Key Points
Question
Among a cohort of US frontline and essential workers infected with the original strain or the Delta or Omicron variants of SARS-CoV-2, is there a difference in COVID-19 symptoms or viral RNA load among those receiving mRNA vaccines compared with being unvaccinated?
Findings
In this prospective cohort study that included 1199 participants with SARS-CoV-2 infection, receipt of 2 or 3 mRNA vaccine doses before Delta infections and 3 mRNA vaccine doses before Omicron infections was significantly associated with milder COVID-19 (less frequently symptomatic, febrile, or medically attended or shorter duration of illness) compared with being unvaccinated. Receipt of 2 mRNA vaccine doses 14 to 149 days prior to either Delta or Omicron infection was significantly associated with lower viral RNA load.
Meaning
Among a cohort of US frontline and essential workers, recent vaccination with 2 or 3 mRNA vaccine doses, compared with being unvaccinated, was associated with attenuated COVID-19 symptoms and lower viral RNA load for Delta and Omicron variants of SARS-CoV-2 in some comparisons.
Abstract
Importance
Data on the epidemiology of mild to moderately severe COVID-19 are needed to inform public health guidance.
Objective
To evaluate associations between 2 or 3 doses of mRNA COVID-19 vaccine and attenuation of symptoms and viral RNA load across SARS-CoV-2 viral lineages.
Design, Setting, and Participants
A prospective cohort study of essential and frontline workers in Arizona, Florida, Minnesota, Oregon, Texas, and Utah with COVID-19 infection confirmed by reverse transcriptase–polymerase chain reaction testing and lineage classified by whole genome sequencing of specimens self-collected weekly and at COVID-19 illness symptom onset. This analysis was conducted among 1199 participants with SARS-CoV-2 from December 14, 2020, to April 19, 2022, with follow-up until May 9, 2022, reported.
Exposures
SARS-CoV-2 lineage (origin strain, Delta variant, Omicron variant) and COVID-19 vaccination status.
Main Outcomes and Measures
Clinical outcomes included presence of symptoms, specific symptoms (including fever or chills), illness duration, and medical care seeking. Virologic outcomes included viral load by quantitative reverse transcriptase–polymerase chain reaction testing along with viral viability.
Results
Among 1199 participants with COVID-19 infection (714 [59.5%] women; median age, 41 years), 14.0% were infected with the origin strain, 24.0% with the Delta variant, and 62.0% with the Omicron variant. Participants vaccinated with the second vaccine dose 14 to 149 days before Delta infection were significantly less likely to be symptomatic compared with unvaccinated participants (21/27 [77.8%] vs 74/77 [96.1%]; OR, 0.13 [95% CI, 0-0.6]) and, when symptomatic, those vaccinated with the third dose 7 to 149 days before infection were significantly less likely to report fever or chills (5/13 [38.5%] vs 62/73 [84.9%]; OR, 0.07 [95% CI, 0.0-0.3]) and reported significantly fewer days of symptoms (10.2 vs 16.4; difference, −6.1 [95% CI, −11.8 to −0.4] days). Among those with Omicron infection, the risk of symptomatic infection did not differ significantly for the 2-dose vaccination status vs unvaccinated status and was significantly higher for the 3-dose recipients vs those who were unvaccinated (327/370 [88.4%] vs 85/107 [79.4%]; OR, 2.0 [95% CI, 1.1-3.5]). Among symptomatic Omicron infections, those vaccinated with the third dose 7 to 149 days before infection compared with those who were unvaccinated were significantly less likely to report fever or chills (160/311 [51.5%] vs 64/81 [79.0%]; OR, 0.25 [95% CI, 0.1-0.5]) or seek medical care (45/308 [14.6%] vs 20/81 [24.7%]; OR, 0.45 [95% CI, 0.2-0.9]). Participants with Delta and Omicron infections who received the second dose 14 to 149 days before infection had a significantly lower mean viral load compared with unvaccinated participants (3 vs 4.1 log10 copies/μL; difference, −1.0 [95% CI, −1.7 to −0.2] for Delta and 2.8 vs 3.5 log10 copies/μL, difference, −1.0 [95% CI, −1.7 to −0.3] for Omicron).
Conclusions and Relevance
In a cohort of US essential and frontline workers with SARS-CoV-2 infections, recent vaccination with 2 or 3 mRNA vaccine doses less than 150 days before infection with Delta or Omicron variants, compared with being unvaccinated, was associated with attenuated symptoms, duration of illness, medical care seeking, or viral load for some comparisons, although the precision and statistical significance of specific estimates varied.
This study uses a prospective cohort of essential and frontline workers in 6 US states with SARS-CoV-2 infection to examine associations between 2 or 3 doses of mRNA COVID-19 vaccine and attenuation of symptoms and viral RNA load across SARS-CoV-2 viral lineages.
Introduction
There are limited data on the epidemiology of asymptomatic SARS-CoV-2 infections or mild to moderately severe COVID-19 disease, which constitute most SARS-CoV-2 infections and are believed to trigger and fuel SARS-CoV-2 outbreaks.1,2,3,4 Although early reports from prospective community cohorts observed that COVID-19 vaccines attenuated COVID-19 symptoms, duration, and viral RNA shedding,5,6,7,8 results have been mixed in recent studies,9,10,11 which may be due to the waning of vaccine-associated immunity,12,13 changes in virologic features and heightened immune evasion by the Delta14 and Omicron variants,15 or a combination of factors. Data are also limited from studies that conduct routine weekly testing for SARS-CoV-2 in the community or compare multiple SARS-CoV-2 strains within the same population.
This study describes a prospective cohort of essential and frontline workers in 6 US states with SARS-CoV-2 infection identified through weekly surveillance testing irrespective of symptoms and has 3 objectives. First, the study described COVID-19 by characterizing clinical and virologic features of infections with the original strains (origin), B.1.617.2 and all AY lineages (Delta), and B.1.1.529 and BA1 lineages (Omicron) and assessing associations between clinical characteristics and viral RNA load. Second, the study assessed the association of second and third doses of mRNA COVID-19 vaccines with the occurrence of symptomatic disease, symptoms, and viral load. Third, the study examined the potential transmissibility of COVID-19 by clinical presentation and vaccination status assessing viral viability by culture for the origin and Delta variant viruses.
Methods
Study Design and Participants
The HEROES-RECOVER network consists of large prospective cohorts of health care personnel, first responders, and other essential and frontline workers, defined as those who work at least 20 hours per week in an occupation involving regular contact within 3 feet of others, such as workers in the following sectors: education, agriculture, food processing, transportation, solid waste collection, utilities, government services, childcare, environmental services, and hospitality. The study was conducted in 6 US states (Arizona, Florida, Minnesota, Oregon, Texas, and Utah) with participants recruited through ongoing occupational cohort studies, health management organization membership, and university or hospital system employment using a stratified enrollment approach according to age and occupational category (eMethods in Supplement 1).16,17 All participants provided written informed consent, and their common protocols and procedures were approved by the institutional review boards at participating sites. The data for these analyses were collected from December 14, 2020, (after authorization of mRNA vaccines) to April 19, 2022, with follow-up until May 9, 2022, reported. The analysis was conducted among participants who tested positive for SARS-CoV-2, excluding those with SARS-CoV-2 infections prior to the study start date, infected prior to completion of a primary mRNA COVID-19 vaccine series (<14 days after dose 2), infected either less than 7 days or at greater than or equal to 150 days after receipt of dose 3, or who received the Ad26.COV2.S (Janssen/Johnson & Johnson) vaccine (eFigure 1 in Supplement 1).
