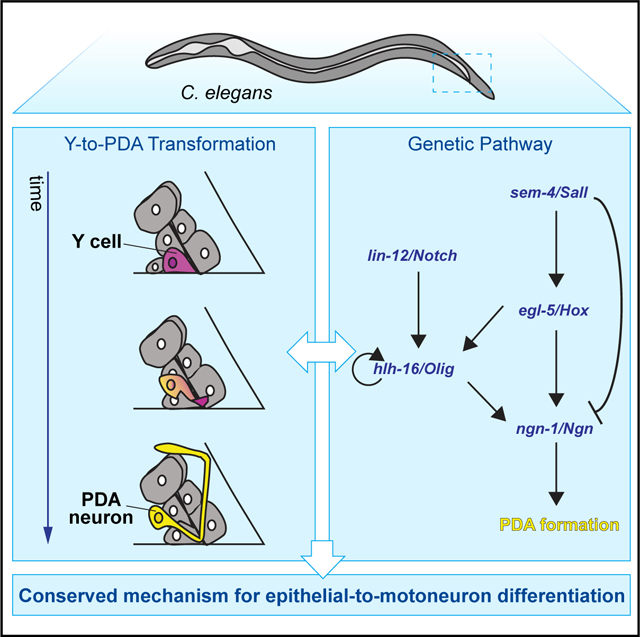
SUMMARY
Motoneurons and motoneuron-like pancreatic β cells arise from radial glia and ductal cells, respectively, both tube-lining progenitors that share molecular regulators. To uncover programs underlying motoneuron formation, we studied a similar, cell-division-independent transformation of the C. elegans tube-lining Y cell into the PDA motoneuron. We find that lin-12/Notch acts through ngn-1/Ngn and its regulator hlh-16/Olig to control transformation timing. lin-12 loss blocks transformation, while lin-12(gf) promotes precocious PDA formation. Early basal expression of ngn-1/Ngn and hlh-16/Olig depends on sem-4/Sall and egl-5/Hox. Later, coincident with Y cell morphological changes, ngn-1/Ngn expression is upregulated in a sem-4/Sall and egl-5/Hox-dependent but hlh-16/Olig-independent manner. Subsequently, Y cell retrograde extension forms an anchored process priming PDA axon extension. Extension requires ngn-1-dependent expression of the cytoskeleton organizers UNC-119, UNC-44/ANK, and UNC-33/CRMP, which also activate PDA terminal-gene expression. Our findings uncover cell-division-independent regulatory events leading to motoneuron generation, suggesting a conserved pathway for epithelial-to-motoneuron/motoneuron-like cell differentiation.
In brief
Rashid et al. report on a conserved epithelial-to-motoneuron transformation pathway in C. elegans requiring ngn-1/Ngn and hlh-16/Olig. lin-12/Notch regulates transformation timing through these genes, while ngn-1/Ngn and hlh-16/Olig expression levels are regulated by sem-4/Sall and egl-5/Hox. Unexpectedly, the cytoskeleton organizers UNC-119, UNC-44, and UNC-33, which are ngn-1/Ngn targets, promote motoneuron terminal identity.
Graphical Abstract
INTRODUCTION
During animal development, neurons and neuron-like cells are generated from progenitor cells that often line tubes. For example, in the developing spinal cord, basal processes of radial glial stem cells line the fluid-filled spinal canal. NOTCH/DELTA signaling is implicated in the differentiation of these cells into motoneurons through control of their proliferative state (de la Pompa et al., 1997; Appel et al., 2001; Tan et al., 2016). Downregulation of the NOTCH-dependent HES1 and HES5 transcriptional inhibitors in differentiating cells induces expression of the proneural basic helix-loop-helix (bHLH) transcription factors OLIG2 and NGN2 (Sagner et al., 2018; Hatakeyama et al., 2004; Ishibashi et al., 1995; Ohtsuka, 1999), leading to adherens junction loss, delamination from the epithelium, cell migration, and neuronal maturation (Ma et al., 1998; Fode et al., 1998; Mizuguchi et al., 2001; Lee et al., 2005; Novitch et al., 2001). A similar sequence of events characterizes the formation of pancreatic insulin-secreting β cells, which are innervated (Nicolls, 2004) and express genes also active in motoneurons (Thor et al., 1991; Ericson et al., 1992; Polak et al., 1993; Cirulli et al., 1994; Naya et al., 1995; Sommer et al., 1996; Sussel et al., 1998; Gradwohl et al., 2000; Muñoz-Bravo et al., 2013). Here, epithelial progenitor cells lining pancreatic ducts express high levels of NOTCH, activating the HES1 repressor, which, in turn blocks NGN3 expression (Apelqvist et al., 1999; Jensen et al., 2000; Murtaugh et al., 2003; Shih et al., 2012; Ninov et al., 2012). Downregulation of NOTCH drives delamination, cell migration, and subsequent differentiation by lifting inhibition of NGN3 (Zhou et al., 2007; Rubio-Cabezas et al., 2011; Gradwohl et al., 2000; Bankaitis et al., 2018) and by allowing NOTCH to directly activate NGN3 expression (Cras-Méneur et al., 2009; Shih et al., 2012). The OLIG family bHLH factor bHLHb4 is expressed in delaminating/migrating cells and may be involved in β cell differentiation (Bramblett et al., 2002).
The dynamics of gene expression and the regulatory interactions among genes driving motoneuron formation have been extensively explored (Dasen and Jessell, 2009; Jessell, 2000; Sagner and Briscoe, 2019). Nonetheless, key questions remain. Specifically, the roles of motoneuron fate regulators are usually assessed through effects of mutants on the expression of differentiation markers. Such defects could arise from a failure to generate sufficient numbers of target cells or from bona fide perturbation of cell-fate determination. Distinguishing between these possibilities can be challenging. To address this issue, we have studied a similar transformation process in the genetically amenable nematode, Caenorhabditis elegans.
The C. elegans Y cell is one of six epithelial cells that line the rectal tube in first-larval-stage (L1) animals (Figure 1A). In L2 animals, Y loses apical junctions with neighboring cells, migrates anterodorsally, and transforms into the PDA motor neuron (Sulston et al., 1983; Jarriault et al., 2008), which innervates the intestinal and anal depressor muscles (White et al., 1986). Remarkably, this transformation takes place without cell division (Sulston et al., 1983). Forward and reverse genetic studies identified a number of genes required for the Y-to-PDA transition, including lin-12/Notch, sem-4/Sall, egl-5/Hox, and several chromatin remodeling genes (Jarriault et al., 2008; Richard et al., 2011; Kagias et al., 2012; Zuryn et al., 2014). Thus, the Y-to-PDA transition is an excellent setting in which to distinguish whether regulators affect motoneuron progenitor cell differentiation or cell division.
Figure 1. Three morphological stages characterize Y-to-PDA transformation.
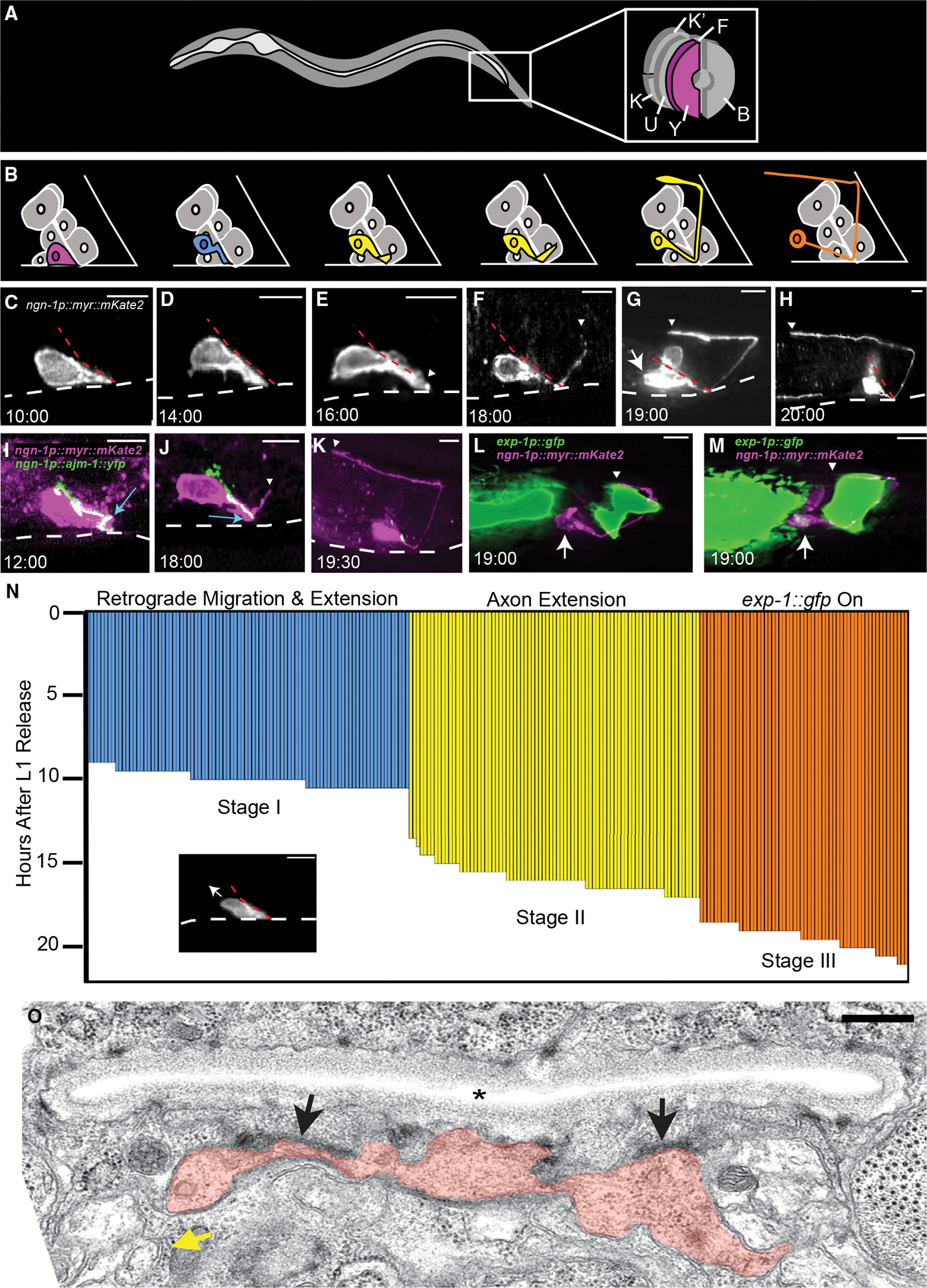
(A) Cellular anatomy of rectal tube in an L1 animal. The Y cell is at the opening. K, K’, F, U, and B cells also line the rectal tube.
(B) Schematic of the Y-to-PDA transformation time course shown in (C–H). Y/PDA is pseudocolored to match (N).
(C–M) Time stamps, hours after L1 arrest exit. Scale bars, 5 μm. Dashed white lines, ventral surface of animal. Dashed red lines, rectal slit. Arrows, cell body. Arrowheads, axon tip. (C) The Y cell before transformation begins. (D) Anterodorsal cell body movement with cell tip anchored at rectal slit. (E) Axon outgrowth initiation. (F–H) Stages of axon extension. (I–K) Apical junctions along the cell length marked by AJM-1:YFP (green) expressed in a Y cell (magenta) are maintained during axon outgrowth (I, J) and disappear afterward (K). Apical junctions at the rectal slit base (blue arrows) are initially present in the Y cell (I) but disappear during axon outgrowth (J). (L and M) exp-1p::gfp expression onset occurs after axon outgrowth.
(N) Each vertical bar represents the Y-to-PDA stage of a single animal. Image shows a representative animal at the onset of retrograde migration and extension stage (arrow, direction of cell migration). Stage I, n = 89 animals, mean = 9.95 ± 0.47 h; stage II, n = 81 animals, mean = 15.93 ± 0.8 h; stage III, n = 58 animals, mean = 19.43 ± 0.73 h.
(O) Electron micrograph of an animal staged as in (F). Orange, Y/PDA; asterisk, rectal slit; black arrows, putative adhesion junctions; yellow arrow, neural bundle into which the PDA process enters. Scale bar, 500 nm. See also Figures S1 and S2; Table S2.
Here, we report our identification of regulators of Y-to-PDA transformation and explore the interactions among them and with previously identified genes. We find that loss of lin-12/Notch blocks PDA neuron formation, and gain of lin-12/Notch function generates a precocious PDA neuron. Thus, lin-12/Notch acts as a timing rheostat for motoneuron generation, acting independent of cell division. lin-12/Notch functions, at least in part, by regulating ngn-1/Ngn, through control of the bHLH gene hlh-16/Olig. ngn-1/Ngn basal expression levels are set early on by sem-4/Sall, egl-5/Hox, and hlh-16/Olig. Coincident with the onset of Y cell morphological changes and migration, we find an increase in ngn-1/Ngn gene expression, governed by sem-4/Sall and egl-5/Hox, but not hlh-16/Olig. Y cell migration is accompanied by retrograde extension of a process that remains anchored at the rectal slit. The tip of this process serves as a growth point for the PDA axon. Following axonal growth, we identify the axonal cytoskeleton genes unc-119, unc-44/Ank, and unc-33/Crmp as targets of hlh-16/Olig and ngn-1/Ngn and demonstrate a previously unappreciated role for these genes in regulating the expression of motoneuron terminal differentiation genes.
Our results highlight intriguing similarities with spinal-cord motoneuron and pancreatic islet formation, suggesting a conserved module for the differentiation of tube-lining cells into motoneuron/motoneuron-like progeny.
