Abstract
The Runt-related transcription factor (Runx) family plays various roles in the homeostasis of cartilage. Here, we examined the role of Runx2 and Runx3 for osteoarthritis development in vivo and in vitro. Runx3-knockout mice exhibited accelerated osteoarthritis following surgical induction, accompanied by decreased expression of lubricin and aggrecan. Meanwhile, Runx2 conditional knockout mice showed biphasic phenotypes: heterozygous knockout inhibited osteoarthritis and decreased matrix metallopeptidase 13 (Mmp13) expression, while homozygous knockout of Runx2 accelerated osteoarthritis and reduced type II collagen (Col2a1) expression. Comprehensive transcriptional analyses revealed lubricin and aggrecan as transcriptional target genes of Runx3, and indicated that Runx2 sustained Col2a1 expression through an intron 6 enhancer when Sox9 was decreased. Intra-articular administration of Runx3 adenovirus ameliorated development of surgically induced osteoarthritis. Runx3 protects adult articular cartilage through extracellular matrix protein production under normal conditions, while Runx2 exerts both catabolic and anabolic effects under the inflammatory condition.
Subject terms: Osteoarthritis, Cartilage, Transcriptional regulatory elements, Osteoarthritis
Possible distinct contributions of Runx 2 and Runx3 in osteoarthritis have not been clarified. Nagata et al. show that Runx3 protects adult articular cartilage by extracellular matrix protein production in normal conditions, while Runx2 exerts both catabolic and anabolic effects during inflammation.
Introduction
Osteoarthritis (OA) is the most prevalent form of the joint disease characterized by cartilage degeneration. Progression of OA eventually leads to disability in older adult patients, which imposes a great socioeconomic burden1,2. OA, a multifactorial disease, is caused by aging, inflammation, heavy weight, hard work, and other factors3,4. In the early stage of OA, cartilage degeneration is obvious in the superficial zone (SFZ) of articular cartilage, which has a specific role for joint lubricity by producing lubricin, encoded by the proteoglycan 4 (Prg4) gene5,6. Prg4-knockout (KO) mice display irregular surfaces at 2 months and subsequently exhibit non-inflammatory hyperplastic synovium6. The deeper zone (DZ) of articular cartilage consists of chondrocytes and extracellular matrix7. Chondrocytes maintain the equilibrium of extracellular matrix and cartilage homeostasis by producing cartilage matrix proteins such as type II collagen α1 (Col2a1) and proteoglycans including Aggrecan (Acan)8,9. Transcription of Col2a1 is strongly induced by sex-determining region Y-box 9 (Sox9) through Sox9 consensus sites in introns 1 and 68,10–13. Notably, SOX9 overexpression was shown to alleviate the progression of experimental OA14.
In contrast to such anabolic factors, matrix metalloproteinase 13 (Mmp13) plays essential roles in cartilage degeneration and OA development through the degradation of Col2a115–19. MMP13 expression was low in normal and early degenerative cartilage, but strongly upregulated in late-stage OA specimens20. Increased MMP13 protein was observed in chondrocytes under inflammatory conditions, e.g., stimulation by interleukin-1-beta (IL-1β)14. Previous studies have revealed multiple transcription factors upstream of Mmp13 expression, including nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB)-associated factors21, hypoxia-inducible factor 2-alpha (HIF-2α)20–24, and Runt-related transcription factor 2 (Runx2)17–19.
The Runx family (Runx1/Runx2/Runx3) plays central roles in the development and homeostasis of cartilage and bone via the DNA-recognition consensus motif TG(T/C)GGT25–27. Runx2 was identified as the master regulator of chondrocyte maturation and endochondral ossification28,29. Runx2 directly induces many kinds of chondrocyte hypertrophy-related genes, such as Mmp13, and positively regulates hypertrophic differentiation of chondrocytes and ossification27,30. In Runx2 heterozygous-knockout mice, OA development was significantly reduced, and expression of MMP13 was decreased31. Similar results were observed in tamoxifen-inducible chondrocyte-specific Runx2-knockout mice, in which mRNA levels of Runx2 in cartilage were decreased by about half32. In addition, chondrocyte-specific Runx2 overexpression led to post-traumatic OA progression33. These in vivo data and other experimental findings consistently indicate the catabolic roles of Runx2 in articular cartilage34,35. Previous studies have also implicated Runx2 in skeletal development. Runx2 homozygous-knockout embryos display remarkable impairment of endochondral ossification28,29. Runx2 is regarded as indispensable during the late stages of endochondral ossification, hypertrophic differentiation of chondrocytes, and osteoblast generation35. However, Kimura et al. showed that Runx1 and Runx2 cooperatively induce Sox5 and Sox6, leading to Col2a1 induction during skeletal growth36. Liao et al. reported that chondrocyte-specific knockout of Runx2 during skeletal growth leads to decreased expression of Col2a1 and Mmp13 in temporomandibular joints37. Nevertheless, these anabolic roles of Runx2 in chondrocytes have not been displayed in adult articular cartilage.
In contrast to the complexity of Runx2 effects, previous studies congruously suggest chondrogenic effects of Runx138. Runx1 enhances cartilage matrix production, whereas Runx1 deficiency accelerates OA progression38. Although Runx3 has been shown to promote aggrecan expression39 and is involved in regulation of chondrocyte proliferation and differentiation40, roles of Runx3 in articular cartilage have not been shown by in vivo experiments. Taken together, we hypothesized that Runx2 and Runx3 may have important roles in the maintenance of adult articular cartilage.
In this study, we examined the roles of Runx2 and Runx3 in OA development using conditional-knockout mice. We employed two surgical OA models in which inflammatory responses are deeply involved, as well as an aging model exhibiting more natural disease progression, in Col2a1-CreERT2 and Prg4-CreERT2 mice. We further analyzed mechanisms underlying the regulation of articular chondrocytes by Runx2 and Runx3 using chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq). We found that Runx3 contributes to the maintenance of articular cartilage through the induction of extracellular matrix, while Runx2 exerts both catabolic and anabolic effects in a biphasic manner under the inflammatory condition.
Results
Expression of Runx2 and Runx3 in articular cartilage during the development of surgically induced OA
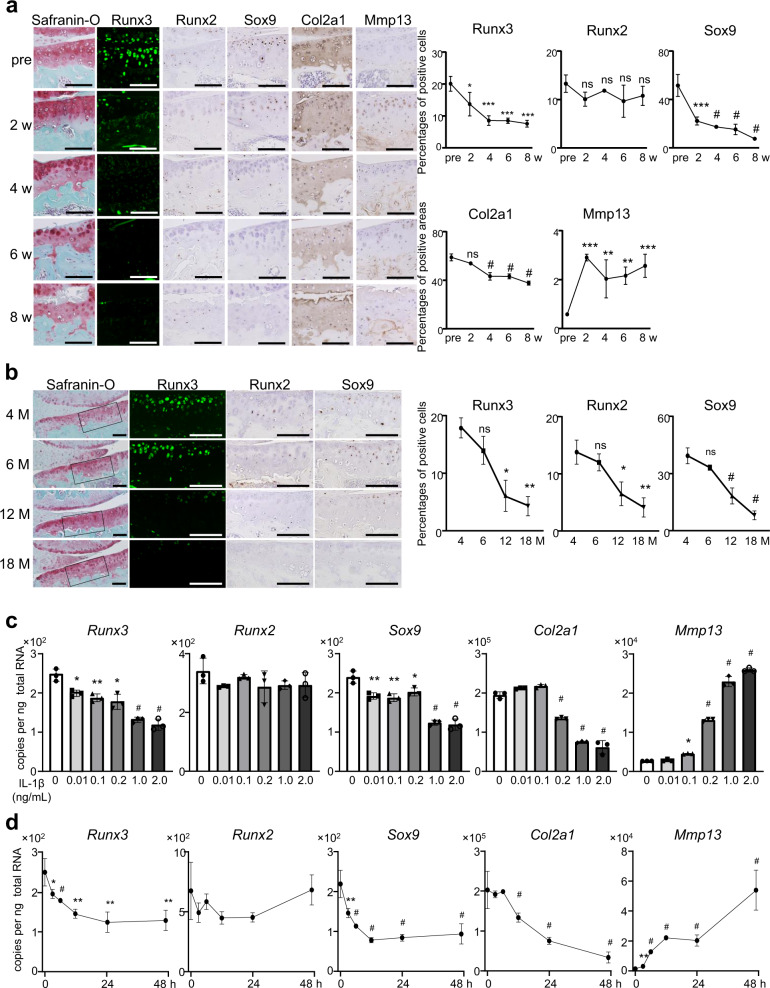
To examine Runx2, Runx3, and Sox9 expression in articular cartilage during OA development, we prepared an 8-week-old mouse OA model by surgical resection of the medial collateral ligament and medial meniscus (medial model)41. Immunohistochemistry of normal cartilage showed that Runx2 protein was predominantly located in the DZ of preoperative articular cartilage, while Runx3 and Sox9 were found in all layers (Fig. 1a). After surgery, Runx3 and Sox9 started to decrease in surgically treated knee joints, accompanied by Col2a1. Runx2 was continuously detected in the DZ. Immunohistochemistry revealed enhanced expression of Mmp13, especially 2 weeks after surgery and predominantly in the DZ of articular cartilage at 4, 6, and 8 weeks after surgery (Fig. 1a). We additionally examined changes in Runx2, Runx3, and Sox9 expression in articular cartilage with aging. In the knee joints of WT mice, their expression decreased over time and was markedly weakened at 12 months of age (Fig. 1b).
Fig. 1. Expression of Runx3 and Runx2, during osteoarthritis (OA) development.
a Immunofluorescence of chondrocyte markers during mouse OA development following resection of the medial meniscus and medial collateral ligament. Scale bars, 100 μm. Percentages of immunofluorescence-positive cells or areas in the Safranin-O-positive area are shown in right panels. b Immunohistochemistry of Runx3, Runx2, and Sox9 in knee joints of 4-, 6-, 12-, and 18-month-old mice. Scale bars, 100 μm. c, d mRNA levels of marker genes in mouse primary chondrocytes exposed to interleukin-1-beta (IL-1β) at concentrations ranging from 0.01 to 2 ng/mL for 24 h (c), or 1 ng/mL IL-1β for 0 − 48 h (d). Data are expressed as mean ± standard deviation (SD) of three biologically independent mice (a, b) or three experiments (c, d). *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001; ns, not significant; ordinary one-way ANOVA with Dunnett’s multiple comparisons test (a, b), one-way ANOVA test with Turkey’s post hoc test (c, d).
We performed in vitro analyses to examine roles of Runx2 and Runx3 under inflammation using IL-1β, which is widely employed to model chondrocyte degradation14,42–44. Primary chondrocytes45 were exposed to IL-1β at different concentrations for 24 h (Fig. 1c); or 1 ng/mL IL-1β for 0−48 h (Fig. 1d). Runx3 was downregulated by IL-1β exposure in dose- (Fig. 1c) and time-dependent manners (Fig. 1d), but levels of Runx2 mRNA were unchanged following IL-1β exposure (Fig. 1c, d). Conversely, exposure to IL-1β elicited dose- and time-dependent upregulation of Mmp13 and suppression of Sox9, followed by a decrease of Col2a1 (Fig. 1c, d). These results indicate that exposure of in vitro chondrocyte to 1 ng/mL IL-1β for 24 h reproduces similar expression patterns of marker genes and proteins observed in vivo surgical OA models, including stable expression of Runx2, reduced Runx3, Sox9, and Col2a1, and increased Mmp13.
