Abstract
Generation of bone marrow (BM) from embryonic stem cells (ESCs) promises to accelerate the development of future cell therapies for life-threatening disorders. However, such approach is limited by technical challenges to produce a mixture of functional BM progenitor cells able to replace all hematopoietic cell lineages. Herein, we used blastocyst complementation to simultaneously produce BM cell lineages from mouse ESCs in a rat. Based on fluorescence-activated cell sorting analysis and single-cell RNA sequencing, mouse ESCs differentiated into multiple hematopoietic and stromal cell types that were indistinguishable from normal mouse BM cells based on gene expression signatures and cell surface markers. Receptor–ligand interactions identified Cxcl12-Cxcr4, Lama2-Itga6, App-Itga6, Comp-Cd47, Col1a1-Cd44, and App-Il18rap as major signaling pathways between hematopoietic progenitors and stromal cells. Multiple hematopoietic progenitors, including hematopoietic stem cells (HSCs) in mouse–rat chimeras derived more efficiently from mouse ESCs, whereas chondrocytes predominantly derived from rat cells. In the dorsal aorta and fetal liver of mouse–rat chimeras, mouse HSCs emerged and expanded faster compared to endogenous rat cells. Sequential BM transplantation of ESC-derived cells from mouse–rat chimeras rescued lethally irradiated syngeneic mice and demonstrated long-term reconstitution potential of donor HSCs. Altogether, a fully functional BM was generated from mouse ESCs using rat embryos as ‘bioreactors’.
Research organism: Rat
Introduction
The bone marrow (BM) is a remarkably complex organ consisting of multiple mesenchymal, immune, endothelial, and neuronal cell types which together comprise a highly specialized microenvironment required to support c blood regeneration or hematopoiesis (Baccin et al., 2020; Baryawno et al., 2019; Rowe et al., 2016; Tikhonova et al., 2019; Vo and Daley, 2015). Hematopoiesis occurs in a stepwise manner and is initiated by a heterogeneous, multipotent, population of hematopoietic stem cells (HSCs), located at the apex of the hematopoietic differentiation tree. Long-term HSCs (LT-HSCs) remain quiescent to maintain their undifferentiated state within the BM niche. When necessary, LT-HSCs can either undergo differentiation or self-renewal, to maintain the HSC pool. Conversely, short-term HSCs (ST-HSCs) are restricted in their self-renewal capacity and primed for differentiation into multipotent progenitors (MPPs), initiating the process of blood cell development. MPPs further differentiate into common myeloid progenitors (CMPs), lymphoid-primed multipotent progenitors (LMPPs), and common lymphoid progenitors (CLPs) that become increasingly lineage restricted with subsequent cell divisions, ultimately yielding all mature blood cell types (Haas et al., 2018). The complexities of the hematopoietic system have been studied extensively in vitro, utilizing paired-daughter and colony-forming unit (CFU) assays (Rowe et al., 2016; Vo and Daley, 2015). Fluorescence-activated cell sorting (FACS) has allowed for precise isolation and characterization of HSCs and progenitor populations based on cell surface markers. Classically, the most biologically relevant way to test HSC function remains to be through serial transplantation and hematopoietic reconstitution of irradiated recipient mice (Purton and Scadden, 2007; Rowe et al., 2016; Vo and Daley, 2015). Recent advances in single-cell RNA sequencing (scRNAseq) have made it possible to further explore heterogeneity of the BM niche (Baryawno et al., 2019; Tikhonova et al., 2019), and identify gene expression signatures of hematopoietic progenitor cells as they differentiate into mature blood cell types (Baccin et al., 2020; Nestorowa et al., 2016).
Generation of functional BM from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) promises to provide new therapeutic opportunities for hematologic and autoimmune disorders. However, this approach is limited by technical challenges to produce functional HSCs or the mixture of hematopoietic progenitors capable of replacing all mature blood cell types after cell transplantation. HSC-like cells have been generated from mouse and human ESCs and iPSCs using in vitro differentiation protocols (Amabile et al., 2013; Doulatov et al., 2013; Grigoriadis et al., 2010; Kitajima et al., 2011; Ledran et al., 2008; Sugimura et al., 2017; Vodyanik et al., 2006). Likewise, ESCs and iPSCs have been used to produce myeloid and lymphoid progenitor cells as well as differentiated hematopoietic cells, including neutrophils, monocytes, erythroid cells, and T and B lymphocytes (Doulatov et al., 2013; Elcheva et al., 2014; Galic et al., 2006; Kennedy et al., 2012; Montel-Hagen et al., 2019; Nafria et al., 2020; Vodyanik et al., 2005). When transplanted into irradiated animals, ESC/iPSC-derived hematopoietic progenitor cells undergo differentiation and engraft into the BM niche, providing an important source of renewal and regeneration for various blood cell lineages (Rowe et al., 2016; Sugimura et al., 2017; Vo and Daley, 2015). While ESC/iPSC-derived hematopoietic cells often express appropriate cell markers, gene expression and functional studies indicate significant differences between ESC/iPSC-derived cells and endogenous cells that have undergone normal morphogenesis in the BM (Lin et al., 2019; Lu et al., 2016; Sugimura et al., 2017).
In vivo differentiation of ESCs into multiple cell lineages can be achieved using blastocyst complementation, in which donor ESCs are injected into blastocysts of recipient animals to create chimeras. Fluorescently labeled ESCs undergo differentiation in recipient embryos that serve as ‘biological reactors’ by providing growth factors, hormones, and cellular niches to support ESC differentiation in the embryo. In mouse and rat apancreatic Pdx1−/− embryos, donor ESCs formed an entire pancreas in which both exocrine and endocrine cells were almost entirely derived from ESCs or iPSCs (Kobayashi et al., 2010; Yamaguchi et al., 2017). Mouse ESC/iPSC-derived β-cells from mouse–rat chimeras were fully differentiated and successfully rescued syngeneic diabetic mice (Yamaguchi et al., 2017). ESCs generated pancreatic cell lineages in apancreatic pigs (Matsunari et al., 2013), kidney in Sall1-deficient rats (Goto et al., 2019), endothelial cells in Flk1−/− mice (Hamanaka et al., 2018), lymphocytes in immunodeficient mice (Muthusamy et al., 2011), and neuronal progenitors in mice with forebrain-specific overexpression of diphtheria toxin (Chang et al., 2018). Recently, mouse ESCs were used to generate lung and thyroid tissues in embryos deficient for Fgf10, Nkx2-1, Fgfr2, or β-catenin (Kitahara et al., 2020; Mori et al., 2019; Wen et al., 2021). Using blastocyst complementation, mouse ESCs effectively produced hematopoietic cells in mice deficient for Kit or Flk1 (Hamanaka et al., 2018; Jansson and Larsson, 2010). ESC-derived endothelial progenitor cells from mouse–rat chimeras were indistinguishable from endogenous endothelial progenitor cells based on gene expression signatures and functional properties (Wang et al., 2021), indicating that ESC/iPSC-derived progenitors can be used for tissue regeneration (Bolte et al., 2020a; Bolte et al., 2018; Dharmadhikari et al., 2015; Kolesnichenko et al., 2021). While all these studies support the effectiveness of blastocyst complementation for differentiation of multiple cell types from ESCs/iPSCs in vivo, generation of functional BM from ESCs in interspecies chimeras has not yet been achieved.
Herein, we used blastocyst complementation to produce mouse BM in a rat. ESC-derived cells from multiple hematopoietic and stromal cell lineages were indistinguishable from normal mouse BM cells based on gene expression signatures and cell surface markers. Transplantation of ESC-derived BM cells into lethally irradiated syngeneic mice prevented mortality and resulted in a long-term contribution to BM and mature blood cell types. Our data demonstrate that interspecies chimeras can be used as ‘bioreactors’ for in vivo differentiation and functional studies of ESC-derived BM hematopoietic and stromal cells.
Results
Generation of BM from pluripotent ESCs in interspecies mouse–rat chimeras
To determine whether mouse ESCs can differentiate into multiple hematopoietic cell lineages in the BM of a rat, blastocyst complementation was performed by injecting GFP-labeled mouse C57BL/6 ESCs (ESC-GFP) into rat SD blastocysts to create interspecies mouse–rat chimeras. Chimeric embryos were transferred into surrogate female rats for subsequent development in utero (Figure 1A). While mouse–rat chimeras were viable, they were smaller than age-matched rats (Figure 1B). Consistent with the presence of mouse ESC-derived cells (black) in the skin tissue (Wang et al., 2021), mixed black and white pigmentation distinguished the mouse–rat chimeras from juvenile rats (Figure 1B). The average body weight of mouse–rat chimeras was smaller than rats, but larger than mice of similar age (Figure 1C). ESC-derived cells were abundant in femur and tibia bones of the chimeras as evidenced by GFP fluorescence (Figure 1D). FACS analysis of BM cells obtained from juvenile mouse–rat chimeras revealed that the percentage of ESC-derived cells was 15–50% (Figure 1E, F). Thus, ESCs contribute to the BM of mouse–rat chimeras.
Figure 1. Mouse embryonic stem cells (ESCs) contribute to hematopoietic stem cells (HSCs) in the bone marrow (BM) of mouse–rat chimeras.
(A) Schematic shows blastocyst complementation of rat embryos with mouse ESCs to generate interspecies mouse–rat chimeras. GFP-labeled mouse ESCs (mESCs) were injected into rat blastocysts, which were implanted into surrogate rat females to undergo embryonic development in utero. Femur and tibia bones of the chimeras were used to obtain BM cells. (B) Photographs of mouse–rat chimeras are taken at postnatal (P) days P3, P8, P13, and P28. Mixed black and white pigmentation distinguishes the mouse–rat chimeras from juvenile rats and mice. (C) Weights of mouse–rat chimeras are shown at different time points and compared to rats and mice of similar ages. Chimeras are significantly smaller than rats, but larger than mice (n = 7–18 in each group), **p < 0.01, see also Source data 1. (D) Fluorescence microscopy shows GFP and bright-field images of femur and tibia bones from P4 rat, mouse, and mouse–rat chimera. (E) Fluorescence-activated cell sorting (FACS) analysis of mouse ESC-derived (GFP-positive) cells in the BM of P10 mouse–rat chimeras. Lineage-negative (Lin−), LSK, short-term HSC (ST-HSC), and long-term HSC (LT-HSC) cell subsets were identified in the BM of mouse–rat chimeras (n = 10) and control mice (n = 8), see also Figure 1—figure supplement 1A, B. (F) Histograms show GFP fluorescence of BM cells from chimeras and control mice. (G–H) FACS analysis shows increased percentages of mouse LSKs and LT-HSCs in BM of mouse–rat chimeras (n = 10) compared to control mice (n = 8), **p < 0.01, N.S. indicates no significance. (I) FACS analysis shows increased numbers of HSCs (ST-HSCs + LT-HSCs) in BM of mouse rat chimeras (n = 10) compared to control mice (n = 8), **p < 0.01.
