ABSTRACT
Intestinal microenvironment dysbiosis is one of the major causes of diseases, such as obesity, diabetes, inflammatory bowel disease, and colon cancer. Microbiota-based strategies have excellent clinical potential in the treatment of repetitive and refractory diseases; however, the underlying regulatory mechanisms remain elusive. Identification of the internal regulatory mechanism of the gut microbiome and the interaction mechanisms involving bacteria-host is essential to achieve precise control of the gut microbiome and obtain effective clinical data. Gut bacteria-derived extracellular vesicles (GBEVs) are lipid bilayer nanoparticles secreted by the gut microbiota and are considered key players in bacteria-bacteria and bacteria-host communication. This review focusses on the role of GBEVs in gut microbiota interactions and bacteria-host communication, and the potential clinical applications of GBEVs.
KEYWORDS: Bacteria-bacteria interaction, bacteria-host interaction, gut bacterial extracellular vesicles, intestinal dysbiosis, intestinal microenvironment
Introduction
The intestinal microenvironment is the most important and complex micro-ecological host system, maintained by the microbial community and the host immune system. In addition to microorganisms, such as bacteria, archaea, protists, fungi, and viruses and their metabolites, components of the intestinal microenvironment include intestinal secretions, intestinal epithelial cells (IECs), and cytokines released during host immune responses.1,2 The intestinal microenvironment plays key roles in nutrient digestion, absorption, and metabolism in the intestine, development of organs, such as gut and brain, and synergistically regulating the host immune system.3 Dysbiosis of the intestinal microenvironment is closely associated with the initiation and progression of diseases such as common digestive system diseases,4 inflammatory bowel disease (IBD), 5 functional gastrointestinal disorders,6 and chronic gastritis.7 Additionally, it is also associated with distal organ lesions, such as those observed in cancer,8 and diseases of the cardiovascular system,9 brain,10 kidney,11 liver,12 and pancreas.13 Intestinal microenvironment dysbiosis mainly manifests as a change in the proportion or number of colonization sites of intestinal microbes. Particularly, an imbalance of the intestinal microbiota occurs, resulting in a loss of host functions including energy metabolism, signal transduction, pathogen resistance, and immune regulation, and finally inducing or worsening a disease. Such changes are mainly attributed to a decrease in the abundance and diversity of commensal microflora coupled with an increase in the abundance of pathogenic bacteria. In a nutshell, the intestinal microenvironment is inextricably associated with the optimal functioning of the gut. Effective intervention strategies to restore homeostasis of the intestinal microenvironment include fecal microbiota transplantation (FMT) and the use of microecologics.14,15
The gut microbiota is predominantly composed of bacteria. More than 1000 bacterial species are commonly found in the intestinal tract.1 The composition of the gut microbiota is influenced by host factors such as intestinal environment, diet, and age.16–18 On the other hand, the physiological and psychological states of the host are influenced by gut bacterial communities.19,20 Studies in mice have demonstrated that up to 70% of the intestinal chemicals are regulated by gut bacterial communities. Moreover, such chemical regulation is observed in organs distant from the gut, such as uterus or brain, 20% of the compounds are regulated by gut bacteria.21
Although gut bacteria are critical for the regulation of the intestinal microenvironment and host health, the mechanisms underlying this regulation have been largely unexplored.22,23 However, recently, bacterial extracellular vesicles (BEVs) secreted by intestinal commensal bacteria, probiotics, and pathogenic bacteria were identified to modulate intestinal microenvironment and host health.24–26 Extracellular vesicles (EVs) are defined as nano-vesicles that are secreted by cell membranes of live cellular organisms such as eukaryotes, bacteria, archaea, and mycoplasma. EVs are mostly spherical in shape and 20–400 nm in diameter.27 The vesicle membrane is a lipid bilayer and may encapsulate molecules such as DNA, RNA, proteins, peptidoglycan, and lipids (Figure 1A).28,29 The development of omics technology has helped elucidate the chemical composition of EVs. However, most of the current studies on cargo mainly focus on structural and functional analysis of protein components, with few studies considering other EV components such as lipids, nucleic acids, and polysaccharides.
Figure 1.
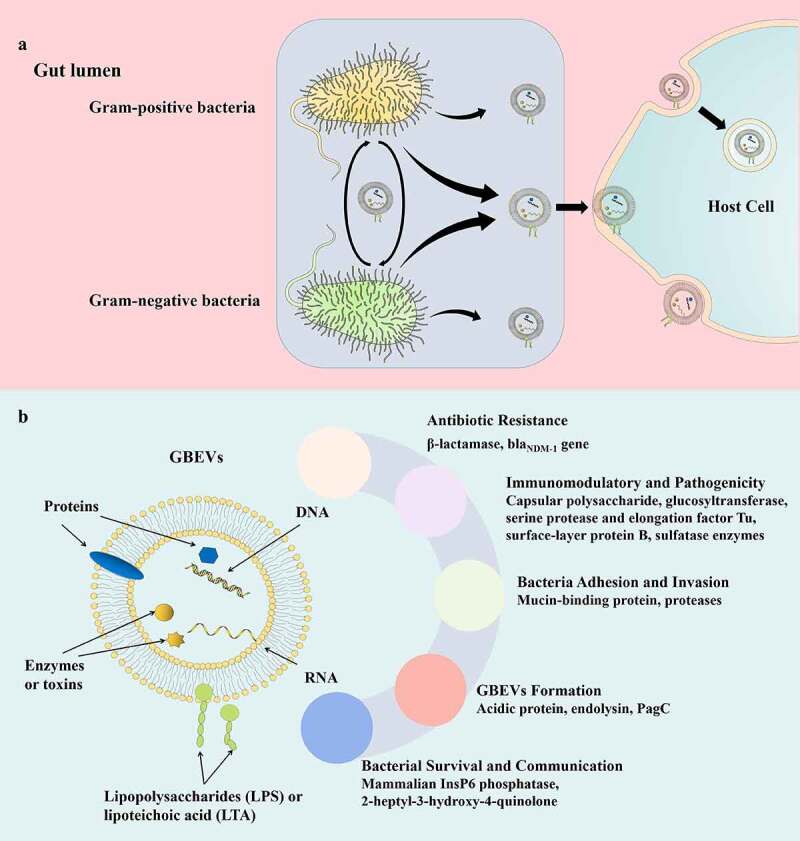
(a) Proposed critical pathophysiological functions of GBEV cargos. (b) Schematic representation of bacteria-host and bacteria-bacteria GBEVs interactions. GBEVs: gut bacteria-derived extracellular vesicles.
In 1981, Wensink and Witholt speculated that excessive production of the bacterial outer membrane was the leading cause of BEV generation;30 however, this hypothesis failed to explain the formation of BEVs under normal growth conditions. In 1998, Leah et al. proposed that gram-negative BEVs result from cell wall turnover, and peptidoglycan turnover causes the outer membrane to bulge and finally bleb, followed by mechanical motion of the cell wall to release BEVs.31 Although this was demonstrated in the formation of Porphyromonas gingivalis EVs, further research revealed that the biogenesis of BEVs is a complex process, and various factors affect their secretion. So far, a consensus mechanism underlying BEV biogenesis applicable to all bacteria has not been identified.32,33 BEVs secreted by gram-negative bacteria are also referred to as outer membrane vesicles (OMVs).34 Gut bacteria-derived extracellular vesicles (GBEVs) were first identified in the electron microscopic images of Escherichia coli by Work et al. in 1966,35 and a similar structure was found in sections of Vibrio cholerae by Chatterjee et al. in 1967.36 The concept of BEVs was explicitly proposed by Dorward et al. in 1990.37 Subsequently, research surrounding gram-positive bacteria, gram-negative bacteria, and archaea in the intestinal tract confirmed the presence of GBEVs.28 Since then, the structure, biogenesis, and interactions within the intestinal flora and with the host of GBEVs has attracted tremendous research interest. Regulation of GBEVs in the intestinal microenvironment and host health has also become an emerging research field in the study of intestinal microbial interactions and bacteria-host interactions (Figure 1B). A variety of commensal bacteria as well as enteric pathogens are present in different stages of the gut (in diseased states or physiological metabolic disorders). To better illustrate the role of GBEVs in the intestinal microenvironment, in addition to GBEVs secreted by intestinal commensal bacteria, GBEVs secreted by enteric pathogens are included in this review. Herein, we describe the regulatory role of GBEVs in the intestinal microenvironment played by bacteria-bacteria and bacteria-host communication, and discuss their possible clinical implications.
GBEVs-mediated bacteria-bacteria interactions
Currently, the regulation of gut microbiota, such as through FMT and probiotic intervention, to restore homeostasis of the intestinal microenvironment is an important therapeutic strategy for recurrent and refractory diseases38 and exhibit excellent clinical therapeutic effects. Nonetheless, the mechanisms underlying these strategies are still unclear. Intestinal bacteria can communicate with the surrounding environment by secreting chemicals such as toxins, quorum sensing (QS) molecules, and nucleic acids. GBEVs, as carriers of these signaling molecules, play an important role in regulating the in vivo balance of intestinal bacteria (summarized in Table 1).
Table 1.
The roles of GBEVs in communication cross-talk between gut bacteria.
| GBEVs source | Cargos of GBEVs | Function of GBEVs | Refs |
|---|---|---|---|
| Bacteroides fragilis | Acidic lipoproteins | Degradation of various polysaccharides. | 39 |
| Bacteroides thetaiotaomicron | 1. β-lactamases; 2. Acidic lipoproteins |
1. Protect commensal bacteria and enteric pathogens against β-lactam antibiotics; 2. Degradation of various polysaccharides. |
39,40 |
| Bifidobacterium longum | Mucin-binding proteins | Promote bifidobacterial colonization. | 24 |
| Escherichia coli | Unknown | Protection of parental bacteria by adsorption of colistin. | 41 |
| Escherichia coli O104:H4 | blaCTX-M-15 | Transmission of antibiotic resistance gene. | 42 |
| Pseudomonas aeruginosa | 2-heptyl-3-hydroxy- 4-quinolone (PQS) |
1. Communicate and coordinate social activities; 2. Promotes biofilm diffusion; 3. Aids in the absorption of iron molecules by bacteria. |
43–45 |
| Vibrio cholerae C6706 | PrtV protease | Helps bacteria resist host defense peptides. | 46 |
| Vibrio cholerae V:5/04 non-O1 non-O139 clinical isolate | Unknown | Inhibit innate immune response of host and promoting bacterial colonization. | 47 |
GBEVs: gut bacteria-derived extracellular vesicles; PQS: Pseudomonas quinolone signal; Refs: references.
GBEVs promote bacterial colonization and growth
In the intestinal microenvironment, GBEVs can both help the colonization of intestinal bacteria and promote their proliferation. As an example, GBEVs are loaded with adhesion factors that promote intestinal bacterial colonization and carry polysaccharide-degrading enzymes which breakdown complex polysaccharides and provide a source of carbohydrates for the intestinal bacteria.24,48
Bacterial surface adhesion molecules, such as surface-exposed human plasminogen receptor,49 enolase,50 sialidase,51 fimbriae,52 and transaldolase,53 are critical for mediating bacteria-host interactions. Recently, Nishiyama et al. identified that the GBEVs secreted by Bifidobacterium longum NCC2705 play a key role in the colonization process.24 Biomolecular interaction analysis has revealed that GBEVs contain a large number of cytoplasmic proteins that bind to porcine gastric mucin such as chaperone GroEL, elongation factor Tu, phosphoglycerate kinase, Tal, and heat shock protein 20. In addition, these proteins were recombinantly expressed in E. coli, immobilized on microbeads, and were found to adhere to the gastrointestinal tract of mice.24 These findings suggest that GBEVs can effectively assist the colonization of B. longum by transporting bacterial adhesion factors into the extracellular environment.
GBEVs may also promote enteric pathogen colonization by reducing the host innate immune response. Bitar et al. coincubated V. cholerae V:5/04 non-O1 non-O139 clinical isolate (a highly motile gram-negative bacterium that colonizes the small intestine and causes cholera) secreted GBEVs with T84 cells for 2 h.47 The expression of microRNAs was determined using quantitative real-time polymerase chain reaction, and a significant increase in miR-146a expression level was observed. Elevated expression levels of miR-146a inhibit the host’s innate immune response. Interestingly, no changes in mRNA expression levels of pro-inflammatory factors, including interleukin (IL)-8, tumor necrosis factor alpha (TNF-α), and IL-1β, were observed. This effect prevents V. cholerae from being immediately cleared by the host immune system, thereby allowing V. cholerae to colonize the host gut.47 Although research exploring the role of GBEVs in colonization of intestinal bacterial is limited, GBEVs are presumed to exert some specific advantages in bacterial colonization because they play an important role in host-bacteria communication. In the future, GBEVs may play a central role in strategies aimed at regulating the host gut bacterial ecological balance. For instance, GBEVs could be used to enhance probiotic colonization, which is critical for the therapeutic effect of probiotics, or GBEVs could be disrupted to reduce the colonization of pathogenic bacteria.
GBEVs can provide nutrients to other bacteria by delivering a variety of enzymes involved in the degradation of complex polysaccharides. Valguarnera et al. analyzed the protein composition of Bacteroides thetaiotaomicron EVs using mass spectrometry and identified several glycosidases and proteases, which were mainly encoded by the polysaccharide utilization loci. When carried by GBEVs and released into the intestinal lumen, enzymes break down polysaccharides present in ingested food or host-synthesized sugar adducts into mono- or oligo-saccharides, thus promoting the growth of other bacteria.48 Researchers further investigated the envelope mechanism and found that it is mediated by a lipoprotein export sequence (LES, a negatively charged rich amino acid motif). Alpha-amylase starch utilization system (Sus) G is mediated by LES from the periplasmic surface of outer membranes to the extracellular environment, which in turn is preferentially encapsulated by GBEVs. On co-culturing SusG-containing GBEVs with the ΔSusG strain (SusG gene knockout strain, which cannot grow in media containing starch as the sole carbon source), the growth of the ΔSusG strain was found to be similar to that of the wild-type.48 Therefore, uncontrolled intestinal flora can be regulated by GBEVs carrying specific effectors, and thus effectively balance the host intestinal microenvironment. In addition to B. thetaiotaomicron EVs, polysaccharide-degrading enzymes were also detected in Bacteroides fragilis EVs, indicating that the encapsulation behavior is not specific to B. thetaiotaomicron.39
GBEVs protect bacteria from antibiotics and host defense peptides
The greatest threat to the existence of host intestinal bacteria are antibiotics and host defense peptides (HDPs). Consequently, bacteria have evolved various effective defense strategies against antibiotics and HDPs, and GBEVs play vital roles in executing these strategies. GBEVs produce a protective effect by adsorbing antibiotics. For example, to protect antibiotic/HDPs-sensitive strains GBEVs adsorb antibiotics and deliver β-lactamase or antibiotic resistance gene (ARG) (Figure 2A).
Figure 2.
