Abstract
We previously identified a protein that was stimulatory for malignant Sézary T cells, termed Sézary T-cell activating factor (SAF). However, the identity of this protein has not been fully elucidated, nor has it’s role been determined in the pathogenesis of cutaneous T-cell lymphoma (CTCL). The basis for epidermotropism and proliferation of malignant cells in the skin of patients with CTCL is unknown. Using a monoclonal antibody inhibitory for SAF activity, we demonstrated that SAF is present in the skin of 16 of 27 samples from patients with mycosis fungoides, the predominant form of CTCL. In this report, the SAF determinant is demonstrated to be associated with Chlamydia pneumoniae bacteria by immunohistochemistry, immunoelectron microscopy, and culture analysis. Reactivity of antibodies against an outer membrane protein of C. pneumoniae or against the lipopolysaccharide of Chlamydiae spp. demonstrated that these determinants are coexpressed in 90% of the SAF-positive samples. We confirmed the presence of C. pneumoniae DNA and RNA in the skin by PCR and reverse transcription-PCR and by sequence analysis of the PCR products. The expression of the C. pneumoniae antigens and SAF appears to be associated with active disease in that C. pneumoniae antigens were absent or greatly diminished in the skin of three patients examined after Psoralen and long-wave UVA radiation treatment. Our results suggest that SAF is a Chlamydia-associated protein and that further investigation is warranted to determine whether SAF and C. pneumoniae play a role in the pathogenesis of CTCL.
The concept that cutaneous T-cell lymphoma (CTCL) could be related to chronic stimulation has been postulated for many years (42). Recent provocative data published by the Duvic group (24) provide information that suggests an association of Staphylococcus aureus with CTCL. That report proposes that S. aureus provides a chronic stimulant in CTCL patients and thus could at least exacerbate the disease process. A bacterial infection in the epidermis would lead to the production of inflammatory cytokines, resulting in lymphocytic infiltration and release of gamma interferon (IFN-γ), followed by IP-10 production, and then to clonal expansion of epidermotropic T cells (37). These studies provide a rational mechanism by which a bacterium could stimulate lymphocytic infiltration and provide a chronic stimulus in CTCL patients. Our investigation supports and extends the concept that a bacterium may provide an important stimulus in the pathogenesis of CTCL, although we implicate a much different bacterial genus and species, viz., Chlamydia pneumoniae.
C. pneumoniae is an obligate intracellular pathogen that replicates within the cytoplasm of the cells in which it infects. C. pneumoniae was originally described as a respiratory pathogen (30). However, the organism has been implicated in several nonpulmonary diseases, such as meningoencephalitis and atherosclerosis (8, 15, 28). In addition, we recently reported an association of C. pneumoniae with Alzheimer’s disease (AD) (4). Epidemiological studies indicate that infection of adults is common in all populations examined (16, 23, 30). The detection of significant anti-C. pneumoniae antibody titers rises with increasing age, peaking in the sixth to seventh decades in most populations (16, 30).
Immunopathology is a general feature of Chlamydia-induced disease. Because C. pneumoniae is an intracellular pathogen, the immune system has difficulty clearing the infection. Thus, persistent chlamydial infections are common and result in chronic inflammation and the presence of Th1/Th2 CD4+ T cells, as well as CD8+ cytotoxic or suppressor T cells, macrophages, and, in some cases, B cells (44). An example of the result of persistent chlamydial infection is described in the synovia of Chlamydia trachomatis-induced reactive arthritis (39). Interestingly, persistent chlamydial infection may be maintained, in part, by host products such as interferons, whose production is induced by the organism (5, 39). Thus, a balance appears to develop between host tissue survival and organism replication. It has been suggested that such a state of semilatency can last for decades (5, 27).
Since it has been shown that C. pneumoniae can traffic to numerous areas of the body (4, 8, 15, 28, 30), we investigated whether C. pneumoniae antigens could be detected in cells within the epidermis in patients with mycosis fungoides, the primary form of CTCL, or in the peripheral blood mononuclear cells (PBMC) of patients with Sézary syndrome, the leukemic variant of CTCL. CTCL represents a malignant clonal amplification of mature, memory (CD45RO+), epidermotropic (CTLA+), helper (CD4+), and T (CD3+) cells (9, 11, 20, 22, 33, 40). These T cells predominantly produce a Th2 cytokine profile (43). Our previous investigations have centered on identifying the growth requirements for malignant cells in CTCL. We identified and characterized a stimulatory factor capable of inducing proliferation of malignant Sézary cells. We termed this factor the Sézary T-cell activating factor (SAF) (1). SAF was originally described as being produced by the PBMC of certain patients with Sézary syndrome (3) and was found to be a potent T-cell mitogenic factor for malignant as well as nonmalignant T cells (1–3). In fact, we used SAF to establish cell lines from patients with Sézary syndrome, some of which contained the predominant malignant clone (3). SAF enabled establishment of T-cell lines from CTCL patients more readily than other methods (13, 14); however, the role SAF plays in the development of CTCL remains to be elucidated. When we first described SAF, we postulated that it was an autocrine growth factor (1); we now have evidence, as described herein, that SAF may not be a eukaryotic product at all but rather a mitogenic bacterial protein.
In this investigation, skin biopsy specimens of individuals with CTCL were tested for reactivity with a variety of immunologic probes, including an anti-C. pneumoniae major outer membrane protein (OMP) monoclonal antibody (MAb), a genus-specific MAb for chlamydial lipopolysaccharide (LPS), and a MAb against the stimulatory SAF protein. In addition, the expression of these antigens was examined in several CTCL patients after Psoralen and long-wave UVA radiation (PUVA) treatment. Furthermore, the presence of C. pneumoniae in the biopsied samples was tested by using nested PCR to detect ompA DNA. In addition, reverse transcription (RT)-PCR was used to determine whether transcriptional activity for C. pneumoniae ompA could be detected in the same samples. Immunoreactivity of the anti-SAF and anti-OMP MAbs against C. pneumoniae was determined by using immunoelectron microscopy. We specifically examined whether antichlamydial antibodies reacted with C. pneumoniae cultured within keratinocytes to determine if epidermal infection is feasible. The overall objective of these investigations was to examine the relationship between the SAF protein and CTCL and to clarify whether C. pneumoniae was a potential source of this mitogenic determinant.
MATERIALS AND METHODS
Preparation of SAF.
Protein with SAF-like activity was recovered and purified from the SZ-4 cell line (2) for the generation of MAbs inhibitory for SAF bioactivity. Conditioned medium (CM) from the SZ-4 cell line was generated as follows: SZ-4 cells were cultured for 18 h in RPMI 1640 media containing combinations of 10% fetal bovine serum (FBS) alone, 10% FBS and 1 μg of phytohemagglutinin (PHA) (Sigma, St. Louis, Mo.) per ml, or 10% FBS and phorbol 12-myristate 13-acetate (TPA) (Sigma) and 1 mM ionomycin (Sigma). The cells were then centrifuged, rinsed in media, and cultured for up to 72 h in serum-free RPMI 1640 containing Neutradoma HU (Boehringer Mannheim, Chicago, Ill.).
To characterize fractions for SAF-like activity and to screen the MAb clones for their ability to neutralize SAF activity, SAF bioassays were performed as previously described (1). Various concentrations of CM from the SZ-4 cell line were added to 105 PBMC recovered from the blood of healthy donors (American Red Cross, Miami, Fla.) along with 10 U of recombinant interleukin 2 (rIL-2; Boehringer Mannheim) per ml in 10% FBS–RPMI 1640 medium. The samples from these healthy donors were determined by the Red Cross to be seronegative for human immunodeficiency virus type 1, hepatitis B virus, and human T-cell leukemia virus. The cells were cultured for 72 h in a humidified 5% CO2 incubator, with the addition of 1 μCi of tritiated thymidine ([3H]TdR) (Amersham, Arlington Heights, Ill.) for the final 4 h in 96-well flat-bottom plates (Costar, Rochester, N.Y.). Cultures were terminated by harvesting them onto filter mats (Schleicher & Schuell, Keene, N.H.) and processed for liquid scintillation counting by drying and adding 3 ml of liquid scintillation fluid (Econofluor; Dupont, NEN, Wilmington, Del.).
Preparation of anti-SAF MAb.
