Abstract
Background & Aims
Nonalcoholic fatty liver disease (NAFLD) is a major cause of liver-related morbidity and mortality whereas the pathogenic mechanism remains largely elusive. DNA N6-methyladenosine (6mA) modification is a recently identified epigenetic mark indicative of transcription in eukaryotic genomes. Here, we aimed to investigate the role and mechanism of DNA 6mA modification in NAFLD progression.
Methods
Dot blot and immunohistochemistry were used to detect DNA 6mA levels. Liver-specific AlkB homolog 1 (ALKBH1)-knockout mice and mice with ALKBH1 overexpression in liver were subjected to a high-fat diet or methionine choline-deficient diet to evaluate the critical role of ALKBH1-demethylated DNA 6mA modification in the pathogenesis of hepatic steatosis during NAFLD. RNA sequencing and chromatin immunoprecipitation sequencing were performed to investigate molecular mechanisms underlying this process.
Results
The DNA 6mA level was increased significantly with hepatic steatosis, while ALKBH1 expression was down-regulated markedly in both mouse and human fatty liver. Deletion of ALKBH1 in hepatocytes increased genomic 6mA levels and accelerated diet-induced hepatic steatosis and metabolic dysfunction. Comprehensive analyses of transcriptome and chromatin immunoprecipitation sequencing data indicated that ALKBH1 directly bound to and exclusively demethylated 6mA levels of genes involved in fatty acid uptake and lipogenesis, leading to reduced hepatic lipid accumulation. Importantly, ALKBH1 overexpression was sufficient to suppress lipid uptake and synthesis, and alleviated diet-induced hepatic steatosis and insulin resistance.
Conclusions
Our findings show an indispensable role of ALKBH1 as an epigenetic suppressor of DNA 6mA in hepatic fatty acid metabolism and offer a potential therapeutic target for NAFLD treatment.
Keywords: DNA 6mA, ALKBH1, Demethylation, Fatty Liver, Lipid Metabolism
Abbreviations used in this paper: Ad, adenovirus; AKO, Alkbh1 knockout; Akt, thymoma viral proto-oncogene 1; ALKBH1, AlkB homolog 1; ALT, alanine aminotransferase; AOE, ALKBH1 overexpression; AST, aspartate aminotransferase; ChIP-seq, chromatin immunoprecipitation sequencing; FATP, fatty acid transport protein; FFA, free fatty acid; GSK3β, glycogen synthase kinase 3 beta; HFD, high-fat diet; MCD, methionine choline-deficient; mRNA, messenger RNA; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NCD, normal chow diet; PA, palmitic acid; qPCR, quantitative polymerase chain reaction; RNA-seq, RNA sequencing; TG, triglyceride; UPR mt, mitochondria unfolded protein response; 6mA, N6-methyladenosine
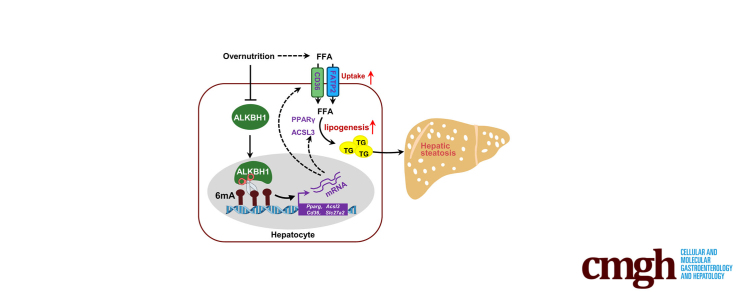
Graphical abstract
Summary.
The pathogenic mechanism of nonalcoholic fatty liver disease and the functions of DNA N6-methyladenosine (6mA) modification in metabolism remain elusive. In this study, we uncovered that DNA 6mA modification regulated by 6mA demethylase AlkB homolog 1 promotes nonalcoholic fatty liver disease progression and insulin resistance via modulating fatty acid metabolism.
Nonalcoholic fatty liver disease (NAFLD) is a leading chronic liver disease and constitutes disorders from simple steatosis, through nonalcoholic steatohepatitis (NASH), to fibrosis and cirrhosis.1,2 Hepatic steatosis is the hallmark of NAFLD and the progression to fibrosis also occurs in patients with steatosis alone,3 although the overall mortality rates are increased in advanced stages and in associated diseases such as NASH, hepatocellular carcinoma, and cardiovascular disease.4 In addition, hepatic steatosis is associated with insulin resistance, dyslipidemia, and cardiovascular disease, and hepatic fat accumulation seems to be more related to these metabolic dysfunctions than obesity status.5, 6, 7 Thus, exploring the molecular mechanisms regulating hepatic lipid homeostasis may be instrumental in the development and progression of NAFLD.
Lipid droplet accumulation results from an imbalance of hepatic lipid homeostasis: acquisition through fatty acid uptake and de novo lipogenesis surpasses the disposal via fatty acid oxidation and export of lipids.4 The uptake of fatty acid in liver is mediated predominately by fatty acid transport proteins (FATPs) and the fatty acid translocase CD36, which facilitates the transportation of fatty acids and drives hepatosteatosis onset, contributing to NAFLD progression.8,9 De novo lipogenesis converts acetyl coenzyme A (acetyl-CoA) to triglycerides (TGs), which involves increased expression of genes such as Fasn, Scd1, Pparg, and Acsl.10, 11, 12 Evidence indicates that the pathogenesis of NAFLD likely is shaped by dynamic interactions between genes and environmental factors such as overnutrition, with epigenetics serving as a mechanistic bridge.2,13 However, how overnutrition triggers epigenetic mechanisms to influence the initiation and progression of NAFLD is largely unknown. Understanding the mechanism by which epigenetic modifiers participate in the progress of NAFLD would provide novel therapeutic strategies for clinic treatment.
DNA N6-methyladenosine (6mA) recently was identified as an epigenetic mark in eukaryotic genomes.14, 15, 16, 17 Different from the genomic distribution of 5-methylcytosine, 6mA localization patterns are species- and tissue-specific, likely reflecting different functions and mechanisms of regulation.16,18,19 The deposition of DNA 6mA on mammal genomes was reported to correlate with epigenetic silencing during early development,16 and its abundance was underactive regulation of pathophysiological changes such as fertilization,15 environmental stress,20 and malignant diseases.21 In hepatocellular carcinoma, DNA 6mA down-regulation correlates with increased tumorigenesis,19 but its contribution to benign diseases, especially hepatic steatosis, is not well characterized.
AlkB homolog 1 (ALKBH1), one member of the AlkB family of oxygenases with known biological functions on nucleotide demethylation,22 was found to act as a demethylase for DNA 6mA.17,19 Deletion of the Alkbh1 gene increases DNA 6mA levels and leads to transcriptional silencing or activation in multicellular species.16,23 Because ALKBH1 whole-body knockout mice were embryonic lethal or showed neural development defects and sex-ratio distortion,24, 25, 26 tissue-/cell-specific ALKBH1-knockout/knockdown mice were used and showed the important role of ALKBH1-demethylated DNA 6mA modification in regulating osteogenic differentiation,27,28 myogenesis,29 hippocampal atrophy,30 axon regeneration,31 vascular calcification,32 and cancer cell growth.17 However, the role of ALKBH1-mediated DNA 6mA modification in diet-induced hepatic metabolic changes and the underlying molecular basis remains unclear.
In this study, we identified that the DNA 6mA level was increased significantly, whereas the expression of ALKBH1, a critical DNA 6mA demethylase, was decreased in fatty liver. Liver-specific ALKBH1 knockout enhanced DNA 6mA modification and lipogenic gene expression, and promoted diet-induced hepatic steatosis and insulin resistance. In contrast, overexpression of ALKBH1 in liver suppressed lipid biosynthesis and protected diet-induced fatty liver and metabolic dysfunction. Notably, we provide evidence supporting that ALKBH1 directly binds to and demethylates DNA 6mA modification of genes involved specifically in fatty acid uptake and lipogenesis, leading to reduced gene transcription and subsequently hepatic lipid accumulation. Therefore, our study shows that ALKBH1-demethylated DNA 6mA modification is a key epigenetic regulator of hepatic TG homeostasis in response to overnutrition.
Results
DNA 6mA Level Is Increased in Liver of Obese Mice
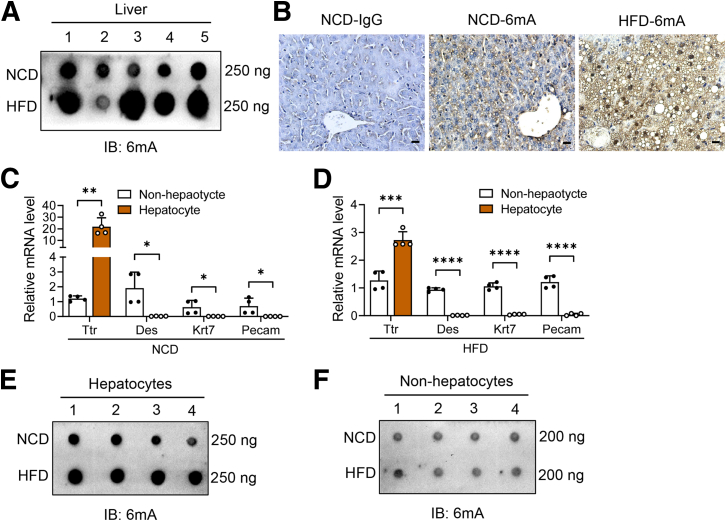
To explore the association of DNA 6mA modification with fatty liver, we examined the DNA 6mA levels in the liver of mice fed with a high-fat diet (HFD), which induced hepatic steatosis. Dot blot analysis showed that the levels of DNA 6mA in the liver of HFD-fed mice was higher than that of normal chow diet (NCD)-fed mice (Figure 1A). To further investigate which liver cell type contributes to the increased DNA 6mA modification in fatty liver, immunohistochemistry staining was performed on liver slice and increased signals of DNA 6mA modification were detected mainly within the nucleus of hepatocytes derived from HFD-fed mice (Figure 1B). Furthermore, hepatocytes and nonhepatocytes were isolated from the mice under NCD and HFD feeding conditions (Figure 1C and D). Results showed that DNA 6mA levels were increased in hepatocytes from HFD-fed mice compared with that from NCD-fed mice (Figure 1E), while no significant changes were observed in nonhepatocytes between NCD- and HFD-fed mice (Figure 1F). Thus, these data showed that DNA 6mA modification was enhanced in the hepatocytes of diet-induced fatty liver.
Figure 1.
DNA 6mA level increases in the liver of obese mice. (A) Dot blot of 6mA levels on genomic DNA from liver of NCD- and HFD-fed mice. A total of 250 ng DNA was loaded for each sample (n = 5). (B) Representative immunohistochemical staining of 6mA levels in the liver sections from NCD- and HFD-fed mice. Scale bar: 10 μm. The mRNA levels of Ttr (hepatocyte marker), Des (HSC marker), Krt7 (cholangiocyte marker), and Pecam (endothelial cell marker) in hepatocytes and nonhepatocytes isolated from livers of (C) NCD-fed lean mice and (D) HFD-fed obese mice (n = 4). Normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA. (E) Dot blot of 6mA levels on genomic DNA extracted from hepatocytes of NCD- and HFD-fed mice. A total of 250 ng DNA was loaded for each sample (n = 4). (F) Dot blot of 6mA levels on genomic DNA extracted from nonhepatocytes of NCD- and HFD-fed mice. A total of 200 ng DNA was loaded for each sample (n = 4). (C and D) Data are presented as the means ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. IB, immunoblotting.
