ABSTRACT
Children are prone to bloodstream infections (BSIs), the rapid and accurate diagnosis of which is an unmet clinical need. The T2MR technology is a direct molecular assay for identification of BSI pathogens, which can help to overcome the limits of blood culture (BC) such as diagnostic accuracy, blood volumes required, and turnaround time. We analyzed results obtained with the T2Bacteria (648) and T2Candida (106) panels in pediatric patients of the Bambino Gesù Children’s Hospital between May 2018 and September 2020 in order to evaluate the performance of the T2Dx instrument with respect to BC. T2Bacteria and T2Candida panels showed 84.2% and 100% sensitivity with 85.9% and 94.1% specificity, respectively. The sensitivity and specificity of the T2Bacteria panel increased to 94.9% and 98.7%, respectively, when BC was negative but other laboratory data supported the molecular result. T2Bacteria sensitivity was 100% with blood volumes <2 mL in neonates and infants. T2Bacteria and T2Candida provided definitive microorganism identification in a mean time of 4.4 and 3.7 h, respectively, versus 65.7 and 125.5 h for BCs (P < 0.001). T2 panels rapidly and accurately enable a diagnosis of a pediatric BSI, even in children under 1 year of age and for very small blood volumes. These findings support their clinical use in life-threatening pediatric infections, where the time to diagnosis is of utmost importance, in order to improve survival and minimize the long-term sequalae of sepsis. The T2 technology could be further developed to include more bacteria and fungi species that are involved in the etiology of sepsis.
KEYWORDS: T2 magnetic resonance, T2 panels, bloodstream infections, molecular diagnosis, pediatric population
INTRODUCTION
Bloodstream infections (BSIs) and sepsis are major causes of death and disability across the age spectrum, including in infants and children. Neonates and young children under the age of five are particularly vulnerable to severe or lethal BSI (1, 2). In 2017, almost half of all sepsis cases worldwide occurred in children, and 2.9 million of those under the age of five died (3–5).
The clinical onset of pediatric sepsis can be insidious and difficult to readily identify (6). In May 2017, the World Health Organization (WHO) adopted the need to improve sepsis recognition as a global health priority. The current gold standard for proven BSI definition requires the isolation of bacteria or yeasts from blood cultures (BCs), a procedure burdened by a prolonged laboratory turnaround time, suboptimal performance for low-volume samples (common in children), and frequent contamination by commensal flora (7–11). More sensitive, rapid, and versatile diagnostic tools are thus needed in this vulnerable population (12, 13).
One of the most innovative and promising technologies for pathogen detection is T2 magnetic resonance (T2MR, T2 Biosystems, Lexington MA, USA), whose nanodiagnostic T2Candida and T2Bacteria panels can detect clinically relevant pathogens from a single fresh whole blood specimen with no need for previous BC, and in a fully automated process requiring few hours of laboratory turnaround time. The T2Candida panel is designed to detect the presence of five Candida species accounting for >90% of candidemia cases in both adult and pediatric populations (14, 15). The T2Bacteria panel enables multiplex detection of the ESKAPEc pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Escherichia coli), which represent the major health care threat (16–18). Both panels have been recently cleared by the Food and Drug Administration (FDA) and have received the Conformité Européenne (CE) mark.
In adults, validation studies have shown per-assay sensitivities of 90% for T2Bacteria and 89–91% for the T2Candida panel, while specificity was 98% for both panels (19–21). These results have been successfully confirmed in a small number of real-life studies in adults but never in children (19, 22–27). Notably, the pediatric population has specific epidemiological (type of microorganisms involved in BSIs), clinical (rapid progression of clinical conditions, particularly in immunocompromised patients), and technical (smaller volume of blood collected) characteristics that preclude the tout court translation of the results obtained in the adult population.
In this study, we sought to verify the usefulness of the T2Bacteria and T2Candida panels of the T2Dx instrument in diagnosing BSI in hospitalized pediatric patients. We evaluated the reduction in the response time (turnaround time) and diagnostic accuracy of the molecular test with respect to blood culture, in light of the characteristics of this age group.
MATERIALS AND METHODS
Study design and participants.
In 2018, Bambino Gesù Children’s Hospital adopted the T2MR as a routine diagnostic tool for assessment of suspected BSIs in association with traditional BC. This retrospective single-center diagnostic accuracy study included consecutive patients aged 0–21 years with a diagnostic BC and a T2MR test requested simultaneously during their hospitalization, from 1 May 2018 to 30 September 2020. No specific criteria were used to define suspected BSI; BC requests and antimicrobial treatments were at the discretion of the treating physicians, and based on local internal protocols. Exclusion criteria comprised previous inclusion of the same patient, age >21 years, and an invalid T2MR result. The institutional review board approved the study protocol (2439_OPBG_2021).
Measurements.
At least one aerobic BC, and whole blood samples for T2MR, were drawn in this order from the same vein or central venous line. BCs were performed in accordance with hospital practices and international recommendations for pediatric populations (8, 9, 28). Each BC bottle was inoculated with 1–3 mL (Bactec Peds Plus/F medium) or 8–10 mL (Bactec Plus Aerobic/F medium, Bactec Lytic/10 Anaerobic/F medium vials, Mycosis IC/F Bactec) of whole blood and incubated at 35°C in a Bactec 9240/70FX BC system (BD Diagnostics). MALDI-TOF Mass Spectrometry (Bruker Daltoniks) or Vitek 2 (bioMérieux) were used for microorganism identification. BCs that did not yield a pathogen were incubated for at least 5 (bacteria) or 10 days (fungi).
When the blood volume was below 2 mL (minimum volume for direct loading), the samples were diluted with General Purpose Buffer (T2 Biosystems) and then loaded, according to the manufacturer’s instructions.
Reference standard.