Conditioned medium containing SAF was semipurified by anion-exchange chromatography with DE-52 anion-exchange resin (Whatman, Clifton, N.J.). Anion-exchange-purified SZ-4 CM (50 U in 25 μl) was mixed with loading buffer containing 2% sodium dodecyl sulfate (SDS) and 5% 2-mercaptoethanol, heated to 55°C for 10 min, and electrophoresed according to the method of Laemmli (29) in a 12% polyacrylamide gel for 4 h at a constant current of 25 mA. Three-millimeter slices were cut from the gel, and proteins from each gel slice were passively eluted into media containing 10% FBS to prevent nonspecific loss of protein. The SDS was removed by dialysis against N,N-bis(2-hydroxyethyl)-2-aminoethanesulfonic acid (5 mM), and the dialysate was tested at various concentrations for SAF activity as described elsewhere (1). The relative mobility (Rf) value of each protein fraction was used to determine the apparent molecular mass of SZ-4 SAF. The 28- to 30-kDa SAF active fraction was immunized into BALB/c mice and hybridomas were made. MAbs reactive with the immunogen were screened for SAF inhibitory activity against both the PBMC-derived SAF and cell line-derived SAF. One clone (58.19), an immunoglobulin G3, called anti-SAF in the present work, was selected and recloned. An anti-SAF assay was performed as follows: various concentrations (0 to 100 μg/ml) of protein G purified MAb immunoglobulin G were added to various concentrations of PBMC-derived SAF and SZ-4 CM for 1 h, and then 105 PBMC were added per well, followed by a standard SAF bioassay (1). Specificity of the inhibition was examined for the MAb to inhibit PHA (1 μg/ml; Sigma)-induced or TPA (10 ng/ml; Sigma) plus ionomycin (1 mM; Sigma)-induced T-cell proliferation.
Infection of cell cultures with C. pneumoniae.
Keratinocyte cell cultures were established on glass chamber well slides (Lab-Tek) or in flasks. Cultures were allowed to reach 40 to 50% confluence, after which 200 inclusion-forming units at 50% confluence (IFU50) per cm2 of culture area of C. pneumoniae (American Type Culture Collection, Rockville, Md.) was added to the media. The flasks (Corning, Cambridge, Mass.) or slide wells (Falcon, Frankline Lakes, N.J.) were then centrifuged at 500 × g for 30 min to increase contact of the organism with the cells. The total volumes were 2 ml for T-25 flasks and 200 μl for the 2-cm2 or 400 μl for the 4-cm2 chambers. (All materials centrifuged were placed in sealed bags to prevent spread of the bacteria in the centrifuge in case of product failure.) After centrifugation, additional media were added to the cultures to bring the 2-cm2 wells to 1 ml, the 4-cm2 wells to 2 ml, and the T-25 flasks to 7 ml.
The nonadherent monocyte cell line THP-1 (gift of David Bernard, Medical College of Pennsylvania and Hahnemann University [MCP-HU]) was centrifuged with the bacteria in 15 ml of conical centrifuge tubes in a 1-ml volume and then brought to 2 × 105 cells/ml in T-25 standing flasks. Cultures were terminated after 72 h with individual T-25 flasks extracted for either RNA or DNA while the chamber slides were prepared for immunohistochemistry. For THP-1 cells, slides were prepared by cytocentrifugation by using a Cytospin III (Shandon) at 800 rpm for 5 min onto ProbeOn Plus slides (Fisher Scientific, Pittsburgh, Pa.). Media were removed from the chamber slides, and cells were rinsed with Hanks buffer twice, after which the cells were fixed with Streck tissue fixative (STF; Streck, Omaha, Nebr.). One milliliter of STF was added to each chamber and allowed to fix at room temperature for 24 h. Cytospun cells were fixed with STF in a Coplan jar for at least 1 h.
Immunohistochemistry.
Skin tissue obtained for diagnostic purposes from 27 patients with CTCL was used for these studies. In addition, sections from two different brains from patients with AD, obtained from autopsy, were used in our study. Five micrometers of formalin-fixed paraffin embedded sections were deparaffinized with xylenes and rehydrated in automation buffer (Biømeda, Foster City, Calif.). For immunohistochemistry on the sections, a modified avidin-biotin complex (ABC) technique was used (21). The modification entails the use of capillary gap ProbeOn Plus slides (Fisher) that have painted surfaces that create a 15-μm gap between the slides when placed face to face. This protocol uses reagents designed for capillary gap slides that includes antibody diluting buffer (Biømeda).
Detection of C. pneumoniae OMP and LPS in keratinocytes.
After fixation, cells were exposed to 1× automation buffer (Biømeda) followed by removal of endogenous peroxidases by using 3% hydrogen peroxide in water (Fisher). The anti-C. pneumoniae OMP MAb (clone RR402; DAKO, Carpinteria, Calif.) and the genus specific anti-Chlamydia LPS MAb from the Imagen kit (DAKO) were used as primary antibodies, both diluted 1:10 in antibody diluting buffer (Biømeda). Cells were incubated with antibody for 30 min, followed by five rinses with 1× automation buffer. Primary antibody binding was detected by using a super ABC kit (Biømeda) following the manufacturer’s instructions, with five rinses between the 30-min incubation with the secondary antibody and ABC reagent. After the final rinse, peroxidase enhancer (Biømeda) was added for 1 min and antibody binding was visualized following incubation with diaminobenzidine cobalt chromagen (Biømeda kit) for 6 to 8 min. Chambers and slides were rinsed with distilled water and counterstained for 1 min with aqueous hematoxylin (Biømeda). Slides were covered with crystal amount (Biømeda) and allowed to dry for 2 h. Slides were photographed with an Olympus BX-40 microscope and an Olympus SC-35 camera.
RNA extraction.
RNA extraction from cultures was performed following the removal of media from the cultures by using Ultraspec RNA (Biotecx, Houston, Tex.) as previously described (35). After extraction, RNA in RNase-free water was treated with RNase-free DNase (9 μg/ml; Sigma) (final concentration of 2 μg/ml) and incubated at 37°C for 2 h. The DNA-free RNA was then extracted through the addition of 100 ml of Ultraspec RNA extraction buffer (Biotecx) followed by 0.2 volumes of chloroform, as described for the original RNA. All RNA preparations were run on a 1% SeaKem agarose gel (FMC, Rockland, Maine) and examined by ethidium bromide incorporation for 28S and 18S rRNA to ensure that the RNA was not degraded.
RT-PCR.
To produce double-stranded cDNA for PCR analysis (RT-PCR), a 1- to 2-μg sample of RNA or the entire DNase-free RNA preparation was mixed with 1 μl of oligo(dT) (0.5 mg/ml; Perkin-Elmer, Foster City, Calif.), 6 μl of 5× first-strand buffer (Life Technologies, Grand Island, N.Y.), 1.5 μl of each deoxynucleoside triphosphate (Perkin-Elmer), 0.6 μl of RNasin (Promega, Madison, Wis.), and 2 μl of Moloney murine leukemia virus (MMLV) reverse transcriptase (Life Technologies) and brought to a final volume of 30 μl. RNA was heated to 70 to 80°C for 3 min followed by a 5-s centrifugation to ensure that the RNA was in the tube bottom. The reaction mixture was added to the RNA, and the incubation lasted for 1.5 h at 37°C, followed by inactivation of the enzyme at 95°C for 3 min. cDNA was stored at −20°C with 2 to 4 μl of cDNA used for each PCR.
Preparation of DNA for PCR.
For DNA extraction from cultures, the cultures were first washed with 5 ml of Hanks balanced salt solution containing 5 mM HEPES to remove debris. Five milliliters of DNA lysis buffer (25 mM Tris [pH 8.0], 100 mM NaCl, 10 mM EDTA, and 0.5% SDS) was added per flask. For DNA extraction from tissues, frozen specimens were homogenized in lysis buffer. DNA from cultures and tissues were then treated identically. The lysates were kept at 37°C for 1 h followed by the addition of 12 ml of proteinase K (20 mg/ml; Life Technologies) and incubation for 2 h at 37°C followed by centrifugation at 1,500 × g for 10 min. After transfer to a fresh tube, DNA was extracted by the hot phenol method as previously described (4). The aqueous phase was precipitated with 2.5 volumes of 100% ethanol and 0.1 volumes of 3 M sodium acetate overnight. The material was pelleted at 10,000 × g for 30 min, dried, suspended in TE buffer (0.05 M Tris, 0.005 M EDTA [pH 8.5]), and stored at −20°C. Following quantitation by A260/A280 on a spectrophotometer 610 (Milton Roy), we used 0.1 μg of DNA in subsequent PCRs.
PCR.
