Abstract
Litchi fruit is consumed across the globe for its high nutritional value and taste. The qualitative profiling of litchi fruit has been carried out by using ultra-high-performance liquid chromatography with QExactive high-resolution accurate mass spectrometry. Acidified water: methanol: acetonitrile (1:1:1) extracts from individual parts (skin, pulp, and seed) of matured litchi, were subjected to LC–MS analysis with electrospray ionization in full MS-ddMS2 mode as a non-target approach. The data was processed through compound discoverer software by the use of mzCloud and ChemSpider databases, for compound identification. We identified 77 compounds with protonated or deprotonated forms based on the polarity and their characteristic fragments are within ± 4 ppm mass error and retention time ± 0.1 min for parent and fragments. Hypoglycin B is the first time reported in litchi fruit along with hypoglycin A. Further, we verified the distribution of the identified components and differentiation of three different parts of litchi through principal component analysis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13197-022-05577-z.
Keywords: Litchi, QExactive, Orbitrap, Non-target, Polyphenols, Amino acids
Introduction
Litchi chinensis belongs to the soapberry family, Sapindaceae. The small fleshy litchi (Lychee) fruit contains a single seed with juicy, sweet pulp similar to the grape taste. Litchi fruits are initially roundish, green and on maturation, turn reddish to pinkish. The fruit of lychee has a thin and rigid peel that comes off quickly exposing a pearl-white, jelly-like edible pulp that has an excellent flavor due to the combination of acids and sugars. It is a native tropical tree of Fujian and Guangdong in China, which is the largest producer of lychees, followed by India, South Africa, and other countries in Southeast Asia. Lychee has limited distribution in India due to specific climatic and soil requirements. It is grown in the states of Bihar, Tripura, West Bengal, Uttar Pradesh, Punjab, and Haryana in India. Out of the total production in India, 74% contribution is from Bihar, with West Bengal being the second-largest lychee-producing state, followed by Tripura and Assam (Singh et al. 2002).
Litchi has been used in traditional Chinese medicine for the treatment of neural pain, swelling, and cardiovascular diseases (Zhu et al. 2019). Apart from its edible part, this fruit draws attention due to beneficial compounds (antioxidant, cancer preventive, antimicrobial, and anti-inflammatory functions) present in the pulp, skin, and seed. Further, various investigations have demonstrated it contains a high amount of bioactive compounds, exhibiting antioxidant, cancer preventive, anti-inflammatory, and antimicrobial activities (Ibrahim and Mohamed 2015; Emanuele et al. 2017). Upadhyaya and Upadhyaya (2017) have discussed the bioactive compounds present in litchi fruits and their medicinal properties. Similarly, Sarni-Manchado et al. (2000) have reported anthocyanins, proanthocyanidins, phenolic acids, and coumarins as significant components of fresh litchi pericarp extract which were linked with high antioxidant activity and a lower incidence of cancers. Epicatechin, and proanthocyanidins (B2, B4) have been reported in litchi pericarp whereas individual phenolic compounds, i.e., gallic acid, chlorogenic acid, (+)-catechin, caffeic acid, epicatechin, and rutin have also been reported in litchi pulp (Ibrahim and Mohamed 2015; Miranda-Hernández et al. 2019; Lin et al. 2014). Further, Zhang et al. (2013) have followed ethanol-based extraction for isolation of polyphenols and reported catechin and epicatechin availability in different forms (free and bounded) in litchi pulp. Alternate extraction/isolation strategies based on the type of matrix can affect the extraction efficiency of natural components. The extraction and identification of polyphenols have also been reported in litchi fruits (Qiang et al. 2015; Kessy et al. 2016).
Outbreaks of hypoglycemic encephalopathy have been reported in Asian countries near litchi-growing areas, including India, Bangladesh, and Vietnam (John and Das 2014). Various researchers have also reported a > 30% mortality rate in young children due to hypoglycemic encephalopathy, and this outbreak happened during the litchi harvesting period (Shrivastava et al. 2015; Mathew and John 2017). Few researchers reported that the presence of Methylenecyclopropyl-glycine (MCPG) in litchi fruit (seed and edible arils) was a probable agent of this illness (John and Das 2014; Shrivastava et al. 2015; Mathew and John 2017; Pulla 2015). Apart from MCPG, hypoglycin A (HGA), which is commonly found in ackee fruit, is also present in litchi and is known to be the causative agent of Jamaican Vomiting Sickness (Tanaka and Ikeda 1990; Barceloux 2009; Isenberg et al. 2016). National Centre for Disease Control, India (NCDC) and the United States (US) Centers for Disease Control and Prevention (US CDC) have investigated the relation between hypoglycin-A/MCPG present in litchi and acute encephalopathy in Muzaffarpur, Bihar. These works pointed to the presence of MCPG and hypoglycin A in litchi (Mathew and John 2017; Bowen-Forbes and Minott 2011; Shrivastava et al. 2014). Further, Shrivastava et al. (2014) have reported a high level of hypoglycin A and MCPG in unripened litchi fruits as compared to the ripen/matured fruits.
In this study, a generic extraction and acquisition were applied by using the UHPLC-QExactive Focus Orbitrap system for the identification of natural compounds from litchi fruits. The data was acquired by using Xcalibur software in the full MS-ddMS2 mode which offered simultaneously full MS (R = 70,000) as well as MSMS (R = 17,500) spectra in both (positive and negative) polarity. The data was processed through the compound discoverer software to identify the maximum number of compounds in litchi through the non-target approach.
Experimental
Chemicals, reagents, and apparatus
Methanol, acetonitrile (Optima LC–MS grade) and formic acid (88%) were procured from Fisher Scientific (Mumbai, India). Quercetin (> 95% purity) was purchased from Sigma-Aldrich, Bengaluru, India. Types of equipment were used in sample preparation were included a mixer and grinder (Maharaja India Pvt. Ltd., Delhi, India), vortex mixer (Fisher Scientific, Mumbai, India), analytical and precision balance (Aczet, CY2202, San Diego, CA), refrigerated centrifuge (Thermo Scientific™ Sorvall™ ST8 ventilated benchtop centrifuge), ultrasonic bath (Thermo Scientific, Mumbai, India). A water purification system from Thermo Scientific, Mumbai was used to generate HPLC-grade water for utilization in sample preparation, dilutions, as well as for UHPLC mobile phase preparation. Hypersep retain SPE cartridge (Thermo Scientific™, Mumbai, India).
