Abstract
Introduction
Previous studies have demonstrated that exposed to the initial suboptimal intrauterine environment of gestational diabetes mellitus (GDM) may increase risk of cardiovascular disease in adulthood.
Methods
In order to investigate the underlying mechanisms involved in the increased risk of cardiovascular diseases (CVDs) in the offspring of GDM, we applied a high-throughput proteomics approach to compare the proteomic expression profile of human umbilical vessels of normal and GDM offspring.
Results
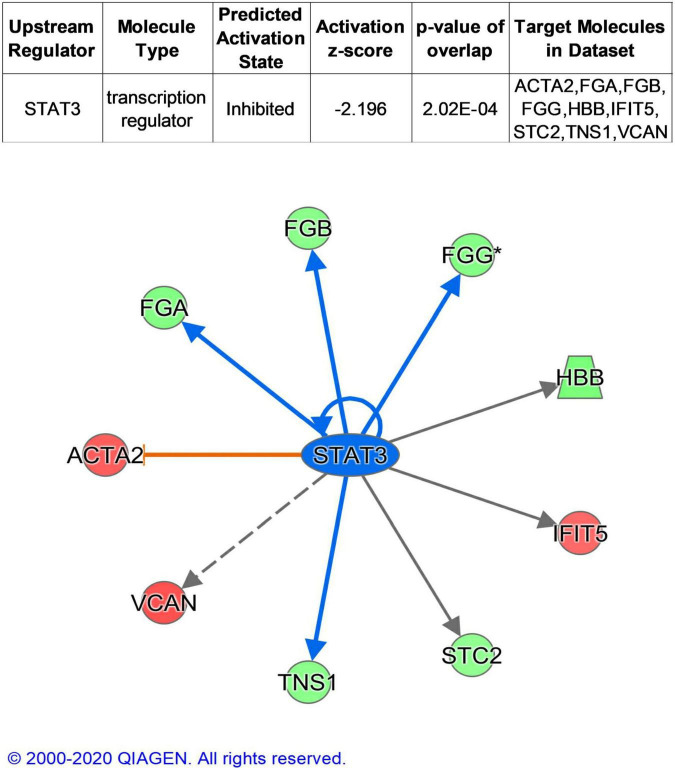
A total of significantly different 100 proteins were identified in umbilical vessels from GDM group compared with normal controls, among which 31 proteins were up-regulated, while 69 proteins were down-regulated. Differentially expressed proteins (DEPs) are validated using Western blotting analysis. The analysis of these differently expressed proteins (DEPs) related diseases and functions results, performed by Ingenuity Pathway Analysis (IPA) software. Based on “Diseases and Disorders” analysis, 17 proteins (ACTA2, ADAR, CBFB, DDAH1, FBN1, FGA, FGB, FGG, GLS, GSTM1, HBB, PGM3, PPP1R13L, S100A8, SLC12A4, TPP2, VCAN) were described to be associated with CVD, especially in Anemia, Thrombus and Myocardial infarction. Functional analysis indicated that DEPs involved in many cardiovascular functions, especially in “vasoconstriction of blood vessel” (related DEPs: ACTA2, DDAH1, FBN1, FGA, FGB, and FGG). Upstream regulator analyses of DEPs identifies STAT3 as inhibitor of ACTA2, FGA, FGB, and FGG.
Conclusion
The results of this study indicate that intrauterine hyperglycemia is associated with an elevated risk of cardiovascular risk in the offspring.
Keywords: Gamete and Embryo-Fetal Origins of Adult Diseases, gestational diabetes mellitus (GDM), offspring, umbilical vessel, iTRAQ
Introduction
A growing number of epidemiological and experimental studies suggest that exposure to adverse intrauterine environment during fetal development can be associated with chronic disease in later life, such as CVD, obesity, type 2 diabetes and cognitive disorder (1–6). The theory of Gamete and Embryo-Fetal Origins of Adult Diseases is used preferentially to describe these associations. Gestational diabetes mellitus (GDM) is defined as any degree of glucose intolerance with onset or first diagnosis during gestation. The prevalence of GDM ranged from 9.3 to 25.5% among 15 collaborating centers using the International Association of the Diabetes and Pregnancy Study Group (IADPSG) criteria (7). GDM is a serious health risk for both pregnant women and their offspring.
Emerging evidence suggests that the vasculature of women with a prior case of GDM is permanently altered, predisposing them to CVD. GDM also increases the offspring’s risk of developing hypertension and CVD. Elevated systolic blood pressure (SBP) and diastolic blood pressure (DBP) (4, 8–11), increased intima-media thickness (IMT) (12), increased cardiac septal hypertrophy (13), and vascular endothelial dysfunctions were observed in the offspring of GDM mother (14, 15). Consistent with epidemiological results, studies in animal models also showed that diabetes during pregnancy affected the development of fetal cardiovascular system (16, 17). In one of our previous works, we also have indicated that intrauterine hyperglycemia could induce IGT (impaired glucose tolerance) and abnormal blood insulin levels in both F1 and F2 offspring (18). In studies of the mechanisms of cardiovascular dysfunction caused by GDM, numbers of potential pathways have been implicated in endothelial cell, including reduced adenosine transport (14), impaired angiogenesis (15) and redox signaling (19). Although many previous studies have been conducted, its precise mechanism involved in the association between intrauterine hyperglycemia and a higher risk of cardiovascular anomalies has yet to be established.
