Abstract
Cells adopt a size that is optimal for their function, and pushing them beyond this limit can cause cell aging and death by senescence or reduce proliferative potential. However, by increasing their genome copy number (ploidy), cells can increase their size dramatically and homeostatically maintain physiological properties such as biosynthesis rate. Recent studies investigating the relationship between cell size and rates of biosynthesis and metabolism under normal, polyploid, and pathological conditions are revealing new insights into how cells attain the best function or fitness for their size by tuning processes including transcription, translation, and mitochondrial respiration. A new frontier is to connect single-cell scaling relationships with tissue and whole-organism physiology, which promises to reveal molecular and evolutionary principles underlying the astonishing diversity of size observed across the tree of life.
INTRODUCTION
Cell size is determined by the relative rates of cell growth and division, and in the past two decades, impressive progress has been made in understanding different mechanisms of size regulation (Campos et al., 2014; Pan et al., 2014; Adiciptaningrum et al., 2015; Ho and Amir, 2015; Schmoller et al., 2015; Taheri-Araghi et al., 2015; Harris and Theriot, 2016; Varsano et al. 2017; Cadart et al., 2018; Garmendia-Torres et al., 2018; Ginzberg et al., 2018; Liu et al., 2018; Micali et al., 2018a,b; Facchetti et al., 2019; Barber et al., 2020; Xie and Skotheim, 2020; Zatulovskiy et al., 2020; Tan et al., 2021). For example, unlike yeast and bacteria, animal cells modulate growth rate in addition to cell cycle duration in order to maintain size homeostasis (Kafri et al., 2013; Cadart et al., 2018; Ginzberg et al., 2018). Further complexity has emerged as different size parameters including mass, volume, and protein levels appear to be regulated by distinct pathways (Delarue et al., 2018; Demian et al., 2019; Knapp et al., 2019) whose uncoupling can be pathological. For example, work in budding yeast revealed the existence of a cell size threshold beyond which biosynthesis does not scale with cell volume, causing dilution of cell contents and premature senescence (Neurohr et al., 2019). In contrast, fission yeast cells display variations in density during the cell cycle under physiological conditions, indicating that variation in intracellular concentrations can also occur normally (Odermatt et al., 2021). In mammals, enlarged cell size is a feature of aging cells (Biran et al., 2017) and was shown to reduce stem cell potential in mice (Lengefeld et al., 2021), providing further evidence that size regulation is important for proper cell function. These findings highlight a fundamental question: how do cellular physiological processes such as biosynthesis or metabolism scale with cell size? Previous reviews have emphasized mechanisms of cell size control (Willis and Huang, 2017; Schmoller, 2017; Ho et al., 2018; Jonas et al., 2018; Zatulovskiy and Skotheim, 2020), the coupling of growth with different size parameters (Cadart et al., 2019; Xie et al., 2022), and the scaling of organelles with cell size (Levy and Heald, 2016; Miller et al., 2020). This review focuses on scaling of biosynthesis and metabolism with cell size and the consequences of disrupting this scaling. For multicellular organisms, size-dependent relationships between cellular physiology and function at the level of an organ or whole organism are poorly understood but have important implications in areas as diverse as ecology (Liedtke et al., 2018) and cancer (Schoenfelder and Fox, 2015).
TRANSCRIPTION, TRANSLATION, AND GROWTH RATE INCREASE WITH CELL SIZE BUT SHOW DISTINCT PATTERNS
At the most basic level, maintaining intracellular homeostasis during cell growth requires that a cell produce its components in the correct amounts, thereby maintaining macromolecular concentrations as cell size increases (Schmoller and Skotheim, 2015). Many cell types grow exponentially in vitro (Cadart et al. 2019), dictating that synthesis rates scale accordingly. Escherichia coli provides the simplest case, with cells growing at a constant exponential rate that scales both with cell size (Si et al., 2017) and with the protein synthesis machinery (ribosome mass fraction) (Scott et al., 2010). The growth rate of budding yeast cells also correlates directly with the ribosome mass fraction (Kafri et al., 2016) and is thought to be exponential (Talia et al., 2007) but other studies have suggested that growth rate may vary across the cell cycle (Ferrezuelo et al., 2012). Growth of Schizosaccharomyces pombe was recently shown to be exponential (Pickering et al., 2019) over a large range of sizes (including mutants that were up to fivefold the size of wild-type cells [Knapp et al., 2019]), contradicting a long-standing belief that these cells follow a bilinear mode of growth (Horváth et al., 2013). New techniques allowing high-throughput single-cell growth measurement in animal cells have revealed that they deviate more dramatically from monoexponential growth and show complex mass (Mu et al., 2020) and volume (Cadart et al., 2018) fluctuations, with a growth rate that is 15% higher in S-G2 than in G1 (Cadart et al., 2022). The origin of cell cycle–dependent changes in growth rate is mysterious and cannot be explained by variation in transcription rate, because transcript amounts scale linearly with cell size (Padovan-Merhar et al., 2015; Sun et al., 2020; Swaffer et al., 2021), independent of genome doubling during S phase. Across diverse organisms, relationships between cell size and growth are still being characterized with the help of newer and more accurate single-cell measurement methods. In many cases, cell size, transcription, translation and volume growth rates show distinct patterns across the cell cycle. Fundamentally, the mechanisms that coordinate biosynthetic processes remain poorly understood, but studies in budding yeast are providing new insight, with recent studies demonstrating that the linear scaling between cell size and transcript levels involved regulation of both RNA polymerase II (Sun et al., 2020; Swaffer et al., 2021) and mRNA stability (Swaffer et al., 2021).
