Abstract
The cerebrovascular-specific molecular mechanism in Alzheimer’s disease (AD) was investigated by employing comprehensive and accurate quantitative proteomics. Highly purified brain capillaries were isolated from cerebral gray and white matter of four AD and three control donors, and examined by SWATH (sequential window acquisition of all theoretical fragment ion spectra) proteomics. Of the 29 ribosomal proteins that were quantified, 28 (RPLP0, RPL4, RPL6, RPL7A, RPL8, RPL10A, RPL11, RPL12, RPL14, RPL15, RPL18, RPL23, RPL27, RPL27A, RPL31, RPL35A, RPS2, RPS3, RPS3A, RPS4X, RPS7, RPS8, RPS14, RPS16, RPS20, RPS24, RPS25, and RPSA) were significantly upregulated in AD patients. This upregulation of ribosomal protein expression occurred only in brain capillaries and not in brain parenchyma. The protein expression of protein processing and N-glycosylation-related proteins in the endoplasmic reticulum (DDOST, STT3A, MOGS, GANAB, RPN1, RPN2, SEC61B, UGGT1, LMAN2, and SSR4) were also upregulated in AD brain capillaries and was correlated with the expression of ribosomal proteins. The findings reported herein indicate that the ribosome complex, the subsequent protein processing and N-glycosylation-related processes are significantly and specifically upregulated in the brain capillaries of AD patients.
Keywords: Ribosome, protein processing, N-glycosylation, Alzheimer’s disease, blood-brain barrier
Introduction
Alzheimer's Disease (AD) is a well known type of dementia and is associated with the progressive decline in cognitive function. Beta-amyloid deposition and neurofibrillary changes in the cerebral cortex are prominent underlying pathophysiological causes. The underlying etiology of AD is not known, but cerebrovascular disease and diabetes are considered to be risk factors. 1 Conversely, physical activity and diet are considered to be protective factors for the development of AD. 1 Thus, multiple factors, including disease and lifestyle can contribute to the development of AD.
In the past decade, the association between cancer and Alzheimer's disease has attracted growing interest among researchers. 2 Multiple epidemiological studies and meta-analyses indicate that AD patients have a lower risk of developing cancer and cancer survivors have a lower risk of developing AD.3–5 Surprisingly, the risk of developing cancer in AD patients was reduced by 50% and the risk of Alzheimer's among cancer patients was reduced by 35%. 3 This suggests that there may be a common molecular mechanism between cancer and AD that is affected by the treatment. Several studies have reported an association between chemotherapy for cancer and a reduced risk of AD.6,7 In addition, compounds and foods such as turmeric (curcumin) and coffee (caffeine) have been reported to reduce the disease risk of AD and cancer.8,9 Although the anti-inflammatory and antioxidant effects of these compounds are thought to be protective against both diseases,10–13 interestingly, these compounds have also been shown to inhibit ribosome biosynthesis. 14 Donepezil and Tacrine, which are clinically used AD drugs, and chemotherapy, a common treatment for cancers, have also been reported to inhibit ribosome biosynthesis. 14 In general, cancer cells are characterized by a higher rate of ribosome biosynthesis than normal cells, which allows for cell growth (increased cell volume) and subsequent cell proliferation (increased cell number) in cancer patients. 15 Therefore, the inhibition of ribosome biosynthesis is thought to be an effective treatment against cancer. This common efficacy of ribosome biosynthesis inhibitors in cancer and AD suggests that ribosome biosynthesis is associated with both the development of cancer and AD.
Increased ribosome biosynthesis is thought to increase protein translation, but surprisingly, several studies have reported a decrease in the rate of protein translation in AD patients 16 and in mild dementia disorder (MCI) patients 17 in the cerebral cortex. The cerebral cortex is composed of glial, neuronal, and endothelial cells in a ratio of 5:4:218,19 which may mask changes in less abundant cells. A snRNA-seq study has revealed an association between increased ribosomal genes in microglia and the progression of AD pathology, 20 but microglia account for only about 6% of all glial cells. 21 Therefore, when considering the brain cortex as a whole, an increase in protein synthesis in AD brain microglia may have been masked by the decrease in protein synthesis in AD brain neurons and other cells that are present in high numbers. 22 The ribosomal inhibitor action of the chemotherapeutic drug mentioned above 23,24 may target Microglia. However, the percentage of microglia in the CNS is quite small, and the permeabilities of these drugs to the CNS are limited due to the blood-brain barrier. 25 Therefore, it is unlikely that these drugs can reach the microglia. As a result, the issue of which cells in the CNS benefit from the effect of ribosome biogenesis inhibitors against AD remains unclear.
It has been reported that angiogenesis, which occurs actively in tumors, occurs in the brains of AD patients, 26 and the results of transcriptomics studies support this finding. 27 Ribosome biogenesis increases with protein demand in actively proliferating cells during angiogenesis, and a protein co-expression network analysis in AD brains has raised the possibility that the expression of ribosome-related proteins is altered in vascular endothelial cells. 28 In addition, curcumin 29 and tanshinone IIA, 30 which have the ability to inhibit ribosome biosynthesis, 14 have been shown to exert anti-AD effects, and also show anticancer effects by inhibiting angiogenesis in cancer.31,32 This suggests that ribosome biosynthesis is enhanced in the AD vascular endothelium, thus also making it a potential therapeutic target for AD as well as cancer.
The SWATH method, a recently developed comprehensive quantitative proteomics method, is superior to shotgun proteomics in terms of quantitative accuracy. 33 Multiple specific peptides derived from a single protein are used for quantification, and the levels of protein expression are then calculated using the average amounts of these peptides that had been quantified. In addition, in previous studies, we applied our own criteria for selecting appropriate peptides in silico. The criteria include (1) less likely to be miscleaved by trypsin, (2) not including regions with low digestion efficiency such as transmembrane regions, (3) not including unstable/inaccurate sequences, and (4) not including regions with post-translational modifications.34–36 These criteria enhance the quantitative accuracy of the conventional SWATH method.
We hypothesized that the protein expression of a ribosome complex and the associated protein translation pathway is upregulated in the vascular endothelium of AD patients. The objective of this study was to confirm this hypothesis by applying the SWATH method to the highly purified brain capillaries derived from AD patients.
Materials and methods
Human brain tissues
The age at death, gender, brain region and weight used, the diagnosis, and prescription medication prior to death are all summarized in Table 1. The brains used were left cerebrum for AD3 and the right cerebrum for the other donors. The frontal/temporal lobe used was located at distance of 5 to 6 cm from the front of the right or left cerebrum, and the temporal/parietal lobe used was located at a distance of 7 to 9 cm from the front of the right cerebrum. The protocols for the present study were approved by the Ethics Committees of the Mihara Memorial Hospital (protocol code 095-06, approved on January 16th 2019) and the Graduate School of Pharmaceutical Sciences, Tohoku University (protocol code 18-03, approved on December 20th 2018), based on the Helsinki Declaration of 2013. Written informed consent was obtained from all subjects involved in the study.
Table 1.
Clinical characteristics of the human subjects described in the present study.
| Human donor No. | Age at death | Gender | Brain region; tissue weight used (g) | Clinical diagnosis | Neuropathological diagnosis | NIA-AA neuropathology | Prescription | Dementia level |
|---|---|---|---|---|---|---|---|---|
| (years) | ||||||||
| Control 1 | 71 | Male | 1. Neocortex (Gray matter) in parietal and temporal lobe; 0.380 | Diabetes, cervical malignant lymphoma | Unremarkable | None | None (previously unknown) | None |
| 2. Corona radiata in White matter under parietal and temporal lobe; 0.486 | ||||||||
| Control 2 | 75 | Male | 1. Neocortex (Gray matter) in parietal and temporal lobe; 0.372 | Hepatic encephalopathy, alcoholic cirrhosis | Hepatic encephalopathy | None | None | None |
| 2. Corona radiata in White matter under parietal and temporal lobe; 0.548 | ||||||||
| Control 3 | 69 | Male | 1. Neocortex (Gray matter) in frontal and temporal lobes; 0.403 | Emphysema | Unremarkable | None | None (previously unknown) | None |
| 2. Corona radiata in White matter under frontal and temporal lobe; 0.572 | ||||||||
| AD 1 | 86 | Male | 1. Neocortex (Gray matter) in parietal and temporal lobe; 0.414 | Pneumonia | AD, small ischemia (medulla) | AD definite | Silodosin, Bromhexine Hydrochloride-Bromhexine, Aspirin, Lansoprazole, Rosuvastatin Calcium, Donepezil Hydrochloride | Advanced |
| 2. Corona radiata in White matter under parietal and temporal lobe; 0.502 | ||||||||
| AD 2 | 86 | Female | 1. Neocortex (Gray matter) in parietal and temporal lobe; 0.437 | Dementia, AMI, hypertension | AD | AD definite | Ranitidine Hydrochloride, Nitrendipine, Magnesium oxide | Advanced |
| 2. Corona radiata in White matter under parietal and temporal lobe; 0.564 | ||||||||
| AD 3 | 83 | Male | 1. Neocortex (Gray matter) in frontal and temporal lobe; 0.405 | Cholangiocarcinoma | AD, ICH (CAA) | AD definite | Ursodeoxycholic Acid, Ifenprodil Tartrate, Donepezil Hydrochloride | Advanced |
| 2. Corona radiata in White matter under frontal and temporal lobe; 0.543 | ||||||||
| AD 4 | 84 | Female | 1. Neocortex (Gray matter) in parietal and temporal lobe; 0.421 | Pneumonia | AD | AD definite | Ranitidine Hydrochloride, Valproic acid | Advanced |
| 2. Corona radiata in White matter under parietal and temporal lobe; 0.545 |
Alzheimer’s disease (AD) was diagnosed based on the neuropathological and National Institute on Aging-Alzheimerʼs Association (NIA-AA) criteria. Autopsy frozen brain tissues were provided from the brain bank of Mihara memorial hospital at Tohoku University with the approval of the ethics committees of Mihara memorial hospital and Tohoku University. The weight of brain tissue described in this Table was used for the isolation of brain capillaries and preparation of whole tissue lysate. AMI: acute myocardial infarction; ICH (CAA): lobar intracerebral hemorrhage (cerebral amyloid angiopathy).
