Summary
Bacterial spores can preserve cellular dormancy for years, but still hold the remarkable ability to revive and recommence life. This cellular awakening begins with a rapid and irreversible event termed germination; however, the metabolic determinants required for its success have been hardly explored. Here, we show that at the onset of the process of sporulation, the metabolic enzyme RocG catabolizes glutamate, facilitating ATP production in the spore progenitor cell, and subsequently influencing the eventual spore ATP reservoir. Mutants displaying low RocG levels generate low ATP-containing spores that exhibit severe germination deficiency. Importantly, this phenotype could be complemented by expressing RocG at a specific window of time during the initiation of sporulation. Thus, we propose that despite its low abundance in dormant spores, ATP energizes spore germination, and its production, fueled by RocG, is coupled with the initial developmental phase of spore formation.
Subject areas: Natural sciences, Biological sciences, Microbiology, Bacteriology
Graphical abstract
Highlights
-
•
The enzyme RocG catabolizes glutamate, producing ATP in the spore progenitor cell
-
•
Mutants with low RocG levels generate low ATP-containing spores
-
•
Low ATP in dormant spores correlates with germination deficiency
-
•
ATP level in spores can be elevated by expressing RocG at sporulation onset
Natural sciences; Biological sciences; Microbiology; Bacteriology
Introduction
When confronted with nutrient limitation, the Gram-positive bacterium Bacillus subtilis (B. subtilis) and its relatives initiate a developmental process, called sporulation, which concludes with the production of a highly resilient dormant spore (Higgins and Dworkin, 2012; Piggot and Hilbert, 2004; Stragier and Losick, 1996). Sporulation commences with the formation of a polar septum, asymmetrically dividing the progenitor cell into a small future spore compartment, termed forespore, and a larger mother cell, which nurtures spore development. Subsequently, the forespore is engulfed by the mother cell, and through the sequential activation of cell type specific sigma factors, acquires its protective shells. A thick layer of peptidoglycan, termed the cortex, as well as inner and outer proteinaceous coats, are deposited around the forespore, conferring spore robustness (Driks and Eichenberger, 2016; Henriques and Moran, 2007; McKenney et al., 2013; Popham and Bernhards, 2015; Stragier and Losick, 1996). The forespore core then undergoes dehydration, mainly by accumulating pyridine-2, 6-dicarboxylic acid [dipicolinic acid or (DPA)] in complex with Ca2+ (Setlow, 2006). Eventually, the phase-bright spore is liberated by the lysis of the mother cell, but intriguingly can still undergo significant molecular remodeling, at least for a few days post-sporulation (Korza et al., 2016; Segev et al., 2012, 2013).
The spore can remain quiescent for years; yet, dormancy is rapidly ceased upon nutrient resumption, culminating in vegetative life renewal. This remarkable revival process comprises three key consecutive phases: (1) Germination, during which the spore undergoes a transition from a phase-bright spore to a phase-dark cell, as manifested by light microscopy (Moir, 2006; Setlow, 2003); (2) Ripening, a lag phase, exploited by the spore for molecular reorganization (Segev et al., 2013; Sinai et al., 2015); (3) Outgrowth, through which the spore converts into a vegetative cell that raises from the breaching spore shells (Moir, 2006; Setlow, 2003). Germination, the most critical revival event, is triggered by the binding of nutrients (e.g., amino acids, sugars, purine nucleosides), termed germinants, to multiple germination receptors embedded in the spore membrane (Paredes-Sabja et al., 2011; Ramirez-Guadiana et al., 2017; Setlow, 2003, 2014; Shah et al., 2008). In B. subtilis, the GerA receptor binds L-alanine (L-Ala), whereas GerB and GerK receptors are jointly stimulated by a mixture of asparagine, glucose, fructose, and potassium ions (AGFK) germinants. These ligand-receptor interactions activate downstream signaling events, instructing the spore to initiate multiple processes, including core rehydration, the release of DPA, cortex hydrolysis, and coat disassembly (Moir, 2006; Setlow, 2003, 2014; Setlow et al., 2017).
Germination is considered independent of macromolecule synthesis (Setlow, 2003; Steinberg et al., 1965; Vinter, 1970); however, in recent years, we have provided evidence that germination necessitates the activation of both transcription and translation for its completion (Sinai and Ben-Yehuda, 2016; Sinai et al., 2015; Zhou et al., 2019, 2022). This view is not collectively accepted, and other laboratories suggested alternative interpretations (Korza et al., 2016; Sinai and Ben-Yehuda, 2016; Swarge et al., 2020). The spore-orchestrated awakening was found to be mediated by the arginine phosphatase YwlE that dephosphorylates the housekeeping sigma factor A (SigA) and the ribosome-associated chaperone Tig, which reactivate the transcriptional and the translational machineries, respectively (Sinai et al., 2015; Zhou et al., 2019). These findings are in accord with previous studies showing that RNA and protein synthesis occurs very early in reviving spores, at a time coinciding with germination [e.g. (Armstrong and Sueoka, 1968; Balassa and Contesse, 1965; Torriani and Levinthal, 1967; Zhou et al., 2022)]. Still, there are gaps in our understanding of the intricate germination pathways, such as the signal transmission from germinant sensing to YwlE activation, and from the stimulated YwlE to its downstream targets.
Accumulating data signify that germination is influenced by sporulation conditions, which in turn impact the spore molecular cargo (Bressuire-Isoard et al., 2018). Furthermore, factors such as spore age and temperature of incubation were found to affect the spore RNA content and to ultimately determine spore germination and revival capacities (Segev et al., 2012, 2013). Interestingly, spores have been shown to contain low levels of ATP, which are considered negligible for germination (Christie and Setlow, 2020; Setlow and Christie, 2020; Setlow et al., 2017). Here, we provide evidence that a key enzyme in glutamate catabolism, RocG, which catalyzes the formation of 2-oxoglutarate (2-OG), a tricarboxylic acid (TCA) cycle component (Stannek et al., 2015), is crucial for spore germination. Surprisingly, we discovered that the timing of RocG activity, specifically at the beginning of sporulation, impacts the future spore ATP level and concomitantly its germination capacity.
Results
The YwlE-interactome revealed arginine utilization regulators to be crucial for spore germination
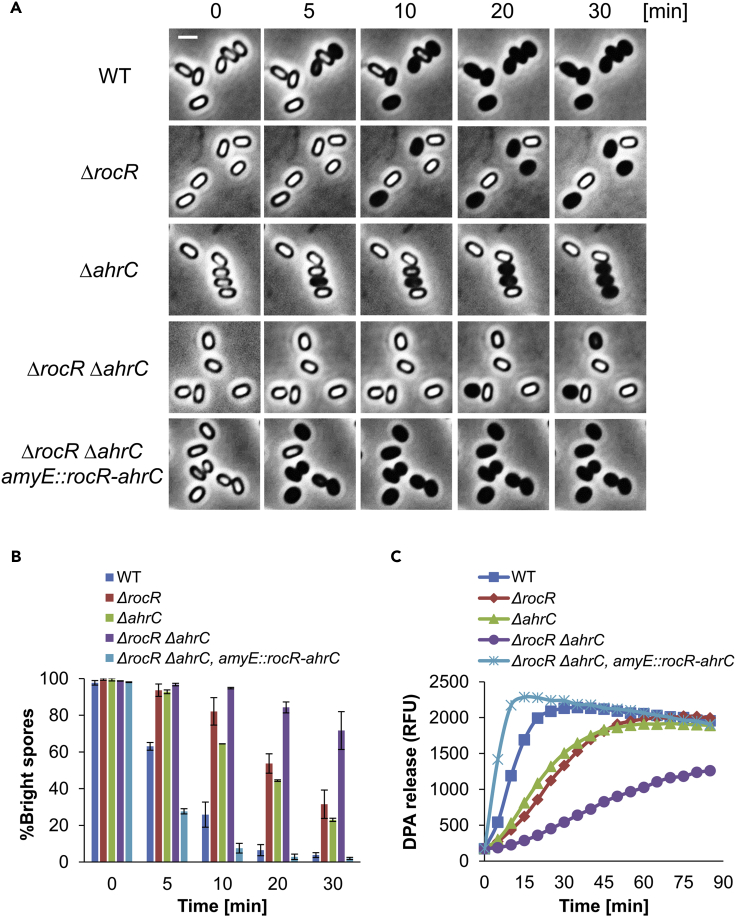
As the YwlE arginine phosphatase is central for germination induction (Zhou et al., 2019), we used YwlE as bait to reach new germination factors. Accordingly, we generated a functional form of YwlE fused to a promiscuous biotin ligase, TurboID, enabling biotin labeling of proximal proteins (Branon et al., 2018) (Figure S1A). Spores carrying TurboID-YwlE were induced to germinate by L-Ala or AGFK, without the addition of other nutrients, in the presence of biotin. Western blot analysis revealed a specific biotin-labeling pattern that varied between the germinants (Figure S1B). The biotinylated proteins were purified and analyzed by mass spectrometry (MS), and 23 factors shared by both L-Ala and AGFK proteomes were chosen for further characterization (Figure S2). Among the identified proteins were those involved in translation (3), metabolism (4), cellular processes (6), and a group of proteins (10) of unknown function. The corresponding mutant spores were consequently tested for their capacity to germinate with either L-Ala or AGFK by monitoring DPA release (Figure S2 and data not shown). Using this approach, we noted that the mutant lacking rocR gene, encoding a transcriptional activator of the arginine utilization operons (Gardan et al., 1997), exhibits a profound germination deficiency (Figures 1A, 1B, and S2).
Figure 1.
Arginine utilization regulators facilitate spore germination
(A) Spores of PY79 (WT), LR31 (ΔrocR), LR30 (ΔahrC), LR32 (ΔrocR ΔahrC), LR205 (ΔrocR ΔahrC, amyE::rocR-ahrC) strains were induced to germinate on an agarose pad supplemented with L-Ala (10 mM) (t = 0) and followed by time-lapse microscopy. Shown are phase contrast images captured at the indicated time points from a representative experiment out of three independent biological repeats. Scale bar, 1 μm.
