Abstract
Opioid prescribing for postoperative pain management is challenging because of inter‐patient variability in opioid response and concern about opioid addiction. Tramadol, hydrocodone, and codeine depend on the cytochrome P450 2D6 (CYP2D6) enzyme for formation of highly potent metabolites. Individuals with reduced or absent CYP2D6 activity (i.e., intermediate metabolizers [IMs] or poor metabolizers [PMs], respectively) have lower concentrations of potent opioid metabolites and potentially inadequate pain control. The primary objective of this prospective, multicenter, randomized pragmatic trial is to determine the effect of postoperative CYP2D6‐guided opioid prescribing on pain control and opioid usage. Up to 2020 participants, age ≥8 years, scheduled to undergo a surgical procedure will be enrolled and randomized to immediate pharmacogenetic testing with clinical decision support (CDS) for CYP2D6 phenotype‐guided postoperative pain management (intervention arm) or delayed testing without CDS (control arm). CDS is provided through medical record alerts and/or a pharmacist consult note. For IMs and PM in the intervention arm, CDS includes recommendations to avoid hydrocodone, tramadol, and codeine. Patient‐reported pain‐related outcomes are collected 10 days and 1, 3, and 6 months after surgery. The primary outcome, a composite of pain intensity and opioid usage at 10 days postsurgery, will be compared in the subgroup of IMs and PMs in the intervention (n = 152) versus the control (n = 152) arm. Secondary end points include prescription pain medication misuse scores and opioid persistence at 6 months. This trial will provide data on the clinical utility of CYP2D6 phenotype‐guided opioid selection for improving postoperative pain control and reducing opioid‐related risks.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
The management of postoperative pain is challenging because of the interpatient variability in opioid response. CYP2D6 phenotype, based on genotype and drug interactions, may contribute to response to tramadol, hydrocodone, and codeine, given the role of the CYP2D6 enzyme in transforming these drugs to potent opioid metabolites.
WHAT QUESTION DOES THIS STUDY ADDRESS?
The primary objective of this prospective, multicenter, randomized pragmatic trial is to determine the effect of postoperative CYP2D6‐guided opioid prescribing on pain control and opioid usage.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This trial will provide data on the clinical utility of CYP2D6 phenotype‐guided opioid selection for postoperative pain management.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Positive findings would support CYP2D6 genotyping in practice, to be considered in combination with CYP2D6 inhibitor use, to inform individualized opioid prescribing to improve postoperative pain control.
INTRODUCTION
Despite a decline in opioid dispensing rates over the past decade, opioid use remains high in the United States, comprising 43 prescriptions per 100 individuals in 2020. 1 Approximately 10% of all opioid prescriptions originate from surgeons, with hydrocodone and tramadol accounting for about 60% of opioid prescriptions in the peri‐operative setting. 2 , 3 Hydrocodone and tramadol were among the top 200 most commonly prescribed drugs in the United States in 2019, ranking number 15 and 35, respectively. 4 Similar to codeine, hydrocodone and tramadol undergo O‐demethylation via the CYP2D6 enzyme to a metabolite that has greater affinity than the parent compound for the μ‐opioid receptor; hydrocodone is metabolized to hydromorphone, tramadol to O‐desmethyltramadol, and codeine to morphine. 5 , 6 , 7
The CYP2D6 gene is highly polymorphic, and up to 30% of individuals, depending on ancestry, have two nonfunctional CYP2D6 alleles and no active enzyme (i.e., poor metabolizers [PMs]) or one nonfunctional and one reduced function allele and significantly impaired enzyme activity (i.e., intermediate metabolizers [IMs]). 8 Through a phenomenon referred to as phenoconversion, use of moderate to strong CYP2D6 inhibitors may also reduce or abolish CYP2D6 activity, akin to having reduced to nonfunctional alleles. Individuals with absent or reduced CYP2D6 activity, secondary to their genotype or use of medications causing phenoconversion, have lower concentrations of active opioid metabolites. 6 In the case of hydrocodone, tramadol, and codeine, this may compromise pain control. 6 , 9 CYP2D6 ultra‐rapid metabolizers (UMs) have increased enzyme activity secondary to gene duplication or multiplication and are at risk for toxic concentrations of active opioid metabolites from these drugs. 10 , 11 , 12 Whereas oxycodone is also metabolized by CYP2D6, data on the effect of CYP2D6 phenotype on oxycodone response are mixed. 6 , 9
Pharmacogenetic guidelines recommend avoiding codeine and tramadol in PMs and UMs because of the potential for reduced effectiveness or toxicity, respectively. 6 No recommendations are provided for hydrocodone because of the limited evidence available on CYP2D6 associations with hydrocodone response. However, evidence of impaired analgesia when hydrocodone is administered with CYP2D6 inhibitors suggests that reduced CYP2D6 activity secondary to genotype may also impair hydrocodone effectiveness. 13 Data from a pilot trial of CYP2D6‐guided management of chronic pain further support important effects of CYP2D6 phenotype on hydrocodone response, but showed no benefit of a CYP2D6‐guided approach in oxycodone‐treated patients. 9
In a single center pilot trial of CYP2D6‐guided opioid prescribing for postoperative pain, we showed that integrating CYP2D6 testing into clinical practice to guide postoperative pain management was feasible. 14 Moreover, compared to usual care, a CYP2D6‐guided approach led to less opioid consumption without compromising pain control. Based on these data, the Implementing Genomics in Practice (IGNITE) Network, funded by the National Human Genome Research Institute, is conducting a multicenter pragmatic trial to test the hypothesis that CYP2D6‐guided opioid selection for postoperative pain management leads to improved pain control or similar pain control with less opioid use. The objective of the study is to determine the effect of CYP2D6‐guided opioid prescribing on pain control and use of Drug Enforcement Administration (DEA) schedule II opioids in postsurgical participants.
