Abstract
Inhibitory G proteins (GNAI/Gαi) bind to the scaffold G protein signaling modulator 2 (GPSM2) to form a conserved polarity complex that regulates cytoskeleton organization. GPSM2 keeps GNAI in a guanosine diphosphate (GDP)-bound state, but how GPSM2-GNAI is generated or relates to heterotrimeric G protein signaling remains unclear. We find that RGS12, a GTPase-activating protein (GAP), is required to polarize GPSM2-GNAI at the hair cell apical membrane and to organize mechanosensory stereocilia in rows of graded heights. Accordingly, RGS12 and the guanine nucleotide exchange factor (GEF) DAPLE are asymmetrically co-enriched at the hair cell apical junction, and Rgs12 mouse mutants are deaf. GPSM2 and RGS12 share GoLoco motifs that stabilize GNAI(GDP), and GPSM2 outcompetes RGS12 to bind GNAI. Our results suggest that polarized GEF/GAP junctional activity might dissociate heterotrimeric G proteins, generating free GNAI(GDP) for GPSM2 at the adjacent apical membrane. GPSM2-GNAI(GDP), in turn, imparts asymmetry to the forming stereocilia to enable sensory function in hair cells.
Heterotrimeric G protein signaling sculpts the motion sensor compartment of hair cells in the inner ear.
INTRODUCTION
Hair cells (HCs) in the inner ear detect sound, body position, and head movements through their apical stereocilia bundle, or hair bundle. Previous work showed that guanine nucleotide–binding proteins (G proteins) of the inhibitory alpha class (GNAI1, GNAI2, and GNAI3; collectively GNAI or Gαi) are essential for hair bundle morphogenesis and for sensory function. In this role, GNAI bind GPSM2 and polarize the apical HC cytoskeleton along the epithelial plane. What upstream regulators drive GPSM2-GNAI assembly and how GPSM2-GNAI relates to heterotrimeric G protein signaling remain unknown.
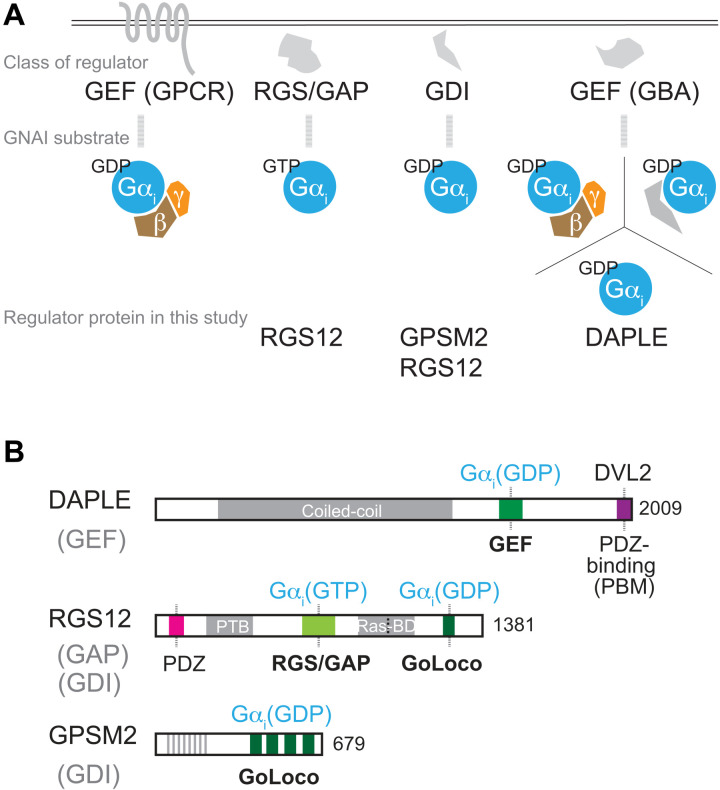
Heterotrimeric G proteins (Gαβγ) are best known as membrane-associated switches that transduce extracellular signals (1). In this context, ligand-bound G protein–coupled receptors (GPCRs) act as guanine nucleotide exchange factors (GEFs) by catalyzing the exchange of guanosine diphosphate (GDP) for guanosine triphosphate (GTP) on the Gα subunit (Fig. 1A). This allows Gα(GTP) and Gβγ to dissociate and signal to effectors to influence cell behavior. Regulator of G protein signaling (RGS) proteins accelerate the intrinsic guanosine triphosphatase (GTPase) activity of Gα, acting as GTPase-activating protein (GAP) to reform the inactive heterotrimer and limit signaling (2, 3). A third type of regulators specific to Gαi/GNAI are guanine nucleotide dissociation inhibitors (GDIs). GDIs bind GNAI via their GoLoco motifs and sequester GNAI in a GDP-bound, inactive state (4, 5).
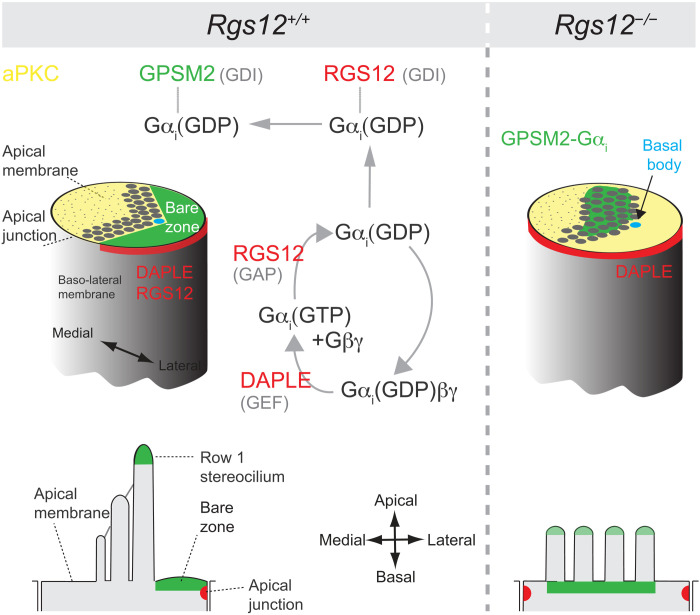
Fig. 1. Overview of GNAI/Gαi regulators and structure of the DAPLE, RGS12, and GPSM2 proteins.
(A) Diagram of the main classes of GNAI regulators and their substrates. (B) Diagram of DAPLE, RGS12, and GPSM2 proteins and their main functional motifs. The GNAI/Gαi species bound by each protein is indicated next to the functional motif involved.
The GDI protein G protein signaling modulator 2 (GPSM2) is a conserved partner of GNAI during oriented cell division (6, 7). In dividing cells, the cortical GPSM2-GNAI(GDP) polarity complex orients the mitotic spindle by recruiting effectors that exert force on astral microtubules (8–12). It remains uncertain to what extent nucleotide-based GNAI signaling is involved, but N-terminal acylation of GNAI is critical to anchor GPSM2 and polarize effectors at the cell membrane (13, 14).
GPSM2-GNAI is also part of the molecular blueprint that polarizes the apical cytoskeleton in postmitotic, differentiating HCs. GPSM2-GNAI first defines the bare zone, a subregion of HC apical membrane deprived of microvilli (15–17). In the auditory epithelium, the GPSM2-GNAI–positive bare zone localizes on the lateral/abneural side of the HC (see Discussion). Microvilli neighboring the bare zone bulk up with the addition and elongation of actin filaments, forming stable membrane protrusions known as stereocilia. GPSM2-GNAI enrichment at the bare zone ensures its specific trafficking to the first row of stereocilia abutting the bare zone. In row 1, GPSM2-GNAI is a module of the elongation complex otherwise comprising the myosin MYO15A, the adapter whirlin (WHRN), and the actin bundling/capping protein EPS8 (18–21). The elongation complex is considered to promote actin nucleation at stereocilia tips, and GPSM2-GNAI confers row 1 its tallest identity by boosting MYO15A-EPS8 levels there (19). The slanted organization of stereocilia by row confers the hair bundle its directional sensitivity, a central property of mechanoelectrical transduction in HCs (22). Mouse models that lack GPSM2 or GNAI function have stunted stereocilia that lack row identity, are profoundly deaf (18–21), and model hereditary hearing loss in families where GPSM2 is mutated (23, 24). The upstream cues assembling and polarizing the GPSM2-GNAI complex at the bare zone remain unknown, clouding how hair bundles acquire their asymmetric architecture.
A first clue emerged after identifying the nonreceptor GEF DAPLE as required for apical HC morphogenesis (25, 26). DAPLE is a GNAI-binding and activating (GBA) protein (Fig. 1A) (27, 28), and Daple mutants show a combination of severe cytoskeleton patterning and HC orientation defects. As DAPLE directly binds GNAI and dishevelled homolog (DVL2) (Fig. 1B) (29), DAPLE was proposed to couple the distinct planar polarity mechanisms that regulate cytoskeleton asymmetry in single HCs (via GPSM2-GNAI) and HC orientation along the epithelial plane [via DVL2 and other planar cell polarity (PCP) proteins ensuring intercellular communication] (25, 30–32). Many questions remain unanswered, however. DAPLE is polarized at the apicolateral HC junction and is adjacent to, but does not coincide with, GPSM2-GNAI at the apical membrane (see Discussion) (25). While GPSM2-GNAI remains enriched at the apical membrane in Daple mutants, GPSM2-GNAI is not properly polarized laterally, resulting in severe stereocilia placement defects. The origin of the GPSM2-GNAI complex and the interplay between the lateral junction and the adjacent bare zone thus remain unknown. Similarly, it remains unclear whether and how exactly GPSM2-GNAI(GDP) is related to heterotrimeric G protein signaling.
A yeast-two-hybrid screen identified RGS12 as a binding partner of GNAI3 in an inner ear cDNA library (25). RGS12 and RGS14 are unique among ~30 RGS proteins because they carry a GoLoco motif for GDI activity (33). RGS12 can thus bind GNAI via (i) its eponymous RGS domain for GAP activity (34) and (ii) its GoLoco motif for GDI activity (35). It follows that both RGS12 and GPSM2 (36, 37) can bind and sequester GNAI(GDP) (Fig. 1B). The largest protein in the family, RGS12 is also characterized by a multidomain architecture. Besides RGS/GAP and GoLoco, RGS12 has a PDZ domain, a phosphotyrosine binding (PTB) domain (38), and tandem Ras-binding domains (39). RGS12 is thus ideally placed to integrate GNAI and other signaling pathways (40). RGS12 was implicated previously in cell differentiation including myogenesis, neurogenesis, and bone formation (39, 41–45). The Drosophila RGS12 homolog Loco regulates oriented cell division with the GPSM2 homolog Pins (46), but the RGS12 protein is not known to be polarized or to affect HC differentiation. It is also unclear how RGS12 and RGS14 cellular functions are served by the “complex coordination of G protein signaling” (35) afforded by having both an RGS and GoLoco motif to bind GNAI.
Here, we find that RGS12 colocalizes with DAPLE at the lateral HC junction and is critical to generate and polarize the GPSM2-GNAI complex at the adjacent apical membrane. RGS12 is a new deafness protein and a previously missing link connecting the apical membrane with the apical junction and GPSM2-GNAI with heterotrimeric G protein signaling in HCs.
RESULTS
Loss of RGS12 causes hair bundle defects reminiscent of Gpsm2 and Gnai inactivation
RGS12 was identified as a GNAI3 binding partner in a yeast-two-hybrid screen with an inner ear library (25). To study the role of RGS12 in HCs, we obtained a mouse strain where an ~11-kb Rgs12 region including exons 2 and 3 was deleted and replaced by a LacZ reporter followed by a neomycin cassette (C57BL/6N-Rgs12tm1(KOMP)/Vlcg; fig. S1, A to C). We used CMV-Cre to delete neomycin, generating the Rgs12tm1.1(KOMP)/Vlcg allele (hereafter Rgs12−; fig. S1, B and C). X-Gal labeling of LacZ expression showed that Rgs12 is broadly transcribed in the cochlear floor from embryonic day 16.5 (E16.5) to postnatal day 20 (P20) and is enriched in HCs (fig. S1D). At P4, F-actin labeling with phalloidin conjugates revealed HCs with dysmorphic hair bundles in an otherwise largely normal auditory epithelium in Rgs12−/− homozygotes [mid-cochlea (50% position): Fig. 2A; cochlear base (15%) and apex (75%): fig. S2, A and B]. In contrast, hair bundles in Rgs12+/− heterozygotes were comparable to Rgs12+/+ wild-type littermates (Fig. 2A). To quantify defects, we first measured the kinocilium index in outer HCs (OHCs), i.e., the length ratio between the two halves (x, y) of the bundle defined by the kinocilium at the bundle vertex (Fig. 2B) (47). We also measured the angle formed by the two bundle halves (Fig. 2C). In each case, Rgs12−/− showed significant deviation from Rgs12+/− and Rgs12+/+ results: specifically, more variable kinocilium indexes and reduced bundle angles. However, quantifying the position of the kinocilium at P4 did not reveal defects in HC orientation in Rgs12−/− (fig. S2C). HCs were also correctly oriented at E17.5 (fig. S2D), ruling out that early defects in orientation are corrected with time in Rgs12 mutants.
