Abstract
Karimabad virus (KARV) is an arthropod-borne viral agent originally found in the Mediterranean region that can cause human infection via sandfly as the main vector. The KARV virion has been only detected from sandfly in western Asian countries and specific antibody has been detected from Rhombomys opimus and human in countries in Africa, Western and Central Asia. In this study, by next-generation sequencing (NGS) on a high variety of wild small animals in Xinjiang Autonomous Region in China, we obtained a complete sequence of KARV from Rhombomys opimus. An expanded epidemiological investigation was subsequently performed on 1713 small wild mammals that were widely collected from seven bioclimatic distinct sites in China by applying KARV specific RT-PCR and sequencing. Positive results were only obtained from 8 (2.29%) of the Rhombomys opimus captured in Xinjiang Autonomous Region, while not in 57 rodent species that were captured in other six provinces. Sequence analysis revealed the currently identified KARV was clustered with Gabek Forest virus, and they shared 79.1–93.9% identity with Iranian KARV that differed for L, M and S segments. Phylogenetic analysis based on eight partial L gene sequences demonstrated the separation of two lineages of the current KARV sequences. The first report of KARV in Rhombomys opimus in China expanded the currently known geographic scope, reservoirs types and the genetic heterogeneity of KARV. Our results show a new host, Rhombomys opimus, for KARV and highlight potential zoonotic transmission of KARV in humans.
Keywords: Sandfly-borne phlebovirus, Rhombomys opimus, Sandfly, Karimabad virus, China
1. Introduction
The genus Phlebovirus of the Phenuviridae Family (order Bunyavirales) contained a group of pathogens associated with human diseases with severe public health concern [1]. According to the most recent classification of the International Committee on Taxonomy of Viruses (ICTV), 66 viral species had been grouped into the genus Phlebovirus [2], which were widely distributed in tropical areas of the New World and semi-arid and temperate areas of the Old World, including southern, southeastern and central Europe, Africa, central and western Asia [2,3]. The predominant part of known phleboviruses is transmitted by sandfly, while a few others are transmitted by mosquitoes (e.g., Rift Valley fever virus, Gouleak virus) and ticks (e.g., Mukawa virus) [4]. According to their antigenic relationships, Old World sandfly-borne phleboviruses could be classified into three groups by serological evidence, Naples phlebovirus serocomplex, Salehabad phlebovirus serocomplex, and Sicilian phlebovirus serocomplex [1,5].
At present, fourteen viruses have been classified within the Sandfly fever Naples phlebovirus serocomplex, including Naples phlebovirus (SFNV), Massilia phlebovirus (MASV), Karimabad phlebovirus (KARV), Tehran phlebovirus (THEV), Toscana virus (TOSV), Gordil phlebovirus (GORV), Punique phlebovirus (PUNV), Granada (GRAV), Fermo phlebovirus (FERV), Saint-Floris phlebovirus (SAFV), Arrábida phlebovirus (ARRV), Saddaguia phlebovirus (SADV), Balkan phlebovirus (BALKV), and Zerdali phlebovirus (ZERV) [1,6]. Among these viruses, SFNV could induce a transient febrile illness known as the “three-day-fever”, “phlebotomus fever”, or “pappataci fever”, which is endemic in the Mediterranean region and in western and central Asia. TOSV, which was isolated in Italy in 1971 and widely distributed in European countries around the Mediterranean basin and Northern Africa, could cause meningitis and encephalitis in humans [1,6]. KARV was firstly isolated from an unidentified pool of Iranian sandflies in 1959 [7], and subsequently from Phlebotomus papatasi, the presumed vector, in Iran in 1975 [7]. Its potential threats to human health were featured by serologic evidence showing an overall prevalence of KARV antibodies among 66% of the human population, and 32% of Rhombomys opimus in an endemic area of sandfly fever in Iran in 1977 [8]. Subsequently, positive antibodies against KARV were detected in serum of rodents (Rattus raftus, 5.5%; Rattus norvegicus, 9.6%) collected in Pakistan in 1981, and in serum of rodents (Tatera indica, 2.5%; Meriones hurrianae, 1.3%) and sheep (0.6%) collected from The Nile Delta region of Egypt in 1983 [9]. A serological survey also demonstrated the antibody prevalence rate in humans in western and central Asia (1%–11% in Sudan, 2% in Egypt, and 1%–62% in Iran and Russia) [7,10].