METHODS
Trial design
This is one of three trials under the umbrella of A Depression and Opioid Pragmatic Trial in Pharmacogenetics (ADOPT‐PGx; registered in ClinicalTrials.gov Identifier: NCT04445792). Each trial is assessing the efficacy of a pharmacogenetic‐guided approach to drug therapy selection. The other two ADOPT‐PGx trials are focused on chronic pain and depression and will be described elsewhere. Participants in the Acute Pain Trial are randomized to CYP2D6‐guided selection of postoperative opioid therapy or usual postoperative pain management, with patient reported outcomes assessed prior to surgery and 10 days and 1, 3, and 6 months after surgery (Figure 1).
FIGURE 1.
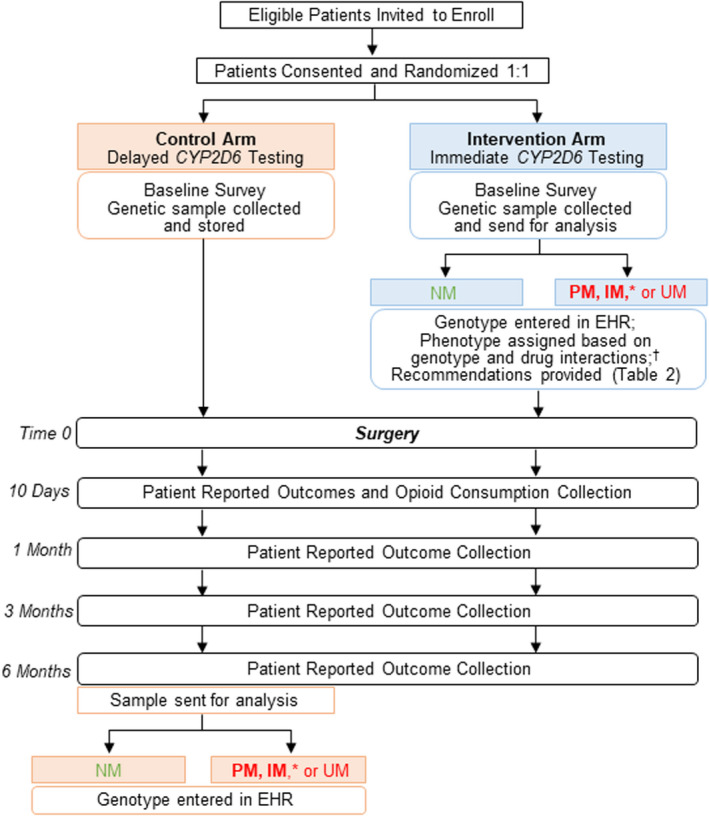
Trial design. *Primary outcomes will be compared between the subgroup of PMs and IMs in the intervention versus control arm. †IM, NM, and PM phenotypes are based on CYP2D6 genotype and use of moderate to strong CYP2D6 inhibitors. EHR, electronic health record; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultrarapid metabolizer.
Study population, location, and personnel
Participants are enrolled from nine health systems: University of Florida (UF) Health; Nemours Children's Health; Indiana University (IU) Health; Eskenazi Health; Duke University Health System (DUHS); Vanderbilt University Medical Center (VUMC); Sanford Health; Nashville General Hospital and Meharry Medical Group affiliated with Meharry Medical College; and Mount Sinai Health System (MSHS). Inclusion criteria are age ≥8 years, which is the minimum age at which the outcome measures are validated without parent proxy; English or Spanish speaking; and having an upcoming scheduled elective or planned surgery. Patients are being enrolled from surgical practices in which the drugs of interest (i.e., hydrocodone, tramadol, or codeine) are usually prescribed for postsurgical pain management. Based on data from the pilot trial mentioned above, it is expected that the intervention will be most effective in patients with a higher postoperative pain burden. 14 Therefore, patients undergoing procedures for which there is a reasonable expectation of pain during the majority of the 10 day period following surgery are prioritized for enrollment. Examples of surgery types that may be included are orthopedic surgeries (e.g., arthroplasty, spine, etc.), open abdominal surgery, and cardiothoracic surgery. Exclusion criteria include chronic opioid use, defined as opioid use, per patient report, on most days of the week for over the past 3 months; life expectancy <12 months; cognitive impairment or illness that may interfere with study participation; history of allogeneic stem cell or liver transplant; and having prior clinical CYP2D6 genotype results available. An important goal of the IGNITE PTN is to include a diverse patient population. 15 To this end, many of the clinics from which patients are being recruited traditionally treat underserved patients, including those of diverse backgrounds and those living in rural areas.
The structure of the IGNITE‐PTN has been described and includes Executive and Steering Committees, a Coordinating Center (Duke University), Protocol and Implementation Teams (PITs), and five Clinical Groups (UF Health, IU Health, DUHS, VUMC, and MSHS). 15 The ADOPT‐PGx PIT is composed of lead investigators across sites and representatives from the National Human Genome Research Institute (NHGRI), Coordinating Center, Clinical Groups, and other enrolling sites. Prior to protocol implementation, the PIT maintained weekly teleconferences to develop the research protocol, documents for institutional review board (IRB) submission, case report forms, and the manual of operations. Smaller working groups held separate teleconferences to establish genotyping procedures, develop clinical decision support, address pediatric specific issues, and establish processes for laboratory reporting of genotype results and providing recommendations to prescribers. The PIT continues to meet via weekly or biweekly teleconferences to discuss study progress and troubleshoot problems that arise. Additional members of the research team are pharmacists; physicians; nurses; researchers with expertise in opioid pharmacogenetics, postsurgical pain management, and pediatrics; clinical bioinformaticists; clinical pathologists; statisticians; and research coordinators. An independent Data and Safety Monitoring Board was established to monitor protocol development, trial data, and patient safety and consists of seven scientists with expertise spanning human and statistical genetics, pharmacogenetics, and clinical trial design.
