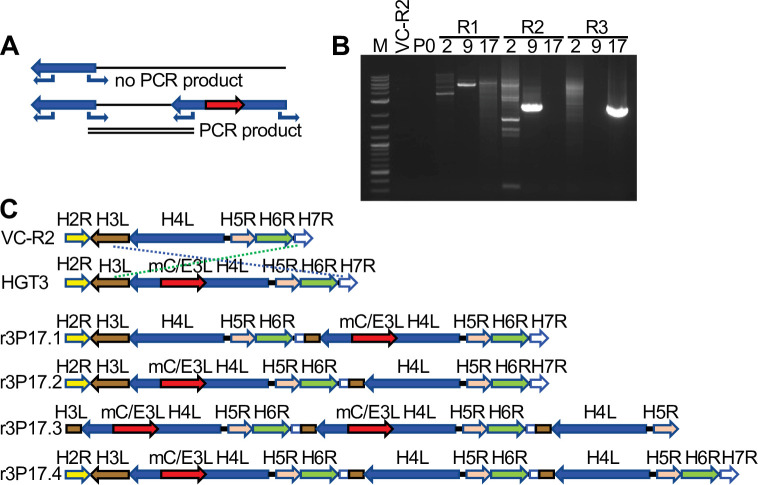
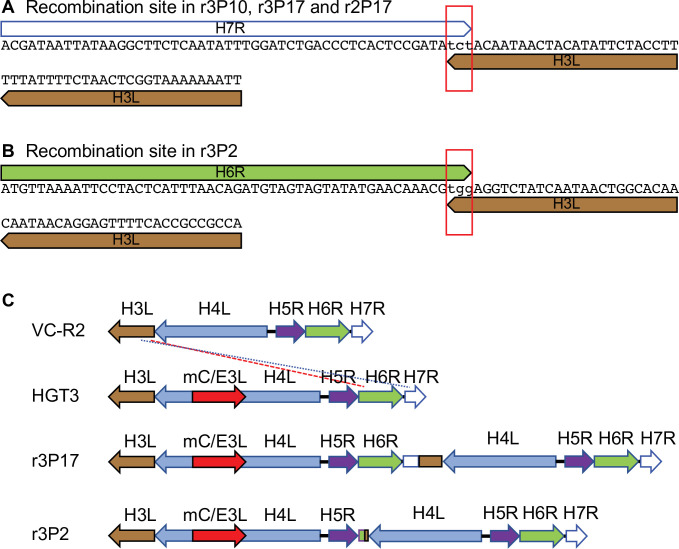
Figure 5. Recombination of complementing viruses after serial passaging.
(A) PCR strategy to detect gene arrays with outward-facing primers. (B) Representative PCR products amplified with outward-facing primers for VC-R2 and three independent HGT3 passages. (C) Recombination of complementing viruses resulted in tandem arrays of the H4L locus as determined by long-read sequencing.