Abstract
Human enterprises through the solar system will entail long-duration voyages and habitation creating challenges in maintaining healthy diets. We discuss consolidating multiple sensory and nutritional attributes into microorganisms to develop customizable food production systems with minimal inputs, physical footprint, and waste. We envisage that a yeast collection bioengineered for one-carbon metabolism, optimal nutrition, and diverse textures, tastes, aromas, and colors could serve as a flexible food-production platform. Beyond its potential for supporting humans in space, bioengineered microbial-based food could lead to a new paradigm for Earth’s food manufacturing that provides greater self-sufficiency and removes pressure from natural ecosystems.
Subject terms: Applied microbiology, Metabolic engineering, Synthetic biology
Long-duration human space travel creates challenges for maintaining healthy diets. Here the authors discuss using synthetic biology approaches to modify yeast into an optimal, and enjoyable, food production platform.
Working up an appetite for food produced in space
Long-duration human space ventures, such as returning to the Moon for extended periods, visiting asteroids, and traveling to Mars, will require self-sufficiency and maximizing life support systems. Reducing dependence on initial launch cargo and resupply from Earth improves autonomy, mitigates risks, and is essential to the economic and logistic viability of long-term human endeavors in space. Biotechnology can play an integral role in achieving this goal1,2. Living systems are self-replicating, self-repairing, and self-organizing and can be engineered to harness available resources for the cost-effective production of desired outputs (e.g., nutrients, pharmaceuticals, biomaterials), thus having great potential to reduce cargoes and resupplies. In particular, the application of biotechnology for food-production off-Earth will be instrumental in supporting human travel and habitation through the solar system3–5. Here, we outline the motivation for developing microbial-based food-production systems. We examine the use of synthetic biology to consolidate multiple sensory and nutritional food attributes into engineered autotroph or waste carbon utilizing microorganisms to contribute to this effort. We also discuss the prospect of the approach in conjunction with three-dimensional (3D) printing to achieve direct consumption of fully personalized microbial food and maximize food-production with minimal waste.
Microorganisms as space food
On short-duration missions, sufficient food can be transported for the entire mission, and regular resupplies keep the International Space Station stocked with food3. However, achieving food self-sufficiency will be essential for future space travelers, as the demand for food increases greatly when the duration of space endeavors lengthens1. By way of example, it is estimated that a mission with six crew for ~500 days of residence on Mars will require approximately five tons of food, plus another eight to ten tons for interplanetary transits and contingencies5. Besides the enormous logistical costs of transporting and storing large quantities of food, stored food is susceptible to deterioration, spoilage, and reductions in nutrient levels or bioavailability, which could lead to food shortages or undernutrition, compromising the crew’s health and physical and cognitive performance. Hence, other than transporting all food required or relying on Earth’s resupplies, the best approach for sustaining extended human space ventures is to produce food on-site. Ideal food systems should be capable of reliably producing appealing nutrient-rich food on-demand, fast, and with minimal inputs and physical footprint.
Different alternatives, including plants, algae, insects, cultured meat, and microbes, have been considered to produce food off-Earth1,6–10. Microorganisms require comparatively fewer inputs, double their biomass more rapidly, and are generally more amenable to bioengineering interventions, all of which are critical advantages that justify developing them as food-production systems. Also, besides being consumed in food and beverages like bread, yogurt, cheese, beer, and wine for millennia and, more recently, as dietary probiotics, microorganisms are increasingly used to produce food additives and are considered a sustainable food source for the future11–13. If microorganisms can be grown on available or waste carbon and nitrogen to produce appealing and nutritious food, then they can not only extend the duration of extraterrestrial human operations but also decrease the environmental impact of current Earth-based agriculture.
The yeast Saccharomyces cerevisiae is a food-grade microorganism with thousands of years of usage in baking, brewing, and winemaking14. Yeast cells are a nutritious food source and have the potential to form a more significant part of a human’s diet, as they have a macronutrient profile similar to soy flour, with ~40.4% protein, ~34.6% carbohydrates, ~1.5% lipids, and ~13 kJ per gram of dry cell weight11,13. The amino acid profile of yeast proteins is also suitable for human nutrition, containing all the essential amino acids that humans cannot produce and must obtain from dietary sources (i.e., histidine, isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine)15. S. cerevisiae is also fast-growing (doubling time of ~90 min under optimal conditions), highly genetically malleable, and one of the most thoroughly characterized organisms16. Furthermore, studies have shown that microgravity conditions do not seem to significantly affect its growth or viability17, and work conducted by NASA has highlighted the potential for engineered S. cerevisiae as a useful source of essential nutrients for human health and nutrition (e.g., NASA’s BioNutrients project). Recent estimates of the nutritional value of a vitamin-prototrophic yeast strain in the context of human space travel suggest that all vitamins and macronutrients necessary for a balanced human diet could be provided for 50–100 people per day from a single 3000 L fermentation18. All this makes S. cerevisiae a promising candidate for developing into a microbial food-production system (Fig. 1).