Enrollment and randomization
Potential participants are identified when they present to clinic for surgery evaluation and scheduling or through electronic health record (EHR) review of scheduled surgeries. They are approached by a research team member through a clinic intercept or telephone call about study participation. Participants who provide informed consent are randomized in a 1:1 allocation to immediate (on enrollment) pharmacogenetic testing and genotype‐guided therapy (intervention arm) or delayed testing (6 months after surgery) and usual postoperative pain management (control arm). Randomization is stratified by enrollment site, with a random block size within each site. The randomization scheme was generated by an unblinded statistician with assignment generated in real time in REDCap at the Coordinating Center. Randomization assignments are not blinded to the participants or their providers but are masked to call center personnel administering follow‐up surveys described below.
Intervention
The intervention was designed to reflect practices and procedures that would be likely with clinical integration of pharmacogenetic testing. To this end, genotype results are returned to the participant's EHR, and clinical decision support is provided. Providers across sites received education on CYP2D6 phenotype, its effects on opioid response, and phenotype‐based treatment recommendations prior to trial launch and periodically thereafter at some sites. However, in line with the pragmatic nature of the trial, the content of the education and mode of delivery was left to the individual site (Table 1).
TABLE 1.
Clinical site CDS and provider education comparison
| UF Health | Nemour Children's Health | Mount Sinai | IFH | VUMC | Sanford Health | Meharry Medical Group | DUHS | IU Health | Eskenazi Health | |
|---|---|---|---|---|---|---|---|---|---|---|
| Clinical decision support | ||||||||||
| Automated alerts based on opioid Rx + PGx | X | X | X | X | X | X | X | |||
| Automated alerts based on opioid Rx + CYP2D6 inhibitor + PGx | X a | X a | ||||||||
| Consult note placed based on CYP2D6 inhibitors + PGx | X | X | X | X | X | X | X | X | ||
| Provider education strategy | ||||||||||
| Presentations in person or via videoconference prior to enrollment | X | X | X | X | X | X | X | X | X | |
| One‐on‐one or small group meetings | X | X | X | X | X | X | ||||
| Emails | X | X | X | X | X | X | ||||
| Refresher presentations periodically after trial launch | X | |||||||||
| Laminated cards of enrollment criteria and how to refer patients into the trial for posting at work stations | X | |||||||||
Abbreviations: CDS, clinical decision support; DUHS, Duke University Health System; IFH, Institute for Family Health at Mount Sinai Health System; IU Health, Indiana University Health; MSHS, Mount Sinai Health System; PGx, pharmacogenetics; Rx, prescription; UF, University of Florida; VUMC, Vanderbilt University Medical Center.
Recommendations are provided solely through automated alerts because of insufficient pharmacy support to provide consult notes.
Following informed consent, a biological sample for genotyping is collected from each participant through venipuncture, saliva collection, or buccal swab, depending on the site and participant preferences, and sent to a Clinical Laboratory Improvement Amendments (CLIA)‐certified laboratory at one of the study sites for immediate genotyping (for the intervention arm) or storage for genotyping after the participant completes study procedures (for the control arm). Once results are available for participants in the intervention arm, recommendations for postoperative pain management are provided as described below, with the prescribing decision ultimately left to the provider. After trial participation is complete, pharmacogenetic results are returned to participants in both the intervention and control arms by the study teams.
Participants in both arms are asked to complete baseline surveys to collect demographic data, medical history, and patient‐reported outcomes prior to randomization and within 45 days before surgery (Table 2). Data are collected by a study coordinator in person or via telephone or email. Follow‐up surveys are administered at 10 (±3) days, 1 month (±7 days), and 3 and 6 months (±14 days) after the date of surgery. Follow‐up surveys for adult participants are administered via telephone by clinical research assistants from a centralized call center, housed in the UF College of Pharmacy, or, if the participant prefers, via an emailed or texted link to a web survey. In the event that a member of the call center cannot reach the participant, a local site coordinator may contact the participant for survey completion. Surveys for pediatric participants are sent to the parent and are completed by the child or parent depending upon the survey. Surveys include assessment of pain intensity, prescription medication misuse, mobility, and overall well‐being via Patient Reported Outcomes Measurement Information System (PROMIS) measures. 16 , 17 Additional surveys (see Supplementary Material) assess opioid consumption, side effects, and continued use (i.e., opioid persistence). The 6‐month survey for the intervention arm asks participants if they became aware of their results over the course of the study. Survey data are gathered using REDCap electronic data tools hosted at Duke University. 18 , 19 The study is approved by the Duke University IRB, as the single IRB.
TABLE 2.
Data collection schedule
| Outcome | Data source/instrument | Baseline a | 10 ± 3 days postsurgery | 1 month ± 7 days postsurgery | 3 month ± 14 days postsurgery | 6 month ± 14 days postsurgery |
|---|---|---|---|---|---|---|
| Pain intensity | PROMIS pain intensity scale | X | X | X | X | X |
| Pain intensity | PROMIS NPRS | X | X | X | X | X |
| Daily opioid dose and type of opioid | Average daily MMEs use of opioids since discharge | X | X | |||
| Opioid use disorder | PROMIS – Prescription pain medication misuse subscale | X | X | X | ||
| Mobility | PROMIS mobility | X | X | X | X | X |
| Opioid side effects | Adapted medication side effect survey (SPACE) 44 , 45 | X | X | X | X | X |
| Opioid persistence | Custom survey | X | ||||
| Well‐being | PROMIS 43/PROMIS Peds37 | X | X | X | X | |
| Awareness of genotype results (Intervention arm only) | Custom survey | X |
Abbreviations: MMEs, morphine milligram equivalents; NPRS, Numeric Rating Scale – Pain Intensity; PROMIS, Patient Reported Outcomes Measurement Information Systems.
Baseline surveys are administered within 45 days of surgery.
Pharmacogenetic testing and phenotype assignment
Six clinical sites are conducting genotyping. Sites without local genotyping send samples to one of these sites for testing. Each site is interrogating the CYP2D6 *2, *3, *4, *5, *6, *8, *9, *10, *17, *29, and *41 alleles in addition to copy number variation, as recommended by the Association of Molecular Pathology. 20 Results are entered into the EHR as discrete variables at all sites.