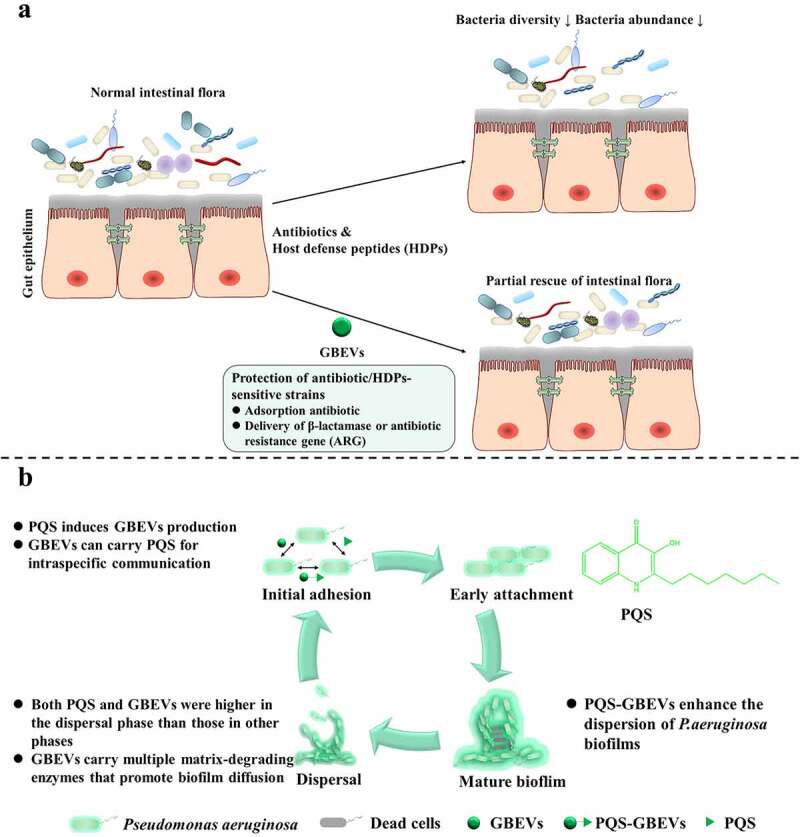
GBEVs mediate cross-talk between gut bacteria. (a) GBEVs can protect strains sensitive to antibiotics/HDPs. Treatment with antibiotics or HDPs disrupts the normal intestinal microbiota, leading to a reduction in both the diversity and abundance of intestinal bacteria, resulting in dysbiosis. GBEVs, through direct antibiotic adsorption41 and intra- and interspecies delivery of β-lactamase40 or antibiotic resistance gene42 can partially recovery. (b) PQS-GBEVs are involved in the developmental cycle of Pseudomonas aeruginosa biofilms.43 PQS can promote the secretion of GBEVs in P. aeruginosa.54 Moreover, PQS can coordinate population behavior through intra-species communication via GBEVs.45 GBEVs: gut bacteria-derived extracellular vesicles; PQS, Pseudomonas quinolone signal; PQS-GBEVs: PQS containing GBEVs.
Colistin is an amphiphilic polypeptide antibiotic that exerts bactericidal or antibacterial effect by electrostatically binding to lipopolysaccharide (LPS) on the outer membrane of gram-negative bacteria, causing Ca2+ and Mg2+ to move out of the phosphate group of LPS, resulting in disruption of the bacterial outer membrane and efflux of cell contents.55 The main cause of colistin resistance in gram-negative bacteria is the presence of mobilized colistin resistance (mcr) gene (mcr-1 to mcr-10).56 For example, mcr-1 gene expression results in phosphoethanolamine (pEtN) modification of phospholipid A on the outer membrane of E. coli, which reduces the negative charge of LPS and lowers its affinity for colistin.56 Interestingly, GBEVs secreted by E. coli with positive and negative mcr-1 expression showed differences in protective effect from parental bacteria. You and Zhang’s team found that both mcr-1 positive or negative E. coli EVs protected colistin-sensitive E. coli strains from the effects of colistin.41 They also found that GBEVs secreted by mcr-1-negative E. coli provide better protection than those secreted by mcr-1-positive E. coli. The main reason for this phenomenon is that the protective effect developed by E. coli EVs is primarily due to their adsorption to colistin. The lipid A on the surface of GBEVs secreted by mcr-1-positive E. coli is the same as that of the parental bacteria and is also modified by pEtN, leading to a weakened binding of mcr-1-positive E. coli EVs to colistin, ultimately reducing the protective effect.41
In addition to antibiotic adsorption to protect sensitive strains, GBEVs can serve as a delivery platform for dissemination of antibiotic-degrading enzymes and ARGs. Among gut bacteria, anaerobic Bacteroides spp. are the most resistant to penicillins and cephalosporins.57 Bacteroides degrade penicillin by secreting β-lactamase58 or excrete antibiotics through Resistance-Nodulation-Division and multidrug and toxic compound extrusion efflux pumps.57 Stentz et al. determined that GBEVs secreted by B. thetaiotaomicron carry β-lactamases that degrade cefotaxime.40 Following proteinase K treatment, 84% of the β-lactamase enzymatic activity was lost, indicating that most of the β-lactamase contained in B. thetaiotaomicron EVs was located on the GBEVs surface. The team further studied GBEVs secreted by other bacteria of the genus Bacteroides (B. fragilis and B. stercoris) and detected high degradation activity for cefotaxime, thus protecting themselves from antibiotics. Furthermore, after adding B. thetaiotaomicron EVs to the media of Salmonella Typhimurium and Bifidobacterium breve, both grew normally in 1 mg/L of cefotaxime or ampicillin. Thus, GBEVs loaded with β-lactamase protect the parental as well as other bacteria from antibiotics.40 A homologous system was also observed in pathogenic E. coli ST131.59 Horizontal gene transfer (HGT) is a unique mechanism by which bacteria rapidly evolve and adapt to complex environments. In addition to transformation, transduction, and conjugation, GBEVs are often used as a fourth mode of HGT among bacteria.60 Bielaszewska et al. found that the diarrhea-causing enteric pathogen E. coli O104:H4 can deliver multiple ARGs (such as blaCTX-M-15 and blaTEM-1) to other bacteria via GBEVs.42 The investigators determined that, compared to that of the untreated group, the transfer frequency of blaCTX-M-15 in simulated intraintestinal environment (simulated ileal environment and colonic environment medium) and under the stress of ciprofloxacin (a drug commonly used to treat diarrhea caused by bacterial pathogens) increased approximately 100-fold.42 This transfer behavior is extremely beneficial for the intestinal bacteria and protects them from antibiotics; however, it possibly poses a major problem for patients with bacterial infections.
Apart from antibiotics, the most frequently encountered antimicrobial agents by gut bacteria in vivo are HDPs. HDPs are short peptides that exert antimicrobial effects primarily by interacting with bacterial membranes, causing membrane cleavage.61 HDPs are often used as alternatives to antibiotics because they can target multiple bacterial sites to exert antibacterial effects, making it difficult for bacteria to develop resistance. Veldhuizen’s team added CATH-2 (chicken-derived cathelicidin),62 PMAP-36 (pig-derived cathelicidin),63 and LL-37 (the only human cathelicidin)64 to E. coli medium and found that E. coli EVs effectively protected the bacteria from HDPs, and this protection was positively correlated with the amount of GBEVs.65 In addition, CATH-2 and PMAP-36 would further stimulate E. coli to secrete more GBEVs and enhance its defense against HDPs. Fortunately for the host, although LL-37 was not effective as a bactericide under the protection of GBEVs, the administration of LL-37 also did not induce the production of more GBEVs thereby enhancing the protective effect on the bacteria. Additionally, the GBEVs secreted by E. coli induced with 2.5 mM CATH-2 (2.5C GBEVs) not only failed to protect E. coli, but even enhanced the bactericidal effect of LL-37. This may be due to the continued presence of active CATH-2 in 2.5C GBEVs, which can exert a synergistic antibacterial effect with LL-37.65 GBEVs from V. cholerae C6706 can also protect parental bacteria from HDPs. Rompikuntal et al. added sublethal concentrations (25 μg/mL) of LL-37 to Luria Bertani medium containing V. cholerae C6706. The treated group, without the addition of V. cholerae C6706 EVs, exhibited significant growth lag compared with that of the control group, whereas the addition of V. cholerae C6706 EVs ameliorated the inhibitory effect of LL-37 on V. cholerae growth.46 Here, the protective effect was attributed to the PrtV protease loaded in GBEVs (PrtV-GBEVs), which protected the bacteria by degrading LL-37. However, replacement of PrtV-GBEVs with GBEVs secreted by the ΔprtV mutant strain was also protective, although the effect was weaker than that with the PrtV overexpression strain and the wild-type strain. Thus, the protective mechanism may not only rely solely on the PrtV protease, but may also be due to the adsorption of some HDPs by GBEVs.
In summary, some GBEVs can effectively help the defend the gut bacteria against antibiotics and HDPs and significantly improve their survival in the host. However, bacterial resistance is a crucial concern and is a major challenge for the global public health system. A comprehensive understanding of how GBEVs influence the interactions of gut bacteria with antibiotics and HDPs will help facilitate the clinical applicability of GBEVs. Furthermore, although only ARG transfer by intestinal bacteria through GBEVs has been reported, HGT via GBEVs is undoubtedly an effective strategy for the evolution of intestinal bacteria and resistance to external environmental pressures. For example, using engineered GBEVs to deliver exogenous genes of interest to dysfunctional host intestinal flora, thereby exerting regulatory effects may be another effective strategy to regulate intestinal microenvironment homeostasis using GBEVs.
GBEVs deliver QS molecules to coordinate bacterial population behavior
QS molecules are signaling substances used by bacteria to coordinate community behavior, such as regulating bacterial symbiosis and competition, influencing the secretion of bacterial metabolites and virulence factors, and regulating biofilm formation(Figure 2B).66 Currently, few studies have investigated the delivery of QS molecules by GBEVs. Of those studies, most have focused on the GBEVs secreted by the opportunistic pathogen Pseudomonas aeruginosa and the production of 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas quinolone signal, PQS). These studies have suggested that the interaction between PQS and P. aeruginosa EVs can be considered “synergistic”. Cooke et al. found that most wild-type P. aeruginosa strains require induction by PQS to secrete GBEVs.67 Further, Bala et al. observed that GBEV production was significantly reduced in the pqs gene mutants of P. aeruginosa, and that exogenous supplementation with PQS induced a significant release of GBEVs by this strain again.54 The mechanism underlying PQS-containing GBEV (PQS-GBEV) production in P. aeruginosa broadly involves the insertion of PQS into the bacterial outer membrane, which leads to the expansion of the outer leaflet, resulting in the bending of the outer membrane and the eventual formation of PQS-GBEVs.68 Subsequently, PQS can enter other P. aeruginosa cells via GBEVs, thereby regulating the behavior of the P. aeruginosa population.
PQS-GBEVs can be used to regulate the behavior of P. aeruginosa. Bacterial biofilms are a major contributor to chronic infections in humans (>65%)69 and antibiotic-resistant biofilms formed by P. aeruginosa can cause persistent infections at surgical sites and burn wounds.70 PQS-GBEVs have been closely associated with the virulence of P. aeruginosa. Addition of PQS-GBEVs to pqs gene mutants of P. aeruginosa has been observed to cause a significant increase in the production of virulence factors.54 Cooke et al. have revealed that PQS-GBEVs enhanced the dispersion of P. aeruginosa biofilms.43 The four different stages of P. aeruginosa biofilm formation are as follows: attachment stage, wherein P. aeruginosa attachment transitions from reversible to irreversible; maturation stage, wherein P. aeruginosa growth gradually changes from three-dimensional microcolonies to mushroom-like clusters; dispersion stage, wherein P. aeruginosa degrades the outer polymer matrix to expand the living space, and spreads further outward to reach a new attachment surface and begin a new cycle.70 Cooke et al. determined that the contents of PQS and GBEVs were significantly higher during the dispersion stage than those during the attachment and maturation stages. During dispersion, GBEVs not only functioned as PQS carriers but also exported various matrix degrading enzymes, including proteases, lipases, and DNAases, to the biofilm to promote the degradation of biofilm matrix components and facilitate P. aeruginosa dispersal.43 Although PQS-GBEVs may enhance the expansion of P. aeruginosa biofilms and further exacerbate the disease in patients infected with P. aeruginosa, the use of PQS-GBEVs to precisely regulate biofilm expansion may be an effective therapeutic strategy for converting chronically drug-resistant biofilm infections from recalcitrant protectors to dispersions that are more amenable to antibiotic treatment.
PQS-GBEVs also function in iron transfer. Iron is an essential micronutrient for many living cells and a cofactor for many cellular enzymes.71 In the host, iron usually forms high-affinity complexes with proteins to achieve sequestration, thereby enabling the host with resistance to pathogenic infestation since pathogens need iron for metabolism and maintaining virulence.72 The main routes of iron uptake by P. aeruginosa include the pyoverdine73 and pyochelin74 iron carriers, both of which bind iron with different affinities and transfer it into P. aeruginosa cells via TonB receptors, and the Feo system,75 which is the most widely distributed ferrous iron transport system in bacteria. Lin et al. constructed a P. aeruginosa mutant strain defective in these three iron uptake pathways. Interestingly, they found that this mutant strain could grow in minimal medium containing 0.5 μg/mL iron chelator ethylenediamine-N,N’-bis(2-hydroxyphenylacetic acid).44 Subsequently. through genetic screening, the crucial role of TseF, a gene close to the type VI secretion system H3 (H3-T6SS), was elucidated. The TseF gene encodes a 178-amino acid protein, TseF, which binds directly and specifically to PQS. Moreover, TseF binds to PQS with stronger affinity in the presence of Fe3+. TseF is packaged along with PQS into GBEVs, resulting in the formation of TseF-containing GBEVs, which interact with the cell surface Fe (III)-pyochelin receptor FptA and porin OprF, thereby delivering iron molecules into the bacterial cells.44
In addition to inducing GBEV secretion by P. aeruginosa cells, PQS can also mediate communication across species. Tashiro et al. noticed that the addition of PQS to E. coli K12 medium significantly increased the production and size of E. coli EVs. For instance, addition of 20 μM PQS increased the size of GBEVs by approximately 20 nm relative to the uninduced group. This effect was also observed in the gram-positive bacterium Bacillus subtilis 168, which seldom produces GBEVs under laboratory conditions.76
Besides PQS system, three other QS systems for bacteria-bacteria communication have been discovered: the LuxI/LuxR type among gram-negative bacteria,77 the oligopeptide-two-component type among gram-positive bacteria,78 and the autoinducer 2 (AI-2) type among both gram-negative and gram-positive bacteria.79 Each of these QS systems has its own unique regulatory mechanism. In the LuxI/LuxR QS system, acyl-homoserine lactone (AHL) serves as a signal that can only be recognized by related species. In the oligopeptide-two-component QS system, for signaling, autoinducer peptides must be delivered by a dedicated oligopeptide transporter, which is usually an ABC transporter. In the AI-2 QS system, AI-2 is predominantly mediates interspecific communication.80 Detection of QS molecules in the host is difficult due to the high volatility and dynamics of QS molecules in the complex intestinal environment. To the best of our knowledge, the phenomena of intestinal bacteria using GBEVs to deliver the three aforementioned QS molecules has not been reported in the literature. Morinaga et al. found that BEVs released by Paracoccus denitrificans, a gram-negative, inactive spherical soil bacterium, were able to take up long-chain AHLs secreted by other bacteria from the environment and use it as a signal to control biofilm formation and cell aggregation. This allows P. denitrificans to rapidly reach QS threshold concentrations in complex microbial communities with relatively low populations of their own.81 In summary, QS molecules can regulate population behavior through their enrichment by BEVs, rather than using the classical diffusion pathway. Therefore, we hypothesize that the use of GBEVs carrying specific QS molecules can regulate bacterial population behavior more precisely. Currently, an important topic in clinical research is the treatment of pathogenic bacterial infections using QS system interfering agents, such as meta-bromo-thiolactone82 (inhibits the production of virulence factor pyocyanin and biofilm formation) and cladodionen (inhibits the production of elastase and rhamnolipid, biofilm formation and diffusion, and down-regulates QS-related mRNA expression).83 In addition, competitive inhibition of GBEVs carrying pathogenic QS molecules using engineered GBEVs or influencing the formation of GBEVs through interventions (such as the use of bicyclic compounds including 4-hydroxyindole, isatin, 1-naphthol and 8-quinolinol to significantly inhibit the production of P. aeruginosa EVs and the synthesis of PQS84) are both potential new strategies for the treatment of pathogenic bacteria.
