ABSTRACT
A vast amount of antimicrobial susceptibility test (AST) data is generated from routine testing in diagnostic laboratories for the primary purpose of guiding clinicians in antimicrobial therapy decisions for their patients. However, there is additional value for these data when they are compiled at the local, regional, national, and global levels. Cumulative AST data can be used to prepare antibiograms at the individual health care facility level. These reports can be used to gain insight into appropriate empirical therapy options prior to the availability of AST results on an individual patient’s isolate. Different types of cumulative AST data reports can also be compiled at the regional, national, and global levels to estimate susceptibility rates in geographic regions, document trends in evolving microbial populations, and recognize the appearance and spread of emerging antimicrobial resistance threats. The first CLSI M39 Guideline for Analysis and Presentation of Cumulative AST Data was published in 2000. Since that time, there have been changes to AST and reporting recommendations as well as the introduction of advanced informatics technologies to analyze and present data. The 5th edition of M39 has taken into consideration these changes to assist those who analyze, present, and utilize routine antibiograms and other types of cumulative AST data reports as well as those who design information systems for the capturing and analyzing of AST data. Furthermore, antimicrobial stewardship programs (ASPs) have expanded considerably, and uses of the antibiogram by ASPs have been addressed. This minireview will remind users of the basic recommendations for analysis and presentation of antibiograms and provide new suggestions to enhance these reports.
KEYWORDS: cumulative antimicrobial susceptibility testing report, M39, antibiogram, antimicrobial susceptibility test
INTRODUCTION
Since the fourth edition of M39, Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test (AST) Data released in January 2014, many new trends in antimicrobial stewardship, public health, rapid diagnostics, and informatics have occurred (1). In particular, antimicrobial stewardship programs (ASPs) have expanded considerably. There are several tools available for clinicians to prescribe antimicrobial agents prudently, including individual isolate susceptibility reports from diagnostic specimens (i.e., the antimicrobial susceptibility profile of an organism) as well as cumulative susceptibility reports (e.g., antibiogram) generated from the compilation of individual isolate data. The antibiogram is not only useful for clinicians at the facility level but also for ASPs and public health where there is an increasing need to better understand what is collectively happening with antimicrobial susceptibility to address emerging antimicrobial resistance threats.
Furthermore, there have been changes in public health and medical microbiology laboratories with the introduction of rapid diagnostic tools (e.g., multiplex molecular panels) and advanced informatics. Novel tools have expanded accessibility to AST data and analysis capabilities. Many of the changes in M39 5th edition take into consideration these and other newer programs and technologies to address current needs of those who rely on cumulative AST data reports, including antibiograms, for various applications.
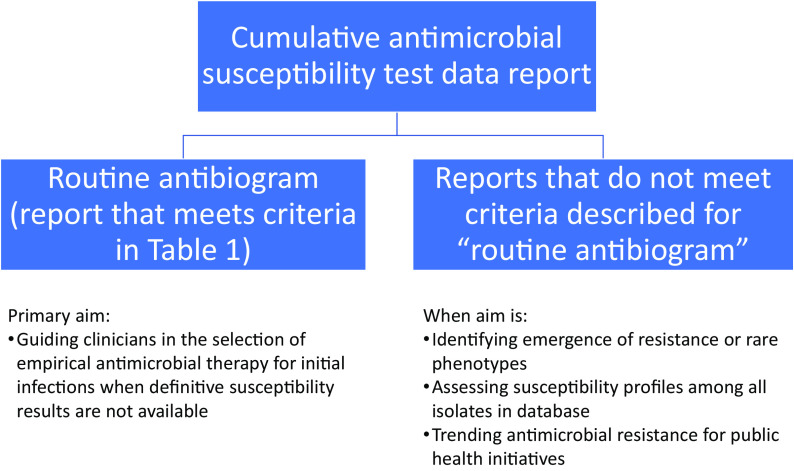
As illustrated in Fig. 1, the recommendations in previous and current editions of M39 for developing a routine antibiogram have been made with the primary aim of guiding clinicians in the selection of empirical antimicrobial therapy for initial infections before definitive susceptibility results become available or when definitive susceptibility results are not available. The term antibiogram should be reserved to describe the annual, cumulative AST report generated from a particular institution(s) following M39 guidance. For cumulative AST reports intended for purposes other than guiding empirical therapy (e.g., identifying emergence of resistance, trending antimicrobial resistance for public health initiatives), alternative analyses may be more appropriate, as discussed briefly in M39 5th edition.
FIG 1.
Different types of cumulative AST data reports.
The basic recommendations for antibiogram preparation remain the same as in previous M39 versions and are summarized in Table 1 (2). Therefore, the focus of this minireview is to summarize changes introduced to the M39 5th edition document to address current trends in medical microbiology laboratories, informatics systems, and ASPs.
TABLE 1.
Key recommendations for routine antibiogram development
| Recommendation |
|---|
| Antibiogram reports should be analyzed and presented at least annually. |
| Only diagnostic (not surveillance) isolates should be included. |
| Only final, verified test results should be included. |
| Duplicates should be eliminated by including only the first isolate of a species, patient, and/or analysis period, regardless of specimen source or antimicrobial susceptibility profile. |
| Only species with testing data for ≥30 isolates should be included. |
| Only antimicrobial agents routinely tested against the population of isolates to be analyzed should be included, and the %S should be calculated from results reported as well as those that may be suppressed on patient reports for which selective or cascade reporting rules have been applied. |
| To decrease biases in susceptibility estimates, laboratorians should refrain from including results for supplemental antimicrobial agents selectively tested on resistant isolates only. |
| Laboratorians should report the %S but exclude the %I and %SDD in the %S statistic. |
CONTENT ELIMINATED FROM M39
The intrinsic resistance appendix and glossaries that are managed by the Clinical and Laboratory Standards Institute (CLSI) AST Subcommittee and published in CLSI M100 (3) were removed from the 5th edition since M100 is updated annually and M39 is on a 5 (or more)-year revision cycle. Listing tables from M100 in M39 that may become obsolete a year or two after publication of M39 was not ideal. M100 is now available for free as a read-only version online (4) with easy access to up-to-date versions of the intrinsic resistance appendix and glossaries.
