Abstract
Alterations in brain cholesterol homeostasis have been broadly implicated in neurological disorders. Notwithstanding the complexity by which cholesterol biology is governed in the mammalian brain, excess neuronal cholesterol is primarily eliminated by metabolic clearance via cytochrome P450 46A1 (CYP46A1). No methods are currently available for visualizing cholesterol metabolism in the living human brain; therefore, a non-invasive technology that quantitatively measures the extent of brain cholesterol metabolism via CYP46A1 could broadly impact disease diagnosis and treatment options using targeted therapies. Here we describe the development and testing of a CYP46A1-targeted positron emission tomography (PET) tracer. 18F-CHL-2205(18F-Cholestify). Our data show that PET imaging readouts correlate with CYP46A1 protein expression and with the extent to which cholesterol is metabolized in the brain, as assessed by cross-species post-mortem analyses of specimens from rodents, non-human primates and humans. Proof-of-concept of in vivo efficacy is provided in the well-established 3xTg-AD murine model of Alzheimer’s disease (AD), where we show that the probe is sensitive to differences in brain cholesterol metabolism between 3xTg-AD mice and control animals. Further, our clinical observations point towards a considerably higher baseline brain cholesterol clearance via CYP46A1 in women, as compared to age-matched men. These findings illustrate the vast potential of assessing brain cholesterol metabolism using PET and establish PET as a sensitive tool for non-invasive assessment of brain cholesterol homeostasis in the clinic.
One-sentence Summary:
Molecular imaging with positron emission tomography now provides a tool to non-invasively assess cholesterol homeostasis in the living human brain.
Introduction
Brain cholesterol homeostasis is tightly regulated by a molecular machinery that orchestrates the biosynthesis, transport, metabolism and clearance of cholesterol from the mammalian central nervous system (CNS) (1–4). Despite the various players involved in neuronal cholesterol regulation, exchange of plasma and brain cholesterol is precluded by the blood-brain barrier (5). Along this line, brain cholesterol is synthesized in situ, namely, by astrocytes and neurons (1). 3-hydroxy-3-methyl-glutaryl-CoA (HMG-CoA) reductase catalyzes the rate-limiting step in the biosynthesis of cholesterol, whereas cytochrome P450 46A1 (CYP46A1) facilitates the clearance of cholesterol from the CNS (6–8). Indeed, CYP46A1 is highly expressed in neurons and constitutes the primary cholesterol clearance pathway by mediating the conversion of cholesterol to 24-hydroxycholesterol – a metabolite that readily penetrates the blood-brain barrier and can be eliminated from the CNS (5, 9).
Several lines of evidence point towards a disturbed brain cholesterol homeostasis in a variety of neurodegenerative, inflammatory, and vascular brain diseases across the lifespan, however, underlying mechanisms are not fully understood (10–12). Indeed, a balanced brain cholesterol turnover is required for key physiological functions including synaptic plasticity, learning and memory function in the mammalian brain (13). Of note, cholesterol metabolism is impaired in Huntington’s disease (HD), with attenuation of CYP46A1 expression in the striatum across different species (14–16). Whereas the knockdown of CYP46A1 in the mouse striatum triggers neurodegeneration, CYP46A1 overexpression reverses disease pathology in the R6/2 and zQ175 mouse models of HD (14, 16). Further, polymorphisms in the gene encoding for CYP46A1 have been linked to Alzheimer’s disease (AD) (17). Given that CYP46A1 is responsible for the clearance of excess cholesterol from neurons, dysfunction of CYP46A1 may ultimately contribute to an enhanced neuronal accumulation of cholesterol esters – a storage form of cholesterol that was shown to be detrimental in AD (18, 19). Indeed, there is emerging evidence suggesting that an impaired balance between cholesterol biosynthesis and clearance from the CNS leads to the formation of cholesterol ester deposits, thereby promoting amyloidogenic processing of the amyloid precursor protein (APP) to amyloid β (Aβ) (12, 19–25). CYP46A1-mediated reduction of cholesterol esters was shown to independently attenuate Aβ and tau pathology in isogenic induced pluripotent stem cell (iPSC)-derived AD neurons (19). Hence, accounting for changes in cholesterol clearance via CYP46A1 is critical to elucidate underlying causes of the impaired brain cholesterol homeostasis in neurodegenerative disorders.
Attempts to assess the plasma concentration of 24-hydroxycholesterol as a surrogate measure for CYP46A1 activity in the brain have yielded conflicting results (26–28). Some studies suggested that patients with mild cognitive impairments and early stages of AD present with augmented concentrations of 24-hydroxycholesterol in the plasma, whereas others found no differences between healthy individuals and individuals with AD (27, 28). Beyond AD, 24-hydroxycholesterol has been suggested as a potential biomarker for HD (29), autism (30), epilepsy (31), depression (32) and Parkinson’s disease (PD) (33). A critical consideration is that 24-hydroxycholesterol is highly susceptible to metabolism in the liver (34, 35). As such, the correlation of 24-hydroxycholesterol plasma concentrations with CYP46A1 function is confounded by downstream metabolic processing of 24-hydroxycholesterol in the periphery, raising substantial concerns about the reliability of 24-hydroxycholesterol as a plasma biomarker of brain cholesterol metabolism (34). Overall, a reliable and non-invasive procedure to assess brain cholesterol clearance via CYP46A1 is currently lacking, which constitutes an unmet need in research and contemporary clinical practice.
Given the key role of cholesterol homeostasis in mammalian aging and brain diseases, a non-invasive technology that quantitatively measures the extent of brain cholesterol metabolism could broadly impact disease diagnosis as well as treatment options using targeted therapies. Here, we developed and tested in animal models and in humans a CYP46A1-targeted positron emission tomography (PET) tracer. We demonstrate that the developed probe provides accurate non-invasive quantification of CYP46A1 abundancy and cholesterol metabolism across different regions of the rodent, non-human primate (NHP) and human brain. In a proof-of-concept study, we show that the tracer is sensitive to differences in brain cholesterol metabolism between 3xTg-AD mice and wild-type animals. Further, we provide preliminary evidence for considerable differences in brain cholesterol metabolism between healthy women and men undergoing CYP46A1-targeted PET imaging.
Results
PET tracer co-localizes with CYP46A1 and 24-hydroxycholersterol in the rodent brain
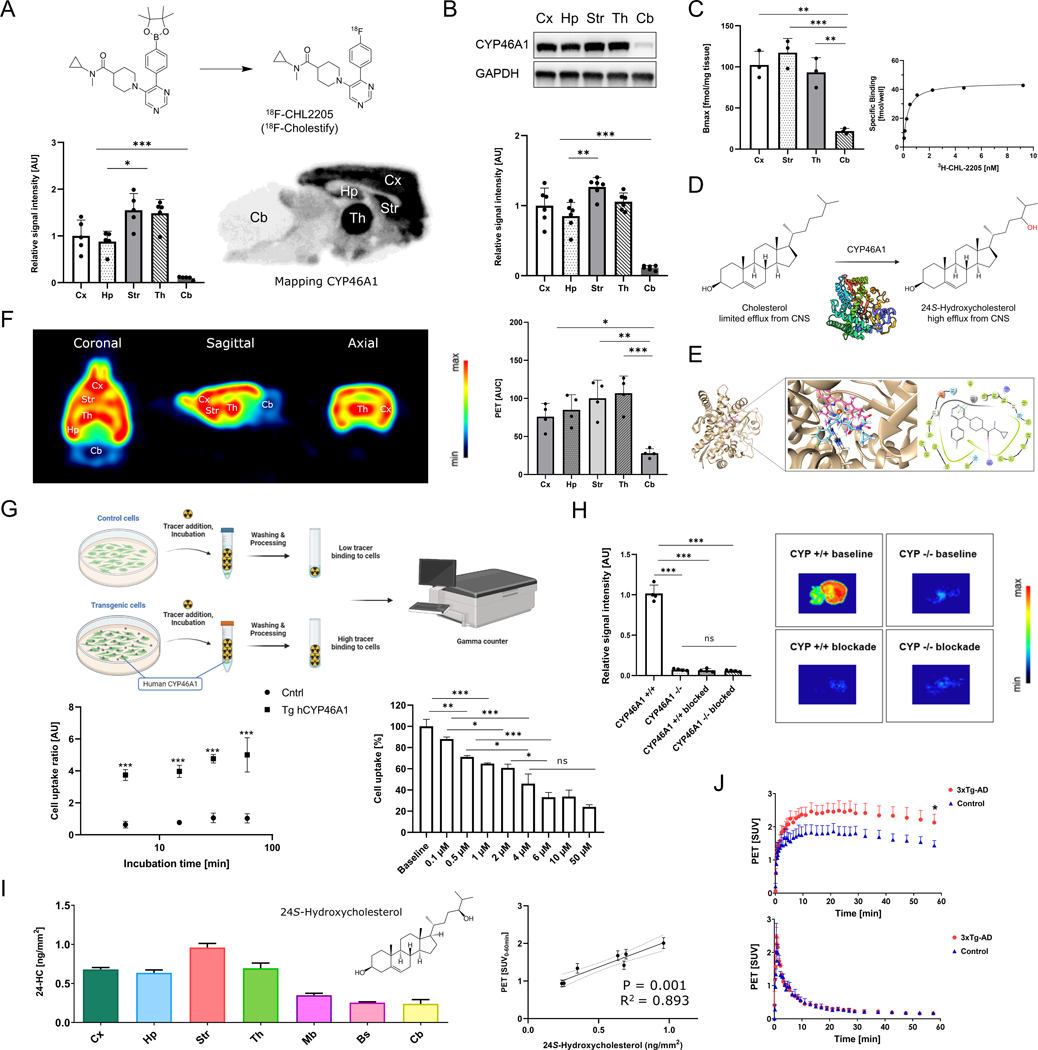
We hypothesized that a targeted PET imaging probe would allow the quantification of cholesterol metabolism in the living brain, ultimately paving the way for mechanistic studies on brain cholesterol metabolism in humans. As such, structure-activity relationship studies (Fig. S1) were performed and led to the discovery of a highly potent ligand for CYP46A1, which was labeled with fluorine-18 via the boronic pinacol ester precursor (Fig. 1A and Fig. S2–S3). The resulting PET tracer, 18F-CHL-2205 (named 18F-Cholestify), exhibited a selectivity for CYP46A1-rich areas of the rodent brain by in vitro autoradiography (Fig. 1A). Highest tracer binding was observed in the CYP46A1-rich cortical, hippocampal, striatal, and thalamic structures, whereas lower abundancy was detected in the cerebellum. The tracer distribution matched closely with the Allen Brain Atlas distribution of CYP46A1 expression data in the mouse brain (36). Quantification of CYP46A1 by Western blot analysis in the same brain regions confirmed that tracer binding patterns were indeed in concert with CYP46A1 expression across all tested brain regions (Fig. 1B). Prior to conducting in vivo experiments, saturation binding studies were performed to determine the dissociation constant (KD) of the probe as well as to assess the amount of available CYP46A1 protein (Bmax) in selected brain regions. Bmax values ranged from 117.3 ± 17.7 fmol/g in the striatum to 21.9 ± 3.3 fmol/g in the cerebellum (p<0.001, Fig. 1C), and a subnanomolar KD value of 0.36 ± 0.06 nM was obtained using striatal homogenates (Fig. S4). Further, pharmacological screening on major CNS targets, including common GPCRs, ion channels, enzymes and transporters at a testing concentration of 10 μM revealed no off-target activity of target compound CHL-2205 (Fig. S5).
