Abstract
We reveal the fundamental understanding of molecular doping of DNAs into organic semiconducting tris (8-hydroxyquinoline) aluminum (Alq3) crystals by varying types and numbers of purines and pyrimidines constituting DNA. Electrostatic, hydrogen bonding, and π-π stacking interactions between Alq3 and DNAs are the major factors affecting the molecular doping. Longer DNAs induce a higher degree of doping due to electrostatic interactions between phosphate backbone and Alq3. Among four bases, single thymine bases induce the multisite interactions of π-π stacking and hydrogen bonding with single Alq3, occurring within a probability of 4.37%. In contrast, single adenine bases form multisite interactions, within lower probability (1.93%), with two-neighboring Alq3. These multisite interactions facilitate the molecular doping into Alq3 particles compared to cytosines or guanines only forming π-π stacking. Thus, photoluminescence and optical waveguide phenomena of crystals were successfully tailored. This discovery should deepen our fundamental understanding of incorporating DNAs into organic semiconducting crystals.
Subject terms: Bioinspired materials, DNA and RNA
DNAs exhibit a unique molecular doping structure, unseen in conventional hybrid assemblies, when doped into light-emitting organic materials. Here authors unveil doping mechanism of DNAs into organic semiconducting crystals and tailor their photoluminescence and optical waveguide phenomena.
Introduction
Since their inception as a typical genetic information carrier, nucleic acids have become a member of the material field and are widely used1–4. The unique physical and chemical properties make nucleic acid-associated materials the focus of numerous studies. For example, a nucleic acid molecule is generally complexed with π-conjugated organic semiconductors and serves as (i) an efficient receptor element for recognizing biological/chemical targets5–7, (ii) a template for the assembly and polymerization of organic semiconductors8–10, (iii) a walking component in a light-driven artificial nanomachine11,12, (iv) a wide-bandgap material in organic light-emitting diodes enhancing their luminescence efficiency2,13,14, (v) a molecular gadget for tuning organic semiconductor crystals bio-active when properly hybridized15, and (vi) a biological moiety of organic hybrid crystals for remote sensing via optical waveguide effects16.
Hybrid assemblies have become important in the field of self-assembly17,18. Binary or ternary hybrid assemblies have been prepared through molecular doping between organic semiconducting components19, involving noncovalent intermolecular interactions, such as van der Waals force, π-π stacking, and hydrogen bonding20–22. The forms of hybrid assemblies can be classified into hetero structures23,24 and uniform-25,26 or gradient-doped27 structures. However, deoxyribonucleic acids (DNAs) doped into light-emitting organic crystals exhibit distinctly different structures of molecular doping that has been unseen in conventional hybrid assemblies3,15. To date, studies have focused on the application of DNA-hybrid assemblies; however, little attention has been paid to how these nucleic acids interface with organic components at the molecular level. A fundamental understanding of the intermolecular interactions between nucleic acids and light-emitting organic molecules can lead to the rational design of these hybrid assemblies, playing a key role in the further development of biophotonics.
This study proposes an interpretation of the molecular doping of oligonucleotide DNA molecules in light-emitting crystals at the molecular level. A typical light-emitting organic material, tris (8-hydroxyquinoline) aluminum (Alq3), was chosen as the model molecule. Alq3 crystals doped with nucleic acid molecules were assembled through a simple reprecipitation method by varying the length of DNA chains and base moieties type. The intermolecular interfacing behavior between nucleic acids and Alq3 is interpreted in terms of electrostatic, hydrogen bonding, and π-π stacking interactions.
Results
Non-uniform molecular distribution of DNA in light-emitting organic particles
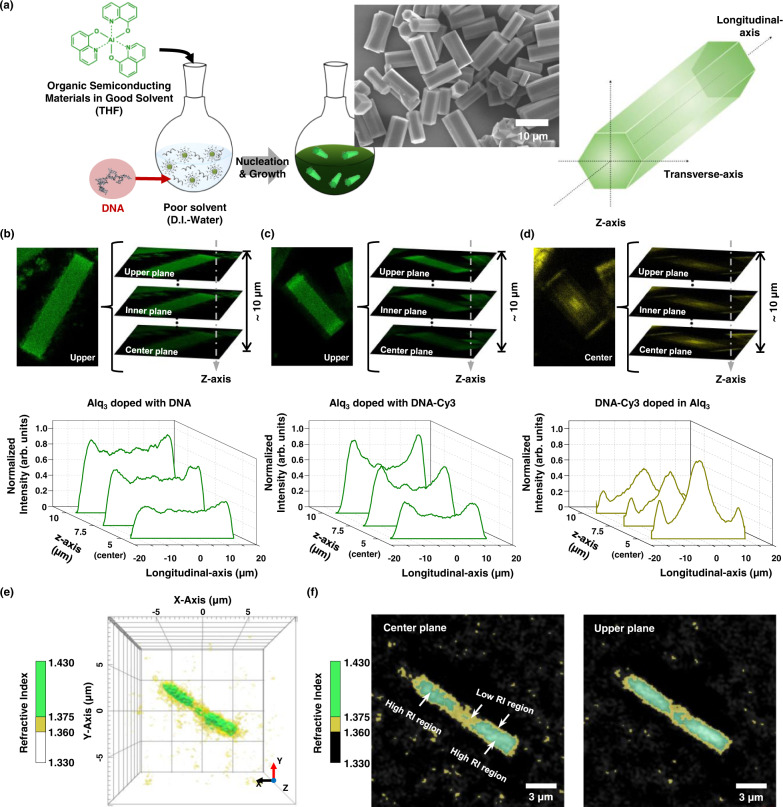
Figure 1a presents a schematic of the bio-functional Alq3 microparticles doped with DNA and illustrates the axial direction of the hexagonal particles. We used confocal laser scanning microscopy (CLSM) to quantify the internal fluorescence distribution of Alq3 and DNA molecules in particles. The Alq3 and DNA molecules, labeled with Cy3 fluorescent dye, are excited using a 405 and 555 nm laser, respectively. The main PL peaks of the Alq3 particles and Cy3 dyes are observed at ~512 and ~572 nm (Supplementary Fig. 1a), in agreement with the known characteristics28. Additionally, the PL spectrum of Alq3 particles overlaps with the absorption spectrum of DNA-Cy3 molecules. Thus, DNA molecules labeled with Cy3 generate Förster resonance energy transfer (FRET) effects from Alq3 to a fluorescent dye with a FRET efficiency of 7.9% and slightly lower the emission intensity of Alq3 molecules compared to Alq3 doped with DNA (Supplementary Fig. 1b). Examples of confocal technical data related to individual z-stack images, three-dimensional (3D) rendered images, and average profiling analysis in the longitudinal direction of Alq3 doped with anthrax lethal factor 27-mer DNA, Alq3 doped with DNA-Cy3, and DNA-Cy3 molecules are provided in Fig. 1b, c, and d, respectively. The Alq3 molecules are distributed into 20 μm particles, while