T2MR was considered positive if one or more targeted microorganisms were detected, and negative otherwise. Positive T2Bacteria results were reported as E. faecium, S. aureus, K. pneumoniae, A. baumannii, P. aeruginosa, and E. coli. Positive T2Candida results were reported as C. albicans/C. tropicalis, C. krusei/C. glabrata, and C. parapsilosis.
A proven BSI was defined when a T2-targeted microorganism was detected in at least one BC bottle (9). By adapting a previous classification proposed by Nguyen and colleagues, BC-negative/T2MR-positive results were defined as probable BSIs if the T2-detected microorganism was isolated within 10 days in a nonconcurrent BC (19), or in specimens collected from other sites or biological fluids compatible with a plausible source of infection (i.e., abdomen, urine, lungs). Rectal surveillance swabs were taken into account only in the presence of laboratory abnormalities suggestive of BSI. Following Nguyen and colleagues, BC-negative/T2MR-positive results were always defined as possible BSIs if the T2-detected microorganism was a plausible cause of disease, according to altered clinical parameters and/or response to ongoing antimicrobial therapy (19, 29, 30). Patients were considered to be receiving an active treatment if they received at least one dose in the 2 days before the sample’s collection.
Statistical analysis.
The 95% CIs for sensitivity and specificity were calculated using the exact Clopper Pearson method. A multivariable logistic regression model with interaction between age group and whole blood volume was used to evaluate T2 overdetection probabilities within age classes by volume (6). To avoid problems due to small numbers and consequent perfect class separation, a Firth penalized logistic regression was used (31). The analyses were conducted using SPSS v.23 and R 4.0.5.
RESULTS
Patient enrollment and sampling.
The final study population consisted of 754 patients: 648 samples from 648 patients for the T2Bacteria panel, and 106 samples from 106 patients for the T2Candida panel (Figure A1in the supplemental material). Table 1 reports the characteristics of the study population. The median (interquartile range, IQR) age was 6.0 (2.2–12.8) years. Younger age groups (<5 years) accounted for 48.9% of the overall population (n/N = 369/754), with 27 suspected BSIs investigated in neonates, 108 in infants, and 234 in toddlers.
TABLE 1.
Baseline characteristics of the study population
| Characteristic | T2Bacteria pediatric population, N = 648 | T2Candida pediatric population, N = 106 |
|---|---|---|
| Pediatric age groups, n (%) | ||
| Neonate (1 wk to 1 mo) | 24 (3.7) | 3 (2.8) |
| Infant (1 mo to 1 yr) | 96 (14.8) | 12 (11.3) |
| Toddler and preschool (2–5 yrs) | 204 (31.5) | 30 (28.3) |
| School age child (6–12 yrs) | 146 (22.5) | 30 (28.3) |
| Adolescent (13–18 yrs) | 148 (22.8) | 29 (27.4) |
| Young adult (19–21 yrs) | 30 (4.6) | 2 (1.9) |
| Hematological malignancy, n (%) | 309 (47.7) | 74 (69.8) |
| Admitted to the intensive care unit, n (%) | 85 (13.1) | 13 (12.3) |
| White blood cell count, n (%) | ||
| Within normal limits | 240 (37.0) | 29 (27.4) |
| Leukocytosis | 95 (14.7) | 8 (7.5) |
| Leukopenia | 313 (48.3) | 69 (65.1) |
| Body temp ≥37.5°C, n (%)a | 399 (88.5) | 61 (82.4) |
| C-reactive protein (mg/dL), n (%)b | ||
| ≤0.5 | 88 (13.6) | 13 (12.3) |
| >0.5 | 559 (86.4) | 93 (87.7) |
| Procalcitonin (ng/mL), n (%)c | ||
| ≥0.5 | 239 (50.7) | 43 (50.0) |
| <0.5 | 232 (49.3) | 43 (50.0) |
| Central venous catheter, n (%) | 329 (50.8) | 66 (62.3) |
| Initial whole blood vol sampled (mL), Median (IQR)d | 2.5 (2.0–3.0) | 2.5 (2.0–3.0) |
| <2 mL | 115 (17.7) | 15 (14.1) |
| 2–3 mL | 292 (45.1) | 55 (51.9) |
| ≥3 mL | 241 (37.2) | 36 (34.0) |
| Initial whole blood vol sampled ≥2 mL, n (%) | 532 (82.1) | 91 (85.8) |
Information available for 451 patients in T2Bacteria population, and 74 patients in T2Candida population.
Information available for 647 patients in T2Bacteria population, and 106 patients in T2Candida population.
Information available for 471 patients in T2Bacteria population, and 86 patients in T2Candida population.
IQR, interquartile range.
For the purpose of T2MR processing, 3 mL of whole blood was successfully obtained only in 37.2% (241/648) of the T2Bacteria and 34.0% (36/106) of the T2Candida population, independent of age. In both populations, blood volumes most frequently comprised between 2 and 3 mL (45.1% [292/648] and 51.9% [55/106], respectively). Even smaller volumes (<2 mL) for T2Bacteria andT2Candida were drawn in 37.5% (9/24) to 66.7% (2/3) of neonates, 34.4% (33/96) to 41.6% (5/12) of infants, and 19.6% (40/204) to 3.3% (4/30) of toddlers/preschoolers, respectively, as well as, unexpectedly, in a minority of school-age children, adolescents, and young adults (Table A1).
Performance of T2Bacteria panel.