Sample preparation
Mature litchi fruits, free from visible damage or disease, were obtained from the local area of Patna (Bihar, Latitude 25° 36′ 45.6372″ N and longitude 85° 9′ 31.9500″ E). After peeling the ripe fruits, the pericarp (skin), pulp (aril) and seed were separated from each other and were kept in cold condition (0 °C) to minimize the degradation of natural compounds for further analysis. Individual fresh samples (pericarp, pulp, and seed) were thoroughly homogenized by using mixer and grinder at room temperature. The skin was dried under controlled temperature (40 °C). After drying the skin, the sample was homogenized to get a uniform fine powder. The seed samples were cut into small pieces and homogenized with water (1:1) to get a uniform particle size. In this study, the extraction protocol reported by Koley et al. (2020) with modification and with the addition of acetonitrile was used. The homogenized samples (10 g Pulp, 2 g skin, and 2 g seed) were weighed separately into a 50 mL polypropylene centrifuge tube, immediately extracted with 10 mL acidified water (1% formic acid), acetonitrile (10 mL), and methanol (10 mL). The samples were vortexed at 3000 rpm for 2 min, followed by centrifugation at 5000 rpm for 5 min at 10 °C. The supernatant was diluted with acidified water (1:1, v/v), filtered through 0.2 µm PTFE membrane filters, and finally injected (5 µL) into the LC–MS (QExactive Focus Orbitrap).
UHPLC-QExactive analysis
The analysis was performed on ultra-high-performance liquid chromatography (Vanquish UHPLC Thermo Scientific™), coupled with a QExactive Focus (Orbitrap, Thermo Scientific, Bremen, Germany). The chromatographic separation was performed on Accucore aQ™ C18 (100 × 2.1 mm, 2.6 μm, Thermo Scientific™, Bremen, Germany) column which was operated at 40 °C. The mobile phase consisted of phase A [water: methanol (90:10, v/v) + 0.2% HCOOH] and phase B [water: methanol (10:90, v/v) + 0.2% HCOOH] with a constant flow rate (0.4 mL/min). A gradient program was used as follows: 0–1. min, 2% B, 1–11 min, 2–100% B, 11–16 min, 100% B, 16–17 min 2% B, 17–22 min, 2% B.
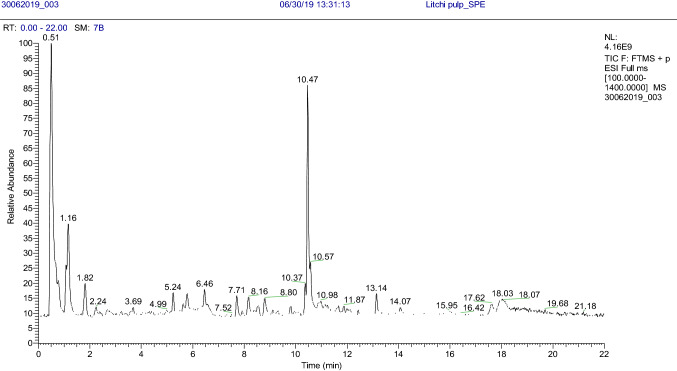
The Orbitrap was operated with heated electrospray ionization (HESI) in positive and negative polarity at the resolving power of 70,000 (full MS) and 17,500 (ddMS2 discovery). The ESI source parameters include ion spray voltage 3.5 kV; capillary temperature 320 °C; S-lens RF level 50 V; auxiliary gas (N2, > 95% purity); aux gas heater temperature 300 °C, sheath gas flow rate 40 au, aux gas flow 10 au, and sweep gas flow rate 1 au (arbitrary units). The automatic gain control was (AGC) set at a target value of 1 × 106. The data was acquired in full MS-ddMS2 mode by using Xcalibur 4.1 software. The full MS-ddMS2 mode offered a full MS with MS/MS spectrum simultaneously in a single LC run. For ddMS2, the normalized collision energy ramped from 10 to 60 v. The full MS spectrum provided information about the intact molecular ion [e.g., M+, M + H + , M-H−], whereas the ddMS2 discovery generates the product ion spectra with ramped collision energy. The mass spectrometer was calibrated weekly by using the multi-component mix to ensure mass accuracy. This data acquisition, as well as processing, was performed without any lock mass. The total ion chromatograms (TICs) for the acquired samples, i.e., pericarp (skin), the pulp (aril), and seed are given in Fig. 1.
Fig. 1.
Total ion chromatogram for litchi pulp sample acquired in full scan ddMS2 mode
Data analysis
The acquired data (n = 6) was processed by using compound discoverer 3.2 v as well as TraceFinder 5.1v software (Thermo Fisher Scientific). The peaks retention time alignment, extraction blanks subtraction, and MS/MS spectra were matched with the database. For identification and confirmation, the precursor ion and product ion mass error (± 5 ppm) criteria were fixed based on the SANTE guideline (2022). This automated screening provided the identification of compounds in comparison with databases. Structure prediction was performed, setting a mass error of ± 5 ppm, and compound prediction based on the fragmentation pattern of the mass spectra of the compounds was performed using “m/zCloud, ChemSpider databases. Through the database, the compound-specific information, i.e. chemical structure, molecular formula, molecular mass, isotopic pattern, and fragments with the mass error were obtained. Identification of the compounds was made according to the given analytical strategies:
Retention time alignment.
Evaluation of the mass error and isotopic pattern between the observed and the theoretical values.
The mass error criteria were set at ± 5 ppm for parent ion with a minimum of two fragments ions.
Retention time criteria were ± 0.1 min for the parent as well as fragments.
The MS/MS spectra comparison between the observed against theoretical fragmentation.
MS/MS spectra were confirmed through the comparison with the mzCloud and ChemSpider databases.
The mzCloud library score should be higher than 65% along with ± 5 ppm mass error.
Statistical analysis
With the help of a compound discoverer, hierarchical cluster analysis (HCA) and principal component analysis (PCA) were employed to investigate the distribution of polyphenols in different parts of litchi fruit. The typical statistical approaches used in chemometrics including PCA, and HCA for cluster analysis in food research, as demonstrated by Yi et al. (2015) were employed. Total identified components in different parts of litchi fruit samples were utilized as a measurement to obtain an HCA heat map.
Results and discussion
The chromatographic conditions were optimized such that most of the compounds would have a uniform peak width and symmetrical peaks. Also, these conditions were feasible for isomer analysis with epimer separation in reverse phase chromatography. The optimized conditions could provide comprehensive coverage including highly polar analytes (amino acids), and mid-polar analytes like quercetin, procyanidin derivatives, and free fatty acids. The samples were extracted with acidified water, methanol, and acetonitrile to achieve better extraction efficiency for a wider range of compounds i.e. polar to non-polar analytes.
Additionally, the diluted supernatant was passed through the pre-conditioned Hypersep retain SPE cartridge. The final extract was eluted with acidified methanol and acetonitrile (1 mL each). In SPE, early eluting compounds i.e. amino acids are not retained on the cartridge due to their highly polar nature. Acetonitrile extraction resulted in improved elution of procyanidins. The optimized chromatographic conditions offered better peak shapes for all the identified compounds. The acidic mobile phase provided stability to the phenolic compounds. Methanol in combination with acidified water used in the mobile phase provided improved ionization efficiency to the compounds identified in litchi. The supernatant was diluted with acidified water and acquired in full MS with ddMS2 scan mode, as mentioned above. A total ion chromatogram is presented in Fig. 1.
