Abstract
Recent studies have shown that probiotic supplementation has beneficial effects on bone metabolism. In a randomized controlled trial (RCT) we demonstrated that supplementation of Lactobacillus reuteri ATCC PTA 6475 reduced bone loss in older women with low bone mineral density. To investigate the mechanisms underlying the effect of L. reuteri ATCC PTA 6475 on bone metabolism, 20 women with the highest changes (good responders) and the lowest changes (poor responders) in tibia total volumetric BMD after one-year supplementation were selected from our previous RCT. In the current study we characterized the gut microbiome composition and function as well as serum metabolome in good responders and poor responders to the probiotic treatment as a secondary analysis. Although there were no significant differences in the microbial composition at high taxonomic levels, gene richness of the gut microbiota was significantly higher (P < 0.01 by the Wilcoxon rank-sum test) and inflammatory state was improved (P < 0.05 by the Wilcoxon signed-rank test) in the good responders at the end of the 12-month daily supplementation. Moreover, detrimental changes including the enrichment of E. coli (adjusted P < 0.05 by DESeq2) and its biofilm formation (P < 0.05 by GSA) observed in the poor responders were alleviated in the good responders by the treatment. Our results indicate that L. reuteri ATCC PTA 6475 supplementation has the potential to prevent a deterioration of the gut microbiota and inflammatory status in elderly women with low bone mineral density, which might have beneficial effects on bone metabolism.
Subject terms: Microbiome, Microbiota
Introduction
Osteoporosis is a highly prevalent bone disease in the elderly population, and is characterized by decreased bone mineral density (BMD), deteriorated bone microarchitecture, reduced bone strength and increased susceptibility to fragility fractures. The risk of fracture can be reduced by pharmacological treatment, but treatment rates in patients with osteoporosis remain low, possibly due to low osteoporosis awareness, costs for medication and fear or rare side effects of presently available drugs1,2. Thus, there is an urgent need to develop a novel and effective intervention for the prevention and treatment of osteoporosis.
Towards this goal, the gut microbiota has been revealed to play pivotal roles in bone metabolism, potentially by regulating the immune system and osteoclast formation in mice3–6. This suggests that modulating the gut microbiome by nutritional supplements with probiotics may provide novel strategies for the prevention and treatment of osteoporosis. In earlier experimental studies, the supplementation of probiotic strain Lactobacillus reuteri ATCC PTA 6475 has been demonstrated to reduce bone loss and increase bone density in mice with estrogen deficiency or increased inflammation7,8. In our previous randomized controlled trial, we observed that supplementation of L. reuteri ATCC PTA 6475 could reduce bone loss by ~50% in older women with low BMD9. Thus, L. reuteri ATCC PTA 6475 may be a potential therapeutic strategy to prevent postmenopausal bone loss. However, we also observed that part of the subjects responded poorly to the treatment with L. reuteri ATCC PTA 6475 (i.e., poor responders with less relative changes in tibia total volumetric BMD)9.
Initial reports suggest that the effects of probiotic supplementations on bone metabolism might involve the modulation of the composition and function of the gut microbiota10–12. Specifically, different probiotics were reported to expand intestinal butyrate-producing bacteria10, indicating that probiotics may indirectly increase production of short chain fatty acids (SCFAs) by the gut microbiota. Moreover, the probiotic Lactobacillus rhamnosus GG has been suggested to increase bone formation by stimulating the production of microbial metabolite butyrate, which induced T cell-produced Wnt10b in eugonadic young mice11.
To gain mechanistic insight into the effect of L. reuteri ATCC PTA 6475 on bone metabolism and identify factors important for a good response to the probiotic, here we characterized the metagenomic profile of the fecal microbiota and the serum metabolome of good responders and poor responders, who had the highest and lowest changes in tibia total volumetric BMD in our randomized controlled trial9, respectively. After a one-year probiotic supplementation, we found decreased inflammation and significantly increased gene richness of the gut microbiota in the good responders, while altered microbial composition and function, including enrichment of E. coli and its biofilm formation in the poor responders. Our study reveals new possible mechanisms contributing to the regulation of bone loss in older women, which could be crucial for the development of novel osteoporosis treatments.
Results
Probiotic supplementation decreases inflammation and increases bone mineral density in the good responders
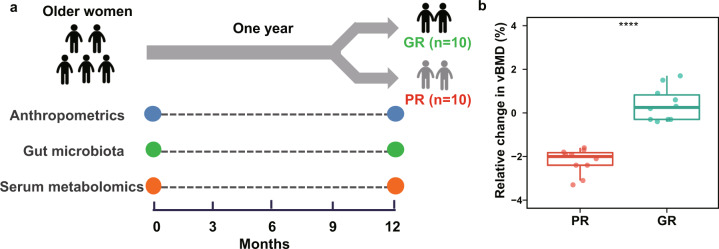
To investigate differences in subjects with different responses to supplementation with the probiotic strain L. reuteri ATCC PTA 6475, we selected 20 elderly women by identifying ten women with a good response (GR group) and ten women with a poor response (PR group) from the per protocol (PP) population as previously described9 (Fig. 1a). The demographic and clinical characteristics of these 20 subjects at baseline and 12 months were shown in Table 1. There were no significant differences in these characteristics between the GR and PR groups at both time points, but subjects in the GR group (27.5 ± 3.6 kg/m2 (baseline) and 27.7 ± 3.7 kg/m2 (12 months)) had slightly higher body mass index (BMI) than the PR group (24.0 ± 2.8 kg/m2 (baseline) and 24.1 ± 3.1 kg/m2 (12 months)) (P < 0.05 by the t-test). After one-year treatment with L. reuteri ATCC PTA 6475, the relative change in tibia total volumetric BMD exhibited significantly increased levels in the GR group (0.39 ± 0.77) compared to the PR group (−2.22 ± 0.58; P < 0.001 by the t-test; Fig. 1b). In addition, relative changes in cortical thickness and cortical volumetric BMD differed between the two groups (P < 0.001 using a t-test; Supplementary Table 1). Moreover, levels of ultrasensitive c-reactive protein (usCRP) in the GR group decreased after one-year treatment (P < 0.05 by the Wilcoxon signed-rank test), which indicates that inflammation in the GR group is possibly treated by supplementation with the probiotics. To further evaluate effects of supplementation with L. reuteri ATCC PTA 6475 on the gut microbiota and metabolomic profiles of the subjects, we collected fecal samples and serum samples from these 20 subjects at baseline and 12 months, respectively, as illustrated in Fig. 1a.
Fig. 1. The scheme diagram of experimental design for elderly women with differential responses to the supplementation with L. reuteri ATCC PTA 6475.
a Women with a good response (GR group, n = 10) and with a poor response (PR group, n = 10) were recruited. Serum samples and fecal samples were collected from the older women with bone loss at baseline and 12 months. The complete metabolomic profiling for these women were performed. However, for metagenomic profiling, one woman in the GR and PR group lost one fecal sample, respectively. Therefore, nine women had time points-paired fecal samples in each group. b The boxplot shows the relative change of tibia total volumetric BMD in the GR and PR groups after one-year treatment. ‘****’ denotes P < 0.001. The upper quartile, median and lower quartile are shown in the box plot.
