosurfactant and spread on the surface, but MS (M with Pfl01_2211 deleted) and MS* (M with both Pfl01_3834 and Pfl01_2211 deleted) failed to do so, as did the WT with an unaltered rsmE gene. These results confirmed that the Pfl01_2211-2213 cluster encodes the biosynthetic genes of the biosurfactant, which is now known to be gacamide A.
Both the mucoid polymer and biosurfactant confer a competitive advantage.
We previously showed that the rsmE knockout mutant outcompetes the WT strain in cocultured colonies but not in liquid cocultures, which indicates that RsmE-regulated products provide benefit exclusively in a structured population (23). To assess the contributions of the RsmE-regulated mucoid polymer and the biosurfactant, we independently competed M, MS, M*, and MS* against the WT in cocultured colonies and assessed their fitness relative to the WT. All four strains outcompeted the WT throughout the duration of the experiments (Fig. 4), with M and MS being nearly equal in fitness and M* and MS* exhibiting decreased fitness at day 4. However, we observed reduced fitness in all secretion mutants compared to M by day 7, with MS and M* being comparable and MS* exhibiting a further reduction. Such stepwise decreases in fitness indicate that each secreted product independently confers a competitive advantage and the two secretions also likely function in an additive manner. Furthermore, the fact that MS* retains the ability to outcompete the WT indicates that there are additional RsmE-regulated genes that contribute to M’s dominance over the WT.
FIG 4.

Competitions of M, with or without mucoid polymer and/or biosurfactant production, against WT showed varying levels of relative fitness over time. WT was chromosomally tagged with streptomycin resistance and all mutants were tagged with kanamycin resistance, and these resistance markers produced neutral relative fitness in P. fluorescens Pf0-1 (23). Error bars represent the standard deviations of the mean relative fitness (mutant over WT) calculated from three independent populations after 4 and 7 days of incubation. Data from each time point were analyzed by ANOVA (P < 0.0001), and Tukey’s honest significant difference test showed that all pairwise comparisons were significantly different (P < 0.05) except for those indicated as nonsignificant (n.s.). A relative fitness (W) of 1 indicates equal fitness of the mutant and WT, and a W value of >1 indicates that the mutant outcompeted the WT. Both the mucoid polymer and the biosurfactant provided a competitive advantage. However, MS* (rsmE mutant with biosynthesis genes of both secretions deleted) still outcompeted the WT, suggesting that additional RsmE-regulated products contribute to the competitive advantage of M (rsmE mutant) against the WT.
The mucoid polymer creates space and the biosurfactant prevents diffusion of the mucoid polymer at the colony surface.
The temporal differences in the relative fitness between MS and M* (Fig. 4) suggest that the mucoid polymer plays a more significant role early in the competition. Importantly, MS*, the double secretion mutant, exhibited an equal growth profile compared to both the WT and M as monoculture in either liquid or colonies (see Fig. S2). The M data here recapitulate the results from our previous study, which also demonstrated that the competitive advantage of rsmE mutants specifically required the formation of spatial structures that decreased local density and provided greater access to oxygen (23).
To explore the functional role of the RsmE-regulated mucoid polymer and biosurfactant in spatial structure formation, we carried out epifluorescence and confocal microscopy analyses of our collection of secretion mutants compared to the WT. We first introduced a constitutively expressed gfp gene into the chromosome of WT, M, MS, M*, and MS* strains. Each green fluorescent protein (GFP) strain was mixed with red fluorescence-labeled WT at a respective ratio of 10−5:1 to best visualize isolated spatiogenetic structures in colonies after 5 days. Epifluorescence imaging of entire colonies showed isolated green fluorescent patches emerging from mostly red fluorescent WT colonies, with M and MS producing consistently bigger patches than M* and MS* (Fig. 5A). Each coculture also produced red fluorescent mucoid patches, which represented de novo rsmE mutants naturally emerging from the red fluorescent WT cells (23); however, no green fluorescent patches were observed in the WT:WT colonies. With epifluorescence imaging at a higher magnification, the green fluorescence signal in the smaller patches formed by M* and MS* was much more intense than in patches formed by M, and MS patches produced the least intense fluorescence signal (Fig. 5B; see also Fig. S3). Individual patches formed by both M and MS typically merged together with nearby patches through continuous expansion over time, but we consistently observed MS patches to be much more amorphous in structure with less-defined individual boundaries. In contrast, green fluorescent WT patches were rarely observed and appeared to comprise only a few cells.
FIG 5.
