Abstract
The Chromosome Passenger Complex (CPC) generates chromosome autonomous signals that regulate mitotic events critical for genome stability. Tip60 is a lysine acetyltransferase that is a tumor suppressor and is targeted for proteasomal degradation by oncogenic papilloma viruses. Mitotic regulation requires the localization of the CPC to inner centromeres, which is driven by the Haspin kinase phosphorylating histone H3 on threonine 3 (H3T3ph). Here we describe how Tip60 acetylates histone H3 at lysine 4 (H3K4ac) to block both the H3T3ph writer and the reader to ensure that this mitotic signaling cannot begin before prophase. Specifically, H3K4ac inhibits Haspin phosphorylation of H3T3 and prevents binding of the Survivin subunit to H3T3ph. Tip60 acetylates H3K4 during S/G2 at centromeres. Inhibition of Tip60 allows the CPC to bind centromeres in G2 cells, and targeting of Tip60 to centromeres prevents CPC localization in mitosis. The H3K4ac mark is removed in prophase by HDAC3 to initiate the CPC localization cascade. Together, our results suggest that Tip60 and HDAC3 temporally control H3K4 acetylation to precisely time the targeting of the CPC to inner centromeres.
INTRODUCTION
Posttranslational modifications (PTMs) of histones have emerged as a central mechanism that regulates both gene expression and other cellular events. During mitosis, there is a global increase in histone tail acetylation that controls chromosome condensation and an increase in histone phosphorylation that controls mitotic signaling (Zhiteneva et al., 2017). Histone phosphorylation drives mitotic signaling by recruiting the Chromosome Passenger Complex (CPC) to the inner centromere of every mitotic chromosome from which it regulates events that ensure faithful chromosome segregation, including spindle checkpoint signaling, sister chromosome cohesion, and the correction of improper kinetochore-microtubule attachments (Trivedi and Stukenberg, 2016). While we understand how histone phosphorylation is used to spatially identify the location to target the CPC to the chromosome region between kinetochores, it is unclear how these events are temporally restricted to the middle of prophase. Moreover, since the CPC and its histone phosphorylation network are among the most overexpressed pathways in highly aneuploid breast tumors (Pfister et al., 2018), it is important to understand how the events are dysregulated in cancer.
The acetylation of histone H3 on lysine 4 (H3K4ac) is a poorly understood histone PTM. In fission yeast, this mark is generated in late S phase as part of a chromodomain switch that epigenetically transfers heterochromatin to the next generation (Xhemalce and Kouzarides, 2010). First, the Chp1/Clr4 complex recognizes H3K9 methylated on an existing histone and methylates H3K9 on neighboring histones deposited during DNA replication. To remove the Chp1/Clr4 methyltransferase, the Tip60 homolog, Mst1, acetylates H3K4 to disrupt the binding of Chp1/Clr4 to the adjacent H3K9 methylated sites. Whether this mechanism is conserved in vertebrates and whether the H3K4 mark has additional cell cycle functions are not known.
The CPC is a four-protein complex consisting of the Aurora-B kinase and three regulatory subunits: the scaffold inner centromere protein (INCENP), Survivin, and Borealin, which are required for chromatin targeting (Carmena et al., 2012; Hindriksen et al., 2017). The CPC regulates mitotic events by generating chromosome autonomous signals that control the spindle checkpoint and kinetochore-microtubule attachments to each mitotic chromosome (Trivedi and Stukenberg, 2016; Hindriksen et al., 2017). The majority of the CPC (∼75%) is localized to the inner centromere (Cooke et al., 1987; Mahen et al., 2014), which is located between sister kinetochore on mitotic chromosomes during prometaphase and metaphase. There are additional pools of CPC at inner kinetochores and on mitotic chromatin (Broad et al., 2020; Hadders et al., 2020; Liang et al., 2020). The CPC locates to the inner centromere during prophase by binding two distinct histone phosphorylation marks that intersect the mitotic chromosome along two orthogonal axes. In the first axis, a Haspin kinase phosphorylates histone H3 at threonine 3 (H3T3ph), which directly associates with the Survivin subunit (Dai and Higgins, 2005; Kelly et al., 2010; Wang et al., 2010). Because Haspin is recruited by cohesin through Pds5 (Goto et al., 2017), the H3T3ph mark is initially found along a central “long” axis between sister chromatids (Yamagishi et al., 2010). A second axis is generated across chromosomes from kinetochore to kinetochore by a Bub1 kinase, which phosphorylates histone H2A on threonine 120 (H2AT120ph) (Yamagishi et al., 2010; Storchova et al., 2011; Liu et al., 2013). This indirectly recruits the CPC through interaction of the Borealin subunit of the CPC with Sgo1 that in turn directly binds histone H2AT120ph. These events are temporally controlled so that the CPC is enriched at noncentromeric chromatin in late G2 and early prophase; however, by the end of prophase the CPC is highly enriched at inner centromeres. The events that temporally control this transition are not known but recently the acetylation of histones at H2AK118 by Tip60 was shown to inhibit the phosphorylation of H2A on T120 by Bub1 (Lee et al., 2018).