Runx3 knockout accelerated OA development through Prg4 and Acan suppression
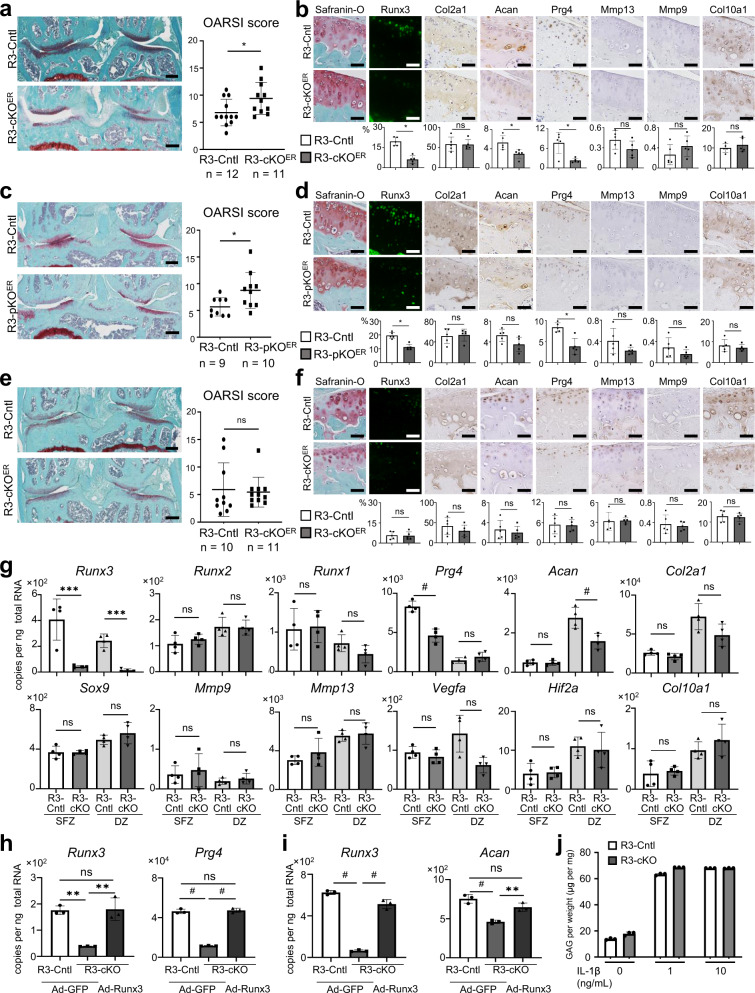
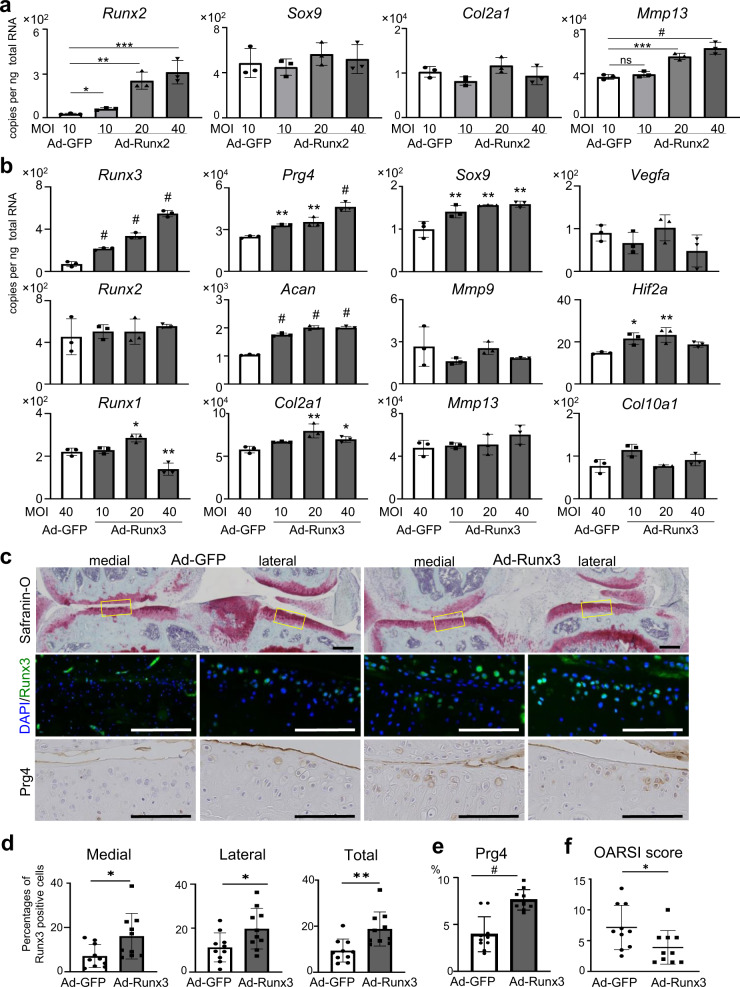
To reveal the roles of Runx3 in whole or SFZ articular cartilage after skeletal growth, we compared Runx3fl/fl littermates46 (R3-Cntl) with Col2a1-CreERT2; Runx3fl/fl (R3-cKOER)47 and Prg4-CreERT2; Runx3fl/fl (R3-pKOER)48 mice. We prepared three distinct OA models (Supplementary Fig. 1a–c). As a result of the medial model41, OA development was significantly accelerated in R3-cKOER knee joints compared with R3-Cntl joints (Fig. 2a). In the articular cartilage of sham-operated R3-cKOER joints, Runx3-positive cells decreased by 70% (Fig. 2b). Positive areas of Acan and Prg4 decreased with Runx3 knockout (Fig. 2b). Similarly, OA development was significantly accelerated in R3-pKOER joints (Fig. 2c). In the articular cartilage of sham-operated R3-pKOER joints, Runx3-positive cells decreased by 40% and the positive area of Prg4 was decreased (Fig. 2d). In addition, 8 weeks after destabilizing the medial meniscus (DMM)49 as a minor injury-induced OA model, OA development was also significantly accelerated in 24-week-old R3-pKOER mice compared with R3-Cntl mice (Supplementary Fig. 2). In the aging model (Supplementary Fig. 1c), OA development was unchanged in R3-cKOER mice compared with R3-Cntl mice (Fig. 2e, f). In the articular cartilage of 18-month-old joints, the rates of Runx3-positive cells were not different for either genotype (Fig. 2f), probably because Runx3 expression decreased with aging, as shown in Fig. 1b. Expression of the marker proteins in both mice was similar as well (Fig. 2f). By Runx3 knockout in the whole articular cartilage, chondrocyte apoptosis was not significantly changed in the sham side, the surgical OA model, or with aging, as well as by Runx3 knockout in the SFZ articular cartilage of the sham side (Supplementary Fig. 3a–d).
Fig. 2. Development of OA in cartilage-specific Runx3-knockout mice.
Development of OA in Runx3fl/fl (R3-Cntl) and Col2a1-CreERT2; Runx3fl/fl (R3-cKOER) mice (a) and in R3-Cntl and Prg4-CreERT2; Runx3fl/fl (R3-pKOER) mice (c) by the medial model, and in R3-Cntl and R3-cKOER mice by the aging model (e). Scale bars, 200 μm (a, c, e). Immunofluorescence of marker proteins in sham-operated knee joints of 16-week-old R3-Cntl and R3-cKOER mice (b) and 16-week-old R3-Cntl and R3-pKOER mice (d), into which tamoxifen was injected at 7 weeks of age. Immunofluorescence of marker proteins in knee joints of 18-month-old R3-Cntl and R3-cKOER mice (f), into which tamoxifen was injected at 2, 6, and 12 months of age. Scale bars, 50 µm (b, d, f). Rates of Runx3-positive cells and the 3,3’-diaminobenzidine (DAB)-positive area in the Safranin-O-positive area are shown in the lower panels b, d, f. g mRNA levels of marker genes in superficial zone (SFZ) and DZ chondrocytes obtained from R3-Cntl and Col2a1-Cre; Runx3fl/fl (R3-cKO) mice. h, i mRNA levels of Runx3 and Prg4 or Acan in the R3-Cntl and R3-cKO SFZ chondrocytes (h) or DZ chondrocytes (i) overexpressing GFP (Ad-GFP) or Runx3 (Ad-Runx3) by adenoviral induction at a multiplicity of infection (MOI) of 20. j Dimethylmethylene blue (DMMB) assays. Amount of proteoglycan released into culture medium from R3-Cntl and R3-cKO mouse femoral heads cultured with or without IL-1β (1, 10 ng per mL) for 3 days. Data are expressed as means ± SD of biologically dependent mice (n = 5 for b, d, f, n = 4 for g, and n = 3 for h–i). *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001, n.s., not significant; ordinary one-way ANOVA test with Turkey’s post hoc test (g, h, i), and two-tailed Mann–Whitney U test (a–f, j).
We then examined expression of OA-associated genes in SFZ and DZ cells from Runx3-knockout and control mice. To enhance knockout efficiency, we prepared Runx3fl/fl (R3-Cntl) and Col2a1-Cre; Runx3fl/fl (R3-cKO) mice, which displayed normal skeletal development (Supplementary Fig. 4a–g). Prg4 was decreased in R3-cKO SFZ cells and Acan was also decreased in R3-cKO DZ cells (Fig. 2g). Other genes were not changed in SFZ cells or DZ cells (Fig. 2g). We then prepared SFZ and DZ cells from R3-pKOER and R3-Cntl littermates, and treated them with water-soluble tamoxifen. Only Prg4 was decreased in Runx3-deficient SFZ cells (Supplementary Fig. 5). We further performed Runx3 overexpression in Runx3-knockout cells using adenoviral transduction. Downregulation of Prg4 in R3-cKO SFZ cells and Acan in R3-cKO DZ cells was recovered to control levels by Runx3 overexpression (Fig. 2h, i). Additionally, we performed dimethylmethylene blue (DMMB) assays50 to investigate the catabolic effect of Runx3. Runx3 knockout did not significantly change the released glycosaminoglycan (GAG) content (Fig. 2j).
Development of surgically induced OA was inhibited by heterozygous Runx2 knockout, but accelerated by homozygous knockout
To investigate Runx2 loss-of-function in articular cartilage after skeletal growth in vivo, we generated a mutant mouse strain expressing a floxed Runx2 allele (Supplementary Fig. 6a). Alizarin red and Alcian blue staining of skeletal preparations was performed in E18.5 embryos. CAG-Cre; Runx2fl/lf mice exhibited a complete absence of mineralized matrix in developing skeletal bones, similar to global knockout mice (Supplementary Fig. 6b)28,29. Hypoplastic clavicles and open fontanelles were observed in CAG-Cre; Runx2fl/+ mice, which indicates a cleidocranial dysplasia phenotype, as reported in general heterozygous-knockout mice (Supplementary Fig. 6b)19.
