Abstract
Purpose
Neoadjuvant therapy modality can increase the operability rate and mitigate pathological risks in locally advanced cervical cancer, but treatment response varies widely. It remains unclear whether genetic alterations correlate with the response to neoadjuvant therapy and disease-free survival (DFS) in locally advanced cervical cancer.
Materials and Methods
A total of 62 locally advanced cervical cancer (stage IB–IIA) patients who received neoadjuvant chemoradiation plus radical hysterectomy were retrospectively analyzed. Patients’ tumor biopsy samples were comprehensively profiled using targeted next generation sequencing. Pathologic response to neoadjuvant treatment and DFS were evaluated against the association with genomic traits.
Results
Genetic alterations of PIK3CA were most frequent (37%), comparable to that of Caucasian populations from The Cancer Genome Atlas. The mutation frequency of genes including TERT, POLD1, NOS2, and FGFR3 was significantly higher in Chinese patients whereas RPTOR, EGFR, and TP53 were underrepresented in comparison to Caucasians. Germline mutations were identified in 21% (13/62) of the cohort and more than half (57%) had mutations in DNA damage repair genes, including BRCA1/2, TP53 and PALB2. Importantly, high tumor mutation burden, TP53 polymorphism (rs1042522), and KEAP1 mutations were found to be associated with poor pathologic response to neoadjuvant chemoradiation treatment. KEAP1 mutations, PIK3CA-SOX2 co-amplification, TERC copy number gain, and TYMS polymorphism correlated with an increased risk of disease relapse.
Conclusion
We report the genomic profile of locally advanced cervical cancer patients and the distinction between Asian and Caucasian cohorts. Our findings highlight genomic traits associated with unfavorable neoadjuvant chemoradiation response and a higher risk of early disease recurrence.
Keywords: Uterine cervical neoplasms, Neoadjuvant therapy, Pathologic response, Disease-free survival, DNA damage repair
Introduction
Cervical cancer is the fourth most common cancer diagnosed among females and every year leads to more than half-million new cases as well as over 300,000 deaths worldwide [1]. Despite recent advances in prevention, diagnosis and treatment, clinical outcome of cervical cancer patients remains poor in the developing countries [2]. While the incidence of cervical cancer in developed countries has more than halved over the past decades, a surge in cervical cancer incidence was recently reported in China [3]. On the macro level, insufficient pap smear screening and human papillomavirus (HPV) vaccination are the major culprits of this international disparity [4]. On the molecular level, there may also exist differences in the mutational landscape of cervical cancer between Chinese and the Western populations, which may reveal clues of carcinogenesis mechanism and susceptibility among different ethnic groups [5].
The primary treatment strategy for patients with early-stage cervical cancers, particularly stage IA–IB1, is radical hysterectomy with or without radiation or chemotherapy [6]. Multiple treatment regimens have been actively explored and proposed for high-risk early-stage (stage IB–IIA) cervical cancer patients [7,8]. Neoadjuvant brachytherapy and chemotherapy followed by radical surgery showed an efficacy non-inferior to standard chemoradiation treatment and a more favorable toxicity profile in stage IB2–IIA cervical cancer [7]. Despite a high three-year disease-free survival (DFS) rate of 90%, there was a portion of patients who failed to respond to the therapy. Identification of potential biomarkers predicting poor treatment response in these patients is much needed.
In this study, we compared the genetic landscape of cervical cancer between Chinese and the Western populations to understand the differences in potential tumorigenesis mechanisms and identified associations between specific genetic alterations and poor treatment response to neoadjuvant therapy.
Materials and Methods
1. Study design and patients
This was a single-institution retrospective study that enrolled a total of 62 patients who were diagnosed of cervical cancer from 2016 to 2019 and received treatment at Shandong Cancer Hospital, Jinan, Shandong, China. The study was approved by the Institutional Review Board/Ethics Committee of Shandong Cancer Hospital. All patients provided written informed consent prior to sample collection.
Patients were included for analysis according to the following criteria: (1) cervical cancer patients with histologically confirmed International Federation of Gynecology and Obstetrics (FIGO) stage IB1–IIA (FIGO 2009) [9]; (2) age ≥ 18 years old; (3) pathological subtypes were squamous cell carcinoma (SQCC), adenocarcinoma (ADC) or adenosquamous carcinoma (ASC), excluding special types of tumors, such as clear cells carcinoma; (4) Eastern Cooperative Oncology Group performance status score of 0-2. Patients voluntarily joined this study, signed informed consent and provided diagnosis and treatment data after cancer diagnosis before entering the group, good compliance, and cooperation with follow-up visits.
Patients were excluded for analysis when (1) potential radiation field overlap caused by previous radiotherapy; (2) patients could not undergo routine imaging examination; (3) any signs of severe or uncontrolled systemic diseases that the researchers believe may significantly affect the patient’s risk/benefit balance, including hepatitis B, hepatitis C and human immunodeficiency virus.