In the present study, we aimed to investigate the cardiovascular risk proteins in offspring exposed to GDM. A proteomics analysis was conducted in umbilical vessels from newborns of mothers with GDM and normal controls using the isobaric tag for relative and absolute quantitation (iTRAQ)-labeling technique to compare the proteomic expression profile. We analyzed the related diseases and functions using ingenuity pathway analysis (IPA) software. The results of the present study may provide valuable information for further investigation of the mechanisms underlying the cardiovascular dysfunction induced by intrauterine hyperglycemia.
Materials and methods
Patients and umbilical cords
Umbilical cords from newborns of 25 mothers with mild GDM and 25 controls were collected by obstetricians in Shaoxing Maternity and Child Health Care Hospital, China. The Ethics Committee of Shaoxing Maternity and Child Health Care Hospital approved the study. All the participants enrolled in this study were with singleton pregnancy and ceased pregnancies with Cesarean section at full term. Tissue samples were stored snap frozen at −80°Cuntil use. The clinical characteristics of the proteomic participants included in this study are outlined in Table 1.
TABLE 1.
Clinical characteristic of the participants.
| Characteristics | Control | GDM | P-value |
| Cases | 25 | 25 | |
| Gestational age, wk | 39.06 ± 0.52 | 38.85 ± 0.62 | 0.108 |
| Birth weight, g | 3439.57 ± 421.29 | 3468 ± 456.14 | 0.241 |
| Maternal age, y | 30.21 ± 3.51 | 31.42 ± 3.11 | 0.071 |
| BMI of pre-pregnancy, kg/m2 | 22.12 ± 2.74 | 22.79 ± 4.12 | 0.058 |
| Glycated hemoglobin, % | 4.94 ± 0.27 | 5.10 ± 0.41 | 0.041* |
| Pregnancy weight gain, kg | 15.65 ± 5.15 | 14.10 ± 8.59 | 0.187 |
| OGTT 75 g, 0 h, mmol/L | 4.47 ± 0.36 | 5.02 ± 0.68 | < 0.001** |
| OGTT 75 g, 1 h, mmol/L | 8.01 ± 0.93 | 10.92 ± 1.68 | < 0.001** |
| OGTT 75 g, 2 h, mmol/L | 6.56 ± 1.02 | 9.38 ± 1.60 | < 0.001** |
| Placenta weight,g | 598.35 ± 89.92 | 603.28 ± 116.46 | 0.097 |
| TC in first-trimester, mmol/L | 4.84 ± 0.87 | 4.78 ± 0.72 | 0.765 |
| TG in first-trimester, mmol/L | 1.23 ± 0.41 | 1.11 ± 0.14 | 0.108 |
| HDL in first-trimester, mmol/L | 1.67 ± 0.34 | 1.61 ± 1.48 | 0.772 |
| LDL in first-trimester, mmol/L | 3.12 ± 0.41 | 3.271 ± 0.37 | 0.692 |
| Diagnosis | NS | GDM | |
| Presence of other major cardiovascular risk factor(s) in the mothers | None | None |
Data are showed as means ± SD, *P < 0.05, **P < 0.001 compared with control.
GDM were diagnosed between gestational weeks 24 and 28 after overnight fasting (for 8–12 h) by an oral glucose tolerance test (OGTT). According to the IADPSG diagnostic criteria, GDM was defined as fasting venous plasma glucose concentration ≥ 5.1 mM and ≥ 10.0 mM at 1-hand/or ≥ 8.5 mM at 2-h after drinking a solution with 75 g glucose. All women with GDM enrolled in the present study experienced dietary management without insulin treatment. Participants with maternal obesity factor (high pre-pregnancy BMI, excessive weight gain during pregnancy, abnormal lipid level in first-trimester) and macrosomia were excluded. The work described in the present study has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki). The study protocols were reviewed and approved by the Research and Ethics Committee of the Women’s Hospital, School of Medicine, Zhejiang University, China, and informed consents were provided by all participants.
Isobaric tag for relative and absolute quantitation analysis
iTRAQ analysis was performed as previously described (20). Briefly, protein was extracted from umbilical artery and measured by BCA assay (Pierce, Rockford, IL, USA) according to the manufacturer’s protocol. Protein digestion was performed according to the FASP procedure, as described by Wisniewski et al. (21). Briefly, 200 μg of total-protein samples were diluted in 30 μL of solution including 4% SDS, 100 mM Tris-HCl pH 8.0 and 100 mM dithiotreitol, and were heated at 95°C for 5 min. After each sample was cooled to room temperature, the sample was loaded onto an ultrafiltration filter (cutoff 10 kDa, Sartorius, Goettingen, Germany). We added 200 μL UT buffer (8 M Urea and 150 mM Tris-HCl, pH 8.0) to the filter and centrifuged it at 14,000 g at 20°C for 30 min. Subsequently, 100 μL of iodoacetamide solution (50 mM iodoacetamide in UT buffer) was added for blocking reduced cysteines, and, the samples were incubated for 20 min in darkness. Then the filters were centrifuged at 14,000 g at 20°C for 20 min. The filters were washed with 100 μL UT buffer at 14,000 g for 20 min. This step was repeated 2 times. Then, 100 μL dissolution buffer (AB Sciex, Framingham, MA, USA) was added to the filter, and it was centrifuged at 14,000 g at 20°C for 30 min, and, this step was repeated twice. Finally, 40 μL of trypsin (Promega, Madison, WI, USA) buffer (2 μg trypsin in 40 μL dissolution buffer) were added, and, the samples were digested overnight at 37°C. Each filter unit was transferred to a new tube and centrifuged at 14,000 g at 20°C for 30 min. The concentration of resulting peptides was determined by UV light spectral density at OD280 (22).