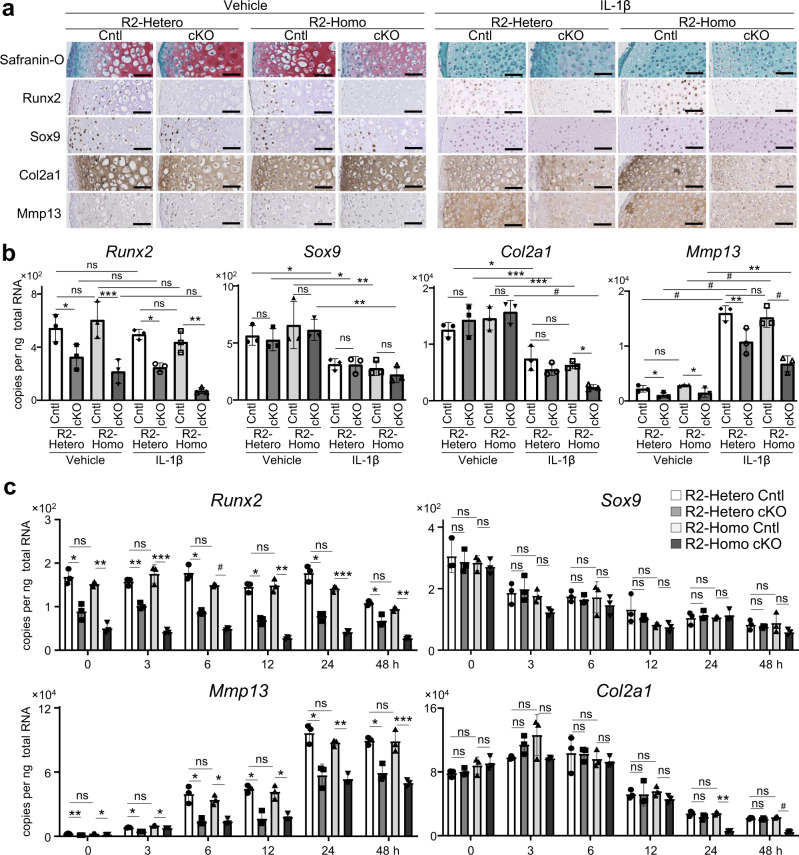
We next mated Runx2fl/fl mice with Col2a1-CreERT2 mice47 and generated Runx2fl/+(R2-Hetero Cntl), Col2a1-CreERT2; Runx2fl/+(R2-Hetero cKO), Runx2fl/fl (R2-Homo Cntl), and Col2a1-CreERT2; Runx2fl/fl (R2-Homo cKO) male littermates. In the medial model41, OA development was significantly inhibited in knee joints of R2-Hetero cKO mice and accelerated in R2-Homo cKO mice compared with their respective controls (Fig. 3a). In the DMM model49, OA development was significantly inhibited in R2-Hetero cKO knee joints, similar to previous reports31,32, but accelerated in R2-Homo cKO mice compared with R2-Cntl joints (Fig. 3a).
Fig. 3. Osteoarthritis development in chondrocyte-specific homozygous and heterozygous Runx2-knockout mice.
a Safranin-O staining and Osteoarthritis Research Society International (OARSI) scores of mouse knee joints of Runx2fl/+ (R2-Hetero Cntl), Col2a1-CreERT2; Runx2fl/+ (R2-Hetero cKO), Runx2fl/fl (R2-Homo Cntl), and Col2a1-CreERT2; Runx2fl/fl (R2-Homo cKO) mice 12 weeks after surgery to destabilize the medial meniscus (DMM) or 8 weeks after resection of the medial meniscus and medial collateral ligament (medial model). Tamoxifen induction was performed at 15 weeks in the DMM group and 7 weeks in the medial model group. Experimental groups consisted of n = 13, 14, 20, and 12 in the DMM model, respectively, and n = 16 per genotype in the medial model. Scale bars, 200 μm. Safranin-O staining and immunohistochemistry of Runx2, Sox9, Col2a1, and Mmp13 in sham (b) and surgical (c) joints of the medial model group. n = 6 biologically independent experiments. Scale bars, 100 μm. Data are expressed as dot plots and mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001; ns, not significant; one-way ANOVA test with Turkey’s post hoc test.
Immunohistochemistry showed that Runx2-positive cells were reduced by approximately 40% in R2-Hetero cKO cartilage and 80% in R2-Homo cKO cartilage of sham and OA joints (Fig. 3b, c), respectively. Protein levels of Sox9 and Col2a1 were unchanged in the sham joints of both genotypes (Fig. 3b), but Col2a1 was significantly suppressed in OA joints of R2-Homo cKO mice (Fig. 3c). Mmp13 was significantly reduced both in sham and OA joints of R2-Hetero and R2-Homo cKO mice (Fig. 3b, c). In the aging model, we further compared OA development between R2-Homo Cntl and R2-Homo cKO mice. At 18 months of age, OA development with aging was unchanged in R2-Homo cKO mice compared with R2-Cntl mice, as well as immunohistochemistry of Runx2, Sox9, Col2a1, or Mmp13 (Supplementary Fig. 7a, b).
To examine chondrocyte apoptosis, we performed TdT-mediated dUTP nick-end labeling (TUNEL) staining of medial and DMM model knee joints. TUNEL staining revealed upregulated chondrocyte apoptosis in the sham joints of R2-Homo KO mice (Supplementary Fig. 8a). Cell numbers in articular cartilage were decreased in R2-Homo cKO mice (Supplementary Fig. 8a). To examine short-term effects of Runx2 knockout, we injected tamoxifen into 7-week-old male littermate mice of the four genotypes daily for 5 days, and sacrificed mice 1 week after injection without any surgery. Chondrocyte apoptosis was enhanced in the articular cartilage of R2-Homo cKO mice with decreased cell numbers (Supplementary Fig. 8b).
Col2a1 expression was suppressed in homozygous Runx2-knockout chondrocytes under inflammation
Surgical induction differently altered OA development in R2-Hetero and Homo cKO mice, but had no effect on aging-model mice, indicating that Runx2 may be involved in cartilage degeneration by severe inflammation rather than aging with mild inflammation. To reveal the roles of Runx2 in chondrocytes under inflammation, we performed ex vivo and in vitro analyses using IL-1β14,40–42. Safranin-O staining of femoral heads cultured for 2 weeks with or without IL-1β displayed consistent decreases in proteoglycans for each genotype (Fig. 4a). Immunohistochemistry confirmed efficient knockdown of Runx2 in R2-Hetero and R2-Homo cKO femoral heads (Fig. 4a). Col2a1 protein levels were unchanged in normal cultures for both genotypes, but decreased following IL-1β exposure, especially in the R2-Homo cKO group (Fig. 4a). qRT-PCR using mRNA from the homogenized femoral heads confirmed that Runx2 was decreased according to genotype, but unchanged by IL-1β exposure (Fig. 4b). Mmp13 increased in all genotypes following IL-1β exposure, and the expression pattern of Mmp13 between genotypes was similar to that of Runx2 (Fig. 4b). Sox9 and Col2a1 were decreased by IL-1β and unchanged between genotypes with a marked decrease in Col2a1 in R2-Homo cKO femoral heads following IL-1β exposure (Fig. 4b).
Fig. 4. Homozygous and heterozygous Runx2-knockout femoral heads and primary chondrocytes with or without interleukin-1-beta (IL-1β).
a Safranin-O staining and immunohistochemistry of Runx2, Sox9, Col2a1, and Mmp13 in femoral heads obtained from Runx2fl/+ (R2-Hetero Cntl), Col2a1-CreERT2; Runx2fl/+ (R2-Hetero cKO), Runx2fl/fl (R2-Homo Cntl), and Col2a1-CreERT2; Runx2fl/fl (R2-Homo cKO) P1 neonates, cultured with or without 1 ng per mL IL-1β for 2 weeks. Scale bars, 100 μm. b mRNA levels of Runx2, Sox9, Col2a1, and Mmp13 in femoral heads. c Time-course of Runx2, Sox9, Col2a1, and Mmp13 mRNA levels in primary chondrocytes obtained from R2-Hetero Cntl, R2-Hetero cKO, R2-Homo Cntl, and R2-Homo cKO mice exposed to 1 ng per mL IL-1β. n = 3 biologically independent experiments. Data are expressed as dot plots and mean ± SD *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001; ns, not significant; one-way ANOVA test with Turkey’s post hoc test.
We next examined temporal changes in mRNA levels of marker genes in chondrocytes from the four genotypes exposed to 1 ng/mL IL-1β from 0 to 8 h. The mRNA level of Runx2 was efficiently decreased in R2-Hetero and R2-Homo cKO chondrocytes, and stable in each genotype following IL-1β exposure (Fig. 4c). Mmp13 increased with time and was suppressed in R2-Hetero and R2-Homo cKO chondrocytes compared with their respective controls at each time point. Sox9 gradually decreased, and there was no difference between genotypes at any point examined. Col2a1 was significantly suppressed in R2-Homo cKO more than 24 h after IL-1β exposure (Fig. 4c).
Genome-wide analysis of Runx2 and Runx3 association profiles in chondrocytes determined by ChIP-seq and RNA-seq
To investigate mechanisms underlying chondrocyte regulation by Runx3 and Runx2, we planned ChIP-seq with an anti-FLAG antibody. For Runx3, we prepared SFZ cells from WT mice transfected with the FLAG-tagged Runx3 expression vector. For the ChIP-seq, 21,730 raw peaks met the peak calling criterion, and ~50% of all peaks mapped to an interval between ±50 and 500 kb from the transcriptional start sites (TSS) (Fig. 5a). Genomic Regions Enrichment of Annotations Tool (GREAT) Gene Ontology (GO) analysis51 indicated that the extracellular structure organization and collagen fibril organization terms were the most significantly enriched in the gene set (Fig. 5a). De novo motif analysis of the top 1000 specific peaks using MEME-ChIP52 identified a previously predicted Runx motif, TG(T/C)GG(T/C) (Fig. 5b). ChIP-seq data around genes encoding Prg4 and Acan showed several peaks for Runx3 binding (Supplementary Fig. 9a–d). Relative luciferase activities of the regions around Prg4 and Acan were increased by Runx3 overexpression (Supplementary Fig. 9b, d). To further investigate the alterations of gene expression profiles by Runx3 knockout, we performed RNA-seq using SFZ and DZ cells from R3-cKO and R3-Cntl mice (Supplementary Fig. 10a–d and Supplementary Tables 1–4). Among 18 genes up- or downregulated by more than twofold in the R3-cKO SFZ cells, nine were the top one-third peak nearest genes as shown in the ChIP-seq, including Prg4 and Mmp9 (Fig. 5c).