The mucoid polymer and biosurfactant function together in the formation of a dominant spatial structure. Each indicated strain was chromosomally tagged with GFP, heavily underrepresented in a mixture with DsRed Express-tagged WT, and representative cocultured colonies were imaged 5 days later. (A) Epifluorescence microscopy images that captured the entire colony. Scale bar, 10 mm. Each sample showed the natural emergence of red mucoid patches that are characteristic of de novo rsmE mutants, stemming from the red fluorescent WT cells. (B) Epifluorescence microscopy images focusing on the surface of individual patches. Scale bar, 50 μm. MS* produced unique patches that appeared to be mixed with red WT cells. (See also Fig. S3 in the supplemental material, which shows the green and red channels separated.) (C) Confocal microscopy images at a higher magnification, focused on the boundaries between the mutant and WT. Scale bar, 10 μm. The mucoid polymer is solely responsible for creating the space of low cell density (black space is devoid of cells), and the biosurfactant appears to physically hold the mucoid polymer and producing cells from flowing out from the newly created space. MS* produces the smallest patches that are densely filled, as reflected by vertically aligned cells (spheres) similar to the WT:WT spatial organization (left panel). However, MS* maintains the ability to form an organized structure that excludes WT cells, suggesting that additional RsmE-regulated products contribute to the spatial dominance of M. See also Fig. S4, in which the green and red channels are separated.
Confocal imaging using an air-corrected 100× Plan Apo objective provided a clear view of individual green fluorescent cells and their spatial arrangement within a given patch surrounded by red fluorescent WT cells (Fig. 5C; see also Fig. S4). M cells were present at a strikingly lower density compared to the neighboring WT cells, with the characteristic black space that was devoid of cells (23). In addition, M patches were defined by a clear boundary formed with a thin layer of M cells, which appeared to exclude the encroachment of WT cells into the black space. In contrast, MS patches lacked a clear exclusionary boundary, with MS cells appearing to flow over the WT cells. This interpretation was also reflected in the lower-magnification observations of MS patches being more mucoid and amorphous (Fig. 5A) and producing less intense fluorescent signal (Fig. 5B and Fig. S3) compared to patches formed by M. M* and MS* both formed much more densely packed patches with clear boundaries against the WT cells, but MS* cells appeared to be even more densely packed, as indicated by the uniquely vertical arrangement of cells (Fig. 5C and Fig. S4) and much smaller sizes of individual patches (Fig. 5A). These observations collectively suggested that the mucoid polymer is the primary driver of creating space, while the biosurfactant spatially sequesters the mucoid polymer to prevent their diffusion. However, MS* retains the ability to produce a spatiogenetic structure that contrasts greatly from the green fluorescent WT cells, which form small clusters of only a few cells without any organized structure (Fig. 5C and Fig. S4), likely representing daughter cells stemming from initially a single mother cell. As reflected by our relative fitness data (Fig. 4), there appear to be additional RsmE-regulated genes that specifically promote spatial competition in a densely populated colony.
DISCUSSION
Several members of the Gammaproteobacteria, including Pseudomonas spp., possess multiple paralogs of CsrA/Rsm proteins, and their corresponding genes are also present in diverse plasmids and bacteriophages (27). We previously showed that mutations in rsmE are exclusively selected as mucoid patches in colonies of P. fluorescens Pf0-1 (23), suggesting that RsmE’s function is not entirely redundant from that of its paralogs, RsmA and RsmI. In this study, we showed that the functions of all three paralogs are accessible to evolutionary selection, and mucoid patches consistently emerge in both rsmA and rsmI knockout colonies. We have also demonstrated that knocking out rsmE results in the production of two visible extracellular secretions, a mucoid polymer and a biosurfactant, but neither is produced in rsmA or rsmI knockouts. Thus, RsmE appears to either directly repress the production of these secretions or modulate the activity of other regulators that directly govern their production. The 5′-untranslated regions of the biosurfactant biosynthesis genes include a potential Rsm-binding site (44). However, it is difficult to predict genes that are directly regulated by Rsm proteins solely through a bioinformatics approach, since the Rsm-binding consensus sequence overlaps with the Shine-Dalgarno sequence (31–34). A recent study in Pseudomonas putida demonstrated that Rsm paralogs directly bind to both overlapping and unique mRNA, and RsmE appears to specifically regulate multiple regulators and extracellular products (30). In addition, Pseudomonas syringae possesses five Rsm paralogs, and three of them have been demonstrated to function in a nonoverlapping fashion to differentially regulate the production of diverse extracellular products and virulence genes (47).