(B) Quantification of the experiment described in (A). Data are presented as percentages of the initial number of the phase bright spores. Shown are average values and SD obtained from three independent biological repeats (n ≥ 300 for each strain).
(C) Spores of PY79 (WT), LR31 (ΔrocR), LR30 (ΔahrC), LR32 (ΔrocR ΔahrC), LR205 (ΔrocR ΔahrC, amyE::rocR-ahrC) strains were incubated with L-Ala (10 mM) to trigger germination. DPA release was determined by monitoring the relative fluorescence units (RFU) of Tb3+-DPA. Shown is a representative experiment out of three independent biological repeats.
Arginine utilization operons are co-regulated by the transcriptional factors RocR and AhrC (Czaplewski et al., 1992); we, therefore, tested the germination ability of spores derived from ΔahrC mutant. Monitoring germination by time-lapse microscopy revealed the ΔahrC spores to display germination defect similar to that of the ΔrocR spores (Figures 1A and 1B). Intriguingly however, the deletion of both rocR and ahrC resulted in an additive and severe germination deficiency (Figures 1A and 1B), suggesting that the two proteins collaborate to execute their function in spore germination. Similar results were obtained when monitoring germination by DPA release, decrease in optical density, or heat resistance assays (Figures 1C and S3C-S3E), deficiencies that were entirely complemented by reintroducing rocR and ahrC into the double mutant strain (Figures 1A-1C and S3F). A germination defect was also evident in ΔrocR ΔahrC mutant spores following the direct activation of germination receptors using high pressure (Figures S4A-S4C), even though the level of the receptors was similar to that of the wild-type (WT) spores (Figure S4D). Consistently, no effect on vegetative growth or sporulation was detected in ΔrocR ΔahrC mutant (Figures S3A and S3B). Collectively, these results indicate that the produced ΔrocR ΔahrC spores are intact, but harbor a germination-specific deficiency.
Glutamate catabolism by RocG is required for spore germination
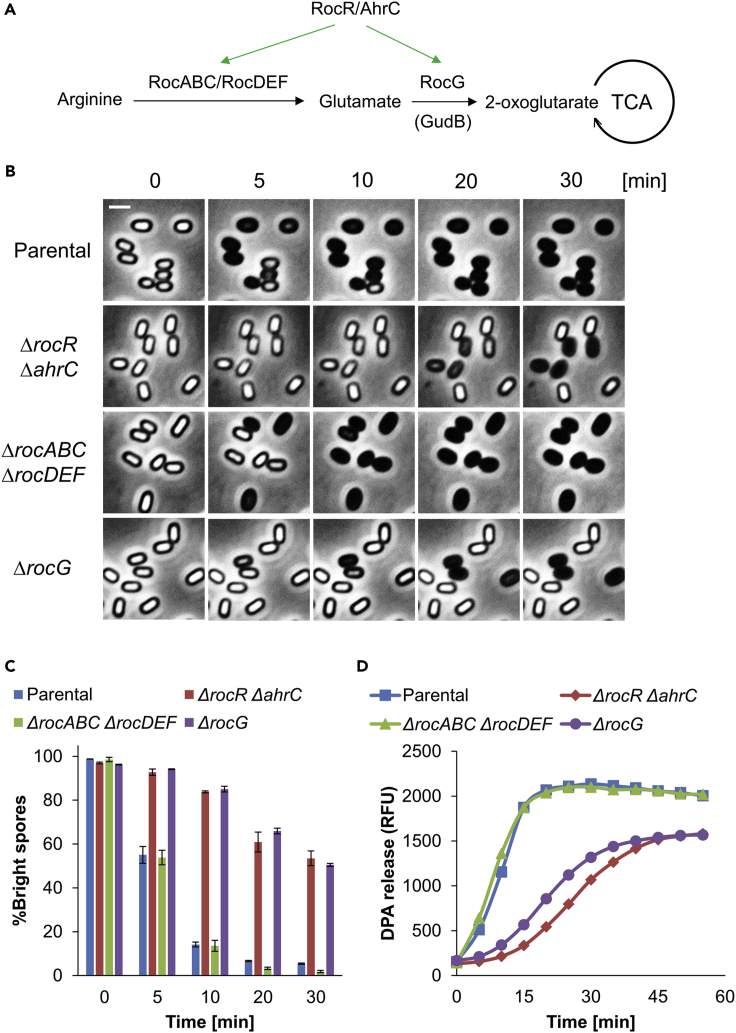
The need for both RocR and AhrC to facilitate germination raised the question of how arginine metabolism impacts the process. RocR-AhrC regulated arginine metabolism consists of two main stages: (i) arginine is converted to glutamate by RocABC and RocDEF (RocA-F), and (ii) RocG dehydrogenase catabolizes glutamate to 2-OG, which participates in the TCA cycle (Figure 2A) (Belitsky and Sonenshein, 1998; Stannek et al., 2015). As both glutamate production and the TCA cycle are vital pathways (Sonenshein, 2007) that can conceivably impact germination, we tested the requirement of each stage for the process. We accordingly investigated whether the knockout of the downstream-regulated genes rocA-F or rocG affect germination. For simplicity, all tested strains were also deleted for glutamate dehydrogenase gudB that might be activated when rocG is inactivated (Belitsky and Sonenshein, 1998; Commichau et al., 2008), a mutation that had no effect on growth, sporulation, or germination (Figure S5). Whereas the ΔrocA-F mutant showed no significant effect on germination, ΔrocG spores exhibited substantial germination perturbation, similarly to that observed for the ΔrocR ΔahrC spores (Figures 2B-2D and S6A-S6C). These results suggest that germination deficiency of ΔrocR ΔahrC is largely attributed to the absence of the downstream RocG enzyme, converting glutamate to 2-OG (Commichau et al., 2007). Although a reduced sporulation efficiency was detected for the ΔrocG mutant, the produced spores were intact as indicated by their capacity to withstand heat (Figures S6D and S6E). Nevertheless, ΔrocR ΔahrC strain, displaying low RocG expression (Figure 4A) but normal sporulation efficiency (Figure S3B), was chosen for further understanding of the RocG role in germination.
Figure 2.
RocG plays a key role in spore germination
(A) The pathway of RocR-AhrC regulated arginine metabolism in B. subtilis is illustrated. It consists of two stages: (1) Arginine is converted to glutamate by RocABC and RocDEF (RocA-F) enzymes, and (2) RocG dehydrogenase catabolizes glutamate to 2-OG, which participates in the tricarboxylic acid cycle (TCA cycle) (Stannek et al., 2015).
(B) Parental LR33, LR38 (ΔrocR ΔahrC), LR203 (ΔrocABC ΔrocDEF), LR137 (ΔrocG) strains, lacking gudB, were induced to germinate on an agarose pad supplemented with L-Ala (10 mM) (t = 0) and followed by time-lapse microscopy. Shown are phase contrast images captured at the indicated time points from a representative experiment out of three independent biological repeats. Scale bar, 1 μm.
(C) Quantification of the experiment described in (B). Data are presented as percentages of the initial number of the phase bright spores. Shown are average values and SD obtained from three independent biological repeats (n ≥ 300 for each strain).
(D) Spores of LR33 (Parental), LR38 (ΔrocR ΔahrC), LR203 (ΔrocABC ΔrocDEF), LR137 (ΔrocG) strains, lacking gudB, were incubated with L-Ala (10 mM) to trigger germination. DPA release was determined by monitoring the relative fluorescence units (RFU) of Tb3+-DPA. Shown is a representative experiment out of three independent biological repeats.
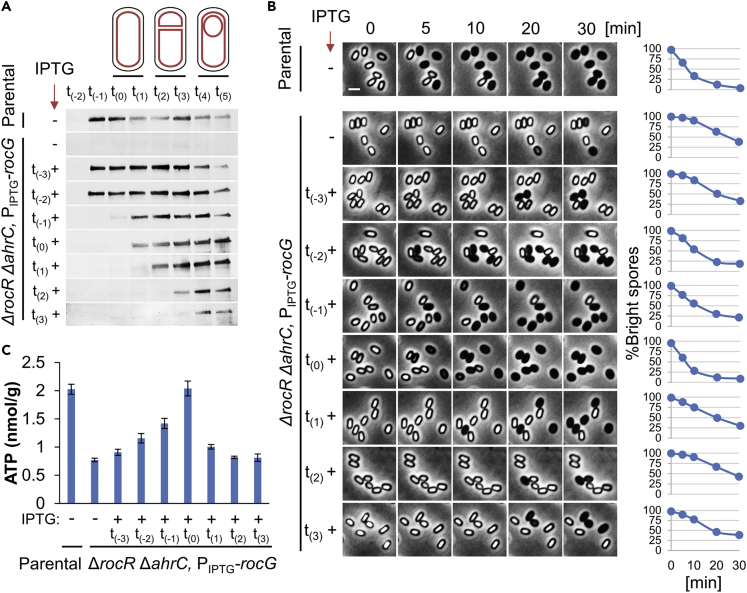
Figure 4.
