Abstract
Staphylococcus aureus is one of the most prominent nosocomial, community and farm acquired bacterial infections among animals and human populations. The main purpose of our study was to identify and characterize antimicrobial resistance (AMR) among Staphylococcus aureus isolated from livestock, poultry and humans and to further identify the associated genes. Staphylococcus aureus isolates from human, bovine, swine and poultry were collected from different laboratories across the United States collected between 2003 and 2016. Antimicrobial susceptibility testing for 13 antimicrobials was performed and conventional PCR was used to detect the presence of the nuc gene, mec gene, and to detect int1 gene. Associations between the presence of mec and intl and specific AMR profiles were determined. Antimicrobial resistance was detected in all four host categories, with the highest overall rates found in swine, 100% resistant to tetracycline, 88% to penicillin and 64% clindamycin. The next highest was found among humans with 81.6% of isolates resistant to penicillin followed by 44% to clindamycin and 43% to erythromycin. Among beef cattle isolates, 63.2% were resistant to penicillin, 15.8% resistant to clindamycin and 15.8% to erythromycin. No isolates from any of the hosts were resistant to linezolid. Among poultry isolates, the highest AMR was found to clindamycin, followed by erythromycin and penicillin. Among dairy cattle, highest resistance was found to penicillin, followed by chloramphenicol and gentamicin. Dairy cattle were the only host category with isolates that are resistant to trimethoprim-sulfamethoxazole. Of the 220 isolates detected by latex agglutination, 217 were confirmed to be S. aureus via PCR of the nuc gene, 21.4% were positive for the mecA gene. Swine had the highest prevalence of the mecA gene, followed by humans, poultry and beef cattle. This study has demonstrated a high occurrence of penicillin resistance among all S. aureus isolates. There were differences observed between host species with tetracycline resistance being the highest among swine isolates and clindamycin being highest in poultry isolates. No detection of oxacillin resistance was found in isolates from dairy cattle but was found in isolates from all of the other host species, 94% of which contained the mecA gene.
Keywords: Antimicrobial resistance, Staphylococcus aureus, MRSA, Livestock, One health, Human, Environment
Highlights
-
•
High occurrence of penicillin resistance in Staphylococcus aureus isolates collected from livestock, poultry and humans.
-
•
Tetracycline resistance was the highest among swine isolates and clindamycin was the highest in poultry isolates.
-
•
Oxacillin resistance was not detected among dairy cattle isolates but was found in isolates from other host species.
-
•
Ninety four percent of the S. aureus isolates were resistant to oxacillin contained the mecA gene.
1. Introduction
Emerging and existing antimicrobial drug resistance (AMR) is a major public health concern with global relevance to overall animal health, specifically livestock. AMR is often only detected following treatment failure, during which time the potential to spread disease to susceptible species is high. Although the development and spread of AMR is multifactorial, it has been shown that AMR definitively increases the severity of foodborne illness [1,2] and other clinical infections in people [[3], [4], [5]] as well as animals.
Among all staphylococci, S. aureus is the most invasive species and an etiological agent of diverse human and animal maladies, including skin infections, abscesses, food poisoning, toxic shock syndrome, septicemia, endocarditis, and pneumonia [[6], [7], [8]]. Additionally, S. aureus is one of the most prominent causes of nosocomial- and community- acquired bacterial infections worldwide [[9], [10], [11]] enhancing the need to determine if the AMR of isolates from humans and other species is significant.
Penicillin was the first antibiotic mass produced for use in humans. Although initially it was highly effective for treatment of S. aureus infections, today over 90% of human S. aureus strains are resistant to this antibiotic. Antimicrobial resistance genes, including those genes encoding penicillin resistance, can be found on mobile genetic elements such as plasmids, transposons, integrons [12,13]. Mobile genetic elements (MGEs) constitute only 25% of the staphylococcal genome [14], and they encode many putative virulence factors and antimicrobial resistance mechanisms. Thus, MGEs have an important role in bacterial adaptability and survival [15].
The main objective of our study was to phenotypically and genotypically identify AMR in Staphylococcus aureus isolates from livestock, poultry, livestock environment (swine and poultry) and humans. The findings from this study increases our understanding of host differences so surveillance for AMR bacteria in humans and food-producing animals could be improved.
As per the U.S. Department of Health and Human Services [16], investing in advanced diagnostics and increasing surveillance for antibiotic-resistant zoonotic and animal pathogens is needed, especially as a means to strengthen detection and control of antibiotic resistance. This study directly addresses the White House AMR initiative by classifying disease-causing Staphylococcus aureus infecting both humans and livestock according to their AMR patterns and molecular markers. The results of this study could be used to address dangerous AMR infections, including S. aureus, originating from livestock as well as from the human hospitals.
In the past decades, indiscriminate use and abuse of existing antibiotics has led to proliferation of antibiotic resistance in microbes [17] and consequently an increasing number of clinical failures in bacterial mediated diseases [[18], [19], [20]]. Several genetic carriers are responsible for the emergence and prevalence of AMR, such as plasmids and transposons. One way of detecting AMR is through detection and characterization of integrons. Integrons are genetic elements that acquire and incorporate gene cassettes that code for antibiotic resistance. Integrons are linked to mobile DNA elements (i.e. transposons and/or conjugative plasmids) and are horizontally transmissible [18,[21], [22], [23], [24]]. Integrons associated with AMR have been detected in many bacterial species, including methicillin-resistant S. aureus (MRSA) [25]. In addition to MRSA, methicillin susceptible S. aureus (MSSA) have been found to contain integrons and can be resistant to at least 6 other antibiotics [26]. MSSA strains are capable of acquiring the methicillin resistance coding mecA gene cassette from MRSA [27].
There are five different classes of integrons, each encoding a distinct integrase gene [28], class 1 integrons being the most common type in clinical isolates of the Enterobacteriaceae [29]. The S. aureus strains that carried class 1 integrons was highest in those found to be multi-drug resistant S. aureus (MDR-SA) [30]. They were found in 72.6% of S. aureus clinical isolates [31].