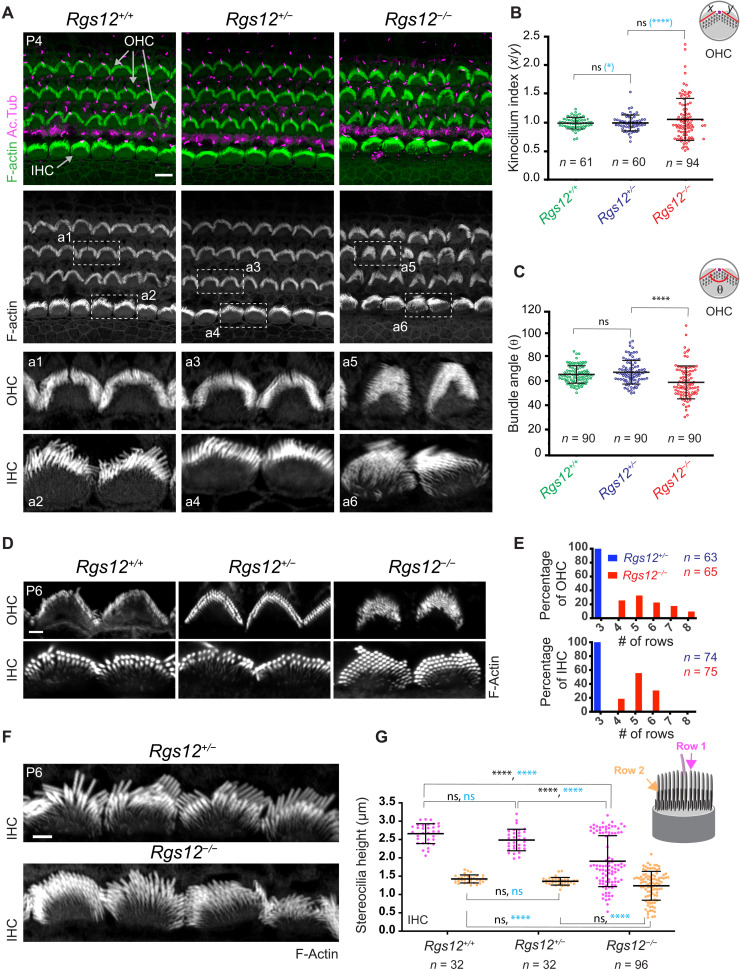
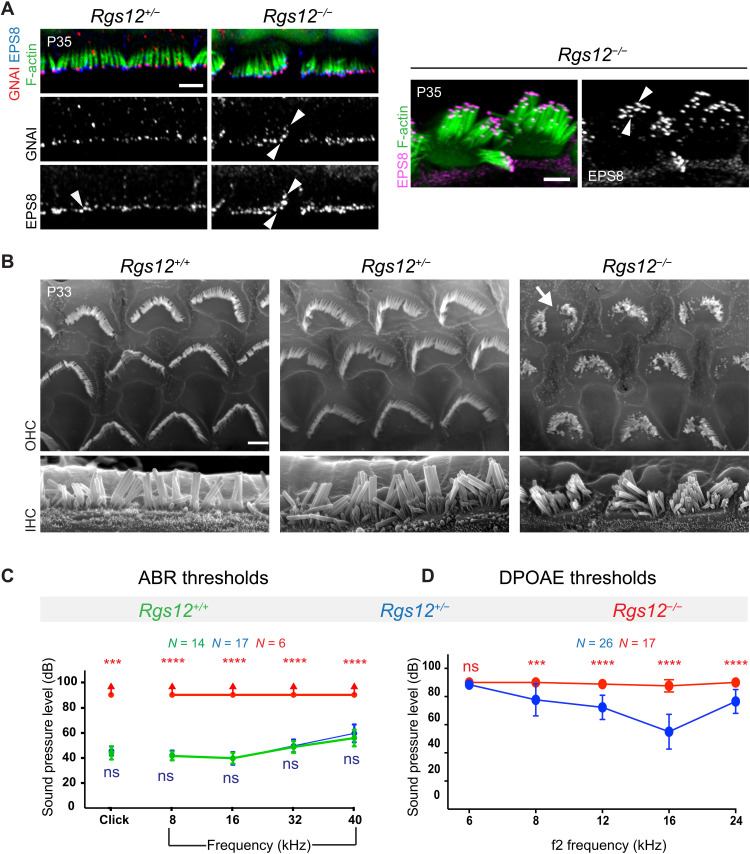
Fig. 2. Defective hair bundle morphogenesis in Rgs12 mutants.
(A) Surface views of the auditory epithelium (cochlear middle-turn, 50% position). F-actin is labeled with phalloidin, and acetylated tubulin (Ac.Tub) is immunolabeled to reveal primary cilia/HC kinocilia. Boxed regions are magnified at the bottom. Cochlear base and apex positions are shown in fig. S2 (A and B). (B and C) Kinocilium index (x/y ratio) (B) and angle (θ) (C) in OHC bundles. n = HCs, N = 3 animals. Means ± SD. Kruskal-Wallis with Dunn’s multiple comparisons to compare data distribution in black [ns (nonsignificant), P > 0.9999; ****P < 0.0001]. F test to compare variance in blue (B) (*P = 0.0157 and ****P < 0.001). (D and E) F-actin labeling shows extra stereocilia rows in Rgs12−/− OHCs and IHCs, as quantified by frequency distribution in (E). n = HCs, N = 3 animals. (F and G) F-actin labeling in IHCs (F) shows reduced stereocilia height in Rgs12−/− mutants as quantified in (G). n = stereocilia, N = 3 (Rgs12+/+; Rgs12+/−) or N = 4 (Rgs12−/−) animals, 8 (Rgs12+/+; Rgs12+/−) or 24 IHCs (Rgs12−/−), and 4 stereocilia per IHC per row. Means ± SD. Two-way analysis of variance (ANOVA) with Tukey’s multiple comparison for data distribution in black (row 1: ns, P = 0.2917; ****P < 0.0001; row 2: Rgs12+/+ versus Rgs12+/−, ns, P = 0.8438; Rgs12+/+ versus Rgs12−/−, ns, P = 0.1176; Rgs12+/− versus Rgs12−/−, ns, P = 0.3936). F test to compare variance in blue (row 1: ns, P = 0.6578; ****P < 0.0001; row 2: ns, P = 0.828; ****P < 0.0001). For all images in the study, the lateral/abneural side of the auditory epithelium is on top. Scale bars, 5 μm (A), 0.5 μm (D), and 1 μm (F).
At P6, Rgs12−/− hair bundles often showed up to eight (OHC) or six [inner HC (IHC)] stereocilia rows instead of three rows in Rgs12+/− and Rgs12+/+ littermates (Fig. 2, D and E). Extra medial rows in embryonic hair bundles are normally resorbed at postnatal stages and indicate a lack of maturation when retained (48). Extra rows were reported in mutant strains where GPSM2 or GNAI are inactivated (18, 21). Next, we measured stereocilia height in IHCs for row 1 (tallest) and row 2. The marked difference in height observed in wild-type hair bundles between row 1 and 2 was preserved in Rgs12+/− but blurred in Rgs12−/− (Fig. 2, F and G). Row 1 stereocilia height showed a bimodal distribution in Rgs12−/−: Some stereocilia had normal height, whereas many were severely stunted, adopting a height close to row 2 (Fig. 2G). Row 2 height was variably reduced in Rgs12−/− (Fig. 2G). As Rgs12+/− and Rgs12+/+ were indistinguishable, Rgs12+/− samples were used as controls for Rgs12−/− mutants in some subsequent experiments.
In summary, much like GPSM2-GNAI, the GAP protein RGS12 is required for hair bundle maturation, including proper stereocilia elongation and resorption of extra rows but not for normal HC orientation. In contrast, HCs lacking the GEF protein DAPLE present distinct HC defects. In Daple mutants, bundles showed early stereocilia degeneration instead of extra rows, and HCs were misoriented (25).
RGS12 is required to deploy the GPSM2-GNAI complex at the bare zone
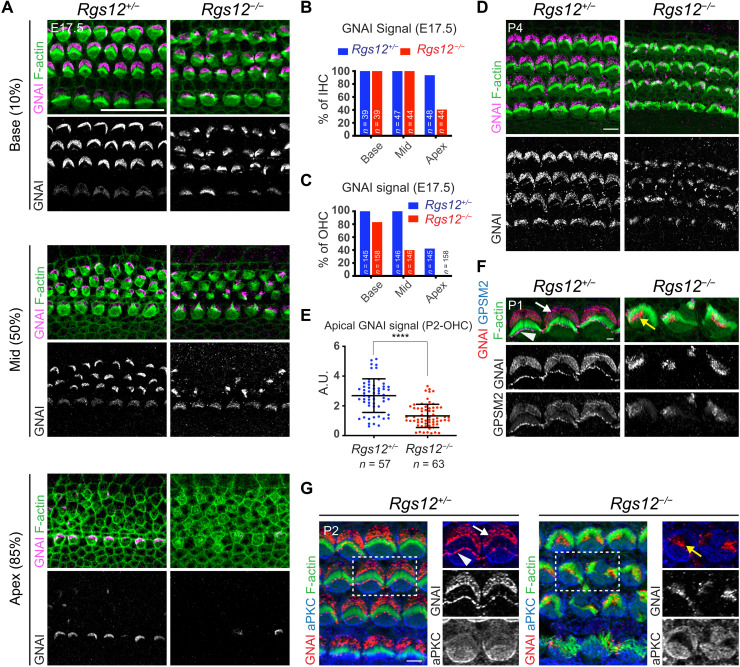
GPSM2-GNAI and atypical protein kinase C (aPKC) segregate to complementary lateral and medial apical domains in HCs, respectively, a molecular blueprint that regulates cytoskeleton polarization (15–17, 49). In Daple mutants, GPSM2-GNAI is maintained at the HC apical membrane but the GPSM2-GNAI versus aPKC blueprint is severely disorganized (25). To ask whether RGS12 is involved in regulating GPSM2-GNAI localization, we immunolabeled GNAI at E17.5 (Fig. 3A). GNAI enrichment at the bare zone observed in controls was largely missing in Rgs12 mutants at the cochlear apex (85% position), only present in a subset of HCs and partially labeling the bare zone in OHCs at the cochlear mid (50%), and partially labeling the bare zone in OHCs and IHCs at the cochlear base (10%; Fig. 3, A to C). As HC differentiation is more advanced at the cochlear base than the apex, these results indicate a delay in GNAI enrichment and polarization, as well as a failure to occupy the entire bare zone when finally enriched there. At P0, GNAI similarly failed to occupy the entire bare zone in Rgs12 mutants and instead concentrated near row 1 stereocilia (fig. S3A). At P4, GNAI was absent at the bare zone in Rgs12 mutants and instead aberrantly enriched at the position of the dysmorphic hair bundle (Fig. 3D). We quantified total GNAI signals at the HC apical membrane at P2 and observed a significant reduction in Rgs12 mutants (Fig. 3E). As expected, GPSM2 colocalized with GNAI at aberrant positions (Fig. 3F). aPKC lost its medial restriction and spread to the entire flat apical membrane in Rgs12 mutants (Fig. 3G), as observed previously when GPSM2 or GNAI were directly inactivated (15, 17). GNAI and aPKC colabeling clarified that GNAI strictly occupied aPKC-negative regions in Rgs12 mutants (Fig. 3G). Together, these results indicate that RGS12 is an upstream regulator required for timely, complete, and sustained deployment of GPSM2-GNAI at the bare zone. RGS12 apparently ensures that GPSM2-GNAI is trafficked in normal amounts and properly polarized at the HC apical membrane. RGS12 is also essential to define a bare zone devoid of aPKC.
Fig. 3. Disruption of the apical blueprint proteins GPSM2-GNAI and aPKC in Rgs12 mutant HCs.
(A) GNAI immunolabeling at E17.5 at different cochlear positions (% of distance from the cochlear base). In Rgs12 mutants, GNAI is missing (apex, 85% position), delayed (mid, 50%), and fails to encompass the whole bare zone (base, 10%) in progressively more mature HCs. (B and C) Proportion of E17.5 IHCs (B) and OHCs (C) where an apical GNAI signal is detected by cochlear position. n = HCs, N = 3 animals. (D) GNAI immunolabeling. (E) Total GNAI signals at the apical HC surface. N = 3 animals. Means ± SD. t test, ****P < 0.0001. (F) GNAI and GPSM2 coimmunolabeling in OHC2. GNAI and GPSM2 at the bare zone (arrow) and stereocilia tips (arrowhead) are absent in Rgs12 mutants and instead match subregions of the hair bundle (yellow arrow). (G) GNAI and aPKC coimmunolabeling. Boxed OHCs are magnified on the right. In Rgs12 mutants, aPKC extends laterally to encompass the entire flat HC surface, and GNAI is found at the position of the aPKC-negative hair bundle (yellow arrow). Unlike indicated differently, in all figures, arrows indicate protein enrichment at the bare zone and arrowheads indicate enrichment at stereocilia tips. A.U., arbitrary units. Scale bars, 20 μm (A), 5 μm (D and G), and 0.5 μm (F).
To characterize more precisely the aberrant distribution of GNAI in Rgs12 mutants, we first colabeled GNAI with WHRN. WHRN and GPSM2-GNAI colocalize at stereocilia tips, and WHRN is also enriched at the base of stereocilia but not at the bare zone with GPSM2-GNAI (19, 50–52). Although GNAI and WHRN occupied the bare zone and stereocilia base in controls, respectively, they colocalized at the position of the hair bundle in Rgs12 mutants (fig. S3B). Next, we colabeled GNAI and TRIO and F-actin binding protein (TRIOBP), an actin-bundling protein in stereocilia rootlets (53). Aberrant GNAI signals in Rgs12 mutants spanned a subregion of the rootlet landscape (Fig. 4A). GNAI accumulation near rootlets appeared to encompass multiple stereocilia rows, suggesting that GPSM2-GNAI loses specificity for row 1 in the absence of RGS12.
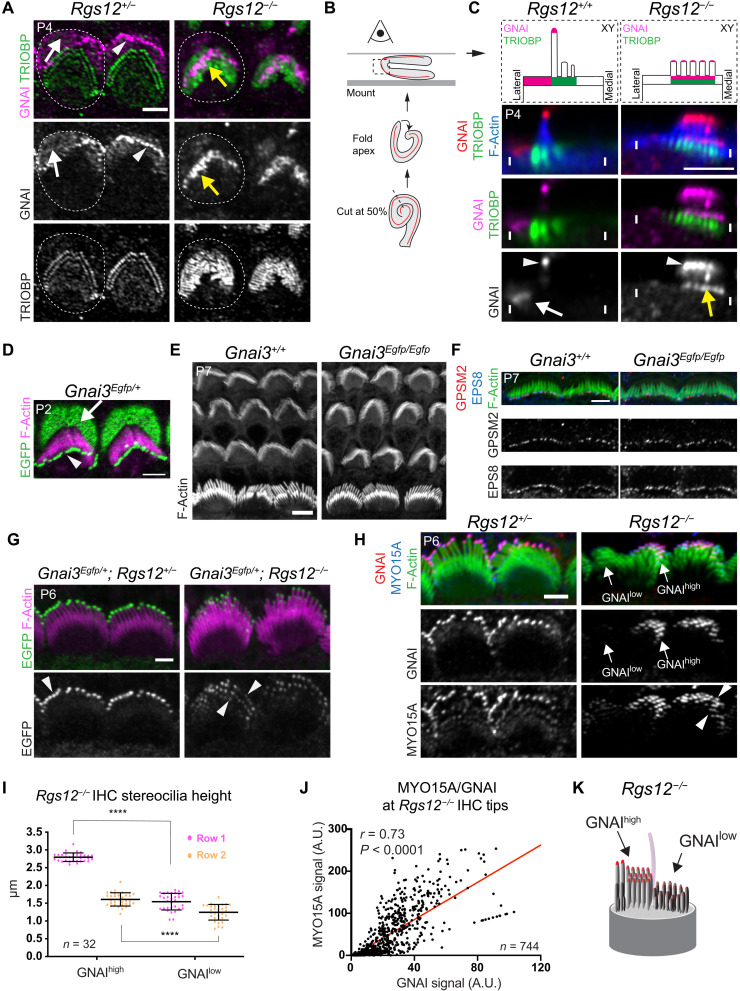
Fig. 4. GNAI loses stereocilia row specificity and accumulates near rootlets in the absence of RGS12.
