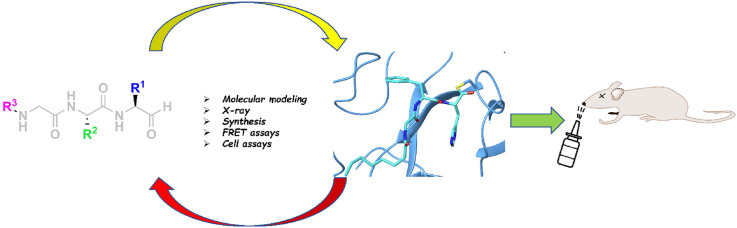
Abstract
Although vaccines are greatly mitigating the worldwide pandemic diffusion of SARS-Cov-2, therapeutics should provide many distinct advantages as complementary approach to control the viral spreading. Here, we report the development of new tripeptide derivatives of AT1001 against SARS-CoV-2 Mpro. By molecular modeling, a small compound library was rationally designed and filtered for enzymatic inhibition through FRET assay, leading to the identification of compound 4. X-ray crystallography studies provide insights into its binding mode and confirm the formation of a covalent bond with Mpro C145. In vitro antiviral tests indicate the improvement of biological activity of 4 respect to AT1001. In silico and X-ray crystallography analysis led to 58, showing a promising activity against three SARS-CoV-2 variants and a valuable safety in Vero cells and human embryonic lung fibroblasts. The drug tolerance was also confirmed by in vivo studies, along with pharmacokinetics evaluation. In summary, 58 could pave the way to develop a clinical candidate for intranasal administration.
Keywords: SARS-CoV-2, Molecular modeling, X-ray crystallography, Antiviral, Covalent inhibitors
Graphical abstract
1. Introduction
The spreading of the virus continues despite an aggressive vaccination campaign with more than 11 billion doses administered worldwide [1]. These reported statistics highlight the need for drug discovery and development of antiviral treatments in combination with an aggressive vaccination campaign to combat the pandemic. While vaccines are a central pillar of our grater efforts to fight against ongoing COVID-19 pandemic [2], small-molecule therapeutics should provide many distinct advantages maintaining a complementary approach. Most of anti-COVID therapeutics, resulting from several repurposing campaigns [3], are penalized by the requirement of specific administration protocols intended exclusively for hospital practices and applicable only to the most severe cases of infection. In this field, several progresses have been reached with monoclonal antibodies (mABs) [4], which now represent the most promising therapy for hospitalized patients. The U.S. Food and Drug Administration (FDA) authorized, under Emergency Use Authorization (EUA), 3 mAB combinations: Tixagevimab plus Cilgavimab specific for Covid-19 prophylaxis [5]; Bamlanivimab plus Etesevimab [6]; Casirivimab plus Imdevimab [7] acting as Spike protein inhibitors, and one single drug, Sotrovimab [8], sharing the same mechanism of the other antibodies. Moreover, monoclonal antibodies Tocilizumab [9] and Baricitinib [10] are used under EUA as immune system modulators in COVID-19 infection. Despite mABs are an attractive approach with potential utility in both COVID-19 prophylaxis and treatment, their use is strongly limited by the economic costs, the problematic production practice, and the global shortage in supply [4,5].
For these reasons small molecule discovery could represent a valid alternative approach to expand anti-SARS-CoV-2 therapeutic arsenal [11]. Guided by the evidence reached in the field of other coronaviruses, such as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), several viral proteins have been prioritized as SARS-CoV-2 antiviral drug targets: the spike protein, the RNA-dependent RNA polymerase (RdRp), the main protease (Mpro), and the papain-like protease (PLpro) [12,13]. The SARS-CoV-2 RdRp inhibitor Remdesivir was EUA granted from FDA on May 2020 [14]. Additional RdRp inhibitors under investigation for SARS-CoV-2 include EIDD-2801, favipiravir (T-705), ribavirin and galidesivir [15,16]. Other two drugs, namely molnupinavir and nirmatrelvir [17] are currently employed to treat COVID-19 infection, but under EUA (https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization#coviddrugs). The first one is a RpRd inhibitor, originally commercialized as anti-flu therapeutic, useable for mild infection and non-hospitalized patients [18]. The latter is the only drug specifically developed against SARS-CoV-2, not resulting from repurposing strategy. It is commercialized by Pfizer as Mpro inhibitor and it is used in combination with remdesivir [[19], [20], [21]].
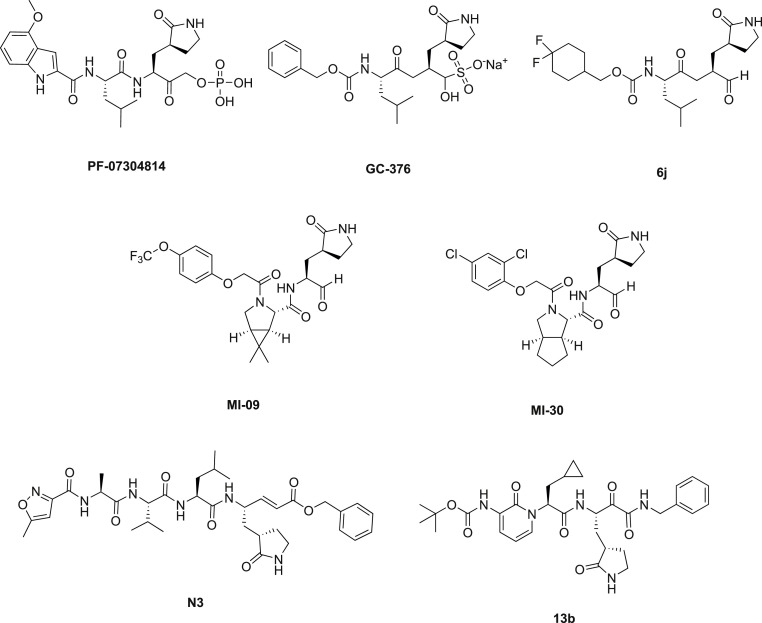
The fusion inhibitor EK1C4, which was designed based on the H2 peptide in the S2 domain of the HCoV-OC43 spike protein, showed promising broad-spectrum antiviral activity against SARS-CoV-2, SARS-CoV, and MERS-CoV, as well as other human coronaviruses HCoV-229E, HCoV-NL63 and HCoV-OC43 [22,23]. Finally, the main protease Mpro has been extensively explored as a drug target (Fig. 1 ) not only for SARS-CoV-2 but also for enteroviruses, rhinoviruses, as well as noroviruses [24].
Fig. 1.
SARS-CoV-2 Mpro inhibitors reported in literature: PF-07304814; GC-376 and analogue 6j, MI-09 and analogue MI-30; peptidomimetic Mpro inhibitors N3 and 13b.
Also known as 3CLpro, Mpro is one of coronaviral non-structural protein that cleaves the viral polyproteins including itself (Nsp5) and generates twelve non-structural proteins (Nsp4-Nsp16) [25]. Thus, pharmacological inhibition of Mpro affects viral protein maturation, preventing the viral replication and therefore represents one of the most exploited anti-coronavirus strategies [26].
Recently, we demonstrated that the zonulin inhibitor AT1001 [27] (1, Fig. 2a) binds Mpro catalytic domain [28], starting from the observation that it shares a similar structural pattern to the peptidomimetic Mpro inhibitors N3 [29] and 13b [30]. These led to the development of a new rational and ambitious research program aimed to investigate AT1001 as a potential new inhibitor of Mpro enzyme. Indeed, these outcomes were exploited to develop a chemical workflow leading to the synthesis of five AT1001 derivatives endowed with cap groups and different sequence length [31]. These modifications increased lipophilicity, while preserving the binding properties towards the Mpro, thus facilitating the penetration of peptides across biological membranes and improving pharmacokinetic properties. Only peptide 2 (Ac-GGVLVQPG-NH2) showed a micromolar range anti-SARS-CoV-2 activity (EC50 = 17.6 ± 2.4 μM), like the reported value of N3 (EC50 = 16.77 ± 1.70 μM) [29] but lower then calpeptin (EC50 = 0.072) [32] and remdesivir (0.89 ± 0.44) [31].
Fig. 2.
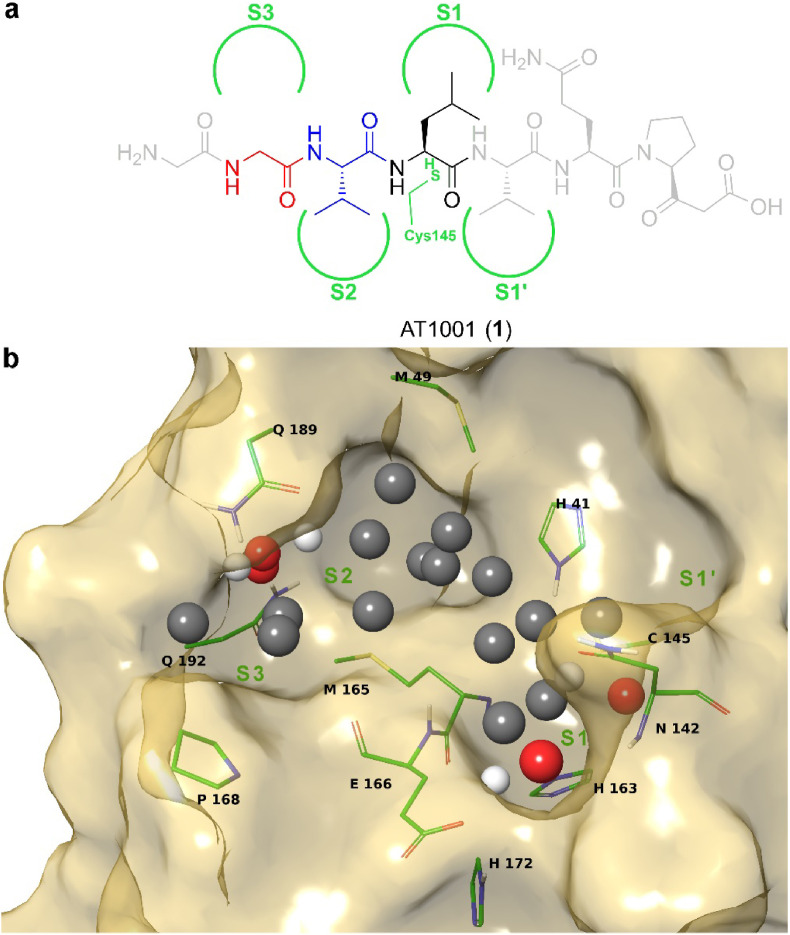
Design diagram of new tripeptide Mpro inhibitors. a) AT1001 chemical structure interacting with enzyme subpockets (green semicircles) S1′ and S1–S3, derived from our previous molecular docking studies [28]. The coloured moieties represent the tripeptide scaffold used for our design at positions P1 (black), P2 (blue) and P3 (red), whereas the grey portions show the residues removed (see main text). b) Extracted feature points for Mpro (Model A, PDB ID: 6M0K): potential hydrogen bond acceptor and donor positions are respectively shown as red and white spheres, while hydrophobic positions are represented as grey spheres.
