Abstract
Actin bundles constitute important cytoskeleton structures and enable a scaffold for force transmission inside cells. Actin bundles are formed by proteins, with multiple F-actin binding domains cross-linking actin filaments to each other. Vasodilator-stimulated phosphoprotein (VASP) has mostly been reported as an actin elongator, but it has been shown to be a bundling protein as well and is found in bundled actin structures at filopodia and adhesion sites. Based on in vitro experiments, it remains unclear when and how VASP can act as an actin bundler or elongator. Here we demonstrate that VASP bound to membranes facilitates the formation of large actin bundles during polymerization. The alignment by polymerization requires the fluidity of the lipid bilayers. The mobility within the bilayer enables VASP to bind to filaments and capture and track growing barbed ends. VASP itself phase separates into a protein-enriched phase on the bilayer. This VASP-rich phase nucleates and accumulates at bundles during polymerization, which in turn leads to a reorganization of the underlying lipid bilayer. Our findings demonstrate that the nature of VASP localization is decisive for its function. The up-concentration based on VASP’s affinity to actin during polymerization enables it to simultaneously fulfill the function of an elongator and a bundler.
INTRODUCTION
In a cell, a wide variety of different actin cytoskeleton structures are necessary to uphold morphological shape, motility, or force transmission (Blanchoin et al., 2014). Actin is predominantly organized into branched mesh works or in aligned filament bundles (Blanchoin et al., 2014). The latter can be found in filopodia, cilia, or stress fibers (Bartles, 2000; Faix et al., 2009). Tight control over the polymerization or bundling of actin by actin-binding proteins is necessary for enabling the cell to dynamically build or maintain actin bundles (Rottner et al., 2017). Cross-linking proteins like α-actinin or fascin can bind formed F-actin filaments into long and stable filament bundles (Schmoller et al., 2011; Winkelman et al., 2016), whereas the long filaments are formed by the continuous barbed-end polymerization of actin (Chesarone and Goode, 2009; Siton-Mendelson and Bernheim-Groswasser, 2017). The most prominent barbed-end elongators are the formin and the Ena/VASP (vasodilator-stimulated phosphoprotein) family of proteins.
VASP consists of a C-terminal coiled-coil domain which forms homotetramers. The actin elongation is facilitated by an F-actin binding domain (FAB) and a G-actin binding domain (GAB) (Krause et al., 2003). Like the formin family, VASP is also able to incorporate profilin bound actin with its proline-rich intrinsically disordered region (IDR) (Kursula et al., 2008; Winkelman et al., 2014). This enables VASP to elongate filaments, whereas spontaneous polymerization or branched actin structures are inhibited by profilin (Rotty et al., 2015). The combination of GAB as well as binding of F-actin enables VASP to both elongate actin filaments and bundle filaments (Winkelman et al., 2014). Thus VASP can be found localized at multiple different cellular actin structures (Benz et al., 2009; Barzik et al., 2014; Gateva et al., 2014; Tojkander et al., 2015). Control over the localization of VASP within cells relies on different proteins being able to bind to the N-terminal EVH1 domain of VASP.
From a cytosolic distribution, VASP can be recruited toward the membrane by the I-BAR protein IRSp53 (Vaggi et al., 2011; Disanza et al., 2013) or lamellipodin (Barzik et al., 2014; Hansen and Mullins, 2015), which leads to enhanced lamellipodium speed and filopodia formation. IRSp53 binds the VASP IDR via SH3 domain, and lamellipodin binds to the EVH1 domain with FPPPP-motifs (Renfranz and Beckerle, 2002). Apart from decorating stress fibers in the cell, VASP plays a critical role in focal adhesions and adherens junctions as well (Krause et al., 2003; Zaidel-Bar et al., 2007; Horton et al., 2015). It is especially involved in the formation and maturation of the adhesion complex (Horton et al., 2015; Tojkander et al., 2015; Livne and Geiger, 2016; Damiano-Guercio et al., 2020). In these multiprotein complexes, there are several proteins present with FPPPP-motifs to recruit VASP (Renfranz and Beckerle, 2002). Zyxin (Hoffman et al., 2006; Cheah et al., 2021), palladin (Gateva et al., 2014), and vinculin (Reinhard et al., 1996) are all situated at the actin-regulating area in the focal adhesion complex and can localize VASP to the forming actin filaments at the adhesion sites (Kanchanawong et al., 2010). Therefore VASP can in biological systems be recruited by multiple different proteins from the cytosol toward the membrane to exert its function on actin.
As a model for clustered barbed-end elongation in filopodia, VASP bound to a solid support elongates nonbundled actin away from the solid surface in a capping protein (CP), a barbed-end polymerization inhibitor, resistant manner (Breitsprecher et al., 2008; Siton and Bernheim-Groswasser, 2014; Winkelman et al., 2014; Brühmann et al., 2017). This is in contrast with its behavior in solution, where it forms thin and unconnected bundles of actin filaments (Hüttelmaier et al., 1999; Barzik et al., 2005; Schirenbeck et al., 2006). Additionally, VASP-mediated polymerization in solution is mostly inhibited by the presence of CP (Schirenbeck et al., 2006; Breitsprecher et al., 2008). VASP bound to a solid support is unable to reconstitute the bundling effect or the mobility expected for VASP in a cellular context. In a localization method incorporating these properties, VASP anchored to fluid lipid bilayers induces the formation of a network of curved and connected actin bundles (Bleicher et al., 2021). It remains unresolved how the localization of VASP on a bilayer determines this effect on the actin structure formation.
Here we demonstrate that the attachment of VASP to a fluid substrate enables the simultaneous elongation, capturing, and alignment of growing barbed ends along existing filaments. Wide bundles are formed by an alignment via polymerization with continued actin elongation at the bundle. The fluidity of the lipid bilayer is crucial for VASP accumulation due to its affinity to actin filaments. On a less mobile lipid monolayer, polymerization is hindered by the slower mobility of VASP, so no bundling occurs. The accumulation of a VASP-enriched phase on bilayers in turn leads to a reorganization within the lipid bilayer by the exclusion of unbound lipids underneath the emerging actin bundles.
RESULTS
VASP forms bundles by capturing and aligning individual filaments
Mobile anchoring of VASP to a surface was achieved by binding His-tagged VASP to a supported lipid bilayer (SLB) via nickel-nitrilotriacetic acid (Ni-NTA) lipids. Using total internal reflection fluorescence (TIRF) microscopy, we observed the formation of a connected bundled meshwork during actin polymerization by using actin monomers labeled with Atto488 (Figure 1A). To ensure polymerization only occurs localized to the bilayer, profilin and CP were added to inhibit bulk actin polymerization (Dürre et al., 2018). Indeed, in absence of VASP on the bilayer, no polymerization was visible under the experimental conditions (Supplemental Figure S1, A–C).
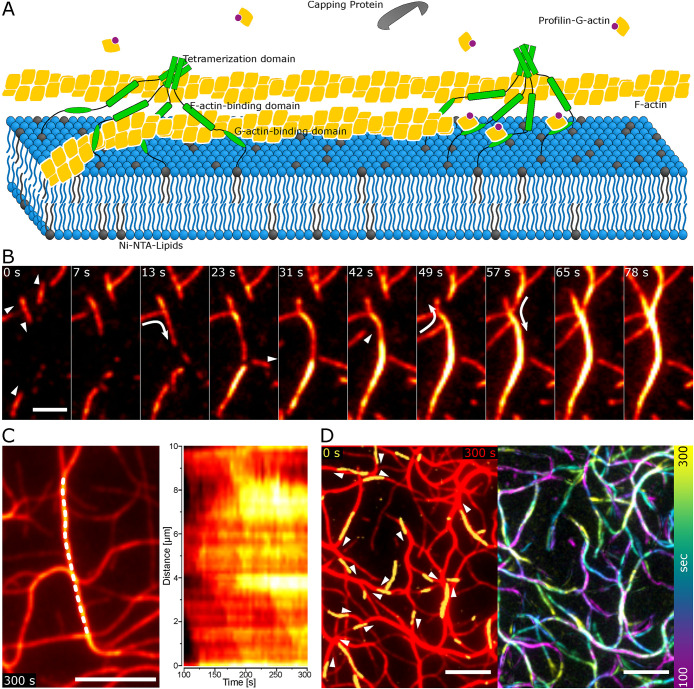
FIGURE 1:
Alignment during polymerization of actin elongated by VASP. (A) Experimental setup of actin polymerized by VASP on supported lipid bilayers. Actin is visualized with 12.5% of actin labeled with Atto488. (B) TIRFM images show actin filament alignment to bundles by 50 nM VASP (scale bar 2 µm). The triangles indicate the direction of polymerization and the arrows display alignment events to a bundle. (C) Kymograph (right) of a newly formed bundle marked by the dashed line at 5 min of polymerization in the left panel (scale bar 5 µm) shows mixed orientation of filaments during polymerization. (D) The first frame (yellow) and the direction of polymerization indicated by the triangles, overlaid over the network at 5 min (red) on the left and the temporal color-coded actin intensity increase on the right, illustrate the bundle network formation from actin seeds and subsequent elongation at the existing filaments (scale bar 5 µm).