(A) GNAI and TRIOBP coimmunolabeling in OHCs. In Rgs12 mutants, GNAI occupies a subregion of the enlarged rootlet space (yellow arrow). Dashed lines, apical circumference of one OHC. (B) Folding of the cochlear floor to image the OHC surface sideways. (C) GNAI and TRIOBP coimmunolabeling in a sideway capture of a single OHC as in (B) (75% cochlear position). Distinct GNAI signals can be resolved near rootlets (TRIOBP; yellow arrow) and at tips (arrowhead) in mutant stereocilia. (D) EGFP signals in Gnai3Egfp/+. The EGFP moiety does not interfere with GNAI3 localization. (E) F-actin labeling. (F) GPSM2 and EPS8 colabeling in IHCs. Hair bundle morphology (E) and GPSM2/EPS8 enrichment at row 1 stereocilia tips (F) are normal in Gnai3Egfp homozygotes. (G) EGFP in Gnai3Egfp/+; Rgs12+/− controls and Gnai3Egfp/+; Rgs12−/− mutant IHCs. Signal at tips encompasses all stereocilia rows in mutants. (H) GNAI and MYO15A colabeling in IHCs. GNAIhigh and GNAIlow indicate stereocilia regions with different GNAI enrichment. (I) Stereocilia height in Rgs12−/− IHCs plotted separately for GNAIhigh and GNAIlow stereocilia. n = stereocilia, 8 IHCs, N = 3 animals, 4 stereocilia per row. Means ± SD. Two-way ANOVA with Tukey’s multiple comparison (****P < 0.0001). GNAI amount is predictive of stereocilia height in Rgs12 mutants. (J) Correlation of GNAI versus MYO15A signal intensity at tips in (H). n = stereocilia, 17 IHCs, N = 3 animals. Pearson’s correlation. (K) Diagram of an Rgs12 mutant IHC. Scale bars, 5 μm (A, C, and E) and 2 μm (D and F to H).
Loss of GNAI row specificity and variable enrichment at stereocilia tips in the absence of RGS12
Abnormal accumulation of GPSM2-GNAI near stereocilia rootlets might reflect impaired trafficking to distal tips, which, in turn, would be expected to limit stereocilia elongation. Shortened hair bundles in Rgs12 mutants (Fig. 2, F and G) made it challenging to address whether GNAI still occupied the tip of stereocilia in addition to their base. To begin to solve this question, we folded the cochlear floor to image P4 HCs at the bending point so that protruding stereocilia are captured in the XY plane offering better resolution than the Z axis (Fig. 4B). These preparations confirmed that in Rgs12 mutants, GNAI was enriched near rootlets in multiple rows (Fig. 4C, yellow arrow). GNAI was also present at stereocilia tips as distinct signals (Fig. 4C, arrowheads). Similar to rootlet level, signals at stereocilia tips were not limited to row 1 and appeared to span multiple rows.
To independently confirm the loss of polarized GPSM2-GNAI in the absence of RGS12, we generated a Gnai3Egfp knock-in reporter strain (fig. S4A). The enhanced green fluorescent protein (EGFP) moiety at GNAI3 C-terminal position did not prevent polarization of the fusion protein at the bare zone or at row 1 stereocilia tips, recapitulating HC signals obtained with immunolabeling (Fig. 4D; compare with Fig. 3F, for example). Hair bundle morphology and GPSM2 and EPS8 distribution at stereocilia tips appeared normal in Gnai3Egfp/Egfp homozygotes at P7 (Fig. 4, E and F), suggesting that EGFP does not interfere with GPSM2-GNAI3 function. When Gnai3Egfp was bred with Rgs12 mutants, GNAI immunolabeling and EGFP signals colocalized in both Rgs12+/− and Rgs12−/− samples (fig. S4B). GNAI3-EGFP could still be enriched at stereocilia tips in P6 Rgs12 mutants, albeit in variable amounts and without restriction to row 1 as seen with GNAI immunolabeling (Fig. 4G for IHCs; fig. S4C for OHCs). Elongation complex partner MYO15A was also variably enriched at tips and more uniformly enriched across stereocilia rows in P6 Rgs12−/− stereocilia, in contrast to its graded enrichment by row in controls at this stage (Fig. 4H). We conclude that abnormal GNAI enrichment near rootlets in all rows leads to inefficient upward trafficking of the elongation complex in all rows as well. This provides new evidence that bare zone enrichment is the mechanism by which GPSM2-GNAI is normally limited to row 1 (see Discussion).
In Rgs12−/− IHCs, some stereocilia showed very low GNAI and MYO15A amounts at tips, whereas others had signals close to controls (Fig. 4H). Signal variability at tips likely originates from GNAI enrichment near a subset of rootlets only (Fig. 4A). We measured stereocilia height in row 1 and 2 in P6 Rgs12 mutant IHCs, tracking whether individual stereocilia enriched high or low levels of GNAI at their tips. By plotting height separately, we could establish that Rgs12−/− row 1 stereocilia with low GNAI signals were significantly shorter than stereocilia with higher GNAI signals (Fig. 4I). A similar trend was observed for row 2. As expected if the elongation complex was generally affected, we found a significant correlation between tip amounts for GNAI and MYO15A in P6 Rgs12−/− stereocilia (Fig. 4, H and J). Stereocilia with low GNAI and MYO15A amounts at their tip must thus represent the stunted fraction of the Rgs12−/− stereocilia population in Fig. 2G.
Immature hair bundles and profound hearing loss in adult Rgs12 mutants
The GNAI enrichment profile observed at stereocilia tips in Rgs12 mutant neonates was maintained in young adults (~P35). GNAI signals were variable in intensity and present at all rows in mutant IHCs (Fig. 5A). Uniform row distribution was also observed with elongation complex partner EPS8, a departure from its specific enrichment at row 1 in P35 control IHCs (Fig. 5A, arrowheads). Along with more uniform MYO15A across rows at P6 (Fig. 4H), these results suggest that stereocilia row identity is at least partially disrupted in the absence of RGS12, as when GPSM2-GNAI is directly inactivated (19). GNAI signals were detectable at tips in highly dysmorphic mutant OHCs (fig. S5). Scanning electron microscopy micrographs clarified that adult Rgs12−/− retained an excess of stereocilia rows in both HC types and highly variable stereocilia height in IHCs (Fig. 5B). There were signs of stereocilia degeneration in Rgs12−/− OHCs, and the bundle outline and the whole HC surface had a round shape compared to the cashew nut shape of Rgs12+/− and Rgs12+/+ controls.
Fig. 5. Adult hair bundle defects and hearing loss in the absence of RGS12.
(A) GNAI and EPS8 coimmunolabeling in IHCs at the middle-turn. GNAI and EPS8 colocalize at stereocilia tips but lack specificity for row 1 in Rgs12−/− (arrowheads). Left: Stereocilia tilted medially. Right: Stereocilia tilted laterally to illustrate hair bundle morphology and EPS8 signal at multiple rows in Rgs12 mutants. (B) Scanning electron microscopy at the middle-turn. IHC hair bundles are disorganized in Rgs12−/−, with extra stereocilia rows showing variable heights, as in neonates (Fig. 2, F and G). OHC bundles are similarly poorly developed and show signs of stereocilia degeneration (arrow). Rgs12+/+ and Rgs12+/− HCs are indistinguishable. (C and D) ABR and DPOAE thresholds at P33 to P38. X axis indicates broadband (click) and pure tone frequency stimuli for ABR (C), or f2 for 2f1-f2 emissions for DPOAE (D), in kilohertz. Y axis indicates the threshold of sound pressure eliciting a response (C) or generating DPOAEs above noise floor (D) in decibel. Means ± SD. N = animals. In (C), red arrows indicate a lack of ABR at 90 dB. ABR click, Kruskal-Wallis test with Dunn’s multiple comparisons. Rgs12+/+ versus Rgs12+/− ns, P > 0.9999; Rgs12+/+ versus Rgs12−/−, ***P = 0.0003. ABR pure tones and DPOAE: two-way ANOVA with Tukey’s multiple comparisons. ABR pure tone: Rgs12+/+ versus Rgs12+/− ns, P = 0.9695 (8 kHz), P = 0.9945 (16 kHz), P = 0.8535 (32 kHz), and P = 0.0685 (40 kHz); Rgs12+/+ versus Rgs12−/−, ****P < 0.0001 for all frequencies. DPOAE: ns, P = 0.5237; ***P = 0.0004; and ****P < 0.0001. Scale bars, 5 μm.
Severe HC apical defects coincided with profound hearing loss in Rgs12 mutants. An auditory brainstem response (ABR) could not be recorded in ~P35 Rgs12−/− animals with broadband (click) or 8-, 16-, 32-, and 40-kHz pure tone stimuli up to 90 dB (Fig. 5C). In contrast, ABR thresholds in Rgs12+/− animals were indistinguishable from Rgs12+/+ littermates. We recorded distortion product otoacoustic emissions (DPOAEs) produced by OHCs and observed higher thresholds for 8, 12, 16, and 24 kHz in Rgs12−/− compared to Rgs12+/− animals (Fig. 5D). Severe hair bundle morphogenesis defects and stereocilia degeneration in OHCs probably prevent HC sensory activity, in turn, precluding auditory function. We cannot rule out, however, that RGS12 also has another role in the auditory cascade downstream of hair bundle function.
RGS12 colocalizes with DAPLE at the lateral HC junction
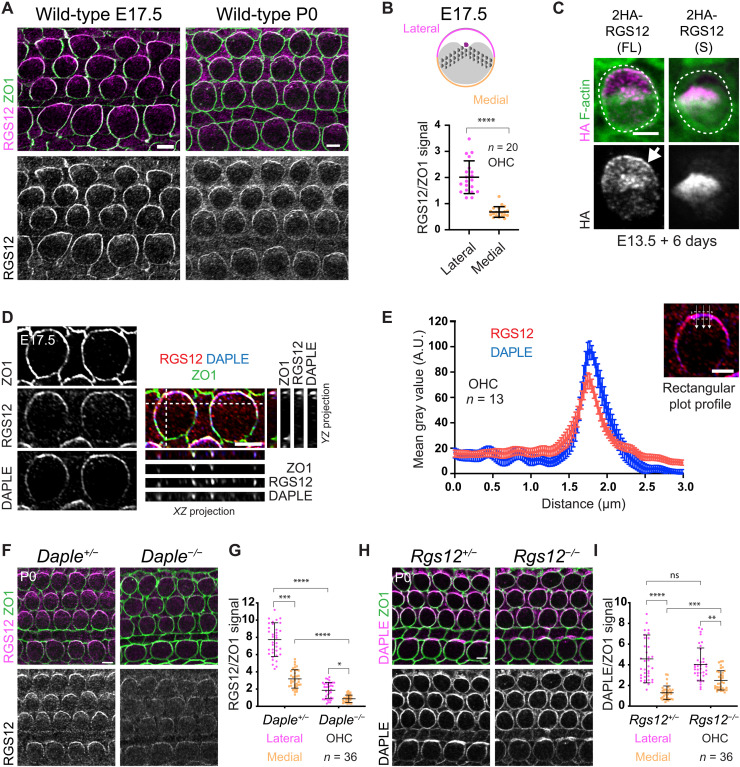
To understand how RGS12 affects GPSM2-GNAI, hair bundle morphogenesis, and hearing, we first used an antibody raised against RGS12 very C-terminal region. At E17.5, the bulk of RGS12 immunosignal was at apical junctions, with higher enrichment laterally in HCs (Fig. 6A). Using ZO1 as a junctional marker, we quantified RGS12 signals at the lateral and medial HC junction and normalized to ZO1 to confirm lateral polarization (Fig. 6B). Lateral polarization was still observed but less robust at P0 (Fig. 6A) and largely absent at P4 (fig. S6A). Polarized signals were not observed in Rgs12 mutants and thus specifically reflected RGS12 distribution (fig. S6B). To obtain independent evidence, we electroporated 2HA-Rgs12 constructs in E13.5 cochlear explants and cultured for 6 days before hemagglutinin (HA) immunodetection. Full-length RGS12 did recapitulate lateral junction enrichment in HCs (Fig. 6C). In contrast, a short isoform lacking the N-terminal PDZ and PTB domains (54, 55) was not enriched or polarized at the HC junction (Fig. 6C). These results suggest that full-length RGS12 is planar-polarized at the lateral HC junction, where it may influence GPSM2-GNAI at the adjacent bare zone. The short RGS12 isoform could still be produced in Rgs12−/− HCs because the corresponding transcript was still detected in Rgs12−/− cochleae (fig. S6, C and D). The short unpolarized RGS12 isoform might account for delayed, incomplete, and transient GNAI enrichment at the bare zone (Fig. 3, A to D, and fig. S3A) and remaining yet unpolarized RGS12 immunosignals (fig. S6B) in the Rgs12−/− model.
Fig. 6. RGS12 and DAPLE colocalize at the apicolateral HC junction.