The iTRAQ labeling of digested peptide samples was performed following the manufacturers protocol with 8-plex isobaric tags for relative and absolute quantitation (iTRAQ) labeling kit (AB Sciex, Framingham, MA, USA). Three umbilical arteries from the control group (C) were labeled with mass 114, 115 and 116 isobaric iTRAQ tags, the other three umbilical arteries from the GDM group were labeled with mass 117, 118, and 119 isobaric iTRAQ tags. Identical quantities of peptide mixtures from the 6 peptides mentioned above were labeled with reagent 113 and served as sample IS (internal standard). According to the manufacturers protocol, The labeling reactions were incubated for 2 h at room temperature before further analysis.
After iTRAQ-labeling the peptide samples were combined and subsequently purified using a strong cation exchange (SCX)-cartridge: Polysulfoethyl 4.6 × 100 mm column (5 μm, 200 Å, Poly LC Inc., Columbia, MD, USA). For LC-MS/MS analysis of the resulting peptides, we followed a previously described method (20). Protein identification and quantification for iTRAQ analysis data was carried out using the MASCOT search engine (version 2.2.1; Matrix Science, London, UK) embedded into Proteome Discoverer 1.3 (Thermo Electron, San Jose, CA, USA), searching against the Uniport database of human protein sequences (03-2013, 133549 entries, downloaded from: http://www.uniprot.org/) and the concatenated target-decoy database. The parameters were set as follows: Trypsin as digestion enzyme, cysteine carbamidomethylation as a fixed modification, Oxidation (M), Gln→Pyro-Glu (N-term Q), iTRAQ 8 plex (K), iTRAQ 8 plex (Y), and iTRAQ 8 plex (N-term) as the variable modification.
Western blotting analysis
Human umbilical arteries were lysed in 1 × RIPA buffer with 1μg/mL leupeptin and 1μg/mL phenylmethylsulfonyl fluoride. Aliquots containing 30 μg of protein samples were separated by 12% SDS-PAGE and transferred electrophoretically to a nitrocellulose transfer membrane (Bio-Rad, Hercules, CA, USA). After blocked with 5% BSA in TBS containing 0.01% Tween 20 (TBST) for 1 h at room temperature, the membrane was incubated overnight at 4°C with primary antibodies against FGA (1:1,000, Abcam, Cambridge, MA, USA), ACTA2 (1:1,000, Abcam, Cambridge, MA, USA), IDH3A (1:1,000, Abcam, Cambridge, MA, USA), GAPDH (1:1,000, Abcam, Cambridge, MA, USA). Subsequently, membranes were washed 10 min for three times with TBST, then each membrane was incubated for 1 h at room temperature with the appropriate secondary antibody (anti-rabbit IgG, anti-mouse IgG; 1:5,000; Abcam). The protein intensities were determined and analyzed using Odyssey® Imager (LI-COR Biosciences, Lincoln, NE, USA).
Bioinformatics analysis of differentially expressed proteins
Proteins exhibiting at least a ± 20% fold change in expression were determined as significantly different. The Cluster 3.01 and Java Tree view software2 were further used to evaluate the capability of the resulting feature proteins in differentiating the two groups of samples. IPA software (QIAGEN, Redwood 185 City, CA) was employed for functional analysis of the identified DEPs between the two groups. In the functional network analysis, the DEPs were represented as nodes, and the biological relationship between two proteins was represented as an edge (line), which was supported by the published articles or the canonical information embedded in the IPA database. Downstream biological processes analysis, which was used to predict the downstream effects of the DEPs based on the observed gene expression changes, was also on the strength of the canonical information embedded in the IPA database. The calculated z-score can be used to infer the activation states (“increased” or “decreased”) of implicated biological processes. Fisher’s exact test was used to calculate a p-value to determine the probability that the association between proteins in the dataset, and the biological process could be explained by chance alone.
Statistical analysis
All analysis was performed with the software Statistical Analysis Software (SPSS 17.0 software, SPSS Inc., USA). Data are displayed as means ± SEM. Statistical evaluation was conducted using an unpaired Student’s t-test, and P < 0.05 was considered statistically significant.
Results
The clinical characteristics of the participants
Twenty five pregnancies with GDM and 25 normal controls were enrolled in the present study, and the specific clinical characteristics of the participants and delivery data were shown in Table 1. None of them experienced major cardiovascular risk factors, such as cardiovascular system diseases, history of type 2 diabetes, hyperthyroidism, assisted reproductive technology treatment, smoking, congenital defect, and any other pregnancy complications. There was no significant difference between GDM group and the controls about the maternal age, gestational weeks, pre-pregnancy BMI, weight gain during pregnancy, lipid level in first-trimester, birth weight and gender distribution of newborns (all P > 0.05). The serum glucose concentration at 0,1, and 2 h of OGTT in the GDM group were significantly higher than that in the normal pregnancies (p < 0.001, respectively). Three pairs of the collected umbilical vessels were selected to compare the proteome profiles between GDM and controls by iTRAQ-LC-MS/MS.