Identification of compounds
The acquired samples were processed through a non-target approach using Compound Discoverer software. This software has a unique feature in which an automatic peak picking and retention time alignment is performed based on accurate mass and mass accuracy. Also, the full MS and MS/MS spectra matching was confirmed with an online database like ChemSpider, mzCloud, etc. A total of 77 compounds identified in litchi fruit, including amino acids and their derivatives (12), phenolic/organic acid compounds (8), flavonols (41), and others (16) are presented in Table 1.
Table 1.
List of identified compounds with their accurate mass, formula, retention time, mass error (ppm), and product ions
| Sr. No | Name of compound | Molecular formula | Theoretical molecular weight | RT [min] | Adduct | Observed molecular ion | Mass error (ppm) | Productions/fragments | mzCloud score |
|---|---|---|---|---|---|---|---|---|---|
| Amino acids | |||||||||
| DL-Arginine | C6H14N4 O2 | 174.11179 | 0.52 | [M + H] + | 175.11913 | − 0.65 | 158.09244, 130.09749, 116.07096, 70.06600, 60.05656, 112.08692 | 93 | |
| DL-Tryptophan | C11H12N2O2 | 204.09010 | 1.72 | [M + H] + | 205.09732 | − 1.07 | 159.09167, 170.06001, 146.06009, 132.08078, 117.06544 | 95 | |
| Hypoglycin A | C7H11NO2 | 141.07911 | 0.75 | [M + H] + | 142.08640 | − 0.97 | 125.02332, 96.08135, 74.02448, 69.07070, 56.05046 | 80 | |
| Hypoglycin B | C12H19N2O5 | 270.121571 | 1.80 | [M + H] + | 271.12897 | 0.44 | 208.09708, 142.08640, 96.08141, 74.02452 | Chemspider | |
| L-Glutamic acid | C5H9NO4 | 147.05322 | 0.54 | [M + H] + | 148.06052 | − 0.43 | 84.04513, 102.05547, 131.05336, | 86 | |
| L-Glutathione (reduced) | C10H17N3O6S | 307.08383 | 0.57 | [M + H] + | 308.09116 | − 0.09 | 233.05875, 179.04848, 162.02194, 130.0500, 116.01664, 84.04507, 76.02235 | 96 | |
| L-Phenylalanine | C9H11NO2 | 165.07913 | 1.036 | [M + H] + | 166.08644 | − 0.90 | 131.04919, 120.08105, 103.05468, 79.05493 | 90 | |
| L-Pyroglutamic acid | C5H7NO3 | 129.04282 | 0.611 | [M + H] + | 130.05011 | − 1.77 | 85.04775, 84.04516, 56.04935 | 74 | |
| L-Tyrosine | C9H11NO3 | 181.07410 | 0.644 | [M + H] + | 182.08136 | − 1.14 | 165.05464, 147.04408, 136.07578, 123.04427, 119.04948, 91.05489, 95.04971 | 91 | |
| L-Valine | C5H11NO2 | 117.07937 | 0.573 | [M + H] + | 118.08661 | − 3.33 | 101.06020, 72.08160, 59.05008, 55.05519 | 68 | |
| Methionine | C5H11NO2S | 149.05122 | 0.603 | [M + H] + | 150.05833 | − 1.13 | 133.03185, 104.05330, 102.05544, 74.02446, 61.01157, 56.05044 | 90 | |
| Prolylleucine | C11H20N2O3 | 228.14755 | 0.572 | [M + H] + | 229.15477 | − 0.67 | 70.06600, 230.15807 | 84 | |
| Phenolic/ other acids derivatives | |||||||||
| 4-Coumaric acid | C9H8O3 | 164.04751 | 0.64 | [M + H] + | 165.05478 | − 1.02 | 119.04951, 147.04414, 91.05488 | 79 | |
| 4-Methoxysalicylic acid | C8H8O4 | 168.04237 | 4.68 | [M + H] + | 169.04953 | − 0.65 | 151.03903, 170.09627 | 66 | |
| 4-Ethoxybenzaldehyde | C9H10O2 | 150.06823 | 4.95 | [M + H] + | 151.07545 | − 0.99 | 123.08070, 105.07026, 95.08609, 67.05510 | 67 | |
| 4-Guanidinobutyric acid | C5H11N3O2 | 145.08524 | 0.55 | [M + H] + | 146.09261 | − 0.76 | 128.08206, 111.05589, 104.07115, 87.04482, 86.06082, 60.05663 | 91 | |
| trans-3-Indoleacrylic acid | C11H9NO2 | 187.06348 | 1.732 | [M + H] + | 188.07077 | − 0.80 | 170.0598, 146.06010, 118.06548, 144.08083, 147.06349 | 88 | |
| α-Eleostearic acid | C18H30O2 | 278.22458 | 11.09 | [M + H] + | 279.23181 | 0.01 | 137.13243, 123.11710, 109.10163, 95.08614, 81.07065, 67.05513, 209.15385 | 85 | |
| γ-Linolenic acid ethyl ester | C20H34O2 | 306.25594 | 11.96 | [M + H] + | 307.26303 | − 0.21 | 261.22083, 243.21005, 123.11713, 109.10153, 95.08602, 81.07057, 67.05509 | 82 | |
| Caffeic acid | C9H8O4 | 180.04226 | 7.71 | [M + H] + | 181.04938 | − 0.15 | 163.03893, 145.04941 | ChemSpider | |
| Phenolic compounds | |||||||||
| Rhamnetin-3-O-neohesperidoside [5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4-oxo-4H-chromen-3-yl 6-O-(6-deoxy-α-L-mannopyranosyl) hexopyranoside] | C28H32O16 | 624.16896 | 5.81 | [M + H] + | 625.17609 | 0.12 | 317.06549, 129.05466, 85.02914, 71.05002, 479.11840 | 79 | |
| Rhamnetin-3-O-neohesperidoside [5,7-Dihydroxy-2-(4-hydroxy-3-methoxyphenyl)-4-oxo-4H-chromen-3-yl 6-O-(6-deoxy-α-L-mannopyranosyl) hexopyranoside] | C28H32O16 | 624.16905 | 6.45 | [M + H] + | 625.17639 | − 0.02 | 480.12292, 479.11832, 317.06549, 147.06502, 129.05487, 85.02911, 71.05003 | 79 | |
| Catechin | C15H14O6 | 290.07921 | 2.06 | [M + H] + | 291.08649 | − 0.59 | 207.06514, 165.05458, 147.04406, 139.03905, 123.04433 | 94 | |