In a study of S. aureus isolated from US meat and poultry samples, 96% were resistant to at least one antibiotic [32] and 78.4% of integrase-positive strains were multi-drug resistant. In the US, livestock-associated S. aureus can also be found in humans; likewise, human strains of S. aureus have also been found in US livestock [33]. Similar findings have been reported in the Netherlands [34]. This indicates a need to assess characteristics shared between S. aureus isolates derived from both humans and animals. Thus, understanding the host differences of AMR as well as AMR gene carriage will be an essential step to effectively manage pathogen transmission, especially within enclosed farm environments and hospitals and thus, reduce the number of new cases among humans, livestock, and poultry.
This work reveals an innovative way of improving detection of antibiotic-resistant zoonotic and animal pathogens nationwide. The diversity of hosts from which S. aureus AMR strains can be isolated suggests that much can be learned to improve our current approaches to mitigate the spread of multi-drug resistant pathogens.
2. Materials and methods
Ethical approval: Live animals were not used for the study.
2.1. Sample collection and culturing methods
Staphylococcus aureus isolates from human, bovine, swine and poultry hosts were collected from various repository laboratories across the United States representing time period between 2003 and 2017. Three-hundred and four isolates were initially collected from the laboratories located in Colorado, Pennsylvania, Washington, Ohio and Iowa. Only 254 isolates indicated appropriate host metadata. Of those, 220 isolates were identified as S. aureus by latex agglutination [35,36]. Single isolates were grown overnight in brain heart infusion broth at 35 °C. After overnight incubation, a 1 ml aliquot was removed, added to 1 ml of glycerol and frozen at −80 °C for further testing.
Molecular amplification of the nuc, mecA and integron cassette genes using multiplex PCR:
If an isolate was tested positive by latex agglutination, PCR of the nuc gene was used to confirm S. aureus [37]. PCRs were also conducted for the methicillin resistance gene (mec) [37] and for class 1 integron gene cassettes [22].
A portion of each S. aureus stock isolate was scraped into a separate microcentrifuge tube, thawed, and centrifuged for 5 min at 5000 xg. The supernatant was removed, and each pellet was resuspended in molecular grade water in a 1:3 ratio (10 μl cell pellet suspended in 30 μl water). A total of 5 μL of each washed, resuspended isolate was used as template and added to the following multiplex PCR master mix reaction to detect nuc gene. A total of 5 μL of each resuspended isolate was used as template in the following multiplex PCR mastermix for a 25 μl total reaction volume: 12.5 μl 2× Qiagen Multiplex PCR master mix (Qiagen, Valencia, CA), 2.0 μM primer mix (containing 2 μM of each primer: nuc forward primer sequence: 5′- AGC CAA GCC TTG ACG AAC TAA −3′; nuc reverse primer sequence: 5′- GCG ATT GAT GGT GAT ACG GTT - 3′; mecA forward primer sequence: 5′- GTA GAA ATG ACT GAA CGT CCG ATA A − 3′; mecA reverse primer sequence: 5′- CCA ATT CCA CAT TGT TTC GGT CTA A - 3′; integron forward primer sequence: 5′- GGC ATC CAA GCA GCA AGC -3′; integron reverse primer sequence: 5′- AAG CAG ACT TGA CCT GAT - 3′), 2.5 μl Q-Solution (Qiagen, Valencia, CA), and 2.5 μl molecular grade water. Each 25 μl reaction was overlaid with 30 μl Chill Out wax (Bio-Rad, Hercules, CA) to prevent evaporation and placed into an MJ Research 60 place thermal cycler (Bio-Rad). Thermal cycling conditions consisted of an initial incubation at 95 °C for 10 min to activate the polymerase, followed by 35 cycles of 94 °C for 30 s, 56 °C for 2 min, 72 °C for 1.5 min, and a final extension incubation at 72 °C for 10 min. Following amplification, samples were run on a 1.5% agarose gel. Samples containing 279 bp products were recorded as nuc gene positive, samples containing 310 bp were recorded as mecA gene positive, and samples containing 1000, 1200, or 1600 bp amplicons were recorded as containing one or more class I integron gene cassette.
Two positive control samples for class I integrons with sizes of 1,000, 1,200, and 1,600 bp were included (5 pg total) with each PCR [22]. Staphylococcus aureus ATCC 25923 was included with each PCR as a positive control for the nuc gene product.
2.2. Antimicrobial susceptibility testing
To demonstrate and characterize antimicrobial resistance, antibiotic susceptibility testing (AST) was conducted on the 220 isolates by the Kirby Bauer (disk diffusion) method according to the Clinical Laboratory Standards Institute (CLSI) guidelines [38]. Isolates were plated on Mueller Hinton II agar. Antibiotics tested included ceftaroline (CPT-30), chloramphenicol (C-30), ciprofloxacin (CIP-5), clindamycin (CC-2), erythromycin (E-15), gentamicin (GM-10), linezolid (LZD-30), cefoxitin (FOX-30; surrogate for oxacillin per CLSI), penicillin G (P-10), rifampin (RA-5), tetracycline (TE-30) and sulfamethoxazole/ trimethoprim (SXT-23.75/1.25). Quality control was performed using a S. aureus positive control for beta-lactamase resistance (ATCC 29213), a S. aureus positive control for methicillin resistance (ATCC 43300), and a general S. aureus AST control for Mueller Hinton agar standardization (ATCC 25923). Antimicrobial resistance patterns were deduced using NARMS (National Antimicrobial Resistance Monitoring System) panels and breakpoints selected according to CLSI recommendations.
In addition, reduced susceptibility to vancomycin was determined by plating three 10 μl drops of a standardized concentration of bacteria (0.5 McFarland), on brain heart infusion agar containing 6 μg/ml vancomycin and counting colonies. Disk approximation testing or D-zone reactions (erythromycin-inducible clindamycin resistance) were also recorded. This was conducted by placing an erythromycin and clindamycin disk 15 mm apart on an agar plate after isolate plating. All erythromycin resistant and clindamycin sensitive isolates were subjected to D-zone test using erythromycin (15 μg) and clindamycin (2 μg) according to Clinical and Laboratory Standards Institute [39]. If there was flattening of the zone of inhibition between the 2 disks and the zone resembles the letter “D,” the test result was interpreted as positive for induction of clindamycin resistance. Enterococcus faecalis (ATCC 51299) was included as a positive control for vancomycin resistance. Reduced susceptibility to vancomycin was recorded if >1 colony per 10 μl drop was observed following 24 h incubation at 37 °C.
