Abstract
Since the onset of the coronavirus pandemic in December 2019, the SARS-CoV-2 virus has accounted for over 6.3 million lives resulting in the demand to develop novel therapeutic approaches to target and treat SARS-CoV-2. Improved understanding of viral entry and infection mechanisms has led to identifying different target receptors to mitigate infection in the host. Researchers have been working on identifying and targeting potential therapeutic target receptors utilizing different candidate drugs. Angiotensin-converting enzyme-2 (ACE2) has been known to perform critical functions in maintaining healthy cardiorespiratory function. However, ACE2 also functions as the binding site for the spike protein of SARS-CoV-2, allowing the virus to enter the cells and ensue infection. Therefore, drugs targeting ACE2 receptors can be considered as therapeutic candidates. Strategies targeting the level of ACE2 expression have been investigated and compared to other potential therapeutic targets, such as TMPRSS2, RdRp, and DPP4. This mini review discusses the key therapeutic approaches that target the ACE2 receptor, which is critical to the cellular entry and propagation of the novel SARS-CoV-2. In addition, we summarize the main advantages of ACE2 targeting against alternative approaches for the treatment of COVID-19.
Keywords: Target receptors, Coronavirus, Receptor binding domain, Smart therapeutics, ACE2, COVID-19, SARS-CoV-2
Introduction
The COVID-19 (CO-corona, VI-virus, D-disease, 19-of 2019) outbreak occurred in 2019, in Wuhan, Hubie, China.59,99 Following the outbreak, the World Health Organization (WHO) declared a global pandemic on March 11, 2020.99 The virus strain was identified to be from the family of coronaviruses known as severe acute respiratory syndrome coronavirus (SARS-CoV).51 As of May 2022, there have been approximately 6.3 million deaths caused globally on account of COVID-19 infections.3 According to the Centers for Disease Control and Prevention (CDC), SARS-CoV-2 transmits through aerosols and respiratory droplets of the infected individuals.57 Due to COVID-19's rapid disease transmission and high fatality rate, an immediate therapeutic intervention is essential to help save lives and prevent further propagation of the virus.41 Current therapeutic strategies seek potential targets to prevent viral entry, survival, and infection.
The etiology of COVID-19 is known to be severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2). Coronavirus is an enveloped non-segmented positive-sense single-stranded ribonucleic acid (ssRNA) virus with a diameter of 80–120 nm.16,61 There are four genera of the coronavirus genus of the Coronaviridae family named alpha-, beta-, gamma-, and delta-coronavirus (α-CoV, β-CoV, γ-CoV, and δ-CoV).61 Among these genera, similar to the known coronaviruses such as MERS-CoV and SARS-CoV, SARS-CoV-2 also belongs to the β-CoV genus.55,70 The genomic size of SARS-CoV-2 is about 29.9 kb.39,55 It has also been found through genomic analysis that despite belonging to the same family and genus as both SARS-CoV and MERS-CoV, SARS -CoV-2 is genetically more similar to SARS-CoV than MERS-CoV.3 However, despite the striking genetic similarities to SARS-CoV, SARS-CoV-2 shares 96% similarity to the horseshoe bat RaTG13 virus found in the bat.93 Understanding the origin, and infection and transmission mechanisms of the SARS-CoV-2 are crucial components in the research and development of potential antiviral therapeutic approaches. Additionally, identifying key therapeutic targets can help in the development of targeted .smart therapies to prevent infection and combat this virus simultaneously.
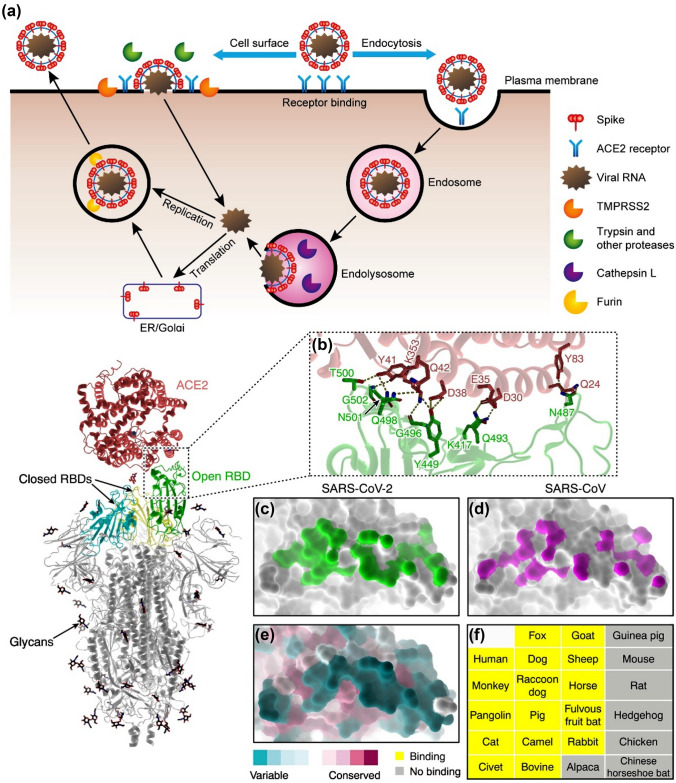
Understanding the viral entry mechanism is essential to the development of targeted therapies. With this objective, researchers investigated the viral entry mechanism of SARS-CoV-2. It is now known that, SARS-CoV-2 depends on cellular proteases to prime the spike protein.65,76 The SARS-CoV-2 virus enters the cell via its spike protein binding to the surface receptors initiating the formation of endosomes and ultimately the fusion of the viral and lysosomal membranes (Fig. 1).73 Once primed, conformational changes occur after the receptor binding and elicit the events leading to integration of the virus with the cell membrane which is subsequently followed by the dissemination of the viral ribonucleoprotein complex within the cytoplasm.65,83 Upon entering the cytoplasm of the host cell, the virus unleashes its RNA which is then translated and cleaved into smaller units by the action of the virus-encoded proteases. The viral polymerase then transcribes the subgenomic mRNAs, which then get translated to form the structural proteins of the virus. These proteins form complexes with the host cell components and form newly assembled viral particles. These particles subsequently get released from the infected cells through the process of exocytosis.65,83
Figure 1.
The mechanism of SARS-CoV-2 viral entry and progression. (a) The SARS-CoV-2 virus utilizes cell surface receptors and proteases to bind to the receptors on the cells. Subsequently the virus enters the cell and replicates and infects the other cells. (b) A close-up representation of the SARS-CoV-2 S protein and ACE2 interface (PDB: 6LZG). The sticks represent the key residues that interact with the ACE2. Dashed lines signify hydrogen bonding and salt bridges. The S-binding footprint for SARS-CoV-2 (c; PDB: 6LZG) and SARS-CoV on ACE2 is shown in (c, d) (D; PDB: 2AJF). More atomic connections are made between SARS-CoV-2 (green) and ACE2 than SARS-CoV (magenta). (e) Maintenance of the S-binding interface of ACE2. ACE2 orthologs from various animal taxa show a significant degree of diversity in the interface for SARS-CoV-2 S binding. (f) A list of examined animals in which ACE2 can bind to the RBD of the SARS-CoV-2 S protein (yellow) or cannot bind to the RBD of the SARS-CoV-2 S protein (grey). (Adapted and modified with permission from Murgolo et al.,54 Shang et al.,33,72 and Peng et al.64).