Expression of DNA 6mA Demethylase ALKBH1 Decreases in Mouse and Human Fatty Liver
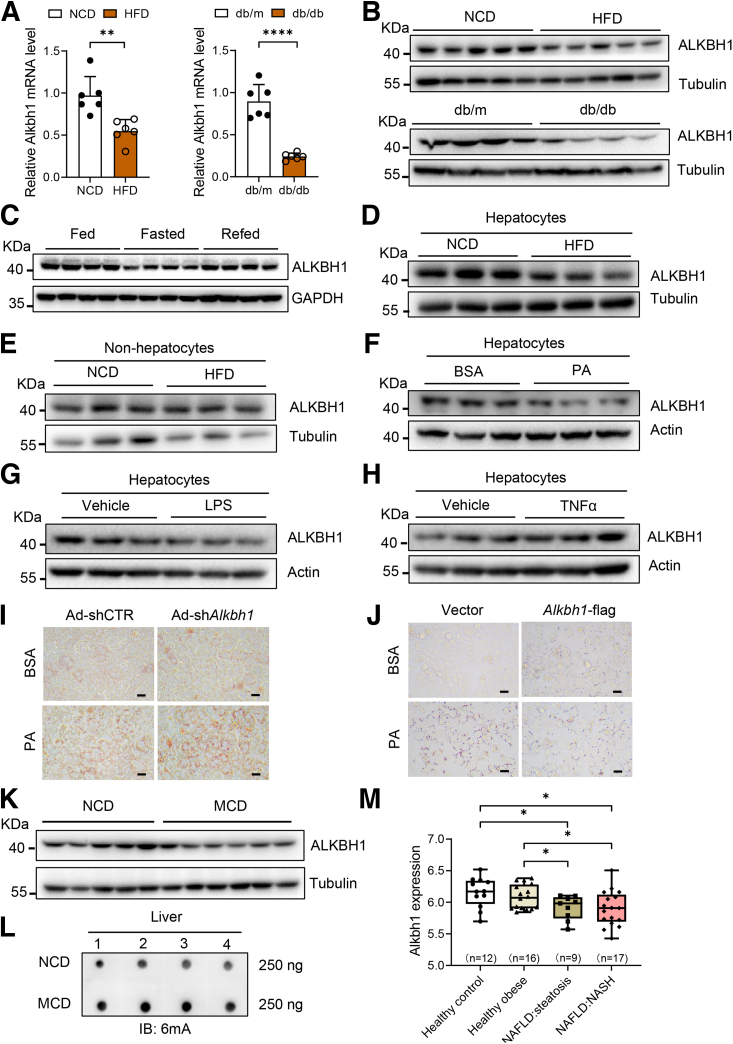
ALKBH1 is a critical DNA 6mA demethylase.19 Consistent with the increased DNA 6mA levels in fatty liver, we found that the messenger RNA (mRNA) and protein levels of hepatic Alkbh1 were down-regulated significantly in livers of obese mice with a HFD compared with mice fed a NCD (Figure 2A and B). In line with this, ALKBH1 expression also was decreased in the liver of db/db genetic obese mice aged 12 weeks (Figure 2A and B), indicating low expression of ALKBH1 in mouse fatty liver. Hepatic lipid accumulation occurs transiently as a metabolic adaptation to fasting.33 Consistent with the negative correlation of ALKBH1 expression with fatty liver status, decreased fasting ALKBH1 expression while refeeding restored ALKBH1 expression (Figure 2C). To investigate the relative contribution of hepatocytes to the decreased hepatic ALKBH1 expression upon obesity, ALKBH1 expression was detected in isolated hepatocytes and nonhepatocytes from NCD-fed and HFD-fed mice. We found up-regulated ALKBH1 expression in hepatocytes but not in nonhepatocytes from HFD-fed mice in comparison with NCD-fed mice (Figure 2D and E), indicating that obesity-regulated hepatic ALKBH1 expression and DNA 6mA modification mainly occurs in hepatocytes. Palmitic acid (PA) was a common chemical used to stimulate excessive influx of free fatty acids (FFAs) into hepatocytes and induce lipid accumulation. We found that ALKBH1 expression also was attenuated after treatment with PA but not lipopolysaccharides or the proinflammatory cytokine tumor necrosis factor α in primary hepatocytes (Figure 2F–H). ALKBH1 knockdown mediated by adenovirus (Ad)-shAlkbh1 promoted, while overexpression of ALKBH1 in hepatocytes alleviated, PA-induced lipid droplet accumulation (Figure 2I and J), suggesting an important role of ALKBH1 in lipid metabolism of hepatocytes. Similar to HFD-induced fatty liver, a methionine choline-deficient (MCD) diet, which was widely used to induce hepatic steatosis together with inflammation and fibrosis, reduced ALKBH1 expression in the fatty liver of wild-type mice (Figure 1K). Correspondingly, MCD-fed wild-type mice showed increased hepatic DNA 6mA levels compared with mice fed with NCD (Figure 1L). More importantly, we analyzed mRNA expression arrays from a data set (GSE48452) containing human liver biopsies of different phases from control to NASH. The results showed that ALKBH1 expression decreased in both liver samples of patients suffering from hepatic steatosis and NASH, with no significant changes in the liver of healthy obese subjects without fatty liver (Figure 1M). Therefore, these results show a decreased level of DNA 6mA demethylase ALKBH1 in hepatocytes of fatty liver, which might exert a critical role on the progression of NAFLD.
Figure 2.
ALKBH1 decreases in the fatty liver of mice and human beings. Alkbh1 (A) mRNA and (B) protein levels in the liver of HFD-induced (HFD, 12 weeks) and genetically obese (db/db, 12 weeks) mice (n = 4–6). The mRNA expression is normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA. (C) ALKBH1 protein levels in the livers of mice under Fed, Fast, and Refed status, as follows: Fed, wild-type mice fed ad libitum; Fasted, wild-type mice fasted for 24 hours; and Refed, wild-type mice fasted for 24 hours and then refed for another 24 hours (n = 4). (D) The protein levels of ALKBH1 in hepatocytes isolated from NCD-fed lean mice and HFD-fed obese mice. (E) Immunoblots of ALKBH1 in nonhepatocytes from NCD- and HFD-fed mice for 12 weeks. (F–H) Immunoblots of ALKBH1 in the mouse primary hepatocytes treated with (F) 0.75 mmol/L PA, (G) 100 ng/mL lipopolysaccharide (LPS), and (H) 20 ng/mL tumor necrosis factor α (TNFα) or vehicle for 24 hours (n = 3). (I) Oil Red O staining of lipid droplets in control (Ad-shCTR) and Alkbh1-silenced mouse primary hepatocytes (Ad-shAlkbh1) treated with 0.75 mmol/L PA or bovine serum albumin (BSA) for 24 hours. Scale bar: 10 μm. (J) Oil Red O staining of lipid droplets in control (vector) and Alkbh1-overexpressing (Alkbh1-flag) hepatocytes treated with 0.75 mmol/L PA or BSA for 24 hours. Scale bar: 10 μm. (K) Immunoblots of ALKBH1 in the livers of NCD- and MCD-fed mice for 4 weeks. (L) Dot blot of 6mA levels on genomic DNA from livers of NCD- and MCD-fed mice for 4 weeks. A total of 250 ng DNA was loaded for each sample (n = 4). (M) Alkbh1 mRNA levels in human liver biopsies of different NAFLD phases (n = 9–17). (A and M) Data are presented as the means ± SD. IB, immunoblotting. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001.
Hepatic ALKBH1 Deficiency Aggravates Diet-Induced Hepatic Steatosis and Insulin Resistance
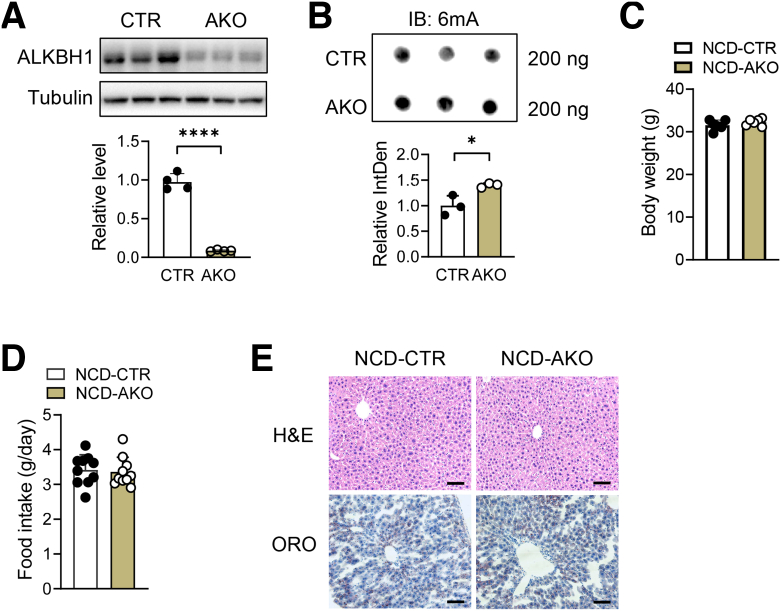
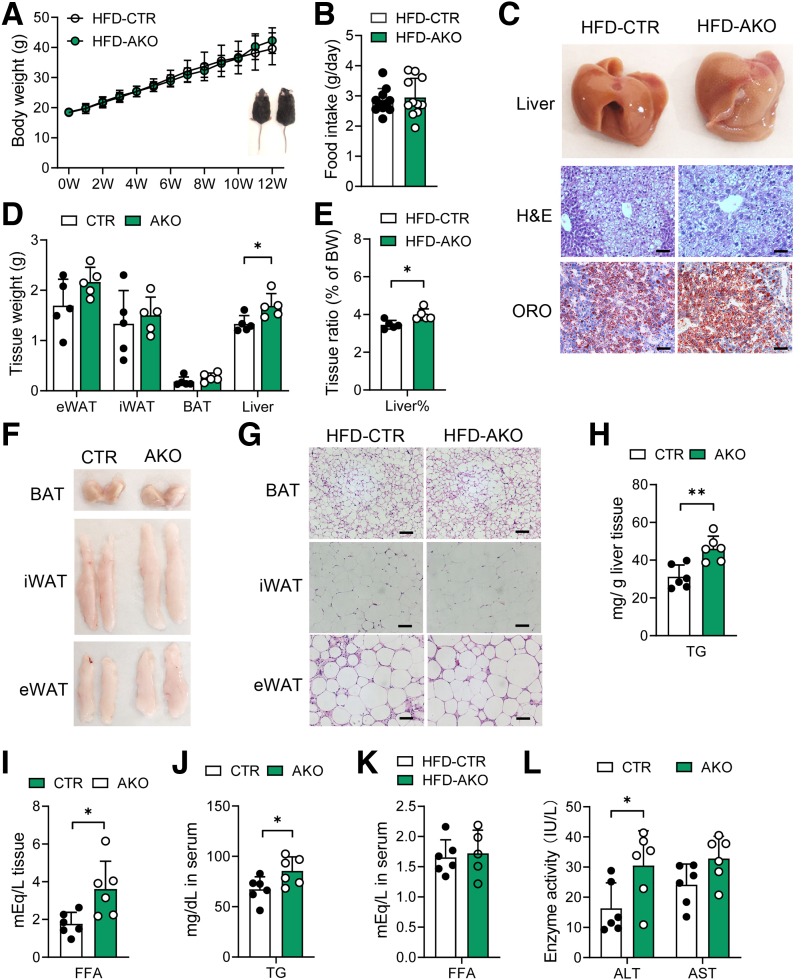
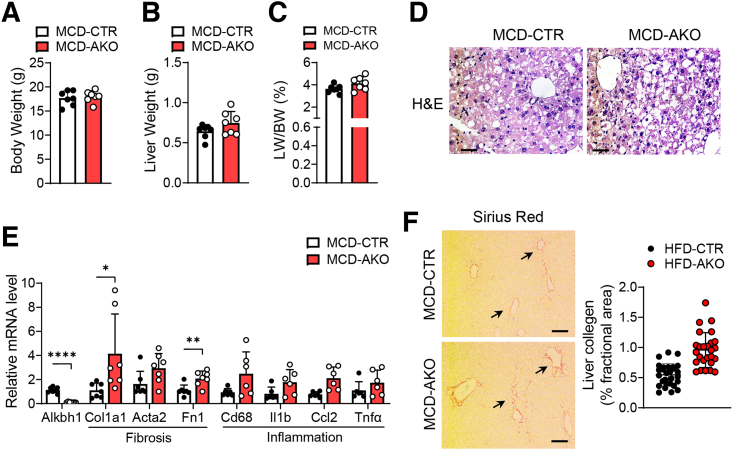
