Abstract
Background & Aims
Covalently closed circular DNA (cccDNA) of hepatitis B virus (HBV), existing as a stable minichromosome in the hepatocyte, is responsible for persistent HBV infection. Maintenance and sustained replication of cccDNA require its interaction with both viral and host proteins. However, the cccDNA-interacting host factors that limit HBV replication remain elusive.
Methods
Minicircle HBV (MC-HBV), a recombinant cccDNA, was constructed based on chimeric intron and minicircle DNA technology. By mass spectrometry based on pull-down with biotinylated MC-HBV, the cccDNA-hepatocyte interaction profile was mapped. HBV replication was assessed in different cell models that support cccDNA formation.
Results
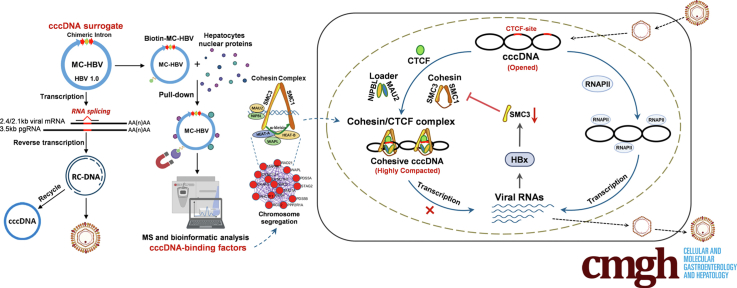
MC-HBV supports persistent HBV replication and mimics the cccDNA minichromosome. The MC-HBV–based screen identified cohesin complex as a cccDNA binding host factor, leading to reduced HBV replication. Mechanistically, with the help of CCCTC-binding factor (CTCF), which has specific binding sites on cccDNA, cohesin loads on cccDNA and reshapes cccDNA confirmation to prevent RNA polymerase II enrichment. Interestingly, HBV X protein transcriptionally reduces structural maintenance of chromosomes complex expression to partially relieve the inhibitory role of the cohesin complex on HBV replication.
Conclusions
Our data not only provide a feasible approach to explore cccDNA-binding factors, but also identify cohesin/CTCF complex as a critical host restriction factor for cccDNA-driven HBV replication. These findings provide a novel insight into cccDNA–host interaction and targeted therapeutic intervention for HBV infection.
Keywords: Cohesin, cccDNA, HBx, SMC3
Abbreviations used in this paper: AFM, atomic force microscopy; ATPase, adenosine triphosphate hydrolysis; BS, binding site; CBP, CREB binding protein; cccDNA, covalently closed circular DNA; ChIP, chromatin immunoprecipitation; Co-IP, co-immunoprecipitation; CTD, C-terminal domain; CTCF, CCCTC-binding factor; DDB1, damage specific DNA binding protein 1; DMEM, Dulbecco’s modified Eagle medium; FBS, fetal bovine serum; GFP, green fluorescent protein; GO, Gene Ontology; HBc, hepatitis B core; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; HBx, HBV X protein; IFN, interferon; MC-HBV, minicircle HBV cccDNA model; HCC, hepatocellular carcinoma; HDAC1, histone deacetylase 1; H3K, histone 3 lysine; MAFG-RE, MAFG responsive element; mBS, mutation at binding site; MAU2, MAU2 chromatid cohesion factor homolog; MCODE, Molecular Complex Detection Algorithm; MS, mass spectrometry; MST, microscale thermophoresis; NIPBL, Nipped-B homolog; NTCP, Na+/taurocholate cotransporting polypeptide; PCR, polymerase chain reaction; PDS5A/Bsister chromatid cohesion protein PDS5 homolog, A/B; pgRNA, pregenomic RNA; PHH, primary human hepatocytes; PP, parent plasmid; qPCR, quantitative polymerase chain reaction; RAD21, double-strand–break repair protein rad21 homolog; RBP, RNA-binding protein; rcccDNA, recombinant covalently closed circular DNA; rcDNA, relaxed circular DNA; RNAPII, RNA polymerase II; RT, reverse-transcription; siRNA, small interfering RNA; SMC, structural maintenance of chromosomes complex; WAPL, wings apart-like protein homolog; WT, wild-type
Graphical abstract
Summary.
The whole cell-based interaction profile identifies cohesin/CTCF complex as the covalently closed circular DNA–interacting factor, which bind and compact covalently closed circular DNA to prevent the recruitment of RNA polymerase II and the consequent hepatitis B virus transcription. However, hepatitis B virus X protein transcriptionally reduces structural maintenance of chromosomes complex 3 expression to evade the cohesin-mediated restriction.
Chronic hepatitis B virus (HBV) infection is one of the major pathogenic causes of liver cirrhosis and hepatocellular carcinoma (HCC).1 HBV is a hepatotropic, partially double-stranded DNA virus. Upon infection, relaxed circular DNA (rcDNA) in the Dane particle is delivered into the nucleus and converted into covalently closed circular DNA (cccDNA), which then serves as the sole transcription template to produce viral RNAs, including pregenomic RNA (pgRNA), driving the production of viral antigens and progeny virions.2 Therefore, cccDNA is recognized as the major obstacle to curing chronic hepatitis B due to its critical role in viral persistence and recurrence after discontinued treatment.3
As the most stable HBV replication intermediate, HBV cccDNA exists in the hepatocyte nucleus as a minichromosome. The stable maintenance and proper functions of the cccDNA minichromosome require its interaction with both viral and host proteins.4 Accumulating evidence has shown that remodeling of the minichromosome is mediated by recruitment of epigenetic regulators (such as CREB binding protein [CBP]/E1A binding protein p300 [p300], histone deacetylase 1 [HDAC1], protein arginine methyltransferase 5 [PRMT5], and sirtuin 3 [Sirt3]) and transcriptional factors (including hepatocyte nuclear factor 1-alpha [HNF1α], HNF4α, zinc fingers and homeoboxes 2 [ZHX2], and farnesoid X-activated receptor [FXR]), which are associated strongly with cccDNA transcriptional activity.5, 6, 7, 8, 9, 10 Furthermore, HBV has evolved multiple strategies to hijack or antagonize host machines for viral replication. HBV X protein (HBx) is the most important viral protein for both HBV replication and minichromosome formation. Through transcriptional transactivation and recruitment of interacting proteins, HBx relieves cccDNA transcriptional repression by altering the acetylation status and methylation status of cccDNA-bound H3/H4.6,11 In addition, HBx activates cccDNA transcription and viral replication by hijacking damage specific DNA binding protein 1 (DDB1)-Cullin 4 (CUL4)-ring finger-containing protein 1 (ROC1) E3 ligase to degrade host proteins such as structural maintenance of chromosomes (SMC)5/6.8,9,12 A complete understanding of host–cccDNA interactions adopted by HBx is mandatory to develop novel strategies to eradicate HBV. However, until now, the host factors responsible for functional maintenance and transcriptional regulation of cccDNA still are less known.
The major obstacles to uncovering the interactome of cccDNA and hepatocyte proteins are the limitation of obtaining enough labeled cccDNA and the lack of appropriate cccDNA labeling methods. The copy number of cccDNA in the nucleus of HBV-infected cells is very low, approximately 1.05 copies per cell,13 which strongly restricts abundant minichromosome acquisition. The recently reported tools producing recombinant cccDNA will bring a new dawn for decrypting cccDNA–host interaction profiles.14,15 The recombinant cccDNA model based on minicircle technology generates high copies of cccDNA-like molecules in vitro, while the inserted attachment site right (attR) sequences cannot be removed from the HBV genome in hepatocytes.15
In the present study, by combining chimeric–intron and minicircle DNA technology,14 we established a recombinant cccDNA model named minicircle HBV (MC-HBV), which supports HBV replication and mimics minichromosome bound with histone and epigenetic factors. Using biotin-labeled MC-HBV, we systemically explored the interaction profile of cccDNA–host factors in hepatocytes by pull-down and mass spectrometry (MS) analysis. Furthermore, we identified cohesin complex, composed of SMC3 and SMC1, as a novel host factor interacting with cccDNA. Mechanistically, cohesin bound and shaped cccDNA conformation to prevent RNA polymerase II (RNAPII) enrichment, leading to inhibition of HBV replication. To get persistent replication, HBx transcriptionally inhibited SMC3 expression. It recently was shown that SMC5/6 inhibits HBV replication but is targeted by the DDB1–CUL4 E3 ligase, hijacked by HBX.9 Although SMC5/6 and cohesin are both members of the SMC complex, they share functional differences and are regulated by distinct mechanisms.16 Our findings strengthen the idea that host chromatin-organizing factors such as SMCs play crucial functions in cccDNA transcription, which different mechanisms can exert. Our data suggest the HBx–cohesin–cccDNA axis as a potential therapeutic target in HBV infection.
Results
Preparation of Recombinant cccDNA Named MC-HBV, Which Supports Epigenetic Modifications and HBV Replication
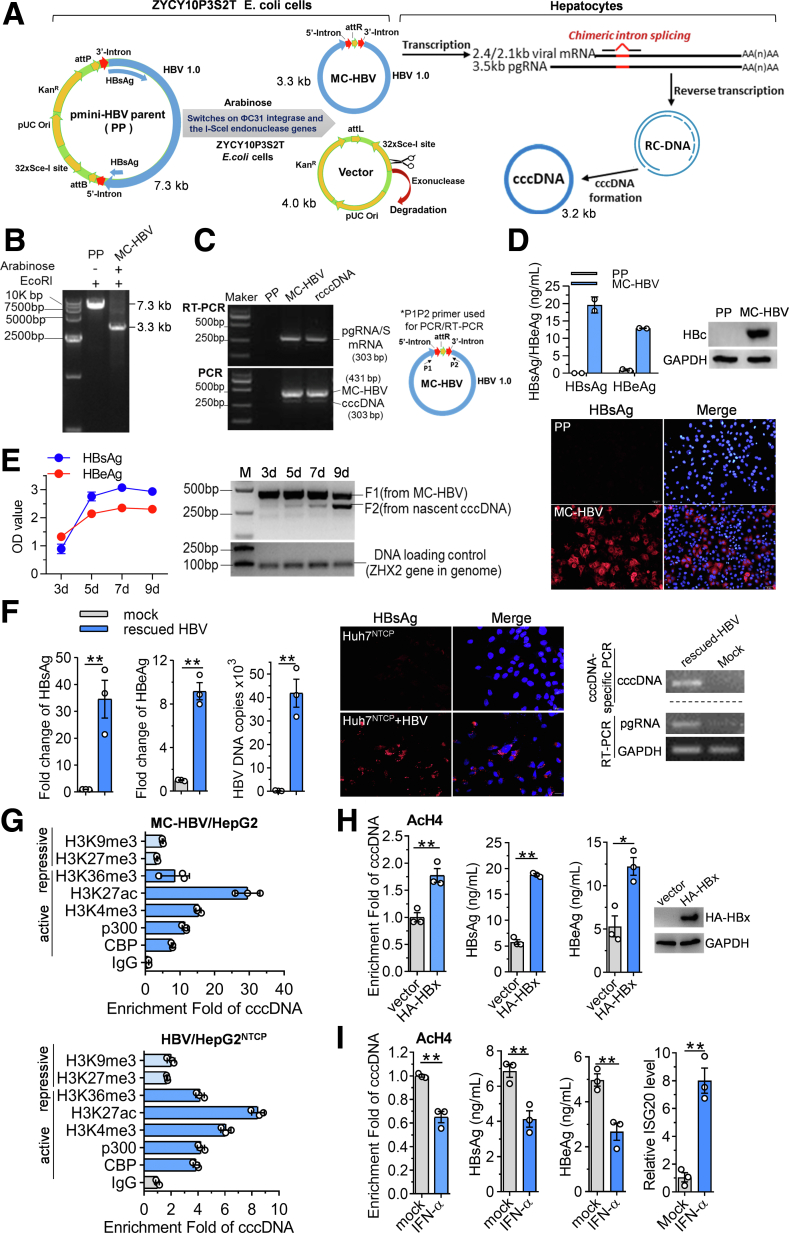
To get sufficient quantity and quality of recombinant cccDNA, MC-HBV was generated from recombination of pmini-HBV parent plasmid (PP) in arabinose-stimulated ZYCY10P3S2T Escherichia coli cells as described in the Methods section (Figure 1A). Results of EcoRI digestion and gel electrophoresis confirmed the complete induction of pmini-HBV (7.3 kb) into MC-HBV (3.3 kb) (Figure 1B). After being transfected, MC-HBV was transcripted and spliced into viral transcripts, including pgRNA, leading to the production of rcDNA and cccDNA in hepatocytes (Figure 1A). Because of the alternative splicing of pgRNA, the chimeric intron (128 bp) was removed from nascent rcDNA and cccDNA. Consequently, as shown in Figure 1C, polymerase chain reaction (PCR) and reverse-transcription (RT)-PCR using P1/P2 primers spanning the splice region produced a 431-bp fragment from MC-HBV and a 303-bp fragment from nascent cccDNA and spliced viral pgRNA/S messenger RNA transcripts in MC-HBV–transfected Huh7 cells, similar to that in the recombinant cccDNA (rcccDNA) model as described.14 In accordance, we also detected the high expression of viral antigens (hepatitis B surface antigen [HBsAg], hepatitis B e antigen [HBeAg], and hepatitis B core antigen [HBc]) in MC-HBV transfectants (Figure 1D), and the expression of HBsAg and HBeAg at least sustained at the peak up to day 9 post-transfection (Figure 1E). Furthermore, HBV particles rescued from MC-HBV–transfected Huh7 cells could infect Huh7Na+/taurocholate cotransporting polypeptide (NTCP) cells, leading to the production of HBsAg, HBeAg, viral HBV DNA, cccDNA, and pgRNA (Figure 1F). Collectively, these results suggest that MC-HBV fully recapitulates the HBV replication cycle, including the generation of viable cccDNA and high-level and long-term HBV antigen expression.
