Abstract
Listeria monocytogenes is an important foodborne intracellular pathogen. The pathogen is the primary cause of human Listeriosis. The main source of human Listeriosis is through consumption of contaminated food products. Other modes of transmission include zoonotic and vertical transmission. The disease often presents in a mild form, but severe and fatal cases, as well as outbreaks, may occur. Despite these challenges, there has been little attempt at enumerating the burden of the disease in countries of Southeast Asia (SEA) and some developing countries. Thus, this study investigated the prevalence of L. monocytogenes in SEA using one health approach through a systematic review and meta-analysis (SR&MA) of the existing literature. In accordance with the PRISMA guidelines, an a priori protocol for the SR&MA was developed and registered in PROSPERO (ID=CRD42021288903). A systematic search of four electronic databases was performed for relevant citations. The identified publications were screened, and 17 studies were included in the review from where data was extracted. The pooling of the prevalence estimate (with the 95% confidence interval [CI]) was done using the random effect model by employing the double transformed arcsine method using MetaXL software. The overall determined prevalence for L. monocytogenes in SEA (in food, animal, and environmental sources) was 16% (95% confidence interval [CI]: 10–23). Further subgroup analysis revealed ready-to-eat food of vegetable origin with the highest prevalence of 21% (CI: 6–41). Also, seven virulence genes were identified to be prevalent in the subregion. The commonest identification method was found to be the polymerase chain reaction (PCR). The knowledge of the high prevalence of L. monocytogenes in SEA is relevant for informed decision making by clinicians, public health practitioners, and policymakers. To achieve the goal of the effective control and prevention of the disease in the subregion.
Keywords: Listeria monocytogenes, Listeriosis, Foodborne pathogen, One health concept, Systematic review, Meta-analysis
Highlights
-
•
Using one-health approach, the prevalence of Listeria monocytogenes in the Southeast Asia was systematically evaluated.
-
•
Estimate prevalence for food, environment and animal sources were pooled using the random effect model.
-
•
L. monocytogenes was reported to be hyper-endemic in SEA with food being the predominant source.
-
•
Prevalent L. monocytogenes virulent genes, high risk food sources, and commonly used detection methods were identified.
1. Introduction
Listeria monocytogenes is an invasive foodborne pathogen of humans and animals that causes the disease known as Listeriosis [1]. The organism is a ubiquitous Gram-positive, non-spore-forming, non-capsulated (unencapsulated), facultative anaerobic, rod-shaped bacterium [2]. There are 13 identified serotypes of L. monocytogenes, but only 1/2a, 1/2b, 1/2c, and 4b are of human importance [3]. In contrast to most other foodborne pathogens, Listeria monocytogenes can grow in food with reasonably low moisture content and high salt concentration [3]. Most importantly, L. monocytogenes thrives in refrigeration temperature in contrast to many other foodborne pathogens [3]. This capacity to persist and multiply in the food environment makes it especially difficult to control.
Listeriosis often presents as a mild disease but can cause severe infection in ‘at-risk groups’ such as young and old individuals, pregnant women, and the immunocompromised [3]. The fatality rate of Listeriosis, particularly in pregnant women, can be as high as 30% [4], exceeding that of Salmonella and Clostridium species. The most common transmission mode is through the consumption of food and/or feed contaminated by the organism [4]. Also, vertical transmission from mother to fetus is another possibility [4]. Other likely but less frequent modes of transmission include from animal-to-human (zoonotic), by contact or nosocomial transmission as seen in cutaneous lesions among veterinarians [5].
Listeriosis is termed as either silage disease or circling disease manifesting commonly as meningoencephalitis in animals. Infections in animals are usually sub-clinical, but severe forms can also occur, leading to abortion in pregnant animals, food poisoning, and death [6]. Nevertheless, it can cause fatal disease in some animals, birds, fishes, and crustaceans, causing septicaemia and encephalitis predominantly [6].
Throughout the world, Listeriosis occurs in sporadic or epidemic forms [6,7]. Many studies have been reported on the prevalence of L. monocytogenes and Listeriosis in different parts of the world [8] and in humans and from food products of animal origin [[9], [10], [11]]. However, the true incidence of foodborne infection is unknown, and there has been little attempt in ascertaining the magnitude of the problem. This is due to the paucity of studies on the epidemiology and surveillance of the disease in some developing countries, including Southeast Asia (SEA).
Additionally, the complex nature of the relationship between animals and man is on the rise and factors responsible include; chemical, physical, biological and social. Furthermore, the newly emerging zoonotic infections have made bold headlines and heightened awareness of the role of wild and domestic animals in spreading diseases to man. Globally, the rapid explosion in the human population and unprecedented increase in numbers and density of animals raised for food production play an important role in spreading zoonotic diseases. Also, the transportation, rearing, marketing, and processing of these animals significantly affect the occupational health of the human beings working and rearing the animals. Thus, raising the risk of zoonotic transmission to the workers due to increased contact with the animals and their products (e.g., dead carcases, blood, and other discharges). The implication is also applicable to the environment where the animals are kept and the entire ecosystem. Thus, emphasising the pivotal role of the ‘one health’ concept in combating the threat of emerging zoonotic infections. Therefore, the importance of Listeria monocytogenes cannot be underemphasised as this may lead to substantial public health consequences in addition to economic losses in the livestock industry. Hence, this SR&MA was conducted to comprehensively investigate the prevalence of L. monocytogenes in SEA.
2. Methods
2.1. Study design
This systematic review and meta-analysis (SR&MA) study was designed according to the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) guidelines (S1 File). In addition, the study was preceded by a priori protocol (S2 File) following the PRISMA protocol (PRISMA-P) guidelines (S3 File). The protocol was then registered on the National Institute for Health Research International Prospective Register of Systematic Reviews (PROSPERO (ID=CRD42021288903) available at: https://www.crd.york.ac.uk/prospero/displayrecord.php?ID=CRD42021288903.