| Epicatechin | C15H14O6 | 290.07912 | 3.63 | [M + H] + | 291.08652 | − 0.30 | 207.06497, 147.04406, 165.05464, 139.03900, 123.04431 | 94 | |
| Flavanone | C15H12O2 | 224.08385 | 9.21 | [M + H] + | 225.09114 | − 0.54 | 197.09612, 165.06966, 93.03346 | 68 | |
| Isoleucine | C6H13NO2 | 131.09484 | 0.78 | [M + H] + | 132.10216 | − 1.60 | 86.09714, 70.07398 | 83 | |
| Isorhamnetin 3-rhamnosyl-(1- > 4)-rhamnosyl-(1- > 6) glucoside | C34H42O20 | 770.22694 | 5.77 | [M + H] + | 771.23364 | − 0.74 | 625.17554, 479.11795, 317.06525, 147.06518, 129.05467 | ChemSpider | |
| Kaempferol-7-O-glucoside | C21H20O11 | 448.10063 | 5.64 | [M + H] + | 449.10797 | − 0.15 | 287.05469, 153.01775, 137.13284, 85.02903 | 76 | |
| Luteolin [I] | C15H10O6 | 286.04765 | 5.64 | [M + H] + | 287.05490 | 0.29 | 153.01816, 288.05820 | 93 | |
| Luteolin [II] | C15H10O6 | 286.04757 | 6.31 | [M + H] + | 287.05496 | 0.58 | 153.01821, 288.05826 | 93 | |
| Naringenin [I] | C15H12O5 | 272.06848 | 4.82 | [M + H] + | 273.07581 | − 0.03 | 171.02220, 153.01830, 147.04402, 119.04933 | 96 | |
| Naringenin [II] | C15H12O5 | 272.06849 | 5.05 | [M + H] + | 273.07584 | − 0.07 | 153.01833, 147.04405, 119.04939 | 97 | |
| Naringenin [III] | C15H12O5 | 272.06852 | 5.97 | [M + H] + | 273.07593 | − 0.16 | 171.02882, 153.01833, 147.04408, 119.04961, | 95 | |
| Naringin [I] | C27H32O14 | 580.17924 | 5.49 | [M + H] + | 581.18634 | − 0.07 | 435.12933, 419.13428, 273.07547, 171.02853, 171.02880, 147.04419, 129.05470, 85.02910 | 75 | |
| Naringin [II] | C27H32O14 | 580.17924 | 5.97 | [M + H] + | 581.18616 | − 0.06 | 435.12857, 419.13327, 315.08621, 273.07556, 219.02855, 195.02865, 153.01820, 147.04398, 85.02914 | 86 | |
| Naringin dihydrochalcone | C27H34O14 | 582.19487 | 5.59 | [M + H] + | 583.20215 | − 0.03 | 437.14398, 421.14905, 317.10144, 275.09109, 169.04976, 129.05462, 107.04958, 85.02910 | 71 | |
| Nictoflorin (Kaempferol 3-O-rutinoside) [I] | C27H30O15 | 594.15840 | 5.64 | [M + H] + | 595.16571 | 0.11 | 449.10770, 287.05490, 147.06519, 129.05481, 85.02841, 71.04914 | 83 | |
| Nictoflorin (Kaempferol 3-O-rutinoside) [II] | C27H30O15 | 594.15848 | 6.32 | [M + H] + | 595.16577 | − 0.02 | 450.11069, 449.10757, 287.05481, 288.05823, 145.04919, 129.05462, 85.02911, 71.05003 | 83 | |
| Phloretin [I] | C15H14O5 | 274.08409 | 5.82 | [M + H] + | 275.09128 | 0.12 | 169.04942, 107.04958, 108.05281 | 92 | |
| Phloretin [II] | C15H14O5 | 274.08412 | 5.59 | [M + H] + | 275.09143 | 0.02 | 170.05321, 149.05942, 107.04964 | 89 | |
| Pinocembrin | C15H12O4 | 256.07360 | 7.14 | [M + H] + | 257.08078 | − 0.16 | 153.01830, 131.04929, 103.05470, 67.01885 | 80 | |
| Procyanidin tetramer | C60H50O24 | 1154.26920 | 3.88 | [M-H] + | 1153.25842 | − 2.08 | 863.17987, 575.11780, 407.07617, 289.07135, 287.05554,161.02299, 125.02283 | ChemSpider | |
| Procyanidin C1 (Procyanidin trimer) | C45H39O18 | 866.20581 | 3.85 | [M + H] + | 867.21263 | − 0.53 | 715.16541, 579.14886, 409.09079, 289.07043, 291.08653, 139.03891, 163.03886, 123.04420 | ChemSpider | |
| Procyanidin C1 (Procyanidin trimer) | C45H39O18 | 866.20581 | 3.85 | [M-H]− | 865.19666 | − 1.10 | 713.14771, 577.13464, 407.07632, 287.05542, 289.0742, 161.02298, 125.02280 | ChemSpider | |
| Procyanidin C1 (Procyanidin trimer) | C45H39O18 | 866.20581 | 2.97 | [M-H]− | 865.19659 | − 0.97 | 575.11914, 713.15167, 407.07654, 287.05530, 289.07040, 161.02293, 125.02277 | ChemSpider | |
| Procyanidin B type trimer | C45H36O18 | 864.19016 | 3.52 | [M-H]− | 863.18073 | − 1.23 | 711.13470, 573.10266, 411.07138, 289.07111, 287.05525, 243.02890, 125.02282 | ChemSpider | |
| Procyanidin B2 | C30H26O12 | 578.1424 | 3.18 | [M-H]− | 577.12592 | − 0.54 | 109.02802, 125.02276, 151.03868, 161.02312, 245.08130, 289.07101, 407.07611, 425.08888, 451.10266 | ChemSpider | |
| Procyanidin A2 | C30H24O12 | 576.12678 | 4.71 | [M-H]− | 575.11804 | − 0.62 | 449.0889, 423.07117, 285.03967, 289.07015, 163.00226, 125.02283, 109.02793 | ChemSpider | |
| Procyanidin A type tetramer | C60H48O24 | 1152.25355 | 3.62 | [M-H]− | 1151.24330 | − 1.63 | 863.1970, 711.14633, 575.12941, 423.07780, 285.04372, 449.8835 | ChemSpider | |
| Pyridoxine | C8H11NO3 | 169.07396 | 0.597 | [M + H] + | 170.08122 | − 0.42 | 152.07069, 134.06009, 153.07437 | 94 | |
| Quercetin | C15H10O7 | 302.04261 | 5.24 | [M + H] + | 303.04993 | 0.15 | 257.04443, 229.04922, 165.01845, 153.01834, 137.02342 | 92 | |
| Quercetin-3-β-D-glucoside [I] | C21H20O12 | 464.09559 | 5.24 | [M + H] + | 465.10297 | − 0.25 | 303.04984, 229.04947, 145.04958, 127.03898, 97.02841, 69.03439 | 86 | |
| Quercetin-3-β-D-glucoside [II] | C21H20O12 | 464.09563 | 5.77 | [M + H] + | 465.10278 | − 0.34 | 419.17548, 303.04947, 304.05292, 97.02890, 91.03960, 85.02908 | 82 | |