2. Clinical data and samples
Patients’ clinical data were carefully reviewed, including age, pathological grade, imaging examination (computed tomography, magnetic resonance imaging or positron emission tomography–computed tomography, etc.) with or without lymph node metastasis, tumor stage, immunohistochemical results, course of disease, location and size of lesions, performance status score, family history. Paraffin samples of tumors were biopsied before and after radiotherapy and chemotherapy for next generation sequencing and pathological response assessment, respectively. Ten millileters of venous blood was collected from each patient after chemo-radiotherapy and was kept in the purple lid EDTA anticoagulant blood collection tube (BD, Franklin Lakes, NJ). The white blood cell or normal tissue adjacent to tumor was used as control of tumor samples.
3. Treatment
All patients received one cycle of chemotherapy (paclitaxel plus cisplatin) and brachytherapy ([500–700] cGy×[1–2] fraction) before the radical cervical cancer resection (extensive hysterectomy and pelvic lymph node dissection and salpingo-oophorectomy or abdominal para-aortic lymphadenectomy). The radical surgery was followed by adjuvant chemotherapy (three cycles), brachytherapy and irradiation (5,040 cGy/28 fraction). A detailed treatment regimen of each patient’s neoadjuvant and adjuvant chemotherapy was provided in S1 Table. DFS was defined as the time from neoadjuvant chemoradiotherapy until the time of tumor relapse or the date of the last follow-up.
4. Pathological assessment
The tumor samples were taken and subject to hematoxylin and eosin (H&E) staining protocol after chemoradiotherapy to evaluate their pathologic response to treatment. H&E slides of sections of tumors after treatment were evaluated by pathologists blinded to the patient information. At least 1 section was taken every centimeter of tumor along its greatest diameter. About 5 to 30 slides were examined for each patient. The percentage of residual viable tumor was determined by dividing the estimated cross-sectional area of viable tumor foci by total cross-sectional areas evaluated on each slide [10,11]. An average (mean) value of the percent of residual viable tumor was determined for each patient. Histologic parameters analyzed include inflammation, necrosis, fibrosis, giant cell reaction, foamy macrophages, and cholesterol cleft granuloma.
5. DNA extraction and library preparation
Sample processing and genomic profiling were performed in a Clinical Laboratory Improvement Amendments (CLIA)- and the College of American Pathologists (CAP)-accredited laboratory (Nanjing Geneseeq Technology Inc., Nanjing, China) as previously described [12,13]. In brief, genomic DNA from tumor specimen and control samples were extracted and quantified by Qubit 3.0. Library preparations were performed with KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA). Target enrichment was performed using customized xGen lockdown probes (Integrated DNA Technologies, Coralville, IA) targeting 474 cancer- and radiotherapy response-relevant genes (Radiotron gene panel, Nanjing Geneseeq Technology Inc.) (S2 Table). The hybridization capture reaction was performed with Dynabeads M-279 (Life Technologies, San Diego, CA) and xGen Lockdown hybridization and wash kit (Integrated DNA Technologies) according to manufacturer’s protocols. Captured libraries were on-beads polymerase chain reaction (PCR) amplified with Illumina p5 and p7 primers in KAPA HiFi HotStart ReadyMix (KAPA Biosystems), followed by purification using Agencourt AMPure XP beads. Libraries were quantified by quantitative real-time PCR using KAPA Library Quantification kit (KAPA Biosystems). Library fragment size was determined by Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA).
6. Targeted next generation sequencing and data processing
Sequencing was performed on the Illumina HiSeq4000 platform (Illumina, San Diego, CA) followed by data analysis as previously described [12,13]. In brief, sequencing data were analyzed by Trimmomatic [14] to remove low-quality (quality < 15) or N bases, and then mapped to the human reference genome hg19 using the Burrows-Wheeler Aligner (https://github.com/lh3/bwa/tree/master/bwakit). PCR duplicates were removed by Picard (available at: https://broadinstitute.github.io/picard/). The Genome Analysis Toolkit (GATK) (https://software.broadinstitute.org/gatk/) was used to perform local realignments around indels and base quality reassurance. Single nucleotide polymorphisms (SNPs) and indels were analyzed by VarScan2 [15] and HaplotypeCaller/UnifiedGenotyper in GATK, with the mutant allele frequency cutoff as 0.5% for tissue samples, 0.1% for cell-free DNA samples, and a minimum of three unique mutant reads. Common SNPs were excluded if they were present in > 1% population frequency in the 1000 Genomes Project or the Exome Aggregation Consortium (ExAC) 65000 exomes database. The resulting mutation list was further filtered by an in-house list of recurrent artifacts based on a normal pool of whole blood samples. Gene fusions were identified by FACTERA [16].
Tumor mutation burden (TMB) was calculated based on the number of non-silent somatic mutations per megabase coding region sequenced. Microsatellite (MS) status of tumor sample was determined on the overall stability of MS loci covered by the sequencing panel (Radiotron, Nanjing Geneseeq Technology Inc.) using a proprietary in-house developed microsatellite instability (MSI) analysis pipeline. Briefly, a total of 108 mononucleotide repeats were evaluated and a subset of 52 loci with a minimum of 15-bp repeats were eventually identified as the MSI determination sites in the targeted sequencing region, including those conventional MSI detection sites such as BAT-25, BAT-26, NR-21, NR-24, and MONO-27. A site is considered qualified for analysis only if > 100× coverage depth. A sample was reported as microsatellite instable (“MSI”) if ≥ 40% of the qualified MS loci display instability, or as “MSS (microsatellite stable)” if < 40% of the qualified MS loci display instability, as previously described [17].