Figure 1: PET tracer co-localizes with CYP46A1 and 24-hydroxycholersterol in the rodent brain.
A) Radiosynthesis and autoradiographic distribution of the probe, 18F-CHL-2205 (named 18F-Cholestify), on the healthy rat brain (n=4) B) Western blot analysis of CYP46A1 expression in selected regions of the rat brain (n=6). C) Representative probe saturation binding kinetics and Bmax determinations in selected rat brain regions. D) CYP46A1-mediated hydroxylation of cholesterol to 24S-hydroxycholesterol in the central nervous system (CNS). E) Molecular docking of CHL-2205 (blue) to CYP46A1 (PDB:3MDM). Binding within the cavity was primarily composed of hydrophobic interactions, as well as one T-shaped pi-pi stacking with the protoporphyrin ring (pink) and a hydrogen bond between the carbonyl group and R226 (arrow). F) Representative PET images of the rat brain reflecting CYP46A1-rich brain regions, averaged from 0–60 min post tracer injection and presented in coronal, sagittal and axial view. Quantitative data is depicted as area under the curve (AUC) from respective time activity curves (n=4). G) Cell uptake studies in transfected human embryonic kidney (HEK) cells overexpressing human CYP46A1 and respective controls at the incubation time of 5, 15, 30 and 60 min, respectively (n=4). Dose-response study was conducted with escalating concentrations of soticlestat. Data is presented as percentage, whereas the average of the baseline (no soticlestat) samples was used as a reference. H) Autoradiographic assessment of 18F-CHL-2205 binding to the brains of Cyp46A1 knock out mice and respective controls (n=4). I) Left: Regional quantification of major cholesterol metabolite, 24-hydroxycholesterol, in the CNS. Right: correlation of PET signal with the extent to which cholesterol was metabolized to 24-hydroxycholesterol. J) PET scans of the validated Alzheimer’s disease mouse model, 3xTg-AD, and respective control animals (n=4). Upper panel shows time-activity curves (TACs) under baseline conditions, where only the tracer is administered. Lower panel shows TACs under blockade conditions, where the tracer is administered together with an excess of non-radioactive CHL-2205 to diminish the number of enzymes that are available for tracer-CYP46A1 interactions. Abbreviations: Bs, brain stem; Cb, cerebellum; Cx, cortex; Hp, hippocampus; Mb, midbrain; Str, striatum; Th, thalamus; AUC, area under the curve; CYP +/+, wild-type mouse; CYP−/−, Cyp46A1 knock out mouse, SUV, standardized uptake value. * p<0.05, ** p<0.01, *** p<0.001.
Cholesterol interacts with the orthosteric CYP46A1 binding site, triggering its conversion to 24S-hydroxycholesterol in the mammalian brain (Fig. 1D). To define the molecular interactions between CHL-2205 and CYP46A1, we conducted docking studies, thereby harnessing previously reported crystal structures of CYP46A1. Of the available crystal structures, PDB:3MDT (36), PDB:7LRL (37), and PDB:3MDM (36) were used because of co-crystallization of CYP46A1 with voriconazole, soticlestat and thioperamide, respectively (38). When CHL-2205 was docked to CYP46A1, the binding pose resembled that of voriconazole, thioperamide, and soticlestat, indicating that CHL-2205 constitutes an orthosteric ligand, which is in accordance with experimental findings showing competitive displacement of 18F-CHL-2205 by voriconazole and soticlestat (Fig. S6). Fig. 1E summarizes the interactions of CHL-2205 observed within the CYP46A1 cavity for the crystal structure PDB:3MDM. Similar docking results were obtained with PDB:3MDT and PDB:7LRL (Fig. S7–S8).
18F-CHL-2205 was stable against radiodefluorination when incubated with mouse, rat, NHP and human liver microsomes (Fig. S9). Based on these encouraging in vitro findings, PET imaging studies were conducted by intravenous injection of 18F-CHL-2205 to rats. Systemic administration resulted in a rapid tracer uptake into the brain – with similarity between in vitro and in vivo tracer binding patterns (Fig. 1F). In line with these observations, we found that >99% of the brain signal was attributed to the intact parent tracer and that the brain signal was virtually devoid of interference by radiometabolites (Fig. S10). By ex vivo studies, we observed that > 90% of the signal in the brain was specific at 15, 30 and 45 min post injection (Fig. S10). Dosimetry experiments revealed an effective dose of 0.012 mSv/MBq, and PET studies in mice corroborated the probe selectivity for the CYP46A1-rich striatum (Fig. S11–S12). These results indicated that 18F-CHL-2205 PET can be used to visualize CYP46A1 in vivo. Overall, in vitro autoradiography and ex vivo biodistribution studies suggested an excellent specificity of 18F-CHL-2205 towards CYP46A1 (Figs. S6 and S10).
In a next step, cell uptake studies were conducted with transfected human embryonic kidney (HEK) cells over expressing human CYP46A1 (hCYP46A1) and respective controls, as depicted in Fig. 1G. Transfected cells displayed a 5-fold increase in cell uptake, as compared to the controls, which was observed at early incubation time points and did not change over time (Fig. 1G). Further, the tracer signal of the transfected cell population was attenuated in a dose-dependent manner by co-incubation with escalating doses of soticlestat.
A key consideration in CNS-targeted PET constitutes the ability of the tracer to selectivity bind to a target protein while lacking interactions with other abundant brain proteins. To assess whether 18F-CHL-2205 exhibited such selectivity, we compared the brain binding patterns between CYP46A1 knock-out (Cyp46A1−/−) and respective control (Cyp46A1+/+) mice (Fig. 1H). High tracer binding was observed in the control brain, whereas the signal diminished in the brains of Cyp46A1−/− animals. A blockade experiment with excess dose of the CYP46A1 inhibitor, soticlestat (38), showed that the tracer binding in control brains was reduced to the signal observed in Cyp46A1−/− animals, corroborating the tracer selectivity for CYP46A1.
To assess whether PET imaging findings not only reflected the abundancy of CYP46A1, but also the extent to which neuronal cholesterol was metabolized to 24-hydroxycholesterol in distinct brain regions, the amount of 24-hydroxycholesterol was measured by advanced mass spectrometry, (39). There was a strong correlation between the PET signal in vivo and amount of 24-hydroxycholesterol ex vivo across all tested brain regions (R2=0.893, p=0.001), suggesting that the PET scan can be used to quantify the metabolic activity of CYP46A1 in the living brain (Fig. 1I). When the same experiment was conducted with a related brain cholesterol metabolite, 24.25-epoxycholesterol, that is formed via CYP46A1-dependent and CYP46A1-independent enzymatic reactions, the correlation was lost (R2=0.07, p=0.58, Fig. S13). These results indicated that the PET signal might serve as a non-invasive surrogate measure for the extent of cholesterol turnover to 24-hydroxycholesterol across different brain regions.
Given the potential importance of CYP46A1 in neurodegenerative diseases such as AD, we assessed the tracer binding in the validated 3xTg-AD mouse model. This model is widely used and exhibits some similarities with histopathological and behavioral features of clinical AD (40, 41). Hippocampal uptake of the CYP46A1 tracer was compared between 3xTg-AD mice and respective controls. Following intravenous administration of the CYP46A1 tracer, time-activity curves were consistently higher in the hippocampus of 3xTg-AD mice than in controls (Fig. 1J, upper panel), pointing towards an increased metabolic clearance of brain cholesterol by CYP46A1 in the AD model. When CYP46A1 binding sites were presaturated (blocked) with excess non-radioactive CHL-2205, tracer uptake in 3xTg-AD and control mice was reduced to the same baseline signal (Fig. 1J, lower panel), supporting the notion that differences in tracer uptake were indeed attributed to distinct bioavailability of CYP46A1.