GBEVs-mediated bacteria-host interactions
The intestinal microenvironment is primarily maintained by a combination of host commensal microbiota, host cells, and their metabolites. The commensal microbiota provides a variety of benefits to the host, such as obtaining nutrition and energy from food, promoting immune system development, and defense against pathogens.85 Previous studies have proved the existence of complex interactions between gut microbiota and the host immune system;86 nonetheless, we know that a thick mucus layer between the gut microbiota and intestinal cells limits their direct contact (except in some diseases, such as ulcerative colitis, where damage to the intestinal barrier allows bacteria to cross the mucus layer and make direct contact with IECs). Communication and interaction between gut microbiota and intestinal cells is a rising concern among researchers. GBEVs can directly enter the mucus layer and be endocytosed by host cells (Figure 1B, 3, and 4). Most of the signaling molecules contained within GBEVs are microbe-associated molecular patterns, which can bind to host cell membrane receptors or intracellular receptors, such as LPS (TLR4 ligand), lipoprotein (TLR2 ligand), peptidoglycan (NOD1/NOD2 ligand), DNA, and RNA (TLR7/TLR9 ligand), to regulate host immune responses or promote disease development.93 In addition, studies have found that GBEVs of 20–100 nm size enter host cells preferentially through the caveolin-mediated endocytosis pathway, whereas GBEVs of 90–450-nm size enter host cells through the macropinocytosis and endocytosis pathways.94 Overall, GBEVs are excellent information carriers that connect bacterial and host cells (Figure 3). Notably, the regulatory effects of GBEVs are strongly dependent on characteristics of the parent bacteria, the cargo loaded, and the host physiology. In this section, we describe the effects of GBEVs on the host (summarized in Table 2). Additionally, some proposed mechanisms underlying GBEV-mediated bacteria-host cross-talk are presented in Figure 4.
Figure 3.
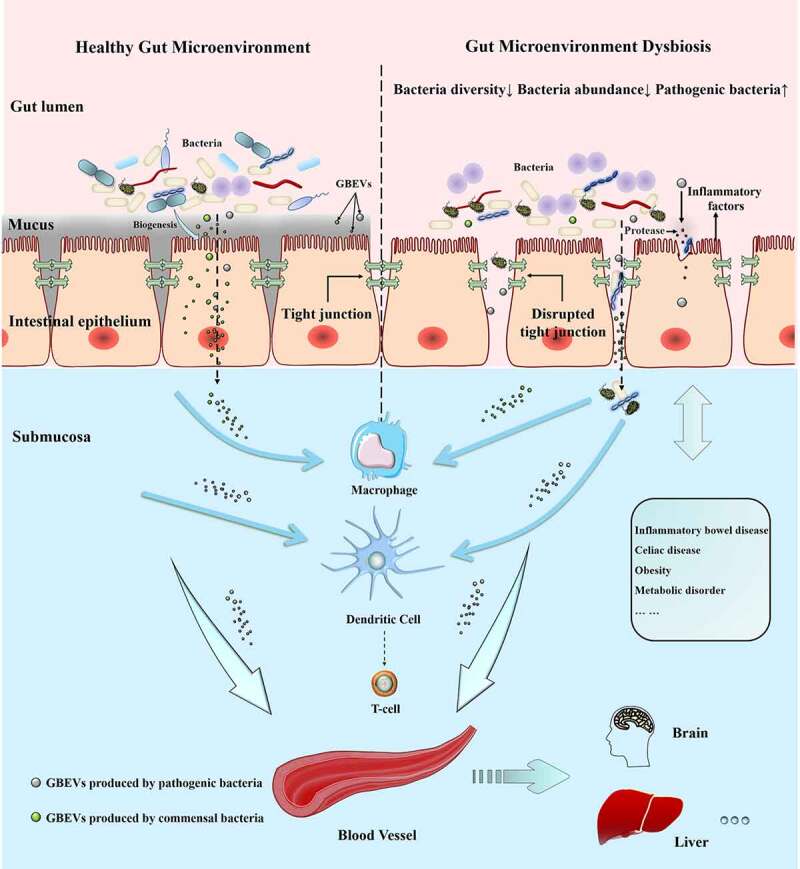
GBEVs mediate cross-talk between gut bacteria and host cells. GBEVs in the gut lumen can access the submucosa. Dysbiosis can lead to the disruption of tight junctions in the intestinal epithelium leading to the liberation of gut bacteria and GBEVs into the underlying submucosa, where GBEVs can activate the immune system and disseminate via the circulatory system. GBEVs, gut bacterial extracellular vesicles. GBEVs: gut bacteria-derived extracellular vesicles.
Figure 4.
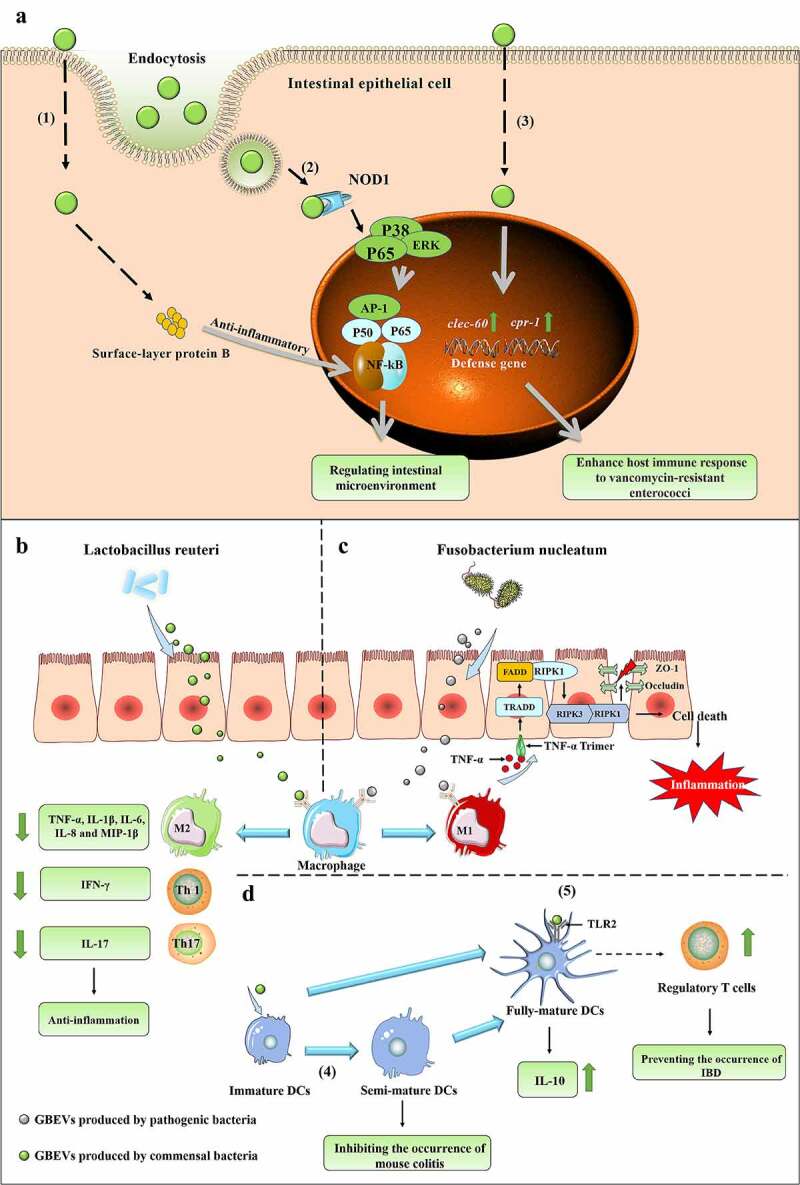
(a) GBEVs mediate communication cross-talk between gut bacteria and intestinal epithelial cells. (1) Propionibacterium freudenreichii CIRM-BIA 129 EVs contain surface-layer protein B, which reduces inflammation by regulating the NF-κB pathway in intestinal epithelial cells.87 (2) GBEVs (secreted by Escherichia coli Nissle 1917) can be internalized by the endocytosis of intestinal epithelial cells and stimulate the intracellular NOD1 receptor, thus regulating the intestinal microenvironment.88 (3) GBEVs produced by Lactobacillus plantarum WCFS1 up-regulates the expression of host defense genes (CTSB and REG3) in intestinal epithelial cells and enhances host immune responses to vancomycin-resistant enterococci.89 (b) The proposed mechanism of Lactobacillus reuteri EV-mediated bacteria-host cross-talk to drive intestinal immune homeostasis against pathogen-induced inflammation in a chicken model.107 L. reuteri EVs can suppress the pro-inflammatory mediators produced by macrophages. (C) The proposed mechanism of Fusobacterium nucleatum EV-driven intestinal mucosal barrier dysfunction in ulcerative colitis.25 The release of GBEVs by F. nucleatum promotes macrophages to secrete pro-inflammatory factors that activate RIPK1-mediated cell death signals in intestinal epithelial cells, leading to the disruption of intercellular tight junctions.25 (D) GBEVs mediate communication cross-talk between gut bacteria and dendritic cells. (4) GBEVs secreted by Bacteroides vulgatus can induce DC semi-maturation and enhance the immune system silencing caused by this strain, thus inhibiting the occurrence of murine colitis.90 (5) Bacteroides thetaiotaomicron and Bacteroides fragilis EVs interact with toll-like receptor-2 on dendritic cells (DCs), enhance the production of regulatory T cells and anti-inflammatory cytokines, and induce immune regulation to prevent the occurrence of inflammatory bowel disease.91,92
Table 2.
The roles of GBEVs in communication cross-talk between gut bacteria and host cells.
| GBEVs source | Cargos | Function | Mechanism | Refs |
|---|---|---|---|---|
| Akkermansia muciniphila | Unknown | 1. Protect the progression of dextran sulfate sodium-induced colitis; 2. Increase 5-HT levels in the host intestine. |
Unknown | 95–97 |
| Bacteroides fragilis | PSA | Induction of immunomodulation and prevention of experimental colitis. | TLR2 | 91 |
| Bacteroides thetaiotaomicron | Unknown | Promote the regulatory DC responses of healthy individuals. | Unknown | 92 |
| Sulfatase enzymes | Promote inflammatory immune stimulation in genetically susceptible hosts. | Sulfatase-dependent manner | 98 | |
| MINPP | Promote intracellular Ca2+ signaling. | Unknown | 99 | |
| Bacteroides vulgatus mpk | MAMPs | Induce semi-maturation of DCs cells and enhance that immune system silence caused by the strain | TLRs | 90 |
| Campylobacter jejuni | Proteases | Triggering the IL-8, IL-6, hBD-3 and TNF-α responses of T84 intestinal epithelial cells and are cytotoxic to Caco-2 IECs and Galleria mellonella larvae. | Unknown | 100 |
| Clostridium difficile | Translation-associated proteins | Inducing a pro-inflammatory response and inducing cytotoxicity of colonic epithelial cells. | Unknown | 101 |
| Escherichia coli BL21 | LPS | Activate caspase-11. | TLR4-TRIF-GBPs | 102 |
| Escherichia coli C25 | Unknown | It causes mild pro-inflammatory response in intestinal epithelial cells. | TLRs | 93 |
| Escherichia coli Nissle 1917 | Unknown | 1. Regulation of innate immunity of intestinal epithelial cells; 2. Up-regulation of tight junction protein expression in intestinal epithelial cells. |
1. NOD1 2. Unknown |
88,103 |
| Escherichia coli W3110 msbB-mutant strain | Unknown | Induces massive production of IFN-γ and promotes T cell-mediated immune responses. | Unknown | 104 |
| Faecalibacterium prausnitzii | Unknown | Increase 5-HT levels in the host intestine. | Unknown | 96 |
| Fusobacterium nucleatum | 1. Autotransporter proteins and other virulence factor (FadA, MORN2 and YadA); 2. Unknown; 3. Unknown |
1. Destroy intestinal epithelial barrier; 2. Promote colon cancer development; 3. Regulation of innate immunity of intestinal epithelial cells. |
1. FADD-RIPK1-caspase 3; 2. Unknown; 3. TLR2 |
25,105–107 |
| Lacticaseibacillus paracasei | Unknown | Inhibit colon cancer development. | PDK1/AKT/Bcl-2 | 108 |
| Lactobacillus plantarum | Unknown | Up-regulate the expression of host defense genes to provide protection for the host | Unknown | 89 |
| Lactobacillus reuteri | Glucosyltransferase, serine protease and elongation factor Tu | Enhancing immunomodulatory cell-mediated immunosuppression by activating macrophages to inhibit Th1 and Th17-mediated inflammatory responses | Unknown | 109 |
| Pediococcus pentosaceus | Unknown | Promote that M2-type polarization of macrophages | TLR2 | 110 |
| Propionibacterium freudenreichii CIRM-BIA 129 | SlpB | Modulate inflammatory responses | NF-κB | 87 |
| Pseudomonas aeruginosa | Unknown | Induces mitochondrial dysfunction in macrophages and inhibits host protein synthesis, ultimately inducing an inflammatory response | Unknown | 111 |
| Salmonella Typhimurium ATCC 14028 | Unknown | Inhibit colon cancer development | Unknown | 112 |
| Vibrio cholerae O395 | Unknown | Activate innate immune response of host | NOD1 | 113 |
Notes: DCs, dendritic cells; HFD, high-fat diet; GBPs, guanylate-binding proteins; MAMPs, microbe-associated molecular patterns; MINPP, mammalian InsP6 phosphatase; OMV, outer membrane vesicles; PSA, polysaccharide; SlpB, surface-layer protein B; TLRs, Toll-like receptors; TRIF, TIR domain-containing adaptor-inducing interferon-β; GBEVs: gut bacteria-derived extracellular vesicles; Refs: references.
GBEVs effects on intestinal barrier function
The intestinal barrier is essential for maintaining the balance of the intestinal microenvironment; it regulates the flow of water, ions, and macromolecules in the lumen.114 The intestinal epithelium is firmly bound using tight junction proteins, forming a mechanical barrier to the intestine, which is the main location for digestion and nutrient absorption, allowing the body to achieve a surface area in contact with the external environment of approximately 200 m2.115 Impaired intestinal barrier function, particularly the intestinal mechanical barrier, can lead to the direct contact of the host cells with intestinal lumen contents, intestinal commensal bacteria, and foreign pathogens, ultimately leading to the onset or development of IBD.114 Recent studies have found that the effect of GBEVs on the intestinal barrier varies based on their origin. Most of the GBEVs produced by pathogens impair intestinal barrier function, a property closely related to the parent bacteria. In contrast, GBEVs secreted by intestinal commensal and probiotic bacteria mostly improve intestinal barrier function and suppress the level of intestinal inflammation. Therefore, studying the effects of GBEVs on intestinal barrier function can help design effective interventions aimed at preventing and treating IBD.