| Quercetin 3-(2Gal-rhamnosyl-robinobioside) | C33H40O20 | 756.21129 | 5.22 | [M + H] + | 757.21824 | − 0.43 | 303.04962, 465.10260, 611.16040, 129.05453, 147.06506, 111.04434 | ChemSpider | |
| Rhamnetin | C16H12O7 | 316.05830 | 5.81 | [M + H] + | 317.06561 | 0.01 | 302.04169 | 84 | |
| Robinin (Kaempferol-3-O-rhamninoside) | C33H40O19 | 740.21639 | 5.64 | [M + H] + | 741.22363 | − 0.01 | 595.16516, 449.10959, 433.11264, 287.05490, 147.06512, 71.05003, 85.02841 | 86 | |
| Rutin [I] | C27H30O16 | 610.15348 | 5.243 | [M + H] + | 611.16077 | − 0.15 | 465.10324, 303.04987, 304.05316, 147.06549, 129.05478, 85.02914, 71.05002 | 92 | |
| Rutin [II] | C27H30O16 | 610.15346 | 5.77 | [M + H] + | 611.16071 | − 0.13 | 465.10303, 466.10660, 303.04987, 304.05316, 229.04987, 147.06523, 129.05479, 85.02913, 71.05003 | 93 | |
| Scopoletin | C10H8O4 | 192.04245 | 4.52 | [M + H] + | 193.04970 | − 0.99 | 175.14775, 137.09599, 133.02849 | 79 | |
| Taxifolin | C15H12O7 | 304.05852 | 3.045 | [M + H] + | 305.06583 | − 0.71 | 287.05453, 259.06009, 231.06529, 195.02855, 167.03389, 153.01830, 149.02339, 123.04433 | 90 | |
| Trigonelline | C7H7NO2 | 137.04778 | 0.537 | [M + H] + | 138.05510 | − 0.75 | 110.06063, 94.06583, 139.05856 | 99 | |
| Other compounds | |||||||||
| 5′-S-Methyl-5′-thioadenosine | C11H15N5O3S | 297.08978 | 1.64 | [M + H] + | 298.09689 | − 0.74 | 136.06183, 163.04205, 61.01157, 163.04234 | 81 | |
| 6-Hydroxy-8-methoxy-3-methyl-3,4-dihydro-1H-isochromen-1-one | C11H12O4 | 208.07375 | 5.19 | [M + H] + | 209.08105 | − 0.90 | 191.07008, 163.07561, 181.08609 | 74 | |
| 5-(1-Hydroxyethyl)-3-(2-hydroxypropyl)-2(5H)-furanone | C9H14O4 | 186.08929 | 4.95 | [M + H] + | 187.09660 | − 0.44 | 151.07550, 169.08635, 123.08073, 125.05986, 95.08619, 79.05504, 81.07069, 67.05515 | 70 | |
| Tavulin (3aR,4aS,5R,7aS,8S,9aR)-5-Hydroxy-4a,8-dimethyl-3-methyleneoctahydroazuleno[6,5-b]furan-2,6(3H,4H)-dione | C15H20O4 | 264.13624 | 4.07 | [M + H] + | 265.14343 | − 0.32 | 247.13263, 229.12231, 201.12739, 175.07521, 173.13248, 161.09596, 131.08571, 105.07038, 93.07038 | 86 | |
| (3aS,10aR,10bR)-6,10a-Dimethyl-3-methylene-3,3a,4,5,7,8,10a,10b-octahydrofuro[3′,2′:6,7]cyclohepta[1,2-b]pyran-2,9-dione | C15H18O4 | 262.12067 | 5.70 | [M + H] + | 263.12790 | − 0.63 | 245.11699, 227.10669, 217.12270, 199.11159, 175.11145, 83.04973 | 70 | |
| 3-(2-Hydroxyethyl) indole | C10H11NO | 161.08418 | 4.93 | [M + H] + | 162.09159 | − 0.74 | 144.08081, 145.08416 | 76 | |
| 3,4-Dihydroxy-benzaldehyde | C7H6O3 | 138.03174 | 3.63 | [M + H] + | 139.03906 | − 0.33 | 111.04454, 93.03410, 65.03947 | 73 | |
| 3-Hydroxy-3,5,5-trimethyl-4-(3-oxo-1-buten-1-ylidene) cyclohexyl β-D-glucopyranoside | C19H30O8 | 386.19423 | 3.92 | [M + H] + | 387.20169 | − 0.42 | 225.14919, 207.13803, 189.12739, 149.09628, 123.08069, 95.08616, 85.06544 | 78 | |
| 4-Benzoylbiphenyl | C19H14O | 258.10447 | 10.99 | [M + H] + | 259.11176 | − 0.03 | 105.03401, 181.06465, 95.04977 | 87 | |
| Adenine | C5H5N5 | 135.05462 | 0.56 | [M + H] + | 136.06189 | − 0.95 | 119.04926, 137.04579 | 85 | |
| Adenosine | C10H13N5O4 | 267.09690 | 0.61 | [M + H] + | 268.10431 | − 0.53 | 136.06190, 137.06627 | 97 | |
| N-Acetylserotonin | C12H14N2O2 | 218.10573 | 3.788 | [M + H] + | 219.11304 | − 0.91 | 202.08621, 160.07573, 132.08095 | 79 | |
| Sedanolide | C12H18O2 | 194.13079 | 8.41 | [M + H] + | 195.13800 | − 0.55 | 167.97124, 149.13252, 135.11682, 93.07043, 79.05492, | 77 | |
| Sedanolide | C12H18O2 | 194.13080 | 8.83 | [M + H] + | 195.13799 | − 0.60 | 177.12741, 149.13528, 107.08592, 79.05496, 97.06478 | 80 | |
| Senkyunolide H | C12H16O4 | 224.10493 | 4.794 | [M + H] + | 225.11218 | − 0.33 | 207.10156, 189.09093, 161.09610, 147.09610, 119.08586, 91.05484, 93.07055 | 76 | |
| Pulegone | C10H16O | 152.12025 | 6.68 | [M + H] + | 153.12755 | − 0.87 | 135.11691, 111.02374, 109.10118, 107.08606, 93.07057, 81.07069, 79.05492, 69.07077, 55.05527 | 81 |
Amino acids
A total of twelve amino acids were identified of which some were in free form (arginine, tryptophan, phenylalanine, tyrosine, isoleucine, and methionine) and their derivatives (pyroglutamic acid, prolylleucine, and glutamylcysteine). Hypoglycin A was also found at trace level, which is a highly toxic amino acid (Shrivastava et al. 2014). Hypoglycin A has been identified with a mass error of 0.04 ppm for a protonated ion m/z 142.08672 [M + H]+ and further confirmed by the product ion spectra compared with the reference spectra from the mzCloud database. The confirmatory ion was m/z 96.08124 (4.80 ppm) which was within a ± 5 ppm mass error. Detailed information related to the identification and confirmation of hypoglycin A is presented in Fig. 2.