the DNA molecules have a strikingly unique distribution in these images. The Alq3 molecules have higher intensity in the upper plane than in the particles’ central plane (10 μm inner plane). The fluorescence intensity of the Alq3 molecules is uniform along the longitudinal axis of particles doped with DNA (Fig. 1b) while that of the sides of particles is slightly higher due to the optical waveguide effects of crystals toward the edges16,29,30. The fluorescence intensity of the Alq3 molecules on the center of particles doped with DNA-Cy3 becomes lower (Fig. 1c). Thus, the difference between the fluorescence intensities on the center and sides of particles doped with DNA-Cy3 increases compared to Alq3 particles doped with DNA. The fluorescence of the DNA-Cy3 molecules is most intense at the center of the particles, and the central plane has a higher intensity than the upper plane (Fig. 1d). It indicates that the DNA molecules, concentrated at the center, generate FRET from Alq3 to the Cy3 fluorescent dye. Besides via the fluorescence distribution, we further confirmed the molecular distribution of doped-DNA molecules in Alq3 particles via the refractive index (RI) distribution. As the RI of the Alq3 is larger than the RI of the DNA3, the distribution of DNA molecules in the Alq3 particles can be visualized through the holotomographic microscopy observing the difference in RI. In the reconstructed 3D RI distributions of DNA doped Alq3 particle (Fig. 1e), yellow and green surfaces represent, respectively, low RI regions ranging from 1.330 to 1.375 and high RI regions ranging from 1.375 to 1.430. The yellow region distributes at the surfaces and center, compared to the rest green region, indicating that DNA molecules with a low RI exist mainly on the surfaces and in the center of the particle. We then obtained cross-sectional images of corresponding 3D RI distribution at the center and upper planes along the z-axis (Fig. 1f). The low RI regions in the particle (yellow-colored) exist at the center region of the particle at both planes; however, the yellow region becomes larger at the center plane, indicating that the DNA molecules are more concentrated, which is consistent with the fluorescence distribution of DNA molecules in Alq3 particles given in Fig. 1d. These results suggest that the DNA molecules are nonuniformly doped into the Alq3 particles.
Fig. 1. DNA as a molecular dopant in light-emitting organic crystals.
a Schematic illustration of the fabrication of DNA-doped Alq3 microparticles (left) and definition of coordination of hexagonal microrods (right). Z-stack confocal laser scanning microscopy images (top) and longitudinal normalized fluorescence profiles averaged from five separate particles from the upper to the center plane (bottom) of (b) DNA doped Alq3 molecules, (c) DNA-Cy3 doped Alq3 molecules, and (d) DNA-Cy3 molecules in a particle. The single-stranded DNA sequence for fabricating the particles is 5′-ATC CTT ATC AAT ATT TAA CAA TAA TCC-3′. The images and profiles of Alq3 molecules are obtained by excitation using a 405 nm laser; a 300‒550 nm filter is used. The images of the DNA-Cy3 molecules are obtained by excitation with a 555 nm laser; a 300‒630 nm filter is used. The fluorescence of DNA-Cy3 molecules is enhanced by 3-fold for clarity. The fluorescence profiles of Alq3 and DNA-Cy3 molecules are normalized by the intensity of Alq3 from sides of particles at upper plane and the intensity of DNA-Cy3 from center of particles at center plane, respectively. e 3D refractive index (RI) distribution of the DNA-doped Alq3 particle. The low RI regions ranging from 1.360 to 1.375 and high RI regions ranging from 1.375 to 1.430 are represented by yellow and green surfaces, respectively. f Cross-sectional images of the corresponding 3D RI distributions on the center plane (left panel) and the upper plane (right panel) of the particle. High and low RI regions were colored as green and yellow, respectively. Low RI regions become larger at the center, especially at the center plane of the particle. Source data are provided as a Source Data file.
Effect of DNA length on molecular doping of DNAs
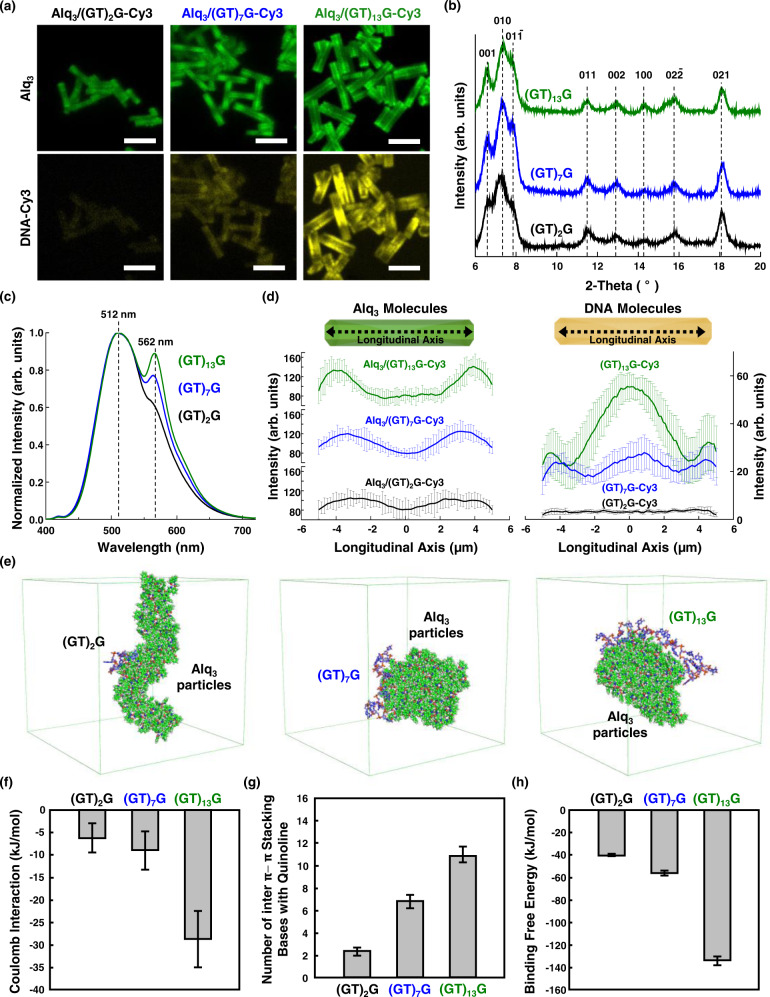
The length of the oligonucleotides of DNAs in the Alq3 microparticles was controlled to observe the molecular doping of DNAs and changes in the optical features of the particles (Fig. 2). In this experiment, DNAs with (GT)2G (5-mer), (GT)7G (15-mer), and (GT)13G (27-mer) nucleotides were labeled with Cy3 fluorescence dye to observe their molecular distribution. The concentration of DNAs used in this experiment was 800 nM. The particle size is independent of the length of the nucleotides doped in the Alq3 microparticles, as determined from the CLSM (Fig. 2a) and scanning electron microscopy (SEM) images (Supplementary Fig. 2). The corresponding X-ray diffraction (XRD) patterns, in Fig. 2b, of DNA-doped Alq3 particles show typical Alq3 α-phase peaks at 11.40° and 12.81°, and δ-phase peak at 11.79°, as observed in earlier studies31,32,33. The normalized photoluminescence (PL) spectra of DNA-Cy3-doped Alq3 microparticles are shown in Fig. 2c. The main PL peaks of the Alq3 particles and Cy3 dyes are observed at ~512 and ~572 nm, respectively28; the peak of Alq3 corresponds to yellowish-green emission from α- and δ-phases, consistent with the observed XRD patterns. Interestingly, the emission ratio calculated by the Gaussian peak area after deconvolution (Α572 nm/Α512 nm) increased from 0.10 to 0.30, as the number of nucleotides increased