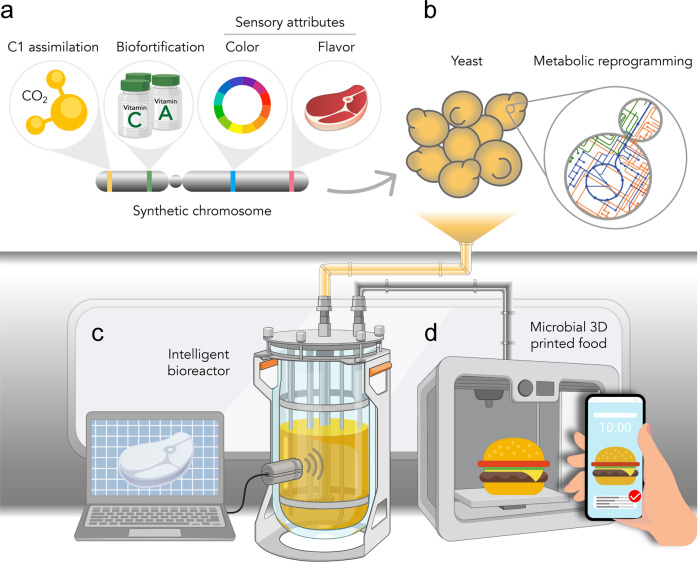
Fig. 1. Bioengineered yeast as a complete food-production system.
Multiple genetic pathways could be consolidated into synthetic chromosomes (a) to reprogram yeast metabolism and bestow it with new engineered traits (e.g., C1-utilization and sensory and nutritional food attributes) (b). Through the use of intelligent bioreactors capable of controlling the expression of specified engineered genetic pathways, yeast cellular physiology could be shaped to tune yeast biomass food properties (c). Microbial 3D-printed food technologies would allow manufacturing food personalized to individual preferences with minimal waste (d).
Alternative microorganisms to S. cerevisiae, including naturally autotrophic ones like the cyanobacterium Arthrospira platensis and the alga Chlorella vulgaris, are already consumed as dietary supplements or food additives, and hence could be used as a food source. However, these and most other organisms are typically less well-characterized and less genetically tractable compared to yeast, meaning that engineered sensory and nutritional attributes can be challenging to implement. Another microbe with a similar depth of characterization and genetic tractability relative to yeast is the bacterium Escherichia coli. However, E. coli is arguably less suitable as a human food source due to the pathogenic nature of some E. coli strains and its lack of history in the human food chain.
Recycling of essential microbial nutrients
On certain extraterrestrial destinations, such as Mars, indispensable nutrients to support microbial growth, like carbon and nitrogen, may be readily available and acquired from the environment (via in situ resource utilization)7,19. However, during space transits and on destinations where essential microbial nutrients are inaccessible, these resources could be partially recovered from crew-generated wastes (via closed-loop systems)1.
It is estimated that an average human requires about 8700 kJ of food and will expel about 740 grams of carbon dioxide (CO2) per day20. If this amount of CO2 could then be used to grow microorganisms in bioreactors with the nutritional content of ~12.5 kJ per gram of cells13, and 0.2–0.5 g of cells made per gram of CO2 per day21,22, then the total energy available from microbial food grown on recycled CO2 would be between 1850 and 4625 kJ per person per day. The potential utilization of different one-carbon (C1) sources using photoautotrophic or chemoautotrophic metabolism in various microorganisms was recently covered in great detail13,23. Assuming that waste or in situ available CO2 can be easily captured and released into a bioreactor, there are numerous options for growing microbial food directly on CO2 or by reducing CO2 to liquid C1 sources, such as formate or methanol. While electrochemical reduction of CO2 to liquid C1’s such as formate and methanol is possible, this technology is not yet mature or efficient due to the low conversion efficiencies and high energy costs24. Alternatively, a more mature technological possibility involves the electrochemical reduction of carbon dioxide with hydrogen to form methanol in a process already commercialized on Earth by the company Carbon Recycling International. Given that the yeast Pichia pastoris can grow efficiently on methanol (growth rate of 0.15/h)25, and there are strains of S. cerevisiae that can grow efficiently on ethanol made from a similar process18, the electrochemical reduction of CO2 with hydrogen poses a promising solution to the recycling of waste carbon to food in a space-travel setting. Once these problems are solved, this process could become very attractive since liquid carbon sources are easier to store and are more easily utilized in a bioreactor.
The major limitation of S. cerevisiae as a food-production system is its requirement for sugars as carbon and energy source. Given that the biomass yield of yeast is ~0.5 g per gram of glucose26, and the fact that glucose is itself a human food source, utilizing sugar-grown yeast would decrease the total calories available for human consumption. A straightforward approach to overcome this limitation would be to develop food-production systems in which yeasts are used in conjunction with autotrophic microorganisms. The simplest configuration in this paradigm would probably be for S. cerevisiae to grow on sugars derived from photosynthetic cyanobacteria CO2 fermentation or cyanobacteria biomass27,28. Strains of cyanobacteria have previously been engineered to secrete sugars into the growth medium29, which could be supplied to a second bioreactor containing sugar-utilizing yeasts, or even utilized within the same vessel through a co-culture system. Furthermore, any CO2 produced by yeast metabolism could theoretically be re-supplied to cyanobacteria. Alternatively, C1 assimilation could conceivably be directly engineered in S. cerevisiae, hence eliminating the requirement for autotrophic partners (Fig. 1).
