Abstract
Simian adenoviruses are in the genus Mastadenovirus of the family Adenoviridae. This family is composed of non-enveloped, double-stranded DNA viruses that infect a wide range of animals. Mastadenoviruses infect mammals, including non-human primates and humans. The close genetic relatedness between simian and human adenoviruses, with its associated potential for the cross-species transmission of zoonotic adenoviruses from monkeys to humans and vice versa, poses important health concerns and thus warrants research. In this study, we performed a molecular survey of adenoviruses in monkeys in Thailand. Most of the monkeys tested here were long-tailed macaques, free-ranging in areas close to human territories across four provinces: Ratchaburi, Kanchanaburi, Lopburi, and Prachuap Khiri Khan. A few fecal samples from captive wild monkeys (a stump-tailed macaque, pig-tailed macaques, gibbons, and a leaf monkey) were also tested. Adenoviruses were detected in 33.3% (70 out of 210) of the fecal or rectal swab samples. The viruses identified in these samples included Simian adenovirus (SAdV)-A, SAdV-B, SAdV-H, Human adenovirus (HAdV)-D, HAdV-G, and a bat adenovirus species. One SAdV-B, SAdV RBR-7-10, was isolated from a long-tailed macaque fecal sample and identified by mass spectrometry. Its full hexon gene and nearly complete DNA polymerase gene were sequenced and analyzed, and the virions were imaged by transmission electron microscopy. The SAdV RBR-7-10 virus was used in a microneutralization assay to identify virus-specific antibodies in monkey plasma and human serum samples collected from the same areas in Prachuap Khiri Khan Province. We detected neutralizing antibodies against SAdV RBR-7-10 in 6.8% (n = 103) of the monkey samples but in none of the 125 human serum samples, suggesting no cross-species transmission of SAdV RBR-7-10 occurred at this study site. Nevertheless, a continuing surveillance of pathogens in monkeys is warranted to quickly identify possible emerging zoonotic outbreaks.
Keywords: Adenovirus, Monkey, Long-tailed macaque, PCR, Seroprevalence
1. Introduction
Simian adenoviruses are non-enveloped, double-stranded DNA viruses of the genus Mastadenovirus in the family Adenoviridae, with icosahedral virions of 70–90 nm in diameter. Adenovirus genomes range from 26 to 48 Kbp in size and encode approximately 40 structural and non-structural proteins involved in host transcriptional modulation, viral DNA replication, virion assembly and maturation, and sabotage of the host immune response. The Adenoviridae family is composed of six genera (Atadenovirus, Aviadenovirus, Ichtadenovirus, Mastadenovirus, Siadenovirus, and Testadenovirus), and viruses in these groups infect mammals, birds, fish, reptiles, and amphibians [1]. Members of the genus Mastadenovirus infect a wide range of mammals, including humans, non-human primates (NHPs), and many others (cattle, dogs, deer, horses, pigs, sheep, rodents, bats, dolphins, sea lions, and polar bears). Adenovirus infections cause diseases that range widely in severity, from asymptomatic to fatal, in mammals. Clinical manifestations can include respiratory symptoms, gastroenteritis, conjunctivitis, meningitis, hepatitis, and systemic infection. A high mortality rate is associated with an immunosuppressive status and other host factors, especially in patients who are particularly young or old [2]. The classification of virus species within the genus depends on several characteristics, including genomic properties, serological properties, host range, and oncogenicity in rodents. Currently, the genus and species demarcation is based mainly on genomic criteria, i.e., phylogenetic distance, but also on genome organization and biological characteristics [1]. Mastadenovirus members currently include >50 species; among them are seven Human mastadenovirus (HAdV-A to -G) and nine Simian mastadenovirus (SAdV-A to —I) species [1]. Several adenoviruses isolated from NHPs are classified as HAdV species. Regarding species denomination, a species containing adenoviruses isolated from NHPs is still named as a Human mastadenovirus if one of its members was isolated from humans [3]. For example, chimpanzee adenoviruses (ChAdVs) such as ChAd-63, SAdV-23, SAdV-24, and SAdV-25, which have been used in the development of viral vectors for gene transfer and vaccine development, are phylogenetically grouped into HAdV-E [4].
Simian adenoviruses identified from a variety of asymptomatic or diseased wild and captive NHPs, including the great apes (chimpanzees, gorillas, bonobos, and orangutans) [3,[5], [6], [7], [8], [9]], old-world monkeys (macaques, mustached monkeys, colobuses, and baboons) [3,7,[10], [11], [12], [13], [14], [15]], and new-world monkeys [7,16,17] have been molecularly identified and characterized. Novel adenoviruses are continually characterized and added to both simian and human virus species. Owing to the genetic relatedness between monkey and human adenoviruses, interspecies transmission is anticipated [18]. To demonstrate the potential for cross-species transmission, phylogenetic and serologic evidence have been described in a number of publications. For example, a genome analysis-based evolutionary study suggested that HAdV-B and HAdV-E members originated in gorillas and chimpanzees, respectively [19]. These viruses have been zoonotically transmitted to humans in the past and are still circulating and causing diseases in humans [19]. The genetically identical nature of HAdV-C viruses identified in humans and gorillas living in the same vicinity suggests that these viruses have also undergone interspecies transmission [3]. Genetic recombination was proposed as a mechanism involved in interspecies co-evolution and transmission [20,21]. Clinical and serological evidence of zoonotic transmission of viruses from monkeys to humans have been noted. As reported in two publications, researchers who were in close contact with colonies of titi monkeys and baboons, in which outbreaks of adenovirus-associated respiratory illnesses occurred, were seropositive for a titi monkey adenovirus and baboon adenovirus, respectively [12,17]. Flu-like symptoms were reported in the human contacts, and possible human-to-human transmission of the monkey-borne virus was indicated [17].
In Thailand, there are areas in which monkeys and humans live in close proximity, increasing the chances of human exposure to monkey viruses and vice versa. Adenovirus transmission can occur via respiratory droplets, the fecal-oral route, the hand-ocular route, and fomites [18]. On the basis of the One Health concept, pathogen surveillance at the animal–human interface is one way to predict and detect an emerging zoonotic outbreak [22,23]. This study conducted a molecular survey performed by using PCR and nucleotide sequence analysis of adenovirus infection in free-ranging monkeys, mainly long-tailed macaques, and some captive monkeys (a stump-tailed macaque, pig-tailed macaques, gibbons, and a leaf monkey) in four Thai provinces: Ratchaburi, Kanchanaburi, Lopburi, and Prachuap Khiri Khan. One simian adenovirus was isolated, characterized, and used in a microneutralization assay to detect virus-specific antibodies in humans and long-tailed macaques that share the same living area in Prachuap Khiri Khan and predict the potential for cross-species transmission. Information about simian adenovirus infection in monkeys and humans is still limited, and we expect that our findings will help address this knowledge gap.
2. Materials and methods
2.1. Ethical statement
The animal study was conducted in accordance with the Ethical Principles and Guidelines for the Use of Animals for Scientific Purposes, the National Research Council of Thailand (NRCT). Our animal study protocol was reviewed and approved by the Animal Care and Use Committee of the Faculty of Tropical Medicine (FTM-ACUC), Mahidol University (No. FTM-ACUC 004/2022E). Our human study protocol was reviewed and approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (No. MUTM 2022–008-01). The human serum samples used in this work were samples leftover from a previous study (EC approval number: MUTM 2019–058-01). During the consent process of that study, the participants had provided permission to use their leftover specimens for further studies.