To unravel the mechanism of the formation of actin bundles on a single filament level, we incubated the lipid bilayers with a low concentration of VASP (50 nM). Growing actin filaments converge on existing filaments, align, and continue elongating. This can best be seen by the subsequent increase in actin intensity on the prior filament after alignment of the growing filaments, as indicated by the arrows in the direction of alignment (Figure 1B; Supplemental Video S1). The arrows indicating the polymerization direction illustrate the elongation of filaments in both directions on a bundle. This apolarity is further demonstrated by a kymograph of the actin intensity on a filament, where the slopes of the increase of actin intensity on the bundle proceed both upward and downward on the filament (Figure 1C). Therefore there is no preferential orientation within the emerging bundles. By overlaying the initially bound filaments with their direction of polymerization and the resulting network at 5 min, it can be seen that the network geometry results from initial filament seeds being elongated and converging upon collision with other filaments. Subsequent elongation then proceeds on the existing actin network. This elongation on the existing filaments is further exemplified by color coding the actin intensity increase over time (Figure 1D). The minimal reorientation of the initial filaments compared with the network at 5 min further indicates minimal force being exerted on the filaments during the alignment process. The random orientation of seeds, followed by polymerization-induced alignment along existing filaments, leads to a broad distribution of filament numbers per bundle.
Movie S1.
Polymerization of labeled actin by 50 nM VASP on a supported lipid bilayer. Scale bar is 5 μm.
Bundle width increases with VASP concentration
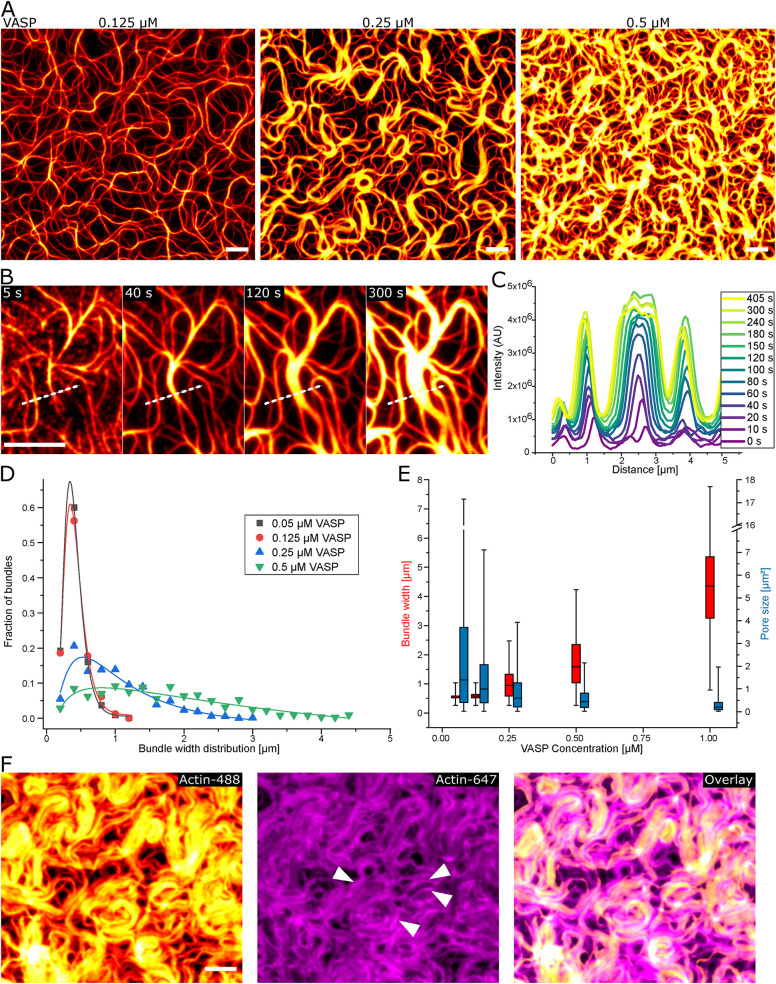
A drastic increase in actin bundle diameter in two dimensions (2D) proportional to the VASP concentration was observed when incubating higher VASP concentrations (Figure 2A; Supplemental Figures S1F and S2, A–E). This result is in contrast with the bundles formed in bulk at comparable VASP concentrations, as these are sparse, thin, and less connected (Supplemental Figure S1D). At 0.25 µM VASP on the bilayer, the observation of the growth of the bundle width is initially diffraction limited and only detectable as an increase of intensity (0–80 s). Afterwards, the bundles grow significantly in thickness up to 3 µm within minutes (Figure 2, B–D). The diffraction limit leads to very small differences in bundle width being observable between 0.05 and 0.125 µM VASP, whereas changes in the pore size through a more interconnected network are already observable (Figure 2, A and D). However, at higher VASP concentrations, the width noticeably increases at 0.25 and 0.5 µM VASP, up to almost complete decoration of the bilayer with actin at 1 µM of VASP (Figure 2, A and D; Supplemental Figure S2, E and G). The complete decoration at 1 µM VASP then prohibited reliable determination of individual bundle widths. The variance in bundle thickness increases at higher VASP concentration as well, showing a more heterogeneous width distribution, while the pore size of the bundle network steadily decreases (Figure 2, A, D, and E). In contrast, varying the actin concentration has comparably little effect on the bundle width (Supplemental Figure S3). Using actin at a concentration of 4 µM, which is double the amount compared with the general conditions, actually leads to a decrease in median bundle width. When a high actin concentration was used and the CP and profilin concentration was kept constant, the actin polymerization kinetics were enhanced (Supplemental Figure S3E). Presumably, the faster polymerization and slightly lower CP to actin ratio leads to more VASP-independent polymerization and thus elongating filaments, which are not bound to the bundles (Supplemental Figure S3, D–F).
FIGURE 2:
Bundle thickening by elongation at the bundle borders. (A) On supported lipid bilayers, actin bundles increase in size in a VASP concentration (0.125, 0.25, 0.5 µM)-dependent manner. Continuous growth of filaments at existing bundles is visible, with swelling of bundles at 0.25 µM VASP (B) and the corresponding intensity profile (C) indicated by the dashed line in (B). (D) Histogram of the bundle width distribution at different VASP concentration with a log-normal fit of the distribution. (E) Box plot of bundle width and pore size showing the median, 25–75th percentile range as boxes and 1–99th percentile range as whiskers. (F) At 1 µM VASP, the exchange of the actin-Atto488 to actin-Atto647 (0.5 µM) after 3 min exhibits the incorporation at the sides of big bundles (indicated by arrows). All scale bars are 5 µm.
The anchoring of the filaments and the capture of the barbed ends by VASP bound to the bundles leads to further processive elongation (Pasic et al., 2008; Winkelman et al., 2014). Continued polymerization supposedly is guided by the existing bundle and thus leads to the thickening of the bundles. This effect is slightly enhanced by the presence of CP. Barbed ends growing upward or away from VASP at the bundles can be capped by CP (Trichet et al., 2008). Without CP, filaments can freely polymerize between VASP processive elongation runs. The free polymerization is not bound to the surface, so it does not contribute to the bundling mechanism. Given that VASP is a potent actin polymerase and most of the polymerization is mediated by VASP, the network morphology only marginally changes, with a slight increase of small filaments polymerizing away from the thick bundles (Supplemental Figure S4, A and C).