PCR was performed with 1× buffer II containing MgCl2 and 1.25 U of AmpliTaq DNA polymerase (Perkin-Elmer) per reaction. Deoxynucleoside triphosphates were at a final concentration of 200 μM each. Twenty picomoles of each primer was used in all reactions. DNA (0.1 μg), cDNA (2 μl), or PCR product (2 μl) for nested PCRs were added and brought to a final volume of 50 μl with water. External and internal nested primers were used for detection of C. pneumoniae DNA and cDNA ompA gene and 16S rRNA gene sequences in cultures infected with TW-183. ompA primer sequences are given in base pair numbers and were obtained from GenBank (accession no. M64064) (32); all are 5′ to 3′ as follows: ompA-EXT plus-strand primers 281 to 300 and minus-strand primers 1073 to 1053; omp-A internal plus-strand primers 390 to 411 and minus-strand primers 730 to 711 (ompA primer set 1,340 bp). Primers and sequences for the 16S rRNA gene were obtained from GenBank (accession no. L06108) (12). 16S rRNA external plus-strand primers are 71 to 90, and external minus-strand primers are 1465 to 1446. Internal plus-strand primers for the 16S rRNA sequence are 243 to 266, and minus-strand primers are 976 to 953. PCR conditions were as follows. For the 16S rRNA gene external primers for C. pneumoniae, the conditions were 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 48°C, and 1 min at 72°C, with termination for 5 min at 72°C. The conditions for the 16S rRNA internal primer were identical to those of the external primer except that the annealing conditions were 68°C for 30 s (product size, 710 bp). For PCR the ompA gene of C. pneumoniae, the external primer conditions were 5 min at 94°C and 35 cycles of 15 s at 94°C, 15 s at 56°C, and 15 s at 72°C, with termination for 7 min at 72°C. The internal primer conditions were identical except that the annealing temperature was set at 59°C instead of 56°C (product size, 340 bp).
For the detection of ompA RNA and DNA in patient samples, the following nested primer system was employed: external plus-strand primers with bp 26 to 43 with minus-strand primers with bp 567 to 548, internal plus-strand primers with bp 115 to 135, and minus-strand primers with bp 462 to 444 (32). PCR conditions were 35 cycles of 30 s at 95°C, 1 min at 52°C, and 1 min at 72°C for extension for both external and internal PCRs (ompA primer set 2,350 bp). Nested PCR products were electrophoresed in 1.8% agarose gels and visualized by ethidium bromide. All other conditions were as described above. PCR products from infected cultures were cloned by using a TA cloning kit (Invitrogen, Carlsbad, Calif.). Sequencing (DNA Sequencing Core Facility, MCP-HU) confirmed the identity of these products to be the ompA gene and 16S rRNA gene sequences.
Ultrastructural analysis.
Negative staining immunoelectron microscopy consisted of absorbing 5 to 10 μl of sample onto carbon-coated copper electron microscopy grids followed by rinsing with phosphate-buffered saline (PBS). The grids were blocked with 0.1% cold-water fish gelatin (Sigma) in PBS for 10 min prior to incubation for 15 to 30 min in the primary antibodies, anti-LPS (1:20), anti-OMP (1:5), and anti-SAF (1:100). The grids were reblocked in 0.1% cold-water fish gelatin prior to rinsing in PBS and subsequently incubated for 30 min in secondary antimouse antibody conjugated to 5 to 10 nm of colloidal gold particles at 1.0 mg/ml (Amersham). The grids were rinsed in double-distilled H2O prior to negative staining with 2.0% uranyl acetate. The grids were then examined at 80 kV on a Zeiss EM-10 electron microscope.
RESULTS
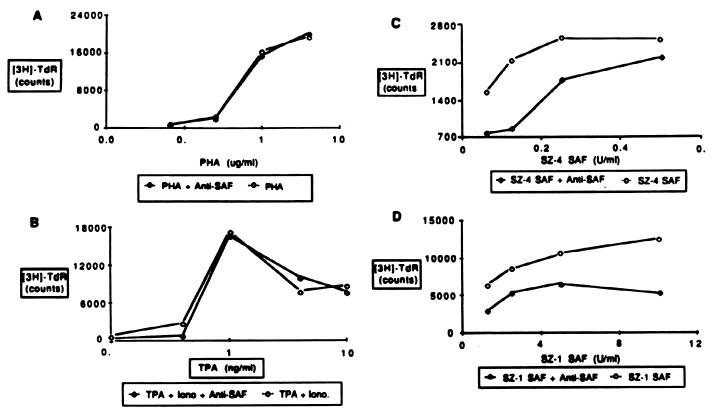
SAF is a 28- to 30-kDa protein with the ability to stimulate CTCL malignant cell growth (1). It has previously been found that SAF can be produced by cells within the PBMC of patients with Sézary syndrome and can be used to establish cell lines containing the predominant T-cell clone (2, 3). One of these lines (SZ-4) was shown to produce detectable amounts of SAF (2). MAbs reactive against both PBMC- and cell line-derived SAF were selected for their ability to neutralize SAF activity. The biological specificity of the selected MAb reactive with SAF is demonstrated in Fig. 1. Anti-SAF was tested against either PHA, TPA plus ionomycin stimulation, cell line-derived SAF, or PBMC-derived SAF (SZ-1). No inhibition of the PHA or TPA plus ionomycin response was observed, demonstrating that the antibody does not inhibit proliferation through toxicity or nonspecific cell surface interactions. However, the antibody inhibits about 50% of both the cell line- and PBMC-derived SAF-induced proliferation. These PBMC and cell lines were derived from patients with Sézary syndrome.
FIG. 1.
Inhibition of SAF bioactivity by anti-SAF. Twenty-five micrograms of anti-SAF (58.19) per milliliter was cultured with PBMC from a healthy donor along with various amounts of either PHA (A), TPA plus ionomycin (B), cell line-derived SAF (SZ-4 SAF) (C), or PBMC-derived SAF (SZ-1 SAF) (D) in the presence of 15 U of recombinant IL-2 per ml for 72 h. Cells were pulsed with tritiated thymidine ([3H]TdR) for the final 6 h and harvested and prepared for liquid scintillation counting. Data are means of triplicate cultures, and the standard error was less than 10% of the mean.
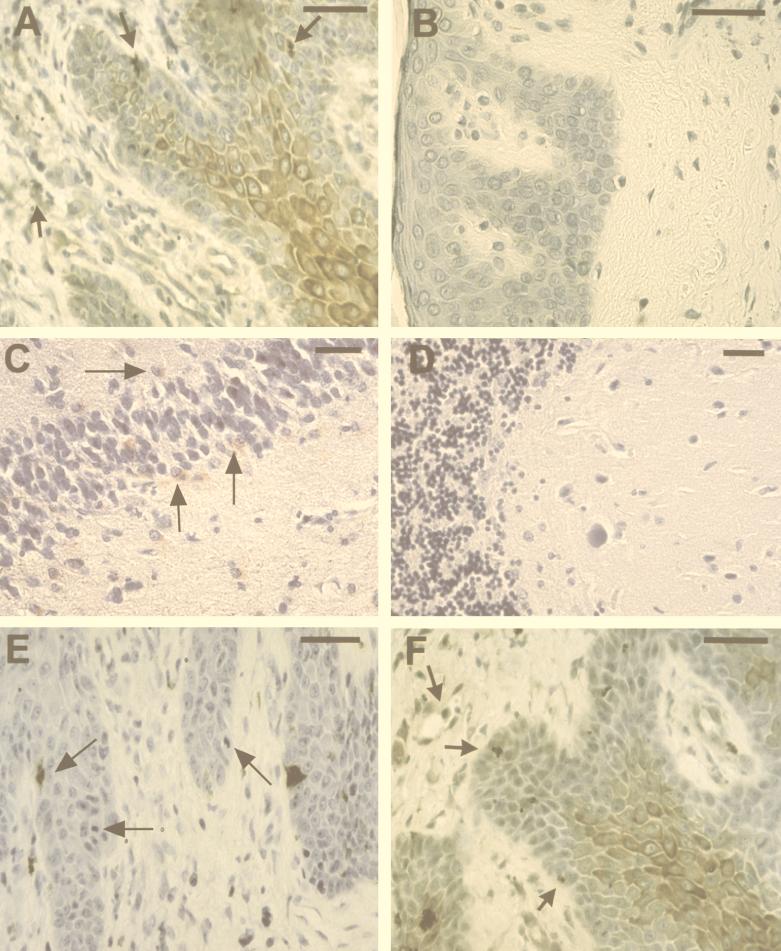
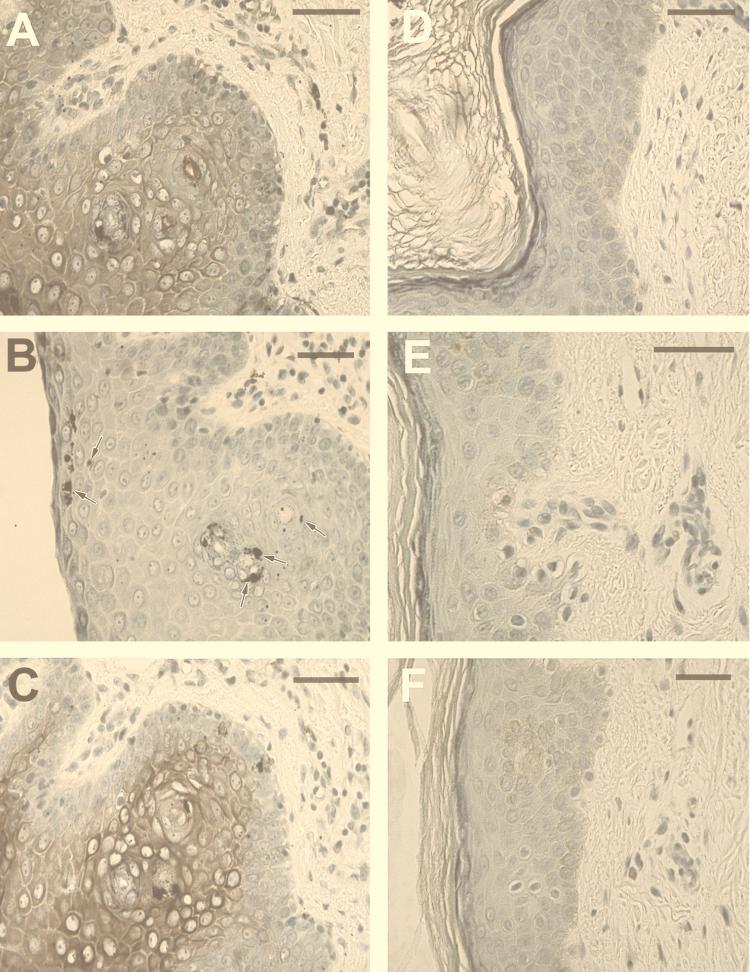
To determine whether SAF expression can be detected within the skin of patients with mycosis fungoides, the predominant form of CTCL, sections from formalin-fixed biopsy specimens obtained from lesions of patients with mycosis fungoides were labeled with anti-SAF (Fig. 2A and B). Sections from 16 of 27 patients were reactive with the anti-SAF antibody. Interestingly, reactivity was found in keratinocytes within the epidermis and in endothelial cells and histiocytes within the dermis, as illustrated by the micrograph in Fig. 2A. Sections from the remaining 11 patients showed little or no immunoreactivity (Fig. 2B), suggesting that the immunolabeling was specific.