The process for assigning CYP2D6 phenotype based on genotype results is shown in Figure 2. An activity value is assigned for each allele. Activity values are then summed to derive the activity score, which is adjusted as needed to account for phenoconversion. The activity score is multiplied by 0.5 for moderate inhibitors and by 0 for strong inhibitors, with inhibitors defined by US Food and Drug Administration (FDA) guidance and shown in Figure 2. 21 An online calculator was designed for use in the trial to enable a standardized approach to phenoconversion calculation. 22
FIGURE 2.
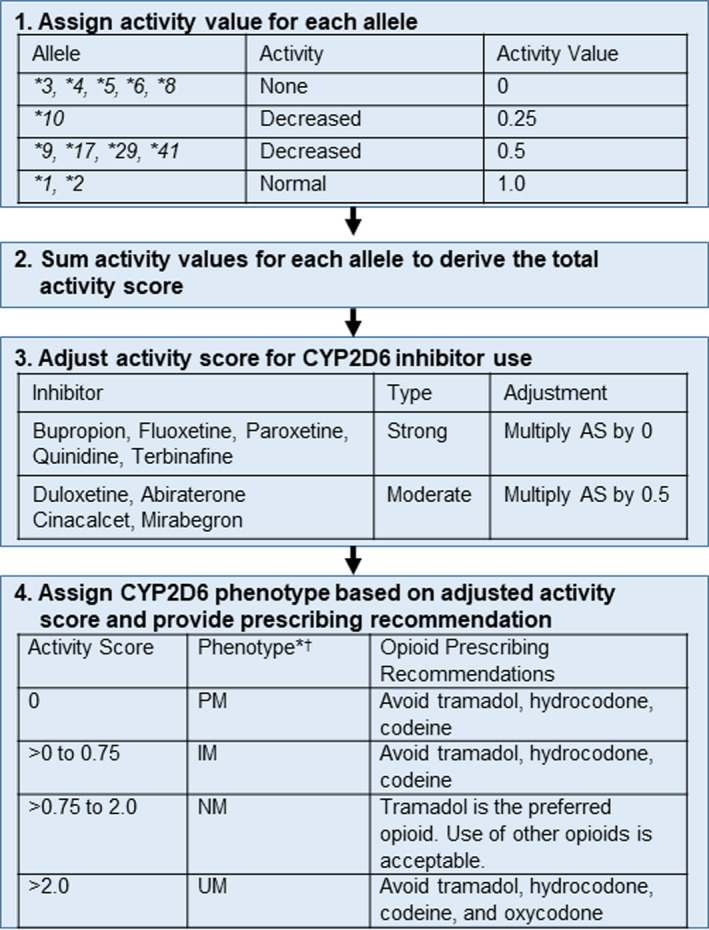
CYP2D6 phenotype assignment and phenotype‐based opioid prescribing recommendations for participants in the intervention arm. *Based on CYP2D6 genotype and use of moderate or strong CYP2D6 inhibitors. Abiraterone, cinacalcet, duloxetine, and mirabegron are moderate inhibitors and reduce the activity score by 50%. Bupropion, fluoxetine, paroxetine, quinidine, and terbinafine are strong inhibitors that reduce the activity score to 0. †Phenotype definitions differs from Clinical Pharmacogenomic Implementation Consortium and Dutch Pharmacogenetics Working Group consensus definitions. Whereas sites detect copy number variation, they do not detect which allele is duplicated or multiplicated. This can result in a ranged phenotype. Given the potential for the IM and/or UM phenotype, recommendations consistent with these phenotypes are provided. In the case of copy number variation leading to ranged phenotypes, recommendations are provided to avoid hydrocodone, tramadol, or codeine for the IM to NM ranged phenotype and to avoid hydrocodone, tramadol, codeine, or oxycodone for the IM to UM and NM to UM ranged phenotypes. AS, activity score; IM, intermediate metabolizer; NM, normal metabolizer; PM, poor metabolizer; UM, ultra‐rapid metabolizer.
The adjusted activity score is used to assign CYP2D6 phenotype (Figure 2). Whereas the Clinical Pharmacogenetics Implementation Consortium (CPIC) assigns the IM phenotype to patients with an activity score of >0 and <1.25, only patients with an activity score >0 to 0.75 are considered IMs for this study given both the significant variability in enzyme activity across the broader IM classification and results from CYP2D6‐guided opioid prescribing pilot studies that defined IMs using the lower score range. 9 , 14 , 23 , 24 The genotyping assays across sites detect allele duplication, but they do not detect which allele is duplicated or multiplicated or the number of allele copies. A ranged phenotype is assigned when the phenotype cannot be definitively assigned based on copy number variation (e.g., IM to UM phenotype assigned for the *4/*17 genotype when duplication of one of the alleles is detected).
Phenotype‐based recommendations
For PMs, IMs, and UMs, a non‐CYP2D6‐metabolized opioid (e.g., hydromorphone and morphine) or non‐opioid analgesic is recommended (Figure 2). Tramadol, a DEA schedule IV opioid, is recommended as the preferred opioid for adult normal metabolizers (NMs) given its opioid and non‐opioid mechanisms and purported lower risk for misuse. 25 , 26 Tramadol is not recommended for pediatric patients (age < 18 years) given contraindications to tramadol use in this population. 27 No recommendations are provided in PMs or IMs treated with oxycodone based on conflicting data regarding the impact of CYP2D6 phenotype on pain response. 6 , 9 , 28 , 29 However, there are data to suggest UMs are at increased risk for toxicity with oxycodone exposure. 30 Thus, although not part of the primary hypothesis, avoidance of oxycodone in UMs is also recommended for safety reasons.
Recommendations are delivered across sites in the form of a written consult note placed in the EHR and/or automated provider alerts within the EHR (Table 1). Alerts are triggered when hydrocodone, tramadol, or codeine is ordered for a participant with the PM or IM phenotype or when hydrocodone, tramadol, codeine, or oxycodone is ordered for a participant with the UM phenotype. One or both mechanisms for delivering recommendations account for phenoconversion. An example of a consult note and alert are shown in Figures 3 and 4.