This mechanism of continued polymerization at the bundles causes a few larger bundles having enhanced growth at their sides and a more heterogeneous width distribution. At 1 µM VASP concentration, it could be shown that the polymerizing filaments are being incorporated at the bundle sides by exchanging the polymerization mix to one with a differently labeled actin (0.5 µM actin-Atto647N) after 3 min. Filaments elongating from actin-Atto647N preferentially grow at the external border of pre-existing bundles, illustrating that the bundle width increases through extended actin polymerization at the bundle edge (Figure 2F). This result further highlights the fact of polymerization being limited in 2D to the surface by way of VASP continuously polymerizing the filaments guided to the sides of bundles.
Effect of tetramerization on bundles
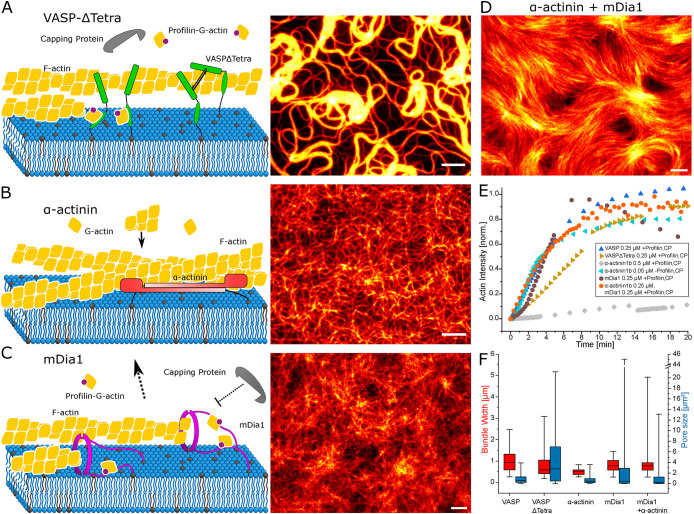
This polymerization-mediated bundling stands in contrast with bundling by traditional cross-linkers, which bind together filaments with multiple FAB domains. Since tetrameric VASP can bind multiple actin filaments, we wanted to determine if multimerization is required for bundle formation. To this end, we use monomeric VASP (VASPΔTetra) as a membrane anchor and elongator. In solution VASPΔTetra is unable to polymerize actin, whereas bound to a solid support it behaves like tetrameric VASP (Breitsprecher et al., 2008). On the fluid membranes it was therefore unclear if VASPΔTetra can polymerize actin or induce any bundling. However, the monomeric VASP was still able to form thick actin bundles. This would be in accordance with the purely polymerization-driven bundle formation model (Figure 3A; Supplemental Video S2). The polymerization of actin by surface bound VASP is solely dependent on the local concentration of VASP-GAB and FAB domains (Brühmann et al., 2017). In the case of VASPΔTetra even without multimerization, it accumulates on the slowly forming actin filaments. Thus there are enough actin-binding domains from unconnected VASP molecules next to each other to enable elongation. Compared with tetrameric VASP, this is demonstrated by slower polymerization kinetics but a similar bundle morphology after polymerization (Figure 3, A and E). Although the median bundle width of 0.584 µm is smaller than for tetrameric VASP (0.942 µm), the resulting bundle thickness is more heterogeneous with a larger variation in width and pore size. This leads to some bundles even exhibiting a large bundle thickness of up to 3.2 µm compared with 3 µm for tetrameric VASP (Figure 3F). However, the thick bundle segments are limited to short lengths or vortices, which also results in a larger pore size (Figure 3, A and F).
FIGURE 3:
Actin network by different elongators and cross-linkers. (A) With monomeric VASPΔTetra (0.25 µM) the network formed is more heterogeneous with filaments more clustered to single vortices or bundles. (B) The nonelongating bundler α-actinin1b only pulls down filaments to the surface without bundling. To be able to visualize and analyze filaments, a concentration of 0.05 µM of α-actinin1b is shown. CP and profilin are emitted from α-actinin1b experiments to enable actin polymerization. C) The nonbundling elongator mDia1 (0.25 µM) is unable to form large bundles or an actin network. (D) mDia1 (0.25 µM) and α-actinin1b (0.25 µM) together on a bilayer form a high-density nematic field. (E) Kinetics of actin polymerization shown by measured actin intensity with TIRFM. To compare α-actinin1b with VASP and mDia1, different concentrations with or without profilin and CP are shown. (F) Box plot of bundle width and pore size showing the median, 25–75th percentile range as boxes, and 1–99th percentile range as whiskers. All scale bars are 5 µm.
Movie S2.
Polymerization of labeled actin by 0.25 μM VASPΔTetra on a supported lipid bilayer. Scale bar is 5 μm.
This width distribution can be attributed to the fact that VASPΔTetra requires a high local concentration of VASP to be able to polymerize actin. On larger bundles, the accumulated VASPΔTetra is able to track and elongate actin-barbed ends. The maximal bundle width being larger than for tetrameric VASP gives insight into the probability of polymerizing filaments being polymerized at the larger bundle at intersecting filaments. The high dynamic local concentration of VASP at the larger bundle increases the likelihood that the growing filament is processively elongated and thus aligned to the bundle. Given that single VASPΔTetra molecules are not able to bind and elongate a growing filament, VASPΔTetra is more dependent on the effect of a high local concentration at large bundles. Therefore it is less likely for filaments to polymerize away from the bundles. The result is the observed formation of isolated thick bundle segments.
Elongation-mediated bundling depends on VASPs unique dynamic actin binding properties
To further prove that polymerization is the essential component in bundle formation, the cross-linker α-actinin1b was used to mimic static bundling without the ability of actin elongation. Even at 0.5 µM α-actinin1b, almost no actin polymerization was visible. Actin polymerization kinetics similar to VASP were observable when profilin and CP were omitted (Figure 3E). The α-actinin1b pulls down F-actin polymerizing by itself from the bulk solution to the membrane, with indiscriminate orientation and no observable bundling. This leads to complete decoration of the bilayer with actin. As a result, a lower α-actinin1b concentration on the bilayer (0.05 µM) had to be used in order to be able to quantify the width of the actin filaments. Polymerization occurs with filaments crossing each other and no visible alignment (Figure 3, B and E; Supplemental Video S3). In a similar way, prepolymerized actin filaments are pulled down by VASP without elongating and bundling the filaments (Supplemental Figure S4, D and E).
Movie S3.
Actin polymerization bound to a supported bilayer by 50 nM α-actinin1b without CP or profilin present. Scale bar is 5 μm.
This in turn raised the question of whether barbed-end elongation without FAB might be enough to form the observed bundles. To test this hypothesis, 0.25 µM of the formin mDia1 was incubated to the membrane instead of VASP. mDia1 is able to polymerize filaments, but these are thin and not connected to one another (Figure 3, C, E, and F). Since mDia1 only binds the barbed end of F-actin during polymerization, the filaments are free to diffuse away from the membrane. This is in contrast to when additional depleting agents force the filaments to the bilayer (Yadav et al., 2019). Additionally, the CP in solution competes with mDia1 for the barbed end, thus causing filaments to detach from the bilayer. The result is an entangled actin mesh fluctuating above the bilayer after polymerization stalls out, as can be seen by the actin intensity decrease in TIRF after 6 min (Figure 3D; Supplemental Video S4).
Movie S4.
Actin polymerization by 0.25 μM mDia1 bound to a supported bilayer. Scale bar is 5 μm.
Combining 0.25 µM mDia1 and 0.25 µM α-actinin1b on the membrane is also not sufficient for forming actin bundles equivalent to those observed with VASP. In these conditions, high actin polymerization bound to the membrane is visible, with no filaments diffusing off (Figure 3, D and E; Supplemental Video S5). Upon collision, the filaments do not immediately align which, coupled to the fast actin polymerization, causes the filaments to be trapped in nematic defects and no continuous elongation bound to bundles. This creates a high-density nematic field on the bilayer (Figure 3, D and F), thus demonstrating that the confinement of actin in 2D through the dynamic binding of polymerizing F-actin to VASP is crucial for the polymerization-mediated alignment. The mechanism of VASP forming large bundles in 2D is based on the combination of its ability to anchor actin filaments, capture barbed ends, and processively elongate them, confirming the role of active actin polymerization and side binding for efficient bundle formation.
Movie S5.