2.2. Monkey sample collection and processing
Fecal (n = 120) and rectal swab (n = 90) samples were collected from 210 monkeys, specifically 194 long-tailed macaques (Macaca fascicularis), 1 stump-tailed macaque (Macaca arctoides), 8 pig-tailed macaques (Macaca nemestrina), 6 white-handed gibbons (Hylobates lar), and 1 leaf monkey (Presbytis melalophos), across four provinces, two in Central Thailand (Lopburi and Ratchaburi) and two in Western Thailand (Kanchanaburi and Prachuap Khiri Khan), from 2013 to 2019.
The fecal samples were mixed with phosphate-buffered saline (PBS) to produce a 30% (wt/vol) fecal suspension. Each rectal swab, submerged in transport media, was mixed by vortexing and pressing the cotton swab to the wall of collection tube prior to discarding the cotton swab. All suspensions were centrifuged at 2000 ×g for 10 min at 4 °C to pellet the fecal debris. The resulting supernatants were collected and stored at −80 °C until further use for RNA extraction or virus culture.
2.3. Nucleic extraction and polymerase chain reaction
Viral nucleic acid was extracted from 200 μl of processed fecal/rectal swab sample or virus culture supernatant with the GenUP™ Virus DNA/RNA kit (Biotechrabbit GmbH, Berlin, Germany) in accordance with the manufacturer's instructions. Nucleic acid was eluted in 60 μl of elution buffer and stored at −80 °C until further use.
For adenovirus molecular detection in fecal and rectal swab samples, nested PCR was performed on the extracted nucleic acid using primers specific to the DNA region encoding the polymerase gene (Table 1). The first-round PCR reaction contained 1× MyTaq HS Red Mix (Bioline, London, UK), 0.4 μM of each forward (Adeno-pol F1) and reverse (Adeno-pol R1) primer, and 5 μl of DNA template in a total volume of 25 μl. The thermal cycling conditions were: 95 °C for 3 min, followed by 35 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 90 s, and a final elongation at 72 °C for 10 min. The nested PCR reaction was prepared with primers Adeno-pol F2 and Adeno-pol R2 using 1 μl of the first-round PCR product. The thermal cycling conditions were: 95 °C for 3 min, followed by 35 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 30 s, and a final elongation at 72 °C for 10 min. PCR products were resolved on a 1.5% agarose gel, stained with gel red, and visualized under a gel documentation system (Molecular Imager Gel Doc XR imaging system, Bio-Rad, Hercules, CA, USA).
Table 1.
PCR primers.
| Primer | Sequence (5′-3′) | Region | Position | Size (bp) |
|---|---|---|---|---|
| Adeno-pol F1 | TGATGCGYTTCTTACCTYTGGTYTCCATGAG | DNA polymerase | 5524–5554 | 1515 |
| Adeno-pol R1 | AGTTYTACATGCTGGGCTCTTACCG | DNA polymerase | 7014–7038 | |
| Adeno-pol F2 | GTGACAAAGAGGCTGTCCGTGTCCCCGTA | DNA polymerase | 5571–5599 | 258 |
| Adeno-pol R2 | TCACGTGGCCTACACTTACAAGCCAATCAC | DNA polymerase | 5799–5828 | |
| Pol5030F | AGAGGAGCTCGGCGAGGGT | Iva2 | 4910–4928 | 769 |
| Pol5030R | CTTCGTCTCAGAGTGGTCCGA | DNA polymerase | 5658–5678 | |
| Pol8626F1 | GGTCTCGCGGATGAGGTCGT | DNA polymerase | 6896–6915 | 905 |
| Pol8626R1 | AGCGGCTCTTTGTCACCTACGA | DNA polymerase | 7779–7800 | |
| Pol8626F2 | CGCTCGGGGGTAAGACAGT | DNA polymerase | 7602–7620 | 1140 |
| Pol8626R2 | TCGAGCAGTTCATGGCCGAGAT | pTP | 8720–8741 | |
| Hex18078F | GGCAGAGCACGCTAAACAGCAT | pVI | 17,935–17,955 | 385 |
| Hex18078R | TTGTACGAGTACGCCGTGTCC | Hexon | 18,299–18,319 | |
| Adeno-hex F | CAGGATGCTTCGGAGTACCTGAG | Hexon | 18,129–18,151 | 1844 |
| Adeno-hex R | TTGGCNGGDATDGGGTAVAGCATGTT | Hexon | 19,947–19,972 | |
| Hex20855F | AGCACGCTGGGTAACGATTTG | Hexon | 19,779–19,799 | 1167 |
| Hex20855R | GGGAAAGCGCTTGTCGAAGGT | Protease | 20,925–20,945 |
Primer binding and PCR product sizes were predicted based on sequence JN880449.1 of SAdV-B.
For adenovirus hexon and DNA polymerase gene amplification, PCR was performed with MyTaq HS Red Mix as described above, using 1 μl of the nucleic acid extracted from a virus culture supernatant and sets of overlapping primers specific to the region in the hexon (Hex18078F/Hex18078R, Adeno-hex F/Adeno-hex R, and Hex20855F/Hex20855R) and DNA polymerase (Pol5030F/Pol5030R, Adeno-pol F1/Adeno-pol R1, Pol8626F1/Pol8626R1, and Pol8626F2/Pol8626R2) genes (Table 1). The thermal cycling conditions were as described above, except that the length of time for the elongation at 72 °C was varied as follows: 120 s for primer set Adeno-hex F/Adeno-hex R; 90 s for primer set Adeno-pol F1/Adeno-pol R1; and 60 s for primer sets Hex20855F/Hex20855R, Pol8626F1/Pol8626R1, and Pol8626F2/Pol8626R2. Annealing at 58 °C and elongation at 72 °C for 60 s were performed for the primer sets Hex18078F/Hex18078R and Pol5030F/Pol5030R.
2.4. Nucleotide sequence analysis
For the sequencing of a partial DNA polymerase gene from monkey samples, nested PCR products (258 bp) were excised from the agarose gel, and the DNA was purified using the QIAquick gel extraction kit (Qiagen, Hilden, Germany) in accordance with the manufacturer's instructions. Purified DNA was sent for Sanger sequencing at Bionics Co., Ltd. (Seoul, Korea), using primers Adeno-pol F2 and Adeno-pol R2. For sequencing the full hexon and DNA polymerase genes from the isolated virus SAdV RBR-7-10, overlapping PCR products of the expected sizes (Table 1) were sent for BTSeq™ sequencing at Celemics, Inc. (Seoul, Korea). The sequencing chromatograms were inspected and processed using BioEdit, version 7.0.9.0. Nucleotide sequences were queried against the NCBI database using BLAST and aligned with published reference sequences using ClustalW in BioEdit. A phylogenetic tree was constructed using Molecular Evolutionary Genetics Analysis (MEGA, version 7.0.21) software. The maximum likelihood method with the Tamura 3-parameter model was applied on the basis of the phylogenetic model analysis results. A bootstrap resampling analysis of 1000 replicates was used. Seventy partial DNA polymerase sequences retrieved from the fecal and rectal swab samples were submitted to the GenBank database and received accession numbers ON081673–ON081742. The full hexon and nearly complete DNA polymerase sequences of SAdV RBR-7-10 received accession numbers ON072488 and ON072489.