GBEVs disrupt the intestinal barrier and induce or aggravate IBD
GBEVs can promote the development and progression of IBD by directly inducing apoptosis in IECs. Fusobacterium nucleatum, a gram-negative bacterium closely associated with preterm birth, periodontal disease, IBD, and colorectal cancer,116 secretes GBEVs that promote macrophage polarization to the M1 type and induce apoptosis in IECs via FADD-receptor interacting protein kinase 1 (RIPK1)-caspase 3 pathway, disrupting the intestinal epithelial barrier (Figure 4C).25 Liu et al. co-incubated F. nucleatum EVs with peripheral blood mononuclear cells and found that it induced a significant increase in the expression levels of pro-inflammatory factors TNF-α and interferon gamma (IFN-γ), inhibited the expression of anti-inflammatory factor IL-10, and polarized macrophages to the M1 type. In the macrophage/colorectal adenocarcinoma (Caco-2) cells co-culture model, treatment of F. nucleatum EVs in the presence of macrophages significantly promoted apoptosis of Caco-2 cells and significantly reduced transepithelial electrical resistance (TEER), resulting in the loss of epithelial barrier function.25 Moreover, the addition of TNF-α neutralizing antibody or necrostatin-1, a phosphorylated RIPK1 kinase inhibitor, significantly increased Caco-2 cell survival and restored intestinal epithelial barrier function (increased TEER values), suggesting that RIPK1 and RIPK3 in IECs may be involved in the apoptotic process. In addition, these findings were verified using a dextran sodium sulfate (DSS)-induced colitis mouse model. The GBEVs-treated group showed shorter colonic lengths, more severe damage to crypt and epithelial structures, and lower survival rates (100 % survival in the DSS model group and only 50 % survival in the 50 μg/d F. nucleatum EVs-treated group) compared with those showed by the DSS model group.25 Another study on F. nucleatum also demonstrated that the bacteria secretes GBEVs that promote intestinal inflammation. Engevik et al. added F. nucleatum subspecies polymorphum EVs to HT29 cell (a human colorectal adenocarcinoma cell line) monolayer, which resulted in the significant promotion of IL-8 production.117 Interestingly, F. nucleatum EVs did not disrupt the intestinal epithelial barrier, although cytokine production and inflammatory responses were observed, which may be strain-dependent. Additionally, clinical isolates from patients with IBD have shown an increased ability to invade Caco-2 cells and induce TNF-α expression than those isolated from healthy populations.118 Thus, the virulence of secreted GBEVs varies among different strains of the same species. Remarkably, the virulence mechanism of GBEVs may not be in good agreement with that of parental bacteria. The protein components of Clostridium difficile EVs are mainly transcription-related proteins, and no toxic proteins, such as TcdA or TcdB, are contained in the parental bacterial. However, co-incubation of C. difficile EVs with Caco-2 cells and HEp-2 cells induces cell death.101
In addition to disrupting the intestinal epithelial barrier by inducing apoptosis in IECs, GBEV cargo can affect intestinal barrier function. Wang et al. incubated E. coli BL21 EVs with Caco-2, HT-29, and NCM460 cells, and found that GBEVs reduced E-cadherin expression in cells in a concentration-dependent manner.119 The team further found that internalization of E. coli BL21 EVs by IECs promotes the recruitment of caspase-5 and PIKfyve (involved in endosome maturation) to early endosomal membranes by sorting nexin 10 (SNX10, a member of the SNX family proteins), while promoting the release of LPS from GBEVs into the cytosol. Activated caspase-5 also leads to the phosphorylation of Lyn, the upstream regulator of Snail/Slug, which promotes nuclear translocalization of Snail/Slug and downregulates E-cadherin expression, ultimately leading to intestinal barrier dysfunction. In addition, knocking out SNX10 using CRISPR/Cas9 or treating with the SnX10 inhibitor DC-SX029 blocks LPS release, caspase-5 activation, and downstream signaling, effectively alleviating intestinal barrier dysfunction induced by E. coli BL21 EVs.119 Although the E. coli BL21 used in this study is not a typical strain in the host gut, these findings are significant for research on IBD pathogenesis. Apart from LPS, GBEVs secreted by some enteropathogenic bacteria contain protein hydrolases. These GBEVs are internalized by IECs and directly cleave intracellular tight junction proteins, thereby facilitating bacterial-host cell interactions. Elmi et al. analyzed protein fractions in the GBEVs of Campylobacter jejuni, the major pathogen of bacterial and foodborne gastroenteritis worldwide, and found three proteases, HtrA, Cj0511, and Cj1365c, responsible for the proteolytic activity of C. jejuni EVs (GBEVs produced by HtrA, Cj0511 and Cj1365c mutant strains as well as pretreatment with serine protease inhibitors reduced the protease activity of C. jejuni EVs). Co-incubation of C. jejuni EVs with T84 monolayers revealed their ability to degrade E-cadherin and occludin in cells in a dose- and time-dependent manner, thereby promoting the adhesion and invasion of C. jejuni.100 In summary, it is clear that the ability of GBEVs to disrupt the intestinal barrier is inextricably linked to the properties of their parental bacteria. Based on this characteristic, the development of anti-vesiculation drugs will be another new potential strategy for the treatment of IBD.
GBEVs modulate host immune system and intestinal barrier to improve IBD
In the intestinal tract, GBEVs secreted by commensal bacteria (including probiotics), may be effective in preventing and treating IBD by modulating the host immune system and intestinal barrier. GBEVs improve IBD by modulating macrophages and dendritic cells (DCs). Pediococcus pentosaceus is an intestinal commensal lactic acid bacterium with potent anti-inflammatory activity.120 Bulut et al. found that P. pentosaceus EVs promote the polarization of marrow-derived macrophages to M2-like macrophages with the involvement of TLR2 receptors, and this anti-inflammatory activity has been validated in both the yeast polysaccharide-induced mouse peritonitis model and the DSS-induced acute colitis mode.110 GBEVs secreted by Bacteroides vulgatus can induce DC semi-maturation and enhance immune system silencing caused by this strain, thereby inhibiting the occurrence of murine colitis (Figure 4D).90 Probiotic E. coli Nissle 1917 EVs activate DCs to initiate a Th1 response essential for fighting pathogen infections, and GBEV programmable DCs secreted by the intestinal commensal bacteria ECOR12 coordinate the production of a T-regulatory response essential for sustained immune tolerance in the intestine.121
Apart from directly interacting with immune cells, GBEVs can also alleviate intestinal inflammation by augmenting the endoplasmic reticulum stress response. Lactobacillus paracasei is an important Lactobacilli, and clinical studies have found a link between a reduction in Lactobacillus spp. in the gut and the increased incidence of IBD.122,123 Choi et al. applied L. paracasei-derived GBEVs to LPS-stimulated HT29 cells and DSS-induced colitis mice model. The results showed that L. paracasei EVs significantly reduce the levels of LPS-induced inflammatory factors (IL-1α, IL-1β, IL-2, and TNF-α), inflammation-related proteins (cyclooxygenase-2, inducible nitric oxide synthase, and nuclear factor kappa B) and nitric oxide (molecules strongly associated with IBD severity), and significantly improve the symptoms of DSS-induced colitis. In contrast, the addition of endoplasmic reticulum stress inhibitors, salubrinal or 4-phenylbutyric acid, or CHOP siRNA block the anti-inflammatory effect of L. paracasei EVs in LPS-stimulated HT29 cells and restore the LPS-induced inflammatory response.124 In addition to exerting immunomodulatory effects, GBEVs can also ameliorate IBD by modulating the structural proteins of IECs. For example, GBEVs secreted by E. coli Nissle 1917 and intestinal commensal strain ECOR63 promote the upregulation of tight junction proteins ZO-1 and claudin-14 in IECs, downregulate claudin-2 levels, enhance the intestinal barrier, and reduce intestinal permeability.103
While some GBEVs can improve IBD by directly stimulating the host immune system or modulating the intestinal mechanical barrier, the ameliorative effect of GBEVs on IBD is majorly achieved by the cargos they carry. Hu et al. found that Lactobacillus reuteri BBC3 EVs carry nucleic acid molecules as well as a variety of bioactive proteins, such as glucosyltransferase (glycosylation of host target proteins, activation of DC-SIGN signaling, induction of IL-10 production),125 serine protease (selective targeting of pro-inflammatory factors to suppress the inflammatory response)126 and elongation factor Tu (involved in intercellular signaling).99 The administration of GBEVs to LPS-stimulated broilers significantly lessened the reduction in body weight and increased food intake and effectively reduced mortality. Measurement of pro- and anti-inflammatory gene expression levels in jejunum tissues revealed that L. reuteri BBC3 EVs significantly inhibit the expression of pro-inflammatory genes (TNF-α, IL-1β, IL-6, IL-8, and IL-17) and promote the expression of anti-inflammatory genes (IL-10 and TGF-β), thereby reducing LPS-induced intestinal inflammation. In vitro cellular experiments revealed similar results, wherein GBEVs were internalized within 6 h of co-incubation with chicken HD11 macrophages. GBEVs pretreatment significantly inhibited NF-kB expression, reduced TNF-α, IL-1β, and IL-6 gene expression, and enhanced IL-10 and TGF-β gene expression in LPS-stimulated HD11 cells (Figure 4B). However, the addition of DNase I (10 U/mL), RNase I (10 U/mL), and proteinase K-agarose (1 mg/mL) significantly attenuated the inhibitory effect of L. reuteri BBC3 EVs on the LPS-induced inflammatory response.109 Although no specific effector substances were identified in this study, identifying substances within GBEVs that are involved in the regulation of host immune responses was possible. Polysaccharide is an immunomodulatory factor produced by B. fragilis, and Shen et al. found that B. fragilis can deliver polysaccharide to immune cells via GBEVs, thereby preventing IBD. B. fragilis EVs are endocytosed by DCs after entering the body and interact with polysaccharide via TLR2 receptors to enhance the production of regulatory T cells and anti-inflammatory cytokines and induce immune regulation (Figure 4D).91 Propionibacterium freudenreichii CIRM-BIA 129 EVs contain a large amount of surface-layer protein B (SlpB), which interacts with the immunomodulatory transcription factor NF-κB through protein-protein interactions. Co-incubation of GBEVs with HT-29 cells revealed that P. freudenreichii CIRM-BIA 129 EVs inhibit LPS-induced IL-8 secretion, and this anti-inflammatory effect is dependent on SlpB, which is not eliminated by the proteinase K-induced hydrolysis of EV surface proteins, suggesting that SlpB is present within GBEVs (Figure 4A).87
Overall, the effects of GBEVs secreted by intestinal pathogenic bacteria, intestinal commensal bacteria, and probiotic bacteria on IBD are all based on their corresponding parental bacteria. However, it is worth noting that we cannot equate the effect of GBEVs on IBD with the effect of parental bacteria on IBD. Kang et al. revealed that the pretreatment of CT26 cells with Akkermansia muciniphila EVs significantly reduced IL-6 levels produced under E. coli EV induction. The oral administration of A. muciniphila EVs was also shown to improve DSS-induced phenotypic changes in IBD in animals. However, the direct treatment of DSS mice with A. muciniphila did not have a therapeutic effect, instead it exacerbated the colitis.95 This suggests that GBEVs may affect the host differently than the parental bacteria, and these effects are not uniquely beneficial or detrimental and need to be judged in the context of the intestinal environment. Collectively, these findings suggest that GBEVs may be a double-edged sword, the consequences of which need to be further explored.
GBEVs regulate colon cancer occurrence and progression
Globally, colon cancer is one of the most common tumors. Statistically, it ranks third and second in incidence and mortality, respectively, among all cancer types.127 Imbalance in the intestinal microenvironment is an important factor in the development and progression of colon cancer.128 According to 16S rRNA sequencing, compared with healthy individuals, patients with colon cancer manifest significantly lower gut flora diversity and abundance.129 In addition, researchers have found that F. nucleatum, genotoxic E. coli, and enterotoxigenic B. fragilis, all promote the development of colon cancer in the animals.128 GBEVs, as important bacteria-bacteria and bacteria-host communication agents, also play an important role in the occurrence and development of colon cancer. Recently, Park et al. performed a metagenomic analysis of GBEVs in the stool samples of 70 patients with colon cancer and 158 control participants and found significant differences in the composition and diversity of GBEVs. The GBEV composition in the stool of patients with advanced colon cancer was significantly altered relative to that of patients with early colon cancer. Among these, GBEVs secreted by bacteria belonging to the genus Alistipes, a member of Bacteroidetes phylum, first reported in 2003, and linked to liver fibrosis, IBD, cancer, and cardiovascular disease130 may be a biomarker for colon cancer staging.131,132###
In addition to the association of GBEVs with colon cancer progression, studies have suggested that F. nucleatum-secreted GBEVs are also involved in colon cancer development. Tai et al. co-cultured F. nucleatum EVs with Caco-2 cells and found that it promotes mitochondrial fusion and invasiveness in cells, while the use of Paris polyphylla, an herb, inhibits the spread of colon cancer.105 Using enzyme-linked immunosorbent assay (ELISA), Lamprinaki et al. determined that LPS-loaded F. nucleatum ssp. animalis EVs could bind to sialic acid-binding immunoglobulin-like lectins-7, which are abundant on innate immune cells and tumor infiltrating T cells and inhibit immune activation upon binding to LPS, and GBEVs induced TNF-α production by human monocyte-DCs, thereby creating a pro-inflammatory environment to promote colon cancer development.133
Some bacteria secrete GBEVs that promote colon cancer progression, whereas some naturally occurring GBEVs induce apoptosis in colon cancer cells to exert antitumor effects. Shi et al. found that Lacticaseibacillus paracasei EVs have an inhibitory effect on colon cancer growth. GBEVs are absorbed by colon cancer cells (HCT116, SW1116, and SW620 cells) and significantly inhibit colon cancer cell proliferation, migration, and invasion in a concentration-dependent manner.108 L. paracasei EVs (200 μg/mL), upon co-incubation with colon cancer cells for 48 h, significantly promote their apoptosis, as determined using Annexin V/PI double staining. The xenograft tumor (seeding HCT116 cells) mice assay has shown extremely slow tumor growth in the GBEVs-treated group. Studies on its antitumor mechanism revealed that L. paracasei EVs significantly inhibit 3-phosphoinositide-dependent protein kinase-1 (serine tryptophan kinase, involved in signal transduction) and AKT (serine-threonine kinase, involved in the regulation of cell survival, proliferation, and apoptosis) genes and decreased the expression of Bcl-2 protein (an anti-apoptotic protein) in colon cancer cells.108 Similar effects have been observed with S. Typhimurium ATCC 14028 EVs.112 Although these studies demonstrated the anti-colon cancer effect of some GBEVs, none of them identified which specific molecules were responsible for the anti-tumor effect. Since the cargos of GBEVs secreted by bacteria vary with time period and environment, strategizing to effectively achieve the quality control of GBEVs with anti-colon cancer effect is critical.
Apart from directly inducing apoptosis in colon cancer cells, GBEVs can also be used as an advanced immune irritant to effectively induce in vivo anti-tumor immune responses for cancer treatment. Kim et al. injected E. coli BL21 and E. coli W3110 msbB-mutant-derived EVs (msbB, the gene encoding lipid A acyltransferase) into the tail vein of colon adenocarcinoma CT26 mice and found that GBEVs could be specifically targeted and enriched in tumor tissue, with extended anti-tumor activity by inducing massive IFN-γ production and promoting T-cell-mediated immune responses. In addition, GBEVs also exhibit therapeutic effects on other tumors; intravenous injection into mice inoculated with 4T1 murine carcinoma and B16BL6 melanoma cells significantly inhibits tumor growth and metastasis.104
Currently, colon cancer research is inclined towards considering its role in terms of EVs of host cell origin, neglecting the large number of GBEVs of intestinal bacterial origin. Moreover, our knowledge of how GBEVs affect colon cancer is extremely limited. For example, it is known that B. fragilis134 and its secreted toxin (BFT)135 can promote colon cancer development; however, regarding B. fragilis EVs, it is only known that they can improve IBD. Whether BFT can be loaded and whether it can affect the occurrence and progression of colon cancer are remain unclear. Recently, some studies have also found that probiotics function as disruptors in cancer development. For instance, Lactobacillus spp. promote the growth of pancreatic cancer tumors in mice.136 However, the role of GBEVs, which act as bacteria-host communicators, therein needs to be further investigated.
Immune modulation by GBEVs
Intestinal bacteria can contribute to the maturation and development of the host immune system through GBEVs. The IEC barrier is an important host defense mechanism against infection by exogenous pathogens and is the first site of interaction between GBEVs and the host. Once GBEVs cross the epithelial barrier into the submucosa, they interact with various immune cells, thereby regulating the host’s immune system. Kaparakis-Liaskos and Ferrero have provided a complete summary of how bacterial GBEVs modulate the host immune system.137 Hence, this section focusses on the recently reported modulation of the host immune system and intestinal microenvironment by intestinal pathogenic bacteria-, intestinal commensal bacteria-, and probiotics-secreted GBEVs.
GBEVs interactions with IECs
The regulatory role of intestinal bacteria on IECs innate immune system (rapid response to intestinal pathogenic microorganisms and slow response to intestinal commensal and probiotic bacteria) is an important factor in maintaining homeostasis of the intestinal microenvironment.138 GBEVs also play an important role in this regard by activating the innate immunity of IECs through various pathways, thus promoting the host defence against pathogenic bacteria and maintaining stability of the intestinal microenvironment.
