Abstract
This study was conducted to produce a recombinant species-specific oocyst wall protein of Cryptosporidium parvum. Antigens unique to C. parvum were identified by gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting of oocyst proteins from several different Cryptosporidium species. Antiserum was then prepared against a 41-kDa antigen unique to C. parvum and used to identify a recombinant DNA clone, designated rCP41. Expression of CP41 mRNA in C. parvum oocysts was confirmed by reverse transcriptase PCR (RT-PCR). Although the CP41 sequence was shown by PCR to be present in the genome of C. baileyi, CP41 mRNA was not detected in this species by RT-PCR. Immunofluorescence staining with antiserum against recombinant CP41 detected native CP41 antigen on the surface of C. parvum oocysts but failed to detect CP41 on C. baileyi oocysts. Immunoelectron microscopy demonstrated that native CP41 was distributed unevenly on the C. parvum oocyst surface and was associated with amorphous oocyst wall material. In an enzyme-linked immunosorbent assay, purified rCP41 performed as well as native C. parvum oocyst protein in measuring the serological responses of young calves and adult cows to experimental and natural C. parvum infections. These results indicate that recombinant CP41 antigen may have potential in the immunodiagnosis of cryptosporidiosis.
The detection of Cryptosporidium parvum oocysts in environmental samples usually relies on one of three different techniques: vital-dye staining (e.g., modified Ziehl-Neelsen acid-fast staining), direct or indirect immunofluorescence assay (IFA), or enzyme immunoassay using Cryptosporidium-reactive antibodies (Abs). Differences in the relative sensitivities of these assays have been noted (5, 6, 10, 14, 31). Confirmatory diagnosis of cryptosporidiosis in patients is often carried out by assaying sera for specific C. parvum antigens (4, 24). Several low-molecular-weight C. parvum oocyst antigens, such as the 15-, 17-, and 23-kDa proteins, appear to be useful for immunodiagnosis of Cryptosporidium infection (26). The immunogenicity of the 15-, 17-, and 23-kDa antigens and somewhat-higher-Mr antigens (e.g., 32 and 47 kDa) has been observed in other mammalian species infected or immunized with C. parvum oocysts (3, 20–23, 27–30, 32). However, laboratory studies have shown that these immunodominant antigens and other oocyst or sporozoite proteins are present in other Cryptosporidium species (27, 33). This cross-reactivity of immunodominant antigens may explain why commercial Ab-based tests cannot differentiate C. parvum from species of Cryptosporidium that are not infectious for humans. The purpose of the present study was to identify a species-specific antigen of C. parvum oocysts and obtain a DNA sequence encoding this antigen for use in immunodiagnosis of human cryptosporidiosis.
MATERIALS AND METHODS
Parasites.
C. parvum (AUCP-1 strain) oocysts were obtained by infecting a 1-day-old calf with 106 oocysts. The calf was obtained at birth from the dairy herd at the Beltsville Agricultural Research Center and housed in a 4- by 6-m concrete-floored pen with cinder block walls in a sanitized masonry building. Feces were collected from days 3 through 10 postinfection, pooled, and passed through a series of sieves of increasingly finer mesh, ending with a no. 325 mesh screen. Sieved fecal material was mixed with 2 M sucrose and subjected to continuous-flow centrifugation (35) followed by CsCl gradient centrifugation (15) for purification of C. parvum oocysts. Clean oocysts were resuspended in distilled H2O, stored at 4°C, and used from 1 to 6 months after collection, depending on the objectives of the experiment. Limited numbers of oocysts of other Cryptosporidium species were obtained from outside sources; the species were C. baileyi (B. Blagburn, Auburn University), C. meleagridis (M. Levy, North Carolina State University), C. serpentis (T. Graczyk, Johns Hopkins University), and C. wrairi (C. E. Chrisp, University of Michigan, Ann Arbor).
Preparation of parasite nucleic acid and protein.
Cryptosporidium oocysts destined for RNA or DNA extraction were treated for 30 min with 2.5% sodium hypochlorite (50% Clorox), washed five times with deionized H2O, resuspended in 1.0 ml of deionized H2O, and immersed dropwise into a mortar containing liquid nitrogen. The oocysts were ground in liquid nitrogen to a fine powder and then transferred to a tube containing either RNA or DNA extraction buffer. TRIZOL reagent was used to prepare total RNA in accordance with the manufacturer’s (Gibco-BRL, Gaithersburg, Md.) directions. A high salt concentration step was incorporated, as per the instructions of the manufacturer, to remove polysaccharide, which appears to exist in large quantities in cryptosporidia (1). DNA was extracted by using proteinase K and sodium dodecyl sulfate (SDS) as previously described (11). RNA and DNA yields were estimated by optical density at 260 nm (OD260)/OD280 readings. Total oocyst protein was prepared by resuspending the parasites in protein extraction buffer (10 mM Tris-HCl [pH 7.3], 1 mM MgCl2) containing phenylmethylsulfonyl fluoride. The oocysts were subjected to five freeze-thaw cycles, using dry ice-ethanol and 37°C water baths.
SDS-PAGE and immunoblotting of native and recombinant C. parvum protein.
Protein extracts of Cryptosporidium oocysts were treated with sample buffer containing 2-mercaptoethanol (19), heated for 3 min in a boiling-water bath, fractionated by 7.5 to 15% gradient SDS-polyacrylamide gel electrophoresis (PAGE), and transblotted to an Immobilon (Millipore, Bedford, Mass.) membrane as described previously (11). The antigen-impregnated membranes were treated briefly with phosphate-buffered saline (PBS), then immersed in PBS containing 2% nonfat dry milk (NFDM) to block nonspecific-Ab binding in subsequent steps. After being subjected to blocking, the membranes were incubated for 2 h with a 1:100 dilution of rabbit antiserum to native or recombinant C. parvum antigen in PBS containing 0.05% Tween 20 (PBS-Tw20). The membranes were then probed for 2 h with biotinylated goat anti-rabbit immunoglobulin G (IgG) (heavy and light [H+L] chain specific; Vector Laboratories, Burlingame, Calif.); this was followed by a 1-h incubation with avidin-peroxidase (Sigma Chemical Co., St. Louis, Mo.) and a final treatment with peroxidase substrate (0.5 mg of 4-chloro-1-naphthol/ml and 0.015% H2O2 in PBS) to visualize Ab binding. The membranes were washed with PBS-Tw20 three times between each incubation step.
