Abstract
The complex, unpredictable nature of pathogen occurrence has required substantial efforts to accurately predict infectious diseases (IDs). With rising popularity of Machine Learning (ML) and Deep Learning (DL) techniques combined with their unique ability to uncover connections between large amounts of diverse data, we conducted a PRISMA systematic review to investigate advances in ID prediction for human and animal diseases using ML and DL. This review included the type of IDs modeled, ML and DL techniques utilized, geographical distribution, prediction tasks performed, input features utilized, spatial and temporal scales, error metrics used, computational efficiency, uncertainty quantification, and missing data handling methods. Among 237 relevant articles published between January 2001 and May 2021, highly contagious diseases in humans were most often represented, including COVID-19 (37.1%), influenza/influenza-like illnesses (9.3%), dengue (8.9%), and malaria (5.1%). Out of 37 diseases identified, 51.4% were zoonotic, 37.8% were human-only, and 8.1% were animal-only, with only 1.6% economically significant, non-zoonotic livestock diseases. Despite the number of zoonoses, 86.5% of articles modeled humans whereas only a few articles (5.1%) contained more than one host species. Eastern Asia (32.5%), North America (17.7%), and Southern Asia (13.1%) were the most represented locations. Frequent approaches included tree-based ML (38.4%) and feed-forward neural networks (26.6%). Articles predicted temporal incidence (66.7%), disease risk (38.0%), and/or spatial movement (31.2%). Less than 10% of studies addressed uncertainty quantification, computational efficiency, and missing data, which are essential to operational use and deployment. This study highlights trends and gaps in ML and DL for ID prediction, providing guidelines for future works to better support biopreparedness and response. To fully utilize ML and DL for improved ID forecasting, models should include the full disease ecology in a One-Health context, important food and agricultural diseases, underrepresented hotspots, and important metrics required for operational deployment.
Keywords: Systematic review, Infectious diseases, Disease prediction, Disease forecast, Machine learning, Deep learning
Highlights
-
•
An exponential increase in use of ML and DL models over the past decade.
-
•
ML/DL can model highly complex systems with a wide variety of input features.
-
•
ML/DL disease models should include the full disease ecology in a One-Health context.
-
•
Important food & agricultural diseases and key disease hotspots are underrepresented.
-
•
Studies must focus on operational deployment needs to better support biopreparedness.
1. Introduction
Infectious disease (ID) events have plagued human and animal populations throughout history, resulting in massive numbers of morbidities and mortalities as well as substantial social and economic impacts across the world [1]. These ID events can take the form of a localized endemic disease outbreak, emergence or reemergence of a disease in a new location, or an epidemic/pandemic affecting multiple countries. ID prediction is a field of epidemiology that is broadly comprised of predicting when (i.e., the temporality of a disease incident), where (i.e., geographical presence along with the extent of spread of a disease), and identifying how an ID event is going to occur (i.e., various factors that influence disease occurrence) in a population based on a variety of information influencing disease presence. The effects of climate change, urbanization, and globalization have rendered these IDs borderless, enabling them to spread easily across regions and inevitably increasing the risk of epidemics and pandemics. Currently, ID prediction is one of the most important operational epidemiological tools with the potential to provide early warning towards actively preventing disease occurrence and spread. By combining robust data collection at the speed of relevance, engineering, and analytic strategies, models can now predict disease event information, such as location, timing, intensity, and the influence of various factors responsible for its occurrence, which can be used to effectively mitigate ID impacts. The timeliness and accuracy of this predicted ID information are crucial for decision makers to adequately mobilize health resources to the area of concern and properly implement control and prevention strategies [2].
Predicting ID is a challenging task mainly due to the complex and unpredictable nature of pathogen ecology and evolutionary dynamics [3]. These inherent complexities demand robust uncertainties quantification systems for better decision making. The challenge of ID prediction is exacerbated due to inadequate and biased disease surveillance initiatives, a lack of disease reporting systems, as well as incomplete and delayed epidemiological data sharing [4,5]. Despite these limitations, significant efforts have been made in the past couple of decades to utilize ID prediction models in operational control and prevention strategies. In particular, the emergence of the coronavirus disease 2019 (COVID-19) pandemic has resulted in accelerated development and integration of ID prediction models in worldwide public health decision making [6].
Machine learning (ML) is a branch of computer science and artificial intelligence that aims to enable computers to autonomously learn and improve in the tasks that they perform using data and experience, without explicit programming by a human [7]. Deep learning (DL) is an advancement in the field of ML that uses an artificial neural network framework (i.e., a biologically-inspired network of artificial neurons that convert multiple input features into a single output) to generate highly accurate predictions [8]. Though ML and DL models are fundamentally governed by statistical, mathematical, and computational theories, they differ greatly from a traditional statistical or mathematical model [9,10]. Traditionally, statistical and mathematical models have been used in epidemiological studies to better understand the nature and dynamics of ID. The statistical approaches mainly utilize regression-based techniques to determine the nature of a relationship between explanatory features and the disease outcome as well as use time series techniques to predict patterns of presence. On the other hand, mathematical approaches are based on compartmental modeling that partitions the population into different groups or “compartments” and models disease outcomes based on known disease dynamics and simulation techniques [11,12]. One of the major advantages that the ML and DL techniques have over traditional approaches is that they do not impose major constraints on the data, such as prior assumptions about data distribution and structure, the number of input feature variables, or a complete understanding of the input features influence on disease presence [13,14]. These advantages can drastically decrease the human resources and time required to build a prediction model. Due to the data-driven nature of the algorithms, ML and DL models can handle large amounts of data and understand hidden connections between the input features, which leads to better predictions. Furthermore, these techniques can accommodate sudden changes in ID transmission while still producing highly accurate predictions, which is key for real-time, operational decision-making [15,16]. A broad conceptual comparison between statistical, compartmental and ML and DL models is presented in Table 1.
Table 1.
A broad comparison between statistical, compartmental, and machine learning and deep learning models.
| Statistical models | Compartment models | Machine learning and deep learning models | |
|---|---|---|---|
| Definition | Probability theory-based models with a set of statistical assumptions concerning the generation of data used to estimate quantitative measures. | Ordinary differential equation-based models that partition the population into different compartments, used to simulate the movement pattern of the population between the compartments. | Data-driven models built to learn and self-improve based on past experiences with the aim of finding patterns in that data and making accurate predictions. |
| Pros |
|
|
|
| Cons |
|
|
|
In recent years, factors, such as an exponential increase in computing power, easy access to large and diverse datasets, and advancements in artificial intelligence, have facilitated extraordinary growth in the field of infectious disease predictions [17]. The ML and DL methods are gaining popularity and are widely being used for a variety of disease intelligence tasks, including temporal, spatial, and risk factor predictions [18]. ML models have been shown to outperform traditional statistical techniques to give more accurate and reliable predictions [19,20]. The popular ML techniques most widely used in the field of ID prediction include tree-based approaches [[20], [21], [22]] and Support Vector Machines (SVM) [[23], [24], [25]] due to their ease of implementation and interpretability. On the other hand, DL techniques, such as feed-forward neural networks (FNN) [26,27] and recurrent neural networks (RNN) [28,29], are popular for their ability to integrate large and complex data into their predictions.