7. Statistical analysis
Categorical variables were compared between mutation carriers and non-carriers using the Fisher exact test. The subgroup analysis of TMB was performed using Student’s t test. The Kaplan-Meier method was used for DFS analysis, and statistical significance was assessed using the log-rank test. A p-value of less than 0.05 was considered statistically significant. All statistical analyses were performed using R ver. 3.4.4 (R Software, R Foundation for Statistical Computing, Vienna, Austria). Gene pathways were analyzed using ReactomePA R package [18].
Results
1. Patient overview
A total of 62 locally advanced cervical cancer patients (stage IB–IIA) who received neoadjuvant chemoradiation plus radical hysterectomy in Shandong Cancer Hospital from 2016 to 2019 were retrospectively reviewed in this study (Table 1). The median age of the cohort was 47 years (range, 26 to 66 years). Approximately 55% of patients were diagnosed of stage IB disease. SQCC accounted for ~82% of the cohort, with the remaining subjects being ADC (~15%) and ASC (~3%). Most patients (44/62, 71%) were classified as high-risk HPV types including 16, 18, 31, 33, 45, 52, and 58 [19], with the HPV type 16 being most common (30/62, 48%), while the remaining 18 patients remained unknown for the HPV type (S1 Table).
Table 1.
Clinical characteristics of cervical cancer patients
| Chinese (n=62) | Caucasiana) (n=82) | |
|---|---|---|
| Age (yr) | ||
| > 44 | 35 (56) | 46 (56) |
| Median (range) | 47 (26–66) | 45 (20–80) |
| Clinical stage | ||
| IB | 34 (55) | 72 (88) |
| IIA | 28 (45) | 10 (12) |
| Histological type | ||
| Squamous cell carcinoma | 51 (82) | 61 (74) |
| Others | 11 (18) | 21 (26) |
| Residual viable tumor (%) | ||
| 0–10 (major response) | 13 (21) | n/a |
| 10–50 (partial response) | 22 (35) | n/a |
| > 50 | 27 (44) | n/a |
Values are presented as number (%) unless otherwise indicated. n/a, not applicable.
The Caucasian cohort data were derived from The Cancer Genome Atlas database.
2. Genomic characteristics of locally advanced cervical cancer
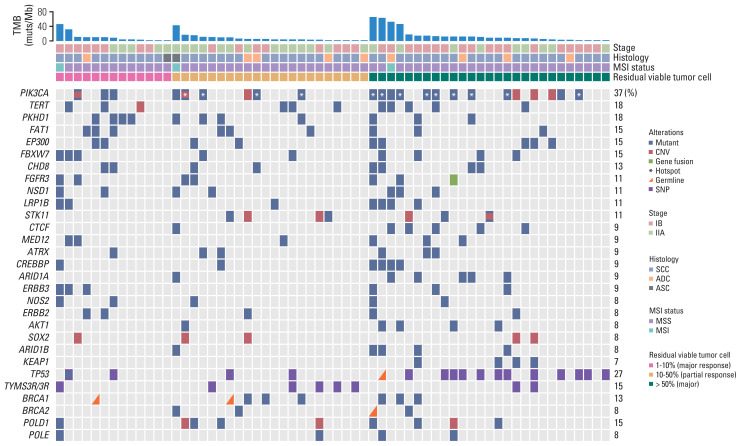
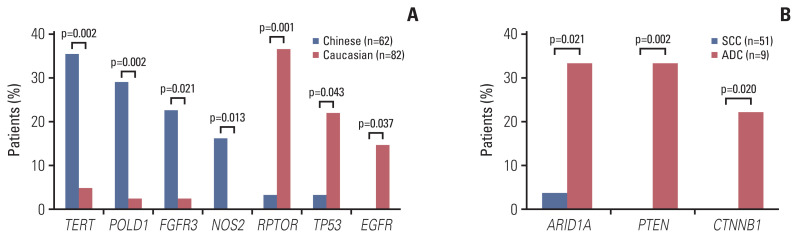
We first characterized the mutational landscape of those 62 locally advanced cervical cancers through comprehensive genomic profiling by using targeted next generation sequencing (see “Materials and Methods”). The median depth of coverage was 974× (range, 322× to 2,159×), and the median coverage depth after removing PCR duplicates was 525× (range, 190× to 1,504×) (S3 Table). As shown in Fig. 1, PIK3CA represented the most frequently mutated gene of which mutations were detected in 37% of the cohort, comparable to what was reported in a Caucasian population of 82 cervical cancer patients from The Cancer Genome Atlas (TCGA) [20] (S4 Table), followed by TERT (18%) and PKHD1 (18%). We acknowledge that there was a difference in the clinical stage between the TCGA dataset and the current cohort (Table 1), but no significant difference of mutation frequencies between IB and IIA patients was observed in either cohort. More than half of mutations identified in PIK3CA were hotspot mutations located in exons 9 and 20, including E542, E545, and H1047, which were involved in inhibitory interaction with regulatory subunit (E542 and E545) and membrane association (H1047) [21]. PIK3CA amplification was also detected in approximately 10% (6/62) of the cohort, and of note, three patients had multiple PIK3CA aberrations (Fig. 1). TERT, POLD1, NOS2, and FGFR3 genes were frequently altered in Chinese cervical cancer, whereas RPTOR, EGFR, and TP53 gene variants were significantly enriched in Caucasian cervical cancer (Fig. 2A). ARID1A, PTEN, and CTNNB1 gene mutations were frequently observed in cervical ADC (Fig. 2B).