Figure 1.
Establishment of recombinant cccDNA mimicking the cccDNA minichromosome. (A) Schematic diagram of MC-HBV recombination in E coli and MC-HBV drove the entire cycle of pgRNA-rcDNA-cccDNA in hepatocytes. The 5'/3'-intron is represented as a red arrow, the HBV genome is represented as a blue box. (B) EcoRI digestion and gel electrophoresis analysis were performed to confirm the recombination efficiency of MC-HBV induced by arabinose in E coli cells (pmini-HBV PP ≈ 7.3 kb, MC-HBV ≈ 3.3 kb). (C) MC-HBV was transfected into Huh7 cells for 72 hours, PP and pCMV-Cre/parent rcccDNA (prcccDNA) as control, pgRNA/S messenger RNA (mRNA) and cccDNA were detected by RT-PCR and PCR, respectively, using primers P1/P2 as indicated with arrows. MC-HBV/unspliced viral transcripts are 431 nt, wild cccDNA/spliced viral transcripts with a chimeric intron are 303 nucleotides (nt). (D) PP and MC-HBV were transfected into Huh7 cells for 72 hours. HBsAg, HBeAg, and HBc expression were measured by enzyme-linked immunosorbent assay (ELISA), Western blot, and indirect immunofluorescence assay (IFA), respectively (n = 2). (E) MC-HBV was transfected into Huh7 cells, and persistent HBV replication was dynamically monitored by detecting HBsAg/HBeAg expression (ELISA) and nascent cccDNA formation (PCR). The ZHX2 gene of undigested whole-cell genomic DNA was amplified as the DNA loading control. (F) MC-HBV was transfected into Huh7 cells for 5 days, then virion particles in the supernatant were concentrated to infect Huh7NTCP cells for 5 days; HBsAg and HBeAg were detected by ELISA and IFA assay; and HBV-DNA, viral pgRNA, and cccDNA were analyzed by PCR (n = 3). (G) HepG2 cells were transfected with MC-HBV, and HepG2NTCP cells were infected with HBV at 200 genome equivalents (Geq) for 4 days, and the enrichment of active makers CBP, p300, H3K4me3, H3K27ac, and H3K36me3, and repressive makers H3K9me3 and H3K27me3, on cccDNA was measured by ChIP assay (n = 3). (H) HA-HBx and MC-HBV were cotransfected into Huh7 cells for 72 hours, or (I) MC-HBV was transfected Huh7 cells for 24 hours and then treated with 2000 U/mL IFN-α for another 48 hours. ChIP assay was performed to evaluate the occupancy of acetylation of histone H4 (AcH4) on cccDNA, and HBsAg/HBeAg expression and pgRNA and interferon stimulated exonuclease gene 20 (ISG20) mRNA levels were detected (n = 3). Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
We then came to verify whether MC-HBV can be occupied and regulated by epigenetic machinery as a cccDNA minichromosome.5, 6, 7, 8,11 As shown in Figure 1G, cccDNA-specific chromatin immunoprecipitation (ChIP) quantitative assays detected a similar accumulation level of active epigenetic modifications (p300, CBP, histone H3 tri-methylation on lysine 4 [H3K4me3], histone H3 acetylation on lysine 27 [H3K27ac], and H3K36me3) and repressive modifications (H3K9me3 and H3K27me3) on cccDNA in both HBV-infected HepG2NTCP cells and MC-HBV–transfected HepG2 cells, consistent with previous reports.6,15 Moreover, as reported that both HBx and interferon (IFN)-α contribute to the epigenetic regulation of the cccDNA minichromosome,6 HBx significantly promoted while IFN-α greatly decreased the enrichment of acetylation of histone H4 (AcH4) in MC-HBV, followed by HBx up-regulated and IFN-α down-regulated expression of HBsAg and HBeAg (Figure 1H and I). All these data support the potential of MC-HBV as a surrogate of the cccDNA minichromosome.
MC-HBV–Based Screen Identifies Cohesin Complex as a Novel cccDNA Binding Factor
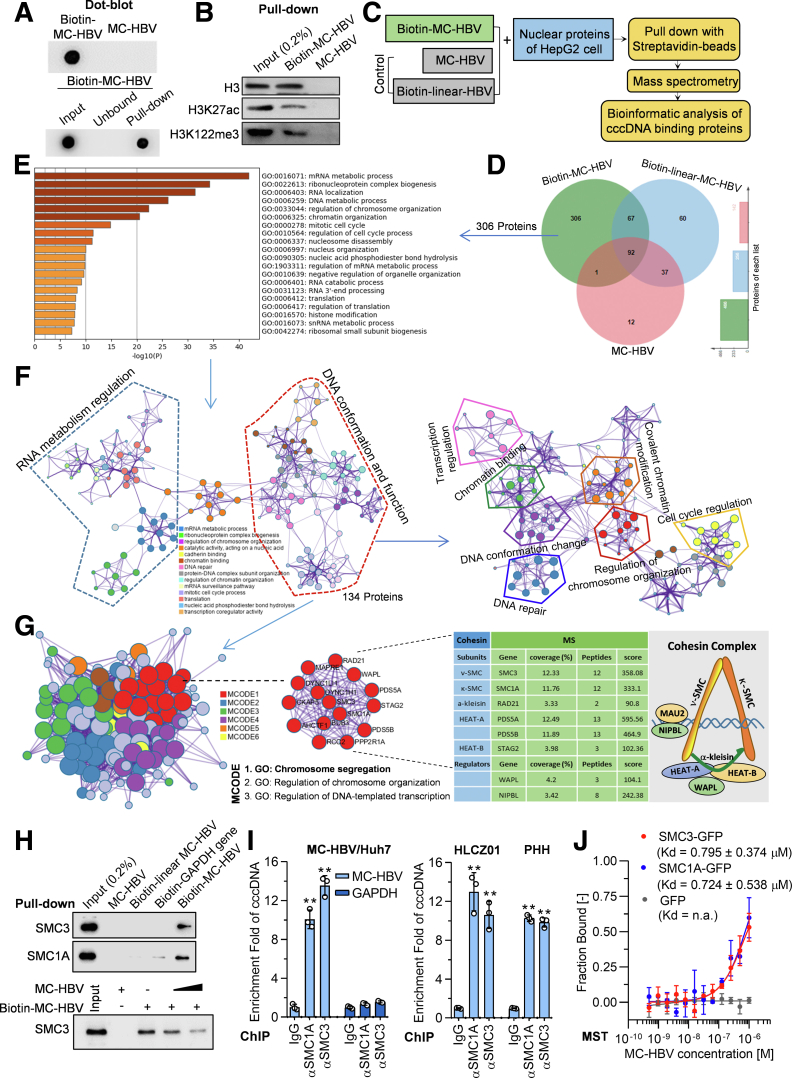
To explore the interaction profile of cccDNA and hepatocyte proteins, MC-HBV was labeled with biotin (biotin-MC-HBV) (Figure 2A). Pull-down assay confirmed the high enriched efficiency of biotin-MC-HBV (Figure 2A) and the interaction of biotin-MC-HBV with the known cccDNA binding factors H3, H3K27ac, and H3K122me3 (Figure 2B), suggesting that biotin-MC-HBV/pull-down is applicable to investigate cccDNA–host interaction. We thus incubated biotin-MC-HBV and nuclear proteins of HepG2 cells, then performed a pull-down assay with streptavidin beads for further Orbitrap MS. To exclude the nonspecific binding factors, unlabeled MC-HBV and biotin-linear-MC-HBV (linearized by the EcoRI enzyme) were taken as negative controls in pull-down and MS analysis (Figure 2C). A total of 306 proteins were identified explicitly in the biotin-MC-HBV group (Figure 2D and Supplementary Table 1) and significantly enriched into chromosome-related pathways by gene ontology (GO) pathway analysis, including chromosome organization, chromatin binding, DNA conformation change, and chromatin modification pathways (Figure 2E). Enriched ontology clusters and visualized network analysis classified these pathways into 2 major clades: DNA conformation/function and RNA metabolism regulation (Figure 2F, left panel). The 134 proteins involved in DNA conformation/function regulation clades were enriched into 7 clusters: DNA conformation change, chromatin binding, chromatin modification, DNA repair, chromosome organization, cell-cycle regulation, and transcription regulation (Figure 2F, right panel). These data indicate that cccDNA interacts with host DNA conformation regulators, which might be important for the formation of the cccDNA minichromosome and viral transcription.
Figure 2.
MC-HBV–based screen identifies cohesin complex as a novel host cccDNA binding factor. (A) MC-HBV was biotinylated and detected by dot blot using horseradish-peroxidase–streptavidin (upper panel); the input, unbound, and streptavidin-beads pull-down DNA was analyzed by dot blot to detect pull-down efficiency (lower panel). (B) Biotin-MC-HBV and MC-HBV were incubated with nuclear protein of HepG2 cells, and the interaction of H3, H3K27Ac, and H3K122me3 with cccDNA was identified by pull-down assay. (C) Schematic diagram of cccDNA-interacting factors screen with biotin-MC-HBV/pull-down/MS/bioinformatic analysis, biotin-linear-MC-HBV (linearized by EcoRI), and unlabeled MC-HBV taken as control. (D) Venn diagram of MS-identified proteins among 3 groups. A total of 306 candidates specifically were identified in the biotin-MC-HBV group and (E) analyzed for GO enrichment, or (F, left panel) were analyzed for enriched ontology clusters and a visualized network, (F, right panel) together with the 134 candidates classified into the DNA conformation regulation set. A circle node represented an enriched cluster, and nodes of the same color belong to the same cluster. (G) The protein–protein interaction network of these 134 proteins was mapped onto 6 pathways by MCODE (left panel), among which the chromosome segregation pathway was highlighted with red. Right: Cohesin composition and the information of 6 cohesin subunits and 2 regulators identified by MS. (H) A total of 1 μg biotin-MC-HBV, biotin-linear MC-HBV, and unlabeled MC-HBV were incubated with 400 μg HepG2 cells nuclear protein for 6 hours, then the interaction of SMC3 and SMC1A with cccDNA was identified by pull-down assay (upper panel); and 1 μg biotin-MC-HBV was incubated with 400 μg HepG2 cells nuclear protein with/without the presence of 1–3 μg unlabeled MC-HBV, then SMC3 were identified by pull-down assay (lower panel). (I) MC-HBV was transfected into Huh7 cells, or HBV-infected HLCZ01 cells and PHH cells at 200 genome equivalents (Geq), the enrichment of endogenous SMC3 and SMC1A on cccDNA was measured by ChIP assay (n = 3). (J) The green fluorescent protein (GFP)-SMC1A and GFP-SMC3 proteins were incubated with increasing concentrations of MC-HBV, the specific interactions were quantified by MST and plotted with the dissociation constant (Kd) equation, GFP protein served as control (n = 3). Data are presented as means ± SD. ∗∗P < .01.
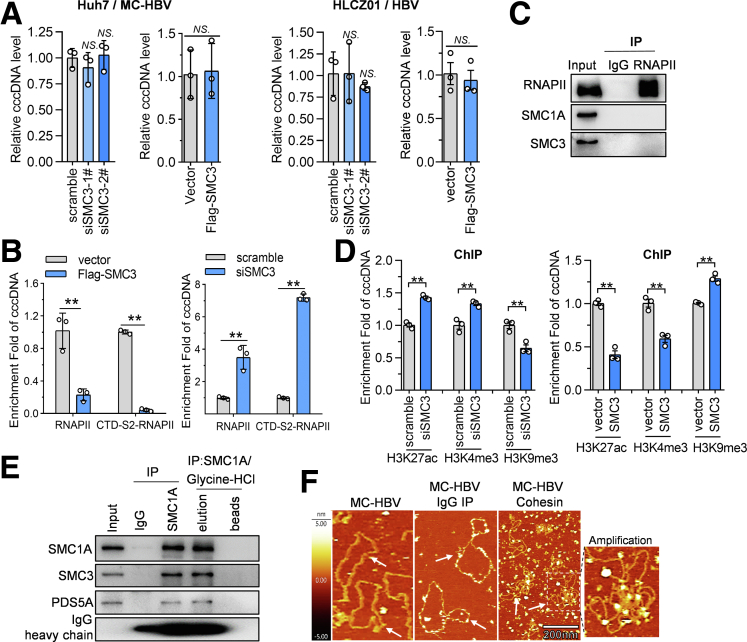
Interestingly, further protein and protein interaction analysis with Cytoscape-based (https://cytoscape.org) Molecular Complex Detection Algorithm (MCODE) uncovered a chromosome segregation interaction network with dense connection (Figure 2G, green nodes, left panel), among which 6 subunits of the cohesin complex (SMC3, SMC1A, double-strand–break repair protein rad21 homolog [RAD21], sister chromatid cohesion protein PDS5 homolog (PDS5)A, PDS5B, and stromal antigen 2 [STAG2]) and its 2 critical regulators (Nipped-B homolog [NIPBL] and wings apart-like protein homolog [WAPL]) were included (Figure 2G, right panel). SMC3 and SMC1A are 2 indispensable arms of the cohesin complex to embrace sister chromatids as a ring,17 contributing to chromatin structure and gene regulation. Consistent with the results of MS, streptavidin beads coupled with biotin-MC-HBV pulled down SMC3 and SMC1A, which were not precipitated with MC-HBV and biotin-linear-MC-HBV (Figure 2H, upper panel), and this interaction was competed efficiently by unlabeled MC-HBV in a dose-dependent manner (Figure 2H, lower panel). Further ChIP assays showed that SMC3 and SMC1A significantly accumulated with cccDNA in MC-HBV–transfected Huh7 cells and HBV-infected HLCZ01 cells and primary human hepatocytes (PHH) cells (Figure 2I). Furthermore, results of microscale thermophoresis (MST), a biophysical assay widely used to quantify the interaction between biomolecules, showed that the recombinant cccDNA molecules interacted with SMC3-green fluorescent protein (GFP) and SMC1A-GFP with the dissociation constant (Kd) values of 0.795 μmol/L (±0.374) and 0.724 μmol/L (±0.538), respectively, while cccDNA showed no association with GFP (Figure 2J). Collectively, these data fully show the cohesin complex composed of SMC3 and SMC1A as a novel HBV cccDNA binding factor.