Fig. 2.
Identification of hypoglycin A in litchi pulp. Extracted ion chromatogram (XIC) in solvent (A), litchi pulp (B), full MS spectra (C), ddMS2 spectra (D) and MS/MS spectra overlay with reference spectra of hypoglycin A from mzCloud library
Along with hypoglycin A, there is a first-time report of hypoglycin B in litchi pulp and seed shown in Fig. 3. Bowen-Forbes and Minott (2011) have reported hypoglycin B in ackee fruit (Blighia sapida soapberry family, Sapindaceae) but there was no report on hypoglycin B in litchi fruit. Identification of hypoglycin B was performed based on the accurate mass m/z 271.12897 (0.60 ppm) and characteristic fragments (m/z 142.08640, 96.08141, 74.02452, 208.09708). Detection of hypoglycin A in the seed is quite high (3X) as compared to the pulp, whereas hypoglycin B is 2X higher in pulp (aril) in comparison with the seed which agrees with Bowen-Forbes and Minott (2011).
Fig. 3.
Identification of hypoglycin B in litchi pulp; Extracted Ion Chromatogram (EIC) for hypoglycin B in extract of seed (A), skin (B) and litchi pulp (C), full MS spectra (D), ddMS2 spectra (E) for hypoglycin B
Apart from amino acids, abscisic acid and 4-methoxysalicylic acid were identified. For abscisic acid, two adducts viz [M + H] and [M + Na] are formed with − 0.26 and − 0.58 ppm errors. As usual, the sodiated adduct had reduced fragmentation as compared to the protonated adduct. Hence, the protonated molecular ion (m/z 265.14337) was further confirmed as product ion spectra in which all the ions are within 1 ppm mass error (Table 1). Caffeic acid was identified with a mass error of − 0.15 ppm for a protonated molecular ion with two confirmatory ions within ± 5 ppm error. A trace level of caffeic acid content was observed (Zhang 2013).
Flavonols
Tentatively identified flavonols included epicatechin, procyanidins, rhamnetin, kaempferol, and quercetin derivatives. All identifications are based on the criteria mentioned earlier. Epicatechin was prominent as compared to catechin (trace level) and identified at 3.68 min and confirmed with the mzCloud library with > 94% score as well as ChemSpider. Identification of epicatechin is presented in supplementary Fig. 1, which includes extracted ion chromatogram (XIC) for epicatechin and the full Scan MS spectra with protonated molecular ion (m/z 291.08636, − 0.07 ppm error). The ddMS2 spectra and the reference spectra from the mzCloud library showed a > 94% match (supplementary Fig. 1). An individual epicatechin detection in litchi pulp due to its free form and catechin in the bound form is available (Zhang 2013).
Quercetin includes the aglycone form of flavonoid-glycosides, such as rutin, quercetin-3-O-glucoside, quercetin-3-(2-Gal-rhamnosyl)-robinobioside, found in litchi pulp. Similarly, kaempferol was found in the form of kaempferol-3-O-rutinoside, kaempferol-7-O-glucoside, and kaempferol-3-O-(2,6-di-O-alpha-L-rhamnopyranosyl)-beta-D-galactopyranoside (Table 1). Quercetin, quercetin-3-(2Gal-rhamnosyl-robinobioside), and quercetin-3-O-b-D-glucoside are eluted at the same retention time (5.24 min). Similarly, kaempferol 3-O-rutinoside and kaempferol 3-O-(2,6-di-O-alpha-L-rhamnopyranosyl)-beta- D-galactopyranoside are eluted at the same retention time 5.63 min. Based on the observed fragmentation pattern, these molecules were probably generated due to in-source fragmentation (Table 1).
Rhamnetin is a monomethoxy-flavone, i.e., quercetin methylated at position 7. It is an antioxidant and an anti-inflammatory agent. Rhamnetin parent compound with derivatives, i.e., rhamnetin-3-O-neohesperidoside, isorhamnetin 3-rhamnosyl-(1- > 4)-rhamnosyl-(1- > 6) glucoside was identified with a mass error − 0.50, − 1.09 and − 0.74 ppm, respectively. Rhamnetin and rhamnetin-3-O-neohesperidoside were detected at 5.79 min due to the possibility of in-source fragmentation of isorhamnetin 3-rhamnosyl-(1- > 4)-rhamnosyl-(1- > 6) glucoside. An identification of isorhamnetin 3-O-rhamnosyl-(1- > 4)-rhamnosyl-(1- > 6) glucoside (Table 1) was determined based on the protonated molecular ion m/z 771.23407 (mass error − 0.19 ppm) with elemental composition C34H43O20. Further, it was confirmed through the characteristic fragment/product ions i.e. m/z 625.17554 (-C6H11O4; one rhamnose molecule), 479.11795 (–C12H22O8; two rhamnose molecule), 317.06525 (removal of two rhamnose molecules, C12H22O8 and one glucose molecule, C6H11O5), 302.04166 (–CH3 from quercetin methyl ether), 147.06511 (C6H11O4, rhamnose) and m/z 129.05467 (C6H9O3). All these characteristic fragment ions, including parent ion, are within ± 1.3 ppm (supplementary Fig. 2A). A possible fragmentation pattern based on the observed ddMS2 spectra is presented in supplementary Fig. 2B.
Procyanidin
The data acquisition in negative polarity showed many high molecular ions with m/z 575.1182, 577.1335, 865.1965, 863.1807, 1151.2429, 1153.2584 with an adduct [M-H]−. All these ions have ddMS2 spectra (MS/MS), in which the common fragment ions m/z 289.07121, 161.02299, 125.02284, 109.02787 were strongly correlated with the epicatechin fragmentation pattern. Also, an addition of epicatechin molecule (m/z 289.07121) forms a dimer which resulted in the m/z 577.1335 fragment ion, hinting at polymerization. Qiang et al. (2015) have reported the degree of polymerization as 2–6, and that epicatechin is dominant in the skin (pericarp) and seed (Qiang et al. 2015).
Procyanidin C1 (Procyanidin trimer) was identified based on the protonated molecular ion m/z 867.21263 observed at 3.95 min in full MS with mass error − 0.53 ppm (C45H38O18) in comparison with theoretical mass m/z 866.20581. Procyanidin C1 is a proanthocyanidin comprising epicatechin (3 units) joined by two successive (4beta- > 8)-linkages. It was further confirmed through MS/MS spectra at the same retention. The observed fragment ions are 715.16541 (C37H31O15; − 0.47 ppm), 579.14886 (C30H27O12; − 1.44 ppm), 409.09079 (C22H17O8; − 2.45 ppm), 289.07043 (C15H13O6; − 0.79 ppm), 163.03886 (C9H7O3; − 0.65 ppm), 139.03891 (C7H7O3; − 0.43 ppm), 123.0440 (C7H7O2; 1.14 ppm). The full MS and MS/MS spectra are presented in Fig. 4A.
Fig. 4.