Targeted PET predicts CYP46A1 abundancy and allows the quantification of drug-CYP46A1 interactions in the brains of non-human primates
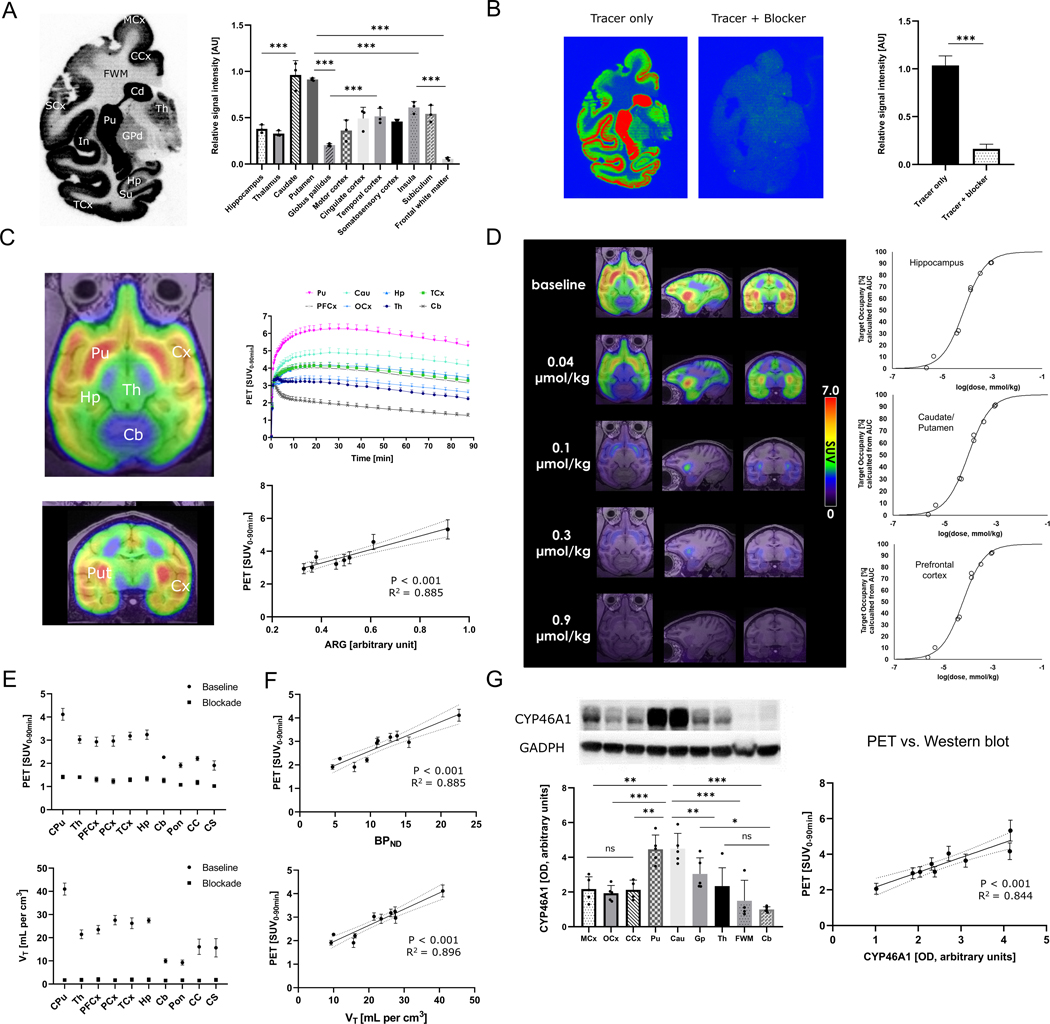
In a next step, PET imaging experiments were conducted in non-human primates. In line with observations from rodent data, the signal intensity of tracer binding was consistent with high abundancy regions for CYP46A1, including the putamen, caudate and limbic cortical regions such as superficial layers of temporal, insular, and cingulate cortex, as evidenced by in vitro autoradiograms of the brain (Fig. 2A). By co-incubation with excess soticlestat (blocker in Fig. 2B), a relative signal reduction of 87.2 ± 0.06% was observed (Fig. 2B), suggesting that the tracer is specific towards CYP46A1 in non-human primates. Next, the tracer was injected intravenously to rhesus monkeys and the time-activity curves in the brain were recorded by PET over a scan duration of 90 min. Regional brain distribution was consistent between in vitro autoradiograms and in vivo PET (Fig. 2C, R2=0.885, p<0.001), thereby showing highest tracer uptake in the putamen, caudate, and cortical regions, followed by the thalamus and hippocampus. Lowest tracer uptake was detected in the cerebellum, brain stem and white matter regions, implying that these regions exhibited poor CYP46A1 abundancy.
Figure 2: Targeted PET predicts CYP46A1 abundancy and allows the quantification of drug-CYP46A1 interactions in the brains of non-human primates.
A) Autoradiographic distribution of the probe, 18F-CHL-2205, on the healthy non-human primate brain (n=3). B) Baseline vs. blockade study with excess non-radioactive CYP46A1 inhibitor, soticlestat (n=3). C) PET images and respective time-activity curves (TACs) in non-human primates (n=4). Correlation between in vitro autoradiography and in vivo PET. D) Target occupancy study in which the probe was used to estimate the extent of target engagement by clinical drug candidate, soticlestat, at different doses and in distinct brain regions. E) Calculation of averaged standardized uptake values from 0–90 min post tracer injection (SUV0–90) and volumes of distribution (VT) from kinetic modeling for different brain regions (n=3). F) Correlation (VT) and binding potentials with PET signal in assessed brain regions. G) left: Western blot analysis of post-mortem non-human primate brain specimens (n=5). Right: correlation of PET signal with Western blot analysis. Abbreviations: PCx, parietal cortex; PFCx, prefrontal cortex; MCx, motor cortex; OCx, occipital cortex; CCx, cingulate cortex; SCx, somatosensory cortex; TCx, temporal cortex; CPu, caudate/putamen; Th, thalamus; In, insula; Su, subiculum; GPd, globus pallidus; Pu, putamen; Cau, caudate; FWM, frontal white matter; Hp, hippocampus; Cb, cerebellum; Pon, pons; CC, corpus callosum; AUC, area under the curve; SUV, standardized uptake value. Dashed lines indicate 95% confidence intervals. * p<0.05, ** p<0.01, *** p<0.001.
The assessment of target engagement constitutes a fundamental component of early-stage drug discovery and development in neuroscience (42, 43). To test whether targeted visualization and quantitative assessment of drug-CYP46A1 interactions in the brain of non-human primates could be obtained with this PET tracer, we performed a dose-response experiment using soticlestat as a model compound. It should be noted that soticlestat and is currently under clinical evaluation (NCT04938427). Employing a broad array of soticlestat i.v. doses, ranging from 0.04 to 0.9 μmol/kg, a consistent dose-dependent PET signal attenuation was observed, which allowed target occupancy modeling with noteworthy model accuracy across different brain regions (Fig. 2D and Fig. S14). D50 values (dose that exhibits 50% target occupancy) were around 0.1 μmol/kg for the hippocampus, striatum (caudate/putamen) and prefrontal cortex, with a target occupancy > 90% achieved at the dose of 0.9 μmol/kg for all regions. These results demonstrate that 18F-CHL-2205 PET serves as a valuable tool to assess the extent to which CYP46A1 inhibitors engage with their target at a given dose regimen.
A major consideration of neuroimaging is that the PET image reflects a balance between multiple dynamic molecular processes that may include tracer delivery to the brain, binding to the desired target protein, potential metabolism or cellular internalization as well as washout from the brain. To assess the extent to which the PET signal reflected tracer binding to CYP46A1, kinetic modeling was performed and tissue volumes of distribution (VT) as well as non-displaceable binding potentials (BPND) were calculated from a two-tissue compartment model. VT constitutes the ratio of tracer concentrations in the target region vs. in the plasma at equilibrium, whereas BPND can be considered a quantitative index of in vivo target abundancy (44, 45). Employing kinetic modeling, we found that VT values across different brain regions revealed a comparable pattern to what was observed from PET images (Fig. 2E). These findings were further corroborated by a strong correlation between the PET signal and kinetic modeling parameters (PET vs. BPND, R2=0.885, p<0.001 and PET vs. VT, R2=0.896, p<0.001, Fig. 2F). Similarly, the PET signal correlated with the expression of CYP46A1, as assessed by Western blot analysis of post-mortem brain tissue (PET vs. Western blot analysis, R2=0.844, p<0.001, Fig 2G). These results suggest that PET imaging reflects the abundancy of CYP46A1 across different brain regions of non-human primates.