Polymerization of labeled actin by 0.25 μM mDia1 and 0.25 μM α-actinin1b both being bound to the membrane. Scale bar is 5 μm.
VASP bundling depends on fluidity-mediated colocalization
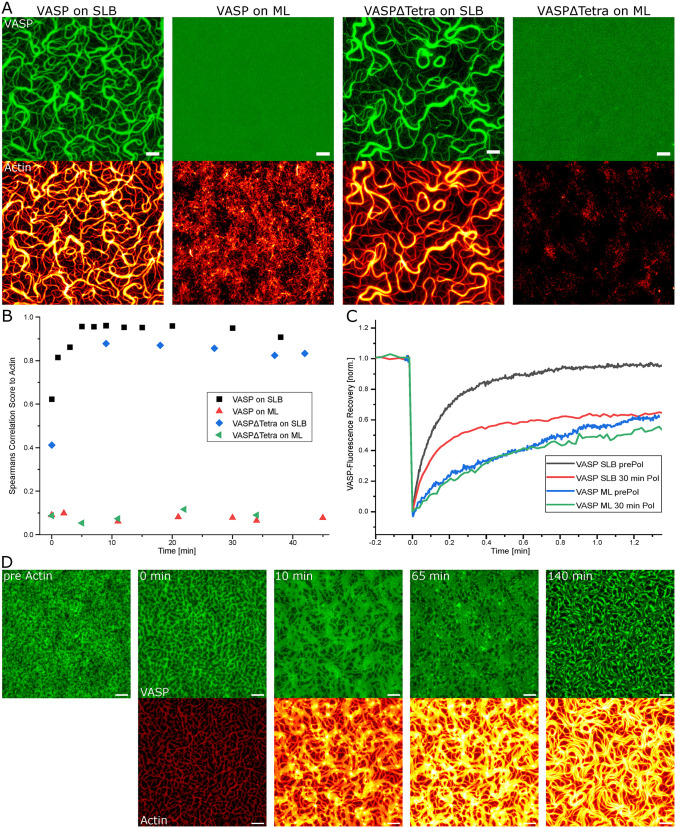
The ability of VASP to create the actin bundles with a heterogeneous width distribution is expected to depend on VASP being able to diffuse on the bilayer and accumulate on the actin filaments. To determine the localization of VASP during polymerization, we labeled it with Alexa532. Immediately upon F-actin being bound to the bilayer, VASP can be observed accumulating on single filaments, on which it was proven to be able to diffuse in a 1D direction (Hansen and Mullins, 2010). VASP up-concentrates on the forming bundles to form a uniformly dense VASP phase (Figure 4A; Supplemental Video S6). This is shown in the almost complete colocalization of VASP to actin as well and is further demonstrated by the fact that the mobile fraction of VASP on a bilayer is reduced from 0.97 to 0.65 after actin polymerization (Figure 4, B and C). VASPΔTetra also accumulates on the bundles and confirms the formation of a high local concentration necessary for the elongation of actin in a manner similar to tetrameric VASP (Figure 4, A and B).
FIGURE 4:
Colocalization of VASP to actin filaments. (A) Images of actin (12.5% Actin-Atto488) and 0.25 µM VASP-Alexa532 or VASPΔTetra-Alexa532 on SLBs or monolayers (ML) exhibit their colocalization to actin. (B) Colocalization score of labeled VASP or VASPΔTetra to actin during polymerization on SLBs or MLs. (C) Fluorescence recovery after photobleaching curves of VASP on SLBs or MLs show a reduced mobility of VASP after polymerization (30 min Pol) compared with before actin polymerization (prePol). (D) VASP forms a pattern of VASP enriched phase before actin addition. After start of polymerization (0 min) the VASP phase accumulates under the actin bundles. All scale bars are 5 µm.
Movie S6.
Polymerization of labeled actin (left) by 0.25 μM VASP-Alexa532 (middle) bound to a supported lipid bilayer, visualized by Cy5-DSPE (right). Scale bar is 5 μm.
A change in VASP distribution is observed at high VASP concentrations (≥0.5 µM) even before the addition of actin. VASP alone performs liquid–liquid phase separation into a protein-enriched phase and a phase with a low concentration on the bilayer. The total area covered by the enriched phase depends on the total VASP concentration (Figure 4D; Supplemental Figure S4, G and H). After the addition of actin, the VASP-enriched phase immediately accumulates around the nucleating filaments. The slow actin polymerization speed at 0.5 µM actin (Supplemental Figure S3E) enabled direct observation of how VASP droplets coalesce and fuse onto the slowly forming actin filaments (Supplemental Video S7). Upon stalling out of the polymerization, VASP slowly moves to the bundle edges to reform clusters. The accumulation of VASP and its mobility demonstrate the partitioning of VASP on the bilayer into an actin-associated enriched phase and an unbound dilute phase. This process depends on the mobility of VASP and, therefore, on the fluidity of the bilayer.
Movie S7.
VASP-Alexa532 droplet fuse together under actin bundles during polymerization of 0.5 μM actin. To visualize the dissolution of actin associated droplets the long time acquisition is speed up compared to beginning of the polymerization. Scale bar is 5 μm.
A lipid monolayer was used as a substrate to bind labeled VASP in order to confirm the role of mobile anchoring of VASP for bundle formation. The mobility of VASP was reduced by a factor of 4 compared with a lipid bilayer anchorage, as determined by fluorescent recovery after photobleaching (FRAP) experiments of the labeled VASP. The recovery on monolayers shows a recovery time of t1/2 = 0.399 min, as compared with the faster recovery time on bilayers with t1/2 = 0.102 min. VASP-mediated actin polymerization needs to polymerize against a diffusion barrier of the friction within the monolayer. This leads to a slowdown of VASP-mediated polymerization to a point where free polymerization is faster and thus more favored. The freely polymerizing barbed ends are then susceptible to capping by CP prior to the diffusive capture by VASP. The decreased mobility of VASP on the monolayer thus results in abrogation of actin polymerization, and no bundle formation is observable. VASP does not colocalize with the actin filaments anymore (Figure 4, A and D). This observation is in agreement with the observed minimal change of the fraction of mobile VASP (0.61 to 0.57 after polymerization). VASP is still able to accumulate on larger prepolymerized actin filaments (Supplemental Figure S4I), which show that the reduced diffusion prevents the VASP-mediated polymerization but not the VASP accumulation on filaments. Given that alignment is dependent on VASP-mediated polymerization and no polymerization is visible, VASP on monolayers is only able to pull down actin seeds from the bulk solution, which barely elongate (Figure 4A). Even by omitting CP, similar to the nonelongating cross-linker α-actinin1b without profilin and CP, the network consists of short filaments which are crossing each other with no alignment (Supplemental Figure S4, B and C).
Formation of a VASP-enriched phase drives lipid reorganization
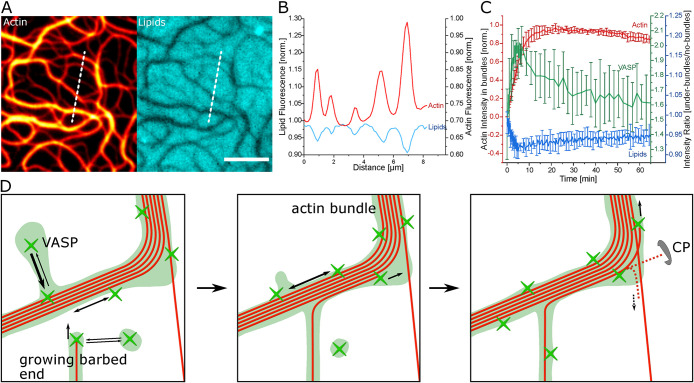
Having established the importance of the mobility and resulting accumulation of VASP underneath the bundles, we then explored how this outcome affects the lipid bilayer. A significant decrease in intensity underneath the actin bundles is visible by observing fluorescently labeled lipids in the membrane (Figure 5, A and B). This negative colocalization to actin proceeds in direct opposition to the accumulation of VASP at the filaments (Figure 5C; Supplemental Video S6). VASP is tethered to the membrane via Ni-NTA lipids, which consequently accumulate underneath the actin bundles. This strong accumulation leads to the volume exclusion of other lipids including the fluorescent lipids. The displacement of fluorescent lipids directly coupled to actin polymerization is visible when observing the intensity underneath the bundles. After the initial displacement coupled to actin polymerization observed under a bundle, the lipid intensity relaxes to around half of initial intensity after 60 min, reciprocally to the VASP intensity under the bundles (Figure 5C). VASP has a higher affinity toward barbed ends than F-actin sides at high actin monomer concentrations (Hansen and Mullins, 2010) and therefore accumulates more during active polymerization. VASP within bundles is less diffusive and equilibrates to forming clusters at actin bundle sides after polymerization (Figure 5C). This result demonstrates that the polymerization-driven up-concentration of VASP is able to reorganize the underlying lipid layer and locally effect the bilayer composition.