Fig. 5. Genome-wide analysis of Runx2 and Runx3 association profiles in chondrocytes.
a Genome-wide distribution of Runx3-associated regions relative to transcriptional start sites (TSS) with Genomic Regions Enrichment of Annotations Tool (GREAT) gene ontology (GO) and MGI expression annotations of Runx3 peaks showing the top five enriched terms. b Enriched motifs determined by Runx3-FLAG ChIP-seq. c Schematic diagram to identify candidate genes by ChIP-seq and RNA-seq using primary SFZ cells obtained from Cntl and cKO mice. d Genome-wide distribution of Runx2-associated regions relative to TSSs, and top five enriched terms identified by GREAT GO of Runx2 peaks. Left upper panel, primary chondrocytes treated with vehicle; left lower panel, inflamed chondrocytes exposed to 1 ng per mL IL-1β; right panels, overlap of Runx2-FLAG ChIP-seq peaks between primary and inflamed chondrocytes. e Enriched motifs determined by Runx2-FLAG ChIP-seq. f Enrichment of TG(T/C)GGT and poly-A motifs recovered de novo from Runx2 and Sox9 peak regions. The upper panels show motif logos, displaying nucleotide frequencies (scaled relative to the information content) at each position. Lower panels show enrichment levels of TG(T/C)GGT and poly-A motifs mapped to Runx2 and Sox9 peaks, whereby x and y axes represent the distance from mapped motifs to the peak center and frequency of mapped motifs, respectively. g Signal intensity plots of ChIP-seq data for Sox9, H3K4me2, H3K27ac, RNA polymerase II (polII), and rabbit immunoglobulin G (IgG) controls in ±1 kb from Runx2-FLAG peaks. Normalized mean signal intensity for the following ChIP-seq reads are shown in the top panels. TES, transcriptional end site. h Schematic of identification of anti-apoptosis-associated candidates among downregulated genes by homozygous Runx2 knockout in primary chondrocytes. i Schematic of identification of candidate transcriptional target genes of Runx2 in inflamed chondrocytes.
For Runx2, we performed ChIP-seq in accordance with previous reports12,53, using chondrocytes treated with vehicle control (primary chondrocytes) or exposed to 1 ng/mL IL-1β (inflamed chondrocytes) derived from Runx2-FLAG mice (Hojo H. et al.54). For the ChIP-seq, 37,163 raw peaks in primary chondrocytes and 14,571 raw peaks in inflamed chondrocytes met the peak calling criteria (Fig. 5d). Peak distributions between primary chondrocytes and inflamed chondrocytes were similar. In both groups, a striking enrichment was observed around the TSS; ~24% of all peaks from two ChIP-seq data are within ±500 bp of the TSS, even though this region represents only 0.001% of the genome51 (Fig. 5d). GREAT GO analysis51 identified “collagen fibril organization” as the fifth and second most significantly enriched term in the gene sets for primary chondrocytes and inflamed chondrocytes, respectively (Fig. 5d). We hypothesized that there were functional differences between the TSS-associated dataset and dataset excluding data ±500 bp from the TSS, similar to Sox912. GREAT GO analyses showed cell cycle-associated terms in TSS-associated Runx2 datasets of both primary and inflamed chondrocytes and terms for a Runx2-regulated skeletal program in the dataset excluding data ±500 bp from TSS (Supplementary Fig. 11a, b). A Venn diagram of GREAT GO analyses indicated that Runx2 was associated with collagen fibril organization in chondrocytes with or without inflammation (Fig. 5d).
We next performed de novo motif analysis of the top 1000 specific peaks using MEME-ChIP52. As expected, primary motifs were similar to the previously predicted Runx motif, TG(T/C)GGT, even in the dataset excluding data ±500 bp from the TSS (Fig. 5e). Interestingly, we observed that poly-A sequence, reported as a SOX9 consensus sequences55, was enriched in the TSS-associated Runx2 dataset. To identify the functional relationship between Sox9 and Runx2, we further analyzed our present dataset combined with previous Sox9 and related ChIP-seq datasets12. The TG(T/C)GGT motif was highly enriched at the predicted center of Runx2 ChIP-seq peaks in primary and inflamed chondrocytes. Interestingly, the TG(T/C)GGT motif was slightly enriched at the predicted center of Sox9 ChIP-seq peaks (Fig. 5f). A poly-A sequence was also recovered, but there was no centering within the Runx2 and Sox9 peaks (Fig. 5f). Thus, the poly-A motif is unlikely to be the preferred primary site for Runx2 or Sox9 engagement in chondrocytes.
To test the integration of multiple regulatory inputs through Runx2- and Sox9-directed enhancer modules, we analyzed peak intensity associations. There were strong associations between Sox9-12 and Runx2-peak regions, which were notably enhanced in inflamed chondrocytes (Fig. 5g). Clear associations of Runx2-peak regions with enhancer signatures were evident, specifically: (1) bi-modal patterns of H3K4 dimethylation (H3K4me2) peaks at Runx2-peak center regions, which indicates both promoters and putative enhancers;56 (2) peaks of H3K27 acetylation (H3K27Ac) flanking Runx2 peaks, which indicate open chromatin57,58, were more evident in inflamed chondrocytes compared with primary chondrocytes; and (3) RNA polymerase II peaks associated with Runx2-peak regions, consistent with active enhancers59 (Fig. 5d). Together, these results indicate that Runx2 enhances transcription of collagen fibrils by binding TG(T/C)GGT target sequences near Sox9-binding site, both in primary chondrocytes and inflamed chondrocytes.
To further investigate the general functions of Runx2 in chondrocytes, we performed RNA-seq analyses of R2-Homo Cntl and cKO chondrocytes with or without IL-1β exposure (Supplementary Fig. 12a–f and Supplementary Tables 5 and 6). The principal component analysis highlighted the effect of Runx2 knockout under inflammation induced by IL-1β. As chondrocyte apoptosis was enhanced in R2-Homo cKO mice (Supplementary Fig. 8b, c), we examined apoptosis-related genes. Among the top 200 genes downregulated by Runx2 knockout in primary chondrocytes, eleven genes (including Ihh, Igf1, and Wnt11) were associated with “negative regulation of apoptosis” (Fig. 5h). Among the top 200 genes downregulated by Runx2 knockout in inflamed chondrocytes, ten (including Col2a1) matched with the top 500 genes nearest to Runx2 peaks detected by ChIP-seq (Fig. 5i).
Transcriptional regulation of Col2a1 and Mmp13 by Runx2 and Sox9
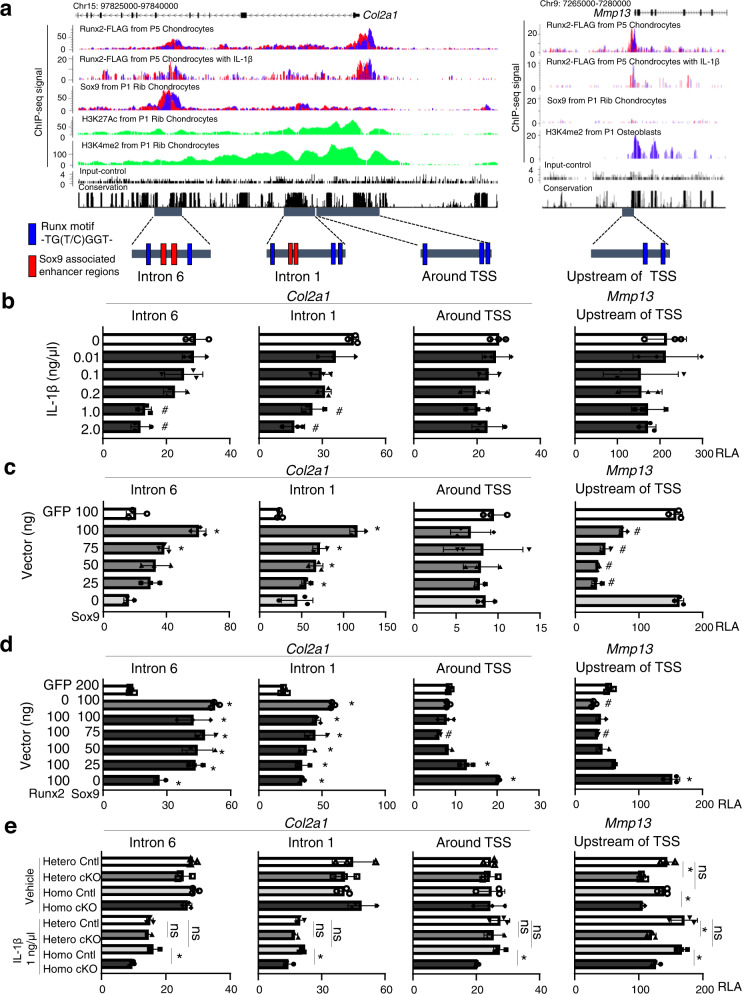
We mapped Runx2-FLAG, Sox912, and active histone marks (H3K4me2 or H3K27Ac in chondrocytes12 or osteoblasts53 from 1-day-old mice) in ChIP-seq data using CisGenome browser. Sox9 and Runx2 sequence data showed similar patterns in introns 1 and 6 of Col2a1; specifically, insignificant peaks at intron 1 and major significant peaks at intron 6 (Fig. 6a). These peaks are identical to the functional enhancers of Col2a1 containing Sox9 motifs8. Magnified views indicated that the peak centers of Runx2 and Sox9, containing their consensus motifs, were 300−400 bp apart (Supplementary Fig. 13). Based on the CisGenome browser, we prepared luciferase reporter vectors containing the Runx2 motifs around Col2a1 and Mmp13 genes shown in Fig. 6a. Relative luciferase activity of Col2a1 intron 1 and 6 fragments decreased in chondrocytes exposed to ≥1 ng/mL IL-1β (Fig. 6b), which exhibited decreased Sox9 mRNA while that of Runx2 remained stable (Fig. 1c, d). However, luciferase activities of the region around the Col2a1 TSS and upstream of the Mmp13 TSS were unchanged compared with controls (Fig. 6b). We next examined the effect of Sox9 on the activity of each reporter. Sox9 overexpression increased the activities of reporters containing the enhancers in Col2a1 introns 1 and 6 in a dose-dependent manner (Fig. 6c), but did not affect the activity of the region around the Col2a1 TSS and decreased activity of the region upstream of the Mmp13 TSS (Fig. 6c). In addition, we co-transfected Runx2 and Sox9 (Fig. 6d). Notably, when the amount of Sox9 was decreased, luciferase activities of the Col2a1 intron 6 enhancer were increased by Runx2 co-transfection (Fig. 6d). In contrast, when Sox9 was overexpressed, luciferase activities of the Col2a1 intron 1 enhancer were not affected by Runx2 transfection (Fig. 6d). Activity of the region around the Col2a1 TSS was increased by Runx2 transfection without Sox9 transfection (Fig. 6d).
