Highlights
-
•
Epibenthic dinoflagellates are abundant on macrophytes at the Swedish west coast.
-
•
The distributions of Prorocentrum lima and Coolia monotis are presented.
-
•
Scanning electron microscopy and metabarcoding revealed a large diversity.
-
•
The phycotoxin producer Prorocentrum lima is a potential risk to human health.
-
•
Artificial substrates are unsuitable for quantitative sampling in this area.
Keywords: Benthic microalgae, Shellfish toxins, Diarrhetic shellfish poisoning, Dinoflagellate diversity, Metabarcoding, Benthic harmful algal blooms (BHABs)
Abbreviations: ED, Epibenthic dinoflagellate; HAB, Harmful Algal Blooms; BHAB, Benthic Harmful Algal Blooms; DSP, Diarrhetic Shellfish Poisoning; ASVs, Amplicon Sequence Variant; DCA, Detrended Correspondence Analysis; PCR, Polymerase chain reaction
Abstract
Epibenthic dinoflagellates occur globally and include many toxin-producing species of concern to human health and benthic ecosystem function. Such benthic harmful algal blooms (BHABs) have been well described from tropical and sub-tropical coastal environments, but assessments from north temperate waters, e.g., northern Europe, and polar regions are scarce. The present study addressed the biodiversity and distribution of potentially toxic epibenthic dinoflagellate populations along the west coast of Sweden (Kattegat-Skagerrak) by morphological and molecular criteria. Morphological analysis conducted by light- and electron-microscopy was then linked by DNA barcoding of the V4 region of 18S rRNA gene sequences to interpret taxonomic and phylogenetic relationships. The presence of two potentially toxigenic epibenthic dinoflagellates, Prorocentrum lima (Ehrenberg) F.Stein and Coolia monotis Meunier was confirmed, along with a description of their spatial and temporal distribution. For P. lima, one third of the cell abundance values exceeded official alarm thresholds for potentially toxic BHAB events (>1000 cells gr–1 of macroalgae fresh weight). The same species were recorded consecutively for two summers, but without significant temporal variation in cell densities. SEM analyses confirmed the presence of other benthic Prorocentrum species: P. fukuyoi complex, P. cf. foraminosum and P. cf. hoffmannianum. Analyses of the V4 region of the 18S rRNA gene also indicated the presence P. compressum, P. hoffmannianum, P. foraminosum, P. fukuyoi, and P. nanum. These findings provide the first biogeographical evidence of toxigenic benthic dinoflagellates along the west coast of Sweden, in the absence of ongoing monitoring to include epibenthic dinoflagellates. Harmful events due to the presence of Coolia at shellfish aquaculture sites along the Kattegat-Skagerrak are likely to be rather marginal because C. monotis is not known to be toxigenic. In any case, as a preliminary assessment, the results highlight the risk of diarrhetic shellfish poisoning (DSP) events caused by P. lima, which may affect the development and sustainability of shellfish aquaculture in the region.
Graphical abstract

1. Introduction
Epibenthic dinoflagellates (EDs) are found primarily along sheltered coasts and attached to a wide range of substrates such as macroalgae, seagrasses, floating detritus or sediment surfaces (Gorbi et al., 2013; Hoppenrath et al., 2014; Durán-Riveroll et al., 2019). Over the past few decades, proliferation of toxigenic EDs has become an emergent phenomenon, and dense cell aggregations of these species are now considered a serious threat to benthic marine ecosystems, human health and seafood industries, particularly for coastal communities (Wells et al., 2015; Berdalet et al., 2016; Vila et al., 2016; Durán-Riveroll et al., 2019). Benthic species do not form classic harmful algal blooms (HABs) like planktonic microalgae, but excessive growth of toxigenic EDs upon benthic substrates may contribute to formation of so-called Benthic Harmful Algal Blooms (BHABs). Toxins accumulated directly from these substrates or after tychoplanktonic release of cells to the surrounding water column can be transferred along marine food chains. When sequestered in shellfish and finfish, these toxins become vectors of human toxicity syndromes, such as diarrhetic shellfish poisoning (DSP) or ciguatera fish poisoning (CFP) when contaminated seafood is consumed (Yasumoto et al., 1987; Toda et al., 2012; Berdalet et al., 2016).
There is a generally high awareness of toxic epibenthic dinoflagellates and associated toxin syndromes in tropical and sub-tropical coastal regions (Durán-Riveroll et al., 2019), where many cases of seafood poisoning are registered annually. Risk assessments in other parts of the world, however, are scarce. Along northern European coasts, for example, the paucity of confirmed human toxicity cases related to BHABs has led to low public awareness of risk and to regulatory laxness in monitoring potentially toxigenic benthic microalgae. Nevertheless, over the past two decades, the diversity and harmfulness of EDs in temperate seas have attracted increasing interest of the scientific community (Berdalet et al., 2016; Berdalet et al., 2017). Unexpected blooms and shifts in biogeographical distribution patterns for species previously considered tropical or sub-tropical, such as Gambierdiscus spp. found in the northeast Atlantic (Fraga et al., 2011; Hoppenrath et al., 2019) and along extensive Japanese coasts (Nishimura et al., 2013), are evidence of potential spreading phenomena. Furthermore, recent reports of Ostreopsis spp. along Japanese coasts (Parson et al., 2012), in the Mediterranean Sea (Penna et al., 2005; Blanfuné et al., 2015; Accoroni et al., 2016), and in Australia (Verma et al., 2016), tend to link ED biodiversity and abundance with environmental changes occurring worldwide.
Establishing a knowledge base on biogeographical distribution and abundance with time-series data on variation in ED trends poses a great challenge for temperate coastal waters, especially where historical data on species is limited and taxonomically selective. As pointed out by Giussani et al. (2017) intensive monitoring efforts for some genera such as Ostreopsis Johs.Schmidt, and the pan-tropical and subtropical association of Gambierdiscus R.Adachi and Y.Fukuyo, andFukuyoa Gómez, D.X.Qiu, R.M.Lopes and Senjie Lin with ciguatera fish poisoning (CFP) has generated concerted scientific and public health interest. Most other recognized potentially toxigenic BHAB genera, e.g., of Prorocentrum Ehrenberg, Coolia A.Meunier, Amphidinium Claperède and Lachmann, etc. have received less scientific attention because they have caused few known incidences of toxin accumulation above regulatory limits or seafood poisoning. With possible exception of P. lima (Ehrenberg) F.Stein, none of these species are confirmed to cause cases of seafood poisoning.
Despite numerous alternative published sampling procedures for investigating EDs (see GEOHAB, 2012; Tester et al., 2014; Jauzein et al., 2016), there is no formal standardized approach that comprehensively addresses the study of all BHAB species. Molecular approaches based primarily on rRNA sequencing and quantitative polymerase chain reaction (qPCR; Penna et al., 2005; Nagahama et al., 2011) have contributed to resolution of taxonomic issues and cryptic species, as well as determining phylogenetic relationships, for many EDs (Durán-Riveroll et al., 2019). Nevertheless, field studies and monitoring programs for BHABs are still based primarily on microscopic analysis of morphotaxonomic characteristics (e.g., as illustrated in Hoppenrath et al., 2014), albeit with increasing supporting evidence from molecular analysis.
Evaluating the potential toxicity risks and magnitude of global distributional changes among EDs requires extensive coverage of these knowledge gaps and the extension of studies to geographical areas not previously investigated, or poorly surveyed, e.g., temperate and high-latitude coastal seas. Such regions may already be affected by BHAB events to some extent, but confirmation of the abundance and distribution of EDs is essential to enhance formal monitoring programs and facilitate the evaluation of potential risks for the aquaculture industry. In most north European countriesthere are no ongoing monitoring programs for assessing ED abundance. Harmful microalgal sampling strategies only focus on the planktonic community; sediments and macroalgal samples for EDs are rarely collected.
These gaps in the biogeographical distribution of EDs encouraged the present survey, which according to the authors’ knowledge, is the first such study of EDs in coastal waters of Sweden. This research had two principal aims: (i) to provide insights into the spatial and temporal distribution of EDs for preliminary toxin risk assessment at selected sites on the west coast of Sweden, focusing on potentially toxic species that may affect shellfish aquaculture, and (ii) to determine the effectiveness of two ED monitoring strategies, comparing natural macrophyte sampling (Hansen et al., 2001; Okolodkov et al., 2007; Reguera et al., 2016) with the artificial substrate method (Tester et al., 2014).
2. Material and methods
2.1. Field collection
The collection of samples was conducted in two sampling campaigns for two consecutive summers (2018–2019). The dinoflagellate samples and associated spatial distribution and environmental data were collected in late summer (August-September 2018) from three locations at the Swedish west coast: Gothenburg (GOT), Tjärnö (TJÄ) and Kristineberg (KRIS) (Fig. 1). Inside each coastal embayment, three randomly chosen stations were sampled in a single campaign over two days (Table 1). During the following summer (June – September 2020), sampling and field data collection was restricted to Hovåsbadet Bay near Gothenburg, where samples were collected twice a month but not at defined stations.
Fig. 1.
Sampling locations along the Swedish west coast. In the left map the rectangle indicates the area shown in the detailed map to the right. The 2018 campaign included three major sites, Tjärnö (TJÄ), Kristineberg (KRIS) and Gothenburg (GOT), whereas the 2019 sampling was exclusively from Hovåsbadet Bay (HOV).