To determine whether ALKBH1-catalyzed DNA 6mA modification plays an essential role in hepatic lipid metabolism, liver-specific Alkbh1 knockout mice (AKO) were established by crossing Alkbh1 floxed mice with albumin-Cre mice (Figure 3A). Consistent with the role of ALKBH1 in DNA 6mA demethylation, hepatocytes from AKO mice showed enhanced DNA 6mA levels (Figure 3B). Compared with control mice, AKO mice showed normal body weight, food intake, and hepatic lipid accumulation when fed with NCD (Figure 3C–E). Under HFD feeding conditions, while the body mass and food intake were not altered between the AKO and control mice (Figure 4A and B), the AKO mice had a pale liver appearance, increased liver weight, and exacerbated HFD-induced hepatic steatosis compared with control mice (Figure 4C–E), without affecting adipose tissue performance (Figure 4F and G). Moreover, hepatic TG, hepatic FFA, and serum TG concentrations also were higher in AKO mice with HFD feeding, although serum FFAs remained unaltered (Figure 4H–K). In addition, ALKBH1 knockout aggravated HFD-induced liver injury, as marked by an increased tendency of serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in AKO mice (Figure 4L). Hepatic steatosis may lead to impaired glucose tolerance and hepatic insulin sensitivity. In line with more severe hepatic steatosis, ALKBH1 knockout markedly attenuated systemic glucose tolerance (Figure 5A). Moreover, aggravated insulin resistance also was observed in AKO mice compared with control mice (Figure 5B). Consistently, the insulin-induced phosphorylation of thymoma viral proto-oncogene 1 (Akt) and glycogen synthase kinase 3 beta (GSK3β) were alleviated in ALKBH1-deficient primary hepatocytes compared with controls (Figure 5C and D). Similar to mice fed with a HFD, MCD-diet–fed AKO mice showed comparable body and liver weights (Figure 6A–C), but developed more hepatic steatosis than control mice (Figure 6D). In addition, AKO mice challenged with MCD diet showed increased fibrotic and inflammatory tendencies (Figure 6E and F). Collectively, these data show that liver-specific ALKBH1 is essential for restraining diet-induced hepatic steatosis and insulin resistance.
Figure 3.
Deletion of hepatic ALKBH1 did not affect body weight, food intake, or liver histology of mice under NCD feeding. (A) Alkbh1 protein (n = 3) and mRNA levels (n = 4) in liver of adult control and AKO mice. Normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA. (B) Dot blot of 6mA levels on genomic DNA of hepatocytes isolated from control and AKO mice, and the statistical analysis of its relative integrated density. A total of 200 ng DNA was loaded for each sample (n = 3). (C) Body weight of control and AKO mice fed with NCD for 12 weeks (n = 6). (D) Food intake (n = 10) of control and AKO mice fed with NCD for 12 weeks. (E) H&E staining and Oil Red O (ORO) staining on liver slices of control and AKO mice fed with NCD for 12 weeks. Scale bar: 10 μm. (A–D) Data are presented as means ± SD. CTR, control mice; IB, immunoblotting; IntDen, Integrated Density. ∗P < .05, ∗∗∗∗P < .0001.
Figure 4.
Hepatic ALKBH1 deficiency promotes HFD-induced hepatic steatosis. For the following studies, AKO and control mice were fed with a HFD for 12 weeks. (A) Body weight gain of HFD-fed control and AKO mice (n = 6). (B) Food intake of HFD-fed control and AKO mice (n = 11). (C) Liver appearance and representative H&E and Oil Red O (ORO) staining of liver slices from HFD-fed control and AKO mice. Scale bar: 10 μm. (D) Tissue weight and the (E) ratio of liver weight to body weight of HFD-fed control and AKO mice (n = 5–6). (F) Tissue appearance and (G) representative H&E staining of brown adipose tissue (BAT), epididymal white adipose tissue (eWAT), and inguinal white adipose tissue (iWAT) from control and AKO mice under HFD feeding condition. Scale bar: 10 μm. Concentration of (H) hepatic TGs and (I) FFAs of HFD-fed control and AKO mice (n = 6). Concentration of (J) TGs and (K) FFAs in the serum of HFD-fed control and AKO mice (n = 6). (L) ALT and AST activity in the serum of HFD-fed control and AKO mice (n = 6). (A, B, D, E, and H–L) Data are presented as the means ± SD. BW, body weight; CTR, control mice. ∗P < .05 and ∗∗P < .01.
Figure 5.
Deletion of hepatic ALKBH1 exacerbates HFD-induced insulin resistance. (A) Glucose tolerance tests (GTTs) and (B) insulin tolerance tests (ITTs) of control and AKO mice fed with a 12-week HFD and their area under the curve (AUC) analysis (n = 6). (C) Immunoblots of factors in insulin signaling pathways in primary hepatocytes. The hepatocytes were isolated from control and AKO mice and transfected with or without Ad-shAlkbh1 for 48 hours, then followed up with 10 nmol/L insulin or vehicle treatment for 15 minutes (n = 3). (D) Statistical analysis of relative protein expression in panel C. (A, B, and D) Data are presented as the means ± SD. Akt, thymoma viral proto-oncogene 1; CTR, control mice; pAkt, phosphorylated Akt; pGSK3β, phosphorylated glycogen synthase kinase 3 beta; shCtr+Veh, hepatocytes transfected with Ad-shCtr and treated with vehicle; shCtr+Ins, hepatocytes transfected with Ad-shCtr and treated with insulin; shAlkbh1+Veh, hepatocytes transfected with Ad-shAlkbh1 and treated with vehicle; shAlkbh1+Ins, hepatocytes transfected with Ad-shAlkbh1 and treated with insulin. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
Figure 6.
Effects of hepatic ALKBH1 on the MCD diet–induced hepatic steatosis, fibrosis, and inflammation. (A) Body weight, (B) liver weight, and the (C) ratio of liver weight to body weight of control and AKO mice fed with MCD diet for 4 weeks. (D) Representative H&E staining of the livers from MCD diet–fed control and AKO mice. Scale bar: 10 μm. (E) qPCR analysis of the mRNA levels of fibrotic genes (col1a1, Acta2, and Fn1) and inflammatory cytokines (Cd68, Il1b, Ccl2, and Tnfα) in the livers from control and AKO mice fed with MCD diet for 4 weeks. Normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA. (F) Sirius Red staining of liver slices from control and AKO mice fed with MCD diet for 4 weeks, the statistical image analysis of the collagen (percentage area) from Sirius red staining using ImageJ software (National Institutes of Health, Bethesda, MD). Black arrows indicate the collagen fibers stained with picrosirius red. Scale bar: 20 μm. (A–C and E) Data are presented as means ± SD. ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001. CTR, control mice; LW/BW, liver weight/body weight.
