Abstract
Light is a factor affecting muscle development and meat quality in poultry production. However, few studies have reported on the role of light in muscle development and meat quality in geese. In this experiment, 10 healthy 220-day-old Zhedong white geese were reared for 60 d under a long photoperiod (15L:9D, LL) and short photoperiod (9L:15D, SL). The gastrocnemius muscles were collected after slaughter to evaluate muscle fiber characteristics and meat color, and RNA-seq analysis. The results showed that compared to the LL group, the SL group had large muscle fiber diameter and cross-sectional area, few muscle fibers per unit area, high meat color a* value, and low L* value at 24 h postmortem. On comparing the 2 groups, 70 differentially expressed genes (DEGs) were identified. Compared to the SL group, the LL group had 25 upregulated and 45 downregulated genes. Gene Ontology (GO) enrichment analysis showed that these DEGs were mainly involved in cell, cell part, binding, cellular processes, and single-organism processes. Several significantly enriched athways were identified in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, such as the calcium and PI3k-Akt signaling pathways. The expression of five randomly selected DEGs was verified using quantitative real-time PCR, and the results were consistent with the RNA-seq data. This study provides a theoretical basis for studying the molecular mechanisms by which light affects muscle development and meat color in geese.
Key words: muscle development, meat color, Zhedong White Geese, photoperiod, RNA-seq
INTRODUCTION
In the global meat market, goose meat is an excellent source of high-quality protein which contains all kinds of amino acids needed by humans (Haraf et al., 2021). The ideal high protein, low fat, and low cholesterol levels makes goose meat a popular nutritional health food among consumers (Buzała et al., 2014; Journal, 2021; Orkusz, 2021). China is the largest producer and consumer of goose meat globally. However, owing to the lack of a scientific breeding method, the breeding cycle of geese is longer than that of other poultry, which seriously restricts the development of the goose industry (Gao et al., 2021a). Therefore, exploring scientific methods to improve muscle development and meat quality of Chinese local geese is a useful way to overcome the current predicament.
Changes in photoperiod affect animal behavior, growth, food intake, and reproductive status (Mariné-Casadó et al., 2018). Atlantic salmon treated with continuous light showed an increased rate of muscle fiber recruitment, a high proportion of fast-twitch fibers, and a rapid increase in the muscle fiber cross-sectional area (Johnston et al., 2003). In addition, photoperiod induced weight gain and insulin-like growth factor-I (IGF-I) levels in the muscle of mice (Chennaoui et al., 2017). Monochromatic green light stimulation promotes the expression of the growth hormone/insulin-like growth factor-I (GH-IGF-I) axis and activates muscle satellite cell proliferation and myofibril formation in chick embryos (Bai et al., 2019). However, there are few reports on whether photoperiod affects muscle development in geese. Exploring a reasonable and effective light regime may be an important research direction for improving the production performance of geese.
Meat color is one of the key indicators for consumers to evaluate the quality of meat and also is one of the most important quality traits influencing consumer purchasing decisions (You et al., 2020). In poultry production, light is a key factor affecting the poultry meat color. Long light exposure reduces the redness of broiler meat (Tuell et al., 2020a). It is better to use intermittent light during duck feeding or only short light in the early stages to maintain a higher redness value of duck meat (Starčević et al., 2021). Dim light reduces the brightness of broiler meat color (Fidan et al., 2017), whereas long light makes the chicken meat color lighter and increases muscle lipid oxidation and protein denaturation (Tuell et al., 2020b). Prolongation of the photoperiod may negatively affect muscle oxidative stability and color by altering muscle metabolites (Tuell et al., 2020a). However, whether light affects geese meat color and the underlying molecular mechanisms by which light rgulates poultry meat color remain to be further investigated.
Since RNA-seq technology was proposed in 2008, it has been widely used in biological research, medical diagnosis and treatment (Ghosh et al., 2018; Kloet et al., 2020). RNA-seq technology has been used to compare the skeletal muscle transcriptomes of Muscovy ducks at different developmental stages and many potential candidate genes have been identified (Hu et al., 2020). In a study of chicken meat color, 1,317 candidate genes related to glycolysis, fatty acid metabolism, and protein metabolism, were screened using RNA-seq technology (Sun et al., 2022). In addition, 397 differentially methylated genes highly correlated with cellular metabolic processes were identified after m6A transcriptome sequencing of the pectoralis muscle in Ding'an geese (Xu et al., 2021). Thus, whole genome sequencing technology plays an important role in exploring the molecular mechanism of poultry traits.
In this study, the Chinese local breed Zhedong White Geese was used as the experimental object. High-throughput sequencing technology and bioinformatics were used to identify differentially expressed genes and signaling pathways present in the gastrocnemius muscles of geese reared under different photoperiod conditions. The results of this study provide a theoretical basis for improving the meat production performance of geese.
MATERIALS AND METHODS
Experimental Animals and Tissues
The Zhedong white geese (Anser cygnoides) used in this study were raised at the East Zhejiang White Geese Original Breeding Farm, Xiangshan County, Zhejiang Province, China. Twenty healthy 220-day-old Zhedong white goose females with similar weights were randomly divided into 2 groups. One group was kept under a long photoperiod (15L:9D, LL) and the other was kept under a short photoperiod under (9L:15D, SL) using artificial light controlled at 30 lx. After 60 d of feeding, 6 geese were dissected from each group and leg muscle samples were collected. All samples were immediately frozen in liquid nitrogen and stored at −80°C until use.
All animal experimental procedures used in this study were approved by the Ethics Committee for Animal Experiments of Zhejiang A&F University and were performed in accordance with the Guidelines for Animal Experimentation of the University (Hangzhou, China).
Total RNA Extraction, Library Construction, and Sequencing
Total RNA was extracted from the gastrocnemius muscle of the legs using the TRIzol method (Puch-Hau et al., 2019). To ensure that the samples qualified, quality testing was performed by the sample testing center of Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). The quality reports are presented in Table S1. All RNA samples met the requirements for Optical density (OD) values (260/280 > 2.0) for further analysis. The RNA-seq transcriptome library was prepared using the TruSeq RNA sample preparation Kit from Illumina (San Diego, CA) using 1 μg of total RNA. Briefly, messenger RNA was isolated according to the polyA selection method using oligo (dT) beads and then fragmented using fragmentation buffer. Second, double-stranded cDNA was synthesized using a SuperScript double-stranded cDNA synthesis kit (Invitrogen, CA) with random hexamer primers (Illumina). The synthesized cDNA was then subjected to end-repair, phosphorylation, and ‘A’ base addition according to Illumina's library construction protocol. Libraries were selected for cDNA target fragments of 300 bp on 2% low range ultra-agarose, followed by PCR amplification using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantification using TBS380, the paired-end RNA-seq sequencing library was sequenced using the Illumina HiSeq xten/NovaSeq 6000 sequencer (2 × 150 bp read length). Goose leg muscles of the SL group were numbered sl 1, sl 2, and sl 3, and those of the LL group were numbered ll 1, ll 2, and ll 3, for subsequent analysis.