Participants reported sociodemographic information, including self-reported race and ethnicity, history of SARS-CoV-2 infection, and chronic medical conditions (using a combination of fixed categories and open-ended text). Race and ethnicity were assessed due to the potential for differential vaccine receipt by race and ethnicity. Active surveillance for symptoms associated with COVID-19 (defined as presence of 1 or more of the following: fever, chills, cough, shortness of breath, sore throat, diarrhea, muscle aches, or a change in smell or taste) was conducted through weekly text messages, emails, and reports obtained directly from the participant.
COVID-19 vaccination status was reported by participants via online surveys and verified through direct upload of images of vaccination cards at all sites. In addition, electronic medical records, occupational health records, or state immunization registries were reviewed by site staff in Minnesota, Oregon, Texas, and Utah.
Laboratory Testing
Participants self-collected midturbinate nasal swabs, irrespective of symptoms, each week and at the onset of COVID-19 symptoms. Specimens were shipped on cold packs and tested by qualitative reverse transcriptase–polymerase chain reaction (RT-PCR) assay at the Marshfield Clinic Research Institute. Virus lineage was determined by whole genome sequencing18 or imputed as the predominant circulating virus (eMethods in Supplement 1). To estimate viral shedding, viral RNA load (hereafter referred to as viral load) was assessed by quantitative RT-PCR assays conducted at the Wisconsin State Laboratory of Hygiene.
To assess virus viability, infectious virus particles were quantified by determination of plaque-forming units (PFU) per milliliter on Vero cells ectopically expressing TMPRSS3 and human ACE219 at St Jude Children’s Research Hospital. Infected cell monolayers were stained and PFUs were counted visually. Specimens were eligible for PFU assessment if the RT-PCR cycle threshold was less than 30, virus lineage had been confirmed, and the viral load was greater than 1 log10 RNA copies/μL (eMethods in Supplement 1). Attempts to culture Omicron variant viruses were not successful.
Exposures
SARS-CoV-2 infection lineage and vaccination status were the primary exposures of interest. Virus lineage was categorized as origin (defined as viruses circulating prior to recognized variants), Delta variant, or Omicron variant. The infection index date was defined as the date of illness onset or specimen collection if asymptomatic. Vaccination status was categorized by doses received relative to the infection index date: 2 doses received within 14 to 149 days prior to infection, 2 doses received at least 150 days prior to infection, and 3 doses received within 7 to 149 days prior to infection. The comparator group was unvaccinated.
Outcomes
Clinical outcomes were presence of symptomatic COVID-19 and, among cases of symptomatic COVID-19, the number of symptoms (0 to 19), presence of fever or chills, illness duration, number of days sick in bed for at least half the day,20 hours of work missed due to illness, and medical care seeking. Clinical outcomes were self-reported via online surveys sent to participants on reporting of COVID-19 symptoms. Additional online surveys were sent after 7 days if symptoms were ongoing and on illness resolution; asymptomatic participants did not receive illness surveys but confirmed absence of symptoms at the time RT-PCR test results were communicated.
Virologic outcomes were viral load and PFU counts from self-collected specimens. When viral load was available from more than 1 specimen per infection, the primary analyses referred to the specimen with the highest viral load. Viral load is reported as overall viral load, peak viral load, time until peak, and time until load was less than 1. Viral load range was 10 to 6 million U/μL. PFU range was 20 to 11 000 U/mL. Both outcomes were analyzed and reported as log10 units.
Statistical Analysis
For COVID-19 clinical outcomes, odds ratios (ORs) were estimated with logistic regression. Mean differences for clinical and virologic outcomes were estimated with linear regression. All models were adjusted for race, ethnicity, and number of daily prescription medications for potential association with vaccination status and outcomes (eTables 3 and 4 in Supplement 1). Occupation was included in adjusted models for the association between virus lineage and missed work, but could not be included in models assessing the association of vaccination status and missed work due to collinearity with vaccine exposure. Analyses of viral load and PFU counts were adjusted for days between index and specimen collection dates. Model assumptions, including linearity, collinearity, and equal variance for linear models, were assessed and met.
Similar to previously published methods,21 viral load by days since onset was estimated using bayesian linear hierarchical models with virus lineage or presence of symptomatic illness as exposures. Time since index date was fitted as a 3-knot natural spline. Models were fit among participants with at least 3 positive RT-PCR specimens and at least 1 specimen with quantifiable load. Viral load was predicted for positive specimens with too little virus to quantify using the linear relationship between the qualitative RT-PCR cycle threshold value and viral load. A sensitivity analysis required at least 2 quantifiable specimens (eMethods in Supplement 1). Models converged with R-hat less than 1.05 and effective sample size greater than 100.
Two-sided 95% CIs for ORs that did not include 1.0 and mean differences that did not include 0 were considered statistically significant. All analyses were conducted with SAS software, version 9.4 (SAS Institute), and R software, version 4.0.2 (R Foundation for Statistical Computing). Because of the potential for type I error due to multiple comparisons, findings should be interpreted as exploratory.
Results
Cohort Characteristics
Of 7223 participants enrolled in active surveillance, 1710 were excluded due to prior SARS-CoV-2 infections; of the remaining 5513 participants, 997 (18.1%) were primary health care personnel (defined as physicians, physician assistants, nurse practitioners, and dentists), 1785 (32.4%) were nurses and other allied health professionals, 1218 (22.1%) were first responders, and 1513 (27.4%) were other essential/frontline workers. Participants completed a median (IQR) of 90.5% (78.1%-97.0%) of weekly surveillance reporting and specimen collection. Ninety-five percent of illness surveys were completed, with a range of 94.0% to 98.5% completion across the vaccination status groups. A total of 1493 infections were identified by RT-PCR testing. After exclusion criteria (eFigure 1 in Supplement 1), the study sample consisted of 1199 unique infections confirmed by RT-PCR testing during the 70-week study period (Table 1); these consisted of 14.0% origin, 24.0% Delta, and 62.0% Omicron infections. Among the 1199 infected participants, the median (IQR) age was 41 (35.0-50.5) years, 714 (59.5%) were women, 232 (19.3%) were Hispanic, 22 (1.8%) were non-Hispanic Asian, 35 (2.9) were non-Hispanic Black, and 871 (72.6%) were non-Hispanic White (eTable 2 in Supplement 1). At least 1 chronic medical condition was reported for 27.4% of infected participants, and 27.7% took at least 2 medications per day. Characteristics of the uninfected participants not included in this study are shown in Table 1.