FIGURE 5:
Reorganization of lipid bilayer through VASP accumulation. (A) Lipid fluorescence (Texas Red-DHPE) inversely correlates with actin bundles formed with 0.125 µM VASP. (B) Intensity profile of the bundle cross-section indicated in by the dashed line in (A). (C) Lipid intensity (Texas Red-DHPE) changes reciprocally to the actin intensity during polymerization at 0.25 µM VASP. Afterward it normalizes more toward the values at the start of polymerization inversely to the way the VASP-Alexa532 intensity behaves under bundles. (D) Schematic of the bundle formation mechanism. On the fluid bilayer, patches of VASP can freely diffuse to actin bundles and alongside the bundles. F-actin bundles have more sites to bind VASP and therefore a higher dynamic VASP concentration than single filaments. Barbed ends growing toward bundles are captured and aligned by localized VASP. Filaments are most likely to be continuously polymerized by the higher VASP concentration on the larger bundles, whereas actin filaments polymerizing away from VASP are inhibited by CP. This leads to an enhanced thickening of larger bundles. All scale bars are 5 µm.
DISCUSSION
The effect of VASP on actin structure formation is highly dependent on the nature of the anchoring of VASP to a substrate. Whereas binding of VASP onto fluid lipid bilayers enables the simultaneous elongation and bundling of actin filaments, immobilized anchoring on nonfluid substrates prevents its bundling activity, and only its elongation effect is detectable. On lipid bilayers, the growing filaments are aligned and bound alongside existing bundles due to capture of growing filaments in 2D and the inhibition of VASP independent polymerization (Figure 5D). It is unlikely that the effect of longer processive elongation of individual VASP molecules on lagging barbed ends in fascin bundles (Winkelman et al., 2014; Harker et al., 2019) plays a role in the presented bundle growth, since VASP and fascin achieve a different filament spacing (Breitsprecher et al., 2008; Winkelman et al., 2016). The enhanced polymerization on the bundles in this case is achieved by a high concentration of VASP on the filaments facilitating the immediate recapture of the barbed end after the processive elongation run of an individual VASP (Hansen and Mullins, 2010). In addition, the adjacent VASP molecules under the bundle can also accelerate polymerization by supplying actin monomers to the barbed end using the high flexibility and reach of the intrinsically disordered region (Brühmann et al., 2017). The transient binding and large distance between actin binding domains should allow the movement of VASP between the filaments in a bundle. This leads to larger bundles having a larger area for VASP acquisition and therefore a higher propensity to bind VASP from the fluid bilayer than a single actin filament. The high local VASP concentration then leads to the capture of barbed ends at the bundles and increases the probability of continuous processive elongation at the large bundles. In effect, this results in a positive feedback loop of increased VASP and actin filament accumulation at thicker actin bundles. Combined with the competition of elongating ends for actin monomers, larger bundles with a heterogeneous width distribution are formed.
The observed growth process resembles the actin alignment recently found for filaments propelled by myosin II motors bound to lipid bilayers. These active transport processes lead to a polar and antipolar alignment of the individual filaments according to their collision angle (Sciortino and Bausch, 2021). For the bundle and vortices formation, the volume exclusion and minimal force between the colliding filaments was essential, even though this was achieved by active propulsion of filaments instead of the guided polymerization in the setup presented here (Jung et al., 2020; Sciortino and Bausch, 2021). Differences in the structure formation process between monomeric and tetrameric VASP seem to be reminiscent of the structure differences found in simulations by differences in steric exclusion strength between filaments (Jung et al., 2020). The monomeric VASP acts analogous to weaker steric exclusion, so there is less chance of filaments diverging from large bundles, creating a more heterogeneous width distribution than tetrameric VASP. The network morphology observed in this case is dependent on continuous elongation, which leads to equivalent but static structures in comparison with the active propelling of filaments by molecular motors in the simulations.
In our setup, it was striking, that VASP is already phase separating on fluid membranes by itself in the absence of actin filaments. The self-affinity leading to the liquid–liquid phase separation of VASP could be based on an interaction between its EVH1 domain and its proline-rich domain. The EVH1 binds the proline-rich FPPPP motif. Although no FPPPP motif is located in the proline-rich domain of VASP, the high concentration of prolines could form a weakly bound nonconventional complex with the EVH1 domain (Acevedo et al., 2017). Moreover, the disordered region of VASP could contribute to a mechanism for minimizing surface area of the protein cluster, as shown for other intrinsically disordered proteins (van der Lee et al., 2014). The formation of the protein-rich phase functions as a nucleation area for the actin filaments, which might turn out to be essential for the localized polymerization of actin filaments within cells. Phase separation was shown to be involved in nucleating actin in cells for various activators of Arp2/3 in the formation of branched actin networks (Dürre et al., 2018; Case et al., 2019; Ditlev et al., 2019; Yan et al., 2021) but so far has not been observed for proteins forming actin bundles.
The subsequent growth-induced accumulation of VASP along the actin filaments induces the up-concentration of VASP-bound lipids. Whereas biotin binding of actin induced a phase separation in supported lipid bilayers on mica (Honigmann et al., 2014), in our study the transient binding of VASP to actin is able to exert an effect on the lipid composition even on a glass surface-supported bilayer. This is notable, because the friction of lipid bilayers on glass surfaces normally does prevent phase separations in lipid bilayers (Goodchild et al., 2019), thus indicating the strong effect of the VASP to actin binding on the lipid organization. These actin growth kinetics-guided phase separation appears to be an efficient coupling of cytoskeletal structure formation and lipid phase separation-induced local lipid up-concentration, which ultimately could induce further signaling and complex formation, as involved in focal adhesion complex formation. Indeed, in vivo the highest accumulation of VASP on actin filaments has been observed in focal adhesions and dorsal stress fibers (Reinhard et al., 1992; Bear and Gertler, 2009). This accumulation is also associated with an important biological function, since knockdown of VASP leads to a decrease in the size and strength of the dorsal stress fibers and focal adhesions (Tojkander et al., 2015; Damiano-Guercio et al., 2020). VASP is one of the most mobile proteins in focal adhesion complexes (Lavelin et al. 2013; Stutchbury et al., 2017; Legerstee et al., 2019). This is consistent with the bundle formation presented herein, in which the VASP mobility is crucial for efficient bundling. The reorganization of the lipids by the VASP bundles suggests that the mechanism may lead to up-concentration and reorganization of the proteins recruiting VASP to the membrane, for example, zyxin, vinculin, or IRSp53.
In contrast with conventional cross-linkers like fascin and α-actinin (Claessens et al., 2008; Falzone et al., 2012) or the recently observed bundling system of talin and vinculin on lipid membranes (Kelley et al. 2020), the polymerization-driven bundle formation mechanism presented in this study forms large bundles at comparatively low concentrations of the bundling protein. These mechanisms, as well as the formin AFH1, which is able to bind at the sides of F-actin as well and continuously nucleates new filaments on the existing filament (Michelot et al., 2006), rely on filament zippering through filament fluctuations to create bundles. Therefore the enlargement of bundles is limited by the reduced Brownian motion of large bundles. In contrast, the feedback mechanism of VASP on large bundles reinforces bundle growth. Furthermore, VASP is able to bundle in the presence of CP and profilin, which can inhibit actin polymerization without an elongator in a cellular context. As talin and vinculin are present in focal adhesions as well (Horton et al., 2015), this suggests an interplay between the elongation and bundling of actin by VASP and the binding together of formed filaments by α-actinin or talin/vinculin.
The mechanism of combined elongating, aligning, and bundling of actin filaments by VASP is unique by virtue of its multiple actin binding sites, self-affinity, and coupling to fluid membranes. The in vitro reconstitution presented makes it possible to identify the interplay of the fluidity of lipid bilayers and the biochemical activity of VASP.