Preparation of highly pure capillaries and whole-tissue lysates of human brain tissues
The highly pure brain capillaries were isolated as described in a previous report, with minor modifications. 37 The cortex (neocortex; gray matter) was collected with a razor less than 3 mm from the surface of the cerebrum, and the white matter was collected from the radial crown just under the cortex used. The weights of collected tissues are listed in Table 1. The collected brain tissue was transferred to a 2 mL screw-cap tube (P000945-LYSK0-A, Bertin Instruments, Paris, France) containing 13 stainless beads (3.2 mm, 1.8 g, TOMY SEIKO, Tokyo, Japan). 1 mL of homogenizing buffer (101 mM NaCl, 4.6 mM KCl, 2.5 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 15 mM HEPES, pH 7.4) was added, and the brain tissue was then homogenized using a bead homogenizer (Precellys Evolution, Bertin Instruments, Paris, France) for 60 s at a speed of 4500 rpm. The homogenate was transferred to a new 2 mL tube and then centrifuged (1000 g, 10 min, 4°C). A portion of the brain homogenate (20 μL) was collected in another tube as a whole tissue lysate. The supernatant was removed carefully and up to 1 mL of the homogenizing buffer was added to the pellet. After preparing a suspension, an equal volume of 45 w/v% dextran/homogenize buffer was added to the 2 mL tube and mixed by inverting. The samples were immediately centrifuged (5800 g, 10 min, 4°C) and the supernatant was collected in a new 2 mL tube, while the pellets were stored on ice. The supernatant was centrifuged (5800 g, 10 min, 4°C) again and the supernatant was discarded. After removing the fat that had adhered to the wall of the tube with a Kimwipe, the pellets were suspended in the suspension buffer (homogenizing buffer containing 25 mM NaHCO3, 10 mM glucose, 1 mM pyruvate, and 5 g/L bovine serum albumin) (200 μL, 2 times), and samples of the two tubes were combined into one tube. The samples were filtered through a cell strainer (70 μm) (pluriStrainer-Mini, pluriSelect, Leipzig, Germany) and the strainer mesh was washed four times with 400 μL of suspension buffer. The samples that passed through the 70 μm mesh were added to a cell strainer filled with 600 mg of glass beads (BN-04, 0.35–0.5 mm, AS ONE, Osaka, Japan) and then washed 20 times with 500 μL of suspension buffer. After washing, the glass beads were transferred to a new tube using a spatula, after which, 1 mL of suspension buffer was added and the resulting suspension was mixed by inverting. The supernatant was then quickly transferred to a new tube. Glass beads were again added to the 500 μL suspension buffer and the resulting suspension was mixed by inverting. The supernatant was quickly transferred to the previous tube. This process of washing the glass beads and collecting the supernatant was repeated again using another tube. The two tubes were centrifuged (1000 g, 5 min, 4°C) and supernatant was removed. The resulting pellet was suspended in 100 μL of homogenizing buffer. A portion of the isolated brain capillary fraction was observed under a microscope. The isolated brain capillary fraction and the whole tissue lysate prepared during the capillary isolation were dissolved in a solubilizer (7 M Guanidine hydrochloride, 0.5 M Tris-HCl, 10 mM EDTA-Na) and sonicated. The protein recovery was measured using a Pierce BCA protein assay kit.
Standard method for isolating human brain capillaries
To compare the purity of the brain capillaries isolated by the above isolation method with the conventional method, brain capillaries were also isolated by the previously reported conventional isolation method. 38
Sample preparation for SWATH-based quantitative proteomics
Protein digestions with LysC/trypsin were performed as described previously. 39 The tryptic digests were cleaned up with a self-packed SDB-XD 200 µL tip (3 M, Minnesota) as previously described. 40
LC-MS/MS measurement for SWATH-based quantitative proteomics
The highly pure peptide samples (1 μg peptide) were injected into an NanoLC Ultra system (Eksigent Technologies, Dublin, CA, USA) coupled with an electrospray-ionization Triple TOF 5600 mass spectrometer (SCIEX, Framingham, MA, USA), which was set up for a single direct injection and analyzed by SWATH-MS acquisition, as previously described.36,41
Data analysis for SWATH-based quantitative proteomics
Spectral alignment and data extraction from SWATH chromatograms (uploaded to the Peptide Atlas website with Identifier PASS01717) were performed with the SWATH Processing Micro App in Peakview (SCIEX) using an in-house spectral library (uploaded to the Peptide Atlas website with Identifier PASS01717) as previously described.35,36 Only data below 1% FDR were selected. According to the previously described procedure, 35 unreliable peaks and peptides were removed based on the criteria of data selection and amino acid sequence-based peptide selection, and the peak areas at the peptide level were calculated as an average of those in transition level after normalizing the differences in signal intensity between different transitions. The details of this procedure have been reported previously. 35 The peak areas of individual proteins were calculated as an average of those at the peptide level, and were compared between the AD and control groups.
Statistical analysis
All statistical analyses were performed under the null hypothesis, assuming that the means of the compared groups were equal. Comparison between two groups was performed by an unpaired two-tailed t-test, and the p value was adjusted by Benjamini-Hochberg (BH) correction in the case of multiple comparisons. For the correlation analysis, Spearman's rank correlation coefficient and p value were calculated using Excel statistical software version 2010. If the p-value was less than 0.05, the difference was considered to be statistically significant and the null hypothesis was rejected. No formal power calculation was performed to estimate the required sample size. No randomization or blinding was performed in this study.
Results
Isolation of highly pure brain capillaries from AD and control donors
In order to clarify the pathological changes in the proteome of vascular endothelial cells that occur in AD, it is necessary to isolate highly purified brain capillaries from human brain tissues. A method for accomplishing this from rodent brain tissues has been reported. 37 We modified this isolation method and then applied it to human brain tissues in the present study. Using this procedure, it was possible to isolate highly pure capillaries (Supplementary figure 1). We also used the conventional standard isolation method, but contamination by parenchymal tissues other than capillaries was observed (Supplementary figure 1). These findings suggest that the purity was higher in the brain capillaries isolated by our modified method than those by the conventional method. Photographs of all 14 brain capillaries that were isolated from two regions (gray and white matters) of 7 donors (4 AD patients and 3 controls) are shown in Supplementary figure 2.
To quantify the purity of the vessels, the levels of marker proteins was quantitatively determined (Supplementary figure 3). The levels of the cerebral vascular endothelial cell markers ABCB1 (P-gp) and SLC2A1 (GLUT1) were 2.45-fold and 9.64-fold higher, respectively, in vessels isolated by the method described here compared to conventional methods. The levels of the neuronal markers NFL and NFM, the astrocyte marker GFAP and the oligodendrocyte marker MOG were 3.90-, 2.96-, 8.30- and 40.7-fold lower, respectively, compared to the values for the conventional method. To compare the ratio of endothelial cells to non-endothelial cells between the present and conventional isolation methods, the levels of endothelial cell markers was divided by the values for the markers of non-endothelial cells: the ABCB1/NFL, ABCB1/NFM, ABCB1/GFAP, ABCB1/MOG, SLC2A1/NFL, SLC2A1/NFM, SLC2A1/GFAP and SLC2A1/MOG ratios in present method were 6.35-, 5.29-, 6.38-, 133-, 26.0-, 21.5-, 23.1- and 533-fold greater than the values for the conventional method, respectively.
Furthermore, for the vessels isolated by present method, there were no statistically significant differences in the levels of the endothelial cell markers ABCB1 (P-gp), PECAM1 (CD31) and ABCG2 (BCRP) between the AD patient and control groups (Supplementary figure 4).
Aβ and APP levels in whole tissue lysates and the capillary fraction from gray and white matter
The total levels of Aβ and APP were quantified by the peak area of the tryptic peptide “LVFFAEDVGSNK” which is shared in Aβ and APP. In AD brains, the total expression of Aβ and APP was found to be significantly up-regulated in both gray and white matter by 31.1- and 3.50-fold, respectively, compared to the values for the controls (Supplementary figure 5). In AD brains, the total expression of Aβ and APP in gray matter was 38.6-fold greater than that in white matter. In the blood vessels isolated from AD brains, significant 17.1-fold and 10.5-fold increases in the total amounts of Aβ and APP in gray and white matters, respectively, were found compared to controls (Supplementary figure 5).