Previous editions of M39 suggested that percentage of isolates with intermediate susceptibility should be listed in addition to percent susceptible (%S) for penicillin with viridans group streptococci. This recommendation was initially added to address treatment of infective endocarditis caused by viridans group streptococci. However, the recommendation was eliminated, as providing %I for penicillin is unlikely to impact empirical therapy (5).
NEW CONTENT IN M39 5TH EDITION
A summary of new content in M39 5th edition is listed in Table 2. In addition, the list of “Contents” in the front of M39 has been expanded to enable the user to locate content related to a specific topic more readily. In addition to the references that link to the sources for some of the statements made in M39, the “Additional Resources” section contains a list of supplemental publications. For the most part, these focus on specific topics covered in M39, for example, combination antibiograms, antimicrobial stewardship, antimicrobial surveillance, etc. Several of the appendices have been expanded, and there are five new appendices (which will be described in further detail below), including one for “Frequently Asked Questions” (new Appendix L in reference 2).
TABLE 2.
New and expanded content in M39 5th editiona
| New and expanded content |
|---|
| Refined definitions of “cumulative AST data report” and “antibiogram”. |
| Practical concerns and advantages and disadvantages in extracting data from various data sources for antibiogram preparation. |
| Combining results from rapid diagnostics and resistance markers with the antibiogram. |
| Developing antibiograms for |
| Yeast and antifungal agents |
| Long-term care facilities |
| Veterinary practices |
| Developing multifacility antibiograms. |
| Use of antibiograms in antimicrobial stewardship programs. |
| Preparing cumulative AST data reports for peer-reviewed publication. |
| Using statistical analysis including percentiles, interquartile ranges, MIC50, and MIC90, to evaluate antibiogram data. |
| Including intermediate^ results in antibiograms for urinary tract agents. |
| Defining antibiogram percent susceptible thresholds related to empirical therapy decisions. |
AST, antimicrobial susceptibility test.
There has been confusion about several terms used in previous editions of M39 to describe cumulative susceptibility reports, especially cumulative AST data reports, cumulative antibiogram, routine antibiogram, and enhanced antibiogram. Therefore, these terms have been further defined in M39 5th edition. “Cumulative AST data reports” is the broadest term used to describe all types of reports generated by analysis of compiled AST results over a defined period of time that reflect the percentage of isolates of a given species or organism group that is susceptible to each of the antimicrobial agents tested. Cumulative AST data reports include antibiograms and other relevant analyses presented in tabular, graphic, and other types of formats. In contrast, the term “cumulative antibiogram” has been eliminated. The routine antibiogram is defined as the report generated by analysis of collective AST results usually from a single health care facility following M39 guidance specifically (Table 1). The enhanced antibiogram is a report where the %S data are further stratified using specific parameters (e.g., specimen source-specific or patient location antibiogram), and multifacility antibiograms aggregate data from multiple facilities.
Every step in M39 assumes that the data available for analysis are accurate. The updated version of M39 places greater emphasis on making certain that all AST results are verified and finalized by the laboratory prior to sending them to the database used for analyses. Laboratories that suppress certain antimicrobial results by applying selective or cascade reporting rules to encourage prudent antimicrobial prescribing should make every effort to capture the suppressed results in the database to avoid biases in susceptibility rate estimates. When applying suppression rules to facilitate cascade reporting (e.g., suppressing results for broader-spectrum agents if narrow-spectrum agents in the same drug class are susceptible), some drugs will only be reported on more resistant isolates (6). Consequently, failure to capture results from suppressed drugs in the antibiogram can result in %S values that are falsely low. It is essential to understand exactly how data are transmitted to the database in order to troubleshoot any inconsistencies in the %S data. One of the new appendices in M39 5th edition emphasizes the need to review the completed antibiogram report before it is distributed, and it includes tips for identifying %S data that might be erroneous (new Appendix D in reference 2). For example, when reviewing routine antibiogram data for Escherichia coli, %S values of 80% for cefepime and 95% for ceftriaxone are unusual and should be investigated.
Other concepts related to AST and reporting that have evolved in the past few years are now addressed in M39 5th edition, including (i) how to handle multiple breakpoints for a single agent such as cefazolin, which has breakpoints for isolates associated with both uncomplicated urinary tract infections and systemic infections; (ii) consideration of the susceptible dose-dependent (SDD) interpretive category; and (iii) consideration of the intermediate versus intermediate^ category, the latter of which relates to isolates associated with uncomplicated urinary tract infections.
There has been considerable activity within the CLSI AST subcommittee to ensure breakpoints used to interpret AST results, particularly for older drugs, are reliable (7, 8). Reanalysis of breakpoints using recent data resulted in updating a number of breakpoints, and it is likely that additional breakpoints will be updated in the near future. Breakpoint changes can have a significant impact on %S results in the antibiogram, and managing breakpoint changes for specific organism/antimicrobial agent combinations is thoroughly described in M39 5th edition.
Following a request to address color coding on the routine or enhanced antibiogram to reflect various levels of %S, M39 suggests green might represent %S values >80%, red for %S ≤60%, and yellow for %S values between these two. The discussion of the optional use of color coding emphasizes, as is done throughout M39, that other factors besides %S values must always be taken into consideration when empirical therapy decisions are made, such as infection type, severity of illness, patient history, standard treatment guidelines, and consequences of treating patients with an ineffective antimicrobial agent.
Other additions to M39 5th edition include recommendations for preparation of an antifungal antibiogram for yeast isolates. Laboratories that perform routine antifungal susceptibility testing should consider generation of such a report and might consider combining more than 1 year of data if the numbers of isolates of certain yeast species are less than 30. Instructions for combining data from multiple years or multiple sources have been expanded in M39 5th edition.
Below, we review the new content in the M39 5th edition (Table 2), which is presented in order of its appearance in the M39 document.