RocG activity is required at the onset of sporulation to allow successful germination
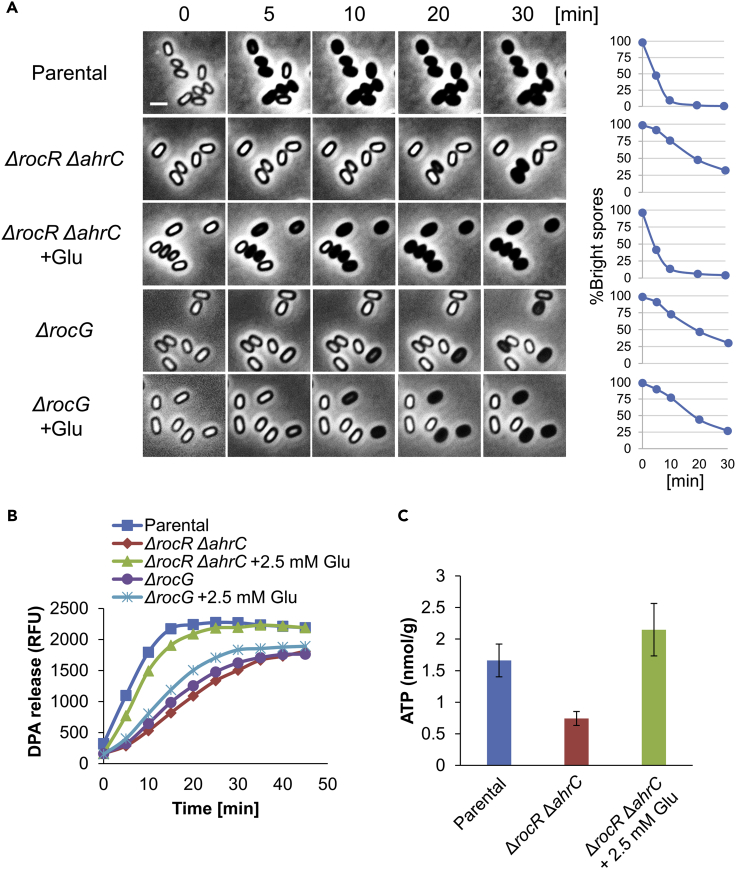
(A) LR33 (Parental) and LR127 (ΔrocR ΔahrC, PIPTG-rocG) strains, lacking gudB, were grown in DSM sporulation medium. IPTG was added to the LR127 cultures at the indicated sporulation time points to induce rocG expression. Cells were collected at each time point for protein extraction. Equal amounts of protein extracts were subjected to Western blot analysis using antibody against RocG.
(B) LR33 (Parental) and LR127 (ΔrocR ΔahrC, PIPTG-rocG) strains, lacking gudB, were grown in DSM sporulation medium. IPTG was added to the LR127 cultures at the indicated sporulation time points to induce rocG expression. After 20 hrs of incubation, spores were purified, induced to germinate on an agarose pad supplemented with L-Ala (10 mM) (t = 0), and followed by time-lapse microscopy. Shown are phase contrast images captured at the indicated time points from a representative experiment, out of three independent biological repeats. Graphs on the right show the quantification of the percentages of the initial number of the phase bright spores (n ≥ 300 for each strain). Scale bar, 1 μm.
(C) LR33 (Parental) and LR127 (ΔrocR ΔahrC, PIPTG-rocG) strains, lacking gudB, were grown in DSM sporulation medium. IPTG was added to the LR127 cultures at the indicated sporulation time points to induce rocG expression. After 20 hrs of incubation, spores were purified from DSM cultures and analyzed for ATP level using ATP Bioluminescence Detection Kit (Promega). Shown are average values and SD obtained from three independent biological repeats.
Expressing RocG at the onset of sporulation suppresses ΔrocR ΔahrC germination deficiency
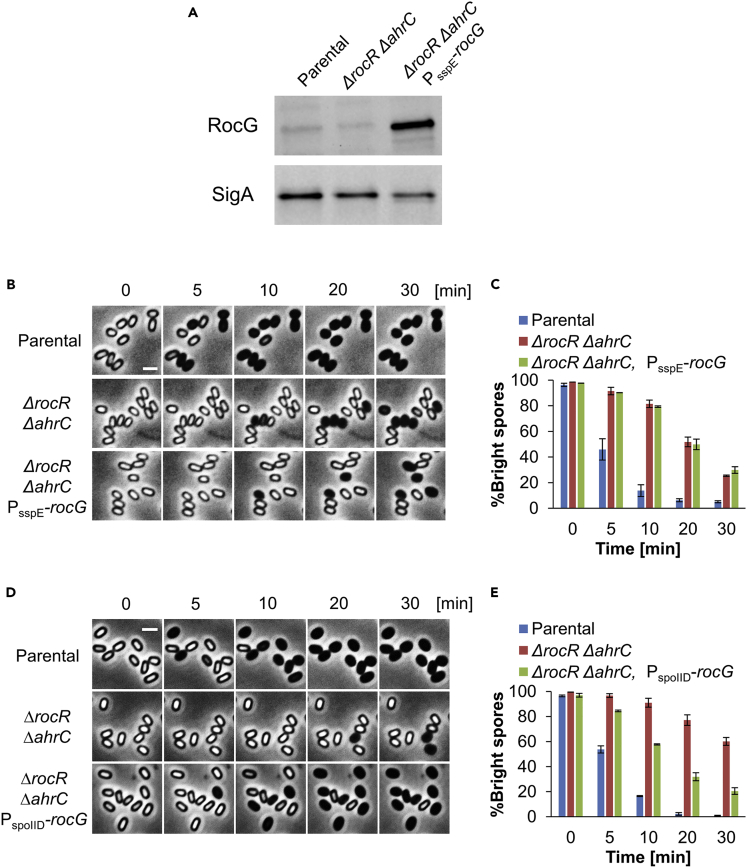
As glutamate and arginine were the only free amino acids significantly detected in dormant spores (Nelson and Kornberg, 1970), we considered the possibility that glutamate catabolism by RocG provides the energy needed to propel spore germination by fueling the TCA cycle. If correct, artificially increasing RocG levels in ΔrocR ΔahrC spores should relieve their germination deficiency. Thus, rocG was expressed from the late sigma G forespore promoter belonging to the sspE gene (Fajardo-Cavazos et al., 1991) in ΔrocR ΔahrC strain. Indeed, RocG levels in dormant spores were boosted (Figure 3A), but surprisingly the germination defect was not concomitantly restored (Figures 3B, 3C, S7A, and S7B). Nevertheless, an earlier expression of RocG from the sigma E mother cell regulated promoter spoIID could largely suppress the ΔrocR ΔahrC germination defect (Figures 3D, 3E, S7C, and S7D). Thus, it seems that a metabolite generated by RocG during sporulation, rather than the enzyme itself, is required at the time of germination.
Figure 3.
Early expression of RocG during sporulation is crucial for germination
(A) Spores of LR33 (Parental), LR38 (ΔrocR ΔahrC), LR134 (ΔrocR ΔahrC, PsspE-rocG) strains, lacking gudB, were disrupted for protein extraction. Equal amounts of protein extracts were subjected to Western blot analysis with antibody against RocG. SigA was monitored in parallel as a loading control using anti-SigA antibody.
(B) Spores of LR33 (parental), LR38 (ΔrocR ΔahrC), LR134 (ΔrocR ΔahrC, PsspE-rocG) strains, lacking gudB, were induced to germinate on an agarose pad supplemented with L-Ala (10 mM) (t = 0) and followed by time-lapse microscopy. Shown are phase contrast images captured at the indicated time points from a representative experiment out of three independent biological repeats. Scale bar, 1 μm.
(C) Quantification of the experiment described in (B). Data are presented as percentages of the initial number of the phase bright spores. Shown are average values and SD obtained from three independent biological repeats (n ≥ 300 for each strain).
(D) Spores of LR33 (Parental), LR38 (ΔrocR ΔahrC), LR133 (ΔrocR ΔahrC, PspoIID-rocG) strains, lacking gudB, were induced to germinate on an agarose pad supplemented with L-Ala (10 mM) (t = 0) and followed by time-lapse microscopy. Shown are phase contrast images captured at the indicated time points from a representative experiment, out of three independent biological repeats. Scale bar, 1 μm.
(E) Quantification of the experiment described in (D). Data are presented as percentages of the initial number of the phase bright spores. Shown are average values and SD obtained from three independent biological repeats (n ≥ 300 for each strain).
To further address this notion, we cloned rocG under an Isopropyl β-D-1-thiogalactopyranoside (IPTG) inducible promoter and tested its ability to complement ΔrocR ΔahrC germination deficiency, when expressed at different time points along the process of sporulation (Figures 4A and S8A-S8C). The corresponding spores were then purified and tested for their capacity to induce germination by time-lapse microscopy, DPA release, and optical density analyses. We revealed that germination deficiency was rescued in spores, when RocG was induced from t(-2) up until t(0) of sporulation, with t(0) appearing as the optimal induction time point (Figures 4B, S8D, and S8E). Taken together, germination deficiency of ΔrocR ΔahrC can be fully complemented by artificially expressing their downstream component RocG enzyme at the onset of sporulation.
Impaired glutamate catabolism during sporulation yields spores with low ATP levels that exhibit germination deficiency
To elucidate whether RocG activity is required to produce an essential metabolite that facilitates germination, we supplied the sporulation medium with glutamate, the RocG substrate (Figure 2A), and assayed the ΔrocR ΔahrC germination capacity. Indeed, the addition of glutamate was highly effective, clearly rescuing the mutant germination defect (Figures 5A, 5B, S9A, and S9B). This suppression was RocG dependent, as it was abolished in the absence of the protein (Figures 5A, 5B, and S9B). Examination of RocG levels following glutamate addition showed no increase in the protein levels (Figure S9C), indicating that a low amount of RocG is sufficient to enhance germination, as long as an adequate amount of glutamate was added to the sporulation medium. The simplest interpretation of these findings is that spores reserve a critical metabolite, influenced by RocG-mediated glutamate catabolism, for germination.
Figure 5.
Glutamate supplementation at the onset of sporulation suppresses ΔrocR ΔahrC germination deficiency
(A) LR33 (Parental), LR38 (ΔrocR ΔahrC), LR137 (ΔrocG) strains, lacking gudB, were grown in DSM sporulation medium with or without 2.5 mM glutamate. After 20 hrs of incubation, spores were purified, induced to germinate on an agarose pad supplemented with L-Ala (10 mM) (t = 0), and followed by time-lapse microscopy. Shown are phase contrast images captured at the indicated time points from a representative experiment, out of three independent biological repeats. Graphs on the right show the quantification of the percentages of the initial number of the phase bright spores (n ≥ 300 for each strain). Scale bar, 1 μm.
(B) LR33 (Parental), LR38 (ΔrocR ΔahrC), LR137 (ΔrocG) strains, lacking gudB, were grown in DSM sporulation medium with or without 2.5 mM glutamate. After 20 hrs of incubation, spores were purified, induced to germinate by L-Ala (10 mM) (t = 0), and germination was followed by DPA release, monitoring the relative fluorescence units (RFU) of Tb3+-DPA. Shown is a representative experiment out of three independent biological repeats.