FIG. 2.
Immunohistochemical analysis of anti-SAF, anti-C. pneumoniae OMP, and anti-Chlamydia LPS reactivity in formalin-fixed skin and brain tissue. Anti-SAF immunoreactivity is shown in panels A to D. (A) Immunolabeling of epidermal cells with the morphology of keratinocytes, as well as inclusion-like structures and perivascular cells (arrows). (B) A section from a mycosis fungoides patient found negative for immunolabeling (negative control). (C) Anti-SAF (67 μg/ml) reactivity on temporal cortex of a patient with AD confirmed positive for C. pneumoniae in other studies (positive control) (4). Immunolabeling of apparent glial cells in the denate gyrus is depicted by arrows. (D) Negative immunoreactivity of anti-SAF is shown on a section from the cerebellum (uninvolved) of the same patient (negative control). (E) Immunolabeling of inclusion-like structures with anti-OMP MAb (1:10) (arrows) of a section from a patient with mycosis fungoides. (F) Immunoreactivity of anti-LPS (1:10) of cells with the morphology of keratinocytes as well as labeling of inclusion-like structures and perivascular cells (arrows). Diaminobenzidine-cobalt (dark brown) chromagen was used for panels A, B, E, and F. 3-Amino-9-ethylcarbazole (red) chromagen was used for panels C and D. Bars = 50 μm.
We considered whether SAF may be a C. pneumoniae-associated protein, since one of the antigenic proteins produced by C. pneumoniae is 30 kDa (17, 18), a molecular mass similar to that of SAF. As a positive control, we used our anti-SAF MAb to perform immunohistochemistry on an OMP- and LPS-positive sample from a brain from an AD patient and found it to be positive for SAF (Fig. 2C). In contrast, a section from the cerebellum of another patient with AD, which did not demonstrate C. pneumoniae infection, was found to be negative for reactivity with the anti-SAF MAb (Fig. 2D). These data compelled us to test whether SAF reactivity observed in the skin is predictive of OMP and LPS reactivity in this tissue. Immunocytochemistry was performed on sections made from the same biopsy specimens used in the anti-SAF experiments by using commercially available MAbs specific for a C. pneumoniae OMP and chlamydial LPS. The anti-OMP reactivity in consecutive sections was highly localized, appearing to specifically stain inclusions within keratinocytes and histiocytes (Fig. 2E). In contrast, a similar reactivity pattern of both diffuse and highly localized staining (Fig. 2A and F) was observed for the anti-LPS (Fig. 2F) in a manner similar to that for the anti-SAF (Fig. 2A). As the photomicrograph depicts, keratinocytes, endothelial cells, and histiocytes demonstrate both diffuse and intense focal immunoreactivity. Interestingly, anti-OMP staining also demonstrated diffuse immunoreactivity in some samples obtained from patients with more advanced disease (data not shown). In a series of 27 specimens tested, immunoreactivity was found in approximately 60% of the specimens with 90% concordance between reactivity of the anti-SAF, anti-LPS, and anti-OMP MAbs; thus, SAF appears to immunolabel C. pneumoniae bacteria.
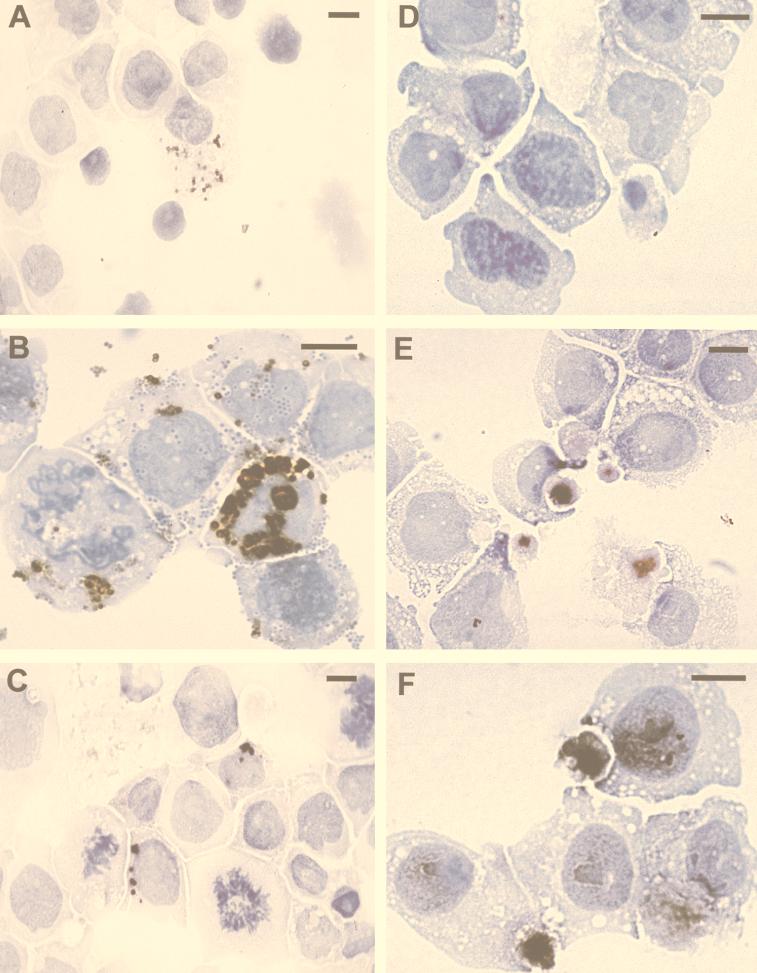
To test directly whether SAF is expressed by C. pneumoniae, we compared the expression of the anti-OMP, anti-LPS, and anti-SAF antibodies on MØ cell line cultures (THP-1) infected with either the laboratory strain of C. pneumoniae (TW-183) or C. pneumoniae isolated from the brains of patients who had suffered from AD (4). The identity of the associated AD bacterium as C. pneumoniae was confirmed by PCR, RT-PCR, and ultrastructural and culture analyses (4). Immunohistochemistry demonstrated that 7-day cultures infected with AD-isolated organisms showed a similar reactivity pattern. Reactivity of the anti-OMP (Fig. 3A), anti-SAF (Fig. 3B), anti-LPS (Fig. 3C), and anti-SAF on uninfected cells (Fig. 3D) clearly demonstrated that these antibodies reacted with the bacteria infecting these cells. The anti-SAF reactivity was specific for organisms that appeared to be the metabolically active larger reticulate bodies but not for the infectious quiescent elementary bodies. Anti-SAF clearly reacted with bacteria but did not show any of the diffuse staining observed in the tissues. In contrast, the THP-1 cell line infected with the TW-183 strain of these bacteria showed a more diffuse staining with the anti-OMP (Fig. 3E) and anti-SAF (Fig. 3F) MAbs on cells infected for 11 days. The anti-LPS had both diffuse and more focused staining (data not shown). These staining patterns presumably represent differences in the antigenic profiles between these two isolates of C. pneumoniae and demonstrate differential labeling for more persistent infections (i.e., 7 days versus 11 days). These results suggest that SAF is a protein directly associated with C. pneumoniae infection.