RESULTS
Y-to-PDA transformation occurs in distinct stages through a transient epithelial-neuronal hybrid intermediate
To image the sequence of events leading to PDA formation, we developed a single fluorescent reporter expressed in both Y and PDA, as this had not been previously published. We leveraged our genetic studies identifying ngn-1 as a relevant gene in Y-to-PDA transformation (see below) to generate an ngn-1 promoter:myr:mKate2 transgene, containing ~4 kb of ngn-1 upstream regulatory sequences, a myristoylation (myr) sequence for membrane localization, and the mKate2 red fluorescent reporter gene. This reporter is strongly expressed in the Y cell of L1 animals (Figures 1C–1H). We synchronized a pool of animals by hatching embryos in the absence of food, resulting in growth arrest at the equivalent of ~2 h of postembryonic development. Separate groups of animals from this pool were imaged every 30 min between 6 and 22 h following food introduction. We found that the Y-to-PDA transition occurs in a stereotypical pattern (Figures 1B–1H and 1N). Between 9 and 10.5 h, the Y cell begins to migrate anteriorly and dorsally, while its posterior tip remains attached to the rectal slit, thereby forming a thick posterior process (Figures 1C, 1D, and 1N). These initial events are reminiscent of retrograde extension of sensory-neuron dendrites and nerve-ring axons (Heiman and Shaham, 2009; Singhal and Shaham, 2017; Fan et al., 2019). Axon outgrowth, initiating from the anchored posterior tip, then begins between 13.5 and 17 h (Figures 1E–1H, and 1N).
Importantly, during the Y-to-PDA transition, the cell does not fully delaminate from the rectal epithelium, as was previously proposed (Richard et al., 2011). Supporting this conclusion, we find that the apical junction marker AJM-1:YFP, expressed specifically in Y, continues to localize along the length of the cell, except at the base of the rectal slit, while the cell extends its axon (Figures 1I and 1J). Furthermore, electron micrographs of Y/PDA with a partially extended axon reveal persistent apical junctions with other rectal epithelial cells (Figure 1O). Once the axon is fully extended, AJM-1 no longer localizes to the rectal slit (Figure 1K). We also observed that the nucleus of the cell retains a single prominent nucleolus, a feature of epithelial cells, and adopts a neuron-like speckled nucleolar morphology only after axon outgrowth is complete (Figure S1) (Sulston and Horvitz, 1977). Thus, Y-to-PDA transformation proceeds through a transitory state exhibiting both epithelial and neuronal characteristics.
The mature PDA neuron expresses the EXP-1/GABRB3 GABA-gated cation channel (Jarriault et al., 2008; Beg and Jorgensen, 2003). Surprisingly, we found that an exp-1 promoter:gfp reporter transgene is only detected starting at 18.5–21 h post hatching, once the PDA axon has passed the dorsal aspect of the anal depressor muscle (Figures 1L–1N). Thus, exp-1 expression is turned on after axon extension has initiated. We believe this conclusion is not confounded by a lag in visualization of GFP, as previous studies demonstrate that GFP should be detected within 30–60 min of gene expression initiation (e.g., Maurer et al., 2007).
Under certain conditions, starvation can alter Y-to-PDA transformation timings and events (Becker et al., 2019). To confirm that this is not the case in our studies, we repeated the time-lapse experiments in non-starved animals. We found that retrograde migration occurs between 10.5 and 12.5 h after hatching, axon extension occurs between 16 and 18 h after hatching, and onset of exp-1 promoter::gfp expression occurs between 19.5 and 22 h after hatching (Figure S2). Since starvation arrests animals at ~2 h after hatching, we conclude that our protocol does not affect the timing or the events of Y-to-PDA transformation.
Taken together, our observations reveal three distinct stages in the transformation of Y to PDA: (I) anterior and dorsal cell-body migration, coupled with posterior process retrograde extension; (II) axon outgrowth initiation; and (III) exp-1 gene expression. These stages are temporally non-overlapping, as cells extending axons always undergo cell migration and retrograde extension (n = 81), and cells expressing EXP-1/GABRB3 always undergo migration, retrograde extension, and axon outgrowth (n = 58; Figure 1N).
NGN-1/NGN is required early during Y-to-PDA transformation
To identify genes required for Y-to-PDA transformation, we performed a forward genetic screen, seeking animals that fail to express the exp-1p:gfp reporter transgene in adults. Such mutants could affect transitions between any of the three stages we defined. We identified 16 such mutants (Table S1), and through a combination of complementation tests and whole-genome sequencing found that three (ns188, ns189, ns192) carry defects in the gene sem-4. sem-4 encodes a SALL-family transcription factor and was previously shown to be required for the Y-to-PDA transition (Jarriault et al., 2008), demonstrating that our screen can indeed uncover Y-to-PDA transformation mutants.
Animals homozygous for the ns185 allele isolated in our screen have a strong defect in exp-1p:gfp expression (PDAp::gfp in Figures 2A–2D and 2O). To determine where PDA formation fails, we examined ns185 mutants carrying an egl-5 promoter:gfp transgene. This reporter is normally detected in the Y cell of L1 animals, but is not expressed in PDA neurons (Figures 2B and 2C) (Jarriault et al., 2008; Zuryn et al., 2014). We found that in ns185 mutants, the Y cell forms properly and undergoes anterodorsal cell-body movement, and a thick posterior process begins to stretch out. However, an axon is not formed, nucleolar morphology remains epithelial, and the cell constitutively expresses the egl-5p:gfp reporter (Figures 2D–2F and 2O). Furthermore, ns185 mutant animals retain apical junctions along the length and base of the cell into adulthood (Figures 2J and 2K). Together, these results suggest that the gene affected in ns185 animals is required for the transition from stage I to stage II of Y-to-PDA transformation (Figure 1N).
Figure 2. Defective Y-to-PDA transformation in ngn-1 and hlh-16 mutants.
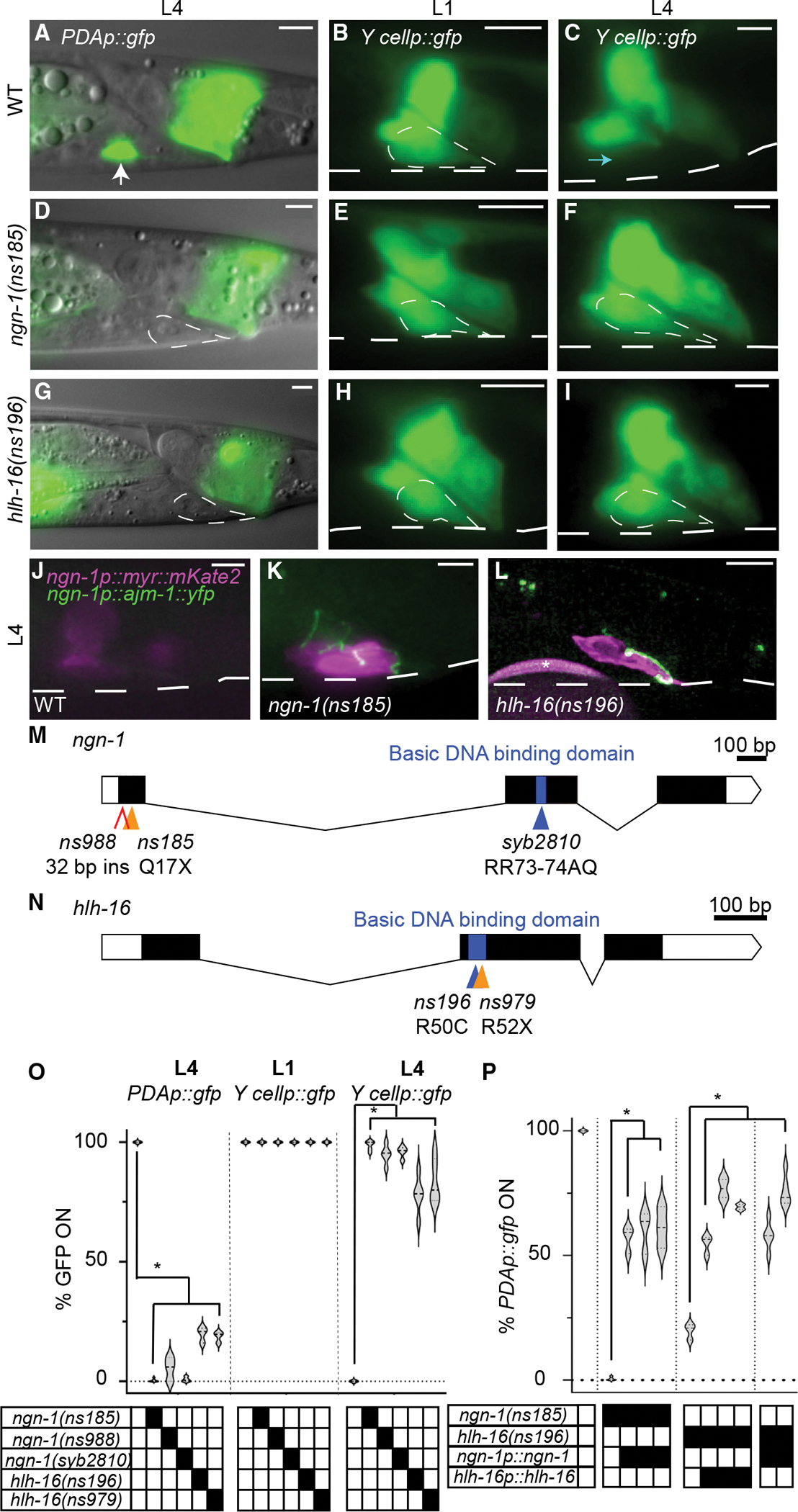
(A–C) Wild-type Y-to-PDA transformation. PDAp:gfp, nsIs131[exp-1p::gfp]; Y cellp::gfp, bxIs7 [egl-5p::egl-5(exon1–3)::gfp] reporter; dashed white circle, outline of Y cell; dashed white line, outline of worm. Scale bars, 5 μm. (A) PDAp::gfp expression in L4 PDA (arrow). (B) The Y cell in an L1 animal. (C) Y cell transformed into PDA neuron in L4 animal, Y cellp::gfp is off. Cyan arrow indicates where Y cell was in L1 animals.
(D–F) Same as (A–C), except in ngn-1(ns185) mutant. PDA is not formed, and Y cellp::gfp is expressed in L4 stage.
(G–I) Same as (D–F), except in hlh-16(ns196) mutant.
(J–L) Persistent Y cell in L4 ngn-1 and hlh-16 mutants maintains apical junctions. Asterisk, bead used for focusing.
(M and N) Amino acid changes are indicated. X, stop codon. (M) The ns988 allele contains a 32 bp insertion with a stop codon between nucleotides 29 and 30.
(O and P) Percentage of animals expressing indicated reporters in indicated genotype. Data are shown as violin plots; n > 100 animals, n > 3 technical replicates for all genotypes. Solid black boxes, presence of a mutation or transgene. (O) Significance at p < 0.01, two-way ANOVA with Dunnett’s post hoc test. (P) Significance at p < 0.01, one-way ANOVA with Dunnett’s post hoc test. See also Tables S1 and S2.
To identify the affected gene, we used SNP mapping to localize the causal lesion to an 885 kb region on linkage group IV, containing the ngn-1 gene. A transgene containing ~4 kb of ngn-1 regulatory sequences, fused to ngn-1 cDNA, restores PDA neuron formation and exp-1p:gfp expression to ns185 mutants (Figure 2P). Incomplete rescue may reflect toxicity of ngn-1 overexpression (which we had previously noticed) or the mosaic nature of the extrachromosomal rescuing array. Sequencing of the gene from ns185 animals revealed a C-to-T nucleotide change predicted to result in a Q17Stop lesion, which would generate a severely truncated protein (Figure 2M). Furthermore, a CRISPR-generated mutant we isolated, ns988, with an early stop codon, phenocopies the defects seen in ns185 animals (Figures 2M and 2O). Thus, ns185 is an allele of ngn-1. An additional allele of ngn-1, ns194, was isolated from the screen and contains a C-to-T nucleotide change predicted to result in a Q29Stop lesion and a severely truncated protein (Table S1).
ngn-1 encodes a protein related to vertebrate neurogenin proteins, a subfamily of bHLH transcriptional regulators that control a variety of developmental processes through DNA binding and helix-loop-helix domain interactions (Sommer et al., 1996; Gradwohl et al., 1996; Ma et al., 1998, 1999; Sun et al., 2001; Grove et al., 2009). To determine whether the DNA-binding domain is required for Y-to-PDA transformation, we used CRISPR-mediated mutagenesis to replace conserved arginine residues (RR) in the NGN-1 basic domain with AQ. This mutation perturbs DNA binding of neurogenin proteins, while leaving other domains functional (Sun et al., 2001). We found that these ngn-1(syb2810) mutants reproduce the defects of ngn-1(ns185) and ngn-1(ns988) mutants, suggesting that the DNA-binding domain of NGN-1 is required for Y-to-PDA transformation (Figures 2M and 2O).
To determine where NGN-1 functions, we generated animals carrying a genomically integrated transgene containing ~4 kb of ngn-1 regulatory sequences fused to myr::mKate2 (Figures 1C–1H). We found that in the rectal area of early L1 larvae, prior to PDA axon extension, this reporter is expressed exclusively in the Y cell. The same promoter was used to drive ngn-1 cDNA in our rescue studies (Figure 2P), suggesting that NGN-1 functions cell autonomously in Y for Y-to-PDA transformation.
HLH-16/OLIG also functions early during Y-to-PDA transformation
ns196 animals also have a strong defect in exp-1p::gfp expression (PDAp::gfp in Figures 2G–2O). Like ngn-1(ns185) mutants, ns196 animals reliably form the Y cell, which initiates migration and retrograde extension. However, axon formation, nucleolar changes, and silencing of the egl-5p:gfp reporter do not take place (Figures 2G–2I and 2O). ns196 mutant animals also retain their apical junctions along the length and base of the cell (Figure 2L), confirming a failure in the transition from stage I to stage II.
SNP mapping of ns196 mutants revealed linkage of the causal lesion to a 335 kb region on chromosome I containing the hlh-16 gene, which encodes a bHLH transcription factor related to vertebrate OLIG2 and bHLHb4 (Reece-Hoyes et al., 2005s; Grove et al., 2009; Bertrand et al., 2011). A transgene containing a 480 bp sequence upstream of the hlh-16 start codon, fused to the hlh-16 cDNA and 1.5 kb of 3′ UTR sequences, rescues the mutant defects (Figure 2P). Thus, ns196 is an allele of hlh-16. Indeed, sequencing of the gene from ns196 mutants revealed a C-to-T nucleotide change leading to a predicted R50C amino acid substitution in the conserved basic DNA-binding domain (Figure 2N). Thus, the DNA-binding domain of hlh-16 is likely required for Y-to-PDA transformation. Furthermore, a CRISPR-generated R52Stop mutation (ns979) phenocopies the defects seen in ns196 (Figures 2N and 2O), confirming that hlh-16 is the relevant gene.