In the present work, we discuss the process leading to the development of a new series of tripeptide derivatives of AT1001 with significantly improved in vitro and in vivo activities. Considering the previous results obtained on AT1001 analogues, we collected pivotal clues to design a new series of more potent Mpro inhibitors. The new molecules developed here as well as the overall strategy have a strong potential to lead to generally applicable anti-COVID-19 therapeutics.
2. Results and discussion
2.1. Design
Our structural studies [28,31] on AT1001 and its analogues revealed that the peptide bond between L4 and V5 of 1 is positioned at ∼3 Å from C145 (Fig. 2a), residue responsible for proteolytic activity of Mpro. Indeed, experimental investigation by FRET assays showed a bell-shaped dose-response profile, due to the enzymatic hydrolysis on small peptides [28,31]. Moreover, molecular dynamics investigations, integrated by MM-GBSA predictions, suggested that the residues G1, Q6, P7 and G8 of 1 (grey residues, Fig. 2a) largely fluctuated over the simulation contributing to a lesser extent to the affinity towards the enzyme. Combining these structural considerations, we designed a new generation of AT1001 analogues, shortening the sequence length to three amino acids and converting the carboxylic group of L4 of 1 into aldehydic warhead able to establish a reversible covalent bond with C145 of the enzyme, increasing the binding affinity over the macromolecule.
Based on this structural scheme, we generated a small library of tripeptides, preserving the H-bond network given by peptide backbone and varying the structural features of the new inserted standard and nonstandard amino acid side chains in terms of size, polarity, donor/acceptor groups of H-bonds, as suggested by binding cavity analysis through AutoSite (Fig. 2b,S1,2) [33]. Specifically, as protein model, we used three available X-ray structures of Mpro (PDB IDs: 6M0K as Model A [34], 6LZE as Model B [34] and 6LU7 as Model C [29]), because structural experiments revealed different spatial rearrangements, upon ligand binding, of residues M49 and Q189 (Fig. S3) rimming subpockets S2 and S3 [35,36]. For S1 subpocket, this analysis on Models A-C (Fig. 2b,S1,S2) suggested the advantageous placement of H-bond donors/acceptors close to residues H163 and E166, as well as hydrophobic substituents to interact with other delimiting residues (F140, N142, H163, E166, L141, H172). For the deep pocket S2 mainly delimited by H41, M49, M165, D187, Q189, hydrophobic groups could be accommodated, and H-bond donor/acceptor groups could also establish interactions with side chain of Q189. Similar accommodations and interactions could be identified for S3 delimited by M165, L167 and Q189. Moreover, the G2 was maintained or substituted with non-standard amino acids, without affecting the H-bonds with E166 backbone. The so designed compound collection (3–56, Table S1 and Scheme 1 ) was screened in silico by CovDock [37] on Models A-C and rescored by MM-GBSA (Table S2). Based on binding energies and visual inspection, including the expected accommodation of side chains (R1-3) into corresponding subpockets (S1-3) and the network of established H-bonds, the docking outcomes of all tested compounds led to a focused library of tripeptides (3–9, Scheme 1), that were useful to provide a minimum information for structure–activity relationship studies. In particular, unlike AT1001, histidine and tyrosine were chosen at P1 position in the filtered compounds (Scheme 1), as both residues looked to better interact with H163 and E166 through H-bonds, mimicking the glutamine side chain of endogenous substrate of Mpro.
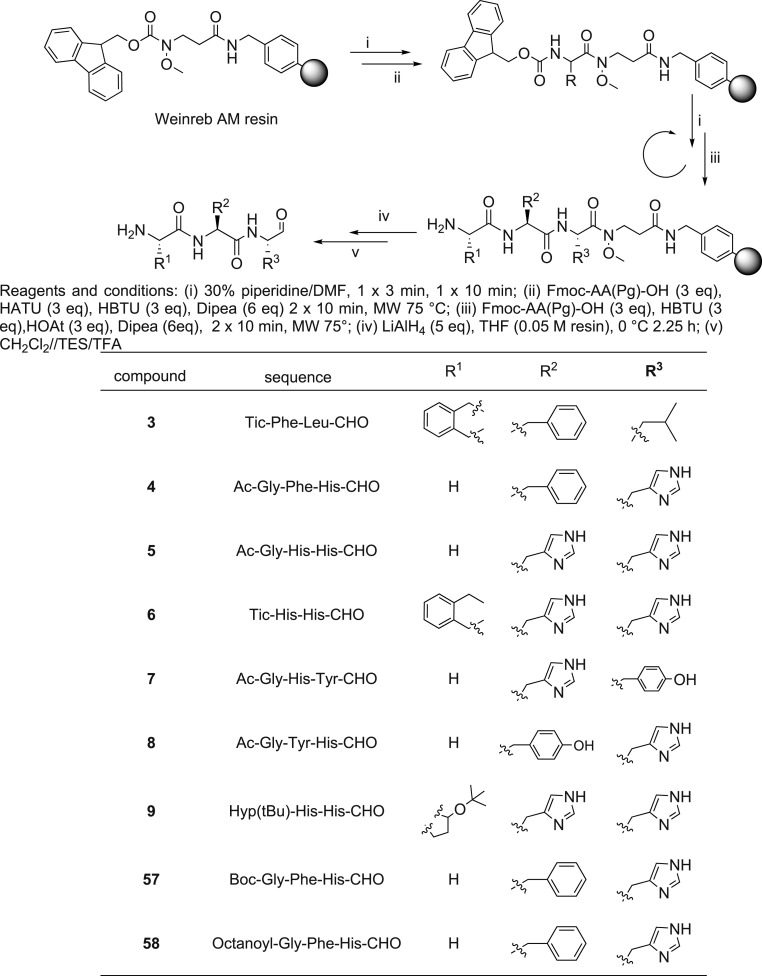
Scheme 1.
General procedure of aldehydic tripeptide (3–9, 57 and 58) synthesis.
2.2. Synthesis
Because of the interest in peptide C-modified aldehydes, several methods for solid phase synthesis of these compounds have been described. Specifically, it is possible to introduce the aldehyde function directly on the peptide, for example by the oxidation of an appropriate peptide alcohol [38,39] Weinreb amide [40,41] reduction of peptide thioesters [42] by a backbone amide linker (BAL) approach [43] or perform a step-wise synthesis using a masked pre-formed aldehyde [44,45]. Although many strategies have been developed for synthesizing peptide aldehydes, the major limitation is the low yield and the purity of these compounds, as epimerization can occur.
In this study we planned to use a common resin-bound Weinreb amide as the starting point for C N assembly of peptides as shown in Scheme 1. The correspondent C-terminal peptide aldehydes were obtained by reduction with lithium aluminum hydride (LiAlH4) in tetrahydrofuran (THF). The Crude peptides were obtained in 30% yield.
2.3. Enzymatic inhibition assay
The synthesized 3–9 compounds were investigated for inhibition of Mpro enzymatic activity by using FRET. The experiments revealed that compound 4 has the highest inhibitory activity (IC50 = 2.51 ± 0.24 μM) against Mpro (Table 1 ). It is worth of note that compound 4 showed an inhibitory activity comparable with that of calpeptin, which was used as reference compound (IC50 = 2.43 ± 0.20 μM). Moreover, compound 4 also presented a similar KD with reference, suggesting the formation of covalent bond with C145 [32] (as confirmed by X-ray experiments, see below). Compound 6 also appreciably interfere with enzymatic activity showing IC50 = 37.84 ± 5.23 μM. Lower enzyme modulation was observed for compound 8 (197.73 ± 23.77 μM), whereas the remaining analogues’ affinity (IC50) was not detectable with significantly less than 50% probe displacement at the highest compound applied concentration.
Table 1.
Enzyme inhibition profiling of 3–9 and calpeptin.
| compound | IC50 (μM) | aKD (μM) |
|---|---|---|
| 3 | >425 | >2.13 E+02 |
| 4 | 2.51 ± 0.24 | 1.26 ± 0.12 |
| 5 | >213 | >1.07 E+02 |
| 6 | 37.84 ± 5.23 | 18.92 ± 2.61 |
| 7 | >425 | >2.13 E+02 |
| 8 | 197.73 ± 23.77 | 98.86 ± 11.89 |
| 9 | >170 | >8.50 E+01 |
| 57 | 6.49 ± 0.73 | 3.26 ± 0.36 |
| 58 | 2.37 ± 0.81 | 1.19 ± 0.40 |
| calpeptin | 2.43 ± 0.20 | 1.22 ± 0.10 |
calculated by using the Cheng Prussoff equation.