Tip60 is a lysine acetyltransferase that is often dysregulated in human tumors. Tip60 acetylates histone tails and other substrates and is a coregulator of transcription factors and a modulator of DNA damage response signals (Sapountzi et al., 2006). Tip60 has also been implicated in mitotic regulation. Tip60 directly acetylates Aurora B, which stimulates kinase activity by preventing T-loop dephosphorylation by PP2A (Mo et al., 2016). Tip60 is part of the RSF1 complex that temporally regulates Sgo1 binding at mitotic centromeres by acetylating histone H2A that prevents Bub1 phosphorylation of H2A T120, until this mark is removed by the HDAC1 protein (Lee et al., 2018).
In this study we show that temporal regulation of CPC localization to the inner centromere is controlled by H3K4ac. Tip60 acetylates H3K4 in centromeres during S/G2 to both prevent the phosphorylation of H3T3 by Haspin (Han et al., 2011) and the association of Survivin with H3T3ph. HDAC3 removes the H3K4ac in the middle of prophase to enable CPC targeting to the inner centromere. We suggest that Tip60 acetylation inhibits CPC localization to inner centromeres at numerous steps to control the proper timing of CPC localization and signaling during mitosis.
RESULTS
It has been shown that H3K4 acetylation (H3K4ac) reduces the ability of the Haspin kinase to phosphorylate H3T3 in vitro (Han et al., 2011); however, neither the importance of this event nor whether H3K4ac controls binding of Survivin to the tail is known. We therefore determined whether H3K4 PTMs might regulate in vitro the binding of histones tails by Survivin using isothermal titrating calorimetry (Table 1). Both acetylation and trimethylation of H3K4 inhibited Survivin binding even when the peptides contained H3T3ph. In contrast, mono- and dimethylation of H3K4 had little effect on Survivin binding to H3T3ph. Trimethylation of H3K4 is associated with active promoters (Santos-Rosa et al., 2002), suggesting that this mark would inhibit CPC binding to transcriptionally active sites. Since the CPC is normally found at pericentric heterochromatin (Ruppert et al., 2018), this mark is unlikely to control CPC at centromeres. Therefore we focused our investigation on H3K4ac mark, which had not been examined previously for its impact on Survivin interaction with H3.
TABLE 1:
Thermodynamic parameters of interaction between hSurvivin and histone H3 peptides at pH 7.2 at 25°C.
| Peptide | N | Kd [µM] | ΔH (cal/mol) | –TΔS (cal/mol) |
|---|---|---|---|---|
| H3T3phK4me1(1-12) | 1.0301 ± 0.005 | 3.10 ± 0.14 | –4692 ± 26 | –2820 |
| H3T3phK4me2(1-12) | 0.873 ± 0.004 | 2.82 ± 0.16 | –6250 ± 60 | –1310 |
| H3T3phK4me3(1-12) | 1.21 ± 0.02 | 11.5 ± 1.0 | –4630 ± 90 | –2820 |
| H3K4me1(1-12) | 1.080 ± 0.012 | 9.1 ± 1.0 | –2680 ± 60 | –4200 |
| H3K4me2(1-12) | 0.899 ± 0.010 | 12.0 ± 0.8 | –2610 ± 40 | –4110 |
| H3K4me3(1-12) | 0.83 ± 0.07 | 87 ± 10 | –6000 ± 700 | –500 |
| H3T3phK4ac(1-12) | 0.965 ± 0.009 | 15.4 ± 0.8 | –7300 ± 100 | –700 |
| H3T3ph(1-12) | 1.03 ± 0.002 | 2.31 ± 0.08 | –4880 ± 30 | –2800 |
| H3(1-12) | 1.21 ± 0.01 | 9.37 ± 0.31 | –2790 ± 20 | –4040 |
Measurements were performed in 25°C in a triple component buffer containing citric acid, HEPES, CHES, and 10 mM β-mercaptoethanol. N, stoichiometry; Kd, dissociation constant; ΔH, entalphy change; ΔS, entropy change; T, temperature. Values for H3T3ph(1-12) and H3(1-12) were taken from Niedzialkowska et al. (2012).