Transcriptomic and Chromatin Immunoprecipitation Analysis Suggest That ALKBH1 Regulates Lipid Metabolism
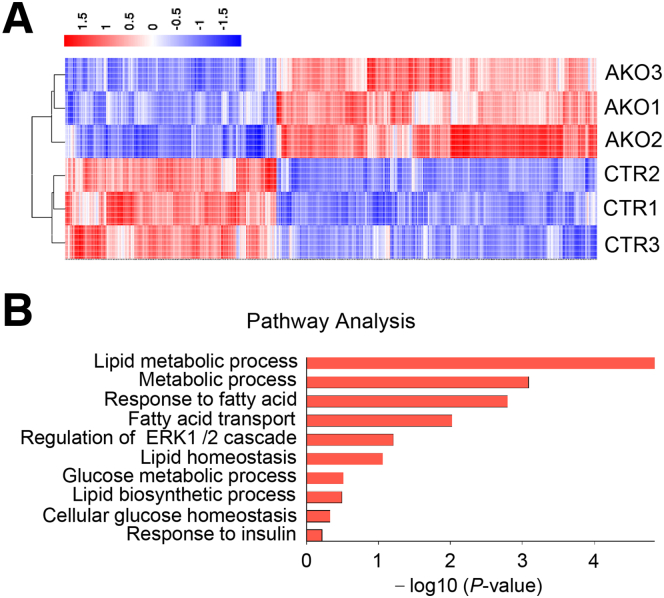
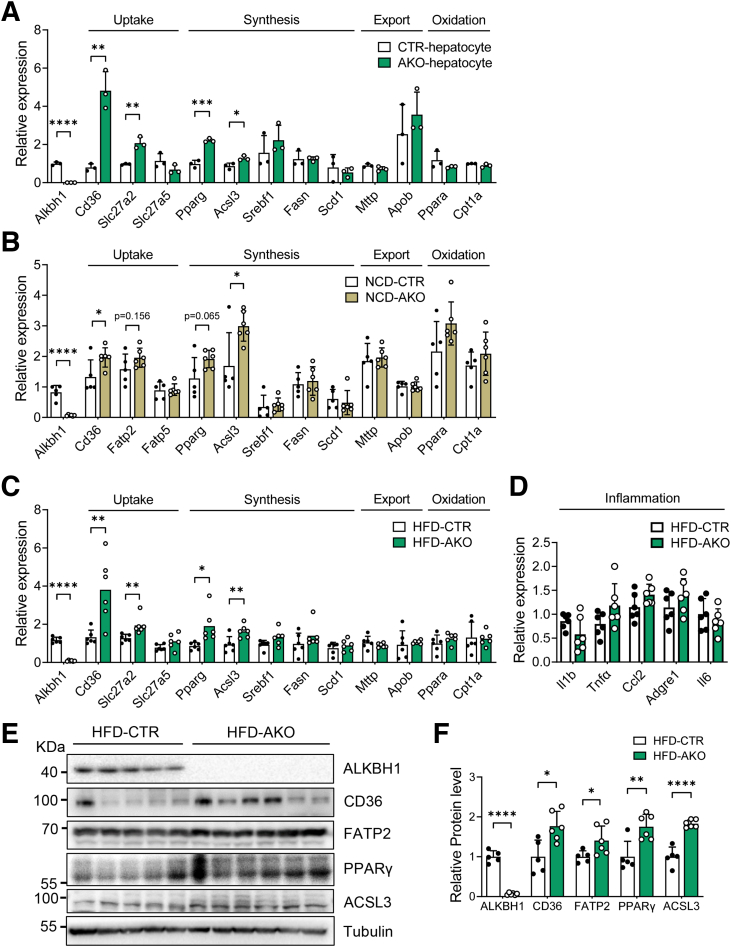
We then asked how ALKBH1 regulates lipid deposits in hepatocytes under metabolic stress. We first performed RNA sequencing on PA-treated primary mouse hepatocytes isolated from control and AKO mice (Figure 7A). Transcriptomic analysis of RNA sequencing (RNA-seq) identified that lipid metabolism–related pathways were enriched significantly in the AKO group (Figure 7B), suggesting a role of ALKBH1 in regulating expression of lipid-metabolic genes. Furthermore, ALKBH1 protein showed more accumulation in the nucleus than in the cytoplasm of primary hepatocytes (Figure 8A), indicating ALKBH1 may function mainly in the nucleus as DNA 6mA demethylase. To further clarify the precise mechanism of ALKBH1 in regulating lipid metabolism in hepatocytes, we investigated global DNA-binding sites of ALKBH1 protein across the hepatocyte genome by performing chromatin immunoprecipitation sequencing (ChIP-seq) within the primary hepatocytes overexpressed with ALKBH1-flag fusion protein (Figure 8B). Overall, 8461 genes and 8404 protein-coding genes were discovered in ALKBH1 IP samples. Of those transcribed genes, more than 50% of ALKBH1 binding sites were distributed in intergenic regions (Figure 8C), which is consistent with previous findings of 6mA peaks in mouse embryonic stem cells and human glioblastomas.16,17 Integrating the analysis of RNA-seq and ChIP-seq data resulted in 437 ALKBH1 potential target genes (Figure 8D). Gene ontology analysis on this set of up-regulated target genes also showed significant enrichment in biological processes including the lipid metabolic process, response to lipids, fatty acid transport, and lipid biosynthesis (Figure 8E). Thus, the expression profiles of genes related in the lipid uptake, synthesis, export, and oxidation were validated in control and AKO primary hepatocytes. Of note, ALKBH1 knockout in hepatocytes markedly increased expression of lipid uptake genes Cd36 and Slc27a2 and lipid synthesis genes Pparg and Acsl3, which are among the overlapped genes between ChIP-seq and RNA-seq, with little effect on the expression of other lipid metabolism–related genes (Figure 9A). In line with this, AKO mice displayed slightly increased transcription of those lipid uptake and synthesis genes compared with control mice when fed with a NCD (Figure 9B). This effect became more pronounced when the mice switched to HFD feeding, manifested as significantly up-regulated both mRNA and protein levels of hepatic Cd36, Slc27a2, Pparg, and Acsl3 genes in AKO mice, despite no significant differences in other lipid metabolic pathways and hepatic inflammation (Figure 9C–F). Taken together, these findings show that ALKBH1 deficiency promotes HFD-induced hepatic steatosis via increasing gene expression in hepatic lipid uptake and synthesis.
Figure 7.
Transcriptomic analysis shows the profile of differentially expressed genes between control and AKO hepatocytes. (A) Heatmap displays differentially expressed genes (log2 fold change >1.3; P < .05) between control and AKO hepatocytes in RNA-seq (n = 3). (B) Representative Gene Ontology analysis with differentially expressed genes annotated in lipid metabolic process under RNA-seq (n = 3). CTR, control mice; ERK, extracellular signal-regulated kinase.
Figure 8.
ChIP analysis suggests that ALKBH1 regulates lipid metabolism. (A) Immunoblots of ALKBH1 in the cytoplasmic and nuclear components of primary mouse hepatocytes. (B) Experimental steps for ChIP analysis of ALKBH1–DNA interactions. (C) The percentage of ALKBH1-bound peaks within various regions of the hepatocyte genome. (D) Venn diagram showing overlapped genes between direct ALKBH1 binding targets from ChIP-seq and differential expression genes from RNA-seq. (E) Representative Gene Ontology analysis with overlapped genes annotated in the fatty acid metabolic processes under ChIP-seq and RNA-seq analysis. ERK, extracellular signal-regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TSS, transcription start site.
Figure 9.
Deletion of hepatic ALKBH1 increases gene expression involved in lipid uptake and synthesis. (A) qPCR analysis of the mRNA levels of genes related to lipid uptake, synthesis, export, and oxidation in primary hepatocytes of HFD-fed control and AKO mice. qPCR analysis of the mRNA levels of genes related to lipid uptake, synthesis, export, and oxidation in the livers of control and AKO mice on a (B) NCD (n = 6) and (C) HFD (n = 6) feeding plan. (D) qPCR analysis of the mRNA level of inflammation-related genes in the livers of HFD-fed control and AKO mice (n = 6). (E) Immunoblots of ALKBH1, CD36, FATP2, peroxisome proliferator-activated receptor (PPAR)γ, and ACSL3 protein levels in the liver of HFD-fed control and AKO mice (n = 5–6). (F) Statistical analysis of relative protein expression in panel E. (A–D and F) Data are presented as the means ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. CTR, control mice.
ALKBH1 Directly Binds to and Demethylates DNA 6mA Modification of Genes Involved in Lipid Uptake and Synthesis
DNA 6mA modification has been reported to be an epigenetic mark indicative of transcription via depositing on promoter regions or nonpromoter regions such as long interspersed element-1 (LINE-1) transposons, the stress-induced DNA double helix destabilization (SIDD) region, and the heterochromatin region.16,17,28 To detect the regions of ALKBH1 binding enrichment by ChIP-seq, we searched for peaks of ALKBH1-ChIP vs input DNA. All the binding sites occupied by ALKBH1 protein in the gene structure of Slc27a2, Acsl3, Cd36, and Pparg then were identified (Figure 10A and B). Furthermore, the ChIP–quantitative polymerase chain reaction (qPCR) assay showed that ALKBH1 precipitated the sequences of Acsl3 at the first 3 intronic regions: Slc27a2 at the first intronic region, Cd36 at the intergenic gene, and Pparg at the intergenic, the first, second, and seventh intronic regions (Figure 10C–F). To examine whether ALKBH1 regulates the expression of these lipid metabolic genes through functioning as DNA 6mA demethylase, 6mA ChIP-qPCR was performed using the 6mA antibody in wild-type and ALKBH1-deficient primary hepatocytes. More 6mA enrichment on the specific binding sites of these genes was observed in AKO hepatocytes than that in control hepatocytes (Figure 10G–J), which lead to increased gene expression (Figure 9A–C). Taken together, these findings suggest that ALKBH1 regulates Slc27a2, Acsl3, Cd36, and Pparg transcription by removing 6mA modifications of these genes at intergenic and/or intronic regions in hepatocytes.
Figure 10.
ALKBH1 directly binds to and demethylates DNA 6mA modification of genes involved in lipid uptake and synthesis. (A) ALKBH1-bound peaks within Acsl3, Slc27a2, Cd36, and Pparg genes. Tracks display ChIP-seq data from Input and ALKBH1-flag IP samples. The X-axis shows the position in gene structure, and the Y-axis shows signal coverage. (B) Schematic representation of primers binding sites around potential ALKBH1 binding sites on DNA fragments of Acsl3 (S1, first intron; S2, second intron; and S3, third intron), Slc27a2 (S1, first intron), Cd36 (S1, intergenic region; S2, fourth intron; S3, fourth exon; S4, second intron; and S5, first intron), and Pparg (S1, intergenic region; S2, first intron; S3, second intron; S4, sixth exon; and S5, seventh intron) genes, respectively. (C–F) ChIP-qPCR was performed using anti-ALKBH1 antibody (ALKBH1 IP) or IgG (IgG IP) with specific primers for binding regions of Acsl3, Slc27a2, Cd36, and Pparg. Signals obtained from each immunoprecipitation are expressed as a percentage of the total input chromatin (n = 3). (G–J) ChIP-qPCR was performed using anti-6mA antibody (6mA) or IgG with specific primers for binding regions of Acsl3, Slc27a2, Cd36, and Pparg in control and AKO primary hepatocytes. The levels of control IgG binding were designated as 1 (n = 3). (C–J) Data are presented as the means ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. CTR, control mice; TSS, transcription start sites.