Table 1. Characteristics of Study Participants at the Start of Observation Period.
| Characteristics | No. (%) | ||
|---|---|---|---|
| SARS-CoV-2 infection | Uninfected | ||
| Unvaccinated | Vaccinated (≥2 doses) | ||
| All participants | 352 | 847 | 4020 |
| Demographics | |||
| Age, median (IQR), y | 41 (34.0-50.1) | 41.3 (35.0-50.5) | 41 (34.0-50.0) |
| Sex | |||
| Female | 180 (51.1) | 534 (63.0) | 2463 (61.3) |
| Male | 170 (48.3) | 313 (37.0) | 1537 (38.2) |
| Racea | |||
| American Indian/Alaskan Native | 2 (0.57) | 5 (0.59) | 31 (0.77) |
| Asian | 7 (2.0) | 21 (2.5) | 149 (3.7) |
| Black or African American | 7 (2.0) | 22 (2.6) | 89 (2.2) |
| White | 313 (88.9) | 765 (90.3) | 3514 (87.4) |
| Multiracial | 4 (1.1) | 6 (0.71) | 46 (1.1) |
| Did not disclose | 19 (5.4) | 28 (3.3) | 191 (4.8) |
| Ethnicitya | |||
| Hispanic | 83 (23.6) | 150 (17.7) | 722 (18.0) |
| Non-Hispanic | 253 (71.9) | 690 (81.5) | 3228 (80.3) |
| Did not disclose | 16 (4.5) | 7 (0.83) | 70 (1.7) |
| HEROES-RECOVER cohort locations | |||
| Phoenix, Arizona | 74 (21.0) | 162 (19.1) | 733 (25.4) |
| Tucson, Arizona | 123 (34.9) | 195 (23.0) | 1269 (31.6) |
| Miami, Florida | 52 (14.8) | 86 (10.2) | 395 (9.8) |
| Duluth, Minnesota | 18 (5.1) | 116 (13.7) | 458 (11.4) |
| Portland, Oregon | 18 (5.1) | 72 (8.5) | 495 (12.3) |
| Temple, Texas | 25 (7.1) | 72 (8.5) | 309 (7.7) |
| Salt Lake City, Utah | 42 (11.9) | 144 (17.0) | 361 (9.0) |
| Occupationb | |||
| Primary health care personnel | 17 (4.8) | 170 (20.1) | 752 (18.7) |
| Nurses and allied health care personnel | 87 (24.7) | 261 (30.8) | 1332 (33.1) |
| First responders | 145 (41.2) | 191 (22.6) | 819 (20.4) |
| Essential and other frontline workers | 103 (29.3) | 225 (26.6) | 1116 (27.8) |
| Medical history | |||
| Chronic conditionsc | |||
| None | 248 (70.5) | 606 (71.5) | 2781 (69.2) |
| 1 or more | 91 (25.9) | 238 (28.1) | 1137 (28.3) |
| No. of medications | |||
| None | 175 (49.7) | 414 (48.9) | 1872 (46.6) |
| 1 | 69 (19.6) | 181 (21.4) | 841 (20.9) |
| 2 or more | 90 (25.6) | 242 (28.6) | 1171 (29.1) |
| Self-rated health | |||
| Excellent/very good | 231 (65.6) | 622 (73.4) | 2688 (66.9) |
| Good/fair/poor | 121 (34.4) | 225 (26.6) | 1325 (33.0) |
| Vaccine product | |||
| BNT162b2 (Pfizer-BioNTech) | 538 (63.5) | 2050 (51.0) | |
| mRNA-1273 (Moderna) | 269 (31.8) | 1048 (26.1) | |
| Mixed dosingd | 40 (4.7) | 110 (2.7) | |
| SARS-CoV-2 lineage | |||
| Origin strains | 165 (46.9) | 3 (0.4) | |
| Delta variant | 78 (22.2) | 210 (24.8) | |
| Omicron variant | 109 (31.0) | 634 (74.9) | |
Abbreviations: HEROES, the Arizona Healthcare, Emergency Response, and Other Essential Workers Surveillance Study; RECOVER, Research on the Epidemiology of SARS-CoV-2 in Essential Response Personnel.
Race and ethnicity determined by participant self-report using closed categories.
Primary health care providers included physicians, physician assistants, nurse practitioners, and dentists; allied health care personnel included nurses, therapists, technicians, medical assistants, orderlies, and all others providing clinical support in inpatient or outpatient settings; first responders included firefighters, law enforcement, corrections officers, and emergency medical technicians; and other essential and frontline workers included teachers and hospitality, delivery, and retail workers, as well as all other occupations that require routine close contact with the public, customers, or coworkers.
Chronic conditions included asthma, chronic lung disease, cancer, diabetes, heart disease or condition, immunosuppression, kidney disease, liver disease, neurologic or neuromuscular disease, and autoimmune disease.
Mixed dosing refers to participants who received a third vaccine dose with a different vaccine manufacturer than the first 2 doses.
Of the 1199 participants with SARS-CoV-2 infections, 352 (29.4%) were unvaccinated, 72 (6.0%) received the second dose 14 to 149 days before infection, 374 (31.2%) received the second dose at least 150 days before infection, and 401 (33.4%) received the third dose 7 to 149 days before infection (Table 2; eTable 1 in Supplement 1). Of those who received the third dose, 11 (2.7%) were immunocompromised by self-report; dose 3 was considered primary series vaccination for these individuals. All participants received the same vaccine product for dose 1 and dose 2: 280 (62.3%) received BNT162b2 (Pfizer-BioNTech) and 166 (37.2%) received mRNA-1273 (Moderna) vaccines. For dose 3, a total of 22 of 401 individuals (5.5%) received a different mRNA vaccine product than their primary series.
Table 2. COVID-19 Characteristics for Delta and Omicron Variants by mRNA COVID-19 Vaccination Status Among Adults With SARS-CoV-2 Infection.