Figure 1—figure supplement 1. Identification of lineage− cells, LSKs, short-term HSCs (ST-HSCs), and long-term HSCs (LT-HSCs) in the bone marrow (BM).
To identify ESC-derived HSCs, we used GFP fluorescence and mouse-specific antibodies recognizing multiple cell surface antigens (Figure 1E and Figure 1—figure supplement 1A, B). First, ESC-derived GFP+ BM cells were subdivided into lineage-positive (Lin+) and lineage-negative subpopulations (Lin−) (Figure 1E and Figure 1—figure supplement 1A, B). The percentage of ESC-derived Lin− cells in the BM of mouse–rat chimeras was similar to the percentage of Lin− cells in the BM of age-matched C57BL/6 mice (Figure 1E, G). Next, we used Sca1 and CD117 (c-KIT) antibodies to identify Lin−Sca1+c-KIT+ cells (LSKs) (Figure 1E). The percentage of LSKs was higher in the BM of mouse–rat chimeras compared to the control (Figure 1G). Based on cell surface expression of CD150 and CD48, the percentage of LT-HSCs among LSKs was also higher in mouse–rat chimeras (Figure 1E, H). While changes in ST-HSCs were not significant (Figure 1H), total numbers of HSCs (LT-HSCs + ST-HSCs) were higher in mouse–rat chimeras compared to mice of the same age (Figure 1I). Thus, mouse ESCs can differentiate into hematopoietic progenitor cells in the BM of mouse–rat chimeras.
Single-cell RNA sequencing identifies multiple subpopulations of ESC-derived hematopoietic cells in the BM of mouse–rat chimeras
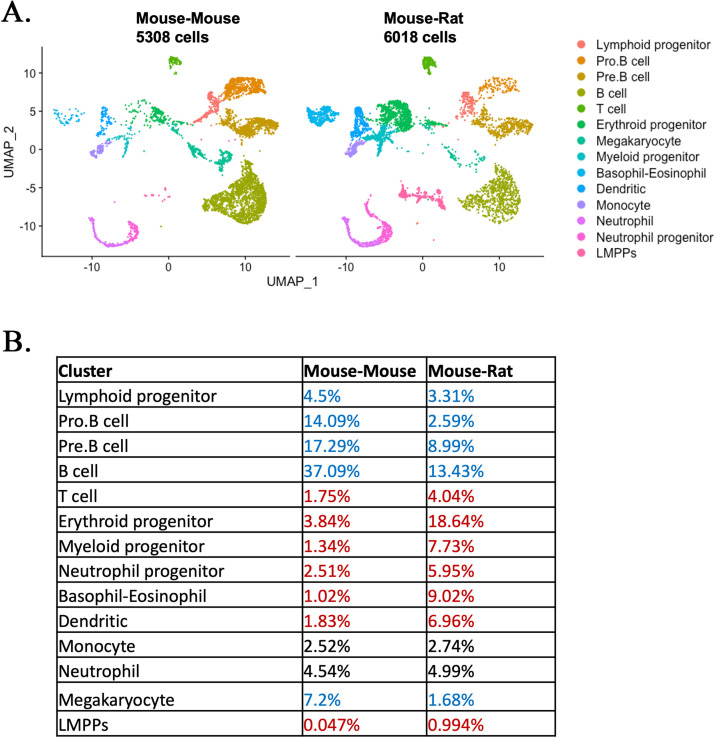
To identify ESC-derived cells in the BM, single-cell RNAseq (the 10× Chromium platform) of FACS-sorted GFP+ BM cells was performed. Mouse ESC-derived cells from P10 mouse–rat chimeras were compared to ESC-derived cells from P10 mouse–mouse (control) chimeras, the latter of which were produced by complementing mouse blastocysts with mouse ESCs from the same ESC-GFP cell line. Based on GFP fluorescence, contribution of ESCs to BM cells in both chimeras was similar (Figure 2—figure supplement 1A, B). Since the numbers of HSCs and other hematopoietic progenitor cells in the BM are low compared to numbers of differentiated hematopoietic cells, we enriched for BM progenitor cell populations prior to single-cell RNA sequencing by combining 90% of FACS-sorted GFP+Lin− cells and 10% of GFP+Lin+ cells in each experimental group. BM cells from 3 animals per group were combined prior to FACS sorting. Based on published gene expression signatures of mouse BM cells (Baccin et al., 2020), 11,326 cells from 14 major cell subtypes were identified: 5308 cells from control mouse–mouse chimeras and 6018 cells from mouse–rat chimeras. These include lymphoid, erythroid, myeloid, and neutrophil progenitors, Pro-B, Pre-B, B and T lymphocytes, megakaryocytes, dendritic cells, neutrophils, basophils/eosinophils, monocytes, and LMPP cells (Figure 2A and Figure 2—figure supplement 2A). Analysis of BM cells from mouse–rat and mouse–mouse chimeras demonstrated similar distributions of hematopoietic cell lineages derived from CMP and CLP (Figure 2A), indicating identical cell types in mouse–rat and control chimeras. For selected genes, we used violin plots to confirm cell specificity and expression levels of Ptprc (Cd45), Pclaf, Vpreb1, Tmpo, Ebf1, Ms4a4b, Vamp5, Elof1, Elane, Ms4a2, Siglech, Ngp, Clec4d, Ctss, and Ftl1-ps1 in the combined dataset (Figure 2—figure supplement 3). Markers of endothelial cells, adipocytes, osteocytes, and neuronal cells were undetectable in BM cell suspensions from both chimeras (Figure 2—figure supplement 2B). Percentages CLP-derived lymphoid progenitors, Pro-B, Pre-B, and B cells were lower in mouse–rat chimeras compared to the control (Figure 2A, B). In contrast, percentages of CMP-derived erythroid, myeloid and neutrophil progenitors, dendritic cells, and basophils/eosinophils were higher (Figure 2B). Monocytes and neutrophils were similar, whereas megakaryocytes were decreased in the BM of mouse–rat chimeras (Figure 2B). The percentage of LMPPs in mouse–rat chimeras was increased compared to the control (Figure 2A, B). HSCs, identified by coexpression of Kit, Ly6a(Sca1), and Flt3 mRNAs (Rowe et al., 2016; Vo and Daley, 2015), clustered together with myeloid and erythroid progenitors (Figure 2—figure supplement 4A, B). The number of ESC-derived HSCs was higher in BM of mouse–rat chimeras compared to the control (Figure 2—figure supplement 4C), findings consistent with FACS analysis (Figure 1H, I). Only 6 out of 6018 BM cells (0.1%) in mouse–rat chimeras contained both mouse and rat mRNA transcripts (Supplementary files 1 and 2), indicating that the fusion of mouse and rat BM cells is rare. Thus, although the cellular composition of ESC-derived hematopoietic BM cells was similar in mouse–rat and mouse–mouse chimeras, mouse–rat BM was enriched in HSCs, LMPPs, and CMP-derived erythroid, myeloid, and neutrophil progenitors.
Figure 2. Single-cell RNAseq analysis identifies embryonic stem cell (ESC)-derived hematopoietic cell lineages in the bone marrow (BM) of mouse–rat chimeras.
(A) Parallel dimension UMAP plots show identical hematopoietic cell clusters in the BM of mouse–mouse chimera (5308 cells) and mouse–rat chimera (6018 cells). ESC-derived BM cells were obtained from the BM of P10 chimeras using fluorescence-activated cell sorting (FACS) for GFP+ cells, see Figure 2—figure supplement 1A, B. Cells from n = 3 animals per group were pooled together prior to FACS sorting. Cell clusters were identified from single-cell RNAseq datasets using Uniform Manifold Approximation and Projection (UMAP) method, see also Figure 2—figure supplements 2A, B and 3. Hematopoietic stem cells (HSCs) were identified by coexpression of Kit, Ly6a (Sca1), and Flt3 (Flk2), see Figure 2—figure supplement 4. Heatmaps and linear regression analysis identified significant similarities in gene expression signatures of lymphoid and myeloid progenitor cells obtained from mouse–rat (R) and mouse–mouse chimeras (M), see Figure 2—figure supplement 5A, B and Figure 2—figure supplement 6A, B. Gene expression profiles of ESC-derived HSCs and lymphoid-primed multipotent progenitor cells are shown in Figure 2—figure supplement 7A, B. (B) Table shows percentages of cells in individual clusters in mouse–mouse and mouse–rat chimeras. Blue color indicates decreased percentages of cells in mouse–rat chimeras compared to mouse–mouse chimeras. Red color indicates increased percentages of cells in mouse–rat chimeras.
Figure 2—figure supplement 1. Purification of mouse embryonic stem cell (ESC)-derived cells from bone marrow (BM) of mouse–rat and mouse–mouse chimeras before scRNAseq.
Figure 2—figure supplement 2. Single-cell RNAseq analysis identifies hematopoietic cell subsets in the bone marrow (BM) of mouse–rat chimeras.
Figure 2—figure supplement 3. Violin plots confirm expression of hematopoietic marker genes in bone marrow (BM) cell clusters.
Figure 2—figure supplement 4. Single-cell RNAseq analysis identifies genes expressed in hematopoietic stem cells (HSCs) in chimeric bone marrow (BM).
Figure 2—figure supplement 5. Embryonic stem cell (ESC)-derived lymphoid cell types in mouse–rat and mouse–mouse chimeras exhibit identical gene expression profiles.
Figure 2—figure supplement 6. Embryonic stem cell (ESC)-derived myeloid cell types in mouse–rat and mouse–mouse chimeras exhibit similar gene expression profiles.