Cohesin Inhibits HBV Transcription and Replication via Shaping cccDNA Conformation to Impede RNAPII Recruitment
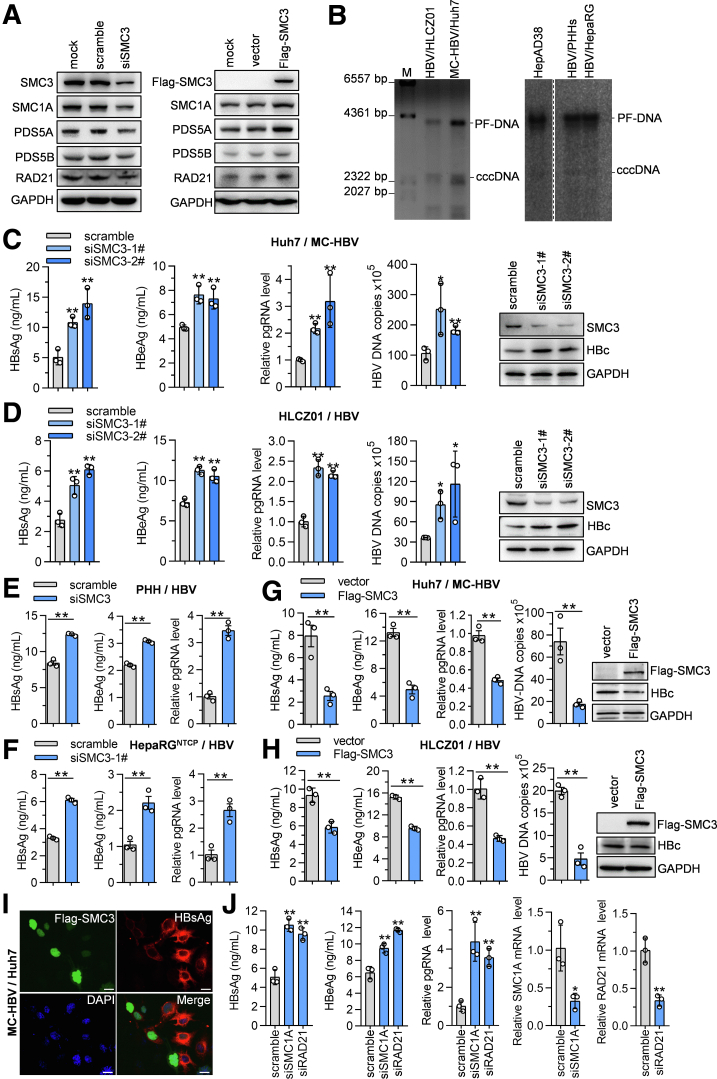
Given the critical role of cccDNA in HBV replication and the interaction of cohesin complex with cccDNA, we thus hypothesized that cohesin complex might participate in the regulation of HBV replication. To address this issue, we interfered with SMC3 expression by small interfering RNA (siRNA). As reported,18,19 SMC3 silence induced an obvious decrease of SMC1A and other cohesin components, including PDS5A, PDS5B, and RAD21, while ectopic SMC3 led to augmented expression of all detected cohesin components (Figure 3A). Therefore, we then focused on manipulating SMC3 expression to investigate whether cohesin complex regulates HBV replication in different cell models that support cccDNA formation, including MC-HBV–transfected Huh7 cells as well as in HBV-infected HLCZ01 cells, HepaRGNTCP cells, and PHH cells (Figure 3B). As shown in Figure 3C–H, SMC3 knockdown significantly increased the level of viral antigens (HBsAg, HBeAg, and HBc), pgRNA, and HBV DNA, while ectopic expression of Flag-SMC3 strongly repressed viral antigens, pgRNA, and HBV-DNA abundance. Immunofluorescence assays also observed an obvious decrease of HBsAg levels in SMC3-overexpressed Huh7 cells (Figure 3I). Furthermore, the knockdown of another 2 crucial cohesin subunits, SMC1A and RAD21, also significantly promoted HBsAg/HBeAg expression and pgRNA transcription (Figure 3J). These data illustrate that the cohesin complex inhibits HBV replication.
Figure 3.
Cohesin inhibits HBV replication. (A) Huh7 cells were transfected with Flag-SMC3 or siSMC3 for 72 hours, the expression of cohesin complex subunits was measured with Western blot. (B) Huh7 cells were transfected with MC-HBV for 3 days, while PHH, HepaRGNTCP, and HLCZ01 cells were infected with HBV at 200 genome equivalents (Geq) for 7 days. Then, cccDNA was extracted using a Hirt protein-free DNA extraction procedure and subjected to Southern blot assay. The cccDNA from the HepAD38 cell line was taken as a positive control. (C and H) In the MC-HBV/Huh7 model, MC-HBV was cotransfected with Flag-SMC3 or siSMC3 into Huh7 cells for 72 hours; in HBV-infected HLCZ01, PHH, and HepaRGNTCP models, cells were infected with HBV at 200 Geq for 24 hours and then transfected with Flag-SMC3 or siSMC3 for another 5 days, HBV replication was analyzed through measuring HBsAg/HBeAg (enzyme-linked immunosorbent assay), pgRNA, HBV DNA, and HBc antigen (HBcAg) (Western blot) (n = 3). (I) Huh7 cells were transfected with Flag-SMC3 and MC-HBV for 72 hours, and immunofluorescence staining was performed to detect Flag-SMC3 and HBsAg expression levels. Scale bar: 20 μm. (J) Huh7 cells were transfected with MC-HBV and siSMC1A or siRAD21 for 72 hours, and viral replication was analyzed (n = 3). Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PF-DNA, protein-free DNA.
Although SMC3 significantly inhibited the production of HBV antigens, HBV DNA, and pgRNA, the cccDNA level was not altered significantly by either SMC3 overexpression or SMC3 knockdown in MC-HBV–transfected Huh7 cells and HBV-infected HLCZ01 cells (Figure 4A), suggesting that cohesin inhibits HBV replication mainly at the transcriptional level. RNAPII and phosphorylation at its C-terminal domain (CTD) were essential for transcriptional initiation and elongation,20 we then assessed whether SMC3 altered the accumulation of RNAPII and its phosphorylated forms CTD-S2-RNAPII on cccDNA. As shown in Figure 4B, the ChIP assay showed that SMC3 overexpression decreased significantly, while SMC3 knockdown increased the enrichment of RNAPII and CTD-S2-RNAPII on cccDNA. Concurrently, the co-immunoprecipitation (Co-IP) assay failed to detect any interaction between RNAPII and SMC3/SMC1A (Figure 4C), suggesting that the SMC3-induced suppression of RNAPII enrichment on cccDNA was not owing to the interaction of RNAPII with cohesin. Epigenetic modification of cccDNA-bound histones is critical for the dynamic epigenetic modulation of cccDNA transcriptional activity, further ChIP assay found that ectopic SMC3 expression decreased the enrichment of active H3K27ac and H3K4me3 modification, but increased the abundance of repressive H3K9me3 modification in cccDNA, and vice versa (Figure 4D). The prominent role of cohesin complex is to embrace sister chromatids as a ring, leading to alteration of chromatin structure and gene regulation.17 Therefore, we hypothesized that the binding of cohesin changed the structure of cccDNA, leading to subsequent epigenetic repression and decreased RNAPII recruitment. To address that, we purified cohesin complex from cultured HepG2 cells with SMC1A-based IP (Figure 4E), and analyzed the interaction of cohesin and cccDNA in vitro using atomic force microscopy (AFM) imaging. As shown in Figure 4F, MC-HBV existed mainly as a relaxed circular formation under mock treatment or incubation with IgG-precipitated cell lysates, but it formed a dense cluster structure after incubation with cohesin complex, suggesting that cohesin extrudes cccDNA loops to shape its organization. Altogether, these data indicate that the cohesin complex inhibits HBV transcription and replication via reshaping cccDNA conformation to impede RNAPII recruitment on cccDNA.
Figure 4.
Cohesin reshapes cccDNA conformation to impede RNAPII recruitment. (A) In MC-HBV–transfected Huh7 and HBV-infected HLCZ01 models, SMC3 was overexpressed or knocked down, and cccDNA levels were detected by qPCR (n = 3). (B) Flag-SMC3 or siSMC3 was cotransfected with MC-HBV into Huh7 cells for 72 hours, enrichment of RNAPII and CTD-S2-RNAPII on cccDNA was measured by ChIP (n = 3). (C) Interaction of RNAPII with cohesin complex was analyzed by detecting SMC1/SMC3 enrichment using Co-IP assay. (D) Flag-SMC3 or siSMC3 was cotransfected with MC-HBV into Huh7 cells for 72 hours, and H3K27ac, H3K4me3, and H3K9me3 enrichment on cccDNA was measured by ChIP (n = 3). (E) Cohesin complex was purified from HepG2 cells with Co-IP using SMC1A antibody and glycine-HCl elution. (F) MC-HBV was incubated with purified cohesin complex at room temperature for 30 minutes, mock treatment and IgG precipitation were taken as control. MC-HBV conformation was analyzed with AFM imaging, and arrows indicate the classic cccDNA structure, the box represents the amplified region. Data are presented as means ± SD. ∗P < .05, ∗∗P < .01.
Cohesin Restricts HBV Transcription in a SMC5/6-Independent Manner
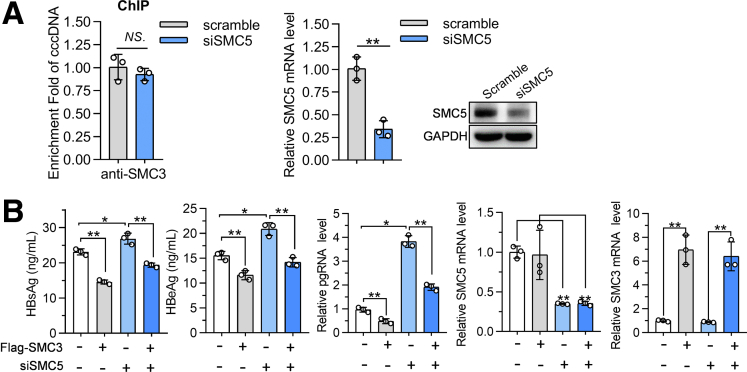
The SMC5/6 complex, another member of SMC complexes, recently was identified as a host HBV restriction factor for repressing cccDNA transcription.9 To evaluate whether the antiviral ability of cohesin couples with SMC5/6 complex, SMC5 was knocked down in MC-HBV–transfected Huh7 cells. The ChIP assay showed that SMC5 silence did not alter the accumulation of SMC3 on cccDNA (Figure 5A). Furthermore, although SMC5 knockdown significantly increased HBsAg and HBeAg expression and pgRNA transcription, which is consistent with a previous report,9 SMC5 knockdown did not affect SMC3-mediated down-regulation of HBsAg/HBeAg and pgRNA (Figure 5B). These results suggest that the inhibitory role of cohesin on HBV replication may be independent of the SMC5/6 complex.
Figure 5.