Sequencing Quality Assessment and Differentially Expressed Gene
Screening
The SeqPrep and Sickle software were used to remove low quality reads from the raw data. Q20, Q30, GC content, and sequence repeat levels were calculated using fastp. All downstream analyses were based on high-quality clean reads. The goose genome sequences were downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/genome/31397?genome_assembly_id=229313). The resulting clean reads were compared with the reference genome of the goose using Hisat2.
To identify differentially expressed genes (DEGs) between the 2 groups, the expression level of each transcript was calculated according to the transcripts per million reads (TPM) method. RSEM (http://deweylab.biostat.wisc.edu/rsem/) (Dewey and Li, 2011) was used to quantify gene abundances. Differential expression analysis was performed using the DESeq2 (Love et al., 2014), DEGseq (Wang et al., 2010), and EdgeR (Robinson et al., 2010) method. Differentially expressed genes with |log2FC|>1 and Q value ≤ 0.05 (DESeq2 or EdgeR) or Q value ≤ 0.001 (DEGseq) were considered to be significantly differentially expressed.
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis
The Goatools based on the Gene Ontology (GO) database (http://www.geneontology.org/) were used to analyze the GO enrichment. Three parts, Biological Process (BP), Molecular Function (MF), and Cellular Component (CC) were used for classification annotation. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/), we used the KOBAS software (2.1, Peking University, China) to test the statistical enrichment of DEGs in the KEGG pathways.
Quantitative Real-Time PCR
Using the NCBI gene library, primers of 5 randomly selected DEGs and the house-keeping gene GAPDH were designed using Primer Premier 5 software (Table 1). Primers were synthesized by Hangzhou Youkang Biotechnology Co., Ltd. Total RNA was used to synthesize cDNA using the 5 × All-In-One MasterMix Kit (ABM, Zhengjiang, China). As the template, cDNA was used for quantitative PCR using the EvaGreen 2 × qPCR MasterMix Kit (ABM). Three replicates were performed for each sample. The relative expression of each gene was calculated using 2−△△CT statistical analysis. All data are shown as mean ± SEM, and unpaired Student's t tests were used to calculate P-values.
Table 1.
Primers used in the real-time PCR.
| Gene name | Sequence (5’-3’) | Product size |
|---|---|---|
| GAPDH | F: TTCCTCCACCTTTGATGCGG R:ACCATCAAGTCCACCACACG |
114bp |
| PHKB | F:CAACAATGGCAGTCCAGAGC R:TTGGGTACAACAGGACACCC |
543bp |
| SOX6 | F:TGGCTGGTGTGGTAGGAG R:AGGCAGATGAGAGGTCGC |
188bp |
| GCK | F:GCAGAAAGTGGAGATGGA R:GTGACACGGGAAAGAGAA |
172bp |
| HSPB1 | F:GTATTTCCGTCTGCTGCC R:TGTTTCTCCTCGTGCTTG |
222bp |
| PHGDH | F:TGAAGGAAGGCAAATGGG R:GGTCTCGGGGGTGATGAT |
164bp |
Analysis of Muscle Structure
Goose leg muscle samples were fixed with 4% paraformaldehyde and embedded in paraffin blocks. After the paraffin block was sectioned (5 μm), it was stained with hematoxylin and eosin solution, and the slides were placed in 70% ethanol for 20 s, 90% ethanol for 20 s, 100% ethanol for 1 min, and xylene for 3 min, and then air-dried and sealed (Wang et al., 2017). Sections were photographed using a fluorescence microscope (Olympus, Tokyo, Japan). Three samples were obtained from each group of geese, and 5 micrographs were randomly selected for each sample. A total of 90 muscle fibers per group were counted using the ImageJ software, and the average of 5 measurements was taken as the fiber cross-sectional area (μm2), diameter (μm), and density (fibers/mm2) (Zhang et al., 2014).
Detection of Meat Color
The goose meat color parameters were recorded on the L*a*b* color scale (Ganz and Pauli, 1995), using a Minolta Chromameter (Minolta Camera Co., Osaka, Japan). Goose meat color was measured 24 h after slaughter. The results are expressed as L* (lightness), a* (redness), and b* (yellowness) and were calculated from the average of 5 different locations on each muscle (Orkusz et al., 2017).
RESULTS
Differences in Muscle Fiber Characteristics of Goose Leg Muscles Under Different Photoperiods
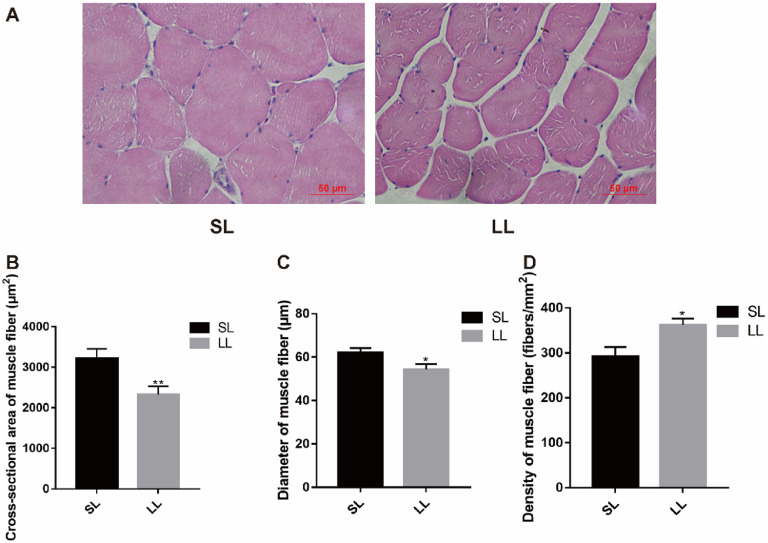
To explore the effect of photoperiod on muscle fiber characteristics in geese, gastrocnemius muscles were sectioned and stained. The results showed that the density of muscle fibers in the SL group was lower than that in the LL group, but the cross-sectional area and diameter of the muscle fibers were larger (Figures 1A–1D). Therefore, it was inferred that SL may promote goose muscle hypertrophy.
Figure 1.
Muscle fiber characteristics of geese under different photoperiods. (A) Representative micrographs of hematoxylin and eosin (H&E) staining of leg muscles of geese reared under different photoperiods. (B) Measurement of cross-sectional area of leg muscle fibers of geese reared under different photoperiods. (C) Measurement of the diameter of leg muscle fibers of geese reared under different photoperiods. (D) Measurement of the number of muscle fibers per unit area of leg muscles of geese reared under different photoperiods. ∗∗ P < 0.01, ∗ P < 0.05, n = 6.