2.2. Eligibility criteria
The following were defined as the eligibility criteria for the studies to be included in this SR&MA:
Inclusion criteria: Studies that meet the requirements outlined below were included in this SR&MA:
-
•
Study type: All observational studies (cross-sectional, cohort, case-control, prevalence surveys) conducted on human, animal, food, and environmental samples were included.
-
•
Study location: All accessible published full articles from studies conducted in South-Eastern Asia countries were included.
-
•
Time period: There was no time limitation on the period/year of publication.
-
•
Age and sex: no restriction
-
•
Language of publication: Only studies published in the English language were included.
-
•
Publication type: Published peer-reviewed articles were included,
Exclusion criteria: The ensuing criteria were used to exclude studies from this SR&MA:
-
•
Studies with incomplete data, no clear study design were excluded.
-
•
Studies outside SEA countries were not considered.
-
•
In silico studies, In vitro studies, Letters, books, dissertations, review articles, opinion papers, reports, and conference papers were excluded.
-
•
Articles published in languages other than the English language were excluded.
2.2.1. Outcomes
Primary outcome: is to determine the overall prevalence of Listeria monocytogenes in humans, animals, food, and the environment in SEA countries.
Secondary outcomes:
-
•
To identify the virulent genes in isolated L. monocytogenes in SEA.
-
•
To determine the prevalent sample types/origin for the isolation of L. monocytogenes in SEA.
-
•
To assess the frequent types of detection methods/techniques used to identify L. monocytogenes in SEA.
2.3. Search and selection strategy
A search strategy with specific search – terms was designed and applied on four selected electronic databases. Also, hand searching of references of selected (review) articles and conference proceedings was done after the electronic search. Additionally, a search was equally conducted on some non-specific internet search engines (Google Scholar and Google search) using specific terms for more literature.
2.4. Databases
The following databases were selected and searched: ProQuest, MEDLINE, PubMed, and Academic Search Complete. The specific search – terms used on each database are outlined in the study protocol (S2 file). First, however, the search algorithm used in the ProQuest database is given as follows; (Prevalence OR Occurrence OR Incidence) AND (Virulent factors OR Virulent genes OR Virulence Agents OR virulent strains) AND (Listeria monocytogenes OR Listeria OR Listeriosis OR Listeria species OR Listeria spp. OR L. monocytogenes) AND (human* OR animal* OR meat OR Vegetable* OR fruit* OR environment) AND (Malaysia OR Indonesia OR Singapore OR Thailand OR Cambodia OR Philippines OR Laos PDR OR Brunei OR Myanmar OR Vietnam OR East Timor).
2.5. Data management and selection process
The citations identified following the search were exported to the Mendeley reference manager for de-duplication. The de-duplicated sources were then transferred to the Rayyan Intelligent Systematic Review software [12] for the title/abstract and full-text screening based on the study inclusion and exclusion criteria. Three independent reviewers undertook the screening process, with a fourth reviewer deciding on areas of dispute between the three reviewers.
2.6. Data collection process
The data extraction process began with creating an a priori data extraction form in a Microsoft Excel (MS) spreadsheet. Afterwards, the characteristics of the studies included, and other relevant data were retrieved and inputted in the data extraction form. Three independent reviewers carried out the complete data extraction procedure, and a fourth reviewer crosschecked to ascertain the accuracy of the extracted data.
2.7. Study quality assessment
Each article that meets the study inclusion and exclusion criteria was subjected to a quality assessment using the appraisal instrument developed for use in systematic reviews addressing questions of prevalence [13]. The appraisal tool has ten questions that were answered as either yes (Y), no (N), unclear (UN), or not applicable (NA). A score of 1 was given to questions with ‘Y’ answers, 0 was awarded to questions with ‘N’, and ‘UC’ answers and no score was awarded to ‘NA’. The total scores were then calculated as percentages. Thus, studies with <50% score were termed low quality, those with >50%-to- < 70% were labelled medium quality, and those with ≥70% score were high-quality studies. Two independent reviewers carried out the critical appraisal and were cross-checked by a third reviewer.
2.8. Meta-analysis
2.8.1. Statistical assessment
MetaXL software (add-in for Microsoft Excel) was used to analyse the extracted data quantitatively. The meta-analysis and pooling of the prevalence estimate (with the 95% confidence interval) were done using the random effect (RE) model to account for heterogeneity. This meta-analysis was done by employing (the transformed) double arcsine method.
2.8.2. Assessment of heterogeneity
Estimation of statistical heterogeneity among the included studies was done using the X2 test, Cochrane Q test, tau and I2 statistics. An I2 value of 0 to ≤40% was considered low heterogeneity, >40% to 60% was considered moderate heterogeneity, >60% to 75% was deemed substantial heterogeneity, and > 75% to 100% was judged as high heterogeneity. Significant heterogeneity was considered for a p < 0.10.
2.8.3. Sensitivity analysis
Sensitivity analysis was done based on the leave-one-out model to identify the study that significantly influenced the meta-analysis result.
2.8.4. Subgroup analysis and meta-regression
Subgroup analysis and meta-regression were conducted to identify the factors that may contribute to heterogeneity among the studies. The factors considered for the analyses include population type studied (food or environment or animals or humans), year of publication, sample size, country of study and study quality. For meta-regression, univariate analysis was done for each of the covariates. Due to the low power of the test (meta-regression) and the fact that this study is considered as the first, 0.25 was considered significant.