from 5 to 27. Furthermore, the emission ratio (Α572 nm/Α512 nm) of Alq3 microparticles doped with (GT)2G-Cy3 (0.11) is still low after increasing the concentration of DNA-Cy3 to 1500 nM (Supplementary Fig. 3) compared to the emission ratio of Alq3 microparticles doped with (GT)7G-Cy3 and (GT)13G-Cy3. The efficiency of energy transfer between the Alq3 donor and Cy3 dye acceptor is expected to be maximal when the distance between the two is minimal34. However, the longest DNA molecule [(GT)13G)] affords the highest energy transfer efficiency among the samples tested. The insight into this unrevealed factor influencing the energy transfer between Alq3 and DNA in these hybrid particles was obtained from the molecular distributions determined by CLSM. The CLSM images (Fig. 2a, bottom panel) indicate that DNA-Cy3 molecule distribution differs depending on the DNA length; the yellow-fluorescence signal from the Cy3 molecules is more intense for longer DNA lengths. The average local fluorescence intensities of Alq3 and DNA molecules are shown in Fig. 2d. The longitudinal fluorescence profile of the DNA molecules (Fig. 2d, right) in the Alq3/DNA[(GT)2G]-Cy3 particle has low intensity across the entire region, that is, little DNA is doped into these particles. The fluorescence of DNA molecules in Alq3/DNA[(GT)7G]-Cy3 is spread across the entire region, while that in Alq3/DNA[(GT)13G]-Cy3 is most intense in the central region, indicating that longer DNAs are more strongly doped into the particles. The DNA distribution in the particles affects energy transfer from the Alq3 molecules. The fluorescence intensity of the Alq3 molecules (Fig. 2d, left) is relatively uniform for particles with the shortest DNA[(GT)2G]-Cy3 except that that the fluorescence intensity of the edges of particles is slightly higher due to the optical waveguide effects. By contrast, the fluorescence intensity of the Alq3 molecules becomes lower at the center of the particles with DNA[(GT)7G]-Cy3 and DNA[(GT)13G]-Cy3 along the longitudinal axis. These results indicate that the energy transfer from the Alq3 donor to the Cy3 dye acceptor is largest at the highest degree of doping.
Fig. 2. Effect of DNA length on molecular doping of DNAs into Alq3 microparticles.
a CLSM images captured at the center plane of Alq3/DNA[(GT)xG]-Cy3 particles; x = 2, 7, and 13 (scale bar, 10 μm). b XRD patterns of all DNA-doped Alq3 particles show α−phase peak for Alq3 at 11.55° and 12.94°, along with a δ-phase peak at 11.79°. c PL spectra with excitation at 365 nm, revealing FRET properties of Alq3/DNA[(GT)xG]-Cy3 particles; x = 2, 7, and 13. d Longitudinal fluorescence profiles quantifying the local intensity of Alq3 (left) and DNA[(GT)xG]-Cy3 molecules (right) at the center plane averaged from six separate particles. CLSM images and profiles were obtained with excitation by a 405 nm laser for the Alq3 molecules and 555 nm laser for the DNA-Cy3 molecules used for doping. The filters used for observing the Alq3 and Cy3 molecules ranged from 300 to 550 nm and from 300 to 630 nm, respectively. The concentration of DNA in this experiment is 800 nM. e Molecular dynamics simulation for the length-dependent molecular doping of DNAs. Final configurations were shown for Alq3 assembly with DNA[(GT)xG] (x = 2, 7, and 13). f Electrostatic interaction between Alq3 particles and DNAs, and (g) averaged number of bases that form inter π-π stacking between bases of DNAs and quinoline of Alq3 assembly during the final 10 ns of simulations. h Calculated free energy change (binding free energy) of the DNAs interacting with Alq3 assembly, obtained from umbrella sampling. Error bars in (d), (f), (g), and (h) represent standard deviation. Source data are provided as a Source Data file.
All atomic molecular dynamics (AA MD) simulations were performed to interpret factors inducing increased doping for longer DNA segments. The final configurations of the three systems with 100 Alq3 molecules and DNA[(GT)xG] (x = 2, 7, and 13) are shown in Fig. 2e. All DNA segments are adsorbed onto the Alq3 particles. The electrostatic interaction (Fig. 2f) and number of π-π stacking structures (Fig. 2g) were obtained from the final 10 ns of the simulations. Longer DNA segments induce stronger electrostatic interactions between the phosphate backbone and Alq3 particles, facilitating the molecular doping of DNAs. In this acidic system, H+ ligands are present around the O cation of Al-O; thus, Alq3 is positively charged via the absorption of H+ by the 8-hydroxyquinoline moiety35. The dissociation constant (pKa) of the phosphodiester group of DNA is approximately 136, thereby maintaining negative charges on the DNA backbone37. This implies that the negative charges of DNA and the positive charges of the Alq3 molecules can engender more intense intermolecular attraction for longer DNA segments. Moreover, larger numbers of π-π stacking structures are observed between the DNA bases and quinoline of Alq3 particles for longer DNA segments. We then performed a series of steered molecular dynamics (SMD) simulations to quantify the free energy change before and after adsorption of different length DNA segments, as shown in Fig. 2h. The binding free energies of DNA[(GT)xG] (x = 2, 7, and 13) adsorbed onto the Alq3 particles are −39.9, −56.8, and −134.4 kJ/mol, respectively. From these results, it is reasonable to deduce that longer DNA molecules experience stronger interaction, including electrostatic attraction and π-π stacking, with Alq3, thereby increasing the extent of doping.