EXTRACTING DATA FROM DIFFERENT SOURCES
Antibiograms are generated using data derived from one or more of the following sources:
-
•
automated or semiautomated AST instrument
-
•
laboratory information system (LIS)
-
•
hospital electronic health record (EHR)
-
•
third-party clinical decision support system (CDSS) used by infection prevention and/or ASP
Data from the AST instrument are a convenient and well-standardized source of AST results. However, valuable patient demographic, patient location, and specimen source details may not be available depending on the completeness of information received through the bidirectional interface with the LIS. Furthermore, the AST instrument typically would not contain results from manual methods such as disk diffusion or gradient diffusion. Data from the AST instrument may include erroneous AST results if steps are not taken to ensure all data to be analyzed for the antibiogram are accurate and “final.” Data from the LIS are generally most complete with regard to patient, location, and specimen details and contain AST results for tests performed manually. However, the LIS may not include results of “suppressed” antimicrobial agents if they are suppressed in the AST instrument as a part of a cascade or selective reporting for antimicrobial stewardship. A general recommendation would be to transfer all antimicrobial results from the AST instrument to the LIS, including those that might be suppressed on the final patient reports. Data derived from the EHR or CDSS are likely to contain more patient, location, and specimen details, which can be parsed out to prepare enhanced antibiograms based on clinical need. For example, when evaluating the prevalence of methicillin (oxacillin)-resistant Staphylococcus aureus (MRSA) in various locations in a health care facility, it would be easier to obtain the oxacillin %S statistic among S. aureus from intensive care unit (ICU) patients when using an EHR or CDSS data source where patient demographics are stored. However, the EHR or CDSS databases may not contain results for all antimicrobial agents tested if cascade or selective reporting is implemented at the AST instrument or LIS levels. Data derived from the AST instrument, LIS, and EHR sources may be combined; however, data quality and uniqueness of data extracted have to be carefully scrutinized before use in antibiograms.
For some data sources, data analysis is performed in the same system where the data are stored. Alternatively, data generated from one or more of these sources can be exported to other third-party data management and analysis tools, such as Microsoft Excel, Microsoft Access, WHONET, or dedicated statistical analysis software provided for antibiogram preparation. Choice of data source utilized depends on the end user and the scope of the program that is using the antibiogram. Although data are usually analyzed annually, some may find it valuable to generate reports on a rolling basis (9). Regardless of the data source, it is important to be attentive to the suitability of each with regard to availability, content, granularity, and formatting of extracted data. Table 3 summarizes the attributes and qualities of the various sources by which data acquisition is possible for generation of the antibiogram.
TABLE 3.
| Attribute or characteristic | Data source: |
||
|---|---|---|---|
| AST instrument | LIS | EHR/CDSS | |
| Data extraction | |||
| Ease of data extraction | Standard data export features with predefined formats, generally simple | Format created by vendor; may need specific module or procedure (simple or difficult depending on the system) | Format created by vendor; may need additional programming or module installation (simple or difficult depending on the system) |
| Availability of patient demographics, patient locations, and specimen source data | Possibly, depending on the AST system and completeness of manual data entry or bidirectional LIS interface | Generally, yes | Yes |
| Availability of suppressed antimicrobial resultsa | Yes | Depends on the configuration. Results from suppressed antimicrobials often missing. | Generally, no |
| Availability of quantitative MIC data | Generally, yes | Generally, yes, if desired. | Generally, yes, if desired |
| Availability of results from offline AST (e.g., gradient diffusion and disk diffusion) | No, unless manually entered | Yes | Yes |
| Capability of removing duplicate AST results | No | Yes | Possibly |
| Data analysis | |||
| Customizability | Low | Moderate to high | Moderate to high |
| Stratification of results by patient details | Possibly, depending on the AST system and completeness of manual data entry or bidirectional LIS interface | Yes | Yes |
| Ability to reanalyze retrospective data using updated breakpoints | No | Possibly, if there are separate fields for MIC or zone value and interpretation. | Possibly, if there are separate fields for MIC or zone value and interpretation. |
| Frequency of data analysis, capability of automation | Generated interactively by the user | Can often be run interactively by the user or automated depending on the system | Can often be run interactively by the user or automated depending on the system |
| Additional support for setup and use | |||
| Additional cost | None, utilizing standard AST system features | Standard features within some systems, but needs creation and customization by others | Standard features within some systems, but needs creation and customization by others |
| Dependency on local informatics resources | No | Yes | Yes |
| Dependence on vendor-defined reports | Yes | Depends on the system capabilities, including predefined and customizable reports. | Depends on the system capabilities, including predefined and customizable reports. |
Applies to AST results available/unavailable based on cascade or selective reporting. AST, antimicrobial susceptibility test; LIS, laboratory information system; EHR, electronic healthcare record; CDSS, clinical decision support system.
INCORPORATING ANTIMICROBIAL RESISTANCE MARKER TEST RESULTS WITH THE ANTIBIOGRAM
Detection of antimicrobial resistance (AMR) markers (e.g., mecA/C, blaCTX-M) or mechanisms mediating AMR (e.g., broad detection of carbapenemase production by a phenotypic method) are becoming more commonplace in medical microbiology laboratories. Tests for the detection of AMR can be performed from recovered isolates, positive blood culture broths, or directly from clinical specimens (e.g., respiratory secretions) for diagnostic or surveillance purposes (10). Often, the results of these tests are available prior to the antimicrobial susceptibility profile of the associated organism(s) and may be helpful to optimize therapy for individual patients.