Fig. 1.
Distribution of gene alterations correlated with pathologic response. Gene alterations and patient clinical characteristics were shown at the top and bottom, respectively. Patients were separated into three groups, of which H&E stains exhibited < 10%, 10%–50% and 50%–100% viable tumor cells. The BRCA1/2, POLD1, and POLE were genes related to targeted therapy or immunotherapy. PIK3CA hotspot mutations on E542, E545, and H1047 were marked by white asterisks. The 0%–10%, 10%–50%, and 50%–100% viable tumor cells represent major, partial and poor pathologic response, respectively. ADC, adenocarcinoma; ASC, adenosquamous carcinoma; CNV, copy number variation; MSI, microsatellite instability; MSS, microsatellite stability; SCC, squamous cell carcinoma; SNP, single nucleotide polymorphism; TMB, tumor mutation burden.
Fig. 2.
Genetic alterations enriched in Chinese, Caucasian patients or cervical adenocarcinoma (ADC). (A) Gene alterations significantly enriched in Chinese (blue) and Caucasian (red) patients were shown on the left and right respectively. (B) Gene mutations associated with cervical adenocarcinoma were shown in red. SCC, squamous cell carcinoma.
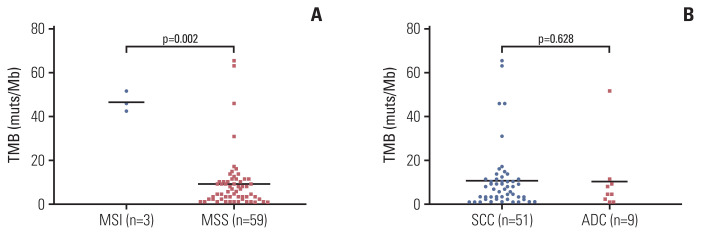
Furthermore, the TMB (median TMB: 46 muts/megabase [Mb]) of microsatellite unstable (MSI) cervical cancer patients (n=3) was significantly higher than MSS patients (median TMB: 9.2 muts/Mb) (Fig. 3A), although the MSI subgroup size was restricted. No significant differences of TMB were observed between histology subgroups as to SQCC or ADC (Fig. 3B).
Fig. 3.
Association of tumor mutation burden (TMB) with microsatellite instability in cervical cancer. Comparison of TMB levels in patients separated by microsatellite instability status (A) and histological types (B). ADC, adenocarcinoma; MSI, microsatellite instability; MSS, microsatellite stable; SCC, squamous cell carcinoma.
In addition, germline mutations were detected in 21% (13/62) of the cohort (Table 2). The median age of the patients who carried germline mutations was 43 years, who were younger than those without germline mutations by an average of 6 years. Most of the patients (77%, 10/13) carried nonsynonymous mutations of genes including TP53, BRCA2, BRIP1, BRCA1, FANCM, MUTYH, FANCE, and PALB2, which play parts in DNA damage repair pathways (Table 2). Particularly, BRCA2, BRIP1, BRCA1, and PALB2 that were involved in homology-directed repair process were detected in four patients (31%) (Table 2).
Table 2.
Germline mutant patient characteristics
| Patient ID | Age (yr) | Stage | Histology | Gene | AA change | Variant type |
|---|---|---|---|---|---|---|
| CC_006 | 35 | IIA | ADC | MPL | W398X | Nonsense variant |
| CC_007 | 44 | IIA | SCC | MMP1 | A330LfsX45 | Frame shift variant |
| CC_015 | 41 | IB | SCC | PMS1 | K894RfsX17 | Frame shift variant |
| CC_019 | 36 | IB | SCC | AXIN2 | R714W | Missense variant |
| CC_028 | 60 | IB | SCC | BRCA1 | c.4358-2A>G | Splice variant |
| CC_028 | 60 | IB | SCC | BRIP1 | S618* | Nonsense variant |
| CC_034 | 66 | IB | SCC | EPCAM | L78R | Missense variant |
| CC_036 | 59 | IIA | SCC | PALB2 | S537L | Missense variant |
| CC_039 | 50 | IB | SCC | MUTYH | Y453C | Missense variant |
| CC_039 | 50 | IB | SCC | TP53 | A86V | Missense variant |
| CC_056 | 43 | IIA | SCC | BRCA2 | S2414* | Nonsense variant |
| CC_072 | 38 | IB | ADC | FANCE | S157Kfs*21 | Frame shift variant |
| CC_079 | 41 | IIA | SCC | BRCA1 | S451Lfs*20 | Frame shift variant |
| CC_081 | 44 | IB | ADC | MLH1 | S295G | Missense variant |
| CC_111 | 42 | IB | SCC | FANCM | L923Cfs*3 | Frame shift variant |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.