Preparation of antiserum to C. parvum 41-kDa protein.
Total oocyst protein extracted from C. parvum, C. baileyi, C. meleagridis, and C. serpentis was electrophoresed in adjacent lanes of a 7.5 to 15% gradient SDS-polyacrylamide gel (107 oocysts/lane) and transblotted to a nitrocellulose membrane (Schleicher and Schuell, Keene, N.H.). The blot was immunostained with rabbit antiserum raised against total C. parvum oocyst protein to identify antigens unique to C. parvum. A 41-kDa protein was identified, excised from adjacent unstained lanes, ground with a miniature mortar and pestle, resuspended in PBS containing ImmunoMax SR adjuvant (Zonagen, The Woodlands, Tex.), and used to immunize New Zealand White rabbits (Covance, Denver, Pa.) by intramuscular injection. The rabbits received subsequent booster immunizations 4 weeks after the primary immunization and were then bled for serum by central auricular artery puncture 2 weeks after the last booster immunization.
Immunoscreening of C. parvum genomic DNA libraries for CP41 clones.
Genomic C. parvum DNA was subjected to partial digestion with Tsp5901 restriction enzyme (recognition site, AATT; New England BioLabs, Beverly, Mass.) to obtain fragments approximately 1 kb in length. The DNA fragments were cloned into the EcoRI site of the lambda Zap II bacteriophage expression system (Stratagene, La Jolla, Calif.). Bacteriophages containing C. parvum genomic DNA were screened at a density of 104 plaques per petri dish, using the directions supplied by the manufacturer (Stratagene). Nitrocellulose filters soaked in 10 mM isopropyl-β-d-thiogalactopyranoside (IPTG) were overlaid on the petri dishes containing phage plaques for 4 h at 37°C. The filters were washed with PBS, treated with PBS-NFDM, and then probed with rabbit antiserum to native CP41 protein. Ab binding was visualized as described above for immunoblotting. Positive phages were picked and subjected to multiple rounds of screening until a clonal population was obtained.
DNA sequencing.
Recombinant pBluescript plasmid DNA was excised from recombinant lambda Zap bacteriophage by using an excision protocol supplied by the manufacturer (Stratagene). The DNA sequence of the recombinant pBluescript clone was obtained by using a 35S-dideoxy sequencing kit (Amersham-Pharmacia Biotech, Piscataway, N.J.) and a PCR fluorescence dye terminator kit (Applied Biosystems, Foster City, Calif.). The nonradioactive sequencing reactions were analyzed on an ABI 373 sequencer (Applied Biosystems). The complete DNA sequence was obtained by performing multiple reactions with both insert-specific and pBluescript-specific primers. Sequence compilation and identification of open reading frames (ORFs) for the recombinant clones were performed by using the GCG sequence analysis package (Genetics Computer Group, Madison, Wis.).
Production of recombinant polyhistidine proteins and immunization of rabbits for production of antisera against recombinant protein.
The Cryptosporidium insert DNA was excised from pBluescript by PstI and HindIII digestion and ligated into the pTrcHis A vector (Invitrogen, Carlsbad, Calif.) cut with the same restriction enzymes. Recombinant plasmid DNA was introduced into Escherichia coli DH5 cells by standard transformation procedures (7). Maintenance of the ORF between pTrcHis and the insert DNA was confirmed by DNA sequencing. A time course study to identify the time of IPTG induction leading to peak expression of recombinant protein indicated that a 4-h induction was optimal. New Zealand White rabbits (Covance) were immunized three times over a 2-month period with recombinant protein impregnated on nitrocellulose paper which had been ground in a miniature mortar and pestle in a manner similar to that described above. The rabbits were bled for serum by central auricular artery puncture 2 weeks after the last booster immunization.
IFA and IEM.
C. parvum sporozoites were first excysted by treating oocysts with 1% sodium hypochlorite (20% Clorox) for 10 min at room temperature, washing them five times with Hanks’ balanced salt solution (HBSS), resuspending them in HBSS, and incubating them for 1 h in a humidified chamber at 37°C in an atmosphere of 5% CO2. For examination by immunofluorescence microscopy, the excysted oocyst-sporozoite mixture was pipetted onto multiwell glass slides at a density of 104 oocysts per well (based on pre-excystation counts) and allowed to air dry at room temperature (RT). The parasites were then treated with PBS containing 1% bovine serum albumin (PBS-BSA) for 1 h at room temperature prior to incubation with a 1:100 dilution of antiserum in PBS-BSA for 2 h at room temperature in a humidified chamber. Ab binding was detected by treatment with fluorescein isothiocyanate-labeled anti-rabbit IgG (H+L chain specific; Sigma Chemical Co.) for 1 h at room temperature in a humidified chamber. The slides were washed after each incubation step by three separate immersions in PBS. After the last incubation, each slide was allowed to air dry prior to being overlaid with several drops of VectaStain antibleaching mounting medium (Vector Laboratories) and a coverslip. The C. parvum oocysts and sporozoites were examined with an epifluorescence microscope.