The process of identifying therapeutic target receptors has led researchers to explore commonly expressed enzymes and their receptors and their possible roles in viral entry and infection. In humans, transmembrane protease serine 2 (TMPRSS2) is an enzyme secreted by the epithelial cells in the gastrointestinal tract and the respiratory system.5 Recent research has revealed that TMPRSS2 also plays a role in the priming of the spike protein of the SARS-CoV-2 S (spike) upon binding to the ACE2 receptors. The TMPRSS2 thus supports the fusion of the virus and the host cell membrane which causes infection.58 This enzyme typically is present on the extracellular surface of the cells.5 To enter the host cell, the spike protein of the SARS-CoV-2 virus first binds to the host cell receptor, driving TMPRSS2 to facilitate S protein priming by cleavage of the S protein sequence proteolytically.67 This cleavage enables a conformational change favorable for the virus to enter the host cell via cell–cell and virus–cell fusion.73 The gastrointestinal tract, urogenital epithelium, and respiratory epithelium express TMPRSS2.33 In particular, type II pneumocytes, nasal goblet secretory cells, and ileal absorptive enterocytes co-express TMPRSS2 and ACE2, making them highly susceptible to viral infection. In comparison to TMPRSS2, dipeptidyl peptidase-4 (DPP4), a serine exopeptidase, is expressed in most human cells including lung, kidney, liver, gut, and immune cells.79 DPP4 has been recognized as a potential binding site for the receptor-binding S protein in SARS-CoV and MERS-CoV viruses but not in SARS-CoV-2.84 With detailed investigation, DPP4 inhibition has thus been ruled out as a potential target for developing COVID-19 therapeutics.84
Recent studies have shown that similar to SARS-CoV, SARS-CoV-2 is able to bind to human Angiotensin Converting Enzyme (hACE2).71,82,98 Angiotensin Converting Enzyme-2 (ACE2) is also a functional receptor on the surface of the cells and is found to be highly prevalent in the kidney, lung and cardiac cells. The Renin–angiotensin–aldosterone system (RAAS) relies on ACE2 as a key regulator.9 Analysis of the crystal structure of SARS-CoV-2 receptor binding domain (RBD) with hACE2 exhibited significant functional differences that allow SARS-CoV-2 RBD to have a higher binding affinity to hACE2 than SARS-CoV RBD.73 In the human body, ACE2 performs the essential function of conversion of angiotensin I and II into angiotensin (1–9) and angiotensin (1–7), respectively. This function is integral to the renin–angiotensin–aldosterone system (RAAS) pathway to regulate healthy cardiorespiratory functions.37 Because of this high binding affinity and critical physiological and functional role, despite other receptors being identified as potential therapeutic targets, research has shown that targeting the ACE2 receptors is one of the most potent, smart, and effective ways of targeting and treating the SARS-CoV-2 viral infection. Therefore, significant efforts focus on developing therapeutic approaches that specifically target ACE2 receptors and expression. In this paper, we summarize different ACE-2 targeting strategies for developing smart COVID-19 therapeutics.
Therapeutic Targets for Treatment of SARS-CoV-2
The viral enzymes that contribute to the process of viral replication can serve as potential targets for smart therapeutic drugs to treat the viral infection.56 To target the novel SARS-CoV-2, various antiviral treatments have been developed for inhibition of the action of the RNA-dependent RNA-polymerase enzyme (RdRP) and two viral proteases PLpro and Mpro, which play a significant role in the infection process.56 The RNA polymerase enzyme of the virus assists RNA replication by catalyzing the formation of phosphodiester bonds which is dependent on the RNA template.22 In viruses, the protease enzymes play an important role in the maturation of the viral protein. It does so by clearing out the protein precursors (proproteins) after the translation process into the cytosol of the host cell.52 Therefore, these viral proteases are frequently looked at as potential drug targets in various therapeutic approaches. Inhibiting the viral proteases can limit the turnout of mature viral particles. In SARS-CoV-2, particularly the Mpro is an essential viral protease for the proteolytic release of the enzymes that induce viral replication which include nonstructural protein 13 (nsp 13) that control the NTPase and RNA helicase activity.52
Despite the fact that this therapeutic approach of targeting viral enzymes has many advantages, targeting the proteins of the host cell has shown to be more practical since the proteins of the host cell are more immune to mutations.90 The drug therapies for chronic viruses, such as the human immunodeficiency virus (HIV), are distinctive from therapies for acute viruses such as the SARS-CoV-2. In chronic viral infection, the immune system cannot completely remove the virus whereas in the acute viral infection the immune system can eventually clear the virus completely.90 The rate of viral replication within the host cell is directly correlated with disease severity; therefore, therapeutic strategies typically aim to decrease the viral replication rate.90 Over the course of the pandemic, the use of kinase inhibitors to ameliorate the symptoms of COVID-19 has been studied. However, utilizing antiviral agents, such as inhibitors targeting viral RNA-dependent RNA polymerase (RdRp), does not suppress the replication rate, leading to development of resistance over time.90 Moreover, the RdRp inhibitors have a very narrow time window during which they can be effective, limiting their usage to the early stages of disease.90 While the therapeutic approaches that target the host cell kinases have some advantages over the conventional direct-acting antiviral drugs, the disadvantage associated with this approach is the lack of specificity to infected cells, which results in undesired toxicity to healthy cells. This limits the scope of using kinase inhibitors as a therapeutic approach for mitigating infection due to SARS-CoV-2.66 Therefore, there is an unmet need to identify more effective treatment targets for the novel coronavirus.
To identify the most potent and effective therapeutic target and approach, researchers have turned to already characterized viral binding targets or drugs that successfully alleviate the COVID-19 symptoms. DPP4 is found in many human tissues and is involved in different physiological processes and immune system ailments. For instance, the DPP4/CD26 receptors are expressed on epithelial and endothelial cells which constitute the vasculature, kidney, lung, small intestine, and cardiac function. Specifically the DPP4 distribution in the human respiratory tract facilitates viral entry in the airway tract and contributes to the development of the cytokine storm immunopathology triggering fatal COVID-19 induced pneumonia.77,86 The spike protein on the SARS-CoV-2 has two domains, S1 and S2, which interact with the ACE2 receptor in human cells. The S1 domain acts as a receptor-binding domain and actively interacts with the DPP4 of the host cell, while the S2 domain is involved in membrane fusion.94 The phylogenetic analysis has shown that MERS-CoV is related to Bat-CoV-HKU4, which targets seven DPP4 residues and is predicted to interact with the SARS-CoV-2.88 In addition, DPP4 inhibitors play a role in decreasing the inflammation associated with severe COVID-19.79 DPP4 can also help to block pathways leading to cytokine storms and help alleviate the severity of the disease during COVID-19 infection.
However, it has been established that DPP4 does not directly function as a receptor in SARS-CoV-2 infection resulting in COVID-19 but it helps alleviate the symptoms of the infection.97 Recent studies suggest that SARS-CoV-2 can bind to ACE2 but not to DPP4 expressing cells.79
An alternative therapeutic target is TMPRSS2, which is expressed in human lung cells.29 SARS-CoV-2 utilizes cathepsin, a protease that plays a vital role in protein degradation and processing for S protein priming in TMPRSS2 expressing cells.29 There are 11 classes of cathepsins.62 Among them, cathepsin B and L (CatB/L) have gained interest as a target for developing new therapeutic agents.45 In addition, the expression of TMPRSS2 is partially dependent on CatB/L activities, making CatB/L an essential protein required to drive transmission of the SARS-CoV-2 virus.29 Therefore, blocking both proteases is necessary for effectively inhibiting the entry of the SARS-CoV-2.29