The effect of electrostatic attraction on the molecular doping of DNAs was further investigated by assembling Alq3 particles with DNAs in aqueous solutions of various pH values. The particle morphologies and molecular distributions of the DNA-doped Alq3 particles at various pH are shown in Supplementary Fig. 4. At pH 4, 7, and 10, the particles have regular prismatic hexagonal shapes, while no defined microstructures are observed at pH 3 and 12. CLSM images captured at the z-centric planes show that the Alq3 (middle) and DNA-Cy3 (bottom) molecules in these particles are distributed differently at various pH (Supplementary Fig. 4). Green fluorescent microparticles were observed at pH 4, 7, and 10. Specifically, efficient doping of DNA molecules into the Alq3 particles is observed at pH 4, and no yellow-fluorescence signals of the DNA-Cy3 molecules are observed in particles fabricated at other pH values. The quantitative histogram of DNA-Cy3 molecules also shows that the DNAs are predominantly incorporated into the Alq3 particles at pH 4 (Supplementary Fig. 5a). The PL spectra of Alq3 microparticles doped with DNA-Cy3 at various pH values are shown in Supplementary Fig. 5b. The emission ratio calculated by the Gaussian peak area (Α572 nm/Α512 nm) is 0.23 at pH 4, indicating significant doping, and ~0.07 at other pH values. It appears that the intermolecular interaction is pH-dependent; the electrostatic attraction between positively charged Alq3 and DNA with a relatively strong negative charge is dominant at pH 4. This is consistent with the report that the binding between aluminum ions and calf thymus DNA is pH dependent38. The binding constant increased in the pH range of 3.5‒4.5, reaching a maximum at pH 4.5. However, at pH above 5.5, the binding constant declined, and no binding of aluminum ions to DNA is observed at pH 6.0 and 7.0. Therefore, the acidity of the aqueous solutions used in the assembly of the Alq3 particles should be controlled to generate strong electrostatic attraction, inducing the incorporation of DNA molecules.
Effect of base units on molecular doping of DNAs
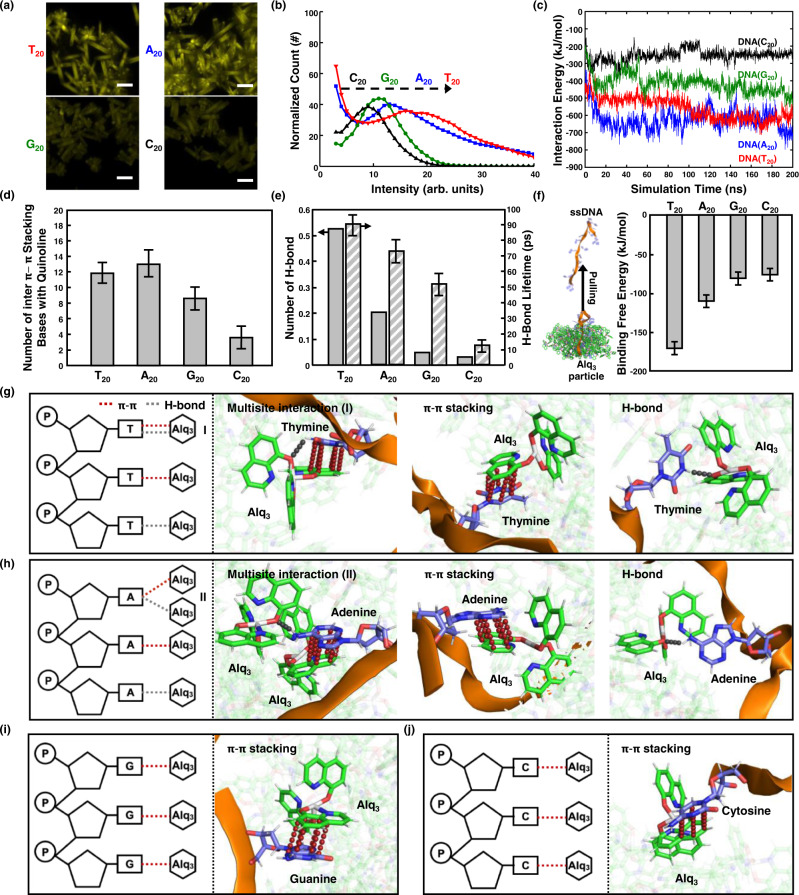
To explore the molecular doping of DNA with respect to the purine and pyrimidine base units, we fabricated Alq3 particles with DNAs having different patterns of the same length. In this experiment, four patterns were considered: poly(T), poly(A), poly(G), and poly(C) with a 20-mer sequence. The morphological features of DNA-Alq3 particles are presented in Supplementary Fig. 6. The Alq3 molecules crystallized similarly when doped with the 20-mer DNAs. Interestingly, the fluorescence signals in the CLSM images indicate that DNA(T20)-Cy3 and DNA(A20)-Cy3 are more effectively doped into the Alq3 particles than the other DNAs (Fig. 3a). The normalized quantitative histogram obtained from CLSM image shows the most intense fluorescence in particles doped with DNA(T20)-Cy3 (Fig. 3b). The PL spectra of the samples are shown in Supplementary Fig. 7. The emission at 572 nm after excitation at 500 nm is most intense for DNA(T20), consistent with the quantitative histogram of CLSM images. AA MD simulations were conducted to identify the binding phenomena, inducing doping between different bases and Alq3 particles. We constructed systems using 100 Alq3 molecules and DNA segments with 20-mer sequences: poly(T), poly(A), poly(G), and poly(C). The final configurations of the four systems are shown in Supplementary Fig. 8. The DNA segments with poly(T) and poly(A) sequences are fully adsorbed onto the Alq3 particles, whereas poly(G) and poly(C) sequences are partially adsorbed. The interaction energy between the DNA segments and Alq3 particles (Fig. 3c) also indicates strong interaction of DNA(T20) and DNA(A20) with the Alq3 particles than the other DNAs, leading to full adsorption on the Alq3 particles. Furthermore, we monitored the π-π stacking interaction (Fig. 3d) and hydrogen bonding (Fig. 3e) between the Alq3 particles and DNA segments to clarify differences in the interaction energy of the DNA molecules. Larger numbers of π-π stacking structures and hydrogen bonds are observed for DNA(T20) and DNA(A20). The average hydrogen bond lifetimes also show that Alq3 forms stronger hydrogen bonds with thymine and adenine than with guanine and cytosine. A series of SMD simulations were performed to calculate the binding free energy of fully adsorbed DNAs. The initial configurations for the calculation were derived from Alq3 with DNA(A20) to ensure that the four DNA segments are fully adsorbed on the Alq3 particles (see details in Methods section). The binding free energies averaged over three replicates of DNAs (T20, A20, G20, and C20) adsorbed on the Alq3 particles are ‒170.5, ‒110.4, ‒81.3, and ‒76.6 kJ/mol, respectively (Fig. 3f), indicating that DNAs (T20 and A20) with relatively large binding free energy tend to be fully adsorbed onto Alq3 particles (Supplementary Fig. 8). By closely inspecting how base units form noncovalent interactions, including π-π stacking and hydrogen bonding during the final 10 ns of simulations, we show key interactions that make DNA(T20 and A20) strongly doped into Alq3 (Fig. 3g and Table 1). Both π-π stacking and hydrogen bonds are frequently formed between the bases (thymine and adenine) and Alq3 molecules. In particular, 56 thymine molecules form multisite interactions with single Alq3 molecules via both π-π stacking and hydrogen bonding, whereas 26 adenine molecules form hydrogen bonds with one of the Alq3 molecules and subsequent π-π stacking interactions with neighboring Alq3 molecules. Considering the higher occurrence probability (4.39%) of multisite interactions and the strongest binding energy (‒170.5 kJ/mol) with Alq3 particles, we conclude that the multisite interaction of π-π stacking and hydrogen bonding between thymine and single Alq3 molecules causes the strongest attraction and intensifies the extent of doping into Alq3 particles. In comparison, few hydrogen bonding interactions with Alq3 molecules are observed for DNA(G20) and DNA(C20). Only π-π stacking interactions are involved in the doping of DNA(G20) (Fig. 3i and Table 1) and DNA(C20) (Fig. 3j and Table 1). The adsorption energies of the DNA(T20, A20, G20, and C20) on the Alq3 crystal surface were also calculated (Supplementary Fig. 9). The adsorption energy is highest for thymine molecules on the Alq3 crystal surface. Combined with experimental results, we confirm that thymine molecules induce stronger attraction with Alq3, suggesting that the oligonucleotide constituents control molecular doping in light-emitting organic crystals.