We have shown that the production of both the mucoid polymer and the biosurfactant significantly boosts competition through spatial structure formation. The two key characteristics associated with the dominant spatial structure formed by the rsmE mutant are creation of space with a low cellular density and exclusion of the neighboring WT cells from this local environment (23). Here, we have demonstrated that the mucoid polymer is solely responsible for creating the space. We had initially interpreted our findings to indicate that the biosurfactant forms the exclusionary boundary, based on the mixed presence of the biosurfactant knockout and WT cells. However, we consistently observed that the WT cells rarely invaded deeply into the areas of low cellular density at high optical magnification. In addition, the borders of individual patches formed by the biosurfactant mutant were less defined and the mutant cells appeared to flow out on top of the neighboring WT cells, akin to lava flowing out from a volcano. However, these observations indirectly contradicted the results of our membrane assay, which showed that the same biosurfactant promoted the spreading of cells on the membrane surface. In fact, we initially referred to the corresponding secretion as a biosurfactant, due to the well-known function of bacterial surfactants in reducing surface tension to promote swarming on semisolid agar surfaces (48).
We identified the biosynthetic genes of the biosurfactant in this study, which were recently characterized by an independent group to produce a cyclic lipopeptide named gacamide A that promotes swarming (44). Pseudomonas spp. produce numerous cyclic lipopeptides that variably contribute to surface spreading and biofilm formation, and this variability potentially depends on discrete interactions with diverse extracellular or cell membrane-associated products (46, 49). The amphiphilic structure of gacamide A likely promotes its interaction with both hydrophilic compounds, like the mucoid polymer, and hydrophobic compounds that coaccumulate within the patches formed by the rsmE mutant. Importantly, removing the production of both the mucoid polymer and gacamide A maintained the respective rsmE mutant’s ability to outcompete the WT, albeit with much-reduced spatial dominance. These observations suggest that there are additional RsmE-regulated products that contribute to the competitive advantage of the rsmE mutant, which is clearly manifested through beneficial structures (23). Pressure likely builds up internally within a localized patch as the accumulating mucoid polymer constantly pushes away the surrounding WT cells to expand space. We thus speculate that gacamide A physically stabilizes the mucoid polymer and additional RsmE-regulated products to prevent their diffusion at the surface of the colony, which is uniquely devoid of neighboring cells and provides much less resistance.
A potential criticism of this study is the utilization of bacterial colonies to explore spatial structure formation, as these colonies lack important mechanical properties that manifest in natural microbial communities (7). However, resolving the problem of space and resource constraints in a densely populated colony likely shares common principles with other organisms in different experimental systems. Extracellular polysaccharides produced by Vibrio cholerae growing in microfluidic device biofilms promote the formation of isogenic structures that exclude the neighboring nonproducers (50), and glycolipid biosurfactants produced by Streptococcus spp. selectively displace competing genotypes on the tooth surface (51). In addition, cyclic lipopeptide production in Bacillus subtilis impacts the structure of fruiting body formation on an agar surface (52), but it is not essential (53). Our study also established a highly tractable experimental pipeline to identify and characterize additional RsmE-regulated products and to explore why RsmA and RsmI are functionally excluded from the formation of spatial structures.
MATERIALS AND METHODS
Strains and culture conditions.
Liquid and solid Lennox LB medium (Fisher) was used for general overnight cultures. Pseudomonas agar F (PAF; Difco) medium was used for all phenotypic screens, competitions, and microscopy. Pseudomonas minimum medium (PMM; 3.5 mM potassium phosphate dibasic trihydrate, 2.2 mM potassium phosphate monobasic, 0.8 mM ammonium sulfate, 100 mM magnesium sulfate, 100 mM sodium succinate) was used to selectively grow P. fluorescens isolates from conjugations with Escherichia coli donors. Routine cloning was carried out in E. coli 10B (Invitrogen) or E. coli JM109 (Promega), and E. coli S17.1λpir (54) was used as the donor strain in conjugations. When required, antibiotics were added to the medium at the following final concentrations: kanamycin (50 μg/mL), streptomycin (50 μg/mL), ampicillin (100 μg/mL), and gentamicin (20 μg/mL). P. fluorescens was cultured at 30°C or at room temperature (~22°C), and E. coli strains were cultured at 37°C. Liquid cultures were incubated with shaking at 250 rpm. All P. fluorescens strains used in this study are listed in Table 1.
TABLE 1.