Associating the cumulative AST data with the collective results from AMR marker tests on an antibiogram can provide further insight into patient management, antimicrobial stewardship, and infection control or serve as a quality indicator in the laboratory. The current edition of M39 provides guidance and examples on how collective AMR marker test results can be included in the antibiogram as either a separate line listing or combined with %S data from follow-up phenotypic AST results. The presentation of AMR results using these approaches yields different types of information. For instance, when AMR data are incorporated as a separate line listing in the antibiogram, the results can be compared to %S from phenotypic results for a given species and serve as a quality indicator for the accuracy of the diagnostic tests performed in the laboratory. Alternatively, one can document the accuracy of antimicrobial resistance markers to predict susceptibility before the availability of phenotypic AST results to patient-facing clinicians. The best example involves detecting the mecA gene in S. aureus. Separate line listings for mecA detection for prediction of oxacillin (e.g., 53% of isolates negative for mecA would predict 53% S to oxacillin) can be compared to the %S based on oxacillin susceptibility testing among the same isolates to demonstrate the correlation between genotype and phenotype.
Alternatively, if the AMR marker test results are combined with the corresponding %S data into a single line on the antibiogram, the combined results will provide insight into an isolate’s susceptibility associated with the presence or absence of an AMR marker. The associated results can provide additional information on the local epidemiology and may benefit patient management. For example, the absence of a resistance marker among Gram-negative organisms in a single case is often inconclusive, as resistance to specific agents may be conferred by mechanisms other than those associated with the targeted AMR marker (e.g., the absence of carbapenemase genes does not rule out carbapenem resistance). However, by combining the cumulative AST data for a given species in the absence of an AMR marker(s), high %S rates may be observed (e.g., high rates of susceptibility to carbapenems in the absence of detecting a carbapenemase marker). Alternatively, information regarding agents from alternative classes not traditionally predicted by the AMR marker may also yield fruitful information. For example, aminoglycoside resistance genes may be harbored on the same plasmid as carbapenemase genes, and therefore, isolates that harbor a carbapenemase might demonstrate a higher association with aminoglycoside resistance (e.g., lower %S) than isolates that do not harbor a carbapenemase. These data may be used to support therapeutic decision-making or may be considered part of antimicrobial stewardship and infection control initiatives.
Thus, combining AMR marker test data with the antibiogram can help guide early therapy and provide information to different stakeholders on the epidemiology and predictive value of the presence and/or absence of the AMR markers for their patient population. There are limitations to incorporating AMR marker results into the antibiogram, which are discussed in detail in the M39 5th edition document (2).
LONG-TERM CARE FACILITY ANTIBIOGRAMS
The Centers for Disease Control and Prevention (CDC) and Agency for Healthcare Research and Quality (AHRQ) recommend that all long-term care facilities (LTCFs) (e.g., nursing homes, skilled nursing facilities, assisted living facilities) develop an annual antibiogram to guide clinicians in the selection of initial empirical antimicrobial therapy (11, 12). The general recommendations for antibiogram development should also be applied to LTCF antibiogram development when possible (Table 1). It is recognized that there are a number of challenges within LTCFs that may preclude the generation of an annual antibiogram according to M39 guidelines, as described below and in further detail in the M39 5th edition (2). Similar to acute care hospitals, a multidisciplinary group (e.g., LTCF leadership, LTCF medical director, LTCF consultant pharmacist, LTCF infection control, LTCF antimicrobial stewardship program, LTCF lab provider, etc.) should formulate a plan for antibiogram development that meets the individual needs of the LTCF.
The accuracy of an antibiogram is highly dependent upon the culturing practices within an individual LTCF. Selective culturing of residents who are likely infected will not provide an accurate representation of the susceptibility rates of bacteria at the LTCF. Therefore, a best practice recommendation would be to perform routine culture and susceptibility of all LTCF patients with suspected infection to improve patient care and the accuracy of the data in the LTCF antibiogram. This recommendation is in agreement with the CDC Core Elements of Antimicrobial Stewardship for Nursing Homes, which recommend LTCFs implement and monitor at least one policy or practice to improve antimicrobial use (12, 13). Additionally, AHRQ has developed a toolkit within the Nursing Home Antimicrobial Stewardship Guide to assist in the identification and diagnosis of common infections in LTCF residents based on signs/symptoms and physical exam findings (13).
Traditionally, LTCFs do not have their own microbiology laboratory and send specimens to one or multiple referral laboratories. Ideally, sending to one laboratory ensures that the same AST method and interpretive breakpoints are utilized. When multiple referral laboratories are utilized, variations in lab practices must be considered when evaluating and analyzing susceptibility data, which may complicate the data analysis and creation of the antibiogram as well as lead to discrepancies in the data. For hospital-based LTCFs that only receive their patient referrals from an attached acute care hospital, the LTCF can consider using the hospital antibiogram for empirical antimicrobial regimen selection if the LTCF is unable to generate an antibiogram. If only small numbers of isolates are available (<30 bacteria) for the time period of the antibiogram, a number of data analysis options can be utilized, such as combining data from multiple years, combining species (if applicable), or using data from other sources (e.g., regional health department data, local/regional published susceptibility data, susceptibility data from other health care facilities in the community, etc.). However, because the susceptibility patterns of isolates encountered in LTCFs are often unique to that facility, use of antibiograms from other sources is not ideal.
Last, all prescribing clinicians within the LTCF should have access to the annual antibiogram for empirical antimicrobial selection. In addition, a copy of the LTCF annual antibiogram should be included in the transfer documentation anytime an LTCF resident is sent to an acute care hospital or other facility.
THE VETERINARY ANTIBIOGRAM
Use of antibiograms is considered one of the core elements of many veterinary ASPs. Although many of the principles of antibiogram development and presentation apply in both human and veterinary medicine, the 5th edition of CLSI M39 contains an entirely new chapter dedicated to unique circumstances surrounding the veterinary antibiogram (2). This new chapter provides guidance to developers of veterinary antibiograms on how to address these unique challenges. A brief synopsis of the chapter discussion on the challenges associated with isolate inclusion, presentation, and use of the antibiogram is provided here.
Veterinary antibiograms are often developed by regional diagnostic laboratories to capture a sufficient number of bacterial isolates (at least 30) to make the antibiogram useful. Therefore, the veterinary antibiogram is a hybrid between multifacility and enhanced antibiograms. In this situation, it is possible that differences in antimicrobial susceptibility across geography or patient subpopulations are masked. If the process behind developing the veterinary antibiogram is transparent to the veterinary prescriber, the applicability of the cumulative antibiogram data to a specific case for which empirical antimicrobial therapy is being considered can be assessed by the clinician.