INCREASING PLOIDY EXTENDS THE RANGE IN WHICH BIOSYNTHESIS SCALES WITH CELL SIZE
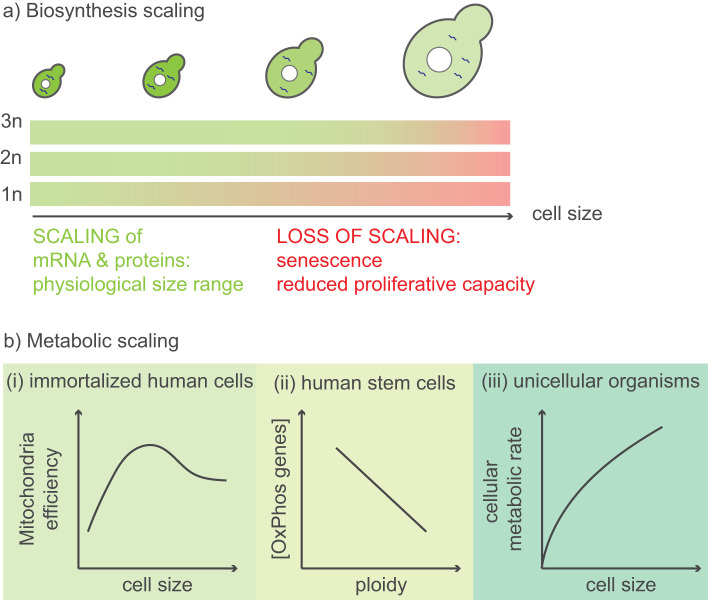
In a landmark study, Neurhor and colleagues analyzed the consequences of extreme cell size in budding yeast by transiently blocking cell cycle progression to obtain haploid cells that were up to sixfold larger than normal (Neurohr et al., 2019). Volume growth of large cells outpaced protein and RNA synthesis rates, leading to cytoplasm dilution (Neurohr et al., 2019). Importantly, uncoupling of volume growth rate and protein synthesis occurred at a larger cell size in diploids, indicating that transcripts become rate-limiting for translation (Lin and Amir, 2018; Metzl-Raz et al., 2020) and that the ploidy sets an upper bound to the range in which biosynthesis scales with cell size (Figure 1a). Polyploidy has long been hypothesized as a mechanism to increase the metabolic capacity of highly synthetically active cells such as Drosophila nurse cells or salivary gland cells (Frawley and Orr-Weaver, 2015) by providing more gene copies and thus increasing rates of biosynthesis. However, measurements of cultured animal cells in suspension spanning ploidies from 2N to 64N revealed a constant mass-normalized growth rate that did not increase with genome copy number (Mu et al., 2020). Interestingly, because these cells were in suspension and had a spherical shape, these findings refute the hypothesis that nutrient import, which is dictated by surface area and scales quadratically with volume for a sphere, is rate-limiting and slows biosynthesis, because cells with volumes three orders of magnitude larger than normal displayed similar growth rates. Thus, increased ploidy boosts the ability of cells to obtain larger sizes with constant cytoplasmic density by maintaining a constant size-normalized biosynthesis rate.
FIGURE 1:
Examples of biosynthesis and metabolism scaling with cell size. (a) In budding yeast, mRNA and protein levels scale with cell size. The upper limit to a physiologically fit size range is thought to be set by ploidy, which can become rate-limiting for transcription. (b) Cellular respiration and metabolic rate appear to decrease with cell size. (i) In immortalized human cells, mitochondrial efficiency is optimal at intermediate cell sizes (Miettinen and Bjorklund, 2016). (ii) In human stem cells, mitochondrial- and nuclear-encoded oxidative phosphorylation genes are expressed at higher levels in haploids compared with diploids (Sagi et al., 2016). (iii) Across unicellular organisms, a power law with an exponent <1 relates cellular metabolic rate and cell size (Glazier, 2009).