For immunoelectron microscopy (IEM), the excysted sporozoite-oocyst mixture was washed several times in PBS, enumerated on a hemocytometer, aliquoted at 108 sporozoites per 1.5-ml microcentrifuge tube, and pelleted by centrifugation. The oocyst-sporozoite pellet was resuspended in 0.1 M sodium cacodylate containing 2% paraformaldehyde and 0.5% glutaraldehyde for 20 min at room temperature. The fixed parasites were washed twice with 0.1 M sodium cacodylate and pelleted by centrifugation. The oocyst-sporozoite pellet was dehydrated in a graded ethanol series, infiltrated overnight in LR White hard-grade acrylic resin (London Resin Company, London, England), and cured at 55°C for 24 h. Thin sections (90 nm thick) were obtained by using a Diatome diamond knife on a Reichert/AO Ultracut microtome and collected on 200-mesh nickel grids. The grids were floated on drops of PBS containing 0.1 M glycine and 1% BSA for 10 min, washed with PBS, floated on drops of PBS-NFDM-Tw20, and floated on drops of PBS containing a 1:100 dilution of normal goat serum. The grids were incubated in a humidified chamber on drops containing a 1:100 dilution of rabbit antiserum in PBS-NFDM-Tw20. The grids were incubated for 2 h at room temperature, washed three times with PBS-Tw20-NFDM, and floated for 1 h at room temperature on drops of a 1:100 dilution of gold particle (10 nm diameter)-labeled goat anti-rabbit IgG (H+L chain specific; Vector Laboratories). The grids were washed three times with PBS-Tw20 and once with distilled water, air dried, stained with 5% uranyl acetate for 30 min, and examined under a Hitachi H500H transmission electron microscope at 75 kV.
RT-PCR analysis of C. parvum oocyst RNA.
C. parvum oocysts stored for 1, 3, or 6 months at 4°C were analyzed for the presence of CP41 mRNA by reverse transcriptase PCR (RT-PCR). First-strand cDNA synthesis from 1 μg of total C. parvum RNA was carried out at 42°C in a reaction buffer containing 20 U of Superscript II RT (Gibco-BRL) and either 2 pmol of a gene-specific CP41-F1 primer (5′ AGCATTAGTAGCAACAGTAG 3′) or 4 pmol of oligo(dT) primer in a total volume of 20 μl as per the manufacturer’s (Gibco-BRL) directions. After RNase H treatment, PCR was carried out with 2 μl of the first-strand reaction mixture, 0.25 U of Taq polymerase (Gibco-BRL), and 50 pmol (each) of CP41-F1 and CP41-R1 (5′ GAGATGGACTATTCTAGG 3′) primers in a final volume of 50 μl. Amplification was performed with a Stratagene Robocycler, using the following PCR amplification protocol: denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min for 30 cycles, followed by a final extension at 72°C for 2 min. To analyze for the presence of the CP41 gene in genomic DNA, a single PCR was performed on DNA equivalent to 103 Cryptosporidium oocysts under reaction conditions identical to those described above. The PCR products were analyzed by PAGE, ethidium bromide staining, and UV transillumination followed by capture to a charge-coupled device camera as described previously (13).
ELISA.
Antisera were obtained weekly, beginning at 1 day of age, for 7 to 10 weeks from four calves that had been given normal bovine colostrum (NBC) at birth and were being housed in a dairy operation that had experienced sporadic outbreaks of cryptosporidiosis (natural infection). The calves were examined daily for diarrhea and excretion of C. parvum oocysts in feces, using standard methods. Antiserum was also obtained from a calf prior to, and every week for 1 month after, being fed NBC at birth and inoculated per os with 106 C. parvum oocysts (experimental infection). Antisera were also obtained from adult cows (3 to 4 years old) before and for several weeks after experimental C. parvum challenge and from two adult cows that had not been exposed to C. parvum. Antisera were tested by enzyme-linked immunosorbent assay (ELISA) for recognition of native C. parvum oocyst protein and recombinant CP41 antigen. In the former, C. parvum oocysts were subjected to multiple freeze-thaw cycles and disruption on a Mini-Bead Beater (BioSpec Products, Bartlesville, Okla.) followed by centrifugation to pellet insoluble material. The oocyst protein supernatant (50 μl), equivalent to 4 × 104 oocysts (150 ng of protein), was pipetted into individual wells of Immulon II microtiter plates (Dynex Technologies, Inc., Chantilly, Va.) and incubated overnight at 4°C. The native C. parvum protein ELISA was performed as previously described (8). Titers are given as the reciprocal of the highest dilution giving an absorbance reading twice that of the average reading for the 1:80 dilution of the negative-control serum.
Recombinant CP41 polyhistidine fusion protein was purified by denaturing Ni-nitrilotriacetic acid (NTA) affinity chromatography in accordance with the manufacturer’s (Invitrogen) directions. Eluates containing peak amounts of purified protein, as indicated by SDS-PAGE and immunoblotting, were pooled and adsorbed to the surface of Immulon II microtiter plates as described above. The wells were washed with PBS to remove unbound recombinant antigen and then treated with PBS containing 2% normal horse serum (Sigma Chemical Co.) for 1 h at room temperature to inhibit nonspecific-Ab binding in subsequent incubation steps. The wells were washed with PBS-Tw20, and then the plates were incubated for 2 h at room temperature with 100-μl volumes of serial dilutions of positive-control bovine serum or a 1:100 dilution of negative-control bovine serum or sera from calves or cows as described above. After the plates were washed three times with PBS-Tw20, 100-μl volumes of a 1:1,000 dilution of peroxidase-labeled goat anti-bovine IgG (H+L chain specific; Sigma Chemical Co.) were added to the wells, and the plate was incubated for 1 h at room temperature. The wells were washed three times with PBS-Tw20 prior to addition of 50-μl aliquots of peroxidase substrate (0.01 mg of o-phenylene diamine/ml and 0.001% H2O2 in PBS) and a 10-min incubation at room temperature. Color development was stopped by the addition of 50 μl of 2% H2SO4 to each well. The absorbance at 492 nm was read on a Bio-Rad model 450 microplate reader. The titer of anti-recombinant CP41 antigen Ab in serum was estimated through the use of a standard curve generated with the positive-control serum by previously described procedures (12).