ACE2 directly affects cardiovascular health and functions of various organs such as the heart, kidney, and lungs via counter-regulation of the renin-angiotensin system (RAS).11,24 ACE2 is a crucial receptor for SARS-CoV-2 entry to the human cells.86 Hence, the majority of the therapeutic approaches are focused on targeting the ACE2 receptor in an attempt to block the viral entry and prevent or significantly reduce the risk of infection.86
ACE2-Targeted Therapeutic Approaches
MicroRNA Based Therapeutics Targeting ACE2
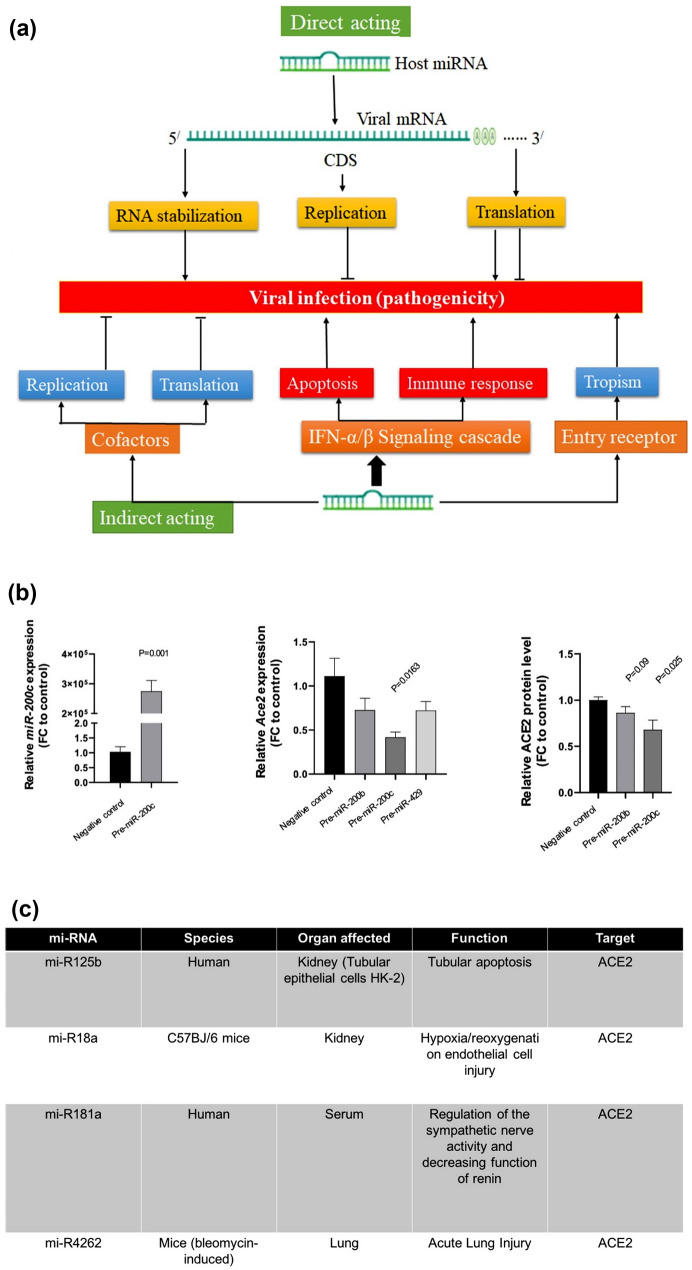
The microRNA (miRNA) is a small noncoding double-stranded RNA molecule that regulates post-transcriptional gene expression and guides molecules in RNA silencing.25,31 An ample number of miRNAs are present in human cells, and their biogenesis is linked to various diseases (Fig. 2).13,78 The miRNA can target the binding sites of ACE2 transcripts in the three prime untranslated regions (3’-UTR), thereby inhibiting ACE2 expression.48 Recent studies revealed the regulation of the ACE2 expression and the RAS system by miRNAs as they can act as autocrine, paracrine and endocrine regulators.21 This finding indicates the important role that miRNA plays in regulating the renin–angiotensin–aldosterone system (RAAS) pathway.18 RAAS is an endocrine system that plays an important role in regulation of blood pressure or hypertension development.18 A study by Dongchao et al. demonstrated the use of miRNA to regulate ACE2 expression in cardiac cells, thus influencing the rate of infection of SARS-CoV-2.48 In this study, human induced pluripotent stem cells (hiPSC)-derived cardiomyocytes were transfected with three therapeutic miRNAs: miR-429, -200b, and -200c. A quantitative PCR was performed utilizing miRNA-specific TaqMan to evaluate the ACE2 mRNA expression levels.48 These miRNAs were selected because they displayed highest 3’UTR binding site conservation among in silico candidates. Western blotting techniques were used to evaluate the ACE2 protein levels in the transfected cells. Of the three therapeutic miRNAs, miR-200c has displayed the most promising results by strongly diminishing the expression of the ACE2.48 This study successfully demonstrated the use of miR-200c to regulate ACE2 expression in cardiomyocytes. However, researchers must acknowledge that the patients with severe cardiovascular diseases (CVD) could be more susceptible to the severe symptoms of COVID-19 infection due to the suppression of the ACE2 expression. An increase in the ACE2 expression could mitigate the CVD symptoms but increase the risk of virus infection. This therapeutic approach for treating COVID-19 should be employed in the patients with cardiovascular diseases cautiously while meticulously monitoring the link between RAAS, ACE2, and miRNA. Angiotensin-converting enzyme inhibitors and angiotensin receptor blockers are typically administered to cardiac patients, reducing cardiac stress and increasing ACE2 expression and activity. The increased levels of ACE2 can make cells more susceptible to infection by SARS-CoV-2. Studying the correlation between ACE2 expression and miRNA-based therapy has garnered much interest in the scientific community and could be an intermediate therapeutic intervention until effective vaccines are available that can keep up immunity against newer emerging variants of SARS-CoV-2.48
Figure 2.
miRNAs play a crucial role in the replication of viruses and can be used by infected cells to regulate viral replication and have therefore been investigated for their potential to control and mitigate viral infection due to SARS-CoV-2. (a) It has been documented in the literature that human miRNAs can indirectly bind to the viral genome in both the coding and noncoding 3′UTR regions, suppressing translation and having an antiviral effect as a result.43 Evidence from the literature has suggested that host-miRNAs bind to viral-RNA, controlling its translation and altering viral pathogenesis.38 Micro-RNA Response Elements (MREs), which are often present in the 5′UTR, coding region (CDS), and 3′UTR of the virus genetic material, assist the underlying mechanism for host miRNA-viral RNA interactions. Viral replication directly affects the following outcomes, which have been used to pinpoint these interactions: (i) Viral mRNA is suppressed when host miRNA attaches to its 3′-NTR. (ii) When host miRNA attaches to the viral mRNA's coding area, the viral genes' ability to translate is suppressed, which stops the virus from replicating. (iii) Viral RNA is stabilized as a result of host miRNA binding to the 5′-NTR of the viral mRNA, which improves viral replication (Fig. 2). (b) mi-R200c show the lowest ACE2 expression and lower the susceptibility of cardiomyocytes to viral infection. (c) A summary of mi-RNA that target ACE2 receptors. (Adapted and modified with permission from Paul et al.,63 Widiasta et al.,91 Abu-Izneid et al.4 and Dongchao et al.48).
Antiviral Drugs Targeting ACE2
ACE2 targeted clinical therapeutic drugs are increasingly utilized to treat COVID-19 patients.6 Two main concerns arise for clinical targeting of ACE2 receptors as treatment for viral infection.6 Many clinically approved medications routinely used for the treatment of COVID-19 symptoms and underlying comorbidities have evidently been known to raise the mRNA expression of ACE2.6 During the early stages of the pandemic, the increase in mRNA expression of ACE2 has raised uneasiness about the harmful implications of ACE2-targeted-drugs on the health of the infected patients with comorbid conditions. Regardless, researchers have confirmed that such medication can help alleviate the symptoms of the disease with no significant increased risk or compounded danger of viral infection. This has led to a speculation that ACE inhibitors (ACEIs) and Angiotensin receptor blockers (ARBs) may not be detrimental to COVID-19 patients and can be explored as a therapeutic alternative to manage the symptoms of the disease.6 Previous studies have exhibited clinically approved drugs targeting ACEI, e.g., Enalapril and Lisinopril and ARB, e.g., Arbidol, Losartan, Olmesartan, and Irbesartan, can increase ACE2 expression in plasma and heart tissues.14
Experimental studies have suggested increased expression of ACE2 mRNA with the use of clinical drugs. ARB drugs such as Losartan was studied in plasma, heart, and kidney of Lewis rats and transgenic Ren2 rats. Olmesartan was analyzed in the aorta, plasma, and urine of spontaneously hypertensive rats (SHR), Lewis rats, and hypertensive human patients. Irbesartan was examined in the heart and aorta of C57BL/6 mice.23,32,35 Likewise, ACEI drugs such as Enalapril were studied in the heart and plasma of Sprague Dawley rats. Lisinopril was analyzed in the heart, kidney, and plasma of transgenic ren2 and Lewis rats.23,31,35,60 ARBs have displayed increased expression of ACE2 mRNA in animal models in the heart, plasma, kidney, and aorta. The quantification of mRNA and protein was performed utilizing RT-PCR. Although cardiac ACE2 mRNA expression did not alter in myocardial infarction (MI) in vehicle-treated rats, Losartan and Olmesartan both increased the ACE2 mRNA by 97 and 42 percent, respectively.32 Similarly, a significant increase in ACE2 expression was displayed in cardiac and kidney transgenic Ren2 rats.40 In a separate study, TaqMan real-time PCR analysis and western blotting demonstrated that utilizing Irbesartan increased the ACE2 mRNA in the heart and aorta of C57BL/6 mice.43 Particularly, studies concerning the Olmesartan drug in aorta suggested a fivefold increase in ACE2 mRNA expression in RT-PCR analysis and an increase in ACE2 antibodies in immunostaining assay in SHR rats.60 ACEIs have also displayed increased ACE2 mRNA expression in animal plasma, the heart, and the kidney; RT-PCR suggested an increase in ACE2 activity was caused by utilizing Enalapril in the heart of Sprague Dawley rats.41 Furthermore, studies utilizing the Lisinopril drug also suggested the similar corresponding increase in ACE2 activity in heart and kidney of the animal models.35 Therefore, the use of ACE2 targeting drugs have opened up a new domain of therapeutics which could help alleviate symptoms of the COVID-19 infection effectively.