The protein expression of ribosome complexes in the brain capillaries of AD patients are increased but are not increased in brain parenchyma
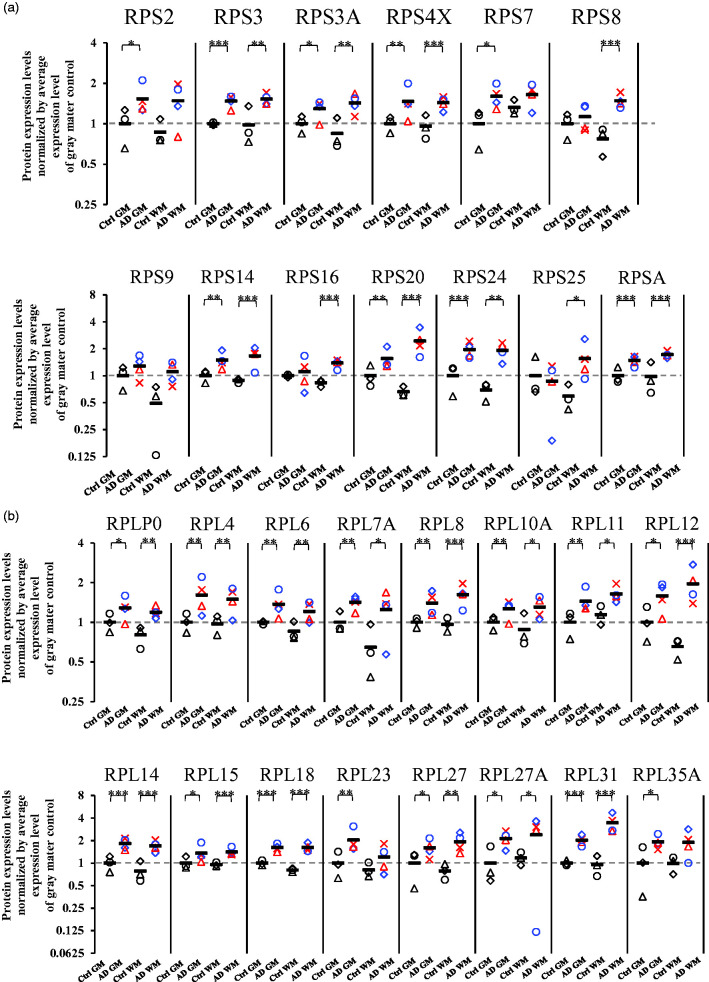
Brain capillaries isolated from gray (neocortex) and white (corona radiata) matter of four AD and three control donors were subjected to a SWATH analysis. A total of 1174 proteins were quantified (Supplementary table 1). Of these 1174 proteins, 164 proteins showed significant differences (BH-adjusted p value <0.05) in the protein expression levels in the brain capillaries of gray matter between AD and control patients, and 269 proteins showed significant differences (BH-adjusted p value <0.05) in protein expression levels in the brain capillaries of white matter between AD and control patients (Supplementary table 1). In order to clarify the molecular mechanisms that are commonly involved in the alteration in the capillaries of gray and white matter, a pathway analysis was performed by using the Kyoto Encyclopedia of Genes and Genomes (KEGG) using 97 molecules that were significantly altered in the capillaries of both gray and white matter in AD patients compared to control donors. Table 2 shows the top four pathways or complexes which were identified by the KEGG analysis. The ribosome complex was significantly enriched with an adjusted p-value of 9.7E-18. In the capillaries of AD gray matter, the protein expression of 25 ribosomal proteins were significantly upregulated when compared to those of the control gray matter (Figure 1; RPS2 (1.53-fold), RPS3 (1.47-fold), RPS3A (1.29-fold), RPS4X (1.46-fold), RPS7 (1.60-fold), RPS14 (1.49-fold), RPS20 (1.55-fold), RPS24 (1.95-fold), RPSA (1.48-fold), RPLP0 (1.28-fold), RPL4 (1.61-fold), RPL6 (1.37-fold), RPL7A (1.42-fold), RPL8 (1.40-fold), RPL10A (1.26-fold), RPL11 (1.44-fold), RPL12 (1.58-fold), RPL14 (1.82-fold), RPL15 (1.35-fold), RPL18 (1.61-fold), RPL23 (2.02-fold), RPL27 (1.59-fold), RPL27A (2.12-fold), RPL31 (2.02-fold), and RPL35A (1.91-fold)). In the capillaries of AD white matter, the protein expression of 24 ribosomal proteins were significantly upregulated when compared to those of the control white matter (Figure 1; RPS3 (1.55-fold), RPS3A (1.69-fold), RPS4X (1.49-fold), RPS8 (1.91-fold), RPS14 (1.87-fold), RPS16 (1.66-fold), RPS20 (3.70-fold), RPS24 (2.77-fold), RPS25 (2.63-fold), RPSA (1.75-fold), RPLP0 (1.49-fold), RPL4 (1.54-fold), RPL6 (1.42-fold), RPL7A (1.93-fold), RPL8 (1.68-fold), RPL10A (1.48-fold), RPL11 (1.44-fold), RPL12 (2.99-fold), RPL14 (2.17-fold), RPL15 (1.47-fold), RPL18 (2.00-fold), RPL27 (2.45-fold), RPL27A (2.04-fold), and RPL31 (3.63-fold)). To clarify whether the upregulation of ribosome complexes is specific to the capillaries, we performed a SWATH analysis on a whole tissue lysate of the parenchyma of four AD and three control donors (Supplementary table 2), and extracted the data for ribosomal proteins. No significant upregulation of ribosomal protein was observed in the AD patients compared to the control donors, in both gray and white matter parenchyma (Supplementary figure 6). This suggests that the increases in the expression of ribosomal proteins in AD patients are specific to the brain capillaries.
Table 2.
KEGG-based identification of molecular pathways/complexes for which the protein expression levels were changed in the isolated brain capillaries of AD patients compared to control donors.
| Pathway/Complex | Number of proteins included in 97 proteins | P-value | Adjusted P-value |
|---|---|---|---|
| Ribosome | 21 | 8.8E-20 | 9.7E-18 |
| Protein processing in endoplasmic reticulum | 5 | 0.065 | 1 |
| N-Glycan biosynthesis | 3 | 0.072 | 1 |
| Amyotrophic lateral sclerosis | 3 | 0.074 | 1 |
Raw data of the SWATH-MS analysis using the highly purified brain capillaries is shown in Supplementary table 1. 97 proteins which showed a significant change (p < 0.05 or p = 0.05) in the protein expression levels between AD and control brain capillaries both in gray and white matters were applied to KEGG-based pathway analysis. Top four pathways or complexes enriched in this analysis were listed in the order of p-value. The p-value and adjusted p-values were obtained from David enrichment analysis website.
Figure 1.
Protein expression levels of ribosomal proteins in highly purified brain capillaries isolated from cerebral gray and white matter in AD patients. Brain capillaries were isolated from gray (neocortex) and white (corona radiata) matter of four AD and three control donors. Tryptic digests were produced and subjected to SWATH analysis. The protein expression levels were normalized by the average protein expression levels in gray matter capillaries of control donors. BH-adjusted p value <0.05 (*), <0.01 (**), and <0.001 (***), significantly different between AD and control groups. (a) Small ribosomal subunit proteins (40S ribosomal protein) and (b) Large ribosomal subunit proteins (60S ribosomal protein). Ctrl GM, gray matter capillaries in control donors; AD GM, gray matter capillaries in AD donors; Ctrl WM, white matter capillaries in control donors; AD WM, white matter capillaries in AD donors. Circle, control 1 or AD 1; Triangle, control 2 or AD 2; Diamond, control 3 or AD 3; Cross, AD 4. Bar, average value in each group. For AD patients, blue plots correspond to males, and red plots to females.