FIG. 3.
Immunolabeling of THP-1 monocyte cell line infected with C. pneumoniae isolated from a brain from a patient with AD and with the laboratory strain of C. pneumoniae (TW-183). Cultured THP-1 cells were placed on slides by a cytospin and fixed with STF. THP-1 cells infected with C. pneumoniae isolated from the brain of a patient with AD are shown in panels A to C: immunolabeling of anti-OMP (A), anti-SAF (B), and anti-LPS (C) of bacterial inclusions within infected cells. (D) Uninfected THP-1 cells that reacted with anti-SAF (negative control). THP-1 cells infected for 10 days with the laboratory strain of C. pneumoniae TW-183 immunolabeled with anti-OMP (E) or anti-SAF (F) contain diffusely stained inclusions. Bars = 10 μm.
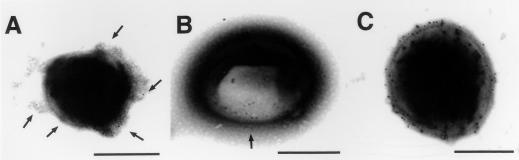
To determine if SAF is localized on C. pneumoniae, immunoelectron microscopy was performed. C. pneumoniae was reacted with anti-SAF or anti-OMP followed by 5 nm of gold-conjugated goat antimouse antibodies. Immunoelectron microscopy confirms that anti-SAF (Fig. 4A), anti-OMP (Fig. 4B), and anti-LPS (Fig. 4C) specifically label C. pneumoniae.
FIG. 4.
Negative staining combined with immunoelectron microscopy of C. pneumoniae. Pellets from culture supernatants of THP-1 cells infected with C. pneumoniae isolated from a patient with AD and cultured for 7 days were absorbed onto carbon-coated copper electron microscopy grids. Grids were exposed to the primary antibodies, incubated, and subsequently incubated with secondary antimouse antibody conjugated to 5 nm colloidal gold particles. (A) Immunogold particles are bound to the anti-SAF-labeled bacterium, examples of which are depicted with arrows. (B) An area of a bacterium apparently rich in OMP (arrow) is exposed and labeled with gold particles. (C) A bacterium reacted with anti-LPS demonstrates heavy labeling with many gold particles bound to its surface. Bars = 0.75 μm (A) and 0.5 μm (B and C).
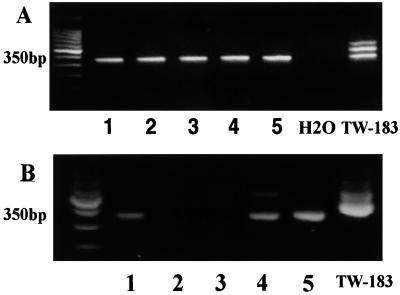
If the detection of C. pneumoniae antigens in skin lesions of CTCL patients is a result of C. pneumoniae infection, we should be able to detect C. pneumoniae-specific DNA sequences by PCR in lesions of CTCL patients as well. DNA was isolated from skin samples of five patients with mycosis fungoides. PCR was performed with our nested ompA-specific C. pneumoniae-specific PCR primer pair (see Materials and Methods). PCR was found positive for all five of these preparations (Fig. 5A). The PCR pattern with the positive control DNA contained some additional bands of unknown origin that were not found in CTCL DNA preparations. These data suggest that our results were not a spurious outcome of DNA contamination from the positive control extractions, a conclusion also supported by water controls that were consistently negative. The PCR data, therefore, are consistent with the immunohistochemical data. If SAF expression is associated with an active C. pneumoniae infection, then C. pneumoniae RNA encoding the ompA gene product should be detectable in at least some patients with CTCL. RNA was isolated from lymph node biopsies from five patients, two with mycosis fungoides and three with Sézary syndrome. RT-PCR was performed with the ompA-specific primers on the DNase-treated RNA. The appropriate 350-bp product was detected in three of five samples, one with mycosis fungoides and two with Sézary syndrome (Fig. 5B). These data suggest that ompA is actively transcribed in certain patients with CTCL. To ensure that these PCR products contain C. pneumoniae-related sequences, we isolated, cloned, and sequenced the 350-bp product and found it to be identical to the predicted sequence, as previously described (32). These results were consistent with our PCR and immunohistochemical data, indicating that at least some patients with CTCL are infected with transcriptionally active C. pneumoniae.
FIG. 5.
C. pneumoniae-specific PCR and RT-PCR results from patients with CTCL. A nested primer set specific for the ompA gene of C. pneumoniae was used. (A) DNA extracted from the skin of five patients with mycosis fungoides (lanes 1 to 5) and subjected to PCR (ompA primer set 2; see Materials and Methods) shows the predicted 350-bp product. The PCR containing only water is negative, while the PCR product with DNA from the positive control TW-183 shows three bands, one at 350 bp. (B) RNA extracted from lymph nodes of patients with tumor stage mycosis fungoides (lanes 1 and 2) or with Sézary syndrome (lanes 3 to 5) were DNase treated, and then 1 to 2 μg of RNA was exposed to MMLV reverse transcriptase. Three microliters of the RT product was subjected to PCR with the same ompA-specific primers as those used for DNA PCR. Positive control DNA from TW-183 has a band at 350 bp as well as two bands at higher molecular masses. The no RT control was negative for all samples.
To initially evaluate the relationship between active disease and the expression of SAF, OMP, and LPS, biopsy specimens from four patients were obtained pre- and post-PUVA treatment from sites in close proximity to one another. Immunocytochemistry was performed with the anti-LPS, anti-SAF, and anti-OMP MAbs on these identically processed specimens. Immunoreactivity for all of these antigens was greatly diminished after therapy. Figure 6 demonstrates pretreatment and posttreatment results from one of these patients. Anti-SAF, anti-OMP, and anti-LPS reactivities are also depicted in Fig. 6. These micrographs clearly demonstrate that the staining in these and the other pretreatment specimens (Fig. 2) is specific and indicate that SAF and the expression of other C. pneumoniae RNA-encoded gene products are associated with active disease.
FIG. 6.
Effect of PUVA therapy on C. pneumoniae antigen expression. Immunolabeling of formalin-fixed sections from a biopsy sample taken from the same patient before and after PUVA therapy. Sections from biopsy specimens obtained before treatment are shown in panels A to C, while sections from biopsy specimens obtained after PUVA treatment are shown in panels D to F. Anti-SAF labeling of epidermal and dermal cells in a diffuse and highly localized pattern is observed in panel A, while little or no labeling is observed in panel D. Anti-OMP immunolabeling of epidermal cells in a highly localized pattern is found in panel B (arrows), while no labeling is observed in the posttreated section (E). Intense diffuse and localized staining is observed in the anti-LPS-reacted section from the pretreated biopsy (C), while virtually no immunoreactivity is observed in the LPS-reacted section obtained posttreatment (F). Sections were stained simultaneously. Bars = 50 μm.