Fig. 3. Effect of base units on molecular doping of DNAs possessing distinct intermolecular interactions.
a Fluorescent analysis of doped DNA(T20, A20, G20, and C20)-Cy3 in Alq3 particles. CLSM images are obtained with excitation by a 555 nm laser (scale bar, 10 μm). b Quantitative histogram of doped DNA-Cy3 using CLSM. Molecular dynamics simulations for the base-dependent incorporation of DNAs into Alq3 microparticles. c Time-traced total interaction energy between Alq3 particles and DNAs. d Number of bases that form inter π-π stacking with the quinoline of Alq3 assembly during simulation. e Number of hydrogen bonds formed between Alq3 molecules and DNAs and their corresponding lifetimes during the final 100 ns simulation. f Binding free energy of the DNAs on Alq3 assembly, obtained from umbrella sampling for three replicates of DNAs fully adsorbed on Alq3 particles. The schematics and molecular views of (g) T20, (h) A20, (i) G20, and (j) C20 doped in Alq3 particles from the final 10 ns of AA MD simulation. Three key interactions, including multisite interaction, stoichiometric π-π stacking, and hydrogen bonding, for doping of T20 and A20 into Alq3 are shown in (g) and (h), respectively. π-π stacking interaction doping G20 and C20 into Alq3 are shown in (i) and (j), respectively. The hydrogen bonds and π-π stacking interaction in schematics and molecular views are depicted in gray and red dashed lines, respectively. Error bars in (d), (e), and (f) represent standard deviation. Source data are provided as a Source Data file.
Table 1.
Summary of the intermolecular interactions between Alq3 and DNA with different bases during the final 10 ns of AA MD simulations
| Numbers for the Occurrence of Interactions (Probability, %) | ||||
|---|---|---|---|---|
| Multisite interaction (Type I) | Multisite interaction (Type II) | H-bond | π-π stacking | |
| T20 | 56 (4.39%) | - | 81 (6.34%) | 1140 (89.3%) |
| A20 | - | 26 (1.93%) | 40 (2.96%) | 1284 (95.1%) |
| C20 | - | - | 1 (0.20%) | 491 (99.8%) |
| G20 | - | - | 1 (0.10%) | 823 (99.9%) |
Tailored bio-specific photonics and optical waveguide effects of DNA-Alq3 hybrid crystals
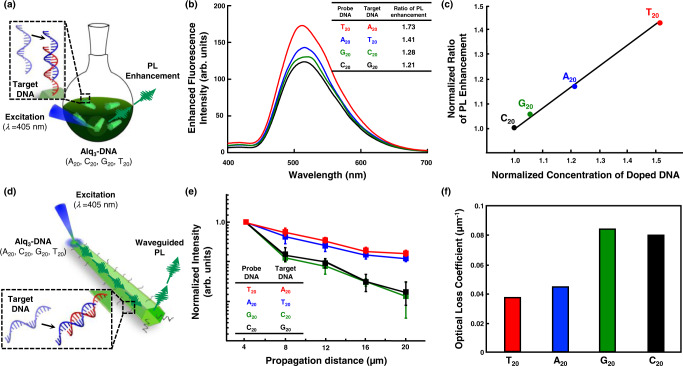
Fluorescence enhancement (Fig. 4a) and change in optical waveguide efficiency (Fig. 4d) of Alq3 particles with DNAs were observed after treatment with specific target DNA (tDNA) molecules to investigate the resulting biophotonics of Alq3 particles. The tDNAs used in this experiment were A20, T20, C20, and G20, for which the hybridization was complementary to single-stranded DNAs (T20, A20, G20, and C20), respectively. As shown in Fig. 4b, the largest fluorescence enhancement is observed when DNA(T20) doped in Alq3 particles interact with tDNA (A20). The relative ratio of PL enhancement is plotted as a function of the concentration of DNA-Cy3 doped into the Alq3 (Fig. 4c). After interacting with complementary tDNAs, the ratio increases linearly up to ~1.5 fold as the amount of DNA-Cy3 doped in Alq3 particles increases, indicating that larger degree of doping enhances bio-recognitive photonics. Specific detection of DNA-doped Alq3 microparticles is also confirmed (Supplementary Fig. 11). Only complementary recognition triggers PL enhancement in the Alq3 particles. We then measured the optical waveguide characteristics of single isolated Alq3 particles with DNAs (T20, A20, G20, and C20) using a house-built laser confocal microscope (LCM) system with a diode laser (λex = 405 nm). PL intensities of all Alq3 particles are decreased along the increasing propagation distance (Supplementary Fig. 12). In particular, the PL intensities have low optical transmission loss for Alq3 doped with DNAs (T20 and A20) (Fig. 4e). The optical loss coefficient of each Alq3 microparticle was calculated by exponentially fitting the emission intensities with respect to the propagation distance to distinguish the optical waveguide efficiency more quantitatively. Loss coefficient values, measured for the Alq3 particles doped with T20 (0.03701 μm−1) and A20 (0.04421 μm−1), are twice lower than those of the other Alq3 particles (Fig. 4f). However, compared to fluorescent enhancement, optical waveguide phenomena reveal similar loss coefficient values for T20 and A20 and G20 and C20. For the optical waveguide phenomena, light propagation is mainly affected by the surface of the Alq3 crystal16, whereas PL enhancement after treatment with complementary DNAs originates from the surface and inner core of the Alq3 particles. The molecular distribution of doped DNA with different base moieties at the Alq3 surface is shown in Supplementary Fig. 13. The fluorescence of the DNA (T20 and A20)-Cy3 molecules exhibits similar distribution, most intense in the central region along the longitudinal axis of the Alq3 surface. In contrast, DNA (G20 and C20)-Cy3 molecules demonstrate a lower intensity over the entire Alq3 surface. Hence, we deduce that a larger doping extent of DNA molecules at the Alq3 particle’s surface reduces the optical loss during light propagation in T20 and A20. Thus, the extent of DNA doping can tailor the biophotonic properties, including the bio-specific photonics and optical waveguide phenomenon, of light-emitting organic crystals.