(C) LR33 (Parental), LR38 (ΔrocR ΔahrC) strains, lacking gudB, were grown in DSM sporulation medium with or without 2.5 mM glutamate. After 20 hrs of incubation, spores were purified and analyzed for ATP level using ATP Bioluminescence Detection Kit (Promega). Shown are average values and SD obtained from three independent biological repeats.
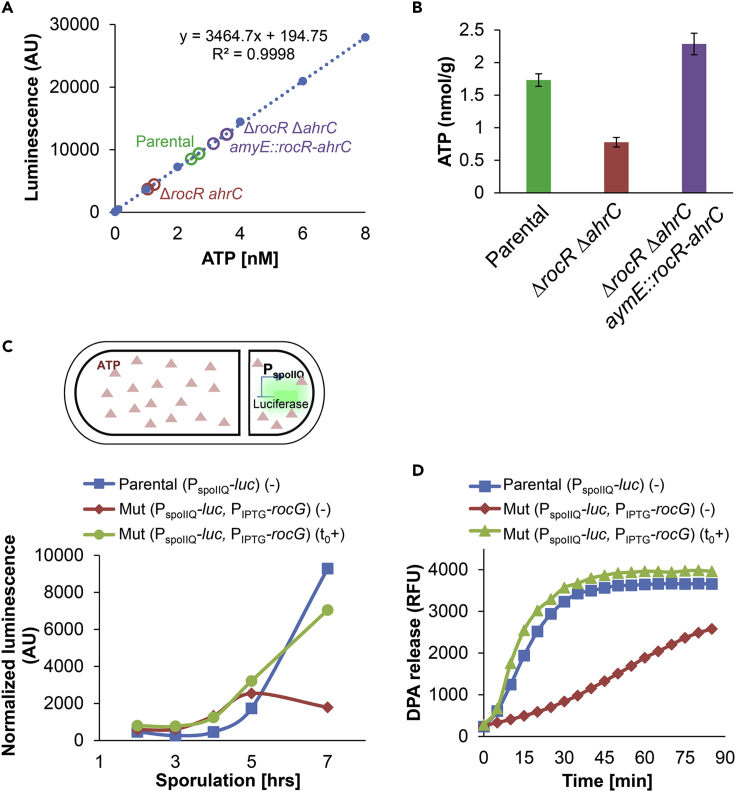
The potential contribution of RocG activity to the TCA cycle (Figure 2A) hints that RocG could facilitate germination by fueling the production of ATP, among other compounds, during sporulation. As such, ATP could be a spore-accumulated component that serves to energize germination. To test this possibility, we attempted to compare the ATP pools in the WT and the ΔrocR ΔahrC mutant dormant spores. Detection of ATP levels from spores (∼2x109 spores per measurement) by the sensitive luciferase that catalyzes luminescence reaction revealed the WT spores to harbor 1.73 ± 0.1 nmol/g ATP (Figures 6A and 6B), in line with previously reported concentration (2 nmol/g) (Setlow and Kornberg, 1970). Remarkably, the ΔrocR ΔahrC mutant spores harbored only 0.77 ± 0.07 nmol/g ATP, whereas the complemented strain harbored levels even higher than the WT (2.28 ± 0.17 nmol/g) (Figures 6A and 6B). Likewise, the addition of glutamate during sporulation recovered the ATP level of ΔrocR ΔahrC mutant spores, along with improving their germination capability (Figure 5C). Furthermore, ATP content examined in dormant spores of ΔrocR ΔahrC mutant expressing rocG under IPTG-inducible promoter correlated with the spore ability to germinate (Figures 4B and 4C). Notably, the production of RocG prior to the initiation of sporulation was insufficient to sustain germination, probably owing to premature glutamate consumption by the enzyme, draining the sporangium from its potential ATP source (Figure 4).
Figure 6.
ATP generated by RocG-mediated glutamate catabolism is stored in spores for future germination
(A and B) Spores of LR33 (Parental), LR38 (ΔrocR ΔahrC), LR207 (ΔrocR ΔahrC, amyE::rocR-ahrC) strains, lacking gudB, were disrupted for ATP extraction. ATP levels were determined using an ATP bioluminescence assay kit (Promega), referring to a standard curve of known ATP concentrations (A). The ATP levels in spores are the total ATP in extracts (nmol)/dry weight of spores (g) (B). Shown are average values and SD obtained from three independent biological repeats.
(C) Upper panel: shows a schematic description of ATP detection in vivo from the forespore compartment during sporulation by luciferase-catalyzed luminescence reaction (Parrell and Kroos, 2020). Lower panel: LR209 [Parental (PspoIIQ-luc)], LR227 [Mut (PspoIIQ-luc, PIPTG-rocG, ΔrocR ΔahrC)] strains were induced to sporulate in DSM medium. At t0 of sporulation, IPTG was added to induce rocG expression, and samples were subjected to luminescence assay at the indicated time points. Shown are luminescence values in arbitrary units (AU) normalized to total luciferase levels measured by Western blot analysis.
(D) Spores derived from the cultures described in (C) were purified, induced to germinate by L-Ala (10 mM) (t = 0), and germination was followed by monitoring the relative fluorescence units (RFU) of Tb3+-DPA. Shown is a representative experiment out of three independent biological repeats.
To substantiate that sporulation-generated ATP, via glutamate catabolism, facilitates germination, we monitored the ATP levels in vivo by measuring the kinetics of luciferase activity in the forespore compartment during sporulation, utilizing the system developed by the Kroos laboratory (Figures 6C, S9C) (Parrell and Kroos, 2020). In accordance with our findings, the ATP level of ΔrocR ΔahrC forespore was significantly lower than that of the WT, a phenotype that was again restored by RocG induction at the onset of sporulation that rescued germination deficiency (Figures 6C and 6D). Hence, we infer that RocG activity during sporulation contributes to the spore ATP reservoir, which correlates with germination efficiency.
Discussion
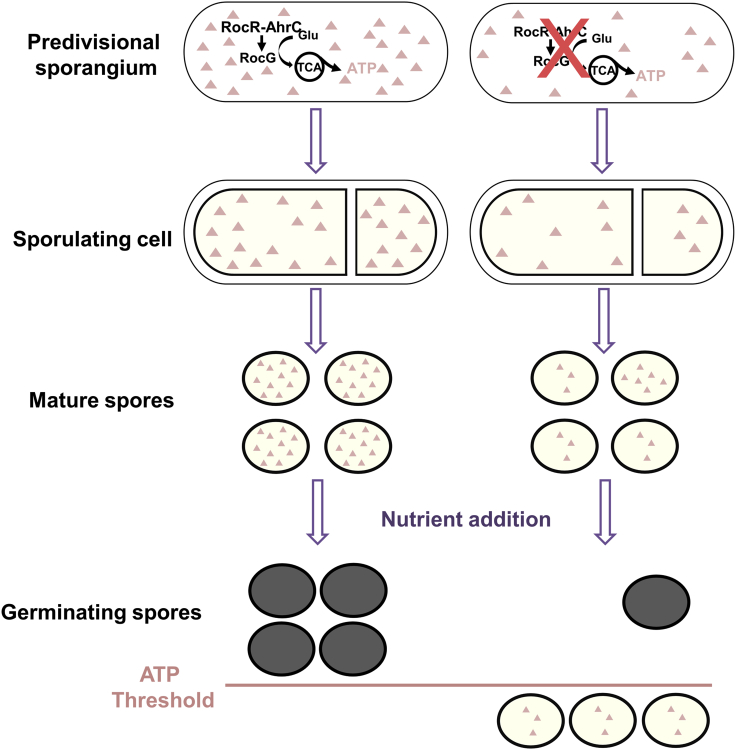
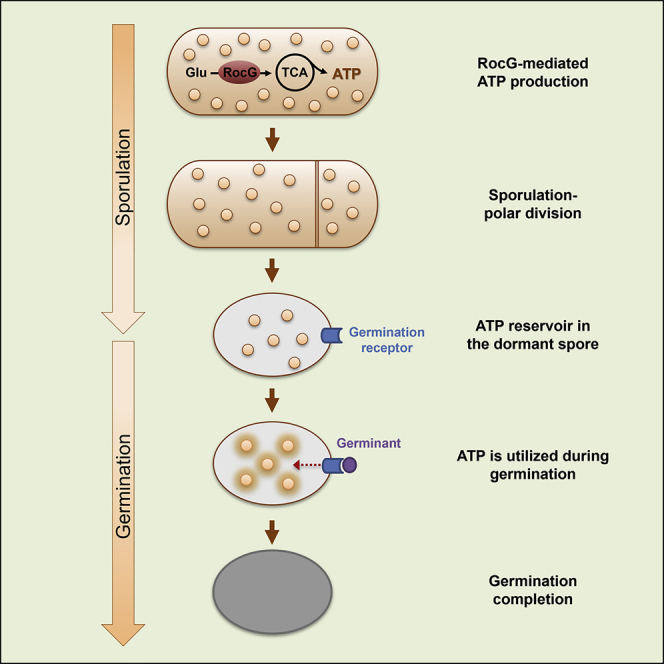
Here we present evidence supporting that the limited spore ATP reservoir associates with germination competence. We describe that glutamate degradation by RocG, at the beginning of sporulation, promotes ATP generation and subsequently its accumulation in the forespore compartment, potentially providing an energy reservoir for fueling future spore germination. Accordingly, mutants producing low RocG levels generate low ATP-containing spores that exhibit a severe germination defect. This phenotype can be rescued by inducing RocG production at the onset of sporulation, which concomitantly restores the wild-type ATP level in spores. The RocG product 2-OG also has an anabolic function acting as a precursor for the synthesis of amino acids and nucleotides, as well as contributing to the NADH pool (Huergo and Dixon, 2015), as such it can also affect the spore molecular content via alternative metabolic pathways. Nevertheless, based on the strong correlation between RocG activity and the spore ATP reservoir, we propose that sporulation-synthesized ATP, mainly via the RocG-glutamate degradation pathway, should be accumulated to a threshold level to enable successful spore germination (Figure 7). Thus, despite its low concentration, we suggest that ATP is a major determinant of spore germination capacity.