2.3. Statistical analysis
The data on AMR results of each antimicrobial were classified into susceptible, intermediate and resistant. The AMR data was considered as a binary variable (resistance or susceptible) for each drug tested. Those isolates which demonstrated an intermediate resistance to antibiotics were considered resistant for analyses. All data was visually represented using a heat map to depict the host variations on the patterns. The AST data on intermediate results was combined with resistance pattern to evaluate host differences using a logistic regression analysis for the comparisons with appropriate number of observations. All analyses were performed using SAS v9.4 (SAS Inc., Cary, NC).
3. Results
3.1. Antimicrobial resistance profiles of Staphylococcus aureus across host species
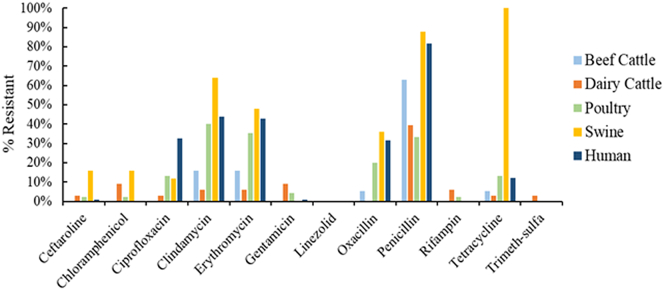
Seventy five percent of all S. aureus isolates were resistant to at least one antimicrobial drug and 40% were resistant to at least three antimicrobial drugs. The percentage of isolates found to be resistant to at least one antimicrobial drug among various hosts were: 68% of beef cattle, 42% of dairy cattle, 86% of human isolates, 64% of poultry, and 100% of swine isolates. Antimicrobial resistance was detected in all 5 host categories, with the highest overall rates found in swine, 100% resistance to tetracycline, 88% to penicillin and 64% clindamycin. The next highest was found among humans with 81.6% of isolates resistant to penicillin followed by 44% to clindamycin and 43% to erythromycin. Among beef cattle isolates, 63.2% were resistant to penicillin, 15.8% resistant to clindamycin and 15.8% to erythromycin. None of the isolates from any of the hosts were resistant to linezolid. Among poultry isolates, the highest AMR was found to clindamycin in 40% of the isolates, 35.6% to erythromycin, and 33.3% to penicillin. Among dairy cattle, 39.4% of isolates were resistant to penicillin, followed by 9.1% resistance to chloramphenicol and 9.1% to gentamicin. Dairy cattle were the only host category with isolates that are resistant to trimethoprim-sulfamethoxazole.
The percentage of S. aureus isolates determined to be resistant to various antimicrobials via susceptibility testing are depicted by host category in Fig. 1. The isolates with intermediate resistance were combined with resistant isolates.
Fig. 1.
Antimicrobial resistance of S. aureus by host category.
Oxacillin resistance was found among 22.7% of total S. aureus isolates, highest among swine (36%), followed by humans (31.6%), poultry (20%) and beef cattle (5.3%). None of the dairy cattle isolates were resistant to oxacillin.
3.2. mecA gene prevalence
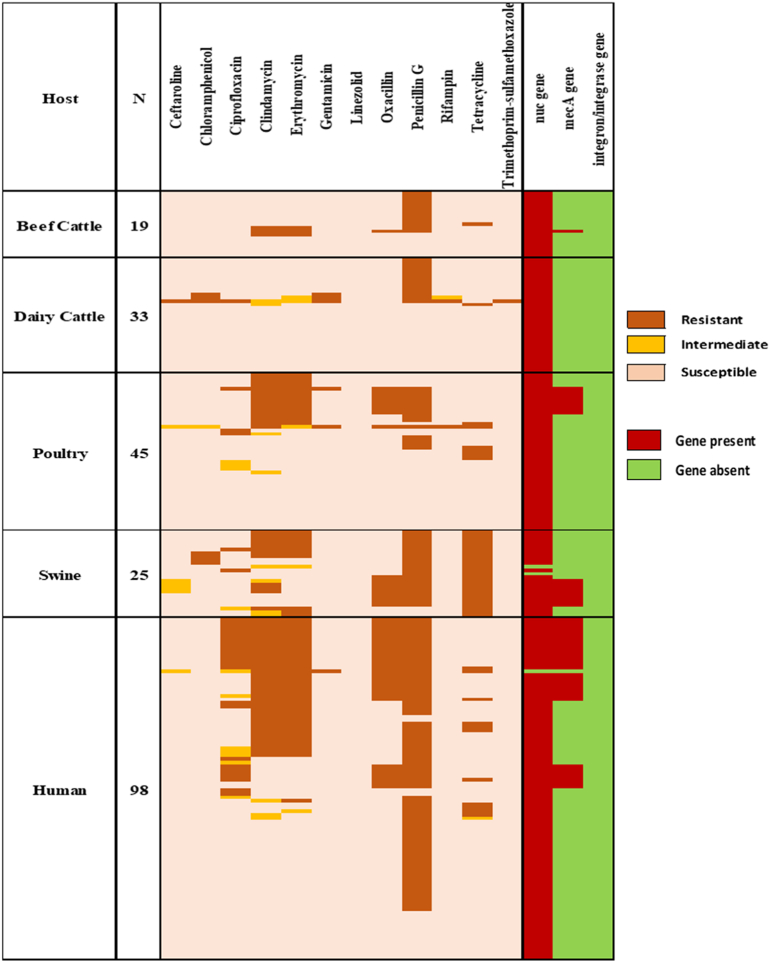
Latex agglutination identified 220 isolates serologically as S. aureus. Fig. 2 depicted AMR patterns of S. aureus with characterization of genes by host category. Briefly, the isolates were distributed in the five host categories: beef cows (n = 19), dairy cows (n = 33), poultry (n = 45), swine (n = 25) and humans (n = 98). Of the 220 isolates, 217 were confirmed to be S. aureus via PCR of the nuc gene. The three isolates negative for the nuc gene but serologically positive for S. aureus were included in the study as described by Hoegh et al. (2014) where nuc-negative clinical isolates of S. aureus were described [40].