Fig. 4. Bio-specific photoluminescence and optical waveguide phenomena of Alq3 microparticles upon complementary DNA-DNA hybridization.
a Schematic illustration for observing bio-recognitive photonics of Alq3 doped with DNAs. b PL spectra of Alq3/DNAs (T20, A20, G20, and C20) after treatment with corresponding complementary DNAs (excitation at 365 nm). The inset chart represents the ratio of PL enhancement compared to PL intensity before complementary DNA treatment (see details in Supplementary Fig. 10). c Normalized ratio of PL enhancement as a function of the normalized concentration of doped DNA-Cy3 molecules. Concentration is derived from the fluorescence intensity of doped DNA-Cy3 molecules. d Schematic illustration for measuring optical waveguide phenomena of Alq3 microparticles doped with DNAs. e Normalized intensity of waveguided PL from Alq3/DNAs (T20, A20, G20, and C20) microparticles after treatment with corresponding complementary DNAs. f Optical loss coefficient of each Alq3 microparticle, calculated by exponentially fitting the emission intensities with respect to the propagation distance. Error bars in (e) represent standard deviation. Source data are provided as a Source Data file.
Discussion
In summary, we proposed a molecular-level interpretation of the doping behavior of DNAs in light-emitting organic particles by varying types and numbers of purine and pyrimidine bases constituting oligonucleotide. Intermolecular interactions with Alq3 particles, which induce molecular doping, originate from three noncovalent interactions: electrostatic, hydrogen bonding, and π-π stacking interactions. For the DNA length effect, stronger electrostatic interactions between the phosphate backbone of nucleic acid and Alq3 for longer nucleic acid molecules induce a higher degree of doping. For the bases effect, adenine and thymine molecules generate stronger attraction with Alq3 molecules due to the multisite interactions combining π-π stacking and hydrogen bonding interactions, in spite of low occurrence probabilities. This strong attraction contributes to a higher degree of doping than cytosine and guanine, which only form π-π stacking interactions with Alq3 molecules. In particular, MD simulations revealed that thymine molecules exert the strongest attraction through multisite interactions with single Alq3 molecules, facilitating the molecular doping into Alq3 particles up to 1.5-fold.
This molecular doping of nucleic acid molecules successfully tailors the resulting biophotonics of hybrid Alq3 particles in terms of (i) enhancing the recognitive photonics for complementary DNA molecules and (ii) reducing optical loss during the optical waveguide phenomena. We expect that rational designs of doped nucleic acids based on the fundamental understanding of intermolecular interactions can impact the technological applications of such DNA-hybrid light-emitting materials with tailored biophotonics.
Methods
Fabrication of DNA-doped Alq3 microparticles
A facile reprecipitation method for fabricating DNA-doped Alq3 microparticles has been previously reported15,28. Commercial Alq3 powder and tetrahydrofuran (≥99%) were purchased from Sigma–Aldrich. The oligonucleotides and oligonucleotides labeled with Cy3 fluorescent dyes used for doping were synthesized and purified by Bioneer Corporation (Daejeon, Korea). The quality of the synthesized oligonucleotides and quantity of labeled Cy3 molecules was verified using the MALDI-TOF mass spectrometer. The Alq3 powder was dissolved in tetrahydrofuran at a concentration of 1 mg mL‒1 to form a stock solution. The stock solution (2 mL) was injected into a series of 20 mL aqueous DNA solutions at a concentration of 0.8 μM with vigorous stirring (~800 rpm) for 2 min. The mixture was stored overnight at 25 °C to form a visible precipitate. The DNA sequences used in this study are listed in Supplementary Table 1. DNA was labeled with cyanine dye (Cy3) derivatives for molecular distribution and energy transfer analysis.
Hybridization of DNA molecules doped into Alq3 microparticles
A method for hybridizing DNA-doped Alq3 microparticles with tDNA has been previously reported15. The DNA-doped Alq3 microparticles with different sequences (T20, A20, G20, or C20) were reacted with 0.8 μM complementary tDNA(A20, T20, C20, or G20) at 52 °C for 30 min and then cooled to 25 °C.
Characterization of Alq3 microparticles
The morphology of the Alq3 particles was analyzed by field-emission SEM (Hitachi, S-4300) at an acceleration voltage of 15 kV. A holotomography microscope (HT-2H, Tomocube Inc., Republic of Korea)39 was performed to visualize 3D RI distributions of DNA doped Alq3 particles. The acquired images were processed via Tomostudio software (Tomocube Inc.). CLSM (Carl Zeiss, LSM700) provides z-sectioning fluorescence images of the Alq3 particles and DNA molecules. The Alq3 molecules were excited at 405 nm, and fluorescence was detected after passing through a 300‒550 nm filter. The DNA molecules labeled with Cy3 dye were excited at 550 nm, and fluorescence was detected after passing through a 300‒630 nm filter. The Alq3 particles were analyzed using a z-stack of images collected at 100 nm intervals. PL spectra were acquired using a fluorescence spectrophotometer (Hitachi, F-7000; excited by a xenon lamp). FRET efficiency between Alq3 particles and Cy3 fluorescence dyes was calculated by comparing the fluorescence intensities of the donor (Alq3) at 512 nm in the presence and absence of the acceptor (Cy3). The emission ratio (Α572 nm/Α512 nm) was calculated after the deconvolution of PL spectra of Alq3 particles by three gaussian peaks. X-ray diffraction (XRD; Rigaku, SmartLab) patterns were captured at a voltage of 40 kV and current of 40 mA using Cu-Kα radiation (λ = 1.540 Å). The optical waveguides of the Alq3 particles were measured using an in-build LCM system comprising a diode laser (λex = 405 nm) for excitation and a spectrometer (Andor, SR-3031-B) for recording the PL spectra. Three to five Alq3 particles from each sample were used for calculating intensities of waveguided PL. We separated the energy irradiation and signal detection points using a piezoelectric stage (Nanofocus Inc., Albatross) to obtain precise waveguide intensity at each propagation point.
AA MD simulations of DNA molecules on Alq3 particles
The all atomic simulation of DNA molecules on the Alq3 particles was performed by GROMACS 5.1.4, using the CHARMM36 force-field40,41. The simulations were conducted using a leapfrog integrator with a time step of 2 fs. PME with a cutoff of 1.2 nm calculated the electrostatic interactions42. Neighbor lists built using the Verlet cutoff scheme with a cutoff range of 1.2 nm were updated at every simulation step. The LINCS algorithm constrains the bond lengths43. All systems were maintained at 298.15 K using a V-rescale thermostat44 and at 1 bar using the Berendsen45 and Parrinello-Rahman46 barostats for the equilibrium and production runs, respectively.