Many, complex factors contribute to and influence the presence of an ID event, such as epidemiologic, geographic, climatic, demographic, behavioral, and sociopolitical. Traditional ID prediction models can only process a limited number of explanatory variables and do not perform well on cross-correlated features. On the other hand, ML and DL models excel at processing large amounts of feature data and finding complex and hidden connections among data sources. ID prediction modeling has, therefore, greatly benefited from the recent “big data” revolution [30]. Remote sensing satellite imagery and census data yield high resolution information about critical disease related factors, such as climate, environment, population density, and demography. With the increase in worldwide internet and mobile phone usage, non-traditional information (e.g., internet searches, social media usage, phone call records, news media trends, and population mobility data) are also readily available. The ML and DL approaches have become highly efficient in utilizing large and complex information gathered through multiple channels to provide a unique opportunity to understand and model ID dynamics like never before [3,31]. However, the utilization of large datasets and increased complexity of the prediction models could lead to an exponential rise in computational requirements. Hence, optimizing the memory and processing requirements of the ML and DL algorithms without compromising their predictive capabilities is crucial.
This study investigates the advances to and quality of ID prediction capabilities, focusing on ML and DL techniques applied over the past two decades. To do this evaluation, we systematically reviewed the scientific literature to identify research that included ML and/or DL models to predict IDs in humans and/or animals. Within the collection, we highlighted specific tasks performed by each prediction model type, input features used for model building, the study spatial and temporal scales, and error metrics applied. We specifically noted if the studies addressed the important issues of uncertainty quantification, computational efficiency, and missing data when building the models. By focusing on the above-mentioned research areas, we identified the best approaches and strategies as well as revealed gaps present in the field of ID prediction modeling. This systematic analysis can be used as a guide to improve future research studies, to better address operational needs for model deployment, and inform areas where public health and veterinary policies can help improve predictive capabilities.
2. Methodology
To assess the application of ML and DL techniques in the field of infectious disease prediction, we conducted a systematic review following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [32]. A diverse set of subject matter experts, spanning infectious diseases, public health, epidemiology, computer engineering, data science, and statistics, formulated the following specific research questions:
-
•
Which IDs are modeled using ML and DL techniques?
-
•
Which global geographic regions are modeled in the ID prediction studies?
-
•
What is the trend and extent of ML and DL types and sub-types used in ID predictions?
-
•
What are the various tasks performed by ID prediction models?
-
•
What are the different input features used for ID predictions?
-
•
What are the spatial (geographic extent) and temporal (duration) scales of the studies?
-
•
What are the error metrics used?
-
•
Is uncertainty quantification, computational efficiency, or missing data handling addressed?
2.1. Eligibility criteria
Specific eligibility criteria were developed based on subject matter expert recommendations. Inclusion criteria required that the study (1) must explicitly include temporal, spatial, and/or risk prediction models of infectious diseases; (2) must utilize ML and/or DL techniques for predictions; (3) must be an original study; and (4) must be published in a peer-reviewed journal in the English-language between Jan 2001 and May 2021. We excluded prediction studies containing diseases that are primarily transmitted by sexual contact, cancer, clinical trials, and only biomarker data (e.g., genomics, proteomics, transcriptomics). In addition, we did not include research that primarily utilized traditional statistics-based regression or classification methods (e.g., linear, non-linear, autoregressive moving average, logistic or Poisson models). Preprints, book chapters, conference presentations, reviews, opinions, commentaries, and dissertations were excluded. We also excluded articles with missing or inaccessible full texts.
2.2. Search strategy
In May 2021, the scientific literature databases of PubMed, Web of Sciences, Embase, Scopus, and Google Scholar were searched to guarantee effective and adequate coverage of targeted studies (Table 2). The literature published between Jan 2001 and May 2021 was searched using the keywords recommended by subject matter experts. We restricted the Google Scholar searches to the first 300 results, which provides an acceptable search coverage of academic literature without excluding useful references [33]. The citation manager Mendeley (https://www.mendeley.com/) was used to manage imported review citations.
Table 2.
Search keywords and scientific literature databases used to identify potentially relevant publications for systematic review.
| Search keywords | Databases |
|---|---|
| (Forecast* OR Predict* OR Distribution OR Estimate) AND (Machine learning OR Deep learning OR Artificial Intelligence OR Random Forest OR regression tree OR extreme gradient boosting OR Neural Network OR Time-Series OR LSTM) AND (Infect* OR zoonos* OR vector borne OR Virus OR bacteria OR parasite) NOT (geno* OR gene* OR protein OR proteomics OR transcript* OR lipidomics OR metabol* OR plant OR imag* OR Biomarker OR cancer) |
|
2.3. Selection strategy
Citations were first de-duplicated before proceeding to the manual screening of abstracts. As the first step, each abstract was evaluated by two independent reviewers for possible eligibility in the systematic review based on defined eligibility criteria. Next, the full texts of potential candidate articles were evaluated in detail by the reviewers to ensure all criteria were met. Articles that passed the two-part screening were included in the final publication list and, ultimately, in the systematic review. Any differences in opinion between the independent reviewers were resolved through internal discussion until consensus was achieved.
2.4. Information extraction
The ML and DL models present in the review literature were classified into broad categories based on the tasks they performed listed below.
Temporal prediction models utilize historic disease information to predict future disease events. These models attempt to answer when the next disease outbreak would occur in the future based on past events.
Spatial prediction models utilize historic disease information to predict the geographic distribution of disease events. These models attempt to answer where the next disease outbreak might occur by imputing the locations where disease occurrence information is not available.
Risk prediction models assess the relationship between disease events and various factors associated with their occurrence. These models attempt to estimate spatial and/or temporal risk factors correlated with the disease event.
During the process of full-text review, the reviewers recorded the following information: model types and subtypes, disease names, primary study hosts, input features or explanatory variables used for predictions, study area, study duration, temporal forecasting distance, error metrics used, uncertainty quantification, missing data handling, and computational efficiency. These groupings are not mutually exclusive. For example, Zhang et al. [a211] (Supplementary material 2) compared the performance of temporal prediction models belonging to FNN and RNN to forecast typhoid fever incidence in China. To evaluate their model performance, they used three error metrics (mean absolute error, mean absolute percentage error, and mean square error). Hence, this citation was placed under multiple prediction model subtype and error metric categories. Similarly, if a publication model performed multiple tasks, such as modeling multiple diseases, geographic locations, or prediction categories, the citation was placed in all relevant categories. Any differences in opinion between the independent reviewers raised during the collection, screening, and information recording processes of the review were resolved through internal discussion until consensus was achieved.