Fig. 6. Transcriptional regulation of Col2a1 and Mmp13 by Runx2 and Sox9.
a CisGenome browser screenshots showing Runx2 and reference engagement around Col2a1 (Chr15: 97,825,000 − 97,840,000) and Mmp13 (Chr9: 7,265,000 − 7,280,000). Lower panels show genomic regions cloned into the pGL4.10[luc] vector. b Luciferase activities of enhancer regions in mouse chondrocytes exposed to various concentrations of IL-1β. c, d Luciferase activities of enhancer regions in mouse chondrocytes transfected with various ratios of Runx2 and Sox9, or GFP control. e Luciferase activities of enhancer regions in chondrocytes from Runx2fl/+ (R2-Hetero Cntl), Col2a1-CreERT2; Runx2fl/+ (R2-Hetero cKO), Runx2fl/fl (R2-Homo Cntl), and Col2a1-CreERT2; Runx2fl/fl (R2-Homo cKO) mice with or without IL-1β exposure. n = 3 (b–e) biologically independent experiments. Data shown mean ± SD. ns not significant. #P < 0.05 vs control (white bars) (b), *P < 0.05 (c–e); one-way ANOVA test with Turkey’s post hoc test. RLA relative luciferase activity.
We further performed luciferase assays using chondrocytes from the four genotypes with or without IL-1β exposure. Relative luciferase activities of Col2a1 intron 1 and 6 enhancers were decreased in inflamed chondrocytes compared with primary chondrocytes for each genotype (Fig. 6e), similar to the results shown in Fig. 6b. In addition, luciferase activities of Col2a1 intron 1 and 6 enhancers, and the region around the Col2a1 TSS, were suppressed in inflamed chondrocytes of R2-Homo cKO mice compared with other genotypes (Fig. 6e). Compared with controls, Runx2 heterozygous or homozygous knockout decreased the activities of reporters containing the region upstream of the Mmp13 TSS in both primary and inflamed chondrocytes (Fig. 6d). Taken together, the present results indicate that Runx2 could activate Col2a1 transcription through the enhancer in intron 6, in addition to the enhancer in intron 1 and region around the Col2a1 TSS.
Enhanced expression of Runx3 inhibited surgically induced OA development
Finally, we investigated the effects of adenoviral Runx2 or Runx3 overexpression in vitro and in vivo. Transduced of a Runx2 adenovirus (Ad-Runx2) at different doses to WT chondrocytes increased Runx2 expression in a dose-dependent manner (Fig. 7a). Although mRNA levels of Sox9 and Col2a1 were unchanged, Mmp13 was upregulated in Runx2 overexpressing chondrocytes (Fig. 7a).
Fig. 7. Effects of Runx2 or Runx3 overexpression.
a mRNA levels of Runx2, Sox9, Col2a1, and Mmp13 in WT primary chondrocytes overexpressing GFP (Ad-GFP) or Runx2 (Ad-Runx2) by adenoviral induction. b mRNA levels of Runx family members and marker genes in primary articular chondrocytes transduced with GFP (Ad-GFP) or Runx3 (Ad-Runx3) by adenoviral vectors. c Safranin-O staining and Runx3/Prg4 expression after OA development in WT knee joints after intra-articular administration of Ad-GFP or Ad-Runx3. Representative images among ten mice in each group were shown. All mice underwent medial model surgery at 8 weeks of age, and the intra-articular administration of 10 µL of 1 × 109 plaque-forming unit per mL Ad-GFP or Ad-Runx3 was performed at 1, 4, and 7 weeks after the surgery. Inset boxes in Safranin-O staining indicate regions of immunofluorescence below. Scale bars indicate 200 μm. d Rates of Runx3-positive cells in medial, lateral, and both sides of knee joints. e Percentage of 3,3’-diaminobenzidine (DAB)-positive area in the Safranin-O-positive area. f OARSI scores for evaluation of OA development between the two groups. Data are expressed as dot plots and means ± SD of biologically dependent mice (n = 3 for a, b, n = 10 for c–e). *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001; ordinary ANOVA with Dunnett’s multiple comparisons test for a, b, and two-tailed Mann–Whitney U test for d, e.
When we transduced Runx3 adenovirus at different doses to primary chondrocytes from WT mice, Prg4 was increased in a dose-dependent manner (Fig. 7b). Acan and Sox9 were also increased by Runx3 overexpression (Fig. 7b). Catabolic factors were not changed, except for slightly increased Hif2a (Fig. 7b). We next introduced GFP or Runx3 adenoviral vectors into knee joints of WT mice that received surgical induction for the medial model at 8 weeks of age. Runx3 expression was efficiently enhanced by the intra-articular injection of adenovirus, resulting in significant suppression of OA progression and enhanced Prg4 expression (Fig. 7c–f).
Discussion
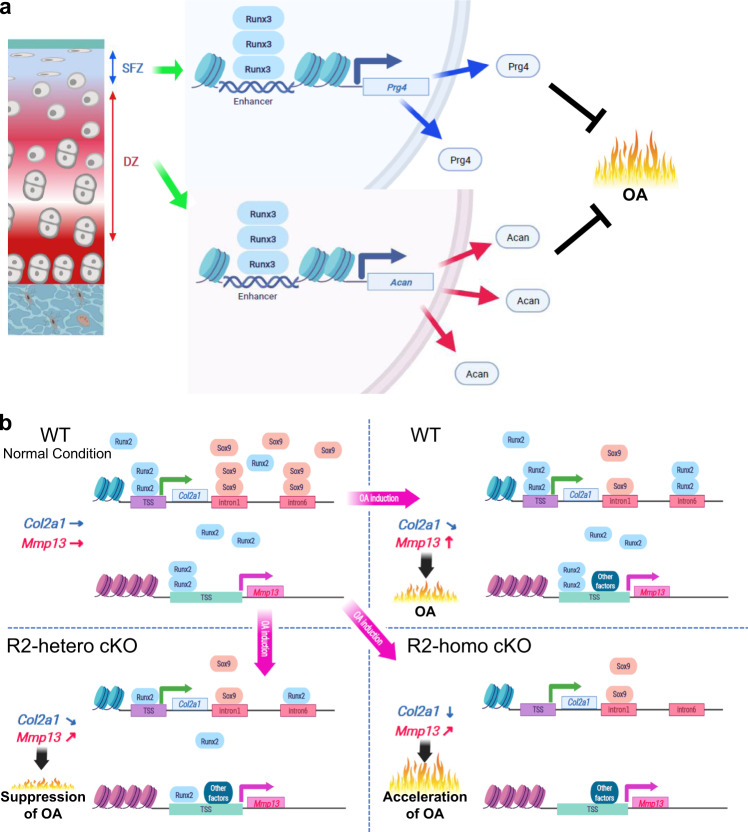
This study revealed that Runx3 protects articular cartilage degradation in mouse surgically induced OA models through extracellular matrix protein production, while Runx2 exerts both catabolic and anabolic effects under inflammatory conditions (Fig. 8). In vitro and comprehensive analyses of transcriptional regulation by Runx3 indicate its association with the organization of extracellular matrix, and identified Prg4 in SFZ as a transcriptional target gene, as well as Acan in DZ as a potential target (Figs. 2 and 5). Consistent with previous studies, chondrocyte-specific heterozygous knockout of Runx2 inhibited OA and decreased Mmp13 expression (Fig. 3). Notably, in contrast, homozygous knockout of Runx2 in articular chondrocytes markedly accelerated OA development, which was accompanied by suppressed Col2a1 protein level. Loss-of-function of Runx2 in vitro decreased Col2a1 only in inflamed chondrocytes in which Sox9 was decreased (Fig. 4), while Mmp13 expression was downregulated in both primary and inflamed chondrocytes. In the presence of IL-1β, de novo Runx2 binding to regions around the known Sox9-binding sites was enhanced (Fig. 5). Luciferase assays indicated that Runx2 can compensate for Col2a1 transcription in inflamed chondrocytes (Fig. 6). Overexpression of Runx2 increased Mmp13, but not Col2a1. In contrast, overexpression of Runx3 increased Prg4 and Acan, and intra-articular administration of Runx3 adenovirus suppressed surgically induced OA (Fig. 7).
Fig. 8. Schematic diagrams representing molecular pathways in which Runx3 and Runx2 regulate articular cartilage during OA development.
a Runx3 contributes to cartilage homeostasis through the induction of Prg4 and Acan. b Runx2 exerts the catabolic effect through Mmp13 induction, while Runx2 sustains Col2a1 expression under the inflammatory condition, in which Sox9 was decreased.
In this study, OA development was enhanced in homozygous Runx2-knockout mice. This finding challenges the results of previous studies which directly or indirectly indicated catabolic effects of Runx2 in articular cartilage31,32,60–62. Compensation of Col2a1 expression by Runx2 was observed under inflammatory conditions in surgically induced OA models employing R2-Homo cKO mice, but not R2-Hetero cKO mice. This effect was not obvious in previous studies of general heterozygous-knockout mice31 or Aggrecan-CreERT2; Runx2fl/fl mice in which the efficiency of Runx2 knockdown was approximately 50%32. Considering the efficiency of Runx2 suppression was ~80% in R2-Homo cKO mice, smaller amounts of Runx2 (compared with Mmp13 transcription) may be able to sustain Col2a1 transcription under inflammatory conditions. Although this anabolic effect of Runx2 is less prominent than its catabolic effect, it may contribute to the homeostasis of articular cartilage under pathological conditions in which Sox9 expression is decreased.
Col2a1 expression was unchanged by Runx2 homozygous knockout in primary chondrocytes, but decreased in inflamed chondrocytes (Fig. 4). These data were also supported by in vivo experiments showing that Col2a1 protein was not decreased in sham joints of R2-Homo cKO mice, but was significantly decreased in OA joints (Fig. 3d, e). A number of previous studies showed that Sox9 is most responsible for transcriptional induction of Col2a18,10–13, while Sox9 expression decreases during the early stage of OA development (Fig. 1). These data evoked our hypothesis that Runx2 may sustain Col2a1 expression under inflammatory conditions where Sox9 is decreased. The results of ChIP-seq and luciferase assays showed that the enhancer in Col2a1 intron 6, which contains both Sox9 and Runx2 consensus motifs, was activated by both transcription factors (Fig. 6c, d). The luciferase activity of the enhancer was tightly regulated by the amount of Sox9 and suppressed in accordance with decreased expression of Sox9 (Fig. 6c). Interestingly, although co-transfection of Runx2 and Sox9 did not enhance the activity of the intron 6 enhancer, Runx2 significantly upregulated its activity when Sox9 was decreased (Fig. 6d). Accordingly, the intron 6 enhancer may be more important for transcriptional regulation of Col2a1 by Sox9 and Runx2 than the intron 1 enhancer. Previous studies also showed that the intron 6 region is a functional enhancer for the Col2a1 gene8, and the transactivity of the intron 6 enhancer by Sox9 is stronger than that of the intron 1 enhancer8,12. All these data are consistent with our hypothesis, however, we could not further evaluate the biological effect sizes of Runx2/Sox9 binding to these enhancers. Considering that luciferase reporter assays often do not match in vivo data, it is necessary to perform OA model experiments using genome-edited mice in which these enhancers are mutated for further understanding.