2.4. Structural analysis of Mpro-ligand complex by X-ray crystallography
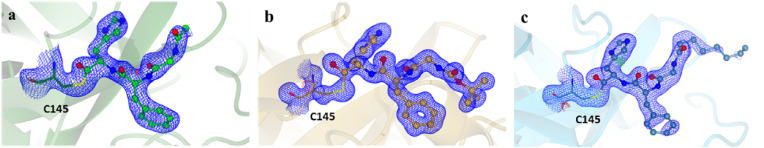
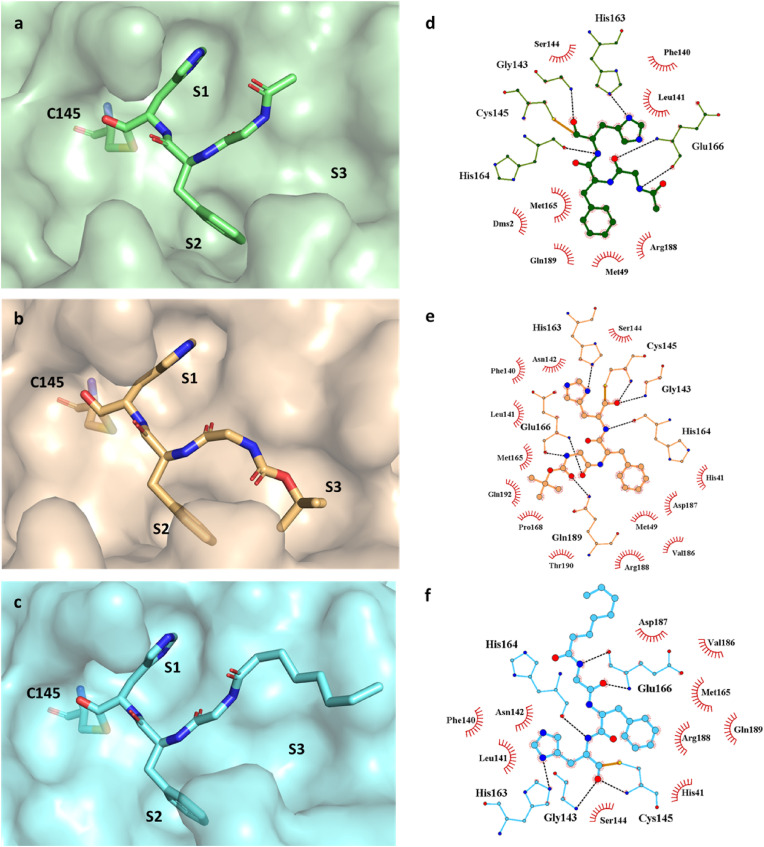
In order to obtain the structural information on Mpro-inhibitor interactions, we performed X-ray crystallographic analysis of Mpro-4 complex. Co-crystal structure of 4 (PDB ID 7ZV5) bound to Mpro were solved at 2.00 Å and crystallographic data collection and refinement statistics are presented in Supplemental Table S3. The Mpro-4 structure corresponds to the canonical SARS-CoV-2 Mpro dimer formed by two identical subunits (Fig. S4) [29,30,46,47]. For simplicity, only one subunit is shown here. The electron density corresponding to 4 can be readily identified and interpreted (Fig. 3 a). As expected, in the structure the inhibitor binds at the Mpro substrate binding pocket between domains I and II. The clear continuous electron density between aldehydic carbon and the sulphur atom of C145 confirms the formation of a covalent bond between the Mpro and 4. In the Mpro-4 complex, the side chain of His at P1 neatly fits Mpro subpocket S1 forming a hydrogen bond with Mpro H163 (Fig. 4a,d), while the carbonyl oxygen of His forms an additional H-bond with the backbone amide of G143 (Fig. 4a,d). The phenylalanine in P2 accommodates into the hydrophobic S2 pocket packing between enzyme M49 and M165 (Fig. 4a,d). Both the carbonyl and amino groups of glycine at position P3 form H-bonds with the backbone amine and carbonyl of Mpro E166, respectively (Fig. 4a,d). This approximates P4 towards subpocket S3. However, the small terminal carbonyl group of 4 at P4 is orientated towards the surface of the protein rather than the S3 pocket ( Fig. 4a,d) with non-direct interactions with Mpro.
Fig. 3.
Co-crystal structures of the covalent inhibitors bound to SARS-CoV-2 Mpro C145. The 2Fo – Fc omit map at a sigma level of 3 is shown in blue. a) Mpro bound to inhibitor 4 (PDB ID 7ZV5) in green, b) Mpro bound to inhibitor 57 (PDB ID 7ZV7) in orange, c) Mpro bound to inhibitor 58 (PDB ID 7ZV8) in cyan.
Fig. 4.
Inhibitors bind to the active site of Mpro and form covalent bound with C145. The ligand accommodation in active site represented in green surface for 4 (a), orange surface for 57 (b), cyan surface for 58 (c). The hydrogen bond interaction (dashed lines) between inhibitors and Mpro presented via LigPlot + for 4 (d), 57 (e), 58 (f).
2.5. Stability, antiviral activity and compound optimization
Before proceeding with cell assays, the stability of the most active compound (4), determined by enzymatic experiments, was evaluated in aqueous solution [48]. Compound 4 was incubated at 25 °C and 37 °C in PBS buffer at pH 7.0. Its stability was monitored by HPLC at different time points (3, 6, 8, and 24 h) post incubation. The peptide is stable in PBS at both tested temperatures for overall time (Fig. S5).
Subsequently, compound 4 was evaluated for its efficacy in inhibiting the replication of three SARS-CoV-2 variants in Vero cells (Table 2 ).
Table 2.
Activity of the synthesized peptides against SARS-CoV-2 in Vero cells.
| compound | Antiviral activity (EC50, μM)a |
Cytotoxicity (μM) |
|||
|---|---|---|---|---|---|
| UC-1074 | RG2674 | NVDBB-2220 | Cell morphology (MCC)b | Cell growth (CC50)c | |
| 4 | 66.1 ± 4.1 | >100 ± 0 | 40.3 ± 6.2 | ≥100 | >100 ± 0 |
| 57 | 50.2 ± 7.8 | 84.9 ± 26.2 | 43.3 ± 4.8 | ≥100 | >100 ± 0 |
| 58 | 5.0 ± 2.2 | 39.9 ± 12.1 | 5.2 ± 4.5 | ≥100 | >100 ± 0 |
Effective concentration required to reduce virus induced cytopathic effect by 50%. Virus input was 100 CCID50.
Minimum cytotoxic concentration that causes a microscopically detectable alteration of cell morphology.
Cytotoxic concentration required to reduce cell growth by 50%.
Compound 4 showed a comparable activity against UC-1074 (Wuhan) and NVDBB-2220 (UK) variants in micromolar range, while resulting ineffective vs. the RG2674 (South African) variant. As these outcomes could be ascribed to low membrane permeability, we tried to improve cell permeability by modifying the cap group at N-terminal as suggested by our computational predictions (Table S4) [49]. In details, we introduced BOC (compound 57) and octanoyl (compound 58) groups at position P4 in the place of acetyl of 4 (Scheme 1). Prior to the antiviral tests, in order to evaluate the effect of newly introduced cap groups to the Mpro binding, we performed X-ray crystallographic analysis of Mpro-57 and Mpro-58 complexes. Similarly to 4, compounds 57 (Fig. 3b) and 58 (Fig. 3c) bind into Mpro substrate binding pocket and form covalent bound between aldehydic carbon of 57 and 58, and the sulphur atom of Mpro C145, also in agreement with enzymatic assays (Table 1). The high-resolution structures of Mpro-57 and Mpro-58 show a similar pattern of interactions around position P1, P2 and P3 as compared to Mpro-4 but with one additional H-bond formed between the backbone carbonyl of P1 group and the amide of Mpro C145 (Fig. 4b–f). However, the three inhibitors differ significantly at P4 (Fig. S6). 57 has a BOC group at P4 which enters to the S3 subpocket. The carbonyl of BOC group establishes an H-bond with the side chain of Q189 of Mpro inducing a sharp twist in the direction of the small molecule backbone. This orients the BOC tert-butyl group towards the S3 pocket filling it completely and where it is stabilized by hydrophobic interactions with Mpro M165, P168 and carbon atoms in the side chain of Q192 (Fig. 4b,e). In 58 the terminal group at P4 is an octanoyl group and is orientated towards the surface of the protein as in 4. The octanoyl chain extends along the sidechain of P168 leaving most of the S3 pocket unoccupied (Fig. 4c,f). The octanoyl group shows partial electron density, indicating a certain degree of flexibility in this part of the molecule as would be expected from its chemical structure and the scarcity of direct interactions with Mpro.
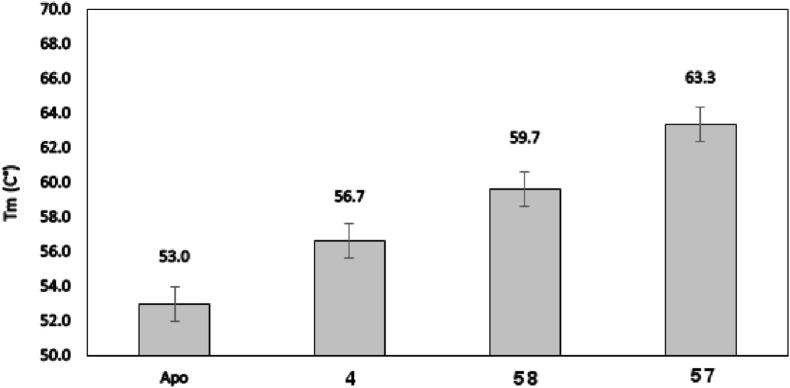
Interestingly, of the three inhibitors, 57 shows the higher level of stabilization in thermal shift assays [50,51] (Fig. 5 ), which is consistent with the observation that 57 exploits optimally interactions with the three Mpro pockets.
Fig. 5.
Evaluation of lead compounds effect of SARS-CoV-2 Mpro stability by thermal shift assay.
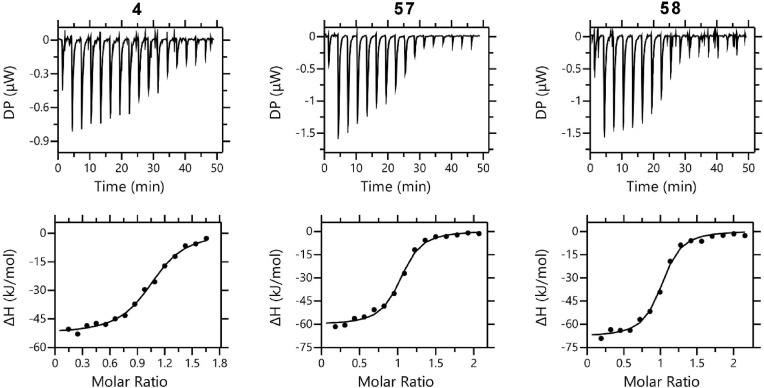
Furthermore, these three inhibitors were evaluated for their binding affinity to the Mpro and to assess their binding thermodynamics using isothermal titration calorimetry (ITC) (Fig. 6 , Table 3 ). The binding to Mpro by all three compounds is primarily driven by enthalpy. 4 binds to Mpro with K D values of 0.640 ± 0.082 μM. However, inhibitors 57 and 58 bind to Mpro with similar affinity, 0.442 ± 0.099 μM and 0.425 ± 0.075 μM, respectively. This finding is not unexpected, as all of them share a high degree of structural similarity and the binding energy is likely dominated by the formation of the covalent bond formation.
Fig. 6.
In vitro binding isotherms for the interaction of 4, 57 and 58 with SARS CoV-2 Mpro.
Table 3.