Overexpression of ALKBH1 in Liver Protecting Against HFD-Induced Hepatic Steatosis and Insulin Resistance
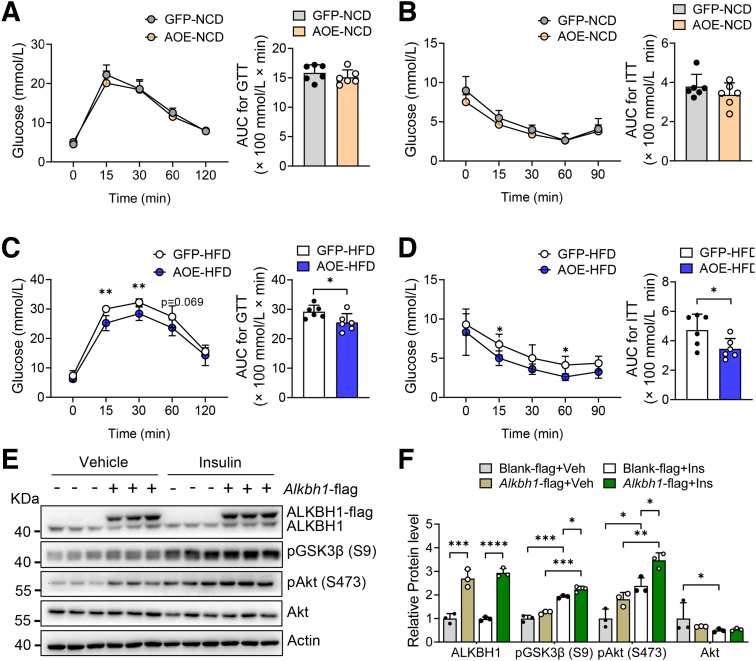
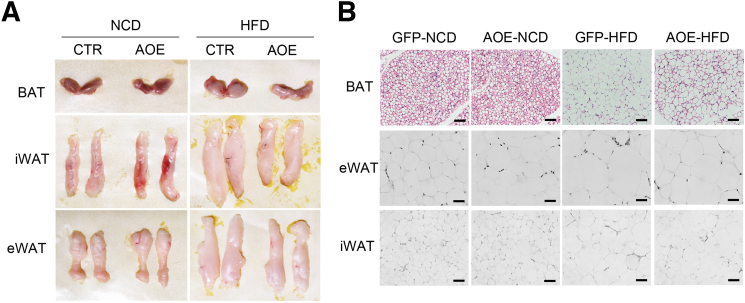
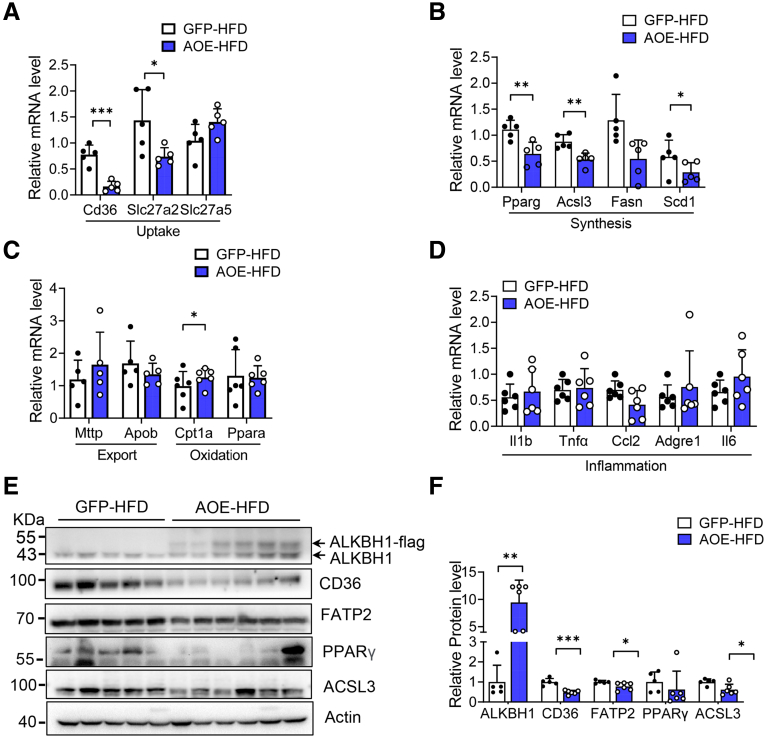
We next performed a gain-of-function analysis via adeno-associated virus-mediated hepatic ALKBH1 overexpression in mice (AOE mice) (Figure 11A). Similar to the ALKBH1-knockout model, AOE mice showed the comparable food intake, body weight, and hepatic lipid accumulation compared with control mice when fed with a NCD (Figure 11B–D). Notably, overexpression of ALKBH1 in liver significantly relieved the HFD-induced body weight gain, hepatic steatosis, and liver weight (Figure 11C–E), without affecting food intake (Figure 11B). In line with this, ALKBH1 overexpression decreased the content of TGs and FFAs in liver (Figure 11F and G), and circulating levels of TGs and FFAs in serum under HFD feeding conditions (Figure 11H and I). The levels of AST and ALT in serum also were lower in HFD-fed AOE mice than that in control mice (Figure 11J and K). Moreover, enhanced ALKBH1 expression in liver did not influence systemic glucose tolerance and insulin sensitivity in NCD conditions (Figure 12A and B), although significantly improved HFD-induced glucose intolerance and insulin resistance (Figure 12C and D). The improved insulin sensitivity as shown by increased insulin-induced phosphorylation of Akt and GSK3β also was observed in ALKBH1-overexpressed primary hepatocytes compared with controls (Figure 12E and F). In addition, consistent with decreased body weight, AOE mice displayed reduced adipose tissue volumes and adipocyte size when compared with control mice under HFD conditions (Figure 13A and B). In contrast to induced fatty acid uptake and lipogenesis by ALKBH1 deletion, the expression of Cd36, Slc27a2, Pparg, and Acsl3 genes was suppressed both at mRNA and protein levels in liver of HFD-fed AOE mice (Figure 14). Taken together, these data showed that enhanced hepatic ALKBH1 expression protects mice from HFD-induced hepatic steatosis and insulin resistance via decreasing lipid intake and biosynthesis.
Figure 11.
Liver-specific overexpression of ALKBH1 alleviates HFD-induced hepatic steatosis. (A) qPCR analysis of Alkbh1 mRNA levels in the liver of control (green fluorescent protein, GFP) and AOE mice under NCD and HFD feeding conditions (n = 6). Normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA. (B) Food intake and (C) body weight gain of control and AOE mice under NCD and HFD feeding conditions (n = 6). (D) Representative liver morphology, Oil Red O staining (ORO), and H&E staining of liver slices from control and AOE mice under NCD and HFD feeding conditions. Scale bar: 10 μm. (E) Tissue weight of epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT), brown adipose tissue (BAT), and liver of HFD-fed control and AOE mice. Concentration of (F) hepatic TGs and (G) FFAs, and serum levels of (H) TGs and (I) FFAs of control and AOE mice under NCD and HFD feeding conditions (n = 6). Measurement of (J) AST and (K) ALT levels in the serum of control and AOE mice under NCD and HFD feeding conditions (n = 6). (A–C and E–K) Data are presented as the means ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001. AOE, ALKBH1 liver-specific overexpression mice; GFP, control mice with GFP expression.
Figure 12.
Liver-specific overexpression of ALKBH1 attenuates HFD-induced insulin resistance. (A) Glucose tolerance tests (GTTs) and its area under the curve (AUC), and (B) insulin tolerance tests (ITTs) and its AUC of control and AOE mice under NCD feeding conditions (n = 6). (C) GTT and its AUC, and (D) ITT and its AUC of control and AOE mice under HFD feeding conditions (n = 6). (E) Immunoblots of factors in insulin signaling pathways in primary hepatocytes, which were transfected with or without Alkbh1-flag for 48 hours, followed with 10 nmol/L insulin or vehicle treatment for 15 minutes. (F) Statistical analysis of the relative protein expression in panel E. (A–D and F) Data are presented as the means ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. Akt, thymoma viral proto-oncogene 1; Alkbh1-flag+Ins, hepatocytes transfected with Alkbh1-flag plasmid and treated with insulin; Alkbh1-flag+Veh, hepatocytes transfected with Alkbh1-flag plasmid and treated with vehicle; AOE, ALKBH1 liver-specific overexpression mice; Blank-flag+Ins, hepatocytes transfected with Blank-flag plasmid and treated with insulin; Blank-flag+Veh, hepatocytes transfected with Blank-flag plasmid and treated with vehicle; GFP, control mice with GFP expression; pAkt, phosphorylated Akt; pGSK3β, phosphorylated glycogen synthase kinase 3 beta.
Figure 13.
Adipose morphology of control and AOE mice under NCD and HFD feeding conditions. (A) Tissue appearance of brown adipose tissue (BAT), epididymal white adipose tissue (eWAT), and inguinal white adipose tissue (iWAT) from control and AOE mice under NCD and HFD feeding conditions. (B) Representative H&E staining of BAT, eWAT, and iWAT from control and AOE mice under NCD and HFD feeding conditions. Scale bar: 10 μm. AOE, ALKBH1 liver-specific overexpression mice; CTR, control mice; GFP, control mice with GFP expression.
Figure 14.
Overexpression of ALKBH1 in liver suppresses gene expression in fatty acid uptake and lipogenesis. qPCR analysis of genes related to (A) lipid uptake and (B) lipid synthesis in the liver of control and AOE mice under HFD feeding conditions (n = 5). Normalized to Gapdh mRNA. qPCR analysis of genes related to (C) lipid export and lipid oxidation and (D) inflammation in the liver of control and AOE mice under HFD feeding (n = 5–6). (E) Immunoblots of protein expression of genes related to lipid uptake and synthesis in the liver of HFD-fed control and AOE mice (n = 5–6). (F) Statistical analysis of the relative protein expression in panel E. (A–D and F) Data are presented as the means ± SD. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. AOE, ALKBH1 liver-specific overexpression mice; GFP, control mice with GFP expression; PPAR, peroxisome proliferator-activated receptor.
Discussion
NAFLD is considered an epigenetics-driven disease. Although the critical role of 5-methylcytosine modification in the regulation of transcription is recognized, the functions of other novel noncanonical DNA modifications in metabolism remain obscure. Here, we found increased DNA 6mA modifications in liver of obese mice and decreased expression of demethylase ALKBH1 in mouse and human fatty liver, implicating a critical role of this epigenetic mark in hepatic metabolism. Furthermore, depletion of ALKBH1 transcriptionally activated fatty acid uptake genes Cd36 and Slc27a2 and lipid synthesis genes Pparg and Acsl3, and thus promoted diet-induced hepatic steatosis. However, overexpression of ALKBH1 resulted in opposite phenotypes. The mechanism of ALKBH1 regulating hepatic steatosis was evidenced further through directly binding to and removing DNA 6mA modifications of Cd36, Slc27a2, Pparg, and Acsl3, therefore reducing the transcription of these genes (Figure 8). Our study uncovers a novel epigenetic regulation, DNA 6mA modification by ALKBH1, and acts as a potential therapeutic target for preventing hepatic steatosis, which expands the repertoire of genetic marks in metabolism.
ALKBH1 was reported to exert multiple effects based on diverse substrates such as DNA,16 RNA,34 and methylated histone H2A,25 and varied cellular distribution including mitochondria, cytoplasm, and nucleus.34, 35, 36 In this study, we observed abundant nuclear locations of ALKBH1 in primary hepatocytes and increased DNA 6mA levels by ALKBH1 deletion, suggesting a potential role of demethylation of ALKBH1 for genomic 6mA modification in hepatocytes. In support of this, both RNA-seq and ChIP-seq were performed and showed enriched pathways related to lipid metabolism in ALKBH1-deficient hepatocytes. Further evidence showed that ALKBH1 mainly regulates the expression of lipid uptake and synthetic genes via directly binding to and demethylating 6mA in the target genes. However, we could not exclude other possible mechanisms of ALKBH1 action. It has been reported that ALKBH1 in mitochondrion is responsible for mitochondrial transfer ribonucleic acid (tRNA) modifications, mainly involved in the mitochondria unfolded protein response pathway (UPRmt).34,35 Although we did not further examine whether ALKBH1 exists in the mitochondria of hepatocytes, we found no significant differences in the expression of UPRmt-related genes such as Lonp1, Clpp, and Hspd1 between the livers from AKO and control mice, suggesting that modulation of the UPRmt may not be a key mechanism by which ALKBH1 regulates hepatic lipid metabolism. Because ALKBH1 also is located in the cytoplasm in hepatocytes, it is possible that cytoplasmic ALKBH1 plays a role in lipid metabolism except for DNA 6mA demethylation, which needs to be investigated further.
Alteration in body weight is a consequence of metabolic disorders in multiple organs. The liver plays a central role in regulating energy balance by interacting with various organ systems through secreting factors. Communication between the liver and adipose tissue plays a critical role in regulating systemic metabolism.37 In our study, hepatic ALKBH1 deletion did not affect mice body weight, while ALKBH1 overexpression in liver significantly decreased fat mass, leading to reduced body weight. It is possible that overexpression of ALKBH1 in liver promotes fat metabolism and subsequently limits body weight gain through liver–fat communication. However, this interorgan communication signal may be much weaker in AKO mice under HFD-fed conditions because a lack of ALKBH1 promotes the development of fatty liver and may disturb hepatokine secretion. In this sense, ALKBH1-knockout mice display comparable phenotypes in fat tissue and body weight compared with control mice.