The system consists of DNA[(GT)xG] (x = 2, 7, and 13) or 20-mer DNA molecules with different sequences (T20, A20, G20, or C20), 100 Alq3 molecules, and 30,000 pre-equilibrated water molecules in a simulation box with dimensions of 10 × 10 × 10 nm. The Alq3 molecule was parameterized based on the parameters developed by Voorhis et al.47. The interaction energies, including electrostatic energy and hydrogen bonding, between the DNAs and Alq3 particles, were calculated using the GROMACS analysis tool40. For the π-π stacking structures, the vertical separation between the base and quinoline was less than 0.45 nm48.
AA MD simulation calculating the binding free energy of DNA molecules on Alq3 particles
Umbrella sampling simulations were conducted for each DNA to compute the binding energies of the DNA molecules. To construct initial configurations for binding free energy, we manually constrained the position of the backbone of DNA (A20), fully adsorbed on the Alq3 particle, and replaced adenine with other bases. Thereafter, the four systems with DNAs (T20, A20, G20, or C20) were equilibrated for 100 ns. To generate the z-directional reaction coordinate, DNA was pulled from the Alq3 particles using a force constant of 1000 kJ mol‒1 nm‒2. Forty windows with a spacing of 0.15 nm were created in each system. Equilibration and production runs of 1 and 10 ns, respectively, were performed for each window. WHAM tools in the GROMACS package measured the mean force potential. The binding energy is the difference between the highest and lowest energy states of the potential of mean force (PMF) curves (Supplementary Fig. 14).
Supplementary information
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF-2021R1A2C3009955 and 2017M3D1A1039421), 111 Project (D18012), and Korea University Grant. We acknowledge Dr. Yeol Kyo Choi for his discussions related to computational simulation and Yedam Lee for her assistance to holotomographic measurements.
Source data
Author contributions
D.J.A. designed and supervised the project. W.H.J., J.H.P., and S.K. contributed equally to this work. J.H.P. and W.H.J. fabricated DNA-organic semiconductor particles and conducted the recognition and optical analyses (confocal molecular profiling and fluorescence microscopy analyses). S.K. performed optical waveguide experiments and holotomographic microscopy analyses. W.H.J. conducted MD simulations to interpret the molecular doping of DNAs. W.H.J., J.H.P., S.K., and C.C. prepared the initial manuscript draft. D.J.A. reviewed and edited the manuscript. All the authors agree with the final content.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Data availability
All data needed to evaluate the conclusions in this study are present within the article and Supplementary Information. Source data for main figure and Supplementary Information are provided with this paper. Additional data related to this paper are available from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Woo Hyuk Jung, Jin Hyuk Park, Seokho Kim.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-33999-y.
References
- 1.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Steckl AJ. DNA–a new material for photonics? Nat. Photonics. 2007;1:3–5. doi: 10.1038/nphoton.2006.56. [DOI] [Google Scholar]
- 3.Cui C, Park DH, Ahn DJ. Organic semiconductor–DNA hybrid assemblies. Adv. Mater. 2020;32:2002213. doi: 10.1002/adma.202002213. [DOI] [PubMed] [Google Scholar]
- 4.Ong LL, et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature. 2017;552:72–77. doi: 10.1038/nature24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson LA, Kowalik J, Josowicz M, Janata J. Label-free DNA hybridization probe based on a conducting polymer. J. Am. Chem. Soc. 2003;125:324–325. doi: 10.1021/ja027929z. [DOI] [PubMed] [Google Scholar]
- 6.Lee K, Rouillard JM, Pham T, Gulari E, Kim J. Signal‐amplifying conjugated polymer–DNA hybrid chips. Angew. Chem. Int. Ed. 2007;46:4667–4670. doi: 10.1002/anie.200700419. [DOI] [PubMed] [Google Scholar]
- 7.Nakatsuka N, et al. Aptamer–field-effect transistors overcome Debye length limitations for small-molecule sensing. Science. 2018;362:319–324. doi: 10.1126/science.aao6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pruneanu S, et al. Self‐assembly of DNA‐templated polypyrrole nanowires: spontaneous formation of conductive nanoropes. Adv. Funct. Mater. 2008;18:2444–2454. doi: 10.1002/adfm.200701336. [DOI] [Google Scholar]
- 9.Wang X, et al. Exciton delocalization in a DNA-templated organic semiconductor dimer assembly. ACS Nano. 2022;16:1301–1307. doi: 10.1021/acsnano.1c09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knudsen JB, et al. Routing of individual polymers in designed patterns. Nat. Nanotechnol. 2015;10:892–898. doi: 10.1038/nnano.2015.190. [DOI] [PubMed] [Google Scholar]
- 11.Kamiya Y, Asanuma H. Light-driven DNA nanomachine with a photoresponsive molecular engine. Acc. Chem. Res. 2014;47:1663–1672. doi: 10.1021/ar400308f. [DOI] [PubMed] [Google Scholar]
- 12.Keya JJ, et al. DNA-assisted swarm control in a biomolecular motor system. Nat. Commun. 2018;9:453. doi: 10.1038/s41467-017-02778-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez EF, Venkatraman V, Grote JG, Steckl AJ. DNA bases thymine and adenine in bio-organic light emitting diodes. Sci. Rep. 2014;4:7105. doi: 10.1038/srep07105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagen JA, Li W, Steckl A, Grote J. Enhanced emission efficiency in organic light-emitting diodes using deoxyribonucleic acid complex as an electron blocking layer. Appl. Phys. Lett. 2006;88:171109. doi: 10.1063/1.2197973. [DOI] [Google Scholar]
- 15.Back SH, Park JH, Cui C, Ahn DJ. Bio-recognitive photonics of a DNA-guided organic semiconductor. Nat. Commun. 2016;7:10234. doi: 10.1038/ncomms10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S, et al. Bio‐photonic waveguide of a DNA‐hybrid semiconductor prismatic hexagon. Adv. Mater. 2020;32:2005238. doi: 10.1002/adma.202005238. [DOI] [PubMed] [Google Scholar]
- 17.Sun J, et al. 2D–Organic hybrid heterostructures for optoelectronic applications. Adv. Mater. 2019;31:1803831. doi: 10.1002/adma.201803831. [DOI] [PubMed] [Google Scholar]
- 18.Yan Y, Zhang C, Yao J, Zhao YS. Recent advances in organic one‐dimensional composite materials: design, construction, and photonic elements for information processing. Adv. Mater. 2013;25:3627–3638. doi: 10.1002/adma.201300325. [DOI] [PubMed] [Google Scholar]