Cohesin inhibits HBV transcription independently on SMC5/6 complex. (A) Huh7 cells were transfected with siSMC3 and MC-HBV for 72 hours, SMC3 enrichment on cccDNA was detected by ChIP (n = 3), RT-qPCR and Western blot measured SMC5 expression. (B) MC-HBV was cotransfected with Flag-SMC3 and siSMC5 into Huh7 cells for 72 hours, and HBsAg, HBeAg, and pgRNA levels were measured as described in Figure 3A (n = 3). Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Cohesin Loading Onto cccDNA Is Necessary for Its Antiviral Role
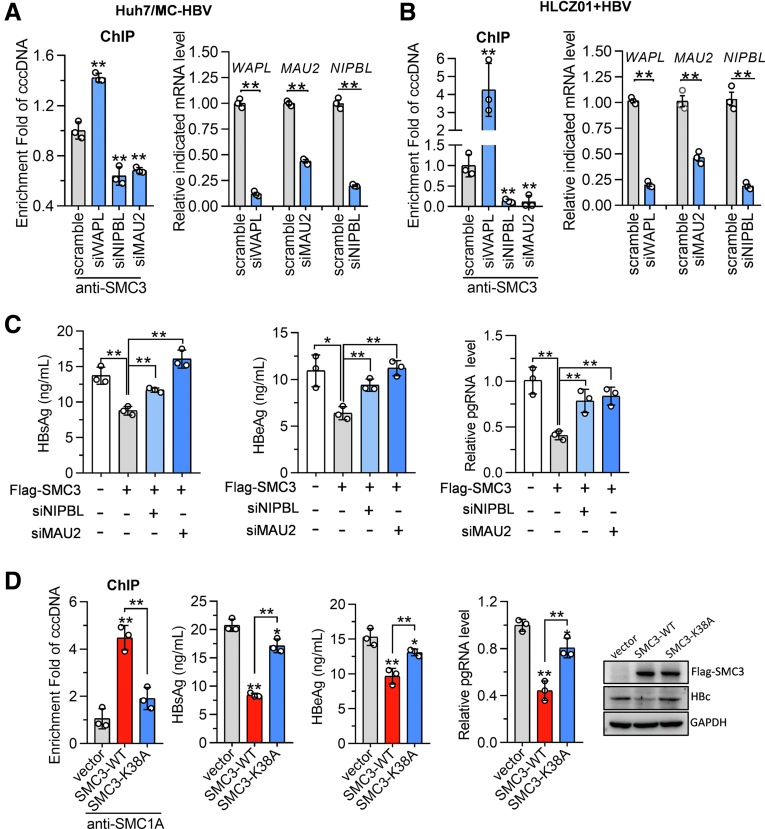
Cohesin is loaded on chromosomes by NIPBL- MAU2 chromatid cohesion factor homolog (MAU2) complex,21 and is released from chromatin by WAPL.22 Both NIPBL and WAPL were detected in our MS data and mapped into a cluster of chromosome segregation pathways (Figure 2G). We therefore came to analyze whether these regulators are involved in the loading of the cohesin complex on cccDNA. As shown in Figure 6A and B, WAPL knockdown increased significantly, while NIPBL and MAU2 silence repressed the enrichment of SMC3 on cccDNA. Moreover, silencing NIPBL or MAU2 significantly restored the SMC3-mediated repression of viral antigen expression and pgRNA transcription in MC-HBV–transfected Huh7 cells (Figure 6C).
Figure 6.
The loading of cohesin onto cccDNA is necessary for its antiviral role. (A) MC-HBV was cotransfected with siWAPL, siNIPBL, or siMAU2 into Huh7 cells for 72 hours, (B) HBV-infected (200 genome equivalents [Geq]) HLCZ01 cells for 24 hours, then siWAPL, siNIPBL, and siMAU2 were transfected for another 48 hours. RT-qPCR detected the knockdown effect and the enrichment of SMC3 on cccDNA was measured by ChIP (n = 3). (C) MC-HBV was cotransfected with Flag-SMC3 and siNIPBL or siMAU2 into Huh7 cells for 72 hours, and HBsAg, HBeAg, and pgRNA levels were measured (n = 3). (D) MC-HBV was cotransfected with WT SMC3 or SMC3-K38A mutant into Huh7 cells for 72 hours, and HBsAg, HBeAg, HBc, and pgRNA levels were measured (n = 3). SMC1A binding on cccDNA was detected by ChIP (n = 3). Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; mRNA, messenger RNA.
The adenosine triphosphate hydrolysis (ATPase) activity of cohesin is required for cohesin loading, cohesion establishment, and DNA compaction.23 To evaluate the role of ATPase activity in cohesin's antiviral effect, a K38A mutation was induced in SMC3 to abolish its ATPase activity.23 ChIP assay found that overexpression of SMC3 but not K38A mutant (SMC3-K38A) significantly promoted the accumulation of cohesin on cccDNA (Figure 6D). In accordance, SMC3-K38A failed to repress HBV antigen expression and pgRNA transcription (Figure 6D). Collectively, these data suggest that the loading of cohesin on cccDNA is necessary for its inhibition on HBV replication, and the ATP activity of SMC3 is required for its enrichment on cccDNA and cohesin-mediated inhibition on HBV replication.
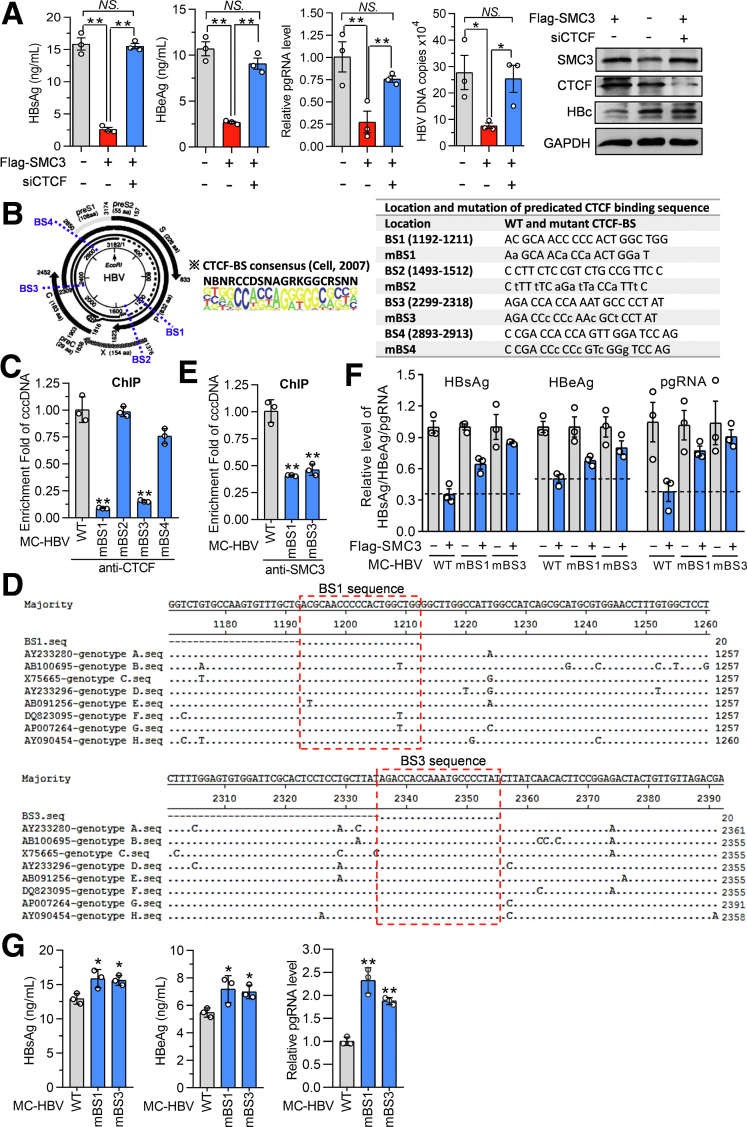
CCCTC-Binding Factor–Mediated Recruitment of Cohesin Complex on cccDNA Is Necessary for Its Inhibition on HBV Replication
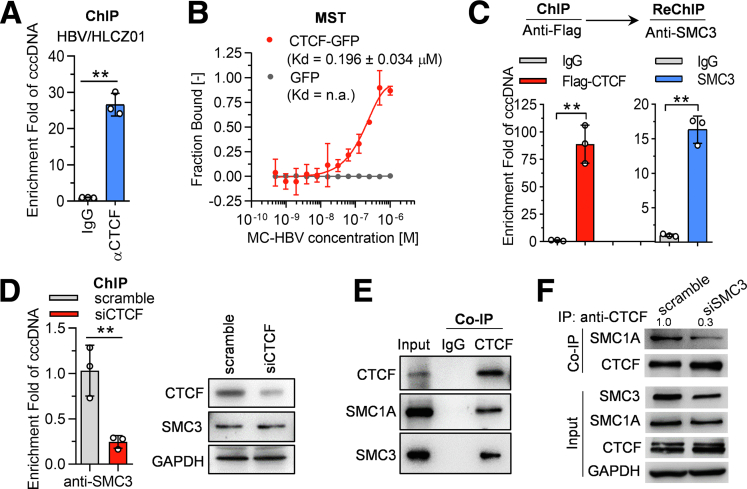
It has been well documented that CCCTC-binding factor (CTCF) mediates the recruitment of cohesin complex onto DNA for transcriptional insulation,24 and CTCF was identified to potentially interact with both circular and linear cccDNA (Supplementary Table 1), which prompted us to explore the role of CTCF in the cohesin-mediated inhibition on HBV replication. In HBV-infected HLCZ01 cells, the ChIP assay showed the occupancy of CTCF on cccDNA (Figure 7A). In accordance, the MST experiment confirmed the binding of GFP-CTCF with MC-HBV at a Kd of 0.196 ± 0.034 μmol/L (Figure 7B). Next, to further evaluate whether CTCF forms a complex to bind with cccDNA, Flag-CTCF and MC-HBV were cotransfected into Huh7 cells, and a Flag-ChIP/SMC3-sequential ChIP (reChIP) assay was performed. As shown in Figure 7C, both SMC3 and CTCF were found to occupy cccDNA concurrently. Moreover, CTCF knockdown led to a significant reduction of SMC3 occupancy on cccDNA without changing SMC3 expression (Figure 7D). Furthermore, Co-IP verified the interaction of CTCF with SMC1A and SMC3 (Figure 7E), and SMC3 knockdown remarkably attenuated the interaction of CTCF and SMC1A (Figure 7F). These data suggest that CTCF recruits cohesin to bind with cccDNA as a complex.
Figure 7.
CTCF mediates cohesin loading onto HBV cccDNA. (A) HLCZ01 cells were infected with HBV (200 genome equivalents [Geq]) for 72 hours, the interaction of CTCF with cccDNA was analyzed by ChIP (n = 3). (B) The green fluorescent protein (GFP)-CTCF protein was incubated with increasing concentrations of MC-HBV, specific interactions were quantified by MST and plotted with the dissociation constant (Kd) equation, GFP protein served as control (n = 3). (C) Huh7 cells were transfected with Flag-CTCF and MC-HBV for 72 hours, binding of CTCF with cccDNA was first estimated with ChIP using anti-Flag antibody, after competitively eluting with Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys (DDDDK) peptide, reChIP assay was performed using an anti-SMC3 antibody to detect the occupancy of SMC3 with CTCF on cccDNA (n = 3). (D) siCTCF was cotransfected into Huh7 with MC-HBV for 72 hours. ChIP assay was performed with an anti-SMC3 antibody to analyze the enrichment of SMC3 on cccDNA (n = 3). (E) Co-IP was performed with an anti-CTCF antibody or IgG to analyze the interaction of CTCF with SMC1A and SMC3. (F) siSMC3 was transfected into Huh7 for 72 hours, and Co-IP was performed with an anti-CTCF antibody to analyze the interaction of CTCF and SMC1A. Relative quantification was performed by analyzing the relative ratio of band density using ImageJ software (National Institutes of Health, Bethesda, MD) as follows: relative SMC1A level interacting with CTCF = band density ratio (immunoprecipitated SMC1A/immunoprecipitated CTCF)/band density ratio (input SMC1A/input CTCF), scramble group set as 1. Data are presented as means ± SD. ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To further determine whether CTCF participates in cohesin-mediated inhibition of HBV replication, we simultaneously silenced CTCF and forced SMC3 expression in MC-HBV–transfected Huh7 cells. As expected, CTCF knockdown almost completely rescued the SMC3-induced reduction of viral proteins (HBsAg/HBeAg/HBc), pgRNA transcription, and HBV-DNA abundance (Figure 8A). Further sequence analysis using the CTCF binding site database (http://insulatordb.uthsc.edu) predicted 4 potential CTCT binding sites (BS)1–4 in the HBV genome (ayw strain, NC_003977) (Figure 8B). The synonymous mutation then was induced at each CTCF-BS in MC-HBV without altering the viral protein-coding sequence (Figure 8B), and cccDNA-specific ChIP analysis found BS1 and BS3 mutation greatly abrogated the CTCT enrichment on cccDNA (Figure 8C). Sequence alignment showed that CTCF-BS1 and BS3 were highly evolutionarily conserved among different HBV genotypes (Figure 8D). ChIP assay showed that, compared with wild-type (WT) MC-HBV, MC-HBV with a mutation at BS1 and BS3 (mBS1 and mBS3) significantly lost the binding with cohesin (Figure 8E). In accordance, SMC3 overexpression greatly suppressed the level of HBV antigens and pgRNA in hepatocytes transfected with WT MC-HBV, but not mBS1 and mBS3 (Figure 8F). These data suggest that the sequence-specific binding of CTCF to the HBV genome is indispensable for the accumulation of cohesin with cccDNA and the cohesin-mediated HBV inhibition. Furthermore, compared with WT MC-HBV, mBS1 and mBS3 mutants showed a higher viral replication level (Figure 8G). Collectively, all these data strongly suggest that CTCF interacts with and recruits cohesin to cccDNA and then insulates against HBV transcription and replication.
Figure 8.