Quantitative assessment of cholesterol metabolism in the living human brain
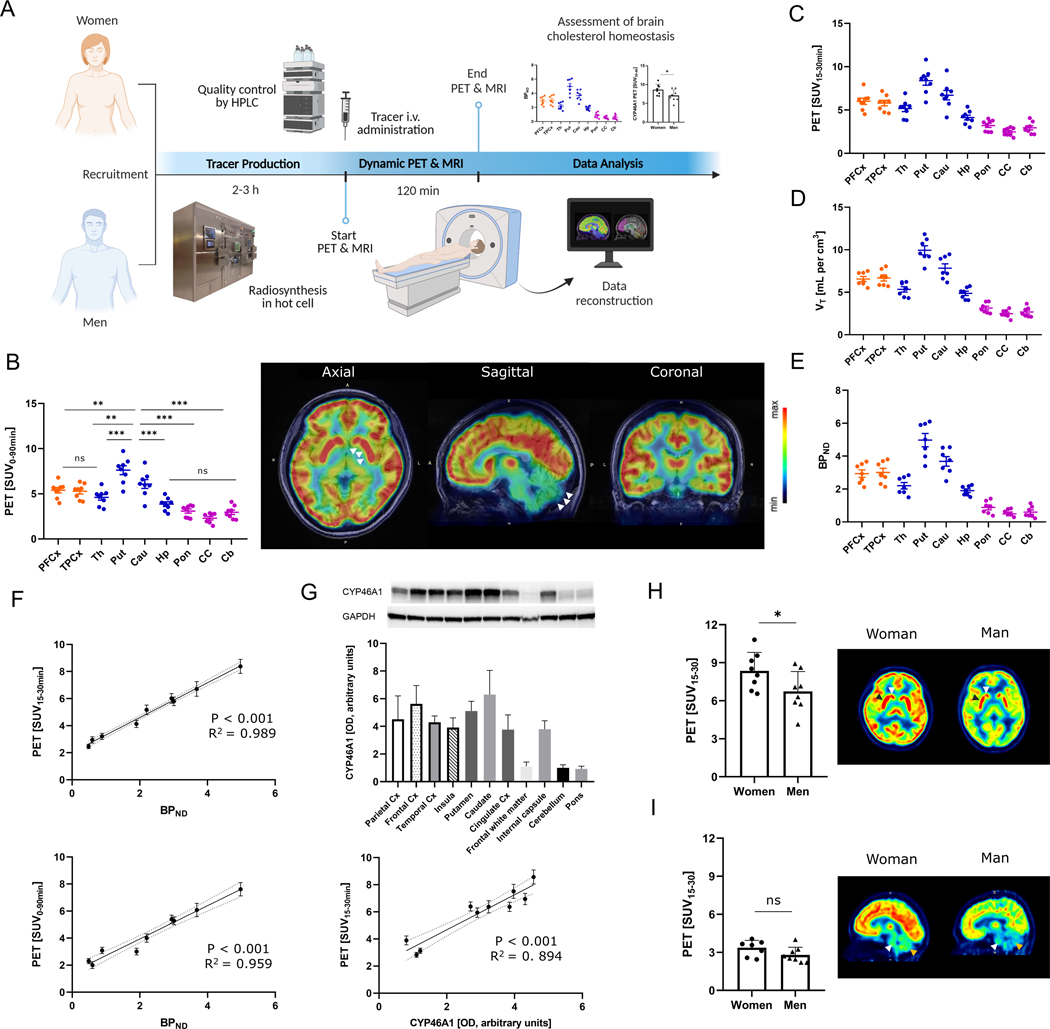
Eight healthy participants (n=4 each sex, female vs. male mean age, 24.5 ± 1.0 y vs. 27.5 ± 2.0 y, p=0.23) underwent a PET scan with 18F-CHL-2205, followed by a magnetic resonance imaging (MRI) scan for anatomical orientation. The study design is outlined in Fig. 3A. Due to the relatively short physical half-life of fluorine-18 (109.8 min), the tracer was produced on the day of the experiment in a designated hot cell that provided shielding from radiation exposure and was equipped with an automated synthesis module. PET scans were performed for a duration of 90 min and the data was subsequently reconstructed to allow the assessment of tracer uptake as a surrogate for CYP46A1 abundancy and cholesterol metabolism. Averaged standardized uptake values for a PET scan duration of 90 min (SUV0–90) revealed a heterogenous tracer accumulation pattern in the human brain, with areas of high radioactivity in the cortical, thalamic and basal ganglia regions (Fig. 3B). In sharp contrast, brain areas with limited CYP46A1 abundancy such as the cerebellum, brain stem and corpus callosum revealed the lowest tracer accumulation (putamen vs. cerebellum, 6.9 ± 0.7 vs. 2.5 ± 0.2, p<0.001; putamen vs. pons, 6.9 ± 0.7 vs. 2.9 ± 0.4, p<0.001; putamen vs. corpus callosum, 6.9 ± 0.7 vs. 2.2 ± 0.3, p<0.001, Fig. 3B). Hence, the distribution and in vivo binding of 18F-CHL-2205 in the human brain closely matched the binding patterns in the rodent and nonhuman primate brain. No adverse events were observed in any of the participants during the entire study.
Figure 3: Quantitative assessment of cholesterol metabolism in the living human brain.
A) Study design. B) Representative PET images of the human brain reflecting CYP46A1-rich brain regions, averaged from 0–90 min post tracer injection and presented in axial, sagittal and coronal view. CPu (CYP46A1-rich region on axial image) and Cb (CYP46A1-poor region on sagittal image) are highlighted with white arrows. Quantitative data is depicted as standardized uptake values (SUVs) from respective individual scans (n=8). C) Distribution of 18F-CHL-2205 in the human brain presented as standardized uptake values from 15 to 30 min post injection (SUV15–30) for the respective individual scans (n=8). D) Kinetic modeling assessment of tissue volumes of distribution (VT) for 18F-CHL-2205 in the human brain. E) Kinetic modeling assessment of non-displaceable binding potentials (BPND) for 18F-CHL-2205 in the human brain. For B to E, cortical regions are depicted in orange, whereas subcortical regions are shown in blue. Regions with low CYP46A1 expression are shown in purple. F) Correlation of PET signal averaged from 15–30 min (upper panel) and 0–90 min (lower panel) post tracer injection with non-displaceable binding potentials in selected brain regions. G) Western blot analysis of post-mortem human brain specimens. Correlation of PET signal with Western blot analysis in selected brain regions. H) Female vs. male brain cholesterol metabolism in the high CYP46A1 regions, caudate (n=4, white arrow) and putamen (n=4, black arrow), respectively. I) Female vs. male brain cholesterol metabolism in the low CYP46A1 regions, cerebellum (n=4, brown arrow) and brain stem (n=4, white arrow), respectively. Results are presented as standardized uptake values averaged from 15 to 30 min post tracer injection (SUV15–30). PFCx, prefrontal cortex; TPCx, temporal cortex; Th, thalamus; Pu, putamen; Cau, caudate; Hp, hippocampus; Cb, cerebellum; Pon, pons; CC, corpus callosum; AUC, area under the curve; SUV, standardized uptake value; PET, positron emission tomography; MRI, magnetic resonance imaging. Dashed lines indicate 95% confidence intervals. * p<0.05, ** p<0.01, *** p<0.001.
For patients undergoing PET imaging, scan duration of 90 min constitutes a major challenge because minor head movements may prompt detrimental effects on the image quality. To shorten the required scan duration in future studies, we next assessed whether averaged standardized uptake values from 15–30 min post tracer injection (SUV15–30) proved useful for quantitative PET. Indeed, a comparison between SUV15–30 (Fig. 3C) with regional tissue volumes of distribution (Fig. 3D) and binding potentials (Fig. 3E) revealed a virtually identical distribution pattern across a broad range of tested brain regions. The latter observation was further supported by the strong relationship between SUV15–30 and BPND (R2=0.989, p<0.001), which was comparable to that of SUV0–90 vs. BPND (R2=0.959, p<0.001, Fig. 3F). These findings indicated that targeted PET accurately predicted the bioavailability of CYP46A1 in humans, with a relatively short scan duration of 15 min.
To provide further evidence corroborating that quantitative PET constitutes an accurate surrogate measure of CYP46A1 tissue density in various brain regions, we performed Western blot analysis with post-mortem specimens of healthy human brains. As depicted in Fig. 3G, quantification of CYP46A1 by Western blot analysis revealed high target protein expression in the caudate, putamen and various cortical regions, whereas lowest CYP46A1 abundancy was found in the cerebellum, pons and white matter. The strong relationship observed between PET and Western blot analysis across various brain areas (R2=0.894, p<0.001, Fig. 3G) further indicates that the PET signal constitutes an accurate non-invasive measure of CYP46A1 protein density, providing a potential valuable tool to assess cholesterol metabolism in the living human brain.
Finally, we assessed whether CYP46A1-targeted PET was sensitive to sex differences in brain cholesterol metabolism. Women generally showed a higher brain PET signal, and the regional values of SUV15–30 in the caudate and putamen (CYP46A1-rich regions) were higher in women than in men (Fig. 3H). Conversely, there was no difference in tracer uptake for low target density regions such as the cerebellum and brain stem (Fig. 3I). These findings imply that considerable sex differences in brain cholesterol metabolism via CYP46A1 exist in humans.
Discussion
Our findings document the non-invasive in vivo quantification of brain abundancy and activity of CYP46A1, the primary enzyme responsible for clearance of cholesterol from the mammalian CNS. We developed a PET tracer, 18F-CHL-2205 (18F-Cholestify) for CYP46A1 and demonstrated that molecular imaging readouts correlate with CYP46A1 protein expression and the extent to which brain cholesterol is metabolized to 24-hydroxycholesterol, as assessed by post-mortem analyses of brain specimens from rodents, non-human primates and humans. Initial data indicate potential differences in CYP46A1-related tracer uptake between a mouse model of AD (3xTg-AD) and wild type animals. Finally, our findings point towards considerable differences in brain cholesterol metabolism via CYP46A1 between women and men.
The proper balance between cholesterol biosynthesis and clearance is a key feature of the healthy mammalian CNS. As such, a wealth of evidence generated over the past decades has linked impaired neuronal cholesterol homeostasis to several common neurological disorders including AD. In spite of the complexity by which cholesterol content seems to be regulated in the mammalian brain, CYP46A1 has been deemed critical to the brain cholesterol homeostasis due to its primary role in eliminating excess cholesterol from the CNS (9, 46, 47). Small-animal PET imaging allowed us to show an enhanced CYP46A1-related signal in the brains of 3xTg-AD mice, as compared to control animals. Consistent with this observation, several previous studies have reported on augmented concentrations of 24-hydroxycholesterol in the cerebrospinal fluid (CSF) of patients with mild cognitive impairments and early AD (48–52). Indeed, it is currently hypothesized that CYP46A1 function is enhanced in early AD, as an attempt to eliminate excess neuronal cholesterol and attenuate further accumulation of amyloidogenic cholesterol esters (19–25, 50). Further, it should be noted that patients with advanced AD tend to present with blunted concentrations of 24-hydroxycholesterol, potentially owing to the loss of neurons in CYP46A1-expressing brain areas (46). Although degeneration of neurons is less pronounced in mouse models of AD compared to patients (53), these observations raise concerns about the interpretation of CYP46A1 protein measurements in AD patients, particularly those with advanced disease stages. As such, future studies comprising CYP46A1-targeted PET in patients with AD must account for neuronal cell loss as a potential confounder.