Guidance for developing antibiograms in human medicine recommends including only the first isolate from a given patient per analysis period to limit the influence of repeat patient isolates on the antibiogram (Table 1). The rationale for this guidance also applies to veterinary antibiograms; however, there are logistical challenges to implementation. First and foremost, a coordinated system of patient identification is not present in veterinary medicine. Veterinary patients are often identified by pet name and owner last name. At the level at which veterinary antibiograms are typically developed (regional diagnostic laboratory), it may be difficult to distinguish patients with similar names (e.g., “Buffy” Smith). Additionally, in production animal settings, animal identification may only be at the herd, pen, or flock level, so determining if isolates from the same premise are similar may be impossible. The veterinary chapter of M39 5th edition poses potential methods to address this challenge.
As one of the components of an antimicrobial stewardship program, antibiograms can be used to convey information to veterinary prescribers beyond the local prevalence of AMR. Antibiograms may be presented in such a way that they highlight other considerations in the antimicrobial selection process. In veterinary medicine, this may include the regulatory status of an antimicrobial, specifically with regard to uses in food-producing animals. Antibiograms may also be developed to reflect clinical practice guidelines. As veterinary interpretive criteria are host-species specific, the veterinary antibiogram should clearly delineate to the clinician if species-specific breakpoints were used to determine the %S and, if not, which breakpoints were used in the calculation. Example antibiograms are presented in the veterinary chapter of M39 5th edition.
PREPARATION AND USE OF MULTIFACILITY ANTIBIOGRAMS
The use of single-facility antibiograms has been well described and may be mandated by antimicrobial stewardship metrics and accreditations (14). While single-facility antibiograms guide empirical therapy decisions and interventions, there are several instances when a multifacility antibiogram may be developed and used separately. There may be times when AST data from a single facility are not available, limited in size or scope, or subject to significant biases. In these situations, aggregating data from multiple external sources may be useful to increase awareness of antimicrobial resistance and emerging resistance in a region and can serve as a benchmark for comparing %S data among individual facilities. Besides the acute care hospital, sources of data for constructing multifacility antibiograms include referral laboratories, long-term care sites, public health departments, and national surveillance laboratories. Susceptibility data from any of these sources can be combined to develop a multifacility antibiogram. The M39 5th edition document describes different approaches and considerations to preparing multifacility antibiograms.
There are three main scenarios for the generation of multifacility antibiograms. The approach used is determined by the study goals, resources available, and agreements between facilities and the network coordinator for sharing isolates, isolate-level data, and/or local antibiograms. The three approaches described in the M39 include (i) collection and testing of clinical samples or isolates by a centralized laboratory, (ii) centralized collection of electronic isolate-level AST data, and (iii) centralized collection and collation of single-facility antibiograms. These approaches to aggregating data to prepare multifacility antibiograms and their strengths and weakness are described in M39 5th edition (see Table 31 in reference 2). Overall, the centralized laboratory isolate collection approach is likely to generate the highest-quality and most consistent data. However, it can be resource intensive, with concerns about sustainability. The centralized collection of single-facility antibiogram approach may be the most feasible, as many facilities already generate annual facility-level antibiograms. However, this approach generates the lowest quality of data at the aggregate level, as variability across many parameters may exist between facilities. It is important to identify any inconsistencies and outliers among the submitted data to assess the effect on the final report. Certain variables may be collected from each facility to better assess their potential effect on the final collated data, such as AST reporting rules (e.g., suppression rules), breakpoints applied to interpret the results, antimicrobial agents tested, and level of organism identification. The results from the centralized laboratory testing and the centralized collection of electronic isolate-level AST data can be analyzed as described for the single-facility antibiogram, whereas special stepwise instructions are provided in the M39 for the centralized collection of single-facility antibiograms approach (see new Appendix H in reference 2).
The presentation of a multifacility antibiogram can be similar to that recommended for a single-facility antibiogram by presenting the %S statistic for each organism/antimicrobial agent combination in a table format. When data are aggregated from a large number of facilities, it is recommended that percentiles or interquartile ranges are included to help identify institution outliers and variability in the data set. Guidance on how to perform these calculations is available in the M39 5th edition.
Multifacility antibiograms may be valuable in certain circumstances to guide local empirical therapy recommendations (e.g., organisms that do not routinely have AST performed, organisms that are infrequently isolated, etc.). However, when such data are applied, it is critical to consider the approach that was used for generating the report, limitations of the data due to sampling biases or protocols and methods used for AST, and relevance of the data used for a particular health care facility’s geographic location, patient population, and clinical setting. When available, facility-level data should be prioritized over multifacility-level data for local empirical therapy recommendations and guidelines.
There are many other uses for multifacility antibiograms, which vary based on the needs of different stakeholders. For example, infection prevention and control programs may use the multifacility antibiogram as a benchmark for local data to help determine whether investigations or interventions in infection prevention practices may be needed. Alternatively, public health departments may monitor susceptibility or resistance profiles across specific jurisdictions using the centralized collection of single-facility antibiograms (15). It is important for various stakeholders to understand the methods used to assemble the data to appreciate the potential limitations of multifacility antibiogram data.
USING THE ANTIBIOGRAM TO GUIDE EMPIRICAL THERAPY OF INITIAL INFECTIONS
Susceptibility data in the annual antibiogram provide current information on the relative prevalence and susceptibility of bacteria at a particular institution/health care center. There are two types of antibiograms, the routine antibiogram where the %S values of all Gram-negative or Gram-positive bacteria from the entire health care system or hospital unit are included (regardless of specimen type or patient characteristic) and the enhanced antibiogram where the %S values of bacteria are evaluated and presented using specific criteria such as infection type (bloodstream isolates only, urinary tract isolates only, etc.), patient type (cystic fibrosis, renal transplant, hematology/oncology, etc.), or patient location (medical floor, intensive care unit, outpatient, etc.).