Figure 7.
ATP is produced at the onset of sporulation to facilitating spore germination
Left panels: At the onset of sporulation, RocG, activated by RocR and AhrC, catalyzes glutamate, feeding the TCA cycle and increasing the ATP pool in the pre-divisional sporanguim. A fraction of the produced ATP is stored in the forespore and subsequently in the mature spore. A threshold level of ATP determines the efficiency of germination. Right panels: show the scheme in the absence of the glutamate catabolic pathway, whereby many of the produced spores have ATP level lower than the threshold.
According to our data and calculations, we estimate the spore ATP reservoir to be around 1 μM, which is within the Km range of ATP binding proteins [e.g. (Knight and Shokat, 2005; Noriega et al., 2008). Such intracellular concentration is likely to propel early molecular events occurring during germination. ATP storage could facilitate germination-induced phosphorylation events, such as the signal transduction triggered by PrkC kinase or processes that are stimulated by the YwlE phosphatase (Shah et al., 2008; Zhou et al., 2019). As germination appears to be promoted by the rapid restoration of transcription and translation (Sinai et al., 2015; Zhou et al., 2019), the preserved ATP pool might enable the reestablishment and initial progression of these processes. ATP level was found to correlate with the resuscitation capacity of viable but nonculturable E. coli cells (Pu et al., 2019), suggesting that a threshold ATP level could be a common factor required in the awakening phase of quiescent bacteria. Remarkably, ATP concentration was shown to be increased by 100-fold during the first minutes of germination (Setlow and Kornberg, 1970). The initial spore ATP reservoir could be necessary to trigger and propel this active ATP synthesis. This boost in ATP levels might be generated by enzymes consuming energy sources such as phosphoglyceric acid (PGA) and malate that reach high concentrations within spores (Nelson and Kornberg, 1970; Sinai et al., 2015). Furthermore, the presence of arginine and glutamate in dormant spores (Nelson and Kornberg, 1970), implies that their utilization might contribute to this ATP rise. In fact, the potential interaction between YwlE and RocR during germination, noted by the TurboID analysis, hints that YwlE could activate RocR to induce arginine catabolism.
The finding that RocG needs to be expressed prior to polar division to optimally complement the mutant phenotype infers that the TCA enzymes, exploiting its product, are abundant at this stage but are scarce at later stages of sporulation. In fact, a shutdown of metabolic enzyme production was shown to typify the forespore compartment following polar septation (Riley et al., 2021), corroborating the temporal importance of the enzyme expression. Hence, our results highlight how a metabolic pathway is tightly connected to cellular differentiation in bacteria. Interestingly, substantial complementation was monitored when RocG was expressed under an early mother cell-specific promoter, suggesting that the mother cell could augment the forespore ATP reservoir. This result coincides with the notion that the mother cell utilizes a feeding tube apparatus to nourish the forespore with small molecules needed for its biosynthetic activities during development (Camp and Losick, 2009; Doan et al., 2009; Meisner et al., 2008; Riley et al., 2021).
Emerging evidence reinforces the view that phenotypic diversity among spores determines their ability to germinate. Broadly, spores formed in a nutrient-poor sporulation medium germinate slower than those formed in the nutrient-rich medium, indicating that their molecular content differs (Ramirez-Peralta et al., 2012). We propose that such difference could emanate, at least in part, from dissimilarity in the ATP levels deposited during sporulation. Variations in gene expression also affect germination capacity, on the one hand causing the formation of non-germinating-super dormant spores owing to low levels of germination receptors, and on the other hand, increasing stochastic germination owing to low expression of a sporulation transcription factor (Chen et al., 2014; Ghosh et al., 2012; Ghosh and Setlow, 2009; Sturm and Dworkin, 2015). Likewise, variation in the metabolic enzyme alanine dehydrogenase expression level during sporulation was shown to impact spore revival kinetics (Mutlu et al., 2018, 2020). Additionally, we have found that the spore RNA profile is modified during aging according to the temperature of residing, a process that significantly alters germination kinetics (Segev et al., 2012, 2013). Thus, it appears that phenotypic diversity acquired during sporulation and post-sporulation periods shapes the spore molecular reservoir, and subsequently impacts its germination and revival capabilities. Here we show that ATP, produced during sporulation commencement, corresponds to both, the dormant spore ATP pool and the success of its future germination.
Limitations of the study
We show the direct impact of RocG activity during sporulation on spore germination efficiency and attribute that to RocG impact on the spore ATP level. However, we did not demonstrate the direct connection between RocG activity and the spore ATP reservoir or show whether any intermediate molecules are involved in this path. Although our study provides evidence that the dormant spore ATP level correlates with germination capacity, we did not demonstrate a direct connection between ATP and germination.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| PY79 (B. subtilis wild type) | (Youngman et al., 1984) | NA |
| BKK30700 (rpmEB::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK22880 (ypfD::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK28470 (lysC::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK40350 (rocR::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK05850 (gmuR::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK29010 (speD::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK05950 (ydiF::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK35940 (rbsA::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK25100 (zur::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK15770 (prkC::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK13400 (ykoU::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK19430 (cdaS::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK00650 (yabS::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK13350 (ykoN::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK00630 (yabR::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK33620 (yvaK::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK02330 (ybfQ::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK29830 (ytpQ::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK13250 (ykoG::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK10000 (yhaH::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK05770 (ydhI::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK31390 (yugI::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK24250 (ahrC::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| BKK37790 (rocG::kan) | B. subtilis Strain Collection (Koo et al., 2017) | NA |
| LR14 (mls-turboID-ywlEHAx2) | This study | NA |
| LR30 (ahrC::kan) | This study | NA |
| LR31 (rocR::mls) | This study | NA |
| LR32 (rocR::mls, ahrC::kan) | This study | NA |
| LR33 (gudB::tet) | This study | NA |
| LR38 (gudB::tet, rocR::mls, ahrC::kan) | This study | NA |
| LR127 (gudB::tet, rocR::mls, ahrC::kan amyE::PIPTG-rocG-spc) | This study | NA |
| LR133 (gudB::tet, rocR::mls, ahrC::kan amyE::PspoIID-rocG-cat) | This study | NA |
| LR134 (gudB::tet, rocR::mls, ahrC::kan amyE::PsspE-rocG-cat) | This study | NA |
| LR137 (gudB::tet, rocG::kan) | This study | NA |
| LR197 (rocABC::spc) | This study | NA |
| LR198 (rocDEF::cat) | This study | NA |
| LR200 (gudB::tet, rocABC::spc) | This study | NA |
| LR201 (gudB::tet, rocDEF::cat) | This study | NA |
| LR203 (gudB::tet rocABC::spc, rocDEF::cat) | This study | NA |
| LR205 (rocR::mls, ahrC::kan amyE::rocR-ahrC-cat) | This study | NA |
| LR207 (gudB::tet, rocR::mls, ahrC::kan amyE::rocR-ahrC-cat) | This study | NA |
| BDP87 (amyE::PspoIIQ-lucH245F-cat) | A gift from Prof. Lee Kroos (Parrell and Kroos, 2020). | NA |
| LR209 (gudB::tet amyE::PspoIIQ-lucH245F-cat) | This study | NA |
| LR211 (gudB::tet, rocR::mls, ahrC::kan amyE::PspoIIQ-lucH245F-cat) | This study | NA |
| LR225 (sacA::PIPTG-rocG-spc) | This study | NA |
| LR227 (gudB::tet, rocR::mls, ahrC::kan amyE::PspoIIQ-lucH245F-cat sacA::PIPTG-rocG-spc) | This study | NA |
| Chemicals, peptides, and recombinant proteins | ||
| Chloramphenicol | Sigma-Aldrich | Cat#: C0378 |
| Tetracycline | Sigma-Aldrich | Cat#: 87128 |
| Kanamycin | US Biological | Cat#: K0010 |
| Lincomycin | Sigma-Aldrich | Cat#: 62143-5G |
| Erythromycin | Sigma-Aldrich | Cat#: E0774 |
| Spectinomycin | Sigma-Aldrich | Cat#: S4014-5G |
| Ampicillin | Sigma-Aldrich | Cat#: A9518-25G |
| L-Alainine | MP BIOMEDICALS | Cat#: 100287 |
| Difco™ Nutrient Broth | BD Biosciences | Cat#: 234000 |
| L-aspargine | Sigma-Aldrich | Cat#:A8381-100G |
| D-glucose | Sigma-Aldrich | Cat#:1632-1KG |
| D-fructose | Sigma-Aldrich | Cat#:F3510-500G |
| Histodenz | Sigma-Aldrich | Cat#:D2158-100G |
| RQ1 DNase | Promega | Cat#:M610A |
| Terbium(III) chloride hexahydrate | Sigma-Aldrich | Cat#:212903 |
| Critical commercial assays | ||
| Q5 High-Fidelity DNA Polymerase | NEW ENGLAND BioLabs | Cat#: M0491S |
| Gibson Assembly Master Mix | NEW ENGLAND BioLabs | Cat#: E2611L |
| Quick Ligation Kit | NEW ENGLAND BioLabs | Cat#: M2200S |
| ENLITEN® ATP Assay System | Promega | Cat#:FF2000 |
| Halt™ Protease Inhibitor Cocktail (100X) | Thermo | Cat#: 78438 |
| iScript™ cDNA Synthesis Kit | BIO-RAD | Cat#:170-8891 |
| iTaq Universal SYBR Green Supermix | BIO-RAD | Cat#: 1725124 |
| FastRNA PRO™ BLUE KIT | MP BIOMEDICALS | Cat#: 116025050 |
| Recombinant DNA | ||
| pDR111 (amyE::PIPTG-spc) | a gift from David Rudner (Harvard Medical School) | NA |
| pDG364 (amyE::cat) | (Guerout-Fleury et al., 1996) | NA |
| pET2 (amyE::gfp-cat) | laboratory stock | NA |
| pLR4 (amyE::PIPTG-rocG-spc) | This study | NA |
| pLR5 (amyE::PspoIID-rocG-cat) | This study | NA |
| pLR6 (amyE::PsspE-rocG-cat) | This study | NA |
| pLR7 (amyE::ProcR-rocR-PahrC-ahrC-cat) | This study | NA |
| Sequence-based reagents | ||
| Primers used in this study are listed in Table S1. | All primers were designed during this study, and synthesized by Integrated DNA Technologies (IDT). | NA |
Resource availability
Lead contact
For additional information about reagents and resources, contact the Lead Contact, Sigal Ben-Yehuda at: sigalb@ekmd.huji.ac.il.