2.8.5. Publication bias
A funnel plot of the double arcsine prevalence against standard error was constructed to examine publication bias. The observed asymmetry on the funnel plot, indicating potential publication bias, was further assessed using the Doi plot to evaluate the degree of the asymmetry observed in the funnel plot. This was followed by Egger's regression test to test for the significance of the confirmed asymmetry.
3. Results
3.1. Characteristics of included studies and study selection process
The literature search from the selected electronic databases returned 1867 citations. Additionally, six studies were identified from other searches conducted (manual search of references and other internet search engines). After de-duplication of the total citations, 1217 publications were screened on title/abstract and 48 were then selected for full-article review (Table S1). Finally, 17 publications were included in this SR&MA (Fig. 1 and S4 File) with a total sample size of 7160 (food, animal, and environmental samples). Of the included studies, 53% (9) were conducted in Malaysia. Other countries include Thailand, Indonesia, and Singapore, with no studies identified from the remaining seven countries of SEA. No study sampled humans in all the included publications. One study sampled food, animals, and environment [14], one sampled food and environment [15] and another [16] worked on animal and environmental samples. All other remaining studies sampled only food. Other characteristics of the included studies are outlined in Table 1.
Fig. 1.
PRISMA flow diagram.
Table 1.
Characteristics of included studies.
| Author | Year of sampling | Country | Study design | Sample size | Studied population |
Overall reported prevalence % |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Animal | Environment | Food | Human | Animal | Environment | Food | |||||
| Minami et al., 2010 | 2006–2007 | Thailand | Prevalence survey | 388 | N | N | N | Y | – | – | – | 6 |
| Arumugaswamy et al., 1994 | 1993 | Malaysia | Prevalence | 234 | N | N | N | Y | – | – | – | 43.2 |
| Wong et al., 2012 | 2009 | Malaysia | Prevalence | 112 | N | N | N | Y | – | – | – | 22.3 |
| Vongkamjan et al., 2016 | 2013 | Thailand | Prevalence | 200 | N | N | N | Y | – | – | – | 7.5 |
| Kuan et al., 2013 | 2010–2011 | Malaysia | Prevalence | 216 | N | N | N | Y | – | – | – | 26.39 |
| Sugiri et al., 2014 | 2012–2013 | Indonesia | Prevalence | 184 | N | N | N | Y | – | – | – | 15.8 |
| Marian et al., 2012 | 2008 | Malaysia | Prevalence | 140 | N | N | N | Y | – | – | – | 16.4 |
| Vongkamjan et al., 2017 | 2013–2014 | Thailand | Prevalence | 595 | N | N | Y | Y | – | – | 3.7 | – |
| Goh et al., 2012 | 2011–2012 | Malaysia | Prevalence | 210 | N | N | N | Y | – | – | – | 20 |
| Jamali et al., 2013 | 2006–2012 | Malaysia | Prevalence | 396 | N | N | N | Y | – | – | – | 11.4 |
| Chau et al., 2017 | 2011–2015 | Singapore | Prevalence survey | 527 | N | N | N | Y | – | – | – | 12.7 |
| Aksono et al., 2020 | Indonesia | Cross sectional | 60 | N | N | N | Y | – | – | – | 5 | |
| Indrawattana et al., 2011 | 2007 | Thailand | Prevalence | 104 | N | N | N | Y | – | – | – | 15.4 |
| Hassan et al., 2001 | Malaysia | Prevalence | 101 | N | N | N | Y | – | – | – | 23.8 | |
| Kanarat et al., 2011 | 2004–2009 | Thailand | Prevalence | 3600 | N | Y | Y | Y | – | – | – | 1.7 |
| Lesley et al., 2016 | 2012 | Malaysia | Prevalence | 30 | N | Y | Y | N | – | 33.3 | – | |
| Kuan et al., 2013b | 2010–2011 | Malaysia | PS | 63 | N | N | N | Y | – | – | – | 33.3 |
CS: cross sectional, N: No, not included, PS: Prevalence survey, Y: Yes, include.
3.2. Risk of bias (quality) assessment
The result of the study quality assessment is provided in Table S2. Of the included studies, 88% (15) were high quality, and the remaining two studies were medium quality. There was no study with low quality.
3.3. Outcomes
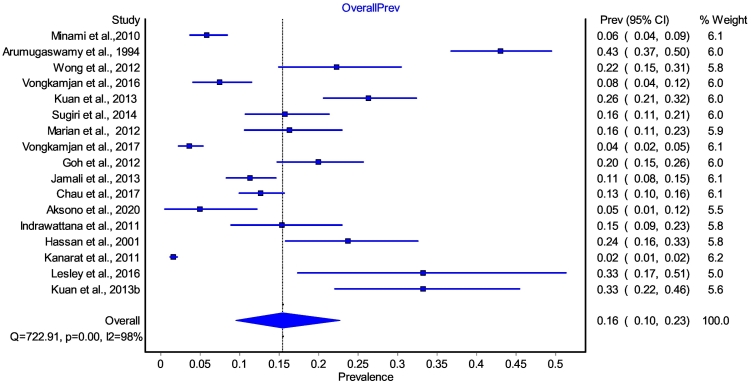
Primary outcome: the primary outcome was to pool the overall prevalence of L. monocytogenes in humans, animals, food, and the environment (one health concept). In this review, prevalence (proportion) is defined as the number of cases (positive L. monocytogenes) in the (sampling) population, divided by the population number (sample size) [17]. So, all the included studies (17) assessed the primary outcome. However, L. monocytogenes prevalence was not determined for humans (samples) in all the included studies. Prevalence results for food (only) samples were determined in 15 [14,[18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31]] of the included studies (Table 1). One study [15] had a result for the environmental (only) sample and one [16] for environmental and animal samples. Thus, all 17 studies were included in the meta-analysis of the primary outcome. The overall (RE pooled) prevalence estimate for L. monocytogenes in SEA was determined to be 16% (95% confidence interval [CI]: 10–23). To test for heterogeneity, the following statistics were computed: Cochrane Q value (Q; =1403.354), X2 p < 0.0001, and I2 = 98.9%. The forest plot (Fig. 2) gives the graphical presentation of the result, and Table S3 summarises the meta-analysis result.