Figure 2—figure supplement 7. Heatmaps identify gene expression profile of embryonic stem cell (ESC)-derived hematopoietic stem cells (HSCs) and lymphoid-primed multipotent progenitor (LMPP) cells from mouse–rat and mouse–mouse chimeras.
Single-cell RNA sequencing identifies close similarities in gene expression signatures between ESC-derived hematopoietic cells in mouse–rat and mouse–mouse chimeras
Comparison of gene expression signatures between mouse–rat and mouse–mouse chimeras revealed significant similarities among ESC-derived hematopoietic cell types. Lymphoid progenitors and pro-B cells isolated from mouse–rat and control chimeras expressed Mif, Rcsd1, and Tspan13, whereas pre-B cells expressed Hmgb2 and Pgls (Figure 2—figure supplement 5A). Cd79a and CD79b transcripts were detected in B cells of mouse–rat and control chimeras, whereas Cd3g and Lck were restricted to T cells (Figure 2—figure supplement 5A). Based on the correlation analysis, gene expression profiles of all lymphoid cell types were similar between mouse–rat and control chimeras (Figure 2—figure supplement 5B). Likewise, gene expression signatures of myeloid, erythroid, and neutrophil progenitors and their derivatives in the BM were similar in both experimental groups (Figure 2—figure supplement 6A, B). Furthermore, single-cell RNAseq identified close similarities in gene expression signatures of ESC-derived HSCs and LMPPs in both chimeras (Figure 2—figure supplement 7A, B). Thus, gene expression signatures of ESC-derived hematopoietic cells were similar in mouse–rat and control mouse–mouse chimeras.
Chimeric BM is enriched in mouse hematopoietic progenitor cells and rat chondrocytes
To examine the composition and origin of stromal cells in mouse–rat chimeras, we used an enzymatic digestion to obtain both hematopoietic and stromal cells from BM of P5 mouse–rat chimeras and compared them to BM cells of mice and rats of the same age. Flow sorting for GFP was performed to separate donor mouse cells (GFP+) and recipient rat cells (GFP−) in the chimeric BM. BM from control P5 mice and rats was also FACS-sorted for GFP– BM cells to ensure similar conditions of cell preparations prior to single-cell RNAseq. Based on published gene expression signatures (Baccin et al., 2020), 6375 mouse and 5495 rat cells were identified in the chimeras, which were compared to 6418 cells from control mice and 7016 cells from control rats. Similar hematopoietic and stromal cell clusters were present in BM of mice, rats, and mouse–rat chimeras (Figure 3A–C). These included stromal cell clusters (endothelial cells, fibroblasts, myofibroblasts, and chondrocytes) and hematopoietic cell clusters with various progenitor and differentiated hematopoietic cell types. Since we did not enrich BM cell populations for Lin− cells, some rare BM cell subsets, such as HSCs, LMPPs, and dendritic cells, were not detected as separate cell clusters. Compared to normal BM from P5 mice, chimeric BM was enriched in mouse ESC-derived hematopoietic progenitor cells, such as myeloid, granulocyte, and erythroid progenitors, whereas mouse-derived B cell lineages were reduced (Figure 3A), findings consistent with single-cell RNAseq comparison of P10 BM from mouse–rat and mouse–mouse chimeras (Figure 2). The percentage of mouse endothelial cells was increased in mouse–rat BM, whereas the percentages of mouse chondrocytes and fibroblasts were reduced compared to mouse control (Figure 3A). In contrast, mouse–rat BM was enriched in rat-derived chondrocytes and fibroblasts, but the percentages of endothelial and most hematopoietic cells were reduced compared to age-matched rats (Figure 3B). Thus, mouse cells preferentially contributed to hematopoietic progenitors and endothelial cells, whereas rat cells contributed to the majority of chondrocytes and fibroblasts.
Figure 3. Single-cell RNAseq analysis shows increased percentages of embryonic stem cell (ESC)-derived hematopoietic progenitors and endothelial cells but decreased percentages of ESC-derived chondrocytes in the bone marrow (BM) of mouse–rat chimeras.
(A, B) Parallel dimension UMAP plots show identical hematopoietic and stromal cell clusters in the BM of P5 mice, rats, and mouse–rat chimeras. BM cells were obtained from P5 animals using an enzymatic digestion (n = 5 animals per group) and pooled prior to single-cell RNAseq. Cell clusters were identified from single-cell RNAseq datasets using Uniform Manifold Approximation and Projection (UMAP) method. Red color in the tables indicates increased percentages of cells in mouse–rat chimeras compared to either mice or rats of the same age. Blue color indicates decreased percentages of cells in mouse–rat chimeras. Gene expression signatures of mouse and rat hematopoietic and stromal cells are shown in Figure 3—figure supplement 1A–D. (C) A bar graph shows relative percentages of ESC-derived mouse cells (green) and endogenous rat cells (blue) in the BM of P5 mouse–rat chimeras.
Figure 3—figure supplement 1. Heatmaps compare gene expression profile in mouse and rat hematopoietic and stromal cells that form bone marrow (BM) in mouse–rat chimeras.
Direct comparison of mouse and rat cells within chimeric BM demonstrated significant similarities between gene expression signatures of hematopoietic and stromal cell lineages (Figure 3—figure supplement 1A–D). To examine cell signaling between hematopoietic progenitors and stromal cells in BM of mouse–rat chimeras, we generated the map of potential ligand–receptor interactions using P5 single-cell RNAseq datasets. There were remarkable similarities in major receptor–ligand interactions between stromal and erythro-myeloid progenitor cells (EMPs) (Figure 4). Regardless of mouse and rat origins of BM cells, endothelial cells interacted with EMPs through the Cxcl12-Cxcr4 receptor–ligand signaling pair. The main signaling circuit between fibroblasts and EMPs was Lama2-Itga6, whereas chondrocytes signaled to EMPs through App-Itga6 and Comp-Cd47 pathways (Figure 4). Major receptor–ligand interactions between granulocyte–monocyte progenitor (GMP) cells and stromal cells were also similar in BM cells of mouse and rat origin (Figure 4—figure supplement 1). These include Cxcl12-Cxcr4 signaling between endothelial cells and GMPs, Col1a1-Cd44 signaling between fibroblasts and GMPs, and App-Il18rap signaling between chondrocytes and GMPs (Figure 4—figure supplement 1). Analysis of expression patterns for several ligands and their receptors revealed no obvious differences between mouse and rat cells (Figure 4—figure supplement 2). These results demonstrate that mouse and rat BM cells use similar signaling pathways between stromal and hematopoietic progenitor cells.
Figure 4. Single-cell RNAseq analysis shows remarkable similarities in major receptor–ligand interactions between erythro-myeloid progenitors and stromal cells of mouse and rat origins.
Bone marrow (BM) cells were obtained from P5 animals using an enzymatic digestion (n = 5 animals per group). Single-cell RNAseq was performed to identify BM stromal and erythro-myeloid progenitor cells (EMPs) based on gene expression signatures. The R package NicheNet was used to analyze the expression of ligands and receptors to identify intercellular communication patterns between EMPs and BM stromal cells. Receptor–ligand interactions between stromal and granulocyte–monocyte progenitor (GMP) cells are shown in Figure 4—figure supplement 1. Violin plots were used to identify expression of ligands and their receptors in hematopoietic and stromal BM cells, see Figure 4—figure supplement 2.
Figure 4—figure supplement 1. Single-cell RNAseq analysis shows similar receptor–ligand interactions between stromal and granulocyte–monocyte progenitor (GMP) cells.
Figure 4—figure supplement 2. Violin plots show expression of ligands and their receptors in hematopoietic and stromal bone marrow (BM) cells.
Mouse HSCs in mouse–rat chimeras develop earlier than rat HSCs
Fetal HSCs emerge from hemogenic endothelium in the aorta–gonad–mesonephros region and later undergo expansion in the embryonic liver (Gao et al., 2018; Weijts et al., 2021). To examine the development of HSCs in mouse–rat chimeras, mouse-derived (GFP+) and rat-derived (GFP−) hemogenic endothelial cells were visualized in the dorsal aorta by colocalization of FLK1 with RUNX1 transcription factor (Figure 5A, B). At E11, mouse embryos were significantly larger than rat and mouse–rat chimeric embryos (Figure 5—figure supplement 1), consistent with previous studies demonstrating that the main stages of mouse embryonic development occur approximately 1.5 days faster compared to embryonic development in the rat (Farrington-Rock et al., 2008; Marcela et al., 2012; Takahashi and Osumi, 2005; Torres et al., 2008). Therefore, we compared E11 mouse embryos with E12.5 rat and chimeric embryos which were in similar developmental stages. In the dorsal aorta of mouse–rat chimeras, the majority of FLK1+RUNX1+ cells expressed GFP, indicating the mouse origin of these cells (Figure 5B). Later in development, percentages of mouse Lin− cells, LSKs, and ST-HSCs were higher in fetal livers of mouse–rat chimeras as demonstrated by FACS analysis for Lin, CD117, Sca1, CD48, and CD150 (Figure 5C and Figure 5—figure supplement 2). The percentage of LT-HSC in fetal livers was unchanged (Figure 5C). Thus, ESC complementation causes the earlier development of donor HSCs in the dorsal aorta and increases percentages of donor-derived Lin− cells, LSKs, and ST-HSCs in the fetal liver.
Figure 5. Mouse hematopoietic stem cells (HSCs) in mouse–rat chimeras develop earlier than rat HSCs.