Fig. 2.
Antimicrobial resistance patterns of S. aureus with gene characterization by host category.
Of the 220 S. aureus isolates tested, 21.4% (47/220) were positive for the mecA gene. Among the host categories studied, swine had the highest prevalence of the mecA gene, with 32% (8/25) of the samples. Humans had the second highest prevalence at 31% (30/98), followed by poultry at 18% (8/45) and beef cattle at 5% (1/19). There was no detection of the mecA gene in the dairy cow isolates. There was one isolate each from poultry, swine and human that were resistant to cefoxitin (surrogate for oxacillin) and did not carry the mecA gene.
3.3. Integron detection
None of the S. aureus isolates contained class 1 integrons, hence no further analysis was performed.
3.4. Association of AMR profiles and host differences
The statistical analyses to find differences between host species was performed for penicillin, clindamycin and erythromycin as there were not adequate number of isolates for analyses that were resistant to the other antimicrobials (Table 1).
Table 1.
Likelihood of clindamycin, erythromycin and penicillin resistance among host species.
| Comparison of host species | Clindamycin |
Erythromycin |
Penicillin |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | ||||
| Beef Cattle vs Dairy Cattle | 2.91 | 0.44 | 19.20 | 0.27 | 2.91 | 0.44 | 19.20 | 0.27 | 2.64 | 0.82 | 8.45 | 0.10 |
| Beef Cattle vs Poultry | 0.28 | 0.07 | 1.11 | 0.07 | 0.34 | 0.09 | 1.35 | 0.12 | 3.43 | 1.12 | 10.50 | 0.031⁎ |
| Swine vs Dairy Cattle | 27.55 | 5.31 | 142.95 | <0.001⁎ | 14.31 | 2.80 | 73.08 | 0.0014⁎ | 11.28 | 2.80 | 45.47 | 0.0007⁎ |
| Swine vs Beef Cattle | 9.48 | 2.16 | 41.61 | 0.0029⁎ | 4.92 | 1.14 | 21.23 | 0.033⁎ | 4.28 | 0.93 | 19.65 | 0.06 |
| Swine vs Poultry | 2.67 | 0.97 | 7.33 | 0.057 | 1.67 | 0.62 | 4.52 | 0.31 | 14.67 | 3.78 | 56.93 | 0.0001⁎ |
| Swine vs Human | 2.27 | 0.92 | 5.64 | 0.077 | 1.23 | 0.51 | 2.97 | 0.64 | 1.65 | 0.45 | 6.12 | 0.45 |
| Poultry vs Dairy Cattle | 10.33 | 2.20 | 48.63 | 0.0031⁎ | 8.55 | 1.81 | 40.47 | 0.0068⁎ | 0.77 | 0.30 | 1.96 | 0.58 |
| Human vs Dairy Cattle | 12.12 | 2.75 | 53.45 | 0.001⁎ | 11.62 | 2.63 | 51.30 | 0.0012⁎ | 6.84 | 2.88 | 16.25 | <0.001⁎ |
| Human vs Beef Cattle | 4.17 | 1.14 | 15.24 | 0.031⁎ | 4.00 | 1.09 | 14.62 | 0.04⁎ | 2.59 | 0.90 | 7.51 | 0.08 |
| Human vs Poultry | 1.17 | 0.57 | 2.40 | 0.66 | 1.36 | 0.66 | 2.82 | 0.41 | 8.89 | 3.98 | 19.85 | <0.001⁎ |
OR = Odds Ratio.
CI = Confidence Interval.
Significant at p-value<0.05
The isolates from swine had a significantly higher likelihood of penicillin, clindamycin and erythromycin resistance individually when compared to isolates from dairy cattle. Additionally, swine isolates were more likely to have clindamycin and erythromycin resistance when compared to isolates from beef cattle. There were no statistically significant differences detected in resistance to the three antimicrobials between dairy and beef cattle isolates.
Penicillin resistance was significantly higher in swine, beef cattle and human isolates when compared to poultry isolates. However, clindamycin and erythromycin resistance were significantly higher in poultry isolates when compared to dairy cattle isolates.
Human isolates were more likely to have clindamycin and erythromycin resistance when compared to dairy and beef cattle isolates, and significantly higher number of penicillin resistant isolates when compared to dairy cattle and poultry isolates.
4. Discussion
Antimicrobial resistance in bacterial infections continues to be a high-priority health concern in humans as well as animals. In humans, there is a high cost burden associated with treating infections caused by AMR bacteria due to longer hospitalization periods and high costs associated with complex treatment plans [41]. Staphylococcus aureus is prevalent in human and animal populations and is known to have developed resistance to different antimicrobials. Humans are the primary reservoir for S. aureus and serve as a hub for host switching events, allowing S. aureus to successfully infect multiple species [[42], [43], [44], [45]]. Confirmed cases of interspecies transmission between pigs, humans and cattle, emphasize the importance of understanding host specific antimicrobial resistance patterns for S. aureus to study transmission to animal and public health [44,46]. This study aimed to identify the prevalence of MRSA isolates and the antimicrobial resistance profiles of S. aureus isolates collected from humans and major food animal groups.
In the current study, swine and human S. aureus isolates had the highest detection of the mecA gene, a finding that was also reported by previous studies [45,[47], [48], [49], [50]]. This study suggests that MRSA was more likely to be isolated from swine populations when compared to poultry or cattle among S. aureus collected for laboratory analysis. The prevalence of methicillin resistance in swine isolates poses a potential risk for public health. In 2005, a swine-adapted lineage of MRSA was detected that could infect humans while having a low pathologic potential in swine [51]. Another study found a near 100% rate of nasal MRSA carriage in Dutch slaughter pigs, posing a potential risk for pig to human transmission [52].