(A) RGS12 and ZO1 coimmunolabeling. (B) RGS12 signal as a ratio of ZO1 at the lateral and medial HC junction. Means ± SD. n = HCs, N = 3 animals. Mann-Whitney test, ****P < 0.0001. (C) HA labeling of representative OHCs expressing the full-length (FL; Rgs12-201) or short (S; Rgs12-204) Rgs12 isoform (see fig. S6, C and D). The cochlea was electroporated at E13.5 and cultured for 6 days. 2HA-Rgs12 (FL) but not (S) is enriched at the lateral HC junction (arrow). Outcome representative of >18 OHCs in >4 explants. (D and E) RGS12 and DAPLE coimmunolabeling along with ZO1 as junctional marker in OHCs. In (D), side projections in the YZ (right) and XZ (bottom) axis are shown along with surface views. RGS12 and DAPLE signals are polarized and colocalize at the lateral HC junction. (E) Plot profile of averaged signal intensity along lines crossing the junction as illustrated. Means ± SEM. N = 3 animals. (F and G) RGS12 immunolabeling. RGS12 signals are reduced and less markedly polarized laterally in Daple mutants (F), as quantified in (G) for OHCs. (H and I) DAPLE immunolabeling. DAPLE junctional amounts are normal but less polarized in Rgs12 mutant OHCs (H), as quantified in (I). (G and I) Means ± SD. N = 3 animals. Kruskal-Wallis test with Dunn’s multiple comparisons. (G) ****P < 0.0001, ***P = 0.0001, and *P = 0.0266. (I) ns, P > 0.9999; ****P < 0.0001; ***P = 0.0004; and **P = 0.0021. Scale bars, 5 μm.
We previously showed that the cytoplasmic GEF DAPLE also localizes to the lateral HC junction (25). We thus coimmunolabeled DAPLE and RGS12 along with ZO1 at E17.5. The localization and medial extent of the DAPLE and RGS12 crescents precisely coincided, as observed in top and side projection views (Fig. 6D) and as quantified in plot profiles of signal intensity along a line crossing the lateral junction (Fig. 6E). To explore the relationship between RGS12 and DAPLE, we immunolabeled and quantified RGS12 in Daple mutants and DAPLE in Rgs12 mutants. In P0 OHCs, RGS12 was still detected at junctions in Daple mutants, but RGS12 enrichment and lateral polarization were severely reduced (Fig. 6, F and G). In contrast, junctional DAPLE amounts were similar to controls in Rgs12 mutant OHCs, but DAPLE polarization was compromised with a reduced ratio of lateral versus medial enrichment (Fig. 6, H and I). RGS12 and DAPLE enrichment in IHCs was overall less affected (Fig. 6, F and H). Next, we interrogated RGS12 distribution after disrupting GNAI function with pertussis toxin catalytic subunit (PTXa). PTXa ADP-ribosylation and uncouples GNAI from GPCRs and other regulators with broad functional consequences (56). In addition to disrupting GPSM2-GNAI and hair bundle morphogenesis (15, 18), PTXa also disrupts the EMX2-GPR156-GNAI cascade that regulates HC orientation (57). In PTXa HCs, polarized enrichment of RGS12 was intact and in register with the cytoskeleton when HCs were misoriented (fig. S6E). RGS12 was also normally enriched in HCs lacking GNAI3 (fig. S6F), a GNAI protein required for normal hair bundle morphogenesis (20, 21). In conclusion, polarized RGS12 distribution does not depend on GNAI, as expected if RGS12 is acting upstream of GNAI. Despite their spatial and functional association, we could not coimmunoprecipitate DAPLE with RGS12 by transfecting 2HA-Rgs12 and Egfp-Daple constructs in human embryonic kidney (HEK) 293 cells (fig. S6G). Cotransfecting untagged GNAI3 did not help (fig. S6G).
In summary, the apicolateral HC junction hosts both a GEF and a GAP, which are expected to regulate guanine nucleotide exchange on GNAI in opposite ways [DAPLE: promote GNAI(GTP); RGS12: hydrolyze GNAI(GTP) back to GNAI(GDP)]. DAPLE and RGS12 are co-dependent for their respective distribution and, without evidence for their interaction, could operate in tandem at the junction.
Evidence that GNAI is a DAPLE/RGS12 substrate at the lateral junction
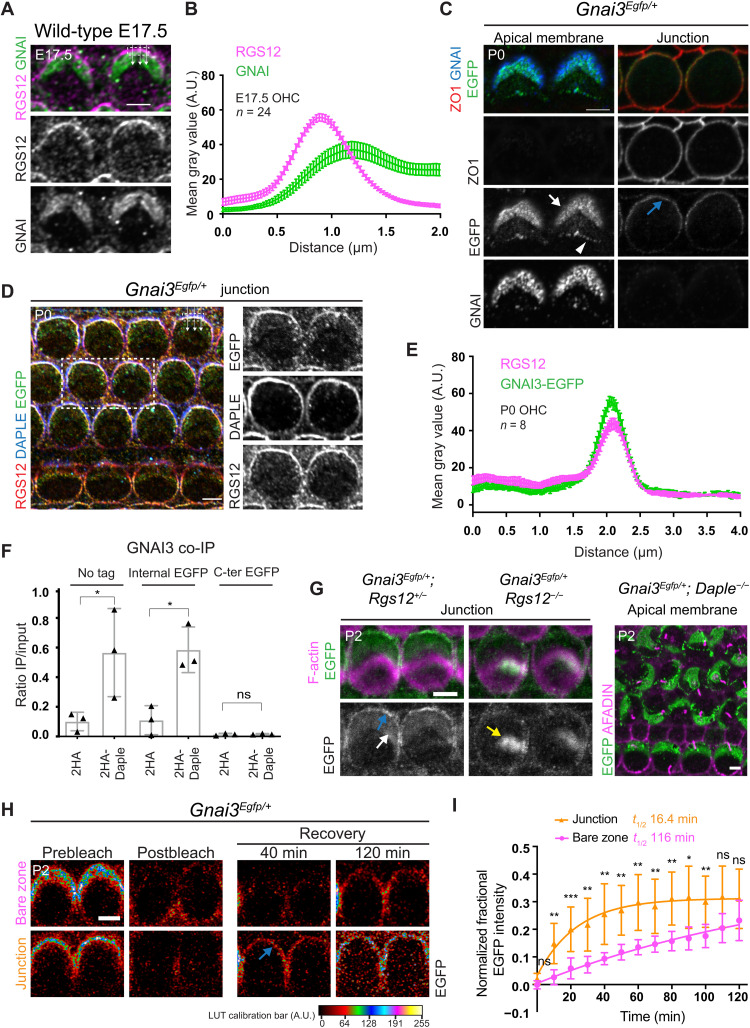
We hypothesized that DAPLE/RGS12 GEF/GAP activities at the lateral HC junction might be the origin of free GNAI needed for the atypical GPSM2-GNAI complex at the adjacent bare zone (see model in Discussion). Notably, however, we have not been able to immunodetect GNAI at the HC junction despite testing multiple antibodies and conditions (25, 57). RGS12 and GNAI coimmunolabeling at E17.5 produced overlapping but nonmatching signals (Fig. 7A). Plot profiles along a line crossing the junction into OHCs showed that RGS12 peaks at the junction whereas GNAI peaks at the bare zone past the junction (Fig. 7B). On the basis of immunodetection alone, RGS12 and GNAI are thus only partially overlapping in the transition zone between the HC apical junction and the HC apical membrane.
Fig. 7. Evidence for GNAI enrichment at the lateral HC junction.
(A) GNAI and RGS12 coimmunolabeling in OHC2. (B) Plot profile of averaged signal intensity across the lateral junction. Means ± SEM. n = OHCs, N = 3 animals. (C) EGFP with GNAI immunolabeling in Gnai3Egfp/+ OHC2. Apical and junctional sections in the same OHC stacks. EGFP but not GNAI is junctional (blue arrow). (D) RGS12 and DAPLE coimmunolabeling with EGFP in Gnai3Egfp/+. (E) Plot profile of averaged signal intensity as in (B). N = 3 animals. (F) GNAI3 amounts coimmunoprecipitated with 2HA or 2HA-DAPLE in HEK293 cells (see fig. S7B). Ratio of immunoprecipitated (IP) GNAI3 over input. Means ± SD, three experiments. One-way ANOVA with Tukey’s multiple comparisons. No tag: *P = 0.0164; internal EGFP: *P = 0.019; C-terminal EGFP: ns, P > 0.9999. (G) GNAI3-EGFP in OHCs. EGFP is lost at the junction (blue arrow) and bare zone (white arrow) but aberrantly enriched at the hair bundle (yellow arrow) in Rgs12 mutants. In contrast, EGFP is retained but mislocalized in Daple mutant. (H) Representative FRAP results for Gnai3Egfp/+ OHCs. Signal recovery is shown at bare zone versus junction in the same stack and first observed at the junction (blue arrow). (I) FRAP quantification. Half-times (t1/2) are based on nonlinear fit curves. Means ± SD. n = 12 OHCs, four experiments. Two-way ANOVA with Tukey’s multiple comparisons (from left: ns, P = 0.0635; **P = 0.0012; ***P = 0.0007; **P = 0.0031; **P = 0.0042; **P = 0.0011; **P = 0.0019; **P = 0.0063; **P = 0.0017; *P = 0.01; **P = 0.0094; ns, P = 0.0714; ns, P = 0.49). Scale bars, 5 μm.
Antibodies might not access GNAI/Gαi in complex with Gβγ, and free GNAI might be present in low amounts at the junction. In contrast, GPSM2 is known to sequester free GNAI(GDP) via its four GoLoco motifs (58, 59), locally increasing GNAI amounts and likely facilitating GNAI immunodetection at the bare zone and at stereocilia tips. To visualize what could be an obscured or fleeting GNAI presence at HC junctions, we leveraged the Gnai3Egfp model. One antibody raised against a GNAI C-terminal region (sc-262; Materials and Methods) largely failed to detect GNAI3-EGFP in Gnai3Egfp/Egfp HCs (fig. S7A), providing a means to distinguish endogenous GNAI and GNAI3-EGFP in the same HC. In Gnai3Egfp/+ HCs, EGFP and GNAI signals colocalized at the bare zone and at stereocilia tips, as shown before (Fig. 7C and fig. S4B). At junctional level, however, EGFP but not GNAI was enriched and polarized laterally (Fig. 7C). Moreover, EGFP colocalized with DAPLE and RGS12 at the lateral junction in Gnai3Egfp/+ HCs (Fig. 7, D and E). Seeking an explanation for GNAI3-EGFP presence at the HC junction, we cotransfected HEK293 cells with 2HA-Daple and various Gnai3 constructs, immunoprecipitated 2HA-DAPLE in cell lysates, and quantified the amount of coimmunoprecipitated GNAI3. Unlike untagged GNAI3 and GNAI3 carrying an internal EGFP reporter, GNAI3 with a C-terminal reporter (GNAI3-EGFP) was poorly coimmunoprecipitated (Fig. 7F and fig. S7B). This suggests that the C-terminal EGFP moiety impairs GNAI3 binding to DAPLE. In contrast, GNAI3-EGFP could be coimmunoprecipitated with 2HA-RGS12, similar to untagged GNAI3 (fig. S7C). We thus hypothesize that GNAI3-EGFP presence at the lateral HC junction could stem from its abnormally stable association with RGS12 there, possibly linked to its defective binding to DAPLE. Accordingly, GNAI3-EGFP was absent at the lateral junction in Rgs12 mutant HCs (Fig. 7G) but faithfully recapitulated aberrant enrichment near stereocilia rootlets observed with the GNAI antibody (Fig. 7G and fig. S4B). GNAI3-EGFP was otherwise polarized normally at the apical membrane, and junctional presence of GNAI3-EGFP did not significantly interfere with apical HC morphogenesis since Gnai3Egfp/Egfp hair bundles were largely normal (Fig. 4, D to F). We thus conclude that DAPLE can still regulate GNAI3-EGFP in HCs despite their defective binding in HEK293 cells. Accordingly, the actual loss of DAPLE severely mislocalized GNAI3-EGFP along with native GNAI at the apical HC membrane (Fig. 7G to fig. S7D). As reported previously (25), the polarized distribution but not the apical enrichment of GNAI thus heavily depends on DAPLE, and this applies to GNAI3-EGFP as well. In summary, by virtue of the EGFP moiety, GNAI3-EGFP can be used as a probe to visualize what may be a low-amount, short-lived enrichment of GNAI at the HC lateral junction without overtly disrupting apical HC morphogenesis.
GPSM2 outcompetes RGS12 for an apical pool of free GNAI
We hypothesized that free GNAI(GDP) might be produced at the lateral HC junction by RGS12 and handed over to GPSM2 to be sequestered and accumulated at the adjacent bare zone (see model in Discussion). If this were true, then new apical GNAI proteins should first occupy the HC junction before diffusing to the bare zone. To begin to challenge this hypothesis, we used fluorescence recovery after photobleaching (FRAP) to quantify and compare recovered GNAI3-EGFP signals at the lateral junction and the bare zone in P2 Gnai3Egfp/+ cochlear explants (Fig. 7H). After photobleaching of the OHC apex, we observed that GNAI3-EGFP signal expressed as a ratio of its original intensity recovered faster at the junction compared to the bare zone (Fig. 7I; junction: estimated half-time, 16.4 min; mobile fraction, 31%; bare zone: half-time, 116 min; mobile fraction, 42%). This result is compatible with a transfer from the junction to the bare zone but is not evidence as the two GNAI3-EGFP populations could also turn over separately at different rates. While GNAI3-EGFP supports normal GPSM2-GNAI function and HC morphogenesis (Fig. 4, D to F), endogenous GNAI protein dynamics might differ from GNAI3-EGFP that does not bind DAPLE effectively (Fig. 7F).
RGS12 has two domains to bind GNAI. Besides the RGS (GAP) domain, a single GoLoco motif specifically binds GNAI in its GDP form (Fig. 1B). RGS12 can thus accelerate hydrolysis of free GNAI(GTP) (GAP activity) and also prevent a fraction of the resulting GNAI(GDP) product from reforming the G protein heterotrimer (GDI activity). As GPSM2 can also bind GNAI(GDP) (Fig. 1B), we hypothesized that GPSM2 and RGS12 might compete for GNAI(GDP) and that four GoLoco motifs might give GPSM2 an advantage.