| Illness characteristics | Unvaccinated, No. (%) | 2 Doses 14-149 d prior to infection | 2 Doses ≥150 d prior to infection | 3 Doses 7-149 d prior to infection | |||
|---|---|---|---|---|---|---|---|
| No. (%) | Difference vs unvaccinated (95% CI)a,b | No. (%) | Difference vs unvaccinated (95% CI)a,b | No. (%) | Difference vs unvaccinated (95% CI)a,b | ||
| SARS-CoV-2 infections by lineage | |||||||
| Origin strain | 165/168 (98.2) | 3/168 (1.8) | 0/168 | 0/168 | |||
| Delta variant | 78/288 (27.1) | 27/288 (9.4) | 165/288 (57.3) | 18/288 (6.3) | |||
| Omicron variant | 109/743 (14.7) | 42/743 (5.7) | 209/743 (28.1) | 383/743 (51.6) | |||
| Total | 352 (29.4) | 72 (6) | 374 (31.2) | 401 (33.4) | |||
| Illness characteristics by lineagec | |||||||
| Delta variant | |||||||
| Symptomatic COVID-19d | 74/77 (96.1) | 21/27 (77.8) | OR, 0.13 (0.0 to 0.6) | 152/163 (93.3) | OR, 0.53 (0.1 to 2.0) | 13/18 (72.2) | OR, 0.13 (0.0 to 0.7) |
| Among symptomatic illnesses | |||||||
| Fever or chillse,f | 62/73 (84.9) | 15/19 (79.0) | OR, 0.66 (0.2 to 2.5) | 100/142 (70.4) | OR, 0.39 (0.2 to 0.9) | 5/19 (38.5) | OR, 0.07 (0.0 to 0.3) |
| Received medical caree,g | 35/74 (47.3) | 5 (26.3) | OR, 0.41 (0.1 to 1.3) | 45/143 (31.5) | OR, 0.51 (0.3 to 1.0) | 3/12 (25) | OR, 0.32 (0.1 to 1.3) |
| No. of symptoms, mean (SD)e,h | 9.0 (3.9) [n = 73] | 7.5 (3.5) [n = 19] | −1.3 (−3.2 to 0.5) | 7.5 (3.7) [n = 143] | −1.4 (−2.5 to −0.3) | 5.2 (3) [n = 13] | −4.3 (−6.5 to −2.1) |
| Duration of symptoms, mean (SD), di | 16.4 (8.6) [n = 74] | 13.6 (11) [n = 21] | −2.5 (−7.3 to 2.2) | 17.1 (10) [n = 151] | 0.6 (−2.2 to 3.4) | 10.2 (5.4) [n = 13] | −6.1 (−11.8 to −0.4) |
| Time spent sick in bed for half the day, mean (SD), dj | 4.9 (4.6) [n = 58] | 2.3 (2.2) [n = 16] | −2.3 (−4.4 to −0.2) | 3.7 (3.9) [n = 120] | −1.1 (−2.3 to 0.2) | 1.8 (1.9) [n = 9] | −3.6 (−6.3 to −0.9) |
| Time missed work, mean (SD), hj | 62.8 (35) [n = 58] | 43.8 (372) [n = 16] | −17.3 (−36.5 to 1.8) | 47.1 (32.3) [n = 120] | −15 (−26.4 to −4.1) | 55.1 (42.8) [n = 9] | −10.3 (−34.5 to 14.0) |
| Omicron variant | |||||||
| Symptomatic COVID-19d | 85/107 (79.4) | 36/41 (87.8) | OR, 2.0 (0.7 to 5.6) | 180/206 (87.4) | OR, 1.8 (1.0 to 3.4) | 327/370 (88.4) | OR, 2.0 (1.1 to 3.5) |
| Among symptomatic illnesses | |||||||
| Fever or chillse,f | 64/81 (79) | 31/36 (86.1) | OR, 1.5 (0.5 to 4.5) | 132/169 (78.1) | OR, 0.85 (0.4 to 1.7) | 160/311 (51.5) | OR, 0.25 (0.1 to 0.5) |
| Received medical caree,k | 20/81 (24.7) | 4/36 (11.8) | OR, 0.36 (0.1 to 1.2) | 33/169 (19.5) | OR, 0.7 (0.4 to 1.4) | 45/308 (14.6) | OR, 0.45 (0.2 to 0.9) |
| No. of symptoms, mean (SD)e,h | 6.9 (3.1) [n = 81] | 7.9 (3.1) [n = 36] | 0.9 (−0.4 to 2.2) | 7.3 (3.5) [n = 169] | 0.2 (−0.7 to 1.1) | 5.9 (3.2) [n = 311] | −1.3 (−2.1 to −0.4) |
| Duration of symptoms, mean (SD), di | 12.3 (9.1) [n = 85] | 14.3 (9.9) [n = 36] | 2.1 (−1.1 to 5.3) | 12.1 (8.2) [n = 180] | −0.1 (−2.3 to 2.0) | 11.8 (7.6) [n = 327] | −0.4 (−2.4 to 1.5) |
| Time spent sick in bed for half the day, mean (SD), dj | 2.6 (3.1) [n = 72] | 2.5 (1.9) [n = 33] | −0.3 (−1.4 to 0.7) | 2.8 (2.6) [n = 149] | 0.1 (−0.7 to 0.9) | 1.9 (2.6) [n = 282] | −0.9 (−1.6 to −0.2) |
| Time missed work, mean (SD), hj | 35.9 (31.5) [n = 72] | 33.5 (23.8) [n = 33] | −3.9 (−15.1 to 7.3) | 33.3 (26.8) [n = 119] | −4.1 (−11.9 to 3.7) | 26.6 (25.3) [n = 282] | −11.1 (−18.4 to −3.9) |
Odds ratios (ORs) estimated using logistic regression with unvaccinated as the referent participant group adjusted for number of daily medications and race and ethnicity.
Mean differences compared with unvaccinated estimated using linear regression and adjusted for number of daily medications and race and ethnicity.
Due to the small number of breakthrough infections associated with origin strains, comparisons are limited to Delta and Omicron variants.
Assessed among participants reporting COVID-19–like symptoms or no symptoms.
Assessed among participants who reported COVID-19 symptoms and responded to the illness survey.
Fever or chills defined as self-reported fever, feverishness, chills, or measured temperature higher than 38 °C.
Among those who received medical care, 14 went to the emergency department and 4 were hospitalized.
Participants were asked about the following: fever or feeling feverish, chills, dry cough, cough that produces phlegm or sputum, sore throat, runny nose or nasal congestion, shortness of breath or difficulty breathing, muscle or body aches, joint pain, abdominal pain, nausea or queasy feeling that you may vomit, vomiting, diarrhea, headache, fatigue, rash, discoloration or blistering of fingers or toes, eye redness, and loss of smell or taste.
Assessed among participants who reported COVID-19 symptoms.
Assessed among participants who reported COVID-19 symptoms and responded to questions on days spent sick in bed for at least half of the day and hours of missed work.
Among those who received medical care, 6 were hospitalized.
Clinical Characteristics of COVID-19 Among Unvaccinated Participants
To describe the clinical characteristics of infection by each viral lineage, the 352 episodes of infection that occurred among unvaccinated individuals (among the 1199 total infection episodes) were examined (Table 3); of these, 44 (12.5%) were asymptomatic and 24 (6.8%) had symptoms not in the COVID-19 case definition. Asymptomatic infections were significantly more commonly associated with Omicron compared with Delta variants (OR, 5.6 [95% CI, 1.6-19.6]). When symptomatic, 64.8% of participants with COVID-19 reported fever or chills; the percentage was significantly higher for Delta compared with origin infection (84.9% vs 71.8%; OR, 2.2 [95% CI, 1.1-4.8]). Medical care seeking was significantly less common among Omicron infections (24.7%) compared with Delta (47.3%) infections (OR, 0.39 [95% CI, 0.2-0.8]), but was not significantly different among Omicron vs origin (36.9%) infections (OR, 1.8 [95% CI, (1.0-3.2)]).
Table 3. COVID-19 Characteristics Among Unvaccinated Participants by SARS-CoV-2 Lineage.