Effect of Different Photoperiods on Meat Color of Geese
To investigate whether light affects geese meat color, meat color measurements were performed of leg muscles of geese reared under different photoperiods (Figure 2). The results showed that SL significantly reduced the L* value (brightness) (P-value = 0.025) while increasing the a* value (redness) (P-value = 0.0315) of goose meat color compared to that under LL (Table 2).
Figure 2.
Gastrocnemius muscle of Zhedong White geese in the two groups. n = 4.
Table 2.
L* (brightness), a* (redness) and b* (yellowness) values of goose leg muscles at 24 h postmortem under different photoperiods (n = 10).
| Light/h | Brightness L* | Redness a* | Yellowness b* |
|---|---|---|---|
| 15h | 26.65 ± 1.21a | 55.91 ± 0.81b | 15.26 ± 0.59 |
| 9h | 23.49 ± 1.60b | 58.30 ± 1.40a | 15.23 ± 1.19 |
| P-value | 0.0250 | 0.0315 | 0.4830 |
In the same column, values with different lowercase superscripts indicate significant differences (P < 0.05).
Sequencing Quality Evaluation
During the sequencing process, the raw data were quality controlled and a quality assessment was provided to ensure the accuracy of the subsequent analysis. The results listed in Table 3 show that the values of Q20 (base quality > 20 and error rate < 0.01)) are all above 98% and those of Q30 (base quality > 30 and error rate < 0.001) are all above 94%. The GC content ranged from 53 to 57%. The results indicated that the data had a low probability of error which met the sequencing requirements.
Table 3.
Quality assessment of sequencing date.
| Sample | Clean reads | Clean bases | Q20(%) | Q30(%) | GC content(%) |
|---|---|---|---|---|---|
| sl_1 | 54,313,760 | 80,26,863,149 | 98.27 | 95.01 | 54.53 |
| sl_2 | 44,389,142 | 6,555,398,259 | 98.35 | 95.26 | 56.73 |
| sl_3 | 48,539,644 | 7,211,649,150 | 98.75 | 95.98 | 54.97 |
| ll_1 | 46,095,306 | 6,861,678,422 | 98.76 | 95.99 | 54.88 |
| ll_2 | 51,839,210 | 7,677,991,815 | 98.25 | 94.95 | 54.92 |
| ll_3 | 49,484,182 | 7,363,615,166 | 98.74 | 95.92 | 53.94 |
sl and ll represent short and long photoperiod groups, respectively. 1/2/3 represent three biological replicates for each group. Clean reads: total number of sequencing data entries after quality control. Clean bases: total sequencing data volume after quality control (i.e., number of clean reads multiplied by the length of reads). Q20 and Q30: the percentage of bases with sequencing quality above 99% and 99.9% of the total bases, respectively. GC content: Quality control (QC) data corresponded to the sum of G and C bases as a percentage of the total bases.
More than 68% of the reads were aligned to the reference genome and 65 to 75% of the clean reads had unique comparison positions on the reference genome (Table 4). This illustrates that the clean reads obtained were available for post-sequencing analysis.
Table 4.
Statistical results of the comparison with the reference genome.
| Sample | Total reads | Total mapped | Multiple mapped | Uniquely mapped |
|---|---|---|---|---|
| sl_1 | 54,313,760 | 40,223,184(74.06%) | 1,160,976(2.14%) | 39,062,208(71.92%) |
| sl_2 | 44,389,142 | 30,206,700(68.05%) | 1,050,449(2.37%) | 29,156,251(65.68%) |
| sl_3 | 48,539,644 | 35,760,428(73.67%) | 1,084,433(2.23%) | 34,675,995(71.44%) |
| ll_1 | 46,095,306 | 34,149,665(74.08%) | 1,158,824(2.51%) | 32,990,841(71.57%) |
| ll_2 | 51,839,210 | 37,800,821(72.92%) | 14,66,913(2.83%) | 36,333,908(70.09%) |
| ll_3 | 49,484,182 | 37,292,580(75.36%) | 11,01,398(2.23%) | 36,191,182(73.14%) |
sl and ll represent short and long photoperiod groups, respectively. 1/2/3 represent three biological replicates for each group. Total reads: total number of sequencing data entries after quality control (i.e., clean reads). Total mapped: number of clean reads localized to the genome. Multiple mapped: number of clean reads with multiple comparison positions in the reference sequence. Uniquely mapped: number of clean reads with a unique comparison position on the reference sequence.
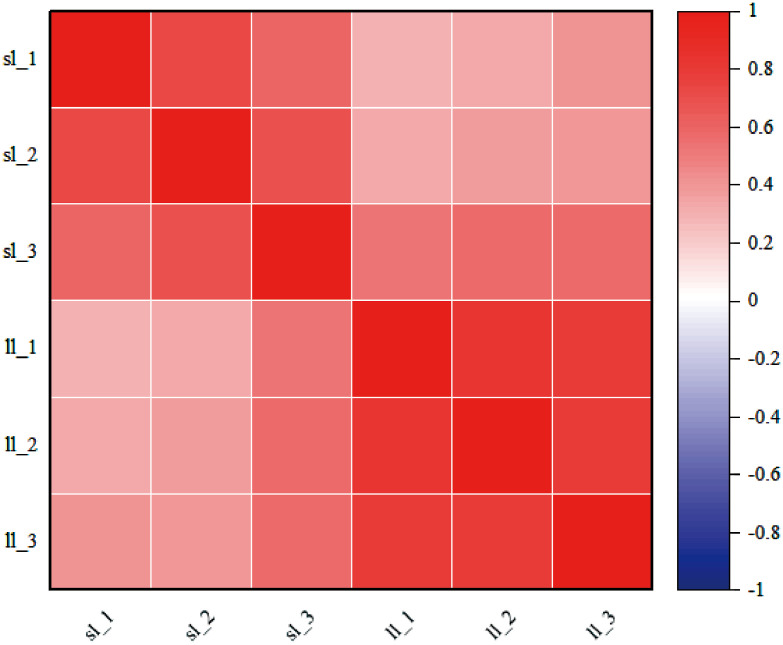
Correlation analysis was performed to assess the quality of repeatability within the group. The results showed that there were differences between the groups and the biological replicates within the groups were good (Figure 3).
Figure 3.
Heat map of intersample correlation.