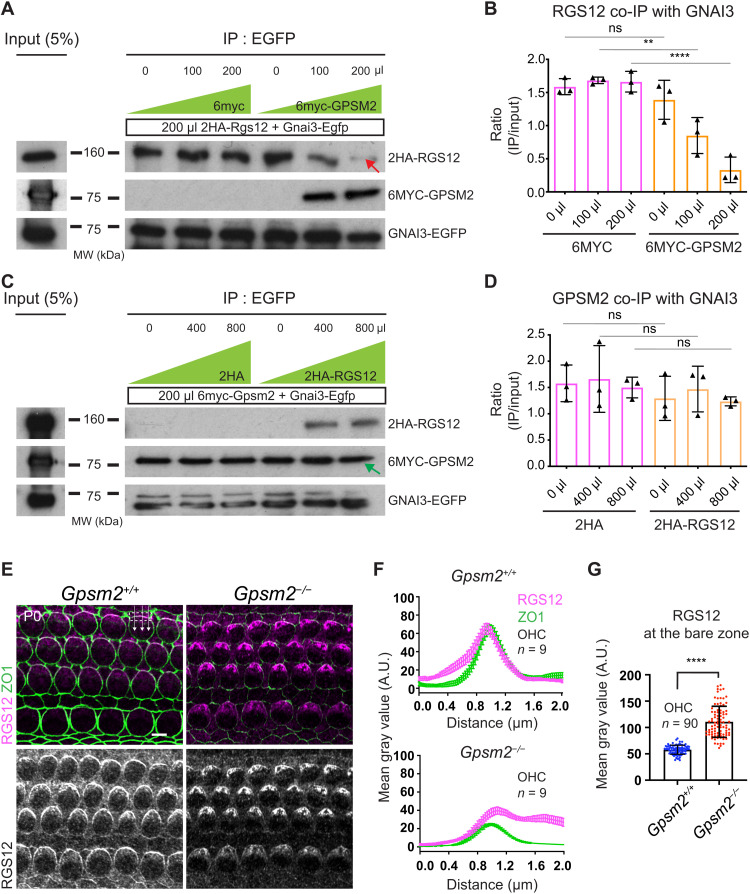
To test whether GPSM2 and RGS12 compete to bind GNAI, we separately transformed HEK293 cells with (i) 2HA-Rgs12 and Gnai3-Egfp constructs and (ii) 6myc or 6myc-Gpsm2 and cultured them for 2 days. We then mixed cell extracts (i + ii) with variable amounts of 6MYC or 6MYC-GPSM2, incubated the mixture for 1 hour, immunoprecipitated GNAI3-EGFP, and quantified the amount of coimmunoprecipitated 2HA-RGS12. Increasing the amount of 6MYC-GPSM2 in the mixture decreased the amount of 2HA-RGS12 coimmunoprecipitated with GNAI3-EGFP (Fig. 8, A and B). This demonstrates that GPSM2 can displace RGS12 as a GNAI3 binding partner. In control experiments, increasing the amount of the 6MYC extract did not affect binding of GNAI3 to RGS12 (Fig. 8, A and B). In a reciprocal experiment, we next separately transformed HEK293 cells with (i) 6myc-Gpsm2 and Gnai3-Egfp and (ii) 2HA or 2HA-Rgs12. In this case, 2HA-RGS12 could not displace GPSM2 as a GNAI3 binding partner, even when higher amounts of the 2HA-RGS12 competing extract were used in the mixture compared to the original design (Fig. 8, C and D). GPSM2 thus outcompetes RGS12 as a GNAI3 partner in a cell extract, likely because it carries four GoLoco motifs compared to one GoLoco in RGS12.
Fig. 8. GPSM2 outcompetes RGS12 for GNAI binding and prevents RGS12 enrichment at the bare zone.
(A to D) Binding competition assays. (A and B) Separate HEK293 cultures were transfected with (i) 2HA-Rgs12 and Gnai3-Egfp and (ii) 6myc or 6myc-Gpsm2. Protein extracts were then mixed (i + ii) with variable amounts of 6myc or 6myc-Gpsm2 extract (0, 100, or 200 μl for 200 μl of 2HA-Rgs12/Gnai3-Egfp extract). Blots in (A) show 2HA-RGS12 coimmunoprecipitation with GNAI3-EGFP, quantified in (B). (C and D) Reciprocal binding competition assays with (i) 6myc-Gpsm2 and Gnai3-Egfp and (ii) 2HA or 2HA-Rgs12. Blots in (C) show 6MYC-GPSM2 coimmunoprecipitation with GNAI3-EGFP, quantified in (D) (0, 400, and 800 μl of 2HA or 2HA-Rgs12 for 200 μl of 6myc-Gpsm2/Gnai3-Egfp). 6MYC-GPSM2 can outcompete 2HA-RGS12 for GNAI3-EGFP binding [(A), red arrow], but 2HA-RGS12 cannot outcompete 6MYC-GPSM2 [(C), green arrow]. (B and D) IP band intensity (ratio of input). Means ± SD, three experiments. One-way ANOVA with Tukey’s multiple comparisons. (B) ns, P = 0.8225; **P = 0.0027; and ****P < 0.0001. (D) ns: P = 0.9439 (0 μl), P = 0.9893 (400 μl), and P = 0.9586 (800 μl). Similar amounts of GNAI3-EGFP are immunoprecipitated across conditions (see fig. S8, A and B). (E) RGS12 immunolabeling. (F) Plot profile of averaged signal intensity along lines crossing the lateral OHC junction in (E). Means ± SEM. n = OHCs, N = 3 animals. (G) RGS12 intensity at the bare zone in (E). Means ± SD. N = 3 animals. Mann-Whitney test. ****P < 0.0001. In the absence of GPSM2, RGS12 signals appear reduced at the lateral junction and extend onto the bare zone, and ZO1 signals are unexpectedly reduced. MW, molecular weight in kilodalton. Scale bar, 5 μm.
As a proxy for testing whether GPSM2 has higher affinity than RGS12 for free GNAI in vivo, we asked how RGS12 behaves when GPSM2 is absent in HCs. In P0 Gpsm2 constitutive mutants, RGS12 enrichment was reduced at the lateral OHC junction but increased at the adjacent bare zone where GPSM2 normally resides (Fig. 8, E to G). Unexpectedly, enrichment of ZO1 at the OHC junction was also reduced in Gpsm2 mutants (Fig. 8, E and F). On the basis of these results, we speculate that free GNAI(GDP) could be transferred from the lateral junction to the apical membrane when GPSM2 outcompetes RGS12 as a GoLoco-based binding partner. In the transfer process, GPSM2 prevents RGS12 enrichment at the bare zone, restricting the bulk of RGS12 to the lateral junction.
DISCUSSION
Planar polarization of the cytoskeleton at the HC apical membrane, including the graded height arrangement of stereocilia, is essential for auditory function and relies heavily on the GPSM2-GNAI complex. Our findings indicate that RGS12 is the source of GPSM2-GNAI. We propose a model where RGS12 at the lateral HC junction limits GNAI(GTP) signaling (via its RGS/GAP activity) and sequesters a pool of the resulting free hydrolyzed GNAI(GDP) (via its GDI activity; Fig. 9). That pool could then be transferred to GPSM2 at the adjacent HC apical membrane because GPSM2 (four GoLoco motifs) outcompetes RGS12 (one GoLoco) to bind GNAI(GDP). It was reported previously that RGS12 can act as an enzymatic scaffold, integrating G protein signaling with other pathways. Notably, the long isoform of RGS12 carries a PDZ domain that may associate with other polarity proteins at the lateral HC junction and ensure a lateral bias in GAP and GDI activities. In contrast, it remained unclear how carrying an RGS and GDI motif that both bind and regulate GNAI can serve RGS12 functions. Our study suggests that this complex coordination of G protein signaling (35) might generate the GPSM2-GNAI complex.
Fig. 9. Summary and working model: RGS12 at the lateral HC junction produces the GPSM2-GNAI/Gαi complex deployed on the neighboring apical membrane (bare zone).
We hypothesize that DAPLE could dissociate the Gαi(GDP)βγ heterotrimer at the lateral junction. The RGS/GAP domain of RGS12 could accelerate hydrolysis of free Gαi(GTP). A fraction of the resulting Gαi(GDP) could be kept free by binding to the GoLoco domain of RGS12, whereas the rest is used to reform the heterotrimer. Gαi(GDP) could then be transferred from RGS12 to GPSM2 at the apical membrane on the basis of favorable GoLoco stoichiometry. In the absence of RGS12, GPSM2-GNAI/Gαi loses its polarization at the bare zone and at the tips of row 1 stereocilia. Instead, GPSM2-GNAI/Gαi accumulates at the base of stereocilia, where variable protein amounts still traffic to tips without row specificity. This leads to stunted stereocilia bundles with defective graded height architecture and row identity.
GEF-GAP activity on GNAI at the lateral HC junction
Two decades of active research only uncovered rare evidence that GPCRs (receptor GEFs; Fig. 1A) regulate GPSM2-GNAI or oriented cell division (6, 60, 61). In invertebrates, the cytoplasmic GEF and GNAI chaperone Ric8a was proposed to work with Rgs7 to generate free GNAI(GDP) for Gpsm2 and to influence the orientation of the mitotic spindle (62–64). Our work in postmitotic HCs suggests that the nonreceptor GEF DAPLE that belongs to the G protein–binding and activating (GBA) family (27, 28) might work with RGS12 to generate free GNAI(GDP) for GPSM2. Similar to GPCRs, GBA proteins can dissociate heterotrimeric G proteins (29, 65), one possible source of free GNAI(GTP) for RGS12 at the HC junction (Fig. 9). Previous results in vitro and in cell culture are compatible with this model. The GBA protein GIRDIN/GIV dissociates Gαiβγ more effectively than RGS12 (66), but the RGS12 GoLoco motif has a higher affinity for GNAI3 than GIRDIN GBA motif (67).
Immunolabeling detects GPSM2-mediated GNAI(GDP) accumulation at the HC apical membrane but not at the apical junction where GNAI is expected to be a substrate for DAPLE and RGS12. We speculate that antibodies may not be able to bind GNAI in the heterotrimeric complex and that low amounts of other GNAI species compared to GPSM2-GNAI(GDP) likely produce signals close to background level. We recently showed that the GPCR GPR156 signals via GNAI at the opposite (medial) junction to reverse HC orientation downstream of the transcription factor EMX2, and GNAI could not be immunodetected at the medial HC junction either (57). In contrast, our Gnai3-Egfp knock-in model provides evidence for GNAI presence at the lateral HC junction (Fig. 7). EGFP at the N- or C-terminal end of Gα proteins is well known to interfere with plasma membrane, Gβγ and GPCR interaction, and functional Gα assays typically use an internal EGFP (68). In our study, however, C-terminal EGFP does not affect the polarized localization of GNAI3 at the bare zone and at stereocilia tips or apical morphogenesis in HCs (Fig. 4, D to F). This is probably because C-terminal EGFP does not prevent GNAI from associating with GPSM2 (Fig. 8, A to D) (15, 59). GNAI3-EGFP can also still bind RGS12 (fig. S7C), and GNAI3-EGFP enrichment at the lateral HC junction depends on RGS12 (Fig. 7G). By contrast, GNAI3-EGFP does not bind DAPLE effectively in HEK293 cells (Fig. 7F), possibly because EGFP imposes a steric hindrance. DAPLE may thus increase turnover and limit junctional enrichment of endogenous GNAI, whereas GNAI3-EGFP would be stabilized there by preferentially binding to RGS12. In this regard, FRAP results obtained with GNAI3-EGFP may not reflect precisely the behavior of endogenous GNAI (Fig. 7, H and I). Despite these limitations, DAPLE and RGS12 depend on each other for normal co-enrichment at the lateral HC junction (Fig. 6, F to I) and likely work in tandem with roles that are distinct and remain to be completely elucidated. Unlike in Rgs12 mutants, GPSM2-GNAI is still enriched at the HC apical membrane in Daple mutants (fig. S7D) (25). However, GPSM2-GNAI does depend on DAPLE for normal polarized distribution, and complementary GPSM2-GNAI versus aPKC apical domains are preserved but severely distorted in Daple mutants (25). Because GNAI3-EGFP distribution is similarly distorted in Daple mutants but not in control HCs (Figs. 4D and 7G and fig. S7D), we conclude that DAPLE can appreciably regulate GNAI3-EGFP in HCs despite their defective interaction in the heterologous context of HEK293 cells.
Coupling HC-intrinsic and intercellular planar polarity mechanisms
Besides generating GPSM2-GNAI(GDP) as a derivative (Fig. 9), DAPLE and RGS12 are expected to drive conventional heterotrimeric G protein signaling at the lateral HC junction (Fig. 9). Emerging evidence suggests that conventional GNAI signaling might more specifically regulate HC orientation by integrating with the PCP machinery. First, DAPLE can bind and influence the PCP protein DVL2 that is also polarized at the lateral HC junction (25, 29, 69). In addition, DAPLE was proposed to respond to Wnt ligands and signal via Gβγ and phosphatidylinositol 3-kinase to regulate HC orientation, although the identity of the Wnt receptor remains unclear (47, 70). RGS12 (at the junction) and GPSM2 (at the bare zone) might create a permissive environment for lateral Gβγ signaling by sequestering free GNAI/Gαi(GDP). In this view, heterotrimeric G protein signaling at the HC lateral junction might be central to coupling HC-intrinsic polarization of the cytoskeleton [via RGS12 and GPSM2-GNAI(GDP)] and intercellular PCP mechanisms regulating HC orientation at cell-cell junctions [via DVL2>DAPLE>Gβγ].
Graded stereocilia height architecture and auditory function
This work lends support to the notion that restricted trafficking of GPSM2-GNAI to the first stereocilia row is secured by its prior polarization at the adjacent bare zone (18). In Rgs12 mutants, loss of GPSM2-GNAI at the bare zone coincides with its ectopic accumulation near stereocilia rootlets without row specificity (Fig. 9). Consistently, variably reduced GPSM2-GNAI enrichment at stereocilia tips also lacks row specificity, and the resulting hair bundles show extra rows and shortened stereocilia reminiscent of Gpsm2 and Gnai mutants. A similar loss of GNAI row specificity and similar structural defects were previously observed in HCs expressing Pertussis toxin (19). Evidence for bare zone–mediated trafficking to row 1 was also observed in Inscuteable and Gpsm2 mutants (18) and in mutants lacking the apical polarity protein MPDZ (49). As GPSM2-GNAI is required at stereocilia tips to confer row 1 its unique tallest identity (19), we conclude that G protein signaling at the HC lateral junction is an upstream cue and a major driver for graded stereocilia height architecture. To our knowledge, RGS12 was not implicated in human hearing loss to date. Wolf-Hirschhorn syndrome caused by hemizygote deletions of the distal end of chromosome 4 includes sensorineural hearing loss in about 15% of cases (71). As larger deletions can include RGS12 (72), RGS12 haploinsufficiency may contribute to hearing loss.