To determine where HLH-16 functions, we first generated animals carrying a genomically integrated transgene containing 480 bp of hlh-16 upstream regulatory sequences fused to myr:gfp. We did not detect any GFP fluorescence in the rectal area. However, animals carrying a similar transgene, with an additional 1.5 kb fragment of the hlh-16 3′ UTR, showed specific GFP expression in the Y cell, demonstrating that the 3′ UTR of the gene is important for Y-specific expression (Figure S3A). A construct with the same promoter and UTR driving hlh-16 cDNA rescues hlh-16 mutant defects (Figure 2P), suggesting that hlh-16 functions cell autonomously in Y for the Y-to-PDA transformation.
hlh-16/Olig controls its own expression and the basal expression of ngn-1/ngn
As ngn-1 and hlh-16 mutants exhibit similar defects, they may function in the same pathway. To address this possibility, we first determined the temporal expression pattern of these genes. We observed animals carrying the hlh-16p::myr::gfp:hlh-16 3′ UTR reporter for a 14 h period following L1 arrest exit, at 2 h intervals (n > 53 per time point; Figure S3B). Only small changes in Y/PDA expression (~20%) were detected over this time interval. A similar experiment, using the ngn-1p:myr:mKate2 reporter, also revealed little change in expression over the first 10 h of imaging. However, from 10 to 14 h, as changes in Y cell morphology become evident, ngn-1 reporter expression levels nearly doubled (from 73 to 145 a.u.; n > 57 per time point; p < 0.0001; Figures 3A and S4A). This suggests that ngn-1 expression is subject to at least two modes of control. Before 10 h, ngn-1 is maintained at a static basal level. Between 10 and 14 h, expression is boosted. We obtained similar results using two different CRISPR-mediated knockin reporters in the endogenous ngn-1 locus, with gfp sequences inserted either directly upstream of the stop codon (translational fusion) or as part of a trans-splicing cassette immediately downstream of the ngn-1 gene (transcriptional fusion) (Figures S4E and S4F). Interestingly, while expression of the transcriptional fusion is seen in several cells in the animal, the translational reporter is not expressed initially (Figures S4F–S4H). At 8–10 h post arrest, NGN-1 protein expression then becomes detectable specifically in the Y cell, suggesting that NGN-1 protein is under differential control in different cell types (n > 40 animals per time point; Figures S4F–S4H).
Figure 3. Time course of and effects of mutants on ngn-1 and hlh-16 reporter expression.
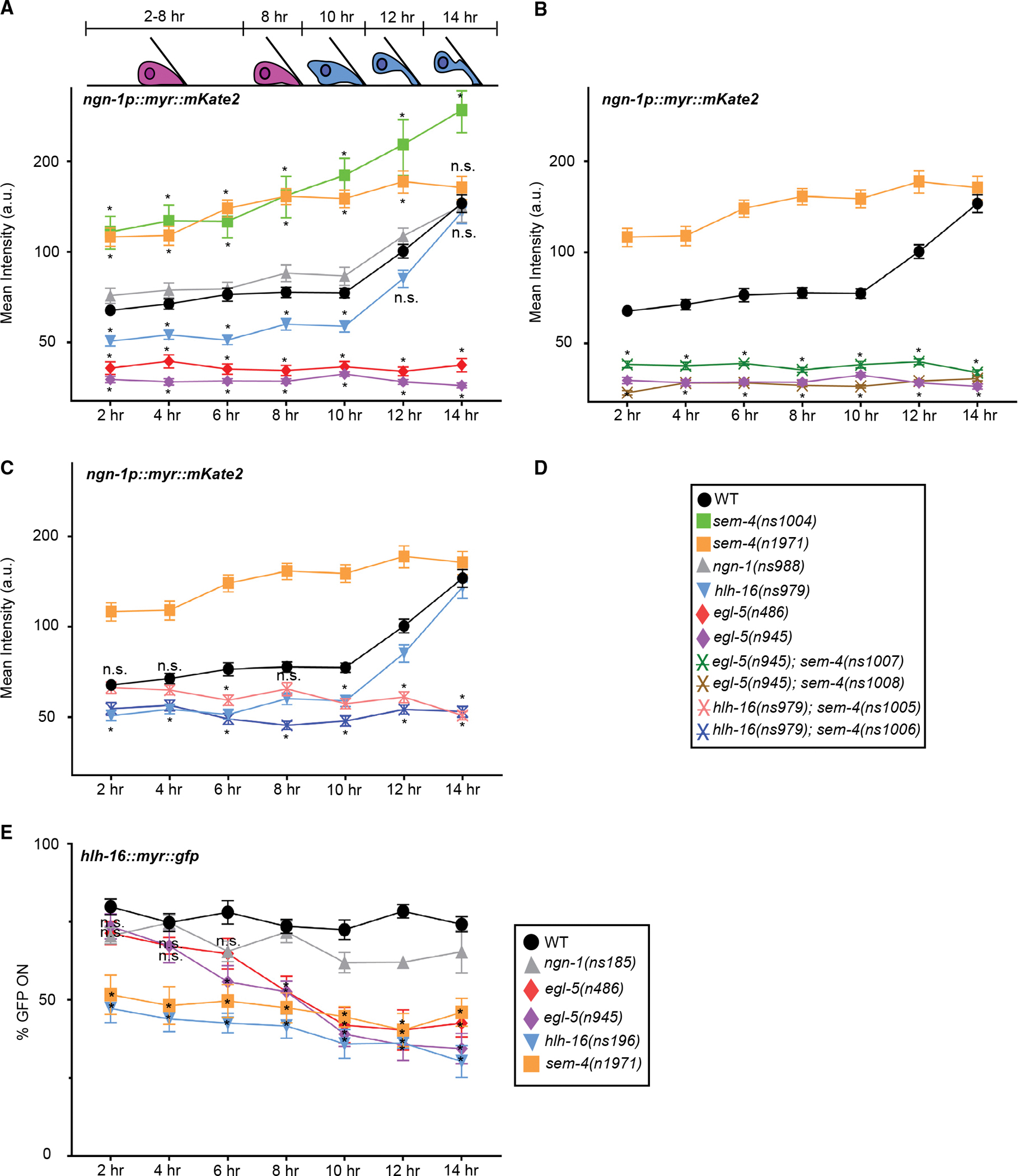
Schematic depicts Y cell morphology at the indicated time points. Horizontal axis, time after L1 arrest exit; n.s., not significant.
(A–C) Y cell fluorescence levels of nsIs913 [ngn-1p:myr:mKate2] in the indicated mutant backgrounds; n > 24 animals per time point. The Y axis is plotted on a log2 scale. (B and C) Significance values are shown only for strains not present in (A).
(D) Key for graphs (A–C).
(E) As nsIs943 [hlh-16p:myr:gfp] reporter expression is faint, signal quantification is unreliable. We therefore scored the percentage of animals for which any fluorescence signal was detected in the Y cell of indicated mutants. Statistics for ngn-1 mutants are not plotted for clarity. ngn-1 mutants do not show statistically significant differences from wild-type (WT) animals; n > 5 technical replicates for all genotypes, n > 15 animals per time point per replicate; two-way ANOVA with Dunnett’s post hoc test. Significance at *p < 0.05. Error bars, SEM. Statistics in Table S2. See also Figures S3 and S4; Table S2.
We next investigated hlh-16 and ngn-1 reporter expression in ngn-1 and hlh-16 mutants, respectively. hlh-16 reporter expression is not perturbed in ngn-1(ns185) mutants (five experiments, n > 20 per time point per experiment; Figure 3E). However, basal ngn-1 reporter levels are significantly reduced in hlh-16 mutants (n > 28 per time point; Figures 3A and S4B; n > 29 per time point; Figure S4E). Of note, the boost in ngn-1 reporter levels occurs normally in hlh-16 mutant animals. Thus, hlh-16 is required for ngn-1 basal expression, but is not required for the increase in ngn-1 expression that follows. Consistent with these observations, we found that a high-copy ngn-1p:ngn-1 cDNA transgene can restore Y-to-PDA transformation to hlh-16 mutants (Figure 2P), supporting the notion that hlh-16 functions to establish ngn-1 basal expression levels.
To assess autoregulation, we examined ngn-1 and hlh-16 multicopy reporter expression in ngn-1 and hlh-16 mutants, respectively. While ngn-1 expression is not perturbed, hlh-16 expression is significantly reduced in hlh-16 mutants (six experiments, n > 15 per time point per experiment; Figures 3A–3E), suggesting that HLH-16 is required for its own expression.
egl-5/Hox functions upstream of ngn-1 and hlh-16
Previous studies showed that egl-5/Hox is required for Y-to-PDA transformation. In egl-5 mutants, a PDA neuron is not generated, and Y epithelial characteristics are constitutively maintained (Jarriault et al., 2008). To uncover possible interactions among egl-5, hlh-16, and ngn-1, we examined the expression of the ngn-1 and hlh-16 reporters in two different egl-5 mutants, egl-5(n486) and egl-5(n945). We found that both egl-5 alleles strongly reduce ngn-1 transcriptional and translational reporter expression at all time points, and induction of ngn-1 expression between 10 and 14 h is entirely blocked (n > 27 per time point; Figures 3A, S4C, S4E, and S4F). Intriguingly, we found that expression of an egl-5p::egl-5(exons1–3)::gfp genomically integrated reporter transgene (Ferreira et al., 1999) is dynamic over the imaging period. Between 0 and 12 h, reporter levels do not vary extensively. However, after the 12 h time point, there is a significant increase in egl-5 expression levels (four experiments, n > 4 per time point per experiment; Figure S3C). This increase correlates with the increase in ngn-1 reporter expression, raising the possibility that a rise in egl-5 expression drives increased ngn-1 expression. We believe that the onset of increased egl-5 reporter expression is likely slightly delayed compared with the ngn-1 reporter strain because of different growth rates of the strains. Our combined results strongly support the notion that EGL-5/HOX functions upstream of ngn-1/Ngn for both basal and induced expression, and suggest that these may be independent functions.
egl-5 mutants also exhibit defects in hlh-16 reporter expression. While initial levels of hlh-16 appear normal, they steadily and significantly decline over the 14 h imaging period (n > 7 experiments, n > 17 per time point per experiment; Figure 3E). These observations suggest several conclusions. First, EGL-5 is not required for the initial expression of hlh-16. Second, EGL-5 is required for maintaining hlh-16 gene expression before and during initiation of Y-to-PDA transformation. Third, the effects of egl-5 on ngn-1 are partly independent of hlh-16, as the levels of hlh-16 at the start of the experiment appear normal, but ngn-1 levels are dramatically reduced even at this time point. This last observation is consistent with our observations that ngn-1 expression levels track the expression levels of egl-5, but not of hlh-16, between the 10 and the 14 h time points (Figures 3A, S3B, and S3C).
SEM-4 can activate and inhibit ngn-1 expression
The sem-4 gene encodes a SALL-family transcription factor previously reported to promote Y-to-PDA transformation, and we identified three alleles of the gene in our screen (Table S1). Like egl-5 mutants, the Y cell in sem-4 mutant animals retains epithelial properties and fails to transform into PDA (Jarriault et al., 2008). We therefore examined the expression of ngn-1, hlh-16, and egl-5 reporter transgenes in sem-4 mutants. Unexpectedly, and unlike in egl-5 mutants, we found that in two different sem-4 mutants (ns1004, n1971), the ngn-1 multicopy reporter is induced to nearly twice the level of wild-type animals during the 0–10 h observation period. This increased expression is subsequently retained, so that ngn-1 expression levels at the 14 h time point are comparable to, or perhaps slightly higher than, wild-type levels (n > 26 animals per time point; Figures 3A and S4D). These observations suggest that at early time points, SEM-4 may repress ngn-1 expression, but this inhibition becomes less pronounced as Y-to-PDA transformation proceeds. Interestingly, however, we found that in sem-4(n1971) mutants, basal expression of our knockin transcriptional and translational ngn-1 reporters is not increased (n > 28 animals per time point; Figures S4E and S4F). This discrepancy could be due to perturbed ngn-1 regulation from inserting sequences into the genomic locus, or to missing regulatory elements on the multicopy transgene. We conclude that early on, SEM-4 can engage repressive elements within the ngn-1 locus, whose physiological significance is unclear.
To further study the effects of sem-4 on ngn-1, we assessed ngn-1 multicopy transgene expression in egl-5(n945) and hlh-16(ns979) mutants also harboring a sem-4 deletion allele. We found that animals with both egl-5 and sem-4 mutations have greatly reduced ngn-1 reporter expression levels throughout the imaging period, comparable to egl-5 single mutants alone (n > 24 per time point; Figure 3B). In sem-4; hlh-16 double mutants, the ngn-1 multicopy reporter expression levels are slightly reduced compared with wild-type levels, as in hlh-16 single mutants, during the 0–10 h imaging period. However, ngn-1 induction, which takes place normally in hlh-16 single mutants, now fails to occur (n > 24 per time point; Figure 3C). Thus, from 0 to 10 h, the increased expression of the ngn-1 multicopy reporter in sem-4 mutants requires functional egl-5 and hlh-16 genes. At later times points, all our reporters suggest that sem-4 promotes induction of ngn-1 expression.
We also examined the effects of sem-4 loss on egl-5 and hlh-16 reporter expression. We find that expression of both reporters is reduced (but not eliminated) at all time points (six experiments, n > 15 per time point per experiment; Figure 3E; n > 22 per time point; Figure S3C). Thus, sem-4 promotes egl-5 and hlh-16 expression at all time points.