FIGURE 3.

Example of an ADOPT PGx Consult Note. The note shown is for a CYP2D6 poor metabolizer as the result of taking a strong CYP2D6 inhibitor. Bulleted text are the required elements per the ADOPT PGx Clinical Decision Support Working Group Guidelines for consult notes. Consult note format and wording varied among sites, but all included these common elements. ADOPT PGx, A Depression and Opioid Pragmatic Trial in Pharmacogenetics; CPIC, Clinical Pharmacogenetics Implementation Consortium; FDA, US Food and Drug Administration; MRN, Medical Record Number.
FIGURE 4.
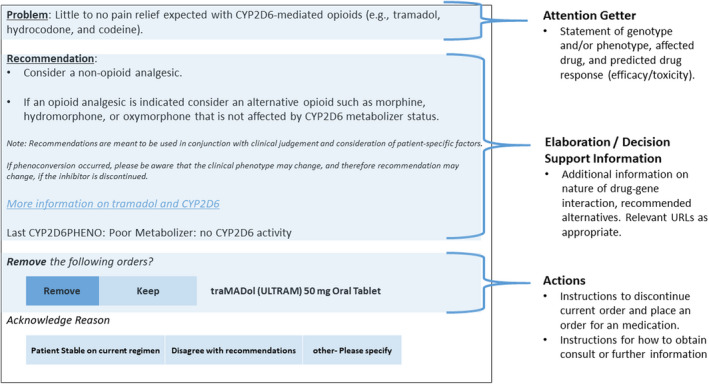
Example of an ADOPT PGx Automated Alert. The alert shown is for a CYP2D6 poor metabolizer prescribed tramadol. A similar alert fires when hydrocodone or codeine is prescribed. Bulleted text are the required elements per the ADOPT PGx Clinical Decision Support Working Group Guidelines for automated alerts. ADOPT PGx, A Depression and Opioid Pragmatic Trial in Pharmacogenetics.
Data analysis
The primary end point is the Silverman Integrated Analgesic Assessment (SIA) score, which is a composite of pain intensity (composite of current pain and worst and average pain in the past 7 days) and opioid usage at 10 days postsurgery. 31 This composite end point was chosen based on data from the pilot trial suggesting that pain intensity would be similar between arms, but that less opioid (in morphine milligram equivalents [MMEs]) would be needed in the intervention arm to achieve a similar pain intensity as in the control arm. 14 Opioid usage is calculated as the difference between participant‐reported opioid tablets prescribed and tablets remaining at the 10‐day timepoint, expressed as MME using standard conversion factors and the strength of the prescribed opioid, 32 and standardized by the number of days since hospital discharge. Secondary end points include individual components of the composite primary outcome; mobility score at 1 month; prescription pain medication misuse at 3 months; opioid persistence, overall well‐being, and subdomains of overall well‐being (pain interference, physical function, sleep disturbance, social role and activities functioning, fatigue, anxiety, and depression) at 6 months; and concordance between metabolizer phenotype and prescribed medication. The analysis is an intent to treat study design, and primary and secondary end points will be compared between the intervention and control arms in the subset of participants with the PM or IM phenotype based on genotype and phenoconversion. End points will be compared between arms using a two‐sided t‐test or Mann–Whitney U test, as appropriate. In addition to the intent to treat analysis, a per‐protocol subgroup analysis is also planned. If there is an imbalance in baseline characteristics, including surgery type, between trial arms, a covariate‐adjusted sensitivity analysis, accounting for differences between groups, will be conducted.
Power calculations were based on data from the pilot trial suggesting provider adherence to CYP2D6‐guided recommendations for intervention arm participants with an actionable phenotype was >70% and a Cohen's D of 0.375. 14 Including 304 IMs or PMs (152 in each arm) is estimated to provide 90% power to detect a standardized effect size of 0.375 for SIA score between arms with a two‐sided type 1 error rate of 0.049 to account for a single interim analysis of the primary end point. 33 Additional assumptions were that 10% would drop out or be lost to follow‐up, and an additional 7% would not proceed to surgery. The total sample size is dependent on the prevalence of the IM or PM phenotype; 1518, 1732, or 2020 participants will need to be enrolled if 24%, 21%, or 18% have the IM or PM phenotype, respectively.
RESULTS
The trial enrolled its first participant in March 2021. Enrollment was initially expected to be completed by December 2022; however, with delays caused by the pandemic, the enrollment period was extended to June 2023, with follow‐up planned through April 2024. As of March 2022, the trial has enrolled 504 participants. The majority of patients enrolled to date were scheduled to undergo total joint arthroplasty (89%) or spinal (6%) surgery. Of 191 participants randomized to the genotype‐guided arm who have undergone surgery to date, 185 (97%) had genotype results available prior to their procedure.
Table 3 summarizes challenges faced to date and actions taken to overcome these. Unanticipated recruitment challenges include temporary holds placed on elective surgeries and limits to clinic access by research staff as a result of the coronavirus disease 2019 (COVID‐19) pandemic, restrictions placed on opioid prescribing in some states in an effort to combat the opioid epidemic, and changes in opioid prescribing practices away from the drugs of interest in the trial. Anticipated challenges as a result of the pragmatic trial design include the inability to completely mask participants to the study arm and to mask patients in the intervention arm to their genotype results. This is because genotyping is done clinically, with results placed in the EHR and accessible through patient portals. Finally, we faced a challenge with quantifying opioid consumption for participants prescribed liquid dosage forms. We ultimately decided to avoid enrolling patients for whom liquid opioid prescriptions were anticipated. In the event that, despite this, a participant was prescribed a liquid dosage form, the investigator team conceded that because of the difficulty with quantifying intake, we would not capture opioid consumption for these patients.
TABLE 3.