Nucleotide sequence accession number.
The nucleotide sequence of the rCP41 clone has been deposited in GenBank under accession no. AF144621.
RESULTS
Antiserum raised against total C. parvum oocyst protein recognized a number of antigens of C. parvum and C. baileyi (Fig. 1). A similar recognition pattern was observed with C. meleagridis and C. serpentis (data not shown). Although the majority of antigens were common to all Cryptosporidium species, a few antigens were unique to C. parvum. In particular, a 41-kDa protein was present in C. parvum but not in the other Cryptosporidium species (Fig. 1). A section of nitrocellulose adjacent to the unique p41 antigen in an unlabeled section of the blot was used to immunize rabbits for monospecific antisera to the p41 protein. These antisera were used to identify a recombinant bacteriophage clone encoding epitopes of the C. parvum 41-kDa antigen. DNA sequencing of this clone, designated rCP41, revealed an insert of 740 nucleotides (nt) in length containing an ORF that was in frame with beta-galactosidase of lambda ZAP/pBluescript (Fig. 2). Two potential ATG start sites were identified, one at nt 139 and the other at nt 196. Both sites conform to the Kozak consensus sequence for translation initiation at the −3 and +4 nt position (18).
FIG. 1.
Immunostaining of SDS-PAGE-fractionated native C. parvum (Cp) or C. baileyi (Cb) oocyst protein or Ni-NTA-purified (P) and unpurified (IP) recombinant CP41 protein (rCp41) with rabbit antiserum to whole C. parvum oocyst protein (R α CpOO) or to native (R α NATIVE Cp41) or recombinant (R α RECOMBINANT Cp41) CP41 antigen or with normal control rabbit serum (NRS). The positions of the 28-, 36-, and 41-kDa proteins are noted. MrS, molecular weight standards.
FIG. 2.
DNA sequence and predicted amino acid sequence of the CP41 DNA clone isolated from C. parvum oocysts. Putative ATG start sites are indicated in boldface.
Oligonucleotide primers (CP41R and CP41F) were prepared based on the rCP41 DNA sequence and used in PCRs to amplify the respective sequence in genomic DNAs from two C. parvum isolates, C. baileyi, and C. wrairi. An amplification product similar in size to the insert DNA was observed, indicating that rCP41 was present in C. parvum (bovine and human isolates), C. baileyi, and C. wrairi genomic DNAs (Fig. 3). RT-PCR of total RNA showed that the CP41 transcript was present in C. parvum oocysts (Fig. 4). Although equivalent amounts of total RNA were used in all reactions, the intensity of the RT-PCR signal appeared to be inversely correlated with the age of the C. parvum oocysts. The RT-PCR signals derived from RNA isolated from oocysts that had been stored at 4°C for 6 or 3 months were about 25 and 50%, respectively, of the signal from 1-month-old oocysts (Fig. 4, lanes CP1+, CP2+, and CP3+, respectively). The control reactions in which RT was not included did not show a PCR product, indicating that the RT-PCR signal was due to the presence of mRNA rather than contaminating DNA (Fig. 3, lanes CP1−, CP2−, and CP3−). In RT-PCR assays on C. baileyi RNA, CP41 mRNA was not detectable (data not shown). RT-PCR could not be performed on C. wrairi because RNA from this species was not available. Attempts to identify the transcript size for C. parvum by Northern blot hybridization assay were unsuccessful.
FIG. 3.
PCR amplification of the CP41 sequence from genomic DNA (equivalent to 103 oocysts) of a bovine isolate of C. parvum (Cp bov), a human isolate of C. parvum (Cp hu), C. baileyi (Cb), and C. wrairi (Cw). The positions of φx174 DNA standards are shown on the left.
FIG. 4.
Molecular analysis of C. parvum for the presence of the CP41 sequence in genomic DNA by PCR and for the presence of CP41 mRNA in total RNA by RT-PCR. The PCR assay was performed on DNA equivalent to 103 C. parvum oocysts (CP DNA). RT-PCR was performed on total RNA equivalent to 5 × 105 C. parvum oocysts stored at 4°C for 6 months (CP1), 3 months (CP2), or 1 month (CP3). +, Superscript RTase added to first-step cDNA synthesis reaction; −, no RT added to first-step cDNA synthesis reaction. The positions of φx174 DNA standards are shown on the left.
The CP41 DNA sequence was inserted into the pTrcHis expression vector to produce a recombinant polyhistidine-CP41 fusion protein. Immunoblots of SDS-PAGE-fractionated protein extracts from E. coli harboring the recombinant pTrcHis-CP41 plasmid contained two unique protein bands, migrating at positions corresponding to 28 and 36 kDa, produced 4 h after IPTG induction (Fig. 1). Protein extracts from E. coli harboring the nonrecombinant pTrcHis plasmid contained neither the 28- nor the 36-kDa recombinant protein (data not shown). Purification of rCP41 by Ni-NTA affinity chromatography showed that only the 36-kDa protein was isolated (Fig. 1). The 28- and 36-kDa recombinant CP41 proteins were excised from unlabeled membranes and subjected to N-terminal amino acid sequencing (Beckman Center, Stanford University Medical Center, Palo Alto, Calif.). The 36-kDa protein appeared to be the actual fusion protein containing pTrcHis-encoded amino acids at its N terminus. The amino acid sequence of the recombinant 28-kDa protein indicated that in E. coli, translation also initiated from the CP41 start codon at nt 196. In an immunoblotting assay, antiserum against the 36-kDa recombinant protein recognized a native 41-kDa C. parvum antigen (Fig. 1). A similar pattern was observed with antiserum against the 28-kDa recombinant protein (data not shown). Antiserum raised against native or recombinant CP41 antigen showed a similar pattern in immunoblots with whole native C. parvum oocyst protein or with purified or unpurified recombinant CP41 protein (Fig. 1, lanes Cp, P, and IP, respectively). Normal control serum showed negligible recognition of native C. parvum protein or purified recombinant CP41 antigen (Fig. 1). A band similar in size to the recombinant CP41 antigen was present in unpurified recombinant protein extracts and recognized by normal control serum (Fig. 1).