Screening of DEGs
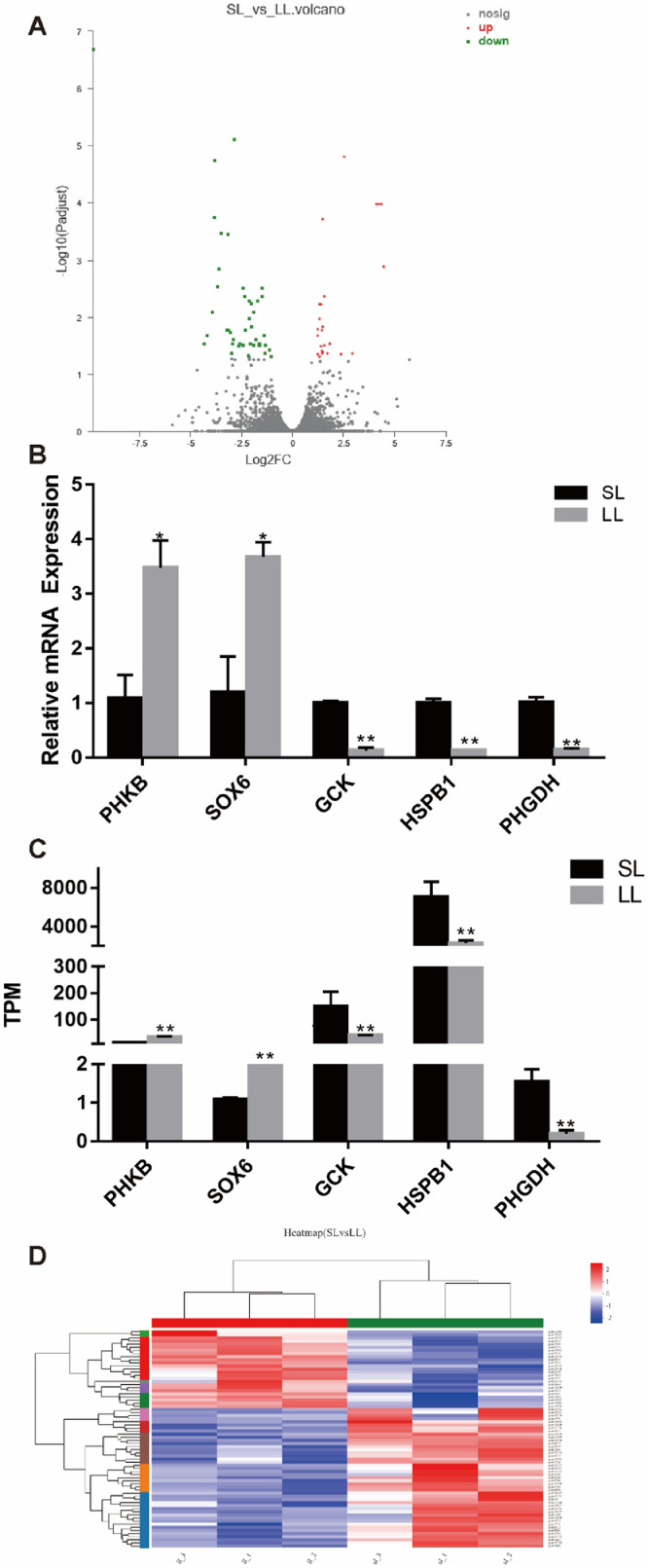
The total number of genes screened by RNA-Seq was 9,858. To display gene expression distribution, a volcano plot was constructed using -log10 (adjusted) as the vertical coordinate and log2 (FC) as the horizontal coordinate (Figure 4A). A total of 70 DEGs were identified. There were 45 upregulated genes and 25 downregulated genes in the SL group compared to the LL group (Figure 4A). The expression levels of 5 randomly selected DEGs, including 3 downregulated genes (GCK, HSPB1, and PHGDH) and 2 upregulated genes (PHKB and SOX6), were verified using quantitative real-time PCR. The results showed that their expression trends were highly consistent with the sequencing results, which proved the reliability of the microarray analysis (Figures 4B and 4C).
Figure 4.
Differential gene expression (DEGs) analysis. (A) Volcano plot for differential gene expression. (B) Validation of DEGs by qRT-PCR. (C) DEGs in RNA-seq results. (D) Heat map of differential gene expression. ** P < 0.01, * P < 0.05, n = 3.
To ensure the accuracy of the analysis, the screened differentially expressed genes were subjected to hierarchical clustering analysis. The horizontal coordinates represent sample clustering. One column represents one sample, and clustering analysis was based on the similarity of gene expression patterns between samples. The closer the distance, the more similar were the samples (Figure 4D). These results suggests that these DEGs clearly divided the leg muscle samples into short and long photoperiod groups, indicating the reliability of our analysis.
GO Annotation and Enrichment Analysis
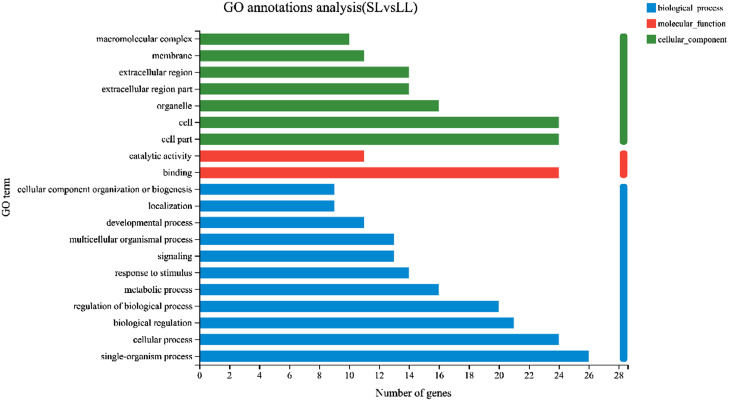
To further reveal the molecular characterization of these DEGs, they were analyzed using GO annotations. The results showed that 70 DEGs were annotated with 20 GO terms, including 11 biological processes, 7 cellular components, and 2 molecular function annotations. The major GO terms were significantly enriched in biological processes, which included positive regulation of biological processes, localization, development process, multicellular organismal process, signaling, response to stimulus, metabolism, regulation of biological processes, biological regulation, cellular processes, and single-organism processes. In addition, macromolecular complexes, membrane, extracellular region, extracellular region part, organelles, and cell part and cells were enriched among cellular components. The main functional term of molecular function were catalytic activity and binding, which were enriched in 11 and 24 genes, respectively (Figure 5).
Figure 5.
GO annotations analysis of the DEGs. Abbreviation: DEGs, differential gene expression.
KEGG Pathway Enrichment Analysis
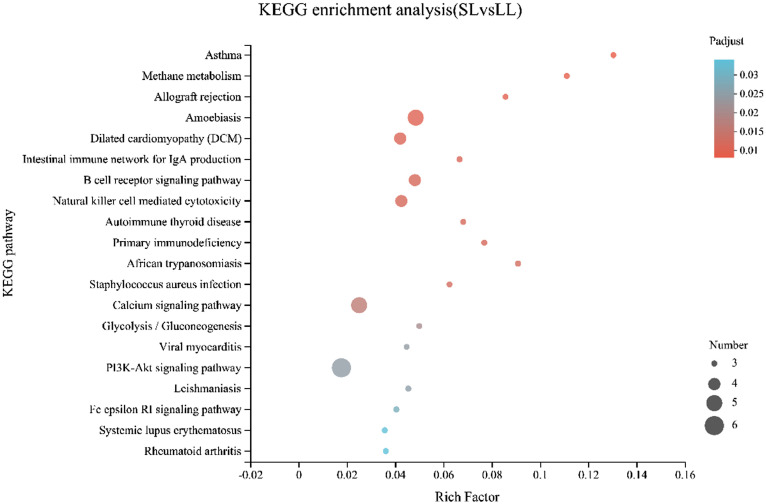
To understand the signaling pathways in which the genes were distributed, a KEGG pathway analysis was performed. A total of 48 DEGs were mapped to 127 KEGG pathways, of which 20 were significantly enriched (P < 0.05). There were several significantly enriched signaling pathways, including amoebiasis, the calcium signaling pathway, and the PI3k-Akt signaling pathway (Figure 6).