The CTCF-mediated cohesin recruitment on cccDNA is necessary for cohesin-initiated inhibition on HBV replication. (A) MC-HBV was cotransfected with Flag-SMC3 and siCTCF into Huh7 cells for 72 hours, and HBsAg, HBeAg, HBc, pgRNA, and HBV-DNA levels were measured (n = 3). (B) The localization of CTCF-BS in the HBV genome and their sequence of WT and synonymous mutants. ※The CTCF-BS consensus sequence. (C) MC-HBV and CTCF-BS mutants were transfected into Huh7 for 72 hours, and the enrichment of CTCF on cccDNA was analyzed by ChIP (n = 3). (D) Sequence alignment of CTCF-BS1 and BS3 with different genotypes of HBV strains using the Clustal W algorithm of DNAstar Megalign software (Madison, WI). The dot represents the identical nucleotide; the red box indicates CTCF-BS1 and BS3 sequences. (E) MC-HBV and CTCF-BS mutants were transfected into Huh7 for 72 hours. The cccDNA binding of SMC3 was analyzed by ChIP (n = 3). (F) Flag-SMC3 was cotransfected with WT MC-HBV and CTCF-BS mutants (mBS1, mBS3) into Huh7 cells for 72 hours, HBsAg/HBeAg expression and pgRNA transcription were measured (n = 3). (G) WT, CTCF-mBS1, and CTCF-mBS3 mutants of MC-HBV were transfected into Huh7 for 72 hours, HBsAg/HBeAg expression and pgRNA transcription were measured (n = 3). Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
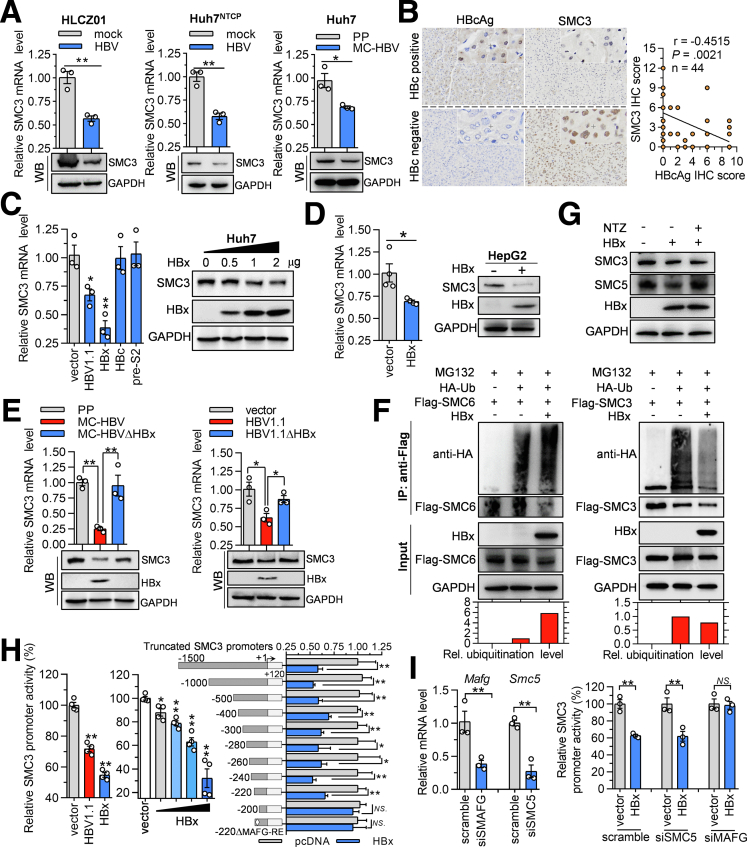
HBx Transcriptionally Represses SMC3 Expression
It is well known that HBV has evolved multiple mechanisms against host antiviral effects.8,9,11,25 Hence, we further investigated whether HBV regulated SMC3 expression. As shown in Figure 9A, SMC3 expression was inhibited in both MC-HBV–transfected Huh7 cells and HBV-infected HLCZ01 and Huh7NTCP cells. To further verify the negative correlation between HBV replication and SMC3 expression in patients, SMC3 abundance was accessed in para-nontumor liver tissues from 44 clinical HCC patients. As shown in Figure 9B, quantification analysis of immunohistochemical staining showed a significant negative correlation between SMC3 and HBc expression level, a well-accepted HBV replication marker. Collectively, these data suggest that HBV suppresses SMC3 expression.
Figure 9.
HBx transcriptionally represses SMC3 expression. (A) HLCZ01 and Huh7NTCP cells were infected with HBV at 200 genome equivalents (Geq) for 5 days, Huh7 cells were transfected with MC-HBV for 3 days, and the messenger RNA (mRNA) and protein abundance of SMC3 were detected by RT-qPCR and Western blot. (B) The correlation analysis of immunohistochemical (IHC) staining of SMC3 and HBc antigen (HBcAg) in consecutive sections of para-nontumor tissue section obtained from HCC patients. (C) Huh7 cells were transfected with HBV1.1, HBc, HBx, and pre-S2 expression plasmids for 72 hours, SMC3 mRNA level was measured with RT-qPCR (n = 3) (left panel), HA-HBx plasmid at a different dose was transfected into Huh7 cells for 72 hours, and SMC3 protein was detected with Western blot (right panel). (D) HBx construct was transfected into HepG2 cells for 48 hours, SMC3 mRNA and protein expression were detected (n = 3). (E) MC-HBV and MC-HBVΔHBx or HBV1.1 and HBV1.1ΔHBx were transfected into Huh7 cells for 72 hours, and SMC3 mRNA and protein expression were detected. (F) Flag-SMC3 or Flag-SMC6 was cotransfected with HBx and HA-Ub into HEK293 cells for 48 hours, and then treated with MG132 for 4 hours. Flag-SMC6 (left) and Flag-SMC3 (right) were immunoprecipitated and immunoblotted with anti-HA antibody to measure their ubiquitylation. (G) HBx was overexpressed in Huh7 for 24 hours and then treated with 20 μmol/L nitazoxanide (NTZ) for 36 hours, SMC3 and SMC5 protein expressions were detected. (H) HBV1.1 and HBx constructs or HBx construct at different doses were cotransfected with SMC3 promoter-reporter, or HBx were cotransfected with a series of truncated SMC3 promoter-reporter constructs into Huh7 cells for 48 hours, the dual-luciferase assay was performed to detect SMC3 promoter activity (n = 3). (I) MAFG-siRNA or SMC5-siRNA were transfected into Huh7 cells for 24 hours and then the cells were cotransfected with SMC3 promoter-reporter and HBx expressing plasmid for 48 hours. The knockdown efficiency was evaluated by RT-qPCR, and SMC3 promoter activity was detected by dual luciferase assay (n = 3). Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
To determine which viral protein is responsible for repressing SMC3 expression, we transfected different HBV constructs into Huh7 cells. Overexpression of HBx and HBV1.1 significantly reduced SMC3 messenger RNA abundance (Figure 9C, left panel), and HBx inhibited SMC3 protein expression in a dose-dependent manner in Huh7 cells (Figure 9C, right panel). Consistently, ectopic HBx expression significantly repressed SMC3 transcription and expression in HepG2 cells (Figure 9D). Moreover, mutation of A at 1376 (ATG→TTG) causing HBx deletion in both MC-HBV and HBV1.1 (MC-HBVΔHBx and HBV.1ΔHBx) nearly destroyed the HBV-induced suppression of SMC3 expression in Huh7 cells (Figure 9E), which further confirmed the critical role of HBx in down-regulating SMC3 expression.
Recently, the SMC5/6 complex was reported to be degraded by HBx through hijacking DDB1-containing E3 ubiquitin ligase,9 and nitazoxanide efficiently can inhibit the HBx-DDB1 interaction by restoring SMC5 protein level.26 After that, HBx overexpression accelerated SMC6 ubiquitylation. However, HBx did not affect SMC3 ubiquitylation (Figure 9F). Similarly, nitazoxanide treatment dampened HBx-induced SMC5 degradation but failed to rescue the HBx-mediated reduction of SMC3 protein (Figure 9G). Consistent with the well-defined transactivation activity of HBx,8 the luciferase reporter assay found that HBV1.1 and HBx significantly suppressed SMC3 promoter activity in a dose-dependent manner (Figure 9H). Furthermore, cotransfection and luciferase reporter assays with a series of truncated SMC3 promoter-reporter constructs further identified -220 to -200 nt as the core region of SMC3 promoter retaining responsiveness to HBx (Figure 9H). Using the JASPAR database (https://jaspar.genereg.net), a responsive element of MAF BZIP transcription factor G (MAFG-RE) was predicted in this core region. To verify whether MAFG-RE is responsible for HBx-mediated suppression of SMC3 transcription, a deletion construct of the SMC3 promoter-reporter plasmid losing the MAFG-RE (-220ΔMAFG-RE) was prepared for cotransfection and dual-luciferase assays. As shown in Figure 9H (right panel), HBx lost the inhibition effect on the promoter activity of -220ΔMAFG-RE. Consistently, knockdown of MAFG abrogated HBx-mediated suppression of SMC3 promoter activity (Figure 9I). Considering that HBx can degrade the SMC5/6 complex and repress the transcription of episomal plasmid DNA,9 we then evaluated whether the SMC5/6 complex is involved in HBx-regulated SMC3 expression. As shown in Figure 9I, SMC5 silence did not affect the HBx-induced decrease of SMC3 promoter activity. Collectively, all these results suggest that HBx inhibits SMC3 transcription in a SMC5/6-independent manner.
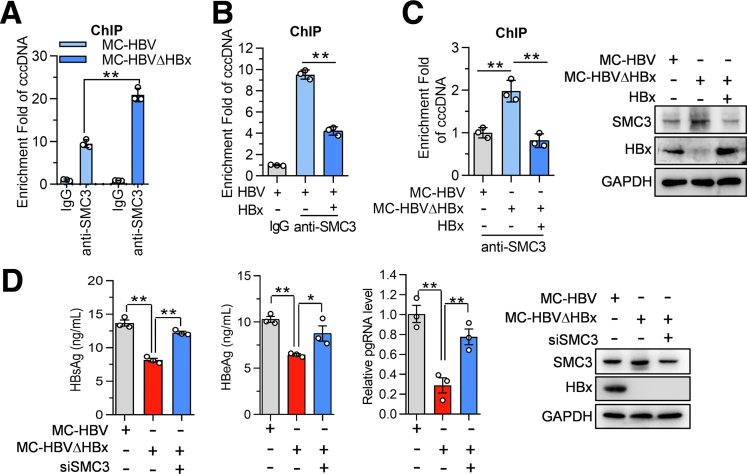
We then came to investigate whether HBx altered SMC3 accumulation on cccDNA. As expected, HBx deletion in MC-HBV (MC-HBVΔHBx) increased the abundance of SMC3 on cccDNA (Figure 10A), and ectopic HBx expression significantly reduced the enrichment of SMC3 on cccDNA in HBV-infected HepG2NTCP cells (Figure 10B). Further rescue assay found that HBx overexpression greatly restored the reduced SMC3 enrichment on cccDNA caused by HBV in Huh7 cells (Figure 10C). In line with the well-characterized critical role of HBx in HBV replication,6 compared with WT MC-HBV, MC-HBVΔHBx drove a lower level of HBsAg/HBeAg expression and pgRNA transcription, accompanied by an increase in SMC3 expression (Figure 10D). More importantly, SMC3 knockdown partially rescued the pgRNA and HBsAg/HBeAg abundance in MC-HBVΔHBx–transfected hepatocytes (Figure 10D). These data imply that HBx transcriptionally reduces SMC3 expression to counteract its antiviral effect.
Figure 10.
HBx promotes viral evasion from cohesin's restriction role. (A) MC-HBV and MC-HBVΔHBx were transfected into Huh7 cells for 3 days, (B) HBV-infected HepG2NTCP cells for 24 hours, and then transfected HA-HBx for another 3 days. ChIP assay was performed to measure the enrichment of SMC3 on cccDNA (n = 3). (C) HBx construct was cotransfected with MC-HBV or MC-HBVΔHBx into Huh7 cells for 3 days, and ChIP assay was performed to measure the enrichment of SMC3 on cccDNA (n = 3) while SMC3 expression was analyzed by Western blot. (D) SMC3 siRNA was transfected into Huh7 for 24 hours, then the cells were transfected with MC-HBV or MC-HBVΔHBx for another 72 hours. HBV replication was analyzed (n = 3) and SMC3 expression was detected by Western blot. Data are presented as means ± SD. ∗P < .05, ∗∗P < .01. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
Chronic HBV infection remains a critical global public health concern, and cccDNA is the major barrier to curative HBV therapy.2 cccDNA in the nuclei of a hepatocyte exists as a minichromosome via interacting with viral and host proteins, and these interactions are crucial for HBV replication.27 However, the host factors involved in the functional regulation of HBV cccDNA remain poorly understood. Here, by a MC-HBV–based screen model, we identified cohesin complex as a novel host factor binding with cccDNA and restricting HBV replication. Mechanistically, with the help of the NIPBL-MAU2 complex and CTCF, cohesin occupies and shapes cccDNA to prevent the recruitment of RNAPII and the consequent transcription of cccDNA. Conversely, HBV antagonized its antiviral role via HBx-mediated SMC3 transcriptional repression, forming a negative feedback loop in HBV infection. Thus, our data link cohesin to HBV transcription/replication control and provide novel insight into HBV therapy.
The interplay between host factors and cccDNA determines minichromosome remodeling and its transcriptional activity,4 but the very low cccDNA copies in HBV-infected hepatocytes and lack of a suitable cccDNA labeling method severely limit the labeling cccDNA–host interaction study. Here, we established an MC-HBV model serving as a cccDNA surrogate, supporting a high level of HBV replication and binding with epigenetic regulators in HBV-infected HepG2NTCP cells. With the help of a biotinylated MC-HBV/pull-down/MS–based screening approach, we described the interplay profile between cccDNA and host cell nuclear proteins. A total of 306 candidates clustered mainly into DNA conformation/function regulation, and RNA metabolism regulation pathways, were identified. In particular, 134 candidates were enriched into DNA conformation and function regulation clusters, consistent with accumulating evidence showing the close association of epigenetic regulation of cccDNA with HBV transcription and replication.5,7,28 The candidates identified in our study include some reported epigenetic regulators of cccDNA such as HDAC1, HDAC2, and KDM2A. In addition to the cohesin complex, we found several chromatin remodeling complexes in our MS data, including the minichromosome maintenance (MCM) complex, the tailless complex polypeptide 1 ring complex (TRiC)/the ATP-dependent chaperone chaperonin containing tailless complex polypeptide 1 (CCT) complex, the LCR-associated remodeling complex (LARC), the RuvB like AAA ATPase 1/2 (RUVBL1/RUVBL2) complex, and the Switch/sucrose non-fermentable (SWI-SNF) chromatin remodeling-related breast cancer type 1 susceptibility protein (BRCA1) complex. These complexes have been reported to regulate viral replication,29, 30, 31, 32 but it needs to be investigated further whether they participate in cccDNA minichromosome maintenance and HBV replication.