LIN-12/NOTCH determines Y-to-PDA transformation timing through ngn-1/ngn and hlh-16/Olig-dependent and independent mechanisms
Our results suggest that a temporal switch in Y cell state, manifested by changes in the functions of the HLH-16 and SEM-4 transcription factors, correlates with initiation of Y-to-PDA transformation. We sought, therefore, to identify regulators of this cell-state change. A previous study reported that lin-12(n676n930) homozygous mutants, carrying a temperature-sensitive partial-loss-of-function mutation in the lin-12/Notch gene, fail to generate a Y cell at 25°C (Jarriault et al., 2008). This allele was originally generated by reverting the lin-12(n676) gain-of-function allele to generate a lin-12 allele containing the original gain-of-function mutation and a new loss-of-function mutation (Greenwald et al., 1983; Sundaram and Greenwald, 1993). We confirmed this result, showing that, while a few newly hatched L1 animals in this mutant at 25°C express hlh-16 or ngn-1 reporters in a rectal cell with the morphology of the Y cell (Figure 4C), most do not (n > 70, Figure 4E). Curiously, in some animals, while a Y cell could not be detected, a PDA neuron expressing the hlh-16 or ngn-1 reporter was apparent. We wondered whether this precocious formation of PDA (>20 h early) might reflect residual lin-12/Notch gain-of-function activity of this strain. We therefore examined newly hatched animals at 15°C, the permissive temperature for the loss-of-function defects, and found that almost all contain a precocious PDA neuron, and rare animals possess a normal Y cell (n > 80, Figures 4D and 4F). These results suggest that at 15°C, lin-12(n676n930) animals have an appreciable gain of lin-12 function in Y. This is consistent with previous work showing that at 15°C, this allele has some gain-of-function activity in other contexts (Sundaram and Greenwald, 1993).
Figure 4. lin-12/Notch promotes Y-to-PDA transformation.
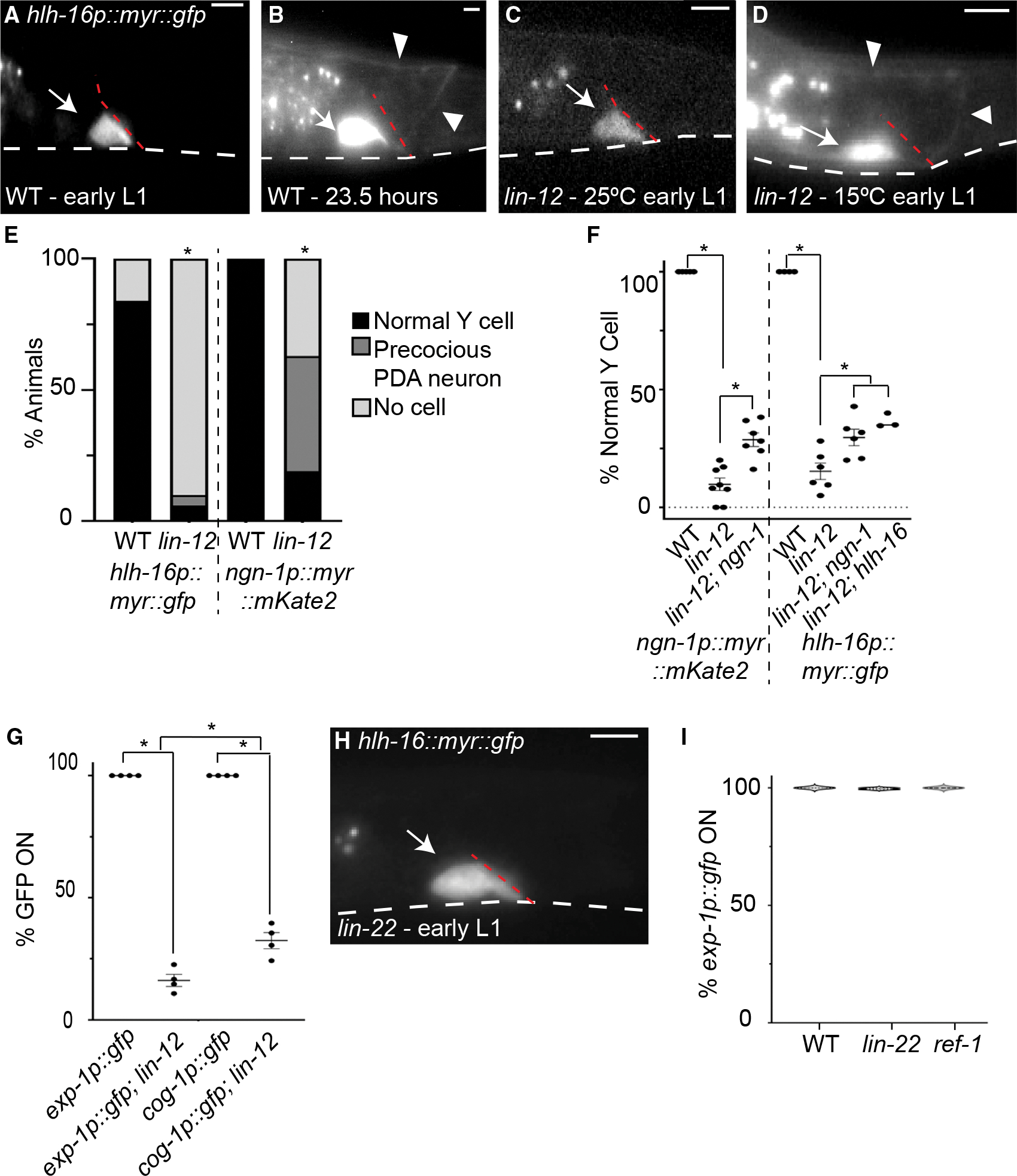
(A) WT Y cell in early L1 at 15°C.
(B) WT Y cell, 23.5 h after release from L1 arrest at 15°C.
(C) Early lin-12(n676n930) mutant L1 at 25°C either has normal Y cell morphology (shown) or does not express nsIs943 [hlh-16p:myr:gfp].
(D) Early lin-12(n676n930) mutant L1 at 15°C displays precocious PDA.
(E) Quantification of Y/PDA in early L1 animals at 25°C using nsIs943 and nsIs913 [ngn-1p:myr:mKate2] in WT and lin-12(n676n930) mutant animals. Fisher’s exact test, significance at *p < 0.01; n > 70 animals.
(F) Quantification of presence of a precocious PDA in early L1 animals at 15°C using nsIs913 and nsIs943 reporter in WT, lin-12(n676n930), lin-12(n676n930); ngn-1(ns988), lin-12(n676n930); ngn-1(ns185), and lin-12(n676n930); hlh-16(ns196) mutants. For nsIs943, percentages were quantified from animals with fluorescence expression. One-way ANOVA with Tukey’s post hoc test. Significance at *p < 0.05; n > 50 animals for all genotypes. Error bars, SEM.
(G) Quantification of nsIs130 [exp-1p:gfp] and syIs63 [cog-1p:gfp] expression in WT and lin-12(n676n930) L4 animals raised at 15°C. ANOVA with Tukey’s post hoc test. Significance at *p < 0.01; n > 100 animals for all genotypes. Error bars, SEM.
(H) Early lin-22(n372) mutant L1 at 20°C with normal Y cell.
(I) nsIs131 [exp-1p:gfp] expression in WT, lin-22(n372), and ref-1(mu220) mutant L4 animals raised at 20°C. Data are shown as violin plots. One-way ANOVA; n > 350 animals. Scale bars, 5 μm. Dashed white lines, ventral surface of animal; dashed red lines, rectal slit; arrows, cell body; arrowheads, axon.
To examine whether these precociously formed PDA neurons are properly differentiated, we examined the expression of exp-1p::gfp and cog-1/Nkx6–3 promoter::gfp, which is expressed slightly earlier (Zuryn et al., 2014), in lin-12(n676n930) animals grown at 15°C. We found that only 18% of L4 animals express exp-1p::gfp and only 33% of L4 animals express cog-1p::gfp (n > 68; Figure 4G). Therefore, the Y cell in most lin-12(n676n930) animals executes stages I and II of Y-to-PDA transformation precociously but fails to reach stage III.
Is ngn-1 required for the precocious formation of PDA? To address this question, we assayed PDA formation in lin-12; ngn-1 double mutants. We found that loss of ngn-1 can suppress precocious PDA formation, suggesting that lin-12/Notch acts in part through ngn-1/neurogenin (n > 79; Figure 4F). Supporting this notion, a previous study reported that lin-12/Notch promotes hlh-16 expression in other C. elegans motoneurons (Bertrand et al., 2011), raising the possibility that NOTCH acts through hlh-16 to affect ngn-1 activity in Y/PDA. Indeed, loss of hlh-16 can also partially suppress precocious PDA formation in lin-12(n676n930) mutants (n > 50; Figure 4F). Importantly, however, a substantial fraction of lin-12(n676n930) mutants, also carrying ngn-1 or hlh-16 mutations, still exhibit precocious PDAs, suggesting that a pathway independent of ngn-1 must also exist.
In spinal cord radial glia, NOTCH inhibits Olig2 and Ngn2 gene expression through HES-family transcriptional repressors (Sagner et al., 2018; Hatakeyama et al., 2004; Ishibashi et al., 1995; Ohtsuka, 1999). Our findings suggest that the opposite mechanism is at play during Y-to-PDA transformation, as lin-12/Notch promotes hlh-16 and ngn-1 activities. To understand this difference in more detail, we tested whether lin-22, encoding the only C. elegans protein containing all HES-protein domains (Wrischnik and Kenyon, 1997; Alper and Kenyon, 2001; Neves and Priess, 2005), or ref-1, a distantly related protein (Alper and Kenyon, 2001; Neves and Priess, 2005; Chou et al., 2015), are required for Y-to-PDA transformation. We found that neither lin-22(n372) nor ref-1(mu220) mutants display precocious PDA formation (Figure 4H) and properly express the exp-1p:gfp reporter in PDA neurons of L4 animals (n > 350; Figure 4I). Thus, lin-22 and ref-1 are not required for Y-to-PDA transformation, consistent with an alternative mechanism by which NOTCH regulates this epithelial-to-neuronal transition. Such a mechanism may share features with that employed in pancreatic islet development, where NOTCH promotes NGN3 expression (Cras-Méneur et al., 2009; Shih et al., 2012). The C. elegans Y/PDA may be a good system to study this elusive interaction.
Previous characterization of the lin-12(n676) strain argues that this allele increases wild-type LIN-12/NOTCH function (Greenwald et al., 1983; Sundaram and Greenwald, 1993). Taken together, our studies therefore suggest that lin-12/Notch activity levels determine when Y-to-PDA transformation takes place. Low levels of NOTCH result in no transformation, which can be viewed as an infinite delay in transformation onset; wild-type levels result in transformation initiation at 10 h post L1 arrest; and excess lin-12/Notch expression drives precocious PDA formation.
Cytoskeletal organizers UNC-119, UNC-44/ANK, and UNC-33/CRMP function downstream of HLH-16 and NGN-1
Our findings that exp-1 reporter expression turns on after the onset of PDA axon extension, and that lin-12(gf) results in formation of a precocious axon-bearing PDA neuron defective in exp-1 expression, prompted us to investigate how the transition from stage II to stage III of Y-to-PDA transformation takes place. To address this, we went back to the mutants collected in our screen. We found that, while ns200 mutants exhibit a defect in exp-1p:gfp expression, they generate a normal Y cell in L1 animals (Figure 5A) and silence the egl-5p:gfp reporter normally. Thus, ns200 mutants appear to be defective in transitioning from stage II to stage III of the Y-to-PDA transformation (Figure 5A). ns200 mutants also exhibit pronounced locomotion defects. SNP mapping uncovered linkage of the causal lesion to chromosome III, and whole-genome sequencing revealed a G-to-T mutation in the unc-119 gene (Figure 5C). This mutation is predicted to cause a G79Stop mutation, truncating all isoforms of the UNC-119 protein. Introducing the G79Stop lesion into the genomic unc-119 locus of wild-type animals via CRISPR (allele ns913) reproduces the exp-1 expression defect of ns200 mutants, although the penetrance of the defect is lower (Figure 5A), suggesting that unc-119(ns200) mutants also carry enhancer mutations in the background. A transgene containing 1 kb of unc-119 upstream regulatory sequences fused to the unc-119 cDNA from the related nematode C. briggsae restores both exp-1 expression and wild-type locomotion to transgenic animals (Figure 5B). Furthermore, an ngn-1p:unc-119 cDNA transgene fully restores exp-1/Gabrb3 expression, but only weakly rescues the locomotion defects of unc-119 mutants (Figure 5B), suggesting that unc-119 acts cell autonomously in Y/PDA.
Figure 5. Cytoskeleton organizers UNC-119, UNC-44, and UNC-33 promote exp-1p::gfp expression in the PDA neuron.