3. Genomic traits related to poor neoadjuvant chemoradiation response and higher disease relapse risk
The pathologic response to neoadjuvant chemoradiation was evaluated by quantifying the percent of residual viable tumor following the neoadjuvant chemoradiation (see “Materials and Methods”). Twenty-two patients (35%) showed partial pathologic response to neoadjuvant therapy (< 50% residual viable tumor), and thirteen patients (21%) demonstrated major pathologic response (< 10% viable tumor cells) [10,11], including two patients who showed complete pathologic response (Table 1, S1 Table). Univariate analysis showed that Kelch-like ECH-associated protein 1 (KEAP1) mutations (p=0.031), TMB-high (TMB-H) (p=0.011) and TP53 polymorphism (rs1042522 P72R, p=0.007) were significantly associated with poor pathologic response to neoadjuvant chemoradiation in those patients (S5 Table). The PIK3CA mutations had a trend to associate with poor pathologic response (S5 Table). Multivariate analysis showed that TP53 polymorphism was an independent factor that correlated with poor pathologic response (p=0.014) but not necessarily with poor DFS (hazard ratio [HR], 1.8; 95% confidence interval [CI], 0.2 to 20.8; p=0.616).
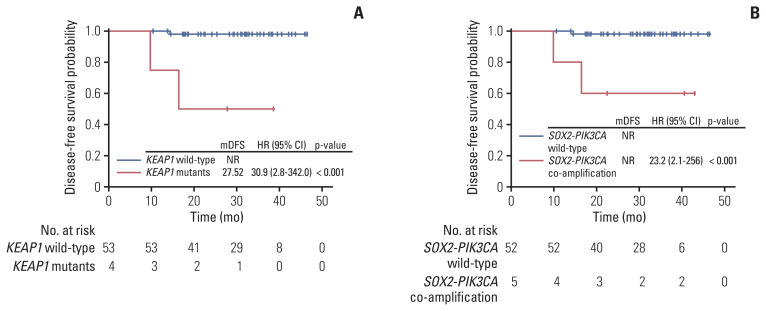
As of manuscript writing, the median follow-up time of the cohort was 31 months. Most of the patients (95%) remained relapse-free up till the data cutoff date. Five patients were lost to follow up after surgery. Firstly, major pathologic response (MPR) patients demonstrated better DFS than non-MPR patients although not significantly (S6 Fig). Secondly, we were able to identify four genomic alterations that were significantly associated with poor DFS, including KEAP1 mutations (n=4; HR, 30.9; 95% CI, 2.8 to 342; p < 0.001) (Fig. 4A), SOX2-PIK3CA co-amplification (n=5; HR, 23.2; 95% CI, 2.1 to 256; p < 0.001) (Fig. 4B), thymidylate synthase triple repeats (3R/3R) polymorphism (n=8; HR, 12.8; 95% CI, 1.2 to 142; p=0.007) (S7A Fig.), and TERC copy number gain (n=3; HR, 45.6; 95% CI, 4.1 to 507; p < 0.001) (S7B Fig.).
Fig. 4.
Association of KEAP1 mutation and SOX2-PIK3CA co-amplification with high cervical cancer recurrence risk. Poor disease-free survival was observed in patients harbouring KEAP1 mutation (A) or SOX2-PIK3CA co-amplification (B). CI, confidence interval; HR, hazard ratio; mDFS, median disease-free survival; NR, not reported.
Discussion
In this study, we characterized genetic alteration of 62 cervical cancer cases in China and compared their molecular profile with that of Caucasian cervical cancer patients in TCGA database. PIK3CA was the most frequently mutated gene in cervical cancer regardless of racial groups, suggesting a universal dependence of cervical cancer on phosphoinositide 3-kinase (PI3K)/AKT signal pathway. Both datasets highlighted three mutation hotspots in PIK3CA gene, including E542, E545, and H1047 which accounted for half of mutation sites. Interestingly, several genes’ mutation frequency differed significantly in Chinese and Caucasian cervical cancer. The three genes predominantly mutated in Caucasian population were RPTOR, EGFR, and TP53, all associated with PI3K/AKT pathway. The genes mainly mutated in Chinese patients were TERT, POLD1, NOS2, and FGFR3. TERT and POLD1 were associated with telomere maintenance in cells. It has been reported that HPV type 16 E6 could activate TERT gene transcription [22], suggesting a close relationship between HPV infection and TERT expression. Gene amplification, rearrangement and protein expression of TERT were associated with poor clinical outcome in human cancers including thyroid cancer, glioma, and neuroblastoma [23]. Further clinical investigation is needed to evaluate the effect of TERT promoter mutation on survival of cervical cancer patients.