Table 1.
Description of the sampling campaigns performed in 2018 and 2019.
| Location | Date | Station Coordinates | Macrophytes selected | Location Typology | |
|---|---|---|---|---|---|
| Latitude N | Longitude E | ||||
| Gothenburg | 20–21 Aug 2018 |
57º33.775′ | 11º54.963′ | Ceramium spp., Polysiphonia spp. | Natural rocky shore, shallow and sheltered |
| 57º33.339′ | 11º53.038′ | Polysiphonia spp. | |||
| 57º33.539′ | 11º54.475′ | Polysiphonia spp. | |||
| Tjärnö | 28–29 Aug 2018 |
58º52.133′ | 11º 09.222′ | Polysiphonia spp. | Natural rocky/sandy shore, shallow and sheltered |
| 58º52.217′ | 11º 09.131′ | Ceramium spp., Polysiphonia spp. | |||
| 58º52.016′ | 11º 08.321′ | Ceramium spp., Polysiphonia spp. | |||
| Kristineberg | 8–9 Sep 2018 |
58º14.902′ | 11º27.074′ | Ceramium spp. | Modified rocky/muddy shore, with some seagrass beds and sheltered |
| 58º14.867′ | 11º26.760′ | Ceramium spp., Sphacelaria spp. | |||
| 58º14.868′ | 11º27.301′ | Ceramium spp., Polysiphonia spp. | |||
| Hovåsbadet | 24 June – 25 Sep 2019 |
58º37.081′ | 11º55.340′ | Polysiphonia spp. | Rocky shore and sheltered |
Both summer campaigns followed the general protocols and guidelines for the study of benthic microalgae (Moreira and Tester, 2016), with modifications as specified below. The ambient seawater parameters salinity and temperature (ºC) were measured with a multiparameter probe (YSI Inc., 30 M/25 FT, Yellow Springs, OH 45,387 USA). GPS coordinates were also recorded for each site.
2.1.1. Macrophyte sampling and dinoflagellate cell preparation
Macrophyte sampling in both 2018 and 2019 campaigns followed previously established methods for epibenthic dinoflagellates (Hansen et al., 2001; Okolodkov et al., 2007). Macroalgal samples (n = 5 per station) of the rhodophytes Ceramium spp. and Polysiphonia spp. were collected at < 1 m depth and placed with the ambient surrounding seawater in 250 mL plastic jars. On the same day, each plastic jar was vigorously shaken (1 min) to suspend epiphytic dinoflagellates in the seawater, and macroalgae were weighed after separation. The seawater volume was measured, and the suspension gravity-filtered through two nested nylon sieves: 200 µm (top) and 20 µm (bottom). The cells retained were collected by backwashing with filtered seawater into 50 mL Falcon conical plastic centrifuge tubes. The final volume (50 mL) was divided into two subsamples; one was preserved with neutral Lugol's iodine solution for morphological analysis and cell enumeration by microscopy, whereas the second subsample was prepared for DNA metabarcoding.
2.1.2. Artificial substrate sampling and dinoflagellate cell preparation
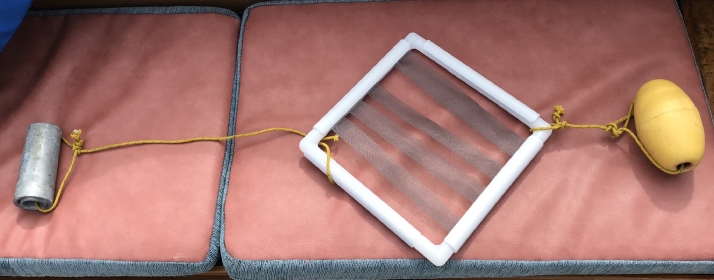
The artificial substrate (AS) sampling method (Tester et al., 2014) was only deployed during the 2018 sampling campaign. Each device (Fig. 2) consisted of four rectangular pieces of plastic screen (21.0 ± 1.5 cm × 3 cm) attached to a rigid frame (27.5 cm x 27.5 cm). The mesh size of the screen was 1.5 mm x 1.5 mm. The frame was connected by a rope to a weight and a subsurface float to keep the device vertical in the water column. Three devices were placed separately at each site for 24 h, suspended at a depth of <1 m below surface. Afterwards, each screen piece was collected underwater and transferred to a 250 mL plastic jar. The preparation procedure was identical to that for macrophyte samples (2.1.1), with subsampling for DNA metabarcoding and quantitative cell analysis by microscopy.
Fig. 2.
Photograph of the artificial substrate (AS) sampling device used in part of this study, shown connected with a weight and float. .
2.2. Quantitative analyses of spatial and temporal distribution of epibenthic dinoflagellates
Lugol's iodine-fixed subsamples (20–25 mL) were prepared for ED cell counting and preliminary identification in Utermöhl sedimentation chambers (Utermöhl, 1958) and examined with an inverted microscope (Carl Zeiss AB, Axiovert 200, Göttingen Germany) fitted with an epifluorescence module. Thecae of dinoflagellate cells were stained according to the calcofluor method (Fritz and Triemer, 1985) with fluorescent brightener 28 (Sigma-Aldrich F3543, St Louis, Mo, USA) for more critical taxonomic analysis by epifluorescence microscopy (excitation: 300–395, centered at 365 nm, Beam Splitter: 395 nm, emission: Long Pass 420 nm) at magnifications 100, 200 and 400x. Cell enumeration followed the guidelines by Olenina et al. (2006) and in the reference manual for HELCOM (Anonymous, 2017), with application of counting software (Plankton Toolbox, v1.3.1; Karlson et al., 2015, available at http://nordicmicroalgae.org/tools). The cell abundance of each species was defined as the number of cells per unit of fresh weight of macroalgae (cells g–1 of FW MA) or per unit of artificial substrate area (cells cm–2 of AS).
The final cell abundance of each ED species per sample was calculated by the following equations (Moreira and Tester, 2016):
2.3. Scanning electron microscopy of epibenthic dinoflagellates
Species identification was confirmed by scanning electron microscopy (SEM). Each subsample containing EDs from artificial and natural substrates was filtered under low vacuum pressure (< 5 cm Hg) through a 5 µm pore-size filter (MF-Millipore™, SMWP04700). Filtered cells were rinsed twice with distilled water and subsequently dehydrated through a graded ethanol concentration series (30, 50, 70, 85, 90, 100%; 10 min each), followed by drying with hexamethyldisilazane at room temperature. Dry samples were mounted on an SEM stub and sputter-coated with gold-palladium (SCD 050 Bal-Tec). Cells were observed under a Tescan VEGA3 electron microscope (Elekronen-Optik-Service GmbH, Dortmund, Germany) at 15 kV.
Terminology of Prorocentrum cell orientation, designation of thecal plates and platelets, and ornamentation follows Hoppenrath et al. (2013), but includes some additions and modifications as suggested by Tillmann et al. (2019).
2.4. rRNA gene metabarcoding of epibenthic dinoflagellates
Subsamples containing ED cells harvested from natural macrophytes and artificial substrates were filtered upon 3 µm pore-size polycarbonate membrane filters (MF-Millipore™, SSWP04700). The filtered aggregate was detached by vortex-mixing in 700 µL prewarmed (60 ºC) lysis Buffer SL1 from the NucleoSpin Soil kit (Machery-Nagel, Düren, Germany). Each sample was stored in a cryovial at -80 ºC until DNA extraction.
2.4.1. DNA extraction and sequencing
DNA extraction was performed with the NucleoSpin Soil kit (Macherey-Nagel, Germany) in SL1 buffer according to the manufacturer's protocol. The 18S rRNA gene V4 region was targeted for amplification using forward and reverse primers (Bradley et al., 2016) with overhang adapters attached. The Illumina overhang nucleotide sequences were added to the 18S amplicon PCR forward and reverse primer. The workflow for preparation of 18S ribosomal RNA gene amplicons for the Illumina MiSeq system was derived from the document 16S metagenomic sequencing library preparation distributed by Illumina (https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf) with modifications for preparation of 18S ribosomal gene amplicons. Paired end Illumina sequencing (MSC 2.5.0.5/RTA 1.18.54, 2 × 300 bp) of 152 benthic plankton samples was performed on a MiSeq platform (Illumina, United States). Raw reads have been deposited in the European Nucleotide Archive via the GFBio portal.
2.4.2. Bioinformatic and statistical analysis
2.4.2.1. Processing of amplicon sequence data
The sequence data were processed with the DADA2 algorithm for detecting the inference of the amplicon sequence variants (ASVs) in a sample from the library of noisy reads generated by amplicon sequencing (Rosen et al., 2012; Callahan et al., 2016) as described in Elferink et al. (2020).
2.4.2.2. Phylogenetic analyses
A reference alignment was created for the phylogenetic analysis of the ASV reads. A set of 18S rDNA sequences was downloaded from DinoRef (https://doi.org/10.6084/m9.figshare.5568454) and NCBI (https://www.ncbi.nlm.nih.gov/genbank/), representing most of the known published diversity sequences of the dinoflagellates Prorocentrum and Coolia. The sequences were aligned with MAFFT and the L-INSI settings and calculated with RAxML (v8.2.12) with the GTRGAMMA model and 1000 Bootstrap analyses. This tree served as a reference for the phylogenetic assignment of the ASV sequences to Prorocentrum (20,292 reads) or Coolia (27,143 reads). The ASV sequences were inserted as Fasta files into the alignment with MAFFT and the "—add fragments –reorder" option in the second step. Subsequently, a Maximum Likelihood phylogenetic tree was generated in the RAxML program (v8.2.12) with 1000 bootstrap replications.