Fig. 2.
Forest plot of overall meta-analysis of L. monocytogenes (for the animal, environment, and food) in SEA.
3.4. Sensitivity analysis
A sensitivity analysis was done by removing the study with the largest weight [14] to determine its influence on the overall pooled prevalence. The removed study had no much impact on the pooled result (Fig. 3), indicating the stability of the meta-analysis.
Fig. 3.
Forest plot of sensitivity analysis of L. monocytogenes in SEA.
3.5. Subgroup analysis and meta-regression
Subgroup analysis was used to explore the observed high heterogeneity in order to determine the predictors (source(s) of high heterogeneity). The factors examined in the subgroup analysis were sampled population (food, environment, and animals), country of study, year of publication, detection methods, and sample size. Table 2 gives the summary result of the subgroup analysis for each of the factors while the forest plots are presented in the S5 File. In other to determine the effect of the covariates as moderators of the cumulative prevalence, a univariable meta-regression was conducted. The covariates used in the subgroup analysis were used as the moderators in the meta-regression. Table 3 shows the proportion (R2) of each of the moderators' effect on heterogeneity with their corresponding p values. Country of study (study location) is the only predictor with significant value, and it accounted for 64% of the detected heterogeneity. A multivariable meta-regression was not done because only one variable was significant.
Table 2.
Summary of subgroup analysis result.
| Subgroups | Number of studies | Pooled prevalence |
Heterogeneity |
||
|---|---|---|---|---|---|
| % | 95% CI | I2 | p | ||
| Food-environment-animal | 18.0 | ||||
|
15.0 | 16.0 | 9.0–24.0 | 98.0% | <0.00001 |
|
2.0 | 15.0 | 0.0–60.0 | 87.0% | <0.00001 |
|
1.0 | 32.0 | 14.0–53.0 | – | – |
| Country of study | 17.0 | ||||
|
2.0 | 10.0 | 2.0–22.0 | 81.0% | <0.00001 |
|
9.0 | 25.0 | 17.0–33.0 | 91.0% | <0.00001 |
|
5.0 | 5.0 | 2.0–10.0 | 94.0% | <0.00001 |
|
1.0 | 13.0 | 10.0–16.0 | – | – |
| Sample size | 17.0 | ||||
|
3.0 | 22.0 | 2.0–46.0 | 90.0% | <0.00001 |
|
11.0 | 18.0 | 12.0–25.0 | 94.0% | <0.00001 |
|
3.0 | 5.0% | 0.0–12.0 | 98.0% | <0.00001 |
| Year of publication | 17.0 | ||||
|
2.0 | 33.0 | 15.0–53.0 | 92% | <0.00001 |
|
10.0 | 15.0 | 7.0–25.0 | 98.0% | <0.00001 |
|
5.0 | 10.0 | 4.0–17.0 | 91.0% | <0.00001 |
| Detection methods | 17.0 | ||||
|
2.0 | 10.0 | 2.0–20.0 | 88.0% | <0.00001 |
|
3.0 | 11.0 | 7.0–16.0 | 64.0% | <0.00001 |
|
3.0 | 19.0 | 0.0–61.0 | 99.0% | <0.00001 |
|
7.0 | 16.0 | 9.0–24 | 94.0% | <0.00001 |
|
2.0 | 23.0 | 17.0–30.0 | 59.0% | 0.12 |
Table 3.
Univariate meta-regression.
| Covariates | R2 (%) | p value |
|---|---|---|
| Country of study | 64.00 | 0.011 |
| Detection methods | 18.30 | 0.775 |
| Population type | 18.80 | 0.423 |
| Sample size | 28.00 | 0.029 |
| Study quality | 0.633 | 0.762 |
| Year of publication | 67.90 | 0.401 |
R2: explains the proportion of between study variance (the effect of covariates on heterogeneity).
3.5.1. Secondary outcomes
Virulence gene profile: one of the secondary outcomes was to assess the different virulent genes identified from L. monocytogenes isolates across SEA. A total of seven (hly, [hly A-F, and hly A-R, LLO] prs, prfA, actA, flaA, iap, inlA, B,) different virulence genes (Table 4) were identified from 14 (out of the 17) included studies in this review. In addition, virulence genes were reported from Malaysia (in seven studies), Thailand (in four studies), Indonesia (in two studies), and in one study from Singapore. The hly gene is the most frequently reported virulence gene, reported in 86% of the 14 studies.
Table 4.
Methods for the detection of L. monocytogenes and virulent genes targeted.