P. fluorescens strains used in this study
| Strain | Relevant genotype | Relevant phenotype | Source |
|---|---|---|---|
| Pf0-1 | WT | Nonmucoid, no biosurfactant | 66 |
| Pf0-1S | WT (Tn7-SmR) | Streptomycin resistance | 23 |
| Pf0-1R | WT (Tn7-DsRed Express) | Red fluorescence | 23 |
| ΔrsmE | WT (ΔPfl01_1912) | Mucoid, biosurfactant | 23 |
| ΔrsmA | WT (ΔPfl01_4273) | Nonmucoid, no biosurfactant | This study |
| ΔrsmI | WT (ΔPfl01_4104) | Nonmucoid, no biosurfactant | This study |
| M | WT (126th nucleotide deleted in rsmE) | Mucoid, biosurfactant | 23 |
| MK | M (Tn7-KmR) | Kanamycin resistance | 23 |
| MG | M (Tn7-Gfpmut2) | Green fluorescence | 23 |
| M* | M (ΔPfl01_3834) | Nonmucoid, biosurfactant | 42 |
| M*K | M* (Tn7-KmR) | Kanamycin resistance | This study |
| M*G | M* (Tn7-Gfpmut2) | Green fluorescence | 42 |
| MS | M (ΔPfl01_2211) | Mucoid, no biosurfactant | This study |
| MSK | MS (Tn7-KmR) | Kanamycin resistance | This study |
| MSG | MS (Tn7-Gfpmut2) | Green fluorescence | This study |
| MS* | M (ΔPfl01_2211 ΔPfl01_3834) | Nonmucoid, no biosurfactant | This study |
| MS*K | MS* (Tn7-KmR) | Kanamycin resistance | This study |
| MS*G | MS* (Tn7-Gfpmut2) | Green fluorescence | This study |
RT-qPCR.
Total RNA was isolated from colonies grown for 3 days at room temperature on PAF plates by using the TRIzol reagent (Thermo Fisher) following the manufacturer’s protocol. Total RNA quality and concentration were assessed using a NanoDrop spectrometer. First-strand cDNA synthesis was carried out using the High-Capacity RNA-to-cDNA kit (Applied Biosystems) with 1 μg of RNA and random hexamers, following the manufacturer’s protocol. Quantitative PCR (qPCR) optimized primers were obtained from Integrated DNA Technologies (see Table S1 in the supplemental material), and their quality was assessed through PCR with genomic DNA, cDNA, and no-reverse transcriptase (no-RT) cDNA reactions. qPCR was performed using SYBR green (Thermo Fisher) on the StepOnePlus instrument (Applied Biosystems). Each reaction was analyzed to ensure only one amplicon was amplified, by using dissociation curves. Gene expression was calculated using the 2−ΔΔCT method with the 16S rRNA gene as an internal reference and quantified relative to rsmI expression (55).
Biosurfactant assay.
Nuclepore Track-Etch polycarbonate membranes (Whatman; 0.4-μM pore size, 90-mm diameter) were used for assessing biosurfactant production. As previously described (23), one side of the membrane is shiny and the other is dull due to the manufacturing process. The dull side’s surface contains gaps and ridges that physically trap cells, but the biosurfactant permeates to produce a visible ring around colonies. The shiny side’s surface is smooth, which allows biosurfactant-producing cells to spread out through growth. Sterile forceps were used to overlay the membrane on the PAF agar surface, and 20 μL of overnight culture was spotted directly on the membrane and allowed to fully dry before the plates were inverted and incubated overnight at room temperature.
Identification of biosurfactant biosynthesis genes by transposon mutagenesis.
Random transposon mutagenesis, using the plasmid pUT-miniTn5-KmlacZ2 (56, 57) in E. coli S17.1λpir as the donor, was carried to identify the biosynthesis genes of the biosurfactant, as previously described to identify the biosynthesis genes of the mucoid polymer (53). Briefly, overnight cultures of the donor and M* strains were washed in PMM, mixed at the relative ratio of 1:6, spotted on solid LB to conjugate, incubated at 30°C for 3 h, harvested, and plated out on solid PMM supplemented with kanamycin. Over 20,000 transconjugant colonies were picked and rearrayed using the QBot system (Genetix) into 384-well plates containing kanamycin-supplemented PMM and then incubated at 30°C. Surfactant assays on overnight cultures were conducted on PMM plates overlaid with the dull side of the polycarbonate membrane, as described above, with a disposable 384-pin replicator (Scinomix). Mutants that were defective in biosurfactant production (dull side) were rearrayed into 96-well plates containing kanamycin-supplemented PMM and then incubated at 30°C. Overnight cultures were retested for biosurfactant production as described above using a disposable 96-pin replicator (Scinomix). Mutants that failed to produce the biosurfactant ring were selected, ignoring those that had obvious growth defects. The transposon insertion sites were identified by arbitrary primed PCR as previously described (53).