2.5. Statistical analyses of spatio-temporal distribution and ASV data
A nested one-way analysis of variance (ANOVA) followed by a multiple comparisons Student Newman-Keuls (SNK) test were employed to detect differences in the spatial distribution of epibenthic dinoflagellate species. A value of 1 was added to every measurement prior to log transformation - a necessary step since some values = 0. Statistical tests were performed with the R software and the packages “outliers”, “nlme” and “agricolae”. Cell abundances of epibenthic dinoflagellate species collected with the alternative sampling macrophyte and artificial substrate were compared by linear regression analysis (Microsoft Excel, v16.16.1) of the log-transformed abundances from the data recorded from the same stations in summer 2018, following methods from Tester et al. (2014). Again, a value of 1 was added to all cell abundances. The temporal distribution of epibenthic species was analysed from samples collected at Hovåsbadet Bay in 2019. Since the sampling design was not nested, the statistical analyses consisted of a one-way ANOVA followed by a multiple comparisons Student Newman-Keuls test (Microsoft Excel, v16.16.1).
3. Results
3.1. Species identification
Examination by light-, epifluorescence- and scanning electron microscopy revealed the presence of several known ED species upon both macrophyte and artificial substrates. The most abundant and frequently found EDs were referable to Prorocentrum lima (Ehrenberg) F.Stein and Coolia monotis Meunier, but other potentially toxigenic Prorocentrum taxa, including P. compressum (Bailey) T.H.Abé ex J.D.Dodge (not truly epibenthic), P. fukuyoi complex, P. cf. foraminosum and P. cf. hoffmannianum, were also identified.
Prorocentrum lima cells (n = 20) were ovoid, 32–50 µm long and 20–28 µm deep, with smooth thecal plates with scattered large pores (center devoid of pores) and a row of marginal pores (Fig 3A-C). On the right lateral thecal plate, a wide V-shaped periflagellar area was observed (Fig 3A, B). Platelets were not investigated in detail, but platelet lists were visible in right thecal view (Fig 3A, B). A collar was not obvious. A golden-brown chloroplast with a central pyrenoid was visible in the light microscope.
Fig. 3.
SEM micrographs of Prorocentrum lima (A-C) and Prorocentrum compressum (D-H) cells from field samples. A, B) P. lima cells in right lateral view; note the ovoid cell shape, the pore pattern and the wide V-shaped periflagellar area; C) P. lima cells in left lateral view; note the ovoid cell shape and the pore pattern; D, E) P. compressum cells in left lateral view; note the ornamentation and pores, and the apical spine-like wings; F) P. compressum in posterior dorsal or ventral view showing the intercalary band and sagittal suture; note the smooth posterior thecal surface with pore cluster; G) Periflagellar area in right lateral view showing some platelets, especially the spine-like wings on platelets 1 and 4. H) Antapical to lateral view showing the smooth posterior thecal surface with antapical pore cluster. Scale bars = 10 µm, except for G = 2 µm.
Coolia monotis cells (n = 20) were globular to lens-shaped, slightly anterior-posteriorly compressed, with an oblique axis, 30–50 µm of dorsoventral depth, and 24–40 µm wide. The thecal surface was smooth, covered with scattered round to oval pores (Fig. 4) of a wide size range (0.15–0.43 µm in diameter, n = 20). The epitheca was slightly smaller than the hypotheca. The observed tabulation in Kofoidian notation was: APC 3′ 7′′ ?c ?s 5′′′ 2′′′′ (Fig. 4); with an apical pore plate (Po) about 6 to 8 µm in length (Fig. 4D); with a narrow, oblong first apical plate (1′, Figs 4A-C, H) left of the center; a small seventh precingular plate (7′′, Figs 4A-C, G, H); and the second postcingular plate (2′′′) not in contact with the second antapical plate (2′′′′), such that plates 3′′′ and 1′′′′ were touching (Figs 4E, F, large arrow).
Fig. 4.
SEM micrographs of Coolia monotis cells from field samples. A-C) Apical views of the epitheca; note the narrow elongated first apical (1′) plate and the small seventh precingular (7″) plate; D) Detail of the left dorsal part of the epitheca showing the apical pore complex (APC) with the apical pore plate (Po) and the slit-like apical pore. E, F) Antapical view of the hypotheca; note the suture between first antapical and third postcingular plate (large arrow); G,H) Right lateral view showing the large sixth (6″) and the small seventh (7″) precingular plate. Scale bars = 10 µm, except D) = 5 µm.
A third species P. compressum was also abundant in macrophyte and artificial substrate samples. The cells were broadly round-ovate with two spine-like extensions (Figs 3D-H) that were also visible in the light microscope, 35–50 µm in length and 25–30 µm in depth (n = 20). The two lateral thecal plates had a reticulate-foveate ornamentation with pores in and between the depressions (Figs 3D, E). The marginal area of the posterior plate part was nearly smooth with scattered pores and a characteristic cluster of densely arranged pores at the antapical end (Figs 3D, F, H). Some of the (likely nine) platelets in the nearly straight periflagellar area (very narrow excavation) were observed (Fig. 3G), platelets 1, 3, 4, 5, 6a, 6b. Platelets 1 and 4 possessed spine-like wings (Fig. 3G).
SEM analyses confirmed the presence of other benthic Prorocentrum species: P. fukuyoi Murray and Nagahama complex, P. cf. foraminosum M.A.Faust and P. cf. hoffmannianum M.A.Faust emend S.Fraga (supplementary material, Fig. S1). These species were not included in the quantitative analysis because they occurred only sporadically and in low cell abundance.
3.2. Spatial distribution of ED species along the Swedish west coast
The comparison among ED species between settlement upon macroalgal versus artificial substrates collected in summer 2018 at Gothenburg, Kristineberg, and Tjärnö stations yielded no significant differences. Linear regression analyses showed an absence of significant correlation (p > 0.05) between the abundance of P. lima, C. monotis and P. compressum cells for the alternative sampling methods. This result was confirmed again by the pooled data, which revealed no discernible patterns between the average cell abundances upon macroalgal and artificial substrates among locations. In general, the artificial substrates retained only very low cell densities; in some cases ED species were absent.
3.2.1. Quantitative species distribution upon macrophytes
Three major potentially toxigenic species were found at every location studied and during both sampling campaigns (2018 and 2019; Fig. 5; Table 2). The cell abundances of P. lima and C. monotis in macrophyte samples did not vary significantly (ANOVA, SNK-test, p>0.05; n = 15 per station) among the three locations sampled at the west coast. In contrast, cell abundance of P. compressum at Tjärnö differed significantly from the other locations (p<0.05), by the near absence of this species at Tjärnö (Fig. 5). Within each site, there were significant variations (p<0.05) in cell abundances of the three species. The highest cell abundances for P. lima and C. monotis were recorded from Tjärnö, 1766 and 805 cells g–1 fresh weight (FW) of macroalgae, respectively. For P. compressum, the highest abundance was observed at Gothenburg with 168 cells g–1 FW of macroalgae (Table 2). No significant correlation was found between the ED cell number variability and the macroalgal genus as substrate.
Fig. 5.
Spatial distribution of epibenthic dinoflagellates on macroalgae (2018). Log- transformed abundances of cells from macroalgae for P. lima, P. compressum and C. monotis collected at Gothenburg (GOT), Kristineberg (KRIS) and Tjärnö (TJÄ-) stations. Each black circle represents the value of an individual sample. The ends of the boxwhiskers represent the maximum and minimum of the log cell abundances from that location.
Table 2.
Cell abundances of P. lima, C. monotis and P. compressum collected from MA (cells g–1 FW MA) and ASs cells cm–2 AS) from the three sampling stations at Gothenburg, Tjärnö and Kristineberg. The seawater temperature and salinity were recorded during sampling at 1 m depth. The symbol “*” indicates for P. lima which stations present abundance values above official alarm threshold (> 1000 cells g–1 FW MA).
| P. lima | C. monotis | P. compressum | Temp (ºC) | Salinity (PSU) | ||||
|---|---|---|---|---|---|---|---|---|
| Cells g–1 | Cells cm–2 | Cells g–1 | Cells cm–2 | Cells g–1 | Cells cm–2 | |||
| Gothenburg | 182 – 1112* | 0 – 33 | 22 – 155 | 0 – 2 | 0 – 10 | 0 – 1 | 18.6 | 23.8 |
| 50 – 366 | 0 – 16 | 8 – 47 | 0 – 1 | 2 – 168 | 0 – 1 | 19.2 | 23.7 | |
| 121 – 389 | 0 – 1 | 47 – 209 | 0 – 1 | 4 – 19 | 0 – 1 | 19.3 | 23.9 | |
| Tjärnö | 83 – 1521* | 12 – 17 | 64 – 805 | 8 – 22 | 0 – 8 | 0 | 16.2 | 29.0 |
| 189 – 319 | 2 – 8 | 146 – 326 | 2 – 10 | 0 | 0 | 16.2 | 28.9 | |
| 410 – 1766* | 2 – 17 | 170 – 421 | 1 – 17 | 0 | 0 | 16.8 | 29.2 | |
| Kristineberg | 82 – 181 | 0 – 1 | 15 – 36 | 0 – 1 | 5 – 10 | 12 – 14 | 17.7 | 26.6 |
| 132 – 539 | 0 – 1 | 73 – 371 | 0 – 1 | 4 – 42 | 10 – 22 | 17.6 | 26.5 | |
| 245 – 724 | 0 – 1 | 114 – 309 | 0 – 1 | 14 – 18 | 22 – 34 | 18.4 | 26.7 | |
3.2.2. Quantitative species distribution upon artificial substrates
Due to the high heterogeneity of variances for C. monotis and P. lima cell abundances among AS samples, a p-value threshold of 0.001 was selected to fit the parametric requirements of the statistical test. A less stringent p-value threshold of 0.05 was applied for analysis of P. compressum cell abundance data collected with the artificial substrate method.