| Study | Sample type | Mode of sample collection | Detection methods |
Virulence gene (target gene) | |||
|---|---|---|---|---|---|---|---|
| Culture (type of media) | Biochemical | Serology | Molecular | ||||
| Minami et al., 2010 | Meat and seafood | Vendors' bare hand | PALCAM, CHROMagar, BHI, TSA | Rhamnose, Mannitol, Dextrose | Seroagglutination | PCR | hly |
| Arumugaswamy et al., 1994 | Raw foods, RTE foods | Purchased | PALCAM | MR, VP, Rhamnose, Xylose, Mannitol | – | – | – |
| Wong et al., 2012 | Raw burger patties | Purchase | PALCAM, TSA | – | – | PCR | hlyA, 16S rRNA |
| Vongkamjan et al., 2016 | RTE foods | Purchase | BHI, CHROMaga | – | – | PCR, sigB sequencing | Prs, hly |
| Kuan et al., 2013 | Raw chicken offal | Purchase | – | – | – | PCR, | hlyA, 16S rRNA |
| Sugiri et al., 2014 | Raw chicken carcass | N/S | PALCAM, CHROMagar, BHI, TSA | CAMP-test | – | PCR | Prs, prfA, ORF2819 |
| Marian et al., 2012 | Raw foods, RTE foods | Purchase | PALCAM, TSA | – | – | PCR | hlyA, 16S rRNA |
| Vongkamjan et al., 2017 | Food, Environmental | Sponge-stick swab | CHROMagar, BHI | – | – | PCR | hly |
| Goh et al., 2012 | Raw chicken meat | N/S | N/S | – | – | PCR | hlyA, 16S rRNA |
| Jamali et al., 2013 | RTE foods | N/S | PALCAM, TSA, BHI, CHROMagar | MR-VP, catalase, oxidase. Urea, SIM, and TSI | – | PCR | 16S rRNA, LLO |
| Chau et al., 2017 | RTE foods | Purchase | PALCAM, OAA | – | – | PCR, MLST | Prs, inlA |
| Aksono et al., 2020 | Raw chicken meat | N/S | PALCAM, | MR-VP, SIM, and TSIA, CAMP | – | PCR, PGA | hlyA |
| Indrawattana et al., 2011 | Raw meat | N/S | Chrom agar, PALCAM, TSA, TSYEA | CAMP, | Listeria antisera, for O and H antigen | PCR, | hlyA, actA, flaA, iap, inl A, B, prf A |
| Hassan et al., 2001 | Frozen beef or meat | Purchase | TSYEA, | TSI, MR-VP, catalase, oxidase, CAMP | – | – | – |
| Kanarat et al., 2011 | Soil litter, chicken feed, water, meat, RTE | Swab with sterile, gauze | ALOA, PALCAM, TSA | Catalase, Oxidase, CAMP | – | – | – |
| Lesley et al., 2016 | Bats, birds, rodents, shrew, feces, water & sediment | Mist nets, cage traps, by scooping & dipping, anal & coacal by cotton bud swab | PALCAM | – | – | PCR | hly A-F, hly A-R |
| Kuan et al., 2013b | Raw beef offal | Purchased | TSA | – | – | PCR | 16S Rrna, LLO |
BHI; Brain Heart Infusion, CAMP; Christie Atkins-Munch-Peterson, CHROMagar™; Trade name, MLST; Multilocus Sequence Typing, MR-VP; Methyl Red and Vogues-Proskauer, NS; Not stated, PALCAM; Trade name for listeria culture agar. PCR; Polymerase chain reaction, PGA; Phylogenetic Analyses, RTE; Ready to Eat, SIM; Sulfide Indole Motility, TSA; Tryptic Soy Agar, TSATE; Tryptic Soy Agar Yeast Extract, TSI; Triple Sugar Iron. 16S – 16 sub-units, actA; – actin polymerization protein, FlaA; – Flagellin A, – hlyA; – Hemolysin O, iap; – associated protein, InL; – internalin A, LLO; – Listeriolysin O, ORF; – Open Reading Frame, plcA; Phosphotidylinositol A. PrfA; – perforin A, prs; – pyrophosphokinase, rRNA; – ribosomal, Ribonucleic Acid.
Prevalent sample types: Food being the predominant sampled source of L. monocytogenes in all the included studies, a further meta-analysis was done by classifying the food sources into different classes. Details of classification are outlined in the Table S4. The obtained pooled prevalence estimates for the various food classifications are ready-to-eat (RTE) foods of aquatic origin (16% CI: 6–29), RTE-food products of animal origin (18% CI: 2–43), RTE-foods of vegetable origin (21% CI: 6–41), and other products (25% CI: 16–35). Also, meta-analysis was done for raw foods of aquatic origin (13% CI: 1–30), raw food products of animal origin (19% CI: 9–31). All results are graphically presented in S6 File.
Detection techniques: this outcome was set to outline the various methods used to isolate and identify L. monocytogenes. Different detection techniques were used; culture, biochemical, serology, and molecular (Table 4). The polymerase chain reaction (PCR) is the most common method used to identify L. monocytogenes. The PCR was used in 82% of the included studies. However, most of the included studies used a combination of two or more methods for identifying L. monocytogenes. Combination of culture and biochemical methods was used in 18% of the studies. Three (18%) studies combined culture, biochemical, and PCR methods. An additional 12% combined four methods (culture, biochemical, serology, and PCR), and PCR alone was used in two studies. While 41% of the included studies used culture and PCR to isolate and identify L. monocytogenes.
3.6. Publication bias
A lack of symmetry was observed from the constructed funnel plot, which illustrates potential publication bias (Fig. 4). To this effect, a Doi plot was constructed to determine the level of asymmetry. The LFK index of 4.9 (Fig. 5) observed from the Doi plot indicates the presence of major asymmetry. To quantify the level of asymmetry Egger's regression test was conducted which was not significant (p = 0.052).
Fig. 4.
Funnel plot of double arcsine prevalence against standard error showing observed asymmetry.
Fig. 5.
Doi plot of double arcsine prevalence against Z-score showing evidence of major asymmetry.
4. Discussion
One Health is defined as an integrated, unifying strategy with the goal of enhancing the health of people, animals, and ecosystems in a sustainable manner [32]. This concept of One Heath is increasingly gaining recognition as a viable tool in strengthening and supporting global health security. The acceptance of the One Health concept further buttresses the interconnectivity of human health to animal health and the common environment. [33]. In recent years the world has seen disease outbreaks of emerging and resurging pathogens due to increased human-animal-environment interaction [34]. This close interaction has provided more opportunities for the emergence of zoonotic diseases. One such pathogen is the L. monocytogenes transmitted directly from infected animals and contaminated food products to humans [35]. This organism has a unique ability to withstand extreme food preservation conditions, thus making it a serious food safety threat [36]. Thus, addressing the threat of this pathogen will require robust data (from human, animal, food, and environment sources) on the prevalence of this pathogen at a regional level.