2.5. Virus isolation
Virus isolation was performed using Vero cells (ATCC CCL81). The cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1× Antibiotic-Antimycotic solution, and 1× GlutaMAX (Gibco™, Thermo Fisher Scientific, Waltham, MA, USA) to a confluency of 80%–90%. The supernatants of 30% (wt/vol) fecal solutions were filtered through a 0.45 μm pore-sized membrane using a syringe filter and then diluted five-fold with viral growth medium (DMEM supplemented with 2% FBS, 1× Antibiotic-Antimycotic solution, and 1× GlutaMAX). The Vero cell monolayer in a 96-well tissue culture plate was inoculated with 50 μl/well of the diluted filtered samples (3 wells/sample). Virus adsorption was achieved by centrifugation of the plate at 25 °C for 20 min prior to incubation at 37 °C under a 5% CO2 atmosphere for 20 min. The centrifugation step was repeated before viral growth medium (150 μl/well) was added, and the cells were incubated at 37 °C under a 5% CO2 atmosphere. The cells were observed microscopically for cytopathic effect (CPE) daily for 5 days. The supernatants and cells from CPE-positive wells were collected and stored at −80 °C. Two blind passages were performed.
2.6. Viral protein identification by mass spectrometry
The supernatant (50 ml) from virus cultured in Vero cells was collected, and cell debris was removed by centrifugation at 1000 ×g for 15 min at 4 °C. The viruses in the supernatant were concentrated by ultracentrifugation in polycarbonate centrifuge bottles (no. 355603, Beckman Coulter, Brea, CA, USA) using a Beckman L7–65 ultracentrifuge (rotor 70.1 Ti) set at 35,000 rpm for 1.5 h at 4 °C. The resulting pellets were mixed with lysis buffer containing 1% NaCl, 1% sodium dodecyl sulfate (SDS), and 1% Triton-X to produce a virus protein lysate, which was then processed for mass spectrometric analysis via LC-MS/MS by a MicroToF Q II mass spectrometer (Bruker, Germany) coupled to an Ultimate 3000 nano-LC system (Dionex, Sunnyvale, CA, USA) as previously described [24].
2.7. Hemagglutination assay
Hemagglutination assays were performed as previously described [25]. Equal amounts of two-fold serially diluted virus and red blood cells (RBCs) were mixed and incubated at room temperature. Goose and chicken RBCs were used at a concentration of 0.5%, whereas sheep and human RBCs were used at a concentration of 0.75%.
2.8. Transmission electron microscopy
Virus-infected Vero cells were collected by centrifugation at 1000 ×g for 15 min at 4 °C. The resulting cell pellets were fixed with 2.5% glutaraldehyde for 1 h and subsequently fixed with 1% osmium tetroxide for 1 h, at room temperature. The fixed pellets were dehydrated with a graded ethanol series, infiltrated and embedded in LR white resin (EMS, Hatfield, PA, USA), polymerized in an oven set to 65 °C for 24–48 h, cut into 100-nm-thick sections, and stained with uranyl acetate and lead citrate. The sections were examined under a transmission electron microscope (Hitachi; model HT7700, Tokyo, Japan), to identify the ultrastructural changes of a cell during viral infection.
2.9. Virus neutralization assay
The CPE-based microneutralization (NT) assay was performed on Vero cells in 96-well micro-culture plates. Test specimens (125 human serum and 103 long-tailed macaque plasma samples) were heat inactivated at 56 °C for 30 min and serially two-fold diluted with maintenance medium (Minimum Essential Medium (MEM) supplemented with 1× Antibiotic-Antimycotic solution, and 1× GlutaMAX) starting from a dilution of 1:20. The diluted serum or plasma samples were mixed with an equal volume of a test virus suspension (prepared at a concentration of 2000 TCID50/ml) and incubated at room temperature for 1 h. The virus-serum mixture was transferred into quadruplicate wells (50 μl/well) containing a Vero cell monolayer in 150 μl of maintenance medium, and the mixture-containing plates were incubated at 37 °C under a 5% CO2 atmosphere. CPE was assessed at day 3 post-inoculation. The neutralizing antibody titer was defined as the reciprocal of the serum or plasma dilution that inhibited 50% (NT50) of virus infectivity, as determined by the absence of CPE on Vero cells.
3. Results
3.1. Molecular detection and identification of adenoviruses in monkey feces and rectal swabs
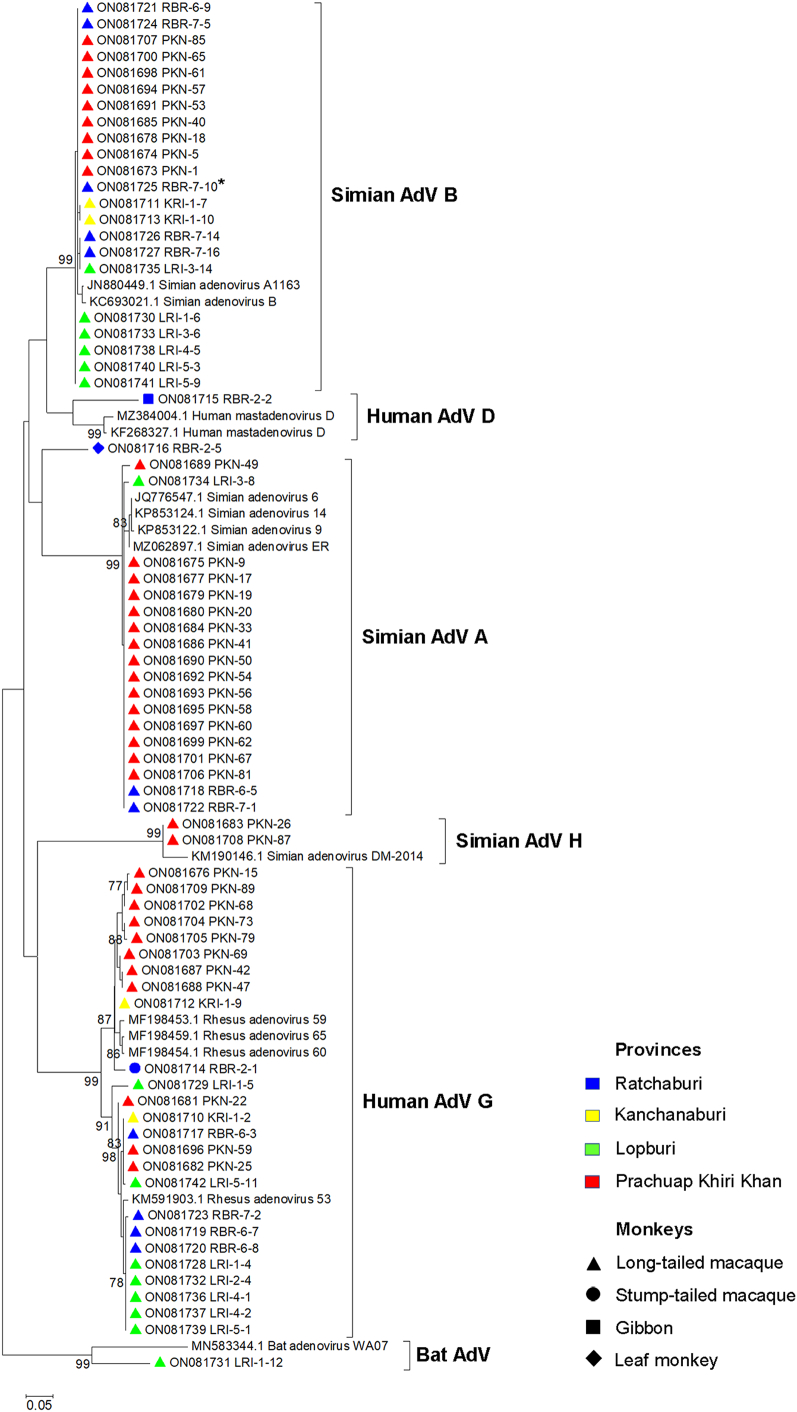
Partial DNA polymerase gene nested PCR was used to detect adenovirus in monkey fecal and rectal swab samples. Of the 210 total samples, 70 (33.3%) were found to be adenovirus positive. Of the 194 samples from long-tailed macaques (M. fascicularis), 67 (34.5%) were adenovirus positive. Adenoviruses were also detected in the stool samples from a stump-tailed macaque (M. arctoides), gibbons (H. lar), and a leaf monkey (P. melalophos). Table 2 shows the adenovirus infection prevalence in monkeys from different sample collection sites. All 70 PCR products were sequenced and phylogenetically analyzed to identify adenovirus species. An analysis of the partial DNA polymerase gene (227 bp) revealed that the viruses detected in long-tailed macaques can be grouped into SAdV-A, SAdV-B, SAdV-H, and HAdV-G, with a nucleotide identity of >94% (Fig. 1); a virus detected in the stump-tailed macaque was closely related to HAdV-G (nucleotide identity: 96%), and one detected in gibbons was related to HAdV-D (nucleotide identity: 86%). Two viruses, one each from a long-tailed macaque and a leaf monkey, were found to be separate from the groups; a BLAST analysis against the NCBI database revealed that they were related to an unclassified bat adenovirus (nucleotide identity: 81%) and HAdV-D (nucleotide identity: 87%), respectively. Regarding sampling locations, SAdV-B and HAdV-G detections were distributed across all four sample collection sites, whereas SAdV-A was detected in three sites, with a high prevalence in Prachuap Khiri Khan (Fig. 2). SAdV-H was detected only in samples from Prachuap Khiri Khan, whereas HAdV-D was detected only in samples from Ratchaburi. The bat AdV-like virus was detected in a sample from Lopburi Province.