The relationship between native and recombinant CP41 proteins was unknown except for the fact that these antigens have at least one epitope in common. The size discrepancy between the native CP41 protein and either the 36-kDa or the 28-kDa recombinant CP41 protein did not appear to be due to the presence of glycan residues on the native protein. Glycosidase treatment was performed on total oocyst extracts to determine if CP41 was glycosylated. In addition, untreated SDS-PAGE-fractionated C. parvum protein impregnated on an Immobilon membrane was treated with sodium perodiate to remove glycan residues. No differences in molecular weight or immunoreaction intensity were observed between the treated and untreated protein samples (data not shown). The results indicated that native CP41 was not heavily glycosylated and that epitopes recognized by serum against native or recombinant CP41 protein did not involve glycan moieties.
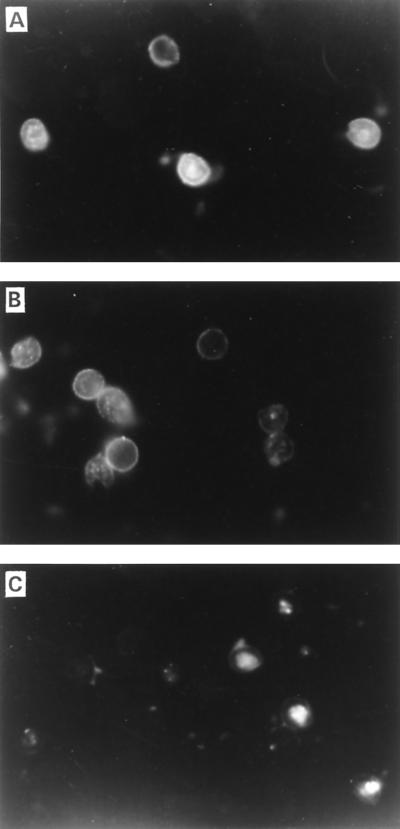
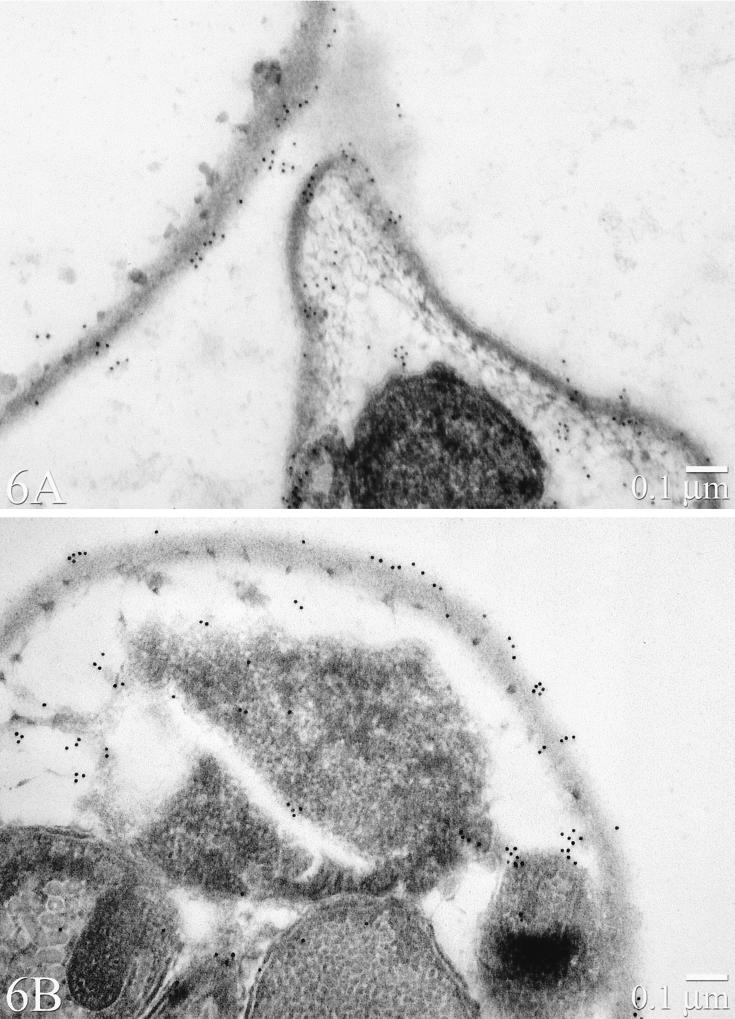
As shown by IFA, antiserum to native or recombinant CP41 antigen bound a surface antigen of C. parvum oocysts (Fig. 5). Although not evident from the IFA figures, the target antigen appeared to be distributed unevenly on the surface of the oocyst. This surface location and distribution was confirmed by IEM (Fig. 6). The antigen was present on both external and internal regions of the oocyst wall and was also associated with amorphous material on the oocyst outer surface. Negligible staining of C. baileyi oocysts was observed with antiserum to native or recombinant CP41 antigen (data not shown).
FIG. 5.
IFA staining of C. parvum oocysts with rabbit antiserum prepared against native CP41 protein (A) or recombinant CP41 antigen (B) or with normal control rabbit serum (C).
FIG. 6.
IEM staining of C. parvum oocysts with rabbit antiserum prepared against native CP41 protein (A) or recombinant CP41 antigen (B).