Recent studies have shown that DNA 6mA modification is associated strongly with introns and intergenic regions, and influencing the regulation of gene expression. For example, increased 6mA modification epigenetically silences LINE-1 transposons, together with their neighboring enhancers and genes during mouse embryonic stem cell differentiation,16 and represses SIDD-special AT-rich sequence binding protein 1 (SATB1) interactions to regulate gene expression during trophoblast development.38 Xie et al17 found that DNA 6mA modification is enriched in heterochromatin in human glioblastoma, and increasing DNA 6mA modification by ALKBH1 depletion inhibited transcription of oncogenic genes through decreasing chromatin accessibility. Our study indicated that ALKBH1 in hepatocytes demethylates the 6mA modification at the intergenic regions and introns on the Cd36, Slc27a2, Acsl3, and Pparg gene structure, subsequently inhibits the expression of these genes, and suppresses diet-induced hepatic steatosis. The common presence of disease-associated loci in intergenic and intronic regions usually is attributed to potential regulatory functions of DNA sequence.39 However, how 6mA modification at these genomic regions could be recognized and then subsequently affect gene transcription in hepatocytes still needs further investigation.
Insulin resistance and lipotoxicity are considered to act synergistically to induce NAFLD development and progression, which makes it likely that the treatment for NAFLD will require either a single drug with multiple functions or combination therapy. In our study, combined analysis of RNA-seq and ChIP-seq showed close correlation of ALKBH1-mediated DNA 6mA modification and transcriptional level of genes involved in fatty acid uptake and lipogenesis. Among them, CD36 and FATP2 are important fatty acid transporters to promote the intake of exogenous fatty acid and contribute to the progression of NAFLD.8,9 Peroxisome proliferator-activated receptor-γ induces lipogenic factors, which are implicated in fatty acid synthesis, leading to lipid droplet deposition within hepatocytes.10,11 Acyl-CoA synthetase long chain family member 3 (ACSL3) mediates transcriptional control of hepatic lipogenesis and is the only long-chain fatty acyl-CoA synthetase found consistently on growing and mature lipid droplets.12,40 These results indicate multiple direct targets of ALKBH1. In summary, we showed DNA 6mA demethylase ALKBH1 acting as an epigenetic suppressor to regulate diet-induced hepatic steatosis. Targeting ALKBH1 to inhibit hepatic lipid accumulation by inactivating fatty acid uptake and lipid synthesis pathways would be a promising approach for the treatment of NAFLD and its related metabolic diseases.
Materials and Methods
Animals
All animal protocols for this study were reviewed and approved by the Animal Care and Use Committee of the Laboratory Animal Research Center at Xiangya Medical School of Central South University. C57BL/6N-Alkbh1tm1a(EUCOMM)Hmgu/Ieg (strain HEPD0751_6_G08) and albumin-cre mice (Alb-cre) were purchased from the EMMA repository (Neuherberg, Germany) and Jackson Laboratory (Bar Harbor ME,) separately. Heterozygous female C57BL/6N-Alkbh1tm1a(EUCOMM)Hmgu/Ieg mice were crossed with FLPe recombinase-expressing males (Jackson Laboratory), and then C57BL/6J males to remove the Flp driver and generate Alkbh1tm1c allele mice with exon 2 floxed mice (Alkbh1 f/f mice). Alkbh1 f/f mice then were backcrossed with wild-type (C57BL/6J) males for more than 5 generations onto a C57BL/6J background. Alkbh1 liver-specific knockout mice (Alkbh1 f/f; Alb-cre) and control mice (Alkbh1 f/f) were generated by crossing Alkbh1 f/f mice with Alb-cre mice. Wild-type mice and leptin receptor-mutated (db/db) mice aged 8 weeks were purchased from Hunan Laboratory Animal Company (SJA, Hunan, China). Mice were fed with NCD or HFD containing 60% kcal from fat, 20% kcal from carbohydrates, and 20% kcal from protein (D12492; Research Diets) for 12 weeks at 6 weeks of age. Mice were fed a MCD diet (A02082002B; Research Diets) for 4 weeks at 12 weeks of age. All mice were kept at the Laboratory Animal Research Center with specific pathogen-free standards at Central South University, and housed on a 12-hour light/dark cycle at 22°C–24°C, with free access to water and diet.
Primary Hepatocytes and Nonhepatocyte Isolation and Treatment
Primary hepatocytes were isolated from C57BL/6J mice and cultured as described in a previous study.41 Briefly, mice were deeply anesthetized with isoflurane and perfused with digestion buffer containing 100 CDU/mL (CDU = collagen digestion units) type IV collagenase through the inferior vena cava. Once the digestion was done, the liver cells were extracted and filtered through a 70-μm membrane. After centrifugation, the supernatant containing nonhepatocytes and the remaining hepatocyte pellet were transferred, washed 3 times, and collected separately. For the treatment of hepatocytes, the isolated primary hepatocytes continued to be plated into 12-well culture dishes coated with collagen at 2.8 × 105 cells/well. Hepatocytes were cultured in MEMα (Minimum Essential Medium α) with 10% fetal bovine serum (Gibco, Waltham, MA) plus 100 U/mL penicillin and 100 mg/mL streptomycin (Gibco, Waltham, MA) at 37°C with a humidified atmosphere of 5% CO2 for 4 hours, and then kept in serum-free medium until the primary hepatocytes were treated with serum-free medium containing 0.75 mmol/L PA (P0500; Sigma, Darmstadt, Germany), 100 ng/mL lipopolysaccharide (L4391; Sigma, Darmstadt, Germany), and 20 ng/mL recombinant tumor necrosis factor α (AF-300-01A; Peprotech, Cranbury, NJ) for 24 hours.
Nuclear and Cytoplasmic Protein Extraction
Fresh primary mouse hepatocytes were transfected with or without Alkbh1-flag plasmids with Lipofectamine 2000 (Waltham, MA), and were washed and scraped in cold phosphate-buffered saline. Cells then were collected by centrifugation at 4°C and immediately used for isolation of nuclear and cytoplasmic protein with a commercial kit (P0027; Beyotime, Jiangsu, China), according to the manufacturer’s protocol.
Gene Knockdown and Overexpression
For generation of liver-specific overexpression of ALKBH1 mice, green fluorescent protein (GFP)-tagged Alkbh1 with thyroxine binding globulin (TBG) promoter adeno-associated virus (AAV) and control AAV were purchased from OBiO Technology (Shanghai, China). To silence endogenous Alkbh1 gene expression, short hairpin RNA adenovirus (Ad-shAlkbh1) that target Alkbh1 gene and its control adenovirus (Ad-shCtr) was obtained from OBiO Technology. The target sequence of short hairpin RNAs was as follows: Ad-shAlkbh1, 5’-GGACAGATCTGAGCTAGAT-3’, and Ad-shCTR, 5’-TTCTCCGAACGTGTCACGT-3’. To overexpress ALKBH1 in hepatocytes, the plasmids expressing a C-terminal flag tag on the protein of ALKBH1 (Alkbh1-flag) or blank vectors (Blank-flag) were generated from OBiO Technology. The transfection of plasmid was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. PA or other treatment was administered 2 days after the transfection. Cells then were collected for further analysis.
Dot Blotting
Genomic DNA was isolated from tissue or cells with RNase treatment and denatured at 95°C for 10 minutes, and then immediately ice-cooled. No more than 2 μL DNA samples were spotted on a positively charged nylon membrane (FFN10; Beyotime Biotechnology), air-dried for 5 minutes, baked at 80°C for 1 hour, and then UV cross-linked for 30 minutes. Membranes then were blocked with 5% milk diluted in Tris-buffered saline (20 mmol/L Tris base and 150 mmol/L NaCl, pH 7.6), and incubated with primary antibodies against 6mA (202-003, 1:2000; Synaptic Systems, Goettingen, Germany) overnight at 4°C. Subsequently, membranes were washed in Tris-buffered saline with 0.1% Tween 3 times and incubated with horseradish-peroxidase–linked secondary anti-rabbit IgG antibody for 1 hour at room temperature. Membranes were washed and incubated with chemiluminescence substrate (SB-WB011; Share-Bio) and imaged using the ChemiDoc XRS+ system (Bio-Rad).
Histology Analysis and Immunohistochemistry Staining
Liver and adipose tissue including brown adipose tissue, inguinal white adipose tissue, and epididymal white adipose tissue samples were fixed in 4% paraformaldehyde after harvesting, dehydrating, and embedding in paraffin. For H&E staining and Sirius Red staining, paraffin-embedded tissues were cut into 3- to 4-μm–thick sections, dewaxed, hydrated, and then stained in H&E and picro-sirius red solution. For immunohistochemical staining, the sections were cut at 4 μm, dried, deparaffinized, and incubated within a boiling citrate buffer for 10 minutes. The sections subsequently were treated with 3% H2O2 for 10 minutes, penetrated with 0.3% Triton X-100 (Darmstadt, Germany) in phosphate-buffered saline for 30 minutes, treated with RNase for 1 hour to remove RNA contamination, and blocked with 5% bovine serum albumin for 1 hour. Liver sections were incubated with antibody against 6mA overnight at 4°C and horseradish-peroxidase–conjugated anti-rabbit secondary antibodies, then developed with 3,3'-Diaminobenzidine substrates (AR1022; Boster, Wuhan, China), and counterstained with hematoxylin. Nuclear 6mA staining was imaged with a SOPTOP RX50 microscope (Shunyu, Ningbo, China). For Oil Red O staining, fresh liver samples were snap-frozen in Tissue-Tek® O.C.T. Compound (Sakura, Torrance, CA). The slices (8 μm) were cryo-sectioned and stained with Oil Red O solutions (Sigma).
Glucose Tolerance Tests and Insulin Tolerance Tests
Glucose tolerance tests were performed on mice by intraperitoneal injection of D-glucose (2 g/kg; Sigma-Aldrich) after a 16-hour fast. Insulin tolerance tests were performed on mice injected intraperitoneally with biosynthetic human insulin Novolin R (0.75 U/kg; Novo Nordisk, Bagsvaerd, Denmark) after a 6-hour fast. Blood glucose level was measured using a portable blood glucose meter (Accu-Chek Performa; Roche) at 0, 15, 30, 60, 90, and 120 minutes after injection.
Quantification of Total TGs, FFAs, ALT, and AST in the Liver and Serum
According to the manufacturer’s instructions, TGs from liver and serum were measured using commercial kits (BC0265; Solarbio, Beijing, China). FFAs in serum were determined using a LabAssay NEFA kit (294-63601; Wako, Osaka, Japan). ALT and AST levels in serum were examined with commercial kits (E-BC-K235-M and E-BC-K236-M; Elabscience, Wuhan, China).
RNA Isolation and Real-Time qPCR
Total RNA was isolated from liver tissues and cell lysates using the TRIzol reagent (15596026; Life Technologies Corporation, Carlsbad, CA) according to the manufacturer’s instructions. The complementary DNA was synthesized from 1-μg aliquots of each RNA sample using an Evo M-MLV RT Kit with genomic DNA (gDNA) Clean for qPCR (AG11705; Accurate Biology, Hunan, China). Real-time qPCR was performed using the SYBR Green Premix Pro Taq HS qPCR Kit (Rox Plus, AG11718; Accurate Biology ) in a QuantStudio 3 real-time PCR system (Applied Biosystems). The sequences of primers used are listed in Table 1. Relative gene expression was calculated using the 2−ΔΔCt method by normalization with mRNA levels of the Gapdh housekeeping gene and presented as the fold increase relative to control.
Table 1.