The connection between genome size and cell size has long been recognized, though its basis is not understood. Ploidy and cell volume have been shown to scale linearly in budding yeast (Jorgensen et al., 2007; Yahya et al., 2021) and fission yeast (Neumann and Nurse, 2007). Importantly, the correlation between ploidy and cell size within a species (Gillooly et al., 2015) differs from the scaling relationship between genome size and average erythrocyte size observed across species (Gregory, 2001), suggesting that chromosomal mass alone does not completely account for effects of ploidy on cell size, with mechanisms still currently debated (Gregory, 2001; Cantwell and Dey, 2021). The few available quantifications in animals seem to tell a story different from that in unicellular organisms because, for example, the volume of muscle fibers in vivo scales sublinearly with ploidy in adult humans and adult and developing mice (Cramer et al., 2020; Hansson et al., 2020). Different cell types in humans also display a sublinear scaling relationship between average cell size and ploidy (Gillooly et al., 2015). It is therefore possible that other factors limit the increase in cell size enabled by polyploidization. Conversely, a recent study suggests that a minimal cell size is required to sustain polyploidy. Gemble and colleagues showed that cultured RPE1 cells induced to undergo whole-genome duplication in a single cell cycle grew a similar extent during the following G1 phase compared with their diploid counterparts (Gemble et al., 2022). The newborn tetraploids then accumulated high rates of DNA damage during the subsequent S phase, a phenomenon that was rescued if tetraploid cells were induced to spend a longer time growing in G1. Thus, proteome scaling with ploidy, including all the factors necessary for DNA replication, is necessary for cells to remain healthy while undergoing polyploidization. To date, the mechanisms leading to an increase in cell size following polyploidization remain largely unknown (Frawley and Orr-Weaver, 2015) and it is becoming increasingly clear that the proper coordination of ploidy, cell volume, and mass has important implications for cell physiology.
INCREASED CELL SIZE IS ASSOCIATED WITH BIOSYNTHESIS SCALING DEFECTS AND CAN LEAD TO SENESCENCE
Several recent studies provided evidence that increases in cell size are not a consequence, but a cause, of senescence. Budding yeast cells induced to reach large sizes with more dilute cytoplasm showed physiological defects leading to stress pathway activation, decreased proliferation rate, and premature senescence (Neurohr et al., 2019). In an elegant study, Lengefeld and colleagues brought further evidence for a causal link between cell size and senescence in mouse hematopoietic stem cells (HSCs) in vivo (Lengefeld et al., 2021). In mice, radiation treatment caused HSCs to enlarge and become senescent, a phenotype that could be rescued by treatment with rapamycin to maintain their size small. Conversely, inhibition of cell cycle proliferation caused up to a 15% increase in cell size, led to DNA damage, and impaired the ability of treated HSCs to reconstitute the hematopoietic lineage in recipient mice in a manner that depended on cell size.
The mechanisms causing senescence when cell size increases remain unclear, but a defect in proteome scaling with cell size has emerged as an important factor. A novel triple-SILAC approach to analyze subcellular compartment-specific scaling of the proteome with cell size revealed that in cultured human cells, components typically associated with cell senescence such as lysosomes, β-galactosidase, and metalloproteases were up-regulated with enlarged cell size (Lanz et al., 2021). Thus, contrary to the simple view that all proteins and organelles adapt in the same way to cell size (Levy and Heald, 2012), several processes appear to deviate from a linear scaling pattern (Cheng et al., 2021; Lanz et al., 2021; Liu et al., 2021). Furthermore, when cells reach sizes beyond their physiological range, overall proteome content no longer scales with size and the cytoplasm becomes diluted, presumably due to defective coordination of cell volume growth and biosynthesis. The consequences of cytoplasm dilution are an active area of research, with recent evidence demonstrating effects on reaction rates (Jin et al., 2022; Molines et al., 2022) and phase separation (Delarue et al., 2018) that could negatively impact cell function. New techniques enabling cellular density measurement with unprecedented precision (Miettinen et al., 2022; Oh et al., 2022), as well as theoretical approaches applying principles of colloidal physics to study cytoplasmic crowding (Maheshwari et al., 2019), are likely to provide exciting new insights.
OXIDATIVE PHOSPHORYLATION IS THOUGHT TO CHANGE WITH CELL SIZE AND PLOIDY
While many studies have focused on the importance of cytoplasmic density and scaling of biosynthesis for cell viability (Neurohr and Amon, 2020), characterization of enlarged senescent mouse HSCs also revealed metabolic defects, including a decrease in mitochondrial concentration and lower levels of reactive oxygen species (Lengefeld et al., 2021). Although the metabolic consequences of enlarged cell size remain poorly understood, one important set of results was provided by Miettinen and colleagues, who showed that hepatocytes remodeled their metabolome as their size increased, decreasing mitochondrial oxidative phosphorylation (Miettinen et al., 2014). They also reported that although the concentration of mitochondria remained constant over a range of cell sizes, mitochondrial membrane potential reached a peak at an intermediate cell size (Miettinen and Bjorklund, 2016) (Figure 1b). For a review on the relationship between mitochondria and cell size, see Miettinen and Björklund (2017). Reinforcing the idea that oxidative phosphorylation decreases with increasing cell size, studies of polyploid yeast revealed down-regulation of several proteins involved in mitochondrial respiration (Yahya et al., 2021). Furthermore, in human embryonic stem cells, expression levels of both mitochondrial and nuclear genes involved in mitochondrial respiration were up-regulated in haploid cells (Sagi et al., 2016) (Figure 1b). New approaches to quantify ATP production (Yaginuma and Okada, 2021) and mitochondrial function at the single-cell level (Papagiannakis et al., 2017; Kang et al., 2020) will help elucidate connections between expression of mitochondrial proteins and cellular respiration rate.