Mutant construction, complementation, and tagging.
Gene deletion mutants were constructed by the gene splicing by overlap extension method (58), using the plasmid pMQ30 (59) or pSR47s (60) as previously outlined (23, 42). PCR primers used to construct and confirm each mutation are listed in Table S2. Briefly, for each targeted gene, approximately 500 bp of its flanking upstream and downstream regions were individually amplified, joined together, first cloned into the pGEM-T Easy vector system (Promega), then subcloned into pMQ30 or pSR47s, and transformed into E. coli S17.1λpir as the donor strain. Overnight cultures of the donor and target strains were washed in PMM and mixed at an equal ratio, spotted on solid LB, incubated at 30°C overnight, harvested, and plated out on solid PMM supplemented with gentamicin (pMQ30) or kanamycin (pSR47s). Transformants were grown on solid LB supplemented with sucrose (5% [wt/vol]) overnight, and the resulting colonies were screened using primers that bind outside the two flanking fragments for the expected reduction in amplicon size. To confirm the gene deletions, we isolated genomic DNA from overnight cultures using the DNeasy UltraClean microbial kit (Qiagen) following the manufacturer’s protocol, and whole-genome sequencing was conducted at the Microbial Genome Sequencing Center (MiGS; Pittsburgh, PA). Kanamycin-resistant and streptomycin-resistant strains used in competitions and GFP-tagged and DsRed Express-tagged strains used in microscopy were constructed using the mini-Tn7 chromosomal insertion system (61) as previously described (23, 42). The same set of the kanamycin and streptomycin resistance markers utilized in this study has been demonstrated to be neutral for conducting competition experiments in P. fluorescens Pf0-1 (23). The ΔrsmE mutant was complemented by integrating the native rsmE locus into its chromosome by using the mini-Tn7 system (pHRB2) via conjugation as previously described (62). The native rsmE locus, including the 500-bp upstream and 500-bp downstream sequences from the open reading frame, was PCR amplified from P. fluorescens Pf0-1 using primers RsmE1 (5′-CGCTGGCATCCTTGATGACG) and RsmE2 (5′-TCTGGATCCGGTGAGGTCGC). The amplified product was cloned into pGEMT-Easy, then subcloned into pHRB2 using the ApaI and EcoRI sites, and introduced into the ΔrsmE mutant as described above. Chromosomal integration of the native rsmE locus into the noncoding Tn7 insertion site was confirmed by whole-genome sequencing.
Measurement of monoculture growth.
For the measurement of growth in colonies, overnight cultures were resuspended in PMM and 20 μL was spotted on PAF plates and incubated at room temperature. To enumerate the initial population size, each cell suspension in PMM was serially diluted and plated on LB plates, and resulting colonies were counted on the following day. Three spotted colonies were scraped on each day over 7 days and resuspended in 5 mL of PMM using a sterilized bent glass Pasteur pipette. Cell suspensions were vortexed until clumps were no longer visible and then serially diluted and enumerated as described above. For the measurement of growth in liquid, overnight cultures were diluted into a manually formulated PAF without agar (63) in six replicates, and optical density at 600 nm was measured every 30 min over 48 h (30°C, constant shaking) in the Bioscreen C MBR system (Oy Growth Curves Ab Ltd.).
Competition assay.
Competitions between kanamycin-resistant mutant strains and the streptomycin-resistant WT strain were conducted as previously described (23). Briefly, overnight cultures (1.5 mL) were washed in fresh PMM and resuspended in 1.0 mL PMM, and the mutant strain suspension was serially diluted to 10−3 in PMM and mixed with equal volumes of the undiluted WT strain suspension. Twenty microliters of each competition mixture was spotted in triplicate on a PAF plate and incubated at room temperature. To enumerate the initial population sizes of the competing strains, each competition mixture was serially diluted and plated on LB plates supplemented with either kanamycin or streptomycin, and resulting colonies were counted on the following day. Four or seven days later, the spotted colonies were scraped and resuspended in 5 mL of PMM, serially diluted, plated, and counted as for the initial competition mixture. The results of the competitions were analyzed by calculating the relative fitness (W) of each competing strain against the WT (64), as follows: [ln(CFU of mutant at time of sampling/CFU of mutant at time zero)]/[ln(CFU of WT at time of sampling/CFU of WT at time zero)].