The %S data tables for either type of antibiogram include information on the type of bacteria tested, the number of isolates (first isolate per patient per reporting period) tested during the time frame of the antibiogram, the antimicrobials tested for susceptibility, and the %S to each antimicrobial agent (see Table S1 in the supplemental material). The %S data from either the routine or enhanced antibiogram can be used to guide empirical antimicrobial therapy while waiting for culture and antimicrobial susceptibility profile from a patient’s clinical specimen.
When selecting empirical antimicrobial therapy, the %S information from the antibiogram will be evaluated along with the site and severity of infection (e.g., systemic infections with high morbidity and/or early mortality versus those that are local and/or easily eradicated), likely infecting organism(s) based on the clinical scenario and possibly Gram strain findings, infection origin (e.g., community versus hospital versus LTCF), and past infections/antimicrobial therapy for a particular patient. Additional factors that may be considered include relevant regional treatment guideline recommendations, public health consequences of treating patients with an ineffective antimicrobial agent (e.g., additional transmission of infection to the community), and antimicrobial stewardship objectives to limit the use of priority reserve antimicrobial agents (e.g., broad-spectrum antimicrobial agents [meropenem] or antimicrobial agents used for multidrug-resistant infections [ceftazidime-avibactam]).
PERCENT SUSCEPTIBILITY THRESHOLD TO GUIDE EMPIRICAL ANTIMICROBIAL THERAPY
There are limited published data on the %S threshold for an antimicrobial that is considered acceptable to be selected for empirical therapy. For the treatment of patients with bacterial meningitis caused by Streptococcus pneumoniae, the World Health Organization (WHO) recommends penicillin %S of ≥90% for penicillin to be considered an empirical treatment option to minimize the risk of death (16). In contrast, a trimethoprim-sulfamethoxazole %S of ≥80% is a suitable threshold for consideration of this agent in the treatment of acute, uncomplicated cystitis (17, 18). A %S of 80% to 90% is also endorsed by other infection treatment guidelines (19, 20). Based on this limited published information, antimicrobial agents with %S of at least 90% or 95% should be empirically selected for the treatment of infections where the risk of mortality or significant morbidity is high (e.g., meningitis, sepsis, patients in the ICU). However, for patients with infections where there is no significant concern for mortality within 24 to 48 h (e.g., uncomplicated urinary tract infections [UTIs], simple community-acquired infections), antimicrobial agents with a lower %S threshold of 80% to 85% may be appropriate. If there are no antimicrobial agents with %S of >80%, the antimicrobial agent with the highest %S or combination therapy should be considered. Local institutional antimicrobial stewardship formulary policies and guidelines should also be considered when selecting empirical therapy.
USING ANTIBIOGRAMS TO GUIDE EMPIRICAL ANTIMICROBIAL THERAPY WHEN THE CAUSATIVE ORGANISM AND SUSCEPTIBILITY ARE UNKNOWN
When selecting empirical antimicrobial therapy in patients with infections where the causative organism and AST results are not yet known, the antibiogram can be used to guide selection of antimicrobial therapy targeting the most common causative organisms based on the infection type and criteria listed above. For example, if the antibiogram shown in Table S1 in the supplemental material is used to select an empirical antimicrobial agent for the treatment of a patient with Gram-negative bacteremia and no known drug allergies, cefepime (78% to 99% susceptibility) or meropenem (80% to 100% susceptibility) would be suitable choices because most Gram-negative species commonly associated with bloodstream infections demonstrate a high %S for these agents. If an enhanced antibiogram displaying the %S for bloodstream isolates from hospitalized patients is available, it should be preferentially utilized to guide empirical antimicrobial therapy selection for patients with Gram-negative bacteremia.
USING ANTIBIOGRAMS TO GUIDE EMPIRICAL ANTIMICROBIAL THERAPY WHEN THE ORGANISM IS KNOWN BUT THE SUSCEPTIBILITY IS UNKNOWN
When selecting empirical antimicrobial therapy in patients with infections where the causative organism is known but the susceptibility results are pending, the antibiogram can be used to select more targeted empirical antimicrobial therapy. For example, if a patient with hospital-acquired pneumonia has Klebsiella pneumoniae detected in their respiratory sample using a molecular method, the antibiogram shown in Table S1 in the supplemental material can be used to select an empirical antimicrobial agent with good activity against K. pneumoniae, such as ceftriaxone (91% susceptible), and avoid the use of other broad-spectrum or antipseudomonal antimicrobials.
ANTIMICROBIAL STEWARDSHIP PROGRAMS AND THE ANTIBIOGRAM
With Joint Commission regulations and endorsements from the CDC, the number of ASPs has increased rapidly with 85% of acute care hospitals reporting implementation of core elements of antimicrobial stewardship compared to only 41% in 2014 (14). The antibiogram plays a pivotal role in many initiatives implemented by ASPs to optimize antimicrobial use (Table 4).
TABLE 4.
Uses of the routine and enhanced antibiogram by antimicrobial stewardship programs
| Use of antibiogramsa |
|---|
| Routine antibiograms |
| Selection of empirical antimicrobial therapy for individual patients |
| Development of empirical antimicrobial treatment guidelines/algorithms based on infection type (e.g., febrile neutropenia, community-acquired and/or hospital/ventilator-associated pneumonia, sepsis, urinary tract infection, diabetic foot infection, etc.) |
| Decisions regarding the antimicrobial formulary |
| Enhanced antibiograms |
| Assist with more targeted empirical therapy based on infection type, patient type, organism, isolate source, patient location, etc. Examples include Gram-negative bacilli antibiograms for inpatients, ICU patients, burns patients, outpatients, and patients with cystic fibrosis |
| Urinary tract isolate antibiograms for inpatients, ICU patients, outpatients, patients in an emergency department, and renal transplant patients |
| Blood isolate antibiograms for inpatients, ICU, patients with hematologic/oncologic disorders, burn patients |
| Streptococcus pneumoniae antibiograms for patients with community-acquired pneumonia or meningitis |
| Staphylococcus aureus antibiograms based on patient location (e.g., inpatient, outpatient/ED, ICU patients, etc.) |
| Antimicrobial agent combinations antibiograms to indicate the increased coverage with the combination over the individual antimicrobial agents alone |
ICU, intensive care unit; ED, emergency department.