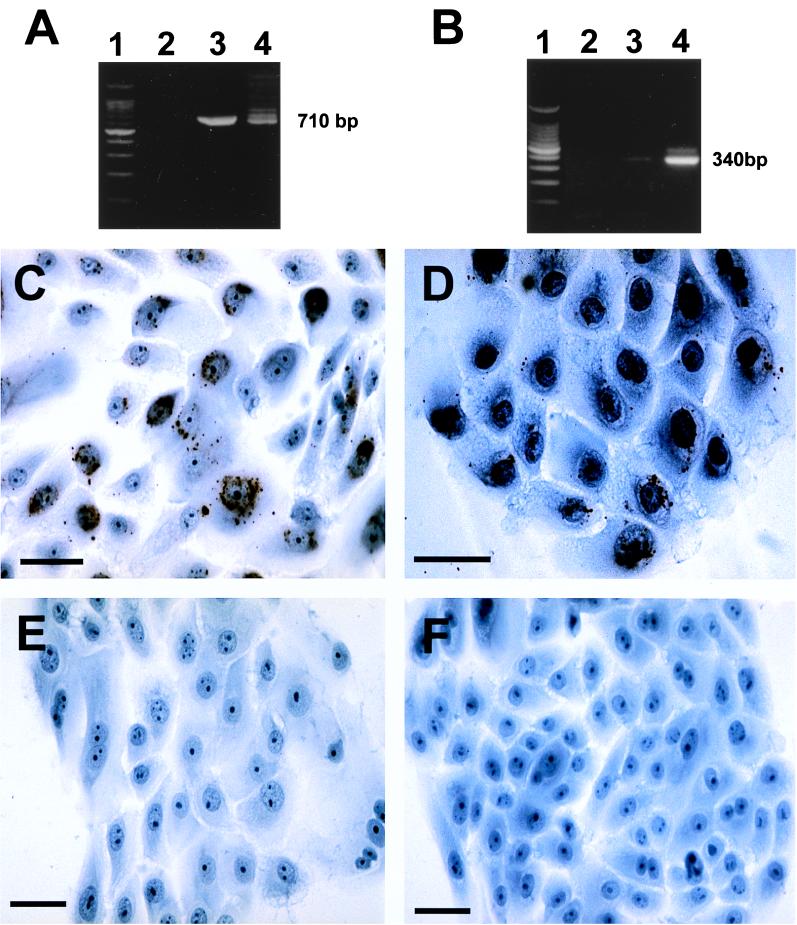
Infection of C. pneumoniae in the epidermis has not been previously described. Thus, immunostaining of keratinocytes from CTCL patients with the anti-SAF- and anti-OMP-specific MAbs was intriguing. Although the data presented here are consistent with keratinocyte infection, we wanted to determine to what extent we could experimentally demonstrate infection of keratinocytes with the laboratory strain of C. pneumoniae. Normal human keratinocytes grown in T-25 flasks or in chamber slides were incubated for 3 days with 200 IFU50 per cm2 of TW-183. T-25 flasks were harvested, and DNA and RNA were isolated and subjected to PCR or RT-PCR analysis with primers independent from those used on the biopsied samples to diminish the possibility of contamination. The chamber slides were fixed with STF and then stained with anti-OMP or LPS, as before. We observed that keratinocytes can be productively infected with C. pneumoniae. RT-PCR results using 16S rRNA and ompA primers were found positive (Fig. 7A and B), and LPS (Fig. 7C)- and OMP (Fig. 7D)-positive organisms were found within the cultured keratinocytes with immunocytochemistry. These data indicate that keratinocytes in vitro can be infected with C. pneumoniae, thereby supporting our observation of the immunolabeling of infected keratinocytes in the epidermis from CTCL patients in situ.
FIG. 7.
Infection of cultured keratinocytes with C. pneumoniae TW-183. Normal human neonatal keratinocytes were infected with 5,000 IFU50 and cultured for 3 days in T-25 flasks. RNA was extracted followed by DNase treatment, and then cDNA was prepared with 1 to 2 μg of RNA by using MMLV reverse transcriptase. cDNA was subjected to PCR with primers annealing with DNA of the 16S rRNA gene of C. pneumoniae or the ompA gene (ompA primer set 1; see Materials and Methods). (A) Lane 1, DNA ladder; lane 2, cDNA from uninfected cells; lane 3, PCR product for 16S rRNA gene from infected cells; lane 4, DNA from TW-183 (positive control). (B) Lane 1, DNA ladder; lane 2, cDNA from the uninfected culture; lane 3, PCR product for the ompA gene from the infected culture (light 340-bp band); lane 4, PCR product for ompA gene from TW-183. Keratinocytes in chamber slides were infected simultaneously with the T-25 flasks with an equal concentration of C. pneumoniae (200 IFU50 per cm2) (C and D), and uninfected cultures were established (E and F). After 3 days, cells were washed and fixed with STF and then reacted with anti-LPS (C and E) or anti-OMP (D and F). Bars = 50 μm (C to F).
DISCUSSION
Cell lines of malignant Sézary cells generated by using a combination of SAF and irradiated allogenic PBMC suggested that SAF may be an autocrine growth factor for these cells (3). Since our previous studies were restricted to patients with Sézary syndrome, we wanted to determine whether SAF could be detected within the skin of patients suffering from mycosis fungoides, the predominant subset of CTCL. While our results indicating that SAF is present in the skin of patients with CTCL were striking, we were surprised that the SAF-positive cells did not include infiltrating lymphocytes. Instead, we detected SAF staining of endothelial cells, histiocytes, and keratinocytes. These data suggest that SAF was present in vivo and found in cells resident to the skin rather than in the infiltrating lymphocytes. The identity of the SAF-positive cells within the skin and the ability of SAF to stimulate malignant cell growth suggests that SAF could directly contribute to the pathogenesis of CTCL and potentially help explain the epidermotropism of this disease.
The decision to examine C. pneumoniae as the biologic source of SAF came about following the determination that SAF appeared to have a cell-associated form and that this form of SAF was found to be complexed with RNA and DNA in the cytoplasm. These observations indicated that SAF could be associated with a large macromolecular complex. Originally, anti-SAF was found to be reactive intracellularly with the SZ-4 cell line and was thought to be associated with a retrotransposon, but we could not find evidence to support this. For reasons not established at that point, the cell line over time lost its immunoreactivity and ability to produce soluble SAF. We now consider the use of antibiotics during routine passaging of the cultures as the reason for loss of SAF activity. Results from our recently reported experiments, in which C. pneumoniae was identified and cultured from patients with AD (4), led us to investigate whether SAF could be associated with this bacterium, as several features of C. pneumoniae lend themselves to serious consideration of such. These include its intracellular location (30), its production of a 30-kDa protein with T-cell stimulatory activity (17, 18), and its ability to infect a variety of cell types, including cell lines derived from macrophages (U937) (36) and from larynx epithelium (Hep-2) (7). Initially, anti-SAF reactivity was examined in a site previously established as C. pneumoniae positive, that being in sections from the brains of patients who had suffered from AD (4). Results in this study showed SAF staining within glial cells, in a fashion similar to that observed for LPS in the same brain (4). These results mandated the further study of the skin of patients with CTCL for C. pneumoniae antigens.
Anti-OMP immunoreactivity was found in approximately 60% of the patients tested, and overall there was nearly 90% concordance between anti-SAF, LPS, and anti-OMP staining. Negative results may relate to treatment received before evaluation or to disease unrelated to the observed infection. OMP-specific primers were found to amplify a 350-bp C. pneumoniae amplimer in both DNA and RNA preparations, supporting the immunohistochemical results. These data indicate that C. pneumoniae was present in the skin of many patients with CTCL. However, the question remained whether SAF was a chlamydial antigen or induced by C. pneumoniae infection. To address this issue, we infected THP-1 monocyte cells with C. pneumoniae isolates and immunolabeled them with antichlamydial reagents and anti-SAF. Striking results indicated clearly that SAF immunoreactivity was associated with the AD-isolated bacterium in these cultures and was expressed in inclusions in a diffuse pattern in TW-183-infected cultures. The small bodies that were not reactive with either antibody in the AD isolate appear to represent the infectious elementary bodies or may be a persistent form of the bacteria that has been observed in other disorders (27). To further confirm these studies, ultrastructural analysis by using a combination of negative staining and immunoelectron microscopy indicated that anti-SAF labeled the AD-isolated bacterium.
If SAF is a chlamydial protein, then protein extracts from the bacteria should contain specific bands reactive with the anti-SAF MAb. Immunoblotting with the anti-OMP and anti-SAF MAbs was performed (data not shown) and indicated that anti-SAF reacted to the predicted 30-kDa SAF band. In addition, a few other higher-molecular-mass bands showed some reactivity with anti-SAF. However, because the identity of chlamydial antigens is still not complete, we cannot fully interpret whether these bands represent forms of the SAF or cross-reactivity to homologous proteins. It is clear that anti-SAF, an antibody raised against a soluble factor, is reacting with protein bands made from lysates containing C. pneumoniae. Further experiments, however, are required to clarify this issue.