SCALING OF CELLULAR RESPIRATION AND METABOLIC RATE WITH CELL SIZE
Why should cellular respiration decrease with cell size? Since the famous ¾ power law between body size and metabolic rate initially observed in animals by Kleiber was extended to unicellular organisms (Savage et al., 2004; Glazier, 2009), cellular metabolic rate was assumed to scale sublinearly with cell size across species (Figure 1b). However, in multicellular organisms, cell type and whole-organism metabolism impact this relationship and add a layer of complexity, which may lead to a trade-off between average cell size and cellular metabolic rate. Highly proliferative and biosynthetically active cells such as epithelial cells maintain a constant size and appear to scale their metabolic rate to that of the whole body. In contrast, other cells maintain a more constant metabolic rate while their size tends to increase with body size—storage cells like adipocytes seem to fall in this category (Savage et al., 2007). Quantitative experiments in planarians recently provided evidence that cell size, body size, and body metabolic rate are connected and showed that a nonlinear increase in average cell size, not cellular metabolic rate, accounted for the conservation of Kleiber’s law as body size increased over several orders of magnitude (Thommen et al., 2019). However, while cross-species studies have established that scaling between cellular metabolic rate and cell size occurs, we lack a clear understanding of this relationship, and direct quantification of in vivo cellular respiration in multicellular animals is needed to bring more conclusive answers.
Novel investigations of cellular bioenergetics (Yang et al., 2021) provide a promising approach to understanding how cellular metabolism changes with cell size by helping identify 1) the key energy-producing components that could be affected by changes in cellular geometry and 2) the energetic cost of central cellular processes whose relative usage could change with cell size or ploidy. One example in the first category is the proposition that the decrease in surface area to volume ratio that occurs as E. coli cells grow larger leads to saturation of membrane space available for respiratory proteins, triggering activation of fermentation pathways (Zhuang et al., 2011; Szenk et al., 2017). Reminiscent of this idea, in animal cells, it has been proposed that changes in mitochondrial surface area-to-volume ratio and activity could emerge with changes in cell size due to network remodeling, although scant experimental evidence supports this hypothesis (Miettinen and Björklund, 2017). Moreover, how changes in mitochondrial morphology might relate to decreased expression of mitochondrial genes observed with increasing ploidy in animal cells and budding yeast (Sagi et al., 2016; Yahya et al., 2021) is unclear. Is it possible that overall cellular energy demand changes with cell ploidy or size? The development of approaches deconvolving the energetic cost of key cellular processes, including nutrient import, membrane synthesis, energy production, and each step of the central dogma, similarly to what has been done in E. coli (Belliveau et al., 2021), will likely bring insights. In zebrafish embryos, for example, energy expenditure scales with cellular plasma membrane synthesis (Rodenfels et al., 2020), not cell volume, thus demonstrating that cell number, size, and geometry relate to whole-organism metabolism.
CONCLUSION
Recent studies provide compelling evidence that enlarged cell size decreases the ability to thrive (fitness) by contributing to cellular senescence (Neurohr et al., 2019; Lanz et al., 2021) and loss of replicative potential (Lengefeld et al., 2021). The emerging view is that defective scaling between biosynthesis and cell size, which is limited by ploidy (Neurohr et al., 2019), is a more important factor than decreased nutrient import rates or a suboptimal surface area-to-volume ratio, even at very large cell sizes (Mu et al., 2020). However, the mechanisms underlying scaling relationships of cell size and ploidy at each step of the central dogma are only beginning to be elucidated (Swaffer et al., 2021). Another underlying explanation for changes in cell fitness likely relates to how cellular metabolism scales with cell size, as several independent observations suggest that oxidative respiration decreases with size (Miettinen and Bjorklund, 2016) or ploidy (Sagi et al., 2016). Investigating links among cell size, cellular physiology, and tissue physiology is likely to prove extremely fruitful as examples of connections between cell size or ploidy and organ metabolism are rife. For example, polyploidization accompanied by increases in cell size can affect the metabolic demand of organs such as the liver (Donne et al., 2020) while across evolutionary timescales, the well-described scaling relationship between genome size and cell size has been hypothesized to affect overall metabolic demand of the organism (Liedtke et al., 2018; Gardner et al., 2020). Exploring connections between size and fitness at the cellular and organismal levels will undoubtedly yield exciting new principles of physiology, as well as how evolution has enabled life to exist at sizes that vary over 20 orders of magnitude.