Interestingly, some RNA-binding proteins (RBPs), such as several DEAD-box RNA helicases (DDXs) and RNA binding motif proteins (RBMs), are identified as the candidate interaction partners of cccDNA. Recent large-scale RBP ChIP sequencing analysis showed that widespread RBPs present inactive chromatin regions in the human genome, and RBPs show a strong preference for hotspots in the genome, particularly in gene promoters. RBPs show extensive co-association with transcription factors, which are important in splicing regulation and chromatin binding, DNA looping, and transcription.33 Thus, we propose that the presence of these RBPs in cccDNA might enhance the interaction network of transcription factors, regulatory RNAs, and epigenetic chromosome remodeling. Altogether, although the MC-HBV model carries bacterial dam methylation and cannot simulate the cccDNA-host interaction under HBV infection circumstances well, we provide whole cell–based interplay profiling of cccDNA with host nuclear factors, which should be beneficial for understanding the molecular mechanisms of cccDNA maintenance and function.
The other important finding is that we identified cohesin complex as a novel host restriction factor of HBV. Cohesin originally was identified and named for its role in controlling sister chromatid cohesion. Increasing evidence indicates that cohesin participates in DNA looping, transcriptional regulation, and chromosome stability.34 Interestingly, for Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) with the circular genome, cohesin is shown to regulate viral replication through mediating DNA looping of viral episome and controlling viral gene transcription.35,36 The conformation of cccDNA plays a vital role in HBV transcription.7,27,28 Our data indicate that cohesin represses pgRNA transcription via compacting cccDNA to pause RNAPII recruitment on HBV cccDNA. Pull-down, ChIP, and MST assays validated the binding of cohesin with cccDNA.
Furthermore, AFM analysis observed that the closed cccDNA looping was formed from the relaxed circular formation after incubation with purified cohesin complex, suggesting that cohesin complex is an important cccDNA conformation reconstruction factor. Cohesin's loading on chromosomes depends on the NIPBL-MAU2 loader.21 Either NIPBL silence or MAU2 knockdown reduced the cohesin-cccDNA binding and impaired the inhibitory role of cohesin on HBV replication. The ATPase activity of cohesin also is necessary for cohesin loading and cohesion establishment.23 SMC3-K38A mutation abolished its ATPase activity23 and successively compromised its cccDNA loading and antiviral ability. Although another SMC complex, SMC5/6, has been identified as a novel host restriction factor of cccDNA, its antiviral mechanism and cccDNA loading regulation are unclear.9 Otherwise, the inhibitory role of cohesin on HBV was independent of the SMC5/6 complex. Our findings extend the understanding of cohesin complex assembly on cccDNA to form a closed conformation and suggest the conservative role of SMC complexes in determining cccDNA conformation and viral replication.
It has been well documented that CTCF is critically involved in cohesin-mediated DNA looping of both human and viral genomes.24,35,36 It also has been reported that CTCF can be co-immunoprecipitated with the cohesin complex.37 In this study, the reChIP and Co-IP assays confirmed that CTCF and cohesin composed a complex to co-localize to cccDNA. CTCF fostered cohesin's loading on cccDNA and is required for cohesin-mediated inhibition of HBV replication. Nevertheless, in the mammal genome, CTCF is dispensable for cohesin to load onto DNA, which mainly favors enrichment of cohesin at specific binding sites.24 The small size of cccDNA may cause this difference, and CTCF is in favor of positioning cohesin. HBV mutagenesis assays found that CTCF binding sites in the HBV genome (BS1 at the enhancer I (EnhI) region and BS3 at the HBc coding region) were responsible for CTCF and cohesin's binding on cccDNA. Similarly, D'Arienzo et al38 reported that CTCF accumulated on cccDNA through binding to the BS1 to repress HBV transcription. Here, we identified an extra CTCF binding site, BS3, in the HBV genome, which is located in the HBc gene and served as another evolutionarily conserved CTCF-BS among genotype A–H strains. Functionally, CTCF is indispensable for recruiting cohesin complex to insulate viral transcription.
HBx, a multifunctional regulator, is required for the viral life cycle and can hijack host factors for sustaining viral replication.39 HBV manipulates HBx to degrade SMC5/6 by hijacking DDB1-containing E3 ubiquitin ligase, relieving the inhibition and allowing HBV replication.9 Here, we showed the HBx-mediated repression of SMC3. Moreover, HBx down-regulated SMC3 expression via transcriptional repression, and SMC3 knockdown partially rescued viral replication in the MC-HBVΔHBx model. These findings suggest a common feature of HBx-mediated functional inhibition of SMC complexes to maintain cccDNA-driven viral replication. Considering the well-known protumoral role of HBx, HBx-induced SMC3 down-regulation may be a new pathway contributing to HBV-related HCC. SMC3 deficiency triggers genomic instability and p53-dependent apoptosis,34 and mutations in genes encoding cohesin subunits recently were identified in several types of tumors.40 However, further effort is needed to uncover the exact role of HBx-mediated SMC3 repression in HCC.
HBx-induced degradation of the SMC5/6 complex is one of the most important mechanisms for HBV to relieve host restriction. Here, we found that SMC3 silence also partially rescued the replication of HBVΔHBx. The cohesin and SMC5/6 complexes have been reported to interplay with each other during chromosome segregation and DNA repair. Human SMC5/6 complex promotes DNA double-strand break repair by facilitating the recruitment of cohesin to double-strand breaks.41 In addition, cohesin retention defects also have been described in Saccharomyces cerevisiae with Smc5 mutation during aberrant meiotic divisions.42 More importantly, SMC5/6 dynamics are very similar to those of cohesin in time and space.43,44 In Schizosaccharomyces pombe, SMC5/6 locations also significantly overlap with cohesin distribution, and the SMC5/6 complex is required for timely removal of cohesin from chromosome arms.45,46 This evidence indicate the functional crosstalk of these 2 complexes in chromosomal structure maintenance, although little is known about the mechanisms by which this occurs. Both cohesin and SMC5/6 complexes occupy HBV cccDNA and restrict its transcription, suggesting that cohesin also may interplay with SMC5/6 complex on cccDNA and this interplay might further lead to the failure of the SMC5/6 complex in restricting HBVΔHBx transcription when SMC3 was knocked down. The potential functional interplay of cohesin and SMC5/6 complexes on HBV cccDNA transcription will be investigated further in future studies.
In conclusion, this study established a cccDNA-host interaction screening model and provided a comprehensive view of the cellular proteins associated with cccDNA functional regulation. The cohesin complex binds to cccDNA to restrict HBV transcription and replication in a CTCF-dependent manner. These findings expand our understanding of HBV and host interaction and provide new insights into controlling HBV replication via targeting the cohesin complex.
Materials and Methods
Regents and Cell Culture
The human HCC cell lines Huh7, HepG2, HepG2NTCP, and Huh7NTCP47 were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The HepG2.2.15 cell line stably transformed with 2 copies of the HBV genome was maintained in minimum essential medium (MEM) supplemented with 10% FBS, 200 μg/mL G418, and 2 mmol/L glutamine. The HLCZ01 cell line was kindly gifted by Professor Zhu (Hunan University) and cultured with DMEM/F12 medium supplemented with 10% FBS, 40 ng/mL dexamethasone, and 10 ng/mL epidermal growth factor (EGF) in collagen-coated tissue culture plates as described previously.48 HepaRGNTCP cells49 were kindly gifted by Professor Yuchen Xia (Wuhan University) and Professor Stephan Urban (University Hospital Heidelberg) and cultured with Williams' medium E, supplied with 10% FBS, 1% penicillin-streptomycin (PS), insulin, 5 μg/mL hydrocortisone, and 80 μg/mL gentamicin. Before HBV infection, HepaRGNTCP cells were cultured with medium containing 1.8% dimethyl sulfoxide for 48 hours. PHH cells were cultured on rat tail collagen-coated plates with Advanced DMEM/F12 (Thermo Scientific, Waltham, MA) plus 500 ng/mL R-spondin1 (Peprotech, Rocky Hill, NJ), B27 (minus vitamin A), 1.25 mmol/L N-acetylcysteine (Sigma, St. Louis, MO, USA), 10 mmol/L nicotinamide (Sigma), 50 ng/mL epidermal growth factor (Peprotech), 10 nmol/L gastrin (Sigma), 3 μmol/L CHIR99021 (Sigma), 50 ng/mL hepatocyte growth factor (HGF) (Peprotech), 100 ng/mL fibroblast growth factor-7 (FGF7) (Peprotech), 100 ng/mL FGF10 (Peprotech), 20 ng/mL transforming growth factor α (Peprotech), and 2 μmol/L A83-01 (Tocris Bioscience, Bristol, UK).
The commercial antibodies used for Western blot in this study were as follows: rabbit anti-SMC3 monoclonal antibody (1:2000, 5696; Cell Signaling Technology, Danvers, MA), rabbit anti-SMC1A monoclonal antibody (1:10,000, ab109238; Abcam, Cambridge, MA), rabbit anti-CTCF polyclonal antibody (1:4000, ab188408; Abcam), rabbit anti-HBc polyclonal antibody (1:2000, B0586; Dako, Copenhagen, Denmark), rabbit anti-HBx polyclonal antibody (1:2000, ab39716; Abcam), mouse anti-PDS5B monoclonal antibody (1:50, SC-81635; Santa Cruz Biotechnology, Santa Cruz, CA), mouse anti-PDS5A monoclonal antibody (1:50, SC-515263; Santa Cruz Biotechnology), mouse anti-RAD21 monoclonal antibody (1:50, SC-271601; Santa Cruz Biotechnology), mouse anti–β-actin monoclonal antibody (1:5000, 66009-I-Ig; Proteintech, Chicago, IL), mouse anti–glyceraldehyde 3-phosphate dehydrogenase (GAPDH) monoclonal antibody (1:5000, 66004-I-Ig; Proteintech), mouse anti-Flag-tag monoclonal antibody (1:1000, M2; Sigma), and mouse anti-HA-tag monoclonal antibody (1:2000, ab130275; Abcam). The commercial antibodies used for ChIP assay in this study were as follows: anti-H3K4me3 antibody (ab8580; Abcam), anti-H3K27me3 (ab6002; Abcam), anti-H3K27ac (ab4729; Abcam), anti-H3K36me3 (ABE435; Sigma), anti-H3K122ac (ab33309; Abcam), anti-H3K9me3 (ab8898; Abcam), anti-KAT3A/CBP (ab2832; Abcam), anti-RNAPII C-terminal domain (CTD) repeat YSPTSPS (ab5095; Abcam), and anti-RNAPII antibody (05-623-Z; Sigma).
The minicircle DNA vector system was purchased from System Biosciences, LLC (Palo Alto, CA), In-Fusion HD Cloning Kit was purchased from Takara Bio, Inc (Kyoto, Japan), commercial enzyme-linked immunosorbent assay kit for HBsAg and HBeAg was purchased from Dade Behring (Marburg, Germany), Label IT nucleic acid labeling kit was purchased from Mirus Bio LLC (Madison, WI), NE-PE Nuclear and Cytoplasmic Extraction Reagents was purchased from Thermo Fisher Scientific, ChIP kit was purchased from Millipore (Merck KGaA, Darmstadt, Germany), plasmid-safe DNase was purchased from Epicentre (Madison, WI), PEG8000, 3-aminopropyltriethoxy-silane and horseradish-peroxidase–streptavidin all were purchased from Sigma, and care HBV PCR Assay V3 kits were purchased from QIAGEN Divisions (Shenzhen, China).
Establishment of MC-HBV
MC-HBV was constructed by combining minicircle DNA technology and the HBV rcccDNA model.14 Briefly, a HBV 1.0 copy genome-containing chimeric intron was amplified from parent rcccDNA plasmid, and minimized minicircle vector retaining the bacterial attachment site (attB)/the phage attachment site (attP) was amplified from pCMV.MC plasmid by high-fidelity PCR. The fragments were extracted and ligated with the In-Fusion HD Cloning Kit (Takara Bio) to form a 7.3-kb pmini-HBV PP. The PP then was transformed into E coli strain ZYCY10P3S2T and the transformed E coli then was induced with 0.2 mg/mL arabinose (Sigma-Aldrich, St. Louis, MO) at 30°C, pH 7.0, to generate MC-HBV (Figure 1A). The induction efficiency was estimated with EcoRI digestion and gel electrophoresis analysis. MC-HBV contains a 1.0 copy HBV genome (3.2 kb) and a chimeric intron (128 bp). As theoretically calculated, MC-HBV only produced a single band of approximately 3.3 kb after total recombination and degradation of PP plasmid (Figure 1B). In hepatocytes, because of the alternative splicing of pgRNA, the chimeric intron was removed from nascent rcDNA and cccDNA, which can be identified by PCR amplification using P1/P2 primer spanning the chimeric intron region in Table 1, and produced a 431-bp fragment in MC-HBV and a shorter fragment (303 bp) in rcDNA and nascent cccDNA (Figure 1C).