Overview of the proteomics analysis
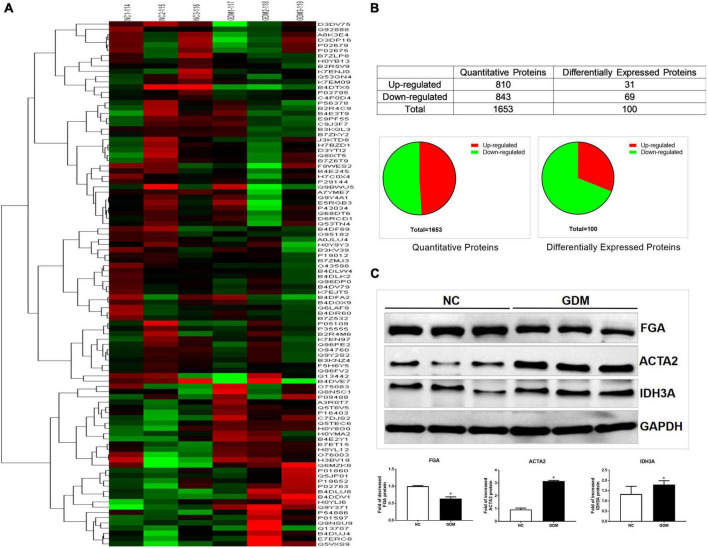
To identify the differentially expressed proteins (DEPs) in the umbilical arteries between GDM and controls in genome-wide level, iTRAQ-LC-MS/MS was performed in 3 GDM samples and 3 control samples. In the present study, a total of 1,653 proteins were quantified in the umbilical arteries from both GDM and the controls (Supplementary Table 1). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (23) with the dataset identifier PXD024892. Using the screening criteria of fold change over ± 1.2, 100 proteins were regarded as differentially expressed in umbilical arteries from GDM group, of which, 31 proteins were up-regulated and 69 proteins were down-regulated (Supplementary Table 2 and Figure 1B). Hierarchical clustering analysis was further performed with the identified 100 DEPs mentioned above. After unsupervised clustering, the GDM group were significantly distinguished from the controls (Figure 1A), indicating the significant effect of intrauterine hyperglycemia on the expression of proteins in umbilical vessels of newborn.
FIGURE 1.
iTRAQ analysis results for the GDM umbilical vessels. (A) Hierarchical clustering of DEPs. (B) The statistic of the proteins identified in umbilical vessels. (C) Validation of the differential expression of the selected proteins in umbilical vessels by western blotting: FGA, ACTA2, IDH3A. Data are presented as mean ± SE (n = 3). *p < 0.01.
Western blotting validation
To validate the results carried out by iTRAQ-LC-MS/MS, Western blotting was conducted in additional three umbilical vessels from GDM and three normal umbilical vessels. Based on the significant expression difference and the biological function in the cardiovascular system, three proteins, including FGA, ACTA2, and IDH3A, were selected for further investigation. As shown in Figure 1C, the change trends of these three proteins within Western blotting analysis were in accordance with iTRAQ analysis results.
Bioinformatics analysis of differentially expressed proteins
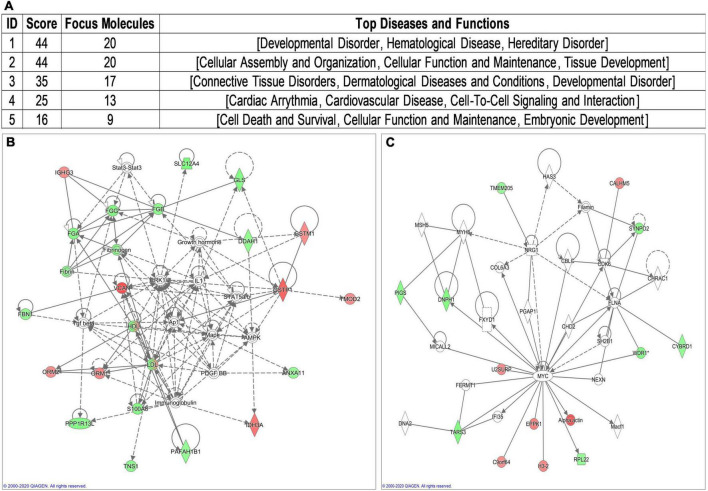
To assist functional interpretation of the DEPs, the bioinformatics analysis was performed based on the 100 DEPs. These genes were loaded into IPA database (IPA®, QIAGEN)3 for pathway, disease and function, and network analysis. Three cardiovascular signaling pathways identified by IPA software (Supplementary Table 3 and Supplementary Figure 1) including “Atherosclerosis Signaling” (related gene: ORM1, ORM2, and S100A8). According to overlapping p-values, these 73 subcategories showed in “Disease and Disorder” analysis, including “Developmental Disorder” and “CVD” (Supplementary Table 4 and Supplementary Figure 2). In addition to the gene ontology enrichment, 22 subcategories involed in “Physiological System Development and Function,” especially in “Cardiovascular System Development and Function” (Supplementary Table 5 and Supplementary Figure 3). Network analysis identified five biological networks (Figure 2A), which included developmental network (Figure 2B) and cardiovascular network (Figure 2C). The most related network emerged comprising 20 of those DEPs. It is associated with the IPA functions “Developmental Disorder” (Figure 2B).
FIGURE 2.
Visual representation of the principal network generated by ingenuity pathway analysis (IPA). (A) Principal network generated by IPA. (B) Developmental Disorder associated network. (C) Cardiovascular Disease associated network.