| Characteristica | No. (%) | Difference (95% CI) | ||||
|---|---|---|---|---|---|---|
| Origin strains | Delta | Omicron | Delta vs origin | Omicron vs origin | Omicron vs Delta | |
| Asymptomatic SARS-CoV-2b | 19/165 (11.5) | 3/78 (3.9) | 22/109 (20.2) | OR, 0.36 (0.1 to 1.1)c | 1.8 (0.9 to 3.6) | 5.6 (1.6 to 19.6) |
| Symptoms, mean (SD)d,e | 7.7 (3.4) [n = 142] | 9 (3.9) [n = 73] | 6.9 (3.1) [n = 81] | 1.3 (0.3 to 2.3)f | −0.60 (−1.5 to 0.4) | −1.9 (−3.0 to −0.8) |
| Fever or chillsd,g | 102/142 (71.8) | 62/73 (84.9) | 64/81 (79) | OR, 2.2 (1.0 to 4.8)c | 1.6 (0.8 to 3.1) | 0.71 (0.3 to 1.7) |
| Received medical care for COVID-19 d,h | 52/141 (36.9) | 35/74 (47.3) | 20/81 (24.7) | OR, 1.8 (1.0 to 3.2)c | 0.69 (0.4 to 1.3) | 0.39 (0.2 to 0.8) |
| Duration of COVID-19 symptoms, mean (SD)i | 15.6 (9.4) [n = 144] | 16.4 (8.6) [n = 74] | 12.3 (9.1) [n = 85] | 0.80 (−1.9 to 3.4)f | −2.9 (−5.5 to −0.3) | −3.6 (−6.6 to −0.7) |
| Time spent sick in bed for at least half a day, mean (SD), dj | 4.3 (4.3) [n = 120] | 4.9 (4.6) [n = 58] | 2.6 (3.1) [n = 71] | 0.8 (−0.5 to 2.1)f | −1.2 (−2.5 to 0.0) | −2.0 (−3.4 to −0.6) |
| Time missed work due to COVID-19, mean (SD), hj | 54.5 (41.4) [n = 120] | 62.8 (35.0) [n = 58] | 35.9 (31.5) [n = 72] | 6.3 (−5.0 to 17.6)k | −19.2 (−29.8 to −8.6) | −25.5 (−8.0 to −13.0) |
| Viral RNA load, mean (SD), log10 copies/μL | 3.9 (1.7) [n = 165] | 4.0 (1.8) [n = 78] | 3.5 (1.8) [n = 109] | 0.18 (−0.31 to 0.68)f | −0.30 (−0.77 to 0.16) | −0.49 (−1.03 to 0.06) |
| Plaque-forming units, mean (SD), log10 copies/mL | 3.5 (1.0) [n = 9] | 4.8 (1.4) [n = 36] | 1.7 (0.7 to 2.7)f | |||
Illness characteristics are limited to unvaccinated participants to describe the natural history of infection without the vaccine-induced changes to immunity or the potential attenuation from vaccination.
Asymptomatic compared with COVID-19–like illness symptoms or non-COVID-19–like illness symptoms.
Odds ratios estimated using logistic regression with Ancestral strains as the referent participant group adjusted for number of daily medications and race and ethnicity.
Assessed among participants who reported COVID-19–like symptoms and responded to the illness survey.
Participants were asked about the following: fever or feeling feverish, chills, dry cough, cough that produces phlegm or sputum, sore throat, runny nose or nasal congestion, shortness of breath or difficulty breathing, muscle or body aches, joint pain, abdominal pain, nausea or queasy feeling that you may vomit, vomiting, diarrhea, headache, fatigue, rash, discoloration or blistering of fingers or toes, eye redness, and loss of smell or taste.
Mean difference estimated with linear regression adjusted for number of daily medications and race and ethnicity.
Fever or chills defined as self-reported fever, feverishness, chills, or measured temperature higher than 38 °C.
Among 296 unvaccinated participants who received medical care, including 273 who received outpatient care, 15 who went to the emergency department, and 8 who were hospitalized; 8 participants who did not report whether medical care was received during illness are excluded.
Assessed among participants who reported COVID-19–like symptoms.
Assessed among participants who reported COVID-19–like symptoms and responded to questions on days spent sick in bed for at least half a day and hours of missed work.
Mean difference estimated with linear regression adjusted for number of daily medications, race and ethnicity, and occupation.
Among symptomatic participants, COVID-19 symptoms associated with infection due to the Omicron variant lasted a mean (SD) of 12.3 (9.1) days, which was 2.9 (95% CI, −5.5 to −0.3) days fewer than origin (mean of 15.6 days) infection and 3.6 (95% CI, −6.6 to −0.7) days fewer than Delta (mean of 16.4 days) infection. Participants with Omicron infection spent a mean (SD) of 2.6 (3.1) days sick in bed, which was a mean of 1.2 (95% CI, −2.5 to −0.0) days fewer than those with origin infection and 2.0 (95% CI, −3.4 to −0.6) days fewer than those with Delta infection. Further findings regarding the associations between clinical and participant characteristics are in Table 3 and eTable 3 in Supplement 1.
Comparison of Delta and Omicron Clinical Characteristics by Vaccination Status
Vaccinated and unvaccinated participants were examined to assess potential vaccine-associated attenuation of disease. Among participants with Delta infections, illness was milder among those who were vaccinated, although the precision of these estimates varied (Table 2). Participants who received the second dose 14 to 149 days before Delta infection compared with those unvaccinated were significantly less likely to be symptomatic (77.8% vs 96.1%; OR, 0.13 [95% CI, 0.0-0.6]) and spent significantly fewer days sick in bed (2.3 vs 4.9; difference, −2.3 [95% CI, −4.4 to −0.2] days). Symptomatic participants who received dose 2 at least 150 days prior to Delta infection compared with those who were unvaccinated were significantly less likely to report fever or chills (70.4% vs 84.9%; OR, 0.39 [95% CI, 0.2-0.9]) or seek medical care (31.5% vs 47.3%; OR, 0.51 [95% CI, 0.3-1.0]) and missed significantly fewer hours of work (47.1 vs 62.8; difference, −15.2 [95% CI, −26.4 to −4.1] hours). Participants who received the third vaccine dose 7 to 149 days prior to Delta infection compared with those who were unvaccinated were significantly less likely to have symptomatic illness (72.2% vs 77.8%; difference, 0.13% [95% CI, 0.0%-0.7%]) and, when symptomatic, were significantly less likely to report fever or chills (38.5% vs 84.9%; OR, 0.07 [95% CI, 0.0-0.3]) and spent significantly fewer days symptomatic (mean [SD] of 10.2 [5.4] days vs 16.4 [8.6]; difference, −6.1 [95% CI, −11.8 to −0.4] days).
Significant differences in clinical presentation between unvaccinated individuals and those who received 2 vaccine doses were not observed among Omicron variant infections. However, symptomatic participants who received the third vaccine dose 7 to 149 days prior to Omicron infection compared with those who were unvaccinated were significantly less likely to report fever or chills (51.5% vs 79.0%; OR, 0.25 [95% CI, 0.1-0.5]) or receive medical care (14.6% vs 24.7%; OR, 0.45 [95% CI, 0.2-0.9]) and missed significantly fewer hours of work (mean [SD] of 26.6 [25.3] vs 35.9 [31.5]; difference, −11.1 [95% CI, −18.4 to −3.9] hours), although guidance on postinfection isolation and returning to work changed during the Omicron wave. Participants who received the third vaccine dose 7 to 149 days prior to Omicron infection were more likely to be symptomatic than those who were unvaccinated (88.4% vs 79.4%; OR, 2.0 [95% CI, 1.1-3.5]).
Viral Load
For 886 participants (73.8%), the specimen with the highest viral load was their first positive specimen collected. Mean viral load did not differ by demographic characteristics (eTable 4 in Supplement 1). Symptomatic infections compared with asymptomatic infections had a significantly higher mean viral load (mean difference, 1.3 log10 copies/μL [95% CI, 0.3-1.6]; Figure 1); this difference occurred consistently for Delta and Omicron infections (eTable 5 in Supplement 1). For all studied lineages, mean viral load was significantly higher for infections with fever or chills (3.8 vs 3.4 log10 copies/μL; mean difference, 0.4 [95% CI, 0.2-0.7]), those with 8 or more symptoms (3.9 vs 3.0 log10; mean difference, 0.9 [95% CI, 0.6-1.3] log10 copies/μL), and those with symptoms lasting at least 14 days (4.0 vs 3.5; mean difference, 0.6 [95% CI, 0.3-0.9] log10 copies/μL) (Figure 1 and eTable 5 in Supplement 1).