Monoclonal Antibody Therapy Targeting ACE2
Another alternative therapeutic approach is utilizing passive immunotherapy to treat and prevent viral infection.75 The humoral immune response actively contributes to viral infection recovery.34 Of the two innate and adaptive immune system response mechanisms, the humoral immune response is a part of the adaptive immune defense mechanisms mediated by antibodies. Apart from antibody production, many other processes could concurrently occur in response to antigen or virulent particle exposure. The initial step to the humoral immune response is neutralizing the toxin, which slows down the replication of viruses. Simultaneously the complement system which comprises of an assembly of proteins found in the blood and other fluids in the body and surfaces of the cells also gets activated, in response to the antigen.50 The soluble parts of the complement system initiate a proteolytic cascade which in turn triggers the complement effectors that can target the cells to elicit an immune response.50 The activation of these complement effectors enhances the antibody activity to remove damaged cells and assist inflammation.34 Lastly, opsonins, which are extracellular proteins that can bind to antigens can induce phagocytosis by acting as a tag.45 Neutralizing antibodies targeting RBD, induced by SARS-CoV-2, positively correlate with disease outcomes.12,25 There is a multitude of antibody-neutralization mechanisms to block viral entry and prevent infection.48–50 Antibodies can cluster viruses to block their entry and prevent binding to cell surface receptors. Moreover, antibodies can bind to the host receptor further stopping the entry of the virus into the cell. Antibodies can also hinder conformational changes required for cell membrane fusion. Other antibody-neutralization mechanisms include the antibody impairing the virus, preventing the removal of the viral capsid, and inhibiting viral replication and release.48–50
Monoclonal antibodies (mAbs) constitute a crucial class of biotherapeutics (Fig. 3). The mAbs are hybridoma white blood cells produced from spleen and myeloma cells.39 Studies investigating the role of mAbs to neutralize SARS-CoV and MERS-CoV viruses have shown promising results in preventing viral escape.8,80,85,87 The majority of SARS-CoV treatments consist of antibodies that target the N and C terminals of the RBD. For neutralizing SARS-CoV-2 via monoclonal antibody studies, anti-ACE2 monoclonal antibodies (mAb) were utilized.75 Analysis revealed two neutralization epitopes, 201, which binds to the receptor binding domain, and 68, which binds to the outer surface of the domain. Immunoprecipitation and western blotting were performed to detect the membrane-bound S protein using the monoclonal antibody (mAb).26 Previous research has revealed that the catalytic site and the RBD-binding site of ACE2 do not overlap.19 It was therefore hypothesized that the RBD binding site on ACE2 with antibodies can effectively block the entry of all ACE2 dependent viruses while preserving the physiological functions of the ACE2.19 This approach that uses monoclonal antibodies to target ACE2 can be a promising therapeutic avenue to mitigate the SARS-CoV-2 infection.
Figure 3.
The use of monoclonal antibody therapy for preventing binding of SARS-CoV-2 to the ACE2 receptor as potential treatment of SARS-CoV-2. (a) Schematic representation for the neutralization mechanism of SARS-CoV-2. The SARS-CoV-2 spike protein-targeting monoclonal antibodies may limit the ability of the virus to attach to its cellular receptor, thereby preventing the virus from entering the cell. (b) SARS-CoV-targeting neutralizing monoclonal antibodies and their mode of action. These spike protein-targeting monoclonal antibodies showed promising results both in vitro and in vivo and could be potentially effective in combating SARS-CoV-2. (Adapted and modified with permission from Shanmugaraj et al.75).
On November 12, 2020, the FDA issued an Emergency Use Authorization (EUA) for imdevimab, and casirivimab, humanized monoclonal antibodies (IgG1) to be administered intravenously for the treatment of mild to moderate COVID-19 in adults and pediatric patients with positive results of direct SARS-CoV-2 viral testing.1 The pediatric patients were stipulated to be 12 years of age or older and weighing at least 40 kg, to be eligible for the intravenous treatment. Those at high risk for progressing to severe COVID-19 were also eligible for treatment which included those who were 65 years of age or older or who had certain chronic medical conditions. In the clinical trial of patients with severe COVID-19, casirivimab and imdevimab were administered. They showed a decrease in COVID-19-related hospitalization and emergency room visits in high-risk patients within 4 weeks after treatment compared to the placebo. The safety and effectiveness of this experimental therapy for the treatment of COVID-19 continues to be evaluated.1 It was found in a study by Chen et al that the monoclonal antibody 3E8 binds to human ACE2 with moderate affinity. The results of this study also reflected 3E8 was able to successfully block the SARS-CoV-2 infection both in vitro and in vivo within a prophylactic COVID-19 infected mouse model.19 For clinical development, full evaluation must be performed in non-human primates before this approach could advance into clinical trials. The results provided a potential broad spectrum and potent strategy to combat all coronaviruses that can bind to ACE2 for entry and is revealed as an anti-coronavirus epitope on human ACE2.19
Cell-Based Therapies Targeting ACE2
There has been an increase in cell therapy based clinical investigations utilized to treat the coronaviruses.38 Cell therapies use cell replacement to repair the pathways that initiate the illness and promote its progression.15 Mesenchymal (stromal) stem cells (MSCs) have been widely used for cell-based therapy for the treatment of respiratory ailments caused by COVID-19 infection.20,92 The majority of the MSCs are present in the blood vessels through capillary endothelial cell interactions.38 Many anti-inflammatory mediators such as anti-inflammatory cytokines, antimicrobial peptides, angiogenic growth factors, and extracellular vesicles are released during virus-induced lung damage and regulation of immune inflammation is mediated through receptors expressed in MSCs.30,38,40,43
Currently, there are two types of therapeutic approaches in which the MSCs are utilized in the treatment of COVID-19. The first therapeutic strategy constitutively increases the interferon (IFN)-stimulating gene (ISGs), acts as a mediator and secondary response to IFN, triggering ISG which generates resistance against the respiratory virus, including the coronavirus.38 The alternative approach is comprised of both the intrinsic and inducible innate defense against viruses.38 Studies suggest that the activation of various receptors such as Toll-like receptors (TLRs) 3 and 4, interleukin (IL)-6, IL-8, and CXCL10, mediates the release of anti-inflammatory mediators, specific to the cellular environment.46,89 Furthermore, the angiopoietin-1 secreted by MSCs helps repair the disrupted alveolar-capillary barriers.42 ACE2 receptors are widely distributed in human cells and are primarily present in the capillary endothelium and the alveolar type II cells (AT2).27 In the recent studies, ACE2 has also been suggested to be an ISG in nasal epithelial cells.100 SARS-CoV-2 may utilize the overexpression of ACE2 to increase viral infection. Therefore, ACE2 must be monitored as the increase in expression of the anti-inflammatory mediators directly correlates with the expression of the ACE2.
In a separate study, Leng et al. investigated patients infected with coronavirus after the bone marrow-derived MSCs (BM-MSCs) transplantation in a clinical trial.44 Seven patients with confirmed COVID-19 diagnosis, including one critically severe type (patient 1), four severe types (patients 2, 3, 6, 7), and two common types (patients 4, 6) were candidates for MSCs transplantation. Simultaneously, three severe infected types were listed to receive the placebo control. Prior to the MSC transplantation, it was reported that patients had symptoms of very high fever, weakness, shortness of breath, and low oxygen saturation levels.
Results demonstrated that just within 2–4 days post-transplantation, these symptoms were alleviated entirely in all of the patients. Additionally, there was no acute allergic reaction or infusion-related or allergic reactions post transplantation. Moreover, no delayed secondary infections or hypersensitivity was observed after the treatment.44 These findings show a strong therapeutic promise to treat patients in severe stages of the disease.