In this study, we screened a great variety of wild small mammals in seven ecological regions in China, determining the presence of KARV in Rhombomys opimus from Xinjiang Autonomous Region for the first time.
2. Material and methods
2.1. Study site and sample collection
From September 2013 to June 2021, wild small mammals were captured with snap traps from seven provinces (Autonomous Region), i.e., Heilongjiang, Xinjiang, Inner Mongolia, Henan, Jiangsu, Guangdong, Yunnan during an ongoing program of identifying pathogens in wild small mammals in China. The spleen samples were collected and stored at −80 °C until further use. The species of wild small mammals were initially identified by morphology, and further confirmed with PCR and sequencing [11].
2.2. Extraction of viral RNA and next-generation sequencing (NGS)
Twelve randomly selected spleen samples from four dominant wild small animal species (Mus musculus, Rhombomys opimus, Meriones libycus, and Spermophilus erythrogenys) in Xinjiang Autonomous Region were subjected to metagenomic analysis by NGS as previously described [12]. Briefly, total viral nucleic acids were extracted by using AllPrep DNA/RNA Mini Kit (Qiagen, Germany), from which rRNA was depleted using MGIEasy rRNA Depletion Kit (BGI, China) according to the manufacturer's instructions. A high-throughput sequencing library was constructed using an MGIEasy RNA Library Prep Kit (BGI) and then sequenced on the MGI2000 platform (BGI) to generate 150 bp paired-end reads. After filtering, trimming, and error removal for the original data, high quality reads were obtained and mapped to reference KARV genome sequences (KF297912-KF297914) using BWA (Version: 0.7.15). De novo assembly was performed using MEGAHIT (v1.1.2) software. The assembled contigs were blasted against the reference KARV genome sequences (KF297906-KF297914) to identify virus-related contigs. KARV specific primers were designed based on successfully assembled viral genomic sequences for PCR confirmation.
2.3. Reverse transcription-PCR (RT-PCR) and sequencing for KARV
We performed KARV screening by PCR amplification of a 475-bp fragment of the large (L) gene by using specific primers (KA1F 5′-ATAGAATCAAGAGGRGCAATG-3′ and KA1R 5′-CRTGGTATGCAGARAAYAAYT-3′). PrimeScript™ One Step RT-PCR Kit was used according to the manufacturer's instructions, briefly, the PCR mix was in a volume of 25 μl containing 12.5 μl of one Step Buffer (2×), 1 μl of PrimeScript one Step Enzyme Mix, 1 μl of PCR primer mix (10 μM of sense and antisense each), total RNA 2 μl and RNase free dH2O (8.5 μl). RT-PCR was carried with one cycle of 50 °C for 30 min and 94 °C for 2 min, followed by 40 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s in PCR System 9700 (Applied Biosystems, USA). The amplified products were determined by agarose gel electrophoresis, and the bands with expected size were subjected to Sanger sequencing. All steps of the nucleic acid extraction and RT-PCR test were conducted in parallel with positive and negative controls.
2.4. Phylogenetic analysis
The nucleotide and amino acid sequences from the currently determined KARV and representative species belonging to family Phenuiviridae that were downloaded from GenBank were aligned by ClustalW method using MEGA-X. Phylogenetic trees were constructed by using Gouleako goukovirus in the genus Goukovirus of the family Phenuiviridae as an outgroup. Phylogenetic trees were constructed using the maximum likelihood (ML) method and the robustness of each node was tested by 1000 bootstrap replications.
2.5. Nucleotide sequence accession numbers
The KARV sequences generated in this study were submitted to GenBank under the accession numbers ON456601-ON456603 (full-length genome sequence), OP150887-OP150891 and OP150893-OP150894 (partial L gene).
3. Results
3.1. Identification of KARV in Rhombomys opimus
Of the 12 samples subject to NGS, KARV specific sequences were determined from Rhombomys opimus captured in Xinjiang Autonomous Region in 2019. Totally 1213 reads were obtained, and 78% of reads were annotated to KARV. Based on 945 reads, complete genome sequences of KARV (strain DLT97) were obtained. To confirm the genome sequence of KARV, we designed 27 primer pairs for Sanger sequencing, which had produced consistent sequences with those from NGS.