BC resulted positive in 75/648 (11.6%) of patients, of whom 53/75 (70.7%) were classified as proven BSI episodes and 22/75 (29.3%) as contaminations. T2Bacteria-targeted microorganisms (ESKAPEc) accounted for 71.7% (38/53) of all proven BSI episodes. T2Bacteria yielded positive results in 120/648 of patients (18.5%), and it was concordant with the corresponding BC in 85.8% of cases (556/648). The overall spectrum of bacteria identified according to the result of concurrent BC is reported in Fig. 1a. The combined performance of T2Bacteria and BC in the diagnosis of proven BSIs caused by T2-targeted bacteria is reported in Table 2. We identified 38 proven BSIs by 39 T2-targeted bacteria. The most frequently isolated pathogen was S. aureus (26.3%, 10/38), followed by K. pneumoniae and P. aeruginosa (23.7% each, 9/38), E. faecium and E. coli (13.2% each, 5/38), and A. baumannii (2.6%, 1/38). In 11 samples, additional T2Bacteria-targeted organisms were identified by T2Bacteria assay, but they were not identified in concurrent blood cultures (Table A2).
FIG 1.
T2 magnetic resonance results in pediatric patients with suspected bloodstream infections, in comparison with concurrent blood culture. Microorganisms identified by T2Bacteria (Panel a) and T2Candida (Panel b) are shown. aTwelve microorganisms identified in 9 patients. bInstead of Enterococcus faecium isolated in blood culture. cInstead of Klebsiella pneumoniae isolated in blood culture. dIsolation of Staphylococcus aureus (n = 2), Enterococcus faecium (n = 1), and Klebsiella pneumoniae (n = 1) in blood cultures.
TABLE 2.
Performance of T2Bacteria and T2Candida panels in diagnosis of bloodstream infections episodes by T2-targeted microorganismsa
| Panel | n | Prevalence (95% CI) | Mean time to blood culture positivity in hours (SD) | Mean time to identification after positive blood culture in hours (SD) | Mean time to T2 positivity in hours (SD) | T2 panels performance |
|||
|---|---|---|---|---|---|---|---|---|---|
| Sensitivity (95% CI) | Specificity (95% CI) | LR+ | LR− | ||||||
| T2Bacteria panel | |||||||||
| Proven BSIs | 38 | 5.9% (4.2–8) | 10.9 (5.5) | 65.7 (24.5) | 4.4 (0.7) | 84.2% (81.1–86.9) | 85.9% (82.9–88.4) | 6.0 | 0.2 |
| Microorganisms responsible for proven BSIs, N (%)b | |||||||||
| Staphylococcus aureus | 10 | 1.5% (0.8–2.9) | 12.3 (6.8) | 54.6 (16.6) | 4.4 (0.7) | 80.0% (76.7–83.0) | 98.7% (97.5–99.4) | 63.8 | 0.2 |
| Klebsiella pneumoniae | 9 | 1.4% (0.7–2.7) | 6.9 (4.3) | 64.2 (21.3) | 4.2 (0.8) | 77.8% (74.3–80.9) | 95.9% (94.0–97.3) | 19.1 | 0.2 |
| Pseudomonas aeruginosa | 9 | 1.4% (0.7–2.7) | 13.4 (6.5) | 77.7 (31.0) | 4.3 (0.7) | 100% (99.3–100) | 95.9% (94.0–97.3) | 24.6 | 0.0 |
| Enterococcus faecium | 5 | 0.8% (0.3–1.9) | 12.0 (4.6) | 44.6 (16.4) | 4.5 (0.7) | 60% (56.1–63.8) | 97.8% (96.3–98.8) | 27.6 | 0.4 |
| Escherichia coli | 5 | 0.8% (0.3–1.9) | 9.5 (2.0) | 76.0 (27.1) | 4.7 (1.0) | 100% (99.3–100) | 97.2% (95.5–98.3) | 35.7 | 0.0 |
| Acinetobacter baumannii | 1 | 0.2% (0–1.0) | 12.2 | 62.3 | 3.9 | 100% (99.3–100) | 97.7% (96.1–98.6) | 43.1 | 0.0 |
| Proven and probable BSIs | 66 | 10.2% (8.1–12.9) | 4.4 (0.9) | 90.9% (88.4–93) | 90.0% (87.3–92.1) | 9.1 | 0.1 | ||
| Proven, probable and possible BSIs | 117 | 18.2% (15.3–21.4) | 4.5 (0.9) | 94.9% (92.8–96.4) | 98.7% (97.4–99.4) | 71.4 | 0.1 | ||
| T2Candida panel | |||||||||
| Proven BSIs | 4 | 3.8% (1.2–9.9) | 36.9 (37.1) | 125.5 (20.8) | 3.7 (0.7) | 100% (95.6–100) | 94.1% (87.3–97.5) | 17 | 0.0 |
| Candida species responsible for proven BSIs, N (%) | |||||||||
| Candida parapsilosis | 2 | 1·9% (0.3–7.3) | 17.8 (20.1) | 121.2 (29.7) | 4.1 (0.7) | 100% 95.6–100) | 99% (94.1–99.9) | 104.0 | 0.0 |
| Candida albicans/Candida tropicalis | 2 | 1.9% (0.3–7.3) | 56.0 (47.7) | 129.8 (18.6) | 3.4 (0.5) | 100% (95.6–100) | 96.2% (90–98.7) | 26.0 | 0.0 |
| Proven, probable and possible BSIs | 10 | 9.4% (4.9–17.1) | 3.7 (0.7) | 100% (95.6–100) | 100% (95.6–100) | ∞ | 0.0 | ||
BSIs, bloodstream infections; CI, confidence interval; LR+, positive likelihood ratio; LR−, negative likelihood ratio; SD, standard deviation.
In 1 patient, 2 T2Bacteria-targeted organisms were identified in the same blood culture. Therefore, 39 microorganisms were identified in 38 proven infection cases. In 11 samples, additional T2Bacteria-targeted organisms were identified by T2Bacteria assay but they were not identified in concurrent blood cultures (Table A2).
The per-patient sensitivity of T2Bacteria for proven BSIs caused by T2-targeted bacteria was 84.2% (95% CI:81.1–86.9). The per-organism sensitivity ranged from 60% (95% CI:56.1–63.8) for E. faecium to 100% (95% CI:99.3–100) for E. coli, A. baumannii, and P. aeruginosa. The identification results were provided in a mean of 4.4 (SD: 0.7) hours, a time significantly shorter than that required for BC to turn positive (mean [SD] = 10.9 [5.5] hours; P < 0.001), and to allow the final microorganism identification (mean [SD] = 65.7 [24.5] hours; P < 0.001).