Recent advances have allowed the functional integration of autotrophic carbon-fixing enzymes in S. cerevisiae30,31 and enabled the yeast P. pastoris and E. coli to grow directly on CO2, with reducing power provided by the C1 substrate formate in E. coli32,33. C1 substrates, such as formate and methanol, may also serve as carbon sources for microbial food production in space, as they can be derived readily from CO2 using additional oxidation and reduction reactions that are becoming inexpensive and efficient34. These advances make the recent engineering of formate35 and methanol utilization in S. cerevisiae21,22,36 relevant to microbial food production in space. While the growth rates and biomass yields of these synthetic C1-utilizing systems are still far from optimal, and co-substrates are still required, it is inevitable that they will be improved using the tools of laboratory evolution, synthetic biology, and systems biology. Although strains of S. cerevisiae with synthetic C1 fixation pathways may be the ultimate solution for engineered microbial food, the more immediate deployment of waste C1-utilizing microbes in space or on Earth would require the use of naturally occurring autotrophs such as hydrogenotrophic, methanotrophic, or photosynthetic microorganisms, which have been covered recently1,23,27,28,37.
In addition to carbon, microorganisms require a source of nitrogen. The necessary amount of nitrogen in microbial growth media is often comparable to carbon in terms of weight, making it another important consideration for long-duration space ventures and resource recycling. Human urine is one potential source of waste nitrogen that could be used for microbial fermentation, as it has high concentrations of urea (~10 g/L). Urea is a readily assimilable nitrogen source for many microorganisms, including yeast38. The yeast Yarrowia lipolytica has been grown successfully on both synthetic and natural urine39. Although S. cerevisiae can utilize urea as a sole nitrogen source, we were unable to find any reports of its growth in urine. However, given the similarity of nitrogen metabolism to Y. lipolytica, it is likely that S. cerevisiae growth can be supported in the urine-based medium. In addition to urea, human urine has other components, such as phosphates and salts, that would form part of a normal microbial growth medium, making its use particularly well-suited to support microbial food production.
It is worth noting that, although nutrient recovery from wastes to support microbial growth is feasible, bioengineered waste-reclamation systems will always be accompanied by inherent biological inefficiencies and associated energy costs. Furthermore, it is important to acknowledge that while waste recycling through synthetic microorganisms could theoretically extend a space mission, reduce the launch weight, or provide an emergency food supply, it could not do so indefinitely as there would be nonrecoverable losses of nutrients and carbon at each recycling stage.
Developing yeast into a complete food-production system
In addition to enabling altered carbon source utilization, current synthetic biology capabilities make it possible to propose engineering multiple traits for the nutritional and sensory enhancement of S. cerevisiae to repurpose it for producing edible microbial biomass. We envisage that a collection of yeast strains engineered for optimal nutrition and a range of textures, tastes, aromas, and colors would help sustain a healthy diet and enable food customization to individual preferences to increase food acceptability and prevent menu fatigue, critical challenges for space food systems3. Such a yeast collection could be air-dried preserved (air-drying has been shown to maximize long-term yeast cell viability in space40) without the requirement for refrigeration and would minimize size and weight, thus being convenient for transportation and storage purposes. Selected yeast strains grown in microbial bioreactors on C1 sources or photosynthesis-derived sugars could rapidly generate edible biomass for human consumption as needed (Fig. 1).
Many strains of the proposed yeast collection could be generated by native gene knockouts and the addition of relatively small numbers of heterologous genes. However, engineering multiple genetic pathways comprising many genes and consolidating them in a minimum number of yeast strains may be challenging to achieve through classic genetic engineering approaches. One way to overcome this limitation would be to combine numerous genetic pathways of interest into synthetic neochromosomes41,42 designed to implement many new dietary features into yeast simultaneously (Fig. 1). We envision these dietary-dedicated neochromosomes as versatile platforms on which genetic pathways for different textures, tastes, odors, pigments, and nutrients could be added as modules depending on the characteristics of the desired strain. Designing these multi-gene modules as ‘synthetic eukaryotic operons’ (e.g., exploiting 2A peptide- or tobacco etch virus protease-based polycistronic constructs43,44) would allow minimized neochromosome sizes and would simplify their construction by dramatically reducing the number of promoters and terminators required.
We also foresee that strain engineering could be integrated with the development of intelligent bioreactors capable of dynamically controlling yeast biomass properties by adjusting culture conditions and cellular physiology45,46. Through the use of synthetic biology approaches, such as biosensors, optogenetics, and electrogenetics, these bioreactors could be programmed to orchestrate the expression of specified genes to customize yeast sensory and nutritional characteristics and produce edible microbial biomass personalized to specific requirements on demand (Fig. 1). To mitigate potential growth penalties derived from any metabolic burden caused by the activity of engineered pathways, yeast could be grown to optimal cell density only then to induce the expression of food-attributes-conveying pathways before biomass harvesting. Furthermore, once reaching the desired cell density, yeast biomass could be split into separate batches, each one induced to display particular characteristics to enable a varied menu (e.g., different meals for breakfast, lunch, and dinner) and the preparation of elaborate multi-component food (e.g., a layer cake, a burger).