Cardiovascular risk proteins analysis
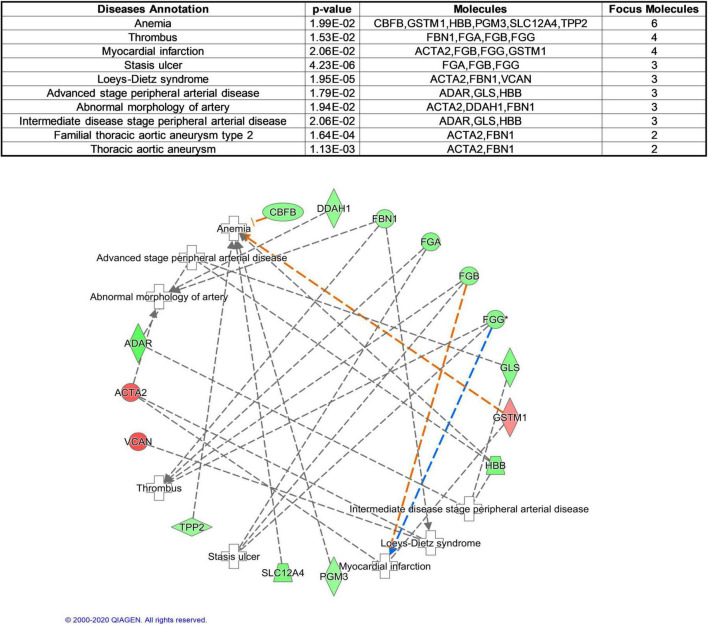
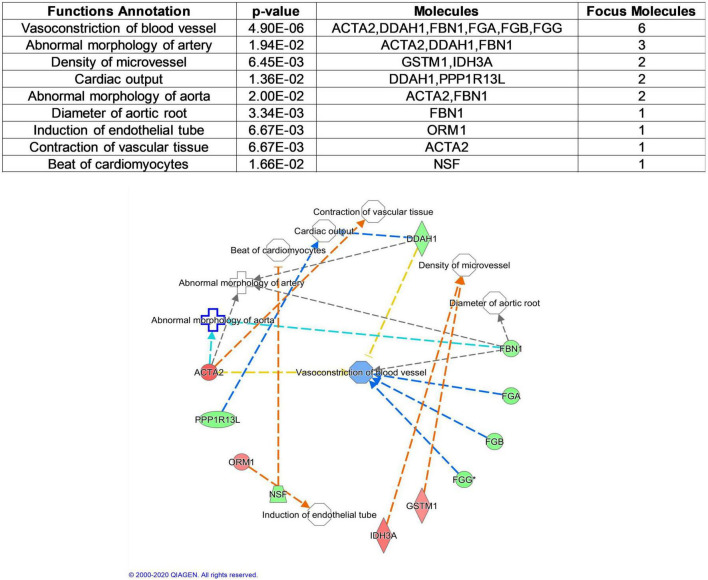
To better understand cardiovascular risk proteins in offspring exposed to GDM, these DEPs were further analyzed in CVDs and functions. Based on “Diseases and Disorders” analysis, 17 proteins (ACTA2, ADAR, CBFB, DDAH1, FBN1, FGA, FGB, FGG, GLS, GSTM1, HBB, PGM3, PPP1R13L, S100A8, SLC12A4, TPP2, VCAN) were described to be associated with CVD, especially in Anemia, Thrombus and Myocardial infarction (Figure 3). Based on “Physiological System Development and Functions” analysis, 11 proteins (ACTA2, DDAH1, FBN1, FGA, FGB, FGG, GSTM1, IDH3A, NSF, ORM1, PPP1R13L) were described to be associated with Cardiovascular System Development and Function, especially in the function of “vasoconstriction of blood vessel” (Figure 4).
FIGURE 3.
Cardiovascular Disease analysis of differentially expressed proteins between normal and GDM umbilical vessels. For this developmental disease network, genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge. All edges are supported by at least one publication as stored in the Ingenuity Knowledge database. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation.
FIGURE 4.
Cardiovascular Functional analysis of differentially expressed proteins between normal and GDM umbilical vessels. For this cardiovascular function network, genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge. All edges are supported by at least one publication as stored in the Ingenuity Knowledge database. The intensity of the node color indicates the degree of up- (red) or down- (green) regulation.
The term “upstream regulator” as used in IPA refers to any molecule that can affect the expression of another molecule. In upstream regulator analysis, STAT3 was predicted in inhibited state (z-score = −2.196) and regulated ACTA2, FBN1, FGA, FGB, and FGG (Figure 5).
FIGURE 5.
Upstream analysis of DEPs. Colorized nodes represented our input proteins. Green, down-regulated proteins. Red, up-regulated proteins.
Discussion
Cardiovascular and metabolic disorders often present in adult life, but may have their origins in changes to the intrauterine environment during fetal development. Previous studies including epidemiological investigations and experimental projects have demonstrated the alteration of vascular function and cardiovascular system in the offspring of GDM (8, 24). The current study firstly focus on the proteomics of umbilical vessels from GDM and normal controls, of normal birth weights, which excluded maternal obesity factors (high pre-pregnancy BMI, excessive weight gain during pregnancy, abnormal lipid level in first-trimester). Our results demonstrated that the intrauterine hyperglycemia indeed individually affected the expression patterns of proteins and vessel function.
Several studies implicated that the association between maternal hyperglycemia and childhood metabolic outcomes was attenuated after adjusting for covariates including maternal BMI and weight gain during pregnancy (11), overall, existing evidence suggested gestational diabetes as an independent element. To date, intensified treatment during pregnancy of maternal hyperglycemia reduce maternal weight gain and macrosomia at birth, whether similar associations still exist between maternal hyperglycemia and offspring cardiovascular outcomes is few studied. In the current study, participants with no maternal obesity factors (high pre-pregnancy BMI, excessive weight gain during pregnancy, abnormal lipid level in first-trimester) were recruited, and macrosomia was excluded.