The cell abundances of P. compressum (p<0.05) and C. monotis (p<0.001) showed significant variation between locations (summarized in Fig. 6). The absence of P. compressum cells at Tjärnö reveals this high heterogeneity between locations, as was pointed out for the macroalgal samples (Table 2). For P. lima, no significant variation in its cell abundance was found between locations (p>0.001; p = 0.045, respectively). Nevertheless, within each location, the cell abundance varied significantly for P. compressum (p<0.05) and P. lima (p<0.001). The highest cell abundances recorded by the surface area of artificial substrate were for P. lima at Gothenburg, C. monotis at Tjärnö and for P. compressum at Kristineberg (Table 2).
Fig. 6.
Spatial distribution of epibenthic dinoflagellates retained on artificial substrate (2018). Log-transformed abundances of cells found on artificial substrate (AS) for P. lima, P. compressum and C. monotis collected at Gothenburg (GOT), Kristineberg (KRIS) and Tjärnö (TJÄ-) stations. Each black circle represents the value of an individual sample. The ends of the boxwhiskers represent the maximum and minimum of the log cell abundances from each location.
3.3. Temporal distribution of EDs in Hovåsbadet Bay
Cell abundances of P. lima and C. monotis in macroalgal samples collected at Hovåsbadet Bay did not vary significantly during the period June – September 2019 (Fig. 7.). Even though these species were consistently present during the whole season, their respective cell abundances were three orders of magnitude lower than from macroalgal samples collected in 2018. The highest cell abundances for P. lima and C. monotis were recorded in July, with 22 and 16 cells g–1 FW MA, respectively. Prorocentrum compressum was rarely observed at Hovåsbadet, with just few cells present in samples from June 2019.
Fig. 7.
Temporal distribution of log-transformed cell abundances found in 2019 for P. lima and C. monotis from macrophyte samples collected at Hovå-sbadet Bay, near Gothenburg. Each black circle represents the value of an individual sample. The ends of the boxwhiskers represent the maximum and minimum of the log cell abundances from that location.
During this sampling period, the average surface salinity was 21.3 with a minimum of 18.3 in August and a maximum of 25.8 in July. The average seawater temperature was 21.3 ºC, with a minimum of 14.1 ºC in September and a maximum of 21.6 ºC in July.
3.4. Metabarcoding of V4 region 18S rRNA gene
Phylogenetic analyses supported nine species assignments, but with different degrees of confidence at various taxonomic levels (Fig. 8). From the total 48 ASVs of Coolia and Prorocentrum, 16 were assigned to Coolia and 32 to Prorocentrum, respectively. All 13 Coolia ASVs clustered well- supported with C. monotis. But 13 Prorocentrum ASVs could only be assigned down to genus level, whereas 19 taxa could be assigned to species. ASVs clustered well with reference sequences of Prorocentrum lima and P. hoffmannianum M.A.Faust P. hoffmannianum, represented by eight ASVs. One ASV was assigned to P. compressum with 10 reads matching in the metabarcoding samples. The phylogenetic analyses according to DADA2 with PR2 also indicated the presence of P. levis, P. foraminosum, P. nanum, P. cordatum and P. fukuyoi with good to moderate bootstrap table (BT) support.
Fig. 8.
Phylogenetic tree displaying the phylogenetic analysis of the ASV reads for a taxonomic assignation to Prorocentrum (20,292 reads) and Coolia (27,143 reads). A set of 18S rDNA sequences was downloaded from DinoRef and NCBI representing most of the known and published diversity of the dinoflagellates Prorocentrum and Coolia. The sequences were aligned with MAFFT and the l-INSI settings and calculated with RAxML and 1000 Boostraps analyses.
The presence of P. leve M.A.Faust, Kibler, Vandersea, Tester and Litaker, P. foraminosum M.A.Faust, P. nanum J.Schiller, P. cordatum (Ostenfeld) J.D.Dodge and P. fukuyoi Shauna Murray and Y.Nagahama was confirmed with good (>85%) to moderate BT support. Thirteen ASVs fitting within Prorocentrum could not be reliably assigned to any clade with reference sequences.
The taxonomic classification and absolute number of reads per ASV in each sample are presented in Table S1. Most of the ASVs could be assigned to one major microeukaryotic taxonomic rank (superphylum to subphylum). The remaining ASVs were assigned as unclassified eukaryotes. The supergroup Alveolata dominated the benthic communities in richness and abundance at all sampling stations. Dinoflagellates were the most abundant phylum within the alveolates, accounting for 71% in samples collected from macroalgae in 2018 and 90% in 2019. In comparison, in samples collected from artificial substrate in 2018 dinoflagellates represented 53% of the total ASVs and 84% in planktonic water samples collected in 2019.
The percentage of assignable ASVs of the whole dinoflagellate community collected from macroalgae samples (MA) in 2018 was 6% for genus Coolia and 5% for Prorocentrum. In 2019, the ASV percentages for genera Coolia and Prorocentrum were 6% and 3%, respectively. The ASV percentage of the genera Coolia and Prorocentrum of the whole dinoflagellate community collected from artificial substrate (AS) samples in 2018 was 5% and 4%, respectively, whereas in planktonic samples from 2019 no representatives of Coolia and only 2% of Prorocentrum were detected. Coolia sp. was found at every sampling station, with the overall most abundant targeted species detected in 2018 at Gothenburg and Tjärno, both from samples extracted from MA and ASsubstrate. The most abundant Prorocentrum species was P. lima. At Kristineberg station the target species richness was more diverse, showing Coolia and P. lima at nearly equal relative ASV abundance from macroalgae, but both species only in low relative abundances from artificial substrate. P. foraminosum was slightly lower in relative abundance, but detected at high relative abundance from AS.
In 2019 Coolia was the targeted genus with the highest detected numbers of reads in macroalgae samples, followed by P. foraminosum and P. lima (Fig. 9). The planktonic samples showed P. cordatum with high number of ASV reads and to a lower extent other Prorocentrum, but which could not be assigned to species level (Fig. 9). Sixty-nine sequencing reads of the class Dinophyceae could be taxonomically assigned to genus level. Moreover, 104 reads could be assigned to species level, comprising 19 epiphytic species. The Coolia ASVs could only be assigned to the genus level. The light microscopic analysis documented only Coolia monotis in the samples. Some Prorocentrum species could not be assigned to species level and therefore the light microscopic analysis of Prorocentrum compressum could not be confirmed by sequencing analysis.
Fig. 9.
Target species richness of amplicon sequence variants at the different sampling stations in 2018 and 2019 extracted from macroalgae (MA) artificial substrates (AS) and planktonic samples (PL), respectively.
4. Discussion
Benthic microalgae are considered as likely group beneficiaries of climate change, whereby a warmer ocean and shifts in current patterns could promote invasive colonization and range expansion into colder seas. For in situ resident populations, a temperature-dependent increase in the growth potential and overall cell abundance is also expected for this dinoflagellate assemblage (Tester et al., 2020), as well as enhanced substrate availability due to increasing macroalgal and seagrass biomass and benthic biofouling. For Ostreopsis ovata, for example, an expansion in distribution due to climate change effects is expected (Drouet et al., 2021). Nevertheless, the lack of ecophysiological and toxicological field data for EDs from diverse global habitats complicates evaluation of the effects of climate change on their toxicity and bloom dynamics (Durán-Riveroll et al., 2019), and hence assessment of the risk to human and ecosystem health via climate-mediated global dispersion remains an open challenge.
This first investigation of the spatial and temporal distribution of EDs along the west coast of Sweden yielded the presence of several epibenthic species, with morphological characteristics conforming perfectly with recent published morphospecies concepts for the respective taxa (Nagahama and Fukuyo, 2005; Nagahama et al., 2011; Laza-Martinez et al., 2011; Hoppenrath et al., 2014; Karafas et al., 2015). Among these species, P. lima poses the greatest potential risk as a source of polyether shellfish toxins associated with DSP in Sweden because of its frequent occurrence in shellfish-growing areas, at relatively high cell abundance, and known consistent toxigenicity. A global survey of published reports on the toxin composition of EDs (Duran-Riveroll et al., 2019) indicates that production of DSP toxins (DSTs) is a ubiquitous characteristic of this species. Although presence of DSTs in P. lima from Swedish coastal waters has not been confirmed, these populations are likely to be toxigenic. Prorocentrum compressum is frequently found in samples from the Swedish west coast but is not considered to be a toxin producer (Rhodes and Syhre, 1995; Duran-Riveroll et al., 2019). The rarer species P. cf. foraminosum and P. cf. hoffmannianum are, however, potentially toxigenic, as producers of dinophysistoxin-1 (DTX1), other DTXs (Cembella et al., 2021), unknown “fast-acting toxin” (perhaps prorocentrolide) and okadaic acid (OA) (Aikman et al., 1993; Kameneva et al., 2015).