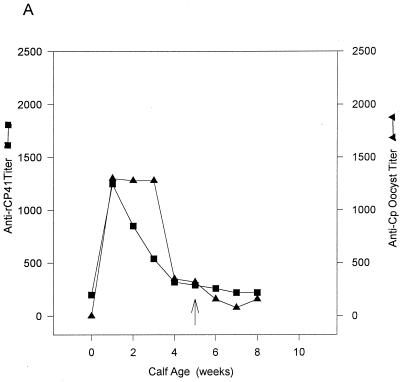
In the ELISA, recombinant CP41 antigen showed levels of binding similar to those of the native C. parvum oocyst antigen when serum from adult cows that were exposed to C. parvum was used as the probe (Table 1). The preinfection titers of Abs to the native and recombinant antigens were high and did not appear to increase appreciably after experimental challenge. Perhaps boosting of the Ab response was not observed because the oocyst challenge did not result in patent infection due to the age-related resistance of adult cows to C. parvum infection (9). In young calves exposed to a natural C. parvum infection, titers of Abs to recombinant CP41 antigen were high at the first postcolostral bleeding, possibly due to the presence of anti-CP41 antibodies in normal colostrum, and then decreased (Fig. 7A to D). No increase in titers of Ab to rCP41 was noted after exposure to C. parvum. The titers of Ab to native C. parvum oocyst antigen were variable. In two calves (Fig. 7B and D), the highest titers of Ab against native antigen occurred at the time of oocyst shedding. In the other two calves, peak anti-C. parvum antigen Ab titers were observed immediately after colostrum feeding (Fig. 7A and C). A similar pattern was observed after an experimental C. parvum infection of a 1-day-old calf (Fig. 7E). Titers of Abs to rCP41 and native C. parvum oocyst antigen were highest after colostrum feeding and natural C. parvum oocyst inoculation (Fig. 7E). Although anti-rCP41 Ab titers decreased with time, the response to native C. parvum oocyst titers was variable (Fig. 7E).
TABLE 1.
Anti-recombinant CP41 and anti-native C. parvum (nCP) oocyst antigen Ab titers in sera from adult cows after experimental cryptosporidiosis infection or in unexposed control adult cows
| Study group and cow no. | Time postinfection (wk) | ELISA titer
|
|
|---|---|---|---|
| Anti-rCP41 | Anti-nCP oocyst antigen | ||
| Experimental infections | |||
| 1 | 0 | 1,100 | 2,560 |
| 1 | 1,310 | NDa | |
| 2 | 1,200 | 1,280 | |
| 3 | 1,200 | 2,560 | |
| 4 | 1,110 | 2,560 | |
| 2 | 0 | 900 | 2,560 |
| 1 | 1,020 | ND | |
| 2 | 1,260 | 1,280 | |
| 3 | 1,070 | 2,560 | |
| 3 | 0 | 890 | ND |
| 1 | 850 | 1,280 | |
| 2 | 900 | 2,560 | |
| 3 | 1,200 | 5,120 | |
| Unexposed controls | |||
| 1 | <100 | <100 | |
| 2 | <100 | 320 | |
ND, not done.
FIG. 7.
Serological titers of Abs against recombinant CP41 (■) and native C. parvum oocyst protein (▴) in four calves (A to D) exposed to a natural C. parvum infection and in one calf (E) exposed to an experimental natural C. parvum infection, as revealed by ELISA. The arrows indicates the first day of C. parvum oocyst shedding.
DISCUSSION
This study represents initial stages in the development of reagents for serodiagnosis of C. parvum infection and for detecting this parasite in environmental samples. Although the CP41 gene sequence is present in the genome of Cryptosporidium species that do not infect humans and calves (C. baileyi and C. wrairi), RT-PCR and IFA (for C. baileyi) indicated that CP41 is expressed only in C. parvum. It is possible, however, that CP41 mRNA and/or CP41 oocyst antigen is less stable in C. baileyi. Further testing of other C. baileyi isolates, C. wrairi, and other Cryptosporidium species is required before it can be concluded that CP41 is C. parvum specific. The results also indicate that the CP41 primers used in the present study may be useful for species-specific RT-PCR analysis of total Cryptosporidium RNA but probably will not be useful for a species-specific PCR test based on genomic DNA. The presence of CP41 antigen on the surface of C. parvum oocysts, as revealed by IFA testing and IEM observations, indicates that polyclonal sera or monoclonal antibodies to CP41 can be a valuable tools for species-specific immunodetection of the parasite in environmental samples.
The differences in the apparent molecular masses of the recombinant CP41 proteins (36 and 28 kDa) and the native C. parvum protein (41 kDa) do not appear to be due to the presence of glycan moieties on the native protein. Glycosidase treatment did not affect the molecular weight of the native 41-kDa C. parvum protein. These findings were corroborated by experiments using sodium periodate. Perhaps rCP41 is not a full-length clone, i.e., is missing upstream and/or downstream coding sequences. The absence of a stop codon in the DNA sequence supports this hypothesis. The predicted size of the recombinant protein, based on the pTrcHis fusion peptide (4 kDa) and the CP41 DNA sequence, is 31 kDa. The predicted size of the recombinant protein initiating from the ATG at nt 196 is 19.5 kDa. The size discrepancy between either the predicted size of the full-length fusion protein (31 kDa) or the predicted truncated protein initiating at the internal ATG start site (19.5 kDa) and the observed 36- and 28-kDa recombinant proteins may be due to any of several phenomena. One is the observed aberrant migration of E. coli-expressed recombinant proteins during SDS-PAGE for proteins with atypical amino acid compositions (1a, 16, 17, 34). The CP41 amino acid sequence contains a disproportionate number of Asn (21%) and Thr (11%) residues, representing almost one-third of the total. The recombinant protein initiating from the internal ATG start site contains even higher percentages of Asn (27%) and Thr (14%).