GBEVs modulate the innate immunity of IECs by interacting with TLR2 receptors. Martin-Gallausiaux et al. exposed Caco-2 cells in a non-inflammatory environment to F. nucleatum EVs (10 μg/mL). Epithelial resistance of the cells was measured and found that F. nucleatum EVs did not disrupt the barrier of IECs and had no effect on the expression levels of the tight junction proteins occludin and claudin-2. In contrast, after the co-culture of F. nucleatum EVs with T84 cells, GBEVs induced activation of the NF-kB pathway and induced production of the inflammatory factor IL-8 (neither Caco-2 nor HT-29 cells showed NF-kB activation).106 FomA, a porin which is immunogenic and is involved in bacterial adhesion,139,140 is a key substance central to this phenomenon. The investigators used anti-FomA binding peptide to pretreat GBEVs, which effectively reduced their activity, while NF-kB expression was also significantly reduced after TLR2 was blocked in T84 cells using siRNA. This suggests that both FomA and TLR2 are involved in the activation of NF-kB pathway by F. nucleatum EVs.106 In previous studies, F. nucleatum EVs were generally found to have adverse effects on the host, such as exacerbating IBD or promoting colon cancer development. However, when the IECs were placed under steady-state conditions, F. nucleatum EVs did not cause pathological inflammatory responses and activated the host innate immunity by interacting with TLR2. Thus, the effects of GBEVs on the host cannot be categorized as “good” or “bad”, but are inextricably linked to the host’s physiological state.
In addition to recognizing extracellular TLR receptors, GBEVs can enhance innate immunity by activating intracellular nucleotide oligomerization domain 1 (NOD1) receptors in IECs through the peptidoglycan they carry. The NOD1 receptor is a member of the NOD-like receptor family of innate immune proteins, which is expressed in a variety of cell types and is particularly highly expressed in IECs.141 The NOD receptor consists mainly of the N-terminal caspase-activated recruitment domain (involved in protein interactions), the central NACHT domain (involved in receptor dimerization), and the C-terminal leucine rich repeat domain (which recognizes specific ligands).142 It can be activated by bacterial peptidoglycan fragments, thereby acting as an intracellular sensor of bacterial infection. Canas et al. incubated E. coli Nissle 1917 and GBEVs secreted by the intestinal commensal bacterium ECOR12 with IECs and determined, through confocal fluorescence microscopy, that GBEVs could be internalized by IECs through endocytosis. Internalized GBEVs further trigger the formation of NOD1 aggregates in IECs, which activates the NF-κB signaling pathway and promotes the expression of inflammatory factors IL-6 and IL-8 (Figure 4A).88 While the sustained stimulation of NOD1 by GBEVs from both sources results in an inflammatory response, GBEVs secreted by E. coli Nissle 1917 and Commensal ECOR63 promote the expression of tight junction proteins in IECs, thereby improving IBD.103 Therefore, the biological effects of GBEVs should be evaluated from multiple perspectives, and should not be judged by a single indicator. Thus, NOD1 receptors in IECs can activate GBEVs produced by probiotic and intestinal commensal bacteria; GBEVs secreted by V. cholerae O395 can also mediate the host innate immune response via NOD1 receptors. Chatterjee and Chaudhuri co-cultured 50 μg/mL V. cholerae O395 EVs with Int407 cells (the most common cell model for studying V. cholerae-host interactions) and HEK293 cells and found that the levels of pro-inflammatory IL-8 and granulocyte-macrophage colony-stimulating factor (GM-CSF) were significantly elevated.113 The investigators also observed that NOD1 expression was significantly upregulated in HEK293 cytoplasm, while NOD1, IL-8, and GM-CSF expression levels were significantly reduced after treatment with NOD1-specific siRNA. In the co-culture system of HT-29 and DCs, the expression of co-stimulatory molecules (HLA-DR, CD80, and CD83) on the surface of DCs was significantly upregulated by adding V. cholerae O395 EVs to stimulate HT-29 cells, indicating that V. cholerae O395 EVs can activate DCs. Based on this, the team added naive CD4+CD45RA+ T cells purified from peripheral blood to the above culture system for 5 d. The results of ELISA indicated that IL-4, IL-13, and IL-17 levels were significantly upregulated and IFN-γ expression levels were significantly downregulated, indicating that the T cell population was stimulated by V. cholerae O395 EVs to Th2/Th17 and the responses were polarized.113 Overall, this study demonstrated, at the in vitro cellular level, that V. cholerae O395 EVs induce inflammatory responses and activate host innate immune responses via the NOD1 pathway. Notably, the regulation of host innate immune responses by GBEVs secreted by different serotypes of V. cholerae may also differ; for example, the secreted GBEVs of V. cholerae V:5/04 non-O1 non-O139 clinical isolate,47 fail to activate host innate immune responses and promote strain colonization by suppressive effects.
GBEVs can protect the host from pathogens by activating the innate immune system of IECs and upregulating the expression of host defense genes in IECs. Li et al. evaluated the protective effect of Lactobacillus plantarum WCFS1 EVs using Caenorhabditis elegans and Caco-2 cells attacked by Enterococcus faecium (VRE) as a model. The solid killing assay revealed that nematodes pretreated with L. plantarum WCFS1 EVs survived the VRE attack for approximately 4 d longer than that by the control, and their defense genes cpr-1 (expressing gut-specific cysteine protease) and clec-60 (expressing C type lectin) were upregulated approximately 3-fold and 9-fold, respectively. Similarly, L. plantarum WCFS1 EVs co-incubated with Caco-2 cells for 24 h did not show cytotoxicity and result in a significant upregulation of CTSB (cathepsin B) and REG3 (regenerating islet-derived protein 3-gamma), which are functionally similar to cpr-1 and clec-60 genes (Figure 4A).89
Effects of GBEVs on neutrophils
Neutrophils are an important part of the host’s innate immune system. During bacterial infection, neutrophils are rapidly recruited to the infection site, and they swiftly clear the infection by secreting hydrolytic enzymes, proteinases, reactive oxygen species (ROS), chemotactic agents, and other antimicrobial substances.143 GBEVs are involved in regulating memory-like inflammatory responses of neutrophils, with dose-dependent variations in immune responses. Hudalla et al. co-incubated fecal-derived GBEVs with mouse bone marrow-derived neutrophils and found that GBEVs directly mediate adaptive immune responses.144 Pro-inflammatory sensitivity of neutrophils induced by low doses of GBEVs (1 ng/mL) is significantly enhanced, and TNF-α, IL-6, and ROS levels are significantly upregulated after the second hit of LPS. In contrast, increasing the dose of GBEVs to 28.1 μg/mL leads to an immune tolerance phenotype (diminished response to LPS restimulation) in neutrophils and promotes the expression of the anti-inflammatory factor IL-10.144 Of note, since the source of GBEVs in this study was feces, it may be difficult to separate host cell EVs and GBEVs in fecal samples by centrifugation and qEV columns alone.
Modulation of macrophages and DCs by GBEVs
Except for the above mentioned GBEVs that can improve IBD by modulating macrophages and DCs, not many studies have been conducted on GBEVs interacting with macrophages and DCs to affect the host immune system. In terms of macrophages, Deo et al. found that P. aeruginosa EVs activate macrophages-mediated inflammatory responses. The team co-cultured P. aeruginosa EVs with macrophages and found that they induced mitochondrial dysfunction and inhibited host protein synthesis, ultimately activating intrinsic apoptosis and inflammatory responses.111 In a study of the interaction between GBEVs and DCs, Durant et al. found that B. thetaiotaomicron EVs are involved in regulating DCs-mediated immune responses. They co-incubated DCs from the colonic mucosa and blood of healthy volunteers with B. thetaiotaomicron EVs and measured the cytokines secreted by the DCs. Interestingly, B. thetaiotaomicron EVs produced different effects on DCs from different sources, stimulating significant expressions of regulatory IL-10 and protective IL-6 for DCs of colonic and peripheral blood origin, respectively (Figure 4D). Thus, B. thetaiotaomicron EVs balance DCs-mediated immune responses when the host is in a healthy state.92 The investigators also used B. thetaiotaomicron EVs to stimulate DCs in patients with IBD (Crohn’s disease and ulcerative colitis); however, the above effects completely disappeared. This suggests that the different physiological states of the host may also affect the response of DCs to GBEVs.
GBEVs regulate serotonin levels and affect intestinal microenvironment stability
Serotonin (5-HT) is an important signaling molecule in the body, which transmits signals from the intestinal lumen to the lateral neurons, thus participating in synaptic signaling in the nervous system. In addition, 5-HT is a key molecule in maintaining the stability of the intestinal environment by regulating the secretory and motor functions of the gut.145 Approximately 95 % of 5-HT in humans is localized in the gastrointestinal tract and is mainly synthesized by enterochromaffin cells in the intestinal mucosa with the help of the enzyme tryptophan hydroxylase 1 (TpH1).146
Recently, Yaghoubfar et al. found that GBEVs can alter 5-HT levels in the host intestine.96 By co-incubating bacteria with GBEVs, Yaghoubfar et al. revealed that the pretreatment of CT26 cells with A. muciniphila EVs and Faecalibacterium prausnitzii EVs significantly increased 5-HT levels, expression of Slc6a4 (involved in serotonin transport) and Tph1 (rate-limiting enzyme associated with serotonin biosynthesis), while decreasing the expression of Mao gene (associated with serotonin metabolism). Curiously, the parental strains of both only affected the expression of Slc6a4 (significantly increased) and Mao (significantly decreased) genes, with no effect on serotonin levels in cells and Tph1 expression.96 Another study by the team on A. muciniphila and its GBEVs found that 5-HT levels in both the hippocampus and colon of C57BL/6 J mice significantly increase after 5 weeks of administering A. muciniphila and its EVs. The mRNA levels of the Tph1 gene in the colon are also significantly increased after treatment with both A. muciniphila and its GBEVs.97 Although the effects of A. muciniphila differed in the two studies, the effects of its secreted GBEVs were similar in both cases. Unfortunately, neither of these studies further investigated the biological effects in the host after the rise in 5-HT caused by GBEVs. As 5-HT is both a hormone and a neurotransmitter, it has multiple effects on both the intestinal microenvironment and the central nervous system, and the role of GBEVs in the regulation of 5-HT needs to be further explored.
In addition to regulating host intestinal microenvironment via the above pathways, GBEVs can also affect lesions involving distal host organs via the gut-brain axis, gut-liver axis, and other pathways (Figure 3). This part of the content is not the focus of this review. We recommend that readers interested in the topic of “GBEV-mediated gut-distal organ communication” to refer to an in-depth review of the gut-brain axis147 and gut-liver axis.148 In the following section, we describe the potential medical applications of GBEVs.
Repurposing GBEVs in medical applications
GBEVs as vaccines
The activation of host innate and adaptive immune responses by GBEVs secreted by pathogenic intestinal bacteria allows the development of GBEVs as a low-toxicity, highly effective vaccine or adjuvant. S. typhimurium is the main acute gastroenteritis-causing pathogen. Schetters et al. found that S. typhimurium EVs can induce the maturation of human monocyte-derived DCs, mouse bone marrow-derived DCs, and CD11c+ splenic DCs, which in turn activate CD8+T cell responses. The CD8+T cell response is currently a key clinical treatment strategy for tumors and intracellular viruses. Therefore, S. typhimurium EVs could be developed as an effective vaccine to inhibit viral replication and tumor growth.149 Enteric bacterial infections are a leading cause of diarrhea. Statistically, in developing countries, up to 10% of deaths in children under five years of age are caused by diarrhea.150 Shigella boydii is a major diarrhea-causing pathogen in children, and approximately 165 million cases have been reported annually worldwide. Although antibiotics are an effective strategy to control the outbreak of shigellosis, they are accompanied by the continuous emergence of antibiotic-resistant Shigella. Mitra et al. found that S. boydii EVs can significantly inhibit S. boydii-mediated inflammation and enable prolonged survival of infected mice.151
Introducing and expressing heterologous genes into non-pathogenic engineered E. coli is also a novel strategy for preparing a low-toxicity, well-controlled, and high-yield GBEV-based vaccine. In 2004, Kesty et al. first demonstrated that heterologously expressed outer membrane and periplasmic proteins can be incorporated into GBEVs.152 These findings enable the production of “customized” GBEVs. Price et al. introduced a plasmid containing a gene encoding the structure of glycan (derived from C. jejuni) into E. coli for expression, and the secreted GBEVs contained a large amount of glycan molecules identical to the pathogenic bacterial surface glycan (C. jejuni heptasaccharide N-glycan).153 The test in a C. jejuni attack model revealed that the numbers for colonization by C. jejuni in chicken intestines inoculated with GBEVs was nearly 104-fold lower than that in a model group.153 Guido et al. introduced a heterologous antigen gene with a fused lipoprotein leader sequence into E. coli, successfully synthesized the heterologous antigens by lipid modifications, and realized high-level accumulation in GBEVs (accounting for 5–20% of the total protein content of GBEVs) at the same time. In addition, the expression of lipidated xenoantigens also interfered with the acylation of lipid A, so as to significantly reduce LPS-mediated reactivity. This preparation strategy was verified using five Staphylococcus aureus antigens, and mice immunized with the engineered GBEVs could ultimately tolerate pathogenic attacks by S. aureus.154
GBEVs as delivery vehicles
Owing to their small size and natural lipid bilayer, GBEVs can accommodate a variety of biomolecules with different properties, such as lipids, nucleic acids, and proteins, making them an ideal drug delivery system.
Modified GBEVs play a therapeutic role
Surface modification of GBEVs improves their targeting ability and reduces their toxic effects. Peng et al. introduced a plasmid containing recombinant human tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) gene into engineered bacteria (E. coli) to construct GBEVs containing the active TRAIL protein.155 To reduce LPS toxicity in GBEVs, the investigators co-incubated the GBEVs with 2 mg/mL lysozyme for 1.5 h, allowing lysozyme to bind to LPS efficiently. Meanwhile, to achieve tumor targeting, the investigators modified the αvβ3 integrin targeting ligand (RGP) on GBEVs. After applying the modified GBEVs to melanoma sites in mice, the GBEVs fused with the skin through the nano-size effect, thereby efficiently penetrating the dermal stratum corneum of the mice. Upon entry into the skin, GBEVs specifically recognize melanoma cells with the help of RGP, thereby releasing the TRAIL protein and promoting the apoptosis of melanoma cells.155 In addition, in order to circumvent antibody-dependent clearance of GBEVs and the high toxicity caused by intravenous injection of GBEVs secreted by E. coli BL21, Qing et al. covered the surface of GBEVs with a pH-sensitive calcium phosphate shell, which could neutralize the acidic microenvironment of tumors and cause macrophages to become highly and effectively polarized from M2 to M1 types, thereby improving the antitumor effects of the GBEVs.156
GBEVs play a therapeutic role as a drug carrier
Exploiting the similarity between GBEVs secreted by pathogenic bacteria and parental bacteria as well as the natural structure of the lipid bilayer, researchers wrapped antibiotics with GBEVs, effectively realizing antibiotic camouflage and intracellular delivery, successfully solving the clinically-difficult problem of intracellular bacterial infection. For example, Gao et al. used S. aureus EVs as carriers and wrapped poly (lactic-co-glycolic acid) nanoparticles containing antibiotics by membrane-coating technique (NP@EV).157 The efficiency of NP@EV uptake by murine Ana-1 macrophage was examined using a fluorescence microscope and fluorimeter. Subsequently, Gao et al. discovered that Dil-labeled NP@EV was efficiently taken up by macrophages, and relatively more NP@EV was taken up by macrophages infected with S. aureus. The investigators also used PEGylated lipid bilayer-coated nanoparticles and E. coli EV-coated nanoparticles as controls and found that both were internalized less efficiently by macrophages than that observed for the NP@EV group. To evaluate the in vivo bacteriostatic effect of NP@EV, Gao et al. injected NP@EV into S. aureus bacteremia-bearing mice via the tail vein. NP@EV was significantly enriched in the kidney, lung, spleen, and heart of the mice model compared with that in the normal controls. Moreover, NP@EV reduced bacterial burden in the kidney and lungs by approximately 1–2 orders of magnitude compared with the untreated group, indicating effective alleviation of S. aureus metastatic infection.157
Some GBEVs can stimulate the host immune system. These GBEVs can be combined with anticancer drugs or formulations by virtue of their characteristics, thereby exerting synergistic therapeutic effects. Chen et al. used polyethylene glycol and tumor-targeting ligand Arg-Gly-Asp peptide to surface-modify GBEVs isolated from attenuated Salmonella to improve the circulatory stability and tumor-targeting of GBEVs.158 Additionally, micelles loaded with tegafur, a prodrug of fluorouracil that triggers direct apoptosis of tumor cells and also sensitizes tumor cells to cytotoxic T lymphocytes, were encapsulated within GBEVs (ORFT) to improve its inhibitory effect on tumor cells. Studies on in vitro cellular uptake of ORFT revealed that both murine macrophages RAW264.7 and B16F10 cells achieved rapid and high levels of ORFT uptake. Studies on in vivo efficacy of ORFT have revealed that ORFT injection effectively delays tumor development (tumors appeared at day 13 in the saline group, day 17 in the tegafur-loaded micelles group, and day 21–24 in the ORFT group), prolongs the survival of melanoma mice (from 31 to 39 d), and effectively inhibits tumor metastasis to the lung (significant reduction of pulmonary metastatic melanoma).158 Li et al. used E. coli Trans T1 EVs to mask cisplatin nanoparticles, exploiting its pathogen-associated molecular pattern, which is similar to that of the parental bacteria, resulting in effective recognition and internalization of these EVs by neutrophils, which were subsequently transferred to the site of tumor inflammation.159 Under the stimulation of inflammation, neutrophils rapidly released cisplatin nanoparticles, which were then absorbed by tumor cells and exerted anti-cancer effects.159
GBEVs as diagnostic biomarkers
Diseases are strongly correlated with biological imbalances involving intestinal microbiota. Diagnosing the existence and development of diseases by detecting changes in gut flora is a crucial area of clinical research. Current studies surrounding biomarkers are dominated by detection of changes in microbiota and metabolites in fecal samples; however, in fecal samples, in addition to live bacteria, a significant number of dead bacteria are also present. The possible reasons for bacterial death are numerous and may involve drug-induced inactivation or prolonged exposure of bacteria to air during sampling. The existence of these partially-inactivated bacteria will undoubtedly interfere with the search for disease-related biomarkers, making it difficult to translate research findings into clinical settings.