An association between CTCL and an infection of cells resident to the epidermis is an attractive hypothesis. The fact that C. pneumoniae expresses heat shock proteins, LPS, and the SAF determinant indicates that this bacterium certainly could induce a potent immune response resulting in lymphocytic infiltration of the skin. CTCL is a disease with a high degree of complexity whose pathogenesis is assuredly a result of multiple interactions. These include the production of cytokines and chemokines by host skin cells, infiltrating nonmalignant cells, and infiltrating malignant clonal T cells, in addition to the status of putative stimuli initiating the inflammatory process (10). Although the Th1/Th2 balance has not been directly studied in chlamydial responses, the induction of proinflammatory cytokines, such as tumor necrosis factor alpha, IL-1β, IL-6, IFN-γ, and IFN-α, suggests that a Th1 response would normally predominate (18, 26, 39). However, this may not be the case in elderly individuals or in the skin of patients with CTCL, and there is evidence indicating that chlamydial infections can also induce a Th2 response (17). The Th2-like cytokine pattern observed in CTCL lesions (43) may create a permissive environment for C. pneumoniae infection. This concept is supported by studies demonstrating that the skin of aged mice has been shown to be highly susceptible to infection by intracellular pathogens (41). Although data in reactive arthritis generally support a Th1 response, Th2 activation in chlamydial immunity also is supported by production of IL-4 and IL-10 by chlamydia-specific T-cell clones (39). In fact, detection of IL-10 is considered a sign of chronic C. pneumoniae infection (26). Likewise, high concentrations of IFN-γ and IFN-α inhibit growth but promote persistence of chlamydial infections purportedly by a lessening of exogenous tryptophan (38), thereby reducing the apparent infection. Thus, the relationship between increased CD8+ cells and improved prognosis in CTCL might be explained by their IFN production rather than solely by the proposed increase in an anti-tumor cell-mediated response (45). Furthermore, the therapeutic value of IFN-α in CTCL (25) could be attributed to the bacteriostatic effects of this cytokine (38).
Considering recent evidence demonstrating that C. pneumoniae is associated with inflammatory diseases throughout the body, such as reactive arthritis (15), atherosclerosis (8), and AD (4), it is understandable how this bacterium also could be found in the skin of patients with CTCL. Interestingly, the related species C. trachomatis has been shown to cause lymphomatus granulosum venereum, a disorder involving lymphoproliferation (6) and shown to become systemic through infection of monocytes. Studies have not yet been conducted to examine whether C. pneumoniae infection of the skin is common. In addition, under the correct permissive circumstances, many organisms that commonly infect humans can cause serious illness, for example, group A streptococcus and necrotizing fascitis (31). C. pneumoniae could reasonably contribute to the heterogeneity of inflammatory cells that infiltrate CTCL lesions (10, 45). In addition, this bacterium could induce the immunoregulatory events that occur during CTCL (3, 19) and the histologic changes that include the expression of HLA-DR (46), consistent with a bacterial infection (34). We and others (24, 42) hypothesize that chronic epidermal stimulation leads to clonal T-cell expansion, followed by conversion of that clone to a malignant entity that can potentially lead to tumor development and, in some cases, death of the patient.
In this report, we have shown that our biologically and biochemically characterized stimulatory factor SAF is present on C. pneumoniae organisms, that SAF and C. pneumoniae antigens are present in lesions of certain CTCL patients, and that DNA and RNA of C. pneumoniae can be detected in those CTCL patients. We also demonstrate that following effective PUVA therapy, SAF and other C. pneumoniae antigens are greatly reduced within the skin, perhaps as a result of UV damage to the C. pneumoniae bacteria. Furthermore, experiments in vitro demonstrated that keratinocytes were capable of being infected with C. pneumoniae, supporting the morphologic identification of the infected cells in situ. It is premature to claim any casual and/or risk factor relationship between the observed infection and the pathogenesis of CTCL; however, this possibility should be considered based on the fact that SAF is a C. pneumoniae-associated determinant.
ACKNOWLEDGMENTS
We thank the Departments of Dermatology and Pathology and Laboratory Medicine for supporting this work and the Foundation for Research into Diseases of Aging and PCOM for supporting the publication of this article.
We dedicate this work to all the individuals suffering from CTCL. We thank Subrata K. Ghosh for his support and helpful comments throughout this project.
REFERENCES
- 1.Abrams J T, Ghosh S K, DeFreitas E. Sezary T-cell-activating factor induces functional interleukin 2 receptors on T cells derived from patients with Sezary syndrome. Cancer Res. 1993;53:5501–5506. [PubMed] [Google Scholar]
- 2.Abrams J T, Lessin S, Ghosh S K, Ju W D, Vonderheid E C, Nowell P C, Murphy G, Elfenbein B, DeFreitas E. A clonal CD4 positive T cell line established from the blood of a patient with Sézary syndrome. J Investig Dermatol. 1991;96:31–37. doi: 10.1111/1523-1747.ep12514693. [DOI] [PubMed] [Google Scholar]
- 3.Abrams J T, Lessin S, Ghosh S K, Nowell P C, Ju W, Vonderheid E C, Rook A, DeFreitas E. Malignant and non-malignant T cell lines from human T cell lymphotropic virus type I-negative patients with Sézary syndrome. J Immunol. 1991;146:1455–1462. [PubMed] [Google Scholar]
- 4.Balin B J, Gérard H C, Arking E J, Appelt D M, Branigan P J, Abrams J T, Whittum-Hudson J A, Hudson A P. Identification and localization of Chlamydia pneumoniae in the Alzheimer’s brain. Med Microbiol Immunol. 1998;187:23–42. doi: 10.1007/s004300050071. [DOI] [PubMed] [Google Scholar]
- 5.Beatty W L, Byrne G I, Morrison R P. Morphologic and antigenic characterization of interferon g-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci USA. 1989;90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgoyne R A. Lymphogranuloma venereum. Prim Care. 1990;17:153–157. [PubMed] [Google Scholar]
- 7.Campbell J F, Barnes R C, Kozarsky P E, Spika J S. Culture-confirmed pneumonia due to Chlamydia pneumoniae. J Infect Dis. 1991;164:411–413. doi: 10.1093/infdis/164.2.411. [DOI] [PubMed] [Google Scholar]
- 8.Campbell L A, O’Brien E R, Cappuccia A L, Kuo C C, Wang S P, Stewart D, Patton D L, Cummings P K, Grayston J T. Detection of Chlamydia pneumoniae TWAR in human coronary atherectomy tissues. J Infect Dis. 1995;172:585–588. doi: 10.1093/infdis/172.2.585. [DOI] [PubMed] [Google Scholar]
- 9.Edelson R L. Pathogenesis of T-cell lymphoma of skin. J Am Acad Dermatol. 1983;9:957–960. doi: 10.1016/s0190-9622(83)80083-8. [DOI] [PubMed] [Google Scholar]
- 10.Fermand J P, Mitjavila M T, Le Couedic J P, Tsapis A, Berger R, Modigliani R, Seligmann M, Brouet J C, Vainchenker W. Role of granulocyte-macrophage colony-stimulating factor, interleukin-3 and interleukin-5 in the eosinophilia associated with T cell lymphoma. Br J Haematol. 1993;83:359–364. doi: 10.1111/j.1365-2141.1993.tb04657.x. [DOI] [PubMed] [Google Scholar]
- 11.Fivenson D P, Hanson C A, Nickoloff B J. Localization of clonal T cells to the epidermis in cutaneous T-cell lymphoma. J Am Acad Dermatol. 1994;31:717–723. doi: 10.1016/s0190-9622(94)70231-4. [DOI] [PubMed] [Google Scholar]
- 12.Gaydos C A, Palmer L, Quinn T C, Falkow S, Brooks G F, Eiden J J. Phylogenetic relationship of Chlamydia pneumoniae to Chlamydia. Int J Syst Bacteriol. 1993;43:610–612. doi: 10.1099/00207713-43-3-610. [DOI] [PubMed] [Google Scholar]