T2Bacteria was discordant with BC in 15.8% of proven BSIs caused by T2-targeted bacteria (6/38), due to four false-negative results and two different identifications, detailed in Table A3.
Non-T2Bacteria-targeted microorganisms accounted for 28.3% (15/53) of all proven BSI episodes and included coagulase-negative staphylococci (n = 5), Enterococcus faecalis (n = 3), Enterobacter cloacae (n = 2), Streptococcus pneumoniae (n = 1), Streptococcus spp. (n = 1), Achromobacter xylosoxidans (n = 1), Acinetobacter junii (n = 1), and Salmonella spp. (n = 1). There were another 22 BCs positive for coagulase-negative staphylococci and other non-ESKAPEc bacteria that were classified as contaminants upon in-depth clinical analysis as per the hospital and laboratory protocols.
Among the 610 patients with negative BC or BC negative for T2-targeted bacteria, T2Bacteria excluded the presence of a proven BSI caused by the ESKAPEc with 85.9% (95% CI:82.9–88.4) specificity, and 0.2 negative likelihood ratio.
T2Bacteria identified 28 probable BSIs and 51 possible BSIs in samples defined negative by BC, in a mean of 4.5 (SD: 1.1) and 4.6 (SD: 1.0) hours, respectively. Only seven putative false-positive results were observed, with an excellent positive likelihood ratio for proven and probable and possible BSIs diagnosis (71.4). As detailed in Table 2, the per-patient sensitivity of T2Bacteria for proven and probable BSIs was 90.9% (95% CI:88.4–93); it increased to 94.9% (95% CI:92.8–96.4) when proven, probable, and possible BSIs were analyzed together. Similarly, the per-patient specificity was 90.0% (95% CI:87.5–92.3) for proven and probable BSIs, and 98.7% (95% CI:97.4–99.4) for proven, probable, and possible BSIs, with 0.1 negative likelihood ratios in all cases.
Patients with probable BSIs (18.5%, 5/27) and with possible BSIs (24.5%, 12/49), all with available data, had been receiving a potentially active antimicrobial treatment for >48 h at the time of testing, in addition to the 25% of patients with proven BSIs (9/36 with available data).
Role of age and blood volumes on T2Bacteria performance.
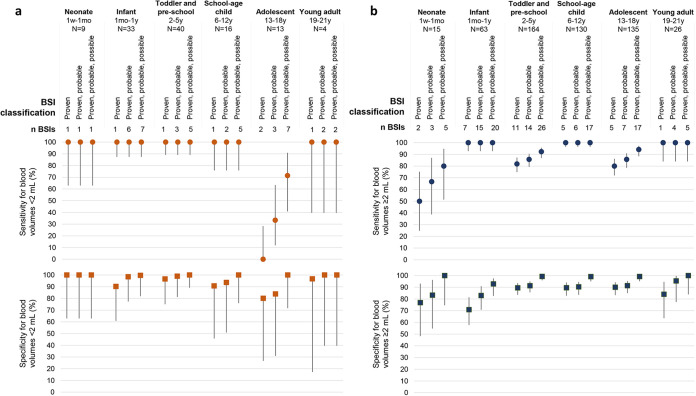
Both sensitivity and specificity of T2Bacteria were consistently high across the spectrum of age and blood volumes analyzed (Fig. 2; Table A4). Notably, initial blood volumes <2 mL allowed a stable sensitivity of 100% for BSIs diagnosis in all age groups, with the exception of adolescents.
FIG 2.
T2Bacteria panel performance in specific age groups and for different initial blood volume processed. Sensitivity (dots) and specificity (squares) values and 95% confidence intervals for T2Bacteria panel in proven, proven and probable, and proven, probable, and possible BSIs are reported, classified for initial blood volume samples for T2Bacteria panel performance lower (Panel a, orange dots) or higher (Panel b, blue dots) than 2 mL.
The rate of T2MR overdetection was higher in 13 adolescents from whom initial blood volumes <2 mL were drawn (38.5% [5/13], Fig. 3). According to the penalized logistic regression, the interaction between blood volume and age group was borderline statistically significant (P = 0.069), supporting the observed differential association between T2MR overdetections and blood volumes within the different age groups.
FIG 3.
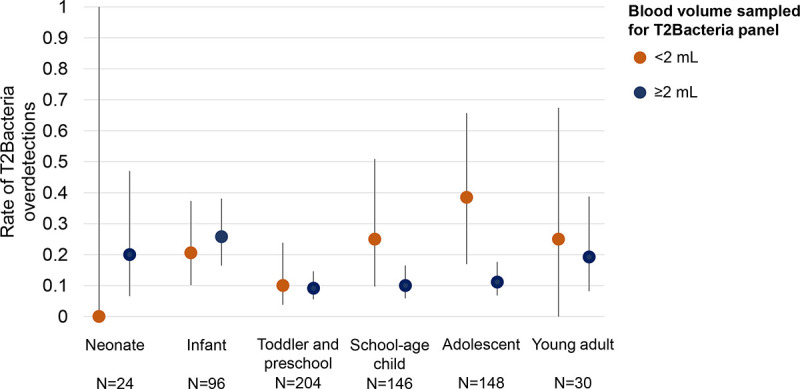
Rate of T2Bacteria panel overdetections compared to concurrent blood culture. Rates (and 95% confidence intervals) of samples in which T2Bacteria panel provided additional microorganisms identifications compared to standard blood culture are reported for each age group, classified for initial blood volume samples for T2Bacteria panel performance lower (orange) or higher (blue) than 2 mL.