MATERIALS AND METHODS
Animal strain and care
The Rgs12 strain used in this study is a derivative of C57BL/6N-Rgs12tm1(KOMP)Vlcg (MGI:4399678) created by the KOMP/VelociGene consortium. In Rgs12tm1(KOMP)Vlcg, targeted insertion of the ZEN-Ub1 cassette created a 10,452–base pair (bp) deletion between positions 35122222 to 35132673 on chromosome 5 (GRCm38). We removed the neo selection cassette with CMV-Cre (B6.C-Tg(CMV-cre)1Cgn/J; MGI: 75204) and segregated CMV-Cre in subsequent crosses with C57BL/6J animals to obtain Rgs12tm1.1(KOMP)Vlcg (here Rgs12−; fig. S1). Other mouse strains in the study are “wild-type” [animals of mixed background between C57BL6/J (MGI:3028467) and FVB(MGI:2163709)], Daple/Ccdc88c KO [Ccdc88ctm1(KOMP)Mbp, MGI:1915589], R26-LSL-PTXa [Gt(ROSA)26Sorem1(ptxA)Btar, MGI:6163665 (18)], FoxG1-Cre [B6.129P2(Cg)-Foxg1tm1(cre)Skm/J, MGI:1932522], Gpsm2 knockout [KO; Gpsm2tm1a(EUCOMM)Wtsi, MGI:4441912; (15)], and Gnai3 KO (Gnai3tm1Lbi; MGI:3652743). Gnai3Egfp was engineered to generate a fusion protein between GNAI3 and EGFP (C-terminal) using CRISPR-Cas9. A donor vector included the following fragments in order: a 2004-bp 5′ homology arm, a GCGGCCGCG linker, Egfp followed by a stop codon, and a 1880-bp 3′ arm [arms were cloned by polymerase chain reaction (PCR) using C57BL/6J genomic template DNA]. No alterations to the Gnai3 genomic locus were introduced besides the linker/Egfp insertion. Two guide RNAs (GGAATGTGGGCTTTATTGAG and TGTGGGCTTTATTGAGAGGA) were designed to insert Egfp in frame at the C-terminal end of the Gnai3 coding sequence in exon 8. Guide RNAs and Cas9 protein were microinjected in fertilized C57BL6/J oocytes. Following identification of a founder animal using PCR, the allele was bred with C57BL6/J for two generations to segregate other potential genomic alterations. Experimental mice in the study ranged in age between E13.5 and P38 as indicated in each figure. Males and females were both included, but sex was not recorded except for ABR/DPOAE hearing tests since there is no published evidence that sex influences HC development. The initial characterization of the Rgs12 mutant strain was carried out with Rgs12+/+, Rgs12+/−, and Rgs12−/− littermates (Figs. 2 and 5 and fig. S2). Because physiological, anatomical, and molecular results were similar in Rgs12+/+ and Rgs12+/−, Rgs12+/− animals were used as controls in subsequent experiments to limit animal production. Animals were maintained under standard housing conditions (14-hour light/10-hour dark cycle, ambient temperature, and normal humidity). All animal work was reviewed for compliance and approved by the Animal Care and Use Committees of The Jackson Laboratory (Animal Use Summary AUS no. 14012).
Immunolabeling of cochlear tissue
Inner ears from embryonic (E14 to E17) or postnatal (P0 to P6) mice were extracted from the skull and either (i) fixed with 10% trichloroacetic acid (TCA; T8657, Thermo Fisher Scientific) on ice for 10 min or (ii) further dissected to expose the auditory epithelium and then fixed in 4% paraformaldehyde (PFA; 15710, Electron Microscopy Sciences) for 1 hour at 4°C. The auditory epithelium was exposed (TCA), and the tectorial membrane was removed (PFA) before permeabilization and blocking in 0.5% Triton X-100 and 1% bovine serum albumin in phosphate-buffered saline (PBS) for 30 min at room temperature. Next, primary antibodies were added and the tissue was incubated at 4°C overnight. The next day, samples were washed in PBS + 0.05% Triton X-100 (3× 15 min) and conjugated secondary antibodies and phalloidin were added for a second 4°C overnight incubation. Following washes, samples were postfixed in 4% PFA for 1 hour at room temperature. Sensory epithelia were detached from the modiolus and mounted on slides (M1021, Denville) with 10% Mowiol (475904, EMD/Millipore; in 0.1 M tris-Cl, pH 8.5) either compressed by the cover glass (48366-045, VWR) or using office tape as spacer to avoid compression. Primary antibodies were chicken anti-RGS12 (GW21317, Sigma-Aldrich; TCA, 1:500; raised against the 75 most C-terminal amino acids of the human protein), mouse anti-acetylated Tubulin (23950, Santa Cruz Biotechnology; PFA, 1:500), rat anti-ZO1 (R26.4C, Developmental Studies Hybridoma Bank; PFA and TCA, 1:200), chicken anti-GNAI3 (Fig. 4, A, C, and H and fig. S3B; GW22489, Sigma-Aldrich; PFA, 1:400; human amino acids 345 to 353), rabbit anti-GNAI3 (Figs. 3, A, D, F, and G; 5A; and 7, A and C ; and figs. S3A, S4B, S5 and S7, A and D; sc-262, Santa Cruz Biotechnology; PFA and TCA, 1:400; rat undisclosed C-terminal peptide), goat anti-GPSM2 (PA5-18646, Thermo Fisher Scientific; PFA, 1:250), mouse anti-EPS8 (610143, BD Biosciences; PFA, 1:250), mouse anti-aPKC/PRKCZ (sc-216, Santa Cruz Biotechnology; PFA, 1:100), rabbit anti-DAPLE/CCDC88C (25769-1-AP, Proteintech; TCA, 1:400), rabbit anti-WHRN (gift from J. Yang Laboratory, University of Utah; 1:500), rabbit anti-MYO15A short isoform (PB48, gift from J. Bird Laboratory, University of Florida; PFA, 1:500), and sheep anti-Afadin (AF7829, R&D Systems; PFA, 1:400; this antibody labels apical junctions as well as primary cilia and HC kinocilia; cilia labeling might be unspecific). Alexa Fluor (AF)–conjugated antibodies from Thermo Fisher Scientific were donkey anti-mouse (AF488: A21202; AF555: A-31570; AF647: A-31571), donkey anti-rabbit (AF555: A-31572; AF647: A-31573), donkey anti-goat (AF647: A-21447), donkey anti-rat (AF488: A-21208), and goat anti-chicken (AF546: A-11040). Conjugated phalloidin was used to reveal F-actin (AF488: A12379; AF555: A34055, Thermo Fisher Scientific; CF405: 89138-126, Biotium). Wheat germ agglutinin was used to visualize the hair bundle (W11261, Thermo Fisher Scientific; 1:2000).
Image acquisition
Most images were acquired using an LSM 800 line scanning confocal microscope using Zen blue 2.3 or Zen 2.6 software (Carl Zeiss AG). Most images were captured in regular confocal mode using a 63×/1.4 NA (numerical aperture) oil objective (420782-9900-799, Carl Zeiss AG), the Airyscan detector and sequential channel capture. Some specific experiments were imaged in Airyscan mode (Figs. 2D, 3F, 4A and C, 5A, and 7H) and raw images were processed with Zen software’s automated settings for two-dimensional (2D) or 3D processing, standard processing strength, and auto filtering. Acquisition for fixed tissue was done at 2500 × 2500 pixel resolution and a z-spacing of 0.15 μm. Capture settings (laser intensity and gain) were kept constant when imaging samples of different genotypes (littermates in the same cohort). Wide-field images of X-Gal stainings in fig. S1D were captured with a Leica DM5500B microscope using the LAS X 3.7.2 software.
Image analysis and quantification
All quantifications unless mentioned were performed in Fiji (2.3.0) (73). For quantification of the kinocilium index and hair bundle angle (Fig. 2, B and C), methods by Landin Malt. et al. (47) were adopted. Briefly, for kinocilium index in Fig. 2B, the length of the two halves of each hair bundle (left and right to the kinocilium) were measured and their ratio (x/y) plotted. For Fig. 2C, the angle between the lines used for kinocilium index was plotted. To quantify the number of stereocilia rows in IHC and OHC in Fig. 2E, the number of stereocilia along a lateral-to-medial line at the position of the kinocilium was counted and plotted in a frequency distribution histogram. Quantification of HC orientation in fig. S2C was done as previously (57). Briefly, at the 50% cochlear position, we used the position of the kinocilium labeled with acetyl-tubulin antibody relative to the center of the HC apical surface to draw one vector. The angle between this vector and a vector parallel to the apical surface of inner Pillar cells was measured. Either right or left cochleae were used with the field reversed (“Flip Canvas Horizontal” in Adobe Photoshop) as needed so that 0° (“3 p.m.”) on circular histograms consistently pointed toward the cochlear base. 90° (“12 p.m.”) indicated the lateral/abneural direction (cochlear periphery). To quantify stereocilia height in IHCs in Figs. 2G and 4I, confocal Z-stacks were analyzed in Imaris (8.2.4, Oxford Instruments). The height of an individual stereocilium was measured from root to tip in a 3D environment. To quantify the percentage of cells positive for GNAI in Fig. 3 (B and C), we acquired images at the cochlear base (10%), middle-turn (50%), and apical-turn (85%) and counted HCs where any apical GNAI signal was visible in a 260-μm longitudinal window. The number of positive cells was then divided by the total number of outer or inner HCs in each field to obtain percentages shown in the plots. For signal plot profiles in Figs. 6E, 7 (B and E), and 8F, we used the rectangle (averaging) plot profiling function in Fiji. A rectangular region of interest (ROI) was drawn at the lateral junction and lateral-to-medial signals on the short side of the ROI were averaged by the Fiji function to obtain one value per HC [edge crossing junction: 3 μm (Fig. 6E), 2 μm (Figs. 7B and 8F), and 4 μm (Fig. 7E); edge parallel to junction: 4 μm for all].
To measure protein enrichment, we used the mean gray value function in Fiji. Background was calculated using the same size ROI by averaging three measurements lateral and medial to HCs where no signal enrichment was visible. This average was subtracted from every HC signal value measured in the same image. To quantify GNAI signals in Fig. 3E, the F-actin channel was used to outline each HC and total signal at the apical membrane was measured. To quantify GNAI and MYO15A signals at stereocilia tips in Fig. 4J, a circular ROI (radius 0.18 μm) was placed using the F-actin channel and GNAI and MYO15A signals were measured for the same ROI. To quantify protein enrichment at junctions in Fig. 6 (B, G, and I), a 0.6 μm–by–0.2 μm ROI was placed at the lateral and medial junction in each HC. ZO1 signals were quantified in the same ROI, and enrichment of the protein of interest was normalized to averaged lateral and medial ZO1 signals. To quantify signals at the bare zone in Fig. 8G, a 0.6 μm–by–0.6 μm ROI was drawn inside the bare zone and signal measured. Image processing was done in Adobe Photoshop (CC 2020) applying the same treatment to samples of different genotypes (littermates in the same cohort). Figures were assembled in Adobe Illustrator (CC 2020).
Scanning electron microscopy
Temporal bones from P33 to P38 animals were fixed in 2.5% glutaraldehyde (16200, Electron Microscopy Sciences) + 4% PFA in 0.1 M sodium cacodylate with 1 mM MgCl2 and 20 mM CaCl2. After three washes in PBS, samples were decalcified in 0.11 M EDTA overnight. Decalcified tissue was then dissected to obtain three pieces (base, mid, and apex) of the cochlear floor. The samples were dehydrated with an ethanol series (30-50-70-80-90-100%) with 15-min incubation steps. Sample dehydration was performed with hexamethyldisilazane (HMDS; 16700, Electron Microscopy Sciences). After three 10-min HMDS changes, HMDS was removed, and samples were left to dry in a fume hood. Samples were then mounted on aluminum stubs (75200, Electron Microscopy Sciences) using double-sided carbon tape (77816, Electron Microscopy Sciences). Mounted samples were sputter-coated with gold-palladium and imaged with a Hitachi S-3000N VP microscope at 20 kV.
Cochlea electroporation and culture
Inner ears from wild-type animals were collected at E13.5 in Hanks’ balanced salt solution + 25 mM Hepes buffer (14065-056 and 15630-080, respectively; Gibco). Plasmid DNA was mixed with Fast Green FCF (final, 0.05%; F7252, Sigma-Aldrich) and injected at 2 μg/μl into the cochlear duct using the Wiretrol plunger system (53507-426, Drummond). Plasmids carried the mouse coding sequence for Rgs12 full-length (ENSMUST00000030984.14, Rgs12-201) and short (ENSMUST00000114281.8, Rgs12-204) isoforms with two N-terminal HA tags under the control of the caggs promoter (CMV early enhancer element fused to chicken beta-actin promoter). Next, the whole inner ear was electroporated (27 V, 27 ms, and six square pulses at 950-ms intervals; BTX ECM 830), and the membranous labyrinth was dissected away from the condensed mesenchyme and embedded in Matrigel [8 μl drop of 50% Matrigel in Dulbecco’s modified Eagle’s medium (DMEM); CB40234, Corning]. The explants were cultured for 6 days in DMEM with 10% fetal bovine serum and ciprofloxacin (10 μg/ml; 17850, Sigma-Aldrich) and then fixed in 4% PFA for 15 min at room temperature before being processed for immunolabeling as described above.
Live imaging, FRAP, and quantification
Gnai3Egfp/+ cochleae were dissected at P2. The intact cochlear duct was separated from the modiolus and cut into three segments at the 25 and 75% position from the base. Only the middle-turn segment was used. The Reissner membrane was opened, and the lateral wall still attached to the cochlear floor was flipped open to expose the auditory epithelium. This “open-book” preparation was then explanted on a tissue culture–grade 35-mm dish (1228K66, CellTreat) coated with poly-d-lysine (10 μg/ml; ICN10269410, MP Biomedicals) and cultured in DMEM/F12 (SH3002301, HyClone) + ciprofloxacin (10 μg/ml; 17850, Sigma-Aldrich). A plastic ring (PVC; inner diameter, 12.7 mm; outer diameter, 19 mm; thickness, 0.32 mm; 98090A225, Precision Brand) was used as spacer. A 22-mm cover glass (10026-140, Matsunami) was placed on top of the ring to close the chamber and sealed with silicone grease (Z273554, Dow Corning). The chamber was placed on a microscope incubator (130-800 005, PECON) to keep the tissue at 37°C, 5% CO2, and 10% humidity. Live stacks (range: 4.5 to 6 μm at 0.15-μm intervals) were captured every 10 min in Airyscan mode with an LSM 800 confocal microscope and a 40×/1.1 NA water immersion lens (421867-9970-000, Carl Zeiss AG). Photobleaching was carried out using the timed-bleaching application in Zen 2.6 and by successively photobleaching the bare zone and junction Z planes. Regions to be photobleached were hand-drawn in Zen, and EGFP bleaching was done with the 405 laser (intensity 25%), a speed of 3, and six repetitions within a cycle for a total of three cycles to achieve complete darkening of the selected regions. Laser power for EGFP capture (488) was 0.6%.