3. Results
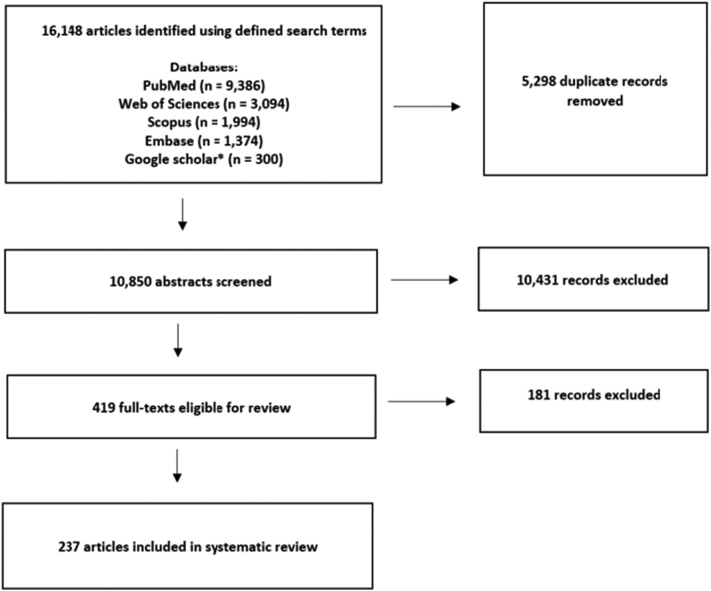
We identified 16,148 articles that were published in peer-reviewed journals between January 2001 and May 2021 (Fig. 1). After removing the duplicates and screening the records for inclusion and exclusion criteria, 237 articles were selected for the final systematic review. The complete list of articles that were included in this systematic review is provided in Supplementary material 2 [a1–a237].
Fig. 1.
PRISMA flow diagram. The illustration of the overall selection process.
* Google Scholar searches were restricted to the first 300 results.
3.1. ML and DL modeling for infectious disease prediction
A large and diverse number of IDs and prediction models applying ML or DL methods were found in the literature based on our search criteria. Out of the 37 unique diseases identified, 37.8% were human-only, 8.1% were animal-only, and 51.4% were zoonotic in nature. Among them, COVID-19 undoubtedly received the most attention and was studied in 88 (37.1%) articles. Influenza and influenza-like illnesses were modeled in 22 (9.3%) articles followed by dengue and malaria in 21 (8.9%) and 12 (5.1%) articles, respectively. The complete list of all the infectious diseases identified in the literature review along with their citations is presented in Table 3.
Table 3.
Citations categorized by infectious disease and study host.
| Disease | Study host | No. of articles |
|---|---|---|
| COVID-19 | Humana1-a88 | 88 |
| Influenza and influenza-like illnesses | Humana89-a110 | 22 |
| Dengue | Humana111-a130, vectora131 | 21 |
| Malaria | Humana132-a142, vectora143 | 12 |
| Tuberculosis | Humana138, a144-a153 | 11 |
| Other mosquito-borne diseases⁎ | Humana113, a155, a156, a158, a159, wild birdsa154, a157, horsea157, vectora157 | 8 |
| Avian influenza | Poultrya160, a165-a167, humana162-a164, wild birds a160-a161 | 8 |
| Tick-borne diseases | Humana168–173, vectorsa172-a175 | 8 |
| Non-specific diseases⁎⁎ | Humansa182–188, livestock a184, wildlifea184 | 7 |
| Brucellosis | Humana176-a181 | 6 |
| Hand foot and mouth disease (HFMD) | Humansa189–193 | 5 |
| Hepatitis A, B, or E | Humansa176, a194-a197 | 5 |
| Leishmaniasis | Humansa200-a202, dogsa198, vectorsa199 | 5 |
| Hemorrhagic fever with renal syndrome (HFRS) | Humansa176, a203-a205 | 4 |
| Plague | Wild animalsa207–209, humansa206, domestic animals a209, vectorsa208 | 4 |
| Typhoid | Humans a120, a138, a176, a210 | 4 |
| Anthrax | Humansa211-a213, livestocka211-a213, wild animalsa212-a213 | 3 |
| Zika | Humansa218-a220 | 3 |
| Ebola and Marburg | Humansa214-a215, wild animalsa214 | 2 |
| Hantavirus | Humansa216-a217 | 2 |
| Others⁎⁎⁎ | Humansa105, a135, a138, a176, a221-a231, domestic animalsa232–236, wild animalsa237 | 21 |
Other mosquito-borne diseases included West Nile fever, lymphatic filariasis, yellow fever, onchocerciasis, chikungunya.
Non-specific diseases included antibiotic resistance, infectious diarrhea, emerging zoonotic diseases, foodborne disease, acute respiratory infectious disease.
Others included scarlet fever, chickenpox, bacillary dysentery, cholera, cryptosporidiosis, schistosomiasis, whooping cough, porcine reproductive and respiratory syndrome, salmonella infection, E. coli infection, leptospirosis, porcine epidemic diarrhea, African swine fever, rabies, peste des petits ruminants. Note: if an article included models for multiple diseases and primary study hosts, it was placed in each respective category.
A large majority (205, 86.5%) of articles focused on modeling only humans followed by only domestic animals (9, 3.8%), only wildlife (6, 2.5%), and only vectors (6, 2.5%) (Fig. 2). There were only a few articles (12, 5.1%) that used more than one host species for modeling IDs.
Fig. 2.
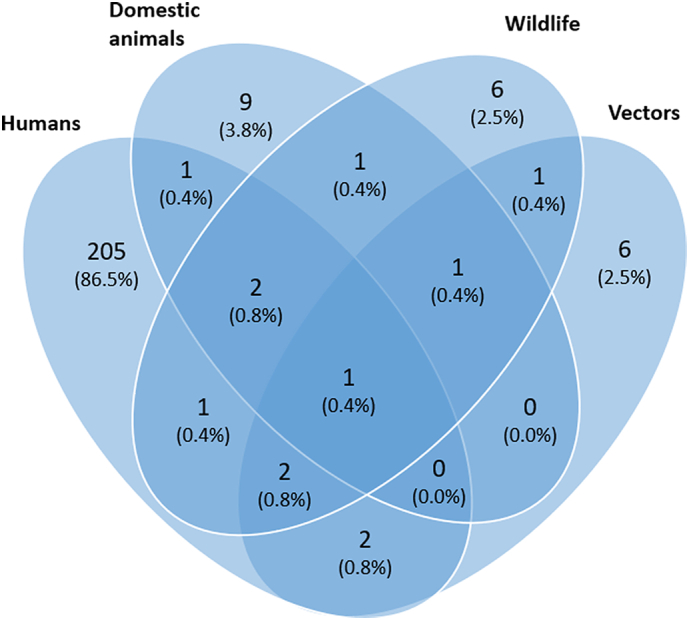
Venn diagram of articles grouped by host species included in infectious disease modeling using machine learning and deep learning techniques. Domesticated animals include livestock and companion animals; wildlife includes wild animals and birds.