The ELISA data suggest that natural C. parvum infections in young calves do not elicit a strong Ab response against the 41-kDa protein. It is unclear whether the high anti-rCP41 Ab titers observed in the experimental infection were due to the presence of anti-C. parvum antibodies in NBC or were stimulated by the C. parvum oocyst inoculation. These observations are consistent with other studies showing that natural and experimental C. parvum infections in young calves elicit a strong serological Ab response to a single low-Mr protein but much weaker Ab responses to other antigens, including the 41-kDa protein (20, 23, 25, 32, 36). The immunoblotting data reported by others, and our unpublished observations during experiments using sera from C. parvum-infected calves (natural and experimental infections), are in contrast with the finding of one study, that calves infected with the parasite exhibit high titers of Abs to a wide-Mr range of C. parvum oocyst antigens (29). The present study also showed high titers of Abs to native and recombinant C. parvum antigen in adult cows previously exposed to the parasite. Similar to the findings of others, adult cows with high anti-C. parvum Ab titers are resistant to challenge infection, neither exhibiting clinical signs of cryptosporidiosis nor shedding C. parvum oocysts in feces (2, 9). The absence of patent infection may explain why titers against native oocyst antigen or recombinant CP41 antigen did not appreciably increase after C. parvum challenge. The ELISA data also indicated that NBC has Ab to the C. parvum 41-kDa protein because serum from calves fed colostrum contained high anti-rCP41 titers at 1 week of age.
REFERENCES
- 1.Abrahamsen, M. Personal communication.
- 1a.Alano P, Premawansa S, Bruce M C, Carter R. A stage specific gene expressed at the onset of gametocytogenesis in Plasmodium falciparum. Mol Biochem Parasitol. 1991;46:81–88. doi: 10.1016/0166-6851(91)90201-g. [DOI] [PubMed] [Google Scholar]
- 2.Casemore D P, Wright S E, Coop R L. Cryptosporidiosis—human and animal epidemiology. In: Fayer R, editor. Cryptosporidium and cryptosporidiosis. Boca Raton, Fla: CRC Press; 1997. pp. 65–92. [Google Scholar]
- 3.Enriquez F J, Riggs M W. Role of immunoglobulin A monoclonal antibodies against P23 in controlling murine Cryptosporidium parvum infection. Infect Immun. 1998;66:4469–4473. doi: 10.1128/iai.66.9.4469-4473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frost F J, de la Cruz A A, Moss D M, Curry M, Calderon R L. Comparisons of ELISA and Western blot assays for detection of Cryptosporidium antibody. Epidemiol Infect. 1998;121:205–211. doi: 10.1017/s0950268898008991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia L S, Shimizu R Y. Evaluation of nine immunoassay kits (enzyme immunoassay and direct fluorescence) for detection of Giardia lamblia and Cryptosporidium parvum in human fecal specimens. J Clin Microbiol. 1997;35:1526–1529. doi: 10.1128/jcm.35.6.1526-1529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graczyk T K, Cranfield M R, Fayer R. Evaluation of commercial enzyme immunoassay (EIA) and immunofluorescent antibody (FA) test kits for detection of Cryptosporidium oocysts of species other than Cryptosporidium parvum. Am J Trop Med Hyg. 1996;54:274–279. doi: 10.4269/ajtmh.1996.54.274. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D. Studies on the transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 8.Harp J A, Woodmansee D B, Moon H W. Effects of colostral antibody on susceptibility of calves to Cryptosporidium parvum infection. Am J Vet Res. 1989;50:2117–2119. [PubMed] [Google Scholar]
- 9.Harp J A, Woodmansee D B, Moon H W. Resistance of calves to Cryptosporidium parvum: effects of age and previous exposure. Infect Immun. 1990;58:2237–2240. doi: 10.1128/iai.58.7.2237-2240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ignatius R, Eisenblatter M, Regnath T, Mansmann U, Futh U, Hahn H, Wagner J. Efficacy of different methods for detection of low Cryptosporidium parvum oocyst numbers or antigen concentrations in stool specimens. Euro J Clin Microbiol Infect Dis. 1997;16:732–736. doi: 10.1007/BF01709253. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins M C, Fayer R, Tilley M, Upton S J. Cloning and expression of cDNA encoding epitopes shared by 15- and 60-kilodalton proteins of Cryptosporidium parvum sporozoites. Infect Immun. 1993;61:2377–2382. doi: 10.1128/iai.61.6.2377-2382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins M C, Wouda W, Dubey J P. Serological response over time to recombinant Neospora caninum antigens in cattle after a neosporosis-induced abortion. Clin Diagn Lab Immunol. 1997;4:270–274. doi: 10.1128/cdli.4.3.270-274.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins M C, Trout J, Fayer R. Development and application of an improved semi-quantitative technique for detecting low level Cryptosporidium parvum infections in mouse tissue using polymerase chain reaction. J Parasitol. 1998;84:182–186. [PubMed] [Google Scholar]
- 14.Kehl K S, Cicirello H, Havens P L. Comparison of four different methods for detection of Cryptosporidium species. J Clin Microbiol. 1995;33:416–418. doi: 10.1128/jcm.33.2.416-418.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilani R T, Sekla L. Purification of Cryptosporidium oocysts and sporozoites by cesium chloride and Percoll gradients. Am J Trop Med Hyg. 1987;36:505–508. doi: 10.4269/ajtmh.1987.36.505. [DOI] [PubMed] [Google Scholar]