Table 1.
siRNA and Primers in This Study
| Gene and primer | Primer sequence, 5’ to 3’ |
|---|---|
| P1 | GTATTTCCCTGCTGGTGGC |
| P2 | GGTGAGTGATTGGAGGTTG |
| pgRNA-fwd | CTCAATCTCGGGAATCTCAATGT |
| pgRNA-rev | AGGATAGAACCTAGCAGGCATAAT |
| cccDNA-fwd | TTCTCATCTGCCGGACCG |
| cccDNA-rev | CACAGCTTGGAGGCTTGAAC |
| SMC3-fwd | GTTTCAACCCAGCTGGCCCGTG |
| SMC3-rev | CGATGGCTGACTTGGTCACCTTCCA |
| GAPDH-fwd | GGAGTCCACTGGCGTCTTCAC |
| GAPDH-rev | GAGGCATTGCTGATGATCTTGAGG |
| SMC1A-fwd | GTTGCATGGTGCTGGTGCGCAGACC |
| SMC1A-rev | GCGCGGGACGTCCTTTGTCTACG |
| RAD21-fwd | GTGGAAAGAGACAGGAGGAGTAG |
| RAD21-rev | AGGTCTTCTGGTACAAGCGGTG |
| NIPBL-fwd | AAGCAGTGGCTGGTATGAAG |
| NIPBL-rev | TGAACACAAAGCGCTAGAGG |
| MAU2-fwd | CACTTGAGCCCAGGAGATTG |
| MAU2-rev | TGCCTTTCTGCGACCTTG |
| WAPL-fwd | GTAGGCAAAGCAGTGGAGGACT |
| WAPL-rev | CTGTGCCTATGAGACCGTCCTG |
| SMC5-fwd | TGTCACTGTGGACCTAGAGCAG |
| SMC5-rev | GTCTTTGTGCTCCAGATGTTTGC |
| ZHX2-fwd | GGTTCGGACATCACAAGTAGTAG |
| ZHX2-rev | GGTGTGCCGATTCCTTTCTCT |
| siRNA | siRNA sequence, 5’ to 3’ |
| SMC3-siRNA-1 | GGACCAAGUAGAACAGGAATT |
| SMC3-siRNA-2 | AUCGAUAAAGAGGAAGUUUTT |
| CTCF-siRNA-1 | GGAGCCUGCCGUAGAAAUUTT |
| CTCF-siRNA-2 | UUGGUUCGGCAUCGUCGUUTT |
| SMC1A-siRNA | GCAAUGCCCUUGUCUGUGATT |
| RAD21-siRNA | GGUGAAAAUGGCAUUACGGTT |
| NIPBL-siRNA | GCAUCGGUAUCAAGUCCCAUUTT |
| MAU2-siRNA | CCCGCAGUUCGAAGAUGUUTT |
| WAPL-siRNA | CGGACUACCCUUAGCACAATT |
| SMC5-siRNA | GGCAUUAUGUGAAGGCGAAAUAAUU |
fwd, forward; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; rev, reverse.
MC-HBV Biotinylation, Pull-Down, and MS Analysis
MC-HBV was labeled with Label IT biotin reagent (Mirus Bio, LLC) at 37°C for 1 hour and purified with a G50 microspin column according to the manufacturer's protocol. A total of 10 μg biotin-MC-HBV, unlabeled MC-HBV, and biotin-linear-MC-HBV were incubated with 10 mg HepG2 cell nuclear protein at 4°C for 6 hours under constant agitation, and then incubated for another 2 hours with an extra 50 μL of Dynabeads M-280 Streptavidin (Thermo Fisher Scientific, Waltham, MA). After careful wash with phosphate-buffered saline with Tween 20 (PBST) 3 times and PBS with 0.2% Nonidet P-40 (NP40) 2 times, boiling with 25 μL 1 × sodium dodecyl sulfate loading buffer, protein samples then were sent to PTM Biolabs, Inc (Hangzhou, Zhejiang, China) for LC-MS/MS analysis.
MST
Cell lysates from Huh7 cells with SMC3-GFP, SMC1A-GFP, or CTCF-GFP overexpression were incubated with MC-HBV at different concentrations in buffer containing 20 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 0.5% Triton X-100 (Sigma, St. Louis, MO), 0.3% bovine serum albumin, and 0.5 mmol/L ATP for 20 minutes at room temperature; GFP-overexpressed cell lysates were used as control. Then, all MST measurements were performed at 27°C using Monolith NT standard capillaries (NanoTemper Technologies, Munich, Germany) and the Monolith NT.115T device (NanoTemper Technologies) with the laser being on for 5 seconds using 40% power. All curves were plotted with the MO Affinity Analysis V10 software (NanoTemper Technologies), and the thermophoresis signals were fitted with the Kd model and normalized to the fraction bound (X), where X = (Y[c] - minimum)/(maximum - minimum), error bars (SD) were normalized by SD norm = SD(c)/(maximum - minimum).
AFM Imaging
For typical AFM imaging, muscovite mica was freshly cleaved and pretreated with volatile 3-aminopropyltriethoxy-silane in a closed tank for 30 minutes to enhance adhesive capacity. A total of 100 ng MC-HBV was mixed with purified cohesin complex on ice in PBS buffer with 112 mmol/L MgCl2, and incubated for 30 minutes at 32°C in the presence of 0.5 mmol/L ATP, with IgG purified product as control. A 50-μL sample was deposited onto the mica for 10 minutes and the sample area then was washed with 200 μL 1 × Tris-EDTA (TE) buffer with 112 mmol/L MgCl2 5 times. Commercial silicon nitride cantilevers with integrated sharpened tips (SNL-10; Bruker Corporation, Santa, Barbara, CA) were used. The topographic images were captured by peak force tapping mode experiments on a Multimode VIII system (Bruker Corporation) in liquid.
MS, Enriched Ontology Clusters. and Protein–Protein Interaction Network Analysis
Identified proteins by MS in the biotin-MC-HBV group were first compared with those in the unlabeled MC-HBV group and the biotin-linear-MC-HBV group to exclude the nonspecific binding proteins, then the remaining proteins in 2 repeats were overlaid and their peptides/scores were averaged (Supplementary Table 1). We then identified all statistically enriched terms based on GO terms, the Kyoto Encyclopedia of Genes and Genomes (KEGG) terms, and canonical analysis with Metascape (http://metascape.org), accumulative hypergeometric P values and enrichment factors were calculated and used for filtering. The top 20 clusters were listed with their representative enriched terms. Count refers to the number of genes in the user-provided lists with membership in the given ontology term. Percent refers to the percentage of all the user-provided genes that are found in the given ontology term. Log10(P) refers to the P value in log base 10. Log10(q) refers to the multitest adjusted P value in log base 10.
The remaining significant terms then were clustered hierarchically into a tree based on κ statistical similarities among their gene memberships. Then, a 0.3 κ score was applied as the threshold to cast the tree into enriched term clusters. Afterward, we selected a subset of representative terms from this cluster and converted them into a network layout. More specifically, the circle nodes represent enriched terms analyzed based on GO terms and KEGG terms, where their size is proportional to the number of input genes that fall into that term, and the color represents its cluster identity (nodes of the same color belong to the same cluster). Terms with a similarity score >0.3 are linked by an edge (the thickness of the edge represents the similarity score). The network is visualized with Cytoscape (v3.1.2) with force-directed layout and with the edge bundled for clarity. One term from each cluster is selected to have its term description shown as the label.
The protein–protein interaction network was analyzed with MCODE in Cytoscape to identify neighborhoods where proteins are densely connected as previously described.50 Each MCODE network is assigned a unique color. GO enrichment analysis was applied to each MCODE network to assign meanings to the network component. Each MCODE network is assigned a unique color.
Dot-Blot Assay
Biotin-MC-HBV (10 ng) was spotted onto a nitrocellulose membrane with positive charge at the center of the grid, the membrane was allowed to dry, and then blocked by soaking in 5% bovine serum albumin in Tris Buffered Saline with Tween 20 (TBST) for 1 hour. Then, it was incubated with horseradish-peroxidase–streptavidin for 30 minutes and exposed using the enhanced chemiluminescence (ECL) reagent to detect the biotinylation of MC-HBV.
Enzyme-Linked Immunosorbent Assay
HBsAg and HBeAg secreted into cell culture supernatant were measured using commercially available HBsAg or HBeAg Enzyme-Linked Immunosorbent Assay kits (InTec, Inc, Xiamen, China) as protocol. The antigen levels were quantitated in triplicates and represented with OD450/630 optical density.
ChIP and ReChIP Assay
Huh7 cells were transfected with MC-HBV alone or with other plasmids for 72 hours, HBV cccDNA ChIP assay then was performed as previously described.10 Briefly, cells were fixed with 1% formaldehyde for 10 minutes at room temperature quenched by 0.125 mol/L glycine. The cell nucleus was isolated and sonicated at 25% amplitude, 10 seconds on, 10 seconds off, for 14 cycles to shear DNA. The protein–DNA complexes were immunoprecipitated with the indicated antibodies or control IgG. For ReChIP, bound material in the first ChIP with anti-Flag antibody was eluted with 50 μL 0.1 mg/mL Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys (DDDDK) peptide, 0.1% sodium dodecyl sulfate, and 30 mmol/L dithiothreitol in TE for 1 hour at 37°C under constant agitation. The eluent was adjusted to 1.2 mL with dilution buffer and subjected to a second round of ChIP with anti-SMC3 antibody and eluted as described earlier. The retrieved DNA of ChIP and ReChIP were analyzed by quantitative PCR (qPCR) with the cccDNA detecting primers (cccDNA-fwd/rev in Table 1). The qPCR results were presented as relative enrichment fold changes as control.
Cohesin Complex Purification
Huh7 cells (3 × 107) were harvested to lyse with RIPA buffer and then incubated with 4 μg SMC1A antibody (ab109238; Abcam) for 8 hours at 4°C under constant agitation. A total of 20 μL protein A/G magnetic beads was added for another 2 hours of incubation and washed 4 times with PBST buffer. Cohesin/SMC1A antibody/protein A/G beads were treated with 20 μL 0.1 mol/L glycine-HCl (pH 3.0) for 15 seconds to dissociate the binding of antibody and protein A/G and rapidly neutralized with 0.5 μL 1 mol/L Tris-HCl (pH 9.0), protein A/G magnetic beads then were discarded and the purified cohesin complex was stored at -80°C. The purification effect of the cohesin complex was evaluated through measuring subunits of cohesin using Western blot.
Immunohistochemical Staining
Immunohistochemical staining was performed in the adjacent nontumor sections from 44 HCC patients (Table 2) who underwent surgery between January 1, 2013, and May 1, 2014, at Qilu Hospital, Shandong University. None of the patients were positive for hepatitis C virus or human immunodeficiency virus. The study was approved by the Shandong University Medical Ethics Committee in accordance with the Declaration of Helsinki. Immunohistochemical staining using anti-SMC3 antibody (5696; Cell Signaling Technology) and anti-HBc antibody (B0586; Dako) was performed, scored, and analyzed as described previously.10,51 Briefly, 8 fields of approximately 1000 cells from each section were counted and HBc and SMC3 staining were reported separately according to the German semiquantitative scoring system. Each sample was scored according to staining intensity (no staining, 0; weak staining, 1; moderate staining, 2; and strong staining, 3) and the number of stained cells (0%, 0; 1%–25%, 1; 26%–50%, 2; 51%–75%, 3; and 76%–100%, 4). Final immunoreactive scores were determined by multiplying the staining intensity by the number of stained cells, with minimum and maximum scores of 0 and 12, respectively.
Table 2.
Clinical Characteristics of Enrolled Subjects
| Cases | 44 |
|---|---|
| Age, ya | 54.0 ± 4.5 |
| HBeAg positive | 23 (52%) |
| HBsAg positive | 31 (72.1%) |
| HBsAb positive | 13 (30.2%) |
| HBeAb positive | 9 (20.9%) |
| HBcAb positive | 42 (95.5%) |
HBcAb, antibodies to HBc; HBeAb, antibodies to HBeAg; HBsAb, antibodies to HBsAg.
Means ± SD.
RNA Interference, Transfection, and RT-qPCR
siRNAs targeting indicated genes were synthesized chemically from GenePharma (Shanghai, China) (Table 1). Cells grown overnight were transfected with siRNA using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's protocol. Total RNA was extracted from cells using TRIzol reagent (Thermo Fisher Scientific), and 1 μg RNA was used to synthesize complementary DNA using a PrimeScript RT Reagent Kit with genomic DNA Eraser (Takara, Kyoto, Japan). RT-qPCR analysis of gene expression was performed using SYBR Premix Ex Taq (Takara) according to the manufacturer's protocol by the indicated primers (Table 1). Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. Relative gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase using the 2 to the power of minus delta delta Ct (2–ΔΔCt) method.
Statistical Analysis
All data are presented as the means ± standard deviation (SD). Statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, San Diego, CA). Statistical differences between groups were assessed using the Student unpaired 2-tailed t test, 1-way analysis of variance, or 2-way analysis of variance. P values <.05 were considered significant.
Acknowledgments
The authors thank professor Qiang Deng (Institute Pasteur of Shanghai, Chinese Academy of Sciences) for the Cre/parent rcccDNA system. The authors thank the Translational Medicine Core Facility of Shandong University for consultation and instrument availability that supported this work.