Information within the routine antibiogram is useful for many common stewardship processes such as prior authorization (e.g., ordering restricted antimicrobial agents based on certain indications as deemed by the stewardship program) and prospective audit with real-time feedback (e.g., monitoring the use of particular antimicrobial agents by the ASP with active feedback to the clinical team if changes are needed). Both of these practices require review and understanding of the institutional antibiogram and %S patterns, which are considered when selecting empirical antimicrobial therapy. The antibiogram is an integral part of the antimicrobial drug selection for individual patients and provides the framework for trainee/prescriber education since it provides data on local rates of susceptibility to guide appropriate use of antimicrobial agents. Guidance on how best to distribute and communicate antibiogram data are summarized in chapter 11 of M39. In addition, ASPs often use local antibiogram data to develop empirical antimicrobial therapy recommendations based on infection type in their institution-specific treatment guidelines. If the annual antibiogram demonstrates activity of an antimicrobial agent is declining and there are other antimicrobial agents on the formulary that may provide broader coverage, a change in empirical therapy guidelines may be warranted.
As described above, enhanced antibiograms are refined reports where the susceptibility data are analyzed to answer specific clinical questions or to guide empirical antimicrobial therapy in certain infections and/or patient types. If an enhanced antibiogram is available for a particular scenario (e.g., respiratory tract isolates only from ventilated ICU patients with pneumonia), it can be utilized to select more targeted empirical therapy based on the clinical situation. Supplemental analyses and presentation options for multidrug-resistant organisms (MDROs) are addressed in the M39, and these can include creating enhanced antibiograms for specific MDROs and including additional agents not reported on the routine antibiogram (e.g., novel beta-lactam-beta-lactamase inhibitor combination agents for carbapenem-resistant Enterobacterales). Additionally, antimicrobial combination susceptibility reports can also be constructed to examine the percentage of isolates susceptible to one or both agents in relevant combinations for empirical therapy against certain organisms or infection types where combination therapy may be warranted (e.g., MDROs). Several examples of antimicrobial agent combination antibiograms are provided in M39 (see new Appendix F in reference 2).
Last, ASPs are often the gatekeepers of the antimicrobial formulary at local institutions. Information from the routine and/or enhanced antibiogram can be reviewed annually to help guide formulary decisions. For example, if a significant decrease in %S is observed for a particular antimicrobial agent, it may be prudent to remove the agent from empirical therapy treatment guidelines as well as from the institution’s formulary. Alternatively, if an increase in the prevalence of Gram-negative organisms with lowered %S values to multiple agents is observed (e.g., suggesting an increase in MDR), the ASP may consider adding additional agents such as the novel beta-lactam-beta-lactamase inhibitor combinations to the formulary to help combat these difficult-to-treat organisms.
CONSIDERATIONS FOR PEER-REVIEWED PUBLICATION OF CUMULATIVE AST DATA
Data from cumulative AST results can be published as part of peer-reviewed literature. Such publications should include complete details on the methods in which the AST data were generated and analyzed. In the new M39, guidance is provided for the content to include in each section of a publication (i.e., introduction, materials and methods, results, and discussion) that examines cumulative AST data from isolates obtained from a single facility or multiple facilities. These suggestions should be considered together with the instructions to authors required for the intended journal.
One of the critical components to ensure reproducibility and comparisons between studies is to have a well-defined materials and methods section. In the new M39, an extensive table covering the components to consider, including in the material and methods section of a peer-reviewed publication, is provided (see Table 35 in reference 2).
The cumulative AST data in peer-reviewed publications are generally presented in multiple tables and/or graphs. The data can be presented in various formats, including a summary of %S, percent intermediate, and percent resistant together with the range of MIC results obtained from testing each organism-antimicrobial agent combination. More descriptive statistics can be provided by summarizing the frequency and distribution of MIC, MIC50, and MIC90 values. Examples are provided on different presentation formats in the M39 (new Appendix I in reference 2). Furthermore, as the MIC50 and MIC90 values are new terms in the M39 document, a section was added to the statistical considerations on how to calculate the MIC50 and MIC90 values. Finally, the discussion section of the peer-reviewed publication should highlight the overall findings of the study in the context of previous studies reported in the literature. Limitations and biases of the data should also be included.
SUMMARY
In summary, the M39 Guideline for Analysis and Presentation of Cumulative AST Data was recently updated with the publication of the 5th edition. The basics of standardized routine antibiogram preparation to guide empirical antimicrobial therapy of initial infections remain unchanged. For this revision, new trends in antimicrobial stewardship, public health initiatives, rapid diagnostics, and informatics have been taken into consideration with the goal of providing additional suggestions to improve the value of cumulative AST data, and antibiograms in particular, in multiple settings.
Footnotes
Supplemental material is available online only.
Contributor Information
Patricia J. Simner, Email: psimner1@jhmi.edu.
Romney M. Humphries, Vanderbilt University Medical Center
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2014. Analysis and presentation of cumulative antimicrobial susceptibility test data, 4th ed. Approved guideline. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2022. Analysis and Presentation of cumulative antimicrobial susceptibility test data, 5th ed. Approved guideline. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2022. Performance standards for antimicrobial susceptibility testing; 32nd informational supplement. CLSI M100-S32. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2022. Performance standards for antimicrobial susceptibility testing; 32nd informational supplement (online version). http://em100.edaptivedocs.net/Login.aspx?_ga=2.16063088.755778026.1648416253-1304901391.1638386189&_gac=1.220515178.1647057379.CjwKCAiAg6yRBhBNEiwAeVyL0DX_L3o6I8oqmAifI6F5XlndKDWWDc9tXChSUHHpJZj4_OfEjm37AhoCVkMQAvD_BwE. Retrieved 7 April 2022.