3.2. Genome characterization of KARV
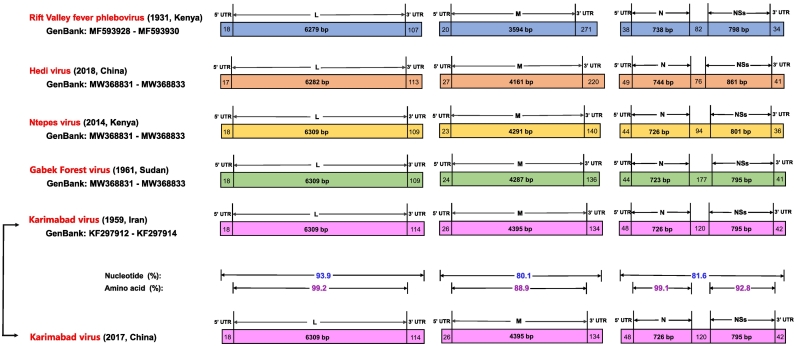
The genome of the obtained KARV comprised a similar negative-sense single-stranded tripartite structure, similar with other closely related members in the family Phenuiviridae, such as Rift Valley fever phlebovirus that was firstly identified in Kenya in 1931, Hedi virus that was determined in China in 2018, Ntepes virus (NPV) that was determined in Kenya in 2014 and Gabek Forest virus (GFV) that was determined in Sudan in 1961 (Fig. 1). The genome comprised of the L segment (6441 nt) encoding RNA polymerase; the medium (M) segment (4555 nt) encoding membrane glycoprotein; the small (S) segment (1729 nt) encoding nucleoprotein (N) and a non-structural protein (NSs) (Fig. 1). The pairwise sequence comparisons of the L, M, and S nucleotide sequences showed 93.9%, 80.1%, and 81.6% identity between the current KARV strain (GenBank accession numbers ON456601-ON456603) and that of Iranian KARV strain I-58 in 1958 (accession numbers KF297912-KF297914) (Fig. 1, Table S1). Amino acid sequences of L, M, N and NSs demonstrated 99.2%, 88.9%, 99.1% and 92.8% similarity with Iranian KARV strain I-58 (Fig. 1, Fig. S1–4, Table S2).
Fig. 1.
Genome organization of Karimabad virus, Ntepes virus, Gabek Forest virus, Hedi virus and Rift Valley fever phlebovirus. Nucleotide identities sequences between Iranian KARV and the current KARV are marked in blue. Amino acid identities sequences between Iranian KARV and the current KARV are marked in purple. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. KARV screening in wild small mammals
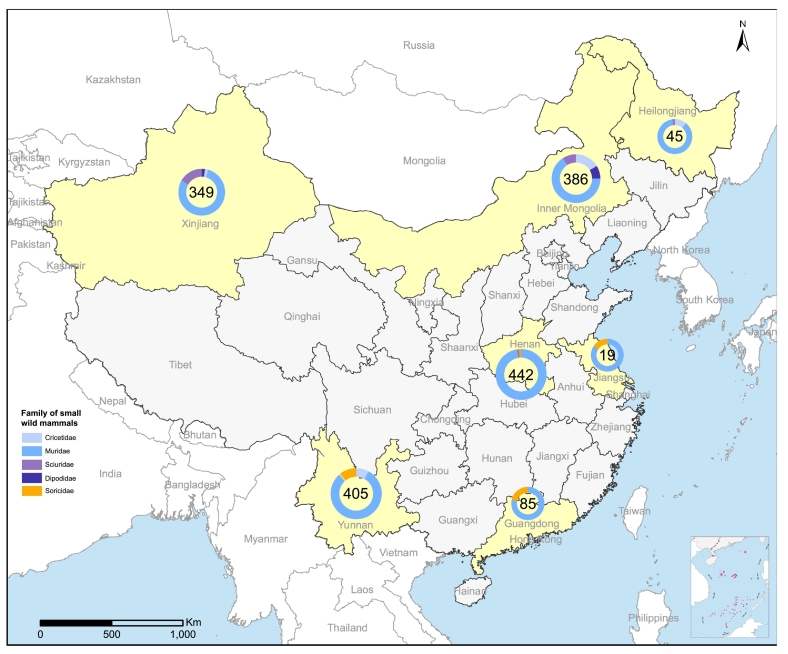
A total of 1713 wild small mammal collected and examined for the presence of KARV by reverse transcription PCR during the study course. The tested samples represented 58 species in 5 families and 2 orders, including 50 species of Cricetidae, Dipodidae, Muridae and Sciuridae family of Rodentia, 8 species of Soricidae family of Soricomorpha (Fig. 2 and Table 1). The highest number of samples were collected from Henan (442/1713, 25.53%), followed by Yunnan (405/1713, 23.40%), Inner Mongolia (386/1713, 22.30%), Xinjiang (349/1713, 20.16%), Guangdong (85/1713, 4.91%), Heilongjiang (45/1713, 2.60%), and Jiangsu (19/1713, 1.10%) (Fig. 2). Positive results of for a 475-bp fragment in the L gene of KARV were only obtained in Rhombomys opimus (8/77, 10.39%) captured in Xinjiang Autonomous Region (Table 1). No positive result for KARV was obtained from any of the other wild small mammal samples (Table 1).