MATERIAL AND METHODS
Request a protocol through Bio-protocol.
Protein purification
Rabbit skeletal muscle actin was purified from acetone powder (Spudich and Watt, 1971). No rabbits were directly involved in this study. Monomeric actin was stored at 4 °C in G-buffer (2 mM Tris, 0.2 mM ATP, 0.2 mM CaCl2, 0.2 mM DTT [dithiothreitol] and 0.005 % NaN3, pH 8.0). For fluorescent actin, surface-exposed lysines were labeled with Atto-488 (Jena Bioscience, FP-201-488) or Atto-647N NHS-ester (Jena Bioscience, FP-201-647N).
All recombinantly purified proteins were expressed in Escherichia coli BL21 CodonPlus DE3-RIPL (Agilent Technologies, 230280).
Human VASP containing the high affinity GAB of Dictyostelium discoideum VASP was expressed as a His(6x)–tagged protein in pET28a vector (Novagen, 69864) (Breitsprecher et al., 2011). Expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 21 °C for 12 h. The bacteria were harvested and lysed by freeze–thaw and French pressing in lysis buffer containing 30 mM HEPES, pH 7.4, 150 mM KCl, 2 mM EGTA, 20 mM imidazole, 1 mM DTT, 5 % (vol/vol) glycerol, 40 µg/ml lysozyme, one tablet of cOmplete protease inhibitor (Roche, 4693132001), and 2 U/ml Benzonase (Sigma, E1014). The protein was subsequently purified from bacterial extracts by affinity chromatography using cOmplete His-Tag purification resin (Roche, 5893801001) and standard procedures. The eluted protein fractions were further purified by size exclusion chromatography using a Superdex 200 16/600 column (GE Healthcare, GE28-9893-35) and a Bio-Rad NGC chromatography system. To label VASP, it was incubated with Alexa532-maleimide (Invitrogen, A10255), and the dye was removed with a NAP-25 size-exclusion chromatography column (GE Healthcare, 17-0852-02). The purified His-VASP was concentrated and stored at –20 °C in storage buffer (30 mM HEPES, pH 7.4, 150 mM KCl, 1 mM DTT, 50 % [vol/vol] glycerol) for later measurements. VASP with deletion of the tetramerization domain (VASPΔTetra) was created by inserting a stop codon instead of proline at amino acid 270 using QuikChange Lightning site-directed mutagenesis (Agilent Technologies, 210518) and purified the same way as VASP.
CP was expressed as a heterodimer of mouse α1- and human β2 subunit cloned into pRFSDuet-1 (Novagen, 71341). Expression was induced with 0.5 mM IPTG and incubation at 26 °C overnight. Bacteria were harvested and lysed by French pressing in a 20 mM Tris, pH 8.0, 10 mM imidazole, 250 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5 tablet of cOmplete protease inhibitor, 5 % glycerol, and 2 U/ml Benzonase buffer. After centrifugation, the supernatant was incubated with Anti-FLAG M2 affinity gel (Sigma, A2220) at 4 °C for 90 min. After washing, 140 µl FLAG peptide (25 mg lyophilized powder per 1.4 ml buffer) were added and incubated at 4 °C for 1 h. The beads were spun down and the supernatant was dialyzed overnight at 4 °C against 10 mM Tris, pH 8.0, 50 mM KCl, and 1 mM DTT. The protein was then frozen in liquid nitrogen and stored at –80 °C.
Expression of Ca2+-independent human α-actinin1b was induced with 1 mM IPTG at 26 °C for 12 h. The pellet was resuspended in lysis buffer containing 30 mM Tris, pH 8.0, 250 mM NaCl, 20 mM imidazole, 5 % glycerol, 2 mM DTT, 40 µg/ml lysozyme, 0.5 tablet of cOmplete protease inhibitor, and 2 U/ml Benzonase. The supernatant after lysis was loaded on a HisTrap-FF column (GE Healthcare, 17525501). After washing, the proteins were eluted with 500 mM imidazole. The protein was then further purified with a Superdex200 Increase 10/300 size exclusion column (GE Healthcare, 28990944) with a buffer consisting of 20 mM TES, pH 8.0, 150 mM KCl, and 1 mM DTT. The protein was then concentrated, frozen in liquid nitrogen, and stored at –80 °C.
Mouse profilin 2a was expressed as a glutathione S-transferase (GST) fusion protein. The protein was expressed using auto-induction medium: 928 ml ZY (12 g/l tryptone, 24 g/l yeast extract), 1 ml 1 M MgSO4, 20 ml 50× 5052 (0.5 % glycerol, 0.05 % D-glucose, 0.2 % α-lactose) and 50 ml 20× NPS (0.5 M (NH4)2SO4, 1 M KH2PO4, 1 M Na2HPO4). The bacteria were incubated at 37 °C for 6 h and further incubated at 15 °C for 60 h. The cells were resuspended in 50 mM Tris, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 40 µg/ml lysozyme, one tablet of cOmplete protease inhibitor, and 2 U/ml Benzonase and then were lysed by french pressing. The supernatant was incubated with 3 ml of glutathione sepharose 4B (GE Healthcare, GE17-0756-01) at 4 °C for 2 h. After washing, the protein was cleaved off using 15 µl of PreScission protease (Cytiva, GE27-0843-01) overnight at 4 °C. The eluted protein was then dialyzed overnight at 4 °C against 20 mM Tris, pH 7.0, 150 mM NaCl, 1 mM DTT, flash-frozen in liquid nitrogen, and stored at –80 °C.
The murine formin mDia1 in a pET28a vector was expressed using auto-induction medium and purified using Ni-NTA resin, as previously described (Bleicher et al., 2020).
Precleaning of glass slides
The coverslips for the supported lipid bilayers were first cleaned by sonication for 30 min in 3 M NaOH. After washing with double-distilled H2O, the coverslips were etched in piranha solution (2:1 H2SO4/H2O2) for 5 min. The coverslips were then extensively washed and stored in double-distilled H2O. The glass slides were cleaned by sonication in 2 % Hellmanex III (Hellma, Z805939-1EA) for 30 min, washed in double-distilled H2O, and stored in ethanol. The cleaned coverslips and glass slides were used within 1 wk after cleaning.
Formation of the supported lipid bilayers
The lipid mix consisted of 87.45 % egg-PC (L-α-phosphatidylcholine from chicken egg) (Sigma, P3556), 10 % DOGS-Ni-NTA (1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt) (Avanti Polar Lipids, 790404), 2.5 % DOPE-PEG2000 (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]) (Avanti Polar Lipids, 880130P), and 0.05 % DHPE-Texas Red (1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine) (Thermo Fisher Scientific, T1395MP). For the measurements with labeled VASP, the DHPE-Texas Red was switched out for DSPE-Cy5 (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[Cyanine 5]) (Avanti Polar Lipids, 810345C). The lipid mix was prepared to 20 mM in chloroform. The lipid mix was dried under a nitrogen stream and left under a vacuum overnight. The dried lipids were resuspended to 1 mM in phosphate-buffered saline (PBS), pH 7.4, vortexed for 30 s, and sonicated for 30 min. To obtain small unilamellar vesicles (SUVs), the solution was extruded 10 times using an Avanti Mini Extruder (Avanti Polar Lipids, 610000-1EA) with a 100 nm pore size Whatman Nuclepore membrane (Sigma, WHA111105). The SUVs were stored at 5 °C and used within 1 wk. For the bilayer formation, the precleaned slides were dried under a nitrogen stream. Immediately afterward, a flow chamber of 50 µl was created with parafilm, and a 1:5 dilution of the SUVs was incubated in the channel for 20 min at room temperature. The chamber was then washed with 40× the chamber volume with PBS. Correct bilayer formation was assessed by FRAP, and the bilayer was immediately used for assays.
Formation of monolayers
The same lipid mix used for bilayers was also used for the monolayers. 0.5 mg of the lipid mix was dried under a nitrogen stream and left under a vacuum overnight. The dried lipids were resuspended in 0.5 ml of 30 % isopropanol in ddH2O and vortexed for 30 s. The monolayer was formed in ibidi µ-slide I luer uncoated (ibidi, 80171); 100 µl of the lipid in isopropanol solution was incubated to the channel and incubated for 10 min at room temperature. The channel was then washed with 0.5 ml of ddH2O and 0.5 ml of PBS afterward. The monolayers were assessed by FRAP and immediately used for assays.