In any case, reports of “toxic Prorocentrum species”, especially early citations of toxicity, type of toxins and species assignments must be interpreted cautiously. The genus has undergone frequent recent taxonomic revisions, leading to species reassignment of strains previously analysed for toxin content (Chomerat et al. 2019; Cembella et al., 2021). Furthermore, early reports of cell toxicity were based on non-specific bioassays (“DSP-like symptoms”) or fluorescence-derivatization methods for DSP toxins but capable of detecting only OA and DTX1.
Only a single species of Coolia, C. monotis was found on macroalgae or artificial substrate sampled over two years at sites on the Kattegat-Skagerrak coast of Sweden. Some Coolia species are reported as toxigenic, but the genus is undergoing frequent taxonomic revisions, i.e., with the addition of new possibly toxigenic species C. palmyrensis Karafas, Tomas & York and C. santacroce Karafas, Tomas & York (Karafas et al., 2015; Karafas and Tomas, 2015). The IOC-UNESCO Taxonomic Reference List of Harmful MicroAlgae (Lundholm et al., 2009 onwards), recognizes only C. tropicalis M.A.Faust as a confirmed toxin-producing Coolia species. The same caveats as applied to “toxic Prorocentrum” must be considered for Coolia. Certain Amphidinium species, some of which can also produce biologically active polyketides (“toxins”), may have been previously misidentified as C. monotis and C. malayensis Leaw, P.-T.Lim and Usup (Duran-Riveroll et al., 2019). Coolia monotis was identified as the source of cooliatoxin (likely a monosulfated analog of yessotoxin) (Holmes et al., 1995, Rhodes and Thomas, 1997), but Mohammad-Noor et al. (2013) later demonstrated that the original strain producing cooliatoxin belonged to C. tropicalis, not C. monotis. In summary, the potential risk to human health and the shellfish production industry posed by the presence of Coolia at aquaculture sites in Sweden is likely to be rather marginal – C. monotis is not known to be toxigenic. Many studies have failed to detect toxicity in any Coolia species (Holmes et al., 2014) and no human poisoning cases clearly linked to Coolia have been reported.
The molecular diagnostic approach was proven effective to identify and confirm Prorocentrum and Coolia as the two major genera of BHAB taxa posing a potential toxicity risk to shellfish harvest and human health on the Swedish west coast. Unfortunately, variation in the 18S rRNA gene does not offer very high resolution for establishing taxonomic and phylogenetic relationships among Prorocentrum species, such that some taxa could not be assigned to species in the reference tree. Exact assignment was, unfortunately, not possible for P. hoffmannianum, although association with the reference sequences was indicated. Furthermore, for genus Prorocentrum reliable reference sequences were not found for all species. For Coolia, there is a similar lack of diversity in the 18S rRNA gene in the reference data. But beyond these caveats, most ED ASVs could be assigned to species and were supported at least by the DADA2 annotation.
Both P. lima and C. monotis were abundant on macroalgae and present along the Swedish west coast as surveyed. The cell densities of both species had a high spatial variability (Fig. 5 and Table 2), which fluctuated greatly over small distances within each location (<10 m). This condition is noted in previous investigations, where the inherently patchy distribution of EDs is described (Cohu et al., 2011, 2013; Mangialajo et al., 2011; Tester et al., 2014). Apparently arbitrary spatial distribution of EDs is also dependent upon the composition and biogeographical density of the macroalgal community. In this study, Kristineberg presented the lowest cell densities recorded for P. lima and C. monotis. This location was characterized by a poor coverage and low diversity of macroalgal substrate, especially, filamentous forms, which may explain the low ED abundances (Maranda et al., 2007; Armi et al., 2010; Totti et al., 2010; Fricke et al., 2016).
Despite being considered a planktonic species, P. compressum was included in the quantitative analyses because of the significantly higher number of cells of this dinoflagellate compared to other planktonic species found in MA samples. The possibility that P. compressum cells were attached to macroalgae suggests the existence of an epiphytic phase in its life cycle, based on inter- and intra-species interactions (e.g. competition, mating, and protection against predators). In that case, P. compressum cells would behave similarly to Ostreopsis spp. which, independent from hydrodynamic conditions, alters between a planktonic stage and an epiphytic phase attached to different substrates (Mangialajo et al., 2011; Accoroni et al., 2016; Giussani et al., 2017). This study does not provide any direct observations of P. compressum attachment to macroalgae substrate. However, as for certain other Prorocentrum species, the poor natatorial performance of P. compressum and the production of mucus may correlate with this transient epiphytic nature.
As reported by Hoppenrath et al. (2013), the available ecological data concerning epiphytic Prorocentrum species is limited. Only a few records have described P. compressum from the plankton, mainly for taxonomic purposes (Steidinger et al., 1997). It should be noted that P. compressum has been renamed for nomenclatural reasons and therefore the name Prorocentrum bidens J.Schiller should be adopted (Cowan and Huisman, 2015). As this has not yet widely been recognized, the more familiar name has been retained herein.
Gaarder (1954) recorded P. compressum (as Exuviaella compressa) and mentioned that specimens were most often found in twos or fours of cells embedded in a jelly-mass. Taylor (1976) recorded the species from the Indian Ocean, Steidinger and Williams (1970) from the Gulf of Mexico, and Munir et al. (2013) and Gul and Saifullah (2011) from the Arabian Sea. All of these reports are from warm water habitats and may not be representative of behavior and life style in temperate waters, such as the west coast of Sweden. These findings highlight the requirement for further investigation of the ecology of P. compressum.
This research monitored ED spatial distribution using two novel sampling methods previously tested for the investigation of epiphytic microalgae. Consistent with the findings of Parsons et al. (2017), comparison of these sampling procedures showed poor relationships and no conclusive results for the west Swedish coast. The AS did not provide consistent estimates for the cell abundances of P. lima, C. monotis or P. compressum on macroalgae. In contrast, the results of Tester et al. (2014) and Jauzein et al. (2018) both support the efficiency and validity of the AS cell estimates. Nevertheless, these previous studies were performed in the warm Mediterranean Sea or in tropical/sub-tropical areas where EDs are extensively studied and high cell density BHABs are frequent. In such cases, the cell density in the water column tends to be higher and consequently the number of cells captured by the mesh of the AS (Mangialajo et al., 2011). This results herein question the deployment of AS for investigating new locations in the study of EDs or in environments where their natural cell abundance is moderate to low; under these circumstances underestimates of the cells attached to macrophytes are likely. This is consistent with similar statements reported previously (Maranda et al., 2007; Giussani et al., 2017). Furthermore, until now no studies have validated the effective functioning of the AS technique for monitoring EDs, other than for Ostreopsis or Gambierdiscus species. Thus, the standardization of sampling methods for other ED species involves some ongoing controversy. The strictly “benthic behavior” of some dinoflagellates may affect the precision of this method (e.g., for P. lima in Giussani et al., 2017 and P. lima and C. monotis in this paper). However, the “natural” MA collection approach has negative aspects as well, such as destruction of the macroalgal community or benthos disruption. The difficulty in standardization and comparison between methods highlights the crucial requirement to develop and implement appropriate sampling strategies that allow for valid comparison of results between ED studies (Berdalet et al., 2016; Giussani et al., 2017).
As mentioned by Levasseur et al. (2003), the temporal abundance of ED attached to macroalgae displays a strong seasonal pattern. In 2019 at Hovåsbadet, this study observed the same ED species as in 2018 along the Swedish west coast. Nevertheless, the cell abundances collected for P. lima and C. monotis were two to three orders of magnitude lower than the 2018 results. The dinoflagellate P. compressum appeared in few samples, but was frequently absent. For 2019 summer, intense wind conditions interrupted the sampling actions between late August and early September. Due to these storm conditions, this study missed sampling during a potential late bloom period for P. lima and C. monotis, such as observed in 2018 data. The degree of vertical mixing caused by this strong hydrodynamic activity may have disrupted the ED summer bloom in this year (Simoni et al., 2004; Totti et al., 2010). Even though this study did not detect significant temporal changes in the cell abundances of EDs throughout the 2019 summer at Hovåsbadet, C. monotis and P. lima cell numbers presented a slight increase during July and August.
Monitoring ED in the marine environment is becoming increasingly critical in locations where shellfish aquaculture is carried out in northern Europe. For Sweden, the culture and consumption of blue mussels has been an important industry in the Skagerrak area since early 1971 (Ackefors and Haamer, 1987). Toxic syndromes, such as DSP, are one of the main economic threats for shellfish farms and consumers (Ciminiello et al., 2003) and concerning toxin levels are frequently reported in shellfish from Swedish waters (Rehnstam-Holm and Hernroth, 2005; Karlson et al., 2007). The risk is especially elevated at mussel farms settled in natural coastal embayments, where ED resuspension likely occurs due to disruptive climate-related conditions (e.g., high wind, waves or tides). For P. lima, a key producer of DSP toxins (Murakami et al., 1982; Lee et al., 1989; Jackson et al., 1993; Morton and Tindall, 1995; Bouaïcha et al., 2001; Hoppenrath et al., 2013), cells are ingested by suspension-feeders such as bivalve mollusks and some undigested cells can still remain active and grow after passing through their guts (Bauder and Cembella, 2000; Bauder et al., 2001).