- 19.Zhao YS, et al. Construction and optoelectronic properties of organic one-dimensional nanostructures. Acc. Chem. Res. 2010;43:409–418. doi: 10.1021/ar900219n. [DOI] [PubMed] [Google Scholar]
- 20.Zhao YS, et al. Single crystalline submicrotubes from small organic molecules. Chem. Mater. 2005;17:6430–6435. doi: 10.1021/cm051949t. [DOI] [Google Scholar]
- 21.Kapadia PP, et al. Semiconducting organic assemblies prepared from tetraphenylethylene tetracarboxylic acid and bis (pyridine) s via charge-assisted hydrogen bonding. J. Am. Chem. Soc. 2011;133:8490–8493. doi: 10.1021/ja203001z. [DOI] [PubMed] [Google Scholar]
- 22.Yee N, Dadvand A, Hamzehpoor E, Titi HM, Perepichka DF. Hydrogen bonding versus π-stacking in charge-transfer co-crystals. Cryst. Growth Des. 2021;21:2609–2613. doi: 10.1021/acs.cgd.1c00309. [DOI] [Google Scholar]
- 23.Lei Y, Sun Y, Liao L, Lee S-T, Wong W-Y. Facet-selective growth of organic heterostructured architectures via sequential crystallization of structurally complementary π-conjugated molecules. Nano Lett. 2017;17:695–701. doi: 10.1021/acs.nanolett.6b03778. [DOI] [PubMed] [Google Scholar]
- 24.Zhao YS, Zhan P, Kim J, Sun C, Huang J. Patterned growth of vertically aligned organic nanowire waveguide arrays. ACS Nano. 2010;4:1630–1636. doi: 10.1021/nn901567z. [DOI] [PubMed] [Google Scholar]
- 25.Liao Q, Fu H, Wang C, Yao J. Cooperative assembly of binary molecular components into tubular structures for multiple photonic applications. Angew. Chem. Int. Ed. 2011;50:4942–4946. doi: 10.1002/anie.201006681. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C, Zheng JY, Zhao YS, Yao J. Self‐modulated white light outcoupling in doped organic nanowire waveguides via the fluctuations of singlet and triplet excitons during propagation. Adv. Mater. 2011;23:1380–1384. doi: 10.1002/adma.201003829. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C, et al. One‐dimensional organic photonic heterostructures: rational construction and spatial engineering of excitonic emission. Adv. Mater. 2012;24:1703–1708. doi: 10.1002/adma.201104326. [DOI] [PubMed] [Google Scholar]
- 28.Cui C, Park DH, Kim J, Joo J, Ahn DJ. Oligonucleotide assisted light-emitting Alq3 microrods: energy transfer effect with fluorescent dyes. Chem. Commun. 2013;49:5360–5362. doi: 10.1039/c3cc41255e. [DOI] [PubMed] [Google Scholar]
- 29.Zheng JY, et al. Wire-on-wire growth of fluorescent organic heterojunctions. J. Am. Chem. Soc. 2012;134:2880–2883. doi: 10.1021/ja209815f. [DOI] [PubMed] [Google Scholar]
- 30.Xu G, et al. Facile solution synthesis without surfactant assistant for ultra long Alq3 sub-microwires and their enhanced field emission and waveguide properties. J. Mater. Chem. 2010;20:3006–3010. doi: 10.1039/b924459j. [DOI] [Google Scholar]
- 31.Cölle M, Brütting W. Thermal, structural and photophysical properties of the organic semiconductor Alq3. Phys. Status Solidi. 2004;201:1095–1115. doi: 10.1002/pssa.200404341. [DOI] [Google Scholar]
- 32.Suzuki F, Fukushima T, Fukuchi M, Kaji H. Refined structure determination of blue-emitting tris (8-hydroxyquinoline) aluminum (III)(Alq3) by the combined use of cross-polarization/magic-angle spinning 13C solid-state NMR and first-principles calculation. J. Phys. Chem. C. 2013;117:18809–18817. doi: 10.1021/jp404430v. [DOI] [Google Scholar]
- 33.Cölle M, Gmeiner J, Milius W, Hillebrecht H, Brütting W. Preparation and characterization of blue‐luminescent tris (8‐hydroxyquinoline)‐aluminum (Alq3) Adv. Funct. Mater. 2003;13:108–112. doi: 10.1002/adfm.200390015. [DOI] [Google Scholar]
- 34.Förster T. 10th Spiers memorial lecture. Transfer mechanisms of electronic excitation. Discuss. Farady Soc. 1959;27:7–17. doi: 10.1039/DF9592700007. [DOI] [Google Scholar]
- 35.Li T, et al. Preparation and properties of 8‐hydroxyquinoline aluminum quinoline/polyaniline core–shell composite. Polym. Compos. 2015;36:272–277. doi: 10.1002/pc.22940. [DOI] [Google Scholar]
- 36.Thaplyal P, Bevilacqua PC. Experimental approaches for measuring pKa’s in RNA and DNA. Methods Enzymol. 2014;549:189–219. doi: 10.1016/B978-0-12-801122-5.00009-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M, Kempaiah R, Huang P-JJ, Maheshwari V, Liu J. Adsorption and desorption of DNA on graphene oxide studied by fluorescently labeled oligonucleotides. Langmuir. 2011;27:2731–2738. doi: 10.1021/la1037926. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, et al. Thermodynamics of the interaction of aluminum ions with DNA: implications for the biological function of aluminum. J. Inorg. Biochem. 2005;99:1145–1154. doi: 10.1016/j.jinorgbio.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Park Y, Depeursinge C, Popescu G. Quantitative phase imaging in biomedicine. Nat. Photonics. 2018;12:578–589. doi: 10.1038/s41566-018-0253-x. [DOI] [Google Scholar]
- 40.Abraham MJ, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 41.Hart K, et al. Optimization of the CHARMM additive force field for DNA: improved treatment of the BI/BII conformational equilibrium. J. Chem. Theory Comput. 2012;8:348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darden T, York D, Pedersen L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 43.Hess B, Bekker H, Berendsen HJ, Fraaije JG. LINCS: a linear constraint solver for molecular simulations. J. Comput Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 44.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 45.Berendsen HJ, Postma JV, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 46.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Apply. Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 47.Difley S, Wang L-P, Yeganeh S, Yost SR, Voorhis TV. Electronic properties of disordered organic semiconductors via QM/MM simulations. Acc. Chem. Res. 2010;43:995–1004. doi: 10.1021/ar900246s. [DOI] [PubMed] [Google Scholar]
- 48.Xu Z, et al. Dynamic cooperative of hydrogen binding and π stacking in ssDNA adsorption on graphene oxide. Chem. Eur. J. 2017;23:13100–13104. doi: 10.1002/chem.201701733. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in this study are present within the article and Supplementary Information. Source data for main figure and Supplementary Information are provided with this paper. Additional data related to this paper are available from the corresponding author upon request. Source data are provided with this paper.