Acknowledgments
R. H. is supported by National Institutes of Health Grant R35GM118183 and the Flora Lamson Hewlett Chair in Biochemistry.
Abbreviations used:
- ATP
adenosine triphosphate
- DNA
deoxyribonucleic acid
- HSCs
hematopoietic stem cells
- RNA
ribonucleic acid
- RPE1
retinal pigment epithelial cells
- SILAC
stable isotope labeling by amino acids in cell culture.
Footnotes
REFERENCES
- Adiciptaningrum A, Osella M, Charl Moolman M, Lagomarsino MC, Tans SJ (2015). Stochasticity and homeostasis in the E. coli replication and division cycle. Sci Rep 5, 18261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber F, Amir A, Murray AW (2020). Cell-size regulation in budding yeast does not depend on linear accumulation of Whi5. Proc Natl Acad Sci USA 117, 14243–14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliveau NM, Chure G, Hueschen CL, Garcia HG, Kondev J, Fisher DS, Theriot JA, Phillips R (2021). Fundamental limits on the rate of bacterial growth and their influence on proteomic composition. Cell Syst 12, 924–944.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biran A, Zada L, Karam PA, Vadai E, Roitman L, Ovadya Y, Porat Z, Krizhanovsky V (2017). Quantitative identification of senescent cells in aging and disease. Aging Cell 16, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadart C, Monnier S, Grilli J, Sáez PJ, Srivastava N, Attia R, Terriac E, Baum B, Cosentino-Lagomarsino M, Piel M (2018). Size control in mammalian cells involves modulation of both growth rate and cell cycle duration. Nat Commun 9, 3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadart C, Venkova L, Piel M, Lagomarsino MC (2022). Volume growth in animal cells is cell cycle dependent and shows additive fluctuations. eLife 11, e70816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadart C, Venkova L, Recho P, Cosentino Lagomarsino M, Piel M (2019). The physics of cell-size regulation across timescales. Nat Phys 15, 993–1004. [Google Scholar]
- Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C ( 2014). A constant size extension drives bacterial cell size homeostasis. Cell 159, 1433–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell H, Dey G (2021). Nuclear size and shape control. Semin Cell Dev Biol, doi: 10.1016/j.semcdb.2021.10.013. [DOI] [PubMed] [Google Scholar]
- Cheng L, Chen J, Kong Y, Tan C, Kafri R, Björklund M (2021). Size-scaling promotes senescence-like changes in proteome and organelle content. BioRxiv, 455193. [Google Scholar]
- Cramer AAW, Prasad V, Eftestøl E, Song T, Hansson KA, Dugdale HF, Sadayappan S, Ochala J, Gundersen K, Millay DP (2020). Nuclear numbers in syncytial muscle fibers promote size but limit the development of larger myonuclear domains. Nat Commun 11, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Brittingham GP, Pfeffer S, Surovtsev IV, Pinglay S, Kennedy KJ, Schaffer M, Gutierrez JI, Sang D, Poterewicz G, et al. (2018). MTORC1 controls phase separation and the biophysical properties of the cytoplasm by tuning crowding. Cell 174, 338–349.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demian WL, Persaud A, Jiang C, Coyaud É, Liu S, Kapus A, Kafri R, Raught B, Rotin D (2019). The ion transporter NKCC1 links cell volume to cell mass regulation by suppressing MTORC1. Cell Rep 27, 1886–1896.e6. [DOI] [PubMed] [Google Scholar]
- Donne R, Saroul-Aïnama M, Cordier P, Celton-Morizur S, Desdouets C (2020). Polyploidy in liver development, homeostasis and disease. Nat Rev Gastroenterol Hepatol 17, 391–405. [DOI] [PubMed] [Google Scholar]
- Facchetti G, Knapp B, Flor-Parra I, Chang F, Howard M (2019). Reprogramming Cdr2-dependent geometry-based cell size control in fission yeast. Curr Biol 29, 350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Colomina N, Palmisano A, Garí E, Gallego C, Csikász-Nagy A, Aldea M (2012). The critical size is set at a single-cell level by growth rate to attain homeostasis and adaptation. Nat Commun 3, 1012. [DOI] [PubMed] [Google Scholar]
- Frawley LE, Orr-Weaver TL (2015). Polyploidy. Curr Biol 25, R353–R358. [DOI] [PubMed] [Google Scholar]
- Gardner JD, Laurin M, Organ CL (2020). The relationship between genome size and metabolic rate in extant vertebrates. Philos Trans R Soc B 375, 20190146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmendia-Torres C, Tassy O, Matifas A, Molina N, Charvin G (2018). Multiple inputs ensure yeast cell size homeostasis during cell cycle progression. eLife 7, e34025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemble S, Wardenaar R, Keuper K, Srivastava N, Nano M, Macé A, Tijhuis AE, Bernhard SV, Spierings DCJ, Simon A, et al. (2022). Genetic instability from a single S phase after whole-genome duplication. Nature 604, 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly JF, Hein A, Damiani R (2015). Nuclear DNA content varies with cell size across human cell types. Cold Spring Harb Perspect Biol 7, a019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginzberg MB, Chang N, D’Souza H, Patel N, Kafri R, Kirschner MW (2018). Cell size sensing in animal cells coordinates anabolic growth rates and cell cycle progression to maintain cell size uniformity. eLife 7, e26947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier DS (2009). Metabolic level and size scaling of rates of respiration and growth in unicellular organisms. Funct Ecol 23, 963–968. [Google Scholar]