Binding affinities of covalent inhibitors 4, 57 and 58 towards Mpro determined by isothermal titration calorimetry.
| compound | KD (μM) | Stoichiometry (N) | ΔG (kJ/mol) | ΔH (kJ/mol) | -TΔS (kJ/mol) |
|---|---|---|---|---|---|
| 4 | 0.640 ± 0.082 | 1.040 ± 0.012 | −35.7 | −53.2 ± 0.8 | 17.4 |
| 57 | 0.442 ± 0.099 | 0.991 ± 0.015 | −36.7 | −60.8 ± 1.9 | 24.2 |
| 58 | 0.425 ± 0.075 | 0.966 ± 0.013 | −36.8 | −68.5 ± 1.3 | 31.8 |
Inhibitor 57 showed an improved antiviral activity against Wuhan (UC-1074) and South African (RG2674) variants, as compared to compound 4, while maintaining a similar profile of the progenitor vs. UK (NVDBB-2220) variant (Table 2). The peptide 58 was able to inhibit the replication of the Wuhan (UC-1074) and UK (NVDBB-2220) variants with 50% inhibitory concentrations of about 5 μM. It is worth of note that the activity against the South African variant also improved, even though is about 8-fold less effective with respect to the other investigated variants. The three peptides altered cell morphology only at concentrations above ≥100 μM and did not inhibit Vero cell growth up to a concentration >100 μM (Table 2).
Compounds 4, 57 and 58 were also evaluated against two herpesviruses (varicella-zoster virus and human cytomegalovirus) in human embryonic lung fibroblasts (Table S5). The three peptides lacked antiviral activity against these two DNA viruses. It is worth of note that the compounds showed a very low cytotoxicity also against embryonic lung fibroblasts (Table S5).
2.6. Preliminary in vivo PK studies
As compound 58 showed the best antiviral activity, it was selected for an early screening to estimate plasma concentrations and pharmacokinetic parameters obtained after oral (PO) and intranasal (IN) single administration to male C57BL6 mice. Specifically, two different doses for both routes were evaluated: 0.5 and 1.25 mg/kg for IN; 10 and 25 mg/kg for PO. Moreover, based on solubility limit, we used a much higher concentration (Table 4 ) for both administration routes than the obtained antiviral EC50 (Table 2), to also get a preliminary safety profile of 58. For all animals treated with compound 58, concentrations were measured between 0 and 8 h (all the concentrations from 8 to 24 h were not quantified), demonstrating that all animals were indeed exposed to the treatment. Concentrations increased with increasing doses. The concentrations obtained for oral administration were 10 times lower than for intranasal administration. The Cmax of compound 58 was determined at 177.97 ± 26.15 ng/mL for IN dose of 0.5 mg/kg, whereas 325.95 ± 42.45 ng/mL for IN dose 1.25 mg/kg. For PO doses of 10 and 25 mg/kg, the Cmax values were 22.72 ± 4.42 and 33.67 ± 3.42 ng/mL, respectively. The compound was rapidly excreted after 8 h from administration. The exposure AUCt/dose (the relative bioavailability) was calculated: intranasal dose 0.5 mg/kg with: 188.05 ± 32.52 h*kg*ng/mL/mg; intranasal dose 1.25 mg/kg with 150.62 ± 23.80 h*kg*ng/mL/mg; oral dose 10 mg/kg with 3.07 ± 0.92 h*kg*ng/mL/mg; oral dose 25 mg/kg with 2.00 ± 0.51 h*kg*ng/mL/mg. The highest exposure was observed with the intranasal administration dose 0.5 mg/kg.
Table 4.
PK analysis of plasma concentrations, with ± SD, after intranasal (IN) and oral (PO) administration in male C57BL6 mice.
| Route | Dose (mg/kg) | Concentration (mg/mL) | Number of animals | Blood collection times (h) | Cmax (ng/mL) | Tmax (h) | AUCt/dose (h*kg*ng/mL/mg) |
|---|---|---|---|---|---|---|---|
| IN | 0.5 | 1.0 | 3 | 0.25, 1, 2, 4, 8, 24 | 177.97 ± 26.15 | 0.25 ± 0.00 | 188.05 ± 32.52 |
| IN | 1.25 | 2.5 | 3 | 325.95 ± 42.45 | 0.25 ± 0.00 | 150.62 ± 23.80 | |
| PO | 10 | 1.0 | 3 | 22.72 ± 4.42 | 0.75 ± 0.43 | 3.07 ± 0.92 | |
| PO | 25 | 2.5 | 3 | 33.67 ± 3.42 | 0.5 ± 0.43 | 2.00 ± 0.51 |
Animals were examined for general health and welfare before administration and after each sampling. The administered compound was well tolerated at two doses for both IN and PO administration routes, as no adverse effects were observed during experiments and all animals survived.
3. Conclusions
In the present work we report the design of the second generation of peptide derivatives of zonulin inhibitor larazotide (AT1001). Specifically, driven by our structural investigations, we demonstrate that, simplifying the structure of the octapeptide progenitor by shortening the sequence length to three amino acids and by introducing at the same time an aldehyde warhead, the enzymatic inhibition activity was improved from partial inhibition (about 30%) to efficient inhibition at very low micromolar range. Thus, we identified the minimum sequence requirement to ensure tighter affinity Mpro binders, also reducing time and costs for their production. Moreover, despite other reported SARS-CoV-2 Mpro inhibitors endowed with imidazole [52,53], we show that introducing a histidine residue at P1 position is an effective strategy to mimic the glutamine residue of the natural substrate of Mpro. Incidentally, this is in line with work of Akaji et al. [54], reporting that His-aldehyde is suitable as P1 moiety for SARS-CoV-1 Mpro binding. X-ray structural studies confirmed that histidine at P1 position is able to establish H-bonds with H163 and E166 (water-mediated), while reducing the entropic loss upon binding thanks to imidazole rigidity. It is noteworthy that most of the reported SARS-CoV-2 Mpro inhibitors present in this position a γ-lactam as surrogate of glutamine, which requires additional and complex synthetic steps for its production. Thus, the use of histidine is more time and cost effective as it avoids the use of complex synthetic procedures. Moreover, unlike many reported Mpro inhibitors, our structural analysis shows that compound 57 interacts with the deep subpocket S3, beyond the hydrogen bond with side chain of Q189. These experimental observations provide new structural elements for drug design. The superimposition of ligand-Mpro complexes showed very similar binding mode and ITC experiments reveal comparable K D for 4, 57 and 58. The tests on SARS-CoV-2 infected cells confirmed that compound 58 is endowed with the right balance of affinity and cell permeability leading to higher antiviral activities. Interestingly, the tripeptides were incubated for five days in infected cells, showing a considerable antiviral activity, despite the long incubation time that could be detrimental for the bioactivity due to potential peptidase-mediated hydrolysis. Thus, this indicates the ligands have good stability. The antiviral activity of 4, 57 and 58 showed a certain variability against the three investigated SARS-CoV-2 strains. The use of Vero cells to evaluate the antiviral effects of compounds against SARS-CoV-2 is being questioned due to drug efflux and the endocytosis entry pathway of the virus. This can explain, at least in part, the variation observed for the EC50 values of compounds 4, 57 and 58 in inhibiting three different strains of SARS-CoV-2 (Table 2). Future studies will require the evaluation of these compounds in more relevant cells like Calu-3 and Caco-2. Moreover, drug design of compounds that directly act on conserved enzymes (such as the main protease or 3C-like protease (Mpro or 3CLpro), are expected to deliver compounds with broad-spectrum and effective anti-SARS-CoV-2 activity. However, SARS-CoV-2 evolution led to the emergence of a distinct pattern of mutations in each clade that exert a different impact on the viral molecular properties. Several studies have focused on the mutational impact on spike protein because of the critical role of this protein in receptor binding and antibody evasion. In contrast, less attention was given to the mutational impact on other (non)structural proteins. Therefore, it is conceivable that mutations, in less studied positions and/or proteins characteristic of certain clades, could influence protein structure and function and potential variations in the efficacy of antiviral drugs [55].
The absence of anti-cytomegalovirus and anti-varicella-zoster virus profiles by 4, 57 and 58 could suggest a specific antiviral activity against SARS-CoV-2. In particular, the anti-cytomegalovirus and anti-varicella-zoster virus assays were carried out on infected human embryonic lung cells, showing the lack of toxicity by the small molecules tested. This strong safety profile was also demonstrated by animal studies that showed the absence of adverse effects during the experiments and the survival of all mice. The pharmacokinetic studies suggest that the highest exposure is obtainable by intranasal administration dose of 0.5 mg/kg. These results lay foundation to develop a nasal spray that can be used 2–3 times per day in an easy way incrementing general applicability and patient compliance. This formulation could complement the vaccine campaign, be effective against new potential variant scaping acquired immunity and should be well-tolerated by the general population, including non-vaccinated people.
The drugs currently approved, such as Remdesivir, have limitations. In fact, remdesivir is approved by the USA-FDA to treat patients infected by COVID-19 but its clinical efficacy remains debatable. The Paxlovid administration is limited to patients that could develop severe symptoms and it presents many adverse effects such as diarrhoea, vomit, dysgeusia. Moreover, it cannot be administered to pregnant or breastfeeding women.
Our work provides key insights for the discovery of new drugs endowed with potent anti-SARS-CoV-2 activity, while presenting a low cytotoxic profile.
4. Experimental section
4.1. Computational details
The 3D structures of ligands were constructed through the Build Panel of Maestro (version 11), and successively their geometries refined applying: OPLS3 force field [56], Polak-Ribière conjugate gradient algorithm (maximum derivative <0.001 kcal/mol), GB/SA (generalized Born/surface area) [57] solvent treatment of H2O. Then, the tripeptides were processed by LigPrep [58] accounting for the protonation states at pH of 7.0 ± 1.0. Protein Preparation Wizard [59,60] was employed to process the X-ray structures of Mpro (PDB IDs: 6M0K, 6LZE and 6LU7): bond order assignment and hydrogen addition; missing side chain and loop check; check of alternate positions of the residues, side chain charge assignment at pH 7.0 ± 1.0; H-bond network improvement through the optimize preference. The H2O molecules were removed. Molecular docking predictions were carried out by CovDock [37]. The docking protocol was validated by redocking [61,62] the co-crystallized 11a and 11b with Mpro and overlapping the docked and experimental poses (Fig. S7, RMSD = 0.334 Å for 11a; Fig. S8, RMSD = 0.497 Å for 11b). The receptor grid was sized as 10 Å inner and 15.37 Å outer boxes, with a coordinate center: 11.61 (x), 11.76 (y), 68.59 (z) for Model A; −11.02 (x), 13.03 (y), 69.30 (z) for Model B; −10.80 (x), 12.53 (y), 68.70 (z) for Model C. For the nucleophilic addition reaction to aldehyde a custom SMARTS pattern was defined (see supporting information). As docking mode, the Pose Prediction (Thorough) was used, with an energy cut-off and maximum number of poses to retain for further refinement of 2.5 kcal/mol and 999, respectively. The output poses per ligand reaction site was set to 100, while the maximum number of top-scoring ligands to 1000. Reference position for docking option was applied. QuikProp [63] of Schrödinger suite was used to calculate the Predicted apparent Caco-2 cell permeability (QPPCaco), applying default parameters and Caco-2 cells as model. Maestro (version 11) was utilized for theoretic study and to generate all depictions.