Nutritional enhancement of yeast
As discussed above, yeast is a nutritious food source for humans, having been consumed in food and beverages for thousands of years. While the extent to which a healthy human diet can consist of yeast is uncertain, dairy cattle have been fed diets with up to 40% liquid yeast cells (11% dry matter) without any negative performance or health effects47. Here, we propose that the nutritional value of yeast could be further enhanced to develop microbial foods for optimal human nutrition (Fig. 1).
One of the most obvious deficiencies in the nutritional profile of S. cerevisiae is its relatively low content of lipids (∼1.5%)13. A healthy human diet requires ~20–35% fats, meaning that the metabolic profile of S. cerevisiae would need to be modified to meet this demand if it were to comprise a significant portion of a human diet. Considerable progress has been made in increasing the lipid content of S. cerevisiae by combining adaptive laboratory evolution with metabolic engineering to develop yeast with high levels of fatty acids48. These strategies and others could be employed to contribute to the mouthfeel for microbial food and to a balanced diet, including the provision of adequate amounts of essential (i.e., linoleic acid and alpha-linolenic acid49) and health-promoting (e.g., omega-3 fatty acids50,51) fats. Alternatively, microbial species that naturally accumulate lipids, such as the oleaginous yeast Y. lipolytica, which is also extensively used for the commercial production of foods52, could be used to increase dietary fats. Y. lipolytica dry weight can comprise ~50% lipids53, and many modern synthetic biology tools are available to facilitate its metabolic manipulation52.
Yeast has been successfully engineered to produce many nutraceuticals54 and essential nutrients that humans must obtain from dietary sources, including vitamin A55, vitamin C56, and 7-dehydrocholesterol, which is directly converted into vitamin D3 in the skin upon ultra-violet (UV) light exposure57. Yeast production of other essential vitamins―such as vitamins B, E, and K―has not yet been fully achieved, but recent research strides in this field indicate that this may be realized in the foreseeable future58. Until yeast production of all essential and desired human nutrients in sufficient amounts becomes a reality, other natural or engineered microorganisms that already synthesize them58 could be used to supplement yeast biomass to formulate microbial food for optimal nutrition.
Engineering food sensory attributes in yeast
Besides fulfilling human nutritional needs, if we are to develop microbial-based foods, it will be essential that edible microbial biomass also enables the production of appealing foods. In this section, we discuss the bioengineering of sensory attributes of food into microorganisms (Fig. 1).
The taste and smell sensations of food are intimately entwined and critical to our enjoyment while eating. Therefore, the engineering of microorganisms to provide a repertoire of food flavors and odors would be crucial for the development of microbial food. Fittingly, widespread applications in the food, feed, cosmetic, and pharmaceutical industries have motivated significant advances in generating taste and scent compounds in multiple yeasts59–61. Examples range from fruit smells, such as S. cerevisiae engineered to produce a ketone that imparts raspberry aroma62, and flavoring agents like vanillin―the key constituent of the natural vanilla flavor―produced in S. cerevisiae and Schizosaccharomyces pombe63, all the way through to meat flavor and aroma by the production of soy leghemoglobin in P. pastoris64.
Another key attribute of food is texture, which is highly determined by both composition and structure. Therefore, engineering S. cerevisiae in ways that can provide textural elements will be an important aspect for the development of yeast-based foods. Several molecules that could be used to this end have already been recombinantly produced in microbes. Examples include cellulose and starch, which are major structural and textural components of plant matter, and collagen and gelatin, which are used as textural agents in food products like yogurt and marshmallows65–67. In addition, as mentioned above, different approaches have also been successfully implemented to alter the lipid content of yeast48. Lipids not only contribute to the nutrition and taste of food, but also to texture, so altering yeast’s lipid content could also serve to modify the textural properties of yeast-based food products. Finally, there is a repertoire of biological compounds, such as pectin, lecithin, transglutaminases, and xanthan gum, that could be conceivably exploited to achieve additional textural manipulations in microbial food products.
Color is also an important sensory attribute of foods, and many microbial-derived pigments are widely used as colorants to make food products more attractive68. Thus, engineered pigmentation could be employed to improve the visual appeal of microbial foods. While yeast is normally a whitish beige, very different colored yeast strains can be generated by genetic manipulation. For instance, carotenoid genes can be used to impart color in the yellow to red range, while the violacein pathway and genes from bacteria and coral can be used to confer purple, green, blue, and magenta colorations69–72. The usage of genes for chromoprotein variants further expands the possibilities for engineering many different pigmentations73.
From the bioreactor to the table
The simplest way to consume food-grade microbial biomass could probably be as a liquid or pureed homogenous food or in the form of solid food products such as noodles or snack bars. However, 3D-food printing technologies make it possible to manufacture more complex food in terms of sensory properties and to create aesthetically pleasing custom meals74. Hence, the development of 3D-printing methods for microbial cells could enable the realization of a plethora of different appealing food from microbial biomass. This approach could also allow maximum food output with minimal waste, as the whole edible microbial biomass harvested from bioreactors could potentially be used for printing food.