Identification of the molecular mechanisms responsible for the increased expression of ribosomal proteins
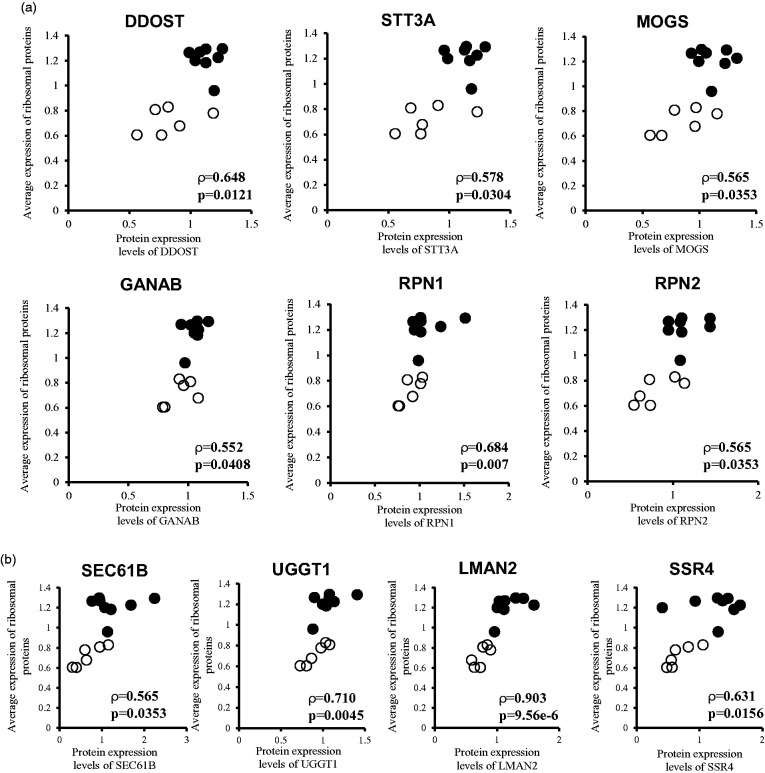
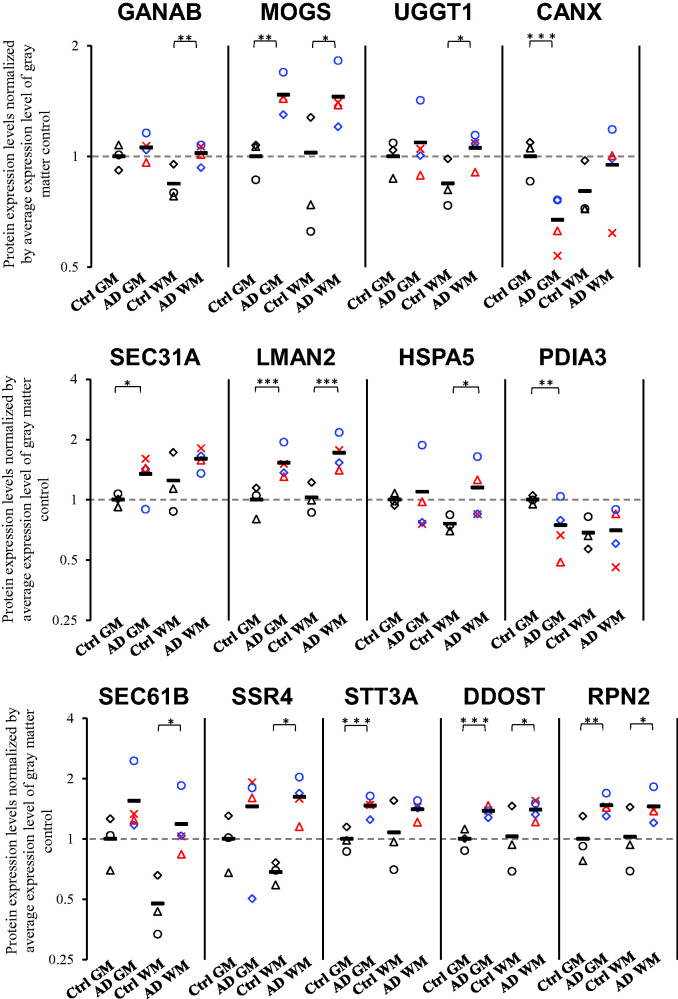
We performed Spearman's rank correlation analysis to identify the molecular mechanisms that are correlated with the increased expression of ribosomal proteins in the brain capillaries of AD patients. To accomplish this, the average of expression levels of 21 ribosomal proteins that were significantly up-regulated in both gray and white matters of AD brain capillaries were calculated for each donor. A Spearman's rank correlation coefficient analysis was performed between the average expression level of 21 ribosomal proteins and the level of expression of individual proteins for the gray and white matter capillaries of four AD and three control donors (n = 14). A pathway analysis was performed by KEGG using 138 molecules that showed a significant (p < 0.05) positive correlation with the average of 21 ribosomal protein expressions. Among the pathways/complexes detected, protein processing in the endoplasmic reticulum pathway and the N-Glycan biosynthesis pathway showed significant correlations with the adjusted p values of 0.012 and 0.013, respectively (Table 3). In the N-Glycan biosynthesis pathway, six molecules (STT3A, DDOST, GANAB, MOGS, RPN1, and RPN2) consisting of enzymes that transfer glycans to protein precursors in the ER, showed a significant positive correlation with ribosomes (Figure 2). In the protein processing in the endoplasmic reticulum pathway, 10 molecules (including SEC61B, UGGT1, LMAN2, SSR4 and the 6 N-Glycan biosynthesis proteins above) showed a significant positive correlation with ribosomes (Figure 2). In addition to the above proteins, we identified proteins related to N-Glycan biosynthesis and protein processing in the ER that were significantly upregulated in either the gray or white matter (Figure 3). In the capillaries of AD gray matter, SEC31A (1.34-fold) was significantly upregulated in addition to the above-mentioned molecules; MOGS (1.26-fold), LMAN2 (1.53-fold), STT3A (1.47-fold), RPN2 (1.47-fold), and DDOST (1.38-fold). In contrast, CANX (0.671-fold) and PDIA3 (0.746-fold) were significantly downregulated (Figure 3). In the capillaries of AD white matter, HSPA5 (1.52-fold) was significantly upregulated in addition to the above-mentioned molecules; GANAB (1.21-fold), MOGS (1.36-fold), UGGT1 (1.25-fold), LMAN2 (1.67-fold), SEC61B (2.49-fold), SSR4 (2.37-fold), DDOST (1.36-fold), and RPN2 (1.42-fold) (Figure 3).
Table 3.
Top 5 pathways/complexes significantly correlated with ribosomal protein expression.
| Pathway/Complex | Number of proteins included in 138 proteins | P-value | Adjusted P-value |
|---|---|---|---|
| Protein processing in endoplasmic reticulum | 10 | 0.000082 | 0.012 |
| N-Glycan biosynthesis | 6 | 0.00018 | 0.013 |
| Fatty acid degradation | 4 | 0.011 | 0.37 |
| Peroxisome | 5 | 0.013 | 0.37 |
| Valine, leucine and isoleucine degradation | 4 | 0.014 | 0.37 |
The average of expression levels of 21 ribosomal proteins that were significantly up-regulated in both gray and white matter of AD brain capillaries were calculated for each donor. Spearman's rank correlation coefficient analysis was performed between the average expression level of 21 ribosomal proteins and the protein expression level of individual proteins for the gray and white matter capillaries of four AD and three control donors (n = 14). A pathway analysis was performed by KEGG using 138 molecules that showed a significant (p < 0.05) positive correlation with the average of 21 ribosomal protein expressions. Top five pathways or complexes enriched in this analysis were listed in the order of p-value. The p-value and adjusted p-values were obtained from David enrichment analysis website.
Figure 2.
Endoplasmic reticulum proteins that are positively correlated with ribosomal proteins by Spearman's rank correlation analysis. The average of expression levels of 21 ribosomal proteins that were significantly up-regulated in both gray and white matter of AD brain capillaries were calculated for each donor. Spearman's rank correlation coefficient analysis was performed between the average expression level of 21 ribosomal proteins and the protein expression level of individual proteins for the gray and white matter capillaries of four AD and three control donors (n = 14). Pathway analysis was performed by KEGG using 138 molecules that showed a significant (p < 0.05) positive correlation with the average expression of 21 ribosomal proteins. It found the N-Glycan biosynthesis pathway (a) and protein processing in endoplasmic reticulum pathway (a and b). (a) Six molecules of N-Glycan biosynthesis pathway (STT3A, DDOST, GANAB, MOGS, RPN1, and RPN2) showed a significant positive correlation with ribosomes. (a and b) Ten molecules of the protein processing in endoplasmic reticulum pathway (including SEC61B, UGGT1, LMAN2, SSR4 and the 6 N-Glycan biosynthesis proteins above) showed a significant positive correlation with ribosomes. ρ, correlation coefficient; p < 0.05, significant correlation; Open symbol, control donors; Closed symbol, AD patients.
Figure 3.
Protein expression levels of N-glycosylation and ER processing related proteins in highly purified brain capillaries isolated from cerebral gray and white matter in AD patients. Brain capillaries were isolated from gray (neocortex) and white (corona radiata)Continued.matter of four AD and three control donors. Tryptic digests were produced and subjected to SWATH analysis. The data for the N-glycosylation and ER processing related proteins was extracted and shown in this figure. The protein expression levels were normalized by the average protein expression levels in gray matter capillaries of control donors. BH-adjusted p value <0.05 (*), <0.01 (**), and <0.001 (***), significantly different between AD and control groups. Ctrl GM, gray matter capillaries in control donors; AD GM, gray matter capillaries in AD donors; Ctrl WM, white matter capillaries in control donors; AD WM, white matter capillaries in AD donors. Circle, control 1 or AD 1; Triangle, control 2 or AD 2; Diamond, control 3 or AD 3; Cross, AD 4. Bar, average value in each group. For AD patients, blue plots correspond to male, and red plots to females.