List of Primer Sequences for qPCR
| Gene name | Forward primer 5’-3’ | Reverse primer 5’-3’ |
|---|---|---|
| Alkbh1 | GGTGGTCAGGTTTCCTCTGAA | GGAGGAAGGGGTTTGGAATGAA |
| Ttr | GTCCTCTGATGGTCAAAGTC | CTCCTTCTACAAACTTCTCATCTG |
| Des | CTTCTCTGCTCTCAACTTCC | CTGACAACCTCTCCATCCC |
| Krt7 | AGGAGATCAACCGACGCAC | GTCTCGTGAAGGGTCTTGAGG |
| Pecam | CTGGTGCTCTATGCAAGCCT | AGTTGCTGCCCATTCATCAC |
| Cd36 | CCTGCAAATGTCAGAGGAAA | GCGACATGATTAATGGCACA |
| Slc27a5 | TTCGAAAGAACCAACCCTTCCT | GCGTCGTACATTCGCAACAA |
| Slc27a2 | CTTCGGGAACCACAGGTCTTC | CATAGCAAGGCCTGTCCCATAC |
| Acsl3 | GGGACTTGGGAGGAGCTGTGTAAC | CTCAATGTCCGCCTGGTAATGTGT |
| Pparg | CCGAAGAACCATCCGATT | CGCAGATCAGCAGACTC |
| Srebf1 | GCTTCTCTTCTGCTTCTCT | GCTGTAGGATGGTGAGTG |
| Fasn | CTCATCCACTCAGGTTCAG | AGGTATGCTCGCTTCTCT |
| Scd1 | CAGCCGAGCCTTGTAAGTTC | GCTCTACACCTGCCTCTTCG |
| Mttp | CTCTTGGCAGTGCTTTTTCTCT | GAGCTTGTATAGCCGCTCATT |
| Apob | AAGCACCTCCGAAAGTACGTG | CTCCAGCTCTACCTTACAGTTGA |
| Pparα | TGCCTTAGAACTGGATGAC | ATCTGGATGGTTGCTCTG |
| Cpt1a | AGCCAGACGAAGAACATC | CCTTGACCATAGCCATCC |
| Il1β | ATGGCAACTGTTCCTGAACTCAACT | CAGGACAGGTATAGATTCTTTCCTTT |
| Tnfα | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| Ccl2 | AGGTCCCTGTCATGCTTCTG | TCTGGACCCATTCCTTCTTG |
| Il6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| Adgre1 | CCCCAGTGTCCTTACAGAGTG | GTGCCCAGAGTGGATGTCT |
| Cd68 | GCTCCAAGCCCAAATTCAAATCCG | CCCTGGACCTTGGTTTTGTTGGG |
| Col1a1 | TGACTGGAAGAGCGGAGAGTACT | TTCGGGCTGATGTACCAGTTC |
| Acta2 | TGCCGAGCGTGAGATTGTC | CGTTCGTTTCCAATGGTGATC |
| Fn1 | GTGCCTGGGCAACGGA | CCCGACCCTGACCGAAG |
| Gapdh | CGTGTTCTACCCCCAATGT | TGTCATCATACTTGGCAGGTTTCT |
Western Blot
Proteins from tissues and cells were obtained using RIPA buffer (Pierce, Rockford, IL) supplemented with a protease inhibitor cocktail (Roche, Mannheim, Germany), and quantitated for concentrations using the Bradford method (23238; Pierce). The protein lysates were heated at 95°C for 5 minutes in sample buffer containing 2% sodium dodecyl sulfate and 1% 2-mercaptoethanol, separated on 10% or 12% sodium dodecyl sulfate–polyacrylamide gels, and transferred to polyvinylidene difluoride membranes using a wet transfer apparatus (Bio-Rad). The membranes were blocked with 5% bovine serum albumin for 1 hour and then incubated with antibodies against ALKBH1 (ab195376, 1:1000; Abcam, Waltham, MA), CD36 (NB400-144SS, 1:1000; Novus Biologicals, Centennial, CO), and phospho-GSK3β-Ser9 (5558, 1:1000; Cell Signaling, Danvers, MA). Phospho-Akt-Ser473 (9271, 1:1000; Cell Signaling, Danvers, MA), Akt (4691, 1:1000; Cell Signaling, Danvers, MA), FATP2 (14048-1-AP, 1:2000; Proteintech, Rosemont, IL), ACSL3 (161226, 1:1000; ZEN BIO, Beijing, China), histone-H3 (17168-1-AP, 1:1000; Proteintech, Rosemont, IL), α-tubulin (11224-1-Ap, 1:4000; Proteintech, Rosemont, IL), β-actin (BM0627, 1:2000; Boster, Wuhan, China), or glyceraldehyde-3-phosphate dehydrogenase (TA802519, 1:8000; Origene, Wuxi, China) overnight followed by a horseradish-peroxidase–conjugated anti-rabbit or anti-mouse IgG. Signals were detected with ChemiDoc XRS+ (Bio-Rad) with Image Lab Software (Hercules, CA).
RNA-seq Analysis
Total RNA was isolated and verified using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Then, the libraries were constructed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA) according to the manufacturer’s instructions. The transcriptome sequencing and analysis were conducted by OE Biotech Co, Ltd (Shanghai, China). Briefly, the libraries were sequenced on an Illumina Nova seq 6000 platform and 150-bp paired-end reads were generated. Raw Reads were processed using Trimmomatic42 to obtain the clean reads and mapped to the mouse genome (Genome Reference Consortium Mouse Build 38, GRCm38) using HISAT2.43 Fragments Per Kilobase of transcript per Million mapped reads (FPKM)44 and read counts of each gene were obtained by Cufflinks45 and HTSeq-count46 separately. Differential gene expression analysis was performed using DESeq. Differentially expressed genes (DEGs) were identified by a P value less than .05 and fold change larger than 2 or fold change less than 0.5.
ChIP-seq Analysis
Mouse primary hepatocytes were isolated and transfected with plasmids expressing ALKBH1 protein with a flag tag for 48 hours. Then, ChIP experiments were performed using a SimpleChIP Enzymatic Chromatin IP Kit (9003; Cell Signaling) with an antibody against flag (14793; Cell Signaling) or the control normal rabbit IgG, according to the manufacturer’s procedures. DNA sequencing libraries were prepared using the VAHTS Universal DNA Library Prep Kit for Illumina V3 (ND607; Vazyme, Nanjing, China). High-throughput sequencing and analysis were conducted by Seqhealth Technology Co, Ltd (Wuhan, China). Briefly, the libraries were sequenced on a Novaseq 6000 sequencer (Illumina) with the PE150 model. Raw data were filtered by Trimmomatic (version 0.36). Then, clean reads were mapped to the mouse genome (GRCm38) using STAR software (version 2.5.3a). RSeQC (version 2.6) was used for read distribution analysis. MACS2 software (version 2.1.1) was used for peak calling. Bedtools (version 2.25.0) was used for peak annotation and peak distribution analysis. The differentially binding peaks were identified by a python script, using the Fisher test. A P value less than .05 was judged statistically significant enrichment.
ChIP–qPCR Assay
ChIP experiments were performed on mouse primary hepatocytes using a SimpleChIP Enzymatic Chromatin IP Kit (9003; Cell Signaling) according to the manufacturer’s procedures, with additional antibodies against ALKBH1 (ab195376; Abcam) and N6-methyladenosine (202 003; Synaptic Systems). Cells were fixed with formaldehyde to covalently cross-link protein to DNA to conserve their interactions. After immunoprecipitation, pulled-down DNA and the input DNA were subjected to qPCR analysis with the primers (Table 2) for the indicated genomic sequence. The results are presented as the percentage of input DNA.
Table 2.
List of Primer Sequences for ChIP-qPCR
| Gene name | Forword sequence 5’-3’ | Reverse sequence 5’-3’ |
|---|---|---|
| Cd36-S1 | AGAGCCATTCATAGGCTTTCAAC | CTCTGCCCAGGTGCTAACTT |
| Cd36-S2 | ACTTCTTCCCATTACCTGCACT | TGTGTCATCTATCTGCGTGCT |
| Cd36-S3 | AGCAGTGAATCCCGTAGCAG | GCATAACTGCGATTGCTCCC |
| Cd36-S4 | AGCCCAATGAATGAAGTGTTCTTG | CGAGACCTCCGTGGGAAATC |
| Cd36-S5 | AATAGCCATCACTCGTTTCCA | TGTAAGCAAACACAAACATTTTACC |
| Acsl3-S1 | GGCAACTGGAGGGCTTGTAT | CAAGGCCATGGACCACTTCT |
| Acsl3-S2 | GGCTGGATCGGCTTTGAACT | ACATAAGGTCTGTGAGTGCCA |
| Acsl3-S3 | ATCCGAGCCCTTGACTTTGG | CAGCCCTGGTCTCACAGAAG |
| Slc27a2-S1 | CTACTTTCCCTGGCCACCTC | GAGGTCCTCTGGACTGGGAA |
| Pparg-S1 | TCAGTGTGCTTGCAGTTTGC | CACAACCAAGCTGATTGCCC |
| Pparg-S2 | CATCAGACACCCTCCGACTG | GCTGAGAGATCTAGGTGTGGC |
| Pparg-S3 | CTCAGGGAGGGCTCCAATTTT | ACTGCAGTAGTATTCCAATGGTTG |
| Pparg-S4 | TTAGGGAGAGCACAATGCGG | AACAGCTTCTCCTTCTCGGC |
| Pparg-S5 | CCCCTAGGGAGCTCTCAACT | TCCTAGCTTCCCCTGTCTCC |
Statistics
All experiments were repeated at least 3 times and expressed as the means ± SEM. Data were analyzed using the t test with GraphPad prism 8 software from GraphPad (La Jolla, CA). A P value less than .05 was considered statistically significant.
Acknowledgments
Data Availability Statement
The data sets generated for this study can be found in the Gene Expression Omnibus (RNA-seq, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193497. Secure token for reviewers: yzafmiyqzpahfuz. ChIP-seq, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197185. Secure token for reviewers: gdorokiyznsxlqp).
CRediT Authorship Contributions
Liping Luo (Conceptualization: Equal; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Software: Equal; Writing – original draft: Lead)
Ya Liu (Formal analysis: Equal; Investigation: Equal; Software: Equal)
Paul Nizigiyimana (Data curation: Equal; Investigation: Equal; Resources: Equal)
Mingsheng Ye (Data curation: Equal; Methodology: Equal)
Ye Xiao (Data curation: Lead; Formal analysis: Equal)
Qi Guo (Formal analysis: Lead; Methodology: Equal)
Tian Su (Data curation: Equal; Methodology: Supporting)
Xianghang Luo (Funding acquisition: Supporting; Project administration: Lead; Supervision: Supporting)
Yan Huang (Funding acquisition: Equal; Project administration: Supporting; Supervision: Equal)
Haiyan Zhou (Funding acquisition: Equal; Project administration: Lead; Supervision: Lead; Writing – original draft: Supporting)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by grants from the National Natural Science Foundation of China (82100914, 82170866, 82000811, 92149306, 82120108009, 81930022, 81900732, and 81900810), the Postdoctoral Science Foundation of China (2021M703639 and 2022M713514), and the Natural Science Foundation of Hunan Province, China (2022JJ40806 and 2022JJ30990).