- Gregory TR (2001). Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol Rev 76, 65–101. [DOI] [PubMed] [Google Scholar]
- Hansson KA, Eftestøl E, Bruusgaard JoC, Juvkam I, Cramer AW, Malthe-Sørenssen A, Millay DP, Gundersen K (2020). Myonuclear content regulates cell size with similar scaling properties in mice and humans. Nat Commun 11, 6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LKK, Theriot JAA (2016). Relative rates of surface and volume synthesis set bacterial cell size. Cell 165, 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PY, Amir A (2015). Simultaneous regulation of cell size and chromosome replication in bacteria. Front Microbiol 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PY, Lin J, Amir A (2018). Modeling cell size regulation: from single-cell-level statistics to molecular mechanisms and population-level effects. Ann Rev Biophys 47, 251–271. [DOI] [PubMed] [Google Scholar]
- Horváth A, Rácz-Mónus A, Buchwald P, Sveiczer Á (2013). Cell length growth in fission yeast: an analysis of its bilinear character and the nature of its rate change transition. FEMS Yeast Res 13, 635–649. [DOI] [PubMed] [Google Scholar]
- Jin M, Tavella F, Wang S, Yang Q (2022). In itro cell cycle oscillations exhibit a rovbust and hysteretic response to changes in cytoplasmic density. Proc Natl Acad Sci USA 119, e2109547119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas F, Soifer I, Barkai N (2018). A visual framework for classifying determinants of cell size. Cell Rep 25, 3519–3529.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B (2007). The size of the nucleus increases as yeast cells grow. Mol Biol Cell 18, 3523–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri M, Metzl-Raz E, Jona G, Barkai N (2016). The cost of protein production. Cell Rep 14, 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Levy J, Ginzberg MB, Oh S, Lahav G, Kirschner MW (2013). Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature 494, 480–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JHo, Katsikis G, Li Z, Sapp KM, Stockslager MA, Lim D, Heiden MGV, Yaffe MB, Manalis SR, Miettinen TP (2020). Monitoring and modeling of lymphocytic leukemia cell bioenergetics reveals decreased ATP synthesis during cell division. Nat Commun 11, 4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp BD, Odermatt P, Rojas ER, Cheng W, He X, Huang KC, Chang F (2019). Decoupling of rates of protein synthesis from cell expansion leads to supergrowth. Cell Syst 9, 434–445.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz MC, Zatulovskiy E, Swaffer MP, Zhang L, Ilerten I, Zhang S, You DS, Marinov G, McAlpine P, Elias JE, Skotheim JM (2021). Increasing cell size remodels the proteome and promotes senescence. BioRxiv, 1–50. 10.1101/2021.07.29.454227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengefeld J, Cheng C-W, Maretich P, Blair M, Hagen H, McReynolds MR, Sullivan E, Majors K, Roberts C, Kang JH, et al. (2021). Cell size is a determinant of stem cell potential during aging. Sci Adv 7, eabk0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R (2012). Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol 28, 113–135. [DOI] [PubMed] [Google Scholar]
- Levy DL, Heald R (2016). Biological scaling problems and solutions in amphibians. Cold Spring Harb Perspect Biol 8, a019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke HC, Gower DJ, Wilkinson M, Gomez-Mestre I (2018). Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nat Ecol Evol 2, 1792–1799. [DOI] [PubMed] [Google Scholar]
- Lin J, Amir A (2018). Homeostasis of protein and MRNA concentrations in growing cells. Nat Commun 9, 4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ginzberg MB, Patel N, Hild M, Leung B, Li Z, Chen Y-C, Chang N, Wang Y, Tan C, et al. (2018). Size uniformity of animal cells is actively maintained by a P38 MAPK-dependent regulation of G1-length. eLife 7, e26947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Tan C, Melo-Gavin C, Mark KG, Ginzberg MB, Blutrich R, Patel N, Rape M, Kafri R (2021). Large cells activate global protein degradation to maintain cell size homeostasis. BioRxiv, 2021.11.09.467936. [Google Scholar]