Up-regulated expression of N-glycosylated proteins in AD brain capillaries
The ribosome complex, the subsequent protein processing and N-glycosylation-related processes were significantly and specifically upregulated in the brain capillaries of AD patients. To further confirm that these functions are, in fact, enhanced, we investigated the issue of whether the expression levels of N-glycosylated proteins are increased in the brain capillaries of AD patients. A search for N-glycosylated proteins was conducted using the Uniprot database. Based on the results of this search, N-glycosylated proteins were extracted from the SWATH data. For gray matter capillaries, of the 36 proteins that showed significant expression changes between AD patients and controls, 75% (27 proteins) were up-regulated in the AD patients (Supplementary figure 7). In white matter capillaries, 67.4% (29 proteins) of the 43 proteins that showed significant expression changes between AD patients and controls were up-regulated in AD patients (Supplementary figure 7). These results indicate that the expression levels of N-glycosylated proteins tend to be increased in cerebrovasculature of AD patients.
Discussion
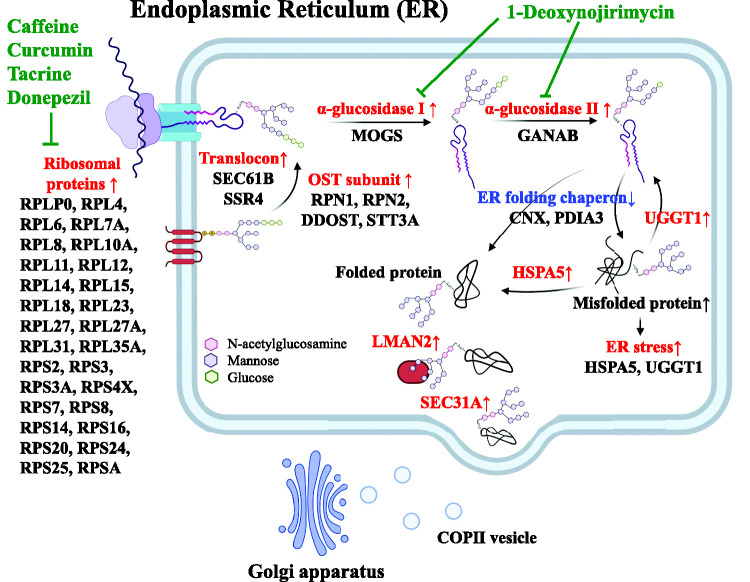
The SWATH analysis was used to reveal cerebrovascular-specific molecular variations in the brains of AD patients. Of the 29 ribosomal proteins that were quantified, 28 (RPLP0, RPL4, RPL6, RPL7A, RPL8, RPL10A, RPL11, RPL12, RPL14, RPL15, RPL18, RPL23, RPL27, RPL27A, RPL31, RPL35A, RPS2, RPS3, RPS3A, RPS4X, RPS7, RPS8, RPS14, RPS16, RPS20, RPS24, RPS25, and RPSA) were significantly up-regulated (Figure 1). This upregulation of ribosomal protein expression was observed only in the brain capillaries and not in whole brain regions. In the vascular endothelium of the AD brains, the protein expression of Protein Processing and N-glycosylation-related proteins in the ER (DDOST, STT3A, MOGS, GANAB, RPN1, RPN2, SEC61B, UGGT1, LMAN2, and SSR4) was upregulated and this upregulation was correlated with the expression of ribosomal proteins (Figure 2). Other molecules related to protein processing in the ER were also significantly upregulated; SEC31A in gray matter and HSPA5 in white matter (Figure 3). However, some were downregulated; CANX and PDIA3 in gray matter (Figure 3). Taking these collective findings into consideration, they suggest that ribosome function is enhanced in the cerebral blood vessels of AD patients and that the subsequent protein translation network is abnormal, as illustrated in Figure 4.
Figure 4.
Hypothetical molecular mechanism in the brain capillary endothelial cells of AD patients. Based on the results in the present study, we illustrate the features of pathological molecular mechanisms that occur in brain capillary endothelial cells of AD patients. The protein expression levels for most of the ribosomal proteins were upregulated. The downstream protein processing mechanisms such as N-glycosylation and folding were also upregulated in the ER. The upregulated molecules and processes are shown in red. The downregulated molecules are shown in blue. The potential drugs and inhibitors are shown in green. The ribosomes represent a potential target of caffeine, curcumin, tacrine and donepezil, as discussed in the main text. 1-Deoxynojirimycin, an α-glucosidase inhibitor, has been shown to improve cognitive function and reduce Aβ deposition in senescence-accelerated mice. 85 We therefore hypothesize that the molecular mechanisms shown in this figure would be promising drug targets for the treatment of AD.
Most of the ribosomal proteins were significantly up-regulated in the vascular endothelium of AD brains. Ribosomes are intracellular organelles consisting of multiple rRNAs and two subunits, the 60S large subunit and the 40S small subunit, which are responsible for protein translation in the cell. 42 The increased expression of these ribosomal proteins in the vascular endothelium of AD brains indicates that ribosome biosynthesis in the endothelium is increased, which can be interpreted to mean that protein synthesis in the AD cerebrovascular endothelium is increased. Ribosomes perform protein translation on the rough-surfaced endoplasmic reticulum (RER) membrane. Protein translation begins with the insertion of a polypeptide into the lumen of the RER (called ER protein translocation) and the resulting polypeptide is transported into the RER by a protein complex on the RER membrane called the translocon, where it undergoes N-glycosylation. ER protein translocation and N-glycosylation are conjugated events. 43 The translocons are formed by the Sec61 complex, the oligosaccharyltransferase complex (OST) and the translocon-associated protein complex (TRAP) (Figure 4). The protein expression of these complexes were up-regulated in AD brain capillaries (Figure 3). OST is a membrane protein complex that is composed of multiple subunits (RPN1, RPN2, DDOST, STT3A) 44 and catalyzes the addition of glycans (G3M9-type glycans) to Asn residues in nascent polypeptides that are inserted by ER protein translocation (Figure 4): 45 STT3A is the catalytic subunit of OST, 46 and the deletion of STT3A reduces OST activity. 45 DDOST is essential for the formation of the OST complex, and gene knockout suppresses N-glycosylation by 70%. 47 It has been reported that RPN1 and RPN2 promote N-glycosylation in a substrate-specific manner.48,49 The increased expression of these proteins suggests that the addition of glycan chains to nascent polypeptides in the ER is enhanced in the vascular endothelium of AD brains (Figure 4).
Two glucose residues are removed from the glycan-added nascent polypeptide in the ER by successive reactions of glucosidase 1 (α-glucosidase I (MOGS)) and glucosidase 2 ((1) thus converting G3M9 to a G2M9-type glycan; (2) from a G2M9 to a G1M9-type glycan) (Figure 4). This reaction allows the glycan to be recognized by the lectin chaperone, allowing the glycoprotein to fold and proceed to the Golgi secretory pathway.50,51 The inhibition of glucosidase 1 inhibits the secretion of glycoproteins in human liver cancer cells. 52 Glucosidase II is a soluble heterodimeric protein composed of an alpha-subunit and a beta-subunit, and GANAB, which is its catalytic subunit, has been reported to catalyze reactions without the beta-subunit. 53 The increased expression of these glucosidases suggests that the above reactions are enhanced in the ER of AD cerebrovascular endothelium (Figure 4).
Polypeptides (G1M9-type glycans) are specifically recognized and folded by chaperones such as CANX, CRT and PDIA3. 54 In the gray matter in the AD brain vascular endothelium, the expression of CANX and PDIA3 was decreased, while the level of expression of UGGT1 was increased in correlation with ribosome expression and the expression level of HSPA5 appeared to be increased, but this increase was not significant (Figures 2 and 3). In the vascular endothelium white matter of AD brains, the expression of HSPA5 and UGGT1 was increased (Figure 3). CANX and PDIA3 are important for the correct folding of nascent polypeptides,55,56 and a decreased expression of these proteins increases the levels of unfolded or misfolded proteins in the ER, leading to an unfolded protein reaction (UPR) (Figure 4). HSPA5 functions as a molecular chaperone to promote folding, and UGGT1 functions as a checkpoint to reapply glucose to the N-glycans of misfolded proteins (Figure 4). 57 It has been reported that the expression of these proteins is induced under conditions of ER stress.58,59 After folding is complete, the glucose unit is removed from the protein by the action of glucosidase 2 (M9-type glycans). At this point, if the protein is correctly folded, it is transported to the Golgi apparatus (Figure 4). If the protein is not in the correct 3D conformation, it is trapped by the folding sensor enzyme UGGT1, which reattaches one glucose moiety and, if deemed irreparable, it is passed to the degradation pathway (Figure 4). 60 LMAN2 can move between the ER and Golgi by recognizing and binding to M9-type glycans (Figure 4). 61 SEC31 is a COPII coat protein that transports proteins from the ER to the Golgi 62 and performs the GTP hydrolysis required for the secretion of COPII from the ER (Figure 4).63,64 Taken together, these findings suggest that AD cerebral blood vessels in the gray matter are prone to occur the UPRs which causes the upregulation of molecules that repair them, with subsequent protein processing also being enhanced (Figure 4).