The physiological and pathological regulation of cardiovascular function derives mainly from the collaboration between vascular endothelial cells and VSMCs (Vascular smooth muscle cells). Previous studies stated that characteristics and alterations of umbilical vessels could offer valuable information since umbilical vessel cells experienced the effect of same intrauterine environment (20, 25–27). In consideration of the ethics limited, umbilical vessels is thought to be the best tissue that could be collected in clinical.
Based on the identified DEPs in umbilical arteries between GDM and normal controls, our results suggested that the “Cardiovascular System Development and Function” was impaired exposure to intrauterine hyperglycemia. In this content, the top related function was “Vasoconstriction of blood vessel,” this is consistent with a large number of epidemiological results. These suppressed physiological processes in the umbilical vessels from GDM might lead to impaired vascular repair under stress or diabetic vessel pathological damage.
In this study, we found 20 DEPs (ACTA2, ADAR, CBFB, DDAH1, FBN1, FGA, FGB, FGG, GLS, GSTM1, HBB, PGM3, PPP1R13L, S100A8, SLC12A4, TPP2, VCAN, IDH3A, NSF, ORM1) related with Cardiovascular system. ACTA2 gene mutations in adults associated with thoracic aortic aneurysm and dissection (TAAD) (28). Transgenetic mouse model study proposes that sufficient ADAR2 enzyme activity might play a vital role in preventing cardiovascular defects (29). CBFB may impair the primitive hematopoiesis (30). FBN1 is a gene with a well characterized role in the pathogenesis of thoracic aortic aneurysm (TAA) in the context of Marfan syndrome (31). Inferring that variation in genomic sequences that regulate the fibrinogen genes (FGA, FGB, and FGG) may affect hepatic fibrinogen production and perhaps CVD risk (32). MicroRNA-200c exacerbates the ischemia/reperfusion injury of heart through targeting the glutaminase (GLS)-mediated glutamine metabolism (33). Combined GSTM1*0/GSTA1*A genotypes might be considered as genetic markers for cardiovascular death risk in ESRD patients, which may permit targeting of preventive and early intervention (34). HBB identified to be related to Abdominal aortic aneurysm (AAA) (35). PPP1R13L affecting NFkB activity may be candidate genes in the study of human CVD (36). Levels of S100A8/A9, a proinflammatory and prothrombotic protein complex, are increased in several diseases, and high levels predispose to CVD (37). Total absence of the VCAN gene halts heart development at a stage prior to the heart’s pulmonary/aortic outlet segment growth (38).
In summary, in the present study, we discovered that the expression profile of proteins in umbilical vessels of newborns from GDM group was different from that in normal controls. The bio-informatics analysis suggested that some DEPs might play important roles in cardiovascular dysfunction of GDM children. Our findings would contribute to the exploration of the potential mechanism underlying the dysregulated balance of apoptosis and autophagy in vessels, angiogenesis and endothelial cell dysfunction in the offspring of GDM pregnancies. However, deeper analyses will still mostly need to be performed to explore the key factors and potential preventive and therapeutic strategies of cardiovascular dysfunction in GDM offspring.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Shaoxing Maternity and Child Health Care Hospital. The patients/participants provided their written informed consent to participate in this study.
Author contributions
H-TP and Y-MX: writing—original draft. H-DZ: methodology. X-LS: formal analysis. BY: visualization. H-GD: validation. R-JX and J-LD: data curation. JZ and TZ: project administration. All authors contributed to the article and approved the submitted version.
Abbreviations
- GDM
Gestational Diabetes Mellitus
- DEPs
Differentially Expressed Proteins
- FDR
False Discovery Rate
- IPA
Ingenuity Pathway Analysis
- SCX
Strong Cationic-exchange Chromatography
- SBP
Systolic Blood Pressure
- IMT
Intima-Media Thickness
- FGA
Fibrinogen Alpha Chain
- ACTA2, Actin
aortic smooth muscle
- IDH3A
Isocitrate dehydrogenase (NAD) subunit alphamitochondrial.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (82071729), the China Postdoctoral Science Foundation (2020M681336), the Science Technology Department of Zhejiang Province, China (LGD20H040001, LGF21H040003, and LGF21H040004), the Health Commission of Zhejiang Province, China (2022RC277, 2022KY412, 2022KY413, and 2021KY375), and the Science Technology Department of Shaoxing, China (2020A13034 and 2020A13035).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor JQ declared a shared parent affiliation with the authors Y-MX and H-TP at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2022.1021112/full#supplementary-material
Cardiovascular related pathways.
Disease and disorder analysis.
Functional analysis.
A total of 1,653 proteins quantified in the umbilical arteries from both GDM and the controls.
A total of 100 differentially expressed proteins (DEPs) in umbilical vessels.
Ingenuity canonical pathways analysis.
Disease and disorder analysis.
Functional analysis.