This study found two potentially toxic ED species, P. lima and C. monotis, along the Swedish west coast from Gothenburg to the Norwegian border. The maximum abundances observed for P. lima and C. monotis were 1766 cells g–1 and 805 cells g–1 FW MA, respectively (at Tjärnö). National food agencies have not defined an official alarm threshold for Coolia spp. since the toxicity of this genus still is uncertain (Ben-Gharbia et al., 2016). On the contrary, for P. lima ICES (2015) noted that certain European countries have adopted thresholds of 100 cells L–1 (in Ireland and the UK) or 50 cell L–1 (in the Spanish Mediterranean). These cell concentrations represent a level of concern for possible DSP events, establishing that P. lima abundances above these limits might cause toxic events. The units used to establish these thresholds are defined by the sampling method applied, corresponding in both cases mentioned to water samples. Previous researchers, who applied the same sampling strategies (MAs), have defined for P. lima an alarm threshold of 1000 cells g–1 FW (Foden et al., 2005; CEFAS, 2012; Tester et al., 2014).
In the present study, P. lima exceeded this limit in 5 of 15 samples collected in Tjärnö with MAs (Table 2). This location, as well as other areas along the west coast of Sweden, has a well-developed shellfish industry. Several studies agree that shellfish aquaculture provides a good environment for macroalgae growth and, as a consequence, a beneficial habitat for epiphytic Prorocentrum spp. (Lawrence et al., 2000; Bravo et al., 2001; Maranda et al., 2007). Thus, P. lima constitutes at least a potential risk for phycotoxin contamination of bivalve shellfish in Sweden with high economic consequences for the aquaculture and harvest industries.
5. Conclusions
This study has shown that epibenthic dinoflagellates are abundant on macrophytes along the Swedish west coast. The presence of two potentially toxigenic epibenthic dinoflagellates, Prorocentrum lima and Coolia monotis was confirmed along with a description of their spatial and temporal distribution. A comparison between two sampling methods, macrophyte sampling and sampling using artificial substrates, indicates that artificial substrates are unsuitable for quantitative sampling of epibenthic dinoflagellates in this area. Scanning electron microscopy and metabarcoding of the V4 region of the 18S rRNA gene revealed a large diversity of benthic dinoflagellates. The presence of the DSP-toxin producer Prorocentrum lima is a potential risk to human health and may affect aquaculture negatively since bivalve molluscs may be contaminated with toxins produced by P. lima.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The late Professor Anna Godhe, University of Gothenburg, took part in the design and early phases of this study and we, therefore, dedicate this article to her memory. This study is a component of the EU project CoCliME as part of ERA4CS, an ERA-NET initiated by JPI Climate, and funded by EPA (IE), ANR (FR), DLR (DE), UEFISCDI (RO), RCN (NO) and FORMAS (SE), with co-funding by the European Union (Grant 690462). CoCliME funding from Swedish Research Council Formas was administered from grant number 2017-1737.
Edited by Christopher J. Gobler
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.hal.2022.102318.
Appendix. Supplementary materials
Data Availability
Data will be made available on request. Sequence data have been deposited in the European Nucleotide Archive via the GFBio portal. The GFBio Submission ID is: daf47116-bfab-47d7-9ff9-38e9d34ccc57.
References
- Accoroni S., Romagnoli T., Penna A., Capellacci S., Ciminiello P., Dell'Aversano C., Tartaglione L., Abboud-Abi Saab M., Giussani V., Asnaghi V., Chiantore M. Ostreopsis fattorussoi sp. nov. (Dinophyceae), a new benthic toxic Ostreopsis species from the eastern Mediterranean Sea. J. Phycol. 2016;52(6):1064–1084. doi: 10.1111/jpy.12464. [DOI] [PubMed] [Google Scholar]
- Ackefors H., Haamer J. International Council for the Exploration of the Sea (ICES); Copenhagen: 1987. A New Swedish technique For Culturing Blue Mussel. [Google Scholar]
- Aikman K.E., Tindall D.R., Morton S.L. In: Toxic Phytoplankton Blooms in the Sea. Smayda T.J., Shimizu Y., editors. Elsevier; Amsterdam: 1993. Physiology and potency of the dinoflagellate Prorocentrum hoffmannianum (Faust) during one complete growth cycle; pp. 463–468. [Google Scholar]
- Anonymous, 2017. Manual for marine monitoring in the COMBINE Programme of HELCOM http://www.helcom.fi/Documents/Action%20areas/Monitoring%20and%20assessment/Manuals%20and%20Guidelines/Manual%20for%20Marine%20Monitoring%20in%20the%20COMBINE%20Programme%20of%20HELCOM.pdf. HELCOM, Helsinki.
- Armi Z., Turki S., Trabelsi E., Maiz N.B. First recorded proliferation of Coolia monotis (Meunier, 1919) in the North Lake of Tunis (Tunisia) correlation with environmental factors. Environ. Monit. Assess. 2010;164:423–433. doi: 10.1007/s10661-009-0903-z. [DOI] [PubMed] [Google Scholar]
- Bauder A.G., Cembella A.D. Viability of the toxic dinoflagellate Prorocentrum lima following ingestion and gut passage in the bay scallop Argopecten irradians. J. Shellfish Res. 2000;19:321–324. [Google Scholar]
- Bauder A.G., Cembella A.D., Bricelj V.M., Quilliam M.A. Uptake and fate of diarrhetic shellish poisoning toxins from the dinoflagellate Prorocentrum lima in the bay scallop Argopecten irradians. Mar. Ecol. Prog. Ser. 2001;213:39–52. [Google Scholar]
- Ben-Gharbia H., Yahia O., Amzil Z., Chomérat N., Abadie E., Masseret E.…Laabir M. Toxicity and growth assessments of three thermophilic benthic dinoflagellates (Ostreopsis cf. ovata, Prorocentrum lima and Coolia monotis) developing in the southern Mediterranean basin. Toxins (Basel) 2016;8(10):297. doi: 10.3390/toxins8100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdalet E., Fleming L.E., Gowen R., Davidson K., Hess P., Backer L.C.…Enevoldsen H. Marine harmful algal blooms, human health and wellbeing: challenges and opportunities in the 21st century. J. Mar. Biolog. Assoc. UK. 2016;96(1):61–91. doi: 10.1017/S0025315415001733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanfuné A., Boudouresque C.F., Grossel H., Thibaut T. Distribution and abundance of Ostreopsis spp. and associated species (Dinophyceae) in the northwestern Mediterranean: the region and the macroalgal substrate matter. Environl. Sci. Pollut. Res. Int. 2015;22(16):12332–12346. doi: 10.1007/s11356-015-4525-4. [DOI] [PubMed] [Google Scholar]
- Bouaïcha N., Chézeau A., Turquet J., Quod J.P., Puiseux-Dao S. Morphological and toxicological variability of Prorocentrum lima clones isolated from four locations in the south- west Indian Ocean. Toxicon. 2001;39:1195–1202. doi: 10.1016/s0041-0101(00)00258-0. [DOI] [PubMed] [Google Scholar]
- Bravo I., Fernández M.L., Ramilo I., Martínez A. Toxin composition of the toxic dinoflagellate Prorocentrum lima isolated from different locations along the Galician coast (NW Spain) Toxicon. 2001;39(10):1537–1545. doi: 10.1016/s0041-0101(01)00126-x. [DOI] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample: illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cembella A.D., Durán-Riveroll L.M., Tarazona-Janampa U.I., Okolodkov Y.B., García-Sandoval R., Krock B., Hörstmann C., John U. Phylogeography and diversity among populations of the toxigenic benthic dinoflagellate Prorocentrum from coastal reef systems in Mexico. Front. Mar. Sci. 2021;8 [Google Scholar]
- Centre for Environment, Fisheries and Aquaculture Science (CEFAS) In Annual report on the results of the Biotoxin and Phytoplankton Official Control Monitoring Programmes for England and Wales 2012; Lowestoft, UK; 2012. [Google Scholar]
- Chomérat N., Bilien G., Derrien A., Henry K., Ung A., Viallon J., Darius H.T., iti Gatti C.M., Roué M., Hervé F. Ostreopsis lenticularis Y. Fukuyo (Dinophyceae, Gonyaulacales) from French Polynesia (South Pacific Ocean): a revisit of its morphology, molecular phylogeny and toxicity. Harmful Algae. 2019;84:95–111. doi: 10.1016/j.hal.2019.02.004. [DOI] [PubMed] [Google Scholar]
- Ciminiello P., Dell'Aversano C., Fattorusso E., Forino M., Magno S. Toxins from Adriatic blue mussels: a decade of studies. J. Macromol. Sci. Pure. Appl. Chem. 2003;75(2–3):325–336. [Google Scholar]