Figure 6.
Top 20 significantly enriched KEGG pathways.
DISCUSSION
Light plays an important role in goose productions and metabolism. Reasonable and effective use of light can greatly affect the production performance of geese (Wang et al., 2009; Liu et al., 2020). The present study showed that the Zhedong white geese in the SL group had a larger muscle fiber diameter and cross-sectional area, but had a smaller number of muscle fibers per unit area than those in the LL group. Melatonin levels in the blood of geese increase under dark conditions (Zawilska et al., 2003). Melatonin can promote proliferation of skeletal muscle satellite cells (Lamosová et al., 1998). Long-term exogenous addition of melatonin effectively prevents muscle atrophy (Lee et al., 2012). Therefore, melatonin may be a key hormone in light-induced muscle hypertrophy in geese. Our study also found that the meat color of geese under SL had a higher a* parameter and lower L* parameter than those under LL conditions, thus having a high commercial value. Glucocorticoid downregulation in birds is caused by prolonged light exposure (House et al., 2021). In muscle cells, increased glucocorticoid levels reduce cellular respiration and shift the cellular metabolic phenotype to glycolysis (Torres-Velarde et al., 2021). Studies have shown that glycolysis is an important metabolic pathway that affects meat color, and meat with low glycolytic dehydrogenase activity has poor color stability (Xin et al., 2018; Gao et al., 2021b). Through RNA-seq analysis of leg muscles of geese reared under different photoperiods, 70 DEGs were identified, among which 5 DEGs were randomly selected for further verification of expression levels using qRT-PCR. Although the expression levels of the genes selected by the 2 methods were different, their expression trends were highly consistent, which proved the reliability of the sequencing analysis results.
The basic processes of muscle development in geese include cellular and biological processes and biological regulation. GO annotation analysis showed that 20 DEGs were enriched in the GO term: regulation of biological processes (Table S2). Among these DEGs, FMOD, HSPB1, and STC1 were upregulated in the goose leg muscles under SL condtions. FMOD promotes chicken myogenic cell differentiation and alleviates muscle atrophy (Yin et al., 2020). Inhibition of HSPB1 during myogenesis represses myotube formation (Kim et al., 2018). STC1 can accelerate the fusion of mouse muscle cells to form myotubes (Jiang et al., 2000). In addition, NOV, PHGDH, and TNNT2 were also identified as upregulated DEGs in goose leg muscles under SL conditions. Loss of the NOV gene in mice causes muscle wasting (Heath et al., 2008). Our previous study indicated that PHGDH promoted the proliferation of chicken skeletal muscle satellite cells (Wang et al., 2022). TNNT2 is an important gene that induces grouper myoblasts differentiation (Kong et al., 2021). Polymorphism in the TNNT2 gene was closely associated with the pork color b* parameter (Ropka-Molik et al., 2018), which provided a theoretical basis for our subsequent study on the role of TNNT2 in light-influenced muscle development and meat color chage in geese. In contrast, KCNJ2 was significantly downregulated under SL conditions. Mutations in KCNJ2 cause malformations in muscle development (Andelfinger et al., 2002). Therefore, we speculated that these genes may play a key role in the regulation of goose myoblast differentiation and muscle hypertrophy.
The energy metabolism of muscle after slaughter affects meat color (Men et al., 2017). GO annotation analysis revealed that 16 DEGs were enriched in metabolic process-related GO terms (Table S2). Inhibition of glycolysis in the skeletal muscle has been reported to increase glycogen synthesis (Clark et al., 1987; Ren et al., 1993). Research has shown that animals with high glycogen levels tend to have worse meat color at 24 h postmortem (Choe et al., 2008). ACSL6, AGL, and GPD2 were upregulated in the LL group. Inhibition of ACSL6 reduces glucose uptake in the skeletal muscle (Jung and Bu, 2020), which in turn reduces muscle glycogen synthesis (Ryder et al., 1999). Cancer cells with knockout of AGL expression were more metabolically active and dependent on glycolysis than cells that express AGL normally (Lew et al., 2015). GPD2 can inhibit glycolysis in tumor cells (Wu et al., 2020; Zhao et al., 2021). In our study, the KEGG pathway enrichment analysis results indicated that the 2 most significantly enriched pathways were related to the energy metabolism in muscles (Table S3). ALDOC is active during the second stage of glycolysis (Wang et al., 2007). Knockdown of ALDOC significantly inhibits MUC16C-induced glycolysis in tumors (Fan et al., 2020). Thus, we inferred that these DEGs were activated during the process of light exposure, affecting goose meat color.
After animal death, mitochondria continue to metabolize oxygen in the skeletal muscle (Tang et al., 2005). Mitochondrial oxygen depletion establishes anaerobic conditions that promote the reduction of MetMb by transferring electrons from NADH or succinate to oxygen (Watts et al., 2010), which improves meat color stability. HSPB1 was upregulated in the SL group and was significantly enriched in GO term metabolic processes (Table S2). HSPB1 was found to restore the mitochondrial respiratory impairment caused by cellular stress (Mule et al., 2021). Pyruvate, a product of glycolysis, supports mitochondrial respiration in postmortem muscles (Mohan et al., 2010). GCK was significantly enriched in the glycolysis/gluconeogenesis pathway (Table S3) and highly expressed under SL conditions. During normal hepatocellular carcinogenesis, GCK expression can link the central carbon metabolism and stimulate mitochondrial respiration (Perrin-Cocon et al., 2021). Thus, these DEGs may be involved in the process of light affecting goose meat color through mitochondrial respiration.
KEGG enrichment analysis showed that the DEGs were significantly enriched in the calcium signaling pathway (Table S3). The intracellular calcium signaling pathway is an important factor that influences meat color (Lonergan et al., 2003; Ropka-Molik et al., 2017). Among the genes upregulated in the LL group, PHKB was significantly enriched in the calcium signaling pathway (Table S3). PHKB is a key regulatory enzyme in glycogen metabolism (Wüllrich-Schmoll and Kilimann, 1996), and mutations in PHKB can lead to the accumulation of muscle glycogen (Burwinkel et al., 1997). Muscles with high glycogen levels usually show a poor meat color at 24 h postmortem (Choe et al., 2008). In addition, the present study found that DEGs were significantly enriched in the PI3K-Akt signaling pathway (Table S3). FIGF was upregulated in the LL group. A C>G mutation in the bird FIGF gene results in a dramatic increase in the number of muscle fibers (Chen et al., 2013). The number of muscle fibers per unit area was higher in the LL group, which may be correlated with the function shown in FIGF.