Full scan MS and MS/MS spectra for procyanidin Trimer (A) and tetramer (B)
Procyanidin tetramer was identified based on the deprotonated molecular ion m/z 1153.25842 observed at 3.90 min in full MS with mass error − 2.08 ppm (C60H49O24) against the theoretical mass m/z 1154.26920. This tetramer comprises four units of epicatechin joined through inter-flavonoid (4beta- > 8)-linkages. Tetramer was further confirmed through MS/MS spectra at the same retention. The observed fragment ions are 865.19635 (C45H37O18; − 0.91 ppm), 863.17987 (C45H35O18; − 2.22 ppm), 575.11780 (C30H23O12; − 1.05 ppm), 407.07617 (C22H15O8; 0.06 ppm), 289.07043 (C15H13O6; − 0.90 ppm), 287.05554 (C15H11O6; 1.83 ppm), 161.02299 (C9H5O3; − 2.02 ppm). This fragmentation pattern provided the common difference of m/z 289.07043 between m/z 1153.25842, 865.19635, 575.11780, and 289.07043 (Fig. 4B). Similarly, other proanthocyanidins (procyanidin dimers, trimers, and tetramers) were identified and are presented in Table 1 (Qiang et al. 2015, Lin et al. 2014).
Other compounds
In addition to amino acids, phenolic acids, and flavonols, seventeen compounds were tentatively identified based on spectral matching with the mzCloud database by using compound discoverer software. The spectral matching score for all the compounds was always greater than 70% in comparison with the mzCloud library spectra (Table 1).
Multivariate analysis
The relative changes in the polyphenols composition in litchi fruit are shown in the heat map (Supplimentary Fig. 3A). Different parts (skin, pulp, and seed) of litchi fruits were screened to evaluate the difference. The PCA score plot based on the polyphenol composition showed the three parts significantly separated from each other. The principal component 1 (PC 1) and principal component 2 (PC 2) showed 61.8 and 34.0% data variance in polyphenols composition (Supplimentary Fig. 3B). The markers identified and reported in this study for litchi fruit were possible due to an advanced analytical approach. The use of high‐resolution accurate mass spectrometry permitted the large-scale screening and identification of highly polar, mid-polar to non-polar compounds. Amino acid (L-glutamic acid), purine nucleotide base (adenosine), and phenolic compounds (catechin, epicatechin, taxifolin) were significantly present in high amounts in seed in comparison with pulp and skin. Naringenin and flavanone were three times higher in seed and skin. Hypoglicine A, luteolin, and robinin were three times higher in seed than pulp. Rutin is significantly higher (> 10X) in pulp than in skin and seed. The marker compounds were reported based on the area with a standard deviation (n = 5) observed in all three parts. and found to be significantly different as shown in Fig. 5. Several biological activities (antioxidant, antitumor, antiviral, antibacterial, anti-inflammatory, antiadipogenic, and cardioprotective effects) have been reported to these polyphenol compounds (Salehi et al. 2019).
Fig. 5.
Comparison of the mean values of area (± standard deviation) of marker compounds observed in litchi pulp (aril), skin (pericarp), and seed
Distribution of identified compounds
The relative distribution of the total identified compounds in three parts of litchi fruit is presented in the form of significant classes (Supplementary Fig. 4). The significant contribution was by phenolic compounds (53%), followed by other compounds (21%), amino acids (16%), and phenolic/other acids (10%) in extracts of litchi fruits.
Conclusions
In this study, a quick and fast extraction procedure was optimized to cover the maximum possible analytes in a single analysis. This study has revealed the analytical capability of the QExactive Orbitrap accurate mass analyzer in terms of detection and identification of phytochemicals in litchi fruit. Using the compound discoverer software, which includes the mzCloud database and direct link with the ChemSpider online database followed by the spectral comparison of MS as well as MS/MS spectra, allowing for accurate identification of phytochemicals. Based on the spectral comparison, a mzCloud library score is provided for every identified compound. A total of 77 compounds, including amino acids, phenolic acid derivatives, phenolic compounds, and others were tentatively identified with the help of libraries. Along with hypoglycin A, hypoglycin B is the first time reported in litchi fruit seed and skin. Earlier this was reported in ackee fruits. Both databases allowed a quick automated search of the analytes present in the sample based on the pre-defined criteria set. Apart from screening, multivariate analysis is possible in the same workflow. The Orbitrap allows retrospective analysis and compounds not included in the developed database, which could be identified through the help of ChemSpider and mass frontier software. Hence, for a non-target approach, the HRMS QExactive Orbitrap could be a better choice for the mass analyzer. As seed and skin are not consumed and discarded but it contains a lot of polyphenols (Flavone and flavo-3-ols) in higher quantities as compared to pulp except for rutin.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
All authors have equally contributed to this work. All authors thank FSSAI and the FSSC for permission to publish this work.
Abbreviations
- LC-MS
Liquid chromatography mass spectrometer
- MCPG
Methylenecyclopropyl-glycine
- UHPLC
Ultra high performance liquid chromatography
- HCA
Hierarchical cluster analysis
- PCA
Principal component analysis
- AGC
Automatic gain control
- HESI
Heated electrospray ionization
- TIC
Total ion chromatogram
- ddMS2
Data dependent MS2
Author contributions
The corresponding author states that, all authors contributed equally to finish this work. We all authors declare that– (1) the work described has not been published before (except in the form of an abstract, a published lecture or academic thesis), (2) it is not under consideration for publication elsewhere, (3) its submission to JFST publication has been approved by all authors (4) if accepted, it will not be published elsewhere in the same form, in English or in any other language, including electronically without the written consent of the copyright holder, and (5) JFST will not be held legally responsible should there be any claims for compensation or dispute on authorship.
Funding
This work was supported by the FSSAI-Thermo Fisher Food Safety Solution Center (FSSC) and the FSSAI, New Delhi.
Availability of data and material
All the data provided in manuscript is correct and also available with us if required. The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there are no conflicts of interest.
Consent to participate
None.