Multiple epidemiological studies and meta-analyses have demonstrated that AD patients have a lower risk of developing cancer, while cancer patients have a lower risk of developing AD.3–5 Increased ribosome biosynthesis in the cerebrovascular endothelium of AD subjects may explain the inverse association between cancer and the risk of developing the disease in Alzheimer's patients. It has been reported that chemotherapy, a common treatment for cancer, 65 and the clinically used anti-AD drugs donepezil and tacrine inhibit ribosome biosynthesis (Figure 4). 14 Increased ribosome biosynthesis is a recognized feature in many types of cancers and has been found to be associated with a poor prognosis. 66 Recent studies suggest that an increased number of ribosomes promote tumorigenesis, 67 and the inhibition of ribosome biosynthesis may lead to cancer suppression.65,68 The results presented here suggest that ribosome biosynthesis is also increased in AD cerebrovascular tissue, although the issue of whether it acts to exacerbate this condition is unknown. This suggests that increased ribosome function is a common phenomenon in cancer and Alzheimer's disease, and that drug treatment of each disease is inversely correlated with the inhibition of ribosome biosynthesis in each disease. Indeed, several studies have reported that chemotherapy for cancer is associated with a reduced risk of AD.6,7 In addition, turmeric (curcumin) and coffee (caffeine) which inhibit ribosome biosynthesis have been reported to reduce the risk of developing AD and cancer (Figure 4).8,9,11,14,69,70 It is thought that turmeric (curcumin) and coffee (caffeine) have anti-AD effects due to their antioxidant and inflammatory properties.10,13 However, given the increased ribosome biosynthesis in the vascular endothelium of AD subjects (although it is not known whether this phenomenon is involved in this exacerbation), these compounds may exert their anti-AD effects through a mechanism that involves the inhibition of ribosome biosynthesis (Figure 4).
Unlike animal models, the use of human brain tissue in studies of molecular mechanisms clinically, very meaningful, but individual differences must be taken into account. In the present study, the AD patients were two male and two female donors, whereas all three donors in the control group were males (Table 1). In addition, the mean age of the AD patients was approximately 10 years greater than that of the control donors. This points out the challenges that are encountered with a small number of cases for each gender and age, and the lack of gender and age matches between the AD patients and the control group. Although it is not easy to obtain human brain tissue, it is clear that future additional experiments with a larger number of specimens will be necessary. However, results of this study are, nevertheless important, even though a limited number of specimens was available, since these findings will be important in terms of future studies. Because, in a proteomic study of brain tissue from Parkinson's disease, one of the same CNS diseases, using brain tissue from only five cases, it was found that the expression of the protein mortalin was reduced in Parkinson's disease, and that this mortalin protein was associated with Parkinson’s toxicity via a pathway that includes oxidative stress, mitochondrial and proteasomal dysfunction. 71 The results reported herein show that, even the results from a small number of cases, based on preliminary findings, can still be useful.
First, with regard to gender imbalance, as shown in Figures 1, 3, Supplementary figures 5, and 6, two male cases (blue plot) and two female cases (red plot) of AD showed almost identical expression changes, suggesting no significant gender differences for these groups of molecules. Secondly, regarding the fact that AD patients were older than the controls, the following interpretation remains a possibility. Decreased expression of ribosomal proteins has been reported in the aged mouse brain tissue (hippocampus). 72 In addition, several cell and animal studies have reported reduced expressions of ribosomal genes and proteins due to ageing.73–78 These findings suggest that ribosomal protein expression decreases with age, and therefore the elevated ribosomal protein expression found for the AD samples used in this study is not due to age but, rather, to the pathology of AD. Although this possible interpretation is based on the preliminary data reported in this study, future analyses using samples from larger numbers of donors that are matched for gender and age between the AD and control groups are needed.
In the isolated vessels, in addition to endothelial cells, brain parenchymal cells are present as contaminants. However, as shown in Supplementary Figure 3, the isolation method established in this study reduced the extent of contamination by brain parenchymal cells such as astrocytes, neurons and oligodendrocytes compared to conventional methods. Astrocytes are particularly prone to contamination because they are in contact with endothelial cells. However, it has been reported that ribosomal expression levels are not altered in astrocytes in AD brains. 28 Therefore, in the present study, the elevated expression of ribosomal proteins in the isolated vessels is suggested to be of vascular endothelial cell origin.
The expression of ribosomal proteins and N-linked glycosylation-related proteins in the ER was increased, but the issue of their functions are enhanced remains uncertain. If these functions are enhanced, the synthesis of proteins that are undergoing N-glycosylation would also be increased. To confirm this, the expression levels of proteins undergoing N-glycosylation were investigated and the results showed that a greater proportion of proteins were up-regulated in the cerebral vessels of AD donors compared to controls (Supplementary figure 7). Thus, not only the expression of ribosomal proteins and N-linked glycosylation-related proteins but also their biological functions may be increased. Further detailed expression and functional analyses will be needed to validate these conclusions.
ABCB1 (P-gp) and SLC2A1 (GLUT1) are important transporters at the BBB; ABCB1 has been reported to be down-regulated in cerebral vessels in AD by an antibody-based analysis. 79 In a subsequent quantitative proteomic analysis, it was reported that ABCB1 expression at the BBB of AD patients is unchanged. 80 Antibody-based analyses lack quantitative accuracy. Given the fact that ABCB1 is a membrane protein, the accuracy of antibody-based quantitative analysis would be even more questionable. The SWATH method that was used in this study provides higher quantitative accuracy than the quantitative proteomics described above. The reason for this is that the above proteomics analysis used a single peptide sequence within the ABCB1 protein, whereas the SWATH analysis in this study used peptide sequences from multiple locations of the ABCB1 protein to quantify it. The present SWATH study showed that there is no significant difference in the protein expression level of ABCB1 in cerebral vessels between the AD and control groups (Supplementary table 1). For the reasons stated above, this result could be closer to the truth.
The PET signal intensity of 18F-fluorodeoxyglucose (FDG) accumulation is reported to be positively regulated by the activity of Hexokinase (HK), but not that of SLC2A1. 81 This suggests that HK is a rate limiting step in the signal intensity related to FDG accumulation; HK activity was reduced in brain capillaries isolated from AD patients. 82 These findings suggest that the cause of the reduced FDG-PET signal in the AD patients is due to reduced HK activity in cerebral vessels, rather than a reduced activity or a reduced expression of SLC2A1 in cerebral vessels. On the other hand, endogenous glucose concentrations (not FDG) are elevated in the brains of AD patients, 83 which could be attributed to an increase in SLC2A1 transport activity in the cerebral vessels of AD patients, rather than a decrease. A recent quantitative proteomic analysis reported that SLC2A1 expression levels do not vary significantly in the cerebral vessels of AD patients and are significantly increased in Lewy body dementia. 80 The degree of increased expression of SLC2A1 in the present study was intermediate between those two. Therefore, SLC2A1 expression and transport activity may be slightly increased at the BBB of AD patients.
In conclusion, we report herein, for the first time, that ribosomal proteins and N-linked glycosylation-related proteins in the ER are significantly upregulated in the brain capillaries of AD patients (Figure 4). We also clarified that these upregulations occur only in the brain capillaries and not in the parenchymal regions. The issue of whether these upregulations act to exacerbate or protect AD is currently unclear, and therefore further study will be needed in future to confirm this. Finally, an RNAseq analysis was recently reported on the brain microvessels of AD patients. 84 However, the mRNA expression levels of ribosomal proteins in the brain microvessels of AD patients were not significantly different from those of normal vessels. Protein levels do not necessarily correlate with mRNA levels. This result is a good example highlighting the need for proteome analysis.
Supplemental Material
Supplemental material, sj-xlsx-1-jcb-10.1177_0271678X221111602 for Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer's disease patients by Masayoshi Suzuki, Kenta Tezuka, Takumi Handa, Risa Sato, Hina Takeuchi, Masaki Takao, Mitsutoshi Tano and Yasuo Uchida in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-2-jcb-10.1177_0271678X221111602 for Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer's disease patients by Masayoshi Suzuki, Kenta Tezuka, Takumi Handa, Risa Sato, Hina Takeuchi, Masaki Takao, Mitsutoshi Tano and Yasuo Uchida in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-3-jcb-10.1177_0271678X221111602 for Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer's disease patients by Masayoshi Suzuki, Kenta Tezuka, Takumi Handa, Risa Sato, Hina Takeuchi, Masaki Takao, Mitsutoshi Tano and Yasuo Uchida in Journal of Cerebral Blood Flow & Metabolism
Acknowledgements
The authors wish to thank Dr Milton S Feather (a scientific editor in the United States of America) for English proofreading the entire manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported, in part, by Grants-in-Aids from the Japanese Society for the Promotion of Science (JSPS) for Scientific Research (B) [KAKENHI: 20H03399], Fostering Joint International Research (A) [KAKENHI: 18KK0446], and Challenging Research (Exploratory) [KAKENHI: 21K19365]. This study was also supported, in part, by Grants-in-Aids from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) for Scientific Research on Innovative Areas [KAKENHI: 20H05495 and 20H04690], and also supported, in part, by the Takeda Science Foundation. This study was also supported, in part, by a grant from the Japan Agency for Medical Research and Development (AMED) [grant number JP21wm0425019] and the intramural fund from the National Center of Neurology and Psychiatry [grant number 3–8].
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions: Masayoshi Suzuki: Conducted experiments, Analysis/acquisition of data, Study design/conception, Drafting of manuscript.