Fig. 2.
The locations for capturing wild small mammals in the current study.
Table 1.
Small wild mammals screened for Karimabad virus in China.
| Location | Year | No. Species (n) | No. total tested | No. (%) of positive |
|---|---|---|---|---|
| Heilongjiang | 2017 | Microtus fortis (5), Rattus norvegicus (22), Apodemus agrarius (16), Tamias sibiricus (1), Mus musculus (1) | 45 | 0 (0) |
| Inner Mongolia | 2019 | Spermophilus dauricus (35), Allocricetulus eversmanni (4), Cricetulus barabensis (2), Cricetulus migratorius (38), Phodopus sungorus (6), Dipus sagitta (5), Allactaga sibirica (28), Mus musculus (2), Phodopus roborovskii (4), Meriones unguiculatus (183), Meriones meridianus (64), Lasiopodomys mandnarinus (15) | 386 | 0 (0) |
| Xinjiang | 2018 | Meriones tamariscinus (4), Spermophilus erythrogenys (31), Eothenomys miletus (2), Rhombomys opimus (77), Sundamys muelleri (4), Rattus norvegicus (20), Meriones libycus (48), Cricetulus migratorius (5), Microtus kikuchii (1), Allactaga sibirica (8), Marmota himalayana (2), Mus musculus (104), Apodemus sylvaticus (15), Spermophilus parryii (25), Meriones meridianus (3) | 349 | 8 (2.29) |
| Henan | 2018, 2021 |
Apodemus agrarius (54), Apodemus draco (2), Berylmys bowersi (2), Mus musculus (59), Niviventer andersoni (2), Niviventer confucianus (3), Niviventer niviventer (18), Rattus norvegicus (23), Rattus tanezumi (268), Callosciurus erythraeus (5), Crocidura tanakae (6) | 442 | 0 (0) |
| Jiangsu | 2021 | Apodemus agrarius (4), Mus musculus (3), Rattus norvegicus (2), Rattus pyctoris (1), Rattus tanezumi (6), Crocidura lasiura (3) | 19 | 0 (0) |