ACE2-RBD Blockers
ACE2 proteins are present on the cell surfaces of many organs such as the kidney, placenta, lung, duodenum, small intestine, and pancreas.49 In SARS-CoV-2, the spike protein is present on the outer surface of the virus.100 The spike domain of the SARS-CoV-2 is comprised of two domains, S1 and S2.100 The S1 domain contains the ACE2 receptor binding protein (RBD), responsible for the virus-receptor interaction.100 In contrast to the S1 domain, the S2 domain comprises the heptad repeat (HR) domain, responsible for viral fusion.100 Earlier sequencing studies reported that in the SARS-CoV-2 genome, the residue 394 (glutamine) in the RBD domain was recognized by the lysine 31 on the human ACE2 receptor, thus establishing binding affinity towards ACE2.44 This interaction is necessary for virus entry and replication. Currently, there are four major categories of targeted therapies for cell entry mechanisms. The four categories for cell entry mechanism is comprised of targeting the virus receptors, the spike protein, the membrane fusion, and the endosomal and non-endosomal virus entry.69 Studies also demonstrate that targeting the viral entry by neutralizing the spike protein could be the most efficient treatment.10,17,49,74
The cell lines used for the in vitro studies of SARS-CoV-2 entry are HEK 293T, BHK-21, Caco-2, Vero, Vero E6, and Calu-3 cells. The majority of these in vitro cell entry mechanism studies target either cell receptors that allow viral attachments, the SARS-CoV-2 spike protein, or enzymes enabling viral fusion. For inhibiting viral attachment, the target can be the spike glycoprotein by neutralization.29 The neutralization of the SARS-CoV-2 spike protein is achieved by various means such as neutralizing antibodies, utilizing recombinant or soluble ACE2, utilizing small peptides and molecules, or by using sera.29 Viral fusion inhibition is accomplished by targeting fusion between the virus and the host cell. Similarly, this could also be achieved by inhibiting the endosome or non-endosome-dependent pathways.29 Mechanisms that target ACE2 receptors on host cells are proving to be an effective way to neutralize the inhibition of viral entry.29 To effectively neutralize the inhibition of viral entry, a polyclonal anti-ACE2 antibody, AF933, against human ACE2 was utilized and successfully inhibited SARS-CoV-2 entry.29 Alternative strategies utilize the recombinant SARS-CoV-2 RBD protein, constructed using the C-terminal Fc of human IgG1.81 Furthermore, this strategy has shown to effectively inhibit the viral attachment onto hACE2-expressing 293T cells in a dose-dependent manner.81 Western blotting and SDS-PAGE analysis of the purified SARS-CoV-2 RBD protein and antisera, had contained SARS-CoV RBD-specific antibodies.28 A flow cytometry analysis was carried out to detect the RBD protein binding to hACE2 in 293T cells. In addition, immunofluorescence staining was performed to detect the binding between RBD and the hACE2 receptor.28 ELISA assays were conducted to detect the binding of SARS-CoV RBD to the sACE2 receptor and binding of SARS-CoV-2 to soluble hDPP4. Subsequently, a cross-titration assay was performed in which two proteins overlap for achieving a higher sensitivity to target the ACE2-RBD interaction. Specifically, the ACE2-His-Avi and RBD-Fc was used as the two proteins in this study. This study utilized a cross-titration assay to determine the concentration of the RBD-Fc and ACE2 and AlphaLISA assay to measure the binding of RBD to ACE2.28 They also performed a qRT-PCR, qPCR for ACE2 mRNA, and ELISA assay for quantifying the corresponding ACE2 expression. Collectively, these analyses presented a simple and rapid approach for therapeutic development by targeting the ACE2-RBD. The results can be used for designing further structure–activity studies and its connection in SARS-CoV-2 infection. Targeting the ACE2-RBD serves as a promising platform for potential drug screening that can inhibit viral infection.28
Inhalable GapmeR and Recombinant ACE2
The messenger RNA (mRNA) is a single-stranded RNA molecule complementary to the gene’s DNA strand.68 Gene silencing is a mechanism which prevents the of expression of a specific gene in a cell.68 This process can be executed by removing the gene of interest, regulating transcriptional gene expression level, or inhibiting gene expression at the translational level.80 Gene silencing can be conducted by utilizing antisense RNA technology. Alternatively, RNA interference (RNAi) can also be used.68 The RNAi is a biological mechanism that uses the DNA sequence of a gene to suppress its own expression, known as silencing.68 The designed antisense RNA is complementary to the targeted mRNA strand. With the help of Riboneuclease H (RNase H) enzyme that catalyzes the process of RNA cleavage, the antisense RNA degrades the targeted mRNA. GapmeR is a chimeric antisense oligonucleotide containing monomers of deoxynucleotide. Studies have shown that using GapmeR, as an antisense oligonucleotide, increased the efficacy of this technology. These single-stranded, antisense oligonucleotides can be easily taken up by the targeted cells indicating that transfection is not needed.22 GapmeR can be used in an aerosol or powdered form, therefore, can be inhalable.47
Studies have shown that when the patient is suffering from severe acute lung failure, the human recombinant soluble ACE2 (hrACE2) prevents the entry of the SARS-CoV-2 virus particles inside healthy cells and thus helps in reducing viral replication.2 SARS-CoV-2 actively targets cells expressing ACE2 such as the alveolar epithelial cells of the lung, cardiac muscle cells, esophagus epithelial cells, urothelial cells, kidney tubules, ocular cells, colon, and epithelial cells of the oral mucosa.28,95,96 This indicates that hrsACE2 can reduce viral replication, but it cannot completely eliminate it. Therefore, it was hypothesized that simultaneous action of the hrsACE2 by blocking the entry of the SARS-CoV-2 and GapmeR, and silencing the genome of the virus particles will synergistically eliminate the viral replication in the host cells.96 This result can be achieved by using novel strategies that employ screening and selection of GapmeR designs, and deliver the potential candidates to the lung using in vitro models and organoids. Subsequently the optimal dosage and selection of the optimal cytotoxicity profile can be performed based on the experimental findings.
A summary for ACE2 targeting COVID-19 therapies is provided in Table 1.
Table 1.
Summary of the in vitro therapeutic approaches for treatment COVID-19 by targeting ACE2 expression.
| Treatment approach | Mechanism | ACE2 level | In vitro techniques |
|---|---|---|---|
| microRNA | Inhibit viral translation | Decreased | RT-PCR |
| Antiviral drugs | Inhibit viral entry | Increased | RT-PCR, immunostaining assay |
| Monoclonal antibody | Neutralize antibody | Decreased | Microtiter assay, virus titration assay, immunoprecipitation, and western blotting |
| MSCs | Mesenchymal stromal cell-based therapy | Decreased | RNA sequencing |
| ACE2-RBDs | Inhibit viral entry | Decreased | qRT-PCR, qPCR for ACE2 mRNA, and ELISA |
| Inhalable GapmeR | Antisense RNA | Decreased | In vitro models and organoids |
Discussion and Future Perspectives
The coronavirus has emerged as a global pandemic and claimed millions of lives. The unmet clinical need for therapeutic approaches to combat this deadly virus are critical to preserve and improve the health of the infected population. There are currently various therapeutic targets in research for COVID-19 treatment. With the advent of vaccines and boosters, gaining control of the severity of the infected patients is expected to improve over time. However, the ever-growing number of variants pose a significant threat to invoking an imminent rise in the cases. Discovering a safe target for therapeutic development is crucial to prevent further viral transmission. Although the high mutation rate of RNA viruses allows them to adapt swiftly to changing environmental conditions, it also renders them vulnerable because there are a limited number of mutations in the essential genes that can accumulate. The Delta variation of the SARS-CoV-2 virus, that caused COVID-19, had a higher infectious transmission rate than earlier variants of the virus.7 The enhanced ACE2 activity would imbalance of RAAS and ACE2-Ang-(1–7) pathways. The more recent growing cases of the Omicron variant still pose an active threat and contribute to the rise in cases all over the globe.53 It is essential and strongly recommended that public health and safety guidelines are strictly obeyed, and vaccines and boosters are encouraged, to help mitigate another crisis wave. Studies have validated that, therapeutic approaches targeting ACE2 displayed promising results in the development of long-term clinical treatments targeting COVID-19. Recent investigations have also ventured into novel ways of targeting ACE2 as a potential therapeutic target such as using decoy ACE2 molecules to trap the virus particles, using pseudoligands to dominate the binding site for SARS-CoV-2, blocking antibodies against ACE2 viral docking sites, using ACE2 inhibitors against the docking sites which have all shown promising results.36 Similarly several approaches to enhance ACE2 shedding and using reagents to promote and inhibit ACE2 internalization have also shown to ease the symptomatic side effects of SARS-CoV-2 infection while limiting the possibility of infection.36 While devising ACE2 targeting strategies, the two basic characteristics of ACE2 must be accounted for: (1) the ACE2 enzymatic activity and S-protein binding both influence viral entry but are independent and (2) both soluble and membrane bound ACE2 share the same viral binding and enzymatic functional properties. Another point of consideration is the stage of the disease and any associated comorbidities, since they would play a critical role in developing an effective therapeutic strategy. For instance, approaches that reduce viral infectivity should be prioritized during the early stage of the infection and strategies to increase ACE2 activity should be considered to treat disease induced inflammation and damage in the advanced stages of disease progression.36 There is a high demand for utilizing ACE2-modulating medications to obtain favorable results in COVID-19 infected patients. The promising potential of ACE2 in controlling viral entry has led to the development of various versatile therapeutic approaches. Future therapies must incorporate efforts to understand how ACE2 expression can be modulated effectively to minimize any side effects of the therapies that elevate or diminish ACE2 levels essential for healthy physiological function.