Technology for 3D printing in microgravity is being developed75, and it would be a powerful approach to use in conjunction with the sensory bioengineering of microorganisms to manufacture foods with a fully customized design. For instance, we imagine that microbial cells with different sensory profiles could be 3D printed to mimic the texture and appearance of vegetables or meats or into new food products of designed shapes that combine multiple types of textures and different flavors (Fig. 1).
Future outlook
A most crucial aspect of future long-duration human space enterprises will be the development of new approaches enabling sustainable food-production off-Earth. In this context, we have considered the bioengineering of microorganisms to develop microbial-based food. We argue that the yeasts, particularly S. cerevisiae, pose advantages that make it attractive to repurpose them into a cornerstone microbial platform for developing a complete food-production system. Although immense hurdles remain, synthetic biology approaches offer a way to overcome yeast’s natural limitations and bestow it with tailored sensory and nutritional food attributes, as discussed above. Eventually, progress in our capacity to thoroughly engineer other microorganisms (e.g., cyanobacteria) and self-replicating synthetic cells that can be programmed to exclusively synthesize specified compounds would allow us to expand the repertoire of cell platforms for producing novel food products. This vision will also require advances in engineering infrastructure for microbial growth (e.g., microgravity conditions may require developing specific bioreactor designs and media composition) and processing (e.g., consumption of microbial biomass may require reducing nucleic acid content since, at too high concentrations, their catabolism in humans leads to accumulation of uric acid, which can cause gout-like symptoms13), as well as printing that interfaces with biology and has a low mass and power footprint. Noteworthy is that microbial food-production systems will not be exempt from risks that could compromise food security, such as bioreactor failure, contamination, and instability of bioengineered traits. These risks could be mitigated by implementing backup microbial stocks and infrastructure, and it is important to note that comparable challenges will likely apply to any food-production system. Experimentation onboard the International Space Station and crewed missions planned to the Moon and Mars in the next three decades1 provide a window of opportunity to prototype and optimize the required technologies, so they mature and become viable for supporting long-duration human presence in space.
Beyond its potential for supporting extended human endeavors in space, bioengineered microbial-based food could also open opportunities to address global food security and immediately impact Earth’s food industry. Global population growth and the associated increasing demands for food, together with the need to reduce the negative environmental impacts of modern agriculture, indicate that radical innovations will be needed to sustainably achieve food security in the coming decades. In addition to a new generation of crop plants with enhanced productivity and resilience to environmental stresses, the development of microbial foods holds enormous potential to address these challenges. The approach envisioned here could find applications in food manufacturing, food services and catering, and food retail, but also increase self-sufficiency in areas of inadequate infrastructure or in need of emergency support where local food-production capacity or availability of external food-sources is compromised (e.g., during crises, conflicts, remote places). Given the cultural importance of food and the fact that it is ingested into the body, conservatism and a high dose of skepticism will likely accompany the development of microbial food. However, the use of microbes for the industrial manufacture of feed and food-grade products is gaining prominence37,76, and the day may arrive soon when supermarkets and restaurants offer microbial food products, and kitchen bioreactors and microbial 3D-food printers are commonplace home appliances.
Acknowledgements
We thank Scott M. Smith (Human Health and Performance Directorate, NASA Johnson Space Center) and Nils J. H. Averesch (Stanford University and Center for the Utilization of Biological Engineering in Space) for valuable discussions on the manuscript. External support for Macquarie University’s Synthetic Biology initiative is acknowledged from Bioplatforms Australia, the New South Wales (NSW) Chief Scientist and Engineer, and the NSW Government’s Department of Primary Industries. Australian Government funding through its investment agency, the Australian Research Council, towards the Macquarie University-led ARC Centre of Excellence for Synthetic Biology is gratefully acknowledged. B.L. is supported by the Gordon and Betty Moore Foundation (GBMF9319, grant 10.37807/GBMF9319), the ARC Centre of Excellence for Synthetic Biology, Twist Bioscience, and the Allen Foundation. Bronte Turner from Serpentine Studio is gratefully acknowledged for the artwork used in the figures. Parts of the figure were created with BioRender.com. We apologize to those colleagues whose relevant work has not been cited due to space limitations.
Author contributions
B.L. conceived, developed, and led the writing of the article. T.C.W., H.D.G., I.S.P., and I.T.P. helped to refine and develop the concept, made key contributions, and co-wrote some sections of the article.
Peer review
Peer review information
Nature Communications thanks Kevin Verstrepen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
T.C.W. is a shareholder and of co-founder Number 8 Bio Pty Ltd. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Berliner AJ, et al. Towards a biomanufactory on Mars. Front. Astron Space Sci. 2021;8:711550. doi: 10.3389/fspas.2021.711550. [DOI] [Google Scholar]
- 2.Berliner, A. J. et al. Space bioprocess engineering on the horizon. Commun. Eng.1, 13 (2022).