3.2. Regional distribution of studies
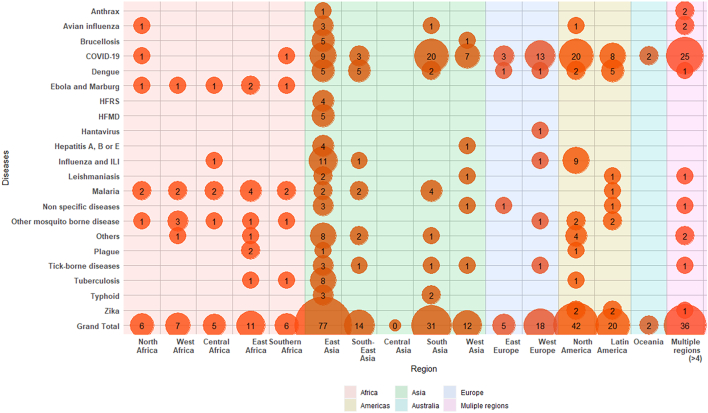
Of the 237 included studies, the majority of them were focused on Eastern Asia (77, 32.5%), followed by North America (42, 17.7%), Southern Asia (31, 13.1%), Latin America (i.e., South and Central America) (20, 8.4%) and Western Europe (18, 7.6%). There were 36 (15.2%) studies that included multiple (more than four) regions which were grouped as a separate category. A complete breakdown of the articles with ID models belonging to each geographical region grouped by diseases is presented in Fig. 3.
Fig. 3.
Distribution of articles with infectious disease models built for each geographical region. If an article included infectious disease models for more than four regions, they were placed in “multiple regions” category. Similarly, if an article included models for multiple diseases, they were placed in each respective disease category.
3.3. Trend and extent of use of ML and DL in infectious disease prediction models
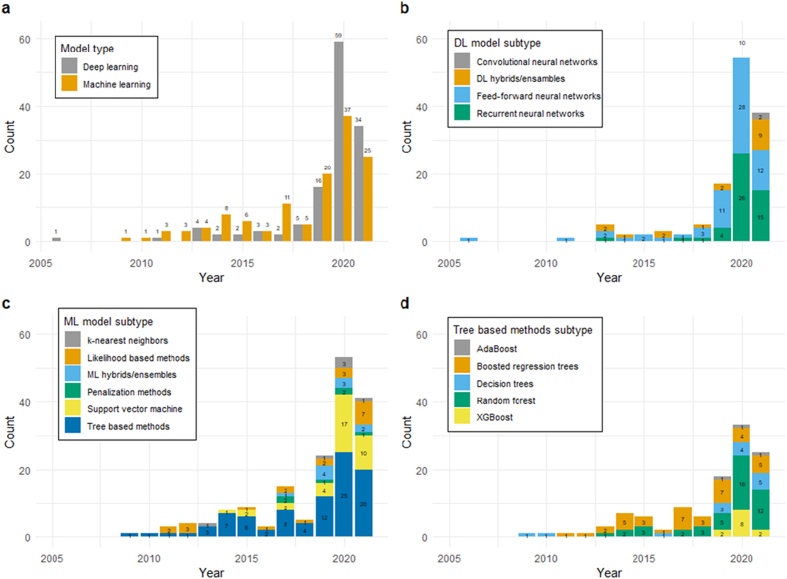
There has been an increasing trend in the use of ML and DL techniques for ID prediction since 2001 with a substantial rise between January 2019 and May 2021 (Fig. 4). Of the 237 articles included in the study, 127 (53.6%) of them applied at least one type of ML approach and 129 (54.4%) used at least one DL technique for disease prediction (Fig. 4a). For the DL models, the FNN (63, 26.6%), RNN (48, 20.3%), and DL hybrids/ensembles (27, 11.4%) were the most common approaches (Fig. 4b). Tree-based methods (91, 38.4%) followed by SVM (36, 15.2%) and then likelihood-based methods (22, 9.3%) were the most common ML approaches (Fig. 4c). Within tree-based ML methods, Random Forest (RF) (44, 18.6%) followed by Boosted Regression Trees (BTR) (30, 12.7%) and Extreme Gradient Boosts (XGB) (12, 5.1%) were most often used (Fig. 4d). More details including the citations grouped by model type and subtype are presented in Supplementary material 1 (S1 Table 1).
Fig. 4.
Trend and extent of ID prediction models published (January 2001–May 2021): Number of citations placed by a) model types (i.e., ML or DL) b) DL model subtypes c) ML model subtypes d) Tree-based ML model subtypes. Note: if an article contained models from different types or subtypes, it was placed in each respective group.
3.4. Utilization of ML and DL approaches for different prediction categories
We grouped the 237 articles into prediction categories based on the tasks they performed. The majority of the articles performed temporal predictions (158, 66.7%) followed by disease risk predictions (90, 38.0%) and spatial predictions (74, 31.2%). COVID-19 was the most frequently modeled disease with the majority being temporal prediction models (Fig. 5). More details, including the citations grouped by prediction categories and model types, are presented in Supplementary material 1 (S1 Table 1).
Fig. 5.
Model prediction categories. The distribution of disease prediction models grouped by model categories and diseases. If an article contained models that performed multiple prediction tasks and for multiple diseases, it was placed in each respective group.
3.5. Spatial and temporal scales of the dataset used in the studies
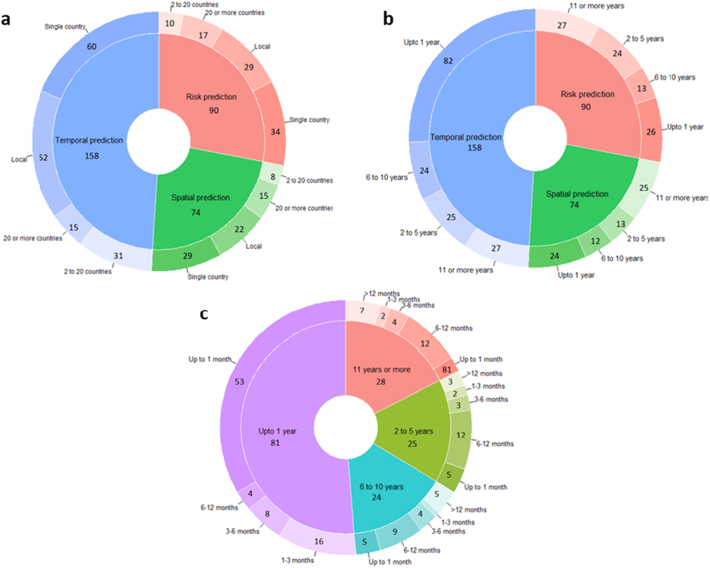
The spatial scale (geographic extent) and temporal scales (duration) of the datasets used to make predictions were identified through the geographic extent/size and duration of the studies, respectively. Overall, for all ID prediction model categories, most articles predicted ID at the country level (123, 51.9%) using only up to one year of data (132, 55.7%) (Fig. 6a, b). Among temporal prediction models, near-term forecasting (up to one month) using one year's worth of data was the most common (53, 33.5%) (Fig. 6c). A complete breakdown of the spatial and temporal scales of the dataset used by each model category for ID prediction is presented in Fig. 6.
Fig. 6.
Spatial and temporal scale of ID prediction models. a) Proportion of the spatial scale (geographic extent) of the models grouped by model categories b) Proportion of temporal scale (duration) of the models grouped by model categories c) Among temporal prediction models, proportion of forecasting distance grouped by temporal scale. An article was placed in its respective groups if it utilized ID models with multiple model categories, spatial and/or temporal scales.