- 13.Gazdar A F, Carney D N, Russel E K, Schechter G P, Bunn P A., Jr In vitro growth of cutaneous T-cell lymphoma. Cancer Treat Rep. 1979;63:587–590. [PubMed] [Google Scholar]
- 14.Golstein M M, Farnarier-Seidel C, Daubney P, Kaplanski S. An OKT4+ T-cell population in Sezary syndrome: attempts to elucidate its lack of proliferative capacity and its suppressive effect. Scand J Immunol. 1986;23:53–64. doi: 10.1111/j.1365-3083.1986.tb01942.x. [DOI] [PubMed] [Google Scholar]
- 15.Gran J T, Hjetland R, Andreassen A H. Pneumonia, myocardidis and reactive arthritis due to Chlamydia pneumoniae. Scand J Rheumatol. 1993;22:43–44. doi: 10.3109/03009749309095111. [DOI] [PubMed] [Google Scholar]
- 16.Grayston J T. Chlamydia pneumoniae, strain TWAR pneumonia. Annu Rev Med. 1992;43:317–323. doi: 10.1146/annurev.me.43.020192.001533. [DOI] [PubMed] [Google Scholar]
- 17.Halme S, Saikku P, Surcel H M. Characterization of Chlamydia pneumoniae antigens using human T cell lines. Scand J Immunol. 1997;45:378–384. doi: 10.1046/j.1365-3083.1997.d01-413.x. [DOI] [PubMed] [Google Scholar]
- 18.Halme S, Surcel H M. Cell mediated immunity to Chlamydia pneumoniae. Scand J Infect Dis Suppl. 1997;104:18–21. [PubMed] [Google Scholar]
- 19.Hanson E R. Immunoregulatory events in the skin or patients with cutaneous T cell lymphoma. Arch Dermatol. 1996;132:554–561. [PubMed] [Google Scholar]
- 20.Haynes B F, Bunn P, Mann D, Thomas C, Eisenbarth G S, Minna J, Fauci A S. Cell surface differentiation antigens of the malignant T cell in Sézary syndrome and mycosis fungoides. J Clin Investig. 1986;67:523–530. doi: 10.1172/JCI110062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu S M, Raine L, Fanger H. The use of antiavidin antibody and avidin-biotin-peroxidase complex in immunoperoxidase techniques. Am J Clin Pathol. 1981;75:816–821. doi: 10.1093/ajcp/75.6.816. [DOI] [PubMed] [Google Scholar]
- 22.Hunt S J, Charley M R, Jegasothy B V. Cutaneous T-cell lymphoma: utility of antibodies to the variable regions of the human T-cell antigen receptor. J Am Acad Dermatol. 1992;26:5552–5558. doi: 10.1016/0190-9622(92)70079-u. [DOI] [PubMed] [Google Scholar]
- 23.Hyman C L, Roblin P M, Gaydos C A, Quinn T C, Schachter J, Hammerschlag M R. Prevalence of asymptomatic nasopharyngeal carriage on Chlamydia pneumoniae in subjectivity healthy adults: assessment by polymerase chain reaction-enzyme immunoassay and culture. Clin Infect Dis. 1995;20:1174–1178. doi: 10.1093/clinids/20.5.1174. [DOI] [PubMed] [Google Scholar]
- 24.Jackow C M, Cather J C, Hearne V, Asano A T, Musser J M, Duvic M. Association of erythrodermic cutaneous T-cell lymphoma, superantigen-positive Staphylococcus aureus, and oligoclonal T-cell receptor V beta gene expansion. Blood. 1997;89:32–40. [PubMed] [Google Scholar]
- 25.Jorg B, Kerl H, Thiers B H, Brocker E B, Burg G. Therapeutic approaches in cutaneous lymphoma. Dermatol Clin. 1994;12:433–441. [PubMed] [Google Scholar]
- 26.Kaukoranta-Tolvanene S S, Teppo A M, Laitinen K, Saikku P, Linnavuori K, Leinonen M. Growth of Chlamydia pneumoniae in cultured peripheral blood mononuclear cells and induction of a cytokine response. Microb Pathog. 1996;21:215–221. doi: 10.1006/mpat.1996.0056. [DOI] [PubMed] [Google Scholar]
- 27.Koehler L, Nettelnbreker E, Hudson A P, Ott N, Gerard H C, Branigan P J, Schumacher H R, Drommer W, Zeidler H. Ultrastructural and molecular analysis of the persistence of Chlamydia trachomatis (serovar K) in human monocytes. Microb Pathog. 1997;22:133–142. doi: 10.1006/mpat.1996.0103. [DOI] [PubMed] [Google Scholar]
- 28.Koskiniemi M, Gancay M, Salonen O, Puolakkainen M, Fakkila M, Saikku P, Vaheri A. Chlamydia pneumoniae associated with central nervous system infections. Eur Neurol. 1996;36:160–163. doi: 10.1159/000117235. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Leinonen M. Pathogenetic mechanisms and epidemiology of Chlamydia pneumoniae. Eur Heart J. 1993;14(Suppl. K):57–61. [PubMed] [Google Scholar]
- 31.Lorber B. Are all diseases infectious? Ann Intern Med. 1996;125:844–851. doi: 10.7326/0003-4819-125-10-199611150-00010. [DOI] [PubMed] [Google Scholar]
- 32.Melgosa-Perez P M, Kuo C C, Campbell L A. Sequence analysis of the major out membrane protein gene of Chlamydia pneumoniae. Infect Immun. 1991;59:2195–2199. doi: 10.1128/iai.59.6.2195-2199.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mielke V, Staib G, Boehncke W H, Duller B, Sterry W. Clonal disease in early cutaneous T-cell lymphoma. Dermatol Clin. 1994;12:351–360. [PubMed] [Google Scholar]
- 34.Nickoloff B J, Mitra R S, Green J, Shimizu Y, Thompson C, Turka L A. Activated keratinocytes present bacterial-derived superantigens to T lymphocytes: relevance to psoriasis. J Dermatol Sci. 1993;6:127–133. doi: 10.1016/0923-1811(93)90002-7. [DOI] [PubMed] [Google Scholar]
- 35.Nolte F S, Thurmond C, Mitchell P S. Isolation of hepatitus C virus RNA from serum for reverse transcription-PCR. J Clin Microbiol. 1994;32:519–520. doi: 10.1128/jcm.32.2.519-520.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Numazaki K, Suzuki K, Chiba S. Replication of Chlamydia trachomatis and C. pneumoniae in the human monocytic cell line U-937. J Med Microbiol. 1995;42:191–195. doi: 10.1099/00222615-42-3-191. [DOI] [PubMed] [Google Scholar]
- 37.Sarris A H, Esgleyes-Ribot T, Crow M, Broxmeyer H E, Karasavvas N, Pugh W, Grossman D, Deisseroth A, Duvic M. Cytokine loops involving interferon-gamma and IP-10, a cytokine chemotactic for CD4+ lymphocytes: an explanation for the epidermotropism of cutaneous T-cell lymphoma? Blood. 1995;86:651–658. [PubMed] [Google Scholar]
- 38.Shener Y, Sarov I. Inhibition of growth by human gamma interferon. Curr Microbiol. 1985;16:9–13. doi: 10.1128/iai.48.2.592-596.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon A K, Seipelt E, Wu P, Wenzel B, Braun J, Seiper J. Analysis of cytokine profiles in synovial T cell clones from chlamydial reactive arthritis patients: predominance of the Th1 subset. Clin Exp Immunol. 1993;94:122–126. doi: 10.1111/j.1365-2249.1993.tb05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sterry W, Mielke V. CD4+ cutaneous T-cell lymphoma show the phenotype of helper/inducer T cells (CD45RA−, Cdw29+) J Investig Dermatol. 1989;93:413–416. [PubMed] [Google Scholar]
- 41.Sunderkotter C, Kalden H, Luger T A. Aging and the skin immune system. Arch Dermatol. 1997;133:1256–1262. doi: 10.1001/archderm.1997.03890460078009. [DOI] [PubMed] [Google Scholar]
- 42.Tan R S H, Butterworth C M, Mclaughlin H, Malka S, Samman P D. Mycosis fungoides—a disease of antigen persistence. Br J Dermatol. 1974;91:607–616. doi: 10.1111/j.1365-2133.1974.tb12449.x. [DOI] [PubMed] [Google Scholar]
- 43.Vowels B R, Cassin M, Vonderheid E C, Rook A H. Aberrant cytokine production by Sezary syndrome patients: cytokine secretion pattern resembles murine Th2 cells. J Investig Dermatol. 1992;99:90–94. doi: 10.1111/1523-1747.ep12611877. [DOI] [PubMed] [Google Scholar]
- 44.Whittum-Hudson J A, Taylor H R, Farazdaghi M, Prendergast R A. Changes in conjunctival lymphocyte populations induced by oral immunization with Chlamydia trachomatis. Curr Eye Res. 1986;5:973–979. doi: 10.3109/02713688608995179. [DOI] [PubMed] [Google Scholar]
- 45.Wood G S, Edinger A, Hoppe R T, Warnke R A. Mycosis fungoides skin lesions contain CD8+ tumor-infiltrating lymphocytes expressing an activated, MHC-restricted cytotoxic T-lymphocyte phenotype. J Cutan Pathol. 1994;21:151–156. doi: 10.1111/j.1600-0560.1994.tb00250.x. [DOI] [PubMed] [Google Scholar]
- 46.Wood G S, Michie S A, Durden F, Hoppe T R, Wanke R A. Expression of class II major histocompatibility antigen by keratinocytes in cutaneous T cell lymphoma. Int J Dermatol. 1994;33:346–350. doi: 10.1111/j.1365-4362.1994.tb01066.x. [DOI] [PubMed] [Google Scholar]