Trial challenges and measures to address challenges
| Challenge | Measure to address challenges |
|---|---|
| Hold on elective surgical procedures as a result of the pandemic | Extended the trial enrollment period by 6 months and seeking additional clinics from which to enroll patients to increase the enrollment rate after holds have ended |
| Institutional restrictions as a result of the pandemic limiting research staff access to surgery clinics | Remote recruitment, consent, and sample collection procedures put into place |
| Opioid prescribing restrictions | Agreement among investigators to focus on enrolling patients undergoing procedures for which the usual opioid prescription is for at least 24 tablets of one of the opioids of interest |
| Changes in opioid prescribing patterns |
Sites conducting EHR data pulls to identify surgeons and surgery practices in which tramadol, hydrocodone, or codeine are commonly prescribed for pain management Periodic assessment of opioids prescribed to ensure that the drugs of interest are being used |
| Quantification of liquid dosage forms for patient reported utilization | Avoid enrolling patients anticipated to be prescribed a liquid dosage form. In the event that a liquid was prescribed, opioid consumption would not be captured |
| Participants not completely masked to trial arm or genotype results |
Leverage data from intervention and control arm participants with a normal metabolizer phenotype to help determine if a placebo effect exists. Query participants in the genotype‐guided arm about whether they became aware of their results over the course of the trial |
Abbreviation: EHR, electronic health record.
DISCUSSION
Data from a single center pilot trial suggested that a CYP2D6‐guided approach to opioid prescribing following total hip or knee arthroplasty reduces opioid consumption without compromising pain control. The current trial aims to examine the efficacy of CYP2D6‐guided opioid prescribing in a larger, multisite cohort of patients undergoing various types of surgery where a high pain burden is expected. If results from the pilot trial are confirmed, they could inform an important change in the approach to postoperative pain management that may result in improved postoperative pain control with decreased opioid use.
Postoperative pain is a major concern for patients. 34 , 35 Indeed, there is significant interpatient variability in response to opioid analgesics, and CYP2D6 PMs and IMs may attain little to no relief from some opioids prescribed for postsurgical pain (i.e., hydrocodone, tramadol, and codeine). Opioid misuse and addiction are additional concerns with postoperative opioid use, and there is evidence that 6% of opioid naïve patients who fill a peri‐operative opioid prescription continue to use opioids 3–6 months following a surgical procedure. 36 , 37 Seeking pain relief is a major reason cited for opioid misuse. 38 , 39 Therefore, optimizing opioid prescribing through precision medicine approaches not only may lead to more effective management of postoperative pain, but in doing so, may minimize the potential for opioid misuse and persistence.
This trial is using a pragmatic clinical trial design, which is expected to more closely align with usual care than a randomized controlled trial, thereby maximizing the generalizability of findings to real world practice. 40 A major pragmatic feature of the trial is that although CYP2D6‐based recommendations are provided, the prescribing decision is ultimately left to the provider. To enhance recommendations’ acceptance, all sites deliver provider education on CYP2D6 phenotype effects on opioid response at baseline and provide treatment recommendations based on phenotype. In our pilot trial of CYP2D6‐guided postoperative pain management, which used a similar design and education strategy, phenotype‐guided recommendations were followed for those with the PM, IM, or UM phenotype 72% of the time, giving us confidence in this trial that recommendations will be followed for a high percentage of the time. The method of communicating phenotype‐based recommendations varies across sites, in line with the pragmatic design. A per‐protocol analysis of participants whose prescribed opioid is consistent with recommendations is planned in the event of variable concordance between the recommended and prescribed opioid across sites. Whereas a limitation of our trial design is that it is not possible to completely mask participants to their study arm or pharmacogenetic results for those in the intervention arm, similar SIA scores between the intervention and control arm participants with a NM phenotype would provide confidence that results were not impacted by a placebo effect. In addition, the final survey administered to those in the intervention arm assess whether participants became aware of their results at any point during the trial, which will provide a mechanism to specifically test any impact of a placebo effect.
We have encountered several challenges in the early stages of the trial. Holds on elective surgeries during periods of pandemic‐related surges have caused halts to enrollment and the need in some cases to reschedule surgeries for patients already enrolled. The team made the decision to expand the original timeline for completing the trial by 6 months and are exploring expansion of enrollment to additional clinics to increase the enrollment rate. Restrictions limiting study staff access to surgery clinics have further impacted trial enrollment and necessitated remote (i.e., via telephone call) recruitment and enrollment procedures.
Restrictions on opioid prescribing in some states, where only a 3‐day supply is allowed, create a challenge in identifying appropriate participants for enrollment. These restrictions make it more difficult to determine which patients are expected to have a higher pain burden and continued opioid use, who we are targeting for the trial. To help provide clarity on this, sites agreed to focus on procedures for which the usual opioid prescription is for at least 24 tablets of one of the opioids of interest (hydrocodone, tramadol, or codeine), as this is the maximum number of tablets that can be prescribed in a 3‐day period per tramadol and hydrocodone labeling. Thus, in states where a 3‐day restriction exists, a prescription for 24 tablets would be an indication that the surgeon is trying to maximize the number of pills in the 3‐day prescription in anticipation of significant pain.
Another concern that arose was the change in opioid prescribing practices away from the drugs of interest at some sites and how this might impact the value of the trial. Many surgeons are now prescribing oxycodone for postoperative analgesia, and based on communication with surgeons, this preference appears to be related, at least in part, to concerns about interpatient variability in response to hydrocodone, tramadol, and codeine, which is precisely what our trial is addressing. Therefore, although it reduces our patient base from which to enroll, it points to the clinical importance of the trial. Minimizing opioid exposure by following a multi‐modal approach, remains of critical importance to the surgery community. 41 Tramadol is the preferred opioid per this approach and is advocated for in the Enhanced Recovery Surgery Pathway given its opioid and non‐opioid mechanisms and purported lower risk of misuse. 25 , 26 , 41 If results from the trial are positive, they may be impactful for influencing future practice as it relates to postoperative pain management.