| Guangdong | 2017, 2021 | Bandicota indica (14), Rattus andamanensis (11), Rattus norvegicus (37), Rattus tanezumi (5), Suncus murinus (17), Crocidura tanakae (1) | 85 | 0 (0) |
| Yunnan | 2013, 2015, 2016 |
Eothenomys cachinus (1), Eothenomys eleusis (8), Eothenomys miletus (16), Eothenomys proditor (8), Apodemus chevrieri (56), Apodemus ilex (80), Berylmys bowersi (1), Chiropodomys gliroides (1), Melomys burtoni (2), Micromys minutus (1), Mus pahari (6), Niviventer andersoni (4), Niviventer coxingi (7), Rattus brunneusculus (7), Rattus steini (7), Rattus tanezumi (144), Rattus yunnanensis (9), Tamiops swinhoei (3), Anourosorex squamipes (15), Blarinella quadraticauda (4), Crocidura horsfieldii (1), Crocidura lasiura (4), Crocidura tanakae (5), Episoriculus caudatus (3), Sorex bedfordiae (8), Suncus murinus (4) |
405 | 0 (0) |
| Total | 1731 | 8 (0.46) |
3.4. Phylogenetic analysis of KARV sequences
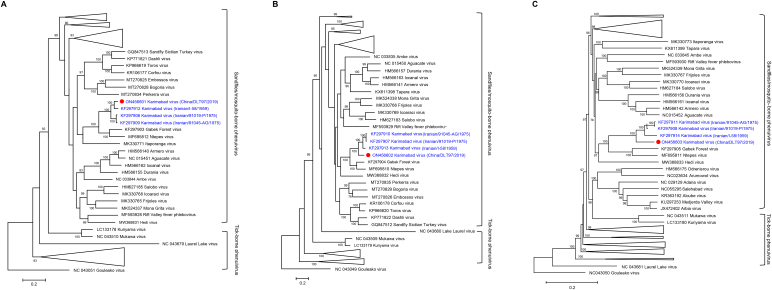
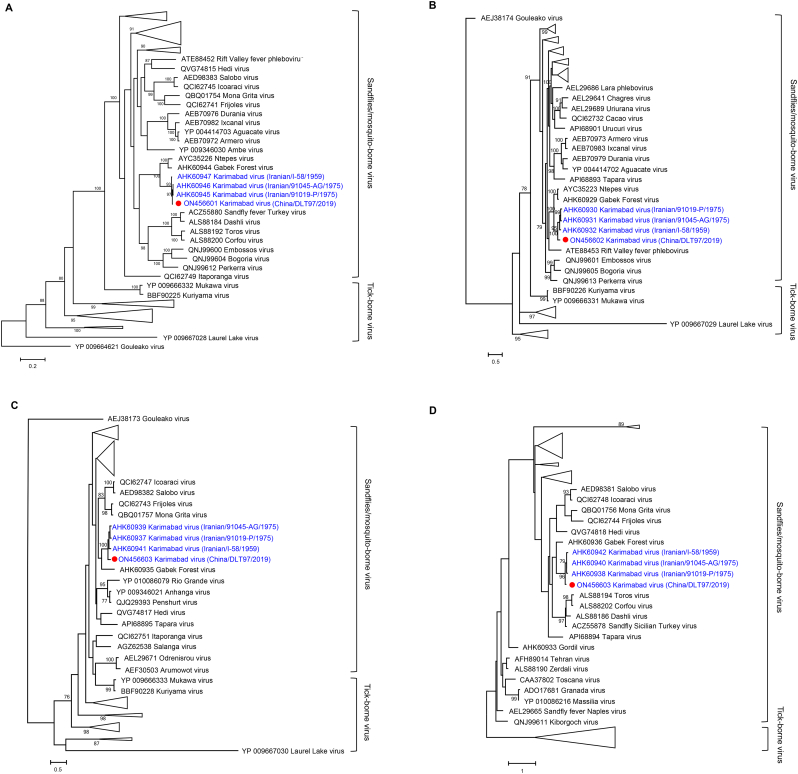
Phylogenetic analyses of L, M, S fragments from all available phlebovirus sequences (Table S3) indicated the current KARV as a novel variant that formed a strongly supported clade with Iranian KARV, more closely related to GFV which were all phleboviruses of sandflies or mosquito sources (Fig. 3). Identical phylogenetic clustering was determined based on the amino acid sequences of the L, M, N, and NSs proteins (Fig. 4). These findings, taken together, suggested KARV as a member of the Karimabad species complex, resembling GFV, its sister taxon.
Fig. 3.