Future research will be directed at determining the optimal photoperiods to utilize these DEGs and signaling pathways to regulate goose muscle development and quality to enable further rapid development of the goose industry.
CONCLUSIONS
The present study demonstrated that short light significantly promoted goose muscle development, resulting in a larger cross-sectional area and diameter of muscle fibers. In addition, short light remarkably improved the redness of goose meat. RNA-seq analysis provided a catalog of mRNAs for the study of the mechanisms by which light affects muscle development and meat color in geese, and screened 3 signaling pathways (glycolysis/gluconeogenesis, calcium signaling pathway, and PI3K-Akt signaling pathway) that may be involved in this regulation. The experimental data provided a theoretical basis for studying the molecular mechanisms by which light affects muscle development and meat color in geese.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NO. 31872397).
DISCLOSURES
The authors declare that there is no conflict of interest regarding the publication of this paper.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2022.102193.
Appendix. Supplementary materials
REFERENCES
- Andelfinger G., Tapper A.R., Welch R.C., Vanoye C.G., George A.L., Jr, Benson D.W. KCNJ2 mutation results in Andersen syndrome with sex-specific cardiac and skeletal muscle phenotypes. Am. J. Hum. Genet. 2002;71:663–668. doi: 10.1086/342360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Cao J., Dong Y., Wang Z., Chen Y.J.P.O. Melatonin mediates monochromatic green light-induced satellite cell proliferation and muscle growth in chick embryo. PLoS One. 2019;14 doi: 10.1371/journal.pone.0216392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwinkel B., Maichele A.J., Aagenaes O., Bakker H.D., Lerner A., Shin Y.S., Strachan J.A., Kilimann M.W. Autosomal glycogenosis of liver and muscle due to phosphorylase kinase deficiency is caused by mutations in the phosphorylase kinase beta subunit (PHKB) Hum. Mol. Genet. 1997;6:1109–1115. doi: 10.1093/hmg/6.7.1109. [DOI] [PubMed] [Google Scholar]
- Buzała M., Adamski M., Janicki B. Characteristics of performance traits and the quality of meat and fat in Polish oat geese. Worlds Poult. Sci. J. 2014;70:531–542. [Google Scholar]
- Chen S., An J., Lian L., Qu L., Zheng J., Xu G., Yang N. Polymorphisms in AKT3, FIGF, PRKAG3, and TGF-β genes are associated with myofiber characteristics in chickens. Poult. Sci. 2013;92:325–330. doi: 10.3382/ps.2012-02766. [DOI] [PubMed] [Google Scholar]
- Chennaoui M., Arnal P., Dorey R., Sauvet F., Ciret S., Gallopin T., Leger D., Drogou C., J.O.M.S. Gomez-merino D.J.I. Changes of cerebral and/or peripheral adenosine A₁ receptor and IGF-I concentrations under extended sleep duration in rats. Int. J. Mol. Sci. 2017;18:2439. doi: 10.3390/ijms18112439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J.H., Choi Y.M., Lee S.H., Shin H.G., Ryu Y.C., Hong K.C., Kim B.C. The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci. 2008;80:355–362. doi: 10.1016/j.meatsci.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Clark A.S., Mitch W.E., Goodman M.N., Fagan J.M., Goheer M.A., Curnow R.T. Dichloroacetate inhibits glycolysis and augments insulin-stimulated glycogen synthesis in rat muscle. J. Clin. Invest. 1987;79:588–594. doi: 10.1172/JCI112851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey C.N., Li B.J.B.B. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan K., Wang J., Sun W., Shen S., Ni X., Gong Z., Zheng B., Gao Z., Ni X., Suo T., Liu H., Liu H. MUC16 C-terminal binding with ALDOC disrupts the ability of ALDOC to sense glucose and promotes gallbladder carcinoma growth. Exp. Cell. Res. 2020;394 doi: 10.1016/j.yexcr.2020.112118. [DOI] [PubMed] [Google Scholar]
- Fidan E.D., Nazlıgül A., Türkyılmaz M.K., Aypak S.Ü., Kilimci F.S., Karaarslan S., Kaya M. Effect of photoperiod length and light intensity on some welfare criteria, carcass, and meat quality characteristics in broilers. Rev. Bras. Zootec. 2017;46:202–210. [Google Scholar]
- Ganz E., Pauli H.K. Whiteness and tint formulas of the Commission Internationale de l'Eclairage: approximations in the Lab color space. Appl. Opt. 1995;34:2998–2999. doi: 10.1364/AO.34.002998. [DOI] [PubMed] [Google Scholar]
- Gao G., Gao D., Zhao X., Xu S., Zhang K., Wu R., Yin C., Li J., Xie Y., Hu S., F.I.G. Wa Q.J. Genome-wide association study-based identification of SNPs and haplotypes associated with goose reproductive performance and egg quality. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.602583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Zhao D., Wang L., Cui Y., Wang S., Lv M., Zang F., Dai R. Proteomic changes in sarcoplasmic and myofibrillar proteins associated with color stability of ovine muscle during post-mortem storage. Foods. 2021;10:2989. doi: 10.3390/foods10122989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Sharma N., Singh A.K., Gera M., Pulicherla K.K., Jeong D.K. Transformation of animal genomics by next-generation sequencing technologies: a decade of challenges and their impact on genetic architecture. Crit. Rev. Biotechnol. 2018;38:1157–1175. doi: 10.1080/07388551.2018.1451819. [DOI] [PubMed] [Google Scholar]
- Haraf G., Wołoszyn J., Okruszek A., Goluch Z., Wereńska M., Teleszko M. The protein and fat quality of thigh muscles from Polish goose varieties. Poult Sci. 2021;100 doi: 10.1016/j.psj.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath E., Tahri D., Andermarcher E., Schofield P., Fleming S., Boulter C.A. Abnormal skeletal and cardiac development, cardiomyopathy, muscle atrophy and cataracts in mice with a targeted disruption of the Nov (Ccn3) gene. BMC Dev. Biol. 2008;8:18. doi: 10.1186/1471-213X-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House G.M., Sobotik E.B., Nelson J.R., Archer G.S. Pekin duck productivity, physiological stress, immune response and behavior under 20L:4D and 16L:8D photoperiods. Appl. Anim. Behav. Sci. 2021;240 [Google Scholar]