To assess probe specificity, we employed validated CYP46A1 inhibitors such as voriconazole and soticlestat. In contrast to voriconazole and soticlestat, efavirenz was shown to trigger CYP46A1 activation at lower concentrations via allosteric modulation mechanism, potentially owing to a conformational change that affects the orthosteric binding site (54). Such conformational change may have affected tracer binding in unpredictable ways, thus rendering it challenging to gain insightful information from PET imaging in the presence of efavirenz. As such, we did not attempt to conduct PET studies in the presence of efavirenz and.
Notwithstanding the compelling evidence linking brain cholesterol dysregulation to neurological disease, the current tools for measuring brain cholesterol homeostasis present major shortcomings. These include the invasive nature of lumbar punctures to study CSF markers, the susceptibility to confounding factors for plasma markers (35), the limited information on affected brain areas, as well as the lack of clinically validated probes for non-invasive molecular imaging. Based on the extensive cross-species work demonstrating the specificity and selectivity of our PET tracer for CYP46A1, the tool for the assessment of CYP46A1 abundancy in humans developed here constitutes a major step forward and enables mechanistic studies on brain cholesterol metabolism in AD and other conditions. Our studies revealed a generally high tracer uptake in the brain, with persistently high uptake only in CYP46A1-rich areas. Although we found that most high expression regions were conserved across species, the hippocampus constituted a notable exception. This area was among the high-expression regions in rodents, whereas lower CYP46A1 expression was noted in non-human primates and humans. The underlying causes are currently unclear, however, we can speculate that attenuated CYP46A1 expression in higher species may ultimately render them more susceptible to excess cholesterol accumulation in hippocampal neurons, thereby contributing to an increased likelihood of spontaneous amyloidogenic processes (19–21). Consistent with this reasoning, positive allosteric modulation of CYP46A1 has recently been suggested as potential treatment approach in AD (55–57). Similarly, hippocampal overexpression of CYP46A1 via adeno-associated vector (AAV) therapy markedly reduced Aß40/42 peptides and Aß oligomers, improved spatial memory deficits and reversed microgliosis as well as astrocytosis in the APP23 transgenic model of AD (58). Despite the solid evidence pointing towards a protective role of CYP46A1 in AD, it remains to be elucidated whether CYP46A1 activation is a viable strategy for treating AD. The effectiveness of CYP46A1-targeted PET as a surrogate marker of cholesterol metabolism opens up the possibility that this probe could become a powerful imaging biomarker for impaired cholesterol homeostasis as well as for assessing response to therapy.
Sex differences in the biology of brain cholesterol have previously been reported. Indeed, several studies suggest that sex affects the AD risk conferred by apolipoprotein E4 (APOE4), which is involved in transporting cholesterol from astrocytes to neurons (59–62). Of note, female carriers of the APOE ε4 allele exhibit a higher risk of AD onset, or conversion from mild cognitive impairment (MCI) to early AD, than male carriers (59–62). Our findings now indicate that differences in cholesterol metabolism via CYP46A1 may further contribute to the observed sex disparities in brain cholesterol biology. However, studies assessing the link between APOE genotype and brain cholesterol metabolism in humans are scarce. The availability of a validated CYP46A1 PET tracer could allow such mechanistic studies in patients without the requirement for an invasive procedure. From an epidemiological point of view, a gender bias has previously been reported in patients with dementia. For instance, AD affects women twice as often as men (63). Further, women tend to convert faster from mild cognitive impairment (MCI) to AD (64). Nonetheless, molecular mechanisms contributing to these disparities are largely unexplored. Considering the emerging role of CYP46A1 as a potential therapeutic target in AD, it is conceivable that sex disparities in CYP46A1 abundancy may constitute a clinically relevant observation. The caveat is that the present findings were observed in healthy individuals and may not necessarily hold true in patients. Future studies in female and male patient populations will shed light on the extent to which CYP46A1 regulation contributes to the gender bias in AD. In addition to gender differences, age-dependent changes in CYP46A1 abundancy have previously been reported (5). By post-mortem analysis, the authors found that human CYP46A1 expression increases from infancy to childhood and remain relatively stable thereafter. Nonetheless, a systematic quantitative assessment of CYP46A1 abundancy in the aging brain is currently lacking. The availability of a non-invasive imaging tool to quantify CYP46A1 abundancy in the living human brain will offer the opportunity to address this question in adequately powered clinical studies.
There are limitations to this study that should be pointed out. Our clinical cohort of healthy individuals was relatively small – with only eight individuals in total. This is, in part, due to the challenge of recruiting healthy people without underlying medical condition to undergo diagnostic medical imaging that involves radiation exposure, notwithstanding the lack of therapeutic interventions. As such, the observed sex differences warrant confirmation in larger clinical studies involving patients. In addition, although the PET signal was reduced to virtually identical intensity between 3xTg-AD and control animals in the absence of free CYP46A1, suggesting that tracer uptake was dependent on available CYP46A1 binding sites, residual confounders that may have resulted from distinct tracer delivery to the brains of 3xTg-AD mice and respective controls were not assessed in the present study. Another limitation of our study is that no data was obtained on 24S-hydroxycholesterol concentrations in the cerebrospinal fluid. Such data would have been useful to further corroborate the utility of CYP46A1-targeted PET to assess brain cholesterol homeostasis in humans.
In conclusion, we believe that the tracer developed here will be of relevance to the neuroscience community where quantification of cholesterol metabolism with traditional approaches such as plasma concentrations of 24-hydroxycholesterol are not optimal. Although neurodegenerative disorders are perhaps the most compelling examples of potential future use of this tracer, it may further be harnessed to better define the role of cholesterol metabolism in other age- and sex-related brain disorders and improve our understanding of existing gender disparities with regard to various neuropathologies. In the era of precision medicine, the use of molecular imaging to track early response to targeted therapies may ultimately facilitate the search for long-sought disease-modifying therapies.
Materials and Methods
Study design
The aim of this study was to develop a translational molecular imaging tool for the assessment of cholesterol turnover in the brain. As such, we targeted CYP46A1 – a brain-specific hydroxylase that is primarily responsible for cholesterol clearance from the CNS – with a structurally optimized PET tracer. We characterized the tracer in vitro using brain tissues and performed in vivo studies in rodents, non-human primates and humans. All biological and animal experiments were performed in accordance with the Institutional Animal Care and Use Committee (IACUC) under protocols approved by the Massachusetts General Hospital or Emory University. Sample sizes for preclinical and clinical investigations were determined empirically, based on previous experience with CNS-targeted PET ligand development, and were indicated in the respective sections. Further, research objectives, subjects and experimental details of the clinical study were described in the respective experimental section. An overview of the clinical study design is provided in Fig. 3A. Clinical studies were conducted in adherence with Good Clinical Practice standards, as well as in accordance with the ethical standards of the 1964 Helsinki declaration and its later amendments. The clinical study was approved by the appropriate Institutional Review Board (IRB). No randomization or blinding was performed as all study participants underwent the same imaging process. Clinical study participants or their legal guardians were informed of the nature of the study and the potential risks involved prior to providing written consent.
Western blot experiments
Rat, non-human primate and human brain tissues were harvested and homogenized in freshly prepared RIPA buffer containing protease and phosphatase inhibitors (Thermo Scientific, A32961) and 1 mM PMSF protease inhibitor (Thermo Scientific, 36978). The supernatant was quantified using the (Bicinchoninic acid) BCA method and lysates were subjected to SDS-PAGE, electrophoretically transferred to PVDF membrane, blocked for 1 h in 5% BSA, and incubated overnight at 4°C with the primary CYP46A1 antibody or GAPDH antibody. Subsequently, the membranes were washed with tris-buffered saline with 0.1% Tween® 20 detergent (TBST) buffer and incubated with corresponding HRP-conjugated polyclonal secondary antibodies for 1 h at ambient temperature. Antibody complexes were visualized by enhanced chemiluminescence using ECL kits (BIO-RAD, 1705060 or 1705062S, USA). The antibodies used in this study include: CYP46A1 antibody (Millipore, MAB2259, 1:1000), GAPDH antibody (Abcam, ab181602, 1:10000), goat anti-rabbit secondary antibody (Invitrogen, A16096,1:1000) and goat anti-mouse secondary antibody (Abcam, ab6789, 1:5000)
In vitro autoradiography with brain tissues
Rodent and NHP post-mortem brain tissue was embedded in Tissue-Tek (O.C.T.). Sections of 10–20 μm thickness were prepared on a cryostat, mounted to glass slides and stored at −80 °C until further use. Prior to in vitro autoradiography experiments, brain sections were initially thawed for 10 min on ice and subsequently preconditioned for 10 min in the assay buffer (pH 7.4) containing 50 mM TRIS and 0.1–1% fatty acid free bovine serum albumin (BSA) at ambient temperature. Upon drying, the tissue sections were incubated with 1 mL of the respective 18F-CHL-2205 solution (0.5–2 nM) for 20–30 minutes at 25 °C. For CYP46A1-blockade, 1 μM of soticlestat was added to the radiotracer solution. The brain sections were washed for 2×2 min in the washing buffer (pH 7.4) containing 50 mM TRIS and further dipped twice in distilled water, subsequently dried and exposed to a phosphor imager plate for 60 minutes. The films were scanned and images were analyzed using ImageQuant TL 8.1.
Saturation binding experiments
To assess the binding affinity of 3H-CHL-2205 towards CYP46A1, radioligand binding assays using 3H-CHL-2205 and the nonradioactive reference were conducted. Briefly, rat or NHP brain membranes were homogenized and suspended in HEPES buffer (50 mM TRIS, 5 mM MgCl2, 0.1 mM EDTA, pH 7.4). Brain membranes (0.23 mg of protein) were incubated in triplicate at 35 °C and with concentrations of 3H-CHL-2205 ranging between 0.1 and 20.4 nM in a total volume of 0.25 mL of HEPES buffer. Nonspecific binding of 3H-CHL-2205 was estimated with 3 μM CHL-2205. After 60 min incubation at 30°C, the samples were filtered, and the filters containing the membranes with bound 3H-CHL-2205 were measured in a β-counter. Bound 3H-CHL-2205 (Bmax, fmol per mg protein) was fitted using GraphPad Prism 8.0.1.