- Maheshwari AJ, Sunol AM, Gonzalez E, Endy D, Zia RN (2019). Colloidal hydrodynamics of biological cells: a frontier spanning two fields. Phys Rev Fluids 4, 1–26. [Google Scholar]
- Metzl-Raz E, Kafri M, Yaakov G, Barkai N (2020). Gene transcription as a limiting factor in protein production and cell growth. G3: Genes, Genomes, Genetics 10, 3229–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali G, Grilli J, Marchi J, Osella M, Cosentino Lagomarsino M (2018a). Dissecting the control mechanisms for DNA replication and cell division in E. coli. Cell Rep 25, 761–771.e4. [DOI] [PubMed] [Google Scholar]
- Micali G, Grilli J, Osella M, Cosentino Lagomarsino M (2018b). Concurrent processes set E. coli cell division. Sci Adv 4, eaau3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen TP, Bjorklund M (2016). Cellular allometry of mitochondrial functionality establishes the optimal cell size. Dev Cell 39, 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen TP, Björklund M (2017). Mitochondrial function and cell size: an allometric relationship. Trends Cell Biol 27, 393–402. [DOI] [PubMed] [Google Scholar]
- Miettinen TP, Ly KS, Lam A, Manalis SR (2022). Single-cell monitoring of dry mass and dry mass density reveals exocytosis of cellular dry contents in mitosis. eLife 11, e76664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen TP, Pessa HKJ, Caldez MJ, Fuhrer T, Kasim Diril M, Sauer U, Kaldis P, Björklund M (2014). Identification of transcriptional and metabolic programs related to mammalian cell size. Curr Biol 24, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KE, Brownlee C, Heald R (2020). The power of amphibians to elucidate mechanisms of size control and scaling. Exp Cell Res 392, 112036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molines AT, Lemière J, Gazzola M, Steinmark IE, Edrington CH, Hsu CT, Real-Calderon P, Suhling K, Goshima G, Holt LJ, et al. (2022). Physical Properties of the cytoplasm modulate the rates of microtubule polymerization and depolymerization. Dev Cell 57, 466–479.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L, Kang JH, Olcum S, Payer KR, Calistri NL, Kimmerling RJ, Manalis SR, Miettinen TP (2020). Mass measurements during lymphocytic leukemia cell polyploidization decouple cell cycle- and cell size-dependent growth. Proc Natl Acad Sci USA 117, 15659–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann FR, Nurse P (2007). Nuclear size control in fission yeast. J Cell Biol 179, 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurohr GE, Amon A (2020). Relevance and regulation of cell density. Trends Cell Biol 30, 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neurohr GE, Terry RL, Lengefeld J, Bonney M, Brittingham GP, Moretto F, Miettinen TP, Vaites LP, Soares LM, Paulo JA, et al. (2019). Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 176, 1083–1097.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt PD, Miettinen TP, Lemière J, Kang JH, Bostan E, Manalis SR, Huang KC, Chang F (2021). Variations of intracellular density during the cell cycle arise from tip-growth regulation in fission yeast. eLife 10, e64901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Lee C, Yang W, Li A, Mukherjee A, Basan M, Ran C, Yin W, Tabin CJ, Fu D, et al. (2022). Protein and lipid mass concentration measurement in tissues by stimulated raman scattering microscopy. Proc Natl Acad Sci USA 119, 629543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padovan-Merhar O, Nair GP, Biaesch AG, Mayer A, Scarfone S, Foley SW, Wu AR, Stirling Churchman L, Singh A, Raj A (2015). Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell 58, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Saunders TE, Flor-Parra I, Howard M, Chang F (2014). Cortical regulation of cell size by a sizer Cdr2p. eLife 3, e02040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakis A, Niebel B, Wit EC, Heinemann M (2017). Autonomous metabolic oscillations robustly gate the early and late cell cycle. Mol Cell 65, 285–295. [DOI] [PubMed] [Google Scholar]
- Pickering M, Hollis LN, D’Souza E, Rhind N (2019). Fission yeast cells grow approximately exponentially. Cell Cycle 18, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenfels J, Sartori P, Golfier S, Nagendra K, Neugebauer K, Howard J (2020). Contribution of increasing plasma membrane to the energetic cost of early zebrafish embryogenesis. Mol Biol Cell 31, 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi I, Chia G, Golan-Lev T, Peretz M, Weissbein U, Sui L, Sauer MV, Yanuka O, Egli D, Benvenisty N (2016). Derivation and differentiation of haploid human embryonic stem cells. Nature 532, 107–111. [DOI] [PubMed] [Google Scholar]