The host groups with the lowest mecA detection in this study were dairy and beef cattle isolates, with no dairy cow isolates positive for the mecA gene. S. aureus is a common cause of contagious mastitis infections in dairy cows and can be difficult to control with antimicrobials alone. Hygiene protocols to prevent contagious mastitis transmission are used extensively in the dairy industry and could possibly explain the low rates in dairy cattle isolates [[53], [54], [55]]. This study's low detection of methicillin resistance in Staphylococcus aureus isolates is similar to a previous study reporting the prevalence in Canadian dairy cows at 0.5% in 2008 [56].
All 220 S. aureus isolates in this study were susceptible to linezolid, including MRSA isolates. Linezolid has been explored as a more effective treatment for MRSA when compared to vancomycin, but linezolid-resistance has been reported in MRSA isolates in human hospitals [[57], [58], [59]]. So far, the majority of linezolid-resistance has been observed in human isolates of S. aureus, but low rates of resistance in swine has been noted by other studies. Similar to this study, no resistance has been reported for cattle or chicken isolates [[60], [61], [62]].
This study found human, dairy cattle and beef cattle isolates to were more likely to be resistant to penicillin. Numerous studies have confirmed high rates of penicillin resistance in S. aureus isolates due to the development of the enzyme penicillinase/ β-lactamase causing inactivation of beta-lactam drugs [[63], [64], [65], [66]]. When comparing dairy and beef cattle isolates, resistance profiles differ in that beef cattle isolates were resistant to erythromycin while no resistant dairy cattle isolates were detected. Dairy cattle isolates were the only isolates to show resistance to ceftaroline and trimethoprim-sulfamethoxazole. Trimethoprim-sulfamethoxazole resistance in dairy cattle isolates has been demonstrated to be found in milk products by Abdeen et al. (2020) [66] and low-level resistance to ceftaroline in the isolates from this study was consistent with results obtained by Abdel-Moein et al. (2019) [60].
One hundred percent of the swine isolates in this study were resistant to tetracyclines, which supports the findings of Dierikx et al. (2016) [52] and Conceição et al. (2017) [67]. This resistance to tetracycline is characteristic of swine isolates of S. aureus and can be linked back to a host-switching event from humans to pigs [44]. It could also be linked to the tetracyclines used as feed additives in swine operations. The tetracycline resistance gene tet(M) has been described to be universally prevalent among livestock-associated MRSA but absent from human-isolates. Tetracyclines are also used at higher amounts in farmed animals when compared against human medicine, which could explain the persistent detection of this resistance [46]. Resistance in swine isolates was detected against penicillin, clindamycin and erythromycin and supporting previous studies [52,67].
Antimicrobial resistance has been attributed to horizontal gene transfer of AMR genes via plasmids, transposons and integrons [68]. Integrons have been detected in S. aureus isolates from poultry litter, human hospitals, and bovine milk, however; this study detected no integrons in the 220 isolates, including MRSA isolates [[69], [70], [71]].
This study was performed on isolates collected from multiple states over the course of 14 years (2003–2017). The sample matrices were not included in the data that was received from any of the laboratories nor was the geographical locations of the sample origins. These limitations of non-availability of enough metadata have the possibility to impact the detection of integrons, as decreasing identification rates of integrons have been noted over time [70]. The isolates were mostly representative of the laboratory submission of clinical as well as surveillance samples.
From the data gathered in this study, it is clear that S. aureus isolates do not rely specifically on integrons for AMR gene transfer, as our study observed antimicrobial resistance in the absence of integron detection.
5. Conclusions
This study has demonstrated a high occurrence of penicillin resistance in Staphylococcus aureus isolates collected from all of the host species. There were differences observed between host species with tetracycline resistance being the highest among swine isolates and clindamycin being highest in poultry isolates. No detection of oxacillin resistance was found in isolates from dairy cattle but was found in isolates from all of the other host species. Ninety four percent of the S. aureus isolates that were resistant to oxacillin contained the mecA gene. Drug resistant S. aureus is medically significant because there is a narrow range of other treatment options in animals or humans.
Funding
This work was supported by the ‘USDA Animal Health & Disease Funding #NI17AHDRXXXXG018’ in 2016.
CRediT authorship contribution statement
Sangeeta Rao: Funding acquisition, Conceptualization, Methodology, Data curation, Formal analysis, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Lyndsey Linke: Funding acquisition, Conceptualization, Methodology, Data curation, Writing – review & editing. Roberta Magnuson: Funding acquisition, Conceptualization, Methodology, Data curation. Linzy Jauch: Validation, Visualization, Writing – original draft, Writing – review & editing. Doreene R. Hyatt: Funding acquisition, Conceptualization, Writing – review & editing.
Declaration of Competing Interest
None.
Acknowledgements
We are thankful to all the institutes who have contributed the Staphylococcus aureus isolates for this study through collaboration: Ohio State University, Kent State University- Ohio, University of Colorado- Health, Washington State University, University of Pennsylvania, and Colorado State University Veterinary Diagnostic Laboratory.