Cell uptake studies
Cell uptake assays were conducted as previously reported (65). Human CYP46A1 Enyzme-HEK cell line (HEK-CYP46A1) and HEK control cells were cultured in DMEM (Gibco, 10566016) supplemented with 10% fetal bovine serum (Hyclone, SH30088.03) as well as antibiotics (0.1 mg/ml streptomycin and 100 IU/ml penicillin). Cells were kept under standard conditions at 37°C and 5% CO2, and subsequently subpassaged using a solution of 0.25% trypsin and 0.02% ethylenediaminetetraacetic acid (EDTA). For cell uptake measurement, HEK-CYP46A1 and HEK control cells were seeded into a 24-well plate at a density of 2×105 cells per well the day prior to the experiment. The medium was replaced with fresh DMEM medium including ca. 74 kBq/well of 18F-CHL-2205 and incubated at 37°C for 15, 30 and 45 min. The supernatant was collected, and the cells were washed twice with cold 1x PBS and then harvested by adding 200 μL of 1 N NaOH followed by additional two times rinsing with 1x PBS. The blocking assay was preformed similarly to the procedure described above with the HEK-CYP46A1 cells, except that the non-radioactive reference compound, soticlestat, was added in various concentrations. The incubation time was 1 h and the washing steps were only repeated once. Supernatant and cell suspensions including 1x PBS from each wash step were collected and then measured using a gamma counter, whereas cell uptake was expressed as the percentage of the added dose (%AD) after decay correction. The data points of cell uptake and blocking studies constitute averages of quadruplicate wells.
Sterol measurements
Sterol measurements were conducted as previously reported (66). Briefly, frozen tissue sections (10 μm thickness) on glass slides were dried in a vacuum desiccator, then a mixture of isotope-labeled standards was sprayed onto the tissue followed by cholesterol oxidase (0.264 units/mL) in 50 mM KH2PO4, pH7 (66). The samples were incubated at 37 °C for 1 h in a damp atmosphere, after which the slides were dried in a vacuum desiccator. [2H5]Girard P (GP) reagent (6.3 mg/mL of bromide salt) in 70% methanol, 5% acetic acid was sprayed on the dried tissue, which was then incubated in a methanol/water atmosphere at 37 °C for 1 h. The slides were removed and dried before analysis by liquid-extraction for surface analysis (LESA)-LC-MS (66).
PET imaging studies in naïve rodents and 3xTg-AD mice
All animal studies were conducted in accordance with the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines as well as the local animal protection law and were approved by the responsible authority. CD1, 3xTgAD mice and respective controls (sex: female, age: 9–13 months, Charles River Laboratories), as well as Sprague Dawley rats were purchased from established commercial vendors and kept under a 12-h light/12-h dark cycle, with ad libitum food and water. Animals were allowed to acclimatize for at least 1 week before the start of the experimental procedures. Mice and rats were anesthetized with isoflurane and scanned in a μPET/CT scanner (Sofie) for 60 min after tail-vein injection of 18F-CHL-2205. Data were reconstructed in user-defined time frames. Time–activity curves were calculated by PMOD, version 4.2 (PMOD Technologies), with predefined regions of interest as previously reported (67). Results are presented as area under the curve (AUC) from the respective time-activity curves (TACs) of standardized uptake values (SUVs), indicating the decay-corrected radioactivity per cm3, divided by the injected dose per gram body weight.
PET/CT and MRI imaging studies in rhesus monkeys
Animal preparation, scan acquisition, and image analysis used were performed as previously reported (68). T1-weighted MR anatomical images were acquired on a 3.0 T scanner. Four rhesus monkeys (4.8–9.3 kg) were anesthetized by intramuscular injection of ketamine (10 mg/kg), and then put on the scanning bed in the supine position. Whole-brain images were acquired with a 3D Bravo T1 sequence. For PET, the subjects were initially anesthetized with ketamine (10 mg/kg; i.m.), put on the scanning bed, and maintained under anesthesia with 2% isoflurane and 98% oxygen. All rhesus monkeys were supine, a stereotactic frame was used to fix the subject’s position of the head. A bolus intravenous injection of 18F-CHL-2205 (104–176 MBq, 0.38–1.49 μg) was performed into the monkey through an intravenous catheter, followed by a dynamic PET scan of the head for 90 min. For target occupancy studies, CYP46A1 inhibitor, soticlestat, was administered intravenously at the doses of either 0.0009, 0.0016, 0.014, 0.016, 0.05, 0.12, 0.32 and 0.34 mg/kg was injected, followed by the injection of 18F-CHL-2205. PET/CT and MR images were co-registered as previously validated.(69, 70). Time-activity curves (TACs) were derived from the respective volumes of interest (VOIs) and were presented as standardized uptake values (SUVs), which were decay-corrected to the time of radioligand injection. Whole-blood samples were collected from an arterial catheter to measure radioactivity in whole-blood and plasma. Indeed, samples were centrifuged, and 100 μL of whole blood and plasma was measured in a gamma counter at 10, 20, 30, 40, 50, 60, 75, 90, 105, 120, 150, 180, 210, 240, 600, 900, 1200, 1800, 2700, 3920, 4500 and 5400 seconds following 18F-CHL-2205 injection. Plasma radiometabolite analysis was performed by collecting additional arterial blood samples (1.0–2.5 mL) at 2, 10, 30, 60 and 90 min post injection. These samples were then centrifuged at 6500 rpm at 4 ºC for 5 min. Portions of the plasma (ca. 0.4 mL) were deproteinated using the same volume of ice-cold acetonitrile, vortexed, and separated by centrifugation for 3 min at 14500 rpm and 4 ºC. The supernatant was mixed with 0.02 mL of CH3CN in which the reference compound CHL-2205 (ca. 0.02 mg) was dissolved. Then, the mixture was injected into a HPLC system equipped with a semipreparative HPLC column (Phenomenex Luna 5 μm C18 100 Å, 250 × 10 mm) and a UV detector with wavelength set at 254 nm. The mobile phase consisted of acetonitrile/water (9/1, v/v) and the flow rate was 2.0 mL/min. The HPLC fractions were collected at 30 sec intervals for 13 min; each fraction was counted with automatic gamma well-counter (Wizard 2480, PerkinElmer). The results were corrected for background radiation and physical decay, whereas unmetabolized 18F-CHL-2205 parent fraction was determined as the ratio of the sum of radioactivity in fractions containing the parent (reference) compound to the total amount of radioactivity collected. Tissue volumes of distribution (VT) and non-displaceable binding potential (BPND) values were determined from a two-tissue compartment model using the PKIN tool in PMOD v.4.2 (PMOD Technologies Ltd.), with metabolite-corrected arterial input function.
Clinical study
An open-label clinical study to evaluate the tolerability, safety and tracer performance characteristics of 18F-CHL-2205 in healthy volunteers (4 men and 4 women, 22 to 31 years of age) was conducted (Fig 3A). Prespecified outcomes of this study included tracer kinetics and distribution volumes of 18F-CHL-2205 in the brain, adverse events for up to 10 d following intravenous bolus administration of the tracer, as well as time-activity curves and binding potentials in prespecified brain regions. All subjects underwent a dynamic 90 min PET and MRI scan following injection of 18F-CHL-2205 (173.9 to 305.8 MBq, 1.57 to 3.63 μg). Individual MRI images were used for anatomical orientation and to delineate volumes of interest. Vital sign and ECG records were taken just before and after the scans. Two-tissue compartment model analyses were conducted using image-derived input functions from the carotid arteries, which provided excellent model fits. Volumes of distribution were further determined by logan graphical analysis and were used to calculate binding potentials. One individual was excluded from kinetic modeling due to limited image quality for the carotid artery. PET data is presented as standardized uptake values, averaged from 0–90 min (SUV0–90) or from 15–30 min (SUV15–30) post injection. SUV data was compared to binding potentials, as well as to regional-dependent CYP46A1 protein expression (Western blot quantification) in brain tissue specimens from previously deceased individuals, which were obtained from the Massachusetts Alzheimer’s Disease Research Center (MADRC).
Statistical analyses
Continuous variables were presented as mean ± standard error of the mean (SEM). Student’s t test and analysis of variance (ANOVA) tests were used for group comparisons of continuous variables as appropriate. For multiple comparisons, Bonferroni correction was employed. Strength and direction of associations were assessed by Spearman’s rank-order correlation (rs). Linear regressions were performed as appropriate. A two-tailed P value of < 0.05 was considered statistically significant and statistical analyses were carried out with (GraphPad Prism version 8.2.1).
Supplementary Material
Acknowledgments:
We thank the staff of the Division of Nuclear Medicine and Molecular Imaging, MGH and Harvard Medical School, and members of the Emory PET Imaging Center & Radiopharmaceutical Discovery Program, Department of Radiology and Imaging Sciences, Emory University, for their expert assistance. We also thank the National Institute of Mental Health’s Psychoactive Drug Screening Program (NIMH PDSP; directed by Bryan L. Roth at the University of North Carolina at Chapel Hill and Jamie Driscoll at NIMH, USA), Emory School of Medicine Endowed Directorship (to SHL), and Gifford Bioscience, Ltd (Dr. Andrew Gifford) for pharmacology screening. For the purpose of open access, the authors have applied a CC BY public copyright license to any Author Accepted Manuscript version arising.