References
- 1.HAPO Study Cooperative Research Group, Metzger BE, Lowe LP, Dyer AR, Trimble ER, Chaovarindr U, et al. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. (2008). 358:1991–2002. 10.1056/NEJMoa0707943 [DOI] [PubMed] [Google Scholar]
- 2.Lawlor DA, Lichtenstein P, Langstrom N. Association of maternal diabetes mellitus in pregnancy with offspring adiposity into early adulthood: sibling study in a prospective cohort of 280,866 men from 248,293 families. Circulation. (2011) 123:258–65. 10.1161/CIRCULATIONAHA.110.980169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boney CM, Verma A, Tucker R, Vohr BR. Metabolic syndrome in childhood: association with birth weight, maternal obesity, and gestational diabetes mellitus. Pediatrics. (2005) 115:e290–6. 10.1542/peds.2004-1808 [DOI] [PubMed] [Google Scholar]
- 4.Krishnaveni GV, Veena SR, Jones A, Srinivasan K, Osmond C, Karat SC, et al. Exposure to maternal gestational diabetes is associated with higher cardiovascular responses to stress in adolescent Indians. J Clin Endocrinol Metab. (2015) 100:986–93. 10.1210/jc.2014-3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu CS, Nohr EA, Bech BH, Vestergaard M, Olsen J. Long-term health outcomes in children born to mothers with diabetes: a population-based cohort study. PLoS One. (2012) 7:e36727. 10.1371/journal.pone.0036727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zitser B. Maternal diabetes and autism in offspring. JAMA. (2015) 314:407. 10.1001/jama.2015.7250 [DOI] [PubMed] [Google Scholar]
- 7.Sacks DA, Hadden DR, Maresh M, Deerochanawong C, Dyer AR, Metzger BE, et al. Frequency of gestational diabetes mellitus at collaborating centers based on IADPSG consensus panel-recommended criteria the hyperglycemia and adverse pregnancy outcome (HAPO) study. Diabetes Care. (2012) 35:526–8. 10.2337/dc11-1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aceti A, Santhakumaran S, Logan KM, Philipps LH, Prior E, Gale C, et al. The diabetic pregnancy and offspring blood pressure in childhood: a systematic review and meta-analysis. Diabetologia. (2012) 55:3114–27. 10.1007/s00125-012-2689-8 [DOI] [PubMed] [Google Scholar]
- 9.Tam WH, Ma RCW, Yang X, Ko GTC, Tong PCY, Cockram CS, et al. Glucose intolerance and cardiometabolic risk in children exposed to maternal gestational diabetes mellitus in utero. Pediatrics. (2008) 122:1229–34. 10.1542/peds.2008-0158 [DOI] [PubMed] [Google Scholar]
- 10.Catalano PM, Farrell K, Thomas A, Huston-Presley L, Mencin P, de Mouzon SH, et al. Perinatal risk factors for childhood obesity and metabolic dysregulation. Am J Clin Nutr. (2009) 90:1303–13. 10.3945/ajcn.2008.27416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam WH, Ma RCW, Ozaki R, Li AM, Chan MHM, Yuen LY, et al. In utero exposure to maternal hyperglycemia increases childhood cardiometabolic risk in offspring. Diabetes Care. (2017) 40:679–86. 10.2337/dc16-2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderwald C, Pfeiler G, Nowotny P, Anderwald-Stadler M, Krebs M, Bischof MG, et al. Glucose turnover and intima media thickness of internal carotid artery in type 2 diabetes offspring. Eur J Clin Invest. (2008) 38:227–37. 10.1111/j.1365-2362.2008.01932.x [DOI] [PubMed] [Google Scholar]
- 13.Aman J, Hansson U, Ostlund I, Wall K, Persson B. Increased fat mass and cardiac septal hypertrophy in newborn infants of mothers with well-controlled diabetes during pregnancy. Neonatology. (2011) 100:147–54. 10.1159/000323741 [DOI] [PubMed] [Google Scholar]
- 14.Westermeier F, Salomón C, González M, Puebla C, Guzmán-Gutiérrez E, Cifuentes F, et al. Insulin restores gestational diabetes mellitus-reduced adenosine transport involving differential expression of insulin receptor isoforms in human umbilical vein endothelium. Diabetes. (2011) 60:1677–87. 10.2337/db11-0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Floris I, Descamps B, Vardeu A, Mitiæ T, Posadino AM, Shantikumar S, et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving MicroRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler Thromb Vasc Biol. (2015) 35:664–74. 10.1161/Atvbaha.114.304730 [DOI] [PubMed] [Google Scholar]
- 16.Koukkou E, Ghosh P, Lowy C, Poston L. Offspring of normal and diabetic rats fed saturated fat in pregnancy demonstrate vascular dysfunction. Circulation. (1998) 98:2899–904. 10.1161/01.cir.98.25.2899 [DOI] [PubMed] [Google Scholar]
- 17.Nehiri T, Van Huyen J-PD, Viltard M, Fassot C, Heudes D, Freund N, et al. Exposure to maternal diabetes induces salt-sensitive hypertension and impairs renal function in adult rat offspring. Diabetes. (2008) 57:2167–75. 10.2337/db07-0780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding GL, Wang F-F, Shu J, Tian S, Jiang Y, Zhang D, et al. Transgenerational glucose intolerance with Igf2/H19 epigenetic alterations in mouse islet induced by intrauterine hyperglycemia. Diabetes. (2012) 61:1133–42. 10.2337/db11-1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng X, Chapple SJ, Patel B, Puszyk W, Sugden D, Yin X, et al. Gestational diabetes mellitus impairs Nrf2-mediated adaptive antioxidant defenses and redox signaling in fetal endothelial cells in utero. Diabetes. (2013) 62:4088–97. 10.2337/db13-0169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan HT, Guo M-X, Xiong Y-M, Ren J, Zhang J-Y, Gao Q, et al. Differential proteomic analysis of umbilical artery tissue from preeclampsia patients, using iTRAQ isobaric tags and 2D nano LC-MS/MS. J Proteomics. (2015) 112:262–73. 10.1016/j.jprot.2014.09.006 [DOI] [PubMed] [Google Scholar]