Table 2.
Numbers of monkey samples that tested positive for adenovirus by nested PCR.
| Site | Sample type | Collection year | Species | No. of samples | No. of positive (%) |
|---|---|---|---|---|---|
| Ratchaburi | Feces | 2018–2019 | M. fascicularis | 38 | 11 (28.9%) |
| M. arctoides | 1 | 1 | |||
| M. nemestrina | 8 | 0 | |||
| H. lar | 6 | 1 (16.7%) | |||
| P. melalophos | 1 | 1 | |||
| Kanchanaburi | Feces | 2018–2019 | M. fascicularis | 12 | 4 (33.3%) |
| Lopburi | Feces | 2013 | M. fascicularis | 54 | 15 (27.8%) |
| Prachuap Khiri Khan | Rectal swab | 2017 | M. fascicularis | 90 | 37 (41.1%) |
| Total | 210 | 70 (33.3%) |
Fig. 1.
Phylogenetic analysis of partial DNA polymerase nucleotide sequences (227 bp) from monkey samples. The unrooted tree was constructed using the maximum likelihood method with a bootstrap value of 1000. The accession numbers of the sequences obtained in this study and of the reference sequences are shown. Bootstrap values of ≥75 are shown at the node. The bar represents nucleotide substitutions per site. The color and symbol labeling used for collection sites (provinces) and monkey types, respectively, are indicated in the figure. Asterisks (*) indicate samples in which an adenovirus was isolated in Vero cell culture.
Fig. 2.
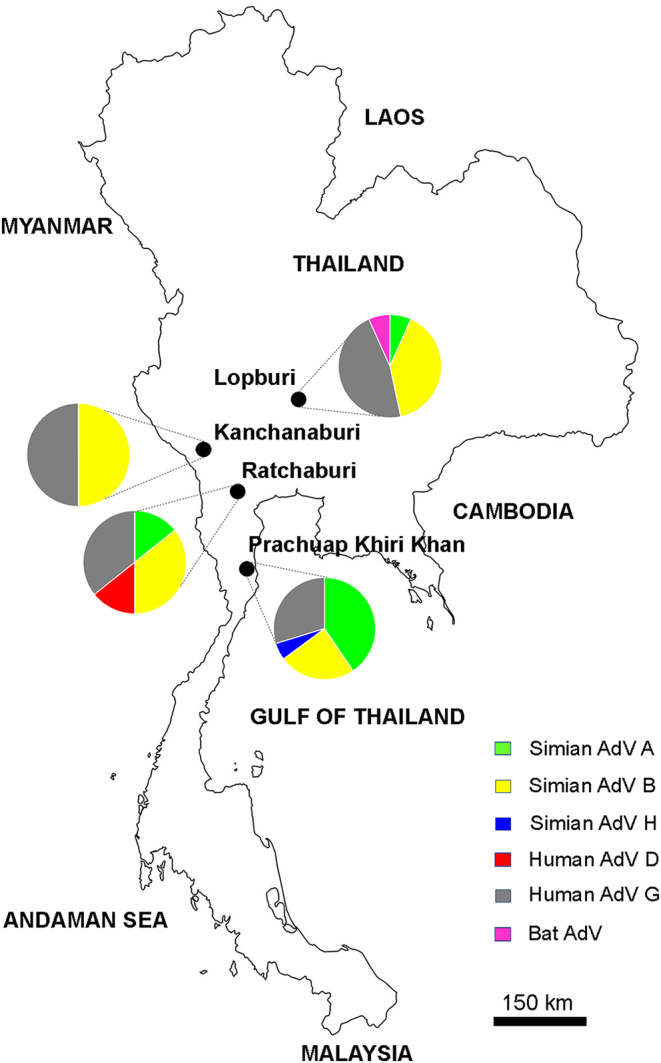
Map of Thailand showing the sample collection sites and proportions of detected adenoviruses. Provinces where the samples were collected (Ratchaburi, Kanchanaburi, Lopburi, and Prachuap Khiri Khan) are indicated by black circles on the map of Thailand. Pie charts show the proportions of adenovirus species that were detected in the adenovirus-positive samples from each area. The color labeling used for adenovirus species is indicated in the figure.