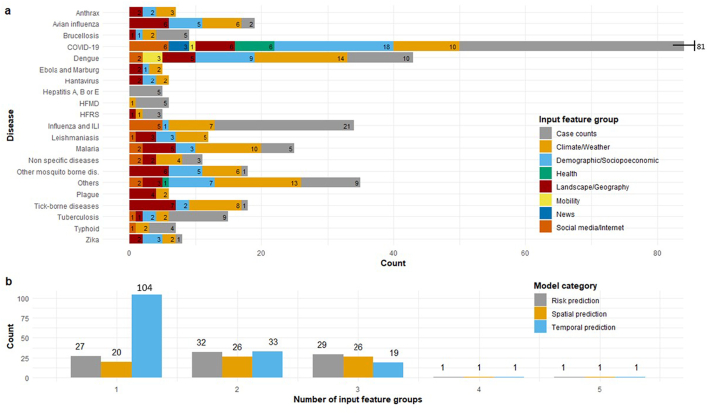
3.6. Input feature groups utilized for disease prediction
The articles included in the study utilized input features that belonged to the following eight groups: case counts (154, 65.0%), climate/weather (98, 41.4%), demographics/socioeconomics (63, 26.6%), landscape/geography (58, 24.5%), social media/internet searches (18, 7.6%), health and comorbidity (7, 3.0%), human mobility (4, 1.7%), and news (3, 1.3%). Each disease modeled has a unique signature of input feature groups used for prediction (Fig. 7a). Focusing on the model prediction type categories, the number of input feature groups used in each category ranged from a minimum of one feature group (n = 151, 63.7%) to a maximum of five groups (n = 3, 1.3%) (Fig. 7b). A complete breakdown of the characteristics of each input feature group utilized for ID prediction is presented in Fig. 7.
Fig. 7.
Characteristics of input feature groups utilized for disease prediction. Articles (n = 237) categorized by a) input feature groups used by disease type b) number of input feature groups utilized by ID prediction model categories. If an article utilized multiple input features, modeled multiple diseases and/or belonged to multiple model categories, the article was counted within each respective grouping.
3.7. Uncertainty quantification, computational efficiency, and missing data
We identified only 21 (8.9%) of the articles to quantify uncertainty in their model predictions. The uncertainty quantification techniques used included frequentist (10, 4.2%) [a46, a67, a68, a91, a107, a123, a145, a152, a193, a195], simulation/sampling based (7, 3.0%) [a26, a53, a156, a200, a213, a214, a219], and Bayesian techniques (3, 1.3%) [a94, a111, a115].
Only 7 (3%) publications [a10, a13, a22, a63, a64, a79, a102, a103] meeting the review criteria included information about computational efficiency while evaluating the performance of their models.
We also noted any missing data handling techniques used in model building. The majority of the articles (220, 84.4%) either did not report any missing data or did not explicitly mention how missing data were handled in their work. For the 18 (7.6%) articles that did discuss this topic, the techniques applied included replacement with mean/median or zeros [a56, a64, a72, a185, a187], moving average [a136, a96, a128], regression [a103, a108, a185], correlation [a220], k-nearest neighbours [a103], multivariate imputation [a111, a136, a139], exclusion [a24], and pixel gap filling [a157].
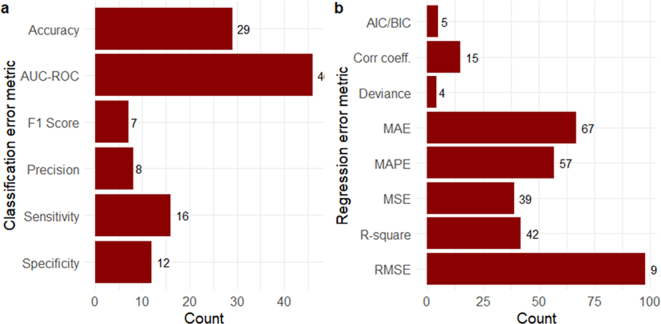
3.8. Common error metrics used in ID prediction modeling
Among classification models that predicted discreet values (e.g., presence or absence of a disease), Area Under the Curve - Receiver Operating Characteristic (AUC-ROC) curve (46, 19.4%), accuracy (29, 12.2%), and sensitivity (16, 6.8%) were the top three error metrics (Fig. 8a). Alternatively, among regression models that predicted continuous values (eg., monthly number of disease cases), Root Mean Square Error (RMSE) (98, 41.4%) followed by Mean Absolute Error (MAE) (67, 28.3%) and Mean Absolute Percentage Error (MAPE) (57, 24.1%) were the most common (Fig. 8b).
Fig. 8.
Error metrics utilized in ID prediction models: Citations grouped by a) Classification error metrics and b) Regression error metrics. If an article used error metrics from different classes, it was placed in each respective group. Abbreviations: AUC-ROC (Area Under the Curve - Receiver Operating Characteristic curve), AIC/BIC (Akaike's/Bayesiasn Information Criteria, corr coeff. (Correlation coefficient), MAE (Mean Absolute Error), MAPE (Mean Absolute Percentage Error), MSE (Mean squared error), RMSE (Root Mean Square Error).
4. Discussion
The ID threat is constantly changing across space and time, hence, an accurate and timely estimation of their occurrence is critical to planning and implementing successful disease preparedness and response strategies [34,35]. To counter these challenges, the landscape of ID prediction is shifting dramatically with the introduction of new disease modeling approaches, especially ML and DL techniques. These techniques are now being extensively applied across a wide range of ID prediction tasks with diverse ecology, transmission pathways, various geographic extents, and temporal scales. This uptick is accompanied by an array of new modeling techniques that cover multiple regression and classification problems. To better characterize these rapidly evolving changes, this systematic review was conducted to understand the current state, trends, and extent of the application of ML and DL algorithms in ID prediction. Our review showed that overall, there was a constant increase in the number of studies that utilized ML and DL to build ID prediction models between 2005 and 2019. Unsurprisingly, we saw an exponential rise in this trend after the COVID-19 pandemic outbreak. The overall global responses to the COVID-19 pandemic by the scientific community, governments, and non-government agencies have been unprecedented. This collective effort has resulted in increased collaboration among health sectors, large-scale disease surveillance, accessible data, and artificial intelligence technology sharing initiatives [36]. The availability of the crucial epidemiological knowledge through these initiatives along with the need for an accurate assessment of the disease dynamics has led to a dramatic increase in the utilization of ML and DL prediction modeling.