GBEVs, as information delivery vectors secreted during the lifespan of living bacteria, contain nucleic acid, protein, and/or lipid molecules derived from parental bacteria. By detecting these molecules contained therein to characterize changes in the intestinal microbiota and metabolites of patients, researchers can ascertain biomarkers more accurately. For example, Kim et al. detected a significant increase in the abundance of Firmicutes phyla, such as Eubacterium, Faecalibacterium, those of the Ruminococcaceae family, and Catenibacterium, by examining intestinal bacteria and GBEVs isolated from the fecal samples of patients with colon cancer.160 Furthermore, Kim et al., showed that GBEVs isolated from patients with severe alcoholic hepatitis have lower α diversity and higher β diversity than those in healthy controls and that the number of GBEVs secreted by Veillonella sp., V. parvula, and Lactobacillales sp. were significantly increased.161 At present, research regarding GBEV-disease-related biomarkers is at a naive stage. Effective isolation and detection of the number and types of GBEVs in biological samples are of the essence, requiring prompt and precise scrutiny.
Conclusions and future perspectives
Overall, our review summarized the role of GBEVs in bacteria-bacteria interactions and bacteria-host communication in the gut, indicating the role of GBEVs in intestinal microenvironment regulation. The research on BEVs has made tremendous progress recently. BEVs are now widely recognized as carriers of information which act as communicative agents between bacteria and hosts. However, to date, most studies on BEVs have focused on BEVs secreted by pathogenic bacteria.
Increase in the knowledge pertaining to GBEVs, a number of issues still need to be further addressed. Currently, methods for separation of GBEVs and host exosomes are generally based on physical factors, such as size and density and clearly differentiating between the two using these methods is difficult. Therefore, the results obtained from studies that have employed such methods need to be further explored. For example, Wei et al. found that GBEVs could increase blood-brain barrier permeability, promote the activation of brain astrocytes and microglia, and induce inflammatory responses and tau hyperphosphorylation by activating the GSK‐3β pathway, resulting in cognitive impairment.162 However, in this experiment, researchers isolated GBEVs directly from feces without considering the removal of exosomes secreted by mammalian cells. Thus, whether the series of effects observed by Wei et al. can completely be attributed to GBEVs remains unclear. Since the outer surface of GBEVs often contains LPS or lipoteichoic acid, which are specific to bacteria, we speculate that efficient separation of GBEVs and host cell exosomes is possible based on these unique properties.
Cargos are the material basis for the pathogenic or regulatory effects of GBEVs on the host, and clarifying the composition of cargos can provide fundamental information and implementation strategies for the development of GBEVs as potential vaccines and microenvironmental regulators. However, putting this theory into practice still remains challenging due to several issues. The cargo that is loaded and carried within GBEVs is closely related to the growth environment and exogenous stimuli of the parental bacteria. For example, Zavan et al. found that Helicobacter pylori secreted different cargos within GBEVs at different growth stages, resulting in different biological effects.163 In 2017, the Tashiro group demonstrated, for the first time that, GBEVs can selectively interact with parental bacterial cells in the microbial community. By labeling GBEVs with fluorescent membrane dyes, they found that GBEVs secreted by Buttiauxella agrestis specifically interacted more with other B. agrestis cells than bacteria of other genera.164 Whether such selective intercellular communication exists in other bacteria remains unclear. Another key issue affecting the application of GBEVs is their targeting behavior once they enter a cell (intestinal bacteria or host cell). Therefore, in view of these uncertainties about naturally occurring GBEVs, customizing GBEVs may be an effective strategy to accelerate their implementation and application. For example, through gene editing techniques, engineered bacteria could be used as “GBEV factories” that secrete GBEVs carrying specific genes, proteins, and polysaccharides, resulting in GBEVs with different bioactivities and targeting properties.165
At present, most studies related to GBEVs are based on experiments that use GBEVs isolated from in vitro cultures. However, GBEV composition is closely related to its growth environment. Whether the physiological characteristics of GBEVs secreted in vitro are the same as those of GBEVs secreted in vivo needs further investigation. In addition, studies on GBEVs have predominantly focused on the effects of GBEVs on the host, neglecting the effects on the intestinal microbiota. Although occasionally reported, the results are mostly invalid and suggestive.166 These unexplored mechanisms pertaining to the interactions between GBEVs and intestinal microbiota still needs to be supplemented with further knowledge. Detailed understanding of the interactions between GBEVs and intestinal microbiota will not only help to advance a better understanding of the interactions between bacteria in the intestine, but also help to better utilize GBEVs to regulate the intestinal microenvironment.
In conclusion, as presented in “125 Questions: Exploration and Discovery” published by “Science” journal,167 through questions such as “How is immune homeostasis maintained and regulated?” and “What role does our microbiome play in health and disease?” the role of GBEVs in the regulation of intestinal microenvironment requires further elucidation.
Acknowledgments
We thank our reviewers and editor for critical feedback that enhanced this work.
Funding Statement
This work was supported, at least in part, by the Natural Science Foundation of China under Grants [number 31770066 and 31470218; Y.W.]; the Natural Science Foundation of China under Grant [number 32000081; K.S.]; the Anhui Provincial Natural Science Foundation under Grant [number 1908085QC120; K.S.]; the Natural Science Foundation of Anhui Higher Education Institutions of China under Grant [number KJ2019A0040; K.S.]; the Open Fund for Discipline Construction, Institute of Physical Science and Information Technology of Anhui University under Grant [Y.W.]; the Outstanding Talents Program of Anhui University, China under Grant [Y.W.]; and the Doctoral Scientific Research Foundation of Anhui University under Grant [K.S.].
Disclosure statement
The authors declare no conflicts of interest.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this review.
References
- 1.Rajilic-Stojanovic M, de Vos WM.. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996–30. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Q, Chen WD, Wang YD. Gut microbiota: an integral moderator in health and disease. Front Microbiol. 2018;9:151. doi: 10.3389/fmicb.2018.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74(16):2959–2977. doi: 10.1007/s00018-017-2509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saffouri GB, Shields-Cutler RR, Chen J, Yang Y, Lekatz HR, Hale VL, Cho JM, Battaglioli EJ, Bhattarai Y, Thompson KJ, et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun. 2019;10(1):2012. doi: 10.1038/s41467-019-09964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14(10):573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 9.Jin M, Qian Z, Yin J, Xu W, Zhou X. The role of intestinal microbiota in cardiovascular disease. J Cell Mol Med. 2019;23(4):2343–2350. doi: 10.1111/jcmm.14195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu S, Jiang Y, Xu K, Cui M, Ye W, Zhao G, Jin L, Chen X. The progress of gut microbiome research related to brain disorders. J Neuroinflammation. 2020;17(1):25. doi: 10.1186/s12974-020-1705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meijers B, Evenepoel P, Anders H-J. Intestinal microbiome and fitness in kidney disease. Nat Rev Nephrol. 2019;15(9):531–545. doi: 10.1038/s41581-019-0172-1. [DOI] [PubMed] [Google Scholar]
- 12.Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15(7):397–411. doi: 10.1038/s41575-018-0011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol. 2020;17(1):53–64. doi: 10.1038/s41575-019-0242-7. [DOI] [PubMed] [Google Scholar]
- 14.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25(5):716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 15.Gupta A, Khanna S. Fecal microbiota transplantation. JAMA. 2017;318(1):102. doi: 10.1001/jama.2017.6466. [DOI] [PubMed] [Google Scholar]
- 16.Kundu P, Blacher E, Elinav E, Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171(7):1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Obata Y, Castano Á, Boeing S, Bon-Frauches AC, Fung C, Fallesen T, de Agüero MG, Yilmaz B, Lopes R, Huseynova A, et al. Neuronal programming by microbiota regulates intestinal physiology. Nature. 2020;578(7794):284–289. doi: 10.1038/s41586-020-1975-8. [DOI] [PubMed] [Google Scholar]
- 18.Ding N, Zhang X, Zhang XD, Jing J, Liu SS, Mu YP, Peng LL, Yan YJ, Xiao GM, Bi XY, et al. Impairment of spermatogenesis and sperm motility by the high-fat diet-induced dysbiosis of gut microbes. Gut. 2020;69(9):1608–1619. doi: 10.1136/gutjnl-2019-319127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanca A, Abbondio M, Palomba A, Fraumene C, Manghina V, Cucca F, Fiorillo E, Uzzau S. Potential and active functions in the gut microbiota of a healthy human cohort. Microbiome. 2017;5(1):79. doi: 10.1186/s40168-017-0293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76(3):473–493. doi: 10.1007/s00018-018-2943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn RA, Melnik AV, Vrbanac A, Fu T, Patras KA, Christy MP, Bodai Z, Belda-Ferre P, Tripathi A, Chung LK, et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature. 2020;579(7797):123–129. doi: 10.1038/s41586-020-2047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng MY, Inohara N, Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10(1):18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama K, Takaki T, Sugiyama M, Fukuda I, Aiso M, Mukai T, Odamaki T, Xiao J-Z, Osawa R, Okada N. Extracellular vesicles produced by Bifidobacterium longum export mucin-binding proteins. Appl Environ Microbiol. 2020;86(19):e01464–20. doi: 10.1128/AEM.01464-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Liang L, Yang C, Zhou Y, Chen Y. Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier through targeting RIPK1-mediated cell death pathway. Gut Microbes. 2021;13(1):1–20. doi: 10.1080/19490976.2021.1902718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz-Garrido N, Badia J, Baldoma L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J Extracell Vesicles. 2021;10(13):e12161. doi: 10.1002/jev2.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 28.Kim JH, Lee J, Park J, Gho YS. Gram-negative and gram-positive bacterial extracellular vesicles. Semin Cell Dev Biol. 2015;40:97–104. doi: 10.1016/j.semcdb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi Y, Obana N, Toyofuku M, Kodera N, Soma T, Ando T, Yoshihiro Fukumori Y, Nomura N, Taoka A. Diversity of physical properties of bacterial extracellular membrane vesicles revealed through atomic force microscopy phase imaging. Nanoscale. 2020;12(14):7950–7959. doi: 10.1039/c9nr10850e. [DOI] [PubMed] [Google Scholar]
- 30.Wensink J, Witholt B. Outer-membrane vesicles released by normally growing Escherichia coli contain very little lipoprotein. Eur J Biochem. 1981;116(2):331–335. doi: 10.1111/j.1432-1033.1981.tb05338.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhou L, Srisatjaluk R, Justus DE, Doyle RJ. On the origin of membrane vesicles in gram-negative bacteria. FEMS Microbiol Lett. 1998;163(2):223–228. doi: 10.1111/j.1574-6968.1998.tb13049.x. [DOI] [PubMed] [Google Scholar]
- 32.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13(10):605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sartorio MG, Pardue EJ, Feldman MF, Haurat MF. Bacterial outer membrane vesicles: from discovery to applications. Annu Rev Microbiol. 2021;75(1):609–630. doi: 10.1146/annurev-micro-052821-031444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol. 2019;17(1):13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 35.Work E, Knox KW, Vesk M. The chemistry and electron microscopy of an extracellular lipopolysaccharide from Escherichia coli. Ann N Y Acad Sci. 1966;133(2):438–449. doi: 10.1111/j.1749-6632.1966.tb52382.x. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee SN, Das J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J Gen Microbiol. 1967;49(1):1–11. doi: 10.1099/00221287-49-1-1. [DOI] [PubMed] [Google Scholar]
- 37.Dorward DW, Garon CF. DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Appl Environ Microbiol. 1990;56(6):1960–1962. doi: 10.1128/aem.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch SV, Pedersen O, Phimister EG. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 39.Elhenawy W, Debelyy MO, Feldman MF, Whiteley M, Greenberg EP. Preferential packing of acidic glycosidases and proteases into bacteroides outer membrane vesicles. mBio. 2014;5(2):e00909–14. doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stentz R, Horn N, Cross K, Salt L, Brearley C, Livermore DM, Carding SR. Cephalosporinases associated with outer membrane vesicles released by Bacteroides spp. protect gut pathogens and commensals against β-lactam antibiotics. J Antimicrob Chemother. 2015;70(3):701–709. doi: 10.1093/jac/dku466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X, Sun L, Li C, Yang X, Wang X, Hu X, Nie T, Zhang Y, You X. The attenuated protective effect of outer membrane vesicles produced by a mcr-1 positive strain on colistin sensitive Escherichia coli. Front Cell Infect Microbiol. 2021;11:701625. doi: 10.3389/fcimb.2021.701625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bielaszewska M, Daniel O, Karch H, Mellmann A. Dissemination of the blaCTX-M-15 gene among Enterobacteriaceae via outer membrane vesicles. J Antimicrob Chemother. 2020;75(9):2442–2451. doi: 10.1093/jac/dkaa214. [DOI] [PubMed] [Google Scholar]
- 43.Cooke AC, Florez C, Dunshee EB, Lieber AD, Terry ML, Light CJ, Schertzer JW, Ellermeier CD. Pseudomonas quinolone signal-induced outer membrane vesicles enhance biofilm dispersion in Pseudomonas aeruginosa. mSphere. 2020;5(6):e01109–20. doi: 10.1128/mSphere.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian PY, Luo ZQ, Shen X. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. 2017;8(1):14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. 2005;437(7057):422–425. doi: 10.1038/nature03925. [DOI] [PubMed] [Google Scholar]