Table 1.
Comparions of characteristics in the GR and PR groups at baseline and 12 months.
| Characteristics | Baseline | 12 months | P1 | P2 | ||
|---|---|---|---|---|---|---|
| GR (n = 10) | PR (n = 10) | GR (n = 10) | PR (n = 10) | |||
| Age (year) | 76.6 ± 1.1 | 76.3 ± 1.0 | 77.6 ± 1.0 | 77.4 ± 1.0 | 0.57 | 0.56 |
| Height (cm) | 162.6 ± 5.1 | 163.1 ± 4.3 | 163.0 ± 5.5 | 163.2 ± 4.3 | 0.82 | 0.94 |
| Weight (kg) | 72.4 ± 8.2 | 63.9 ± 7.7 | 73.3 ± 8.5 | 64.0 ± 8.0 | 0.03 | 0.02 |
| Body mass index (kg/m2) | 27.5 ± 3.6 | 24.0 ± 2.8 | 27.7 ± 3.7 | 24.1 ± 3.1 | 0.03 | 0.03 |
| Bone mineral density (g/cm2) | ||||||
| Lumbar spine | 0.99 ± 0.13 | 0.98 ± 0.1 | 1.01 ± 0.14* | 0.98 ± 0.09 | 0.84 | 0.63 |
| Total hip | 0.82 ± 0.1 | 0.81 ± 0.05 | 0.82 ± 0.09 | 0.8 ± 0.05 | 0.76 | 0.60 |
| Femoral neck | 0.66 ± 0.07 | 0.68 ± 0.07 | 0.66 ± 0.07 | 0.66 ± 0.08# | 0.67 | 0.92 |
| HR-pQCT derived bone variables | ||||||
| Total tibia vBMD (mg/cm3) | 247 ± 39.2 | 231 ± 44.9 | 248 ± 39.8 | 226 ± 44.2# | 0.42 | 0.26 |
| Trabecular BV/TV (%) | 13.1 ± 2.29 | 11.5 ± 1.98 | 13.1 ± 2.34 | 11.4 ± 2.11 | 0.12 | 0.10 |
| Cortical vBMD (mg/cm3) | 787 ± 46.5 | 769 ± 74.5 | 786 ± 44.6 | 758 ± 70.6# | 0.52 | 0.30 |
| Cortical thickness (mm) | 0.84 ± 0.17 | 0.82 ± 0.23 | 0.84 ± 0.17 | 0.79 ± 0.23# | 0.84 | 0.53 |
| Serum markers | ||||||
| N-terminal telopeptide (nM) | 14.8 ± 3.97 | 13.1 ± 2.97 | 14.1 ± 2.35 | 14.8 ± 3.64 | 0.30 | 0.61 |
| BAP (U/L) | 16.4 ± 3.55 | 15.6 ± 2.97 | 17.2 ± 3.91 | 16.2 ± 2.14 | 0.60 | 0.52 |
| usCRP (mg/L) | 2.14 (1.53–3.68) | 0.98 (0.8–2.47) | 1.57 (1.13–1.90)* | 1.36 (0.67–3.19) | 0.25 | 0.91 |
| TNF-α (pg/mL) | 1.28 (1.09–1.57) | 1.23 (0.98–1.47) | 1.24 (1.11–1.59) | 1.15 (1.04–1.35) | 0.63 | 0.47 |
| Body composition (kg) | ||||||
| Total fat mass | 28.9 (27.1–32.0) | 20.8 (19.3–24.5) | 28.2 (24.3–31.7) | 20.0 (18.5–23.7) | 0.04 | 0.04 |
| Total lean mass | 44.2 ± 3.2 | 41.4 ± 3.6 | 45.7 ± 3.6* | 42.3 ± 4.1 | 0.09 | 0.06 |
Note: Mean ± SD. Non-normally distributed variables are presented as median with interquartile range. For comparison between the GR and PR groups at each time point, Student’s t-test or Wilcoxon rank sum test were used as appropriate. For comparison between baseline and 12 months in each group, paired t-test or Wilcoxon signed-rank test were used.
‘*’ and ‘#’ denote significant difference (P < 0.05) between baseline and 12 months in the GR and PR groups, respectively.
P 1 and P 2 values are derived from comparisons between the GR and PR groups at baseline and 12 months, respectively.
The significant differences (P < 0.05) are highlighted in bold.
Total vBMD Total tibia volumetric bone mineral density, Trabecular BV/TV Trabecular bone volume fraction, Cortical vBMD Cortical volumetric bone mineral density, BAP Bone-specific alkaline phosphatase, usCRP ultrasensitive c-reactive protein, TNF-α Tumor necrosis factor alpha.
The overall composition of gut microbiota after one-year supplementation with L. reuteri ATCC PTA 6475
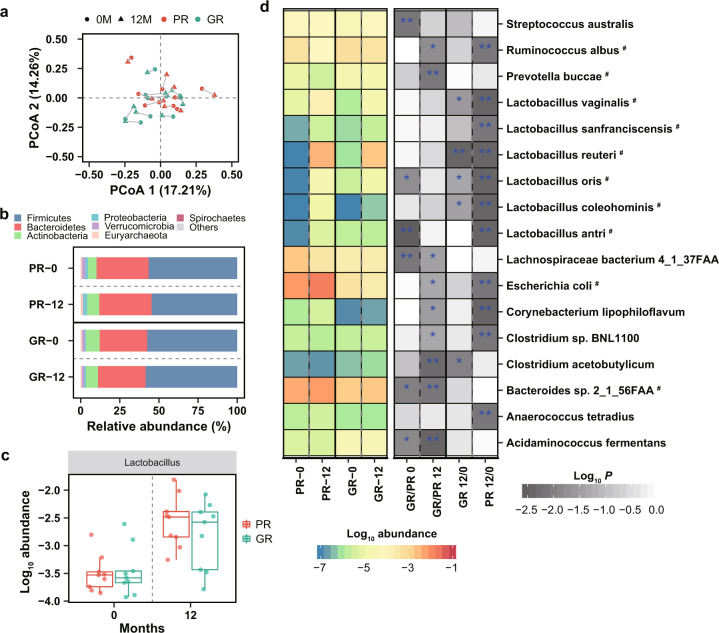
To identify alteration of gut microbiota after L. reuteri ATCC PTA 6475 treatment and potential microbial signatures that are important for a good response to the probiotic, the fecal metagenomes of the selected 20 subjects were characterized using shotgun sequencing. The taxonomic profiles were first calculated using the MEDUSA pipeline13. Principal coordinate analysis (PCoA) and α diversity analysis results showed no significant differences in the overall microbial composition between the GR and PR groups at either baseline or 12 months (Fig. 2a and Supplementary Fig. 1). Also, there were no distinct shifts in the gut microbiota in the two groups after L. reuteri ATCC PTA 6475 treatment (Fig. 2a). Moreover, Fig. 2b showed the overview of microbial species abundances at phylum levels, mainly including Firmicutes, Bacteroidetes and Actinobacteria, which exhibited no significant differences between two groups or time points using the Wilcoxon test (Supplementary Table 2). Furthermore, we performed differential analyses of the taxonomic profiles at levels of class, order, family, genus and species between the GR and BR groups or between the two time points. Genus Lactobacillus and family Lactobacillaceae had elevated abundances after 12 months in the two groups (P < 0.01 by the Wilcoxon signed-rank test; Fig. 2c and Supplementary Fig. 2), but showed no difference between the GR and PR groups at each time point. Therefore, at high taxonomic levels, there were no significant differences in the microbial composition between the two groups or time points in this study, except for genus Lactobacillus and family Lactobacillaceae.