3.2. Isolation and characterization of SAdV-B from long-tailed macaque feces
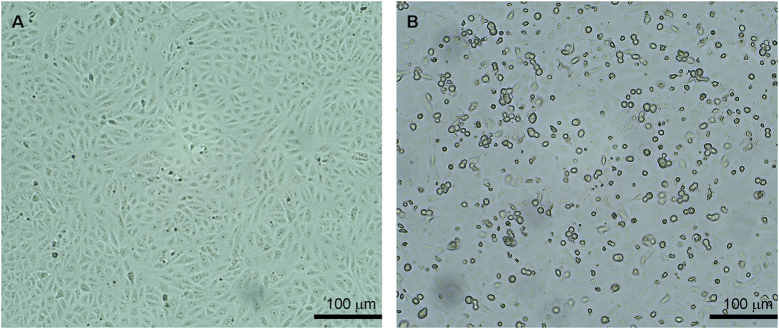
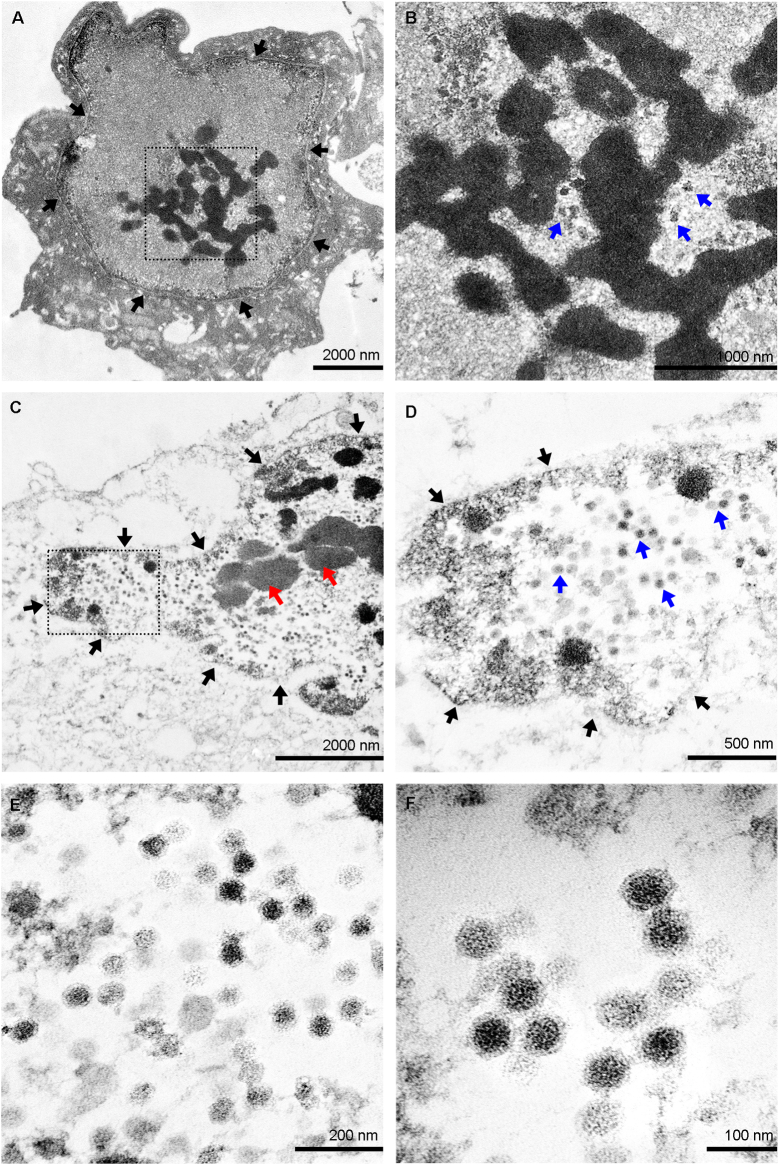
Adenovirus SAdV RBR-7-10 was isolated from a long-tailed macaque fecal sample collected in Ratchaburi (Fig. 1). Microscopically, the virus caused non-syncytial cytopathic effect (CPE) with round-shaped cells in Vero cell culture (Fig. 3). Viral protein identification by mass spectrometry suggested the presence of adenovirus strain A1163-like hexon (protein accession no. gi|379,977,075) in the virus culture sample. In transmission electron micrographs of SAdV RBR-7-10-infected Vero cells, the nuclear membrane was observed on day 1 post-infection, concurrent with early virion production (Fig. 4A and B). As viral replication progressed on day 2 post-infection, nuclear chromatin clumping was observed and increased numbers of viral particles accumulated inside the cell nucleus (Fig. 4C and D). With higher magnification (Fig. 4E and F), the viruses were observed to be non-enveloped icosahedral particles of approximately 70–90 nm in size. Additionally, hemagglutination assays revealed that the virus did not agglutinate human, goose, chicken, or sheep RBCs.
Fig. 3.
Cytopathic effect in SAdV RBR-7-10-infected Vero cells. (A) Uninfected Vero cell. (B) SAdV RBR-7-10-infected Vero cell at day 2 post-infection. Photos were taken under 100× magnification.
Fig. 4.
Transmission electron micrographs of SAdV RBR-7-10-infected Vero cells. Vero cells were infected with SAdV RBR-7-10 for two days and then subjected to transmission electron microscopy (TEM). (A) Electron micrograph of a virus-infected Vero cell at day 1 post-infection. Black arrows indicate the nuclear membrane. The area in the dashed box is enlarged and shown in (B), with blue arrows pointing to viral particles. (C) Electron micrograph of a virus-infected Vero cell at day 2 post-infection. Black arrows indicate the nuclear border. Red arrows point towards clumping of the nuclear chromatin. The area in the dashed box is enlarged and shown in (D), with black arrows indicating the nuclear border and blue arrows pointing to viral particles. (E–F) TEM images showing the non-enveloped icosahedral particles of SAdV RBR-7-10. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The SAdV RBR-7-10 complete hexon (2778 bp) and nearly complete DNA polymerase (3597 bp; lacking 9 bp of the initial sequence) genes were sequenced and submitted to GenBank. Nucleotide BLAST analysis revealed their close relation with the hexon (nucleotide identity: 98.4%) and DNA polymerase (nucleotide identity: 98.9%) genes, respectively, of SAdV-B strain A1163 (JN880449.1) [15]. Furthermore, BLAST analysis of the translated nucleotides against a protein database revealed amino acid identities of 100% and 99.2% for the hexon and DNA polymerase, respectively, compared with those of SAdV-B A1163. Thus, SAdV RBR-7-10 was identified as SAdV-B.
3.3. Assessment of SAdV RBR-7-10-neutralizing antibodies in monkeys and humans
Neutralization assays were performed to evaluate the presence of antibodies against SAdV RBR-7-10 in long-tailed macaque plasma (n = 103) and human serum (n = 125) samples. All samples were collected from the same area in Prachuap Khiri Khan, where monkey habitats are close to human territories. Using a cut-off NT50 titer of ≥20, 7 out of 103 (6.8%) monkey plasma samples but no human serum samples were assessed as positive for SAdV RBR-7-10-neutralizing antibodies (Fig. 5). Among these seven antibody-positive samples, four with an NT50 titer of 20 were PCR negative, one with an NT50 titer of 25.2 was PCR positive for SAdV-A, and one with an NT50 titer of 56.6 was PCR positive for SAdV-B. One “NT50 titer = 20” sample lacked a matching rectal swab.
Fig. 5.
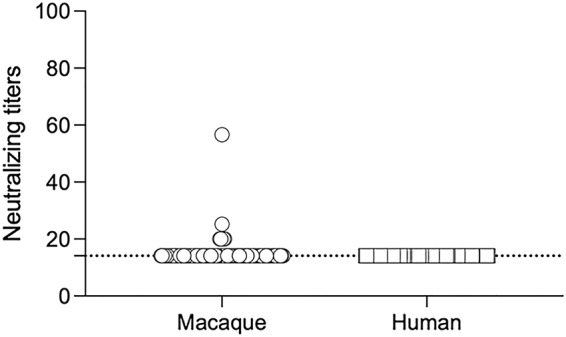
SAdV RBR-7-10 neutralizing antibody detection in long-tailed macaques and humans from the Prachuap Khiri Khan collection site. The initial serum dilution in this assay was 1:20, and seropositivity was defined as an NT50 titer of ≥20. Circles and squares represent monkey plasma and human serum samples, respectively.
4. Discussion
The One Health concept, concerned with human–animal–environment interactions, has been used as a basis for understanding emerging zoonotic human pathogens [22]. It is collaboratively applied in studies of emerging animal-borne viruses that cause outbreaks in humans, such as pandemic influenza, coronavirus, Nipah virus, and Ebola virus [23]. We applied the One Health concept here to study the adenovirus circulation in monkeys and assess the potential of adenovirus cross-species transmission to humans.