Statistical analysis.
Competition experiments were conducted with at least three biological replicates and two technical replicates for each biological replicate. The data were first analyzed with an analysis of variance (ANOVA) to evaluate if the means of the biological replicates differed significantly, and then Tukey’s honest significant difference test (P < 0.05) was applied to make multiple pairwise comparisons within the data set. All comparisons were found to be statistically different or are noted as nonsignificant. Statistical tests were conducted using GraphPad Prism.
Microscopy.
Overnight cultures of GFP-labeled strains and DsRed Express-labeled WT were washed and resuspended in PMM. All GFP-labeled cell suspensions were serially diluted to 10−5 in PMM and mixed with equal volumes of the undiluted DsRed Express-labeled WT suspension. Twenty microliters of each competition mixture was spotted in triplicate on PAF plates and incubated at room temperature. Epifluorescence microscopy was conducted using the Nikon SMZ25 stereo compound microscope with 0.5× and 2× SHR Plan Apo objectives and the NIS Elements software. For confocal microscopy, an agar slice containing the entire colony was placed on a microscope slide and visualized without a coverslip. Confocal microscopy was conducted using a Nikon Ti2 microscope with the air-corrected 100× TU Plan Apo objective and the NIS Elements software. Nonfluorescent imaging of colonies was carried out using the Hayear overhead microscope (HY-2307) or the Canon Rebel EOS T3 DSLR camera. Images were rendered using the NIS Elements and ImageJ software.
Data availability.
All noncommercial plasmids or strains used in this study are available for distribution upon request.
ACKNOWLEDGMENTS
W.K. designed the study; A.E., M.W., J.D., W.M., R.S., A.D., and W.K. performed experiments; A.E., M.W., J.D., and W.K. analyzed data; and A.E., M.W., and W.K. wrote the manuscript. This study was funded by the National Institute of General Medical Sciences of the NIH (1R15GM132856 to W.K.).
Footnotes
Supplemental material is available online only.
Contributor Information
Wook Kim, Email: kimw1@duq.edu.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 2.Sutherland IW. 2001. The biofilm matrix: an immobilized but dynamic microbial environment. Trends Microbiol 9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- 3.Fong JNC, Yildiz FH. 2015. Biofilm matrix proteins. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hobley L, Harkins C, Macphee CE, Stanley-Wall NR. 2015. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev 39:649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branda SS, Vik Å, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol 13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Karygianni L, Ren Z, Koo H, Thurnheer T. 2020. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol 28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, Wingreen NS, Bassler BL, Gitai Z, Stone HA. 2015. The mechanical world of bacteria. Cell 161:988–997. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol Rev 33:206–224. doi: 10.1111/j.1574-6976.2008.00150.x. [DOI] [PubMed] [Google Scholar]
- 9.Xavier JB, Foster KR. 2007. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA 104:876–881. doi: 10.1073/pnas.0607651104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadell CD, Foster KR, Xavier JB. 2010. Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput Biol 6:e1000716. doi: 10.1371/journal.pcbi.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitri S, Xavier JB, Foster KR. 2011. Social evolution in multispecies biofilms. Proc Natl Acad Sci USA 108:10839–10846. doi: 10.1073/pnas.1100292108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. 2016. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA 113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallatschek O. 2011. The noisy edge of traveling waves. Proc Natl Acad Sci USA 108:1783–1787. doi: 10.1073/pnas.1013529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. 2007. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA 104:19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korolev KS, Xavier JB, Nelson DR, Foster KR. 2011. A quantitative test of population genetics using spatiogenetic patterns in bacterial colonies. Am Nat 178:538–552. doi: 10.1086/661897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mavridou DAI, Gonzalez D, Kim W, West SA, Foster KR. 2018. Bacteria use collective behavior to generate diverse combat strategies. Curr Biol 28:345–355.e4. doi: 10.1016/j.cub.2017.12.030. [DOI] [PubMed] [Google Scholar]