- Savage VM, Allen AP, Brown JH, Gillooly JF, Herman AB, Woodruff WH, West GB (2007). Scaling of number, size, and metabolic rate of cells with body size in mammals. Proc Natl Acad Sci USA 104, 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage VM, Gillooly JF, Woodruff WH, West GB, Allen AP, Enquist BJ, Brown JH (2004). The predominance of quarter-power scaling in biology. Funct Ecol 18, 257–282. [Google Scholar]
- Schmoller KM (2017). The phenomenology of cell size control. Curr Opin Cell Biol 49, 53–58. [DOI] [PubMed] [Google Scholar]
- Schmoller KM, Skotheim JM (2015). The biosynthetic basis of cell size control. Trends Cell Biol 25, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller KM, Turner JJ, Kõivomägi M, Skotheim JM (2015). Dilution of the cell cycle inhibitor Whi5 controls budding yeast cell size. Nature 526, 268–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Fox DT (2015). The expanding implications of polyploidy. J Cell Biol 209, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T (2010). Interdependence of cell growth. Science 330, 1099–1102. [DOI] [PubMed] [Google Scholar]
- Si F, Li D, Cox SE, Sauls JT, Azizi O, Sou C, Schwartz AB, Erickstad MJ, Jun Y, Li X, et al. (2017). Invariance of initiation mass and predictability of cell size in Escherichia coli. Curr Biol 27, 1278–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XM, Bowman A, Priestman M, Bertaux F, Martinez-Segura A, Tang W, Whilding C, Dormann D, Shahrezaei V, Marguerat S (2020). Size-dependent increase in RNA polymerase II initiation rates mediates gene expression scaling with cell size. Curr Biol 30, 1217–1230.e7. [DOI] [PubMed] [Google Scholar]
- Swaffer MP, Marinov GK, Zheng H, Jones AW, Greenwood J, Kundaje A, Snijders AP, Greenleaf WJ, Reyes-Lamothe R, Skotheim JM (2021). RNA polymerase II dynamics and MRNA stability feedback determine MRNA scaling with cell size. BioRxiv, 2021.09.20.461005. [Google Scholar]
- Szenk M, Dill KA, Graff AMR (2017). Why do fast-growing bacteria enter overflow metabolism? Testing the membrane real estate hypothesis. Cell Systems 5, 95–104. [DOI] [PubMed] [Google Scholar]
- Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S (2015). Cell-size control and homeostasis in bacteria. Curr Biol 25, 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talia SDI, Skotheim JM, Bean JM, Siggia ED, Cross FR (2007). The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature 448, 947. [DOI] [PubMed] [Google Scholar]
- Tan C, Ginzberg MB, Webster R, Iyengar S, Liu S, Papadopoli D, Concannon J, Wang Y, Auld DS, Jenkins JL, et al. (2021). Cell size homeostasis is maintained by CDK4-dependent activation of P38 MAPK. Dev Cell 56, 1756–1769.e7. [DOI] [PubMed] [Google Scholar]
- Thommen A, Werner S, Frank O, Philipp J, Knittelfelder O, Quek Y, Fahmy K, Shevchenko A, Friedrich BM, Jülicher F, et al. (2019). Body size-dependent energy storage causes Kleiber’s law scaling of the metabolic rate in planarians. eLife 8, e38187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsano G, Wang Y, Wu M, Varsano G, Wang Y, Wu M (2017). Probing mammalian cell size homeostasis by article probing mammalian cell size homeostasis by channel-assisted cell reshaping. Cell Rep 20, 397–410. [DOI] [PubMed] [Google Scholar]
- Willis L, Huang KC (2017). Sizing up the bacterial cell cycle. Nat Rev Microbiol 15, 606–620. [DOI] [PubMed] [Google Scholar]
- Xie S, Skotheim JM (2020). A G1 sizer coordinates growth and division in the mouse epidermis. Curr Biol 30, 916–924.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Swaffer M, Skotheim JM (2022). Eukaryotic cell size control and its relation to biosynthesis and senescence. Annu Rev Cell Dev Biol. 38, doi:10.1146/annurev-cellbio-120219. [DOI] [PubMed] [Google Scholar]
- Yaginuma H, Okada Y (2021). Live cell imaging of metabolic heterogeneity by quantitative fluorescent ATP indicator protein, QUEEN-37C. BioRxiv, 2021.10.08.463131. [Google Scholar]
- Yahya G, Menges P, Ngandiri DA, Schulz D, Wallek A, Kulak N, Mann M, et al. (2021). Scaling of cellular proteome with ploidy. BioRxiv, 2021.05.06.442919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Heinemann M, Howard J, Huber G, Iyer-Biswas S, Le Treut G, Lynch M, Montooth KL, Needleman DJ, Pigolotti S, et al. (2021). Physical bioenergetics: energy fluxes, budgets, and constraints in cells. Proc Natl Acad Sci USA 118, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatulovskiy E, Skotheim JM (2020). On the molecular mechanisms regulating animal cell size homeostasis. Trends Genet 36, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatulovskiy E, Zhang S, Berenson DF, Topacio BR, Skotheim JM (2020). Cell growth dilutes the cell cycle inhibitor Rb to trigger cell division. Science 369, 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang K, Vemuri GN, Mahadevan R (2011). Economics of membrane occupancy and respiro-fermentation. Mol Syst Biol 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]