- 46.Rompikuntal PK, Vdovikova S, Duperthuy M, Johnson TL, Åhlund M, Lundmark R, Oscarsson J, Sandkvist M, Uhlin BE, Wai SN. Outer membrane vesicle-mediated export of processed PrtV protease from Vibrio cholerae. PLoS One. 2015;10(7):e0134098. doi: 10.1371/journal.pone.0134098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bitar A, Aung KM, Wai SN, Hammarström M-L. Vibrio cholerae derived outer membrane vesicles modulate the inflammatory response of human intestinal epithelial cells by inducing microRNA-146a. Sci Rep. 2019;9(1):7212. doi: 10.1038/s41598-019-43691-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valguarnera E, Scott NE, Azimzadeh P, Feldman MF. Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in Bacteroides. mSphere. mSphere. 2018;3(6):e00559–18. doi: 10.1128/mSphere.00559-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, Bergmann S, Hammerschmidt S, Brigidi P. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology (Reading). 2010;156(Pt 6):1609–1618. doi: 10.1099/mic.0.038307-0. [DOI] [PubMed] [Google Scholar]
- 50.Candela M, Biagi E, Centanni M, Turroni S, Vici M, Musiani F, Vitali B, Bergmann S, Hammerschmidt S, Brigidi P. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology (Reading). 2009;155(1):3294–3303. doi: 10.1146/annurev-micro-052821-031444. [DOI] [PubMed] [Google Scholar]
- 51.Nishiyama K, Yamamoto Y, Sugiyama M, Takaki T, Urashima T, Fukiya S, Yokota A, Okada N, Mukai T, Turner MS. Bifidobacterium bifidum extracellular sialidase enhances adhesion to the mucosal surface and supports carbohydrate assimilation. mBio. 2017. doi: 10.1128/mBio.00928-17.;8(5):e00928–17. doi: 10.1128/mBio.00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milani C, Mangifesta M, Mancabelli L, Lugli GA, Mancino W, Viappiani A, Faccini A, van Sinderen D, Ventura M, Turroni F. The sortase-dependent fimbriome of the genus Bifidobacterium: extracellular structures with potential to modulate microbe-host dialogue. Appl Environ Microbiol. 2017;83(19):e01295–17. doi: 10.1128/aem.01295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.González-Rodríguez I, Sánchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, Margolles A. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol. 2012;78(11):3992–3998. doi: 10.1128/aem.08024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bala A, Kumar L, Chhibber S, Harjai K. Augmentation of virulence related traits of pqs mutants by Pseudomonas quinolone signal through membrane vesicles. J Basic Microbiol. 2015;55(5):566–578. doi: 10.1002/jobm.201400377. [DOI] [PubMed] [Google Scholar]
- 55.Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015;31(4):707–721. doi: 10.1185/03007995.2015.1018989. [DOI] [PubMed] [Google Scholar]
- 56.Ling Z, Yin W, Shen Z, Wang Y, Shen J, Walsh TR. Epidemiology of mobile colistin resistance genes mcr-1 to mcr-9. J Antimicrob Chemother. 2020;75(11):3087–3095. doi: 10.1093/jac/dkaa205. [DOI] [PubMed] [Google Scholar]
- 57.Ghotaslou R, Yekani M, Memar MY. The role of efflux pumps in Bacteroides fragilis resistance to antibiotics. Microbiol Res. 2018;210:1–5. doi: 10.1016/j.micres.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 58.Brook I, Pazzaglia G, Coolbaugh JC, Walker RI. In-vivo protection of group A beta-haemolytic streptococci from penicillin by beta-lactamase-producing Bacteroides species. J Antimicrob Chemother. 1983;12(6):599–606. doi: 10.1093/jac/12.6.599. [DOI] [PubMed] [Google Scholar]
- 59.Rangama S, Lidbury I, Holden JM, Borsetto C, Murphy ARJ, Hawkey PM, Wellington EMH. Mechanisms involved in the active secretion of CTX-M-15 β-lactamase by pathogenic Escherichia coli ST131. Antimicrob Agents Chemother. 2021;65(10):e0066321. doi: 10.1128/aac.00663-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soler N, Forterre P. Vesiduction: the fourth way of HGT. Environ Microbiol. 2020;22(7):2457–2460. doi: 10.1111/1462-2920.15056. [DOI] [PubMed] [Google Scholar]
- 61.Drayton M, Deisinger JP, Ludwig KC, Raheem N, Müller A, Schneider T, Straus SK. Host defense peptides: dual antimicrobial and immunomodulatory action. Int J Mol Sci. 2021;22(20):11172. doi: 10.3390/ijms222011172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider VAF, Coorens M, Tjeerdsma-van Bokhoven JLM, Posthuma G, van Dijk A, Veldhuizen EJA, Haagsman HP, Fey PD. Imaging the antistaphylococcal activity of CATH-2: mechanism of attack and regulation of inflammatory response. mSphere. 2017;2(6):e00370–17. doi: 10.1128/mSphere.00370-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scocchi M, Zelezetsky I, Benincasa M, Gennaro R, Mazzoli A, Tossi A. Structural aspects and biological properties of the cathelicidin PMAP-36. FEBS J. 2005;272(17):4398–4406. doi: 10.1111/j.1742-4658.2005.04852.x. [DOI] [PubMed] [Google Scholar]
- 64.Nagaoka I, Tamura H, Reich J. Therapeutic potential of cathelicidin peptide LL-37, an antimicrobial agent, in a murine sepsis model. Int J Mol Sci. 2020;21(17):5973. doi: 10.3390/ijms21175973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balhuizen MD, van Dijk A, Jansen JWA, van de Lest Cha, Veldhuizen EJA, Haagsman HP, van de Lest CHA. Outer membrane vesicles protect gram-negative bacteria against host defense peptides. mSphere. 2021;6(4):e0052321. doi: 10.1128/mSphere.00523-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukherjee S, Bassler BL. Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol. 2019;17(6):371–382. doi: 10.1038/s41579-019-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cooke AC, Nello AV, Ernst RK, Schertzer JW, Hozbor DF. Analysis of Pseudomonas aeruginosa biofilm membrane vesicles supports multiple mechanisms of biogenesis. PLoS One. 2019;14(2):e0212275. doi: 10.1371/journal.pone.0212275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Florez C, Raab JE, Cooke AC, Schertzer JW, Pesci E, Pier GB. Membrane distribution of the pseudomonas quinolone signal modulates outer membrane vesicle production in Pseudomonas aeruginosa. mBio. 2017;8(4):e01034–17. doi: 10.1128/mBio.01034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potera C. Forging a link between biofilms and disease. Science. 1999;283(5409):1837–1839. doi: 10.1126/science.283.5409.1837. [DOI] [PubMed] [Google Scholar]
- 70.Thi MTT, Wibowo D, Rehm BHA. Pseudomonas aeruginosa biofilms. Int J Mol Sci. 2020;21(22):8671. doi: 10.3390/ijms21228671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nairz M, Weiss G. Iron in infection and immunity. Mol Aspects Med. 2020;75:100864. doi: 10.1016/j.mam.2020.100864. [DOI] [PubMed] [Google Scholar]
- 72.Nakashige TG, Zhang B, Krebs C, Nolan EM. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol. 2015;11(10):765–771. doi: 10.1038/nchembio.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schalk IJ, Guillon L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa : implications for metal homeostasis. Environ Microbiol. 2013;15(6):1661–1673. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 74.Kaplan AR, Musaev DG, Wuest WM. Pyochelin biosynthetic metabolites bind iron and promote growth in Pseudomonads demonstrating siderophore-like activity. ACS Infect Dis. 2021;7(3):544–551. doi: 10.1021/acsinfecdis.0c00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau CK, Krewulak KD, Vogel HJ, Bitter W. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev. 2016;40(2):273–298. doi: 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- 76.Tashiro Y, Ichikawa S, Nakajima-Kambe T, Uchiyama H, Nomura N. Pseudomonas quinolone signal affects membrane vesicle production in not only gram-negative but also gram-positive bacteria. Microbes Environ. 2010;25(2):120–125. doi: 10.1264/jsme2.me09182. [DOI] [PubMed] [Google Scholar]
- 77.Sperandio V. SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype O157:H7 in the bovine rumen. Gut Microbes. 2010;1(6):432–435. doi: 10.4161/gmic.1.6.14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ahmed UKB, Ballard JD. Autoinducing peptide-based quorum signaling systems in Clostridioides difficile. Curr Opin Microbiol. 2022;65:81–86. doi: 10.1016/j.mib.2021.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang L, Li S, Liu X, Wang Z, Jiang M, Wang R, Xie L, Liu Q, Xie X, Shang D, et al. Sensing of autoinducer-2 by functionally distinct receptors in prokaryotes. Nat Commun. 2020;11(1):5371. doi: 10.1038/s41467-020-19243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whiteley M, Diggle SP, Greenberg EP. Progress in and promise of bacterial quorum sensing research. Nature. 2017;551(7680):313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morinaga K, Yamamoto T, Nomura N, Toyofuku M. Paracoccus denitrificans can utilize various long-chain N -acyl homoserine lactones and sequester them in membrane vesicles. Environ Microbiol Rep. 2018;10(6):651–654. doi: 10.1111/1758-2229.12674. [DOI] [PubMed] [Google Scholar]
- 82.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc Natl Acad Sci U S A. 2013;110(44):17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang M, Zhao L, Wu H, Zhao C, Gong Q, Yu W. Cladodionen is a potential quorum sensing inhibitor against Pseudomonas aeruginosa. Mar Drugs. 2020;18(4):205. doi: 10.3390/md18040205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tashiro Y, Toyofuku M, Nakajima-Kambe T, Uchiyama H, Nomura N. Bicyclic compounds repress membrane vesicle production and Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2010;304(2):123–130. doi: 10.1111/j.1574-6968.2010.01897.x. [DOI] [PubMed] [Google Scholar]
- 85.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ahern PP, Maloy KJ. Understanding immune-microbiota interactions in the intestine. Immunology. 2020;159(1):4–14. doi: 10.1111/imm.13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodovalho VDR, da Luz BSR, Rabah H, Do Carmo FLR, Folador EL, Nicolas A, Jardin J, Briard-Bion V, Blottière H, Lapaque N, et al. Extracellular vesicles produced by the probiotic Propionibacterium freudenreichii CIRM-BIA 129 mitigate inflammation by modulating the NF-κB pathway. Front Microbiol. 2020;11:1544. doi: 10.3389/fmicb.2020.01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Canas MA, Fabrega MJ, Gimenez R, Badia J, Baldoma L. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front Microbiol. 2018;9:498. doi: 10.3389/fmicb.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol. 2017;17(1):66. doi: 10.1186/s12866-017-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maerz JK, Steimle A, Lange A, Bender A, Fehrenbacher B, Frick J-S. Outer membrane vesicles blebbing contributes to B. vulgatus mpk-mediated immune response silencing. Gut Microbes. 2018;9(1):1–12. doi: 10.1080/19490976.2017.1344810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12(4):509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Durant L, Stentz R, Noble A, Brooks J, Gicheva N, Reddi D, O’Connor MJ, Hoyles L, McCartney AL, Man R, et al. Bacteroides thetaiotaomicron-derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome. 2020;8(1):88. doi: 10.1186/s40168-020-00868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Patten DA, Hussein E, Davies SP, Humphreys PN, Collett A. Commensal-derived OMVs elicit a mild proinflammatory response in intestinal epithelial cells. Microbiology (Reading). 2017;163(5):702–711. doi: 10.1099/mic.0.000468. [DOI] [PubMed] [Google Scholar]
- 94.Turner L, Bitto NJ, Steer DL, Lo C, D’Costa K, Ramm G, Shambrook M, Hill AF, Ferrero RL, Kaparakis-Liaskos M. Helicobacter pylori outer membrane vesicle size determines their mechanisms of host cell entry and protein content. Front Immunol. 2018;9:1466. doi: 10.3389/fimmu.2018.01466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8(10):e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yaghoubfar R, Behrouzi A, Zare Banadkoki E, Ashrafian F, Lari A, Vaziri F, Nojoumi SA, Fateh A, Khatami S, Siadat SD. Effect of Akkermansia muciniphila, Faecalibacterium prausnitzii, and their extracellular vesicles on the serotonin system in intestinal epithelial cells. Probiotics Antimicrob Proteins. 2021;13(6):1546–1556. doi: 10.1007/s12602-021-09786-4. [DOI] [PubMed] [Google Scholar]
- 97.Yaghoubfar R, Behrouzi A, Ashrafian F, Shahryari A, R HR, Choopani S, Hadifar S, Vaziri F, Nojoumi SA, Fateh A, et al. Modulation of serotonin signaling/metabolism by Akkermansia muciniphila and its extracellular vesicles through the gut-brain axis in mice. Sci Rep. 2020;10(1):22119. doi: 10.1038/s41598-020-79171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, et al. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe. 2015;17(5):672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Stentz R, Osborne S, Horn N, W LA, Hautefort I, Bongaerts R, Rouyer M, Bailey P, Shears SB, Hemmings AM, et al. A bacterial homolog of a eukaryotic inositol phosphate signaling enzyme mediates cross-kingdom dialog in the mammalian gut. Cell Rep. 2014;6(4):646–656. doi: 10.1016/j.celrep.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Elmi A, Nasher F, Jagatia H, Gundogdu O, Bajaj-Elliott M, Wren B, Dorrell N. Campylobacter jejuni outer membrane vesicle-associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E-cadherin and occludin. Cell Microbiol. 2016;18(4):561–572. doi: 10.1111/cmi.12534. [DOI] [PubMed] [Google Scholar]
- 101.Nicholas A, Jeon H, Selasi GN, Na SH, Kwon HI, Kim YJ, Choi CW, Kim SI, Lee JC. Clostridium difficile-derived membrane vesicles induce the expression of pro-inflammatory cytokine genes and cytotoxicity in colonic epithelial cells in vitro. Microb Pathog. 2017;107:6–11. doi: 10.1016/j.micpath.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 102.Gu L, Meng R, Tang Y, Zhao K, Liang F, Zhang R, Xue Q, Chen F, Xiao X, Wang H, et al. Toll-like receptor 4 signaling licenses the cytosolic transport of lipopolysaccharide from bacterial outer membrane vesicles. Shock. 2019;51(2):256–265. doi: 10.1097/shk.0000000000001129. [DOI] [PubMed] [Google Scholar]
- 103.Alvarez CS, Badia J, Bosch M, Giménez R, Baldomà L. Outer membrane vesicles and soluble factors released by probiotic Escherichia coli Nissle 1917 and commensal ECOR63 enhance barrier function by regulating expression of tight junction proteins in intestinal epithelial cells. Front Microbiol. 2016;7:1981. doi: 10.3389/fmicb.2016.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kim OY, Park HT, Dinh NTH, Choi SJ, Lee J, Kim JH, Lee SW, Gho YS. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat Commun. 2017;8(1):626. doi: 10.1038/s41467-017-00729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin LT, Shi YC, Choong CY, Tai CJ. The fruits of paris polyphylla inhibit colorectal cancer cell migration induced by Fusobacterium nucleatum-derived extracellular vesicles. Molecules. 2021;26(13):4081. doi: 10.3390/molecules26134081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martin-Gallausiaux C, Malabirade A, Habier J, Wilmes P. Fusobacterium nucleatum extracellular vesicles modulate gut epithelial cell innate immunity via FomA and TLR2. Front Immunol. 2020;11:583644. doi: 10.3389/fimmu.2020.583644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu J, Hsieh CL, Gelincik O, Devolder B, Sei S, Zhang S, Lipkin SM, Chang YF. Proteomic characterization of outer membrane vesicles from gut mucosa-derived fusobacterium nucleatum. J Proteomics. 2019;195:125–137. doi: 10.1016/j.jprot.2018.12.029. [DOI] [PubMed] [Google Scholar]
- 108.Shi Y, Meng L, Zhang C, Zhang F, Fang Y. Extracellular vesicles of Lacticaseibacillus paracasei PC-H1 induce colorectal cancer cells apoptosis via PDK1/AKT/Bcl-2 signaling pathway. Microbiol Res. 2021;255:126921. doi: 10.1016/j.micres.2021.126921. [DOI] [PubMed] [Google Scholar]