Materials availability
Strains and plasmids generated in this study are available upon request.
Experimental model and subject details
Details on bacterial strain construction
B. subtilis strains are derivatives of the wild type PY79 (Youngman et al., 1984). Bacterial strains and plasmids are listed in key resources table, and primers are listed in Table S1. For gene replacement strategy, indicated primer pairs (P1-P4; Table S1) were used to amplify the flanking genomic regions of the corresponding gene. PCR products were used for Gibson assembly (NEB, USA), together with the respective antibiotic resistance gene (Guerout-Fleury et al., 1996) The resultant product was used to transform PY79 to obtain the mutant allele.
LR14 (mls-turboID-ywlEHAx2): turboID fragment was PCR amplified from pLR2 (TurboID-His6_pET21a) (Addgene), and fused to N-terminal region of YwlE harboring a C-terminal region containing 2×HA tag using Gibson assembly kit (NEB, USA).
LR30 (ahrC::kan): PY79 was transformed with genomic DNA of strain BKK24250.
LR31 (rocR::mls): The ORF of rocR was replaced by mls gene using long-flanking-homology PCR with primers rocR-KO-P1-P4.
LR32 (rocR::mls, ahrC::kan): LR30 was transformed with genomic DNA of LR31.
LR33 (gudB::tet): The ORF of gudB was replaced by tet gene using long-flanking-homology PCR with primers gudB-KO-P1-P4.
LR38 (gudB::tet, rocR::mls, ahrC::kan): LR32 was transformed with genomic DNA of LR33.
LR127 (gudB::tet, rocR::mls, ahrC::kan, amyE::PIPTG-rocG-spc): LR38 was transformed with pLR4.
LR133 (gudB::tet, rocR::mls, ahrC::kan, amyE::PspoIID-rocG-cat): LR38 was transformed with pLR5.
LR134 (gudB::tet, rocR::mls, ahrC::kan, amyE::PsspE-rocG-cat): LR38 was transformed with pLR6.
LR137 (gudB::tet, rocG::kan): LR33 was transformed by genomic DNA of strain BKK37790.
LR197 (rocABC::spc): The ORF of rocABC was replaced by spc gene using a long-flanking-homology PCR with primers rocABC-KO-P1-P4.
LR198 (rocDEF::cat): The ORF of rocDEF was replaced by cat gene using a long-flanking-homology PCR with primers rocDEF-KO-P1-P4.
LR200 (gudB::tet, rocABC::spc): LR197 was transformed with genomic DNA of LR33.
LR201 (gudB::tet, rocDEF::cat): LR198 was transformed with genomic DNA of LR33.
LR203 (gudB::tet, rocABC::spc, rocDEF::cat): LR200 was transformed with genomic DNA of LR198.
LR205 (rocR::mls, ahrC::kan amyE::rocR-ahrC-cat): LR32 was transformed with pLR7.
LR207 (gudB::tet, rocR::mls, ahrC::kan amyE::rocR-ahrC-cat): LR38 was transformed with pLR7.
LR209 (gudB::tet, amyE::PspoIIQ-luc H245F-cat): LR33 was transformed with genomic DNA of BDP87.
LR211 (gudB::tet, rocR::mls, ahrC::kan, amyE::PspoIIQ-luc H245F-cat): LR38 was transformed with genomic DNA of BDP87.
LR225 (sacA::PIPTG-rocG-spc): PIPTG-rocG-spc fragment was PCR amplified from genomic DNA of LR127, and cloned into sacA locus using Gibson Assembly Kit (NEB, USA).
LR227 (gudB::tet, rocR::mls, ahrC::kan, amyE::PspoIIQ-luc H245F-cat, sacA::PIPTG-rocG-spc): LR211 was transformed with genomic DNA of LR225.
Details on plasmid construction
Plasmid constructions were performed in E. coli DH5α using standard methods.
pLR4 (amyE::PIPTG-rocG-spc): containing the rocG gene and IPTG inducible promoter with flanking amyE sequences and a spc gene, was constructed by amplifying the rocG gene by PCR using primers PIPTG-rocG-P1-P2. The PCR-amplified DNA was cloned into the HindIII site of pDR111 (amyE::PIPTG-spc) using Gibson Assembly Kit (NEB, USA).
pLR5 (amyE::PspoIID-rocG-cat): containing the rocG gene and spoIID promoter with flanking amyE sequences and a cat gene, was constructed by amplifying the rocG gene by PCR using primers PspoIID-P1-P2 and rocG-P1-P2. The PCR-amplified DNA was cloned into the BamHI site of pDG364 (amyE::cat) using Gibson Assembly Kit (NEB, USA).
pLR6 (amyE::PsspE-rocG-cat): containing the rocG gene and sspE promoter with flanking amyE sequences and a cat gene, was constructed by amplifying the rocG gene by PCR using primers PsspE-P1-P2 and rocG-P1-P2. The PCR-amplified DNA was cloned into the BamHI site of pDG364 (amyE::cat) using Gibson Assembly Kit (NEB, USA).
pLR7 (amyE::ProcR-rocR-PahrC-ahrC-cat): containing the rocR gene (promoter and ORF) and ahrC gene (promoter and ORF) with flanking amyE sequences and a cat gene, was constructed by amplifying the rocR gene by PCR using primers ProcR-rocR-P1-P2, PahrC-P1-P2 and ahrC-P1-P2. The PCR-amplified DNA was cloned into the BamHI site of pDG364 (amyE::cat) using Gibson Assembly Kit (NEB, USA).
Method details
Strains and general methods
B. subtilis strains are derivatives of the wild type PY79 and are listed in key resources table. Plasmids construction is described above and listed in key resources table, and primers are described in Table S1. All general methods for B. subtilis were carried out as described previously (Harwood and Cutting, 1990). Sporulation was induced at 37°C by suspending cells in Schaeffer’s liquid medium (Difco Sporulation Medium, DSM) (Harwood and Cutting, 1990), and mature spores were purified as described previously (Sinai et al., 2015). Sporulation efficiency was evaluated by comparing the number of colony forming units before and after heat treatment (80°C, 30 min) (Zhou et al., 2019). Purified spores were heat activated (75°C, 30 min) prior to germination experiments. Spore germination was induced by L-Ala (10 mM) or AGFK (2.5 mM L-Asparagine, 5 mg/mL D-glucose, 5 mg/mL D-fructose, and 50 mM KCl) at 37°C. Spore revival was induced at 37°C by suspending spores in S7 minimal medium supplemented with L-Ala (Segev et al., 2013).
Spore purification
For all the experiments, spores were purified by combination of water washing and histodenz gradient fractionation. Briefly, a 10 mL of 20 hrs DSM culture was centrifuged and washed 3 times in 10 mL DDW. The pellet was resuspended in 10 mL of DDW and kept in 4°C with constant agitation. On subsequent days, the suspension was centrifuged once and resuspended in DDW. After 7 days washing, the suspension was centrifuged and the pellet was resuspended in 400 μL of 20% histodenz solution for 30 min on ice. Aliquots (200 μL) of spores were then placed on top of 900 μL 50% histodenz. After centrifugation (15,000 RPM, 4°C, 10 min), a pellet was detected at the bottom of the tube. The pellet contains >99% pure spore population, as evaluated by phase contrast microscopy.
Light microscopy
Light microscopy was carried out as described previously (Sinai et al., 2015; Zhou et al., 2019). Briefly, bacterial cells (0.5 mL) were collected by centrifugation and resuspended in 10 μL of PBS×1. FM1-43 was used at a concertation of 1 μg/mL. For time lapse experiments, spore were placed over 1% agarose pads supplemented with L-Ala (10 mM) and incubated in a chamber where temperature was maintained at 37°C with a temperature controller (Pecon-Zeiss). Samples were photographed using Axio Observer Z1 (Zeiss), equipped with a CoolSnap HQII camera (Photometrics, Roper Scientific). System control was performed using MetaMorph software (version 7.7; Molecular Devices), whereas image analysis and processing were conducted using ImageJ 1.52p (National Institutes of Health, USA).
DPA measurements
DPA release was measured as described previously (Yi and Setlow, 2010), with some modifications. Briefly, spores at OD600 of 0.5 were germinated at 37°C with L-Ala or AGFK in a 96-well plate in 200 μL of 25 mM K-Hepes buffer (pH 7.4) supplemented with 50 μM TbCl3. DPA release was measured by quantifying Tb3+-DPA fluorescence at emission of 545 nm and excitation of 270 nm with a Spark 10M plate reader (Tecan, Switzerland).