4.2. AutoSite
For AutoSite [33] analysis, the grid boxes were centred as reported above and sized as 16 × 16 × 16 Å for Models A-C with the grid points spaced of 1.0 Å. For the ligand atom types, the C, HD and OA maps were calculated.
4.3. MM-GBSA
Prime [64,65] module of the Schrödinger suite was used for MM-GBSA predictions applying default parameters. As the covalent bond could not be accounted for this prediction, the C145 was converted into glycine and the aldehyde warhead was rebuilt for ligands.
4.4. Chemistry
4.4.1. Reagents
Nα-Fmoc-protected amino acids, HOBt (1-hydroxybenzotriazole), HATU (2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate), HOAt (1-Hydroxy-7-azabenzotriazole) DIPEA (N,N-diisopropylethylamine), Piperidine and Trifluoroacetic acid (TFA) were purchased from Iris Biotech (Marktredwitz, Germany). Weinreb AM resin was purchased from Novabiochem. Peptide synthesis solvents, reagents, as well as CH3CN for high-performance liquid chromatography (HPLC) were reagent grade and were acquired from commercial sources (Merk Life Science S.r.l. Italy) and used without further purification, unless otherwise noted.
4.4.2. General procedure for solid phase peptide synthesis (SPPS) using the weinreb aminomethyl (AM) resin
The synthesis of tripeptides was performed using an Automated Microwave Peptide Synthesizer from Biotage AB (Initiator + Alstra). Peptides were synthesized on a Weinreb AM resin (0.150 g, loading 0.5 mmol/g) previously Fmoc-deprotected by a 30% piperidine solution in N, N-dimethylformamide (DMF, 1 × 3 min and 1 × 10 min) at room temperature (rt). A Chloranil test was then applied. After a positive Chloranil test (coloured beads), the first amino acid, Nα-Fmoc-Xaa-OH, was linked on to the resin, using as coupling reagents HBTU (3 equiv.), HATU (3 equiv.), and DIPEA (6 equiv.) in N-methyl-2-pyrrolidone (NMP) [66]. The solvent was then filtered off and the procedure was then repeated. Chloranil test was then applied to ensure proper coupling. The peptide resin was washed with DCM (3 × ), DMF (3 × ), and DCM (3 × ) then the Fmoc deprotection protocol, described above, was repeated after each coupling step. The following protected amino acids were then added to the resin using as coupling reagent HBTU (3 equiv.), HOAt (3 equiv.), and DIPEA (6 equiv.) in NMP. All couplings were achieved for 10 min at 75 °C (2 × ) and 2 × 45 min at rt for histidine.
For peptides (4, 5, 7, 8) the N-terminal Fmoc group was removed as described above and the peptides were acetylated adding a solution of Ac2O/DCM (1:3) shaking for 30 min. Acylation of 58 was performed using octanoic acid (6 equiv.); HATU (3equiv.), HBTU (3 equiv.) and DIPEA (6 equiv.) at 10 min at 75 °C (2 × ).
4.4.3. General procedure for cleavage from weinreb AM resin
The on-resin tripeptide (1 equiv.) was swollen in dry THF (0.05 M resin) before use, in a round-bottomed flask equipped with a magnetic stirrer. The flask the flask was flushed with nitrogen, then seal, place in an ice bath at 0 °C for 1 h.
LiAlH4 (5 equiv.) was added portion wise, and the mixture was allowed to stir for 2.25 h. The mixture was again cooled to 0 °C and diluted with ethyl acetate (5 mL). The mixture was then quenched with saturated Rochelle's salt solution (5 mL) and allowed to stir for 15 min to ensure quenching [67]. The mixture was then filtered using a fritted filter to remove any solid particulates. The resulting filtrate was extracted three times using ethyl acetate. The combined ethyl acetate fractions were concentrated in vacuo to yield the desired tripeptide. Finally, protecting group were removed using a cleavage mixture containing 5% TFA, 1% Triisopropylsilane (TIS) and 94% DCM for 30 min.
4.4.4. Purification and characterization
All crude peptides were purified by RP-HPLC on a preparative C18-bonded silica column (Phenomenex Kinetex Biphenyl 100 Å, 100 × 21.2 mm, 5 μm) using a Shimadzu SPD 20 A UV/VIS detector, with detection at 214 and 254 nm. Mobile phase was: (A) H2O and (B) ACN, both acidified with 0.1% TFA (v/v). Injection volume was 5000 μL; flow rate was set to 17 mL/min. The following gradient was employed: 0–18 min, 1–40% B, 18.01–20 min, 40–70% B, 20.01–21 min, 70–90% B, 21.01–23 min, returning to 1% B. Analytical purity and retention time (tr) of each peptide were determined using HPLC conditions in the above solvent system (solvents A and B) programmed at a flow rate of 0.600 ml/min, fitted with C-18 column Phenomenex, Kinetex Biphenyl 100 Å C18 column (100х3.00 mm, 2.6 μm). LC gradient was the following: 0–7 min, 1–40% B, 7.01–8 min, 40–90% B, 8.01–9 min, returning to 1% B, 9–11 min, isocratic for 2 min. All analogues showed ≥97% purity when monitored at 220 nm (Table S6, Figs. S9–S17). Homogeneous fractions, as established using analytical HPLC, were pooled and lyophilized.
Ultra high resolution mass spectra were obtained by positive ESI infusion on a LTQ Orbitrap XL mass spectrometer (Thermo Scientific, Germany), equipped with the Xcalibur software for processing the data acquired (Figs. S9–S17). The sample was dissolved in a mixture of water and methanol (50/50) and injected directly into the electrospray source, using a syringe pump, at constant flow (15 μL/min).
4.5. FRET assay
The recombinant SARS-CoV-2 Mpro (Proteros) (20 nM at a final concentration) was mixed with 85 nL compound in 100% DMSO of tested compounds, by incubating for 15 min at room temperature. Dabcyl-KTSAVLQSGFRKM-E(Edans)-NH2 substrate (5 μM, final concentration) was added appropriate volume of substrate in 10 μL (reaction volume) assay buffer solution (20 mM HEPES, pH 7.5, 1 mM DTT, 1 mM EDTA, 100 mM NaCl, 0.01% Tween-20). Measure reporter displacement after 30 min. Fluorescence signal was monitored every 30 s for 10 min. The IC50 values vs. Mpro were obtained by measuring 12 concentrations per ligand and three independent experiments. The fluorescence signal of the Edans was monitored at an emission wavelength of 500 nm by exciting at 360 nm, by means of Pherastar FSX microplate Reader. Calpeptin was used as reference to set up the experiments. All experimental data was analysed using GraphPad Prism software.
4.6. Stability test of 4 in aqueous solution
The stability of peptide 4 (0.3 mg/mL) was measured in phosphate-buffered saline (PBS, pH 7.0), kept at 25 °C and 37 °C for 24 h and monitored by measuring the peptide peak area by RP-HPLC on a C18-bonded silica column (Kinetex 50 mm x 4.6 mm, 2.6 μm C18 100 Å, Phenomenex) using a Shimadzu SPD 20 UV/Vis detector, with detection at λ 220. The column was perfused at a flow rate of 1.500 mL/min with solvent A (H2O + 0.1% TFA) and a linear gradient 5%–90% of solvent B (CH3CN + 0.1% TFA) over 9 min.
4.7. Protein expression and purification of SARS-CoV-2 Mpro
The expression construct encoding the SARS-CoV-2 Mpro protein was designed as described in Zhang et al., 2020 [30]. Briefly, the SARS-CoV-2 Mpro gene preceded by the Nsp4-Mpro cleavage site (SAVLQ ↓) and with a C-terminal modified 3C cleavage site (SGVTFQ↓GP) and His6-tag was synthesized and codon optimized for expression in E. coli (GeneArt, Regensburg, Germany). The synthetic gene was subcloned into the pGEX-6P1 expression vector via the BamHI/XhoI restriction sites. Upon recombinant protein expression, auto-cleavage of the Mpro will lead to the generation of a native N-terminus. Cleavage with 3C protease during the purification process will produce a native C-terminus. The expression construct was transformed into E. coli BL21(DE3) cells. 10 ml of an overnight starter culture was added to 1 L LB medium supplemented with 100 μg/ml carbenicillin in a 5-L shaking flask. The cultures (6 L in total) were grown at 37 °C until the OD600 was ∼0.8 and induced with 0.5 mM IPTG. After induction of the protein expression, the cultures were grown for an additional 4.5 h at 37 °C. The cells were harvested by centrifugation (30 min, 4 °C, 4600×g). The cell pellets were flash-frozen with liquid nitrogen and stored at −20 °C until further usage. The cell pellet was resuspended into running buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, 20 mM imidazole, 10% glycerol) supplemented with Benzonase® and 2 mM MgCl2. The cells were lysed by 5 consecutive passages through a Microfluidizer device and the cell lysate was cleared by centrifugation (30 min, 4 °C, 30000×g). The cleared lysate was loaded onto a 5 mL Protino® Ni-NTA column (Macherey-Nagel) pre-equilibrated with running buffer. After loading the sample, the Ni-NTA column was washed with running buffer until the UV_280 nm signal returned to baseline and then eluted in a 60 mL linear gradient going from 20 mM imidazole to 500 mM imidazole in the running buffer. The elution fractions containing the recombinant Mpro were pooled and His6-tagged 3C protease was added in a 1:5 (w:w) ratio. The mixture was dialysed overnight at 4 °C against 1 L of dialysis buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM DTT). The next day, the sample was loaded again onto a 5 mL Ni-NTA column and the untagged Mpro protein was collected in the flow through of the column, whereas the His6-tagged 3C protease and any remaining uncleaved Mpro bound to the Ni-NTA column. Finally, the untagged Mpro protein was subjected to a size exclusion chromatography (SEC) step using a HiLoad 16/600 Superdex 75 pg column (Cytiva) pre-equilibrated and run in 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM EDTA, 1 mM DTT. The elution fractions from the SEC containing Mpro were pooled, concentrated to 25 mg/mL, aliquoted, flash-frozen in liquid nitrogen and stored at −80 °C until further usage. The final yielded corresponded to ∼5 mg of Mpro per litre expression culture. The identity of the purified Mpro protein was confirmed by mass spectroscopy (performed by the EMBL Proteomics Core Facility).