In this study, we could not further clarify how Runx2 protein sustains Col2a1 transcription when Sox9 protein decreases. A previous report displayed significant enrichment of TG(T/C)GGT motif in the top 2,000 Sox9 peaks in rib chondrocytes by ChIP-seq12, which indicated (1) direct binding of Sox9 with the TG(T/C)GGT sequence, (2) its surrounding regions, or (3) indirect binding via interaction with Runx2. CisGeonme browser magnified view analysis supported the second idea, as Runx2 peak centers in Col2a1 introns 1 and 6 are about 100 bp away from Sox9-binding sites8,12 (Supplementary Fig. 13). In inflamed chondrocytes, de novo Runx2 binding to the Sox9-binding regions was enhanced (Fig. 5g). Under inflammatory conditions, decreased expression of endogenous Sox9 and subsequent decreased binding of Sox9 with its consensus motif may induce Runx2 to approach Runx motifs located near around the Sox9 motif. This hypothesis may also explain why Runx2 overexpression did not increase mRNA levels of Col2a1 when endogenous Sox9 was sufficiently expressed (Fig. 7a). In contrast to Runx3 and Runx1, Runx2 may support cartilage anabolism only in pathogenic conditions.
ChIP-seq and RNA-seq results of this study displayed the different target genes of Runx2 and Runx3. However, because Runx family members have a common consensus motif, we could not identify the molecular mechanisms underlying how Runx2 and Runx3 distinguish their own target genes. Notably, expression patterns of these two proteins in articular cartilage during OA development are different (Fig. 1). Runx3 is expressed in all cartilage layers and decreased during OA development, whereas Runx2 expression is observed in the DZ and sustained during OA development (Fig. 1). These differences in expression are consistent with our hypothesis, but do not explain differences in their target gene specificity. Various factors, such as epigenomic regulation of genes and cofactors of each Runx protein, probably define transcriptional activation of Runx2- or Runx3-specific target genes.
In articular chondrocytes, Runx3 induces representative cartilage matrix gene, Prg4. Lubricin/Prg4, a mucin-like O-linked glycosylated protein, functions as a boundary lubricant in articular cartilage to decrease wear and friction, and the accumulation of lubricin at the surface of the cartilage is important for joint homeostasis5,63. In this study, both in vivo and in vitro experiments demonstrated that Prg4 is downregulated by Runx3 deficiency and Aggrecan/Acan, working as a shock absorber64 and mainly expressed in DZ65, is also a candidate of Runx3 transcriptional target (Figs. 2 and 5). In contrast to these anabolic effects, Runx3 deficiency did not change the expression of Mmp13 or proteoglycan release from articular cartilage (Fig. 2). Chondrocyte hypertrophy and apoptosis were also not changed by Runx3 knockout (Supplementary Figs. 3 and 4) These data indicate that Runx3 does not directly affect the regulation of cartilage degradation or chondrocyte survival. ChIP-seq and RNA-seq results identified Mmp9 as a target of Runx3 (Fig. 5c); however, consistent data for Mmp9 could not be obtained by qPCR and immunohistochemistry (Fig. 2). Although we did not examine other candidate genes, such as Ccl and Ibsp, they may play some roles in the regulation of articular cartilage as downstream molecules of Runx3.
Mmp13 induction seems to be more straightforward. In the present study, Mmp13 induction was consistently suppressed both by homozygous and heterozygous knockout of Runx2 (Figs. 3 and 4), meanwhile, it was not affected by Runx3. These results indicate that Runx2 directly regulated Mmp13 transcription, as indicated in previous reports17–19,66,67. Although Runx2 activated luciferase activity upstream of the Mmp13 TSS, the Runx2 peak was not as large as we expected (Fig. 6). Obviously, Mmp13 transcription is regulated by various signaling pathways and molecules other than Runx2. Mmp13 was increased by IL-1β exposure in a dose- and time-dependent manner without increasing endogenous Runx2 expression (Fig. 1c, d). We previously reported that NF-κB, HIF-2α, Notch signaling, and its downstream factor Hes1 are potent inducers of Mmp1324,68,69. Runx2 overexpression also indirectly induces MMP13 through mitogen-activated protein kinase pathways17. Considering that Mmp13 suppression by Runx2 knockout was partial (Figs. 3 and 4), Runx2 must be associated with the induction of Mmp13 transcription; however, its effect size remains unknown.
In conclusion, this study revealed that Runx3 protects articular cartilage against post-traumatic OA, while Runx2 exerts both anabolic and catabolic effects on articular cartilage under inflammatory conditions. Prg4 and Acan may be the main mediators of the anabolic effects of Runx3, whereas Runx3 does not affect cartilage catabolism. Runx2 induces Mmp13, whereas it compensates for reduced Col2a1 expression in articular chondrocytes under inflammatory conditions in which Sox9 expression is decreased. Our findings regarding the anabolic roles of Runx3 and the diverse regulation of chondrocytes by Runx2 may contribute to understanding the pathophysiology of OA.
Methods
Animals
All animal experiments were authorized and approved by the Animal Care and Use Committee of The University of Tokyo. We have complied with all relevant ethical regulations. In each experiment, we compared the genotypes of littermates maintained in a C57BL/6J background. Runx2-floxed mice were generated to carry a conditional Runx2 allele with exon 4, which encodes the Runt domain, flanked by loxP sites. Deletion of exon 4 also induced a frameshift and disrupted the Runx2 protein after exon 5. Runx2flox-neo mice were crossed with flippase recombinase target (FRT) transgenic mice to eliminate the neomycin cassette. Positive embryonic stem cells were injected into eight-cell stage embryos to generate chimeric mice for subsequent crossing with C57BL/6 J females. This step provided heterozygous animals for the floxed allele of Runx2 (Runx2flox-neo). Mating these mice with FRT transgenic mice resulted in heterozygous-floxed Runx2 (Runx2fl/+) with the FRT-franked neomycin resistance cassette eliminated. Runx2fl/+ mice were next crossed with Runx2fl/+ to generate Runx2fl/+ or Runx2fl/f mice. We then evaluated the targeting efficiency and specificity of these mice by breeding them with CAG-Cre reporter mice.
Col2a1-Cre mice70, Runx3fl/fl mice46, and Prg4-CreERT2 mice48 were purchased from Jackson Laboratory. Col2a1-CreERT2 mice47 were generously provided by Professor Di Chen (Rush University Medical Center, Chicago, IL, USA). To generate Col2a1-Cre; Runx3fl/fl mice, Runx3fl/fl mice were mated with Col2a1-Cre mice to obtain Col2a1-Cre; Runx3fl/+ mice, which were then mated with Runx3fl/fl mice. Col2a1-CreERT2; Runx3fl/fl mice and Prg4-CreERT2; Runx3fl/fl mice were created in the same manner. To generate Col2a1-CreERT2; Runx2fl/fl mice, Runx2fl/fl mice were mated with Col2a1-CreERT2 mice to obtain Col2a1-CreERT2; Runx2fl/+ mice, which were then mated with Runx2fl/fl mice. After generating Col2a1-CreERT2; Runx2fl/fl mice, they were mated with Runx2fl/+ mice to obtain Runx2fl/+ (R2-Hetero Cntl), Col2a1-CreERT2; Runx2fl/+ (R2-Hetero cKO), Runx2fl/fl (R2-Homo Cntl), and Col2a1-CreERT2; Runx2fl/fl (R2-Homo cKO) mice, which were used in experiments.
Runx2-Biotin-3xFLAG-knockin mice (Runx2-FLAG mice), in which a Biotin-3×FLAG tag was inserted into the C-terminus of Runx2 to produce Runx2-Biotin-3×FLAG fusion protein, were generated by one of the co-authors (Hojo, et al.54). Primer sequences for genotyping are shown in Supplementary Table 7.
OA experiment
We used three models for OA experiments. For the medial model41, tamoxifen (100 μg per gram of body weight) was intraperitoneally injected into 7-week-old male mice daily for 5 days, and surgery was performed at 8 weeks of age. Surgery was then performed at 8 weeks of age. Under general anesthesia, resection of the medial collateral ligament and medial meniscus was performed using a surgical microscope41. For the DMM model45, tamoxifen (100 μg per gram of body weight; Sigma-Aldrich, St Louis, MO, USA) was intraperitoneally injected into 15-week-old male mice daily for 5 days, and surgery was performed at 16 weeks of age, in accordance with a previous report45. Mice were analyzed 8 weeks and 12 weeks after surgery for Runx3-knockout mice or Runx2-knockout mice, respectively. For the aging model, tamoxifen was injected for 5 days at 2, 6, and 12 months, and mice were raised until 18 months of age under physiological conditions. All mice were maintained under identical conditions (three mice per cage). OA severity was quantified by the Osteoarthritis Research Society International (OARSI) system71, which was assessed by two observers blinded to the experimental groups.
Histological analysis
The perfusion fixation was performed soon after euthanasia. Tissue samples were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.4) at 4 °C for 1 day. Specimens were decalcified with 10% ethylenediaminetetraacetic acid (EDTA, pH 7.4) at 4 °C for 2 weeks, embedded in paraffin, and cut into 4-μm-thick sagittal sections. Safranin-O staining was performed according to standard protocols. For immunohistochemistry, sections were incubated with antibodies against Runx3 (ab135248, Abcam, Cambridge, UK), Runx2 (ab192256, Abcam), Runx1 (ab23980, Abcam), Col2a1 (MAB8887, Merck, Darmstadt, Germany), Col10a1 (14-9771-80, Invitrogen, Carlsbad, CA, USA), Acan (13880-1-AP, Proteintech, Rosemont, IL, USA), Prg4 (ab28484, Abcam), Mmp13 (18165-1-AP, Proteintech), Mmp9 (ab38898, Abcam), and Sox9 (ab185230, Abcam). For visualization, 3,3’-diaminobenzidine (DAB; Nakaraitesk, Kyoto, Japan) was used. TdT-mediated dUTP nick-end labeling (TUNEL) staining was performed with an In Situ Cell Death Detection Kit (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Histological analyses were performed at least three times using 3−5 mice per group or genotype for confirmation of results. Images were visualized under a fluorescence microscope (BZ-X710, Keyence, Osaka, Japan). Percentages of positive cells and DAB-positive areas were measured by BZ analyzer software (Keyence) and NDPscan (Hamamatsu Photonics, Hamamatsu, Japan), respectively. The following parameters were used in the software: Pixel Area = 4.86E-08, Hue Value = 0.1, Hue Weight = 0.5, and Color Saturation Threshold = 0.04. We defined the strong intensity threshold as 0 − 100, medium as 100−175, and weak as 175−220 in the algorithm input. After margining the Safranin-O-positive articular cartilage area of lateral and medial sides of tibial and femoral joints by referring to the nearest slide, we evaluated areas strongly positive for Col2a1, and medium to strongly positive areas for Mmp13, respectively. We also counted numbers of Runx2/Sox9 medium/strong-positive cells. For quantification, we selected six samples exhibiting average OA development in each group.