Most of the IDs that were modeled were either zoonotic in nature or diseases solely affecting the human population. Apart from COVID-19, influenza and influenza-like illnesses, dengue, malaria, and tuberculosis received major attention. These diseases have an ability to spread easily among the human community either directly through aerosolization or contact (influenza and influenza-like illnesses, tuberculosis) or propagated by vectors (dengue, malaria). This potential to spread easily and the ability to cause wide-scale mortalities and morbidities most likely led to increased attention from global health communities. Furthermore, almost all recent pandemics and a large proportion of emerging IDs originated from wildlife spillover and involve complex dynamic interactions within human and domesticated animal populations [37]. Our review found the majority of zoonotic diseases modeled had humans as their primary host. More efforts are required to integrate other host species that might significantly affect the transmission and persistence of an ID across time and space. We identified only a very small number of publications on non-zoonotic livestock diseases, which could be due to inadequate livestock disease surveillance and the unavailability of reliable epidemiological data for modeling purposes. More efforts should be made to better predict these economic significance veterinary diseases since many of them, such as African swine fever, are highly contagious transboundary diseases with significant global food security and safety impacts.
Areas of high population and tropical regions of Asia, the Americas, and Africa are known to be the global hotspots for emerging IDs [38]. Our study showed that there is a disparity in the number of studies that used ML and DL techniques in regions of Africa compared to other disease hotspots present in East and South Asia and North America. Although many African nations have put in substantial effort to build strong public health and veterinary infrastructures to tackle major health threats, the ability to harness large quantities of ID-related data and generate new knowledge using cutting-edge disease prediction techniques is lagging [[39], [40], [41]]. Building more robust artificial intelligence and data science capacities along with improved disease surveillance and reporting systems in developing regions of the world like these could help capitalize on the potential of ML and DL techniques in ID prediction for biopreparedness and response.
The articles identified were almost evenly split between ML and DL techniques for ID prediction tasks. Within ML techniques, tree-based methods were popular among all prediction categories. Tree-based methods are often among the best performing types of prediction models [19,42,43]. For instance, XGB and, RF outperformed other traditional modeling approaches in predicting diseases such as brucellosis, avian influenza, and influenza-like illnesses across different regions of the world [a107, a167, a180]. These models are also easy to implement, fast to compute, highly performant, and provide a form of interpretability through input feature importance, which could be the main reason for their popularity in ID modeling [44,45]. Alternatively, FFNs, and RNNs were the most frequently used DL techniques and were mostly used for temporal prediction. The FFNs are artificial neural networks that can learn complex and non-linear patterns without making any prior assumptions concerning data distributions [46,47]. The RNNs are the derivatives of FFNs (e.g., Long Short-Term Memory and Gated Recurrent Unit) and are known to produce strong predictions with time series or other types of sequential data because of their ability to utilize historic information to predict future values [48]. Given that the ID outbreaks generally follow a non-linear and complex pattern, these neural networks are often shown to produce superior predictions compared to other approaches and are hence commonly used in disease forecasting tasks. For example, LSTM models produced better results when compared to conventional statistical techniques in predicting influenza and COVID-19 cases in the United States and Indonesia, respectively [a55, a90]. It is also worthwhile to note that ML and DL hybrids/ensembles have attracted great attention from the ID communities in the past few years, evident by their increased use in publications. Hybrid and ensemble models are information fusion concepts that combine statistical, mechanistic, ML, and/or DL approaches working together (hybrid) or independently (ensemble) to minimize prediction noise and increase accuracy over the individual models, which could be one of the possible explanations for their increased popularity in recent years [18,49]. For instance, combining conventional autoregressive methods with neural networks produced better temporal predictions of tuberculosis, brucellosis, and pertussis in China highlighting the superiority of hybrid and ensemble techniques over the individual models [a144, a179, a227].
A wide variety of input features were used for training ID models. Conventional variables (e.g., previous case counts, climate/weather, demographics/socioeconomics, and landscape/geographic data) were routinely utilized to make disease predictions. However, one of the biggest constraints for building a reliable ID prediction model to accurately estimate the progression of the disease is the timeliness of available, essential outbreak-related data. These constraints are aggravated in cases involving a novel disease outbreak or neglected endemic disease where the spatial and temporal patterns of the pathogen emergence are largely unknown. Furthermore, a major outbreak could lead to a significant shift in population social behavior and movement due to public health efforts and government policies resulting in prediction inaccuracies. Hence, the incorporation of novel data sources that account for these dynamic behaviors is vital for accurate and timely decision making. In our review, we identified studies that utilize news articles [a34, a37], social media or internet search queries [a29, a34, a37, a68, a86, a94, a95, a99, a107, a108, a112, a119, a135, a186, a187], health information collected using phone/wearable devices [a24], and human mobility data [a56, a64, a64, a123, a125]. The ML and DL models used in these studies exploited a large quantity of structured and unstructured data with the goal to produce better ID predictions.
Although methods used in ID prediction are becoming more sophisticated, we also identified consistent concerns in the structure of the analyses that could limit their practical use. First, we found that the data collection duration for a large majority of the studies was less than or equal to one year regardless of the prediction category. Secondly, most articles do not include uncertainty quantification or account for missing data. This was apparent, especially during the early stages of the COVID-19 pandemic where the availability of data was limited and there was a widespread underreporting of the cases. Since each ID is known to show specific occurrence patterns that change over time and space, such short-term predictions could be subject to biases and estimation inaccuracies, which should be carefully accounted for while deploying the algorithms to an operational environment. Though a short turnaround time could be vital for a real-world ID event, we recommend updating the models regularly with new data and retraining them for better and long-term practical usage.
Another serious limitation common to the literature reviewed is a lack of discussion regarding data quality and the functional deployment of an algorithm. While one algorithm may perform the best in terms of overall tested accuracy, it may overstate its confidence, may be unrealistic to implement due to computational efficiency, or may simply fail when in the presence of missing data. Since disease prediction models are meant to provide situational awareness, reliable and near-real-time results are necessary [50]. The fact that so few publications consider the critical aspects of automated algorithm implementation suggests that a greater emphasis should be placed on the operational aspects of epidemiology for biopreparedness and response.
While our systematic review was comprehensive, it still has some limitations. First, we only included peer-reviewed studies that were published in a scientific journal. This could have resulted in a selection bias by excluding important studies disseminated as preprints, conference abstracts, books, dissertations, or theses. Second, we did not include studies that primarily utilized traditional statistics-based regression or classification methods. Considering the amount of literature available about these techniques, they will require a separate literature review of their own.
5. Conclusion
To counter the threats of the ever-increasing risk of ID events, the landscape of disease prediction is also shifting dramatically. Due to the factors such as their high prediction accuracy and effective date handling, ML and DL techniques are increasingly being used in ID prediction tasks. The main purpose of our study was to systematically profile the current state of ID prediction capabilities that utilized ML and DL techniques. We specifically looked for IDs that were modeled, type of the ML and DL techniques utilized, the geographical distribution of the modeling studies, prediction tasks performed, input features utilized, spatial and temporal scale of the studies, error metrics used, the computational efficiency of the models, uncertainty quantification and missing data handling methods adopted. Despite an increase in interest in the field indicated by a diverse number of IDs modeled and a consistent increase in the number of studies that apply ML and DL techniques in ID prediction tasks over the past two decades, there were some major limitations to the literature reviewed. Even with the unique ability of ML and DL models to handle diverse, large amounts of data and uncover connections in the data on their own, studies still include a very limited amount of data related to the full disease ecology. Especially for zoonotic and veterinary diseases, ID prediction models should include important One Health input features to capitalize on the interconnections between human, animal, vector, and environmental factors driving disease presence. Incorporating the assessment of uncertainty in the predictions and computational requirements of the models would enable deployment in an operational environment and the ability for better preparedness and response during an ID emergency by decision makers. Finally, building more robust artificial intelligence and data science capacities in resource-scare settings across regions and diseases could help capitalize on the potential of ML and DL techniques in ID prediction in the future.