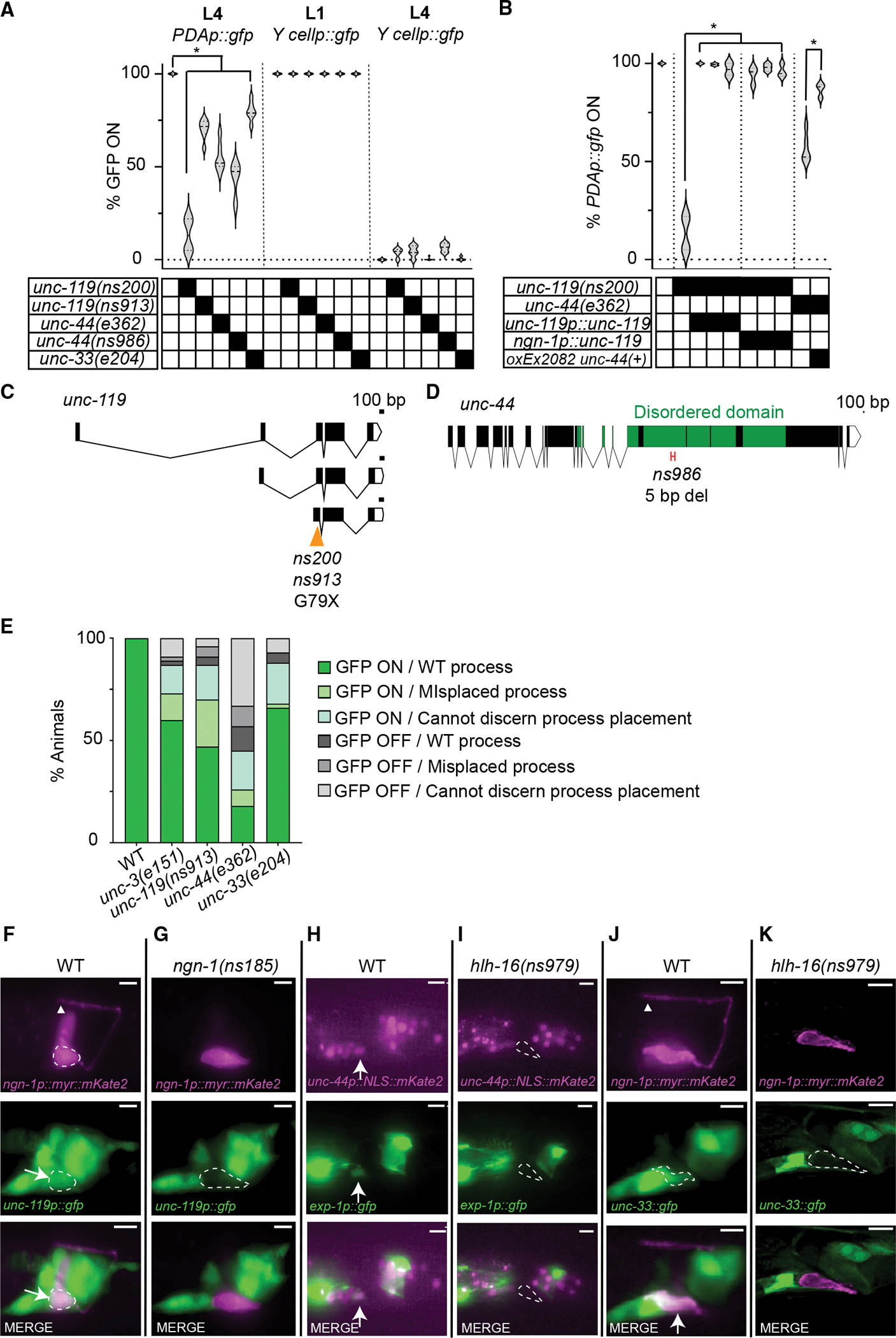
(A) unc-119, unc-44, and unc-33 mutants are defective in PDA maturation. Data are shown as violin plots. Two-way ANOVA with Dunnett’s post hoc test. Significance at *p < 0.01; n > 100 animals.
(B) Similar to Figure 2P. PDAp:gfp, nsIs131[exp-1p:gfp]. A one-way ANOVA with Dunnett’s post hoc test was performed for comparison with unc-119 mutants, and a t test was performed for comparison with unc-44. Significance at *p < 0.01; n > 68 animals.
(C and D) Amino acid changes are indicated. X, stop codon. (D) Only the long neuronal isoform of unc-44 is shown. ns986 has a 5 bp deletion of nucleotides 15,987 to 15,992, leading to an early stop codon.
(E) Defects in PDAp:gfp expression are independent of axon guidance defects; n > 100.
(F–K) Expression of unc-119, unc-44, and unc-33 in wild-type and indicated mutants. Arrows, cell body; arrowhead, axon tip; dashed circle, Y/PDA. Scale bars, 5 μm. See also Figures S5 and S6; Table S2.
When we introduced the ngn-1 and exp-1 reporters together into unc-119(ns913) mutants, we found that, unlike the other mutants we studied, most animals extended a PDA axon. In one-third of the animals, the process is abnormal, and in about 13% of the animals, exp-1 expression is not detected (n > 100; Figures 5E and S5A–S5D). Our results, therefore, demonstrate that the unc-119 gene is required for the transition from stage II to stage III.
UNC-119 is a highly conserved protein that acts as a cytoskeleton organizer in neurons (Maduro et al., 2000; Knobel et al., 2001; Manning et al., 2004; Materi and Pilgrim, 2005; He et al., 2020). A recent study reported that unc-119 functions together with unc-44/Ankyrin and unc-33/Crmp to regulate dendrite polarization, likely through control of actin and microtubule organization, respectively (Hedgecock et al., 1985; Otsuka et al., 1995, 2002; Boontrakulpoontawee and Otsuka, 2002; Tsuboi et al., 2005; Zhou et al., 2008; Maniar et al., 2011; Norris et al., 2014; He et al., 2020; LaBella et al., 2020; Chen et al., 2021). Consistent with this, we found that animals carrying mutations in either unc-44/Ank or unc-33/Crmp exhibit PDA differentiation defects similar to those seen in unc-119 mutants (Figure 5A). A loss-of-function mutation, ns986, specifically affecting the neuronal isoform of unc-44, also displays PDA differentiation defects, and a full-length extrachromosomal unc-44 gene (oxEx2082; LaBella et al., 2020) can rescue unc-44(e362) mutant PDA defects (Figures 5A, 5B, and 5D). Thus, surprisingly, cytoskeletal regulators are important for proper gene expression in stage III of Y-to-PDA transformation.
How might such regulation occur? Although most unc-119, unc-44, or unc-33 mutants have defective PDA axons (Figure 5E), in all three mutants, a population of animals with fully formed, properly placed PDA axons that fail to express the exp-1 reporter was found (Figures 5E and S5A–S5D). This observation suggests that the functions of unc-119/unc-33/unc-44 in axon extension and exp-1 expression are at least partly independent. Expression of cho-1, a choline transporter expressed in mature PDA neurons, is also affected in properly placed axons in unc-44 and unc-33 mutants (n = 5 experiments; Figure S5E).
To understand when unc-119, unc-33, and unc-44 expression begins during Y-to-PDA transformation, we examined the respective reporter constructs. Reporter expression for all three genes is first visible in PDA after the axon begins to grow (Figures 5F, 5H, and 5J). Importantly, in hlh-16 or ngn-1 mutants, these reporters are not expressed at all (Figures 5G, 5I, and 5K). Thus, unc-119, unc-33, and unc-44 are likely to be direct or indirect targets of ngn-1.
We found that mutations in the COE-type transcription factor gene unc-3 have axon extension and exp-1 expression defects highly reminiscent of those seen in unc-119, unc-33, and unc-44 mutants (Figure 5E). This is consistent with previous observations (Richard et al., 2011), and suggests the possibility that NGN-1 might indirectly affect the expression of cytoskeleton-organizing genes through UNC-3. To test this idea, we examined unc-3 expression onset, and found that unc-3 expression in the PDA neuron begins after axon growth initiation, but prior to expression of the cytoskeleton-organizing genes (Figures S6A–S6C). Furthermore, although unc-33::gfp expression is unaltered in unc-3 mutants, unc-119::gfp and unc-44::SL2::NLS::mKate2 reporters show a slight defect (n > 3 experiments; Figure S6D). Thus, UNC-3 may promote cytoskeletal gene expression in the PDA neuron, but this effect would be partially redundant with the activities of other transcriptional regulators (e.g., Stefanakis et al., 2015; Leyva-Díaz and Hobert, 2022).
DISCUSSION
A model for the control of Y-to-PDA transformation
The data presented here are consistent with the model for Y-to-PDA transformation depicted in Figure 6. We propose that NGN-1/NGN plays a pivotal role in the decision of Y to undergo differentiation. Early on, LIN-12/NOTCH activates HLH-16/OLIG. HLH-16/OLIG levels are then maintained by autoactivation and through EGL-5/HOX, which is, in turn, activated by SEM-4/SALL. NGN-1/NGN is positively regulated by HLH-16/OLIG but also potentially inhibited by SEM-4/SALL, keeping its expression at a basal level. At later time points, NGN-1/NGN is released from negative control by SEM-4/SALL and from positive control by HLH-16/OLIG, and its expression levels are boosted through increased EGL-5/HOX expression, requiring SEM-4/SALL. Following the increase in NGN-1/NGN, expression of the cytoskeleton regulators UNC-119, UNC-33/CRMP, and UNC-44/ANK is induced, directly or indirectly by NGN-1. UNC-119, UNC-33/CRMP, and UNC-44/ANK are required for PDA axon extension, but also for transcription of the exp-1 GABA receptor and cho-1 choline transporter genes and perhaps other downstream PDA maturation genes.
Figure 6. A model for Y-to-PDA transformation.
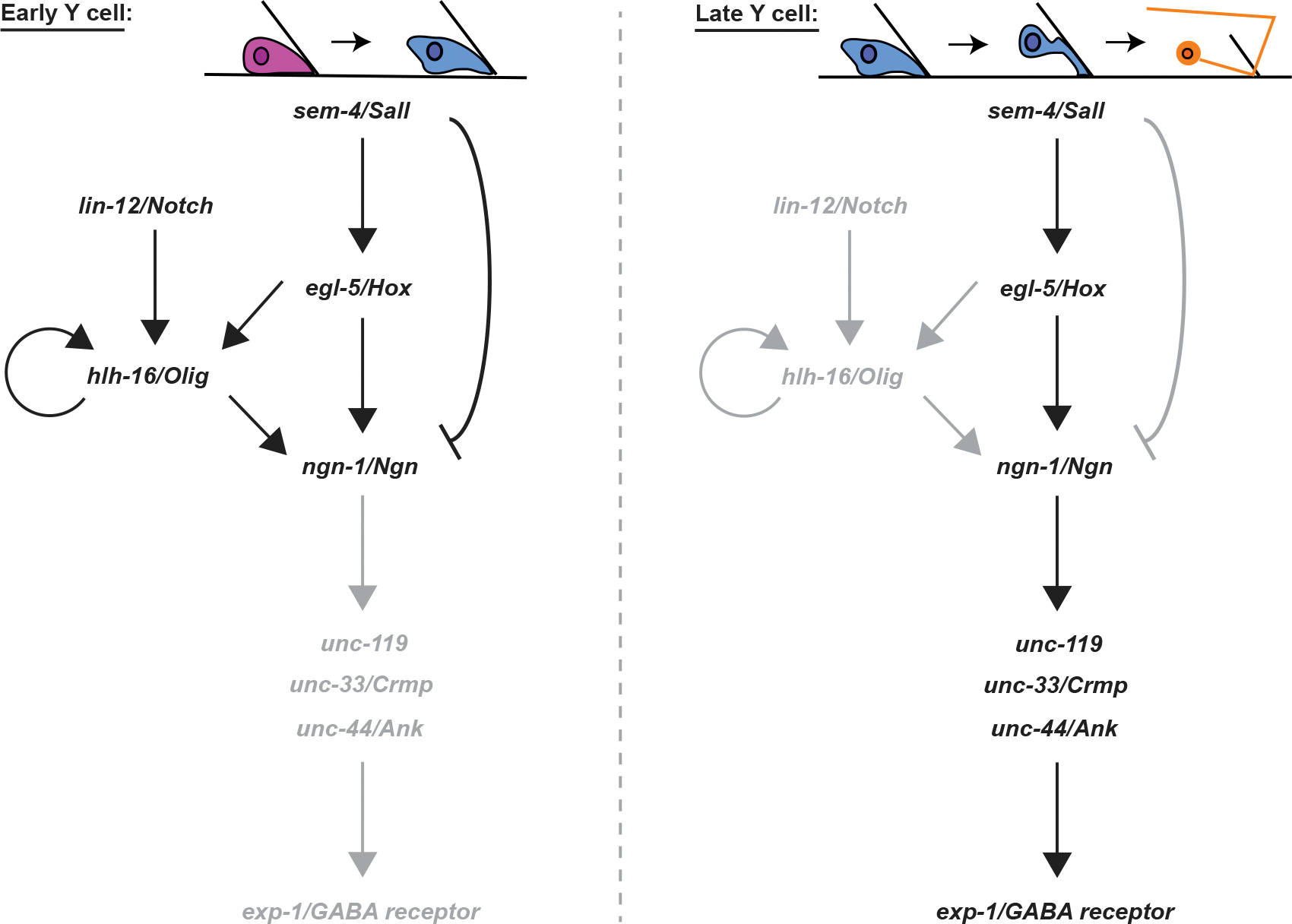
Black and gray letters indicate active or inactive genes/interactions, respectively.
Our model posits that NGN-1/NGN is key for PDA formation in wild-type animals. However, precocious PDA formation in lin-12/Notch mutants is only partially blocked by loss of ngn-1, suggesting that there are likely additional means by which PDA can be produced.
Our model also suggests that SEM-4/SALL has opposing activities as a transcriptional activator and repressor. Previous studies suggest that SEM-4 is part of the NODE transcriptional complex that activates egl-5 in the Y cell (Kagias et al., 2012). In other settings, however, SALL proteins have been suggested to be repressors (Ott et al., 2001; Barembaum and Bronner-Fraser, 2004; Parrish et al., 2004; Sweetman and Münsterberg, 2006). Whether both activities co-exist in the Y cell or whether there is a switch is not clear.
Finally, we point out that the events depicted in Figure 6 occur in the absence of cell division, suggesting that similar cell-division-independent roles for homologous proteins in other animals are likely.
A conserved module for epithelial-to-motoneuron/motoneuron-like fate transitions
The similarities between Y-to-PDA transformation, spinal cord motoneuron formation, and pancreatic islet cell generation are striking. All three events begin with a precursor cell situated within a tube-lining epithelium. Why tubular structures are appropriate sites for progenitor cells is not clear. While pancreatic ducts and the spinal canal could represent milieus that contain signaling molecules for stem cell differentiation (Huang et al., 2010; Lehtinen et al., 2011; Chang et al., 2012; Feliciano et al., 2014), this is unlikely to be the case for the Y cell, where the rectal canal likely transports only intestinal waste. Alternatively, tube-lining epithelial cells may be under specific mechanical stresses that facilitate stem cell division or maintenance (Park et al., 2017; Bogdanova et al., 2018; Guerrero et al., 2019; Legøy et al., 2020).