Cell cultures
Primary articular chondrocytes were isolated from 5-day-old C57BL/6J mice according to a standard protocol using collagenase D (Roche, Basel, Switzerland)45, and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Wako, Osaka, Japan) with 10% fetal bovine serum. The medium was changed every 3 days. SFZ cells were isolated as previously described72. Briefly, the proximal end of the femur and the distal end of the tibia were dissected from 5-day-old mice, and ligaments and tendons were excised. Cartilage tissues were incubated with 0.25% trypsin (Thermo Fisher Scientific, Waltham, MA, USA) for 1 h, followed by 1.5-h digestion with 173 U/mL of type I collagenase (Worthington Biochemical Corporation, Lakewood, NJ, USA). DZ cells were isolated by additional digestion of residual epiphyseal cartilage tissue with 43 U/ml collagenase type I for 5 h. Dissociated cells were seeded on fibronectin-coated culture dishes. Cells were cultured with DMEM with 10% FBS. The cells were cultured as a monolayer in all experiments. To induce Cre recombination in cultured cells, 2 μM 4-hydroxytamoxifen (Sigma-Aldrich) was applied for 48 h. IL-1β was added 24 h after starting this incubation.
Explant cultures
For ex vivo cultures, femoral head cartilage was removed from Hetero Cntl, Hetero cKO, Homo Cntl, and Homo cKO mice at postnatal day 1. Femoral heads were then cultured in DMEM containing 10% fetal bovine serum, with or without 1 ng/μL IL-1β (AF-200-01B, PeproTech, Rocky Hill, NJ, USA). The medium was changed every 3 days, and the total culture period was 2 weeks. Right femoral heads were used for histological analyses and left ones were homogenized using Trizol reagent (15596-018, Life Technologies, Carlsbad, CA, USA) for qRT-PCR.
qRT-PCR
Total RNA was purified with an RNeasy Mini Kit (Qiagen, Hilden, Germany). One microgram of total RNA was reverse transcribed using ReverTraAce qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan). Each PCR reaction contained 1 × THUNDERBIRD SYBR qPCR Mix (Toyobo), 0.3 mM specific primers, and 25 ng of cDNA. The copy number was normalized to rodent total RNA (Thermo-Fisher Scientific, Waltham, MA, USA), with rodent glyceraldehyde-3-phosphate dehydrogenase (Gapdh) used as an internal control. All reactions were run in triplicate. Primer sequences are shown in Supplementary Table 8.
Construction of expression vectors
Full-length cDNA sequences were amplified by PCR and cloned into the pCMV-HA or pShuttle vector (Clontech). Adenovirus vectors were generated using an Adeno-X expression system (Clontech). All vectors were verified by DNA sequencing.
Luciferase assay
Col2a1 enhancer sites (upstream of TSS, intron 1, and intron 6), upstream of the Mmp13 TSS, Prg4, and Acan were amplified by PCR using mouse genomic DNA as the template and cloned into the PGL4.10 [luc2] vector (Promega, Tokyo, Japan). A luciferase assay was performed using primary chondrocytes and the Dual-Luciferase Reporter Assay System (Promega). Results are reported as the ratio of firefly to Renilla activities. Locations of the cloned sequences are shown in Supplementary Table 9.
1, 9-dimethylmethylene blue (DMMB) assay
The GAG content was measured using a 1, 9-dimethylmethylene blue (DMMB) assay50. Lysates (20 μL) were mixed with 200 μL of a DMMB (Sigma-Aldrich) working solution for 30 min at room temperature. The absorbance was then measured at 525 nm. Chondroitin sulfate (Sigma-Aldrich) was used as a standard.
ChIP-seq
Chromatin preparation and ChIP were performed as previously described9,53,73, with minor modifications. Briefly, 5 × 107 primary chondrocytes from Runx2-FLAG mice, expressing 3 × FLAG-tagged Runx2, were cultured for 48 h with or without 1 ng/mL IL-1β for 24 h. For Runx3, we used SFZ chondrocytes from WT mice that were treated by lipofection (Lipofectamine 2000; Thermo Fisher Scientific) with vectors expressing 3xFLAG-tagged Runx3. Briefly, 5 × 107 chondrocytes were harvested and cross-linked with 1% formaldehyde, followed by quenching with glycine. After cell lysis, chromatin was fragmented by sonication (Branson Digital Sonifier 250, Branson Ultrasonics Corporation Danbury, CT, USA) to obtain 100–600 bp DNA fragments. The fragmented DNA lysate was incubated with Dynabeads M-280 sheep anti-mouse (Thermo Fisher Scientific) and antibody complex overnight at 4 °C [anti-FLAG Clone M2 (Sigma-Aldrich) or Ant-mouse IgG (Promega, Madison, WI, USA)]. After washing the beads with radioimmunoprecipitation assay buffer, DNA was eluted with elution buffer at 65 °C for 15 min, and the elution was incubated overnight at 65 °C for reverse crosslinking. DNA was purified using ChIP DNA Clean and Concentrator (ZYMO Research, Irvine, CA, USA). All buffers contained Complete EDTA-Free Protease Inhibitor Cocktail (Roche). Construction of ChIP-seq libraries was performed with a ThruPLEXFD Prep Kit (Takara Bio, Kusatsu, Japan). The library was sequenced on a HiSeq2000 (Illumina, San Diego, CA, USA) platform.
Supercomputing resources were provided by the XXX. DNA sequence information was aligned to the unmasked mouse genome reference sequence mm9 by a bowtie aligner74. Peak calling was performed by two-sample analysis using CisGenome software v2.0 (35) with a P value cut-off of 1 × 10−5 compared with the input control. Peaks with a false discovery rate of <0.01 were incorporated into further analysis. GREAT GO analysis was performed utilizing the online GREAT GO program, version 3.0.052. Each peak category was run against a whole-genome background with assembly mm9. De novo motif recovery was performed with MEME-ChIP52. A 100-bp region surrounding the peak center was extracted from mm9 and used for analyses. Raw and processed data are available in the Gene Expression Omnibus database under accession numbers “GSE157328” and “GSE165086”. Peak data were compared with “GSM1692996”, “GSM1692997”, “GSM1693001”, “GSM1693003”, and “GSM1693005”11.
RNA-seq
For Runx3, RNA samples were collected from SFZ/DZ chondrocytes of 5-day-old Col2a1-Cre; Runx3fl/fl and Runx3fl/fl mice (n = 3, respectively). For Runx2, RNA samples were collected from primary chondrocytes of 5-day-old Col2a1-CreERT2; Runx2fl/fl and Runx2fl/fl mice (n = 2, respectively) and then subjected to 48-h incubation with 4-hydroxytamoxifen with or without 1 ng/μL IL-1β for 24 h. RNA-seq was conducted on a BGIseq-500 (BGI, Hong Kong, China) to produce 100-bp paired-end reads, which were mapped to the mouse genome (mm9). Quantile normalization was applied, and read counts were transformed to log2 counts per million. Linear models were used to test for expression differences between deficient and control samples. Raw and processed data are available in the Gene Expression Omnibus database under accession numbers “GSE157433” and “GSE165084”.
Intra-articular administration of adenoviral vectors
We performed intra-articular administration of an adenoviral vector containing Runx3 or GFP. Injections were performed three times: 1, 4, and 7 weeks after the OA surgery. For each administration, we injected 10 µL of 1 × 109 plaque-forming units of adenoviral vectors solution diluted in PBS.
Statistics and reproducibility
Results were analyzed using Prism version 8.4.1 (GraphPad Software, San Diego, CA, USA). Two-tailed Mann–Whitney U test was used to establish statistical significance between the two groups. One-way ANOVA with Turkey’s post hoc test was used to establish statistical significance in more than three groups. Ordinary one-way ANOVA with Dunnett’s multiple comparisons test was used for dose-dependent or time-course comparisons of multiple groups. P values below 0.05 were considered significant. Biologically independent sample numbers were shown in figures and figure legends. For micrographs, representative images in each group were displayed.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank J. Sugita, R. Homma, A. Ogikubo, and K. Kaneko for technical assistance. We thank Ashleigh Cooper, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This work was supported by JSPS KAKENHI Grant Numbers 19H05654, 19H05565, 18KK0254, 17H04310, 20H03799, 20K09428, and 20K09451.
Source data
Author contributions
Study conception and design: K.N., H.H., S.H.C., and T.S. Data collection: K.N., S.H.C., D.M., Yuma M., M.K., T.K., Y.I., H.N., and Yuji M. Data analysis: K.N., H.H., H.O., R.C., and S.O. Data interpretation: K.N., F.Y., R.C., Y.O., N.T., H.I., J.H., Y.T., U.C., S.T., and T.S. Generation of Runx2-cKO mice: T.S. Generation of Runx2-FLAG mice: H.H., S.O., and U.C. Drafting of the paper: K.N. and T.S. All authors proofread and approved the paper.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Data availability
Raw and processed data are available in the Gene Expression Omnibus database under accession numbers “GSE157328” and “GSE165086” for ChIP-seq. Peak data of “GSM1692996”, “GSM1692997”, “GSM1693001”, “GSM1693003”, and “GSM1693005” were used from the previous report12. “GSE157433” and “GSE165084” for RNA-seq. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-33744-5.