(A, B) Immunostaining for RUNX1 (white) and FLK1 (red) shows that hemogenic endothelium in the dorsal aorta (DA) of mouse–rat chimeras develops mostly from embryonic stem cell (ESC)-derived mouse cells. GFP (green) was used to identify ESC-derived cells, whereas 4′,6-diamidino-2-phenylindole (DAPI, blue) was used to stain cell nuclei. Frozen sections were obtained from E11 mouse embryos and E12.5 embryos from rats and mouse–rat chimeras since these embryos are in similar developmental stages, see also Figure 5—figure supplement 1A–C. DA indicates the lumen of dorsal aorta. Yellow dashed line indicates the luminal surface of DA wall. Inserts show high magnification of hemogenic endothelial cells expressing both RUNX1 and FLK1. Scale bars are: A, 200 μm; B, 20 μm; inserts in B, 5 μm. Abbreviations: DA, dorsal aorta; Li, liver. (C) Fluorescence-activated cell sorting (FACS) analysis shows increased percentages of mouse ESC-derived Lin− cells, LSKs, and short-term HSCs (ST-HSCs) in fetal livers of mouse–rat chimeras (n = 6) compared to control mouse embryos (n = 4), see also Figure 5—figure supplement 2. Fetal livers were obtained from E15.5 mouse–rat chimeras and E14 mouse embryos since these embryos are in similar developmental stages. *p < 0.05, **p < 0.01, N.S. indicates no significance, see also Source data 1.
Figure 5—figure supplement 1. Mouse embryonic development occurs faster than embryonic development in the rat and mouse–rat chimera.
Figure 5—figure supplement 2. Fluorescence-activated cell sorting (FACS) analysis identifies mouse hematopoietic progenitor cells in fetal livers of mouse–rat chimeras.
Transplantation of ESC-derived BM cells from interspecies mouse–rat chimeras rescues lethally irradiated syngeneic mice
To test functional properties of mouse BM hematopoietic progenitor cells derived through a rat, cells were FACS-sorted for GFP from the BM of juvenile mouse–rat chimeras and transferred into the tail vein of syngeneic C57BL/6 adult mice that received the lethal dose of whole-body gamma-irradiation 3 hr prior to the BM transplant (Figure 6A). Consistent with published studies (Rowe et al., 2016; Sugimura et al., 2017; Vo and Daley, 2015), all mice without BM transplant died between 9 and 12 days after irradiation (Figure 6B). In contrast, all 20 mice transplanted with GFP+ BM cells from mouse–rat chimeras survived after lethal irradiation (Figure 6B, C). Histological assessment of femur bones confirmed the presence of GFP+ donor cells in the BM compartment of transplanted mice (Figure 6D). Blood analysis of mice harvested 8 days after irradiation showed significant decreases in white blood cells (WBCs), red blood cells (RBCs), platelets (PLT), hemoglobin (Hb) as well as numbers of granulocytes, monocytes, and lymphocytes (Figure 7A and Figure 7—figure supplements 1 and 2). Transplantation of ESC-derived BM cells from mouse–rat chimeras increased WBC and the numbers of granulocytes, monocytes, and lymphocytes in the peripheral blood at day 8 (Figure 7A and Figure 7—figure supplements 1 and 2). Contribution of ESC-derived BM cells to granulocytes, monocytes, and B cells was higher compared to erythroid and T cells (Figure 7B and Figure 7—figure supplement 3). At 5 months after BM transplantation, ESC-derived cells completely restored blood cell numbers, PLT and Hb in lethally irradiated mice (Figure 7C and Figure 7—figure supplements 1 and 2). Long-term contributions of ESC-derived BM cells to all hematopoietic cell lineages in the peripheral blood were between 49% and 96% (Figure 7C and Figure 7—figure supplement 3). Thus, transplantation of ESC-derived BM cells from mouse–rat chimeras prevented mortality and restored hematopoietic blood lineages in lethally irradiated syngeneic mice.
Figure 6. Transplantation of mouse embryonic stem cell (ESC)-derived bone marrow (BM) cells from interspecies mouse–rat chimeras rescues lethally irradiated syngeneic mice.
(A) Schematic diagram shows transplantation of ESC-derived bone marrow cells (BMCs) into lethally irradiated (IR) mice. ESC-derived cells were obtained from the BM of juvenile mouse–rat chimeras using fluorescence-activated cell sorting (FACS) for GFP+ cells. BM and peripheral blood were harvested 8 days and 5 months after BM transplantation. (B) Kaplan–Meier survival analysis shows a 100% mortality in irradiated mice. Survival is dramatically improved after transplantation of irradiated mice with ESC-derived BM cells obtained from mouse–rat chimeras (IR + BMC). Survival in untreated wild-type (wt) mice is shown as a control (n = 12–20 mice in each group). (C) Photograph shows irradiated C57BL/6 mice 5 months after successful BM transplantation. Untreated C57BL/6 mouse is shown as a control. Gray color of irradiated mice (arrows) is consistent with large doses of whole-body radiation treatment. (D) Hematoxylin and eosin (H&E) staining shows increased amounts of hematopoietic cells in femur bones after BM transplantation into irradiated mice (top panels). GFP+ donor cells (green) are abundant in the BM compartment of transplanted mice (bottom panels). DAPI (blue) was used for counterstaining. Scale bars are: D, 200 μm; inserts in D, 5 μm.
Figure 7. Transplantation of mouse embryonic stem cell (ESC)-derived bone marrow (BM) cells from interspecies mouse–rat chimeras restores hematopoietic cell lineages in the blood and BM of lethally irradiated syngeneic mice.
(A) Blood analysis shows that transplantation with ESC-derived BM cells from mouse–rat chimeras increases white blood cell (WBC) counts and red blood cell (RBC) counts in the peripheral blood of irradiated recipients. Blood samples were obtained from untreated mice (no IR), lethally irradiated mice without BM transplant (IR), and lethally irradiated mice with BM transplant (IR + BMC). BM transplantation was performed using ESC-derived BM cells obtained from juvenile mouse–rat chimeras. Fluorescence-activated cell sorting (FACS) analysis of the peripheral blood to identify granulocytes, B cells, monocytes, T cells, and erythroid cells in shown in Figure 7—figure supplement 1. Concentrations of lymphocytes, monocytes, and neutrophil in the blood were increased after BM transplantation (n = 9–15 mice in each group), **p < 0.01, N.S. indicates no significance, see also Source data 1. BM transplantation also increased concentrations of platelets, hemoglobin, basophils, and eosinophils in the peripheral blood, see Figure 7—figure supplement 2. (B, C) FACS analysis for GFP+ cells in each cell subset shows that ESC-derived BM cells from mouse–rat chimeras contribute to multiple hematopoietic cell lineages in the peripheral blood of lethally irradiated mice (n = 9–16 mice in each group), see also Figure 7—figure supplement 3. (D) FASC analysis shows that transplantation with ESC-derived BM cells from mouse–rat chimeras increases percentages of LSKs, short-term HSCs (ST-HSCs), and long-term HSCs (LT-HSCs) in the BM of irradiated mice 5 months after BM transplantation (n = 9–16 mice in each group), see also Figure 7—figure supplement 4A, B. **p < 0.01, N.S. indicates no significance, see also Source data 1. (E, F) FACS analysis for GFP+ shows that ESC-derived BM cells from mouse–rat chimeras contribute to multiple hematopoietic progenitor cells in the BM of irradiated mice (n = 9–16 mice in each group), see also Figure 7—figure supplement 5. For secondary transplantation of mouse ESC-derived BM cells into lethally irradiated syngeneic mice, see Figure 7—figure supplement 6A–E.
Figure 7—figure supplement 1. Fluorescence-activated cell sorting (FACS) analysis identifies granulocytes, B cells, monocytes, T cells, and erythroid cells in the peripheral blood after bone marrow (BM) transplantation.
Figure 7—figure supplement 2. Transplantation of irradiated mice with embryonic stem cell (ESC)-derived bone marrow (BM) cells from mouse–rat chimeras increases hemoglobin (Hb) concentration and numbers of platelets (PLT), basophils, and eosinophils in the peripheral blood.
Figure 7—figure supplement 3. Identification of embryonic stem cell (ESC)-derived cells in the peripheral blood of irradiated mice after bone marrow (BM) transplantation.
Figure 7—figure supplement 4. Transplantation of mouse embryonic stem cell (ESC)-derived bone marrow (BM) cells from interspecies mouse–rat chimeras results in reconstitution of BM hematopoietic and progenitor cells after irradiation.
Figure 7—figure supplement 5. Identification of embryonic stem cell (ESC)-derived hematopoietic cells in the bone marrow (BM) of irradiated mice after BM transplantation.
Figure 7—figure supplement 6. Secondary transplantation of mouse embryonic stem cell (ESC)-derived bone marrow (BM) cells from interspecies mouse–rat chimeras rescues lethally irradiated syngeneic mice.
Transplantation of ESC-derived BM cells from interspecies mouse–rat chimeras resulted in the long-term contribution of donor cells to hematopoietic progenitor cells
Based on FACS analysis of irradiated mice at day 8, whole-body irradiation decreased the number of hematopoietic progenitor cells in the BM, including LSKs, ST-HSCs, and LT-HSCs (Figure 7D and Figure 7—figure supplement 4A, B). Transplantation of ESC-derived BM cells significantly increased LSKs but did not affect the numbers of ST-HSCs and LT-HSCs in irradiated mice (Figure 7D). Contribution of ESC-derived BM cells to Lin− and LSK cell subsets was high, whereas ESC contribution to ST-HSCs and LT-HSCs at day 8 was low (Figure 7E and Figure 7—figure supplement 5). At 5 months after BM transplantation, percentages of LSKs, ST-HSCs, and LT-HSCs in the BM were increased (Figure 7D and Figure 7—figure supplement 4B). Long-term contribution of ESC-derived BM cells to LSKs, ST-HSCs, and LT-HSCs was between 92% and 95% (Figure 7F and Figure 7—figure supplement 5). Finally, we performed BM transplantation again in secondary recipients to establish the functional potential and self-renewal capacity of the chimeric HSCs (Figure 7—figure supplement 6A). The secondary BM transplantation rescued lethally irradiated mice and resulted in long-term engraftment of ESC-derived HSCs into hematopoietic cell lineages in the BM and peripheral blood (Figure 7—figure supplement 6B–E). Altogether, transplantation of ESC-derived BM cells from mouse–rat chimeras resulted in efficient, long-term contribution of donor cells to the BM and blood of lethally irradiated mice.