References
- 1.Verraes C., Van Boxstael S., Van Meervenne E., Van Coillie E., Butaye P., Catry B., de Schaetzen M.-A., Van Huffel X., Imberechts H., Dierick K., Daube G., Saegerman C., De Block J., Dewulf J., Herman L. Antimicrobial resistance in the food chain: a review. Int. J. Environ. Res. Public Health. 2013;10:2643–2669. doi: 10.3390/ijerph10072643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hashempour-Baltork F., Hosseini H., Shojaee-Aliabadi S., Torbati M., Alizadeh A.M., Alizadeh M. Drug resistance and the prevention strategies in food borne Bacteria: an update review. Adv. Pharm. Bull. 2019;9:335–347. doi: 10.15171/apb.2019.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention Antibiotic Resistance Threats in the United States. 2013. http://www.cdc.gov/drugresistance/threat-report-2013/
- 4.Howard D.H., Scott R.D., 2nd The economic burden of drug resistance. Clin. Infect. Dis. 2005;41:S283–S286. doi: 10.1086/430792. [DOI] [PubMed] [Google Scholar]
- 5.Smith R., Coast J. The true cost of antimicrobial resistance. BMJ. 2013;346:fl493. doi: 10.1136/bmj.f1493. [DOI] [PubMed] [Google Scholar]
- 6.DeLeo F.R., Chambers H.F. Reemergence of antibiotic- resistant Staphylococcus aureus in the genomics era. J. Clin. Invest. 2009;119:2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Belkum A. Staphylococcal colonization and infection: homeostasis versus disbalance of human (innate) immunity and bacterial virulence. Curr. Opin. Infect. Dis. 2006;19:339–344. doi: 10.1097/01.qco.0000235159.40184.61. [DOI] [PubMed] [Google Scholar]
- 8.Weems J.J. The many faces of Staphylococcus aureus infection. Recognizing and managing its life-threatening mani- festations. Postgrad. Med. 2001;110(24–26):29–31. doi: 10.3810/pgm.2001.10.1042. 35–36. [DOI] [PubMed] [Google Scholar]
- 9.Klein E., Smith D.L., Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg. Infect. Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim C.J., Kim H.B., Oh Md., Kim Y., Kim A., Oh S.-H., Song K.-H., Kim E.S., Cho Y.K., Choi Y.H., Park J., Kim B.-N., Kim N.-J., Kim k-h., Lee E.J., Jun J.-B., Kim Y.K., Kiem S.M., Choi H.J., Choo E.J., Sohn K.-M., Lee S., Chang H.-H., Bang J.H., Lee S.J., Lee J., Park S.Y., Jeon M.H., Yun N.R. The burden of nosocomial staphylococcus aureus bloodstream infection in South Korea: a prospective hospital-based nationwide study. BMC Infect. Dis. 2014;14:590. doi: 10.1186/s12879-014-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G., Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Partridge S.R., Kwong S.M., Firth N., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00088-17. e00088–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebmeyer S., Kristiansson E., Larsson D.G.J. A framework for identifying the recent origins of mobile antibiotic resistance genes. Commun. Biol. 2021;4:8. doi: 10.1038/s42003-020-01545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsay J., Holden M. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Malachowa N., DeLeo F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010;67:3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Action plan for Combating Antibiotic Resistant Bacteria . 2020. U.S. Department of Health & Human Services. [Google Scholar]
- 17.Deng Y., Liu J., Peters B.M., Chen L., Miao J., Li B., Li L., Chen D., Yu G., Xu Z., Shirtliff M.E. Antimicrobial resistance investigation on Staphylococcus strains in a local hospital in Guangzhou, China, 2001-2010. Microb. Drug Resist. 2015;21:102. doi: 10.1089/mdr.2014.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Z., Li L., Shi L., Shirtliff M.E. Class 1 integron in staphylococci. Mol. Biol. Rep. 2011;38(8):5261e5279. doi: 10.1007/s11033-011-0676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu Z., Li L., Shirtliff M.E., Peters B.M., Li B., Peng Y., Alam M.J., Yamasaki S., Shi L. Resistance class 1 integron in clinical methicillin-resistant Staphylococcus aureus strains in southern China, 2001-2006. Clin. Microbiol. Infect. 2011;17(5):714–718. doi: 10.1111/j.1469-0691.2010.03379.x. [DOI] [PubMed] [Google Scholar]
- 20.Miao J., Liang Y., Chen L., Wang W., Li B., Li L., Chen D., Xu Z. Formation and development of Staphylococcus biofilm: with focus on food safety. J. Food Saf. 2017;37 [Google Scholar]
- 21.Kargar M., Mohammadalipour Z., Doosti A., Lorzadeh S., Japoni-Nejad A. High prevalence of class 1 to 3 Integrons among multidrug-resistant Diarrheagenic Escherichia coli in southwest of Iran. Osong Public Health Res. Perspect. 2014;5(4):193–198. doi: 10.1016/j.phrp.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao S., Maddox C.W., Hoien-Dalen P., Lanka S., Weigel R.M. Diagnostic accuracy of class 1 integron PCR method in detection of antibiotic resistance in Salmonella isolates from swine production systems. J. Clin. Microbiol. 2008;46:916–920. doi: 10.1128/JCM.01597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solberg O.D., Ajiboye R.M., Riley L.W. Origin of class 1 and 2 integrons and gene cassettes in a population-based sample of uropathogenic Escherichia coli. J. Clin. Microbiol. 2006;44(4):1347–1351. doi: 10.1128/JCM.44.4.1347-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White P.A., McIver C.J., Rawlinson W.D. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 2001;45:2658–2661. doi: 10.1128/AAC.45.9.2658-2661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marathe N.P., Nagarkar S.S., Vaishampayan A.A., Rasane M.H., Samant S.A., Dohe V., Kagal A., Shouche Y.S., Deshpande N. High prevalence of class 1 integrons in clinical isolates of methicillin-resistant Staphylococcus aureus from India. Indian J. Med. Microbiol. 2015;33(2):231–236. doi: 10.4103/0255-0857.154905. [DOI] [PubMed] [Google Scholar]