- Hu Z., Cao J., Liu G., Zhang H., Liu X. Comparative transcriptome profiling of skeletal muscle from black muscovy duck at different growth stages using RNA-seq. Genes (Basel) 2020;11:1228. doi: 10.3390/genes11101228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.Q., Chang A.C., Satoh M., Furuichi Y., Tam P.P., Reddel R.R. The distribution of stanniocalcin 1 protein in fetal mouse tissues suggests a role in bone and muscle development. J. Endocrinol. 2000;165:457–466. doi: 10.1677/joe.0.1650457. [DOI] [PubMed] [Google Scholar]
- Johnston I., Manthri S., Smart A., Campbell P., Nickell D., J.O.E.B. Alderson R.J.T. Plasticity of muscle fibre number in seawater stages of Atlantic salmon in response to photoperiod manipulation. J. Exp. Biol. 2003;206:3425–3435. doi: 10.1242/jeb.00577. [DOI] [PubMed] [Google Scholar]
- W.S.P.S. Journal J.K.J. Goose production and goose products. Worlds Poult. Sci. J. 2021;77:1–12. [Google Scholar]
- Jung Y.H., Bu S.Y. Suppression of long chain acyl-CoA synthetase blocks intracellular fatty acid flux and glucose uptake in skeletal myotubes. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2020;1865 doi: 10.1016/j.bbalip.2020.158678. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Lee J.S., Lee Y., Kim W.S., Peng D.Q., Bae M.H., Jo Y.H., Baik M., Lee H.G. Effect of glutamine on heat-shock protein beta 1 (HSPB1) expression during myogenic differentiation in bovine embryonic fibroblast cells. Food Sci. Biotechnol. 2018;27:829–835. doi: 10.1007/s10068-018-0309-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloet F., Buurmans J., Jonker M.J., Smilde A.K., P.C.B.Westerhuis J.A.J. Increased comparability between RNA-Seq and microarray data by utilization of gene sets. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Wang X., Li M., Song W., Huang K., Zhang F., Zhang Q., Qi J., He Y. Establishment of myoblast cell line and identification of key genes regulating myoblast differentiation in a marine teleost, Sebastes schlegelii. Gene. 2021;802 doi: 10.1016/j.gene.2021.145869. [DOI] [PubMed] [Google Scholar]
- Lamosová D., Zeman M., Juráni M. Influence of melatonin on chick skeletal muscle cell growth. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998;118:375–379. doi: 10.1016/s0742-8413(97)00159-x. [DOI] [PubMed] [Google Scholar]
- Lee S., Shin J., Hong Y., Lee M., Kim K., Lee S.R., Chang K.T., Hong Y. Beneficial effects of melatonin on stroke-induced muscle atrophy in focal cerebral ischemic rats. Lab. Anim. Res. 2012;28:47–54. doi: 10.5625/lar.2012.28.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.J., Chen Z.F., Zhao X.H., Li M.Y., Guo Z.H. Meta-analysis: supplementary artificial light and goose reproduction. Anim. Reprod. Sci. 2020;214 doi: 10.1016/j.anireprosci.2020.106278. [DOI] [PubMed] [Google Scholar]
- Lonergan S.M., Deeb N., Fedler C.A., Lamont S.J. Breast meat quality and composition in unique chicken populations. Poult. Sci. 2003;82:1990–1994. doi: 10.1093/ps/82.12.1990. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew C.R., Guin S., Theodorescu D. Targeting glycogen metabolism in bladder cancer. Nat. Rev. Urol. 2015;12:383–391. doi: 10.1038/nrurol.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariné-casadó R., Domenech-coca C., Delbas J., Bladé C., Arola L., F.I.P. Caimari A.J. The exposure to different photoperiods strongly modulates the glucose and lipid metabolisms of normoweight fischer 344 rats. Front. Physiol. 2018;9:416. doi: 10.3389/fphys.2018.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men X.M., Deng B., Tao X., Ke-ke Q.I., J.O.I.A. Zi-wei X.U.J. Wnt gene expression in adult porcine longissimus dorsi and its association with muscle fiber type, energy metabolism, and meat quality. J. Integr. Agric. 2017;16:144–150. [Google Scholar]
- Mohan A., Hunt M.C., Barstow T.J., Houser T.A., Muthukrishnan S. Effects of malate, lactate, and pyruvate on myoglobin redox stability in homogenates of three bovine muscles. Meat Sci. 2010;86:304–310. doi: 10.1016/j.meatsci.2010.04.030. [DOI] [PubMed] [Google Scholar]
- Mule S.N., Gomes V.M., Wailemann R.A.M., Macedo-da-Silva J., Rosa-fernandes L., Larsen M.R., Labriola L., Palmisano G. HSPB1 influences mitochondrial respiration in ER-stressed beta cells. Biochim. Biophys. Acta Proteins Proteom. 2021;1869 doi: 10.1016/j.bbapap.2021.140680. [DOI] [PubMed] [Google Scholar]
- Orkusz A. Edible insects versus meat-nutritional comparison: knowledge of their composition is the key to good health. Nutrients. 2021;13:1207. doi: 10.3390/nu13041207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkusz A., Haraf G., Okruszek A., Werenska-sudnik M. Lipid oxidation and color changes of goose meat stored under vacuum and modified atmosphere conditions. Poult. Sci. 2017;96:731–737. doi: 10.3382/ps/pew325. [DOI] [PubMed] [Google Scholar]
- Perrin-cocon L., Vidalain P.O., Jacquemin C., Aublin-gex A., Olmstead K., Panthu B., Rautureau G.J.P., Andre P., Nyczka P., Hutt M.T., Amoedo N., Rossignol R., Filipp F.V., Lotteau V., Diaz O. A hexokinase isoenzyme switch in human liver cancer cells promotes lipogenesis and enhances innate immunity. Commun. Biol. 2021;4:217. doi: 10.1038/s42003-021-01749-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puch-hau C., Sánchez-tapia I., Patiño-Suárez V., Olvera-novoa M., Gullian klanian M., Quintanilla-mena M., Zapata-pérez O.J.M. Isostichopus badionotusEvaluation of two independent protocols for the extraction of DNA and RNA from different tissues of sea cucumber. MethodsX. 2019;6:1627–1634. doi: 10.1016/j.mex.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J.M., Marshall B.A., Gulve E.A., Gao J., Johnson D.W., Holloszy J.O., Mueckler M. Evidence from transgenic mice that glucose transport is rate-limiting for glycogen deposition and glycolysis in skeletal muscle. J. Biol. Chem. 1993;268:16113–16115. [PubMed] [Google Scholar]
- Robinson M.D., Mccarthy D.J., Smyth G.K.J.B. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropka-molik K., Bereta A., Żukowski K., Piórkowska K., Gurgul A., Żak G. Transcriptomic gene profiling of porcine muscle tissue depending on histological properties. Anim. Sci. J. 2017;88:1178–1188. doi: 10.1111/asj.12751. [DOI] [PubMed] [Google Scholar]