Acknowledgments
This work was partly supported by the University of Massachusetts Lowell faculty start-up funds and the Transformational Project Award from the American Heart Association (AHA) (19TPA34910111).
Conflict of interest
The authors declare no conflicts of interest.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- ANGI/II
Angiotensin I/II
- CNS
Central Nervous system
- TMPRSS2
Transmembrane protease, serine2
- RdRp
RNA‐dependent RNA polymerase
- DPP4
Dipeptidyl peptidase-4
- PAI-1
Plasminogen activator inhibitor-1
- ARB
Angiotensin II receptor blockers
- ACEI
Angiotensin-converting enzyme inhibitors
- RBD
Receptor binding domain
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coronavirus (COVID-19) Update. FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19. US Food and Drug Administration, 2020.
- 2.Abd El-Aziz TM, Al-Sabi A, Stockand JD. Human recombinant soluble ACE2 (hrsACE2) shows promise for treating severe COVID-19. Signal Transduc. Target Therapy. 2020;5:258–258. doi: 10.1038/s41392-020-00374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdelrahman Z, Li M, Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza a respiratory viruses. Front. Immunol. 2020;11:552909–552909. doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu-Izneid T, AlHajri N, Ibrahim AM, Javed MN, Salem KM, Pottoo FH, Kamal MA. Micro-RNAs in the regulation of immune response against SARS CoV-2 and other viral infections. J. Adv. Res. 2021;30:133–145. doi: 10.1016/j.jare.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguiar JA, Tremblay BJ, Mansfield MJ, Woody O, Lobb B, Banerjee A, Chandiramohan A, Tiessen N, Cao Q, Dvorkin-Gheva A. Gene expression and in situ protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue. Eur. Respir. J. 2020;56:270. doi: 10.1183/13993003.01123-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akhtar S, Benter IF, Danjuma MI, Doi SA, Hasan SS, Habib AM. Pharmacotherapy in COVID-19 patients: a review of ACE2-raising drugs and their clinical safety. J. Drug Target. 2020;28:683–699. doi: 10.1080/1061186X.2020.1797754. [DOI] [PubMed] [Google Scholar]
- 7.Alexandar S, Ravisankar M, Kumar RS, Jakkan K. A comprehensive review on Covid-19 Delta variant. Int. J. Pharmacol. Clin. Res. 2021;5:7. [Google Scholar]
- 8.Berry J. D., K. Hay, J. M. Rini, M. Yu, L. Wang, F. A. Plummer, C. R. Corbett, and A. Andonov. Neutralizing epitopes of the SARS-CoV S-protein cluster independent of repertoire, antigen structure or mAb technology. In: MAbs. Taylor & Francis, 2010, pp. 53–66. [DOI] [PMC free article] [PubMed]
- 9.Beyerstedt S, Casaro EB, Rangel ÉB. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:905–919. doi: 10.1007/s10096-020-04138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee S-S, Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J. Med. Virol. 2020;92:618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bian J, Li Z. Angiotensin-converting enzyme 2 (ACE2): SARS-CoV-2 receptor and RAS modulator. Acta Pharmac. Sin. B. 2021;11:1–12. doi: 10.1016/j.apsb.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisht H, Roberts A, Vogel L, Bukreyev A, Collins PL, Murphy BR, Subbarao K, Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. 2004;101:6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bronze-da-Rocha E. MicroRNAs expression profiles in cardiovascular diseases. BioMed Res. Int. 2014;2014:1–23. doi: 10.1155/2014/985408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooksbank JA, Greene SJ, Harris HM, Stebbins A, Fiuzat M, Whellan DJ, Alhanti B, Kraus WE, Piña IL, O'Connor CM. Beta-blocker and ACE-inhibitor dosing as a function of body surface area: from the HF-ACTION trial. Am. Heart J. 2021;233:1–4. doi: 10.1016/j.ahj.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Buzhor E, Leshansky L, Blumenthal J, Barash H, Warshawsky D, Mazor Y, Shtrichman R. Cell-based therapy approaches: the hope for incurable diseases. Regener. Med. 2014;9:649–672. doi: 10.2217/rme.14.35. [DOI] [PubMed] [Google Scholar]
- 16.Cárdenas-Rodríguez N, Bandala C, Vanoye-Carlo A, Ignacio-Mejía I, Gómez-Manzo S, Hernández-Cruz EY, Pedraza-Chaverri J, Carmona-Aparicio L, Hernández-Ochoa B. Use of antioxidants for the neuro-therapeutic management of COVID-19. Antioxidants. 2021;10:971. doi: 10.3390/antiox10060971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L-J, Xu R, Yu H-M, Chang Q, Zhong J-C. The ACE2/apelin signaling, microRNAs, and hypertension. Int. J. Hypertens. 2015;215:1–6. doi: 10.1155/2015/896861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Zhang Y-N, Yan R, Wang G, Zhang Y, Zhang Z-R, Li Y, Ou J, Chu W, Liang Z, Wang Y, Chen Y-L, Chen G, Wang Q, Zhou Q, Zhang B, Wang C. ACE2-targeting monoclonal antibody as potent and broad-spectrum coronavirus blocker. Signal Transduc. Target. Therapy. 2021;6:315. doi: 10.1038/s41392-021-00740-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connick P, Kolappan M, Crawley C, Webber DJ, Patani R, Michell AW, Du M-Q, Luan S-L, Altmann DR, Thompson AJ, Compston A, Scott MA, Miller DH, Chandran S. Autologous mesenchymal stem cells for the treatment of secondary progressive multiple sclerosis: an open-label phase 2a proof-of-concept study. Lancet Neurol. 2012;11:150–156. doi: 10.1016/S1474-4422(11)70305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fani M, Zandi M, Ebrahimi S, Soltani S, Abbasi S. The role of miRNAs in COVID-19 disease. Fut. Virol. 2021;16:301–306. doi: 10.2217/fvl-2020-0389. [DOI] [Google Scholar]
- 22.Fazil MH, Ong ST, Chalasani ML, Low JH, Kizhakeyil A, Mamidi A, Lim CF, Wright GD, Lakshminarayanan R, Kelleher D, Verma NK. GapmeR cellular internalization by macropinocytosis induces sequence-specific gene silencing in human primary T-cells. Sci. Rep. 2016;6:37721. doi: 10.1038/srep37721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrario CM, Jessup J, Chappell MC, Averill DB, Brosnihan KB, Tallant EA, Diz DI, Gallagher PE. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111:2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- 24.Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong J-C, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glass WG, Subbarao K, Murphy B, Murphy PM. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 26.Greenough TC, Babcock GJ, Roberts A, Hernandez HJ, Thomas WD, Jr, Coccia JA, Graziano RF, Srinivasan M, Lowy I, Finberg RW, Subbarao K, Vogel L, Somasundaran M, Luzuriaga K, Sullivan JL, Ambrosino DM. Development and characterization of a severe acute respiratory syndrome—associated coronavirus—neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 2005;191:507–514. doi: 10.1086/427242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamming I, Timens W, Bulthuis M, Lely A, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanson QM, Wilson KM, Shen M, Itkin Z, Eastman RT, Shinn P, Hall MD. Targeting ACE2–RBD interaction as a platform for COVID-19 therapeutics: development and drug-repurposing screen of an AlphaLISA proximity assay. ACS Pharmacol. Transl. Sci. 2020;3:1352–1360. doi: 10.1021/acsptsci.0c00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffmann M, Kleine-Weber H, Krüger N, Müller M, Drosten C, Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 2020;60:1136. [Google Scholar]
- 30.Hu S, Park J, Liu A, Lee J, Zhang X, Hao Q, Lee J-W. Mesenchymal stem cell microvesicles restore protein permeability across primary cultures of injured human lung microvascular endothelial cells. STEM CELLS Transl. Med. 2018;7:615–624. doi: 10.1002/sctm.17-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Igase M, Strawn WB, Gallagher PE, Geary RL, Ferrario CM. Angiotensin II AT1 receptors regulate ACE2 and angiotensin-(1–7) expression in the aorta of spontaneously hypertensive rats. Am. J. Physiol.-Heart Circ. Physiol. 2005;289:H1013–H1019. doi: 10.1152/ajpheart.00068.2005. [DOI] [PubMed] [Google Scholar]
- 32.Ishiyama Y, Gallagher PE, Averill DB, Tallant EA, Brosnihan KB, Ferrario CM. Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin II receptors. Hypertension. 2004;43:970–976. doi: 10.1161/01.HYP.0000124667.34652.1a. [DOI] [PubMed] [Google Scholar]
- 33.Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janeway C., P. Travers, M. Walport, and M. Shlomchik. Immunologists' toolbox. Immunobiology, 5th ed. New York: Garland Science Publishing, pp. 613–660, 2001.