Kenta Tezuka, Risa Sato, and Hina Takeuchi: Method development of sample preparation.
Takumi Handa: Method development of data analysis.
Masaki Takao and Mitsutoshi Tano: Providing the human brain tissues.
Yasuo Uchida: Drafting of the manuscript, Revision of manuscript, Study design/conception, Analysis/acquisition of data.
ORCID iD: Yasuo Uchida https://orcid.org/0000-0001-9860-1979
Supplemental material: Supplemental material for this article is available online.
References
- 1.Silva MVF, Loures CMG, Alves LCV, et al. Alzheimer's disease: risk factors and potentially protective measures. J Biomed Sci 2019; 26: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanni C, Masi M, Racchi M, et al. Cancer and Alzheimer's disease inverse relationship: an age-associated diverging derailment of shared pathways. Mol Psychiatry 2021; 26: 280–295. [DOI] [PubMed] [Google Scholar]
- 3.Musicco M, Adorni F, Di Santo S, et al. Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study. Neurology 2013; 81: 322–328. [DOI] [PubMed] [Google Scholar]
- 4.Realmuto S, Cinturino A, Arnao V, et al. Tumor diagnosis preceding Alzheimer's disease onset: is there a link between cancer and Alzheimer's disease? J Alzheimers Dis 2012; 31: 177–182. [DOI] [PubMed] [Google Scholar]
- 5.Driver JA, Beiser A, Au R, et al. Inverse association between cancer and Alzheimer's disease: results from the Framingham heart study. Bmj 2012; 344: e1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akushevich I, Yashkin AP, Kravchenko J, et al. Chemotherapy and the risk of Alzheimer's disease in colorectal cancer survivors: Evidence from the medicare system. JCO Oncol Pract 2021; 17: e1649–e1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du XL, Xia R, Hardy D. Relationship between chemotherapy use and cognitive impairments in older women with breast cancer: findings from a large population-based cohort. Am J Clin Oncol 2010; 33: 533–543. [DOI] [PubMed] [Google Scholar]
- 8.Barranco Quintana JL, Allam MF, Serrano Del Castillo A, et al. Alzheimer's disease and coffee: a quantitative review. Neurol Res 2007; 29: 91–95. [DOI] [PubMed] [Google Scholar]
- 9.Ng TP, Chiam PC, Lee T, Chua HC, et al. Curry consumption and cognitive function in the elderly. Am J Epidemiol 2006; 164: 898–906. [DOI] [PubMed] [Google Scholar]
- 10.Tang M, Taghibiglou C. The mechanisms of action of curcumin in Alzheimer's disease. J Alzheimers Dis 2017; 58: 1003–1016. [DOI] [PubMed] [Google Scholar]
- 11.Bohn SK, Blomhoff R, Paur I. Coffee and cancer risk, epidemiological evidence, and molecular mechanisms. Mol Nutr Food Res 2014; 58: 915–930. [DOI] [PubMed] [Google Scholar]
- 12.Shehzad A, Wahid F, Lee YS. Curcumin in cancer chemoprevention: molecular targets, pharmacokinetics, bioavailability, and clinical trials. Arch Pharm (Weinheim) 2010; 343: 489–499. [DOI] [PubMed] [Google Scholar]
- 13.Eskelinen MH, Kivipelto M. Caffeine as a protective factor in dementia and Alzheimer's disease. J Alzheimers Dis 2010; 20: Suppl 1: S167–74. [DOI] [PubMed] [Google Scholar]
- 14.Awad D, Prattes M, Kofler L, et al. Inhibiting eukaryotic ribosome biogenesis. BMC Biol 2019; 17: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res 2012; 72: 1602–1607. [DOI] [PubMed] [Google Scholar]
- 16.Langstrom NS, Anderson JP, Lindroos HG, et al. Alzheimer's disease-associated reduction of polysomal mRNA translation. Brain Res Mol Brain Res 1989; 5: 259–269. [DOI] [PubMed] [Google Scholar]
- 17.Ding Q, Markesbery WR, Chen Q, et al. Ribosome dysfunction is an early event in Alzheimer's disease. J Neurosci 2005; 25: 9171–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrade-Moraes CH, Oliveira-Pinto AV, Castro-Fonseca E, et al. Cell number changes in Alzheimer's disease relate to dementia, not to plaques and tangles. Brain 2013; 136: 3738–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyck L, Santamaria ID, Pakkenberg B, et al. An empirical analysis of the precision of estimating the numbers of neurons and glia in human neocortex using a fractionator-design with sub-sampling. J Neurosci Methods 2009; 182: 143–156. [DOI] [PubMed] [Google Scholar]
- 20.Patel S, Howard D, Man A, et al. Donor-Specific transcriptomic analysis of Alzheimer's Disease-Associated hypometabolism highlights a unique donor, ribosomal proteins and microglia. eNeuro 2020; 7: ENEURO.0255-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelvig DP, Pakkenberg H, Stark AK, et al. Neocortical glial cell numbers in human brains. Neurobiol Aging 2008; 29: 1754–1762. [DOI] [PubMed] [Google Scholar]
- 22.Evans HT, Benetatos J, van Roijen M, et al. Decreased synthesis of ribosomal proteins in tauopathy revealed by non-canonical amino acid labelling. EMBO J 2019; 38: e101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JH. Chemotherapy for colorectal cancer in the elderly. World J Gastroenterol 2015; 21: 5158–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hassan MS, Ansari J, Spooner D, Hussain SA. Chemotherapy for breast cancer (review). Oncol Rep 2010; 24: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 25.Angeli E, Nguyen TT, Janin A, et al. How to make anticancer drugs cross the blood-brain barrier to treat brain metastases. Int J Mol Sci 2019; 21: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jefferies WA, Price KA, Biron KE, et al. Adjusting the compass: new insights into the role of angiogenesis in Alzheimer's disease. Alzheimers Res Ther 2013; 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau SF, Cao H, Fu AKY, et al. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer's disease. Proc Natl Acad Sci U S A 2020; 117: 25800–25809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson ECB, Dammer EB, Duong DM, et al. Large-scale proteomic analysis of Alzheimer's disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat Med 2020; 26: 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra S, Palanivelu K. The effect of curcumin (turmeric) on Alzheimer's disease: an overview. Ann Indian Acad Neurol 2008; 11: 13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Han G, Wu K. Tanshinone IIA alleviates the AD phenotypes in APP and PS1 transgenic mice. Biomed Res Int 2016; 2016: 7631801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Jiang YY, Wang XX, et al. Tanshinone IIA suppresses ovarian cancer growth through inhibiting malignant properties and angiogenesis. Ann Transl Med 2020; 8: 1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang CL, Liu YY, Ma YG, et al. Curcumin blocks small cell lung cancer cells migration, invasion, angiogenesis, cell cycle and neoplasia through janus kinase-STAT3 signalling pathway. PLoS One 2012; 7: e37960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012; 11: O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamiie J, Ohtsuki S, Iwase R, et al. Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm Res 2008; 25: 1469–1483. [DOI] [PubMed] [Google Scholar]
- 35.Uchida Y, Higuchi T, Shirota M, et al. Identification and validation of combination plasma biomarker of afamin, fibronectin and sex hormone-binding globulin to predict pre-eclampsia. Biol Pharm Bull 2021; 44: 804–815. [DOI] [PubMed] [Google Scholar]
- 36.Uchida Y, Sasaki H, Terasaki T. Establishment and validation of highly accurate formalin-fixed paraffin-embedded quantitative proteomics by heat-compatible pressure cycling technology using phase-transfer surfactant and SWATH-MS. Sci Rep 2020; 10: 11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogata S, Ito S, Masuda T, et al. Efficient isolation of brain capillary from a single frozen mouse brain for protein expression analysis. J Cereb Blood Flow Metab 2021; 41: 1026–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida Y, Ohtsuki S, Katsukura Y, et al. Quantitative targeted absolute proteomics of human blood-brain barrier transporters and receptors. J Neurochem 2011; 117: 333–345. [DOI] [PubMed] [Google Scholar]
- 39.Uchida Y, Tachikawa M, Obuchi W, et al. A study protocol for quantitative targeted absolute proteomics (QTAP) by LC-MS/MS: application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood-brain barrier in ddY, FVB, and C57BL/6J mice. Fluids Barriers CNS 2013; 10: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchida Y, Sumiya T, Tachikawa M, et al. Involvement of claudin-11 in disruption of blood-brain, -spinal cord, and -arachnoid barriers in multiple sclerosis. Mol Neurobiol 2019; 56: 2039–2056. [DOI] [PubMed] [Google Scholar]
- 41.Uchida Y, Goto R, Takeuchi H, et al. Abundant expression of OCT2, MATE1, OAT1, OAT3, PEPT2, BCRP, MDR1, and xCT transporters in blood-arachnoid barrier of pig and polarized localizations at CSF- and blood-facing plasma membranes. Drug Metab Dispos 2020; 48: 135–145. [DOI] [PubMed] [Google Scholar]