Fig. 2. Compositional differences of the gut microbiota after supplementation with L. reuteri ATCC PTA 6475.
a Principal coordinate analysis (PCoA) of microbiota community at species level based on Bray–Curtis distance. The overall microbial composition between the GR (n = 9) and PR (n = 9) groups or between the two time points shows no significant difference, estimated by PERMANOVA analysis. The blue and red colors indicate the GR and PR groups, respectively; The dot and triangle indicate the baseline (0 M) and 12 months (12 M), respectively. b Relative profiles of top seven abundant phyla. c Increased abundance of the genus Lactobacillusin in the GR (n = 9) and PR (n = 9) groups at 12 months (P-value < 0.01 by the Wilcoxon signed-rank test). The upper quartile, median and lower quartile are shown in the box plot. The blue and red colors indicate the GR and PR groups, respectively. d The left heatmap shows log-transformed mean abundances of differential species in the two groups at the two time points. The blue and red colors indicate the low and high abundances, respectively; The grey color in the right heatmap indicates P-value of comparative analysis using the Wilcoxon test; ‘*’ denotes P < 0.05; ‘**’ denotes P < 0.01; ‘#’ indicates the differential species were also identified by DESeq2 (adjusted P-value < 0.05 and |log2 fold change | >1.
Detrimental changes of microbial species in poor responders after one-year supplementation with L. reuteri ATCC PTA 6475
After one-year supplementation, species L. reuteri abundances were increased in the GR and PR groups (P < 0.01 by the Wilcoxon signed-rank test and adjusted P < 0.05 & |log2 fold change | > 1 by DESeq2; Fig. 2d and Supplementary Table 2 and 3), but did not show differences between the two groups. In total, we identified 16 species differential between the two groups or between the two time points in total (P < 0.01 by the Wilcoxon test; Fig. 2d). Four species, including Prevotella buccae (adjusted P < 0.05 by DESeq2), Clostridium acetobutylicum, Bacteroides sp. 2_1_56FAA (adjusted P < 0.05 by DESeq2), Acidaminococcus fermentans, had differential abundances between the two groups at 12 months, while three species, including Streptococcus australis, Lactobacillus antri (adjusted P < 0.05 by DESeq2), Lachnospiraceae bacterium 4_1_37FAA, showed differential between the two groups at baseline (P < 0.01 by the Wilcoxon rank-sum test; Fig. 2d). Lactobacillus antri was less abundant in the PR group than the GR group, while Lachnospiraceae bacterium 4_1_37FAA was more abundant in the PR groups at baseline (P < 0.01 by the Wilcoxon rank-sum test). Moreover, Escherichia coli was enriched in the PR group compared to the GR group at 12 months (adjusted P = 0.03 & log2 fold change = −3.8 by DESeq2, Supplementary Table 3).
In addition, 11 differential species, including Escherichia coli, were identified between the two time points in the PR group while only one differential species (L. reuteri) in the GR group (P < 0.01 by the Wilcoxon signed-rank test; Fig. 2d), which suggests that microbial shifts mainly happen in the PR group. E. coli was increased solely in the PR group (adjusted P = 0.04 & log2 fold change = 3.7 by DESeq2, Supplementary Table 3) and meanwhile exhibited different levels between the GR and PR groups at 12 months.
Also, we calculated relative abundances of species using MetaPhlAn2 pipeline14, and performed comparative analyses between the GR and PR groups or the two time points using the Wilcoxon test (Supplementary Table 4). Only four species L. reuteri, Coprococcus catus, Gordonibacter pamelaeae and Erysipelatoclostridium ramosum showed differential abundances between the GR and PR groups or between the two time points (P < 0.01 by the Wilcoxon test; Supplementary Fig. 3). E. coli was enriched, while Akkermansia muciniphila, Ruminococcus bicirculans, Eubacterium_sp_CAG_38 and Butyricimonas virosa were depleted in the PR groups at 12 months, compared to the GR group (P < 0.05 by the Wilcoxon rank-sum test; Supplementary Fig. 3).
Functional alterations of the gut microbiota after supplementation with L. reuteri ATCC PTA 6475
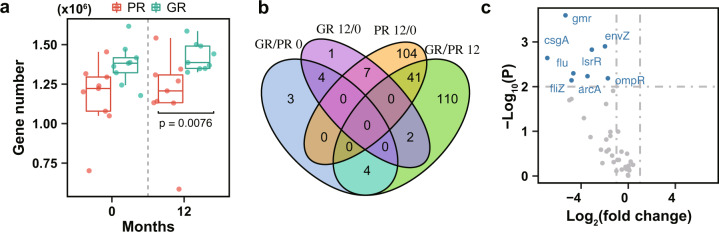
The functional capacities of the gut microbiome in the GR and PR groups were investigated at baseline and 12 months, respectively. We first calculated the gene and KO (KEGG Orthology) profiles using the MEDUSA pipeline13. In comparison to the PR group, gene richness had an increased trend in the GR group at baseline but not statistically significant (Fig. 3a). Interestingly, gene richness was significantly higher in the GR group (1.40 ± 0.11 million) than the PR group (1.18 ± 0.26 million) at 12 months (P < 0.01 by the Wilcoxon rank-sum test; Fig. 3a). By comparative analysis, there were 11 and 157 KOs identified to be significantly differential between the two groups at baseline and 12 months, respectively (Adjusted P < 0.1 by DESeq2; Fig. 3b; Supplementary Table 5). This suggests that functional capacities of the endogenous baseline microbiota show no significant difference between the GR and PR groups. In addition, 152 differential KOs were identified between the two time points in the PR group while only 14 differential KOs in the GR group (Adjusted P < 0.1 by DESeq2; Fig. 3b), indicating that microbial function altered in the PR group over time much more than the GR group.