References
- 1.Younossi Z.M., Golabi P., de Avila L., Paik J.M., Srishord M., Fukui N., Qiu Y., Burns L., Afendy A., Nader F. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. J Hepatol. 2019;71:793–801. doi: 10.1016/j.jhep.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 2.Bayoumi A., Gronbaek H., George J., Eslam M. The epigenetic drug discovery landscape for metabolic-associated fatty liver disease. Trends Genet. 2020;36:429–441. doi: 10.1016/j.tig.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 3.McPherson S., Hardy T., Henderson E., Burt A.D., Day C.P., Anstee Q.M. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. J Hepatol. 2015;62:1148–1155. doi: 10.1016/j.jhep.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 4.Ipsen D.H., Lykkesfeldt J., Tveden-Nyborg P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell Mol Life Sci. 2018;75:3313–3327. doi: 10.1007/s00018-018-2860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hojland Ipsen D., Tveden-Nyborg P., Lykkesfeldt J. Normal weight dyslipidemia: is it all about the liver? Obesity (Silver Spring) 2016;24:556–567. doi: 10.1002/oby.21443. [DOI] [PubMed] [Google Scholar]
- 6.Fabbrini E., Magkos F. Hepatic steatosis as a marker of metabolic dysfunction. Nutrients. 2015;7:4995–5019. doi: 10.3390/nu7064995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ipsen D.H., Tveden-Nyborg P., Lykkesfeldt J. Dyslipidemia: obese or not obese-that is not the question. Curr Obes Rep. 2016;5:405–412. doi: 10.1007/s13679-016-0232-9. [DOI] [PubMed] [Google Scholar]
- 8.Mashek D.G. Hepatic fatty acid trafficking: multiple forks in the road. Adv Nutr. 2013;4:697–710. doi: 10.3945/an.113.004648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson C.G., Tran J.L., Erion D.M., Vera N.B., Febbraio M., Weiss E.J. Hepatocyte-specific disruption of CD36 attenuates fatty liver and improves insulin sensitivity in HFD-fed mice. Endocrinology. 2016;157:570–585. doi: 10.1210/en.2015-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace M., Metallo C.M. Tracing insights into de novo lipogenesis in liver and adipose tissues. Semin Cell Dev Biol. 2020;108:65–71. doi: 10.1016/j.semcdb.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 11.Souza-Mello V. Peroxisome proliferator-activated receptors as targets to treat non-alcoholic fatty liver disease. World J Hepatol. 2015;7:1012–1019. doi: 10.4254/wjh.v7.i8.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bu S.Y., Mashek M.T., Mashek D.G. Suppression of long chain acyl-CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. J Biol Chem. 2009;284:30474–30483. doi: 10.1074/jbc.M109.036665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asif S., Morrow N.M., Mulvihill E.E., Kim K.H. Understanding dietary intervention-mediated epigenetic modifications in metabolic diseases. Front Genet. 2020;11 doi: 10.3389/fgene.2020.590369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greer E.L., Blanco M.A., Gu L., Sendinc E., Liu J., Aristizabal-Corrales D., Hsu C.H., Aravind L., He C., Shi Y. DNA methylation on N6-adenine in C. elegans. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J., Zhu Y., Luo G.Z., Wang X., Yue Y., Wang X., Zong X., Chen K., Yin H., Fu Y., Han D., Wang Y., Chen D., He C. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat Commun. 2016;7 doi: 10.1038/ncomms13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M., Tackett A., Wang G., Hon L.S., Fang G., Swenberg J.A., Xiao A.Z. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U., Zhang C., Dowiak A.V., Lin K., Zeng C., Sui Y., Kim L.J.Y., Miller T.E., Jiang L., Lee C.H., Huang Z., Fang X., Zhai K., Mack S.C., Sander M., Bao S., Kerstetter-Fogle A.E., Sloan A.E., Xiao A.Z., Rich J.N. N(6)-methyladenine DNA modification in glioblastoma. Cell. 2018;175:1228–1243 e1220. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J., Li C., Liu B., Luo Y., Zhu Y., Zhang N., He S., He C., Wang H., Chen D. N6-methyladenine DNA modification in Drosophila. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Xiao C.L., Zhu S., He M., Chen Zhang Q., Chen Y., Yu G., Liu J., Xie S.Q., Luo F., Liang Z., Wang D.P., Bo X.C., Gu X.F., Wang K., Yan G.R. N(6)-methyladenine dna modification in the human genome. Mol Cell. 2018;71:306–318 e307. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Yao B., Cheng Y., Wang Z., Li Y., Chen L., Huang L., Zhang W., Chen D., Wu H., Tang B., Jin P. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun. 2017;8:1122. doi: 10.1038/s41467-017-01195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Wang C. Demethyltransferase AlkBH1 substrate diversity and relationship to human diseases. Mol Biol Rep. 2021;48:4747–4756. doi: 10.1007/s11033-021-06421-x. [DOI] [PubMed] [Google Scholar]
- 22.Fedeles B.I., Singh V., Delaney J.C., Li D., Essigmann J.M. The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290:20734–20742. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ougland R., Jonson I., Moen M.N., Nesse G., Asker G., Klungland A., Larsen E. Role of ALKBH1 in the core transcriptional network of embryonic stem cells. Cell Physiol Biochem. 2016;38:173–184. doi: 10.1159/000438619. [DOI] [PubMed] [Google Scholar]
- 24.Nordstrand L.M., Svard J., Larsen E., Nilsen A., Ougland R., Furu K., Lien G.F., Rognes T., Namekawa S.H., Lee J.T., Klungland A. Mice lacking Alkbh1 display sex-ratio distortion and unilateral eye defects. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ougland R., Lando D., Jonson I., Dahl J.A., Moen M.N., Nordstrand L.M., Rognes T., Lee J.T., Klungland A., Kouzarides T., Larsen E. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem Cells. 2012;30:2672–2682. doi: 10.1002/stem.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan Z., Sikandar S., Witherspoon M., Dizon D., Nguyen T., Benirschke K., Wiley C., Vrana P., Lipkin S.M. Impaired placental trophoblast lineage differentiation in Alkbh1(-/-) mice. Dev Dyn. 2008;237:316–327. doi: 10.1002/dvdy.21418. [DOI] [PubMed] [Google Scholar]
- 27.Cai G.P., Liu Y.L., Luo L.P., Xiao Y., Jiang T.J., Yuan J., Wang M. Alkbh1-mediated DNA N6-methyladenine modification regulates bone marrow mesenchymal stem cell fate during skeletal aging. Cell Prolif. 2022;55 doi: 10.1111/cpr.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C., Liu Y., Li X., Zou J., Zou S. DNA N(6)-methyladenine demethylase ALKBH1 enhances osteogenic differentiation of human MSCs. Bone Res. 2016;4 doi: 10.1038/boneres.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diao L.T., Xie S.J., Yu P.J., Sun Y.J., Yang F., Tan Y.Y., Tao S., Hou Y.R., Zheng L.L., Xiao Z.D., Zhang Q. N(6)-methyladenine demethylase ALKBH1 inhibits the differentiation of skeletal muscle. Exp Cell Res. 2021;400 doi: 10.1016/j.yexcr.2021.112492. [DOI] [PubMed] [Google Scholar]
- 30.Kawarada L., Fukaya M., Saito R., Kassai H., Sakagami H., Aiba A. Telencephalon-specific Alkbh1 conditional knockout mice display hippocampal atrophy and impaired learning. FEBS Lett. 2021;595:1671–1680. doi: 10.1002/1873-3468.14098. [DOI] [PubMed] [Google Scholar]
- 31.Li Q., Qian C., Feng H., Lin T., Zhu Q., Huang Y., Zhou F.Q. N6-methyladenine DNA demethylase ALKBH1 regulates mammalian axon regeneration. Neurosci Bull. 2021;37:809–814. doi: 10.1007/s12264-021-00671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ouyang L., Su X., Li W., Tang L., Zhang M., Zhu Y., Xie C., Zhang P., Chen J., Huang H. ALKBH1-demethylated DNA N6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J Clin Invest. 2021;131 doi: 10.1172/JCI146985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geisler C.E., Renquist B.J. Hepatic lipid accumulation: cause and consequence of dysregulated glucoregulatory hormones. J Endocrinol. 2017;234:R1–R21. doi: 10.1530/JOE-16-0513. [DOI] [PubMed] [Google Scholar]
- 34.Kawarada L., Suzuki T., Ohira T., Hirata S., Miyauchi K., Suzuki T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017;45:7401–7415. doi: 10.1093/nar/gkx354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner A., Hofmeister O., Rolland S.G., Maiser A., Aasumets K., Schmitt S., Schorpp K., Feuchtinger A., Hadian K., Schneider S., Zischka H., Leonhardt H., Conradt B., Gerhold J.M., Wolf A. Mitochondrial Alkbh1 localizes to mtRNA granules and its knockdown induces the mitochondrial UPR in humans and C. elegans. J Cell Sci. 2019;132:jcs223891. doi: 10.1242/jcs.223891. [DOI] [PubMed] [Google Scholar]
- 36.Haag S., Sloan K.E., Ranjan N., Warda A.S., Kretschmer J., Blessing C., Hubner B., Seikowski J., Dennerlein S., Rehling P., Rodnina M.V., Hobartner C., Bohnsack M.T. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–2119. doi: 10.15252/embj.201694885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q., Sharma V.P., Shen H., Xiao Y., Zhu Q., Xiong X., Guo L., Jiang L., Ohta K., Li S., Shi H., Rui L., Lin J.D. The hepatokine Tsukushi gates energy expenditure via brown fat sympathetic innervation. Nat Metab. 2019;1:251–260. doi: 10.1038/s42255-018-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Zhao S., Nelakanti R.V., Lin K., Wu T.P., Alderman M.H., 3rd, Guo C., Wang P., Zhang M., Min W., Jiang Z., Wang Y., Li H., Xiao A.Z. N(6)-methyladenine in DNA antagonizes SATB1 in early development. Nature. 2020;583:625–630. doi: 10.1038/s41586-020-2500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartonicek N., Clark M.B., Quek X.C., Torpy J.R., Pritchard A.L., Maag J.L.V., Gloss B.S., Crawford J., Taft R.J., Hayward N.K., Montgomery G.W., Mattick J.S., Mercer T.R., Dinger M.E. Intergenic disease-associated regions are abundant in novel transcripts. Genome Biol. 2017;18:241. doi: 10.1186/s13059-017-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poppelreuther M., Sander S., Minden F., Dietz M.S., Exner T., Du C., Zhang I., Ehehalt F., Knuppel L., Domschke S., Badenhop A., Staudacher S., Ehehalt R., Stremmel W., Thiele C., Heilemann M., Fullekrug J. The metabolic capacity of lipid droplet localized acyl-CoA synthetase 3 is not sufficient to support local triglyceride synthesis independent of the endoplasmic reticulum in A431 cells. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:614–624. doi: 10.1016/j.bbalip.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Zhou X., Xiao Y., Li C., Huang Y., Guo Q., Su T., Fu L., Luo L. miR-188 promotes liver steatosis and insulin resistance via the autophagy pathway. J Endocrinol. 2020;245:411–423. doi: 10.1530/JOE-20-0033. [DOI] [PubMed] [Google Scholar]
- 42.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated for this study can be found in the Gene Expression Omnibus (RNA-seq, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193497. Secure token for reviewers: yzafmiyqzpahfuz. ChIP-seq, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197185. Secure token for reviewers: gdorokiyznsxlqp).