CRediT Authorship Contributions
Chunhong Ma (Conceptualization: Lead; Supervision: Lead; Writing – review & editing: Lead)
Zhuanchang Wu (Conceptualization: Lead; Methodology: Lead; Resources: Lead; Validation: Lead; Writing – original draft: Lead; Writing – review & editing: Equal)
Liyuan Wang (Methodology: Equal)
Xin Wang (Methodology: Equal; Resources: Equal)
Yang Sun (Methodology: Equal; Project administration: Equal)
Haoran Lo (Investigation: Equal; Methodology: Equal)
Zhaoying Zhang (Methodology: Equal; Project administration: Equal)
Jinghui Lu (Formal analysis: Equal)
Leiqi Xu (Validation: Equal)
Xuetian Yue (Formal analysis: Equal; Writing – review & editing: Supporting)
Yue Hong (Methodology: Equal; Visualization: Supporting)
Qiang Li (Methodology: Supporting; Resources: Supporting)
Haizhen Zhu (Resources: Equal)
Chenjiang Gao (Resources: Equal)
Yaoqin Gong (Writing – review & editing: Supporting)
Lifen Gao (Writing – review & editing: Supporting)
Xiaohong Liang (Writing – review & editing: Supporting)
Caiyue Ren (Investigation: Supporting)
Shuangjie Li (Formal analysis: Supporting)
Huili Hu (Resources: Supporting)
Xiaohui Zhang (Investigation: Supporting; Methodology: Supporting)
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This study was funded by grants from the National Key Research and Development Program (2021YFC2300603), the National Science Foundation of China (Key Program 81830017, 81902051, and 32170157), Taishan Scholarship (tspd20181201), Major Basic Research Project of Shandong Natural Science Foundation (ZR2020ZD12), Collaborative Innovation Center of Technology and Equipment for Biological Diagnosis and Therapy in Universities of Shandong, and the Key Research and Development Program of Shandong (2019GSF108238).
Supplementary Material
References
- 1.Trepo C., Chan H.L., Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–2063. doi: 10.1016/S0140-6736(14)60220-8. [DOI] [PubMed] [Google Scholar]
- 2.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 3.Ganem D., Prince A.M. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 4.Newbold J.E., Xin H., Tencza M., Sherman G., Dean J., Bowden S., Locarnini S. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol. 1995;69:3350–3357. doi: 10.1128/jvi.69.6.3350-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W., Chen J., Wu M., Zhang X., Zhang M., Yue L., Li Y., Liu J., Li B., Shen F., Wang Y., Bai L., Protzer U., Levrero M., Yuan Z. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology. 2017;66:398–415. doi: 10.1002/hep.29133. [DOI] [PubMed] [Google Scholar]
- 6.Belloni L., Pollicino T., De Nicola F., Guerrieri F., Raffa G., Fanciulli M., Raimondo G., Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollicino T., Belloni L., Raffa G., Pediconi N., Squadrito G., Raimondo G., Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Ren J.H., Hu J.L., Cheng S.T., Yu H.B., Wong V.K.W., Law B.Y.K., Yang Y.F., Huang Y., Liu Y., Chen W.X., Cai X.F., Tang H., Hu Y., Zhang W.L., Liu X., Long Q.X., Zhou L., Tao N.N., Zhou H.Z., Yang Q.X., Ren F., He L., Gong R., Huang A.L., Chen J. SIRT3 restricts hepatitis B virus transcription and replication through epigenetic regulation of covalently closed circular DNA involving suppressor of variegation 3-9 homolog 1 and SET domain containing 1A histone methyltransferases. Hepatology. 2018;68:1260–1276. doi: 10.1002/hep.29912. [DOI] [PubMed] [Google Scholar]
- 9.Decorsiere A., Mueller H., van Breugel P.C., Abdul F., Gerossier L., Beran R.K., Livingston C.M., Niu C., Fletcher S.P., Hantz O., Strubin M. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature. 2016;531:386–389. doi: 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 10.Xu L., Wu Z., Tan S., Wang Z., Lin Q., Li X., Song X., Liu Y., Song Y., Zhang J., Peng J., Gao L., Gong Y., Liang X., Zuo X., Ma C. Tumor suppressor ZHX2 restricts hepatitis B virus replication via epigenetic and non-epigenetic manners. Antiviral Res. 2018;153:114–123. doi: 10.1016/j.antiviral.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Riviere L., Gerossier L., Ducroux A., Dion S., Deng Q., Michel M.L., Buendia M.A., Hantz O., Neuveut C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol. 2015;63:1093–1102. doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 12.Murphy C.M., Xu Y., Li F., Nio K., Reszka-Blanco N., Li X., Wu Y., Yu Y., Xiong Y., Su L. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep. 2016;16:2846–2854. doi: 10.1016/j.celrep.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laras A., Koskinas J., Dimou E., Kostamena A., Hadziyannis S.J. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology. 2006;44:694–702. doi: 10.1002/hep.21299. [DOI] [PubMed] [Google Scholar]
- 14.Qi Z., Li G., Hu H., Yang C., Zhang X., Leng Q., Xie Y., Yu D., Zhang X., Gao Y., Lan K., Deng Q. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J Virol. 2014;88:8045–8056. doi: 10.1128/JVI.01024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Z., Zeng J., Yu Y., Xiang K., Hu H., Zhou X., Gu L., Wang L., Zhao J., Young J.A.T., HBVcircle Gao L. A novel tool to investigate hepatitis B virus covalently closed circular DNA. J Hepatol. 2017;66:1149–1157. doi: 10.1016/j.jhep.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Aragon L. The Smc5/6 complex: new and old functions of the enigmatic long-distance relative. Ann Rev Genet. 2018;52:89–107. doi: 10.1146/annurev-genet-120417-031353. [DOI] [PubMed] [Google Scholar]
- 17.Nasmyth K., Haering C.H. The structure and function of SMC and Kleisin complexes. Ann Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 18.Laugsch M., Seebach J., Schnittler H., Jessberger R. Imbalance of SMC1 and SMC3 cohesins causes specific and distinct effects. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y.H., Wang W., Li M., Gao Y., Zhang W., Huang Y.L., Zhuo W., Yan X.Y., Liu W., Wang F.W., Chen D.W., Zhou T.H. NudCL2 is an Hsp90 cochaperone to regulate sister chromatid cohesion by stabilizing cohesin subunits. Cell Mol Life Sci. 2019;76:381–395. doi: 10.1007/s00018-018-2957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asturias F.J., Craighead J.L. RNA polymerase II at initiation. Proc Natl Acad Sci U S A. 2003;100:6893–6895. doi: 10.1073/pnas.1332606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A., Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 22.Murayama Y., Uhlmann F. DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell. 2015;163:1628–1640. doi: 10.1016/j.cell.2015.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ladurner R., Bhaskara V., in't Veld P.J.H., Davidson I.F., Kreidl E., Petzold G., Peters J.M. Cohesin's ATPase activity couples cohesin loading onto DNA with Smc3 acetylation. Curr Biol. 2014;24:2228–2237. doi: 10.1016/j.cub.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J.M. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 25.Lim K.H., Park E.S., Kim D.H., Cho K.C., Kim K.P., Park Y.K., Ahn S.H., Park S.H., Kim K.H., Kim C.W., Kang H.S., Lee A.R., Park S., Sim H., Won J., Seok K., You J.S., Lee J.H., Yi N.J., Lee K.W., Suh K.S., Seong B.L., Kim K.H. Suppression of interferon-mediated anti-HBV response by single CpG methylation in the 5'-UTR of TRIM22. Gut. 2018;67:166–178. doi: 10.1136/gutjnl-2016-312742. [DOI] [PubMed] [Google Scholar]
- 26.Sekiba K., Otsuka M., Ohno M., Yamagami M., Kishikawa T., Suzuki T., Ishibashi R., Seimiya T., Tanaka E., Koike K. Inhibition of HBV transcription from cccDNA with nitazoxanide by targeting the HBx-DDB1 interaction. Cell Mol Gastroenterol Hepatol. 2019;7:297–312. doi: 10.1016/j.jcmgh.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bock C.T., Schwinn S., Locarnini S., Fyfe J., Manns M.P., Trautwein C., Zentgraf H. Structural organization of the hepatitis B virus minichromosome. J Mol Biol. 2001;307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 28.Tropberger P., Mercier A., Robinson M., Zhong W., Ganem D.E., Holdorf M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci U S A. 2015;112:E5715–E5724. doi: 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawaguchi A., Nagata K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 2007;26:4566–4575. doi: 10.1038/sj.emboj.7601881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morwitzer M.J., Tritsch S.R., Cazares L.H., Ward M.D., Nuss J.E., Bavari S., Reid S.P. Identification of RUVBL1 and RUVBL2 as novel cellular interactors of the Ebola virus nucleoprotein. Viruses. 2019;11:372. doi: 10.3390/v11040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K.X., Chu J.J. Antiviral screen identifies EV71 inhibitors and reveals camptothecin-target, DNA topoisomerase 1 as a novel EV71 host factor. Antiviral Res. 2017;143:122–133. doi: 10.1016/j.antiviral.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K., Halfmann P., Oyama M., Kozuka-Hata H., Noda T., Kawaoka Y. DNA topoisomerase 1 facilitates the transcription and replication of the Ebola virus genome. J Virol. 2013;87:8862–8869. doi: 10.1128/JVI.03544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao R., Chen J.Y., Liang Z.Y., Luo D.J., Chen G., Lu Z.J., Chen Y., Zhou B., Li H.R., Du X., Yang Y., San M.K., Wei X.T., Liu W., Lecuyer E., Graveley B.R., Yeo G.W., Burge C.B., Zhang M.Q., Zhou Y., Fu X.D. Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell. 2019;178:107–121.e18. doi: 10.1016/j.cell.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters J.M., Tedeschi A., Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 35.Arvey A., Tempera I., Tsai K., Chen H.S., Tikhmyanova N., Klichinsky M., Leslie C., Lieberman P.M. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012;12:233–245. doi: 10.1016/j.chom.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., Lieberman P.M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen A.S., Pustova I., Cattoglio C., Tjian R., Darzacq X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife. 2017;6 doi: 10.7554/eLife.25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Arienzo V., Ferguson J., Giraud G., Chapus F., Harris J.M., Wing P.A.C., Claydon A., Begum S., Zhuang X., Balfe P., Testoni B., McKeating J.A., Parish J.L. The CCCTC-binding factor CTCF represses hepatitis B virus enhancer I and regulates viral transcription. Cell Microbiol. 2021;23 doi: 10.1111/cmi.13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651–660. doi: 10.1007/s005350170027. [DOI] [PubMed] [Google Scholar]
- 40.Losada A. Cohesin in cancer: chromosome segregation and beyond. Nat Rev Cancer. 2014;14:389–393. doi: 10.1038/nrc3743. [DOI] [PubMed] [Google Scholar]
- 41.Potts P.R., Porteus M.H., Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Copsey A., Tang S., Jordan P.W., Blitzblau H.G., Newcombe S., Chan A.C., Newnham L., Li Z., Gray S., Herbert A.D., Arumugam P., Hochwagen A., Hunter N., Hoffmann E. Smc5/6 coordinates formation and resolution of joint molecules with chromosome morphology to ensure meiotic divisions. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gomez R., Jordan P.W., Viera A., Alsheimer M., Fukuda T., Jessberger R., Llano E., Pendas A.M., Handel M.A., Suja J.A. Dynamic localization of SMC5/6 complex proteins during mammalian meiosis and mitosis suggests functions in distinct chromosome processes. J Cell Sci. 2013;126:4239–4252. doi: 10.1242/jcs.130195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallego-Paez L.M., Tanaka H., Bando M., Takahashi M., Nozaki N., Nakato R., Shirahige K., Hirota T. Smc5/6-mediated regulation of replication progression contributes to chromosome assembly during mitosis in human cells. Mol Biol Cell. 2014;25:302–317. doi: 10.1091/mbc.E13-01-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Outwin E.A., Irmisch A., Murray J.M., O'Connell M.J. Smc5-Smc6-dependent removal of cohesin from mitotic chromosomes. Mol Cell Biol. 2009;29:4363–4375. doi: 10.1128/MCB.00377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tapia-Alveal C., Outwin E.A., Trempolec N., Dziadkowiec D., Murray J.M., O'Connell M.J. SMC complexes and topoisomerase II work together so that sister chromatids can work apart. Cell Cycle. 2010;9:2065–2070. doi: 10.4161/cc.9.11.11734. [DOI] [PubMed] [Google Scholar]
- 47.Wu Z.C., Tan S.Y., Xu L.Q., Gao L.F., Zhu H.Z., Ma C.H., Liang X.H. NgAgo-gDNA system efficiently suppresses hepatitis B virus replication through accelerating decay of pregenomic RNA. Antiviral Res. 2017;145:20–23. doi: 10.1016/j.antiviral.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 48.Yang D., Zuo C., Wang X., Meng X., Xue B., Liu N., Yu R., Qin Y., Gao Y., Wang Q., Hu J., Wang L., Zhou Z., Liu B., Tan D., Guan Y., Zhu H. Complete replication of hepatitis B virus and hepatitis C virus in a newly developed hepatoma cell line. Proc Natl Acad Sci U S A. 2014;111:E1264–E1273. doi: 10.1073/pnas.1320071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ni Y., Urban S. Hepatitis B virus infection of HepaRG Cells, HepaRG-hNTCP cells, and primary human hepatocytes. Methods Mol Biol. 2017;1540:15–25. doi: 10.1007/978-1-4939-6700-1_2. [DOI] [PubMed] [Google Scholar]
- 50.Bader G.D., Hogue C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L., Sun Y., Song X., Wang Z., Zhang Y., Zhao Y., Peng X., Zhang X., Li C., Gao C., Li N., Gao L., Liang X., Wu Z., Ma C. Hepatitis B virus evades immune recognition via RNA adenosine deaminase ADAR1-mediated viral RNA editing in hepatocytes. Cell Mol Immunol. 2021;18:1871–1882. doi: 10.1038/s41423-021-00729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.