All three events also rely on Neurogenin-family members to promote differentiation (Ma et al., 1999; Fode et al., 1998; Mizuguchi et al., 2001; Lee et al., 2005; Zhou et al., 2007; Rubio-Cabezas et al., 2011; Gradwohl et al., 2000; Bankaitis et al., 2018), and Olig-family bHLH proteins are also important. The latter have been directly implicated in vertebrate motoneuron generation and in PDA formation (Takebayashi et al., 2000, 2002; Novitch et al., 2001; Mizuguchi et al., 2001; Lu et al., 2002; Lee et al., 2005). In the pancreas, expression of bHLHb4, an Olig-family bHLH transcription factor, is reported in cells bordering pancreatic ducts (Bramblett et al., 2002); however, no functional studies of the protein in the context of islet formation have been published.
Notch also plays a commanding role in all three differentiation events, although our studies suggest that the effects are not the same in all contexts. In the spinal cord and pancreas, NOTCH, acting through HES transcription factors, inhibits differentiation and/or division of stem cells. In C. elegans, NOTCH activity in Y promotes PDA formation independent of the LIN-22/HES and REF-1/HES-related protein and of cell division. A group of nematode-specific REF-1-family proteins distantly related to HES proteins has been described to function with NOTCH during C. elegans embryogenesis (Alper and Kenyon, 2001; Neves and Priess, 2005; Chou et al., 2015). It is possible that these other family members are relevant NOTCH targets, and that instead of inhibiting gene expression, they promote transcription, explaining the difference with the mammalian settings. Whatever the mechanism, it is likely conserved, as similar NOTCH-dependent, HES-independent induction of NGN3 is seen in the pancreatic islets (Cras-Méneur et al., 2009; Shih et al., 2012). Thus, future investigation of Y-to-PDA transformation could provide additional insight into mammalian pancreatic differentiation.
SALL proteins related to SEM-4 participate in early neural differentiation in the brain, are required for neuronal migration in chick neural crest cells, suppress chick neurogenin, and are expressed in the mouse spinal cord ventricular zone (Ott et al., 2001; Barembaum and Bronner-Fraser, 2004; Parrish et al., 2004; Sweetman and Münsterberg, 2006), suggesting that these proteins may indeed be relevant for spinal cord motoneuron and/or pancreatic islet development. However, their activities in these contexts have not been assessed. Similarly, roles for HOX proteins in neural and pancreatic development have been demonstrated (Aigner and Gage, 2005; Li et al., 2017; Tan et al., 2016; Catela et al., 2016; Larsen et al., 2015; Shi, 2010), but specific interactions with the neurogenin pathway have not been extensively studied.
Cytoskeletal organizers as regulators of gene expression
Our finding that genes controlling cytoskeleton organization are required for transcriptional maturation of the PDA motoneuron is surprising, as it is not immediately clear how these cytoplasmic proteins can affect nuclear events. Recent studies demonstrate that ankyrin proteins, related to C. elegans UNC-44, are required for pancreatic islet function (Healy et al., 2010), and ankyrins have been studied extensively for their roles in neuronal cytoskeleton organization and protein filtering into neuronal processes (Nakada et al., 2003; Jenkins et al., 2015; Sobotzik et al., 2009; Dubreuil and Yu, 1994; Jegla et al., 2016; LaBella et al., 2020). How could these roles be tied to transcriptional control? One possibility is that cellular polarity establishment, which requires cytoskeletal components for localizing proteins that initiate maturation, activates transcription factors that promote expression of terminal-differentiation genes. In neuronal contexts, the cytoskeleton could also monitor synapse maturation (LaBella et al., 2020), setting up a retrograde signal that influences nuclear transcription. Such retrograde signaling has been described in several contexts (Hendry et al., 1974; Feldman et al., 1999; Scheiffele et al., 2000; Asmus et al., 2000; Poo, 2001; Alger, 2002; Baudet et al., 2008). Given the facility with which we identified mutants defective in Y-to-PDA transformation, it is possible that the mechanisms at play could be revealed through additional forward genetic approaches in C. elegans.
Limitations of the study
To examine gene expression, we employed multicopy transgenes containing gene-regulatory sequences fused to fluorescent reporter genes, as well as single-copy fluorescent reporter gene insertions into endogenous loci. Multicopy transgenes may not contain all endogenous regulatory sites, while genomic insertion reporters could perturb chromatin organization, leading to changes in endogenous gene expression. By using both methods, we believe we have exposed relevant gene-regulatory events. Nonetheless, it remains possible that additional complexity in gene control exists.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Shai Shaham (shaham@rockefeller.edu).
Materials availability
All unique strains/stable reagents generated in this study are available from the lead contact without restriction.
Data and code availability
Whole genome sequencing data was deposited to the NCBI Sequence Read Archive (SRA) under BioProjectID PRJNA875470 (SRA: PRJNA875470). All other data reported in this paper will be shared by the lead contact upon request and is available in Table S2.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Caenorhabditis elegans
Experiments were conducted at 20C with C. elegans hermaphrodites unless otherwise indicated. C. elegans were cultivated using standard methods (Brenner, 1974). The wild-type parent for most strains used in this study is the C. elegans Bristol strain N2. Strains generated or used in this study are found in Table S2. The relevant mutations used in this study are: LG I: hlh-16(ns196), hlh-16(ns989), sem - 4(n1971); LG II: ref-1(mu220); LG III: egl-5(n945), egl-5(n486), lin-12(n676n930), unc-119(ns200), unc-119(ns913); LG IV: unc-44(e362), unc-44(ns986), unc-44(syb28170), ngn-1(ns186), ngn-1(syb2810), ngn-1(ns988), ngn-1(syb5802), ngn-1(syb5813), lin-22(n372). Transgenes used in these studies were nsIs131 [3.5 kb exp-1p:gfp::unc-54 3′utr] III, bxIs7 [egl-5p:egl-5(exon1–3):gfp], syIs63 [cog-1:GFP], otIs45 [unc-119:gfp]; otIs118 [unc-33:gfp], nsIs913 [4 kb ngn-1p:myr:mKate2:unc-54 3′utr] IV, nsIs914 [4 kb ngn-1p:myr:mKate2:unc-54 3′utr], nsIs942 [4 kb ngn-1p:myr:mKate2:unc-54 3′utr; 4 kb ngn-1:ajm-1:yfp:unc-54 3′utr], nsIs943 [480 bp hlh-16p:myr:GFP:1.5 kb hlh-16 3′utr], wgIs57 [sem - 4:TY1:EGFP:3xFLAG(92C12) + unc-119(+)], kuIs34 [sem - 4::GFP], oxEx2082 [unc-44 long isoform rescue], otIs591 [unc-3(fosmid)::gfp], otIs354 [cho-1(fosmid)::SL2::NLS::yfp::H2B]. Further information provided in Table S3.
METHOD DETAILS
Electron microscopy
L1 animals with short Y/PDA processes as in Figure 1F were selected for electron microscopy based on ngn-1p:myr:mKate2 fluorescence signals in the tail. Animals were prepared and sectioned using standard methods (Lundquist et al., 2001). Serial images were acquired by using a Titan Themis 200 kV transmission electron microscope with Cs Image Corrector. Image processing and analysis were performed using NIH ImageJ and IMOD software (Schorb et al., 2019).
Plasmid construction
Plasmids were constructed using Gibson cloning (New England Biolabs) into the applicable backbone plasmid.
Mutant analysis and scoring
An EMS screen was performed in the nsIs131 background, seeking F2 L4 animals lacking exp-1p::gfp expression in PDA (Jorgensen and Mango, 2002). Isolates were transferred to individual plates. Penetrance for each mutant was scored for several generations. Once causative mutations were identified in the alleles, bxIs7 [egl-5p:egl-5(exon1–3):gfp] was used to assess the formation of the Y cell in L1 animals and the absence of the Y cell in L4 animals.
SNP mapping
For SNP mapping, mutants were crossed to CB4856 Hawaiian (HA) animals. F2 animals lacking exp-1p:gfp expression were isolated and assessed for the percent penetrance to confirm that it matches the penetrance from the parent strain. F2 recombinants were lysed and analyzed for three SNPs between N2 and HA backgrounds on each chromosome. If a chromosomal region showed linkage to N2 DNA, further SNPs were used to narrow down the chromosomal region of the mutation.
Whole genome sequencing
Whole genome sequencing was performed on animals carrying the alleles sem - 4(ns189, ns192), ngn-1(ns194), unc-119(ns200), and mab-9(ns206). Animals were grown on E. coli strain OP50 until bacteria were depleted, harvested in M9, and resuspended in 1 M pH 7.5 TEN. SDS and proteinase K were added to a final concentration of 0.5% and 0.1 mg/mL respectively and 1 μL of β-mercaptoethanol was also added. The reaction was incubated overnight in a shaking thermocycler at 56C. Phenol/chloroform was added and phase-separated by spinning in a phase-lock tube. The aqueous phase was transferred to 200 proof EtOH. The resulting DNA clot was washed in 70% EtOH, dried, and resuspended in TEN. After rehydration, 0.3 μL of 100 mg/mL RNAse A was added and incubated at 37C for 2 h. Phenol/chloroform extraction was performed again and DNA was rehydrated with EB buffer. The sample was then run on a nanodrop machine and on a 1% gel to confirm that there was no RNA contamination or DNA degradation, respectively. NextSeq High Output 1 × 75 sequencing was performed with Standard Illumina Sequencing primers for gDNA-seq application.
Germline transformation and rescue experiments
Plasmid mixes containing the plasmid of interest, co-injection markers, and pBluescript were injected into both gonads of young adult hermaphrodites at a total of 100 ng/ul. Injected animals were singled onto NGM plates and allowed to grow for two generations. Transformed animals based on co-injection markers were picked onto single plates and screened for stable inheritance of the extrachromosomal array. Only lines from different P0 injected hermaphrodites were considered independent. Lines with stable inheritance of the extrachromosomal array were used to screen for rescues.
CRISPR Cas9 genome editing
Alleles of ngn-1(ns988), hlh-16(ns979), unc-119(ns913), and unc-44(ns986) were generated using a co-CRISPR editing strategy (Farboud et al., 2019). sem - 4(ns1004–1008) mutants were generated using a co-injection strategy (Dokshin et al., 2018). Guide crRNA, repair single-stranded DNA oligos, tracrRNA, and buffers were ordered from IDT. Guide crRNAs used are listed below.
hlh-16: 5′-CATTGTTACTAGCTTCCAATTGG-3′
unc-119: 5′-GCTCTTCCAGGAATCACTCAAGG-3′
unc-44: 5′-GTAGTTTCCATAACATGACGCGG-3′
ngn-1: 5′-TCGCCAGTTTGCAAATGATGCGG-3′
sem - 4: 5′-TGCAAAGACAAATGGCGAGCCGG-3′
5′-ATTGAAAGTGTTTACAGAGAGG-3′
Timing of Y-to-PDA transformation events
To determine the timing of Y-to-PDA events, nsIs913; nsIs131 gravid hermaphrodites were bleached and then incubated on a rotator in M9 overnight for L1 arrest synchronization. L1s were then plated onto agar plates containing NGM medium and E. coli OP50 bacteria. We imaged separate groups from this pool every 30 min between 6 and 22 h after food introduction, and scored for retrograde cell body migration, process extension, and onset of exp-1p::gfp expression until all animals imaged completed all three events.
For experiments in non-starved animals, nsIs913; nsIs131 gravid hermaphrodites were bleached, and the collected embryos were plated onto agar plates containing NGM medium and E. coli OP50 bacteria. As larvae hatched, they were isolated onto separate agar plates containing NGM medium and E. coli OP50 bacteria. We imaged separate groups from this pool every 30 min between 6 and 22 h after food introduction, and scored for retrograde cell body migration, process extension, and onset of exp-1p::gfp expression until all animals imaged completed all three events.
Quantification of fluorescence in the Y cell
To quantify the expression of various reporters before process extension, gravid hermaphrodites were bleached and then incubated on a rotator in M9 overnight for L1 arrest synchronization. L1s were then plated onto agar plates containing NGM medium and E. coli OP50 bacteria. Separate groups were imaged every 2 h until 14 h after release from L1 arrest. nsIs913 [ngn-1p:myr:mKate2], nsIs943 [hlh-16p:myr:gfp:hlh-16 utr], wgIs57 [sem - 4:TY1:EGFP:3xFLAG], kuIs34 [sem - 4:gfp], ngn-1(syb5802), and ngn-1(syb5813) animals were imaged on a Zeiss Imager M2 with an AxioCam r1.4 on a 63x/1.4 NA oil objective at 50, 150, 50, 30, 100, and 100 ms exposure respectively. For nsIs913 and nsIs943, an 80 pixels × 80 pixels square ROI was drawn over the Y cell. The fluorescence intensity was quantified at the z-position at the center of the cell. For wgIs57 and kuIs34, an ROI was drawn around the Y cell nucleus, and the fluorescence intensity was quantified at the z-position at the center of the cell body. For ngn-1(syb5802) and ngn-1(syb5813), a 25 pixels × 25 pixels circular ROI was drawn over the Y cell nucelus, and the fluorescence intensity was quantified at the z-position at the center of the cell body. bxIs7 was imaged on an iSIM with a VisiTech iSIM head and a Leica DMi8 inverted microscope stand with a 100x/1.47 NA oil objective and an Orca sCMOS camera. An ROI was drawn over the entire Y cell, and the fluorescence intensity was quantified at the z-position at the center of the cell.
Lin-12 temperature-sensitive experiments
lin-12(n676n930) mutant animals were raised at 15C. Gravid hermaphrodites were bleached and then incubated on a rotator in M9 at 15C or 20C overnight for L1 arrest synchronization. L1s were plated onto agar plates containing NGM and immediately prepared for imaging.
Cytoskeleton organizers mutant morphology
Gravid animals were bleached and then incubated on a rotator in M9 overnight for L1 arrest synchronization. L1s were then plated onto agar plates containing NGM medium and E. coli OP50 bacteria. 47 h later, animals were prepared for imaging.
UNC-44 expression localization
A CRISPR insertion of SL2:NLS:mKate2 before the stop codon in the long neural isoform of unc-44 was obtained from SUNY Biotech (Fuzhou, China).