- 35.Jessup JA, Gallagher PE, Averill DB, Brosnihan KB, Tallant EA, Chappell MC, Ferrario CM. Effect of angiotensin II blockade on a new congenic model of hypertension derived from transgenic Ren-2 rats. Am. J. Physiol. Heart Circ. Physiol. 2006;291:H2166–H2172. doi: 10.1152/ajpheart.00061.2006. [DOI] [PubMed] [Google Scholar]
- 36.Jia H, Neptune E, Cui H. Targeting ACE2 for COVID-19 therapy: opportunities and challenges. Am. J. Respir. Cell Mol. Biol. 2021;64:416–425. doi: 10.1165/rcmb.2020-0322PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kar M. Vascular dysfunction and its cardiovascular consequences during and after COVID-19 infection: a narrative review. Vasc. Health Risk Manag. 2022;18:105. doi: 10.2147/VHRM.S355410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khoury M, Cuenca J, Cruz FF, Figueroa FE, Rocco PRM, Weiss DJ. Current status of cell-based therapies for respiratory virus infections: applicability to COVID-19. Eur. Respir. J. 2020;55:2000858. doi: 10.1183/13993003.00858-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 40.Krasnodembskaya A, Song Y, Fang X, Gupta N, Serikov V, Lee J-W, Matthay MA. Antibacterial effect of human mesenchymal stem cells is mediated in part from secretion of the antimicrobial peptide LL-37. Stem Cells. 2010;28:2229–2238. doi: 10.1002/stem.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lantigua D. M. C., and G. C. Unal. Point of care platform for detection of SARS-CoV-2/COVID-19. J Infect Dis Ther 2022.
- 42.Lee JW, Krasnodembskaya A, McKenna DH, Song Y, Abbott J, Matthay MA. Therapeutic effects of human mesenchymal stem cells in ex vivo human lungs injured with live bacteria. Am. J. Respir. Critical Care Med. 2013;187:751–760. doi: 10.1164/rccm.201206-0990OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee RH, Pulin AA, Seo MJ, Kota DJ, Ylostalo J, Larson BL, Semprun-Prieto L, Delafontaine P, Prockop DJ. Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell. 2009;5:54–63. doi: 10.1016/j.stem.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leng Z, Zhu R, Hou W, Feng Y, Yang Y, Han Q, Shan G, Meng F, Du D, Wang S, Fan J, Wang W, Deng L, Shi H, Li H, Hu Z, Zhang F, Gao J, Liu H, Li X, Zhao Y, Yin K, He X, Gao Z, Wang Y, Yang B, Jin R, Stambler I, Lim LW, Su H, Moskalev A, Cano A, Chakrabarti S, Min K-J, Ellison-Hughes G, Caruso C, Jin K, Zhao RC. Transplantation of ACE2(-) mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11:216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y-Y, Fang J, Ao G-Z. Cathepsin B and L inhibitors: a patent review (2010-present) Expert Opin. Therap. Patents. 2017;27:643–656. doi: 10.1080/13543776.2017.1272572. [DOI] [PubMed] [Google Scholar]
- 46.Liotta F, Angeli R, Cosmi L, Filì L, Manuelli C, Frosali F, Mazzinghi B, Maggi L, Pasini A, Lisi V, Santarlasci V, Consoloni L, Angelotti ML, Romagnani P, Parronchi P, Krampera M, Maggi E, Romagnani S, Annunziato F. Toll-like receptors 3 and 4 are expressed by human bone marrow-derived mesenchymal stem cells and can inhibit their T-cell modulatory activity by impairing notch signaling. STEM CELLS. 2008;26:279–289. doi: 10.1634/stemcells.2007-0454. [DOI] [PubMed] [Google Scholar]
- 47.Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, Liao P, Qiu J-F, Lin Y, Cai X-F, Wang D-Q, Hu Y, Ren J-H, Tang N, Xu Y-Y, Yu L-H, Mo Z, Gong F, Zhang X-L, Tian W-G, Hu L, Zhang X-X, Xiang J-L, Du H-X, Liu H-W, Lang C-H, Luo X-H, Wu S-B, Cui X-P, Zhou Z, Zhu M-M, Wang J, Xue C-J, Li X-F, Wang L, Li Z-J, Wang K, Niu C-C, Yang Q-J, Tang X-J, Zhang Y, Liu X-M, Li J-J, Zhang D-C, Zhang F, Liu P, Yuan J, Li Q, Hu J-L, Chen J, Huang A-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 48.Lu D, Chatterjee S, Xiao K, Riedel I, Wang Y, Foo R, Bär C, Thum T. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020;148:46–49. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am. J. Pathol. 2007;171:715–727. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marty AM, Jones MK. The novel coronavirus (SARS-CoV-2) is a one health issue. One Health. 2020;9:100123. doi: 10.1016/j.onehlt.2020.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mengist HM, Dilnessa T, Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021 doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohapatra RK, El-Shall NA, Tiwari R, Nainu F, Kandi V, Sarangi AK, Mohammed TA, Desingu PA, Chakraborty C, Dhama K. Need of booster vaccine doses to counteract the emergence of SARS-CoV-2 variants in the context of the Omicron variant and increasing COVID-19 cases: an update. Hum. Vacc. Immunotherap. 2022;18:1–9. doi: 10.1080/21645515.2022.2065824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murgolo N, Therien AG, Howell B, Klein D, Koeplinger K, Lieberman LA, Adam GC, Flynn J, McKenna P, Swaminathan G, Hazuda DJ, Olsen DB. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLOS Pathogens. 2021;17:e1009225. doi: 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naqvi AAT, Fatima K, Mohammad T, Fatima U, Singh IK, Singh A, Atif SM, Hariprasad G, Hasan GM, Hassan MI. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narayanan A, Narwal M, Majowicz SA, Varricchio C, Toner SA, Ballatore C, Brancale A, Murakami KS, Jose J. Identification of SARS-CoV-2 inhibitors targeting Mpro and PLpro using in-cell-protease assay. Commun. Biol. 2022;5:169. doi: 10.1038/s42003-022-03090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nardell EA, Nathavitharana RR. Airborne spread of SARS-CoV-2 and a potential role for air disinfection. JAMA. 2020;324:141–142. doi: 10.1001/jama.2020.7603. [DOI] [PubMed] [Google Scholar]
- 58.Neerukonda SN, Vassell R, Herrup R, Liu S, Wang T, Takeda K, Yang Y, Lin T-L, Wang W, Weiss CD. Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS One. 2021;16:e0248348. doi: 10.1371/journal.pone.0248348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen NN, McCarthy C, Lantigua D, Camci-Unal G. Development of diagnostic tests for detection of SARS-CoV-2. Diagnostics. 2020;10:905. doi: 10.3390/diagnostics10110905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ocaranza MP, Godoy I, Jalil JE, Varas M, Collantes P, Pinto M, Roman M, Ramirez C, Copaja M, Diaz-Araya G. Enalapril attenuates downregulation of angiotensin-converting enzyme 2 in the late phase of ventricular dysfunction in myocardial infarcted rat. Hypertension. 2006;48:572–578. doi: 10.1161/01.HYP.0000237862.94083.45. [DOI] [PubMed] [Google Scholar]
- 61.Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020 doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patel S, Homaei A, El-Seedi HR, Akhtar N. Cathepsins: proteases that are vital for survival but can also be fatal. Biomed. Pharmacotherapy. 2018;105:526–532. doi: 10.1016/j.biopha.2018.05.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paul S, Bravo-Vázquez LA, Reyes-Pérez PR, Estrada-Meza C, Aponte-Alburquerque RA, Pathak S, Banerjee A, Bandyopadhyay A, Chakraborty S, Srivastava A. The role of microRNAs in solving COVID-19 puzzle from infection to therapeutics: a mini-review. Virus Res. 2022;308:198631. doi: 10.1016/j.virusres.2021.198631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng R, Wu L-A, Wang Q, Qi J, Gao GF. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021;46:848–860. doi: 10.1016/j.tibs.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pizzato M, Baraldi C, Sopetto GB, Finozzi D, Gentile C, Gentile MD, Marconi R, Paladino D, Raoss A, Riedmiller I. SARS-CoV-2 and the host cell: a tale of interactions. Front. Virol. 2022 doi: 10.3389/fviro.2021.815388. [DOI] [Google Scholar]