Figure 1. Viral Load and Plaque-Forming Units of SARS-CoV-2 Infection by Virus Lineage, Vaccination Status, and COVID-19 Characteristics.
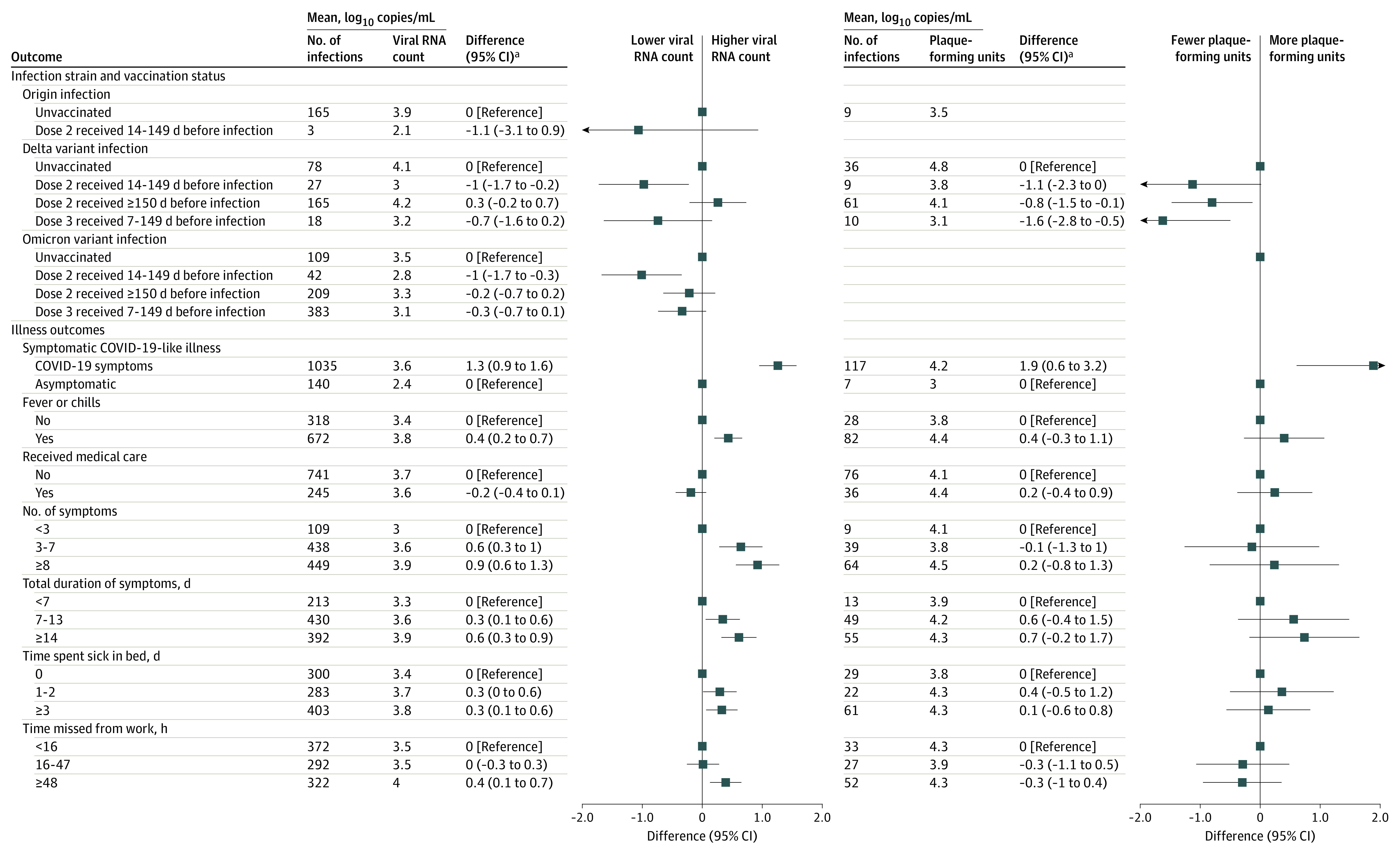
Viral load was measured by quantitative reverse transcriptase–polymerase chain reaction assay to assess viral shedding. Plaque-forming units were quantified on Vero cells ectopically expressing transmembrane serine protease 3 and human angiotensin converting enzyme 2 to assess virus viability.
aMean difference and 95% CIs were estimated using linear regression controlling for time from index to swab collection, number of daily medications reported, and race/ethnicity. Hours of missed work is additionally adjusted for occupation.
Participants with Delta and Omicron infections who received the second vaccine dose 14 to 149 days before infection had a significantly reduced mean viral load compared with unvaccinated participants (3 vs 4.1 log10 copies/μL; mean difference, −1.0 [95% CI, −1.7 to −0.2] log10 copies/μL for Delta; 2.8 vs 3.5 log10 copies/μL; mean difference, −1.0 [95% CI, −1.7 to −0.3] log10 copies/μL for Omicron) (Figure 1).
Viral Load Peak and Decline
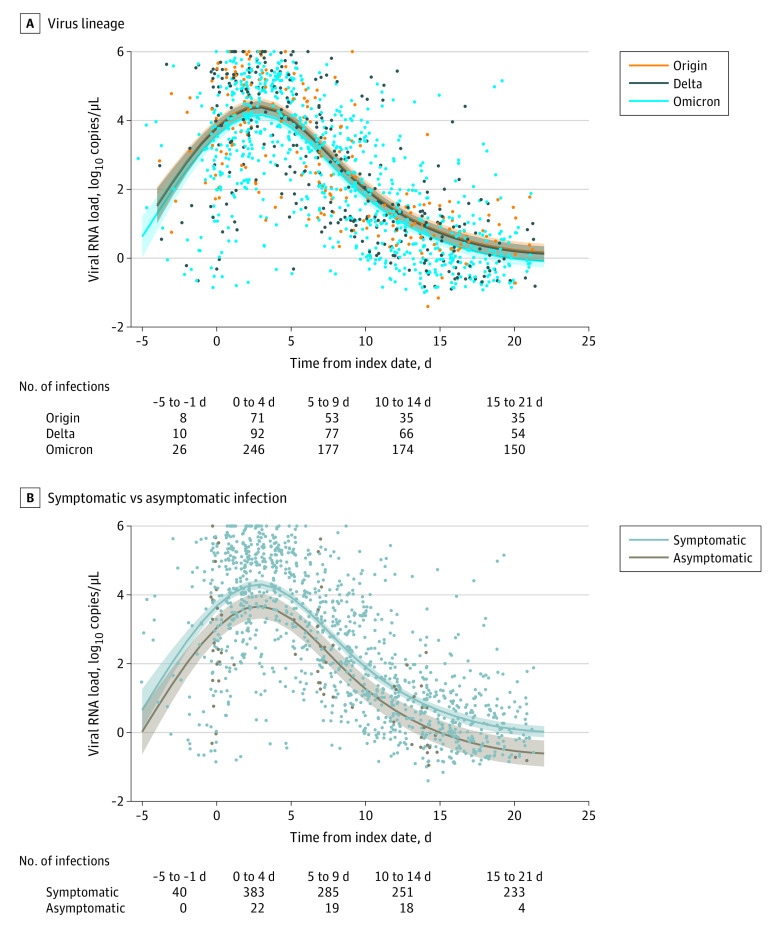
Viral load over the course of infection was estimated using a bayesian hierarchical model among 407 vaccinated and unvaccinated participants who contributed at least 3 specimens positive for SARS-CoV-2 (eTable 6 in Supplement 1). The time from SARS-CoV-2 detection to mean peak viral load was 2.5 days for all 3 virus lineages. Overall mean viral load for origin and Delta infections were higher than for Omicron (mean difference, 0.2 [95% credible interval (CrI), 0.0-0.4] log10 copies/μL for origin infection; mean difference, 0.2 [95% CrI, 0.1-0.5] log10 copies/μL for Delta infection) (Figure 2A). During the estimated peak, viral load did not differ by viral lineage. The estimated time of decline in viral load from peak to a count of less than 1 log10 copies/μL was 11 days for both origin and Delta infection and 10 days for Omicron infection (Figure 2A).