Author contributions
R.K., L.E.C., K.P. and S.D. designed the search strategy, implemented the study protocol, and retrieved articles. R.K. led the screening and data extraction process, performed data analysis and visualization of the results. R.K., L.E.C., K.P., wrote the manuscript. L.E.C and K.P. acquired the funding. All authors reviewed the manuscript and agreed to the published version of the manuscript.
Funding
This work was funded by the Defense Threat Reduction Agency (project number CB11029).
Data availability statement
All data generated or analyzed during this review are included in this article and its supplementary information files.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
The authors wish to thank Nakita Pradhan, Samuel Ortega, and Jaidyn Bryant for their contribution in the initial review process. The authors wish to thank Samantha Erwin for reviewing the manuscript and providing general feedback. R.K. acknowledges the support from the Pacific Northwest National Laboratory (PNNL)-Washington State University (WSU) Distinguished Graduate Research Program (DGRP) Fellowship for facilitating this research collaboration.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2022.100439.
Appendix A. Supplementary data
Systematic Review Citations
Data availability
Supplementary material contains information to access articles reviewed.
References
- 1.Feldmann H., Czub M., Jones S., Dick D., Garbutt M., Grolla A., Artsob H. Emerging and re-emerging infectious diseases. Med. Microbiol. Immunol. 2002;191:63–74. doi: 10.1007/S00430-002-0122-5. [DOI] [PubMed] [Google Scholar]
- 2.Woolhouse M. How to make predictions about future infectious disease risks. Philos. Trans. Royal Soc. B: Biol. Sci. 2011;366:2045–2054. doi: 10.1098/RSTB.2010.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heesterbeek H., Anderson R.M., Andreasen V., Bansal S., DeAngelis D., Dye C., Eames K.T.D., Edmunds W.J., Frost S.D.W., Funk S., Hollingsworth T.D., House T., Isham V., Klepac P., Lessler J., Lloyd-Smith J.O., Metcalf C.J.E., Mollison D., Pellis L., Pulliam J.R.C., Roberts M.G., Viboud C., Arinaminpathy N., Ball F., Bogich T., Gog J., Grenfell B., Lloyd A.L., Mclean A., O’Neill P., Pearson C., Riley S., Tomba G.S., Trapman P., Wood J. Modeling infectious disease dynamics in the complex landscape of global health. Science. 1979;347(2015) doi: 10.1126/SCIENCE.AAA4339/ASSET/8FA31E42-DA90-4C84-BF84-FBB2DA09DB83/ASSETS/GRAPHIC/347_AAA4339_F2.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charles-Smith L.E., Reynolds T.L., Cameron M.A., Conway M., Lau E.H.Y., Olsen J.M., Pavlin J.A., Shigematsu M., Streichert L.C., Suda K.J., Corley C.D. Using social media for actionable disease surveillance and outbreak management: a systematic literature review. PLoS One. 2015;10 doi: 10.1371/JOURNAL.PONE.0139701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keshavamurthy R., Thumbi S.M., Charles L.E. Digital biosurveillance for zoonotic disease detection in Kenya. Pathogens. 2021;10:783. doi: 10.3390/PATHOGENS10070783. 10 (2021) 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker A.D., Grantz K.H., Hegde S.T., Bérubé S., Cummings D.A.T., Wesolowski A. Development and dissemination of infectious disease dynamic transmission models during the COVID-19 pandemic: what can we learn from other pathogens and how can we move forward?, lancet digit. Health. 2021;3:e41–e50. doi: 10.1016/S2589-7500(20)30268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuel A.L. Some studies in machine learning using the game of checkers. II—recent Progress. Comp. Games I. 1988:366–400. doi: 10.1007/978-1-4613-8716-9_15. [DOI] [Google Scholar]
- 8.Lecun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 9.Jordan M.I., Mitchell T.M. Machine learning: trends, perspectives, and prospects. Science. 1979;349(2015):255–260. doi: 10.1126/SCIENCE.AAA8415/ASSET/AB2EF18A-576D-464D-B1B6-1301159EE29A/ASSETS/GRAPHIC/349_255_F5.JPEG. [DOI] [PubMed] [Google Scholar]
- 10.Bi Q., Goodman K.E., Kaminsky J., Lessler J. What is machine learning? A primer for the epidemiologist. Am. J. Epidemiol. 2019;188:2222–2239. doi: 10.1093/AJE/KWZ189. [DOI] [PubMed] [Google Scholar]
- 11.Grassly N.C., Fraser C. Mathematical models of infectious disease transmission. Nat. Rev. Microbiol. 2008;6:477–487. doi: 10.1038/nrmicro1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolhouse M. How to make predictions about future infectious disease risks. Philos. Trans. Royal Soc. B: Biol. Sci. 2011;366:2045–2054. doi: 10.1098/RSTB.2010.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgenstern J.D., Buajitti E., O’Neill M., Piggott T., Goel V., Fridman D., Kornas K., Rosella L.C. Predicting population health with machine learning: a scoping review. BMJ Open. 2020;10 doi: 10.1136/BMJOPEN-2020-037860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bzdok D., Altman N., Krzywinski M. Points of significance: statistics versus machine learning. Nat. Methods. 2018;15:233–234. doi: 10.1038/NMETH.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chae S., Kwon S., Lee D. Predicting infectious disease using deep learning and big data. Int. J. Environ. Res. Public Health. 2018;15:1596. doi: 10.3390/IJERPH15081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chimmula V.K.R., Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos, Solitons Fractals. 2020;135 doi: 10.1016/J.CHAOS.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong Z.S.Y., Zhou J., Zhang Q. Artificial intelligence for infectious disease big data analytics. Infect. Dis. Health. 2019;24:44–48. doi: 10.1016/J.IDH.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Alfred R., Obit J.H. The roles of machine learning methods in limiting the spread of deadly diseases: a systematic review. Heliyon. 2021;7 doi: 10.1016/J.HELIYON.2021.E07371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon S., Keshavamurthy R., Farber D.H., Stevens A., Pazdernik K.T., Charles L.E. A comparison of infectious disease forecasting methods across locations, diseases, and time. Pathogens. 2022;11 doi: 10.3390/PATHOGENS11020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kane M.J., Price N., Scotch M., Rabinowitz P. Comparison of ARIMA and random Forest time series models for prediction of avian influenza H5N1 outbreaks. BMC Bioinform. 2014;15:276. doi: 10.1186/1471-2105-15-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salami D., Sousa C.A., Martins M.R.O., Capinha C. Predicting dengue importation into Europe, using machine learning and model-agnostic methods. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-66650-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrick K.A., Huettmann F., Lindgren M.A. A global model of avian influenza prediction in wild birds: the importance of northern regions. Vet. Res. 2013;44 doi: 10.1186/1297-9716-44-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Zhang T., Young A.A., Li X. Applications and comparisons of four time series models in epidemiological surveillance data. PLoS One. 2014;9 doi: 10.1371/JOURNAL.PONE.0088075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Silva C.C., de Lima C.L., da Silva A.C.G., Silva E.L., Marques G.S., de Araújo L.J.B., Albuquerque Júnior L.A., de Souza S.B.J., de Santana M.A., Gomes J.C., de Barbosa V.A.F., Musah A., Kostkova P., dos Santos W.P., da Silva Filho A.G. COVID-19 dynamic monitoring and real-time spatio-temporal forecasting. Front. Public Health. 2021;9:641253. doi: 10.3389/FPUBH.2021.641253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darwish A., Rahhal Y., Jafar A. A comparative study on predicting influenza outbreaks using different feature spaces: application of influenza-like illness data from early warning alert and response system in Syria. BMC Res. Notes. 2020;13:1–8. doi: 10.1186/S13104-020-4889-5/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mollalo A., Rivera K.M., Vahedi B. Artificial neural network modeling of novel coronavirus (COVID-19) incidence rates across the continental United States. Int. J. Environ. Res. Public Health. 2020;17:4204. doi: 10.3390/ijerph17124204. https://www.mdpi.com/1660-4601/17/12/4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu K., Zhang M., Xi G., Deng A., Song T., Liid Q., Kang M., Yin L. Enhancing fine-grained intra-urban dengue forecasting by integrating spatial interactions of human movements between urban regions. PLoS Negl. Trop. Dis. 2020;14:1–22. doi: 10.1371/journal.pntd.0008924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bomfim R., Pei S., Shaman J. Predicting dengue outbreaks at neighbourhood level using human mobility in urban areas. J. R. Soc. Interface. 2020;17:20200691. doi: 10.1098/rsif.2020.0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santosh T., Ramesh D., Reddy D. LSTM based prediction of malaria abundances using big data. Comput. Biol. Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103859. https://www.sciencedirect.com/science/article/pii/S0010482520302183 [DOI] [PubMed] [Google Scholar]
- 30.Bansal S., Chowell G., Simonsen L., Vespignani A., Viboud C. Big data for infectious disease surveillance and modeling. J. Infect. Dis. 2016;214:S375–S379. doi: 10.1093/INFDIS/JIW400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milinovich G.J., Magalhães R.J.S., Hu W. Role of big data in the early detection of Ebola and other emerging infectious diseases. Lancet Glob. Health. 2015;3:e20–e21. doi: 10.1016/S2214-109X(14)70356-0. [DOI] [PubMed] [Google Scholar]
- 32.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:332–336. doi: 10.1136/BMJ.B2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haddaway N.R., Collins A.M., Coughlin D., Kirk S. The role of Google scholar in evidence reviews and its applicability to Grey literature searching. PLoS One. 2015;10 doi: 10.1371/JOURNAL.PONE.0138237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morse S.S. Public health surveillance and infectious disease detection. Biosecurity Bioterrorism. 2012;10:6–16. doi: 10.1089/bsp.2011.0088. [DOI] [PubMed] [Google Scholar]
- 35.Corley C.D., Pullum L.L., Hartley D.M., Benedum C., Noonan C., Rabinowitz P.M., Lancaster M.J. Disease prediction models and operational readiness. PLoS One. 2014;9 doi: 10.1371/journal.pone.0091989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luengo-Oroz M., Pham K. Hoffmann, Bullock J., Kirkpatrick R., Luccioni A., Rubel S., Wachholz C., Chakchouk M., Biggs P., Nguyen T., Purnat T., Mariano B. Artificial intelligence cooperation to support the global response to COVID-19. Nat. Mach. Intellig. 2020;2:295–297. doi: 10.1038/s42256-020-0184-3. [DOI] [Google Scholar]
- 37.Allen T., Murray K.A., Zambrana-Torrelio C., Morse S.S., Rondinini C., di Marco M., Breit N., Olival K.J., Daszak P. Global hotspots and correlates of emerging zoonotic diseases, Nature. Communications. 2017;8:1–10. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen T., Murray K.A., Zambrana-Torrelio C., Morse S.S., Rondinini C., di Marco M., Breit N., Olival K.J., Daszak P. Global hotspots and correlates of emerging zoonotic diseases, Nature. Communications. 2017;8:1–10. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nkengasong J.N., Maiyegun O., Moeti M. Establishing the Africa Centres for disease control and prevention: responding to Africa’s health threats. Lancet Glob. Health. 2017;5:e246–e247. doi: 10.1016/S2214-109X(17)30025-6. [DOI] [PubMed] [Google Scholar]
- 40.Beyene J., Harrar S.W., Altaye M., Astatkie T., Awoke T., Shkedy Z., Mersha T.B. A roadmap for building data science capacity for health discovery and innovation in Africa. Front. Public Health. 2021;9:1435. doi: 10.3389/FPUBH.2021.710961/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owoyemi A., Owoyemi J., Osiyemi A., Boyd A. Artificial intelligence for healthcare in Africa. Front. Digit. Health. 2020;2:6. doi: 10.3389/FDGTH.2020.00006/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schapire R. 2003. The boosting approach to machine learning: an overview; pp. 141–171. [Google Scholar]
- 43.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016. XGBoost: A scalable tree boosting system; pp. 785–794. [DOI] [Google Scholar]
- 44.James G., Witten D., Hastie T., Tibshirani R. 2021. Tree-Based Methods; pp. 327–365. [DOI] [Google Scholar]
- 45.Kingsford C., Salzberg S.L. What are decision trees? Nat. Biotechnol. 2008;26:1011–1013. doi: 10.1038/nbt0908-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardner M.W., Dorling S.R. Artificial neural networks (the multilayer perceptron)—a review of applications in the atmospheric sciences. Atmos. Environ. 1998;32:2627–2636. doi: 10.1016/S1352-2310(97)00447-0. [DOI] [Google Scholar]
- 47.Eldan R. Vol. 49. 2016. The Power of Depth for Feedforward Neural Networks Ohad Shamir; pp. 1–34. [Google Scholar]
- 48.Che Z., Purushotham S., Cho K., Sontag D., Liu Y. Recurrent neural networks for multivariate time series with missing values. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-24271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ardabili S., Mosavi A., Várkonyi-Kóczy A.R. Vol. 101. 2020. Advances in Machine Learning Modeling Reviewing Hybrid and Ensemble Methods, Lecture Notes in Networks and Systems; pp. 215–227. [DOI] [Google Scholar]
- 50.Broadway K.M., Schwartz-Watjen K.T., Swiatecka A.L., Hadeed S.J., Owens A.N., Batni S.R., Wu A. Operational considerations in global health modeling. Pathogens. 2021;10:1348. doi: 10.3390/PATHOGENS10101348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Systematic Review Citations
Data Availability Statement
All data generated or analyzed during this review are included in this article and its supplementary information files.
Supplementary material contains information to access articles reviewed.