Discussion
Recent single-cell RNA sequencing studies identified remarkable diversity of hematopoietic cell types in the BM (Baccin et al., 2020). Generation of functional BM cells from pluripotent ESCs or iPSCs in a dish or in organoids represents a formidable challenge (Rowe et al., 2016; Vo and Daley, 2015). In the present study, we used blastocyst complementation to generate a diversity of hematopoietic cell types from mouse ESCs in rat embryos. Interspecies mouse–rat chimeras were viable and contained approximately 25% of ESC-derived mouse cells in the BM. It is possible that inactivation of genes critical for hematopoiesis in rat embryos prior to blastocyst complementation can improve the integration of mouse ESCs into the BM of mouse–rat chimeras. This approach was supported by recent studies with mouse–mouse chimeras, in which ESCs contributed to more than 90% of hematopoietic cells in mice deficient for either Kit or Flk1 (Hamanaka et al., 2018; Jansson and Larsson, 2010). While ESCs contributed to all hematopoietic cell lineages in interspecies BM, the percentage of lymphoid progenitors was lower, whereas the percentages of myeloid progenitor cells and HSCs were higher in mouse–rat chimeras compared to control mouse–mouse chimeras. Since both chimeras were produced by complementing blastocysts with mouse ESCs from the same ESC-GFP cell line, it is unlikely that these changes are dependent on donor ESCs. It is possible that the observed differences in BM cellular composition between mouse–rat and mouse–mouse chimeras are due to interactions of donor ESCs with the host embryo. Structural and functional differences between hormones, growth factors, and their receptors in rats and mice can contribute to the efficiency or timing of differentiation of mouse ESCs into hematopoietic cell lineages in BM of chimeras.
Our data demonstrate that chimeric HSCs develop more efficiently from donor mouse cells in the dorsal aorta, fetal liver, and BM, whereas rat cells are less efficient to differentiate into HSCs. Since we observed high numbers of mouse hemogenic endothelial cells in the chimeric dorsal aorta, it is likely that donor hemogenic endothelium undergoes direct transition to functional HSCs in the fetal liver, whereas endogenous (non-GFP+) hemogenic endothelium can be a source of rat HSCs. Since mouse embryos develop faster compared to rat embryos by approximately 1.5 days (Farrington-Rock et al., 2008; Marcela et al., 2012; Takahashi and Osumi, 2005; Torres et al., 2008), it is possible that mouse ESC-derived progenitor cells migrate faster into developing hematopoietic niches in the mouse–rat chimeras, leading to preferential development of HSCs from cells of mouse origin and contributing to increased numbers of mouse-derived hematopoietic progenitors in the BM of mouse–rat chimeras. These data suggest that using donor ESCs from species with less gestational time in interspecies ‘bioreactors’ can lead to larger quantities of ESC-derived hematopoietic progenitors in the chimeric BM. Our single-cell RNAseq analysis enabled us to identify potential signaling pathways and receptor–ligand interactions between hematopoietic progenitors and stromal cells in the BM. These pathways include Cxcl12-Cxcr4 signaling between hematopoietic progenitors and endothelial cells, which plays a critical role in maintenance of HSCs during BM homeostasis and promotes niche regeneration and hematopoietic reconstitution after BM transplantation (Baccin et al., 2020; Singh et al., 2020; Sugiyama et al., 2006). Other pathways identified in our studies, including Lama2-Itga6, App-Itga6, Comp-Cd47, Col1a1-Cd44, and App-Il18rap, have not been extensively studied in the BM microenvironment but are implicated in regulation of cell adhesion, migration, oncogenesis, fibrosis, and inflammatory responses (Kiratipaiboon et al., 2020; Sibin et al., 2019; Rock et al., 2010; Strelnikov et al., 2021; Yang et al., 2017). Notably, our data suggest that some of these signaling pathways can be targeted to modulate the development and expansion of donor ESC-derived hematopoietic progenitor cells in the BM of interspecies chimeras.
Despite mosaicism in interspecies BM, mouse ESC-derived cells from multiple hematopoietic cell lineages were highly differentiated and indistinguishable from the normal mouse BM cells based on gene expression signatures and cell surface proteins. Consistent with functional competency of ESC-derived BM, transplantation of BM cells into lethally irradiated syngeneic mice prevented mortality and resulted in long-term contribution of ESC-derived cells to all hematopoietic cell lineages in the BM and peripheral blood. One of the limitations of our studies is that the functional potential of chimeric HSCs was established from whole BM transplants and not from transplantation of purified HSCs. While these experiments are technically challenging, transplantation of FACS-sorted donor HSCs into lethally irradiated mice will be needed in our future studies to investigate whether chimeric HSCs are fully functional to restore all hematopoietic cell lineages after irradiation. Our results are consistent with recent studies demonstrating the ability of mouse ESCs to generate functional pancreatic, endothelial, and kidney cells in interspecies mouse–rat chimeras (Goto et al., 2019; Wang et al., 2021; Yamaguchi et al., 2017). Interestingly, long-term contribution of donor BM cells to ST-HSCs and LT-HSCs of irradiated mice was high, supporting the ability of donor HSCs to self-renew. In contrast, the short-term contribution of donor BM cells to ST-HSCs and LT-HSCs of irradiated mice was low. Low contribution of donor BM to HSCs at day 8 is not surprising considering an acute hematopoietic deficiency in lethally irradiated mice. It is possible that most donor-derived HSCs undergo rapid differentiation into other hematopoietic cell types to compensate for the loss of injured hematopoietic cells after irradiation.
Generation of intraspecies chimeras through blastocyst complementation creates an interesting opportunity to use patient-derived iPSCs to produce tissues or even organs in large animals, for example, pigs or sheep, which can serve as ‘biological reactors’. However, at this stage of technological advances it is impossible to restrict the integration of ESC/iPSC-derived cells into selected organs or cell types. Off-target integration of ESCs and iPSCs into the brain, testes, and sensory organs raises important ethical concerns for the use of human–animal chimeras in regenerative medicine (Masaki and Nakauchi, 2017; Wu et al., 2016). To improve the selectivity of ESC/iPSC integration into chimeric tissues, various genetic modifications can be introduced into the host embryos to advance the technology. Harvest of tissues from chimeric embryos instead of adult chimeras can alleviate some of the ethical concerns, suggesting a possibility of using chimeric embryos as a potential source of patient-specific hematopoietic progenitor cells.
In summary, blastocyst complementation of rat embryos with mouse ESCs was used to simultaneously generate multiple hematopoietic and stromal cell lineages in the BM. ESC-derived cells in mouse–rat chimeras were indistinguishable from normal mouse BM cells based on gene expression signatures and cell surface markers. Transplantation of ESC-derived BM cells rescued lethally irradiated syngeneic mice and resulted in long-term contribution of donor cells to hematopoietic cell lineages. Thus, the interspecies chimeras could be considered for in vivo differentiation of patient-derived iPSCs into hematopoietic cell lineages for future cell therapies.
Materials and methods
Mice, rats, and generation of mouse–rat and mouse–mouse chimeras through blastocyst complementation
C57BL/6 mice were purchased from Jackson Lab. Interspecies mouse–rat chimeras were generated using blastocyst complementation as described (Li et al., 2021; Wang et al., 2021). Briefly, blastocysts from SD rats were obtained at embryonic day 4.5 (E4.5), injected with 15 GFP-labeled mouse ESC cells (ESC-GFP, C57BL/6 background) (Sun et al., 2021; Wen et al., 2021) and transferred into pseudopregnant SD rat females. Mouse–mouse chimeras were generated by complementing CD1 blastocysts with 15 mouse ESC-GFP cells. For FACS analysis and BM transplantation, BM cells were collected from chimeric pups that were harvested between postnatal day 4 (P4) and P10. For single-cell RNA sequencing, BM cells were prepared from P10 and P5 mice, rats, and chimeras. To perform BM transplantation, BM cells from two tibias and two fibulas of mouse–rat chimeras were collected and FACS-sorted for ESC-derived (GFP+) cells. 500,000 FACS-sorted GFP+ BM cells were intravenously (i.v.) injected into lethally irradiated C57BL/6 male mice (6–8 weeks of age) via the tail vein. Three hours before BM transplantation, whole-body irradiation was performed using 11.75 Gy. Mice were harvested after 8 days or 5 months after BM transplantation. For the second BM transplantation, GFP+ BM cells were FACS-sorted from irradiated mice 5 months after the first BM transplantation and then i.v. injected into new irradiated recipients. Tissue dissection, processing, and preparation of single-cell suspensions were carried out as described (Bolte et al., 2011; Kalin et al., 2008; Kalinichenko et al., 2003; Kim et al., 2005; Wang et al., 2003). Blood analysis was performed in animal facility of Cincinnati Children’s Hospital Research Foundation.
Single-cell RNAseq analysis of ESC-derived BM cells
Prior to scRNAseq (10× Chromium platform), BM cells were pooled from three P10 mouse–rat chimeras and three P10 mouse–mouse (control) chimeras and then FACS-sorted for GFP and the lineage (Lin) marker. Since the numbers of HSCs and other hematopoietic progenitors in BM are significantly low compared to numbers of differentiated hematopoietic cells, the cell mixtures were enriched for BM progenitor cell populations by combining 90% of FACS-sorted GFP+Lin− cells and 10% of GFP+Lin+ cells in each experimental group. This enrichment enabled us to obtain enough progenitor cells for UMAP clustering analysis. In separate scRNAseq experiments, all BM cells (including hematopoietic, vascular, and stromal cells) were prepared from P5 mice, rats, and mouse–rat chimeras using enzymatic digestion and cell purification as described (Baccin et al., 2020). BM cells from five animals were pooled together prior to single-cell RNAseq. All raw data and the processed count matrix of BM datasets were uploaded to the GEO database (accession number GSE184940). Read alignments, quality controls, and false discovery rates were described previously (Guo et al., 2019; Ren et al., 2019; Wang et al., 2022). Identification of cell clusters and quantification of cluster-specific gene expression in BM scRNAseq datasets were performed as described (Baccin et al., 2020; Wang et al., 2021; Wen et al., 2021). To assess the transcriptomic similarity of ESC-derived and endogenous BM cells, the scRNAseq datasets were normalized with SCTransform and then integrated utilizing the canonical correlation analysis. In the integrated scRNAseq datasets, the SelectIntegrationFeatures in Seurat package (version 4.0.0 in R 4.0 statistical environment) was used to identify anchors for integration. The RunPCA function was used for principal component analysis (PCA) of scRNAseq datasets, and the PCElbowPlot function was used to calculate the standard deviations of the principal components (PCs). PCs with standard deviation >3.5 were chosen as input parameters for nonlinear UMAP clustering analysis. Next, the FindNeighbors function was used to compute the k.param nearest neighbors, and BM cell clusters were identified by a shared nearest neighbor modularity optimization clustering algorithm implemented in the FindClusters function with resolution set at 0.4 (Guo et al., 2019; Wang et al., 2021; Wen et al., 2021).