Figure 2. Estimated Viral Load Over the Course of SARS-CoV-2 Infection for Both Vaccinated and Unvaccinated Participants by Virus Lineage and Symptomatic vs Asymptomatic Infections.
Viral load over the time course of infection for vaccinated and unvaccinated participants with at least 3 specimens. Index date is defined as the date of illness onset or the date of specimen collection if the infection was asymptomatic. Negative days indicate infection prior to symptom start. Points are individual positive reverse–transcriptase polymerase chain reaction specimens. Specimens below 1 on the y-axis represent a viral load below the minimum quantifiable level and estimated from cycle threshold values. 106 is the maximum quantifiable by the assay. Curves are predicted mean viral load from bayesian linear models with time from index as a 3-knot natural spline. A, Overall viral count of origin strains and Delta variants are higher than Omicron variants, with mean differences of 0.26 (95% CI, 0.0-0.5) and 0.21 (95% CI, 0.0-0.4), respectively. B. Symptomatic COVID-19 infections had significantly higher viral count than asymptomatic infections (mean difference, 0.70 [95% CI, 0.4-1.1]).
Across viral lineages, the mean viral load for asymptomatic infections was lower than for symptomatic COVID-19 (mean difference, 0.7 [95% CrI, 0.4-1.1] log10 copies/μL) (Figure 2B). Estimated peak viral load was also lower for asymptomatic infections (3.8 [95% CrI, 3.4-4.2] log10 copies/μL) than for symptomatic COVID-19 (4.5 [95% CrI, 4.4-4.6] log10 copies/μL) (eTable 6 in Supplement 1). The estimated time from peak viral load to a count of less than 1 log10 copies/μL was 8 days for asymptomatic infections and 10 days for symptomatic COVID-19 (Figure 2B).
Infectious SARS-CoV-2 Counts
Counts of viable viral particles were only examined for unvaccinated participants infected with origin viruses and among both vaccinated and unvaccinated participants infected with the Delta variant (Figure 1). Mean PFU was significantly higher for participants with symptomatic COVID-19 compared with those with asymptomatic infections (4.2 vs 3; mean difference, 1.9 [95% CI, 0.6-3.2] PFU/mL). Mean PFU was significantly lower among participants who received dose 2 at least 150 days before breakthrough infection or the third dose 7 to 149 days before breakthrough infection compared with unvaccinated participants (4.1 vs 4.8; mean difference, −0.8 [95% CI, −1.5 to −0.1] PFU/mL and 3.1 vs 4.8; mean difference, −1.6 [95% CI, −2.8 to −0.5] PFU/mL) (Figure 1).
Discussion
In this prospective cohort of health care personnel, first responders, and other essential and frontline workers followed up from December 14, 2020, to April 19, 2022, recent receipt of 2 or 3 doses of COVID-19 mRNA vaccine 14 to 149 days prior to Delta or Omicron BA.1 SARS-CoV-2 infection was variably associated with a milder clinical presentation, shorter illness duration, less medical care seeking, and lower viral load, although the precision of specific estimates varied.
The differences between Omicron and Delta infections in clinical and virologic characteristics observed may help explain the previously reported increased transmissibility of the Omicron variant.3,22,23 COVID-19 symptoms associated with Omicron appeared to be milder and of a shorter duration than COVID-19 associated with Delta by many indicators. Additionally, milder symptoms that lasted fewer days and shorter duration of illness was associated with lower viral RNA shedding. In this cohort, 20.2% of infections due to Omicron were asymptomatic, similar to another recent report.24 In contrast, only 3.9% of Delta infections were asymptomatic and were associated with a longer duration of illness. Across all viral lineages in this study, infected participants without symptoms shed viral RNA for about 12 days after peak viral load compared with symptomatic participants with symptoms, who shed viral RNA for about 18 days. During the period of detection, the viral load of Omicron BA.1 infections was similar to Delta infections and higher than infections with origin strains. Although viral RNA shedding cannot be directly attributable to transmission, the relatively high viral load of Omicron infections together with the higher frequency of asymptomatic infection supports previous studies suggesting an association with increased transmission,21,25 particularly during the first 3 to 5 days when viral load peaked.
A study from the HEROES-RECOVER cohort previously reported that participants with origin and other pre-Delta virus infections (from December 2020 to April 2021) after at least 1 dose of mRNA COVID-19 vaccine had lower viral load, were less likely to have febrile symptoms, and had shorter illnesses with fewer days spent sick in bed compared with unvaccinated participants.8 Similarly, in the current study, Delta-associated breakthrough infections 14 to 149 days after dose 2 had lower viral loads compared with those in unvaccinated participants; further, breakthrough infections 14 to 149 days after dose 2 or 7 to 149 days after dose 3 had shorter illnesses, with lower counts of viable and presumed infectious virus. For Omicron infections, only participants with a breakthrough infection after dose 3 showed consistent signs of vaccine attenuation of COVID-19, such as being less likely to have febrile symptoms or seek medical care. It is possible that the recall of immunologic memory that reduces viral replication and accelerates elimination of virally infected cells that may underlie vaccine attenuation of disease26 wanes with time, similar to waning in neutralizing antibodies27 and clinical effectiveness of mRNA vaccines.12
Limitations
This study has several limitations. First, although this was among the largest studies of its kind with routine surveillance testing for SARS-CoV-2, stratification by virus lineage and vaccine exposures resulted in relatively small subgroups, which reduced the precision of estimates. Second, although several relevant confounders were controlled for, unmeasured confounding in this observational study could have occurred. Limitations in sample size and the ability to adjust models for potential confounders made it particularly difficult to interpret unexpected findings, such as a higher percentage of individuals with symptomatic disease among those vaccinated with the third vaccine dose 14 to 149 days before Omicron infection compared with those who were unvaccinated. Third, findings regarding virologic features of COVID-19 are limited to virus present in the nasal cavity, self-collected with midturbinate nasal swabs. Therefore, possible differences between variants in viral RNA present in the full nasopharynx or lower respiratory system could not be examined. Fourth, due to methodological obstacles culturing Omicron variant viruses, virus viability could only be examined with origin and Delta viruses. Fifth, the detection of viral RNA and isolation of viable virus through virus culture are not direct measures of virus transmissibility, although evidence is emerging for SARS-CoV-225 and both are associated with transmission of other viral infections.28,29,30 Sixth, findings on missed work hours should be interpreted with caution because guidance on postinfection isolation and returning to work changed over time and work-related isolation protocols were not considered. Seventh, participant-reported symptoms and duration of illness may be subject to recall and confirmation biases. Eighth, these findings may not generalize to all population in the US, especially given the relatively low percentage of participants who were not White.