- 109.Hu R, Lin H, Wang M, Zhao Y, Liu H, Min Y, Yang X, Gao Y, Yang M. Lactobacillus reuteri-derived extracellular vesicles maintain intestinal immune homeostasis against lipopolysaccharide-induced inflammatory responses in broilers. J Anim Sci Biotechnol. 2021;12(1):25. doi: 10.1186/s40104-020-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alpdundar Bulut E, Bayyurt Kocabas B, Yazar V, Aykut G, Guler U, Salih B, Surucu Yilmaz N, Ayanoglu IC, Polat MM, Akcali KC, et al. Human gut commensal membrane vesicles modulate inflammation by generating M2-like macrophages and myeloid-derived suppressor cells. J Immunol. 2020;205(10):2707–2718. doi: 10.4049/jimmunol.2000731. [DOI] [PubMed] [Google Scholar]
- 111.Deo P, Chow SH, Han ML, Speir M, Huang C, Schittenhelm RB, Dhital S, Emery J, Li J, Kile BT, et al. Mitochondrial dysfunction caused by outer membrane vesicles from Gram-negative bacteria activates intrinsic apoptosis and inflammation. Nat Microbiol. 2020;5(11):1418–1427. doi: 10.1038/s41564-020-0773-2. [DOI] [PubMed] [Google Scholar]
- 112.Aly RG, El-Enbaawy MI, Abd El-Rahman SS, Ata NS. Antineoplastic activity of Salmonella Typhimurium outer membrane nanovesicles. Exp Cell Res. 2021;399(1):112423. doi: 10.1016/j.yexcr.2020.112423. [DOI] [PubMed] [Google Scholar]
- 113.Chatterjee D, Chaudhuri K. Vibrio cholerae O395 outer membrane vesicles modulate intestinal epithelial cells in a NOD1 protein-dependent manner and induce dendritic cell-mediated Th2/Th17 cell responses. J Biol Chem. 2013;288(6):4299–4309. doi: 10.1074/jbc.M112.408302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mehandru S, Colombel J-F. The intestinal barrier, an arbitrator turned provocateur in IBD. Nat Rev Gastroenterol Hepatol. 2021;18(2):83–84. doi: 10.1038/s41575-020-00399-w. [DOI] [PubMed] [Google Scholar]
- 115.Vereecke L, Beyaert R, van Loo G. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol Med. 2011;17(10):584–593. doi: 10.1016/j.molmed.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 116.Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes. 2011;2(5):294–298. doi: 10.4161/gmic.2.5.18603. [DOI] [PubMed] [Google Scholar]
- 117.Engevik MA, Danhof HA, Ruan W, Engevik AC, Chang-Graham AL, Engevik KA, Shi Z, Zhao Y, Brand CK, Krystofiak ES, et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio. 2021;12(2):e02706–20. doi: 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K, Bäumler AJ. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011;79(7):2597–2607. doi: 10.1128/iai.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang X, Ni J, You Y, Feng G, Zhang S, Bao W, Hou H, Li H, Liu L, Zheng M. SNX10-mediated LPS sensing causes intestinal barrier dysfunction via a caspase-5-dependent signaling cascade. EMBO J. 2021;40(24):e108080. doi: 10.15252/embj.2021108080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bian X, Yang L, Wu W, Lv L, Jiang X, Wang Q, Wu J, Li Y, Ye J, Fang D, et al. Pediococcus pentosaceus LI05 alleviates DSS-induced colitis by modulating immunological profiles, the gut microbiota and short-chain fatty acid levels in a mouse model. Microb Biotechnol. 2020;13(4):1228–1244. doi: 10.1111/1751-7915.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Díaz-Garrido N, Bonnin S, Riera M, Gíménez R, Badia J, Baldomà L. Transcriptomic microRNA profiling of dendritic cells in response to gut microbiota-secreted vesicles. Cells. 2020;9(6):1534. doi: 10.3390/cells9061534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bager P, Simonsen J, Nielsen NM, Frisch M. Cesarean section and offspring’s risk of inflammatory bowel disease: a national cohort study. Inflamm Bowel Dis. 2012;18(5):857–862. doi: 10.1002/ibd.21805. [DOI] [PubMed] [Google Scholar]
- 124.Choi JH, Moon CM, Shin T-S, Kim EK, McDowell A, Jo M-K, Joo YH, Kim S-E, Jung H-K, Shim K-N, et al. Lactobacillus paracasei-derived extracellular vesicles attenuate the intestinal inflammatory response by augmenting the endoplasmic reticulum stress pathway. Exp Mol Med. 2020;52(3):423–437. doi: 10.1038/s12276-019-0359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gao L, Song Q, Liang H, Zhu Y, Wei T, Dong N, Xiao J, Shao F, Lai L, Chen X. Legionella effector SetA as a general O-glucosyltransferase for eukaryotic proteins. Nat Chem Biol. 2019;15(3):213–216. doi: 10.1038/s41589-018-0189-y. [DOI] [PubMed] [Google Scholar]
- 126.von Schillde Ma, Hörmannsperger G, Weiher M, Alpert C-A, Hahne H, Bäuerl C, van Huynegem K, Steidler L, Hrncir T, Pérez-Martínez G, et al. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe. 2012;11(4):387–396. doi: 10.1016/j.chom.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 127.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 128.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 129.Liu W, Zhang R, Shu R, Yu J, Li H, Long H, Jin S, Li S, Hu Q, Yao F, et al. Study of the relationship between microbiome and colorectal cancer susceptibility using 16SrRNA sequencing. Biomed Res Int. 2020;2020:7828392. doi: 10.1155/2020/7828392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Parker BJ, Wearsch PA, Veloo ACM, Rodriguez-Palacios A. The genus Alistipes: gut bacteria with emerging implications to inflammation, cancer, and mental health. Front Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park J, Kim NE, Yoon H, Shin CM, Kim N, Lee DH, Park JY, Choi CH, Kim JG, Kim YK, et al. Fecal microbiota and gut microbe-derived extracellular vesicles in colorectal cancer. Front Oncol. 2021;11:650026. doi: 10.3389/fonc.2021.650026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Northrop‐Albrecht EJ, Taylor WR, Huang BQ, Kisiel JB, Lucien F. Assessment of extracellular vesicle isolation methods from human stool supernatant. J Extracell Vesicles. 2022;11(4):e12208. doi: 10.1002/jev2.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lamprinaki D, Garcia-Vello P, Marchetti R, Hellmich C, McCord KA, Bowles KM, Macauley MS, Silipo A, De Castro C, Crocker PR, et al. Siglec-7 mediates immunomodulation by colorectal cancer-associated Fusobacterium nucleatum ssp. animalis. Frontiers in Immunology. 2021;12:744184. doi: 10.3389/fimmu.2021.744184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu -Q-Q, Li C-M, Fu L-N, Wang H-L, Tan J, Wang Y-Q, Sun D-F, Gao Q-Y, Chen Y-X, Fang J-Y. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes. 2020;12(1):1788900. doi: 10.1080/19490976.2020.1788900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, Dejea CM, Fathi P, Chen J, Finard BB, et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe. 2018;23(2):203–214.e5. doi: 10.1016/j.chom.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hezaveh K, Shinde RS, Klötgen A, Halaby MJ, Lamorte S, Ciudad MT, Quevedo R, Neufeld L, Liu ZQ, Jin R, et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity. 2022;55(2):324–340.e8. doi: 10.1016/j.immuni.2022.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kaparakis-Liaskos M, Ferrero RL. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol. 2015;15(6):375–387. doi: 10.1038/nri3837. [DOI] [PubMed] [Google Scholar]
- 138.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535(7610):65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 139.Nakagaki H, Sekine S, Terao Y, Toe M, Tanaka M, Ito H-O, Kawabata S, Shizukuishi S, Fujihashi K, Kataoka K. Fusobacterium nucleatum envelope protein foma is immunogenic and binds to the salivary statherin-derived peptide. Infect Immun. 2010;78(3):1185–1192. doi: 10.1128/iai.01224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Toussi DN, Liu X, Massari P. The FomA porin from Fusobacterium nucleatum is a toll-like receptor 2 agonist with immune adjuvant activity. Clin Vaccine Immunol. 2012;19(7):1093–1101. doi: 10.1128/cvi.00236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mukherjee T, Hovingh ES, Foerster EG, Abdel-Nour M, Philpott DJ, Girardin SE. NOD1 and NOD2 in inflammation, immunity and disease. Arch Biochem Biophys. 2019;670:69–81. doi: 10.1016/j.abb.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 142.Griebel T, Maekawa T, Parker JE. NOD-like receptor cooperativity in effector-triggered immunity. Trends Immunol. 2014;35(11):562–570. doi: 10.1016/j.it.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 143.Németh T, Sperandio M, Mócsai A. Neutrophils as emerging therapeutic targets. Nat Rev Drug Discov. 2020;19(4):253–275. doi: 10.1038/s41573-019-0054-z. [DOI] [PubMed] [Google Scholar]
- 144.Lajqi T, Köstlin-Gille N, Hillmer S, Braun M, Kranig SA, Dietz S, Krause C, Rühle J, Frommhold D, Pöschl J, et al. Gut microbiota-derived small extracellular vesicles endorse memory-like inflammatory responses in murine neutrophils. Biomedicines. 2022;10(2):442. doi: 10.3390/biomedicines10020442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mawe GM, Hoffman JM. Serotonin signalling in the gut—functions, dysfunctions and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2013;10(8):473–486. doi: 10.1038/nrgastro.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Côté F, Thévenot E, Fligny C, Fromes Y, Darmon M, Ripoche MA, Bayard E, Hanoun N, Saurini F, Lechat P, et al. Disruption of the nonneuronal tph1 gene demonstrates the importance of peripheral serotonin in cardiac function. Proc Natl Acad Sci U S A. 2003;100(23):13525–13530. doi: 10.1073/pnas.2233056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Haas-Neill S, Forsythe P. A budding relationship: bacterial extracellular vesicles in the microbiota-gut-brain axis. Int J Mol Sci. 2020;21(23):8899. doi: 10.3390/ijms21238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Villard A, Boursier J, Andriantsitohaina R. Bacterial and eukaryotic extracellular vesicles and nonalcoholic fatty liver disease: new players in the gut-liver axis? Am J Physiol Gastrointest Liver Physiol. 2021;320(4):G485–g495. doi: 10.1152/ajpgi.00362.2020. [DOI] [PubMed] [Google Scholar]
- 149.Schetters ST, Jong WSP, Horrevorts SK, Kruijssen LJW, Engels S, Stolk D, Daleke-Schermerhorn MH, Garcia-Vallejo J, Houben D, Unger WWJ, et al. Outer membrane vesicles engineered to express membrane-bound antigen program dendritic cells for cross-presentation to CD8(+) T cells. Acta Biomater. 2019;91:248–257. doi: 10.1016/j.actbio.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 150.Local Burden of Disease Diarrhoea Collaborators . Mapping geographical inequalities in childhood diarrhoeal morbidity and mortality in low-income and middle-income countries, 2000-17: analysis for the global burden of disease study 2017. Lancet. 2020;395(10239):1779–1801. doi: 10.1016/s0140-6736(20)30114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mitra S, Chakrabarti MK, Koley H. Multi-serotype outer membrane vesicles of Shigellae confer passive protection to the neonatal mice against shigellosis. Vaccine. 2013;31(31):3163–3173. doi: 10.1016/j.vaccine.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 152.Kesty NC, Kuehn MJ. Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J Biol Chem. 2004;279(3):2069–2076. doi: 10.1074/jbc.M307628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Price NL, Goyette-Desjardins G, Nothaft H, Valguarnera E, Szymanski CM, Segura M, Feldman MF. Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci Rep. 2016;6(1):24931. doi: 10.1038/srep24931. doi:ARTN 24931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Irene C, Fantappiè L, Caproni E, Zerbini F, Anesi A, Tomasi M, Zanella I, Stupia S, Prete S, Valensin S, et al. Bacterial outer membrane vesicles engineered with lipidated antigens as a platform for Staphylococcus aureus vaccine. Proc Natl Acad Sci U S A. 2019;116(43):21780–21788. doi: 10.1073/pnas.1905112116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Peng L-H, Wang M-Z, Chu Y, Zhang L, Niu J, Shao H-T, Yuan T-J, Jiang Z-H, Gao J-Q, Ning X-H. Engineering bacterial outer membrane vesicles as transdermal nanoplatforms for photo-TRAIL–programmed therapy against melanoma. Sci Adv. 2020;6(27):eaba2735. doi: 10.1126/sciadv.aba2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Qing S, Lyu C, Zhu L, Pan C, Wang S, Li F, Wang J, Yue H, Gao X, Jia R, et al. Biomineralized bacterial outer membrane vesicles potentiate safe and efficient tumor microenvironment reprogramming for anticancer therapy. Adv Mater. 2020;32(47):e2002085. doi: 10.1002/adma.202002085. [DOI] [PubMed] [Google Scholar]
- 157.Gao F, Xu L, Yang B, Fan F, Yang L. Kill the real with the fake: eliminate intracellular Staphylococcus aureus using nanoparticle coated with its extracellular vesicle membrane as active-targeting drug carrier. ACS Infect Dis. 2019;5(2):218–227. doi: 10.1021/acsinfecdis.8b00212. [DOI] [PubMed] [Google Scholar]
- 158.Chen Q, Bai H, Wu W, Huang G, Li Y, Wu M, Tang G, Ping Y. Bioengineering bacterial vesicle-coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Lett. 2020;20(1):11–21. doi: 10.1021/acs.nanolett.9b02182. [DOI] [PubMed] [Google Scholar]
- 159.Li M, Li S, Zhou H, Tang X, Wu Y, Jiang W, Tian Z, Zhou X, Yang X, Wang Y. Chemotaxis-driven delivery of nano-pathogenoids for complete eradication of tumors post-phototherapy. Nat Commun. 2020;11(1):1126. doi: 10.1038/s41467-020-14963-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kim DJ, Yang J, Seo H, Lee WH, Ho Lee D, Kym S, Park YS, Kim JG, Jang I-J, Kim Y-K, et al. Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Sci Rep. 2020;10(1):2860. doi: 10.1038/s41598-020-59529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kim SS, Eun JW, Cho HJ, Song DS, Kim CW, Kim YS, Lee SW, Kim YK, Yang J, Choi J, et al. Microbiome as a potential diagnostic and predictive biomarker in severe alcoholic hepatitis. Aliment Pharmacol Ther. 2021;53(4):540–551. doi: 10.1111/apt.16200. [DOI] [PubMed] [Google Scholar]
- 162.Wei S, Peng W, Mai Y, Li K, Wei W, Hu L, Zhu S, Zhou H, Jie W, Wei Z, et al. Outer membrane vesicles enhance tau phosphorylation and contribute to cognitive impairment. J Cell Physiol. 2020;235(5):4843–4855. doi: 10.1002/jcp.29362. [DOI] [PubMed] [Google Scholar]
- 163.Zavan L, Bitto NJ, Johnston EL, Greening DW, Kaparakis-Liaskos M. Back Cover: helicobacter pylori growth stage determines the size, protein composition, and preferential cargo packaging of outer membrane vesicles. Proteomics. 2019;19(1–2):e1800209. doi: 10.1002/pmic.201970004. [DOI] [PubMed] [Google Scholar]
- 164.Tashiro Y, Hasegawa Y, Shintani M, Takaki K, Ohkuma M, Kimbara K, Futamata H. Interaction of bacterial membrane vesicles with specific species and their potential for delivery to target cells. Front Microbiol. 2017;8:571. doi: 10.3389/fmicb.2017.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liu Y, Smid EJ, Abee T, Notebaart RA. Delivery of genome editing tools by bacterial extracellular vesicles. Microb Biotechnol. 2019;12(1):71–73. doi: 10.1111/1751-7915.13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lee H, Lee Y, Kim J, An J, Lee S, Kong H, Song Y, Lee C-K, Kim K. Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes. 2018;9(2):155–165. doi: 10.1080/19490976.2017.1405209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.a S B S J T USACPO, 125 questions: Exploration and discovery. https://www.sciencemag.org/collections/125-questions-exploration-and-discovery (14 May 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this review.