We acknowledge that few data are available to support genotype‐guided recommendations for hydrocodone, but believe that the available data justify this approach. In particular, pharmacokinetic studies show lower concentrations of the active hydromorphone metabolite in PMs and IMs. 6 , 42 , 43 In addition, use of CYP2D6 inhibitors, which can mimic the IM or PM genotype, has been shown to reduce the drug's analgesic effectiveness. 13 Moreover, our pilot studies of CYP2D6‐guided opioid therapy support important effects of CYP2D6 phenotype on hydrocodone response. 9 , 14 We also acknowledge differences in CYP2D6 IM phenotype assignment compared to recent consensus definitions by pharmacogenetic guideline groups. 23 Our pilot studies of CYP2D6‐guided therapy began prior to the definition updates; at the time of study initiation, the CPIC definition of IMs conformed with the study definition. 9 , 14 The new definition of an IM includes a much higher percentage of the population, who may have less drug metabolism impairment than with the older, more conservative definition of an IM. This, along with the positive findings from our pilot studies, where we used the more conservative definition of an IM, led the study principal investigators to continue defining IMs as those with a lower activity score (i.e., >0 to ≤0.75).
In summary, this trial will provide data on the clinical utility of CYP2D6‐guided opioid selection. Positive findings would support CYP2D6 genotyping in practice to inform individualized opioid prescribing to improve postoperative pain control and reduce opioid‐related risks.
AUTHOR CONTRIBUTIONS
L.H.C., E.C., and K.W. wrote the manuscript. L.H.C., E.C., K.W., R.B.F., H.C., R.A.M., K.V.B., B.A., J.F.B., W.H.B., K.J.C., E.N.E., C.F.G., Y.G., L.H., J.K., N.K., S.L., K.A.N., A.O.O., V.M.P., H.A.P., M.R., A.S., R.S., M.R., P.S., C.D.T., E.T., P.R.D., C.R.H., L.A.O., J.F.P., T.C.S., S.L.V., S.V., D.V., H.K.P., and J.A.J. designed the research. E.C., B.A., J.F.B., K.J.C., E.N.E., S.L., K.A.N., V.M.P., M.R., A.Z., P.S., and E.T. performed the research. R.A.M. analyzed the data.
FUNDING INFORMATION
This work was supported by grants from the National Institutes of Health (U01 HG007269, U01 HG010232, U01 HG010248, U01 HG010231, U01 HG0010245, and U01 HG010225), and by the NIH IGNITE Network. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
Supporting information
Appendix S1
Cavallari LH, Cicali E, Wiisanen K, et al. Implementing a pragmatic clinical trial to tailor opioids for acute pain on behalf of the IGNITE ADOPT PGx investigators. Clin Transl Sci. 2022;15:2479‐2492. doi: 10.1111/cts.13376
Trial registration: ClinicalTrials.gov Identifier: NCT04445792.
REFERENCES
- 1. Centers for Disease Control and Prevention . Drug Overdose. US Opioid Dispensing Rate Map. https://www.cdc.gov/drugoverdose/rxrate‐maps/index.html. Accessed January 3, 2022.
- 2. Levy B, Paulozzi L, Mack KA, Jones CM. Trends in opioid analgesic‐prescribing rates by specialty, U.S., 2007–2012. Am J Prev Med. 2015;49:409‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ladha KS, Neuman MD, Broms G, et al. Opioid prescribing after surgery in the United States, Canada, and Sweden. JAMA Netw Open. 2019;2:e1910734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ClinCalc DrugStats Database . The top 200 drugs of 2019. https://clincalc.com/DrugStats/. Accessed February 10, 2022.
- 5. Cardia L, Calapai G, Quattrone D, et al. Preclinical and clinical pharmacology of hydrocodone for chronic pain: a mini review. Front Pharmacol. 2018;9:1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crews KR, Monte AA, Huddart R, et al. Clinical pharmacogenetics implementation consortium guideline for CYP2D6, OPRM1, and COMT genotypes and select opioid therapy. Clin Pharmacol Ther. 2021;110:888‐896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Grond S, Sablotzki A. Clinical pharmacology of tramadol. Clin Pharmacokinet. 2004;43:879‐923. [DOI] [PubMed] [Google Scholar]
- 8. CYP2D6 frequency table. https://api.pharmgkb.org/v1/download/file/attachment/CYP2D6_frequency_table.xlsx. Accessed February 28, 2022.
- 9. Smith DM, Weitzel KW, Elsey AR, et al. CYP2D6‐guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet Med. 2019;21:1842‐1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. Codeine, ultrarapid‐metabolism genotype, and postoperative death. N Engl J Med. 2009;361:827‐828. [DOI] [PubMed] [Google Scholar]
- 11. Kelly LE, Rieder M, van den Anker J, et al. More codeine fatalities after tonsillectomy in north American children. Pediatrics. 2012;129:e1343‐e1347. [DOI] [PubMed] [Google Scholar]
- 12. Orliaguet G, Hamza J, Couloigner V, et al. A case of respiratory depression in a child with ultrarapid CYP2D6 metabolism after tramadol. Pediatrics. 2015;135:e753‐e755. [DOI] [PubMed] [Google Scholar]
- 13. Monte AA, Heard KJ, Campbell J, Hamamura D, Weinshilboum RM, Vasiliou V. The effect of CYP2D6 drug‐drug interactions on hydrocodone effectiveness. Acad Emerg Med. 2014;21:879‐885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas CD, Parvataneni HK, Gray CF, et al. A hybrid implementation‐effectiveness randomized trial of CYP2D6‐guided postoperative pain management. Genet Med. 2021;23:621‐628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ginsburg GS, Cavallari LH, Chakraborty H, et al. Establishing the value of genomics in medicine: the IGNITE pragmatic trials network. Genet Med. 2021;23:1185‐1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cella D, Riley W, Stone A, et al. The patient‐reported outcomes measurement information system (PROMIS) developed and tested its first wave of adult self‐reported health outcome item banks: 2005‐2008. J Clin Epidemiol. 2010;63:1179‐1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Patient Reported Outcomes Measurement Information System (PROMIS) HealthMeasures. https://www.healthmeasures.net/explore‐measurement‐systems/promis. Accessed February 11, 2022.