Analysis of potential receptor–ligand interactions using single-cell RNAseq datasets
The R package NicheNet was used to analyze the information about expression of cognate ligands and receptors to identify intercellular communication patterns between hematopoietic progenitors and stromal cells as described (Browaeys et al., 2020). EMP and GMP cells were chosen as potential sources of receptors, whereas BM stromal cell types were chosen as potential sources of ligands. The background expression of genes was specified with default approach used in the NicheNet pipeline, and expressed genes were identified based on >10% detection in specific clusters. To identify ligand–receptor interactions between EMPs/GMPs and stromal cells, we selected the top 20 ligands predicted to drive hematopoietic cell differentiation based on the Pearson correlation coefficient between the ligand–receptor regulatory potential score of each ligand and the target indicator vector. Using the NicheNet pipeline, the Circos plots were generated to show common ligand–receptor interactions between EMPs/GMPs and stromal cells in the BM.
FACS analysis
FACS analysis was performed using cells obtained from the BM and blood. Antibodies for FACS analysis are listed in Supplementary file 3. Immunostaining of cell suspensions were performed as described (Bolte et al., 2017; Xia et al., 2015). Identification of hematopoietic cell types based on multiple cell surface markers is described in Bolte et al., 2020b; Pradhan et al., 2019; Ren et al., 2013; Ren et al., 2010; Sun et al., 2017. To identify ESC-derived HSCs, we used GFP fluorescence and mouse-specific antibodies recognizing multiple cell surface antigens. First, ESC-derived GFP+ BM cells were subdivided into Lin+ and Lin− cell subsets. Second, we used Sca1 and CD117 (c-KIT) antibodies to identify Lin−Sca1+c-KIT+ cells (LSKs). Third, CD150 and CD48 antibodies were used to identify ST-HSCs and LT-HSCs among LSKs. Stained cells were analyzed using a five-laser FACSAria II (BD Biosciences) (Cai et al., 2016; Sun et al., 2021).
Histology and immunostaining
Frozen or paraffin-embedded sections of tissue samples were stained with hematoxylin and eosin (H&E) for histological evaluation (Kalinichenko et al., 2002) or to visualize GFP (Ustiyan et al., 2018; Ustiyan et al., 2016). Frozen sections from embryos were used for immunofluorescent staining as described (Black et al., 2018; Ustiyan et al., 2012; Wang et al., 2010). Primary antibodies for immunostaining are listed in Supplementary file 3. Secondary antibodies were conjugated with Alexa Fluor 488, Alexa Fluor 594, or Alexa Fluor 647 (Invitrogen and Jackson ImmunoResearch Laboratory) to visualize specific staining as described (Bolte et al., 2012; Hoggatt et al., 2013; Milewski et al., 2017a). DAPI (Vector Laboratory) was used to counterstain cell nuclei (Milewski et al., 2017b). Histological and immunofluorescent images were obtained using a Zeiss Axioplan2 microscope (Carl Zeiss Microimaging) as described (Bolte et al., 2015; Kalin et al., 2008; Pradhan et al., 2016).
Statistical analysis
Statistical significance was determined using nonparametric Mann–Whitney U-test, one-way analysis of variance, and Student’s t-test. Multiple means were compared using one-way analysis of variance with the post hoc Tukey test. p ≤ 0.05 was considered statistically significant. Data were presented as mean ± standard error of mean (SEM).
Acknowledgements
We thank Mrs. Erika Smith for excellent secretarial support. This work was supported by NIH Grants HL141174 (to VVK), HL149631 (to VVK), and HL152973 (to VVK and TVK).
Funding Statement
The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Contributor Information
Vladimir V Kalinichenko, Email: Vladimir.Kalinichenko@cchmc.org.
Jalees Rehman, University of Illinois at Chicago, United States.
Didier YR Stainier, Max Planck Institute for Heart and Lung Research, Germany.
Funding Information
This paper was supported by the following grants:
National Heart, Lung, and Blood Institute HL141174 to Vladimir V Kalinichenko.
National Heart, Lung, and Blood Institute HL149631 to Vladimir V Kalinichenko.
National Heart, Lung, and Blood Institute HL152973 to Vladimir V Kalinichenko.
National Heart, Lung, and Blood Institute HL158659 to Tanya V Kalin.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Software, Formal analysis, Supervision, Validation, Investigation, Visualization, Methodology, Writing - original draft, Writing - review and editing.
Resources, Data curation, Software, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing - review and editing.
Data curation, Formal analysis, Validation, Visualization.
Resources, Investigation, Methodology, Writing - original draft, Writing - review and editing.
Investigation, Methodology.
Investigation, Methodology, Writing - review and editing.
Resources, Formal analysis, Funding acquisition, Investigation, Writing - original draft, Writing - review and editing.
Conceptualization, Resources, Data curation, Supervision, Funding acquisition, Validation, Investigation, Methodology, Writing - original draft, Project administration, Writing - review and editing.
Ethics
All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee of the Cincinnati Children's Research Foundation (protocol # IACUC2016-0038).
Additional files
Data availability
Bone marrow single-cell RNA sequencing data have been deposited in GEO under accession number GSE184940.
The following dataset was generated:
Wen B, Wang G, Kalinichenko VV. 2021. The integrated single cell RNAseq analysis of bone marrow cells produced by mouse-mouse intraspecies blastocyst complementation and mouse-rat interspecies blastocyst complementation. NCBI Gene Expression Omnibus. GSE184940
References
- Amabile G, Welner RS, Nombela-Arrieta C, D’Alise AM, Di Ruscio A, Ebralidze AK, Kraytsberg Y, Ye M, Kocher O, Neuberg DS, Khrapko K, Silberstein LE, Tenen DG. In vivo generation of transplantable human hematopoietic cells from induced pluripotent stem cells. Blood. 2013;121:1255–1264. doi: 10.1182/blood-2012-06-434407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccin C, Al-Sabah J, Velten L, Helbling PM, Grünschläger F, Hernández-Malmierca P, Nombela-Arrieta C, Steinmetz LM, Trumpp A, Haas S. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nature Cell Biology. 2020;22:38–48. doi: 10.1038/s41556-019-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno N, Przybylski D, Kowalczyk MS, Kfoury Y, Severe N, Gustafsson K, Kokkaliaris KD, Mercier F, Tabaka M, Hofree M, Dionne D, Papazian A, Lee D, Ashenberg O, Subramanian A, Vaishnav ED, Rozenblatt-Rosen O, Regev A, Scadden DT. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–1932. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black M, Milewski D, Le T, Ren X, Xu Y, Kalinichenko VV, Kalin TV. Foxf1 inhibits pulmonary fibrosis by preventing CDH2-CDH11 cadherin switch in myofibroblasts. Cell Reports. 2018;23:442–458. doi: 10.1016/j.celrep.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Zhang Y, Wang IC, Kalin TV, Molkentin JD, Kalinichenko VV. Expression of FoxM1 transcription factor in cardiomyocytes is required for myocardial development. PLOS ONE. 2011;6:e22217. doi: 10.1371/journal.pone.0022217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Zhang Y, York A, Kalin TV, Schultz JEJ, Molkentin JD, Kalinichenko VV. Postnatal ablation of FoxM1 from cardiomyocytes causes late onset cardiac hypertrophy and fibrosis without exacerbating pressure overload-induced cardiac remodeling. PLOS ONE. 2012;7:e48713. doi: 10.1371/journal.pone.0048713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, Kalin TV, Herring BP, Kalinichenko VV. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. The Journal of Biological Chemistry. 2015;290:7563–7575. doi: 10.1074/jbc.M114.609487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Flood HM, Ren X, Jagannathan S, Barski A, Kalin TV, Kalinichenko VV. Foxf1 transcription factor promotes lung regeneration after partial pneumonectomy. Scientific Reports. 2017;7:10690. doi: 10.1038/s41598-017-11175-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Whitsett JA, Kalin TV, Kalinichenko VV. Transcription factors regulating embryonic development of pulmonary vasculature. Advances in Anatomy, Embryology, and Cell Biology. 2018;228:1–20. doi: 10.1007/978-3-319-68483-3_1. [DOI] [PubMed] [Google Scholar]
- Bolte C, Kalin TV, Kalinichenko VV. Molecular, cellular, and bioengineering approaches to stimulate lung regeneration after injury. Seminars in Cell & Developmental Biology. 2020a;100:101–108. doi: 10.1016/j.semcdb.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Bolte C, Ustiyan V, Ren X, Dunn AW, Pradhan A, Wang G, Kolesnichenko OA, Deng Z, Zhang Y, Shi D, Greenberg JM, Jobe AH, Kalin TV, Kalinichenko VV. Nanoparticle delivery of proangiogenic transcription factors into the neonatal circulation inhibits alveolar simplification caused by hyperoxia. American Journal of Respiratory and Critical Care Medicine. 2020b;202:100–111. doi: 10.1164/rccm.201906-1232OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys R, Saelens W, Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nature Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, Kalinichenko VV. Foxf1 maintains endothelial barrier function and prevents edema after lung injury. Science Signaling. 2016;9:ra40. doi: 10.1126/scisignal.aad1899. [DOI] [PubMed] [Google Scholar]