Therefore, this review included studies of food, environment, and animal samples (as there was no data on human samples) with relatively large sample size. The findings of this SR&MA revealed a high (16%) overall prevalence estimate (for food, environment, and animal samples) of L. monocytogenes in SEA. The pooled prevalence estimate in this study is similar to but slightly lower than the calculated average prevalence of 22.2% recorded from a review study in Africa [37]. The obtained result suggests that L. monocytogenes is highly prevalent in food, environment, and animals in this subregion. Thus, implying that the human population might be at an increased risk of infection by this pathogen. In the presence of a clear indication of increased risk, there is the need for robust prevention and control strategies. Also, improved surveillance and focused research is required to address the threat of this pathogen in the subregion.
Further subgroup analysis equally showed high pooled prevalence for each of the sampled sources (food: 16%, environmental: 15% and animal:32%). There was, however, only one study that reported prevalence from animal origin [16] and only from one country. Hence likely the reason for the high prevalence observed from animal sources in this review. The observed prevalence (32%) from this study [16] is higher than the observed pooled prevalence of 7% from a meta-analysis conducted in Iran [38]. In the cases of environmental and food sources, it might be out of place to compare the observed prevalence in this study to what was obtained in the Iran study. Due to the fact that the Iran study [38] is a country level pooled prevalence. Comparing the result from this meta-analysis with the results of other meta-analyses is challenging because most of the available meta-analyses were conducted at the country level. We are yet to come across any regional level SR&MA that comprehensively pooled L. monocytogenes prevalence from different sampled sources (the One Health approach) as done in this current study.
At country level, this study revealed that the highest prevalence (25%) was observed in Malaysia. The prevalence observed in Malaysia is higher than what was obtained from Indonesia, Singapore, and Thailand. However, the observed prevalence from the other three countries was also relatively high, further confirming that L. monocytogenes is highly prevalent in the subregion. The observed variation in the prevalence between countries could be because Malaysia has the highest number of studies included in this SR&MA. Other possible reasons may include sample size variation, differences in identification methods or seasonal variability. It is also discovered from the meta-analysis that prevalence rises with decreasing sample size. However, the group with the smaller sample sizes (〈100) expectedly has less precision with a higher margin of error. Additional prevalence estimation based on the year of publication showed a decreasing prevalence pattern from 1994 to 2020. The prevalence was highest between 1994-to-2001, and the lowest prevalence was recorded between 2016-to-2020. The observed reduction in the prevalence of L. monocytogenes across the years might not be unconnected with improved sanitary practices and food processing best practices. It was equally observed in this review that studies that used PCR alone had the highest prevalence of 23% (CI: 17–30). Although, this group had the least number of studies.
However, it is important to state that heterogeneity persisted within all the subgroups. Which indicates that the factors evaluated in the subgroup analysis did not completely explain the observed heterogeneity. Nevertheless, the univariate analysis shows that study location (country) can explain 64% of the observed variation in the meta-analysis. Other possible reasons for the observed heterogeneity might be due to some covariates that are not within the scope of this analysis. Such factors may include but not limited to temperature variation during sample processing, sample contamination, sample transportation mode, and sample storage methods and conditions [39].
In terms of the virulence genes, many virulence genes were identified in this study, and the most frequent is the hly gene. The high-level presence of the hly gene in the isolates in the region may have an implication on potential outbreak occurrence and disease severity [40,41]. Also, our findings showed the prevalent sample types from food sources. The highest prevalence was observed in RTE-food of vegetable products followed swiftly by raw-food of animal origin. This information is of clinical and public health significance as it can be used to enumerate high-risk foods for L. monocytogenes infection [42].
In addition, our study provided information on various methods used to identify L. monocytogenes. The results showed that PCR is gaining more prominence as most recent studies used the technique. While culture and biochemical tests that are previously considered as the gold standard, are seldom used in most recent studies. This is probably due to the several limitations associated with these methods [43]. This information is relevant in clinical settings and the food industry. The demand for more sensitive, highly specific, and rapid techniques for identifying the pathogen is on the rise in clinical settings. Whereas in the food industry, there is increased demand for contaminant-free foods [42]. Thus, the need for ultra-rapid, sensitive, and specific detection techniques cannot be overemphasized.
5. Strength and limitation of the study
It is indeed worthy of note that this is the first SR&MA to investigate the prevalence of L. monocytogenes in SEA. The study also comprehensively reviewed the prevalence (comprising food, environment, and animal sources) and related factors of L. monocytogenes in the subregion. This review also provided relevant information on virulence gene profile, prevalent sample types, and widely used identification methods for the pathogen in the subregion. The provided information will improve awareness to understand this essential foodborne pathogen better. Additionally, it will assist in informed – decision making in clinical practice, public health intervention, and policy design for the prevention and control of the disease. However, the limitations of this study include the fact that only articles published in the English language are included in the review. Also, the meta-analysis result showed high heterogeneity between studies even in the subgroup analysis.
6. Conclusion
Addressing the challenges of emerging and resurging pathogens requires intersectoral collaboration, coordination, and communication. The one health approach can help achieve this goal. Using this approach in this study has provided the desired information to address the challenges posed by L. monocytogenes in SEA. We now know that L. monocytogenes is highly prevalent in SEA with this approach. In addition, we are now aware of the high-risk foods, virulence gene profile, and frequently used identification methods for L. monocytogenes. Similarly, there is a need for further research on the human origin of the pathogen in the subregion. Also, more studies are required from countries in the subregion with no reported studies.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its supplementary information files].