Performance of T2Candida panel.
Four proven fungal BSIs were identified in 4/106 children, all caused by T2-targeted microorganisms (C. parapsilosis, n = 2; C. albicans, n = 1; C. tropicalis, n = 1) (Fig. 1b). T2Candida successfully identified all four proven BSIs, thus showing 100% sensitivity, in a significantly shorter time than BC (mean [SD] of 3.7 [0.7] hours versus 125.5 [20.8] hours, respectively; P < 0.001) (Table 2).
The specificity of T2Candida in excluding a proven BSI was 94.1% (95% CI:87.3–97.5). However, T2Candida supported the diagnosis of six additional cases of probable (n = 5) and possible (n = 1) fungal BSIs (Fig. 1b). No false-positive T2Candida results were highlighted. Consequently, per-patient specificity for proven/probable/possible BSIs was 100%, and negative likelihood ratio consistently equal to 0.
DISCUSSION
T2MR is a novel technology for a direct-from-blood rapid identification of the ESKAPEc bacteria and the five Candida species most commonly involved in BSIs. To the best of our knowledge, this is the first study evaluating the clinical performance of both T2Bacteria and T2Candida panels in diagnosing BSIs across the whole spectrum of childhood age.
In our large real-life cohort study, both panels demonstrated a significant advantage over BC (current gold standard) in terms of time to identification, while ensuring consistent sensitivity and specificity. T2Bacteria panel provided definitive microorganism identification in an average time of 4.4 h, compared to 76.6 h for BC, with 84.2% sensitivity and 85.9% specificity for proven BSIs caused by T2-targeted bacteria (n = 38), which accounted for 71.7% (38/53) of all BC-proven BSI episodes. When either probable BSIs (n = 28) or both probable and possible BSIs (n = 79) were classified as true positives missed by concurrent BCs, specificity rose to 90.0% and 98.7%, respectively. The T2Candida panel covered all fungal species associated with BC-proven BSIs in our population, and consistently provided definitive identification in 3.7 h versus 162.4 h for BC, with 100% sensitivity and 94.1% specificity for proven fungal BSIs. Specificity reached 100% when both probable (n = 5) and possible (n = 1) BSIs were considered true positives missed by BC. When the three categories of BSIs were analyzed together, both T2Bacteria and T2Candida demonstrated an excellent ability in excluding a BSI when a negative result was obtained, with negative likelihood ratios of 0.1 and 0.0, respectively.
Overall, our results validate the previous findings and extend them to the pediatric population. In our study, the use of T2MR alongside BC provided significant added values in terms of time to results, support for BSIs exclusion with optimal negative likelihood ratios, and additional identification of BSIs missed by BCs (19–21, 24). It should be noted that the pediatric population is characterized by peculiarities such as the lower availability of adequate blood volumes, which can make an accurate microbiological diagnosis challenging.
T2MR microorganism overdetection compared to BC was the most common reason of discordance (96.3%, 103/107), out of which 89.3% (92/103) was caused by positive T2MR results with negative concurrent BC, and there were very few (n = 7) putative false-positive results. Interestingly, probable and possible BSIs outnumbered proven BSIs by around 2-fold in both T2MR panels. Similar findings have been reported in adults (19, 20). The clinical significance of the frequent situations in which T2MR is positive while BC is negative is still debated, partly because BC, considered the gold standard assay, has well-known limitations, some of which are difficult to account for in retrospective real-life studies as large as ours.
Thanks to a highly efficient assay workflow, T2MR is able to specifically identify circulating live pathogens (either free or white cell encapsulated), thus avoiding false-positive results associated with freely circulating DNA (32, 33). In addition, all T2MR/BC assays were ordered by clinicians on the basis of a strong BSI suspicion. These facts further support the reliability of a positive T2 result even in the presence of a negative BC, in addition to the isolation of the same pathogen in nonconcurrent BCs or other specimens (defining probable BSIs), or the compatibility of laboratory parameters (defining possible BSIs). Notably, the 13.6% (41/301) of BC-negative children under 6 years of age were T2Bacteria positive (51.9% [41/79] of the total cases of BC negative, T2MR overdetections), an interesting finding in light of the known difficulties in collecting adequate blood volumes for a satisfactory BC performance in the youngest children.
Blood volume is widely recognized as the most important factor affecting BC sensitivity and specificity (7, 34). Although there are several recommendations based on weight and age in the pediatric setting, the difficulty of adhering to these standards is well known in clinical practice, responsible for a decreased probability of isolating microorganisms and an extended time to detection (7, 35–37). T2MR protocol usually requires 3 mL of whole blood, and the lowest volume supported by the cartridges is 2 mL. This represents a nonnegligible amount, especially in children under 1 year of age, and in virtue of the necessity of an additional sample for concurrent BC (8, 9). However, we found that 0.5–1.9 mL of blood diluted with General Purpose Buffer up to 2 mL allowed the identification of bacterial BSIs with a sensitivity stable at 100% in children under 13. These results support the feasible use of T2MR with minimum amounts of blood for younger ages, in addition to the possibility of loading 2 to 3 mL samples directly into the cartridges, a method reported to result in a 100% concordance between T2Candida and BC in a small study (38). This result could be extended also to the older age groups, penalized in the present study by an excessive reduction of poststratification numerosity. For example, the reduced sensitivity of T2Bacteria for volumes of <2 mL in adolescents refers to only two proven BSIs caused by T2-targeted bacteria out of 13 patients. In adults, the high 95% CI of sensitivity prevented definite conclusions.
Previous antibiotic treatment is another known confounder for BC performance (39). When administered before BC collection, it can decrease the recovery rate by 45%–50% (8). In our study, similar proportions of patients with proven, probable, or possible BSIs were receiving an active antimicrobial treatment for >48 h at the time of T2MR analysis (18–25%). However, this rate is likely underestimated as most of our patients had chronic (i.e., oncological/hematological) and/or critical conditions (i.e., admission to intensive care unit) associated with frequent and/or prolonged use of broad-spectrum antimicrobials whose history could not be reconstructed. Therefore, it is more plausible that BC failed, rather than that T2MR overdetected the relevant pathogens in patients already undergoing an antibiotic treatment.