- Chang AN, Liang Z, Dai HQ, Chapdelaine-Williams AM, Andrews N, Bronson RT, Schwer B, Alt FW. Neural blastocyst complementation enables mouse forebrain organogenesis. Nature. 2018;563:126–130. doi: 10.1038/s41586-018-0586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmadhikari AV, Szafranski P, Kalinichenko VV, Stankiewicz P. Genomic and epigenetic complexity of the Foxf1 locus in 16q24.1: implications for development and disease. Current Genomics. 2015;16:107–116. doi: 10.2174/1389202916666150122223252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, Li H, Hadland BK, Bernstein ID, Collins JJ, Zon LI, Daley GQ. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva I, Brok-Volchanskaya V, Kumar A, Liu P, Lee JH, Tong L, Vodyanik M, Swanson S, Stewart R, Kyba M, Yakubov E, Cooke J, Thomson JA, Slukvin I. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nature Communications. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrington-Rock C, Kirilova V, Dillard-Telm L, Borowsky AD, Chalk S, Rock MJ, Cohn DH, Krakow D. Disruption of the FlnB gene in mice phenocopies the human disease spondylocarpotarsal synostosis syndrome. Human Molecular Genetics. 2008;17:631–641. doi: 10.1093/hmg/ddm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galic Z, Kitchen SG, Kacena A, Subramanian A, Burke B, Cortado R, Zack JA. T lineage differentiation from human embryonic stem cells. PNAS. 2006;103:11742–11747. doi: 10.1073/pnas.0604244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Xu C, Asada N, Frenette PS. The hematopoietic stem cell niche: from embryo to adult. Development. 2018;145:dev139691. doi: 10.1242/dev.139691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Hara H, Sanbo M, Masaki H, Sato H, Yamaguchi T, Hochi S, Kobayashi T, Nakauchi H, Hirabayashi M. Generation of pluripotent stem cell-derived mouse kidneys in sall1-targeted anephric rats. Nature Communications. 2019;10:451. doi: 10.1038/s41467-019-08394-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis AE, Kennedy M, Bozec A, Brunton F, Stenbeck G, Park IH, Wagner EF, Keller GM. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115:2769–2776. doi: 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Du Y, Gokey JJ, Ray S, Bell SM, Adam M, Sudha P, Perl AK, Deshmukh H, Potter SS, Whitsett JA, Xu Y. Single cell RNA analysis identifies cellular heterogeneity and adaptive responses of the lung at birth. Nature Communications. 2019;10:37. doi: 10.1038/s41467-018-07770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 2018;22:627–638. doi: 10.1016/j.stem.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Hamanaka S, Umino A, Sato H, Hayama T, Yanagida A, Mizuno N, Kobayashi T, Kasai M, Suchy FP, Yamazaki S, Masaki H, Yamaguchi T, Nakauchi H. Generation of vascular endothelial cells and hematopoietic cells by blastocyst complementation. Stem Cell Reports. 2018;11:988–997. doi: 10.1016/j.stemcr.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggatt AM, Kim JR, Ustiyan V, Ren X, Kalin TV, Kalinichenko VV, Herring BP. The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. The Journal of Biological Chemistry. 2013;288:28477–28487. doi: 10.1074/jbc.M113.478974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson L, Larsson J. W41/W41 blastocyst complementation: a system for genetic modeling of hematopoiesis. Blood. 2010;115:47–50. doi: 10.1182/blood-2009-07-235622. [DOI] [PubMed] [Google Scholar]
- Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. American Journal of Respiratory Cell and Molecular Biology. 2008;39:390–399. doi: 10.1165/rcmb.2008-0044OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Zhou Y, Shin B, Stolz DB, Watkins SC, Whitsett JA, Costa RH. Wild-Type levels of the mouse forkhead box F1 gene are essential for lung repair. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2002;282:L1253–L1265. doi: 10.1152/ajplung.00463.2001. [DOI] [PubMed] [Google Scholar]
- Kalinichenko VV, Gusarova GA, Shin B, Costa RH. The forkhead box F1 transcription factor is expressed in brain and head mesenchyme during mouse embryonic development. Gene Expression Patterns. 2003;3:153–158. doi: 10.1016/s1567-133x(03)00010-3. [DOI] [PubMed] [Google Scholar]
- Kennedy M, Awong G, Sturgeon CM, Ditadi A, LaMotte-Mohs R, Zúñiga-Pflücker JC, Keller G. T lymphocyte potential marks the emergence of definitive hematopoietic progenitors in human pluripotent stem cell differentiation cultures. Cell Reports. 2012;2:1722–1735. doi: 10.1016/j.celrep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Kim IM, Zhou Y, Ramakrishna S, Hughes DE, Solway J, Costa RH, Kalinichenko VV. Functional characterization of evolutionarily conserved DNA regions in forkhead box F1 gene locus. The Journal of Biological Chemistry. 2005;280:37908–37916. doi: 10.1074/jbc.M506531200. [DOI] [PubMed] [Google Scholar]
- Kiratipaiboon C, Voronkova M, Ghosh R, Rojanasakul LW, Dinu CZ, Chen YC, Rojanasakul Y. SOX2Mediates carbon nanotube-induced fibrogenesis and fibroblast stem cell acquisition. ACS Biomaterials Science & Engineering. 2020;6:5290–5304. doi: 10.1021/acsbiomaterials.0c00887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitahara A, Ran Q, Oda K, Yasue A, Abe M, Ye X, Sasaoka T, Tsuchida M, Sakimura K, Ajioka Y, Saijo Y, Zhou Q. Generation of lungs by blastocyst complementation in apneumic fgf10-deficient mice. Cell Reports. 2020;31:107626. doi: 10.1016/j.celrep.2020.107626. [DOI] [PubMed] [Google Scholar]
- Kitajima K, Minehata K, Sakimura K, Nakano T, Hara T. In vitro generation of HSC-like cells from murine escs/ipscs by enforced expression of LIM-homeobox transcription factor Lhx2. Blood. 2011;117:3748–3758. doi: 10.1182/blood-2010-07-298596. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Yamaguchi T, Hamanaka S, Kato-Itoh M, Yamazaki Y, Ibata M, Sato H, Lee Y-S, Usui J-I, Knisely AS, Hirabayashi M, Nakauchi H. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787–799. doi: 10.1016/j.cell.2010.07.039. [DOI] [PubMed] [Google Scholar]
- Kolesnichenko OA, Whitsett JA, Kalin TV, Kalinichenko VV. Therapeutic potential of endothelial progenitor cells in pulmonary diseases. American Journal of Respiratory Cell and Molecular Biology. 2021;65:473–488. doi: 10.1165/rcmb.2021-0152TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledran MH, Krassowska A, Armstrong L, Dimmick I, Renström J, Lang R, Yung S, Santibanez-Coref M, Dzierzak E, Stojkovic M, Oostendorp RAJ, Forrester L, Lako M. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 2008;3:85–98. doi: 10.1016/j.stem.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Li E, Ustiyan V, Wen B, Kalin GT, Whitsett JA, Kalin TV, Kalinichenko VV. Blastocyst complementation reveals that NKX2-1 establishes the proximal-peripheral boundary of the airway epithelium. Developmental Dynamics. 2021;250:1001–1020. doi: 10.1002/dvdy.298. [DOI] [PubMed] [Google Scholar]
- Lin Y, Kobayashi M, Azevedo Portilho N, Mishra A, Gao H, Liu Y, Wenzel P, Davis B, Yoder MC, Yoshimoto M. Long-Term engraftment of ESC-derived B-1 progenitor cells supports HSC-independent lymphopoiesis. Stem Cell Reports. 2019;12:572–583. doi: 10.1016/j.stemcr.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YF, Cahan P, Ross S, Sahalie J, Sousa PM, Hadland BK, Cai W, Serrao E, Engelman AN, Bernstein ID, Daley GQ. Engineered murine HSCs reconstitute multi-lineage hematopoiesis and adaptive immunity. Cell Reports. 2016;17:3178–3192. doi: 10.1016/j.celrep.2016.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcela SG, Cristina RMM, Angel PGM, Manuel AM, Sofía DC, Patricia D, Bladimir RR, Concepción SG. Chronological and morphological study of heart development in the rat. Anatomical Record. 2012;295:1267–1290. doi: 10.1002/ar.22508. [DOI] [PubMed] [Google Scholar]
- Masaki H, Nakauchi H. Interspecies chimeras for human stem cell research. Development. 2017;144:2544–2547. doi: 10.1242/dev.151183. [DOI] [PubMed] [Google Scholar]
- Matsunari H, Nagashima H, Watanabe M, Umeyama K, Nakano K, Nagaya M, Kobayashi T, Yamaguchi T, Sumazaki R, Herzenberg LA, Nakauchi H. Blastocyst complementation generates exogenic pancreas in vivo in apancreatic cloned pigs. PNAS. 2013;110:4557–4562. doi: 10.1073/pnas.1222902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski D, Balli D, Ustiyan V, Le T, Dienemann H, Warth A, Breuhahn K, Whitsett JA, Kalinichenko VV, Kalin TV. Foxm1 activates AGR2 and causes progression of lung adenomas into invasive mucinous adenocarcinomas. PLOS Genetics. 2017a;13:e1007097. doi: 10.1371/journal.pgen.1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, Kalinichenko VV, Kalin TV. Foxf1 and foxf2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21Cip1 CDK inhibitor. Oncogene. 2017b;36:850–862. doi: 10.1038/onc.2016.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montel-Hagen A, Seet CS, Li S, Chick B, Zhu Y, Chang P, Tsai S, Sun V, Lopez S, Chen HC, He C, Chin CJ, Casero D, Crooks GM. Organoid-induced differentiation of conventional T cells from human pluripotent stem cells. Cell Stem Cell. 2019;24:376–389. doi: 10.1016/j.stem.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Furuhashi K, Danielsson JA, Hirata Y, Kakiuchi M, Lin CS, Ohta M, Riccio P, Takahashi Y, Xu X, Emala CW, Lu C, Nakauchi H, Cardoso WV. Generation of functional lungs via conditional blastocyst complementation using pluripotent stem cells. Nature Medicine. 2019;25:1691–1698. doi: 10.1038/s41591-019-0635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy N, Chen HC, Rajgolikar G, Butz KG, Frissora FW, Gronostajski RM. Recombination activation gene-2-deficient blastocyst complementation analysis reveals an essential role for nuclear factor I-A transcription factor in T-cell activation. International Immunology. 2011;23:385–390. doi: 10.1093/intimm/dxr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafria M, Keane P, Ng ES, Stanley EG, Elefanty AG, Bonifer C. Expression of RUNX1-ETO rapidly alters the chromatin landscape and growth of early human myeloid precursor cells. Cell Reports. 