Fig. 3. Potential function alterations of the gut microbiota after supplementation with L. reuteri.
a Gene number of the gut microbiota in the PR (n = 9) and GR (n = 9) groups at the two time points. 20 million reads from each sample were sampled in order to rarefy the reads to the same depth of sequencing. The blue and red colors indicate the GR and PR groups, respectively. The upper quartile, median and lower quartile are shown in the box plot. b The Venn diagram shows differential KOs between the GR (n = 9) and PR (n = 9) groups or between baseline and 12 months (Adjusted P-value < 0.1 by DESeq2). ‘GR/PR 0’ and ‘GR/PR 12’ denote the comparative analysis between the GR and PR groups at baseline and 12 months, respectively. ‘GR 12/0’ and ‘PR 12/0’ denote the comparative analysis between the two time points in the GR and PR groups, respectively. c The volcano plot displays the differential genes involved in biofilm formation (Escherichia coli) between the GR (n = 9) and PR (n = 9) groups at 12 months. The blue color indicates the differential genes; The horizontal and vertical dashed lines indicate P-value < 0.01 and |log2 fold change | >1, respectively.
Furthermore, by the gene set analysis (GSA), there were 16 differentially enriched KEGG pathways identified by comparing the GR and PR groups at 12 months or by comparing the two time points in the PR group (P < 0.05 by GSA; Supplementary Table 6). Interestingly, the microbial metabolism related to biofilm formation (E. coli) was enriched in the PR group at 12 months in comparison to baseline (P < 0.05 by GSA; Supplementary Table 6). The abundances of microbial genes involved in biofilm formation, including gmr, arcA, lsrR, rcsC and rcsD, were increased in the PR group (P < 0.01 by DESeq2 and |log2 fold change > 1 | ; Supplementary Fig. 4). Accordantly, we observed difference in the metabolism related to biofilm formation (E. coli) between the GR and PR groups at 12 months (P < 0.05 by GSA; Supplementary Table 6). The genes including arcA, gmr, csgA and envZ involved in biofilm formation, were more abundant in the PR group than the GR group after one-year treatment (P < 0.01 by DESeq2 and |log2 fold change > 1 | ; Fig. 3c). The capacity of microbial phenylalanine metabolism was also increased in the PR group at 12 months compared to the baseline (P < 0.05 by GSA; Supplementary Table 6).
Meanwhile, we calculated the relative profiles of the MetaCyc pathways using the HUMAnN2 tool15, and performed comparative analyses between the GR and PR groups or the two time points (Supplementary Table 7). In agreement with results of the GSA, the enrichment of superpathway of L − phenylalanine biosynthesis was observed in the PR group at 12 months (P < 0.01 by the Wilcoxon signed-rank test; Supplementary Fig. 5). In addition, the capacity of polyisoprenoid biosynthesis (E. coli) was elevated in the PR group at 12 months compared to baseline (P < 0.05 by the Wilcoxon signed-rank test; Supplementary Fig. 6).
Metabolomics changes in response to the supplementation with L. reuteri ATCC PTA 6475
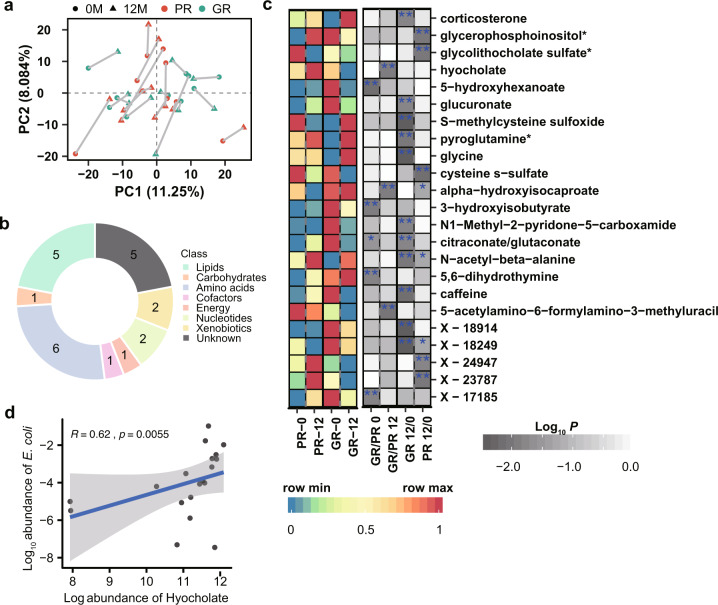
Early studies have revealed that the gut microbiota could impact bone metabolism by producing SCFAs, which can stimulate bone formation11. To investigate the effects of L. reuteri ATCC PTA 6475 on global metabolism of the 20 subjects in the GR and PR groups, we analyzed 40 serum metabolomics profiles collected at baseline and 12 months from these women in our previous study16. In total, the metabolomics dataset comprised of 1,232 metabolites that were measured by liquid chromatography-tandem mass spectroscopy (LC-MS). Principal component analysis (PCA) showed that metabolomic profiles did not exhibit a clear distinction between the GR and PR groups at baseline or 12 months (Fig. 4a).
Fig. 4. Changes of metabolic profiles of the older women after treatment with L. reuteri.
a The score plot of principal components analysis (PCA) based on metabolomic profiles in the GR (n = 10) and PR (n = 10) groups at baseline and 12 months. The blue and red colors indicate the GR and PR groups, respectively; The dot and triangle indicate the baseline (0 M) and 12 months (12 M), respectively. b Biological processes in which the differential metabolites are involved (P < 0.01 by the Wilcoxon test). c The left heatmap shows scaled mean abundances of the differential metabolites in the PR (n = 10) and GR (n = 10) groups at two time points. The blue and red colors indicate the low and high abundances, respectively; The grey color in the right heatmap indicates P-value of comparative analysis; ‘*’ denotes P < 0.05; ‘**’ denotes P < 0.01. d Association between metabolite hyocholate and the species E. coli. The blue line and grey shade indicate the regression line and 95% confidence interval; R denotes Spearman’s correlation coefficient.
Furthermore, we performed differential analysis of metabolite abundances between the two groups or between the two time points (Supplementary Table 8), and a total of 23 metabolites were identified to be differentially abundant (P < 0.01 by the Wilcoxon test), which are mainly involved in biological processes lipids and amino acids (Fig. 4b). Amino acids have been suggested to be associated with bone metabolism17. In accordance, six amino acids derived metabolites were identified to be differential in this study (P < 0.01 by the Wilcoxon test; Fig. 4b). Out of them, three metabolites, including hyocholate, alpha−hydroxyisocaproate, 5−acetylamino−6−formylamino−3−methyluracil, had differential abundances between the two groups at 12 months, while four metabolites, including 5−hydroxyhexanoate, 3−hydroxyisobutyrate, 5,6−dihydrothymine, showed differential between the two groups at baseline (P < 0.01 by the Wilcoxon rank-sum test; Fig. 4c). Hyocholate, involved in secondary bile acid metabolism, had an increased abundance in the PR group at 12 months, compared to the GR group (P < 0.01 by the Wilcoxon rank-sum test).