4.8. Crystallization and X-ray diffraction analysis
The protein solution at 10 mg/mL in 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM EDTA, 1 mM DTT was pre-incubated 2–4 h with 2 mM compounds 4, 57 and 58 (100 mM compounds stock solution in 100% DMSO) and cleared by centrifugation at 12 000 g prior to crystallization experiments. Subsequently, high-throughput crystallization experiments were carried out at HTX facility in EMBL Grenoble using automated protocols [[68], [69], [70], [71], [72]]. Briefly, 672 different crystallization cocktails were screened for each Mpro – compound complexes using commercially available kits from Qiagen (PEG 1 & 2, ComPAS, Classics), Molecular Dimensions (JCSG, PACT), Hampton Research (PEGRx). The crystallization experiments were carried out with a crystallization robot (Mosquito, SPTLabtech). 0.1 μL of protein-compound solution and 0.1 μL of reservoir were mixed to equilibrate against 45 μL reservoir solution using the sitting-drop vapor-diffusion method at 20 °C in 96-well CrystalDirect plates (MiTeGen). Mpro-compound 4 co-crystal appeared within a day in 0.2 M Sodium nitrate, 0.1 M Bis-Tris propane pH 7.5 20% (w/v) PEG 3350. Mpro-compound 57 and Mpro-compound 58 co-crystals appeared within a week in 0.05 M Ammonium sulphate, 0.1 M Sodium Citrate, 15% (w/v) PEG8000, and 0.1 M MES pH 6.5, 12% (w/v) PEG 20000, respectively. The crystals were automatically harvested from 96-well plates using CrystalDirect Harvester [65,[73], [74], [75], [76]] and X-ray diffraction data was collected at an automated beamline ID30A-1/Massif-1 at European Synchrotron Radiation Facility (ESRF) [77]. The analysis of diffraction images, including indexing, determination of accurate cell parameters, integration and scaling were performed using autoPROC [78]. Initial phases were obtained by the molecular replacement method (PHENIX, Phaser) using a search model, PDB 5RGS for Mpro-4 and Mpro-57 complexes, and PDB 6WTM for Mpro-58 [79]. Global Phasing Limited software packages (Buster and Rhofit) were used for structure refinement [80,81]. Real space refinement was carried out with Coot 0.8.9.2 software [82]. The statistics of X-ray data processing and structure refinement are summarized in Table S3. Figures were made with CCP4MG-2.10.11, PyMOL-2.5.2 and LigPlot+ v.2.2.5 software [83,84]. The atomic coordinates and structure factors for the resulting models have been deposited in the https://www.rcsb.org/.
4.9. Thermal shift assay
Thermal shift assay (TSA) was used to identify a positive shift (stabilization of the protein) of the Mpro protein melting temperature (Tm) in presence of compounds. TSA experiments performed with Real-Time qPCR machine (Mx3005P, Agilent) in 96-well plate with final volume of 25 μL [67]. The melting curves were obtained at a protein concentration of 10 μM and 5xSYPRO Orange using buffer containing 50 mM Tris-HCl pH 7.8, 150 mM NaCl, 1 mM EDTA, 1 mM DTT. The small molecules were added to the reaction mix at 1 mM final concentration. The DMSO concentration (1%) was kept identical for all assays, including the control experiment. Scans were measured from 10 °C to 100 °C at a scanning rate of 1 °C/min. All measurements performed in triplicate and single sigmoidal transition curve obtained using Crystallographic Information Management System (CRIMS) thermofluor interface and averaged.
4.10. ITC binding assay
SEC-purified Mpro protein was diluted from the stock solution to 100 μM and dialyzed overnight at 4 °C against 25 mM TRIS-HCl pH 7.6, 20 mM NaCl and 1 mM TCEP (ITC buffer) as in Kneller D. et al. [85]. The concentration of Mpro was measured using UV–Vis spectrophotometer on its 280 nm absorbance and calculated using extension coefficient of ε = 32890. All stock compounds were dissolved in 100% DMSO and freshly diluted in ITC buffer prior to titration. In all experiments, the final DMSO concentration in injectant adjusted to 0.5%. The final DSMO concentration in the cell was also adjusted to 0.5% to avoid potential dilution heat. The titration was performed with between 20 and 25 μM of Mpro in the cell and 160–250 μM compounds in the syringe at 28 °C in a MicroCal PEAQ-ITC instrument (Malvern Panalytical LTD) with continuous stirring. Incremental injections of 2.5 μL were delivered every 180 s. For the control experiments, compounds were titrated to the buffer using the same setting and showed negligible response. Data was processed and plots were generated using the MicroCal PEAQ-ITC Analysis Software provided by Malvern instrument. The stoichiometry was set to 1 and injectant concentrations were adjusted by the software for the data normalization. The data were fitted with a one-site model and the values for enthalpy (ΔH), entropy (ΔS), dissociation constants (K D) were determined mathematically from the fit parameters.
4.11. Biological activity
Vero cells (ATCC-CCL81) were used to evaluate the activity of the peptides against SARS-CoV-2. Cells were grown in Dulbecco's Modified Eagle's Medium (DMEM, ThermoFisher, Belgium) supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate and 10 mM HEPES at 37 °C in a 5% CO2 humidified atmosphere. The SARS-CoV-19 Wuhan strain, denoted UC-1074, was isolated in Vero cells from nasopharyngeal swabs of two COVID-19 patients who had a Ct of 19 for detection of SARS-CoV-2 E protein by RT-qPCR real-time reverse transcription PCR (RT-qPCR). The UC-1074 shares the same genome sequence as the early lineage A sequences (Wuhan/WH04/2020). Two variants of concern, kindly provided by Piet Maes (Laboratory of Clinical and Epidemiological Virology, Rega Institute, KU Leuven, Belgium) were used: NVDBB-2220 (Alpha variant) and RG-2674 (Beta variant). All variants were used after 2–3 passages in cell culture. The infectious virus titer of the different variants was determined in Vero cells and expressed as 50% cell culture infectious dose (CCID50) per mL. For the antiviral assays, Vero cells were seeded in 96-well plates at a density of 1 × 104 cells per well in DMEM 10% FCS medium. After 24 h growth, the cell culture medium was removed, and cells were treated with different compound concentrations in DMEM 2% FCS and mocked-infected or SARS-CoV-2-infected with 100 CCID50/well (final volume 200 μL/well). After 5 days of incubation at 37 °C, viral cytopathic effect (CPE) was recorded microscopically, and the 50% effective concentration (EC50) was calculated for each peptide and remdesivir (reference anti-SARS-CoV-2 compound, Table S7). In parallel, the cytotoxic effects of the derivatives were assessed by evaluating the MCC (minimum cytotoxic concentration that causes a microscopically detectable alteration of cell morphology). The effects of the compounds on cell growth were as well determined by counting the number of cells with a Coulter counter in mock-infected cultures and expressed as cytostatic concentration required to reduce cell growth by 50% (CC50). All SARS-CoV-2-related work was conducted in the high-containment BSL3+ facilities of the KU Leuven Rega Institute (3CAPS) under licenses AMV 30112018 SBB 219 2018 0892 and AMV 23102017 SBB 219 2017 0589 according to institutional guidelines. The peptides were evaluated against two DNA viruses (varicella-zoster virus and human cytomegalovirus as described previously [86].
4.12. In vivo administration and sampling data
12 male C57BL6 mice around 6–8 weeks were used. The in vivo test was located in the rodent area of Eurofins |ADME BIOANALYSES. There was entirely artificial lighting in the room with a controlled cycle of 12 h light, 12 h dark. Animals had free access to food and water before and during the experiment. Process, treatment and euthanasia were conducted according to the current procedures in use at Eurofins|ADME BIOANALYSES and covered by the global project APAFIS#10796–2017072717008661 v8 authorized by the Ministère de l'enseignement supérieur de la recherche et de l'innovation. See also supporting information for further details.