Funding statement
This review was sponsored by Universiti Putra Malaysia (UPM). The funding body has no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript and the decision to publish the study.
Author contributions
Conception of research idea (ZJ), Literature review (GJ and YR), Research protocol design (YR and GJ), Study appraisal (ZJ, SN, and RM), Data extraction (GJ, YR, and SA), Data analysis and interpretation of results (ZJ, SN, RM, GJ, and YR), Manuscript drafting (GJ, SA and YR), and review of the initial and final draft of the manuscript (ZJ, SN, and RM).
The following are the supplementary data related to this article.
PRISMA_2020_checklist
Study protocol
PRISMA-P-Checklist
List of included studies in the SR&MA
Forestplots for subgroup analyses
Supplementary figures
Full articles screened for eligibility
Quality assessment
Meta-analysis summary table
Specific products prevalence
Declaration of Competing Interest
The authors declare that they have no competing interests.
Contributor Information
Garba Gidandawa Jibo, Email: geejeeboo@yahoo.co.uk.
Yakubu Egigogo Raji, Email: rajiegigogoy@ibbu.edu.ng.
Adamu Salawudeen, Email: asalawudeen@gsu.edu.ng.
Syafinaz Amin-Nordin, Email: syafinaz@upm.edu.my.
Rozaihan Mansor, Email: rozaihan@upm.edu.my.
Tengku Zetty Maztura Tengku Jamaluddin, Email: tengkuzetty@upm.edu.my.
References
- 1.Drevets D.A., Bronze M.S. Listeria monocytogenes : epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol. Med. Microbiol. 2008;53(2):151–165. doi: 10.1111/j.1574-695X.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- 2.Roberts B.N., et al. Listeria monocytogenes response to anaerobic environments. Pathogens (Basel, Switzerland) 2020;9(3):210. doi: 10.3390/pathogens9030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Capita R., et al. Characterization of Listeria monocytogenes originating from the Spanish meat-processing chain. Foods (Basel, Switzerland) 2019;8(11):542. doi: 10.3390/foods8110542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J.-Q., et al. Prevalence and methodologies for detection, characterization and subtyping of Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci. Human Wellness. 2017;6(3):97–120. [Google Scholar]
- 5.Mahendra P., et al. Listeria monocytogenes as an emerging global food-borne zoonotic bacterial pathogen. Beverage Food World. 2017;44(3):29–32. [Google Scholar]
- 6.Dhama K., et al. Listeriosis in animals, its public health significance (food-borne zoonosis) and advances in diagnosis and control: a comprehensive review. Vet. Q. 2015;35(4):211–235. doi: 10.1080/01652176.2015.1063023. [DOI] [PubMed] [Google Scholar]
- 7.Desai A.N., et al. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: a review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2019;84:48–53. doi: 10.1016/j.ijid.2019.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.EL-Naenaeey E.-S., et al. Prevalence of Listeria species in dairy cows and pregnant women with reference to virulotyping of Listeria monocytogenes in Egypt. Zagazig Vet. J. 2019;47(3):248–258. [Google Scholar]
- 9.Marina A., et al. 2019. Incidence of Listeria Monocytogenes in Dairy and Food Products of Animal Origin in Central Region of Peninsular Malaysia. [Google Scholar]
- 10.Altekruse S., Cohen M., Swerdlow D. Emerging foodborne diseases. Emerg. Infect. Dis. 1997;3(3):285. doi: 10.3201/eid0303.970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ertaş H.B., Şeker E. Isolation of Listeria monocytogenes from fish intestines and RAPD analysis. Turk. J. Vet. Anim. Sci. 2005;29(4):1007–1011. [Google Scholar]
- 12.Ouzzani M., et al. Rayyan—a web and mobile app for systematic reviews. Syst. Rev. 2016;5(1):1–10. doi: 10.1186/s13643-016-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munn Z., et al. The development of a critical appraisal tool for use in systematic reviews addressing questions of prevalence. Int. J. Health Policy Manag. 2014;3(3):123–128. doi: 10.15171/ijhpm.2014.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanarat S., Jitnupong W., Sukhapesna J. Prevalence of Listeria monocytogenes in chicken production chain in Thailand. Thai J. Vet. Med. 2011;41(2):155. [Google Scholar]
- 15.Vongkamjan K., et al. Longitudinal monitoring of Listeria monocytogenes and Listeria phages in seafood processing environments in Thailand. Food Microbiol. 2017;66:11–19. doi: 10.1016/j.fm.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Lesley M., et al. Detection and antibiotic susceptibility profiles of Listeria monocytogenes in wildlife and water samples in Kubah National Park, Sarawak, Malaysia. Int. Food Res. J. 2016;23(1) [Google Scholar]
- 17.Barendregt J.J., et al. Meta-analysis of prevalence. J. Epidemiol. Community Health. 2013;67(11):974–978. doi: 10.1136/jech-2013-203104. [DOI] [PubMed] [Google Scholar]
- 18.Kuan C., et al. Prevalence and quantification ofListeria monocytogenes in chicken offal at the retail level in Malaysia. Poult. Sci. 2013;92(6):1664–1669. doi: 10.3382/ps.2012-02974. [DOI] [PubMed] [Google Scholar]
- 19.Goh S., et al. Listeria monocytogenes in retailed raw chicken meat in Malaysia. Poult. Sci. 2012;91(10):2686–2690. doi: 10.3382/ps.2012-02349. [DOI] [PubMed] [Google Scholar]
- 20.Minami A., et al. Prevalence of foodborne pathogens in open markets and supermarkets in Thailand. Food Control. 2010;21(3):221–226. [Google Scholar]