Our finding of the enrichment of germline mutations in DNA repair pathway agrees with prior evidence [24]. However, the mutation patterns of cervical cancer differed among studies, likely due to the difference in cohort size and racial/genetic background. In our study, over 60% of the germline mutations were truncation, frameshift, or splicing variants deleterious to protein function, suggesting tumor suppressor role of these genes and importance of inactivation of DNA repair pathway in tumorigenesis. Noteworthy, four patients (31%) carried mutations of genes involved in the homology-directed DNA repair process, yielding a homologous recombination deficiency phenotype, which strongly resembled the results showed in other gynecological cancers including breast, ovarian, and endometrial cancer [25]. So far, several poly(ADP-ribose) polymerase inhibitors (PARPi) have been approved by the US Food and Drug Administration in BRCA1/2-mutant ovarian and breast cancer. Given that, the clinical utility of PARPi in cervical cancer is worth exploration, either alone or in combination with chemotherapy or targeted therapy. The early onset of cancer was found in germline-mutant patients in this study, supporting the critical role of DNA repair gene mutations in carcinogenesis as previously described [26,27].
To date, two randomized phase III trials, NCT00193739 [28] and EORTC Protocol 55994 [29], were designed to compare the neoadjuvant chemotherapy followed by surgery with the standard regimen (concurrent chemoradiation) for FIGO IB–IIA cervical cancer patients, although the latter study has not yet reported its final results. According to both studies, neoadjuvant chemotherapy followed by surgery was not superior to the standard regimen in terms of 5-year DFS or overall survival (OS), while NCT00193739 showed that the neoadjuvant approach had a more favorable safety profile. Nonetheless, the neoadjuvant chemotherapy followed by surgery for FIGO IB–IIA cervical cancer was permitted in National Comprehensive Cancer Network (NCCN) guidelines (ver. 1.2021). Furthermore, the neoadjuvant brachytherapy and chemotherapy to radical hysterectomy was included in the clinical practice guideline in China and has shown promising efficacy [7]. It reduced the size of stage IB2–IIA cervical cancer and enabled radical surgery, achieving an overall survival comparable to standard chemoradiation as well as a more favorable side-effect profile. However, there were still 10% of patients whose tumor progressed after the treatment which may partly be attributed to the poor response to neoadjuvant chemoradiotherapy. Taken together, in view of these limited datasets, further research is warranted to investigate the potential clinical benefit of neoadjuvant chemotherapy in early-stage cervical cancer.
Furthermore, the identification of biomarkers predicting response to neoadjuvant therapy as well as tumor recurrence would enable early detection of recurrence and maximize therapeutic window for patients. KEAP1 mutations have been reported to occur commonly in diverse cancer types including lung cancer (both ADC and SQCC), colon ADC, and endometrial carcinoma [30]. Prior studies have shown that KEAP1 mutations promote cell proliferation in tumors and may also give rise to resistance to chemotherapy [31,32], consistent with what we found in this study that KEAP1 mutations were associated with poor pathologic response to neoadjuvant chemoradiation and an increased risk of early disease relapse in cervical cancer. Furthermore, our data showed that SOX2 and PIK3CA were co-amplified in five patients (four SQCCs and one ADC). SOX2 and PIK3CA are localized in proximity on the chromosome 3q26. This result is consistent with what was recently reported by Voutsadakis [33]. The amplification of 3q26 has also been reported in other cancer types, including head and neck [34], lung [35], and oropharyngeal squamous cell carcinomas [36], further corroborating our findings. Prior studies demonstrated that PIK3CA amplification was associated with shorter survival in lung [37], esophageal [38] and nasopharyngeal SQCC [39], and SOX2 amplification was reported to be associated with clinical progression in squamous lung cancer [35]. Consistently, in this study, we report that in cervical cancer, particularly SQCC, SOX2-PIK3CA co-amplification was significantly associated with poor pathologic response to neoadjuvant chemoradiation and worse disease-free survival.
In addition, according to Li et al. [40], a genome-wide SNPs study of 596 patients with stage IA2–IIIB cervical cancer, four SNPs exhibited strong association with response to neoadjuvant chemotherapy in overall survival (OS) or DFS. In this study, we performed genomic profiling by using targeted next generation sequencing, and through univariate analysis, we found that TP53 polymorphism (P72R, rs1042522), KEAP1 mutations and TMB-H were associated with poor pathologic response to neoadjuvant therapy as measured by the proportion of residual viable tumor, and TP53 remained significantly correlated by a multivariant correction. Though TP53 rs1042522 and wild-type subgroups did not differ significantly in OS or DFS in either cohorts, our data suggest that the association between TP53 polymorphism (P72R) and resistance of chemoradiotherapy also existed in cervical cancer in addition to what has previously been reported for head and neck cancer [41]. In addition, in June 2020, U.S. Food and Drug Administration expanded the approval of pembrolizumab (anti–programmed death-1) to include any cancer with TMB-H. Our previous work has shown that TMB-H (≥ 10 muts/Mb) was associated with favorable response to immune checkpoint blockade in lung cancer [42]. Thus, it seems rational that TMB-H cervical cancer patients can be considered for immunotherapy particularly in view of their poor pathologic response to chemoradiotherapy.