- 21.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. (2009) 6:359–62. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 22.Sheng Q, Dai J, Wu Y, Tang H, Zeng R. BuildSummary: using a group-based approach to improve the sensitivity of peptide/protein identification in shotgun proteomics. J Proteome Res. (2012) 11:1494–502. 10.1021/pr200194p [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Chen T, Wu S, Yang C, Bai M, Shu K, et al. iProX: an integrated proteome resource. Nucleic Acids Res. (2019) 47:D1211–7. 10.1093/nar/gky869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.West NA, Crume TL, Maligie MA, Dabelea D. Cardiovascular risk factors in children exposed to maternal diabetes in utero. Diabetologia. (2011) 54:504–7. 10.1007/s00125-010-2008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Q, Pan H-T, Lin X-H, Zhang J-Y, Jiang Y, Tian S, et al. Altered protein expression profiles in umbilical veins: insights into vascular dysfunctions of the children born after in vitro fertilization. Biol Reprod. (2014) 91:71. 10.1095/biolreprod.114.120659 [DOI] [PubMed] [Google Scholar]
- 26.Xu GF, Zhang J-Y, Pan H-T, Tian S, Liu M-E, Yu T-T, et al. Cardiovascular dysfunction in offspring of ovarian-hyperstimulated women and effects of estradiol and progesterone: a retrospective cohort study and proteomics analysis. J Clin Endocrinol Metab. (2014) 99:E2494–503. 10.1210/jc.2014-2349 [DOI] [PubMed] [Google Scholar]
- 27.Lurbe E, Torro MI. Early vascular phenotypes in the genesis of hypertension. Pediatr Nephrol. (2010) 25:763–7. 10.1007/s00467-009-1357-9 [DOI] [PubMed] [Google Scholar]
- 28.Meuwissen ME, Lequin MH, Bindels-de Heus K, Bruggenwirth HT, Knapen MFCM, Dalinghaus M, et al. ACTA2 mutation with childhood cardiovascular, autonomic and brain anomalies and severe outcome. Am J Med Genet A. (2013) 161:1376–80. 10.1002/ajmg.a.35858 [DOI] [PubMed] [Google Scholar]
- 29.Altaf F, Vesely C, Sheikh AM, Munir R, Shah STA, Tariq A, et al. Modulation of ADAR mRNA expression in patients with congenital heart defects. PLoS One. (2019) 14:e0200968. 10.1371/journal.pone.0200968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, et al. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. (1996) 87:687–96. 10.1016/s0092-8674(00)81388-4 [DOI] [PubMed] [Google Scholar]
- 31.Newell K, Smith W, Ghoshhajra B, Isselbacher E, Lin A, Lindsay ME, et al. Cervical artery dissection expands the cardiovascular phenotype in FBN1-related Weill-Marchesani syndrome. Am J Med Genet A. (2017) 173:2551–6. 10.1002/ajmg.a.38353 [DOI] [PubMed] [Google Scholar]
- 32.Fish RJ, Neerman-Arbez M. A novel regulatory element between the human FGA and FGG genes. Thromb Haemost. (2012) 108:427–34. 10.1160/TH12-04-0274 [DOI] [PubMed] [Google Scholar]
- 33.Liu F, Li Y, Liu G. MicroRNA-200c exacerbates the ischemia/reperfusion injury of heart through targeting the glutaminase (GLS)-mediated glutamine metabolism. Eur Rev Med Pharmacol Sci. (2017) 21:3282–9. [PubMed] [Google Scholar]
- 34.Suvakov S, Damjanovic T, Pekmezovic T, Jakovljevic J, Savic-Radojevic A, Pljesa-Ercegovac M, et al. Associations of GSTM1*0 and GSTA1*A genotypes with the risk of cardiovascular death among hemodialyses patients. BMC Nephrol. (2014) 15:12. 10.1186/1471-2369-15-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan L, Huang J, Ni H, Yu G. Screening key genes for abdominal aortic aneurysm based on gene expression omnibus dataset. BMC Cardiovasc Disord. (2018) 18:34. 10.1186/s12872-018-0766-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson MA, Cook RW, Solanki P, Patton MA, Dennis JA, Crosby AH, et al. A mutation in NFkappaB interacting protein 1 causes cardiomyopathy and woolly haircoat syndrome of Poll Hereford cattle. Anim Genet. (2009) 40:42–6. 10.1111/j.1365-2052.2008.01796.x [DOI] [PubMed] [Google Scholar]
- 37.Lood C, Tydén H, Gullstrand B, Jönsen A, Källberg E, Mörgelin M, et al. Platelet-Derived S100A8/A9 and cardiovascular disease in systemic lupus erythematosus. Arthritis Rheumatol. (2016) 68:1970–80. 10.1002/art.39656 [DOI] [PubMed] [Google Scholar]
- 38.Burns TA, Dours-Zimmermann MT, Zimmermann DR, Krug EL, Comte-Walters S, Reyes L, et al. Imbalanced expression of Vcan mRNA splice form proteins alters heart morphology and cellular protein profiles. PLoS One. (2014) 9:e89133. 10.1371/journal.pone.0089133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cardiovascular related pathways.
Disease and disorder analysis.
Functional analysis.
A total of 1,653 proteins quantified in the umbilical arteries from both GDM and the controls.
A total of 100 differentially expressed proteins (DEPs) in umbilical vessels.
Ingenuity canonical pathways analysis.
Disease and disorder analysis.
Functional analysis.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.