Moreover, 11 and 5 metabolites abundances differed between the two time points in the GR and PR groups, respectively (P < 0.01 by the Wilcoxon signed-rank test; Fig. 4c). The abundance of cysteine s−sulfate decreased in the PR group after one-year probiotic supplementation (P < 0.01 by the Wilcoxon signed-rank test). Cysteine s-sulfate is a structural analog of glutamate and could act as an agonist of N-methyl-D-aspartate receptor (NMDA-R)18, which was reported to be involved in bone resorption19. Interestingly, butyrylcarnitine (C4) showed an elevated trend in the GR group after probiotic supplementation (P < 0.05 by the Wilcoxon signed-rank test; Supplementary Fig. 7 and Supplementary Table 8), which is consistent with our early report that butyrylcarnitine (C4) was increased in older women supplemented with L. reuteri ATCC PTA 6475 compared to placebo16. Butyrylcarnitine (C4) could act as the pool and transporter of butyrate20, which has previously been shown to inhibit bone resorption and stimulate bone formation in mice11,21,22.
To explore the potential links between the differential metabolites and species, we performed correlation analyses at baseline and 12 months, respectively (Supplementary Table 9). At 12 months, we observed associations between 5,6-dihydrothymine and Prevotella buccae (R = 0.61, P = 0.0056, Supplementary Table 9), between citraconate/glutaconate and Ruminococcus albus (R = −0.72, P = 0.00049) and between S-methylcysteine and Lactobacillus.coleohominis (R = −0.58, P = 0.0087). Particularly, metabolite hyocholate showed a positive correlation with species E. coli at 12 months (R = 0.62, P = 0.0055; Fig. 4d). Moreover, at baseline we found associations between glycolithocholate sulfate* and Bacteroides sp.2_1_56FAA (R = 0.71, P = 0.00068, Supplementary Table 9), between pyroglutamine* and Prevotella buccae (R = 0.58, P = 0.01) and between 3-hydroxyisobutyrate and Acidaminococcus fermentans (R = 0.6, P = 0.007).
Discussion
Daily supplementation of L. reuteri ATCC PTA 6475 was reported to substantially reduce bone loss in older women in our recent study9. Here we first selected 20 elderly women with the best or poorest responses to the treatment with L. reuteri ATCC PTA 6475 from the previous PP population. Except for differences in weight and BMI, subjects in the GR and PR groups had no significant differences in the clinical characteristics at baseline, but presented with significant differences in reducing bone loss after one-year probiotic treatment. This indicates that we were successful in sampling a population with similar baseline characteristics, which to some extent eliminates confounding factors for studying the effects of supplementation with L. reuteri ATCC PTA 6475 on bone metabolism and gut microbiota. Intriguingly, in the GR group supplementation with L. reuteri ATCC PTA 6475 decreased levels of the serum inflammation marker ultrasensitive CRP that has been reported to be associated with reduced BMD23,24. In line with this, a previous study suggested that supplementation with L. reuteri ATCC PTA 6475 decreased intestinal inflammation and increased bone density in healthy male mice25.
In this study, significant differences in the microbial composition at high taxonomic levels were not observed between the groups or time points, except for genus Lactobacillus. However, at species levels, there are statistical differences. At baseline, several species (Streptococcus australis, Lactobacillus antri, Lachnospiraceae bacterium 4_1_37FAA, C. catus) and KOs were identified to be differential between the GR and PR groups. The difference in BMI between the GR and PR groups might associate with the discrepancies in the endogenous baseline microbiota, which could be important for the different probiotic effects in good responders versus poor responders. One previous report showed that the baseline microbiota differed for participants who responded with increased insulin sensitivity index after L. reuteri supplementation, and was characterized by higher microbial diversity26.
Multiple studies have suggested that the gut microbiota could be affected by probiotic supplementation10,11, which would regulate bone mass via effects on the immune system3,5,27. Various probiotic strains including Lactobacillus rhamnosus and Bifidobacterium longum have been shown to influence bone formation through the production of SCFAs6,11,28. Here we observed that one-year supplementation with L. reuteri ATCC PTA 6475 resulted in an altered gut microbiome, including a significant increase of gene richness in the GR group. Several SCFAs-producing bacterial species, including C. acetobutylicum, A. fermentans, A. muciniphila, C. catus and R. bicirculans show more abundant in the GR group than the PR group at 12 months. C. acetobutylicum, R. bicirculans and C. catus have been reported to be able to produce SCFAs by fermenting dietary carbohydrates29–32. Moreover, A. fermentans was suggested to be able to ferment glutamate to butyrate33. Previous studies have showed that A. muciniphila has the ability to degrade mucins in the intestine mainly into SCFAs, which have promising probiotic activities34–36. Thus, our results show that probiotic treatment enhances the abundances of these SCFAs-producing bacterial species in the GR group, which might contribute to the reduction of bone loss.
In addition, detrimental changes in the microbial species and function in the poor responders were alleviated in the good responders. E. coli is more abundant in the PR group at 12 months in comparison to the GR group. There is growing evidence that colonization with E. coli could induce expression of pro-inflammatory cytokines and subsequently cause intestinal inflammation and barrier disruption37–39. Moreover, previous studies have implicated that the intestinal dysbiosis potentially contribute to osteoporosis in the elderly via the immune system40,41. Consistently, by functional analysis we found that the capacity of E. coli biofilm formation was increased in the PR group at 12 months. It suggests that E. coli could be more adherent to the gut mucosa in the PR group, which may be more favorable to elicit inflammation42. Therefore, decreased abundance of E. coli and increased abundance of the SCFAs-producing bacterial species in the GR group after treatment with L. reuteri ATCC PTA 6475 indicates that this results in establishment of a more beneficial intestinal ecosystem and barrier function, which could reduce inflammation and improve bone traits. This is in agreement with an early study proposing that probiotic L. reuteri ATCC PTA 6475 could prevent bone loss through reducing intestinal dysbiosis and barrier disruption43.
As previously described, an appropriate sample size enables a microbiome research to discern the differences between groups44. It should be noted that the number of elderly women recruited in this study was small with 20 subjects (ten good responders and ten poor responders), which could have influenced our results. Therefore, future investigations with a larger sample size would further increase the statistical power and improve the generalizability of our results. Moreover, our ability to identify differential metabolites and species could have been affected by multiple comparisons. Another potential limitation is the absence of subjects from the placebo group. Current study only explored the potential differences between the good responders and poor responders. However, the effects of L. reuteri supplementation on the gut microbiota in elder women is still unclear due to the absence of comparison between probiotics group and the placebo group. Therefore, the introduction of subjects of placebo in the comparison will advance our understanding of the effects of probiotics on the gut microbiota related to bone loss.
In conclusion, we observed a significantly increased gene richness, enrichment of SCFAs producing species as well as depletion of E. coli and its biofilm formation in the GR group, by investigating alterations of the gut microbiome during 1 year L. reuteri ATCC PTA 6475 supplementation. This indicates that L. reuteri ATCC PTA 6475 supplementation might promote bone formation by modulating the gut microbiota composition and function, which could be crucial for the development of novel osteoporosis treatments. However, further studies are needed to validate the mechanisms of how the altered gut microbiota is linked to bone metabolism.