Polymerization assay
For the actin polymerization assays on bilayer or monolayer, the respective concentration of VASP, mDia1, or α-actinin1b was incubated for 10 min at room temperature in the flow channel. Unbound protein was then removed by washing with 20 times the chamber volume with PBS. The channel was then washed with 100 µl of KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 20 mM imidazole, pH 7.0). Actin polymerization proceeded in KMEI buffer with 50 nM CP, 10 µM profilin, 1 mM DTT, and 1 mM ATP. 1700 U/ml catalase (Sigma-Aldrich, C40), 26 U/ml glucose-oxidase (Sigma-Aldrich, G2133), and 36 mM glucose were used for bleach protection. Actin was added to a final concentration of 2 µM, of which 12.5% was labeled with Atto488. Directly after the addition of actin, the polymerization mix was flowed onto the bi- or monolayer and the channel was sealed with vacuum grease. Imaging of the chamber was started immediately afterward.
Imaging and data acquisition
TIRF microscopy was conducted using a 100× oil immersion objective with a numerical aperture of 1.47 on a Leica DMi8. Images were captured using a Leica Infinity TIRF HP module and an ORCA-Flash 4.0 CMOS camera (Hamamatsu, C13440-20CU). FRAPs were performed using a Leica Infinity scanner. The FRAP experiments were done by bleaching a circle with a diameter of 4.96 µm at 100% laser power.
Data analysis
Image analysis was performed using FIJI-ImageJ (Schindelin et al., 2012). Fluorescence intensity quantification was done on raw image data with the background intensity subtracted. The average and SD of five bundles were used for the intensity at the bundles. The lipid intensity was measured as an average of 10 frames per time point. For the intensity ratio of VASP or the lipids, the intensity underneath the bundles was divided by the intensity next to the bundles. For segmentation, images were corrected by pseudo-flat-field correction. Segmentation was performed with the machine-learning algorithms provided by ilastik (Berg et al., 2019). The segmented images were then used for further image analysis. The bundle width was determined as the local diameter on the distance ridge using the local thickness plugin in FIJI (Dougherty and Kunzelmann, 2007). Pore size was determined using the particle analyzer in FIJI. The data from triplicate measurements were used for the width and pore size distributions. Depending on the spacing between the filaments in the bundle facilitated by a diameter range from 28 to 70 nm of VASP, we expected a quantity of 14–40 filaments in a 1-µm-thick bundle. We verified this by integrating the intensity over the diameter of filament bundles and used the result to extrapolate the number of filaments for large bundles. This resulted in the expected number of filaments in 1-µm-thick bundles of 45 ± 12, which was in the same order of magnitude as the number calculated from the segmented diameter.
The average ratio and SD of segmented bundle area to total area were used for the decoration of the bilayer. For calculation of the colocalization, an average over 10 frames was pseudo-flat field corrected. Spearman’s rank correlation coefficient was calculated using the plugin Coloc 2 in FIJI. The FRAP data were corrected using bleach correction factor calculated from an unbleached reference region. The data were normalized using the prebleach intensity and first bleach frame (Siggia et al., 2000). The recovery curves were fitted using with the formula y = a∗(1 – e(-t/τ)) + c and the recovery half-time calculated with t1/2 = τ∗ln 2.
Data availability
The data supporting the findings of this study are available from A.R.B. upon request.
Supplementary Material
Acknowledgments
Research was supported by the Deutsche Forschungsgemeinschaft via SFB863 (B1), BA2029 14/1, and by the ERC via the project PoInt (810104). We thank Karin Vogt for the actin preparation and Monika Rusp-Post for the support during CP and profilin purification. We are grateful to Jan Faix (Cytoskeleton Dynamics, Medizinische Hochschule Hannover) for providing the plasmid for VASP and Kristina Djinovic-Carugo (Structural Biology of the Cytoskeleton, Max Perutz Labs, University of Vienna) for providing the plasmid for α-actinin. We also thank Alfredo Sciortino for helpful discussions.
Abbreviations used:
- CP
capping protein
- DTT
dithiothreitol
- FAB
F-actin binding
- FRAP
fluorescence recovery after photobleaching
- GAB
G-actin binding
- IDR
intrinsically disordered region
- IPTG
isopropyl-β-D-thiogalactopyranoside
- Ni-NTA
nickel-nitrilotriacetic acid
- PBS
phosphate-buffered saline
- SLB
supported lipid bilayer
- SUV
small unilamellar vesicle
- TIRFM
total internal reflection fluorescence microscopy
- VASP
vasodilator-stimulated phosphoprotein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0577) on July 13, 2022.
REFERENCES
- Acevedo LA, Greenwood AI, Nicholson LK (2017). A noncanonical binding site in the evh1 domain of vasodilator-stimulated phosphoprotein regulates its interactions with the proline rich region of zyxin. Biochemistry 56, 4626–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartles JR (2000). Parallel actin bundles and their multiple actin-bundling proteins. Curr Opin Cell Biol 12, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA (2005). Ena/vasp proteins enhance actin polymerization in the presence of barbed end capping proteins*. J Biol Chem 280, 28653–28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M, McClain LM, Gupton SL, Gertler FB (2014). Ena/vasp regulates mdia2-initiated filopodial length, dynamics, and function. Mol Biol Cell 25, 2604–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB (2009). Ena/vasp: Towards resolving a pointed controversy at the barbed end. J Cell Sci 122(Pt 12), 1947–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz PM, Blume C, Seifert S, Wilhelm S, Waschke J, Schuh K, Gertler F, Münzel T, Renné T (2009). Differential vasp phosphorylation controls remodeling of the actin cytoskeleton. J Cell Sci 122(Pt 21), 3954–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, et al. (2019). Ilastik: Interactive machine learning for (bio)image analysis. Nat Methods 16, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev 94, 235–263. [DOI] [PubMed] [Google Scholar]
- Bleicher P, Nast-Kolb T, Sciortino A, de la Trobe YA, Pokrant T, Faix J, Bausch AR (2021). Intra-bundle contractions enable extensile properties of active actin networks. Sci Rep 11, 2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleicher P, Sciortino A, Bausch AR (2020). The dynamics of actin network turnover is self-organized by a growth-depletion feedback. Scientific Reports 10, 6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J (2008). Clustering of vasp actively drives processive, wh2 domain-mediated actin filament elongation. EMBO J 27, 2943–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Vinzenz M, Stradal TEB, Small JV, Curth U, Dickinson RB, Faix J (2011). Molecular mechanism of ena/vasp-mediated actin-filament elongation. EMBO J 30, 456–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brühmann S, Ushakov DS, Winterhoff M, Dickinson RB, Curth U, Faix J (2017). Distinct vasp tetramers synergize in the processive elongation of individual actin filaments from clustered arrays. Proc Natl Acad Sci USA 114, E5815–E5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, Rosen MK (2019). Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah JS, Jacobs KA, Lai TW, Caballelo R, Yee JL, Ueda S, Heinrich V, Yamada S (2021). Spatial proximity of proteins surrounding zyxin under force-bearing conditions. Mol Biol Cell 32, 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL (2009). Actin nucleation and elongation factors: Mechanisms and interplay. Curr Opin Cell Biol 21, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens MMAE, Semmrich C, Ramos L, Bausch AR (2008). Helical twist controls the thickness of f-actin bundles. Proc Natl Acad Sci USA 105, 8819–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiano-Guercio J, Kurzawa L, Mueller J, Dimchev G, Schaks M, Nemethova M, Pokrant T, Brühmann S, Linkner J, Blanchoin L, et al. (2020). Loss of ena/vasp interferes with lamellipodium architecture, motility and integrin-dependent adhesion. Elife 9, e55351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Bisi S, Winterhoff M, Milanesi F, Ushakov DS, Kast D, Marighetti P, Romet-Lemonne G, Müller HM, Nickel W, et al. (2013). Cdc42 switches irsp53 from inhibition of actin growth to elongation by clustering of vasp. EMBO J 32, 2735–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Vega AR, Köster DV, Su X, Tani T, Lakoduk AM, Vale RD, Mayor S, Jaqaman K, Rosen MK (2019). A composition-dependent molecular clutch between t cell signaling condensates and actin. eLife 8, e42695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty R, Kunzelmann KH (2007). Computing local thickness of 3d structures with imagej. Microsc Microanal 13, 1678–1679. [Google Scholar]