- Cowan R.A., Huisman J.M. Pyxidicula compressa: diatom or dinoflagellate? Taxon. 2015;64(4):809–810. [Google Scholar]
- Cohu S., Thibaut T., Mangialajo L., Labat J.P., Passafiume O., Blanfuné A., Simon N., Cottalorda J.M., Lemée R. Occurrence of the toxic dinoflagellate Ostreopsis cf. ovata in relation with environmental factors in Monaco (NW Mediterranean) Mar Pollut. Bull. 2011;62:2681–2691. doi: 10.1016/j.marpolbul.2011.09.022. [DOI] [PubMed] [Google Scholar]
- Drouet K., Jauzein C., Herviot-Heath D., Hariri S., Laza-Martinez A., Lecadet C., Plus M., Seoane S., Sourisseau M., Lemée R. Current distribution and potential expansion of the harmful benthic dinoflagellate Ostreopsis cf. siamensis towards the warming waters of the Bay of Biscay, North-East Atlantic. Environ. Microbiol. 2021;23:4956–4979. doi: 10.1111/1462-2920.15406. [DOI] [PubMed] [Google Scholar]
- Durán-Riveroll L.M., Cembella A.D., Okolodkov Y.B. A Review on the biodiversity and biogeography of toxigenic benthic marine dinoflagellates of the coasts of Latin America. Front. Mar. Sci. 2019;6:148. doi: 10.3389/fmars.2019.00148. [DOI] [Google Scholar]
- Elferink S., Wohlrab S., Neuhaus S., Cembella A., Harms L., John U. Comparative metabarcoding and metatranscriptomic analysis of microeukaryotes within coastal surface waters of West Greenland and Northwest Iceland. Front. Mar. Sci. 2020;7:1–20. [Google Scholar]
- Foden J., Purdie D.A., Morris S., Nascimento S. Epiphytic abundance and toxicity of Prorocentrum lima populations in the Fleet Lagoon, UK. Harmful Algae. 2005;4(6):1063–1074. [Google Scholar]
- Fraga S., Rodríguez F., Caillaud A., Diógene J., Raho N., Zapata M. Gambierdiscus excentricus sp. nov. (Dinophyceae): a benthic toxic dinoflagellate from the Canary Islands (NE Atlantic Ocean) Harmful Algae. 2011;11:10–22. [Google Scholar]
- Fricke A., Kopprio G.A., Alemany D., Gastaldi M., Narvarte M., Parodi E.R., Lara R.J., Hidalgo F., Martínez A., Sar E.A., Iribarne O., Martinetto P. Changes in coastal benthic algae succession trajectories and assemblages under contrasting nutrient and grazer loads. Estuar. Coasts. 2016;39:462–477. [Google Scholar]
- Fritz L., Triemer R.E. A rapid simple technique utilizing calcofluor white M2R for the visualization of dinoflagellate thecal plates 1. J. Phycol. 1985;21(4):662–664. [Google Scholar]
- Gaarder K.R. Report on the Scientific Results of the" Michael Sars" North Atlant. Deep-sea Exped. 1910. 1954. Dinoflagellatae from the" Michael Sars" North Atlantic deep-sea expedition 1910; pp. 1–62. [Google Scholar]
- GEOHAB, 2012. Berdalet, E., Tester, P., Zingone, A. (Eds.), GEOHAB Core Research Project: HABs in Benthic Systems. IOC of UNESCO and SCOR, Paris and Newark, 64.
- Giussani V., Asnaghi V., Pedroncini A., Chiantore M. Management of harmful benthic dinoflagellates requires targeted sampling methods and alarm thresholds. Harmful Algae. 2017;68:97–104. doi: 10.1016/j.hal.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Gorbi S., Avio G.C., Benedetti M., Totti C., Accoroni S., Pichierri S.…Regoli F. Effects of harmful dinoflagellate Ostreopsis cf. ovata exposure on immunological, histological and oxidative responses of mussels Mytilus galloprovincialis. Fish Shellfish Immun. 2013;35(3):941–950. doi: 10.1016/j.fsi.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Gul S., Saifullah S.M. The dinoflagellate genus Prorocentrum (Prorocentrales, Prorocentraceae) from the north Arabian sea. Pak. J. Bot. 2011;43(6):3061–3065. [Google Scholar]
- Hansen G., Turquet J., Quod J., Ten-Hage L., Lugomela C., Kyewalyanga M.…Rakotorrinjanahary H. Potentially harmful microalgae of the western Indian Ocean, IOC. Manuals Guides. 2001:41. [Google Scholar]
- Holmes M.J., Lewis R.J., Jones A., Wong-Hoy A. Cooliatoxin, the first toxin from Coolia monotis (Dinophyceae) Nat. Tox. 1995;3:355–362. doi: 10.1002/nt.2620030506. [DOI] [PubMed] [Google Scholar]
- Hoppenrath M., Chomérat N., Horiguchi T., Schweikert M., Nagahama Y., Murray S. Taxonomy and phylogeny of the benthic Prorocentrum Species (Dinophyceae)-A proposal and review. Harmful Algae. 2013;27:1–28. [Google Scholar]
- Hoppenrath M., Kretschmar A.L., Murray S.A., Kaufmann M.J. Morphology and molecular phylogeny of Gambierdiscus excentricus (Dinophyceae) from Madeira Island (NE Atlantic Ocean) Mar. Biodivers. Rec. 2019;12:16. [Google Scholar]
- Hoppenrath M., Murray S.A., Chomérat N., Horiguchi T. Vol. 54. Schweizerbart'sche Verlagsbuchhandlung (Nägele u. Obermiller); Stuttgart, Germany: 2014. Marine benthic dinoflagellates – unveiling their worldwide biodiversity; p. 276. (Kleine Senckenberg-Reihe). [Google Scholar]
- ICES Interim report of the ICES—IOC working group on harmful algal bloom dynamics (WGHABD). 13–18 April 2015; Lisbon, Portugal; 2015. [Google Scholar]
- Jackson A.E., Marr J.C., McLachlan J.L. In: Toxic Phytoplankton Blooms in the Sea. Smayda T.J., Shimizu Y., editors. Elsevier; Amsterdam: 1993. The production of diarrhetic shellfish toxins by an isolate of Prorocentrum lima from Nova Scotia, Canada; pp. 513–518. [Google Scholar]
- Jauzein C., Açaf L., Accoroni S., Asnaghi V., Fricke A., Hachani M.A., Lemée R. Optimization of sampling, cell collection and counting for the monitoring of benthic harmful algal blooms: application to Ostreopsis spp. blooms in the Mediterranean Sea. Ecol. Indic. 2018;91:116–127. [Google Scholar]
- Jauzein C., Fricke A., Mangialajo L., Lemée R. Sampling of Ostreopsis cf. ovata using artificial substrates: optimization of methods for the monitoring of benthic harmful algal blooms. Mar. Pollut. Bull. 2016;107(1):300–304. doi: 10.1016/j.marpolbul.2016.03.047. [DOI] [PubMed] [Google Scholar]
- Kameneva P.A., Efimova K.V., Rybin V.G., Orlaova T.Y. Detection of Dinophysistoxin-1 in clonal culture of marine dinoflagellate Prorocentrum foraminosum (Faust M.A., 1993) from the Sea of Japan. Toxins (Basel) 2015;7:3947–3959. doi: 10.3390/toxins7103947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafas S., Tomas C. Further observations on the genetics and morphometrics of Coolia santacroce (Dinophyceae) Algae. 2015;30(4):275–280. [Google Scholar]
- Karafas S., York R., Tomas C. Morphological and genetic analysis of the Coolia monotis species complex with the introduction of two new species, Coolia santacroce sp. nov. and Coolia palmyrensis sp. nov. (Dinophyceae) Harmful Algae. 2015;46:18–33. [Google Scholar]
- Karlson B., Andreasson A., Johansen M., Mohlin M., Skjevik A.-.T., Strömberg P. Plankton Toolbox – open-source software making it easier to work with plankton data. In: MacKenzie A.L., editor. 16th International Conference on Harmful Algae. Cawthron Institute, Nelson, New Zealand and the International Society for the Study of Harmful Algae (ISSHA); Wellington, New Zealand; 2015. pp. 194–197. [Google Scholar]
- Karlson B., Rehnstam-Holm A., Loo L. Temporal and spatial distribution of diarrhetic shellfish toxins in blue mussels, Mytilus edulis (L.), on the Swedish west coast, NE Atlantic 1988-2005. SMHI Rep. Oceanogr. 2007;35:40. [Google Scholar]
- Lawrence J.E., Grant J., Quilliam M.A., Bauder A.G., Cembella A.D. Colonization and growth of the toxic dinoflagellate Prorocentrum lima and associated fouling macroalgae on mussels in suspended culture. Mar. Ecol. Prog. Ser. 2000;201:147–154. [Google Scholar]
- Laza-Martínez A., Orive E., Irati M. Morphological and genetic characterization of benthic dinoflagellates of the genera Coolia, Ostreopsis and Prorocentrum from south-eastern Bay of Biscay. J. Phycol. 2011;46:45–65. [Google Scholar]
- Lee J.S., Igarashi T., Fraga S., Dahl E., Hovgaard P., Yasumoto T. Determination of diarrhetic shellfish toxins in various dinoflagellate species. J. Appl. Phycol. 1989;1:147–152. [Google Scholar]
- Levasseur M., Couture J.Y., Weise A.M., Michaud S., Elbrächter M., Sauve G., Bonneau E. Pelagic and epiphytic summer distributions of Prorocentrum lima and P. mexicanum at two mussel farms in the Gulf of St. Lawrence, Canada. Aquat. Microb. Ecol. 2003;30:283–293. [Google Scholar]
- Lundholm, N., Churro, C., Fraga, S., Hoppenrath, M., Iwataki, M., Larsen, J., Mertens, K., Moestrup, Ø., Zingone, A. (Eds.), 2009 onwards. IOC-UNESCO taxonomic reference list of harmful micro algae. Accessed at http://www.marinespecies.org/hab on 2021-12-27. doi:10.14284/362.