Etiology and epidemiology of pediatric BSIs are somewhat unique compared to adults, especially in younger ages when the involvement of pathogens not included in T2Bacteria and T2Candida panels, such as coagulase-negative staphylococci, Streptococcus spp., non-E. coli/K. pneumoniae, and Enterobacterales, is not infrequent.
In our cohort, while the T2Candida panel covered all species involved in proven fungal BSIs, T2Bacteria could not identify the 28.3% of the overall bacterial BSIs (15/53) mostly caused by coagulase-negative staphylococci, among others. This rate is not far from the one observed in the adult population (24%) and underlines the need for concomitant BC evaluation, alongside a proper interpretation of negative T2MR results (19).
On the other hand, since coagulase-negative staphylococci are often the main culprits in contamination events (95.4% in our population), blood culture analysis would be vital to establish their clinical significance if the T2Bacteria panel was also able to detect these microorganisms.
The retrospective nature of our study accounted for some limitations, including the high heterogeneity of the population in terms of the clinical status and risk factors, and the low prevalence of BSIs. These aspects emphasize the need for further monitoring of the use of T2MR in pediatric settings, and for more research in different clinical contexts.
In conclusion, the use of T2 panels improved care of hospitalized children by significantly reducing the time to identification compared to BC, and provided a reliable support to confirm or exclude pediatric BSIs. The integration between T2MR and BC could help overcome current diagnostic issues, even in most extreme ages where clinical vulnerability requires a rapid and sensitive approach. In children, it becomes an important first-line tool for a targeted pediatric antimicrobial stewardship and empirical treatment reevaluation, thanks to the high and consistent sensitivity and specificity in identifying the ESKAPEc microorganisms and some species of Candida, which we have shown to be maintained even in patients under 1 year of age and for very small blood volumes.
ACKNOWLEDGMENTS
We thank Marta Argentieri, Laura Pansani, Annamaria Sisto, and Agata Helena Kowalska for their valuable comments on this manuscript. The authors are grateful for the technical staff, Anna Angelaccio, Vittoria Cetra, Francesca Di Leva, Maria Teresa D’Urbano, Giulia Ferri, Gianluca Foglietta, Carmela Parlavecchio, Silvia Tredici, and Ilaria Zullino of the Unit of Microbiology and Diagnostic Immunology, Bambino Gesù Children's Hospital, for their outstanding support in collecting and processing samples and performing laboratory analyses and data management. We also thank Valentina Panetta for support in preparing the study database for statistical analyses.
B.L., V.C., M.A., S.A.-N., L.M., G.M., and M.O. contributed to study design, data collection, analysis and interpretation, as well as drafting and writing the manuscript. F.A. and S.N.M. performed statistical analyses. F.G., L.D.C., T.F., R.B., F.T., C.A., A.D., C.C., S.P., and A.V. supported the study with their role and clinical expertise. M.R. and A.O.M. were responsible for funding acquisition. F.L. followed the patient outcome and critically revised the manuscript. C.F.P. and P.B. supervised the project. The authors approved the final version to be submitted for publication.
The authors have no conflict of interests to declare. This study did not receive any funding.
Footnotes
Supplemental material is available online only.
Contributor Information
Carlo Federico Perno, Email: carlofederico.perno@opbg.net.
Nathan A. Ledeboer, Medical College of Wisconsin
REFERENCES
- 1.Tan B, Wong JJ-M, Sultana R, Koh JCJW, Jit M, Mok YH, Lee JH. 2019. Global case-fatality rates in pediatric severe sepsis and septic shock. JAMA Pediatr 173:352–362. 10.1001/jamapediatrics.2018.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlapbach LJ. 2019. Paediatric sepsis. Curr Opin Infect Dis 32:497–504. 10.1097/QCO.0000000000000583. [DOI] [PubMed] [Google Scholar]
- 3.Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, Colombara DV, Ikuta KS, Kissoon N, Finfer S, Fleischmann-Struzek C, Machado FR, Reinhart KK, Rowan K, Seymour CW, Watson RS, West TE, Marinho F, Hay SI, Lozano R, Lopez AD, Angus DC, Murray CJL, Naghavi M. 2020. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet Lond Engl 395:200–211. 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köstlin-Gille N, Härtel C, Haug C, Göpel W, Zemlin M, Müller A, Poets CF, Herting E, Gille C. 2021. Epidemiology of early and late onset neonatal sepsis in very low birthweight infants: data from the German Neonatal Network. Pediatr Infect Dis J 40:255–259. 10.1097/INF.0000000000002976. [DOI] [PubMed] [Google Scholar]