- 3.Douglas GL, Zwart SR, Smith SM. Space food for thought: challenges and considerations for food and nutrition on exploration missions. J. Nutr. 2020;150:2242–2244. doi: 10.1093/jn/nxaa188. [DOI] [PubMed] [Google Scholar]
- 4.Montague M, et al. The role of synthetic biology for in situ resource utilization (ISRU) Astrobiology. 2012;12:1135–1142. doi: 10.1089/ast.2012.0829. [DOI] [PubMed] [Google Scholar]
- 5.Nangle SN, et al. The case for biotech on Mars. Nat. Biotechnol. 2020;38:401–407. doi: 10.1038/s41587-020-0485-4. [DOI] [PubMed] [Google Scholar]
- 6.Llorente B, Williams TC, Goold HD. The multiplanetary future of plant synthetic biology. Genes. 2018;9:348. doi: 10.3390/genes9070348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menezes AA, Cumbers J, Hogan JA, Arkin AP. Towards synthetic biological approaches to resource utilization on space missions. J. R. Soc. Interface. 2015;12:20140715. doi: 10.1098/rsif.2014.0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Way JC, Silver PA, Howard RJ. Sun-driven microbial synthesis of chemicals in space. Int. J. Astrobiol. 2011;10:359–364. doi: 10.1017/S1473550411000218. [DOI] [Google Scholar]
- 9.Cannon KM, Britt DT. Feeding one million people on Mars. N. Space. 2019;7:245–254. doi: 10.1089/space.2019.0018. [DOI] [Google Scholar]
- 10.Hader DP. On the way to Mars-flagellated algae in bioregenerative life support systems under microgravity conditions. Front. Plant Sci. 2019;10:1621. doi: 10.3389/fpls.2019.01621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi KR, Yu HE, Lee SY. Microbial food: microorganisms repurposed for our food. Micro. Biotechnol. 2022;15:18–25. doi: 10.1111/1751-7915.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Xin F, Alper HS. Bio-synthesis of food additives and colorants-a growing trend in future food. Biotechnol. Adv. 2021;47:107694. doi: 10.1016/j.biotechadv.2020.107694. [DOI] [PubMed] [Google Scholar]
- 13.Linder T. Making the case for edible microorganisms as an integral part of a more sustainable and resilient food production system. Food Security. 2019;11:265–278. doi: 10.1007/s12571-019-00912-3. [DOI] [Google Scholar]
- 14.Samuel D. Investigation of ancient egyptian baking and brewing methods by correlative microscopy. Science. 1996;273:488–490. doi: 10.1126/science.273.5274.488. [DOI] [PubMed] [Google Scholar]
- 15.Onofre SB, Bertoldo IC, Abatti D, Refosco D. Chemical composition of the biomass of Saccharomyces cerevisiae - (Meyen ex E. C. Hansen, 1883) yeast obtained from the beer manufacturing process. Int. J. Environ. Agric Biotechnol. 2017;2:558–562. [Google Scholar]
- 16.Nielsen J. Yeast systems biology: model organism and cell factory. Biotechnol. J. 2019;14:e1800421. doi: 10.1002/biot.201800421. [DOI] [PubMed] [Google Scholar]
- 17.Purevdorj-Gage B, Sheehan KB, Hyman LE. Effects of low-shear modeled microgravity on cell function, gene expression, and phenotype in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2006;72:4569–457. doi: 10.1128/AEM.03050-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell PJL, et al. An electro-microbial process to uncouple food production from photosynthesis for application in space exploration. Life. 2022;12:1002. doi: 10.3390/life12071002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stern JC, et al. Evidence for indigenous nitrogen in sedimentary and aeolian deposits from the Curiosity rover investigations at Gale crater, Mars. Proc. Natl Acad. Sci. USA. 2015;112:4245–4250. doi: 10.1073/pnas.1420932112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meerman R, Brown AJ. When somebody loses weight, where does the fat go? BMJ. 2014;349:g7257. doi: 10.1136/bmj.g7257. [DOI] [PubMed] [Google Scholar]
- 21.Dai Z, et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae. Bioresour. Technol. 2017;245:1407–1412. doi: 10.1016/j.biortech.2017.05.100. [DOI] [PubMed] [Google Scholar]
- 22.Espinosa MI, et al. Adaptive laboratory evolution of native methanol assimilation in Saccharomyces cerevisiae. Nat. Commun. 2020;11:5564. doi: 10.1038/s41467-020-19390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Averesch NJ. Choice of microbial system for in-situ resource utilization on Mars. Front. Astron Space Sci. 2021;8:700370. doi: 10.3389/fspas.2021.700370. [DOI] [Google Scholar]
- 24.Somoza-Tornos A, Guerra OJ, Crow AM, Smith WA, Hodge BM. Process modeling, techno-economic assessment, and life cycle assessment of the electrochemical reduction of CO2: a review. iScience. 2021;24:102813. doi: 10.1016/j.isci.2021.102813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Looser V, et al. Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol. Adv. 2015;33:1177–1193. doi: 10.1016/j.biotechadv.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Verduyn C. Physiology of yeasts in relation to biomass yields. Antonie Leeuwenhoek. 1991;60:325–353. doi: 10.1007/BF00430373. [DOI] [PubMed] [Google Scholar]
- 27.Verseux C, et al. Sustainable life support on Mars—the potential roles of cyanobacteria. Int. J. Astrobiol. 2016;15:65–92. doi: 10.1017/S147355041500021X. [DOI] [Google Scholar]