- 26.Benito D., Gomez P., Aspiroz C., Zarazaga M., Lozano C., Torres C. Molecular characterization of Staphylococcus aureus isolated from humans related to a livestock farm in Spain, with detection of MRSA-CC130 carrying mecC gene: a zoonotic case? Enferm. Infecc. Microbiol. Clin. 2016;34(5):280–285. doi: 10.1016/j.eimc.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Argudín M.A., Argumosa V., Mendoza M.C., Guerra B., Rodicio M.R. Population structure and exotoxin gene content of methicillin-susceptible Staphylococcus aureus from Spanish healthy carriers. J. Mic. Path. 2013;54:26–33. doi: 10.1016/j.micpath.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Mazel D. Integrons: agents of bacterial evolution. Nature. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 29.DeLappe N., O'Halloran F., Fanning S., Corbett-Feeney G., Cheasty T., Cormican M. Antimicrobial resistance and genetic diversity of Shigella sonnei isolates from western Ireland, an area of low incidence of infection. J. Clin. Microbiol. 2003;41:1919–1924. doi: 10.1128/JCM.41.5.1919-1924.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren C., Zhao Y., Shen Y. Analysis of the effect of integrons on drug-resistant Staphylococcus aureus by multiplex PCR detection. Mol. Med. Rep. 2013;7:719–724. doi: 10.3892/mmr.2013.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mostafa M., Siadat S., Shahcheraghi F., Vaziri F., Japoni-Nejad A., Yousefi J.V., Rajaei B., Mood E.H., Zadeh N.E., Moshiri A., Siamdoust S.A.S., Rahbar M. Variability in gene cassette patterns of class 1 and 2 integrons associated with multi drug resistance patterns in Staphylococcus aureus clinical isolates in Tehran-Iran. BMC Microbiol. 2015;15:152. doi: 10.1186/s12866-015-0488-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waters A., Contente-Cuomo T., Buchhagen J., Liu C.M., Watson L., Pearce K., Foster J.T., Bowers J., Driebe E.M., Engelthaler D.M., Keim P.S., Price L.B. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 2011;52:1227–1230. doi: 10.1093/cid/cir181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith T. Livestock-associated Staphylococcus aureus: the United States experience. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Sande-Bruinsma N., Leverstein van Hall M.A., Janssen M., Nagtzaam N., Leenders S., de Greeff S.C., Schneeberger P.M. Impact of livestock-associated MRSA in a hospital setting. Antimicrob. Resist. Infect. Control. 2015;4:11. doi: 10.1186/s13756-015-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tallent Sandra, Hait Jennifer, Bennett (ret.) Reginald W., Lancette Gayle A. AOAC International; Gaithersburg, Md.: 1998. Bacteriological Analytical Manual Chapter 12: Staphylococcus aureus. Content current as of: 12/16/2019. [Google Scholar]
- 36.Dodd C.C., Sanderson M.W., Sargeant J.M., Nagaraja T.G., Oberst R.D., Smith R.A., Griffin D.D. Prevalence of Escherichia coli O157 in cattle feeds in Midwestern feedlots. Appl. Environ. Microbiol. 2003;69(9):5243–5247. doi: 10.1128/AEM.69.9.5243-5247.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velasco V., Sherwood J., Rojas-García P., Logue C. Multiplex real-time PCR for detection of Staphylococcus aureus, mecA and Panton-valentine Leukocidin (PVL) genes from selective enrichments from animals and retail meat. PLoS One. 2014;9(5) doi: 10.1371/journal.pone.0097617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2014. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S24. [Google Scholar]
- 39.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Fifteenth Informational Supplement. CLSI; Wayne, PA: 2005. pp. M100–S14. [Google Scholar]
- 40.Hoegh S.V., Skov M.N., Boye K., Worning P., Jensen T.G., Kemp M. Variations in the Staphylococcus aureus-specific nuc gene can potentially lead to misidentification of methicillin-susceptible and -resistant S. aureus. J. Med. Microbiol. 2014;63:1020–1022. doi: 10.1099/jmm.0.076638-0. [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization Antibiotic Resistance. 2020. https://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance
- 42.Sakwinska O., Giddey M., Moreillon M., Morisset D., Waldvogel A., Moreillon P. Appl. Environ. Microbiol. 2011;77(17):5908–5915. doi: 10.1128/AEM.00238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho P., Chow K., Lai E.L., Law P.Y.T., Chan P., Ho A.Y.M., Ng T., Yam W. Clonality and antimicrobial susceptibility of Staphylococcus aureus and methicillin-Resistant S. aureus isolates from food animals and other animals. J. Clin. Microbiol. 2012;50(11):3735–3737. doi: 10.1128/JCM.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson E.J., Bacigalupe R., Harrison E.M., Weinert L.A., Lycett S., Vrieling M., Robb K., Hoskisson P.A., Holden M.T.G., Feil E.J., Paterson G.K., Tong S.Y.C., Shittu A., van Wamel W., Aanensen D.M., Parkhill J., Peacock S.J., Corander J., Holmes M., Fitzgerald J.R. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018;2:1468–1478. doi: 10.1038/s41559-018-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park S., Ronholm J. Staphylococcus aureus in agriculture: lessons in evolution from a multispecies pathogen. Clin. Microbiol. Rev. 2021;34(2) doi: 10.1128/CMR.00182-20. e00182–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price L.B., Stegger M., Hasman H., Aziz M., Larsen J., Andersen P.S., Pearson T., Waters A.E., Foster J.T., Schupp J., Gillece J., Driebe E., Liu C.M., Springer B., Zdovc I., Battisti A., Franco A., Zmudzki J., Schwarz S., Butaye P., Jouy E., Pomba C., Porrero M.C., Ruimy R., Smith T.C., Robinson D.A., Weese J.S., Arriola C.S., Yu F., Laurent F., Keim P., Skov R., Aarestrup F.M. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 2012;3(1):e00305–e00311. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karikari A.B., Frimpong E., Owusu-Ofori A. Methicillin-resistant Staphylococcus aureus among patients in a teaching hospital in Ghana. Int. J. One Health. 2017;3:46–49. [Google Scholar]
- 48.Abreu R., Roderíguez-Álvarez C., Lecuona M., Castro B., González J.C., Aguirre-Jaime A., Arias A. Increased antimicrobial resistance of MRSA strains isolated from pigs in Spain between 2009 and 2018. Vet. Sci. 2019;6(2):38. doi: 10.3390/vetsci6020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pletinckx L.J., Verhegghe M., Dewulf J., Crombé F., Bleecker Y.D., Arasschaert G., Goddeeris B.M., De Man I. Screening of poultry-pig farms for methicillin-resistant Staphylococcus aureus: sampling methodology and within herd prevalence in broiler flocks and pigs. Infect. Genet. Evol. 2011;11:2133–2137. doi: 10.1016/j.meegid.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 50.Sun J., Yang M., Sreevatsan S., Davies P.R. Prevalence and characterization of Staphylococcus aureus in growing pigs in the USA. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0143670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy A.J., Lindsay J.A., Loeffler A. Are all methicillin-resistant Staphylococcus aureus (MRSA) equal in all hosts? Epidemiological and genetic comparison between animal and human MRSA. Vet. Dermatol. 2012;23(4):267–e54. doi: 10.1111/j.1365-3164.2012.01072.x. [DOI] [PubMed] [Google Scholar]