Western blot analysis
For protein extraction, cells were suspended in PBS×1 supplemented with 0.05% SDS, Halt Protease Inhibitor (Pierce), and lysed using FastPrep (MP, 6.5, 60 s, ×3). Following cell lysis, the supernatant was additionally centrifuged at 4°C and 15, 000 RPM for 10 min, and used for western blot analysis. The extracts were incubated at 100°C for 10 min with Laemmli sample buffer. Proteins were separated by SDS-PAGE 12.5%, and electroblotted onto a polyvinylidene 48 difluoride (PVDF) transfer membrane (Immobilon-P, Millipore). For immunoblot analysis of biotinylated proteins, membranes were blocked for 45 min at room temperature (0.1% Tween-20, 3% BSA, 3% skim milk in TBS × 1), and then incubated for 1 hr at room temperature with Streptavidin-HRP (ab7403, Abcam) (1: 5000 in 0.1% Tween-20 in TBS × 1). For RocG, membranes were blocked for 45 min at room temperature (0.1% Tween-20, 5% skim milk in TBS × 1), and then incubated for 2 hrs at room temperature with anti-RocG antibody (1:10000, 0.1% Tween-20, 3% BSA in TBS × 1) (Commichau et al., 2007). Membranes were incubated for 1 hr at room temperature with anti-rabbit IgG HRP-linked antibody (1:3000 in 0.1% Tween-20, 5% skim milk in TBS × 1). For HA-tagged proteins or SigA detection, membranes were blocked for 45 min at room temperature (0.1% Tween-20, 5% skim milk in TBS × 1), and then incubated for 2 hrs at room temperature with anti-HA or anti-SigA antibody (1:5000 in 0.1% Tween-20, 5% skim milk in TBS × 1). Membranes were incubated for 1 hr at room temperature with anti-rabbit IgG HRP-linked antibody (1:3000 in 0.1% Tween-20, 5% skim milk in TBS × 1). For luciferase detection, membranes were blocked for 45 min at room temperature (0.1% Tween-20, 5% skim milk in TBS × 1), and then incubated for 1 hr at room temperature with goat anti-Luc HRP (200-103-150-0100, Rockland) antibody (1: 10000 in 0.1% Tween-20, 5% skim milk in TBS × 1) (Parrell and Kroos, 2020). For all the experiments, ECL Kit (Advansta, CA, USA) was used for final detection.
Determination of the levels of spore germination proteins
Expression levels of germination receptor subunits (GerAA, GerBC and GerKA) and SpoVAD were detected by Western blot analyses using rabbit antibodies against these proteins and a secondary antibody, as described previously (Ramirez-Peralta et al., 2012). Briefly, 125 ODs of spores were decoated at 70°C for 2 hrs with decoating solution (0.1 M DTT, 0.1 M NaCl, 0.1 M NaOH, 1% SDS), followed by 10 water washes. The decoated spores were lysed with 1 mg lysozyme, 1 mM PMSF, 1 μg RNase, 1 μg Dnase I, and 20 μg of MgCl2 in 0.5 mL TEP buffer (50 mM Tris-HCl pH 7.4, 5 mM EDTA) at 37°C for 5 min, and then incubated on ice for 20 min. The lysed spores were then disrupted using Fastprep (MP) (6.5, 60 seconds, x3), and 100 μL of the lysate was added to 100 μL Laemmli sample buffer containing 55 mM DTT (425 μL BioRad 161-0737 plus 25 μL 1 M DTT) and incubated at 23°C for one hour. Western blot analysis was carried out as described above, using rabbit antibodies against these proteins and a secondary antibody.
High pressure induced germination
Spores (OD600 of 1) were suspended in 1.5 mL of PBS and sealed in sterile flexible plastic bags. Spore suspensions were treated at 150 MPa at 37°C for various times in a FPG7100:9/2C high pressure iso-lab system (Stansted Fluid Power Ltd., Harlow, Exxes, UK) with a 2.5 liter vessel and water as the pressure-transmitting fluid and temperature regulator. The germination of high pressure treated spores was examined by light microscopy and decrease in OD600 assay.
Quantitative Real-Time PCR (qRT-PCR)
B. subtilis strains were induced to sporulate in DSM (Harwood and Cutting, 1990). At indicated time points, 1 mL of sporulating cells were collected and washed 3 times with PBS×1. RNA was extracted using FastRNA Pro Blue Kit (MP Biomedicals) as described previously (Zhou et al., 2019). Extracted RNA (2 μg) was treated with RQ1 DNase (Promega) and subjected to cDNA synthesis using iScript cDNA synthesis kit (Bio-Rad), according to the manufacturer protocol. qRT-PCR reactions were conducted using SYBR-green mix (Bio-Rad), and fluorescence detection was performed using CFX Connect Real-Time PCR Dection System (Bio-Rad), according to manufacturer instructions. qRT-PCR primers (Table S1) were designed using Primer3 software (v.0.4.0). yoxA gene was used to normalize expression data, as its expression was unchanged during vegetative growth and sporulation. The relative gene expression levels were calculated from threshold cycle (CT) values using the 2−ΔΔCT method (Schmittgen and Livak, 2008). Each assay was performed in duplicates with at least two RNA templates prepared from independent biological repeats.
In vitro ATP measurements
ATP measurement was conducted by commercial super sensitive ATP bioluminescence assay kit (Promega) according to the manufacture instructions. ATP was extracted from spores as described previously (Setlow and Kornberg, 1970). Briefly, 150 μL of spores at OD600 of 40 in water were added to 600 μL of boiling 1-propanol, and the suspension was boiled for 5 min, cooled on ice, and flash evaporated to dryness. The dry residue was suspended in 600 μL cold water on ice and the suspension was centrifuged at 4°C for 5 min. ATP was detected by luciferase using a super sensitive ATP bioluminescence assay kit (Promega). ATP concentration was determined using a standard curve of known ATP concentrations.
TurboID-based proximity labelling and MS analysis
TurboID-based proximity labelling was performed as described previously (Branon et al., 2018), with some modifications. Spores harboring TurboID-YwlE at OD600 of 1 were germinated with L-Ala or AGFK at 37°C, in 200 mL PBS × 1 supplemented with 50 μM biotin, 1 mM ATP and 5 mM MgCl2, incubation was proceeded for overnight. The germinated spores were centrifuged and washed with PBS×1. Pellets were resuspended in PBS×1 supplemented with protease inhibitors (Thermo, 78439), lysed using FastPrep (MP) (6.5, 60 s, × 3), and centrifuged. Supernatants containing a mixed population of biotin-labelled and unlabeled proteins were collected. The enrichment of biotin-labeled proteins and trypsin-digestion process were performed as previously described (Dieterich et al., 2007). Briefly, the mixture of proteins was incubated with NeutrAvidin resin (Thermo) for overnight at 4°C with constant agitation, allowing binding of biotinylated proteins to NeutrAvidin. Washing the resin for three times with pH 7.5 PBS×1 containing 1% NP-40 by centrifugation at 2000 g for 5 min at 4°C. Then, washing the resin two times with 50 mM ammonium bicarbonate followed by two washes with PBS×1. For each initial 100 μL NeutrAvidin slurry, resuspend the resin to a total volume of 87 μL in 50 mM ammonium bicarbonate, and heat for 10 min at 70°C with constant agitation. Immediately, add 18 mg of urea beads and vortex to dissolve. After cooling down, add 3.125 mM Tris-(2-carboxyethyl) phosphine (TECP), and incubate for 30 min at 23°C with constant agitation, to reduce disulfide bonds. Then, add 11.2 mM iodoacetamide to the suspension and incubate for 30 min at 23°C in the dark with constant agitation for reducing cysteine residues. Add 0.1 μg endoproteinase Lys-C per 100 μL and incubate for 4 hrs at 37°C for proteinlysis. Finally, add calcium chloride to a final concentration of 0.1 mM, and add 1 μg trypsin per 100 μL of suspension, incubate the reaction mixture overnight at 37°C with constant agitation. Briefly pulse-spin down the suspension, and transfer the supernatant containing tryptic peptides to an empty Eppendorf tube, frozen at −80°C and then subject to lyophilization. The digested samples were analyzed by LC-MS/MS on LTQ-Orbitrap (Thermo). Each presented biotin labelling dataset was based on three independent biological repeats.
Quantification and statistical analysis
Unless stated otherwise, bar charts display a mean ± SD from at least three independent biological experiments. MS Excel was used for all statistical analysis, data processing, and presentation.
Acknowledgments
We are emendable to members of the Ben-Yehuda laboratory, I. Rosenshine and A. Rouvinski (Hebrew U, IL) for valuable discussions and comments. We are grateful to J. Stülke (Gottingen U, DE) and M. Fujita (Houston U, USA) for generously providing antibodies against RocG and SigA, respectively and to Lee Kroos (Michigan State U, USA) for providing B. subtilis strains for in vivo luciferase detection. This work was supported by the National Natural Science Foundation of China (grant no. 32001658) and Agricultural Research Outstanding Talents of China (grant no. 13210317) awarded to L.R., and the Israel Science Foundation (grant no. 774/16, 308/21) awarded to S. B-Y.
Author contributions
S.B-Y., L.R., and L.S. designed the study. L.R., B.Z., R.S., and L.S. designed and performed the experiments. L.R., L.S., R.S., A.M., and S.B-Y. analyzed the data. S.B-Y. and L.S. supervised the project. L.R., L.S., and S.B-Y wrote the article.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105242.
Contributor Information
Lior Sinai, Email: lior.sinai@mail.huji.ac.il.
Sigal Ben-Yehuda, Email: sigalb@ekmd.huji.ac.il.
Supplemental information
Data and code availability
This paper does not report original code.
The data reported in this paper will be shared by the lead contact upon request.
Any additional information required for this paper in available from the lead contact upon request.