Methods
Study population
From the per protocol (PP) population in the placebo-controlled cohort where 68 elderly women had been randomized to supplementation with the probiotic strain L. reuteri ATCC PTA 6475 or placebo9, 20 elderly women with bone loss who supplemented with probiotic L. reuteri ATCC PTA 6475 were selected for the following secondary analysis. Out of these women, ten women had the highest changes (good responders, GR) and the lowest changes (poor responders, PR) in tibia total volumetric BMD after one-year supplementation with L. reuteri ATCC PTA 6475, respectively. These 20 women with different responses to the treatment with L. reuteri ATCC PTA 6475 completed the study and had not used medication in violation of the study protocol. The study was approved by the Regional Ethics Committee in Gothenburg, D-nr 075-15 (26 February 2015) and addendum T229-16 (10 March 2016). The study was registered at Clinicaltrials.gov (number NCT02422082) prior to study start. All participants provided written informed consent to take part in the study.
Serum sample preparation and measurements of serum markers
Serum samples were isolated from whole blood taken from the cubital vein of the selected 20 elderly women at baseline and 12 months as described in our recent study16. These samples were frozen immediately after collection and stored in −80 °C until used in the further measurement assays. A 500 ul aliquot of serum was used for analysis of serum markers. N-telopeptide of type I collagen (NTx; Osteomark®; Alere Scarborough, Inc., Scarborough, ME, USA; intra- and inter-assay variability 4.6 and 6.9%, respectively), bone-specific alkaline phosphatase (BAP; MicrovueTM; Quidel Corporation, Athens, OH, USA; intra- and inter-assay variability 5.0 and 5.9%, respectively), ultrasensitive C-reactive protein (usCRP; BioMerica, Inc., Irvine, CA, USA; intra and inter-assay variability 4.4 and 3.3%, respectively) and tumour necrosis factor-α (TNF-α; Quantikine®; R&D Systems, Inc., Minneapolis, MN, USA; intra- and inter-assay variability 2.0 and 6.7%, respectively) were measured in duplicate by TECO medical AG (Sissach, Switzerland).
Measurements of bone traits and body composition
The lower leg (tibia) of the non-dominant side (based on dominant arm) was measured to determine bone microstructure and geometry, using High Resolution peripheral Computed Tomography (HR-pQCT; Xtreme CT, Scanco Medical AG, Brüttisellen, Switzerland). All measurement procedures were performed according to instructions and protocols from the manufacturer as previously described45,46. A 3D construction of the bone was assembled using 110 slices, obtained from each measurement. The nominal isotropic resolution was 82 μm. After image processing47, the following variables were obtained: total volumetric BMD (total vBMD, mg/cm3), trabecular bone volume fraction (BV/TV, %), cortical thickness (Ct.Th, mm), and cortical volumetric bone mineral density (Ct.vBMD, mg/cm3). Image quality was assessed based on qualitative observations of movement artifacts, including blurring or streaks in reconstructed 2D slices, and was graded from one to five, with a perfect quality was defined as one and a suboptimal quality was five. Only images with a quality level between one and three were used for analysis. Areal BMD (aBMD, g/cm2) was measured at the lumbar spine (L1-L4), total hip and femoral neck and body composition of the total body with a Hologic Discovery A dual x-ray absorptiometer (DXA; Hologic, Waltham, MA, USA).
Fecal sample preparation and metagenomic profiling
We collected 19 fecal samples at baseline and 12 months respectively, from the 20 elderly women in the GR and PR groups. In each group, nine individuals have time points-paired fecal samples. Fecal samples were frozen immediately after collection and stored in −80 °C until analysis. Total fecal genomic DNA was extracted from 100 mg feces using a modification of the IHMS DNA extraction protocol Q48. Briefly, fecal samples were extracted in Lysing Matrix E tubes (MP Biomedicals) containing ASL buffer (QIAGEN), and lysis of cells was obtained, after homogenization by vortexing for 2 min, by two cycles of heating at 90 °C for 10 min each followed by three bursts of bead beating at 5.5 m/s for 60 s in a FastPrep®-24 Instrument (MP Biomedicals). After each bead-beating burst, samples were placed on ice for 5 min. The supernatants containing fecal DNA were collected after the two cycles by centrifugation at 4 °C. Supernatants from the two centrifugations steps were pooled and a 600 µL aliquot from each sample was purified using the QIAamp DNA Mini kit (QIAGEN) in the QIAcube (QIAGEN) instrument using the procedure for human DNA analysis. Samples were eluted in 200 µL of AE buffer (10 mmol/L Tris·Cl; 0.5 mmol/L EDTA; pH 9.0). Libraries for shotgun metagenomic sequencing were prepared by a PCR-free method; library preparation and sequencing were performed at Novogene (Nanjing, China) on a NovaSeq instrument (Illumina Inc.) with 150 bp paired-end reads. The average read count per sample are 24.7 ± 1.4 million.
Tool Kneaddata (https://github.com/biobakery/kneaddata) was first used to perform quality control on the metagenomic sequencing data with parameters –trimmomatic Trimmomatic-0.36 --trimmomatic-options “SLIDINGWINDOW:4:20 MINLEN:50” -t 9 --bowtie2-options “--very-sensitive --dovetail”. The sequencing runs had high quality with almost 96% of the reads passing the quality cut-off. Out of the high-quality reads, on average 0.03% were aligned to the human genome. Then, tools MetaPhlAn2 that relies on unique clade-specific marker genes14, and HUMAnN215 were used to profile the composition and function of the gut microbiota from the quality filtered reads. MetaPhlAn2 and HUMAnN2 were run with default parameters and following databases: mpa_v293_CHOCOPhlAn used for microbial taxon profiling; UniRef90 gene clusters for quantifying genes/proteins; MetaCyc in conjunction with the UniRef90 gene clusters for quantifying pathway abundances. Relative abundances of species at different taxonomy levels and MetaCyc pathways were used for downstream analyses.
In addition, pipeline MEDUSA that was developed to process raw shotgun metagenomics sequence data13, was performed to profile the microbial composition and genes/KEGG Orthology (KOs). Out of the quality filtered no-human reads, average 79% were mapped to the MEDUSA’s software gene catalogue that contains >11 million genes, and average 45% were mapped to genome catalogue that contains 1747 species genomes, using tool Bowtie249. Read counts at different taxonomy levels were normalized by scaling with cumulative sum (i.e. relative abundance). To determine the richness of the microbiota using the gene catalogue, two aligned reads were required to indicate a gene present in a sample as used in the previous study13, and all samples were rarefied to the same number of reads, 20 million, in order to eliminate the effect of different sequencing depth.