2020;31:107691. doi: 10.1016/j.celrep.2020.107691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorowa S, Hamey FK, Pijuan Sala B, Diamanti E, Shepherd M, Laurenti E, Wilson NK, Kent DG, Göttgens B. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–e31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A, Ustiyan V, Zhang Y, Kalin TV, Kalinichenko VV. Forkhead transcription factor Foxf1 interacts with Fanconi anemia protein complexes to promote DNA damage response. Oncotarget. 2016;7:1912–1926. doi: 10.18632/oncotarget.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A, Dunn A, Ustiyan V, Bolte C, Wang G, Whitsett JA, Zhang Y, Porollo A, Hu YC, Xiao R, Szafranski P, Shi D, Stankiewicz P, Kalin TV, Kalinichenko VV. The S52F Foxf1 mutation inhibits STAT3 signaling and causes alveolar capillary dysplasia. American Journal of Respiratory and Critical Care Medicine. 2019;200:1045–1056. doi: 10.1164/rccm.201810-1897OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton LE, Scadden DT. Limiting factors in murine hematopoietic stem cell assays. Cell Stem Cell. 2007;1:263–270. doi: 10.1016/j.stem.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, Kalin TV, Kalinichenko VV. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Molecular and Cellular Biology. 2010;30:5381–5393. doi: 10.1128/MCB.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Shah TA, Ustiyan V, Zhang Y, Shinn J, Chen G, Whitsett JA, Kalin TV, Kalinichenko VV. Foxm1 promotes allergen-induced goblet cell metaplasia and pulmonary inflammation. Molecular and Cellular Biology. 2013;33:371–386. doi: 10.1128/MCB.00934-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Ustiyan V, Guo M, Wang G, Bolte C, Zhang Y, Xu Y, Whitsett JA, Kalin TV, Kalinichenko VV. Postnatal alveologenesis depends on Foxf1 signaling in c-Kit + endothelial progenitor cells. American Journal of Respiratory and Critical Care Medicine. 2019;200:1164–1176. doi: 10.1164/rccm.201812-2312OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock MJ, Holden P, Horton WA, Cohn DH. Cartilage oligomeric matrix protein promotes cell attachment via two independent mechanisms involving CD47 and αvβ3 integrin. Molecular and Cellular Biochemistry. 2010;338:215–224. doi: 10.1007/s11010-009-0355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe RG, Mandelbaum J, Zon LI, Daley GQ. Engineering hematopoietic stem cells: lessons from development. Cell Stem Cell. 2016;18:707–720. doi: 10.1016/j.stem.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibin MK, Harshitha SM, Arati S, Chetan GK, Dhaval PS, Dhananjaya IB. Il18Rap polymorphisms and its plasma levels in patients with lumbar disc degeneration. Clinical Neurology and Neurosurgery. 2019;184:105374. doi: 10.1016/j.clineuro.2019.105374. [DOI] [PubMed] [Google Scholar]
- Singh P, Mohammad KS, Pelus LM. Cxcr4 expression in the bone marrow microenvironment is required for hematopoietic stem and progenitor cell maintenance and early hematopoietic regeneration after myeloablation. Stem Cells. 2020;38:849–859. doi: 10.1002/stem.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelnikov VV, Kuznetsova EB, Tanas AS, Rudenko VV, Kalinkin AI, Poddubskaya EV, Kekeeva TV, Chesnokova GG, Trotsenko ID, Larin SS, Kutsev SI, Zaletaev DV, Nemtsova MV, Simonova OA. Abnormal promoter DNA hypermethylation of the integrin, nidogen, and dystroglycan genes in breast cancer. Scientific Reports. 2021;11:2264. doi: 10.1038/s41598-021-81851-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura R, Jha DK, Han A, Soria-Valles C, da Rocha EL, Lu Y-F, Goettel JA, Serrao E, Rowe RG, Malleshaiah M, Wong I, Sousa P, Zhu TN, Ditadi A, Keller G, Engelman AN, Snapper SB, Doulatov S, Daley GQ. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature. 2017;545:432–438. doi: 10.1038/nature22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Sun L, Ren X, Wang IC, Pradhan A, Zhang Y, Flood HM, Han B, Whitsett JA, Kalin TV, Kalinichenko VV. The FoxM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Science Signaling. 2017;10:eaai8583. doi: 10.1126/scisignal.aai8583. [DOI] [PubMed] [Google Scholar]
- Sun F, Wang G, Pradhan A, Xu K, Gomez-Arroyo J, Zhang Y, Kalin GT, Deng Z, Vagnozzi RJ, He H, Dunn AW, Wang Y, York AJ, Hegde RS, Woods JC, Kalin TV, Molkentin JD, Kalinichenko VV. Nanoparticle delivery of STAT3 alleviates pulmonary hypertension in a mouse model of alveolar capillary dysplasia. Circulation. 2021;144:539–555. doi: 10.1161/CIRCULATIONAHA.121.053980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Osumi N. Identification of a novel type II classical cadherin: rat cadherin19 is expressed in the cranial ganglia and Schwann cell precursors during development. Developmental Dynamics. 2005;232:200–208. doi: 10.1002/dvdy.20209. [DOI] [PubMed] [Google Scholar]
- Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Domínguez Á, Pinho S, Akhmetzyanova I, Gao J, Witkowski M, Guillamot M, Gutkin MC, Zhang Y, Marier C, Diefenbach C, Kousteni S, Heguy A, Zhong H, Fooksman DR, Butler JM, Economides A, Frenette PS, Adams RH, Satija R, Tsirigos A, Aifantis I. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569:222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Weyrauch UM, Sutcliffe R, Dunnett SB. A rat embryo staging scale for the generation of donor tissue for neural transplantation. Cell Transplantation. 2008;17:535–542. doi: 10.3727/096368908785096006. [DOI] [PubMed] [Google Scholar]
- Ustiyan V., Wert SE, Ikegami M, Wang IC, Kalin TV, Whitsett JA, Kalinichenko VV. Foxm1 transcription factor is critical for proliferation and differentiation of Clara cells during development of conducting airways. Developmental Biology. 2012;370:198–212. doi: 10.1016/j.ydbio.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V, Zhang Y, Perl AKT, Whitsett JA, Kalin TV, Kalinichenko VV. Β-catenin and kras/foxm1 signaling pathway are critical to restrict SOX9 in basal cells during pulmonary branching morphogenesis. Developmental Dynamics. 2016;245:590–604. doi: 10.1002/dvdy.24393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustiyan V, Bolte C, Zhang Y, Han L, Xu Y, Yutzey KE, Zorn AM, Kalin TV, Shannon JM, Kalinichenko VV. Foxf1 transcription factor promotes lung morphogenesis by inducing cellular proliferation in fetal lung mesenchyme. Developmental Biology. 2018;443:50–63. doi: 10.1016/j.ydbio.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo LT, Daley GQ. De novo generation of HSCs from somatic and pluripotent stem cell sources. Blood. 2015;125:2641–2648. doi: 10.1182/blood-2014-10-570234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bhattacharyya D, Dennewitz MB, Kalinichenko VV, Zhou Y, Lepe R, Costa RH. Rapid hepatocyte nuclear translocation of the forkhead box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expression. 2003;11:149–162. doi: 10.3727/000000003108749044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Zhang Y, Snyder J, Sutherland MJ, Burhans MS, Shannon JM, Park HJ, Whitsett JA, Kalinichenko VV. Increased expression of FoxM1 transcription factor in respiratory epithelium inhibits lung sacculation and causes Clara cell hyperplasia. Developmental Biology. 2010;347:301–314. doi: 10.1016/j.ydbio.2010.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wen B, Ren X, Li E, Zhang Y, Guo M, Xu Y, Whitsett JA, Kalin TV, Kalinichenko VV. Generation of pulmonary endothelial progenitor cells for cell-based therapy using interspecies mouse-rat chimeras. American Journal of Respiratory and Critical Care Medicine. 2021;204:326–338. doi: 10.1164/rccm.202003-0758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wen B, Deng Z, Zhang Y, Kolesnichenko OA, Ustiyan V, Pradhan A, Kalin TV, Kalinichenko VV. Endothelial progenitor cells stimulate neonatal lung angiogenesis through FOXF1-mediated activation of BMP9/ACVRL1 signaling. Nature Communications. 2022;13:2080. doi: 10.1038/s41467-022-29746-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijts B, Yvernogeau L, Robin C. Recent advances in developmental hematopoiesis: diving deeper with new technologies. Frontiers in Immunology. 2021;12:790379. doi: 10.3389/fimmu.2021.790379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Li E, Ustiyan V, Wang G, Guo M, Na CL, Kalin GT, Galvan V, Xu Y, Weaver TE, Kalin TV, Whitsett JA, Kalinichenko VV. In vivo generation of lung and thyroid tissues from embryonic stem cells using blastocyst complementation. American Journal of Respiratory and Critical Care Medicine. 2021;203:471–483. doi: 10.1164/rccm.201909-1836OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Greely HT, Jaenisch R, Nakauchi H, Rossant J, Belmonte JCI. Stem cells and interspecies chimaeras. Nature. 2016;540:51–59. doi: 10.1038/nature20573. [DOI] [PubMed] [Google Scholar]
- Xia H, Ren X, Bolte CS, Ustiyan V, Zhang Y, Shah TA, Kalin TV, Whitsett JA, Kalinichenko VV. Foxm1 regulates resolution of hyperoxic lung injury in newborns. American Journal of Respiratory Cell and Molecular Biology. 2015;52:611–621. doi: 10.1165/rcmb.2014-0091OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Sato H, Kato-Itoh M, Goto T, Hara H, Sanbo M, Mizuno N, Kobayashi T, Yanagida A, Umino A, Ota Y, Hamanaka S, Masaki H, Rashid ST, Hirabayashi M, Nakauchi H. Interspecies organogenesis generates autologous functional islets. Nature. 2017;542:191–196. doi: 10.1038/nature21070. [DOI] [PubMed] [Google Scholar]
- Yang B, Chen Z, Huang Y, Han G, Li W. Identification of potential biomarkers and analysis of prognostic values in head and neck squamous cell carcinoma by bioinformatics analysis. OncoTargets and Therapy. 2017;10:2315–2321. doi: 10.2147/OTT.S135514. [DOI] [PMC free article] [PubMed] [Google Scholar]