- 28.Verseux C, et al. A Low-Pressure, N2/CO2 atmosphere is suitable for cyanobacterium-based life-support systems on Mars. Front. Microbiol. 2021;12:611798. doi: 10.3389/fmicb.2021.611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ducat DC, Avelar-Rivas JA, Way JC, Silver PA. Rerouting carbon flux to enhance photosynthetic productivity. Appl. Environ. Microbiol. 2012;78:2660–2668. doi: 10.1128/AEM.07901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guadalupe-Medina V, et al. Carbon dioxide fixation by Calvin-Cycle enzymes improves ethanol yield in yeast. Biotechnol. Biofuels. 2013;6:125. doi: 10.1186/1754-6834-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papapetridis I, et al. Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol. Biofuels. 2018;11:17. doi: 10.1186/s13068-017-1001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gassler T, et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO2. Nat. Biotechnol. 2020;38:210–216. doi: 10.1038/s41587-019-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gleizer S, et al. Conversion of Escherichia coli to generate all biomass carbon from CO2. Cell. 2019;179:1255–1263. doi: 10.1016/j.cell.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fabarius JT, Wegat V, Roth A, Sieber V. Synthetic methylotrophy in yeasts: towards a circular bioeconomy. Trends Biotechnol. 2021;39:348–358. doi: 10.1016/j.tibtech.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez de la Cruz J, Machens F, Messerschmidt K, Bar-Even A. Core catalysis of the reductive glycine pathway demonstrated in yeast. ACS Synth. Biol. 2019;8:911–917. doi: 10.1021/acssynbio.8b00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espinosa MI, Williams TC, Pretorius IS, Paulsen IT. Benchmarking two Saccharomyces cerevisiae laboratory strains for growth and transcriptional response to methanol. Synth. Syst. Biotechnol. 2019;4:180–188. doi: 10.1016/j.synbio.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcellin E, Angenent LT, Nielsen LK, Molitor B. Recycling carbon for sustainable protein production using gas fermentation. Curr. Opin. Biotechnol. 2022;76:102723. doi: 10.1016/j.copbio.2022.102723. [DOI] [PubMed] [Google Scholar]
- 38.Godard P, et al. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:3065–3086. doi: 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brabender M, Hussain MS, Rodriguez G, Blenner MA. Urea and urine are a viable and cost-effective nitrogen source for Yarrowia lipolytica biomass and lipid accumulation. Appl. Microbiol. Biotechnol. 2018;102:2313–2322. doi: 10.1007/s00253-018-8769-z. [DOI] [PubMed] [Google Scholar]
- 40.Santa Maria, S. R., Marina, D. B., Tieze, S. M., Liddell, L. C. & Bhattacharya, S. BioSentinel: long-term Saccharomyces cerevisiae preservation for a deep space biosensor mission. Astrobiology20, 1–14 (2020). [DOI] [PMC free article] [PubMed]
- 41.Postma ED, et al. Modular, synthetic chromosomes as new tools for large scale engineering of metabolism. Metab. Eng. 2022;72:1–13. doi: 10.1016/j.ymben.2021.12.013. [DOI] [PubMed] [Google Scholar]
- 42.Kutyna DR, et al. Construction of a synthetic Saccharomyces cerevisiae pan-genome neo-chromosome. Nat. Commun. 2022;13:3628. doi: 10.1038/s41467-022-31305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beekwilder J, et al. Polycistronic expression of a beta-carotene biosynthetic pathway in Saccharomyces cerevisiae coupled to beta-ionone production. J. Biotechnol. 2014;192 Pt B:383–392. doi: 10.1016/j.jbiotec.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 44.Majer E, Llorente B, Rodriguez-Concepcion M, Daros JA. Rewiring carotenoid biosynthesis in plants using a viral vector. Sci. Rep. 2017;7:41645. doi: 10.1038/srep41645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon TA, Williams TC, Pretorius IS. Sensing the future of bio-informational engineering. Nat. Commun. 2021;12:388. doi: 10.1038/s41467-020-20764-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker RSK, Pretorius IS. Synthetic biology for the engineering of complex wine yeast communities. Nat. Food. 2022;3:249–254. doi: 10.1038/s43016-022-00487-x. [DOI] [PubMed] [Google Scholar]
- 47.Besong S, Jackson JA, Hicks CL, Hemken RW. Effects of a supplemental liquid yeast product on feed intake, ruminal profiles, and yield, composition, and organoleptic characteristics of milk from lactating Holstein cows. J. Dairy Sci. 1996;79:1654–1658. doi: 10.3168/jds.S0022-0302(96)76529-3. [DOI] [PubMed] [Google Scholar]
- 48.Yu T, et al. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174:1549–1558.e1514. doi: 10.1016/j.cell.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Yazawa H, Iwahashi H, Kamisaka Y, Kimura K, Uemura H. Improvement of polyunsaturated fatty acids synthesis by the coexpression of CYB5 with desaturase genes in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2010;87:2185–2193. doi: 10.1007/s00253-010-2679-z. [DOI] [PubMed] [Google Scholar]
- 50.Tavares S, et al. Metabolic engineering of Saccharomyces cerevisiae for production of Eicosapentaenoic Acid, using a novel Delta 5-Desaturase from Paramecium tetraurelia. Appl. Environ. Microbiol. 2011;77:1854–1861. doi: 10.1128/AEM.01935-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu X, Hong H, MacKenzie SL. Identification of a Delta 4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 2001;276:31561–31566. doi: 10.1074/jbc.M102971200. [DOI] [PubMed] [Google Scholar]