Funding sources
This research was funded by the projects: Fase 2, studio multicentrico aperto per determinare la sicurezza, tollerabilità ed efficacia della larazotide acetato per l’uso urgente in pazienti. anziani a rischio per la prevenzione di danno acuto polmonare (ali) e la sindrome da distress respiratorio acuto (ards) associate a infezione da covid-19—CUP G58D20000240002—SURF 20004BP000000011 from Regione Campania; X-ray studies of new AT1001 derivatives as basis for rational design of new anti-SARS-CoV-2 Mpro inhibitors with improved pharmacokinetic and pharmacodynamic profiles (PID: 22408 to S.D.M.) from ISIDORe. Financial support was provided by the Horizon Europe EC project ISIDORE ( 101046133). This work benefited from access to the HTX Lab at EMBL Grenoble and was supported by funding from the European CommunityH2020 Program under the projects iNEXT (Grant No 653706) and iNEXT Discovery (Grant No 871037) as well as the Région Auvergne-Rhône-Alpes through the R&D Booster program.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are extremely grateful to M. Brecht Dirix for excellent technical assistance and dedication to evaluate the anti-SARS-CoV-2 activity of the derivatives and to Prof. Piet Maes for providing the SARS-CoV-2 South African variant RG2674 and the UK variant NVDBB-2220. The authors also want to thank the joint EMBL-ESRF Structural Biology Group (JSBG) for support in the use and operation of the ESRF macromolecular beamlines. ITC measurements used the platforms of the Grenoble Instruct-ERIC center (ISBG; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-0005-02) and GRAL, financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). We thank Caroline Mas for assistance and/or access to the Biophysical platform.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2022.114857.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Johns Hopkins University & Meicine, Coronavirus Research Center https://coronavirus.jhu.edu/map.html
- 2.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 3.Giordano A., Forte G., Massimo L., Riccio R., Bifulco G., Di Micco S. Discovery of new erbB4 inhibitors: repositioning an orphan chemical library by inverse virtual screening. Eur. J. Med. Chem. 2018;152:253–263. doi: 10.1016/j.ejmech.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Hwang Y.C., Lu R.M., Su S.C., Chiang P.-Y., Ko S.-H., Ke F.-Y., Liang K.-H., Hsieh T.-Y., Wu H.-C. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed. Sci. 2022;29:1–50. doi: 10.1186/s12929-021-00784-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tixagevimab and Cilgavimab (evusheld) for pre-exposure prophylaxis of COVID-19. JAMA. 2022;327(4):384–385. doi: 10.1001/jama.2021.24931. [DOI] [PubMed] [Google Scholar]
- 6.Dougan M., Nirula A., Azizad M., Mocherla B., Gottlieb R.L., Chen P., Hebert C., Perry R., Boscia J., Heller B., Morris J., Crystal C., Igbinadolor A., Huhn G., Cardona J., Shawa I., Kumar P., Adams A.C., Van Naarden J., Custer K.L., Durante M., Oakley G., Schade A.E., Holzer T.R., Ebert P.J., Higgs R.E., Kallewaard N.L., Sabo J., Patel D.R., Dabora M.C., Klekotka P., Shen L., Skovronsky D.M. Bamlanivimab plus Etesevimab in mild or moderate covid-19. N. Engl. J. Med. 2021;385:1382–1392. doi: 10.1056/NEJMoa2102685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.RECOVERY Collaborative Group Casirivimab and imdevimab in patients admitted to hospital with COVID-19 (RECOVERY): a randomised, controlled, open-label, platform trial. Lancet. 2022;399(10325):665–676. doi: 10.1016/S0140-6736(22)00163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D.R., Sarkis E., Solis J., Zheng H., Scott N., Cathcart A.L., Hebner C.M., Sager J., Mogalian E., Tipple C., Peppercorn A., Alexander E., Pang P.S., Free A., Brinson C., Aldinger M., Shapiro A.E. COMET-ICE investigators. Early treatment for covid-19 with SARS-CoV-2 neutralizing antibody Sotrovimab. N. Engl. J. Med. 2021;385(21):1941–1950. doi: 10.1056/NEJMoa2107934. [DOI] [PubMed] [Google Scholar]
- 9.Banerjee S., Mahapatra A.K. Systematic review on treatment trials of tocilizumab: a repurposing drug against COVID-19. Rev. Recent Clin. Trials. 2021;16(4):381–389. doi: 10.2174/1574887116666210824093611. [DOI] [PubMed] [Google Scholar]
- 10.Lin Z., Niu J., Xu Y., Qin L., Ding J., Zhou L. Clinical efficacy and adverse events of baricitinib treatment for coronavirus disease-2019 (COVID-19): a systematic review and meta-analysis. J. Med. Virol. 2022;94(4):1523–1534. doi: 10.1002/jmv.27482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020;6(50) doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T.C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eastman R.T., Roth J.S., Brimacombe K.R., Simeonov A., Shen M., Patnaik S., Hall M.D. Remdesivir: a review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020;6:672–683. doi: 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Anirudhan V., Du R., Cui Q., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2021;93(1):300–310. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- 16.Sheahan P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., III, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayk Bernal A., Gomes da Silva M.M., Musungaie D.B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martín-Quirós A., Caraco Y., Williams-Diaz A., Brown M.L., Du J., Pedley A., Assaid C., Strizki J., Grobler J.A., Shamsuddin H.H., Tipping R., Wan H., Paschke A., Butterton J.R., Johnson M.G., De Anda C. MOVe-OUT study group. Molnupiravir for oral treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med. 2022;386(6):509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. ACTT-1 study group members. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383(19):1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 20.Mahase E. Covid-19: Pfizer's paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ. 2021;375:n2713. doi: 10.1136/bmj.n2713. [DOI] [PubMed] [Google Scholar]
- 21.Li P., Wang Y., Lavrijsen M., Lamers M.M., de Vries A.C., Rottier R.J., Bruno M.J., Peppelenbosch M.P., Haagmans B.L., Pan Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32:322–324. doi: 10.1038/s41422-022-00618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., Qin C., Sun F., Shi Z., Zhu Y., Jiang S., Yang B., Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.T.K., Wang Q., Du L., Tan W., Wilson I.A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., Wang X., Jochmans D., Neyts J., Młynarski W., Hilgenfeld R., Drag M. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021;17:222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 26.Di Sarno V., Lauro G., Musella S., Ciaglia T., Vestuto V., Sala M., Scala M.C., Smaldone G., Di Matteo F., Novi S., Tecce M.F., Moltedo O., Bifulco G., Campiglia P., Gomez-Monterrey I.M., Snoeck R., Andrei G., Ostacolo C., Bertamino A. Identification of a dual acting SARS-CoV-2 proteases inhibitor through in silico design and step-by-step biological characterization. Eur. J. Med. Chem. 2021;226 doi: 10.1016/j.ejmech.2021.113863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Troisi J., Venutolo G., Terracciano C., Delli Carri M., Di Micco S., Landolfi A., Fasano A. The therapeutic use of the zonulin inhibitor AT-1001 (Larazotide) for a variety of acute and chronic inflammatory diseases. Curr. Med. Chem. 2021;28(28):5788–5807. doi: 10.2174/0929867328666210104110053. [DOI] [PubMed] [Google Scholar]
- 28.Di Micco S., Musella S., Scala M.C., Sala M., Campiglia P., Bifulco G., Fasano A. In silico analysis revealed potential anti-SARS-CoV-2 main protease activity by the zonulin inhibitor larazotide acetate. Front. Chem. 2021;8 doi: 10.3389/fchem.2020.628609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zha Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from 1 COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Micco S., Musella S., Sala M., Scala M.C., Andrei G., Snoeck R., Bifulco G., Campiglia P., Fasano A. Peptide derivatives of the zonulin inhibitor larazotide (AT1001) as potential anti SARS-CoV-2: molecular modelling, synthesis and bioactivity evaluation. Int. J. Mol. Sci. 2021;22(17):9427. doi: 10.3390/ijms22179427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Günther S., Reinke P.Y.A., Fernández-García Y., Lieske J., Lane T.J., Ginn H.M., Koua F.H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Pérez B., Domaracky M., Saouane S., Tolstikova A., White T.A., Hänle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlünzen F., Lourdu Xavier P., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., França B.A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J.J., Knoska J., Peña-Murillo G.E., Mashhour A.R., Hennicke V., Fischer P., Hakanpää J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A.R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T.R., Garcia-Alai M.M., Niebling S., Günther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D.C.F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C.G., Weiss M.S., Schulz E.C., Mehrabi P., Karničar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Günther S., Turk D., Hinrichs W., Chapman H.N., Pearson A.R., Betzel C., Meents A. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372:642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravindranath P.A., Sanner M.F. AutoSite: an automated approach for pseudo-ligands prediction—from ligand-binding sites identification to predicting key ligand atoms. Bioinformatics. 2016;32(20):3142–3149. doi: 10.1093/bioinformatics/btw367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giordano A., del Gaudio F., Johansson C., Riccio R., Oppermann U., Di Micco S. Virtual fragment screening identification of a novel Quinoline-5,8-dicarboxylic acid derivative as selective JMJD3 inhibitor. ChemMedChem. 2018;13:1160–1164. doi: 10.1002/cmdc.201800198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giordano A., Forte G., Terracciano S., Russo A., Sala M., Scala M.C., Johansson C., Oppermann U., Riccio R., Bruno I., Di Micco S. Identification of the 2-benzoxazol-2-yl-phenol scaffold as new hit for JMJD3 inhibition. ACS Med. Chem. Lett. 2019;10:601–605. doi: 10.1021/acsmedchemlett.8b00589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu K., Borrelli K.W., Greenwood J.R., Day T., Abel R., Farid R.S., Harder E. Docking covalent inhibitors: a parameter free approach to pose prediction and scoring. J. Chem. Inf. Model. 2014;54:1932–1940. doi: 10.1021/ci500118s. [DOI] [PubMed] [Google Scholar]
- 38.Pothion C., Paris M., Heitz A., Rocheblave L., Rouch F., Fehrentz J.-A., Martinez J. Tetrahedron Lett. 1997;38:7749–7752. [Google Scholar]
- 39.Paris M., Heitz A., Guerlavais V., Christau M., Fehrenta J.A., Martinez J. Tetrahedron Lett. 1998;39:7287–7290. [Google Scholar]
- 40.Fehrentz J.-A., Paris M., Heitz A., Velek J., Liu C.-F., Winternitz F., Martinez J. Tetrahedron Lett. 1995;36:7871–7874. [Google Scholar]
- 41.Fehrentz J.-A., Paris M., Heitz A., Velek J., Winternitz F., Martinez J. Org. Chem. 1997;62:6792–6796. [Google Scholar]
- 42.Moulin A., Martinez J., Fehrentz J.A. Synthesis of peptide aldehydes. J. Pept. Sci. 2007;13:1–15. doi: 10.1002/psc.787. [DOI] [PubMed] [Google Scholar]
- 43.Jensen K.J., Alsina J., Songster M.F., Vagner J., Albericio F., Barany G. Backbone amide linker (BAL) strategy for solid-phase synthesis of C-terminal- modified and cyclic peptides. J. Am. Chem. Soc. 1998;120:5441–5452. [Google Scholar]