Thailand is home to many species of monkeys living in the wild or free-ranging near human territories. In the provinces where our samples were collected, monkeys are present in public areas, e.g., temples or historic sites, where they share the space with locals and travelers and thereby pose a risk of exposing humans to monkey pathogens. The majority of these monkeys are long-tailed or cynomolgus macaques (M. fascicularis), which are old-world monkeys endemic to Thailand and other Southeast Asian countries such as Myanmar, Malaysia, The Philippines, and Indonesia. Regarding adenovirus surveillance in monkeys, many reported molecular detections and phylogenetic characterizations of such viruses are from African countries, home to several great apes, old-world monkeys, and new-world monkeys [3,5,6,9]. Molecular detection of monkey adenovirus is generally conducted with animal stool samples, or occasionally with organs or blood, using PCR targeting the DNA polymerase and hexon genes. Detected prevalence greatly varies among species and settings. For example, in great apes (chimpanzees, gorillas, and bonobos), the adenovirus prevalence ranges from 36%–100% [3,5,6,9]. Unlike great apes, which are native to only Africa, different species of macaques (Macaca species) are distributed in both Africa and Asia. In addition to living in the wild, colonies of macaques are also present in wildlife sanctuaries, and some, mostly rhesus macaques, are kept for research purposes. An adenovirus survey in non-symptomatic macaques (Macaca mulatta, M. fascicularis, and Macaca sylvanus) from the US, China, and Africa revealed prevalences ranging from 13%–100% [3,9,14]. Limited adenovirus prevalence data from Asia is available, and there is no reported adenovirus surveillance of free-ranging long-tailed macaques in Thailand. The only adenovirus surveillance reported from Thailand was conducted in Macaca assamensis in a wildlife sanctuary; it detected a prevalence of 5.7% [26]. Here, molecular detection revealed an adenovirus prevalence of 33.3%. All but three of our adenovirus-positive samples were from free-ranging long-tailed macaques; the rest were from a captive wild stump-tailed macaque, a leaf monkey, and a white-handed gibbon, all species found in Thailand forests. The data suggest that adenoviruses naturally circulate in monkeys, with different prevalences among various species and locations.
The adenoviruses detected in long-tailed macaques here were tentatively grouped into SAdV-A, SAdV-B, SAdV-H, and HAdV-G species on the basis of their partial DNA polymerase sequences. Macaques are a source of SAdV-A and SAdV-B, which occasionally cause outbreaks of diarrheal and respiratory diseases in monkeys [9,[12], [13], [14]]. Here, SAdV-B was detected in all sampling locations, whereas SAdV-A was detected in three out of four locations; this difference could be due to the small sample size. SAdV-H-like sequences were detected in a few samples from Prachuap Khiri Khan. Members of SAdV-H, recently added to the group of Simian adenovirus species, include an adenovirus isolated from the urine of a simian immunodeficiency virus (SIV)-infected rhesus macaque [27]. Overall, our results suggest that SAdV-A and SAdV-B are endemic to macaques in Thailand. Notably, there was no reported outbreak of disease among monkeys at the time of sample collection, and the monkey colonies assessed were non-symptomatic.
HAdV-G was also detected in all sampling sites, in a higher proportion compared with other adenovirus species, and in the one stump-tailed macaque included in our study. In contrast with HAdV-B, HAdV-C, and HAdV-E, which are widely detected in great apes, HAdV-G is prevalent in old-world monkeys [7]. An adenovirus survey in Assamese macaques in Northeast Thailand found HAdV-G as well [26]. In China, this virus was associated with diarrhea in captive macaques [13]. HAdV-G members include mainly simian adenoviruses identified in macaques. The only well-described human adenovirus is HAdV-52, associated with gastroenteritis in human hosts [28]. HAdV-G likely originated in monkeys and spread to humans via cross-species transmission from old-world monkeys [27]. Although HAdV-E was previously detected in Barbary macaques and other monkeys and apes in Africa [3], it was not detected here. Several studies have suggested a low host species specificity and consequent cross-species transmission potential of HAdV-E from chimpanzees to African people [19]. Cross-species transmission may occur via close contact, contamination with body secretions, or during hunting. Our detection of a high prevalence of HAdV-G, rather than of HAdV-E, in macaques from different sites in Thailand demonstrates the importance of continuing surveillance of this virus species for its zoonosis potential in macaque-endemic areas.
HAdV-D-related adenoviruses were detected in a gibbon and a leaf monkey. HAdV-D may have originated in, and may be specific to, humans; it is isolated mainly from them [3,7,19]. The high levels of sequence identity between human and chimpanzee HAdV-D isolates in Africa may indicate cross-species transmission from humans to chimpanzees [7]. The two animals in our study from which HAdV-D was detected were captured from a forest, and there was some chance of animal contact with humans during their detainment. However, the sequence identities of these two HAdV-D-like viruses with published HAdV-D sequences were < 88%. Therefore, these viruses detected in gibbon and leaf monkey feces might be a HAdV-D variant transmitted from humans to the captive monkeys or they could belong to a new simian virus species in need of characterization.
This study also detected a bat adenovirus-like sequence, in a monkey fecal sample from Lopburi. Notably, we previously found a novel bat reovirus in monkey feces from the same site [24], thus confirming that, in areas where monkeys and bats cohabitate, there is a chance of virus cross-species transmission. Additional studies and evidence are needed to determine whether these results are from cross-species transmission or a cross-contamination of bat virus in the monkey specimen. Nevertheless, all currently available data support conducting zoonotic virus surveys in environments where animals and humans share habitats.
We isolated a simian adenovirus, SAdV RBR-7-10, in one long-tailed macaque stool sample. The results of a nucleotide sequence analysis of full hexon and nearly full DNA polymerase genes indicated that the virus is a SAdV-B species closely related to the SAdV-B strain A1163, which was molecularly characterized after its isolation from the stool of an asymptomatic rhesus macaque from a primate facility in the United States [15]. The identification of this virus from a long-tailed macaque in Thailand suggests the worldwide distribution of this SAdV-B in macaques. Protein identification by spectrometry and virus imaging by transmission electron microscopy confirmed the isolation of simian adenovirus, which propagates inside the cell nucleus and causes chromatin clumping. The clumping of chromatin seen in the infected cell could be a result of interaction between the early viral protein E1A and the host chromatin [29]. The virus was further used in a neutralization assay to identify specific antibodies in plasma/serum samples from monkeys and humans; antibodies against the monkey virus in humans can be an indication of cross-species infection. One limitation of our approach is that the SAdV-B isolate used in the neutralization assay was from the Ratchaburi site, whereas the monkey and human plasma/serum samples were available only from Prachuap Khiri Khan. However, because SAdV-B isolates were detected in monkeys from all sites, and their nucleotide sequences were very closely related, with a sequence identity of 99%–100%, SAdV-RBR-7-10 could appropriately be used as a representative. Herein, a low SAdV-B seroprevalence (6.8%) was detected in monkeys, and no specific antibodies against SAdV RBR-7-10 were detected in humans. The high prevalence of SAdV-B detected by PCR indicates that the macaques are a natural host of the virus without disease. This could be due to a low neutralizing immunity, which would allow the virus to be maintained in the monkey population.
Antibodies generated against a pathogen can be neutralizing or non-neutralizing antibodies. The relevance of this has been well demonstrated in influenza virus infection, where a binding but non-neutralizing antibody can contribute to in vivo protection [30]. The limitation of the neutralization assay used in this study is that it can detect only neutralizing antibodies that inhibit viral infection, represented by the inhibition of virus-induced CPE in cell culture, and cannot detect binding, non-neutralizing antibodies. To clarify this point, more samples and immunological studies are needed. The detection of neutralizing antibodies against simian adenoviruses in humans who live in areas where monkeys are endemic has been reported [31,32]. In the present study, we did not detect antibodies against the SAdV RBR-7-10 in humans, suggesting no cross-species transmission of this monkey virus to humans. Nevertheless, our finding should not impede zoonotic surveillance in monkeys and humans because previous reports and molecular surveys clearly demonstrate that various adenoviruses exist in monkeys and a number of them have zoonotic potential. In addition, the lack of antibodies against SAdV RBR-7-10 in humans suggests that this virus could have potential utility as a viral vector. Adenoviral vectors have displayed their usefulness for gene transfer in vaccine development against HIV, malaria, and other pathogens [31,33,34]. The rising problem for the previously used well-characterized human adenoviral vector is pre-existing immunity against the vector in humans, which compromises vaccine efficacy [31]. Simian adenoviruses, such as chimpanzee adenoviruses, have become a target for vector development owing to their expected low levels of cross-immunity in humans. As such, SAdV RBR-7-10, with its low pre-existing immunity in humans, could be a candidate viral vector, although further study regarding this possibility is needed.