References
- 1.Emery CA, et al. Establishing outcome measures in early knee osteoarthritis. Nat. Rev. Rheumatol. 2019;15:438–448. doi: 10.1038/s41584-019-0237-3. [DOI] [PubMed] [Google Scholar]
- 2.Gupta S, Hawker GA, Laporte A, Croxford R, Coyte PC. The economic burden of disabling hip and knee osteoarthritis (OA) from the perspective of individuals living with this condition. Rheumatology. 2005;44:1531–1537. doi: 10.1093/rheumatology/kei049. [DOI] [PubMed] [Google Scholar]
- 3.Yoshimura N, et al. Prevalence of knee osteoarthritis, lumbar spondylosis, and osteoporosis in Japanese men and women: the research on osteoarthritis/osteoporosis against disability study. J. Bone Min. Metab. 2009;27:620–628. doi: 10.1007/s00774-009-0080-8. [DOI] [PubMed] [Google Scholar]
- 4.Muraki S, et al. Association of occupational activity with radiographic knee osteoarthritis and lumbar spondylosis in elderly patients of population-based cohorts: a large-scale population-based study. Arthritis Rheum. 2009;61:779–786. doi: 10.1002/art.24514. [DOI] [PubMed] [Google Scholar]
- 5.Rhee DK, et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J. Clin. Investig. 2005;115:622–631. doi: 10.1172/JCI200522263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jay GD, et al. Association between friction and wear in diarthrodial joints lacking lubricin. Arthritis Rheum. 2007;56:3662–3669. doi: 10.1002/art.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokuda S, et al. Wwp2 maintains cartilage homeostasis through regulation of Adamts5. Nat. Commun. 2019;10:2429. doi: 10.1038/s41467-019-10177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yasuda H, Oh CD, Chen D, de Crombrugghe B, Kim JH. A novel regulatory mechanism of type II collagen expression via a SOX9-dependent enhancer in Intron 6. J. Biol. Chem. 2017;292:528–538. doi: 10.1074/jbc.M116.758425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldring MB, Marcu KB. Cartilage homeostasis in health and rheumatic diseases. Arthritis Res. Ther. 2009;11:224. doi: 10.1186/ar2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi WM, Deng JM, Zhang ZP, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. J. Bone Miner. Res. 2002;17:S142–S142. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohba S, He X, Hojo H, McMahon AP. Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 2015;12:229–243. doi: 10.1016/j.celrep.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu CF, Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015;43:8183–8203. doi: 10.1093/nar/gkv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouyang Y, et al. Overexpression of SOX9 alleviates the progression of human osteoarthritis in vitro and in vivo. Drug Des. Devel Ther. 2019;13:2833–2842. doi: 10.2147/DDDT.S203974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, et al. MMP13 is a critical target gene during the progression of osteoarthritis. Arthritis Res. Ther. 2013;15:R5. doi: 10.1186/ar4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little CB, et al. Matrix metalloproteinase 13-deficient mice are resistant to osteoarthritic cartilage erosion but not chondrocyte hypertrophy or osteophyte development. Arthritis Rheum. 2009;60:3723–3733. doi: 10.1002/art.25002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tetsunaga T, et al. Regulation of mechanical stress-induced MMP-13 and ADAMTS-5 expression by RUNX-2 transcriptional factor in SW1353 chondrocyte-like cells. Osteoarthr. Cartil. 2011;19:222–232. doi: 10.1016/j.joca.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi A, et al. DNA methylation of the RUNX2 P1 promoter mediates MMP13 transcription in chondrocytes. Sci. Rep. 2017;7:7771. doi: 10.1038/s41598-017-08418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirata M, et al. C/EBPbeta and RUNX2 cooperate to degrade cartilage with MMP-13 as the target and HIF-2alpha as the inducer in chondrocytes. Hum. Mol. Genet. 2012;21:1111–1123. doi: 10.1093/hmg/ddr540. [DOI] [PubMed] [Google Scholar]
- 20.Bau B, et al. Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum. 2002;46:2648–2657. doi: 10.1002/art.10531. [DOI] [PubMed] [Google Scholar]
- 21.Chang SH, et al. Excessive mechanical loading promotes osteoarthritis through the gremlin-1-NF-kappaB pathway. Nat. Commun. 2019;10:1442. doi: 10.1038/s41467-019-09491-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito T, et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat. Med. 2010;16:678–686. doi: 10.1038/nm.2146. [DOI] [PubMed] [Google Scholar]
- 23.Yang S, et al. Hypoxia-inducible factor-2alpha is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 2010;16:687–693. doi: 10.1038/nm.2153. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi H, et al. Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat. Commun. 2016;7:13336. doi: 10.1038/ncomms13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat. Rev. Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- 26.Fukushima-Nakase Y, et al. Shared and distinct roles mediated through C-terminal subdomains of acute myeloid leukemia/Runt-related transcription factor molecules in murine development. Blood. 2005;105:4298–4307. doi: 10.1182/blood-2004-08-3372. [DOI] [PubMed] [Google Scholar]
- 27.Komori T. Runx2, an inducer of osteoblast and chondrocyte differentiation. Histochem. Cell Biol. 2018;149:313–323. doi: 10.1007/s00418-018-1640-6. [DOI] [PubMed] [Google Scholar]
- 28.Komori T, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 29.Otto F, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 30.Wang X, et al. Regulation of MMP-13 expression by RUNX2 and FGF2 in osteoarthritic cartilage. Osteoarthr. Cartil. 2004;12:963–973. doi: 10.1016/j.joca.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Kamekura S, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54:2462–2470. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 32.Liao L, et al. Deletion of Runx2 in articular chondrocytes decelerates the progression of DMM-induced osteoarthritis in adult mice. Sci. Rep. 2017;7:2371. doi: 10.1038/s41598-017-02490-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catheline SE, et al. Chondrocyte-specific RUNX2 overexpression accelerates post-traumatic osteoarthritis progression in adult mice. J. Bone Min. Res. 2019;34:1676–1689. doi: 10.1002/jbmr.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, et al. Runx2 plays a central role in osteoarthritis development. J. Orthop. Transl. 2019;23:132–139. doi: 10.1016/j.jot.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komori T. Molecular mechanism of Runx2-dependent bone development. Mol. Cells. 2020;43:168–175. doi: 10.14348/molcells.2019.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura A, et al. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010;137:1159–1167. doi: 10.1242/dev.045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao L, et al. Deletion of Runx2 in condylar chondrocytes disrupts TMJ tissue homeostasis. J. Cell Physiol. 2019;234:3436–3444. doi: 10.1002/jcp.26761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yano F, et al. Runx1 contributes to articular cartilage maintenance by enhancement of cartilage matrix production and suppression of hypertrophic differentiation. Sci. Rep. 2019;9:7666. doi: 10.1038/s41598-019-43948-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wigner NA, Soung do Y, Einhorn TA, Drissi H, Gerstenfeld LC. Functional role of Runx3 in the regulation of aggrecan expression during cartilage development. J. Cell Physiol. 2013;228:2232–2242. doi: 10.1002/jcp.24396. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Z, Yao B, Zhao D. Runx3 regulates chondrocyte phenotype by controlling multiple genes involved in chondrocyte proliferation and differentiation. Mol. Biol. Rep. 2020;47:5773–5792. doi: 10.1007/s11033-020-05646-6. [DOI] [PubMed] [Google Scholar]
- 41.Kamekura S, et al. Osteoarthritis development in novel experimental mouse models induced by knee joint instability. Osteoarthr. Cartil. 2005;13:632–641. doi: 10.1016/j.joca.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Jenei-Lanzl Z, Meurer A, Zaucke F. Interleukin-1beta signaling in osteoarthritis-chondrocytes in focus. Cell Signal. 2019;53:212–223. doi: 10.1016/j.cellsig.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Z, Du D, Chen A, Zhu L. Circular RNA expression profile of articular chondrocytes in an IL-1beta-induced mouse model of osteoarthritis. Gene. 2018;644:20–26. doi: 10.1016/j.gene.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 44.Arra M, et al. LDHA-mediated ROS generation in chondrocytes is a potential therapeutic target for osteoarthritis. Nat. Commun. 2020;11:3427. doi: 10.1038/s41467-020-17242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gosset M, Berenbaum F, Thirion S, Jacques C. Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 2008;3:1253–1260. doi: 10.1038/nprot.2008.95. [DOI] [PubMed] [Google Scholar]
- 46.Naoe Y, et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J. Exp. Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen M, et al. Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. Genesis. 2007;45:44–50. doi: 10.1002/dvg.20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozhemyakina E, et al. Identification of a Prg4-expressing articular cartilage progenitor cell population in mice. Arthritis Rheumatol. 2015;67:1261–1273. doi: 10.1002/art.39030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthr. Cartil. 2007;15:1061–1069. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 50.Stanton H, et al. Investigating ADAMTS-mediated aggrecanolysis in mouse cartilage. Nat. Protoc. 2011;6:388–404. doi: 10.1038/nprot.2010.179. [DOI] [PubMed] [Google Scholar]
- 51.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hojo H, Ohba S, He X, Lai LP, McMahon AP. Sp7/Osterix is restricted to bone-forming vertebrates where it acts as a Dlx co-factor in osteoblast specification. Dev. Cell. 2016;37:238–253. doi: 10.1016/j.devcel.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hojo H, Saito T, He X, Guo Q, Onodera S, Azuma T. Runx2 regulates chromatin accessibility to direct the osteoblast program at neonatal stages. Cell Rep. 2022;40:111315. doi: 10.1016/j.celrep.2022.111315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhandari RK, Haque MM, Skinner MK. Global genome analysis of the downstream binding targets of testis determining factor SRY and SOX9. PLoS ONE. 2012;7:e43380. doi: 10.1371/journal.pone.0043380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat. Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu X, et al. WNT16 is upregulated early in mouse TMJ osteoarthritis and protects fibrochondrocytes against IL-1β induced inflammatory response by regulation of RUNX2/MMP13 cascade. Bone. 2021;143:115792. doi: 10.1016/j.bone.2020.115793. [DOI] [PubMed] [Google Scholar]
- 61.Dou P, He Y, Yu B, Duan J. Downregulation of microRNA-29b by DNMT3B decelerates chondrocyte apoptosis and the progression of osteoarthritis via PTHLH/CDK4/RUNX2 axis. Aging. 2020;13:7676–7690. doi: 10.18632/aging.103778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen CG, Thuillier D, Chin EN, Alliston T. Chondrocyte-intrinsic Smad3 represses Runx2-inducible matrix metalloproteinase 13 expression to maintain articular cartilage and prevent osteoarthritis. Arthritis Rheum. 2012;64:3278–3289. doi: 10.1002/art.34566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ikegawa S, Sano M, Koshizuka Y, Nakamura Y. Isolation, characterization and mapping of the mouse and human PRG4 (proteoglycan 4) genes. Cytogenet Cell Genet. 2000;90:291–297. doi: 10.1159/000056791. [DOI] [PubMed] [Google Scholar]
- 64.Fraser A, et al. Turnover of type II collagen and aggrecan in cartilage matrix at the onset of inflammatory arthritis in humans: relationship to mediators of systemic and local inflammation. Arthritis Rheum. 2003;48:3085–3095. doi: 10.1002/art.11331. [DOI] [PubMed] [Google Scholar]
- 65.Mori Y, Chung UI, Tanaka S, Saito T. Determination of differential gene expression profiles in superficial and deeper zones of mature rat articular cartilage using RNA sequencing of laser microdissected tissue specimens. Biomed. Res. 2014;35:263–270. doi: 10.2220/biomedres.35.263. [DOI] [PubMed] [Google Scholar]
- 66.Huang J, et al. The microRNAs miR-204 and miR-211 maintain joint homeostasis and protect against osteoarthritis progression. Nat. Commun. 2019;10:2876. doi: 10.1038/s41467-019-10753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishimura R, et al. Osterix regulates calcification and degradation of chondrogenic matrices through matrix metalloproteinase 13 (MMP13) expression in association with transcription factor Runx2 during endochondral ossification. J. Biol. Chem. 2012;287:33179–33190. doi: 10.1074/jbc.M111.337063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hosaka Y, et al. Notch signaling in chondrocytes modulates endochondral ossification and osteoarthritis development. Proc. Natl Acad. Sci. USA. 2013;110:1875–1880. doi: 10.1073/pnas.1207458110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sugita S, et al. Transcription factor Hes1 modulates osteoarthritis development in cooperation with calcium/calmodulin-dependent protein kinase 2. Proc. Natl Acad. Sci. USA. 2015;112:3080–3085. doi: 10.1073/pnas.1419699112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. doi: 10.1002/(SICI)1526-968X(200002)26:2<145::AID-GENE14>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 71.Glasson SS, Chambers MG, Van Den Berg WB, Little CB. The OARSI histopathology initiative—recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthr. Cartil. 2010;18:S17–S23. doi: 10.1016/j.joca.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 72.Yasuhara R, et al. Roles of beta-catenin signaling in phenotypic expression and proliferation of articular cartilage superficial zone cells. Lab Investig. 2011;91:1739–1752. doi: 10.1038/labinvest.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peterson KA, et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data are available in the Gene Expression Omnibus database under accession numbers “GSE157328” and “GSE165086” for ChIP-seq. Peak data of “GSM1692996”, “GSM1692997”, “GSM1693001”, “GSM1693003”, and “GSM1693005” were used from the previous report12. “GSE157433” and “GSE165084” for RNA-seq. Source data are provided with this paper.