Metabolomic profiling
Approximately 500 ul aliquot of serum was used for the metabolome assay. Metabolomics were performed using liquid chromatography-tandem mass spectroscopy (LC-MS) at Metabolon, Inc (Morisville, NC, USA) with their standard and platform method50. The metabolomics dataset comprised a total of 1,232 metabolites with 958 compounds of known identity (named metabolites) and 274 compounds of unknown structural identity (unnamed metabolites).Peak intensities of all metabolites in original scale (provided by Metabolon, Inc) were analyzed. First, we used an imputation method drawing random values from the normal distribution to simulate signals from low abundant metabolites that were assumed to give rise to missing values51. The width of the normal distribution was set as 0.5 times of the standard deviation (SD) of all measured values and the center was down-shifted by 2.5 times of the SD. Second, metabolites with <25% missing values across all samples were selected and analyzed further, which enabled identification of important metabolites with high confidence. Finally, peak intensities of the filtered metabolites within sample i (i = 1, 2, 3…n, n is the total number of samples) were normalized to the total intensity. This normalization method considered the differences of sample volume loaded onto LC-MS and assumes that all samples have equal total intensity52. The normalization process included the following steps: (a) The total intensity of sample i was summed up; (b) Correction factor of sample i was calculated by dividing the total intensity with the lowest total intensity of all samples; (c) Peak areas of all metabolites within sample i were divided by its individual correction factor. After data normalization, we obtained abundances of 993 metabolites with <25% missing values for downstream analyses. The normalized metabolite abundance was log transformed in this study.
Statistical analysis
To visualize and evaluate differences in gut microbiota composition between the two groups at two time points, principal coordinates analysis (PCoA) and PERMANOVA (R function adonis) were performed based on species-level Bray–Curtis distances. The α diversity of the gut microbiota was calculated based on species-levels of each sample using Shannon, Simpson and Invsimpson indices via R package vegan53. To identify the differential KOs and species, multivariate negative binomial generalized linear models were performed by R package DESeq254, using raw read counts from the MEDUSA pipeline. Only genes or species with the sum of counts across all samples ≥10 and existed in at least five samples were considered in the analysis. Raw read counts of KOs and species were normalized using the median of ratios method by DESeq2. To further explore differences in KEGG functions, gene set analysis (GSA) was performed using Piano with the reporter algorithm for KEGG pathways based on fold changes and P-values of KOs55. The differentially enriched KEGG pathways were identified with a distinct directional P-value < 0.05.
To identify differential signatures including metabolites, microbial taxon at different levels and MetaCyc pathways, we used the two-tailed Wilcoxon rank-sum test to compare the GR and PR groups at each time point, and used Wilcoxon signed-rank test to compare the time points-paired samples in each group. The Spearman’s rank correlation analysis was performed to assess the associations between metabolites and species. Principal components analysis (PCA) was performed to visualize differences in serum metabolomic profiles between the GR and PR groups or between the two time points. Adjusted P-values were obtained by using the false discovery rate method (Benjamini–Hochberg) to control for multiple comparisons error.
Supplementary information
Acknowledgements
We thank Lisa Olsson, Valentina Tremaroli and Fredrik Bäckhed for helpful discussions, and Robert Jakubowicz for fecal sample preparation and metagenomic profiling. This work was financially supported by the Novo Nordisk Foundation, Knut and Alice Wallenberg Foundation, Swedish Research Council, BioGaia AB, ALF/LUA grants from the Sahlgrenska University Hospital, and from the IngaBritt and Arne Lundberg Foundation.
Author contributions
J.N. and M.L. conceived and designed the study. P.L. and B.J. performed data analyses and contributed equally as first author in this work. H.L. helped with gut microbiota analysis and graph visualization. D.S. and M.L. collected samples. P.L. wrote the first draft. P.L., B.J., M.L. and J.N. revised the manuscript. All authors critically reviewed and approved the manuscript.
Funding
Open access funding provided by Chalmers University of Technology.
Data availability
The dataset supporting the conclusions of this manuscript is available in the European Nucleotide Archive (ENA) repository under the accession number PRJEB52923.
Code availability
Code and scripts used in the analyses are available upon request.
Competing interests
The authors declare no competing interests. Prof Lorentzon has received lecture or consulting fees from Astellas, Gedeon Richter, Amgen, Lilly, UCB Pharma, Radius Health, Meda, GE-Lunar and Santax Medico/Hologic, all outside the herein presented work. BioGaia AB provided funding for the randomized controlled trial9, which the current report is partially based on. BioGaia AB did not have access to any metabolomics data, and has not participated in the analysis, interpretation or presentation of results of the present study.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Peishun Li, Boyang Ji.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-022-00348-2.
References
- 1.Khosla S, Hofbauer LC. Osteoporosis treatment: recent developments and ongoing challenges. Lancet Diabetes Endocrinol. 2017;5:898–907. doi: 10.1016/S2213-8587(17)30188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorentzon M. Treating osteoporosis to prevent fractures: current concepts and future developments. J. Intern. Med. 2019;285:381–394. doi: 10.1111/joim.12873. [DOI] [PubMed] [Google Scholar]
- 3.Ohlsson C, Sjogren K. Effects of the gut microbiota on bone mass. Trends Endocrinol. Metab. 2015;26:69–74. doi: 10.1016/j.tem.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Medina-Gomez C. Bone and the gut microbiome: a new dimension. J. Lab Precis Med. 2018;3:96. doi: 10.21037/jlpm.2018.11.03. [DOI] [Google Scholar]
- 5.Zhang J, Lu Y, Wang Y, Ren X, Han J. The impact of the intestinal microbiome on bone health. Intractable Rare Dis. Res. 2018;7:148–155. doi: 10.5582/irdr.2018.01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaiss MM, Jones RM, Schett G, Pacifici R. The gut-bone axis: how bacterial metabolites bridge the distance. J. Clin. Invest. 2019;129:3018–3028. doi: 10.1172/JCI128521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britton RA, et al. Probiotic L. reuteri treatment prevents bone loss in a menopausal ovariectomized mouse model. J. Cell Physiol. 2014;229:1822–1830. doi: 10.1002/jcp.24636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins FL, et al. Lactobacillus reuteri 6475 increases bone density in intact females only under an inflammatory setting. PLoS One. 2016;11:e0153180. doi: 10.1371/journal.pone.0153180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson AG, Sundh D, Backhed F, Lorentzon M. Lactobacillus reuteri reduces bone loss in older women with low bone mineral density: a randomized, placebo-controlled, double-blind, clinical trial. J. Intern Med. 2018;284:307–317. doi: 10.1111/joim.12805. [DOI] [PubMed] [Google Scholar]