- Dürre K, Keber FC, Bleicher P, Brauns F, Cyron CJ, Faix J, Bausch AR (2018). Capping protein-controlled actin polymerization shapes lipid membranes. Nat Commun 9, 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TE, Rottner K (2009). Filopodia: Complex models for simple rods. Int J Biochem Cell Biol 41, 1656–1664. [DOI] [PubMed] [Google Scholar]
- Falzone TT, Lenz M, Kovar DR, Gardel ML (2012). Assembly kinetics determine the architecture of α-actinin crosslinked f-actin networks. Nat Commun 3, 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateva G, Tojkander S, Koho S, Carpen O, Lappalainen P (2014). Palladin promotes assembly of non-contractile dorsal stress fibers through vasp recruitment. J Cell Sci 127(Pt 9), 1887–1898. [DOI] [PubMed] [Google Scholar]
- Goodchild JA, Walsh DL, Connell SD (2019). Nanoscale substrate roughness hinders domain formation in supported lipid bilayers. Langmuir 35, 15352–15363. [DOI] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD (2010). Vasp is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol 191, 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD (2015). Lamellipodin promotes actin assembly by clustering ena/vasp proteins and tethering them to actin filaments. Elife 4, e06585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker AJ, Katkar HH, Bidone TC, Aydin F, Voth GA, Applewhite DA, Kovar DR (2019). Ena/vasp processive elongation is modulated by avidity on actin filaments bundled by the filopodia cross-linker fascin. Mol Biol Cell 30, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC (2006). Genetic ablation of zyxin causes mena/vasp mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol 172, 771–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigmann A, Sadeghi S, Keller J, Hell SW, Eggeling C, Vink R (2014). A lipid bound actin meshwork organizes liquid phase separation in model membranes. Elife 3, e01671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton ER, Byron A, Askari JA, Ng DHJ, Millon-Frémillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D, et al. (2015). Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 17, 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S, Harbeck B, Steffens O, Messerschmidt T, Illenberger S, Jockusch BM (1999). Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein vasp. FEBS Lett 451, 68–74. [DOI] [PubMed] [Google Scholar]
- Jung W, Fillenwarth LA, Matsuda A, Li J, Inoue Y, Kim T (2020). Collective and contractile filament motions in the myosin motility assay. Soft Matter 16, 1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM (2010). Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CF, Litschel T, Schumacher S, Dedden D, Schwille P, Mizuno N (2020). Phosphoinositides regulate force-independent interactions between talin, vinculin, and actin. Elife 9, e56110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB (2003). Ena/vasp proteins: Regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol 19, 541–564. [DOI] [PubMed] [Google Scholar]
- Kursula P, Kursula I, Massimi M, Song YH, Downer J, Stanley WA, Witke W, Wilmanns M (2008). High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: The formin homology 1 domain of mdia1 and the proline-rich domain of vasp. J Mol Biol 375, 270–290. [DOI] [PubMed] [Google Scholar]
- Lavelin I, Wolfenson H, Patla I, Henis YI, Medalia O, Volberg T, Livne A, Kam Z, Geiger B (2013). Differential effect of actomyosin relaxation on the dynamic properties of focal adhesion proteins. PLos One 8, e73549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerstee K, Geverts B, Slotman JA, Houtsmuller AB (2019). Dynamics and distribution of paxillin, vinculin, zyxin and vasp depend on focal adhesion location and orientation. Sci Rep 9, 10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livne A, Geiger B (2016). The inner workings of stress fibers - from contractile machinery to focal adhesions and back. J Cell Sci 129, 1293–1304. [DOI] [PubMed] [Google Scholar]
- Michelot A, Derivery E, Paterski-Boujemaa R, Guérin C, Huang S, Parcy F, Staiger CJ, Blanchoin L (2006). A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr Biol 16, 1924–1930. [DOI] [PubMed] [Google Scholar]
- Pasic L, Kotova T, Schafer DA (2008). Ena/vasp proteins capture actin filament barbed ends. J Biol Chem 283, 9814–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Halbrügge M, Scheer U, Wiegand C, Jockusch BM, Walter U (1992). The 46/50 kda phosphoprotein vasp purified from human platelets is a novel protein associated with actin filaments and focal contacts. EMBO J 11, 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard M, Rüdiger M, Jockusch BM, Walter U (1996). Vasp interaction with vinculin: A recurring theme of interactions with proline-rich motifs. FEBS Lett 399, 103–107. [DOI] [PubMed] [Google Scholar]
- Renfranz PJ, Beckerle MC (2002). Doing (f/l)pppps: Evh1 domains and their proline-rich partners in cell polarity and migration. Curr Opin Cell Biol 14, 88–103. [DOI] [PubMed] [Google Scholar]
- Rottner K, Faix J, Bogdan S, Linder S, Kerkhoff E (2017). Actin assembly mechanisms at a glance. J Cell Sci 130, 3427–3435. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE (2015). Profilin-1 serves as a gatekeeper for actin assembly by arp2/3-dependent and -independent pathways. Dev Cell 32, 54–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TE, Schleicher M, Faix J (2006). The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci USA 103, 7694–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller KM, Semmrich C, Bausch AR (2011). Slow down of actin depolymerization by cross-linking molecules. J Struct Biol 173, 350–357. [DOI] [PubMed] [Google Scholar]
- Sciortino A, Bausch AR (2021). Pattern formation and polarity sorting of driven actin filaments on lipid membranes. Proc Natl Acad Sci USA 118, e2017047118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggia ED, Lippincott-Schwartz J, Bekiranov S (2000). Diffusion in inhomogeneous media: Theory and simulations applied to whole cell photobleach recovery. Biophys J 79, 1761–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siton-Mendelson O, Bernheim-Groswasser A (2017). Functional actin networks under construction: The cooperative action of actin nucleation and elongation factors. Trends Biochem Sci 42, 414–430. [DOI] [PubMed] [Google Scholar]
- Siton O, Bernheim-Groswasser A (2014). Reconstitution of actin-based motility by vasodilator-stimulated phosphoprotein (vasp) depends on the recruitment of f-actin seeds from the solution produced by cofilin. J Biol Chem 289, 31274–31286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem 246, 4866–4871. [PubMed] [Google Scholar]
- Stutchbury B, Atherton P, Tsang R, Wang DY, Ballestrem C (2017). Distinct focal adhesion protein modules control different aspects of mechanotransduction. J Cell Sci 130, 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Husain A, Krishnan R, Lappalainen P (2015). Generation of contractile actomyosin bundles depends on mechanosensitive actin filament assembly and disassembly. Elife 4, e06126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trichet L, Sykes C, Plastino J (2008). Relaxing the actin cytoskeleton for adhesion and movement with ena/vasp. J Cell Biol 181, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaggi F, Disanza A, Milanesi F, Di Fiore PP, Menna E, Matteoli M, Gov NS, Scita G, Ciliberto A (2011). The eps8/irsp53/vasp network differentially controls actin capping and bundling in filopodia formation. PLOS Computational Biology 7, e1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, et al. (2014). Classification of intrinsically disordered regions and proteins. Chemical Reviews 114, 6589–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, Kovar DR (2014). Ena/vasp enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled f-actin networks with fascin. Proc Natl Acad Sci USA 111, 4121–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Suarez C, Hocky GM, Harker AJ, Morganthaler AN, Christensen JR, Voth GA, Bartles JR, Kovar DR (2016). Fascin- and α-actinin-bundled networks contain intrinsic structural features that drive protein sorting. Curr Biol 26, 2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan VT, Narayanan A, Jülicher F, Grill SW (2021). A condensate dynamic instability orchestrates oocyte actomyosin cortex activation. bioRxiv.2021.2009.2019.460784. [DOI] [PMC free article] [PubMed]
- Yadav V, Banerjee DS, Tabatabai AP, Kovar DR, Kim T, Banerjee S, Murrell MP (2019). Filament nucleation tunes mechanical memory in active polymer networks. Adv Funct Mater 29, 1905243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B (2007). Functional atlas of the integrin adhesome. Nat Cell Biol 9, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from A.R.B. upon request.