References
- Armstrong R.L., Sueoka N. Phase transitions in ribonucleic acid synthesis during germination of Bacillus subtilis spores. Proc. Natl. Acad. Sci. USA. 1968;59:153–160. doi: 10.1073/pnas.59.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balassa G., Contesse G. Macromolecular synthesis during the germination of spores of B. subtilis. I. Kinetics. Ann. Inst. Pasteur. 1965;109:683–705. [PubMed] [Google Scholar]
- Belitsky B.R., Sonenshein A.L. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 1998;180:6298–6305. doi: 10.1128/jb.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressuire-Isoard C., Broussolle V., Carlin F. Sporulation environment influences spore properties in Bacillus: evidence and insights on underlying molecular and physiological mechanisms. FEMS Microbiol. Rev. 2018;42:614–626. doi: 10.1093/femsre/fuy021. [DOI] [PubMed] [Google Scholar]
- Camp A.H., Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Ray W.K., Helm R.F., Melville S.B., Popham D.L. Levels of germination proteins in Bacillus subtilis dormant, superdormant, and germinating spores. PLoS One. 2014;9:e95781. doi: 10.1371/journal.pone.0095781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie G., Setlow P. Bacillus spore germination: knowns, unknowns and what we need to learn. Cell. Signal. 2020;74:109729. doi: 10.1016/j.cellsig.2020.109729. [DOI] [PubMed] [Google Scholar]
- Commichau F.M., Gunka K., Landmann J.J., Stülke J. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J. Bacteriol. 2008;190:3557–3564. doi: 10.1128/JB.00099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau F.M., Wacker I., Schleider J., Blencke H.M., Reif I., Tripal P., Stülke J. Characterization of Bacillus subtilis mutants with carbon source-independent glutamate biosynthesis. J. Mol. Microbiol. Biotechnol. 2007;12:106–113. doi: 10.1159/000096465. [DOI] [PubMed] [Google Scholar]
- Czaplewski L.G., North A.K., Smith M.C., Baumberg S., Stockley P.G. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol. Microbiol. 1992;6:267–275. doi: 10.1111/j.1365-2958.1992.tb02008.x. [DOI] [PubMed] [Google Scholar]
- Dieterich D.C., Lee J.J., Link A.J., Graumann J., Tirrell D.A., Schuman E.M. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat. Protoc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- Doan T., Morlot C., Meisner J., Serrano M., Henriques A.O., Moran C.P., Jr., Rudner D.Z. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A., Eichenberger P. The spore coat. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.TBS-0023-2016. [DOI] [PubMed] [Google Scholar]
- Fajardo-Cavazos P., Tovar-Rojo F., Setlow P. Effect of promoter mutations and upstream deletions on the expression of genes coding for small, acid-soluble spore proteins of Bacillus subtilis. J. Bacteriol. 1991;173:2011–2016. doi: 10.1128/jb.173.6.2011-2016.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardan R., Rapoport G., Débarbouillé M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 1997;24:825–837. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Setlow P. Isolation and characterization of superdormant spores of Bacillus species. J. Bacteriol. 2009;191:1787–1797. doi: 10.1128/JB.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Scotland M., Setlow P. Levels of germination proteins in dormant and superdormant spores of Bacillus subtilis. J. Bacteriol. 2012;194:2221–2227. doi: 10.1128/JB.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury A.M., Frandsen N., Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Harwood C.R., Cutting S.M. Wiley; Chichester ; New York: 1990. Molecular Biological Methods for Bacillus. [Google Scholar]
- Henriques A.O., Moran C.P., Jr. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Higgins D., Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012;36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huergo L.F., Dixon R. The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol. Mol. Biol. Rev. 2015;79:419–435. doi: 10.1128/MMBR.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight Z.A., Shokat K.M. Features of selective kinase inhibitors. Chem. Biol. 2005;12:621–637. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Koo B.M., Kritikos G., Farelli J.D., Todor H., Tong K., Kimsey H., Wapinski I., Galardini M., Cabal A., Peters J.M., et al. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 2017;4:291–305.e7. doi: 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korza G., Setlow B., Rao L., Li Q., Setlow P. Changes in Bacillus spore small molecules, rRNA, germination and outgrowth after extended sub-lethal exposure to various temperatures: evidence that protein synthesis is not essential for spore germination. J. Bacteriol. 2016;198:3254–3264. doi: 10.1128/JB.00583-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney P.T., Driks A., Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat. Rev. Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner J., Wang X., Serrano M., Henriques A.O., Moran C.P., Jr. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. USA. 2008;105:15100–15105. doi: 10.1073/pnas.0806301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir A. How do spores germinate? J. Appl. Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- Mutlu A., Kaspar C., Becker N., Bischofs I.B. A spore quality-quantity tradeoff favors diverse sporulation strategies in Bacillus subtilis. ISME J. 2020;14:2703–2714. doi: 10.1038/s41396-020-0721-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu A., Trauth S., Ziesack M., Nagler K., Bergeest J.P., Rohr K., Becker N., Höfer T., Bischofs I.B. Phenotypic memory in Bacillus subtilis links dormancy entry and exit by a spore quantity-quality tradeoff. Nat. Commun. 2018;9:69. doi: 10.1038/s41467-017-02477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.L., Kornberg A. Biochemical studies of bacterial sporulation and germination. 18. Free amino acids in spores. J. Biol. Chem. 1970;245:1128–1136. [PubMed] [Google Scholar]
- Noriega C.E., Schmidt R., Gray M.J., Chen L.L., Stewart V. Autophosphorylation and dephosphorylation by soluble forms of the nitrate-responsive sensors NarX and NarQ from Escherichia coli K-12. J. Bacteriol. 2008;190:3869–3876. doi: 10.1128/JB.00092-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes-Sabja D., Setlow P., Sarker M.R. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol. 2011;19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- Parrell D., Kroos L. Channels modestly impact compartment-specific ATP levels during Bacillus subtilis sporulation and a rise in the mother cell ATP level is not necessary for Pro-sigma(K) cleavage. Mol. Microbiol. 2020;114:563–581. doi: 10.1111/mmi.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P.J., Hilbert D.W. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Popham D.L., Bernhards C.B. Spore peptidoglycan. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.TBS-0005-2012. [DOI] [PubMed] [Google Scholar]
- Pu Y., Li Y., Jin X., Tian T., Ma Q., Zhao Z., Lin S.Y., Chen Z., Li B., Yao G., et al. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell. 2019;73:143–156.e4. doi: 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- Ramírez-Guadiana F.H., Meeske A.J., Wang X., Rodrigues C.D.A., Rudner D.Z. The Bacillus subtilis germinant receptor GerA triggers premature germination in response to morphological defects during sporulation. Mol. Microbiol. 2017;105:689–704. doi: 10.1111/mmi.13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Peralta A., Zhang P., Li Y.Q., Setlow P. Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores. Appl. Environ. Microbiol. 2012;78:2689–2697. doi: 10.1128/AEM.07908-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley E.P., Lopez-Garrido J., Sugie J., Liu R.B., Pogliano K. Metabolic differentiation and intercellular nurturing underpin bacterial endospore formation. Sci. Adv. 2021;7:eabd6385. doi: 10.1126/sciadv.abd6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Segev E., Rosenberg A., Mamou G., Sinai L., Ben-Yehuda S. Molecular kinetics of reviving bacterial spores. J. Bacteriol. 2013;195:1875–1882. doi: 10.1128/JB.00093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev E., Smith Y., Ben-Yehuda S. RNA dynamics in aging bacterial spores. Cell. 2012;148:139–149. doi: 10.1016/j.cell.2011.11.059. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spore germination. Curr. Opin. Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- Setlow P. Germination of spores of Bacillus species: what we know and do not know. J. Bacteriol. 2014;196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P., Christie G. Bacterial spore mRNA - what's up with that? Front. Microbiol. 2020;11:596092. doi: 10.3389/fmicb.2020.596092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P., Kornberg A. Biochemical studies of bacterial sporulation and germination. XXII. Energy metabolism in early stages of germination of Bacillus megaterium spores. J. Biol. Chem. 1970;245:3637–3644. [PubMed] [Google Scholar]
- Setlow P., Wang S., Li Y.Q. Germination of spores of the orders Bacillales and Clostridiales. Annu. Rev. Microbiol. 2017;71:459–477. doi: 10.1146/annurev-micro-090816-093558. [DOI] [PubMed] [Google Scholar]
- Shah I.M., Laaberki M.H., Popham D.L., Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L., Ben-Yehuda S. Commentary: changes in Bacillus spore small molecules, rRNA, germination, and outgrowth after extended sublethal exposure to various temperatures: evidence that protein synthesis Is not essential for spore germination. Front. Microbiol. 2016;7:2043. doi: 10.3389/fmicb.2016.02043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai L., Rosenberg A., Smith Y., Segev E., Ben-Yehuda S. The molecular timeline of a reviving bacterial spore. Mol. Cell. 2015;57:695–707. doi: 10.1016/j.molcel.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A.L. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- Stannek L., Thiele M.J., Ischebeck T., Gunka K., Hammer E., Völker U., Commichau F.M. Evidence for synergistic control of glutamate biosynthesis by glutamate dehydrogenases and glutamate in Bacillus subtilis. Environ. Microbiol. 2015;17:3379–3390. doi: 10.1111/1462-2920.12813. [DOI] [PubMed] [Google Scholar]
- Steinberg W., Halvorson H.O., Keynan A., Weinberg E. Timing of protein synthesis during germination and outgrowth of spores of Bacillus cereus strain T. Nature. 1965;208:710–711. [Google Scholar]
- Stragier P., Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu. Rev. Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Sturm A., Dworkin J. Phenotypicdiversity as a mechanism to exit cellular dormancy. Curr. Biol. 2015;25:2272–2277. doi: 10.1016/j.cub.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarge B., Nafid C., Vischer N., Kramer G., Setlow P., Brul S. Investigating synthesis of the MalS malic enzyme during Bacillus subtilis spore germination and outgrowth and the Influence of spore maturation and sporulation conditions. mSphere. 2020;5:004644–e520. doi: 10.1128/mSphere.00464-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani A., Levinthal C. Ordered synthesis of proteins during outgrowth of spores of Bacillus cereus. J. Bacteriol. 1967;94:176–183. doi: 10.1128/jb.94.1.176-183.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinter V. Symposium on bacterial spores: V. Germination and outgrowth: effect of inhibitors. J. Appl. Bacteriol. 1970;33:50–59. doi: 10.1111/j.1365-2672.1970.tb05233.x. [DOI] [PubMed] [Google Scholar]
- Yi X., Setlow P. Studies of the commitment step in the germination of spores of bacillus species. J. Bacteriol. 2010;192:3424–3433. doi: 10.1128/JB.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Perkins J.B., Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Zhou B., Alon S., Rao L., Sinai L., Ben-Yehuda S. Reviving the view: evidence that macromolecule synthesis fuels bacterial spore germination. microLife. 2022;3 doi: 10.1093/femsml/uqac004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Semanjski M., Orlovetskie N., Bhattacharya S., Alon S., Argaman L., Jarrous N., Zhang Y., Macek B., Sinai L., Ben-Yehuda S. Arginine dephosphorylation propels spore germination in bacteria. Proc. Natl. Acad. Sci. USA. 2019;116:14228–14237. doi: 10.1073/pnas.1817742116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code.
The data reported in this paper will be shared by the lead contact upon request.
Any additional information required for this paper in available from the lead contact upon request.