- 52.Dierikx C.M., Hengeveld P.D., Veldman K.T., de Haan A., van der Voorde S., Dop P.Y., Bosch T., van Duijkeren E. Ten years later: still a high prevalence of MRSA in slaughter pigs despite a significant reduction in antimicrobial usage in pigs in the Netherlands. J. Antimicrob. Chemother. 2016;71:2414–2418. doi: 10.1093/jac/dkw190. [DOI] [PubMed] [Google Scholar]
- 53.Taponen S., Dredge K., Henriksson B., Pyyhtiä A.M., Suojala L., Junni R., Heinonen K., Pyörälä S. Efficacy of intramammary treatment with procaine penicillin G vs. procaine penicillin G plus neomycin in bovine clinical mastitis caused by penicillin susceptible, gram-positive bacteria - a double blind field study. J. Vet. Pharmacol. Ther. 2003;26(3):193–198. doi: 10.1046/j.1365-2885.2003.00473.x. [DOI] [PubMed] [Google Scholar]
- 54.Sol J., Sampimon O.C., Barkema H.W., Schukken Y.H. Factors associated with cure after therapy of clinical mastitis caused by Staphylococcus aureus. J. Dairy Sci. 2000;83:278–284. doi: 10.3168/jds.S0022-0302(00)74875-2. [DOI] [PubMed] [Google Scholar]
- 55.Rainard P., Foucras G., Fitzgerald J.R., Watts J.L., Koop G., Middleton J.R. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transbound. Emerg. Dis. 2018;65(1):149–165. doi: 10.1111/tbed.12698. [DOI] [PubMed] [Google Scholar]
- 56.Saini V., McClure J.T., Léger D., Keefe G.P., Scholl D.T., Morck D.W., Barkema H.W. Antimicrobial resistance profiles of common mastitis pathogens on Canadian dairy farms. J. Dairy Sci. 2012;95(8):4319–4332. doi: 10.3168/jds.2012-5373. [DOI] [PubMed] [Google Scholar]
- 57.Wunderink R.G., Rello J., Cammarata S.K., Croos-Dabrera R.V., Kollef M.H. Linezolid vs vancomycin: analysis of two double-blind studies of patients with methicillin-resistant Staphylococcus aureus nosocomial pneumonia. Chest. 2003;124(5):1789–1797. [PubMed] [Google Scholar]
- 58.Azhar A., Rasool S., Haque A., Shan S., Saeed M., Ehsan B., Haque A. Detection of high levels of resistance to linezolid and vancomycin in Staphylococcus aureus. J. Med. Microbiol. 2017;66(9):1328–1331. doi: 10.1099/jmm.0.000566. [DOI] [PubMed] [Google Scholar]
- 59.Gu B., Kelesidis T., Tsiodras S., Hindler J., Humphries R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2013;68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdel-Moein K.A., Zaher H.M. Occurrence of multidrug-resistant methicillin-resistant Staphylococcus aureus among healthy farm animals: a public health concern. Int. J. Vet. Sci. Med. 2019;7(1):55–60. doi: 10.1080/23144599.2019.1689630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang H.Y., Moon D.C., Mechesso A.F., Choi J., Kim S., Song H., Kim M.H., Yoon S., Lim S. Emergence of cfr-mediated linezolid resistance in Staphylococcus aureus isolated from pig carcasses. Antibiotics. 2020;9(11):769. doi: 10.3390/antibiotics9110769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S., Moon D.C., Mechesso A.F., Kang H.Y., Song H., Na S.H., Choi J., Yoon S., Lim S. Nationwide Surveillance on Antimicrobial Resistance Profiles of Staphylococcus aureus isolated from major food animal carcasses in South Korea during 2010–2018. Foodborne Pathog. Dis. 2021;18(6):388–397. doi: 10.1089/fpd.2020.2899. [DOI] [PubMed] [Google Scholar]
- 63.Visciano P., Pomilio F., Tofalo R., Sacchini L., Saletti M.A., Tieri E., Schirone M., Suzzi G. Detection of methicillin-resistant Staphylococcus aureus in dairy cow farms. Food Control. 2014;46:532–538. [Google Scholar]
- 64.Peacock S.J., Paterson G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015;84:577–601. doi: 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 65.Bush K., Bradford P.A. Epidemiology of β-lactamase-producing pathogens. Clin. Microbiol. Rev. 2020;33(2) doi: 10.1128/CMR.00047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdeen E.E., Mousa W.S., Salam S.Y.A., Al-Maary K.S., Mubarak A.S., Moussa I.M., Hemeg H.A., Almuzaini A.M., Alajaji A.I., Alsubki R.A., Elbehiry A. Antibiogram and phylogenic diversity of enterotoxigenic Staphylococcus aureus strains from milk products and public health implications. Saudi. J. Biol. Sci. 2020;27(8):1968–1974. doi: 10.1016/j.sjbs.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conceição T., de Lencastre H., Aires-de-Sousa M. Frequent isolation of methicillin resistant Staphylococcus aureus (MRSA) ST398 among healthy pigs in Portugal. PLoS One. 2017;12(4) doi: 10.1371/journal.pone.0175340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H., Shi L., Li L., Guo S., Zhang X., Yamasaki S., Miyoshi S., Shinoda S. Identification and characterization of class 1 integron resistance gene cassettes among Salmonella strains isolated from healthy humans in China. Microbiol. Immunol. 2004;48:639–645. doi: 10.1111/j.1348-0421.2004.tb03473.x. [DOI] [PubMed] [Google Scholar]
- 69.Nandi S., Maurer J.J., Hofacre C., Summers A.O. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. PNAS. 2004;101(18):7118–7122. doi: 10.1073/pnas.0306466101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng Y., Liu J., Peters B.M., Chen L., Miao J., Li B., Li L., Chen D., Yu G., Xu Z., Shirtliff M.E. Antimicrobial resistance investigation on Staphylococcus strains in a local hospital in Guangzhou China, 2001-2010. Microb. Drug Resist. 2015;21(1):102–104. doi: 10.1089/mdr.2014.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li L., Zhou L., Wang L., Xue H., Zhao X. Characterization of methicillin-resistant and -susceptible staphylococcal isolates from bovine milk in northwestern China. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0116699. [DOI] [PMC free article] [PubMed] [Google Scholar]