- 52.Larroude M, Rossignol T, Nicaud JM, Ledesma-Amaro R. Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol. Adv. 2018;36:2150–2164. doi: 10.1016/j.biotechadv.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beopoulos A, et al. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl. Environ. Microbiol. 2008;74:7779–7789. doi: 10.1128/AEM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan SF, Alper HS. Metabolic engineering of microbial cell factories for production of nutraceuticals. Micro. Cell Fact. 2019;18:46. doi: 10.1186/s12934-019-1096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun L, Kwak S, Jin YS. Vitamin A Production by engineered Saccharomyces cerevisiae from xylose via two-phase in situ extraction. ACS Synth. Biol. 2019;8:2131–2140. doi: 10.1021/acssynbio.9b00217. [DOI] [PubMed] [Google Scholar]
- 56.Branduardi P, et al. Biosynthesis of vitamin C by yeast leads to increased stress resistance. PLoS ONE. 2007;2:e1092. doi: 10.1371/journal.pone.0001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo XJ, et al. Metabolic engineering of Saccharomyces cerevisiae for 7-dehydrocholesterol overproduction. Biotechnol. Biofuels. 2018;11:192. doi: 10.1186/s13068-018-1194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Liu L, Jin Z, Zhang D. Microbial cell factories for green production of vitamins. Front. Bioeng. Biotechnol. 2021;9:66156. doi: 10.3389/fbioe.2021.661562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Wyk N, Kroukamp H, Pretorius IS. The smell of synthetic biology: engineering strategies for aroma compound production in yeast. Fermentation. 2018;4:54. doi: 10.3390/fermentation4030054. [DOI] [Google Scholar]
- 60.Kallscheuer N, Classen T, Drepper T, Marienhagen J. Production of plant metabolites with applications in the food industry using engineered microorganisms. Curr. Opin. Biotechnol. 2019;56:7–17. doi: 10.1016/j.copbio.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 61.Denby CM, et al. Industrial brewing yeast engineered for the production of primary flavor determinants in hopped beer. Nat. Commun. 2018;9:965. doi: 10.1038/s41467-018-03293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee D, Lloyd ND, Pretorius IS, Borneman AR. Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion. Micro. Cell Fact. 2016;15:49. doi: 10.1186/s12934-016-0446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen EH, et al. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker’s yeast (Saccharomyces cerevisiae) Appl. Environ. Microbiol. 2009;75:2765–2774. doi: 10.1128/AEM.02681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shankar, S. & Hoyt, M. A. United States Patent and Trademark Office. Expression constructs and methods of genetically engineering methylotrophic yeast, (ed. USPTO). USA patent (2020).
- 65.Dance A. Engineering the animal out of animal products. Nat. Biotechnol. 2017;35:704–707. doi: 10.1038/nbt.3933. [DOI] [PubMed] [Google Scholar]
- 66.Buldum G, Bismarck A, Mantalaris A. Recombinant biosynthesis of bacterial cellulose in genetically modified Escherichia coli. Bioprocess Biosyst. Eng. 2018;41:265–279. doi: 10.1007/s00449-017-1864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfister B, et al. Recreating the synthesis of starch granules in yeast. Elife. 2016;5:e15552. doi: 10.7554/eLife.15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sen T, Barrow CJ, Deshmukh SK. Microbial pigments in the food industry-challenges and the way forward. Front. Nutr. 2019;6:7. doi: 10.3389/fnut.2019.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keppler-Ross S, Noffz C, Dean N. A new purple fluorescent color marker for genetic studies in Saccharomyces cerevisiae and Candida albicans. Genetics. 2008;179:705–710. doi: 10.1534/genetics.108.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitchell LA, et al. Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res. 2015;43:6620–6630. doi: 10.1093/nar/gkv466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wehrs M, et al. Production efficiency of the bacterial non-ribosomal peptide indigoidine relies on the respiratory metabolic state in S. cerevisiae. Micro. Cell Fact. 2018;17:193. doi: 10.1186/s12934-018-1045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeLoache WC, Russ ZN, Dueber JE. Towards repurposing the yeast peroxisome for compartmentalizing heterologous metabolic pathways. Nat. Commun. 2016;7:11152. doi: 10.1038/ncomms11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liljeruhm J, et al. Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. J. Biol. Eng. 2018;12:8. doi: 10.1186/s13036-018-0100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun J, Peng Z, Yan L, Fuh JY, Hong GS. 3D food printing—An innovative way of mass customization in food fabrication. Int. J. Bioprinting. 2015;1:27–38. [Google Scholar]
- 75.Prater T, et al. 3D printing in zero G technology demonstration mission: complete experimental results and summary of related material modeling efforts. Int. J. Adv. Manuf. Technol. 2019;101:391–417. doi: 10.1007/s00170-018-2827-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Banks M, Johnson R, Giver L, Bryant G, Guo M. Industrial production of microbial protein products. Curr. Opin. Biotechnol. 2022;75:102707. doi: 10.1016/j.copbio.2022.102707. [DOI] [PubMed] [Google Scholar]