There are a few limitations of this study. First, its small sample size may be inadequate for making definitive conclusions regarding the spillover of SAdV-B at the human–animal interface. Second, the study samples were collected from only four study sites, which may not represent the epidemiology of SAdV-B for the entire country. However, the low SAdV-RBR-7-10 seropositivity in monkeys suggests it is unlikely that antibodies against this virus would be detected in the human population, even in individuals from a community in close proximity to monkeys. Nevertheless, the continuous monitoring of viruses in monkeys living in proximity to human populations is crucial for implementing the One Health approach to confront emerging and reemerging zoonotic disease threats.
5. Conclusions
While data of adenovirus survey in monkeys in Southeast Asia is limited, we reported the circulation of adenoviruses in monkeys, mainly long-tailed macaques in the central and western part of Thailand. SAdV-A, SAdV-B and HAdV-G were found in a high prevalence whereas SAdV-H, HAdV-D and bat adenovirus could be detected only in a few monkeys. Antibodies against SAdV-B were surveyed in monkeys and humans in the Prachuap Khiri Khan Province and cross-species transmission was not observed in this study. However, since monkeys are considered a source of zoonotic pathogens, and the types of adenoviruses detected in this study have zoonotic potential, surveillance in monkeys and humans should be continued in more areas by collecting more samples from different monkey species; especially in areas where humans and monkeys live at the interface. The information concerning pathogens that may spillover from animals to human living in a close contact environment is important. It can be used to plan an emerging zoonotic disease control programs following the One Health concept.
Funding
This work was supported by the Research Career Development Grant, Thailand Research Fund (RSA6180052); and the National Science and Technology Development Agency (NSTDA), Thailand, through the eAsia joint research program (P-18-52763).
CRediT authorship contribution statement
Nathamon Kosoltanapiwat: Conceptualization, Validation, Formal analysis, Investigation, Resources, Writing – original draft, Writing – review & editing, Visualization, Supervision, Funding acquisition. Jarinee Tongshoob: Investigation, Writing – review & editing. Sumate Ampawong: Formal analysis, Investigation, Writing – review & editing. Onrapak Reamtong: Formal analysis, Investigation, Writing – review & editing. Luxsana Prasittichai: Investigation, Writing – review & editing. Marnoch Yindee: Investigation, Writing – review & editing. Daraka Tongthainan: Resources, Writing – review & editing. Phitsanu Tulayakul: Resources, Writing – review & editing. Kobporn Boonnak: Conceptualization, Validation, Formal analysis, Resources, Writing – review & editing, Visualization, Supervision, Funding acquisition.
Declaration of Competing Interest
None declared.
Acknowledgements
The authors would like to thank Mr. Banpot Maleehuan, Mr. Songchat Prakalpanont, and Mr. Paitoon Intarabut, the Department of National Parks, Wildlife and Plant Conservation, Ministry of Natural Resources and Environment, Thailand, for their facilitation in sample collection. We also thank Mr. Montee Wijitranon for his technical assistance in monkey sample collection. We thank Divya Lakhotia for her assistance in drafting the manuscript, and Katie Oakley, Ph.D., from Edanz (http://www.edanz.com/ac) for editing this manuscript. We thank the Faculty of Tropical Medicine, Mahidol University, for supporting the cost of English editing.
Contributor Information
Nathamon Kosoltanapiwat, Email: nathamon.kos@mahidol.ac.th.
Jarinee Tongshoob, Email: jarinee.pan@mahidol.ac.th.
Sumate Ampawong, Email: sumate.aum@mahidol.ac.th.
Onrapak Reamtong, Email: onrapak.rea@mahidol.ac.th.
Marnoch Yindee, Email: marnoch.yi@wu.ac.th.
Phitsanu Tulayakul, Email: fvetpnt@ku.ac.th.
Kobporn Boonnak, Email: kobporn.boo@mahidol.ac.th.
Data availability
Data will be made available on request.
References
- 1.Benkő M., Aoki K., Arnberg N., Davison A.J., Echavarria M., Hess M., Jones M.S., Kaján G.L., Kajon A.E., Mittal S.K., Podgorski I.I., San Martín C., Wadell G., Watanabe H., Harrach B., ICTV Report Consortiumet ICTV virus taxonomy profile: Adenoviridae 2022. J. Gen. Virol. 2022;103 doi: 10.1099/jgv.0.001721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lion T. Adenovirus infections in immunocompetent and immunocompromised patients. Clin. Microbiol. Rev. 2014;27:441–462. doi: 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medkour H., Amona I., Akiana J., Davoust B., Bitam I., Levasseur A., Tall M.L., Diatta G., Sokhna C., Hernandez-Aguilar R.A., Barciela A., Gorsane S., La Scola B., Raoult D., Fenollar F., Mediannikov O. Adenovirus infections in African humans and wild non-human primates: great diversity and cross-species transmission. Viruses. 2020;12:657. doi: 10.3390/v12060657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dicks M.D., Spencer A.J., Edwards N.J., Wadell G., Bojang K., Gilbert S.C., Hill A.V.S., Cottingham M.G. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dadáková E., Brožová K., Piel A.K., Stewart F.A., Modrý D., Celer V., Hrazdilová K. Adenovirus infection in savanna chimpanzees (Pan troglodytes schweinfurthii) in the Issa Valley, Tanzania. Arch. Virol. 2018;163:191–196. doi: 10.1007/s00705-017-3576-x. [DOI] [PubMed] [Google Scholar]
- 6.Medkour H., Castaneda S., Amona I., Fenollar F., André C., Belais R., Mungongo P., Muyembé-Tamfum J.J., Levasseur A., Raoult D., Davoust B., Mediannikov O. Potential zoonotic pathogens hosted by endangered bonobos. Sci. Rep. 2021;11:6331. doi: 10.1038/s41598-021-85849-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wevers D., Metzger S., Babweteera F., Bieberbach M., Boesch C., Cameron K., Couacy-Hymann E., Cranfield M., Gray M., Harris L.A., Head J., Jeffery K., Knauf S., Lankester F., Leendertz S.A.J., Lonsdorf E., Mugisha L., Nitsche A., Reed P., Robbins M., Travis D.A., Zommers Z., Leendertz F.H., Ehlers B. Novel adenoviruses in wild primates: a high level of genetic diversity and evidence of zoonotic transmissions. J. Virol. 2011;85:10774–10784. doi: 10.1128/JVI.00810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wevers D., Leendertz F.H., Scuda N., Boesch C., Robbins M.M., Head J., Ludwig C., Kühn J., Ehlers B. A novel adenovirus of Western lowland gorillas (Gorilla gorilla gorilla) Virol. J. 2010;7:303. doi: 10.1186/1743-422X-7-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy S., Vandenberghe L.H., Kryazhimskiy S., Grant R., Calcedo R., Yuan X., Keough M., Sandhu A., Wang Q., Medina-Jaszek C.A., Plotkin J.B., Wilson J.M. Isolation and characterization of adenoviruses persistently shed from the gastrointestinal tract of non-human primates. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Motes C.M., Hundesa A., Almeida F.C., Bofill-Mas S., Girones R. Isolation of a novel monkey adenovirus reveals a new phylogenetic clade in the evolutionary history of simian adenoviruses. Virol. J. 2011;8:125. doi: 10.1186/1743-422X-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lange C.E., Niama F.R., Cameron K., Olson S.H., Nina R.A., Ondzie A., Bounga G., Smith B.R., Pante J., Reed P., Tamufe U., Laudisoit A., Goldstein T., MPassi R.B., Joly D.O. First evidence of a new simian adenovirus clustering with Human mastadenovirus F viruses. Virol. J. 2019;16:147. doi: 10.1186/s12985-019-1248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu C.Y., Yagi S., Lu X., Yu G., Chen E.C., Liu M., Dick E.J., Jr., Carey K.D., Erdman D.D., Leland M.M., Patterson J.L. A novel adenovirus species associated with an acute respiratory outbreak in a baboon colony and evidence of coincident human infection. mBio. 2013;4 doi: 10.1128/mBio.00084-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bányai K., Esona M.D., Liu A., Wang Y., Tu X., Jiang B. Molecular detection of novel adenoviruses in fecal specimens of captive monkeys with diarrhea in China. Vet. Microbiol. 2010;142:416–419. doi: 10.1016/j.vetmic.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Lu J., Wang Q., Wang H., Li G., Gao G. Molecular characterization of adenoviruses in fecal samples of captively bred rhesus macaques in China. Vet. Microbiol. 2011;149:461–466. doi: 10.1016/j.vetmic.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Roy S., Sandhu A., Medina A., Clawson D.S., Wilson J.M. Adenoviruses in fecal samples from asymptomatic rhesus macaques, United States. Emerg. Infect. Dis. 2012;18:1081–1088. doi: 10.3201/eid1807.111665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argüello-Sánchez L.E., de Los Monteros A.E., Santiago-Alarcon D., García-Sepúlveda C.A. Detection and prevalence of adenoviruses from free-ranging black howler monkeys (Alouatta pigra) Virus Genes. 2018;54:818–822. doi: 10.1007/s11262-018-1600-1. [DOI] [PubMed] [Google Scholar]
- 17.Chen E.C., Yagi S., Kelly K.R., Mendoza S.P., Tarara R.P., Canfield D.R., Maninger N., Rosenthal A., Spinner A., Bales K.L., Schnurr D.P., Lerche N.W., Chiu C.Y. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borkenhagen L.K., Fieldhouse J.K., Seto D., Gray G.C. Are adenoviruses zoonotic? A systematic review of the evidence. Emerg. Microbes Infect. 2019;8:1679–1687. doi: 10.1080/22221751.2019.1690953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoppe E., Pauly M., Gillespie T.R., Akoua-Koffi C., Hohmann G., Fruth B., Karhemere S., Madinda N.F., Mugisha L., Muyembe J.J., Todd A., Petrzelkova K.J., Gray M., Robbins M., Bergl R.A., Wittig R.M., Zuberbühler K., Boesch C., Schubert G., Leendertz F.H., Ehlers B., Calvignac-Spencer S. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol. Biol. Evol. 2015;32:2072–2084. doi: 10.1093/molbev/msv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dehghan S., Seto J., Jones M.S., Dyer D.W., Chodosh J., Seto D. Simian adenovirus type 35 has a recombinant genome comprising human and simian adenovirus sequences, which predicts its potential emergence as a human respiratory pathogen. Virology. 2013;447:265–273. doi: 10.1016/j.virol.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehghan S., Seto J., Liu E.B., Walsh M.P., Dyer D.W., Chodosh J., Seto D. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology. 2013;443:197–207. doi: 10.1016/j.virol.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stärk K.D.C., Kuribreña M.A., Dauphin G., Vokaty S., Ward M.P., Wieland B., Lindberg A. One health surveillance - more than a buzz word? Prev. Vet. Med. 2015;120:124–130. doi: 10.1016/j.prevetmed.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Morse S.S., Mazet J.A., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin W.I., Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosoltanapiwat N., Reamtong O., Okabayashi T., Ampawong S., Rungruengkitkun A., Thiangtrongjit T., Thippornchai N., Leaungwutiwong P., Mahittikorn A., Mori H., Yoohanngoa T., Yamwong P. Mass spectrometry-based identification and whole-genome characterisation of the first pteropine orthoreovirus isolated from monkey faeces in Thailand. BMC Microbiol. 2018;18:135. doi: 10.1186/s12866-018-1302-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiriyarat W., Lerdsamran H., Pooruk P., Webster R.G., Louisirirotchanakul S., Ratanakorn P., Chaichoune K., Nateerom K., Puthavathana P. Erythrocyte binding preference of 16 subtypes of low pathogenic avian influenza and 2009 pandemic influenza A (H1N1) viruses. Vet. Microbiol. 2010;146:346–349. doi: 10.1016/j.vetmic.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 26.Sukmak M., Wajjwalku W., Ostner J., Schulke O. A first report of non-invasive adenovirus detection in wild Assamese macaques in Thailand. Primates. 2017;58:307–313. doi: 10.1007/s10329-016-0587-2. [DOI] [PubMed] [Google Scholar]
- 27.Malouli D., Howell G.L., Legasse A.W., Kahl C., Axthelm M.K., Hansen S.G., Früh K. Full genome sequence analysis of a novel adenovirus of rhesus macaque origin indicates a new simian adenovirus type and species. Virol. Rep. 2014;3-4:18–29. doi: 10.1016/j.virep.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones M.S., 2nd, Harrach B., Ganac R.D., Gozum M.M.A., Cruz W.P.D., Riedel B., Pan C., Delwart E.L., Schnurr D.P. New adenovirus species found in a patient presenting with gastroenteritis. J. Virol. 2007;81:5978–5984. doi: 10.1128/JVI.02650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch K.L., Gooding L.R., Garnett-Benson C., Ornelles D.A., Avgousti D.C. Epigenetics and the dynamics of chromatin during adenovirus infections. FEBS Lett. 2019;593:3551–3570. doi: 10.1002/1873-3468.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan G.S., Leon P.E., Albrecht R.A., Margine I., Hirsh A., Bahl J., Krammer F. Broadly-reactive neutralizing and non-neutralizing antibodies directed against the H7 influenza virus hemagglutinin reveal divergent mechanisms of protection. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ersching J., Hernandez M.I.M., Cezarotto F.S., Ferreira J.D.S., Martins A.B., Switzer W.M., Xiang Z., Ertl H.C.J., Zanetti C.R., Pinto A.R. Neutralizing antibodies to human and simian adenoviruses in humans and new-world monkeys. Virology. 2010;407:1–6. doi: 10.1016/j.virol.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 32.Xiang Z., Li Y., Cun A., Yang W., Ellenberg S., Switzer W.M., Kalish M.L., Ertl H.C.J. Chimpanzee adenovirus antibodies in humans, sub-Saharan Africa. Emerg. Infect. Dis. 2006;12:1596–1599. doi: 10.3201/eid1210.060078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchbinder S.P., Mehrotra D.V., Duerr A., Fitzgerald D.W., Mogg R., Li D., Gilbert P.B., Lama J.R., Marmor M., Del Rio C., McElrath M.J., Casimiro D.R., Gottesdiener K.M., Chodakewitz J.A., Corey L., Robertson M.N., Step Study Protocol Team Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the step study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shott J.P., McGrath S.M., Pau M.G., Custers J.H.V., Ophorst O., Demoitié M., Dubois M., Komisar J., Cobb M., Kester K.E., Dubois P., Cohen J., Goudsmit J., Heppner D.G., Stewart V.A. Adenovirus 5 and 35 vectors expressing Plasmodium falciparum circumsporozoite surface protein elicit potent antigen-specific cellular IFN-gamma and antibody responses in mice. Vaccine. 2008;26:2818–2823. doi: 10.1016/j.vaccine.2008.03.080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.