Phylogenetic analysis of KARV RNA segments. (A) The Maximum Likelihood phylogenetic tree constructed based on the full-length nucleotide sequences of the L segment. (B) The Maximum Likelihood phylogenetic tree constructed based on the full-length nucleotide sequences of the M segment. (C) The Maximum Likelihood phylogenetic tree constructed based on the full-length nucleotide sequences of the S segment. The current KARV was labeled with a red dot. Trees were generated using MEGA-X and analyzed included 1000 bootstrap replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Phylogenetic analysis of KARV for deduced amino acid sequences. (A) The Maximum Likelihood phylogenetic tree based on the deduced amino acid sequences of the L segment. (B) The Maximum Likelihood phylogenetic tree based on the deduced amino acid sequences of the M segment. (C) The Maximum Likelihood phylogenetic tree based on the nucleoprotein protein (N). (D) The Maximum Likelihood phylogenetic tree based on the nonstructural protein (NSs). The current KARV was labeled with a red dot. Trees were generated using MEGA-X and analyzed included 1000 bootstrap replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
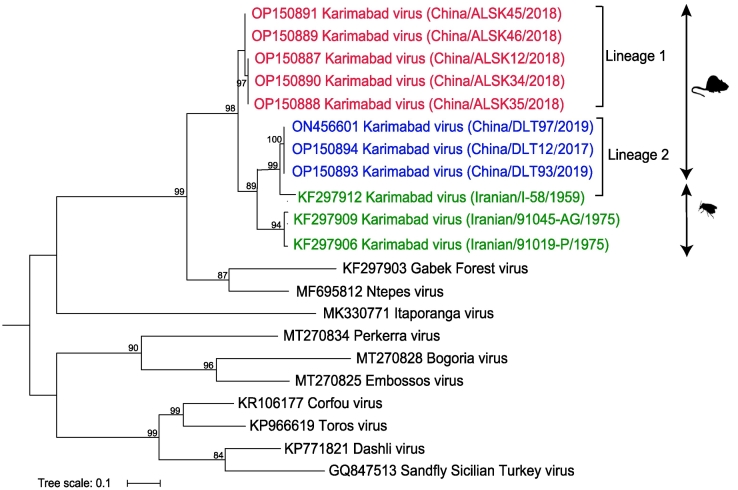
For each of the 8 KARV-positive animals, partial sequences of L gene (475-bp) were obtained for the phylogenetic analyses. The current KARVs were clustered in two major lineages, with mean (± standard deviation) of the nucleotide sequence identity between two lineages as 94.20 ± 5.1%, lower than the identity within each lineage (99.49 ± 0.4% for Lineage 1 and 100% for Lineage 2) (Fig. 5). The clustering of lineage was largely related to their geographic regions. The strains from Alashankou, were clustered into Lineage 1, and the strains from the Dulata Port, were clustered into Lineage 2. In addition, the current three stains of Lineage 2 clustered with the Iranian KARV recorded in 1959 and was distant from other clusters.
Fig. 5.
Phylogenetic analysis of 475-bp KARV positive RNA sequences. The Maximum Likelihood phylogenetic tree constructed based on the 475-bp KARV positive RNA sequences (n = 8) of the L segment. The Iranian KARV sequences were labeled in green. Our KARV sequences were labeled in red (Lineage 1) and blue (Lineage 2). Trees were generated using MEGA-X and analyzed included 1000 bootstrap replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Sandflies are widely distributed around the Mediterranean Basin and transmit a variety of pathogens to humans but are relatively understudied in comparison to mosquitoes and ticks, due to the more difficulty in trapping activity. KARV is maintained in a transmission cycle that involves sandfly and rodents (mainly gerbils) in nature and only occasionally infects humans [8]. Therefore, the investigation in the rodent host might help to understand the epidemiological characteristics of KARV. Here by performing a NGS and subsequent field investigation on the rodent in a wide region in China, we confirmed the presence of KARV in East Asia for the first time, which expanded the known scope of KARV infection that were traditionally limited to the northern Mediterranean basin.
Despite of a wide range screen in 58 rodent species in seven ecological different regions in China, KARV was only determined in one rodent species, Rhombomys opimus captured in Xinjiang. As an animal host of sandfly, Rhombomys opimus may act as a possible mechanical vector for KARV and contribute to human exposure to this pathogen, consistent with Medhat et al. [9]. Rhombomys opimus is known to have a wide distribution throughout Central Asia and some parts of the Middle East, ranging from Iran through Pakistan, Afghanistan via Kazakhstan to China and Mongolia [13,14]. Rhombomys opimus has long been known as a principal natural reservoir of several zoonotic diseases (e.g., plague, leishmaniasis, leptospirosis) [13,15,16]. In China, Rhombomys opimus are mainly distributed in Xinjiang, Inner Mongolia, and Gansu. Sandfly, the important hosts and vectors of KARV, are widely distributed from Inner Mongolia and Jilin in the north to Hainan in the south, from coastal provinces in the east to Xinjiang in the west [17]. Except for Xinjiang where KARV had been currently determined to be prevalent, other regions with Rhombomys opimus warrant investigation to determine their endemic with KARV. Serological investigation of domestic animals, wild animals and humans in Xinjiang and other potential endemic regions should be performed to acquire an enhanced understanding of the transmission cycle and public health significance of KARV infection. In addition, the presence of flea assemblages and sandflies in Rhombomys opimus burrow systems increases their ability to sustain the pathogen and the probability of infectious emergence of vector-borne diseases [13].