Imaging preparation
2% agarose with 6 mM NaN3 was used to make thin pads for mounting animals on microscope slides. Animals were picked into a 2 μL drop of M9 and a 0.17 mm coverslip was placed on top. ngn-1p::myr::mKate2 animals for figure images were imaged on an inverted Olympus IX-70 microscope with a 60X silicone objective, a pco.edge sCMOS camera, and deconvolution using measured PSFs.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism 9.2.0 was used to perform statistical analysis on the data. Specific statistical details can be found in the figure legends. Error bars represent SEM. Statistical significance was determined using p < 0.05 or p < 0.01. Data and statistical results for each figure can be found in Table S2.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli OP50 | CGC | https://cgc.umn.edu/strain/OP50 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Sodium Hypochlorite Solution (4.00–4.99%/Laboratory) | Sigma-Aldrich | Cat# 239305 |
| Cas9-NLS Purified Protein | Berkeley QB3 MacroLab | NA |
| Nuclease Free Duplex Buffer | IDT | Cat#11-01-03-01 |
| Alt-R® CRISPR-Cas9 tracrRNA, 5 nmol | IDT | Cat#1072532 |
| RNAse A | Qiagen | Cat#19101 |
| Q5® High-Fidelity DNA Polymerase | NEB | Cat#M0491L |
| Phusion® High-Fidelity DNA Polymerase | NEB | Cat#M0530L |
| Sibgene Taq DNA Polymerase | Sibgene | Cat#SG-1-500 |
|
| ||
| Deposited data | ||
|
| ||
| Whole Genome Sequencing | This paper | NCBI Sequence Read Archive (SRA) BioProjectID PRJNA875470 (SRA:PRJNA875470) |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| See Table S3 for strains | ||
|
| ||
| Oligonucleotides | ||
|
| ||
| hlh-16 promoter forward primer: GAACATCAGAAATTTGAGACTTC | This paper | NA |
| hlh-16 promoter reverse primer: AACATCTTAGAAGTGACACGTTTG | This paper | NA |
| hlh-16 3′UTR forward primer: AGATGCATTCATTGGTTTGC | This paper | NA |
| hlh-16 3′UTR reverse primer: ATTGAGCTCACCGT CAAAATG | This paper | NA |
| ngn-1 promoter forward primer: AACATTTTGTCGTTGCACC | This paper | NA |
| ngn-1 promoter reverse primer: GTGCAAAACAAAAACACGTGGGTGGGG | This paper | NA |
| myristoyl sequence forward primer: ATGGGATCATGTATTGGAAAAG | This paper | NA |
| mKate2 reverse primer: TTAACGGTGTCCGAGCTTG | This paper | NA |
| GFP/YFP forward primer: ATGAGTAAAGGAGAAGAACTTTTCACTG | This paper | NA |
| GFP/YFP reverse primer: TTATTTGTATAGTTCATCCATGCCAT | This paper | NA |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid pAR22: ngn-1p::myr:: mKate2::unc-54 3′UTR | This paper | NA |
| Plasmid pAR23: hlh-16p::myr:: gfp::hlh-16 3′UTR | This paper | NA |
| Plasmid pAR24: ngn-1p::ajm-1:: yfp::unc-54 3′UTR | This paper | NA |
| Plasmid pAR25: ngn-1p:: unc-119 cDNA (cb):: unc-54 3′UTR | This paper | NA |
| Plasmid pAR28: hlh-16p:: hlh-16 cDNA::hlh-16 3′UTR | This paper | NA |
| Plasmid pMT23: ngn-1p:: ngn-1 cDNA::unc-54 3′UTR | This paper | NA |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| Prism 9.2.0 | GraphPad | https://www.graphpad.com/scientific-software/ |
| Illustrator 26.3.1 | Adobe | https://www.adobe.com/products/illustrator.html |
| ApE | Wayne Davis | https://jorgensen.biology.utah.edu/wayned/ape/ |
Highlights.
ngn-1 and hlh-16/Olig drive epithelial-to-motoneuron transformation in C. elegans
lin-12/Notch controls transformation timing through ngn-1 and hlh-16/Olig
At transformation onset, ngn-1 expression is boosted by sem-4/Sall and egl-5/Hox
Cytoskeleton organizers UNC-33, UNC-44, and UNC-119 promote motoneuron terminal identity
ACKNOWLEDGMENTS
We would like to thank Scott Emmons, Iva Greenwald, Maxwell Heiman, Oliver Hobert, and Erik Jorgensen for strains. Some strains were provided by the CGC, funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). Images were acquired at the Bio-Imaging Resource Center, RRID: SCR_017791, and whole-genome sequencing was performed at the Genomics Resource Center at The Rockefeller University. We would like to acknowledge the Electron Microscope Imaging Facility of CUNY Advanced Science Research Center for instrument use and scientific and technical assistance. Electron microscopy was performed at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR, and NIH (GM103310). This work was funded by a Kavli pilot grant to A.R., a fellowship from the NYSCF to M.T., an NSF predoctoral fellowship to A.R., and NIH grant R35 NS105094 to S.S.
Footnotes
DECLARATION OF INTERESTS
S.S. is a member of the Cell Reports advisory board.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111414.
REFERENCES
- Aigner S, and Gage FH (2005). A small gem with great powers: geminin keeps neural progenitors thriving. Dev. Cell 9, 171–172. 10.1016/j.devcel.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Alger BE (2002). Retrograde signaling in the regulation of synaptic transmission: focus on endocannabinoids. Prog. Neurobiol. 68, 247–286. 10.1016/S0301-0082(02)00080-1. [DOI] [PubMed] [Google Scholar]
- Alper S, and Kenyon C (2001). REF-1, a protein with two bHLH domains, alters the pattern of cell fusion in C. elegans by regulating Hox protein activity. Development 128, 1793–1804. 10.1242/dev.128.10.1793. [DOI] [PubMed] [Google Scholar]
- Apelqvist Å, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, and Edlund H (1999). Notch signalling controls pancreatic cell differentiation. Nature 400, 877–881. 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Appel B, Givan LA, and Eisen JS (2001). Delta-Notch signaling and lateral inhibition in zebrafish spinal cord development. BMC Dev. Biol. 1, 13. 10.1186/1471-213x-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmus SE, Parsons S, and Landis SC (2000). Developmental changes in the transmitter properties of sympathetic neurons that innervate the periosteum. J. Neurosci. 20, 1495–1504. 10.1523/JNEUROSCI.20-04-01495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis ED, Bechard ME, Gu G, Magnuson MA, and Wright CVE (2018). ROCK-nmMyoII, Notch and Neurog3 gene-dosage link epithelial morphogenesis with cell fate in the pancreatic endocrine-progenitor niche. Development 145, dev162115. 10.1242/dev.162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barembaum M, and Bronner-Fraser M (2004). A novel spalt gene expressed in branchial arches affects the ability of cranial neural crest cells to populate sensory ganglia. Neuron Glia Biol. 1, 57–63. 10.1017/S1740925X04000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet C, Pozas E, Adameyko I, Andersson E, Ericson J, and Ernfors P (2008). Retrograde signaling onto Ret during motor nerve terminal maturation. J. Neurosci. 28, 963–975. 10.1523/JNEUROSCI.4489-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker SF, Morin C, Medina Sanchez JD, and Jarriault S (2019). You Are what You Experience: The Impact of Environnment on Transdifferentiation.
- Beg AA, and Jorgensen EM (2003). EXP-1 is an excitatory GABA-gated cation channel. Nat. Neurosci. 6, 1145–1152. 10.1038/nn1136. [DOI] [PubMed] [Google Scholar]
- Bertrand V, Bisso P, Poole RJ, and Hobert O (2011). Notch-dependent induction of left/right asymmetry in C. elegans interneurons and motoneurons. Curr. Biol. 21, 1225–1231. 10.1016/j.cub.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanova M, Kostina A, Zihlavnikova Enayati K, Zabirnyk A, Malashicheva A, Stensløkken KO, Sullivan GJ, Kaljusto M-L, Kvitting JP, Kostareva A, et al. (2018). Inflammation and mechanical stress stimulate osteogenic differentiation of human aortic valve interstitial cells. Front. Physiol. 9, 1635. 10.3389/fphys.2018.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boontrakulpoontawee P, and Otsuka AJ (2002). Mutational analysis of the Caenorhabditis elegans ankyrin gene unc-44 demonstrates that the large spliceoform is critical for neural development. Mol. Genet. Genomics. 267, 291–302. 10.1007/s00438-002-0661-x. [DOI] [PubMed] [Google Scholar]
- Bramblett DE, Copeland NG, Jenkins NA, and Tsai M-J (2002). BHLHB4 is a bHLH transcriptional regulator in pancreas and brain that marks the dimesencephalic boundary. Genomics 79, 402–412. 10.1006/geno.2002.6708. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of caenorhabditis elegans. Genetics 77, 71–94. 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catela C, Shin MM, Lee DH, Liu J-P, and Dasen JS (2016). Hox proteins coordinate motor neuron differentiation and connectivity programs through ret/Gfra genes. Cell Rep. 14, 1901–1915. 10.1016/j.celrep.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JT, Lowery LA, and Sive H (2012). Multiple roles for the Na, K-ATPase subunits, Atp1a1 and Fxyd1, during brain ventricle development. Dev. Biol. 368, 312–322. 10.1016/j.ydbio.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-C, Huang H-R, Hsu C-H, and Ou C-Y (2021). CRMP/UNC-33 organizes microtubule bundles for KIF5-mediated mitochondrial distribution to axon. PLoS Genet. 17, e1009360. 10.1371/journal.pgen.1009360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HT, Vazquez RG, Wang K, Campbell R, Milledge GZ, Walthall WW, and Johnson CM (2015). HES-mediated repression of pten in Caenorhabditis elegans. G3 5, 2619–2628. 10.1534/g3.115.019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli V, Baetens D, Rutishauser U, Halban PA, Orci L, and Rouiller DG (1994). Expression of neural cell adhesion molecule (N-CAM) in rat islets and its role in islet cell type segregation. J. Cell Sci. 107 (Pt 6), 1429–1436. [DOI] [PubMed] [Google Scholar]
- Cras-Méneur C, Li L, Kopan R, and Permutt MA (2009). Presenilins, Notch dose control the fate of pancreatic endocrine progenitors during a narrow developmental window. Genes Dev. 23, 2088–2101. 10.1101/gad.1800209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, and Jessell TM (2009). Chapter six Hox networks and the origins of motor neuron diversity. Curr. Top. Dev. Biol, 169–200. 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, and Conlon RA (1997). Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124, 1139–1148. [DOI] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, and Mello CC (2018). Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics 210, 781–787. 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil RR, and Yu J (1994). Ankyrin and beta-spectrin accumulate independently of alpha-spectrin in Drosophila. Proc. Natl. Acad. Sci. USA 91, 10285–10289. 10.1073/pnas.91.22.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Thor S, Edlund T, Jessell TM, and Yamada T (1992). Early stages of motor neuron differentiation revealed by expression of homeobox gene islet-1. Science 256, 1555–1560. 10.1126/science.1350865. [DOI] [PubMed] [Google Scholar]
- Fan L, Kovacevic I, Heiman MG, and Bao Z (2019). A multicellular rosette-mediated collective dendrite extension. Elife 8, e38065. 10.7554/eLife.38065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farboud B, Severson AF, and Meyer BJ (2019). Strategies for efficient genome editing using CRISPR-Cas9. Genetics 211, 431–457. 10.1534/genetics.118.301775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman DE, Nicoll RA, and Malenka RC (1999). Synaptic plasticity at thalamocortical synapses in developing rat somatosensory cortex: LTP, LTD, and silent synapses. J. Neurobiol. 41, 92–101. [PubMed] [Google Scholar]
- Feliciano DM, Zhang S, Nasrallah CM, Lisgo SN, and Bordey A (2014). Embryonic cerebrospinal fluid nanovesicles carry evolutionarily conserved molecules and promote neural stem cell amplification. PLoS One 9, e88810. 10.1371/journal.pone.0088810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira HB, Zhang Y, Zhao C, and Emmons SW (1999). Patterning of Caenorhabditis elegansPosterior Structures by theAbdominal-BHomolog, egl-5. Dev. Biol. 207, 215–228. 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- Fode C, Gradwohl G, Morin X, Dierich A, LeMeur M, Goridis C, and Guillemot F (1998). The bHLH protein NEUROGENIN 2 is a determination factor for epibranchial placode-derived sensory neurons. Neuron 20, 483–494. 10.1016/s0896-6273(00)80989-7. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Fode C, and Guillemot F (1996). Restricted expression of a novel murineatonal-related bHLH protein in undifferentiated neural precursors. Dev. Biol. 180, 227–241. 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- Gradwohl G, Dierich A, LeMeur M, and Guillemot F (2000). neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. USA 97, 1607–1611. 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, and Horvitz HR (1983). The lin-12 locus specifies cell fates in caenorhabditis elegans. Cell 34, 435–444. 10.1016/0092-8674(83)90377-X. [DOI] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, and Walhout AJM (2009). A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138, 314–327. 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero P, Perez-Carrasco R, Zagorski M, Page D, Kicheva A, Briscoe J, and Page KM (2019). Neuronal differentiation influences progenitor arrangement in the vertebrate neuroepithelium. Development 146, dev176297. 10.1242/dev.176297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, and Kageyama R (2004). Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131, 5539–5550. 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- He L, Kooistra R, Das R, Oudejans E, van Leen E, Ziegler J, Portegies S, de Haan B, van Regteren Altena A, Stucchi R, et al. (2020). Cortical anchoring of the microtubule cytoskeleton is essential for neuron polarity. Elife 9, e55111. 10.7554/eLife.55111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy JA, Nilsson KR, Hohmeier HE, Berglund J, Davis J, Hoffman J, Kohler M, Li L-S, Berggren P-O, Newgard CB, and Bennett V (2010). Cholinergic augmentation of insulin release requires ankyrin-B. Sci. Signal. 3, ra19. 10.1126/scisignal.2000771. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, and Perkins L (1985). Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111, 158–170. [DOI] [PubMed] [Google Scholar]
- Heiman MG, and Shaham S (2009). DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell 137, 344–355. 10.1016/j.cell.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendry IA, Stöckel K, Thoenen H, and Iversen LL (1974). The retrograde axonal transport of nerve growth factor. Brain Res. 68, 103–121. 10.1016/0006-8993(74)90536-8. [DOI] [PubMed] [Google Scholar]