Conclusions
In a cohort of US essential and frontline workers with SARS-CoV-2 infections, recent vaccination with 2 or 3 mRNA vaccine doses less than 150 days before infection with Delta or Omicron variants, compared with being unvaccinated, was associated with attenuated symptoms, duration of illness, medical care seeking, and viral load for some comparisons, although the precision and statistical significance of specific estimates varied.
eResults
Nonauthor collaborators
References
- 1.Meyerowitz EA, Richterman A, Bogoch II, Low N, Cevik M. Towards an accurate and systematic characterisation of persistently asymptomatic infection with SARS-CoV-2. Lancet Infect Dis. 2021;21(6):e163-e169. doi: 10.1016/S1473-3099(20)30837-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science. 2020;368(6490):489-493. doi: 10.1126/science.abb3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iuliano AD, Brunkard JM, Boehmer TK, et al. Trends in disease severity and health care utilization during the early omicron variant period compared with previous SARS-CoV-2 high transmission periods: United States, December 2020-January 2022. MMWR Morb Mortal Wkly Rep. 2022;71(4):146-152. doi: 10.15585/mmwr.mm7104e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipsitch M, Krammer F, Regev-Yochay G, Lustig Y, Balicer RD. SARS-CoV-2 breakthrough infections in vaccinated individuals: measurement, causes and impact. Nat Rev Immunol. 2022;22(1):57-65. doi: 10.1038/s41577-021-00662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strum E, Casagrande Y, Newton K, Unger JB. Healthcare workers benefit from second dose of COVID-19 mRNA vaccine: effects of partial and full vaccination on sick leave duration and symptoms. Public Health Pract (Oxf). 2022;3:100247. doi: 10.1016/j.puhip.2022.100247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadoff J, Gray G, Vandebosch A, et al. ; ENSEMBLE Study Group . Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. 2021;384(23):2187-2201. doi: 10.1056/NEJMoa2101544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Regev-Yochay G, Amit S, Bergwerk M, et al. Decreased infectivity following BNT162b2 vaccination: a prospective cohort study in Israel. Lancet Reg Health Eur. 2021;7:100150. doi: 10.1016/j.lanepe.2021.100150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson MG, Burgess JL, Naleway AL, et al. Prevention and attenuation of Covid-19 with the BNT162b2 and mRNA-1273 vaccines. N Engl J Med. 2021;385(4):320-329. doi: 10.1056/NEJMoa2107058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown CM, Vostok J, Johnson H, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings: Barnstable County, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. 2021;70(31):1059-1062. doi: 10.15585/mmwr.mm7031e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riemersma KK, Grogan BE, Kita-Yarbro A, et al. Shedding of infectious SARS-CoV-2 despite vaccination. medRxiv. 2021;doi: 10.1101/2021.07.31.21261387 [DOI] [PMC free article] [PubMed]
- 11.Puhach O, Adea K, Hulo N, et al. Infectious viral load in unvaccinated and vaccinated patients infected with SARS-CoV-2 WT, Delta and Omicron. medRxiv. 2022;doi: 10.1101/2022.01.10.22269010 [DOI] [PubMed]
- 12.Ferdinands JM, Rao S, Dixon BE, et al. Waning 2-dose and 3-dose effectiveness of mrna vaccines against COVID-19-associated emergency department and urgent care encounters and hospitalizations among adults during periods of Delta and Omicron variant predominance: VISION Network, 10 States, August 2021-January 2022. MMWR Morb Mortal Wkly Rep. 2022;71(7):255-263. doi: 10.15585/mmwr.mm7107e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chemaitelly H, Tang P, Hasan MR, et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N Engl J Med. 2021;385(24):e83. doi: 10.1056/NEJMoa2114114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall EC, Wu M, Harvey R, et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet. 2021;397(10292):2331-2333. doi: 10.1016/S0140-6736(21)01290-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dejnirattisai W, Huo J, Zhou D, et al. ; OPTIC Consortium; ISARIC4C Consortium . SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467-484.e15. doi: 10.1016/j.cell.2021.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards LJ, Fowlkes AL, Wesley MG, et al. Research on the epidemiology of SARS-CoV-2 in essential Response Personnel (RECOVER): protocol for a multisite longitudinal cohort study. JMIR Res Protoc. 2021;10(12):e31574. doi: 10.2196/31574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutrick K, Ellingson KD, Baccam Z, et al. COVID-19 infection, reinfection, and vaccine effectiveness in a prospective cohort of Arizona frontline/essential workers: the AZ HEROES research protocol. JMIR Res Protoc. 2021. doi: 10.2196/28925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paden CR, Tao Y, Queen K, et al. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(10):2401-2405. doi: 10.3201/eid2610.201800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama S, Nao N, Shirato K, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117(13):7001-7003. doi: 10.1073/pnas.2002589117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Searle SD, Mitnitski A, Gahbauer EA, Gill TM, Rockwood K. A standard procedure for creating a frailty index. BMC Geriatr. 2008;8:24. doi: 10.1186/1471-2318-8-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones TC, Biele G, Mühlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373(6551):eabi5273. doi: 10.1126/science.abi5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito K, Piantham C, Nishiura H. Relative instantaneous reproduction number of Omicron SARS-CoV-2 variant with respect to the Delta variant in Denmark. J Med Virol. 2022;94(5):2265-2268. doi: 10.1002/jmv.27560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hay JA, Kissler SM, Fauver JR, et al. Viral dynamics and duration of PCR positivity of the SARS-CoV-2 Omicron variant. medRxiv. 2022;doi: 10.1101/2022.01.13.22269257 [DOI]
- 24.Garrett N, Tapley A, Andriesen J, et al. ; Ubuntu Study Team . High asymptomatic carriage with the Omicron variant in South Africa. Clin Infect Dis. 2022;75(1):e289-e292. doi: 10.1093/cid/ciac237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marks M, Millat-Martinez P, Ouchi D, et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis. 2021;21(5):629-636. doi: 10.1016/S1473-3099(20)30985-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferdinands JM, Thompson MG, Blanton L, Spencer S, Grant L, Fry AM. Does influenza vaccination attenuate the severity of breakthrough infections? a narrative review and recommendations for further research. Vaccine. 2021;39(28):3678-3695. doi: 10.1016/j.vaccine.2021.05.011 [DOI] [PubMed] [Google Scholar]
- 27.Falsey AR, Frenck RW Jr, Walsh EE, et al. SARS-CoV-2 neutralization with BNT162b2 vaccine dose 3. N Engl J Med. 2021;385(17):1627-1629. doi: 10.1056/NEJMc2113468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand BS, Velez M. Assessment of correlation between serum titers of hepatitis C virus and severity of liver disease. World J Gastroenterol. 2004;10(16):2409-2411. doi: 10.3748/wjg.v10.i16.2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacDonald M, Crofts N, Kaldor J. Transmission of hepatitis C virus: rates, routes, and cofactors. Epidemiol Rev. 1996;18(2):137-148. doi: 10.1093/oxfordjournals.epirev.a017921 [DOI] [PubMed] [Google Scholar]
- 30.Tsang TK, Cowling BJ, Fang VJ, et al. Influenza A virus shedding and infectivity in households. J Infect Dis. 2015;212(9):1420-1428. doi: 10.1093/infdis/jiv225 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eResults
Nonauthor collaborators