KARV obtained in the present study differed from the reported KARV (accession numbers KF297912 - KF297914) from Iran over the genome of L (93.9%), M (80.1%), S (81.6%) segments, indicating a high heterogeneity of KARV genomes. Phylogenetic analyses of the L, M and S gene segment sequences of the current KARV confirmed its belonging to the same group cluster as the four previously reported Iranian KARVs [6,10]. As expected, KARV, which forms its own clade along with the previously tentative members GFV, show only distant evolutionary relationships with the other members of the Sandfly fever Naples phlebovirus serocomplex [6]. KARV and GFV appear to constitute a novel species complex within the genus [6].
In conclusion, KARV, the sandflly-borne phlebovirus, was identified for the first time in China, also representing its first determination in eastern Asia. Although with a relatively low prevalence of infection in Rhombomys opimus, our results called for further studies, including active surveillance and a phylogenetic approach to gain better knowledge on the epidemiological characteristics of KARV and guide the diagnosis and management of future human infections.
Funding
This work was supported by grants from the National Science Fund for Distinguished Young Scholars (No. 81825019) and the National Natural Science Foundation of China (81621005).
Ethical approval
The study protocol was approved by the institutional review boards and the ethics committees of the Institute of the Academy of Military Medical Sciences. All animals were treated in accordance with the guidelines in the Regulations for the Administration of Laboratory Animals.
Availability of data
Data are available upon reasonable request.
CRediT authorship contribution statement
Yue Li: Methodology, Software, Validation, Formal analysis, Writing – original draft. Yu-Na Wang: Methodology, Formal analysis, Writing – original draft. Feng Tian: Methodology, Data curation, Investigation. Xiao-Long Zhang: Methodology, Data curation, Investigation. Jing-Tao Zhang: Visualization, Methodology, Software. Shuang Li: Methodology, Software. Hao Li: Methodology, Data curation, Investigation. Xiao-Ai Zhang: Conceptualization, Methodology, Data curation, Formal analysis, Software, Writing – original draft, Writing – review & editing. Wei Liu: Conceptualization, Methodology, Data curation, Formal analysis, Writing – original draft, Writing – review & editing, Funding acquisition, Supervision.
Declaration of Competing Interest
All authors have nothing to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2022.100437.
Contributor Information
Xiao-Ai Zhang, Email: babylovehopi@163.com.
Wei Liu, Email: liuwei@bmi.ac.cn.
Appendix A. Supplementary data
Supplementary material
Data availability
I have shared the link to my data at the Attach File step.