- 17.Crespi BJ. 2001. The evolution of social behavior in microorganisms. Trends Ecol Evol 16:178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- 18.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat Rev Microbiol 4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 19.Smith WPJ, Davit Y, Osborne JM, Kim W, Foster KR, Pitt-Francis JM. 2017. Cell morphology drives spatial patterning in microbial communities. Proc Natl Acad Sci USA 114:E280–E286. doi: 10.1073/pnas.1613007114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. 2007. Evolution of species interactions in a biofilm community. Nature 445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 21.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e00103-13. doi: 10.1128/mBio.00103-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolter R, Greenberg EP. 2006. Microbial sciences: the superficial life of microbes. Nature 441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 23.Kim W, Racimo F, Schluter J, Levy SB, Foster KR. 2014. Importance of positioning for microbial evolution. Proc Natl Acad Sci USA 111:E1639–E1647. doi: 10.1073/pnas.1323632111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lapouge K, Schubert M, Allain FHT, Haas D. 2008. Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol 67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 25.Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J. 2016. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 35:991–1011. doi: 10.15252/embj.201593360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobrero PM, Valverde C. 2020. Comparative genomics and evolutionary analysis of RNA-binding proteins of the CsrA family in the genus Pseudomonas. Front Mol Biosci 7:127. doi: 10.3389/fmolb.2020.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye F, Yang F, Yu R, Lin X, Qi J, Chen Z, Cao Y, Wei Y, Gao GF, Lu G. 2018. Molecular basis of binding between the global post-transcriptional regulator CsrA and the T3SS chaperone CesT. Nat Commun 9:1196. doi: 10.1038/s41467-018-03625-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huertas-Rosales Ó, Romero M, Chan K-G, Hong K-W, Cámara M, Heeb S, Barrientos-Moreno L, Molina-Henares MA, Travieso ML, Ramos-González MI, Espinosa-Urgel M. 2021. Genome-wide analysis of targets for post-transcriptional regulation by Rsm proteins in Pseudomonas putida. Front Mol Biosci 8:50. doi: 10.3389/fmolb.2021.624061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duss O, Michel E, Dit Konté ND, Schubert M, Allain FHT. 2014. Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition. Nucleic Acids Res 42:5332–5346. doi: 10.1093/nar/gku141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romeo T, Babitzke P. 2018. Global regulation by CsrA and its RNA antagonists. Microbiol Spectr 6. doi: 10.1128/microbiolspec.RWR-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol Microbiol 78:158–172. doi: 10.1111/j.1365-2958.2010.07320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker CS, Eöry LA, Yakhnin H, Mercante J, Romeo T, Babitzke P. 2007. CsrA inhibits translation initiation of Escherichia coli hfq by binding to a single site overlapping the Shine-Dalgarno sequence. J Bacteriol 189:5472–5481. doi: 10.1128/JB.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huertas-Rosales Ó, Ramos-González MI, Espinosa-Urgel M. 2016. Self-regulation and interplay of Rsm family proteins modulate the lifestyle of Pseudomonas putida. Appl Environ Microbiol 82:5673–5686. doi: 10.1128/AEM.01724-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramírez-Zapata D, Ramos C, Aguilera S, Bardaji L, Martínez-Gil M, Murillo J. 2020. Two homologues of the global regulator Csr/Rsm redundantly control phaseolotoxin biosynthesis and virulence in the plant pathogen Pseudomonas amygdali pv. phaseolicola 1448A. Microorganisms 8:1536–1538. doi: 10.3390/microorganisms8101536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vakulskas CA, Potts AH, Babitzke P, Ahmer BMM, Romeo T. 2015. Regulation of bacterial virulence by Csr (Rsm) systems. Microbiol Mol Biol Rev 79:193–224. doi: 10.1128/MMBR.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero M, Silistre H, Lovelock L, Wright VJ, Chan K-G, Hong K-W, Williams P, Amara MC, Heeb S. 2018. Genome-wide mapping of the RNA targets of the Pseudomonas aeruginosa riboregulatory protein RsmN. Nucleic Acids Res 46:6823–6840. doi: 10.1093/nar/gky324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris ER, Hall G, Li C, Heeb S, Kulkarni RV, Lovelock L, Silistre H, Messina M, Cámara M, Emsley J, Williams P, Searle MS. 2013. Structural rearrangement in an RsmA/CsrA Ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 21:1659–1671. doi: 10.1016/j.str.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schulmeyer KH, Diaz MR, Bair TB, Sanders W, Gode CJ, Laederach A, Wolfgang MC, Yahr TL. 2016. Primary and secondary sequence structure requirements for recognition and discrimination of target RNAs by Pseudomonas aeruginosa RsmA and RsmF. J Bacteriol 198:2458–2469. doi: 10.1128/JB.00343-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reimmann C, Valverde C, Kay E, Haas D. 2005. Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0. J Bacteriol 187:276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim W, Levy SB, Foster KR. 2016. Rapid radiation in bacteria leads to a division of labour. Nat Commun 7:10508. doi: 10.1038/ncomms10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sieber SA, Marahiel MA. 2003. Learning from nature's drug factories: nonribosomal synthesis of macrocyclic peptides. J Bacteriol 185:7036–7043. doi: 10.1128/JB.185.24.7036-7043.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahanshah G, Yan Q, Gerhardt H, Pataj Z, Lämmerhofer M, Pianet I, Josten M, Sahl H-G, Silby MW, Loper JE, Gross H. 2019. Discovery of the cyclic lipopeptide gacamide A by genome mining and repair of the defective GacA regulator in Pseudomonas fluorescens Pf0-1. J Nat Prod 82:301–308. doi: 10.1021/acs.jnatprod.8b00747. [DOI] [PubMed] [Google Scholar]
- 45.De Bruijn I, De Kock MJD, De Waard P, Van Beek TA, Raaijmakers JM. 2008. Massetolide A biosynthesis in Pseudomonas fluorescens. J Bacteriol 190:2777–2789. doi: 10.1128/JB.01563-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Bruijn I, De Kock MJD, Yang M, De Waard P, Van Beek TA, Raaijmakers JM. 2007. Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species. Mol Microbiol 63:417–428. doi: 10.1111/j.1365-2958.2006.05525.x. [DOI] [PubMed] [Google Scholar]
- 47.Ferreiro M-D, Nogales J, Farias GA, Olmedilla A, Sanjuán J, Gallegos MT. 2018. Multiple CsrA proteins control key virulence traits in Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact 31:525–536. doi: 10.1094/MPMI-09-17-0232-R. [DOI] [PubMed] [Google Scholar]
- 48.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raaijmakers JM, De Bruijn I, Nybroe O, Ongena M. 2010. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev 34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 50.Nadell CD, Bassler BL. 2011. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci USA 108:14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reid G, Younes JA, Van der Mei HC, Gloor GB, Knight R, Busscher HJ. 2011. Microbiota restoration: natural and supplemented recovery of human microbial communities. Nat Rev Microbiol 9:27–38. doi: 10.1038/nrmicro2473. [DOI] [PubMed] [Google Scholar]
- 52.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thérien M, Kiesewalter HT, Auria E, Charron-Lamoureux V, Wibowo M, Maróti G, Kovács ÁT, Beauregard PB. 2020. Surfactin production is not essential for pellicle and root-associated biofilm development of Bacillus subtilis. Biofilm 2:100021. doi: 10.1016/j.bioflm.2020.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon R, Priefer U, Pühler A. 1983. A Broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Lorenzo V, Herrero M, Jakubzik U, Timmis KN. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in Gram-negative eubacteria. J Bacteriol 172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonin M, Quardokus EM, O'Donnol D, Maddock J, Brun YV. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J Bacteriol 182:337–347. doi: 10.1128/JB.182.2.337-347.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 59.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matthews M, Roy CR. 2000. Identification and subcellular localization of the Legionella pneumophila IcmX protein: a factor essential for establishment of a replicative organelle in eukaryotic host cells. Infect Immun 68:3971–3982. doi: 10.1128/IAI.68.7.3971-3982.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lambertsen L, Sternberg C, Molin S. 2004. Mini-Tn7 transposons for site-specific tagging of bacteria with fluorescent proteins. Environ Microbiol 6:726–732. doi: 10.1111/j.1462-2920.2004.00605.x. [DOI] [PubMed] [Google Scholar]
- 62.Kim W, Levy SB. 2008. Increased fitness of Pseudomonas fluorescens Pf0-1 leucine auxotrophs in soil. Appl Environ Microbiol 74:3644–3651. doi: 10.1128/AEM.00429-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kessler C, Mhatre E, Cooper V, Kim W. 2021. Evolutionary divergence of the Wsp signal transduction systems in Beta- and Gammaproteobacteria. Appl Environ Microbiol 87:e01306-21. doi: 10.1128/AEM.01306-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiser MJ, Lenski RE. 2015. A comparison of methods to measure fitness in Escherichia coli. PLoS One 10:e0126210. doi: 10.1371/journal.pone.0126210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 66.Compeau G, Al-Achi BJ, Platsouka E, Levy SB. 1988. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol 54:2432–2438. doi: 10.1128/aem.54.10.2432-2438.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Table S1. Download jb.00285-22-s0001.pdf, PDF file, 0.5 MB (567.9KB, pdf)
Data Availability Statement
All noncommercial plasmids or strains used in this study are available for distribution upon request.