- 5.Zingg W, Hopkins S, Gayet-Ageron A, Holmes A, Sharland M, Suetens C, ECDC PPS study group . 2017. Health-care-associated infections in neonates, children, and adolescents: an analysis of paediatric data from the European Centre for Disease Prevention and Control point-prevalence survey. Lancet Infect Dis 17:381–389. 10.1016/S1473-3099(16)30517-5. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein B, Giroir B, Randolph A, International Consensus Conference on Pediatric Sepsis . 2005. International Pediatric Sepsis Consensus Conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med 6:2–8. 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 7.Huber S, Hetzer B, Crazzolara R, Orth-Höller D. 2020. The correct blood volume for paediatric blood cultures: a conundrum? Clin Microbiol Infect 26:168–173. 10.1016/j.cmi.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 8.O'Hagan S, Nelson P, Speirs L, Moriarty P, Mallett P. 2021. How to interpret a paediatric blood culture. Arch Dis Child Educ Pract Ed 106:244–250. 10.1136/archdischild-2020-321121. [DOI] [PubMed] [Google Scholar]
- 9.Kirn TJ, Weinstein MP. 2013. Update on blood cultures: how to obtain, process, report, and interpret. Clin Microbiol Infect 19:513–520. 10.1111/1469-0691.12180. [DOI] [PubMed] [Google Scholar]
- 10.Dargère S, Cormier H, Verdon R. 2018. Contaminants in blood cultures: importance, implications, interpretation and prevention. Clin Microbiol Infect 24:964–969. 10.1016/j.cmi.2018.03.030. [DOI] [PubMed] [Google Scholar]
- 11.Garcia RA, Spitzer ED, Beaudry J, Beck C, Diblasi R, Gilleeny-Blabac M, Haugaard C, Heuschneider S, Kranz BP, McLean K, Morales KL, Owens S, Paciella ME, Torregrosa E. 2015. Multidisciplinary team review of best practices for collection and handling of blood cultures to determine effective interventions for increasing the yield of true-positive bacteremias, reducing contamination, and eliminating false-positive central line-associated bloodstream infections. Am J Infect Control 43:1222–1237. 10.1016/j.ajic.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 12.Riedel S, Carroll KC. 2016. Early identification and treatment of pathogens in sepsis: molecular diagnostics and antibiotic choice. Clin Chest Med 37:191–207. 10.1016/j.ccm.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 13.Iroh Tam P-Y, Bendel CM. 2017. Diagnostics for neonatal sepsis: current approaches and future directions. Pediatr Res 82:574–583. 10.1038/pr.2017.134. [DOI] [PubMed] [Google Scholar]
- 14.Guinea J. 2014. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 20(Suppl 6):5–10. 10.1111/1469-0691.12539. [DOI] [PubMed] [Google Scholar]
- 15.Warris A, Pana Z-D, Oletto A, Lundin R, Castagnola E, Lehrnbecher T, Groll AH, Roilides E, EUROCANDY Study Group . 2020. Etiology and outcome of candidemia in neonates and children in Europe: an 11-year multinational retrospective study. Pediatr Infect Dis J 39:114–120. 10.1097/INF.0000000000002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng X, Zhou W, Zhu Y, Wan C. 2021. Epidemiology, risk factors and outcomes of bloodstream infection caused by ESKAPEEc pathogens among hospitalized children. BMC Pediatr 21:188. 10.1186/s12887-021-02661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogue JM, Kaye KS, Cohen DA, Marchaim D. 2015. Appropriate antimicrobial therapy in the era of multidrug-resistant human pathogens. Clin Microbiol Infect 21:302–312. 10.1016/j.cmi.2014.12.025. [DOI] [PubMed] [Google Scholar]
- 18.Zhen X, Lundborg CS, Sun X, Hu X, Dong H. 2019. Economic burden of antibiotic resistance in ESKAPE organisms: a systematic review. Antimicrob Resist Infect Control 8:137. 10.1186/s13756-019-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen MH, Clancy CJ, Pasculle AW, Pappas PG, Alangaden G, Pankey GA, Schmitt BH, Rasool A, Weinstein MP, Widen R, Hernandez DR, Wolk DM, Walsh TJ, Perfect JR, Wilson MN, Mylonakis E. 2019. Performance of the T2Bacteria panel for diagnosing bloodstream infections: a diagnostic accuracy study. Ann Intern Med 170:845–852. 10.7326/M18-2772. [DOI] [PubMed] [Google Scholar]
- 20.Mylonakis E, Clancy CJ, Ostrosky-Zeichner L, Garey KW, Alangaden GJ, Vazquez JA, Groeger JS, Judson MA, Vinagre Y-M, Heard SO, Zervou FN, Zacharioudakis IM, Kontoyiannis DP, Pappas PG. 2015. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical trial. Clin Infect Dis 60:892–899. 10.1093/cid/ciu959. [DOI] [PubMed] [Google Scholar]
- 21.Clancy CJ, Pappas PG, Vazquez J, Judson MA, Kontoyiannis DP, Thompson GR, Garey KW, Reboli A, Greenberg RN, Apewokin S, Lyon GM, Ostrosky-Zeichner L, Wu AHB, Tobin E, Nguyen MH, Caliendo AM. 2018. Detecting infections rapidly and easily for candidemia trial, part 2 (DIRECT2): a prospective, multicenter study of the T2Candida panel. Clin Infect Dis 66:1678–1686. 10.1093/cid/cix1095. [DOI] [PubMed] [Google Scholar]
- 22.Voigt C, Silbert S, Widen RH, Marturano JE, Lowery TJ, Ashcraft D, Pankey G. 2020. The T2Bacteria assay is a sensitive and rapid detector of bacteremia that can be initiated in the emergency department and has potential to favorably influence subsequent therapy. J Emerg Med 58:785–796. 10.1016/j.jemermed.2019.11.028. [DOI] [PubMed] [Google Scholar]