- 42.Wilson DN, Doudna Cate JH. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol 2012; 4: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shrimal S, Cherepanova NA, Gilmore R. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin Cell Dev Biol 2015; 41: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 2006; 16: 47R–62R. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Canada C, Kelleher DJ, Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 2009; 136: 272–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsson I, Kelleher DJ, Miao Y, et al. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J Cell Biol 2003; 161: 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roboti P, High S. The oligosaccharyltransferase subunits OST48, DAD1 and KCP2 function as ubiquitous and selective modulators of mammalian N-glycosylation. J Cell Sci 2012; 125: 3474–3484. [DOI] [PubMed] [Google Scholar]
- 48.Honma K, Iwao-Koizumi K, Takeshita F, et al. RPN2 gene confers docetaxel resistance in breast cancer. Nat Med 2008; 14: 939–948. [DOI] [PubMed] [Google Scholar]
- 49.Wilson CM, High S. Ribophorin I acts as a substrate-specific facilitator of N-glycosylation. J Cell Sci 2007; 120: 648–657. [DOI] [PubMed] [Google Scholar]
- 50.Satoh T, Toshimori T, Yan G, et al. Structural basis for two-step glucose trimming by glucosidase II involved in ER glycoprotein quality control. Sci Rep 2016; 6: 20575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barker MK, Rose DR. Specificity of processing alpha-glucosidase I is guided by the substrate conformation: crystallographic and in silico studies. J Biol Chem 2013; 288: 13563–13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasak VW, Ordovas JM, Elbein AD, et al. Castanospermine inhibits glucosidase I and glycoprotein secretion in human hepatoma cells. Biochem J 1985; 232: 759–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilkinson BM, Purswani J, Stirling CJ. Yeast GTB1 encodes a subunit of glucosidase II required for glycoprotein processing in the endoplasmic reticulum. J Biol Chem 2006; 281: 6325–6333. [DOI] [PubMed] [Google Scholar]
- 54.Ellgaard L, Frickel EM. Calnexin, calreticulin, and ERp57: teammates in glycoprotein folding. Cell Biochem Biophys 2003; 39: 223–247. [DOI] [PubMed] [Google Scholar]
- 55.Solda T, Garbi N, Hammerling GJ, et al. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem 2006; 281: 6219–6226. [DOI] [PubMed] [Google Scholar]
- 56.High S, Lecomte FJ, Russell SJ, et al. Glycoprotein folding in the endoplasmic reticulum: a tale of three chaperones? FEBS Lett 2000; 476: 38–41. [DOI] [PubMed] [Google Scholar]
- 57.Molinari M, Eriksson KK, Calanca V, et al. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell 2004; 13: 125–135. [DOI] [PubMed] [Google Scholar]
- 58.Blanco-Herrera F, Moreno AA, Tapia R, et al. The UDP-glucose: glycoprotein glucosyltransferase (UGGT), a key enzyme in ER quality control, plays a significant role in plant growth as well as biotic and abiotic stress in Arabidopsis thaliana. BMC Plant Biol 2015; 15: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato N, Urano F, Yoon Leem J, et al. Upregulation of BiP and CHOP by the unfolded-protein response is independent of presenilin expression. Nat Cell Biol 2000; 2: 863–870. [DOI] [PubMed] [Google Scholar]
- 60.Okada T, Ninagawa S, Mori K. Mannose trimming mechanism in endoplasmic reticulum-associated degradation of glycoproteins. Seikagaku 2016; 88: 257–260. [PubMed] [Google Scholar]
- 61.Kamiya Y, Yamaguchi Y, Takahashi N, et al. Sugar-binding properties of VIP36, an intracellular animal lectin operating as a cargo receptor. J Biol Chem 2005; 280: 37178–37182. [DOI] [PubMed] [Google Scholar]
- 62.D'Arcangelo JG, Stahmer KR, Miller EA. Vesicle-mediated export from the ER: COPII coat function and regulation. Biochim Biophys Acta 2013; 1833: 2464–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bielli A, Haney CJ, Gabreski G, et al. Regulation of Sar1 NH2 terminus by GTP binding and hydrolysis promotes membrane deformation to control COPII vesicle fission. J Cell Biol 2005; 171: 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antonny B, Madden D, Hamamoto S, et al. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol 2001; 3: 531–537. [DOI] [PubMed] [Google Scholar]
- 65.Burger K, Muhl B, Harasim T, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem 2010; 285: 12416–12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer 2018; 18: 51–63. [DOI] [PubMed] [Google Scholar]
- 67.Delloye-Bourgeois C, Goldschneider D, Paradisi A, et al. Nucleolar localization of a netrin-1 isoform enhances tumor cell proliferation. Sci Signal 2012; 5: ra57. [DOI] [PubMed] [Google Scholar]
- 68.Gilles A, Frechin L, Natchiar K, et al. Targeting the human 80S ribosome in cancer: from structure to function and drug design for innovative adjuvant therapeutic strategies. Cells 2020; 9: 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giordano A, Tommonaro G. Curcumin and cancer. Nutrients 2019; 11: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu Y, Wu Y, Wang L, et al. Identification of curcumin as a novel natural inhibitor of rDNA transcription. Cell Cycle 2020; 19: 3362–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin J, Hulette C, Wang Y, et al. Proteomic identification of a stress protein, mortalin/mthsp70/GRP75: relevance to Parkinson disease. Mol Cell Proteomics 2006; 5: 1193–1204. [DOI] [PubMed] [Google Scholar]
- 72.Li Y, Yu H, Chen C, et al. Proteomic profile of mouse brain aging contributions to mitochondrial dysfunction, DNA oxidative damage, loss of neurotrophic factor, and synaptic and ribosomal proteins. Oxid Med Cell Longev 2020; 2020: 5408452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anisimova AS, Meerson MB, Gerashchenko MV, et al. Multifaceted deregulation of gene expression and protein synthesis with age. Proc Natl Acad Sci U S A 2020; 117: 15581–15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ubaida-Mohien C, Lyashkov A, Gonzalez-Freire M, et al. Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. Elife 2019; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marthandan S, Baumgart M, Priebe S, et al. Conserved senescence associated genes and pathways in primary human fibroblasts detected by RNA-Seq. PLoS One 2016; 11: e0154531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peters MJ, Joehanes R, Pilling LC, NABEC/UKBEC Consortium et al. The transcriptional landscape of age in human peripheral blood. Nat Commun 2015; 6: 8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jung M, Jin SG, Zhang X, et al. Longitudinal epigenetic and gene expression profiles analyzed by three-component analysis reveal down-regulation of genes involved in protein translation in human aging. Nucleic Acids Res 2015; 43: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fando JL, Salinas M, Wasterlain CG. Age-dependent changes in brain protein synthesis in the rat. Neurochem Res 1980; 5: 373–383. [DOI] [PubMed] [Google Scholar]
- 79.Wijesuriya HC, Bullock JY, Faull RL, et al. ABC efflux transporters in brain vasculature of Alzheimer's subjects. Brain Res 2010; 1358: 228–238. [DOI] [PubMed] [Google Scholar]
- 80.Al-Majdoub ZM, Al Feteisi H, Achour B, et al. Proteomic quantification of human Blood-Brain barrier SLC and ABC transporters in healthy individuals and dementia patients. Mol Pharm 2019; 16: 1220–1233. [DOI] [PubMed] [Google Scholar]
- 81.Burt BM, Humm JL, Kooby DA, et al. Using positron emission tomography with [(18)F]FDG to predict tumor behavior in experimental colorectal cancer. Neoplasia 2001; 3: 189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marcus DL, de Leon MJ, Goldman J, et al. Altered glucose metabolism in microvessels from patients with Alzheimer's disease. Ann Neurol 1989; 26: 91–94. [DOI] [PubMed] [Google Scholar]
- 83.Mullins R, Reiter D, Kapogiannis D. Magnetic resonance spectroscopy reveals abnormalities of glucose metabolism in the Alzheimer's brain. Ann Clin Transl Neurol 2018; 5: 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang AC, Vest RT, Kern F, et al. A human brain vascular atlas reveals diverse mediators of Alzheimer's risk. Nature 2022; 603: 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen W, Liang T, Zuo W, et al. Neuroprotective effect of 1-Deoxynojirimycin on cognitive impairment, beta-amyloid deposition, and neuroinflammation in the SAMP8 mice. Biomed Pharmacother 2018; 106: 92–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-jcb-10.1177_0271678X221111602 for Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer's disease patients by Masayoshi Suzuki, Kenta Tezuka, Takumi Handa, Risa Sato, Hina Takeuchi, Masaki Takao, Mitsutoshi Tano and Yasuo Uchida in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-xlsx-2-jcb-10.1177_0271678X221111602 for Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer's disease patients by Masayoshi Suzuki, Kenta Tezuka, Takumi Handa, Risa Sato, Hina Takeuchi, Masaki Takao, Mitsutoshi Tano and Yasuo Uchida in Journal of Cerebral Blood Flow & Metabolism
Supplemental material, sj-pdf-3-jcb-10.1177_0271678X221111602 for Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer's disease patients by Masayoshi Suzuki, Kenta Tezuka, Takumi Handa, Risa Sato, Hina Takeuchi, Masaki Takao, Mitsutoshi Tano and Yasuo Uchida in Journal of Cerebral Blood Flow & Metabolism