In conclusion, we report the comprehensive genomic profiles of locally advanced cervical cancer patients and the distinction between Asian and Caucasian populations. Our findings also highlight genomic traits associated with unfavorable neoadjuvant chemoradiation response and increased risk of early disease recurrence. This study has a few limitations. Firstly, a retrospective cohort study design was used, and the cohort size remained limited. Secondly, the study presented a relatively shorter follow-up period in comparison to previous studies. Thirdly, an external dataset with the clinical characteristic of the residual viable tumor would be ideal for the validation of response biomarkers. Future efforts should focus on validating these results in prospectively designed studies of larger patient sample size.
Acknowledgments
We would like to thank the patients and family members who gave their consent on presenting the data in this study, as well as the investigators and research staff involved in this study.
This study was partially funded by Natural Science Foundation of China (NSFC81872475, NSFC81372413, NSFC81972683), National key Research and Development Program (2018YFC1313200), Shandong Natural Science Foundation Joint Fund (ZR2019LZL018), Shandong Key Research and Development Plan (2017CXGC1209 and 2017GSF18164) and the Outstanding Youth Natural Science Foundation of Shandong Province (JQ201423), Jinan Clinical Medicine Science and Technology Innovation Plan (201704095), National Key Research and Development Program of China (2016YFC0904700).
Footnotes
Ethical Statement
This study was approved by the Institutional Review Board/Ethics Committee of Shandong Cancer Hospital and Institute (Reference No. SDTHEC201901007). Written informed consent was obtained from each patient prior to sample collection.
Author Contributions
Conceived and designed the analysis: Wei Y, Yuan S.
Collected the data: Wei Y, Wei C, Chen L, Liu N.
Performed the analysis: Wei Y, Qu Q, Yin JC, Pang J, Fang Z, Wu X, Wang X, Mu D.
Wrote the paper: Wei Y, Qu Q, Yin JC, Pang J, Fang Z.
Study supervision: Shao Y, Yu J, Yuan S.
Conflicts of Interest
Conflict of interest relevant to this article was not reported.
Electronic Supplementary Material
Supplementary materials are available at Cancer Research and Treatment website (https://www.e-crt.org).
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Small W, Jr, Bacon MA, Bajaj A, Chuang LT, Fisher BJ, Harkenrider MM, et al. Cervical cancer: a global health crisis. Cancer. 2017;123:2404–12. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Hu T, Lv W, Zhou H, Li X, Yang R, et al. Changes in prevalence and clinical characteristics of cervical cancer in the People’s Republic of China: a study of 10,012 cases from a nationwide working group. Oncologist. 2013;18:1101–7. doi: 10.1634/theoncologist.2013-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahrami A, Hasanzadeh M, Shahidsales S, Farazestanian M, Hassanian SM, Moetamani Ahmadi M, et al. Genetic susceptibility in cervical cancer: From bench to bedside. J Cell Physiol. 2018;233:1929–39. doi: 10.1002/jcp.26019. [DOI] [PubMed] [Google Scholar]
- 6.Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N, et al. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28:iv72–83. doi: 10.1093/annonc/mdx220. [DOI] [PubMed] [Google Scholar]
- 7.Ma Y, Zhao G, Qi J, Sun P, Liu C, Qu P, et al. Neoadjuvant brachytherapy and chemotherapy followed by radical surgery for stage IB2 and IIA cervical cancer: a retrospective comparison with chemoirradiation. Mol Clin Oncol. 2018;8:617–22. doi: 10.3892/mco.2018.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters WA, 3rd, Liu PY, Barrett RJ, 2nd, Stock RJ, Monk BJ, Berek JS, et al. Concurrent chemotherapy and pelvic radiation therapy compared with pelvic radiation therapy alone as adjuvant therapy after radical surgery in high-risk early-stage cancer of the cervix. J Clin Oncol. 2000;18:1606–13. doi: 10.1200/JCO.2000.18.8.1606. [DOI] [PubMed] [Google Scholar]
- 9.Pecorelli S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int J Gynaecol Obstet. 2009;105:103–4. doi: 10.1016/j.ijgo.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Hellmann MD, Chaft JE, William WN, Jr, Rusch V, Pisters KM, Kalhor N, et al. Pathological response after neoadjuvant chemotherapy in resectable non-small-cell lung cancers: proposal for the use of major pathological response as a surrogate endpoint. Lancet Oncol. 2014;15:e42–50. doi: 10.1016/S1470-2045(13)70334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pataer A, Kalhor N, Correa AM, Raso MG, Erasmus JJ, Kim ES, et al. Histopathologic response criteria predict survival of patients with resected lung cancer after neoadjuvant chemotherapy. J Thorac Oncol. 2012;7:825–32. doi: 10.1097/JTO.0b013e318247504a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu Y, Wu X, Tong X, Wang X, Chang Z, Mao Y, et al. Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep. 2017;7:583. doi: 10.1038/s41598-017-00520-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24:3097–107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 14.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Bratman SV, Stehr H, Lee LJ, Liu CL, Diehn M, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics. 2014;30:3390–3. doi: 10.1093/bioinformatics/btu549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia H, Xue X, Ding H, Ou Q, Wu X, Nagasaka M, et al. Evidence of NTRK1 fusion as resistance mechanism to EGFR TKI in EGFR+ NSCLC: results from a large-scale survey of NTRK1 fusions in Chinese patients with lung cancer. Clin Lung Cancer. 