Funding:
We gratefully acknowledge the financial support from the following funding agencies: NIH (AA028007 to JBD, AG070060 and AG074218 to SHL); BBSRC (BB/S019588/1 and BB/N015932/1 to WJG, BB/L001942/1 to YW), and the European Union, through European Structural Funds (ESF), as part of the Welsh Government funded Academic Expertise for Business project (to WJG and YW); the Swiss National Science Foundation to AH, and Emory Radiology Chair Fund to SHL. We thank David P. Friedman for his support. Further, we thank the Massachusetts Alzheimer’s Disease Research Center (MADRC, grant nr. 1P30AG062421-01 to DHO) for providing postmortem human tissues.
Footnotes
Competing interests: The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: WJG and YW are listed as inventors on the patent “Kit and method for quantitative detection of steroids” US9851368B2. WJG, EY and YW are shareholders in CholesteniX Ltd. SHL, AH and CZ are listed as inventors on the provisional patent application “Novel PET ligands for imaging cholesterol homeostasis” (application number 63/397,463).
Data and materials availability:
All data associated with this study, which were not part of the main manuscript, are available in the Supplementary Material.
References
- 1.Dietschy JM, Turley SD, Cholesterol metabolism in the brain. Curr Opin Lipidol 12, 105–112 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Di Paolo G, Kim T-W, Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nature Reviews Neuroscience 12, 284–296 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mauch DH, Nägler K, Schumacher S, Göritz C, Müller EC, Otto A, Pfrieger FW, CNS synaptogenesis promoted by glia-derived cholesterol. Science 294, 1354–1357 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Barres BA, Smith SJ, Neurobiology. Cholesterol--making or breaking the synapse. Science 294, 1296–1297 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Lund EG, Guileyardo JM, Russell DW, cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proc Natl Acad Sci U S A 96, 7238–7243 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lütjohann D, Breuer O, Ahlborg G, Nennesmo I, Sidén A, Diczfalusy U, Björkhem I, Cholesterol homeostasis in human brain: evidence for an age-dependent flux of 24S-hydroxycholesterol from the brain into the circulation. Proc Natl Acad Sci U S A 93, 9799–9804 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mast N, White MA, Bjorkhem I, Johnson EF, Stout CD, Pikuleva IA, Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain. Proc Natl Acad Sci U S A 105, 9546–9551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lund EG, Guileyardo JM, Russell DW, cDNA cloning of cholesterol 24-hydroxylase, a mediator of cholesterol homeostasis in the brain. Proceedings of the National Academy of Sciences of the United States of America 96, 7238–7243 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Björkhem I, Lütjohann D, Diczfalusy U, Ståhle L, Ahlborg G, Wahren J, Cholesterol homeostasis in human brain: turnover of 24S-hydroxycholesterol and evidence for a cerebral origin of most of this oxysterol in the circulation. J Lipid Res 39, 1594–1600 (1998). [PubMed] [Google Scholar]
- 10.Karasinska JM, Hayden MR, Cholesterol metabolism in Huntington disease. Nature Reviews Neurology 7, 561–572 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Katsuno M, Adachi H, Sobue G, Getting a handle on Huntington’s disease: the case for cholesterol. Nature Medicine 15, 253–254 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Djelti F, Braudeau J, Hudry E, Dhenain M, Varin J, Bièche I, Marquer C, Chali F, Ayciriex S, Auzeil N, Alves S, Langui D, Potier M-C, Laprevote O, Vidaud M, Duyckaerts C, Miles R, Aubourg P, Cartier N, CYP46A1 inhibition, brain cholesterol accumulation and neurodegeneration pave the way for Alzheimer’s disease. Brain 138, 2383–2398 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Kotti Tiina J, Ramirez Denise MO, Pfeiffer Brad E, Huber Kimberly M, Russell David W, Brain cholesterol turnover required for geranylgeraniol production and learning in mice. PNAS 103, 3869–3874 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boussicault L, Alves S, Lamazière A, Planques A, Heck N, Moumné L, Despres G, Bolte S, Hu A, Pagès C, Galvan L, Piguet F, Aubourg P, Cartier N, Caboche J, Betuing S, CYP46A1, the rate-limiting enzyme for cholesterol degradation, is neuroprotective in Huntington’s disease. Brain 139, 953–970 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreilaus F, Spiro AS, McLean CA, Garner B, Jenner AM, Evidence for altered cholesterol metabolism in Huntington’s disease post mortem brain tissue. Neuropathol Appl Neurobiol 42, 535–546 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kacher R, Lamazière A, Heck N, Kappes V, Mounier C, Despres G, Dembitskaya Y, Perrin E, Christaller W, Sasidharan Nair S, Messent V, Cartier N, Vanhoutte P, Venance L, Saudou F, Néri C, Caboche J, Betuing S, CYP46A1 gene therapy deciphers the role of brain cholesterol metabolism in Huntington’s disease. Brain 142, 2432–2450 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Kölsch H, Lütjohann D, Ludwig M, Schulte A, Ptok U, Jessen F, von Bergmann K, Rao ML, Maier W, Heun R, Polymorphism in the cholesterol 24S-hydroxylase gene is associated with Alzheimer’s disease. Molecular Psychiatry 7, 899–902 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Habchi J, Chia S, Galvagnion C, Michaels TCT, Bellaiche MMJ, Ruggeri FS, Sanguanini M, Idini I, Kumita JR, Sparr E, Linse S, Dobson CM, Knowles TPJ, Vendruscolo M, Cholesterol catalyses Aβ42 aggregation through a heterogeneous nucleation pathway in the presence of lipid membranes. Nature Chemistry 10, 673–683 (2018). [DOI] [PubMed] [Google Scholar]
- 19.van der Kant R, Langness VF, Herrera CM, Williams DA, Fong LK, Leestemaker Y, Steenvoorden E, Rynearson KD, Brouwers JF, Helms JB, Ovaa H, Giera M, Wagner SL, Bang AG, Goldstein LSB, Cholesterol Metabolism Is a Druggable Axis that Independently Regulates Tau and Amyloid-β in iPSC-Derived Alzheimer’s Disease Neurons. Cell Stem Cell 24, 363–375.e369 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons M, Keller P, De Strooper B, Beyreuther K, Dotti CG, Simons K, Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc Natl Acad Sci U S A 95, 6460–6464 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puglielli L, Konopka G, Pack-Chung E, Ingano LA, Berezovska O, Hyman BT, Chang TY, Tanzi RE, Kovacs DM, Acyl-coenzyme A: cholesterol acyltransferase modulates the generation of the amyloid beta-peptide. Nat Cell Biol 3, 905–912 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Chan RB, Oliveira TG, Cortes EP, Honig LS, Duff KE, Small SA, Wenk MR, Shui G, Di Paolo G, Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J Biol Chem 287, 2678–2688 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutter-Paier B, Huttunen HJ, Puglielli L, Eckman CB, Kim DY, Hofmeister A, Moir RD, Domnitz SB, Frosch MP, Windisch M, Kovacs DM, The ACAT inhibitor CP-113,818 markedly reduces amyloid pathology in a mouse model of Alzheimer’s disease. Neuron 44, 227–238 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR, The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science 336, 1168–1171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Kulas JA, Wang C, Holtzman DM, Ferris HA, Hansen SB, Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proceedings of the National Academy of Sciences 118, e2102191118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamba P, Giannelli S, Staurenghi E, Testa G, Sottero B, Biasi F, Poli G, Leonarduzzi G, The Controversial Role of 24-S-Hydroxycholesterol in Alzheimer’s Disease. Antioxidants (Basel) 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuliani G, Donnorso MP, Bosi C, Passaro A, Dalla Nora E, Zurlo A, Bonetti F, Mozzi AF, Cortese C, Plasma 24S-hydroxycholesterol levels in elderly subjects with late onset Alzheimer’s disease or vascular dementia: a case-control study. BMC Neurol 11, 121–121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa AC, Joaquim HPG, Nunes VS, Kerr DS, Ferreira GS, Forlenza OV, Gattaz WF, Talib LL, Donepezil effects on cholesterol and oxysterol plasma levels of Alzheimer’s disease patients. European Archives of Psychiatry and Clinical Neuroscience 268, 501–507 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Leoni V, Long JD, Mills JA, Di Donato S, Paulsen JS, Plasma 24S-hydroxycholesterol correlation with markers of Huntington disease progression. Neurobiol Dis 55, 37–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grayaa S, Zerbinati C, Messedi M, HadjKacem I, Chtourou M, Ben Touhemi D, Naifar M, Ayadi H, Ayedi F, Iuliano L, Plasma oxysterol profiling in children reveals 24-hydroxycholesterol as a potential marker for Autism Spectrum Disorders. Biochimie 153, 80–85 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Hanin A, Baudin P, Demeret S, Roussel D, Lecas S, Teyssou E, Damiano M, Luis D, Lambrecq V, Frazzini V, Decavèle M, Plu I, Bonnefont-Rousselot D, Bittar R, Lamari F, Navarro V, Disturbances of brain cholesterol metabolism: A new excitotoxic process associated with status epilepticus. Neurobiology of Disease 154, 105346 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Cenik B, Cenik C, Snyder MP, Brown ES, Plasma sterols and depressive symptom severity in a population-based cohort. PLoS One 12, e0184382-e0184382 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Björkhem I, Patra K, Boxer AL, Svenningsson P, 24S-Hydroxycholesterol Correlates With Tau and Is Increased in Cerebrospinal Fluid in Parkinson’s Disease and Corticobasal Syndrome. Front Neurol 9, 756–756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bretillon L, Lütjohann D, Ståhle L, Widhe T, Bindl L, Eggertsen G, Diczfalusy U, Björkhem I, Plasma levels of 24S-hydroxycholesterol reflect the balance between cerebral production and hepatic metabolism and are inversely related to body surface. Journal of Lipid Research 41, 840–845 (2000). [PubMed] [Google Scholar]
- 35.Norlin M, Toll A, Björkhem I, Wikvall K, 24-hydroxycholesterol is a substrate for hepatic cholesterol 7alpha-hydroxylase (CYP7A). J Lipid Res 41, 1629–1639 (2000). [PubMed] [Google Scholar]
- 36.Allen Institute for Brain Science (2004). Allen Mouse Brain Atlas [dataset]. Available from mouse.brain-map.org.Allen Institute for Brain Science, (2021).