- 21.Arumugaswamy R.K., Ali G.R.R., Hamid S.N.B.A. Prevalence of Listeria monoctogenes in foods in Malaysia. Int. J. Food Microbiol. 1994;23(1):117–121. doi: 10.1016/0168-1605(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 22.Vongkamjan K., et al. Various ready-to-eat products from retail stores linked to occurrence of diverse Listeria monocytogenes and Listeria spp. isolates. J. Food Prot. 2016;79(2):239–245. doi: 10.4315/0362-028X.JFP-15-361. [DOI] [PubMed] [Google Scholar]
- 23.Wong W., et al. Prevalence of Listeria monocytogenes in frozen burger patties in Malaysia. Int. Food Res. J. 2012;19(4) [Google Scholar]
- 24.Sugiri Y.D., et al. Prevalence and antimicrobial susceptibility of Listeria monocytogenes on chicken carcasses in Bandung, Indonesia. J. Food Prot. 2014;77(8):1407–1410. doi: 10.4315/0362-028X.JFP-13-453. [DOI] [PubMed] [Google Scholar]
- 25.Marian M., et al. MPN-PCR detection and antimicrobial resistance of Listeria monocytogenes isolated from raw and ready-to-eat foods in Malaysia. Food Control. 2012;28(2):309–314. [Google Scholar]
- 26.Jamali H., Chai L.C., Thong K.L. Detection and isolation of Listeria spp. and Listeria monocytogenes in ready-to-eat foods with various selective culture media. Food Control. 2013;32(1):19–24. [Google Scholar]
- 27.Chau M.L., et al. Microbial survey of ready-to-eat salad ingredients sold at retail reveals the occurrence and the persistence of Listeria monocytogenes sequence types 2 and 87 in pre-packed smoked salmon. BMC Microbiol. 2017;17(1):1–13. doi: 10.1186/s12866-017-0956-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aksono E.B., et al. Phylogenetic analysis and antibiotics resistance of Listeria monocytogenes contaminating chicken meat in Surabaya, Indonesia. Vet. Med. Int. 2020;2020 doi: 10.1155/2020/9761812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Indrawattana N., et al. Prevalence of Listeria monocytogenes in raw meats marketed in Bangkok and characterization of the isolates by phenotypic and molecular methods. J. Health Popul. Nutr. 2011;29(1):26. doi: 10.3329/jhpn.v29i1.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan Z., et al. Prevalence of Listeria spp and Listeria monocytogenes in meat and fermented fish in Malaysia. Southeast Asian J. Trop. Med. Public Health. 2001;32(2):402–407. [PubMed] [Google Scholar]
- 31.Kuan C.H., et al. Prevalence and quantification of Listeria monocytogenes in beef offal at retail level in Selangor, Malaysia. Braz. J. Microbiol. 2013;44:1169–1172. doi: 10.1590/S1517-83822014005000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adisasmito W.B., et al. One health: a new definition for a sustainable and healthy future. PLoS Pathog. 2022;18(6) doi: 10.1371/journal.ppat.1010537. e1010537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinclair J.R. Importance of a one health approach in advancing global health security and the sustainable development goals. Rev. Sci. Tech. 2019;38(1):145–154. doi: 10.20506/rst.38.1.2949. [DOI] [PubMed] [Google Scholar]
- 34.Mackenzie J.S., Jeggo M. The one health approach-why is it so important? Trop. Med. Infect. Dis. 2019;4(2):88. doi: 10.3390/tropicalmed4020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vázquez-Boland J.A., et al. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001;14(3):584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koutsoumanis K.P., Kendall P.A., Sofos J.N. Effect of food processing-related stresses on acid tolerance of Listeria monocytogenes. Appl. Environ. Microbiol. 2003;69(12):7514–7516. doi: 10.1128/AEM.69.12.7514-7516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dufailu O.A., et al. Prevalence and characteristics of Listeria species from selected African countries. Trop. Dis. Travel Med. Vaccines. 2021;7(1):26. doi: 10.1186/s40794-021-00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranjbar R., Halaji M. Epidemiology of Listeria monocytogenes prevalence in foods, animals and human origin from Iran: a systematic review and meta-analysis. BMC Public Health. 2018;18(1):1–12. doi: 10.1186/s12889-018-5966-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Churchill K.J., et al. Prevalence of Listeria monocytogenes in select ready-to-eat foods—deli meat, soft cheese, and packaged salad: a systematic review and meta-analysis. J. Food Prot. 2019;82(2):344–357. doi: 10.4315/0362-028X.JFP-18-158. [DOI] [PubMed] [Google Scholar]
- 40.Soni D.K., et al. Virulence and genotypic characterization of Listeria monocytogenes isolated from vegetable and soil samples. BMC Microbiol. 2014;14(1):241. doi: 10.1186/s12866-014-0241-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quereda J.J., et al. Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet. Res. 2018;49(1):13. doi: 10.1186/s13567-017-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabit F.T. Contamination, prevention and control of Listeria monocytogenes in food processing and food service environments. Listeria monocytogenes. 2018;71:71–85. [Google Scholar]
- 43.Gasanov U., Hughes D., Hansbro P.M. Methods for the isolation and identification of Listeria spp. and Listeria monocytogenes: a review. FEMS Microbiol. Rev. 2005;29(5):851–875. doi: 10.1016/j.femsre.2004.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA_2020_checklist
Study protocol
PRISMA-P-Checklist
List of included studies in the SR&MA
Forestplots for subgroup analyses
Supplementary figures
Full articles screened for eligibility
Quality assessment
Meta-analysis summary table
Specific products prevalence
Data Availability Statement
All data generated or analysed during this study are included in this published article [and its supplementary information files].