- 23.Lamoth F, Clancy CJ, Tissot F, Squires K, Eggimann P, Flückiger U, Siegemund M, Orasch C, Zimmerli S, Calandra T, Marchetti O, Nguyen MH, Bochud P-Y. 2020. Performance of the T2Candida panel for the diagnosis of intra-abdominal candidiasis. Open Forum Infect Dis 7:ofaa075. 10.1093/ofid/ofaa075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arendrup MC, Andersen JS, Holten MK, Krarup KB, Reiter N, Schierbeck J, Helleberg M. 2019. Diagnostic performance of T2Candida among ICU patients with risk factors for invasive candidiasis. Open Forum Infect Dis 6:ofz136. 10.1093/ofid/ofz136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muñoz P, Vena A, Machado M, Martínez-Jiménez MC, Gioia F, Gómez E, Origüen J, Orellana MÁ, López-Medrano F, Pérez-Granda M-J, Aguado JM, Fortún J, Bouza E, T2MadRid study group . 2018. T2MR contributes to the very early diagnosis of complicated candidaemia: a prospective study. J Antimicrob Chemother 73:iv13–iv19. 10.1093/jac/dky048. [DOI] [PubMed] [Google Scholar]
- 26.Muñoz P, Vena A, Machado M, Gioia F, Martínez-Jiménez MC, Gómez E, Origüen J, Orellana MÁ, López-Medrano F, Fernández-Ruiz M, Merino P, González-Romo F, Frías I, Pérez-Granda M-J, Aguado JM, Fortún J, Bouza E, T2MadRid study group . 2018. T2Candida MR as a predictor of outcome in patients with suspected invasive candidiasis starting empirical antifungal treatment: a prospective pilot study. J Antimicrob Chemother 73:iv6–iv12. 10.1093/jac/dky047. [DOI] [PubMed] [Google Scholar]
- 27.Fortún J, Gioia F, Muñoz P, Graus J, Gómez-García de la Pedrosa E, Martín-Dávila P, Saiz A, Vena A, Moreno S. 2018. T2 magnetic resonance for the diagnosis of deep-seated invasive candidiasis in a liver recipient without candidemia. Rev Iberoam Micol 35:159–161. 10.1016/j.riam.2018.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Miller JM, Binnicker MJ, Campbell S, Carroll KC, Chapin KC, Gilligan PH, Gonzalez MD, Jerris RC, Kehl SC, Patel R, Pritt BS, Richter SS, Robinson-Dunn B, Schwartzman JD, Snyder JW, Telford S, Theel ES, Thomson RB, Weinstein MP, Yao JD. 2018. A guide to utilization of the microbiology laboratory for diagnosis of infectious diseases: 2018 update by the Infectious Diseases Society of America and the American Society for Microbiology. Clin Infect Dis 67:813–816. 10.1093/cid/ciy584. [DOI] [PubMed] [Google Scholar]
- 29.Dursun A, Ozsoylu S, Akyildiz BN. 2018. Neutrophil-to-lymphocyte ratio and mean platelet volume can be useful markers to predict sepsis in children. Pak J Med Sci 34:918–922. 10.12669/pjms.344.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Póvoa P, Coelho L, Almeida E, Fernandes A, Mealha R, Moreira P, Sabino H. 2005. C-reactive protein as a marker of infection in critically ill patients. Clin Microbiol Infect 11:101–108. 10.1111/j.1469-0691.2004.01044.x. [DOI] [PubMed] [Google Scholar]
- 31.Heinze G, Schemper M. 2002. A solution to the problem of separation in logistic regression. Stat Med 21:2409–2419. 10.1002/sim.1047. [DOI] [PubMed] [Google Scholar]
- 32.Pfaller MA, Wolk DM, Lowery TJ. 2016. T2MR and T2Candida: novel technology for the rapid diagnosis of candidemia and invasive candidiasis. Future Microbiol 11:103–117. 10.2217/fmb.15.111. [DOI] [PubMed] [Google Scholar]
- 33.Josefson P, Strålin K, Ohlin A, Ennefors T, Dragsten B, Andersson L, Fredlund H, Mölling P, Olcén P. 2011. Evaluation of a commercial multiplex PCR test (SeptiFast) in the etiological diagnosis of community-onset bloodstream infections. Eur J Clin Microbiol Infect Dis 30:1127–1134. 10.1007/s10096-011-1201-6. [DOI] [PubMed] [Google Scholar]
- 34.Dien Bard J, McElvania TeKippe E. 2016. Diagnosis of bloodstream infections in children. J Clin Microbiol 54:1418–1424. 10.1128/JCM.02919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De SK, Shetty N, Kelsey M. 2014. How to use… blood cultures. Arch Dis Child Educ Pract Ed 99:144–151. 10.1136/archdischild-2013-305197. [DOI] [PubMed] [Google Scholar]
- 36.Mermel LA, Maki DG. 1993. Detection of bacteremia in adults: consequences of culturing an inadequate volume of blood. Ann Intern Med 119:270–272. 10.7326/0003-4819-119-4-199308150-00003. [DOI] [PubMed] [Google Scholar]
- 37.Driscoll AJ, Deloria Knoll M, Hammitt LL, Baggett HC, Brooks WA, Feikin DR, Kotloff KL, Levine OS, Madhi SA, O'Brien KL, Scott JAG, Thea DM, Howie SRC, Adrian PV, Ahmed D, DeLuca AN, Ebruke BE, Gitahi C, Higdon MM, Kaewpan A, Karani A, Karron RA, Mazumder R, McLellan J, Moore DP, Mwananyanda L, Park DE, Prosperi C, Rhodes J, Saifullah M, Seidenberg P, Sow SO, Tamboura B, Zeger SL, Murdoch DR, PERCH Study Group . 2017. The effect of antibiotic exposure and specimen volume on the detection of bacterial pathogens in children with pneumonia. Clin Infect Dis 64:S368–S377. 10.1093/cid/cix101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamula CL, Hughes K, Fisher BT, Zaoutis TE, Singh IR, Velegraki A. 2016. T2Candida provides rapid and accurate species identification in pediatric cases of candidemia. Am J Clin Pathol 145:858–861. 10.1093/ajcp/aqw063. [DOI] [PubMed] [Google Scholar]
- 39.Scheer CS, Fuchs C, Gründling M, Vollmer M, Bast J, Bohnert JA, Zimmermann K, Hahnenkamp K, Rehberg S, Kuhn S-O. 2019. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: a prospective clinical cohort study. Clin Microbiol Infect 25:326–331. 10.1016/j.cmi.2018.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables SA1 to SA4 and Fig. SA1. Download jcm.00292-22-s0001.pdf, PDF file, 0.2 MB (248.4KB, pdf)