Intensity measurements before and after FRAP were done in Fiji with the mean gray value function by drawing ROIs encompassing (i) half the bare zone (freehand selection) on the Z-slice where the bare zone signal was the brightest and (ii) a 0.6 μm–by–0.2 μm region at the lateral junction on the Z-slice where the junctional signal was the brightest. The brightest Z position was selected by measuring and comparing the mean signal intensity of the same ROI at multiple Z positions. Normalized fractional fluorescence intensity was then calculated using the method by Roy and Perrin (74). For every time point, fractional fluorescence was calculated with the formula
where Roblx is the intensity of the bleached region at time x, Roconpre is the intensity of the control region before bleaching, Roconx is the intensity of the control region at time x, Robl0 is the intensity of the bleached region immediately after bleaching, Rocon0 is the intensity of the control region immediately after bleaching, and Roblpre is the intensity of the target region before bleaching. Control regions were from the bare zone and junction of one neighboring HC in the same field whose EGFP signal was not photobleached. To fit curves in Fig. 7I and determine GNAI3-EGFP half-time and mobile fraction at the junction and bare zone, we used nonlinear regression and one-phase association tools in Prims 9 (GraphPad).
Immunoprecipitation, Western blot, and signal quantification
HEK293 cells (CRL-3216, American Type Culture Collection) were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml; 15140122, Gibco) at 37°C. This cell line was not authenticated since acquisition by our laboratory. For transfection, confluent cells were passaged at 1:50 dilution in six-well plates. After 1-day incubation, cells were transfected with a plasmid mixture (total, 2 μg) using jetPRIME transfection reagent (101000046, Polyplus Transfection). Transfected cells were then grown for 2 days. Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer [50 mM tris-Cl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 0.1% NP-40, and proteinase inhibitor cocktail (118361700001, Roche)] and centrifuged to clean up debris. Input fractions (4 to 5% of total lysate) were collected from centrifuged samples. Magnetic Dynabeads (10003D, Invitrogen) were antibody-coated at 4°C for 1 hour and incubated with the cell lysate at 4°C overnight under agitation. The next day, a magnetic stand was used to wash the beads three times with RIPA buffer. Laemmli buffer (6×) was added, and samples were boiled for 10 min at 95°C. Samples were then run on 15% SDS–acrylamide gel. The proteins on the gel were transferred to an activated polyvinylidene difluoride membrane (88520, Bio-Rad). Membranes were blocked in 5% nonfat dry milk in Tris-buffered saline (0.5% Tween 20) for 1 hour. Blocked membranes were incubated with primary antibodies at 4°C overnight in a 0.5% milk/TBST solution under agitation. After three TBST washes, membranes were incubated with horseradish peroxidase (HRP)–conjugated secondary antibodies in a light-safe box. Membranes were washed again and incubated with the ECL substrate (28980926, GE Healthcare). Light signal detection was carried out by exposing the membrane to film (1159T41, HyBlot CL, Denville).
Quantification in immunoprecipitation (Fig. 7F) and protein binding competition analysis (Fig. 8, A to D, and fig. S8) were carried out in Fiji with digital scans of the films. Briefly, protein bands on scans were selected with the rectangle selection tool, and the intensity curve was obtained with the Gel analyzer function. Signals were then normalized to the average of the corresponding inputs (control + experimental inputs). Antibodies used for immunoprecipitation (IP) and immunoblotting (IB) were as follows: rabbit anti-HA for IP (1:500) and IB (1:2000; 3724S, Cell Signaling Technology), rabbit anti-EGFP for IP (1:500) and IB (1:5000; A11122, Molecular Probes), rabbit anti-GNAI2 for IB (1:300; 11136-1-AP, Proteintech; detects GNAI3 equally well), and mouse anti-MYC for IB (1:200; 9e10; The Jackson Laboratory Protein Production Services). HRP-conjugated secondary antibodies are goat anti-rabbit (111-035-144, Jackson ImmunoResearch) and goat anti-mouse (115-035-003, Jackson ImmunoResearch).
Mouse ABR and DPOAE
ABR and DPOAE tests were performed on P33 to P38 mice of both sexes as detailed in data source file S1. Males and females were pooled, as sex did not influence threshold shifts. Mice were anesthetized by intraperitoneal injection of a mix of ketamine and xylazine (10 mg and 0.1 mg per 10 g of body weight, respectively), and body temperature was maintained at 37°C using a heating pad. All tests were conducted in a sound-attenuating chamber. ABR tests used the RZ6 Multi-I/O Processor System coupled to the four-channel Medusa Amplifier (RA4PA, Tucker-Davis Technologies). The BioSigRZ software (4.7.5, Tucker-Davis Technologies) was used to generate specific acoustic stimuli that included broadband clicks and 8-, 16-, 32-, and 40-kHz pure tone bursts. One channel of ABR was recorded after binaural stimulation. Electrodes were placed subdermally. The active electrode was inserted at the vertex; the reference electrode was inserted ventrolateral to the left ear, and the ground electrode was inserted to the right thigh. Auditory thresholds were obtained for each stimulus starting at 90 dB and reducing the sound pressures level (SPL) by 10-dB steps for click and 5-dB steps for pure tones to identify the lowest level at which an ABR could be recognized. This was done by comparing the ABR patterns with two or three suprathreshold ABRs displayed concurrently on the screen. The ABRs were typically identified with 512 stimuli presented at the rate of 21 per second. Arrows for ABR thresholds in Rgs12 mutants (Fig. 5C) indicate that no response could be recorded even at 90 dB. DPOAE tests used the RZ6 Multi-I/O Processor and BioSigRZ software (4.7.5, Tucker-Davis Technologies) to generate and control the stimuli. Pure tone frequencies (f2/f1 ratio = 1.2) at 8, 12, 16, and 24 kHz at equal levels of sound pressure (L1 = L2) were generated by the RZ6 processor and attenuated through PA5 programmable attenuators. Separate drivers were used to route these attenuators to mix acoustically in the ear canal with the help of an earpiece. For each mouse, SPLs from 80 to 20 dB (in 10-dB decrements) were tested in 512 readings. SPLs originating from the ear canal were recorded with a sensitive microphone (ER 10db + Microphone, Etymotic Research). After 10× amplification, the microphone signal was rerouted to the RZ6 processor. This acoustic signal was sampled at 100 kHz and fast Fourier transformations (FFTs) of the signal were averaged. The FFT waveform was used to measure the amplitudes of f1, f2, and the (2f1 − f2) distortion product. Threshold for amplification was determined by comparing the distortion product to the noise floor: If a distortion product peak was higher in magnitude than the noise floor, then it was recognized as a real signal.
Data plotting and statistical analysis
All animals of appropriate age and genotype were included, and no additional inclusion or exclusion criteria were applied. The stage of the tissue analyzed is indicated in the top left corner of each panel group. N indicates the number of animals used for each experiment, and n indicates the number of HCs analyzed, except for Figs. 2G and 4 (I and J) where n indicates the number of stereocilia. N and n values are reported in the figures or figure legends. Data source file S1 contains all the values plotted in all the figures. In experiments where the outcome was not quantified, at least three mutant samples in a minimum of two litters and a similar number of littermate controls were analyzed. In nonquantified experiments, figure panels show a representative outcome observed in all controls and all mutants analyzed. When not quantified (figs. S6G and S7, B and C), immunoprecipitation and Western blots were repeated three times and representative images were shown. All graphs except circular histograms (fig. S2C) were produced in Prism 9 (GraphPad). Circular histograms were generated in R using the coord_polar function in the ggplot2 package in RStudio v4.4.3 (75).
Statistical analyses were performed in Prism 8 or 9 (GraphPad). Error bars indicate SDs, except in plot profile graphs (Figs. 6E; 7, B and E; and 8F) where they indicate SEMs. Normal data distribution was assessed with D’Agostino-Pearson omnibus normality test. F test was used for variance comparison in Fig. 2 (B and G). For pairwise comparisons, unpaired t test and nonparametric unpaired t test (Mann-Whitney) were used for datasets with normal or non-normal distribution, respectively. For >2 comparisons and one factor, one-way analysis of variance (ANOVA) with Tukey’s multiple comparison was used for datasets with a normal distribution. Nonparametric Kruskal-Wallis with Dunn’s multiple comparison was used for datasets where one condition at least did not show normal data distribution. For >2 comparisons and two factors, two-way ANOVA with Tukey’s multiple comparison was used. In Fig. 4J, Pearson’s correlation test was used to correlate GNAI and MYO15A signals at stereocilia tips. In fig. S2C, the distribution of HC orientation in controls and mutants was compared by calculating a normalized difference (ND) value. ND represents the number of circular SDs (CSD) separating the circular means (CM) of the control and mutant distribution. A small ND value thus indicates similar distributions. ND was calculated in R as the angular distance between the control and mutant CMs divided by the average CSD of the two distributions. Circular histograms (fig. S2C) plot the frequency distribution of angles measured in 10° bins; the black bar indicates the CM, and the length of the arc at the tip of the bar indicates the CSD. Detailed information on experimental cohorts and statistics can be found in data source file S1 under the corresponding figure tab.
Acknowledgments
We thank E. Hartig and A. Jarysta for comments on the manuscript. We are grateful to The Jackson Laboratory Genome Engineering Technology and Reproductive Science services for help with generating the Gnai3Egfp strain and cryorecovering the Rgs12tm1(KOMP)/Vlcg strain, respectively.
Funding: This work was supported by a JAX Scholar postdoctoral fellowship (to A.A.) and National Institute on Deafness and Other Communication Disorders grants R01DC015242 and R01DC018304 (to B.T.).