- Ropka-molik K., Bereta A., Żukowski K., Tyra M., Piórkowska K., Żak G., Oczkowicz M. Screening for candidate genes related with histological microstructure, meat quality and carcass characteristic in pig based on RNA-seq data. Asian-Australas. J. Anim. Sci. 2018;31:1565–1574. doi: 10.5713/ajas.17.0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder J.W., Kawano Y., Galuska D., Fahlman R., Wallberg-henriksson H., Charron M.J., Zierath J.R. Postexercise glucose uptake and glycogen synthesis in skeletal muscle from GLUT4-deficient mice. FASEB J. 1999;13:2246–2256. doi: 10.1096/fasebj.13.15.2246. [DOI] [PubMed] [Google Scholar]
- Starčević M., Mahmutović H., Glamočlija N., Bašić M., Andjelković R., Mitrović R., Marković R., Janjić J., Bošković M., A.I.J.O.A.B. Baltić M.J.A. Growth performance, carcass characteristics, and selected meat quality traits of two strains of Pekin duck reared in intensive vs semi-intensive housing systems. Animal. 2021;15 doi: 10.1016/j.animal.2020.100087. [DOI] [PubMed] [Google Scholar]
- Sun J., Tan X., Yang X., Bai L., Kong F., Zhao G., Wen J., Liu R. Identification of candidate genes for meat color of chicken by combing selection signature analyses and differentially expressed genes. Genes (Basel) 2022;13:307. doi: 10.3390/genes13020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Faustman C., Hoagland T.A., Mancini R.A., Seyfert M., Hunt M.C. Postmortem oxygen consumption by mitochondria and its effects on myoglobin form and stability. J. Agric. Food Chem. 2005;53:1223–1230. doi: 10.1021/jf048646o. [DOI] [PubMed] [Google Scholar]
- Torres-velarde J.M., Kolora S.R.R., Khudyakov J.I., Crocker D.E., Sudmant P.H., Vázquez-medina J.P. Elephant seal muscle cells adapt to sustained glucocorticoid exposure by shifting their metabolic phenotype. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2021;321:R413–R428. doi: 10.1152/ajpregu.00052.2021. [DOI] [PubMed] [Google Scholar]
- Tuell J.R., Park J.Y., Wang W., Cooper B., Sobreira T., Cheng H.W., Kim Y.H.B. Effects of photoperiod regime on meat quality, oxidative stability, and metabolites of postmortem broiler fillet (M. Pectoralis major) muscles. Foods. 2020;9:215. doi: 10.3390/foods9020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuell J.R., Park J.Y., Wang W., Cheng H.W., Kim Y.H.B. Functional/physicochemical properties and oxidative stability of ground meat from broilers reared under different photoperiods. Poult. Sci. 2020;99:3761–3768. doi: 10.1016/j.psj.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Yue F., Kuang S. Muscle histology characterization using H&E staining and muscle fiber type classification using immunofluorescence staining. Bio. Protoc. 2017;7:e2279. doi: 10.21769/BioProtoc.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.F., Yuan C.Z., Wang S.H., Zhang H., Hu X.X., Zhang L., Wu C., Li N. Differential gene expression of aldolase C (ALDOC) and hypoxic adaptation in chickens. Anim. Genet. 2007;38:203–210. doi: 10.1111/j.1365-2052.2007.01605.x. [DOI] [PubMed] [Google Scholar]
- Wang C.M., Chen L.R., Lee S.R., Jea Y.S., Kao J.Y. Supplementary artificial light to increase egg production of geese under natural lighting conditions. Anim. Reprod. Sci. 2009;113:317–321. doi: 10.1016/j.anireprosci.2008.05.073. [DOI] [PubMed] [Google Scholar]
- Wang H., Hu M., Ding Z., Zhou X., Yang S., Shen Z., Yan F., Zhao A. Phosphoglycerate dehydrogenase positively regulates the proliferation of chicken muscle cells. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2022.101805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Feng Z., Wang X., Wang X., Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Watts B.M., Kendrick J., Zipser M.W., Hutchins B., O.F.S. Saleh B.J.J. Enzymatic reducing pathways in meat. J. Food Sci. 2010;31:855–862. [Google Scholar]
- Wu S.T., Liu B., Ai Z.Z., Hong Z.C., You P.T., Wu H.Z., Yang Y.F. Esculetin inhibits cancer cell glycolysis by binding tumor PGK2, GPD2, and GPI. Front. Pharmacol. 2020;11:379. doi: 10.3389/fphar.2020.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüllrich-schmoll A., Kilimann M.W. Structure of the human gene encoding the phosphorylase kinase beta subunit (PHKB) Eur. J. Biochem. 1996;238:374–380. doi: 10.1111/j.1432-1033.1996.0374z.x. [DOI] [PubMed] [Google Scholar]
- Xin J., Li Z., Li X., Li M., Li G., Rao W., Zhang D. Meat color stability of ovine muscles is related to glycolytic dehydrogenase activities. J. Food Sci. 2018;83:2432–2438. doi: 10.1111/1750-3841.14321. [DOI] [PubMed] [Google Scholar]
- Xu T., Xu Z., Lu L., Zeng T., Gu L., Huang Y., Zhang S., Yang P., Wen Y., Lin D., Xing M., Huang L., Liu G., Chao Z., Sun W. Transcriptome-wide study revealed m6A regulation of embryonic muscle development in Dingan goose (Anser cygnoides orientalis) BMC Genomics. 2021;22:270. doi: 10.1186/s12864-021-07556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Cui C., Han S., Chen Y., Zhao J., He H., Li D., Zhu Q. Fibromodulin modulates chicken skeletal muscle development via the transforming growth factor-beta signaling pathway. Animals (Basel) 2020;10:1477. doi: 10.3390/ani10091477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You M., Liu J., Zhang J., Xv M., He D.J.I.A. A novel chicken meat quality evaluation method based on color card localization and color correction. IEEE Access. 2020;PP [Google Scholar]
- Zawilska J.B., Berezińska M., Rosiak J., Vivien-roels B., Skene D.J., Pévet P., Nowak J.Z. Daily variation in the concentration of melatonin and 5-methoxytryptophol in the goose pineal gland, retina, and plasma. Gen. Comp. Endocrinol. 2003;134:296–302. doi: 10.1016/s0016-6480(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Guo J., Gao Y., Niu S., Yang C., Bai C., Yu X., Zhao Z. Genome-wide methylation changes are associated with muscle fiber density and drip loss in male three-yellow chickens. Mol. Biol. Rep. 2014;41:3509–3516. doi: 10.1007/s11033-014-3214-6. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhang L., Guo M., Yang H. Taraxasterol suppresses cell proliferation and boosts cell apoptosis via inhibiting GPD2-mediated glycolysis in gastric cancer. Cytotechnology. 2021;73:815–825. doi: 10.1007/s10616-021-00499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.