- 16.Kim K S, Jenkins M C, Lillehoj H S. Immunization of chickens with live Escherichia coli expressing Eimeria acervulina merozoite recombinant antigen induces partial protection against coccidiosis. Infect Immun. 1989;57:2434–2440. doi: 10.1128/iai.57.8.2434-2440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko C, Smith C K, McDonell M. Identification and characterization of a target antigen of a monoclonal antibody directed against Eimeria tenella merozoites. Mol Biochem Parasitol. 1990;41:53–64. doi: 10.1016/0166-6851(90)90096-5. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–239. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lorenzo M J, Ben B, Mendez F, Villacorta I, Ares-Mazas M E. Cryptosporidium parvum oocyst antigens recognized by sera from infected asymptomatic adult cattle. Vet Parasitol. 1995;60:17–25. doi: 10.1016/0304-4017(94)00770-d. [DOI] [PubMed] [Google Scholar]
- 21.Lumb R, Lanser J A, O’Donoghue P J. Electrophoretic and immunoblot analysis of Cryptosporidium oocysts. Immunol Cell Biol. 1988;66:369–376. doi: 10.1038/icb.1988.48. [DOI] [PubMed] [Google Scholar]
- 22.Lumb R, Smith P S, Davies R, O’Donoghue P J, Atkinson H M, Lanser J A. Localization of a 23,000 MW antigen of Cryptosporidium by immunoelectron microscopy. Immunol Cell Biol. 1989;67:267–270. doi: 10.1038/icb.1989.40. [DOI] [PubMed] [Google Scholar]
- 23.McDonald V, Deer R M A, Wright S E, Dyson D A, Chiodini P L, McAdam K P. Comparative study of the antigenic composition of oocyst isolates of Cryptosporidium parvum from different hosts. Parasite Immunol. 1992;14:227–232. doi: 10.1111/j.1365-3024.1992.tb00463.x. [DOI] [PubMed] [Google Scholar]
- 24.McLauchlin J, Casemore D P, Moran S, Patel S. The epidemiology of cryptosporidiosis: application of experimental sub-typing and antibody detection systems to the investigation of water-borne outbreaks. Folia Parasitol. 1998;45:83–92. [PubMed] [Google Scholar]
- 25.Mead J R, Arrowood M J, Sterling C R. Antigens of Cryptosporidium sporozoites recognized by immune sera of infected animals and humans. J Parasitol. 1988;74:135–143. [PubMed] [Google Scholar]
- 26.Moss D M, Chappell C L, Okhuysen P C, DuPont H L, Arrowood M J, Hightower A W, Lammie P J. The antibody response to 27-, 17-, and 15-kDa Cryptosporidium antigens following experimental infections in humans. J Infect Dis. 1998;178:827–833. doi: 10.1086/515377. [DOI] [PubMed] [Google Scholar]
- 27.Nina J M S, McDonald V, Dyson D A, Catchpole J, Uni S, Iseki M, Chiodini P L, McAdam K P W J. Analysis of oocyst wall and sporozoite antigens from three Cryptosporidium species. Infect Immun. 1992;60:1509–1513. doi: 10.1128/iai.60.4.1509-1513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortega-Mora L M, Troncoso J M, Rojo-Vazquez F A, Gomez-Bautista M. Identification of Cryptosporidium parvum oocyst/sporozoite antigens recognized by infected and hyperimmune lambs. Vet Parasitol. 1994;53:159–166. doi: 10.1016/0304-4017(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 29.Peeters J E, Villacorta I, Vanopdenbosch E, Vandergheynst D, Naciri M, Ares-Mazás E, Yvoré P. Cryptosporidium parvum in calves: kinetics and immunoblot analysis of specific serum and local antibody responses (immunoglobulin A [IgA], IgG, and IgM) after natural and experimental infections. Infect Immun. 1992;60:2309–2316. doi: 10.1128/iai.60.6.2309-2316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perryman L E, Jasmer D P, Riggs M W, Bohnet S G, McGuire T C, Arrowood M J. A cloned gene of Cryptosporidium parvum encodes neutralization-sensitive epitopes. Mol Biochem Parasitol. 1996;80:137–147. doi: 10.1016/0166-6851(96)02681-3. [DOI] [PubMed] [Google Scholar]
- 31.Quilez J, Sanchez-Acedo C, Clavel A, del Cacho E, Lopez-Bernard F. Comparison of acid-fast stain and a monoclonal antibody-based immunofluorescence reagent for the detection of Cryptosporidium oocysts in fecal specimens from cattle and pigs. Vet Parasitol. 1996;67:75–81. doi: 10.1016/s0304-4017(96)01023-0. [DOI] [PubMed] [Google Scholar]
- 32.Reperant J M, Naciri M, Iochmann S, Tilley M, Bout D T. Major antigens of Cryptosporidium parvum recognised by serum antibodies from different infected animal species and man. Vet Parasitol. 1994;55:1–13. doi: 10.1016/0304-4017(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 33.Tilley M, Fayer R, Guidry A, Upton S J, Blagburn B L. Cryptosporidium parvum (Apicomplexa:Cryptospordiidae) oocyst and sporozoite antigens recognized by bovine colostral antibodies. Infect Immun. 1990;58:2966–2971. doi: 10.1128/iai.58.9.2966-2971.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomley F M, Bumstead J M, Billington K J, Dunn P J. Molecular cloning and characterization of a novel acidic microneme protein (Etmic-2) from apicomplexan protozoan parasite, Eimeria tenella. Mol Biochem Parasitol. 1996;79:195–206. doi: 10.1016/0166-6851(96)02662-x. [DOI] [PubMed] [Google Scholar]
- 35.Vetterling J M. Continuous-flow differential density flotation of coccidial oocysts and a comparison with other methods. J Parasitol. 1969;55:412–417. [PubMed] [Google Scholar]
- 36.Whitmire W M, Harp J A. Characterization of bovine cellular and serum antibody responses during infection by Cryptosporidium parvum. Infect Immun. 1991;59:990–995. doi: 10.1128/iai.59.3.990-995.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]