- Mangialajo L., Ganzin N., Accoroni S., Asnaghi V., Blanfuné A., Cabrini M., Cattaneo-Vietti R., Chavanon F., Chiantore M., Cohu S., Costa E., Fornasaro D., Grossel H., Marco-Miralles F., Masó M., Reñé A., Rossi A.M., Sala M.M., Thibaut T., Totti C., Vila M., Lemée R. Trends in ostreopsis proliferation along the Northern Mediterranean coasts. Toxicon. 2011;57(3):408–420. doi: 10.1016/j.toxicon.2010.11.019. [DOI] [PubMed] [Google Scholar]
- Maranda L., Corwin S., Hargraves P.E. Prorocentrum lima (Dinophyceae) in northeastern USA coastal waters: I. Abundance and distribution. Harmful Algae. 2007;6(5):623–631. [Google Scholar]
- Moreira A., Tester P.A. In: Guide For Designing and Implementing a Plan to Monitor Toxin-Producing Microalgae. Reguera B., Alonso R., Moreira Á., Méndez S., Dechraoui Bottein M.Y., editors. United Nations Educational, Scientific and Cultural Organization; Paris: 2016. Methods for sampling benthic microalgae; pp. 19–29. [Google Scholar]
- Morton S.L., Tindall D.R. Morphological and biochemical variability of the toxic dinoflagellate Prorocentrum lima isolated from three locations at Heron Island, Australia. J. Phycol. 1995;31:914–921. [Google Scholar]
- Munir S., Naz T., Siddiqui P.J.A., Morton S.L. Morphotaxonomy and seasonal distribution of planktonic and benthic Prorocentrales in Karachi waters, Pakistan Northern Arabian Sea. Chinese J. Oceanol. Limnol. 2013;31(2):267–281. [Google Scholar]
- Murakami Y., Oshima Y., Yasumoto T. Identification of okadaic acid as a toxic component of a marine dinoflagellate Prorocentrum lima. Bull. Jpn. Soc. Sci. Fish. 1982;48:69–72. [Google Scholar]
- Nagahama Y., Fukuyo Y. Redescription of Cryptomonas lima collected from Sorrento, Italy, the basionym of Prorocentrum lima. Plankton Biol. Ecol. 2005;52:107–109. [Google Scholar]
- Nagahama Y., Murray S., Tomaru A., Fukuyo Y. Species boundaries in the toxic dinoflagellate Prorocentrum lima (Dinophyceae, Prorocentrales), based on morphological and phylogenetic characters. J. Phycol. 2011;47:178–189. doi: 10.1111/j.1529-8817.2010.00939.x. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Sato S., Tawong W., Sakanari H., Uehara K., Shah M.…Adachi M. Genetic diversity and distribution of the ciguatera causing dinoflagellate Gambierdiscus spp. (Dinophyceae) in coastal areas of Japan. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0060882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okolodkov Y.B., Campos-Bautista G., Gárate-Lizárraga I., González-González J.A.G., Hoppenrath M., Arenas V. Seasonal changes of benthic and epiphytic dinoflagellates in the Veracruz reef zone, Gulf of Mexico. Aquat. Microbial Ecol. 2007;47(3):223–237. [Google Scholar]
- Parsons M.L., Brandt A.L., Ellsworth A., Leynse A.K., Rains L.K., Anderson D.M. Assessing the use of artificial substrates to monitor Gambierdiscus populations in the Florida Keys. Harmful Algae. 2017;68:52–66. doi: 10.1016/j.hal.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Parsons M.L., Aligizaki K., Bottein M.Y.D., Fraga S., Morton S.L., Penna A., Rhodes L. Gambierdiscus and Ostreopsis: reassessment of the state of knowledge of their taxonomy, geography, ecophysiology, and toxicology. Harmful Algae. 2012;14:107–129. [Google Scholar]
- Penna A., Vila M., Fraga S., Giacobbe M.G., Andreoni F., Riobó P., Vernezi C. Characterization of Ostreopsis and Coolia (Dinophyceae) isolates in the Western Mediterranean Sea based on morphology, toxicity and Internal Transcribed Spacer 5.8S rDNA sequences. J. Phycol. 2005;41:212–225. [Google Scholar]
- Reguera B., Alonso R., Moreira A., Méndez S., Dechraoui-Bottein M.Y. Guide for designing and implementing a plan to monitor Toxin-producing microalgae. 2nd (Eds) Vol. 59. IOC of UNESCO and IAEA; Paris and Vienna: 2016. Methods for sampling benthic microalgae; pp. 19–29. IOC Manuals and Guides. [Google Scholar]
- Rehnstam-Holm A.S., Hernroth B. Shellfish and public health: a Swedish perspective. AMBIO: A J. Human Environ. 2005;34(2):139–145. [PubMed] [Google Scholar]
- Rosen M.J., Callahan B.J., Fisher D.S., Holmes S.P. Denoising PCR-amplified metagenome data. BMC Bioinf. 2012;13:283. doi: 10.1186/1471-2105-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes L., Syhre M. Okadaic acid production by a New Zealand Prorocentrum lima isolate. N.Z.J. MAR. FRESHWATER. RES. 1995;29(3):367–370. [Google Scholar]
- Simoni F., Di Paolo C., Gori L., Lepri L. Further investigation on blooms of Ostreopsis ovata, Coolia monotis, Prorocentrum lima, on the macroalgae of artificial and natural reefs in the Northern Tyrrhenian Sea. Harmful Algae News. 2004;26:5–7. [Google Scholar]
- Steidinger K., Tangen K., Tomas C.R. Academic Press; 1997. Identifying Marine Phytoplankton; pp. 387–584. [Google Scholar]
- Steidinger K.A., Williams J. Florida Department of Natural Resources, Marine Research Laboratory; St. Petersburg, Florida, USA: 1970. Dinoflagellates, In: Memoirs of the Hourglass Cruises; pp. 1–251. [Google Scholar]
- Taylor F.J.R. Dinoflagellates from the International Indian Ocean Expedition. Bibliotheka Botanica. 1976;132:1–234. [Google Scholar]
- Tester P.A., Kibler S.R., Holland W.C., Gires U., Vandersea M.W., Leaw C.P., Litaker R.W. Sampling harmful benthic dinoflagellates: comparison of artificial and natural substrate methods. Harmful Algae. 2014;39:8–25. [Google Scholar]
- Tester P.A., Litaker R.W., Berdalet E. Climate change and harmful benthic microalgae. Harmful Algae. 2020;91 doi: 10.1016/j.hal.2019.101655. [DOI] [PubMed] [Google Scholar]
- Tillmann U., Hoppenrath M., Gottschling M. Reliable determination of Prorocentrum micans Ehrenb. (Prorocentrales, Dinophyceae) based on newly collected material from the type locality. J. Phycol. 2019;54:417–431. [Google Scholar]
- Toda M., Uneyama C., Toyofuku H., Morikawa K. Trends of food poisonings caused by natural toxins in Japan, 1989–2011. Shokuhin eiseigaku zasshi. J. Food Hyg. Soc. Jap. 2012;53(2):105–120. doi: 10.3358/shokueishi.53.105. [DOI] [PubMed] [Google Scholar]
- Totti C., Accoroni S., Cerino F., Cucchiari E., Romagnoli T. Ostreopsis ovata along the Conero Riviera (northern Adriatic Sea): relationships with environmental conditions and substrata. Harmful Algae. 2010;9:233–239. [Google Scholar]
- Verma A., Hoppenrath M., Dorantes-Aranda J.J., Harwood D.T., Murray S.A. Molecular and phylogenetic characterization of Ostreopsis (Dinophyceae) and the description of a new species, Ostreopsis rhodesae sp. nov., from a subtropical Australian lagoon. Harmful Algae. 2016;60:116–130. doi: 10.1016/j.hal.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Vila M., Abós-Herrándiz R., Isern-Fontanet J., Álvarez J., Berdalet E. Establishing the link between blooms and human health impacts using ecology and epidemiology. Sci. Mar. 2016;80:107–115. [Google Scholar]
- Wells M.L., Trainer V.L., Smayda T.J., Karlson B.S., Trick C.G., Kudela R.M.…Cochlan W.P. Harmful algal blooms and climate change: learning from the past and present to forecast the future. Harmful Algae. 2015;49:68–93. doi: 10.1016/j.hal.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto T., Seino N., Murakami Y., Murata M. Toxins produced by benthic dinoflagellates. Biol. Bull. 1987;172:128–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request. Sequence data have been deposited in the European Nucleotide Archive via the GFBio portal. The GFBio Submission ID is: daf47116-bfab-47d7-9ff9-38e9d34ccc57.