- 5.Baddour LM, Wilson WR, Bayer AS, Fowler VG, Jr., Tleyjeh IM, Rybak MJ, Barsic B, Lockhart PB, Gewitz MH, Levison ME, Bolger AF, Steckelberg JM, Baltimore RS, Fink AM, O'Gara P, Taubert KA, American Heart Association Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, Council on Clinical Cardiology, Council on Cardiovascular Surgery and Anesthesia, and Stroke Council. 2015. Infective endocarditis in adults: diagnosis, antimicrobial therapy, and management of complications: a scientific statement for healthcare professionals From the American Heart Association. Circulation 132:1435–1486. 10.1161/CIR.0000000000000296. [DOI] [PubMed] [Google Scholar]
- 6.Liao S, Rhodes J, Jandarov R, DeVore Z, Sopirala MM. 2020. Out of sight-out of mind: impact of cascade reporting on antimicrobial usage. Open Forum Infect Dis 7:ofaa002. 10.1093/ofid/ofaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphries RM, Abbott AN, Hindler JA. 2019. Understanding and addressing CLSI breakpoint revisions: a primer for clinical laboratories. J Clin Microbiol 57:e00203-19. 10.1128/JCM.00203-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simner PC, Martin IW, Sullivan KO, Rhoads D, Rolf R, Souers RJ, Wojewoda C, Humphries RM. 2022. Raising the bar: improving antimicrobial resistance detection by clinical laboratories by ensuring use of current breakpoints. Open Forum Infect Dis 9:ofac007. 10.1093/ofid/ofac007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katzman M, Kim J, Lesher MD, Hale CM, McSherry GD, Loser MF, Ward MA, Glasser FD. 2019. Customizing an electronic medical record to automate the workflow and tracking of an antimicrobial stewardship program. Open Forum Infect Dis 6:ofz352. 10.1093/ofid/ofz352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yee R, Dien Bard J, Simner PJ. 2021. The genotype-to-phenotype dilemma: how should laboratories approach discordant susceptibility results? J Clin Microbiol 59:e00138-20. 10.1128/JCM.00138-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agency for Healthcare Research and Quality. 2016. Help prescribing clinicians choose the right antibiotic. https://www.ahrq.gov/nhguide/toolkits/help-clinicians-choose-the-right-antibiotic/index.html. Retrieved 22 July 2022.
- 12.Centers for Disease Prevention and Control. 2015. The core elements of antibiotic stewardship for nursing homes. http://www.cdc.gov/longtermcare/index.html. Retrieved 22 July 2022.
- 13.Agency for Healthcare Research and Quality. 2017. Toolkit 3. Minimum criteria for common infections toolkit. https://www.ahrq.gov/nhguide/toolkits/determine-whether-to-treat/toolkit3-minimum-criteria.html. Retrieved 22 July 2022.
- 14.Centers for Disease Prevention and Control. 2021. CDC patient safety portal. https://www.cdc.gov/hai/data/portal/AR-Patient-Safety-Portal.html. Retrieved 21 July 2022.
- 15.Los Angeles County Department of Public Health. 2017. Los Angeles County acute care hospital 2017 multi-facility antibiogram. http://publichealth.lacounty.gov/acd/docs/Antibiogram/1AntibiogramFULL.pdf. Retrieved 22 July 2022.
- 16.World Health Organization. 2001. Model prescribing information. Drugs used in bacterial infections. https://apps.who.int/iris/handle/10665/42372. Retrieved 22 July 2022.
- 17.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE, Infectious Diseases Society of America, European Society for Microbiology and Infectious Diseases . 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin Infect Dis 52:e103–e120. 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 18.Warren JW, Abrutyn E, Hebel JR, Johnson JR, Schaeffer AJ, Stamm WE. 1999. Guidelines for antimicrobial treatment of uncomplicated acute bacterial cystitis and acute pyelonephritis in women. Infectious Diseases Society of America (IDSA). Clin Infect Dis 29:745–758. 10.1086/520427. [DOI] [PubMed] [Google Scholar]
- 19.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O'Grady NP, Bartlett JG, Carratala J, El Solh AA, Ewig S, Fey PD, File TM, Jr., Restrepo MI, Roberts JA, Waterer GW, Cruse P, Knight SL, Brozek JL. 2016. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 63:e61–e111. 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solomkin JS, Mazuski JE, Bradley JS, Rodvold KA, Goldstein EJ, Baron EJ, O'Neill PJ, Chow AW, Dellinger EP, Eachempati SR, Gorbach S, Hilfiker M, May AK, Nathens AB, Sawyer RG, Bartlett JG. 2010. Diagnosis and management of complicated intra-abdominal infection in adults and children: guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clin Infect Dis 50:133–164. 10.1086/649554. [DOI] [PubMed] [Google Scholar]
- 21.Forrest GN, Van Schooneveld TC, Kullar R, Schulz LT, Duong P, Postelnick M. 2014. Use of electronic health records and clinical decision support systems for antimicrobial stewardship. Clin Infect Dis 59(Suppl 3):S122–S133. 10.1093/cid/ciu565. [DOI] [PubMed] [Google Scholar]
- 22.Simpao AF, Ahumada LM, Larru Martinez B, Cardenas AM, Metjian TA, Sullivan KV, Galvez JA, Desai BR, Rehman MA, Gerber JS. 2018. Design and implementation of a visual analytics electronic antibiogram within an electronic health record system at a tertiary pediatric hospital. Appl Clin Inform 9:37–45. 10.1055/s-0037-1615787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson G, Badarudeen S, Godwin A. 2010. Real-time validation and presentation of the cumulative antibiogram and implications of presenting a standard format using a novel in-house software: ABSOFT. Am J Infect Control 38:e25–e30. 10.1016/j.ajic.2010.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jcm.02210-21-s0001.pdf, PDF file, 0.04 MB (46.6KB, pdf)