- 44.McConnell R.M., York J.L., Frizzell D., Ezell C. Inhibition studies of some serine and thiol proteinases by new leupeptin analogs. J. Med. Chem. 1993;36:1084–1089. doi: 10.1021/jm00060a016. [DOI] [PubMed] [Google Scholar]
- 45.Ede N.J., Eagle S.N., Wickham G., Bray A.M., Warne B., Shoemaker K., Rosenberg S. Solid phase synthesis of peptide aldehyde protease inhibitors. Probing the proteolytic sites of hepatitis C virus polyprotein. J. Pept. Sci. 2000;6(1):11–18. doi: 10.1002/(SICI)1099-1387(200001)6:1<11::AID-PSC229>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 46.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science (New York, N.Y.) 2003;300(5626):1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 47.Gossen J., Albani S., Hanke A., Joseph B.P., Bergh C., Kuzikov M., Costanzi E., Manelfi C., Storici P., Gribbon P., Beccari A.R., Talarico C., Spyrakis F., Lindahl E., Zaliani A., Carloni P., Wade R.C., Musiani F., Kokh D.B., Rossetti G. ACS Pharmacol. Transl. Sci. 2021;4(3):1079–1095. doi: 10.1021/acsptsci.0c00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sala M., Spensiero A., Scala M.C., Pepe G., Bilotta A., Paduano F., D'Agostino S., Lanzillotta D., Bertamino A., Novellino E., Trapasso F., Gomez-Monterrey I.M., Campiglia P. Design, synthesis, biological activity, and structural analysis of lactam-constrained PTPRJ agonist peptides. ChemMedChem. 2018;13(16):1673–1680. doi: 10.1002/cmdc.201800147. [DOI] [PubMed] [Google Scholar]
- 49.Di Micco S., Pulvirenti L., Bruno I., Terracciano S., Russo A., Vaccaro M.C., Ruggiero D., Muccilli V., Cardullo N., Tringali C., Riccio R., Bifulco G. Identification by Inverse Virtual Screening of magnolol-based scaffold as new tankyrase-2 inhibitors. Bioorg. Med. Chem. 2018;26:3953–3957. doi: 10.1016/j.bmc.2018.06.019. [DOI] [PubMed] [Google Scholar]
- 50.Boras B., Jones R.M., Anson B.J., Aschenbrenner L., Bakowski M.A., Beutler N., Binder J., Chen E., Eng H., Hammond H., Hammond J., Haupt R.E., Hoffman R., Kadar E.P., Kania R., Kimoto E., Kirkpatrick M.G., Lanyon L., Lendy E.K., Lillis J.R., Logue J., Luthra S.A., Ma C., Mason S.W., McGrath M.E., Noell S., Obach R.S., O' Brien M.N., O'Connor R., Ogilvie K., Owen D., Pettersson M., Reese M.R., Rogers T.F., Rosales R., Rossulek M.I., Sathish J.G., Shirai N., Steppan C., Ticehurst M., Updyke L.W., Weston S., Zhu Y., White K.M., García-Sastre A., Wang J., Chatterjee A.K., Mesecar A.D., Frieman M.B., Anderson A.S., Allerton C. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat. Commun. 2021;12:6055. doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir. GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kitamura N., Sacco M.D., Ma C., Hu Y., Townsend J.A., Meng X., Zhang F., Zhang X., Ba M., Szeto T., Kukuljac A., Marty M.T., Schultz D., Cherry S., Xiang Y., Chen Y., Wang J. Expedited approach toward the rational design of noncovalent SARS-CoV-2 main protease inhibitors. J. Med. Chem. 2022;65(4):2848–2865. doi: 10.1021/acs.jmedchem.1c00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stille J.K., Tjutrins J., Wang G., Venegas F.A., Hennecker C., Rueda A.M., Sharon I., Blaine N., Miron C.E., Pinus S., Labarre A., Plescia J., Patrascu M.B., Zhang X., Wahba A.S., Vlaho D., Huot M.J., Schmeing T.M., Mittermaier A.K., Moitessier N. Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CLpro covalent inhibitors. Eur. J. Med. Chem. 2022;229 doi: 10.1016/j.ejmech.2021.114046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akaji K., Konno H., Mitsui H., Teruya K., Shimamoto Y., Hattori Y., Ozaki T., Kusunoki M., Sanjoh A. Structure-based design, synthesis, and evaluation of peptide-mimetic SARS 3CL protease inhibitors. J. Med. Chem. 2011;54(23):7962–7973. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- 55.Sun C., Xie C., Bu G.-L., Zhong L.-Y., Zeng M.-S. Molecular characteristics, immune evasion, and impact of SARS-CoV-2 variants. Signal Transduct. Targeted Ther. 2022;7:202. doi: 10.1038/s41392-022-01039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harder E., Damm W., Maple J., Wu C.J., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L., Kaus J.W., Cerutti D.S., Krilov G., Jorgensen W.L., Abel R., Friesner R.A. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theor. Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 57.Still W.C., Tempczyk A., Hawley R.C., Hendrickson T. Semianalytical treatment of solvation for molecular mechanics and dynamics. J. Am. Chem. Soc. 1990;112:6127–6129. [Google Scholar]
- 58.Schrödinger Release 2017-1, LigPrep. Schrödinger, LLC.; New York, NY, USA: 2017. [Google Scholar]
- 59.Protein Preparation Wizard Schrödinger LLC. Schrödinger LLC.; New York, NY, USA: 2017. [Google Scholar]
- 60.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 61.Di Micco S., Terracciano S., Cantone V., Fischer K., Koeberle A., Foglia A., Riccio R., Werz O., Bruno I., Bifulco G. Discovery of new potent molecular entities able to inhibit mPGES-1. Eur. J. Med. Chem. 2018;143:1419–1427. doi: 10.1016/j.ejmech.2017.10.039. [DOI] [PubMed] [Google Scholar]
- 62.Di Micco S., Masullo M., Bandak A.F., Berger J.M., Riccio R., Piacente S., Bifulco G. Garcinol and related polyisoprenylated benzophenones as Topoisomerase II inhibitors: biochemical and molecular modeling studies. J. Nat. Prod. 2019;82:2768–2779. doi: 10.1021/acs.jnatprod.9b00382. [DOI] [PubMed] [Google Scholar]
- 63.Schrödinger Release 2017-1, QikProp. Schrödinger, LLC.; New York, NY, USA: 2017. [Google Scholar]
- 64.Prime, Schrödinger . Schrödinger, LLC.; New York, NY, USA: 2012. LLC; Prime, Version 3.1. [Google Scholar]
- 65.Knight J.L., Krilov G., Borrelli K.W., Williams J., Gunn J.R., Clowes A., Cheng L., Friesner R.A., Abel R. Leveraging data fusion strategies in multireceptor lead optimization MM/GBSA end-point methods. J. Chem. Theor. Comput. 2014;10:3207–3220. doi: 10.1021/ct500189s. [DOI] [PubMed] [Google Scholar]
- 66.Wang S.S., Tam J.P., Wang B.S., Merrifield R.B. Enhancement of peptide coupling reactions by 4-dimethylaminopyridine. Int. J. Pept. Protein Res. 1981;18(5):459–467. doi: 10.1111/j.1399-3011.1981.tb03007.x. [DOI] [PubMed] [Google Scholar]
- 67.Wilson T.A., Tokarski R.J., Sullivan P., Demoret R.M., Orjala J., Rakotondraibe L.H., Fuchs J.R. Total synthesis of scytonemide A employing Weinreb am solid-phase resin. J. Nat. Prod. 2018;81(3):534–542. doi: 10.1021/acs.jnatprod.7b00912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cornaciu, I.; Bourgeas, R.; Hoffmann, G.; Dupeux, F.; Humm, A. S.; Mariaule, V.; Pica, A.; Clavel, D.; Seroul, G.; Murphy, P.; Márquez, J. A. The automated crystallography pipelines at the EMBL HTX facility in Grenoble. JoVE 2021;(172]. doi: 10.3791/62491. [DOI] [PubMed]
- 69.Münzker L., Petrick J., Schleberger C., Clavel D., Cornaciu I., Wilcken R., Márquez J.A., Klebe G., Marzinzik A., Jahnke W. Fragment-based discovery of non-bisphosphonate binders of Trypanosoma brucei farnesyl pyrophosphate synthase. Chembiochem. 2020;21:3096–3111. doi: 10.1002/cbic.202000246. [DOI] [PubMed] [Google Scholar]
- 70.Dupeux F., Röwer M., Seroul G., Blot D., Márquez J.A. A thermal stability assay can help to estimate the crystallization likelihood of biological samples. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011;67(11):915–919. doi: 10.1107/S0907444911036225. [DOI] [PubMed] [Google Scholar]
- 71.Mariaule V., Dupeux F., Márquez J.A. Estimation of crystallization likelihood through a fluorimetric thermal stability assay. Methods Mol. Biol. 2014;1091:189–195. doi: 10.1007/978-1-62703-691-7_13. [DOI] [PubMed] [Google Scholar]
- 72.Dimasi N., Flot D., Dupeux F., Márquez J.A. Expression, crystallization and X-ray data collection from microcrystals of the extracellular domain of the human inhibitory receptor expressed on myeloid cells IREM-1. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2007;63(3):204–208. doi: 10.1107/S1744309107004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cipriani F., Röwer M., Landret C., Zander U., Felisaz F., Márquez J.A. CrystalDirect: a new method for automated crystal harvesting based on laser-induced photoablation of thin films. Acta Crystallogr. D. 2012;68:1393–1399. doi: 10.1107/S0907444912031459. [DOI] [PubMed] [Google Scholar]
- 74.Zander U., Hoffmann G., Cornaciu I., Marquette J.P., Papp G., Landret C., Seroul G., Sinoir J., Röwer M., Felisaz F., Rodriguez-Puente S., Mariaule V., Murphy P., Mathieu M., Cipriani F., Márquez J.A. Automated harvesting and processing of protein crystals through laser photoablation. Acta Crystallogr. D: Struct. Biol. 2016;72:454–466. doi: 10.1107/S2059798316000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Márquez J.A., Cipriani F. In: Structural Genomics. Methods in Molecular Biology. Chen, Y. Chen Y., editor. Humana Press; Totowa, NJ: 2013. CrystalDirect™: a novel approach for automated crystal harvesting based on photoablation of thin films; pp. 197–203. [DOI] [PubMed] [Google Scholar]
- 76.Healey R.D., Basu S., Humm A.S., Leyrat C., Cong X., Golebiowski J., Dupeux F., Pica A., Granier S., Márquez J.A. An automated platform for structural analysis of membrane proteins through serial crystallography. Cells Rep. Methods. 2021;1(6) doi: 10.1016/j.crmeth.2021.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Svensson O., Monaco S., Popov A.N., Nurizzo D., Bowler M.W. The fully automatic characterization and data collection from crystals of biological macromolecules. Acta Crystallogr. D. 2015;71:1757–1767. doi: 10.1107/S1399004715011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., Hintze B., Hung L.W., Jain S., McCoy A.J., Moriarty N.W., Oeffner R.D., Poon B.K., Prisant M.G., Read R.J., Richardson J.S., Richardson D.C., Sammito M.D., Sobolev O.V., Stockwell D.H., Terwilliger T.C., Urzhumtsev A.G., Videau L.L., Williams C.J., Adams P.D. Macromolecular structure determination using x-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bricogne, G.; Blanc, E.; Brandl, M.; Flensburg, C.; Keller, P.; Paciorek, W.; Roversi, P.; Sharff, A.; Smart, O.; Vonrhein, C.; Womack, T. BUSTER 2011, Version 2.11.8. Global Phasing Ltd, Cambridge, United Kingdom.
- 81.Womack T., Smart O., Sharff A., Flensburg C., Keller P., Paciorek W., Vonrhein C., Bricogne G. Rhofit. Global Phasing Ltd; Cambridge, United Kingdom: 2011. Version 1.2.7. [Google Scholar]
- 82.Emsleya P., Cowtana K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 83.Rigsb R.E., Parker A.B. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 2016;44(5):433–437. doi: 10.1002/bmb.20966. [DOI] [PubMed] [Google Scholar]
- 84.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 85.Kneller D.W., Li H., Phillips G., Weiss K.L., Zhang Q., Arnould M.A., Jonsson C.B., Surendranathan S., Parvathareddy J., Blakeley M.P., Coates L., Louis J.M., Bonnesen P.V., Kovalevsky A. Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nat. Commun. 2022;13:2268. doi: 10.1038/s41467-022-29915-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuan W.Y., Chen X., Liu N.N., Wen Y.N., Yang B., Andrei G., Snoeck R., Xiang Y.H., Wu Y.W., Jiang Z., Schols D., Zhang Z.Y., Wu Q.P. Synthesis, anti-varicella-zoster virus and anti-cytomegalovirus activity of 4,5-disubstituted 1,2,3-(1h)-triazoles. Med. Chem. 2019;15(7):801–812. doi: 10.2174/1573406414666181109095239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.