- 37.Mast N, Charvet C, Pikuleva IA, Stout CD, Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain. J Biol Chem 285, 31783–31795 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishi T, Kondo S, Miyamoto M, Watanabe S, Hasegawa S, Kondo S, Yano J, Watanabe E, Ishi T, Yoshikawa M, Ando HK, Farnaby W, Fujimoto S, Sunahara E, Ohori M, During MJ, Kuroita T, Koike T, Soticlestat, a novel cholesterol 24-hydroxylase inhibitor shows a therapeutic potential for neural hyperexcitation in mice. Scientific Reports 10, 17081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yutuc E, Angelini R, Baumert M, Mast N, Pikuleva I, Newton J, Clench MR, Skibinski DOF, Howell OW, Wang Y, Griffiths WJ, Localization of sterols and oxysterols in mouse brain reveals distinct spatial cholesterol metabolism. Proceedings of the National Academy of Sciences 117, 5749–5760 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billings LM, Oddo S, Green KN, McGaugh JL, LaFerla FM, Intraneuronal Abeta causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice. Neuron 45, 675–688 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Sterniczuk R, Antle MC, Laferla FM, Dyck RH, Characterization of the 3xTg-AD mouse model of Alzheimer’s disease: part 2. Behavioral and cognitive changes. Brain Res 1348, 149–155 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Willmann JK, van Bruggen N, Dinkelborg LM, Gambhir SS, Molecular imaging in drug development. Nature Reviews Drug Discovery 7, 591–607 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Rudin M, Weissleder R, Molecular imaging in drug discovery and development. Nature Reviews Drug Discovery 2, 123–131 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang SC, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE, Consensus nomenclature for in vivo imaging of reversibly binding radioligands. J Cereb Blood Flow Metab 27, 1533–1539 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS, Alexoff DL, Distribution volume ratios without blood sampling from graphical analysis of PET data. J Cereb Blood Flow Metab 16, 834–840 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Petrov AM, Pikuleva IA, Cholesterol 24-Hydroxylation by CYP46A1: Benefits of Modulation for Brain Diseases. Neurotherapeutics 16, 635–648 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lund EG, Xie C, Kotti T, Turley SD, Dietschy JM, Russell DW, Knockout of the cholesterol 24-hydroxylase gene in mice reveals a brain-specific mechanism of cholesterol turnover. J Biol Chem 278, 22980–22988 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Popp J, Meichsner S, Kölsch H, Lewczuk P, Maier W, Kornhuber J, Jessen F, Lütjohann D, Cerebral and extracerebral cholesterol metabolism and CSF markers of Alzheimer’s disease. Biochemical Pharmacology 86, 37–42 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Shafaati M, Solomon A, Kivipelto M, Björkhem I, Leoni V, Levels of ApoE in cerebrospinal fluid are correlated with Tau and 24S-hydroxycholesterol in patients with cognitive disorders. Neuroscience Letters 425, 78–82 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Papassotiropoulos A, Lütjohann D, Bagli M, Locatelli S, Jessen F, Buschfort R, Ptok U, Björkhem I, von Bergmann K, Heun R, 24S-hydroxycholesterol in cerebrospinal fluid is elevated in early stages of dementia. Journal of Psychiatric Research 36, 27–32 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Leoni V, Shafaati M, Salomon A, Kivipelto M, Björkhem I, Wahlund L-O, Are the CSF levels of 24S-hydroxycholesterol a sensitive biomarker for mild cognitive impairment? Neuroscience Letters 397, 83–87 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Besga A, Cedazo-Minguez A, Kåreholt I, Solomon A, Björkhem I, Winblad B, Leoni V, Hooshmand B, Spulber G, Gonzalez-Pinto A, Kivipelto M, Wahlund LO, Differences in brain cholesterol metabolism and insulin in two subgroups of patients with different CSF biomarkers but similar white matter lesions suggest different pathogenic mechanisms. Neuroscience Letters 510, 121–126 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Wirths O, Bayer TA, Neuron loss in transgenic mouse models of Alzheimer’s disease. Int J Alzheimers Dis 2010, 723782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anderson KW, Mast N, Hudgens JW, Lin JB, Turko IV, Pikuleva IA, Mapping of the Allosteric Site in Cholesterol Hydroxylase CYP46A1 for Efavirenz, a Drug That Stimulates Enzyme Activity. J Biol Chem 291, 11876–11886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pikuleva IA, Efavirenz as a cholesterol-targeting drug for AD. Alzheimer’s & Dementia 16, e040603 (2020). [Google Scholar]
- 56.Mast N, El-Darzi N, Petrov AM, Li Y, Pikuleva IA, CYP46A1-dependent and independent effects of efavirenz treatment. Brain Communications 2, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pikuleva IA, Targeting cytochrome P450 46A1 and brain cholesterol 24-hydroxylation to treat neurodegenerative diseases. Explor Neuroprotective Ther 1, 159–172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hudry E, Van Dam D, Kulik W, De Deyn PP, Stet FS, Ahouansou O, Benraiss A, Delacourte A, Bougnères P, Aubourg P, Cartier N, Adeno-associated virus gene therapy with cholesterol 24-hydroxylase reduces the amyloid pathology before or after the onset of amyloid plaques in mouse models of Alzheimer’s disease. Mol Ther 18, 44–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM, Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. Jama 278, 1349–1356 (1997). [PubMed] [Google Scholar]
- 60.Altmann A, Tian L, Henderson VW, Greicius MD, Sex modifies the APOE-related risk of developing Alzheimer disease. Ann Neurol 75, 563–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S, Kim MJ, Kim S, Kang HS, Lim SW, Myung W, Lee Y, Hong CH, Choi SH, Na DL, Seo SW, Ku BD, Kim SY, Kim SY, Jeong JH, Park SA, Carroll BJ, Kim DK, Gender differences in risk factors for transition from mild cognitive impairment to Alzheimer’s disease: A CREDOS study. Compr Psychiatry 62, 114–122 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Neu SC, Pa J, Kukull W, Beekly D, Kuzma A, Gangadharan P, Wang LS, Romero K, Arneric SP, Redolfi A, Orlandi D, Frisoni GB, Au R, Devine S, Auerbach S, Espinosa A, Boada M, Ruiz A, Johnson SC, Koscik R, Wang JJ, Hsu WC, Chen YL, Toga AW, Apolipoprotein E Genotype and Sex Risk Factors for Alzheimer Disease: A Meta-analysis. JAMA Neurol 74, 1178–1189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livingston G, Huntley J, Sommerlad A, Ames D, Ballard C, Banerjee S, Brayne C, Burns A, Cohen-Mansfield J, Cooper C, Costafreda SG, Dias A, Fox N, Gitlin LN, Howard R, Kales HC, Kivimäki M, Larson EB, Ogunniyi A, Orgeta V, Ritchie K, Rockwood K, Sampson EL, Samus Q, Schneider LS, Selbæk G, Teri L, Mukadam N, Dementia prevention, intervention, and care: 2020 report of the Lancet Commission. Lancet 396, 413–446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin KA, Choudhury KR, Rathakrishnan BG, Marks DM, Petrella JR, Doraiswamy PM, Marked gender differences in progression of mild cognitive impairment over 8 years. Alzheimers Dement (N Y) 1, 103–110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haider A, Xiao Z, Xia X, Chen J, Van RS, Kuang S, Zhao C, Rong J, Shao T, Ramesh P, Aravind A, Shao Y, Ran C, Young LJ, Liang SH, Development of a triazolobenzodiazepine-based PET probe for subtype-selective vasopressin 1A receptor imaging. Pharmacol Res 173, 105886 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yutuc E, Angelini R, Baumert M, Mast N, Pikuleva I, Newton J, Clench MR, Skibinski DOF, Howell OW, Wang Y, Griffiths WJ, Localization of sterols and oxysterols in mouse brain reveals distinct spatial cholesterol metabolism. Proc Natl Acad Sci U S A 117, 5749–5760 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haider A, Herde AM, Krämer SD, Varisco J, Keller C, Frauenknecht K, Auberson YP, Temme L, Robaa D, Sippl W, Preclinical evaluation of benzazepine-based PET radioligands (R)-and (S)-11C-Me-NB1 reveals distinct enantiomeric binding patterns and a tightrope walk between GluN2B-and σ1-receptor–targeted PET imaging. Journal of Nuclear Medicine 60, 1167–1173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao Z, Wei H, Xu Y, Haider A, Wei J, Yuan S, Rong J, Zhao C, Li G, Zhang W, Chen H, Li Y, Zhang L, Sun J, Zhang S, Luo H-B, Yan S, Cai Q, Hou L, Che C, Liang SH, Wang L, Discovery of a highly specific 18F-labeled PET ligand for phosphodiesterase 10A enabled by novel spirocyclic iodonium ylide radiofluorination. Acta Pharmaceutica Sinica B 12, 1963–1975 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McLaren DG, Kosmatka KJ, Oakes TR, Kroenke CD, Kohama SG, Matochik JA, Ingram DK, Johnson SC, A population-average MRI-based atlas collection of the rhesus macaque. NeuroImage 45, 52–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nie B, Wang L, Hu Y, Liang S, Tan Z, Chai P, Tang Y, Shang J, Pan Z, Zhao X, Zhang X, Gong J, Zheng C, Xu H, Wey H-Y, Liang SH, Shan B, A population stereotaxic positron emission tomography brain template for the macaque and its application to ischemic model. NeuroImage 203, 116163 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study, which were not part of the main manuscript, are available in the Supplementary Material.