- 66.Raghuvanshi R, Bharate SB. Recent developments in the use of kinase inhibitors for management of viral Infections. J. Med. Chem. 2021;65:893–921. doi: 10.1021/acs.jmedchem.0c01467. [DOI] [PubMed] [Google Scholar]
- 67.Rahman N, Basharat Z, Yousuf M, Castaldo G, Rastrelli L, Khan H. Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecules. 2020;25:2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saurabh S, Vidyarthi AS, Prasad D. RNA interference: concept to reality in crop improvement. Planta. 2014;239:543–564. doi: 10.1007/s00425-013-2019-5. [DOI] [PubMed] [Google Scholar]
- 69.Seyedpour S, Khodaei B, Loghman AH, Seyedpour N, Kisomi MF, Balibegloo M, Nezamabadi SS, Gholami B, Saghazadeh A, Rezaei N. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: a systematic review of in vitro and in vivo studies. J. Cell. Physiol. 2021;236:2364–2392. doi: 10.1002/jcp.30032. [DOI] [PubMed] [Google Scholar]
- 70.Shah VK, Firmal P, Alam A, Ganguly D, Chattopadhyay S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 2020;11:1949. doi: 10.3389/fimmu.2020.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shajahan A, Archer-Hartmann S, Supekar NT, Gleinich AS, Heiss C, Azadi P. Comprehensive characterization of N-and O-glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology. 2021;31:410–424. doi: 10.1093/glycob/cwaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117:202003138. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shang W, Yang Y, Rao Y, Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vacc. 2020;5:18. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shanmugaraj B, Siriwattananon K, Wangkanont K, Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 76.Singh AT, Lantigua D, Meka A, Taing S, Pandher M, Camci-Unal G. based sensors: emerging themes and applications. Sensors. 2018;18:2838. doi: 10.3390/s18092838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Solerte SB, Di Sabatino A, Galli M, Fiorina P. Dipeptidyl peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetol. 2020;57:779–783. doi: 10.1007/s00592-020-01539-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song X, Shan D, Chen J, Jing Q. miRNAs and lncRNAs in vascular injury and remodeling. Sci. China Life Sci. 2014;57:826–835. doi: 10.1007/s11427-014-4698-y. [DOI] [PubMed] [Google Scholar]
- 79.Strollo R, Pozzilli P. DPP4 inhibition: preventing SARS-CoV-2 infection and/or progression of COVID-19? Diabetes/Metab. Res. Rev. 2020;36:e3330. doi: 10.1002/dmrr.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, Moore MJ, Tallarico AS, Olurinde M, Choe H, Anderson LJ, Bellini WJ, Farzan M, Marasco WA. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604:330–336. doi: 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- 83.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Valencia I, Peiró C, Lorenzo Ó, Sánchez-Ferrer CF, Eckel J, Romacho T. DPP4 and ACE2 in diabetes and COVID-19: therapeutic targets for cardiovascular complications? Front. Pharmacol. 2020;11:1161. doi: 10.3389/fphar.2020.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van den Brink EN, Ter Meulen J, Cox F, Jongeneelen MA, Thijsse A, Throsby M, Marissen WE, Rood PM, Bakker AB, Gelderblom HR, Martina BE, Osterhaus AD, Preiser W, Doerr HW, de Kruif J, Goudsmit J. Molecular and biological characterization of human monoclonal antibodies binding to the spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus. J Virol. 2005;79:1635–1644. doi: 10.1128/JVI.79.3.1635-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vankadari N, Wilce JA. Emerging COVID-19 coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg. Microb. Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Walls AC, Xiong X, Park YJ, Tortorici MA, Snijder J, Quispe J, Cameroni E, Gopal R, Dai M, Lanzavecchia A, Zambon M, Rey FA, Corti D, Veesler D. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. 2019;176:1026–1039.e1015. doi: 10.1016/j.cell.2018.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Q, Qi J, Yuan Y, Xuan Y, Han P, Wan Y, Ji W, Li Y, Wu Y, Wang J, Iwamoto A, Woo PCY, Yuen K-Y, Yan J, Lu G, Gao GF. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Waterman RS, Tomchuck SL, Henkle SL, Betancourt AM. A new mesenchymal stem cell (MSC) paradigm: polarization into a pro-inflammatory MSC1 or an immunosuppressive MSC2 phenotype. PLoS ONE. 2010;5:e10088. doi: 10.1371/journal.pone.0010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weisberg E, Parent A, Yang PL, Sattler M, Liu Q, Liu Q, Wang J, Meng C, Buhrlage SJ, Gray N. Repurposing of kinase inhibitors for treatment of COVID-19. Pharmac. Res. 2020;37:1–29. doi: 10.1007/s11095-020-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Widiasta A, Sribudiani Y, Nugrahapraja H, Hilmanto D, Sekarwana N, Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-coding RNA Res. 2020;5:153–166. doi: 10.1016/j.ncrna.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wilson JG, Liu KD, Zhuo H, Caballero L, McMillan M, Fang X, Cosgrove K, Vojnik R, Calfee CS, Lee J-W, Rogers AJ, Levitt J, Wiener-Kronish J, Bajwa EK, Leavitt A, McKenna D, Thompson BT, Matthay MA. Mesenchymal stem (stromal) cells for treatment of ARDS: a phase 1 clinical trial. Lancet Respir. Med. 2015;3:24–32. doi: 10.1016/S2213-2600(14)70291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia X. Domains and functions of spike protein in sars-cov-2 in the context of vaccine design. Viruses. 2021;13:109. doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xin Z, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, Li T, Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang Y, Cai Z, Zhang J. DPP-4 inhibitors may improve the mortality of coronavirus disease 2019: a meta-analysis. PLoS ONE. 2021;16:e0251916. doi: 10.1371/journal.pone.0251916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao X, Chen D, Szabla R, Zheng M, Li G, Du P, Zheng S, Li X, Song C, Li R. Broad and differential animal angiotensin-converting enzyme 2 receptor usage by SARS-CoV-2. J. Virol. 2020;94:e00940. doi: 10.1128/JVI.00940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu H, Wei L, Niu P. The novel coronavirus outbreak in Wuhan, China. Global Health Res. Policy. 2020;5:1–3. doi: 10.1186/s41256-020-00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi L-E, Barbry P, Leslie A, Kiem H-P, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas-Montanes J, Banovich N, Barbry P, Brazma A, Desai T, Duong TE, Eickelberg O, Falk C, Farzan M, Glass I, Haniffa M, Horvath P, Hung D, Kaminski N, Krasnow M, Kropski JA, Kuhnemund M, Lafyatis R, Lee H, Leroy S, Linnarson S, Lundeberg J, Meyer K, Misharin A, Nawijn M, Nikolic MZ, Ordovas-Montanes J, Pe’er D, Powell J, Quake S, Rajagopal J, Tata PR, Rawlins EL, Regev A, Reyfman PA, Rojas M, Rosen O, Saeb-Parsy K, Samakovlis C, Schiller H, Schultze JL, Seibold MA, Shalek AK, Shepherd D, Spence J, Spira A, Sun X, Teichmann S, Theis F, Tsankov A, van den Berge M, von Papen M, Whitsett J, Xavier R, Xu Y, Zaragosi L-E, Zhang K. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]