- Huang X, Liu J, Ketova T, Fleming JT, Grover VK, Cooper MK, Litingtung Y, and Chiang C (2010). Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proc. Natl. Acad. Sci. USA 107, 8422–8427. 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, and Guillemot F (1995). Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 9, 3136–3148. 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Schwab Y, and Greenwald I (2008). A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. USA 105, 3790–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegla T, Nguyen MM, Feng C, Goetschius DJ, Luna E, van Rossum DB, Kamel B, Pisupati A, Milner ES, and Rolls MM (2016). Bilaterian giant ankyrins have a common evolutionary origin and play a conserved role in patterning the axon initial segment. PLoS Genet. 12, e1006457. 10.1371/journal.pgen.1006457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins PM, Kim N, Jones SL, Tseng WC, Svitkina TM, Yin HH, and Bennett V (2015). Giant ankyrin-G: a critical innovation in vertebrate evolution of fast and integrated neuronal signaling. Proc. Natl. Acad. Sci. USA 112, 957–964. 10.1073/pnas.1416544112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J, Pedersen EE, Galante P, Hald J, Heller RS, Ishibashi M, Kageyama R, Guillemot F, Serup P, and Madsen OD (2000). Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36–44. 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jessell TM (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 1, 20–29. 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jorgensen EM, and Mango SE (2002). The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 3, 356–369. 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- Kagias K, Ahier A, Fischer N, and Jarriault S (2012). Members of the NODE (Nanog and Oct4-associated deacetylase) complex and SOX-2 promote the initiation of a natural cellular reprogramming event in vivo. Proc. Natl. Acad. Sci. USA 109, 6596–6601. 10.1073/pnas.1117031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobel KM, Davis WS, Jorgensen EM, and Bastiani MJ (2001). UNC-119 suppresses axon branching in C. elegans. Development 128, 4079–4092. [DOI] [PubMed] [Google Scholar]
- LaBella ML, Hujber EJ, Moore KA, Rawson RL, Merrill SA, Allaire PD, Ailion M, Hollien J, Bastiani MJ, and Jorgensen EM (2020). Casein kinase 1d stabilizes mature axons by inhibiting transcription termination of ankyrin. Dev. Cell 52, 88–103.e18. 10.1016/j.devcel.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen BM, Hrycaj SM, Newman M, Li Y, and Wellik DM (2015). Mesenchymal Hox6 function is required for pancreatic endocrine cell differentiation. Development 142, 3859–3868. 10.1242/dev.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-K, Lee B, Ruiz EC, and Pfaff SL (2005). Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 19, 282–294. 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legøy TA, Vethe H, Abadpour S, Strand BL, Scholz H, Paulo JA, Ræder H, Ghila L, and Chera S (2020). Encapsulation boosts islet-cell signature in differentiating human induced pluripotent stem cells via integrin signalling. Sci. Rep. 10, 414. 10.1038/s41598-019-57305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. (2011). The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron 69, 893–905. 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva-Dáz E, and Hobert O (2022). Robust regulatory architecture of panneuronal gene expression. Curr. Biol. 32, 1715–1727.e8. 10.1016/j.cub.2022.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-J, Hong T, Tung Y-T, Yen Y-P, Hsu H-C, Lu Y-L, Chang M, Nie Q, and Chen J-A (2017). MicroRNA filters Hox temporal transcription noise to confer boundary formation in the spinal cord. Nat. Commun. 8, 14685. 10.1038/ncomms14685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, and Rowitch DH (2002). Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86. 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, and Bargmann CI (2001). Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128, 4475–4488. [DOI] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, and Anderson DJ (1998). neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20, 469–482. 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Fode C, Guillemot F, and Anderson DJ (1999). Neurogenin1 and neurogenin2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes Dev. 13, 1717–1728. 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Gordon M, Jacobs R, and Pilgrim DB (2000). The unc-119 family of neural proteins is functionally conserved between humans, Drosophila and C. Elegans. J. Neurogenet. 13, 191–212. 10.3109/01677060009084494. [DOI] [PubMed] [Google Scholar]
- Maniar TA, Kaplan M, Wang GJ, Shen K, Wei L, Shaw JE, Koushika SP, and Bargmann CI (2011). UNC-33 (CRMP) and ankyrin organize microtubules and localize kinesin to polarize axon-dendrite sorting. Nat. Neurosci. 15, 48–56. 10.1038/nn.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AG, Crawford BD, Waskiewicz AJ, and Pilgrim DB (2004). unc-119 homolog required for normal development of the zebrafish nervous system. Genesis 40, 223–230. 10.1002/gene.20089. [DOI] [PubMed] [Google Scholar]
- Materi W, and Pilgrim D (2005). NovelCaenorhabditis elegans unc-119 axon outgrowth defects correlate with behavioral phenotypes that are partially rescued by nonneural unc-119. Genesis 42, 104–116. 10.1002/gene.20130. [DOI] [PubMed] [Google Scholar]
- Maurer CW, Chiorazzi M, and Shaham S (2007). Timing of the onset of a developmental cell death is controlled by transcriptional induction of the C. elegans ced-3 caspase-encoding gene. Development 134, 1357–1368. 10.1242/dev.02818. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, and Nakafuku M (2001). Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31, 757–771. 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- Muñoz-Bravo JL, Hidalgo-Figueroa M, Pascual A, López-Barneo J, Leal-Cerro A, and Cano DA (2013). GDNF is required for neural colonization of the pancreas. Development 140, 3669–3679. 10.1242/dev.091256. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, and Melton DA (2003). Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA 100, 14920–14925. 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada C, Ritchie K, Oba Y, Nakamura M, Hotta Y, Iino R, Kasai RS, Yamaguchi K, Fujiwara T, and Kusumi A (2003). Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5, 626–632. 10.1038/ncb1009. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Stellrecht CM, and Tsai MJ (1995). Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 9, 1009–1019. 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- Neves A, and Priess JR (2005). The REF-1 family of bHLH transcription factors pattern C. elegans embryos through notch-dependent and notch-independent pathways. Dev. Cell 8, 867–879. 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Nicolls MR (2004). The clinical and biological relationship between Type II diabetes mellitus and Alzheimer’s disease. Curr. Alzheimer Res. 1, 47–54. 10.2174/1567205043480555. [DOI] [PubMed] [Google Scholar]
- Ninov N, Borius M, and Stainier DYR (2012). Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 139, 1557–1567. 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Sundararajan L, Morgan DE, Roberts ZJ, and Lundquist EA (2014). The UNC-6/Netrin receptors UNC-40/DCC and UNC-5 inhibit growth cone filopodial protrusion via UNC-73/Trio, Rac-like GTPases and UNC-33/CRMP. Development 141, 4395–4405. 10.1242/dev.110437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novitch BG, Chen AI, and Jessell TM (2001). Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773–789. 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, and Kageyama R (1999). Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207. 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka AJ, Franco R, Yang B, Shim KH, Tang LZ, Zhang YY, Boontrakulpoontawee P, Jeyaprakash A, Hedgecock E, and Wheaton VI (1995). An ankyrin-related gene (unc-44) is necessary for proper axonal guidance in Caenorhabditis elegans. J. Cell Biol. 129, 1081–1092. 10.1083/jcb.129.4.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka AJ, Boontrakulpoontawee P, Rebeiz N, Domanus M, Otsuka D, Velamparampil N, Chan S, Vande Wyngaerde M, Campagna S, and Cox A (2002). Novel UNC-44 AO13 ankyrin is required for axonal guidance in C. elegans, contains six highly repetitive STEP blocks separated by seven potential transmembrane domains, and is localized to neuronal processes and the periphery of neural cell bodies. J. Neurobiol. 50, 333–349. 10.1002/neu.10036. [DOI] [PubMed] [Google Scholar]
- Ott T, Parrish M, Bond K, Schwaeger-Nickolenko A, and Monaghan AP (2001). A new member of the spalt like zinc finger protein family, Msal-3, is expressed in the CNS and sites of epithelial/mesenchymal interaction. Mech. Dev. 101, 203–207. 10.1016/S0925-4773(00)00552-9. [DOI] [PubMed] [Google Scholar]
- Park MG, Jang H, Lee S-H, and Lee CJ (2017). Flow shear stress enhances the proliferative potential of cultured radial glial cells possibly via an activation of mechanosensitive calcium channel. Exp. Neurobiol. 26, 71–81. 10.5607/en.2017.26.2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish M, Ott T, Lance-Jones C, Schuetz G, Schwaeger-Nickolenko A, and Monaghan AP (2004). Loss of the Sall3 gene leads to palate deficiency, abnormalities in cranial nerves, and perinatal lethality. Mol. Cell Biol. 24, 7102–7112. 10.1128/MCB.24.16.7102-7112.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M, Scharfmann R, Seilheimer B, Eisenbarth G, Dressler D, Verma IM, and Potter H (1993). Nerve growth factor induces neuron-like differentiation of an insulin-secreting pancreatic beta cell line. Proc. Natl. Acad. Sci. USA 90, 5781–5785. 10.1073/pnas.90.12.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo MM (2001). Neurotrophins as synaptic modulators. Nat. Rev. Neurosci. 2, 24–32. 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, and Walhout AJM (2005). A compendium of Caenorhabditis elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 6, R110. 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard JP, Zuryn S, Fischer N, Pavet V, Vaucamps N, and Jarriault S (2011). Direct in vivo cellular reprogramming involves transition through discrete, non-pluripotent steps. Development 138, 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Cabezas O, Jensen JN, Hodgson MI, Codner E, Ellard S, Serup P, and Hattersley AT (2011). Permanent neonatal diabetes and enteric anendocrinosis associated with Biallelic mutations in NEUROG3. Diabetes 60, 1349–1353. 10.2337/db10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagner A, and Briscoe J (2019). Establishing neuronal diversity in the spinal cord: a time and a place. Development 146. 10.1242/dev.182154. [DOI] [PubMed] [Google Scholar]
- Sagner A, Gaber ZB, Delile J, Kong JH, Rousso DL, Pearson CA, Weicksel SE, Melchionda M, Mousavy Gharavy SN, Briscoe J, and Novitch BG (2018). Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. PLoS Biol. 16, e2003127. 10.1371/journal.pbio.2003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, and Serafini T (2000). Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101, 657–669. 10.1016/S0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorb M, Haberbosch I, Hagen WJH, Schwab Y, and Mastronarde DN (2019). Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477. 10.1038/s41592-019-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y (2010). Generation of functional insulin-producing cells from human embryonic stem cells in vitro. In Cellular Programming and Reprogramming, Ding S, ed. (Humana Press; ), pp. 79–85. 10.1007/978-1-60761-691-7_5. [DOI] [PubMed] [Google Scholar]
- Shih HP, Kopp JL, Sandhu M, Dubois CL, Seymour PA, Grapin-Botton A, and Sander M (2012). A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development 139, 2488–2499. 10.1242/dev.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal A, and Shaham S (2017). Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat. Commun. 8, 14100. 10.1038/ncomms14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotzik J-M, Sie JM, Politi C, Del Turco D, Bennett V, Deller T, and Schultz C (2009). AnkyrinG is required to maintain axo-dendritic polarity in vivo. Proc. Natl. Acad. Sci. USA 106, 17564–17569. 10.1073/pnas.0909267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer L, Ma Q, and Anderson DJ (1996). neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8, 221–241. 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- Stefanakis N, Carrera I, and Hobert O (2015). Regulatory logic of panneuronal gene expression in C. elegans. Neuron 87, 733–750. 10.1016/j.neuron.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, and Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156. 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, and Greenberg ME (2001). Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376. 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Sundaram M, and Greenwald I (1993). Genetic and phenotypic studies of hypomorphic lin-12 mutants in Caenorhabditis elegans. Genetics 135, 755–763. 10.1093/genetics/135.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, and German MS (1998). Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development 125, 2213–2221. [DOI] [PubMed] [Google Scholar]
- Sweetman D, and Münsterberg A (2006). The vertebrate spalt genes in development and disease. Dev. Biol. 293, 285–293. 10.1016/j.ydbio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, and Nabeshima Y (2000). Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member. Mech. Dev. 99, 143–148. 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, and Nabeshima Y-I (2002). The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Biol. 12, 1157–1163. 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Tan GC, Mazzoni EO, and Wichterle H (2016). Iterative role of notch signaling in spinal motor neuron diversification. Cell Rep. 16, 907–916. 10.1016/j.celrep.2016.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor S, Ericson J, Brännström T, and Edlund T (1991). The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron 7, 881–889. 10.1016/0896-6273(91)90334-V. [DOI] [PubMed] [Google Scholar]
- Tsuboi D, Hikita T, Qadota H, Amano M, and Kaibuchi K (2005). Regulatory machinery of UNC-33 Ce-CRMP localization in neurites during neuronal development in Caenorhabditis elegans. J. Neurochem. 95, 1629–1641. 10.1111/j.1471-4159.2005.03490.x. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, and Brenner S (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Wrischnik LA, and Kenyon CJ (1997). The role of lin-22, a hairy/enhancer of split homolog, in patterning the peripheral nervous system of C. elegans. Development 124, 2875–2888. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, and Melton DA (2007). A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 13, 103–114. 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Zhou S, Opperman K, Wang X, and Chen L (2008). unc-44 Ankyrin and stn-2 gamma-syntrophin regulate sax-7 L1CAM function in maintaining neuronal positioning in Caenorhabditis elegans. Genetics 180, 1429–1443. 10.1534/genetics.108.091272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuryn S, Ahier A, Portoso M, White ER, Morin M-C, Margueron R, and Jarriault S (2014). Sequential histone-modifying activities determine the robustness of transdifferentiation. Science 345, 826–829. 10.1126/science.1255885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequencing data was deposited to the NCBI Sequence Read Archive (SRA) under BioProjectID PRJNA875470 (SRA: PRJNA875470). All other data reported in this paper will be shared by the lead contact upon request and is available in Table S2.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