2020;21:247–54. doi: 10.1016/j.cllc.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–9. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 19.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 20.Tomczak K, Czerwinska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K pathway in human disease. Cell. 2017;170:605–35. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMurray HR, McCance DJ. Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J Virol. 2003;77:9852–61. doi: 10.1128/JVI.77.18.9852-9861.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan X, Larsson C, Xu D. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019;38:6172–83. doi: 10.1038/s41388-019-0872-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertelsen B, Tuxen IV, Yde CW, Gabrielaite M, Torp MH, Kinalis S, et al. High frequency of pathogenic germline variants within homologous recombination repair in patients with advanced cancer. NPJ Genom Med. 2019;4:13. doi: 10.1038/s41525-019-0087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen L, Martens JW, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11:5584. doi: 10.1038/s41467-020-19406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dite GS, Jenkins MA, Southey MC, Hocking JS, Giles GG, McCredie MR, et al. Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst. 2003;95:448–57. doi: 10.1093/jnci/95.6.448. [DOI] [PubMed] [Google Scholar]
- 27.Yang X, Leslie G, Doroszuk A, Schneider S, Allen J, Decker B, et al. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J Clin Oncol. 2020;38:674–85. doi: 10.1200/JCO.19.01907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta S, Maheshwari A, Parab P, Mahantshetty U, Hawaldar R, Sastri Chopra S, et al. Neoadjuvant chemotherapy followed by radical surgery versus concomitant chemotherapy and radiotherapy in patients with stage IB2, IIA, or IIB squamous cervical cancer: a randomized controlled trial. J Clin Oncol. 2018;36:1548–55. doi: 10.1200/JCO.2017.75.9985. [DOI] [PubMed] [Google Scholar]
- 29.Kenter G, Greggi S, Vergote I, Katsaros D, Kobierski J, Massuger L, et al. Results from neoadjuvant chemotherapy followed by surgery compared to chemoradiation for stage Ib2–IIb cervical cancer, EORTC 55994. J Clin Oncol. 2019;37(15 Suppl):5503. doi: 10.1200/JCO.22.02852. [DOI] [PubMed] [Google Scholar]
- 30.AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017;7:818–31. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong Y, Hoang NT, Lovejoy A, Stehr H, Newman AM, Gentles AJ, et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov. 2017;7:86–101. doi: 10.1158/2159-8290.CD-16-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohta T, Iijima K, Miyamoto M, Nakahara I, Tanaka H, Ohtsuji M, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–9. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 33.Voutsadakis IA. 3q26 amplifications in cervical squamous carcinomas. Curr Oncol. 2021;28:2868–80. doi: 10.3390/curroncol28040251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidson MA, Shanks EJ. 3q26–29 Amplification in head and neck squamous cell carcinoma: a review of established and prospective oncogenes. FEBS J. 2017;284:2705–31. doi: 10.1111/febs.14061. [DOI] [PubMed] [Google Scholar]
- 35.McCaughan F, Pole JC, Bankier AT, Konfortov BA, Carroll B, Falzon M, et al. Progressive 3q amplification consistently targets SOX2 in preinvasive squamous lung cancer. Am J Respir Crit Care Med. 2010;182:83–91. doi: 10.1164/rccm.201001-0005OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kokalj Vokac N, Cizmarevic B, Zagorac A, Zagradisnik B, Lanisnik B. An evaluation of SOX2 and hTERC gene amplifications as screening markers in oral and oropharyngeal squamous cell carcinomas. Mol Cytogenet. 2014;7:5. doi: 10.1186/1755-8166-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawano O, Sasaki H, Okuda K, Yukiue H, Yokoyama T, Yano M, et al. PIK3CA gene amplification in Japanese non-small cell lung cancer. Lung Cancer. 2007;58:159–60. doi: 10.1016/j.lungcan.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 38.Kim HS, Lee SE, Bae YS, Kim DJ, Lee CG, Hur J, et al. PIK3CA amplification is associated with poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Oncotarget. 2016;7:30691–701. doi: 10.18632/oncotarget.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fendri A, Khabir A, Mnejja W, Sellami-Boudawara T, Daoud J, Frikha M, et al. PIK3CA amplification is predictive of poor prognosis in Tunisian patients with nasopharyngeal carcinoma. Cancer Sci. 2009;100:2034–9. doi: 10.1111/j.1349-7006.2009.01292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Huang K, Zhang Q, Zhou J, Sun H, Tang F, et al. Genome-wide association study identifies four SNPs associated with response to platinum-based neoadjuvant chemotherapy for cervical cancer. Sci Rep. 2017;7:41103. doi: 10.1038/srep41103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergamaschi D, Gasco M, Hiller L, Sullivan A, Syed N, Trigiante G, et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell. 2003;3:387–402. doi: 10.1016/s1535-6108(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 42.Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, et al. Comprehensive genomic profiling identifies novel genetic predictors of response to anti-PD-(L)1 therapies in non-small cell lung cancer. Clin Cancer Res. 2019;25:5015–26. doi: 10.1158/1078-0432.CCR-19-0585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.