References
- 1.Bino S., Velo E., Kadriaj P., Kota M., Moureau G., Lamballerie X., et al. Detection of a Novel Phlebovirus (Drin Virus) from Sand Flies in Albania. Viruses. 2019;11(5) doi: 10.3390/v11050469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z., Fan N., Hou X., Wang J., Fu S., Song J., et al. Isolation and identification of a novel phlebovirus, hedi virus, from sandflies collected in China. Viruses. 2021;13(5) doi: 10.3390/v13050772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Özbel Y., Oğuz G., Arserim S.K., Erişöz Kasap Ö., Karaoglu B., Yilmaz A., et al. The initial detection of Toscana virus in phlebotomine sandflies from Turkey. Med Vet Entomol. 2020;34(4):402–420. doi: 10.1111/mve.12450. [DOI] [PubMed] [Google Scholar]
- 4.Calisher C.H., Calzolari M. Taxonomy of phleboviruses, emphasizing those that are sandfly-borne. Viruses. 2021;13(5) doi: 10.3390/v13050918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alwassouf S., Christodoulou V., Bichaud L., Ntais P., Mazeris A., Antoniou M., et al. Seroprevalence of Sandfly-Borne Phleboviruses Belonging to Three Serocomplexes (Sandfly fever Naples, Sandfly fever Sicilian and Salehabad) in Dogs from Greece and Cyprus Using Neutralization Test. PLoS Negl Trop Dis. 2016;10(10) doi: 10.1371/journal.pntd.0005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palacios G., Tesh R.B., Savji N., da Rosa APA Travassos, Guzman H., Bussetti A.V., et al. Characterization of the Sandfly fever Naples species complex and description of a new Karimabad species complex (genus Phlebovirus, family Bunyaviridae) J Gen Virol. 2013;95(Pt 2):292–300. doi: 10.1099/vir.0.056614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tesh R.B., Saidi S., Gajdamovic S.J., Rodhain F., Vesenjak-Hirjan J. Serological studies on the epidemiology of sandfly fever in the Old World. Bull World Health Organ. 1976;54(6):663–674. [PMC free article] [PubMed] [Google Scholar]
- 8.Saidi S., Tesh R., Javadian E., Sahabi Z., Nadim A. Studies on the epidemiology of sandfly fever in Iran. II. The prevalence of human and animal infection with five phlebotomus fever virus serotypes in Isfahan province. Am J Trop Med Hyg. 1977;26(2):288–293. doi: 10.4269/ajtmh.1977.26.288. [DOI] [PubMed] [Google Scholar]
- 9.Darwish M.A., Hoogstraal H., Roberts T.J., Ghazi R., Amer T. A sero-epidemiological survey for Bunyaviridae and certain other arboviruses in Pakistan. Trans R Soc Trop Med Hyg. 1983;77(4):446–450. doi: 10.1016/0035-9203(83)90108-6. [DOI] [PubMed] [Google Scholar]
- 10.Tchouassi D.P., Marklewitz M., Chepkorir E., Zirkel F., Agha S.B., Tigoi C.C., et al. Sand Fly-associated Phlebovirus with evidence of neutralizing antibodies in humans, Kenya. Emerg Infect Dis. 2019;25(4):681–690. doi: 10.3201/eid2504.180750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolas V., Querouil S., Verheyen E., Verheyen W., Mboumba J.F., Dillen M., et al. Mitochondrial phylogeny of African wood mice, genus Hylomyscus (Rodentia, Muridae): implications for their taxonomy and biogeography. Mol Phylogenet Evol. 2006;38(3):779–793. doi: 10.1016/j.ympev.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Chen J.J., Zhang X.A., Fan H., Jiang F.C., Jin M.Z., Dai K., et al. Distribution and characteristics of Beilong virus among wild rodents and shrews in China. Infect Genet Evol. 2020;85:104454. doi: 10.1016/j.meegid.2020.104454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamidi K., Mohammadi S., Ghassemi-Khademi T. Ecological niche modeling of genetic lineages of the great gerbil, Rhombomys opimus (Rodentia: Gerbillinae) PLoS One. 2021;16(9) doi: 10.1371/journal.pone.0257063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamranrashani B., Kia E., Mobedi I., Mohebali M., Zarei Z., Mowlavi G., et al. Helminth Parasites of Rhombomys opimus from Golestan Province, Northeast Iran. Iran J Parasitol. 2013;8(1):78–84. [PMC free article] [PubMed] [Google Scholar]
- 15.Reithinger R., Dujardin J.C., Louzir H., Pirmez C., Alexander B., Brooker S. Cutaneous leishmaniasis. The lancet. Infect Dis. 2007;7(9):581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 16.Wilschut L.I., Addink E.A., Heesterbeek J.A., Dubyanskiy V.M., Davis S.A., Laudisoit A., et al. Mapping the distribution of the main host for plague in a complex landscape in Kazakhstan: An object-based approach using SPOT-5 XS, Landsat 7 ETM+, SRTM and multiple Random Forests. Int J Appl Earth Observ Geoinform. 2014;23(100):81–94. doi: 10.1016/j.jag.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guan L.R., Zhou Z.B., Jin C.F., Fu Q., Chai J.J. Phlebotomine sand flies (Diptera: Psychodidae) transmitting visceral leishmaniasis and their geographical distribution in China: a review. Infectious Diseases of Poverty. 2016;5(15) doi: 10.1186/s40249-016-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data are available upon reasonable request.
I have shared the link to my data at the Attach File step.