Author contributions: Conceptualization: A.A. and B.T. Methodology: A.A. and B.T. Investigation: A.A., M.D., and B.T. Visualization: A.A. and B.T. Writing: A.A. and B.T. Supervision: B.T.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials and are detailed in data source file S1.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Other Supplementary Material for this manuscript includes the following:
Data source file S1
REFERENCES AND NOTES
- 1.Gilman A. G., G proteins: Transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615–649 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Kimple A. J., Bosch D. E., Giguere P. M., Siderovski D. P., Regulators of G-protein signaling and their Gα substrates: Promises and challenges in their use as drug discovery targets. Pharmacol. Rev. 63, 728–749 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neubig R. R., Siderovski D. P., Regulators of G-protein signalling as new central nervous system drug targets. Nat. Rev. Drug Discov. 1, 187–197 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Blumer J. B., Oner S. S., Lanier S. M., Group II activators of G-protein signalling and proteins containing a G-protein regulatory motif. Acta Physiol (Oxf.) 204, 202–218 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Willard F. S., Kimple R. J., Siderovski D. P., Return of the GDI: The GoLoco motif in cell division. Annu. Rev. Biochem. 73, 925–951 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Lechler T., Mapelli M., Spindle positioning and its impact on vertebrate tissue architecture and cell fate. Nat. Rev. Mol. Cell Biol. 22, 691–708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tadenev A. L. D., Tarchini B., The spindle orientation machinery beyond mitosis: When cell specialization demands polarization. Adv. Exp. Med. Biol. 1002, 209–225 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Yu F., Morin X., Kaushik R., Bahri S., Yang X., Chia W., A mouse homologue of Drosophila pins can asymmetrically localize and substitute for pins function in Drosophila neuroblasts. J. Cell Sci. 116, 887–896 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Parmentier M. L., Woods D., Greig S., Phan P. G., Radovic A., Bryant P., O’Kane C. J., Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J. Neurosci. 20, RC84 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaefer M., Shevchenko A., Shevchenko A., Knoblich J. A., A protein complex containing inscuteable and the Gα-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10, 353–362 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Yu F., Morin X., Cai Y., Yang X., Chia W., Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100, 399–409 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Schaefer M., Petronczki M., Dorner D., Forte M., Knoblich J. A., Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107, 183–194 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Grassie M. A., McCallum J. F., Guzzi F., Magee A. I., Milligan G., Parenti M., The palmitoylation status of the G-protein G(o)1 alpha regulates its activity of interaction with the plasma membrane. Biochem. J. 302(Pt. 3), 913–920 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galbiati F., Volonte D., Meani D., Milligan G., Lublin D. M., Lisanti M. P., Parenti M., The dually acylated NH2-terminal domain of gi1alpha is sufficient to target a green fluorescent protein reporter to caveolin-enriched plasma membrane domains. Palmitoylation of caveolin-1 is required for the recognition of dually acylated g-protein alpha subunits in vivo. J. Biol. Chem. 274, 5843–5850 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Tarchini B., Jolicoeur C., Cayouette M., A molecular blueprint at the apical surface establishes planar asymmetry in cochlear hair cells. Dev. Cell 27, 88–102 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Bhonker Y., Abu-Rayyan A., Ushakov K., Amir-Zilberstein L., Shivatzki S., Yizhar-Barnea O., Elkan-Miller T., Tayeb-Fligelman E., Kim S. M., Landau M., Kanaan M., Chen P., Matsuzaki F., Sprinzak D., Avraham K. B., The GPSM2/LGN GoLoco motifs are essential for hearing. Mamm. Genome 27, 29–46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezan J., Lasvaux L., Gezer A., Novakovic A., May-Simera H., Belotti E., Lhoumeau A. C., Birnbaumer L., Beer-Hammer S., Borg J. P., le Bivic A., Nürnberg B., Sans N., Montcouquiol M., Primary cilium migration depends on G-protein signalling control of subapical cytoskeleton. Nat. Cell Biol. 15, 1107–1115 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Tarchini B., Tadenev A. L. D., Devanney N., Cayouette M., A link between planar polarity and staircase-like bundle architecture in hair cells. Development 143, 3926–3932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tadenev A. L. D., Akturk A., Devanney N., Mathur P. D., Clark A. M., Yang J., Tarchini B., GPSM2-GNAI specifies the tallest stereocilia and defines hair bundle row identity. Curr. Biol. 29, 921–934.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beer-Hammer S., Lee S. C., Mauriac S. A., Leiss V., Groh I. A. M., Novakovic A., Piekorz R. P., Bucher K., Chen C., Ni K., Singer W., Harasztosi C., Schimmang T., Zimmermann U., Pfeffer K., Birnbaumer L., Forge A., Montcouquiol M., Knipper M., Nürnberg B., Rüttiger L., Gαi proteins are indispensable for hearing. Cell. Physiol. Biochem. 47, 1509–1532 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Mauriac S. A., Hien Y. E., Bird J. E., Carvalho S. D. S., Peyroutou R., Lee S. C., Moreau M. M., Blanc J. M., Gezer A., Medina C., Thoumine O., Beer-Hammer S., Friedman T. B., Rüttiger L., Forge A., Nürnberg B., Sans N., Montcouquiol M., Defective Gpsm2/Gαi3 signalling disrupts stereocilia development and growth cone actin dynamics in Chudley-McCullough syndrome. Nat. Commun. 8, 14907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudspeth A. J., Corey D. P., Sensitivity, polarity, and conductance change in the response of vertebrate hair cells to controlled mechanical stimuli. Proc. Natl. Acad. Sci. U.S.A. 74, 2407–2411 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh T., Shahin H., Elkan-Miller T., Lee M. K., Thornton A. M., Roeb W., Abu Rayyan A., Loulus S., Avraham K. B., King M. C., Kanaan M., Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet. 87, 90–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doherty D., Chudley A. E., Coghlan G., Ishak G. E., Innes A. M., Lemire E. G., Rogers R. C., Mhanni A. A., Phelps I. G., Jones S. J. M., Zhan S. H., Fejes A. P., Shahin H., Kanaan M., Akay H., Tekin M.; FORGE Canada Consortium, Triggs-Raine B., Zelinski T., GPSM2 mutations cause the brain malformations and hearing loss in Chudley-McCullough syndrome. Am. J. Hum. Genet. 90, 1088–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siletti K., Tarchini B., Hudspeth A. J., Daple coordinates organ-wide and cell-intrinsic polarity to pattern inner-ear hair bundles. Proc. Natl. Acad. Sci. U.S.A. 114, E11170–E11179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozono Y., Tamura A., Nakayama S., Herawati E., Hanada Y., Ohata K., Takagishi M., Takahashi M., Imai T., Ohta Y., Oshima K., Sato T., Inohara H., Tsukita S., Daple deficiency causes hearing loss in adult mice by inducing defects in cochlear stereocilia and apical microtubules. Sci. Rep. 11, 20224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiGiacomo V., Marivin A., Garcia-Marcos M., When heterotrimeric G proteins are not activated by G protein-coupled receptors: Structural insights and evolutionary conservation. Biochemistry 57, 255–257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aznar N., Kalogriopoulos N., Midde K. K., Ghosh P., Heterotrimeric G protein signaling via GIV/Girdin: Breaking the rules of engagement, space, and time. Bioessays 38, 379–393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aznar N., Midde K. K., Dunkel Y., Lopez-Sanchez I., Pavlova Y., Marivin A., Barbazán J., Murray F., Nitsche U., Janssen K. P., Willert K., Goel A., Abal M., Garcia-Marcos M., Ghosh P., Daple is a novel non-receptor GEF required for trimeric G protein activation in Wnt signaling. eLife 4, e07091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarchini B., A reversal in hair cell orientation organizes both the auditory and vestibular organs. Front. Neurosci. 15, 695914 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deans M. R., Conserved and divergent principles of planar polarity revealed by hair cell development and function. Front. Neurosci. 15, 742391 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarchini B., Lu X., New insights into regulation and function of planar polarity in the inner ear. Neurosci. Lett. 709, 134373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soundararajan M., Willard F. S., Kimple A. J., Turnbull A. P., Ball L. J., Schoch G. A., Gileadi C., Fedorov O. Y., Dowler E. F., Higman V. A., Hutsell S. Q., Sundström M., Doyle D. A., Siderovski D. P., Structural diversity in the RGS domain and its interaction with heterotrimeric G protein α-subunits. Proc. Natl. Acad. Sci. U.S.A. 105, 6457–6462 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snow B. E., Hall R. A., Krumins A. M., Brothers G. M., Bouchard D., Brothers C. A., Chung S., Mangion J., Gilman A. G., Lefkowitz R. J., Siderovski D. P., GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain. J. Biol. Chem. 273, 17749–17755 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Kimple R. J., de Vries L., Tronchère H., Behe C. I., Morris R. A., Farquhar M. G., Siderovski D. P., RGS12 and RGS14 GoLoco motifs are Gαi interaction sites with guanine nucleotide dissociation inhibitor activity. J. Biol. Chem. 276, 29275–29281 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Natochin M., Lester B., Peterson Y. K., Bernard M. L., Lanier S. M., Artemyev N. O., AGS3 inhibits GDP dissociation from Gα subunits of the Gi family and rhodopsin-dependent activation of transducin. J. Biol. Chem. 275, 40981–40985 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Kimple R. J., Kimple M. E., Betts L., Sondek J., Siderovski D. P., Structural determinants for GoLoco-induced inhibition of nucleotide release by Gα subunits. Nature 416, 878–881 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Schiff M. L., Siderovski D. P., Jordan J. D., Brothers G., Snow B., de Vries L., Ortiz D. F., Diversé-Pierluissi M., Tyrosine-kinase-dependent recruitment of RGS12 to the N-type calcium channel. Nature 408, 723–727 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Willard M. D., Willard F. S., Li X., Cappell S. D., Snider W. D., Siderovski D. P., Selective role for RGS12 as a Ras/Raf/MEK scaffold in nerve growth factor-mediated differentiation. EMBO J. 26, 2029–2040 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sethakorn N., Yau D. M., Dulin N. O., Non-canonical functions of RGS proteins. Cell. Signal. 22, 1274–1281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S., Li Y. P., Liu T., He X., Yuan X., Li C., Cao J., Kim Y., Mx1-cre mediated Rgs12 conditional knockout mice exhibit increased bone mass phenotype. Genesis 51, 201–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng A. Y. H., Li Z., Jones M. M., Yang S., Li C., Fu C., Tu C., Oursler M. J., Qu J., Yang S., Regulator of G protein signaling 12 enhances osteoclastogenesis by suppressing Nrf2-dependent antioxidant proteins to promote the generation of reactive oxygen species. eLife 8, e42951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gross J. D., Kaski S. W., Schroer A. B., Wix K. A., Siderovski D. P., Setola V., Regulator of G protein signaling-12 modulates the dopamine transporter in ventral striatum and locomotor responses to psychostimulants. J. Psychopharmacol. 32, 191–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroer A. B., Mohamed J. S., Willard M. D., Setola V., Oestreich E., Siderovski D. P., A role for regulator of G protein signaling-12 (RGS12) in the balance between myoblast proliferation and differentiation. PLOS ONE 14, e0216167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J., Chen L., Yao Y., Tang C., Ding J., Fu C., Li H., Ma G., Pivotal role of regulator of G-protein signaling 12 in pathological cardiac hypertrophy. Hypertension 67, 1228–1236 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Yu F., Wang H., Qian H., Kaushik R., Bownes M., Yang X., Chia W., Locomotion defects, together with Pins, regulates heterotrimeric G-protein signaling during Drosophila neuroblast asymmetric divisions. Genes Dev. 19, 1341–1353 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landin Malt A., Hogan A. K., Smith C. D., Madani M. S., Lu X., Wnts regulate planar cell polarity via heterotrimeric G protein and PI3K signaling. J. Cell Biol. 219, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barr-Gillespie P. G., Assembly of hair bundles, an amazing problem for cell biology. Mol. Biol. Cell 26, 2727–2732 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jarysta A., Tarchini B., Multiple PDZ domain protein maintains patterning of the apical cytoskeleton in sensory hair cells. Development 148, dev199549 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ebrahim S., Ingham N. J., Lewis M. A., Rogers M. J. C., Cui R., Kachar B., Pass J. C., Steel K. P., Alternative splice forms influence functions of whirlin in mechanosensory hair cell stereocilia. Cell Rep. 15, 935–943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delprat B., Michel V., Goodyear R., Yamasaki Y., Michalski N., el-Amraoui A., Perfettini I., Legrain P., Richardson G., Hardelin J. P., Petit C., Myosin XVa and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Hum. Mol. Genet. 14, 401–410 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Belyantseva I. A., Boger E. T., Naz S., Frolenkov G. I., Sellers J. R., Ahmed Z. M., Griffith A. J., Friedman T. B., Myosin-XVa is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat. Cell Biol. 7, 148–156 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Kitajiri S., Sakamoto T., Belyantseva I. A., Goodyear R. J., Stepanyan R., Fujiwara I., Bird J. E., Riazuddin S., Riazuddin S., Ahmed Z. M., Hinshaw J. E., Sellers J., Bartles J. R., Hammer J. A. III, Richardson G. P., Griffith A. J., Frolenkov G. I., Friedman T. B., Actin-bundling protein TRIOBP forms resilient rootlets of hair cell stereocilia essential for hearing. Cell 141, 786–798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin-McCaffrey L., Hains M. D., Pritchard G. A., Pajak A., Dagnino L., Siderovski D. P., D’Souza S. J. A., Differential expression of regulator of G-protein signaling R12 subfamily members during mouse development. Dev. Dyn. 234, 438–444 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Snow B. E., Antonio L., Suggs S., Gutstein H. B., Siderovski D. P., Molecular cloning and expression analysis of RatRgs12 and Rgs14. Biochem. Biophys. Res. Commun. 233, 770–777 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Locht C., Coutte L., Mielcarek N., The ins and outs of pertussis toxin. FEBS J. 278, 4668–4682 (2011). [DOI] [PubMed] [Google Scholar]
- 57.Kindt K. S., Akturk A., Jarysta A., Day M., Beirl A., Flonard M., Tarchini B., EMX2-GPR156-Gαi reverses hair cell orientation in mechanosensory epithelia. Nat. Commun. 12, 2861 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Natochin M., Gasimov K. G., Artemyev N. O., Inhibition of GDP/GTP exchange on Gα subunits by proteins containing G-protein regulatory motifs. Biochemistry 40, 5322–5328 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Du Q., Macara I. G., Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell 119, 503–516 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Yoshiura S., Ohta N., Matsuzaki F., Tre1 GPCR signaling orients stem cell divisions in the Drosophila central nervous system. Dev. Cell 22, 79–91 (2012). [DOI] [PubMed] [Google Scholar]
- 61.McDonald W. S., Miyamoto K., Rivera R., Kennedy G., Almeida B. S. V., Kingsbury M. A., Chun J., Altered cleavage plane orientation with increased genomic aneuploidy produced by receptor-mediated lysophosphatidic acid (LPA) signaling in mouse cerebral cortical neural progenitor cells. Mol. Brain 13, 169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Afshar K., Willard F. S., Colombo K., Johnston C. A., McCudden C. R., Siderovski D. P., Gönczy P., RIC-8 is required for GPR-1/2-dependent Gα function during asymmetric division of C. elegans embryos. Cell 119, 219–230 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Hess H. A., Röper J.-C., Grill S. W., Koelle M. R., RGS-7 completes a receptor-independent heterotrimeric G protein cycle to asymmetrically regulate mitotic spindle positioning in C. elegans. Cell 119, 209–218 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Wang H., Ng K. H., Qian H., Siderovski D. P., Chia W., Yu F., Ric-8 controls Drosophila neural progenitor asymmetric division by regulating heterotrimeric G proteins. Nat. Cell Biol. 7, 1091–1098 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Garcia-Marcos M., Ghosh P., Farquhar M. G., GIV is a nonreceptor GEF for Gαi with a unique motif that regulates Akt signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 3178–3183 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia-Marcos M., Complementary biosensors reveal different G-protein signaling modes triggered by GPCRs and non-receptor activators. eLife 10, e65620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Opakua A. I., Parag-Sharma K., DiGiacomo V., Merino N., Leyme A., Marivin A., Villate M., Nguyen L. T., de la Cruz-Morcillo M. A., Blanco-Canosa J. B., Ramachandran S., Baillie G. S., Cerione R. A., Blanco F. J., Garcia-Marcos M., Molecular mechanism of Gαi activation by non-GPCR proteins with a Gα-binding and activating motif. Nat. Commun. 8, 15163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hynes T. R., Hughes T. E., Berlot C. H., Cellular localization of GFP-tagged α subunits. Methods Mol. Biol. 237, 233–246 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Wang J., Mark S., Zhang X., Qian D., Yoo S.-J., Radde-Gallwitz K., Zhang Y., Lin X., Collazo A., Wynshaw-Boris A., Chen P., Regulation of polarized extension and planar cell polarity in the cochlea by the vertebrate PCP pathway. Nat. Genet. 37, 980–985 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Landin Malt A., Clancy S., Hwang D., Liu A., Smith C., Smith M., Hatley M., Clemens C., Lu X., Non-canonical Wnt signaling regulates cochlear outgrowth and planar cell polarity via Gsk3β inhibition. Front. Cell Dev. Biol. 9, 649830 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Battaglia A., Carey J. C., The delineation of the Wolf-Hirschhorn syndrome over six decades: Illustration of the ongoing advances in phenotype analysis and cytogenomic technology. Am. J. Med. Genet. A 185, 2748–2755 (2021). [DOI] [PubMed] [Google Scholar]
- 72.Wieczorek D., Krause M., Majewski F., Albrecht B., Horn D., Riess O., Gillessen-Kaesbach G., Effect of the size of the deletion and clinical manifestation in Wolf-Hirschhorn syndrome: Analysis of 13 patients with a de novo deletion. Eur. J. Hum. Genet. 8, 519–526 (2000). [DOI] [PubMed] [Google Scholar]
- 73.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roy P., Perrin B. J., The stable actin core of mechanosensory stereocilia features continuous turnover of actin cross-linkers. Mol. Biol. Cell 29, 1856–1865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.R Development Core Team, R: A Language and Environment for Statistical Computing, in R foundation for Statistical Computing (R Foundation for Statistical Computing, 2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Data source file S1