- 10.Berni Canani R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J. 2016;10:742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tyagi AM, et al. The microbial metabolite butyrate stimulates bone formation via T regulatory cell-mediated regulation of WNT10B expression. Immunity. 2018;49:1116–1131.e1117. doi: 10.1016/j.immuni.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCabe L, Britton RA, Parameswaran N. Prebiotic and probiotic regulation of bone health: role of the intestine and its microbiome. Curr. Osteoporos. Rep. 2015;13:363–371. doi: 10.1007/s11914-015-0292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson FH, Nookaew I, Nielsen J. Metagenomic data utilization and analysis (MEDUSA) and construction of a global gut microbial gene catalogue. PLoS Comput Biol. 2014;10:e1003706. doi: 10.1371/journal.pcbi.1003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franzosa EA, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, et al. Metabolic alterations in older women with low bone mineral density supplemented with lactobacillus reuteri. JBMR Plus. 2021;5:e10478. doi: 10.1002/jbm4.10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding KH, et al. Amino acids as signaling molecules modulating bone turnover. Bone. 2018;115:15–24. doi: 10.1016/j.bone.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, et al. S-sulfocysteine/NMDA receptor-dependent signaling underlies neurodegeneration in molybdenum cofactor deficiency. J. Clin. Invest. 2017;127:4365–4378. doi: 10.1172/JCI89885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenu C, Serre CM, Raynal C, Burt-Pichat B, Delmas PD. Glutamate receptors are expressed by bone cells and are involved in bone resorption. Bone. 1998;22:295–299. doi: 10.1016/S8756-3282(97)00295-0. [DOI] [PubMed] [Google Scholar]
- 20.Srinivas SR, Prasad PD, Umapathy NS, Ganapathy V, Shekhawat PS. Transport of butyryl-L-carnitine, a potential prodrug, via the carnitine transporter OCTN2 and the amino acid transporter ATB(0,+) Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G1046–G1053. doi: 10.1152/ajpgi.00233.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucas S, et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat. Commun. 2018;9:55. doi: 10.1038/s41467-017-02490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan J, et al. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc. Natl Acad. Sci. USA. 2016;113:E7554–E7563. doi: 10.1073/pnas.1607235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh JM, et al. Higher circulating hsCRP levels are associated with lower bone mineral density in healthy pre- and postmenopausal women: evidence for a link between systemic inflammation and osteoporosis. Osteoporos. Int. 2005;16:1263–1271. doi: 10.1007/s00198-005-1840-5. [DOI] [PubMed] [Google Scholar]
- 24.de Pablo P, Cooper MS, Buckley CD. Association between bone mineral density and C-reactive protein in a large population-based sample. Arthritis Rheum. 2012;64:2624–2631. doi: 10.1002/art.34474. [DOI] [PubMed] [Google Scholar]
- 25.McCabe LR, Irwin R, Schaefer L, Britton RA. Probiotic use decreases intestinal inflammation and increases bone density in healthy male but not female mice. J. Cell Physiol. 2013;228:1793–1798. doi: 10.1002/jcp.24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mobini R, et al. Metabolic effects of Lactobacillus reuteri DSM 17938 in people with type 2 diabetes: A randomized controlled trial. Diabetes Obes. Metab. 2017;19:579–589. doi: 10.1111/dom.12861. [DOI] [PubMed] [Google Scholar]
- 27.Ibanez L, Rouleau M, Wakkach A, Blin-Wakkach C. Gut microbiome and bone. Jt. Bone Spine. 2019;86:43–47. doi: 10.1016/j.jbspin.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Schepper JD, et al. Probiotics in Gut-Bone Signaling. Adv. Exp. Med. Biol. 2017;1033:225–247. doi: 10.1007/978-3-319-66653-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoo M, Nguyen NP, Soucaille P. Trends in Systems Biology for the Analysis and Engineering of Clostridium acetobutylicum Metabolism. Trends Microbiol. 2020;28:118–140. doi: 10.1016/j.tim.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Wegmann U, et al. Complete genome of a new Firmicutes species belonging to the dominant human colonic microbiota (‘Ruminococcus bicirculans’) reveals two chromosomes and a selective capacity to utilize plant glucans. Environ. Microbiol. 2014;16:2879–2890. doi: 10.1111/1462-2920.12217. [DOI] [PubMed] [Google Scholar]
- 31.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rios-Covian D, et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartel U, Buckel W. Sodium ion-dependent hydrogen production in Acidaminococcus fermentans. Arch. Microbiol. 1996;166:350–356. doi: 10.1007/s002030050394. [DOI] [PubMed] [Google Scholar]
- 34.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. The Mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ. Microbiol. 2008;74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 36.Zhou K. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J. Funct. Foods. 2017;33:194–201. doi: 10.1016/j.jff.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellmeyer A, et al. Enterohemorrhagic Escherichia coli suppresses inflammatory response to cytokines and its own toxin. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G576–G581. doi: 10.1152/ajpgi.00050.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kittana H, et al. Commensal Escherichia coli strains can promote intestinal inflammation via differential interleukin-6 production. Front Immunol. 2018;9:2318. doi: 10.3389/fimmu.2018.02318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlowska B, Sobieszczanska BM. Intestinal epithelial barrier: the target for pathogenic Escherichia coli. Adv. Clin. Exp. Med. 2017;26:1437–1445. doi: 10.17219/acem/64883. [DOI] [PubMed] [Google Scholar]
- 40.Ginaldi L, Di Benedetto MC, De Martinis M. Osteoporosis, inflammation and ageing. Immun. Age. 2005;2:14. doi: 10.1186/1742-4933-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ke K, Arra M, Abu-Amer Y. Mechanisms underlying bone loss associated with gut inflammation. Int. J. Mol. Sci. 2019;20:6323. doi: 10.3390/ijms20246323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Medina M, et al. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC) BMC Microbiol. 2009;9:202. doi: 10.1186/1471-2180-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schepper JD, et al. Probiotic Lactobacillus reuteri prevents postantibiotic bone loss by reducing intestinal dysbiosis and preventing barrier disruption. J. Bone Miner. Res. 2019;34:681–698. doi: 10.1002/jbmr.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian XB, et al. A guide to human microbiome research: study design, sample collection, and bioinformatics analysis. Chin. Med. J. 2020;133:1844–1855. doi: 10.1097/CM9.0000000000000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.MacNeil JA, Boyd SK. Improved reproducibility of high-resolution peripheral quantitative computed tomography for measurement of bone quality. Med. Eng. Phys. 2008;30:792–799. doi: 10.1016/j.medengphy.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 46.Sundh D, et al. Increased cortical porosity in women with hip fracture. J. Intern. Med. 2017;281:496–506. doi: 10.1111/joim.12587. [DOI] [PubMed] [Google Scholar]
- 47.Laib A, Hauselmann HJ, Ruegsegger P. In vivo high resolution 3D-QCT of the human forearm. Technol. Health Care. 1998;6:329–337. doi: 10.3233/THC-1998-65-606. [DOI] [PubMed] [Google Scholar]
- 48.Costea PI, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans, A. M. et al. High resolution mass spectrometry improves data quantity and quality as compared to unit Mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics10.4172/2153-0769.1000132 (2014).
- 51.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 52.Veselkov KA, et al. Optimized preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information recovery. Anal. Chem. 2011;83:5864–5872. doi: 10.1021/ac201065j. [DOI] [PubMed] [Google Scholar]
- 53.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset supporting the conclusions of this manuscript is available in the European Nucleotide Archive (ENA) repository under the accession number PRJEB52923.
Code and scripts used in the analyses are available upon request.