- 18. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)‐a metadata‐driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for clinical CYP2D6 genotyping allele selection: a joint consensus recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch pharmacogenetics working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn. 2021;23:1047‐1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Food and Drug Administration . Drug development and drug interactions Table of substrates, inhibitors, and inducers. https://www.fda.gov/drugs/drug‐interactions‐labeling/drug‐development‐and‐drug‐interactions‐table‐substrates‐inhibitors‐and‐inducers. Accessed January 06, 2022.
- 22. Cicali EJ, Elchynski AL, Cook KJ, et al. How to integrate CYP2D6 Phenoconversion into clinical pharmacogenetics: a tutorial. Clin Pharmacol Ther. 2021;110:677‐687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caudle KE, Sangkuhl K, Whirl‐Carrillo M, et al. Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the clinical pharmacogenetics implementation consortium and Dutch pharmacogenetics working group. Clin Transl Sci. 2020;13:116‐124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas CD, Mosley SA, Kim S, et al. Examination of metoprolol pharmacokinetics and pharmacodynamics across CYP2D6 genotype‐derived activity scores. CPT Pharmacometrics Syst Pharmacol. 2020;9:678‐685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adams EH, Breiner S, Cicero TJ, et al. A comparison of the abuse liability of tramadol, NSAIDs, and hydrocodone in patients with chronic pain. J Pain Symptom Manage. 2006;31:465‐476. [DOI] [PubMed] [Google Scholar]
- 26. Preston KL, Jasinski DR, Testa M. Abuse potential and pharmacological comparison of tramadol and morphine. Drug Alcohol Depend. 1991;27:7‐17. [DOI] [PubMed] [Google Scholar]
- 27. Gammal RS, Caudle KE, Quinn CT, et al. The case for pharmacogenetics‐guided prescribing of codeine in children. Clin Pharmacol Ther. 2019;105:1300‐1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Andreassen TN, Eftedal I, Klepstad P, et al. Do CYP2D6 genotypes reflect oxycodone requirements for cancer patients treated for cancer pain? A cross‐sectional multicentre study. Eur J Clin Pharmacol. 2012;68:55‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zwisler ST, Enggaard TP, Mikkelsen S, Brosen K, Sindrup SH. Impact of the CYP2D6 genotype on post‐operative intravenous oxycodone analgesia. Acta Anaesthesiol Scand. 2010;54:232‐240. [DOI] [PubMed] [Google Scholar]
- 30. Samer CF, Daali Y, Wagner M, et al. Genetic polymorphisms and drug interactions modulating CYP2D6 and CYP3A activities have a major effect on oxycodone analgesic efficacy and safety. Br J Pharmacol. 2010;160:919‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dai F, Silverman DG, Chelly JE, Li J, Belfer I, Qin L. Integration of pain score and morphine consumption in analgesic clinical studies. J Pain. 2013;14:767‐777.e8. [DOI] [PubMed] [Google Scholar]
- 32. Dowell D, Haegerich TM, Chou R. CDC guideline for prescribing opioids for chronic pain‐United States, 2016. Jama. 2016;315:1624‐1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeMets DL, Lan KK. Interim analysis: the alpha spending function approach. Stat Med. 1994;13:1341‐1352. discussion 1353‐1346. [DOI] [PubMed] [Google Scholar]
- 34. Hermes D, Matthes M, Saka B. Treatment anxiety in oral and maxillofacial surgery. Results of a German multi‐Centre trial. J Craniomaxillofac Surg. 2007;35:316‐321. [DOI] [PubMed] [Google Scholar]
- 35. Londhe SB, Shah RV, Patwardhan M, Doshi AP, Londhe SS, Subhedar K. Understanding the apprehension and concern haunting patients before a total knee arthroplasty. Art Ther. 2021;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Juprasert JM, Obeid L, Yeo HL. Public perception on opioids & pain management after major surgery. Am J Surg. 2022;223:280‐286. [DOI] [PubMed] [Google Scholar]
- 37. Brummett CM, Waljee JF, Goesling J, et al. New persistent opioid Use after Minor and major surgical procedures in US adults. JAMA Surg. 2017;152:e170504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han B, Compton WM, Blanco C, Crane E, Lee J, Jones CM. Prescription opioid use, misuse, and use disorders in U.S. adults: 2015 National Survey on drug use and health. Ann Intern Med. 2017;167:293‐301. [DOI] [PubMed] [Google Scholar]
- 39. Volkow ND, McLellan AT. Opioid abuse in chronic pain‐misconceptions and mitigation strategies. N Engl J Med. 2016;374:1253‐1263. [DOI] [PubMed] [Google Scholar]
- 40. Dal‐Re R, Janiaud P, Ioannidis JPA. Real‐world evidence: how pragmatic are randomized controlled trials labeled as pragmatic? BMC Med. 2018;16:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wick EC, Grant MC, Wu CL. Postoperative multimodal analgesia pain management with nonopioid analgesics and techniques: a review. JAMA Surg. 2017;152:691‐697. [DOI] [PubMed] [Google Scholar]
- 42. Otton SV, Schadel M, Cheung SW, Kaplan HL, Busto UE, Sellers EM. CYP2D6 phenotype determines the metabolic conversion of hydrocodone to hydromorphone. Clin Pharmacol Ther. 1993;54:463‐472. [DOI] [PubMed] [Google Scholar]
- 43. Stauble ME, Moore AW, Langman LJ, et al. Hydrocodone in postoperative personalized pain management: pro‐drug or drug? Clin Chim Acta. 2014;429:26‐29. [DOI] [PubMed] [Google Scholar]
- 44. Krebs EE, Gravely A, Nugent S, et al. Effect of opioid vs nonopioid medications on pain‐related function in patients with chronic Back pain or hip or knee osteoarthritis pain: the SPACE randomized clinical trial. Jama. 2018;319:872‐882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krebs EE, Jensen AC, Nugent S, et al. Design, recruitment outcomes, and sample characteristics of the strategies for prescribing analgesics comparative effectiveness (SPACE) trial. Contemp Clin Trials. 2017;62:130‐139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