Consent for publication
All authors are agreed for publications.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Analytical quality control and method validation procedure for pesticide residues analysis in food and feed SANTE 11312/2021. https://www.eurl-pesticides.eu/userfiles/file/ EurlALL/SANTE_11312_2021.pdf Accessed on 15.03.2022
- Barceloux DG. Akee fruit and jamaican vomiting sickness (Blighia sapida Köenig) Dis Mon. 2009;55:318–326. doi: 10.1016/j.disamonth.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Bowen-Forbes CS, Minott DA. Tracking hypoglycins A and B over different maturity stages: implications for detoxification of Ackee (Blighia sapida K.D. Koenig) Fruits. J Agric Food Chem. 2011;59:3869–3875. doi: 10.1021/jf104623c. [DOI] [PubMed] [Google Scholar]
- Emanuele S, Lauricella M, Calvaruso G, D’Anneo A, Giuliano M. Litchi chinensis as a functional food and a source of antitumor compounds: an overview and a description of biochemical pathways. Nutrients. 2017;9:992–1006. doi: 10.3390/nu9090992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim SRM, Mohamed GA. Litchi chinensis: medicinal uses, phytochemistry, and pharmacology. J Ethnopharmacol. 2015;174:492–513. doi: 10.1016/j.jep.2015.08.054. [DOI] [PubMed] [Google Scholar]
- Isenberg SL, Carter MD, Hayes SR, Graham LA, Johnson D, Mathews TP. Quantification of toxins in soapberry (Sapindaceae) arils: hypoglycin A and methylenecyclopropylglycine. J Agric Food Chem. 2016;64:5607–5613. doi: 10.1021/acs.jafc.6b02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John TJ, Das M. Acute encephalitis syndrome in children in Muzaffarpur: hypothesis of toxic origin. Curr Sci India. 2014;106:1184–1185. [Google Scholar]
- Kessy H, Hu Z, Zhao L, Zhou M. Effect of steam blanching and drying on phenolic compounds of litchi pericarp. Molecules. 2016;21(729):1–9. doi: 10.3390/molecules21060729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koley TR, Khan Z, Oulkar D, Singh BK, Maurya A, Singh B, Banerjee K. High resolution LC-MS characterization of phenolic compounds and the evaluation of antioxidant properties of a tropical purple radish genotype. Arab J Chem. 2020;13(1):1355–1366. doi: 10.1016/j.arabjc.2017.11.007. [DOI] [Google Scholar]
- Lin LZ, Sun J, Chen P, Monagas MJ, Harnly JM. UHPLC-PDA-ESI/HRMSn profiling method to identify and quantify oligomeric proanthocyanidins in plant products. J Agric Food Chem. 2014;62:9387–9400. doi: 10.1021/jf501011y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew JL, John TJ. Exploration of association between litchi consumption and seasonal acute encephalopathy syndrome. Indian Pediatr. 2017;54:319–325. doi: 10.1007/s13312-017-1095-1. [DOI] [PubMed] [Google Scholar]
- Miranda-Hernández AM, Muñiz-Márquez DB, Wong-Paz JE, Aguilar-Zárate P. Characterization by HPLC–ESI–MS2 of native and oxidized procyanidins from litchi (Litchi chinensis) pericarp. Food Chem. 2019;291:126–131. doi: 10.1016/j.foodchem.2019.04.020. [DOI] [PubMed] [Google Scholar]
- Pulla P. A child-killing toxin emerges from shadows. Science. 2015;348:15–16. doi: 10.1126/science.348.6230.15. [DOI] [PubMed] [Google Scholar]
- Qiang LV, Fenglei L, Xiaoyong Z, Yu L, Guibing H, Chongde S, Xian L, Kunsong C. Identification of proanthocyanidins from litchi (Litchi chinensis Sonn.) Pulp by LC-ESI-Q-TOF-MS and their antioxidant activity. PLoS ONE. 2015;10(3):1–17. doi: 10.1371/journal.pone.0120480.g001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi B, Tsouh Fokou PV, Sharifi-Rad M, Zucca P, Pezzani R, Martins N, Sharifi-Rad J. The therapeutic potential of naringenin: a review of clinical trials. Pharmaceuticals (basel) 2019;11:1–18. doi: 10.3390/ph12010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarni-Manchado P, Le Roux EL, Guernevé CL, Lozano Y, Cheynier V. Phenolic composition of litchi fruit pericarp. J Agric Food Chem. 2000;48:5995–6002. doi: 10.1021/jf000815r. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Kumar A, Thomas JD, Laserson KF, Bhushan G, Carter MD, Chhabra M, Mittal V, Khare S, Sejvar J, Dwivedi M, Isenberg SL, Johnson R, Pirkle JL, Sharer JD, Hall P, Yadav R, Velayudhan A, Papanna M, Singh P, Somashekar D, Pradhan A, Goel K, Pandey R, Kumar M, Kumar S, Chakrabarti A, Sivaperumal P, RameshKumar A, Schier JG, Chang A, Graham LA, Mathews TP, Johnson D, Valentin L, Caldwell KL, Jarrett JM, Harden LA, Takeoka GR, Tong S, Queen K, Paden C, Whitney A, Haberling DL, Singh R, Singh RS, Earhart KC, Dhariwal AC, Chauhan LS, Venkatesh S. Srikantiah P (2017) association of acute toxic encephalopathy with litchi consumption in an outbreak in Muzaffarpur, India. Case-Control Study Lancet Glob Health. 2014;5:458–466. doi: 10.1016/S2214-109X(17)30035-9. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Srikantiah P, Kumar A, Bhushan G, Goel K, Kumar S, Kumar T, Mohankumar R, Pandey R, Pathan P, Tulsian Y, Pappanna M, Pasi A, Pradhan A, Singh P, Somashekar D, Velayudhan A, Yadav R, Chhabra M, Mittal V, Khare S, Sejvar JJ, Dwivedi M, Laserson K, Earhart KC, Sivaperumal P, Ramesh Kumar A, Chakrabarti A, Thomas J, Schier J, Singh R, Singh RS, Dhariwal AC, Chauhan LS. Outbreaks of unexplained neurologic illness-Muzaffarpur, India, 2013–2014. Morb Mortal Wkly Rep. 2015;64:49–53. [PMC free article] [PubMed] [Google Scholar]
- Singh HP, Babita S, FAO RAP (2002) 2002/04, FAO, Rome, 2002, 128p (http://www.fao.org/3/ac684e/ac684e08.htm) Accessed on 10.09.2019
- Tanaka K, Ikeda Y. Hypoglycin and jamaican vomiting sickness. Prog Clin Biol Res. 1990;321:167–184. [PubMed] [Google Scholar]
- Upadhyaya DC, Upadhyaya CP. New bioactive chromanes from litchi chinensis. Lychee Biotechnol. 2017;13:333–361. doi: 10.1007/978-981-10-3644-6_13. [DOI] [Google Scholar]
- Yi L, Dong N, Liu S, Yi Z, Zhang Y. Chemical features of pericarpium citri reticulatae and pericarpium citri reticulatae viride revealed by GC-MS metabolomics analysis. Food Chem. 2015;186:192–199. doi: 10.1016/j.foodchem.2014.07.067. [DOI] [PubMed] [Google Scholar]
- Zhang R, Zeng Q, Deng Y, Zhang M, Wei Z, Zhang Y, Tang X. Phenolic profiles and antioxidant activity of litchi pulp of different cultivars cultivated in Southern China. Food Chem. 2013;136:1169–1176. doi: 10.1016/j.foodchem.2012.09.085. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wang H, Sun J, Yang B, Duan X, Jiang Y. Pericarp and seed of litchi and longan fruits: constituent, extraction, bioactive activity, and potential utilization. J Zhejiang Univ Sci B. 2019;20:503–512. doi: 10.1631/jzus.B1900161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data provided in manuscript is correct and also available with us if required. The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.