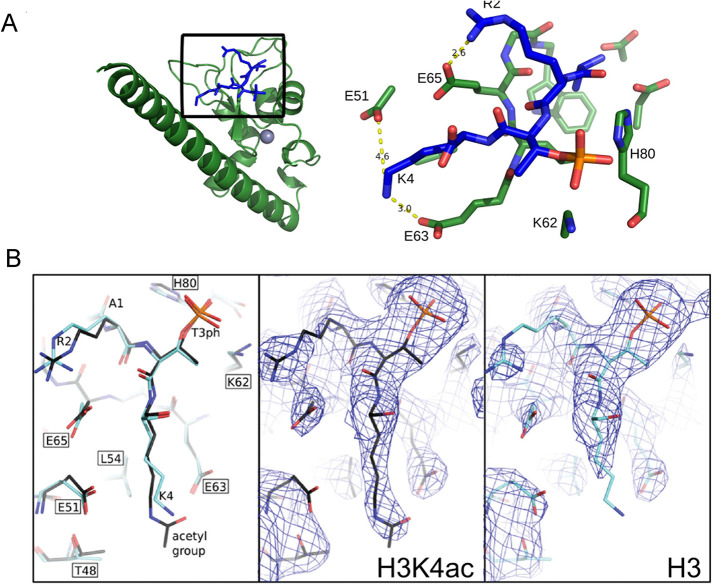
Analysis of the surface of the region interacting with histone H3 tail shows that R2 and K4 form electrostatic interactions with Survivin side chains (Figure 1, A and B). To define what causes the observed differences in binding affinities, we determined structure of Survivin bound to H3T3phK4ac peptide and compared it with the crystal structures of Survivin bound to H3T3ph peptide (Niedzialkowska et al., 2012). Both structures were determined to similar resolution and there were no major conformational changes observed (rmsd 0.199 Å) between the two structures. The peptides used in the crystallization experiments were at saturation concentration; they are refined with occupancy of 1.0 and similar B factors. Therefore the only difference in binding affinities between H3T3ph and H3T3phK4ac peptides can be attributed to H3K4ac and removal of charge around ε nitrogen of H3K4 after acetylation. The positive charge of H3K4 is normally distributed between two salt bridges to negatively charged amino acids (E51 and E63) in the binding pocket of the Survivin subunit. Acetylation of H3K4 disrupts these salt bridges, which can explain the measured effect on the strength of the interaction between Survivin and the H3 tail (Figure 1, A and B). We also determined the structure of Survivin bound to methylated H3K4 and see subtle changes in the conformation of Survivin E51 and E63 in the complex containing the H3T3phK4me3 peptide; however, they are within the error range for the structure resolution and cannot be unambiguously attributed to conformational changes resulting from histone tail binding (Supplemental Figure S1, A–C).
FIGURE 1:
Histone H3K4ac disrupts salt bridges between H3K4 and Survivin E51 and E63. (A) Structure of human Survivin (green) bound to the unacetylated histone H3 peptide (blue). Gray sphere represents zinc atom. Inset shows amino acids involved in interaction between the H3 peptide and the Survivin to highlight the binding interface between Survivin and the histone H3T3phK4 peptide; yellow dashed lines indicate formation of salt bridges between histone H3 lysine 4 side chain and Survivin glutamic acids E51 and E63. Survivin carbon, nitrogen, and oxygen atoms are shown as green, blue, and light red sticks, respectively. Histone atoms are shown in blue; phosphorous and oxygen atoms of phosphorylated T3 are colored orange and red, respectively. (B) Comparison of Survivin bound histone H3 peptide with and without K4 acetylation. Left, stick representation showing unacetylated (light blue) and acetylated (dark blue) peptides; center and right, electron density in the vicinity of Survivin histone H3 peptide binding site with the view on unmodified and acetylated lysine 4. The density around δ and ε carbon of lysine 4 is absent because unmodified K4 side chain can adopt many conformations including salt bridge formation between E51 or E63; therefore it appears disordered in the model. After K4 acetylation there are less possible conformations that can be adopted by acetylated K4 side chain due to steric effects; therefore lysine side chain appears ordered in the model. Map is contoured at 1 sigma.
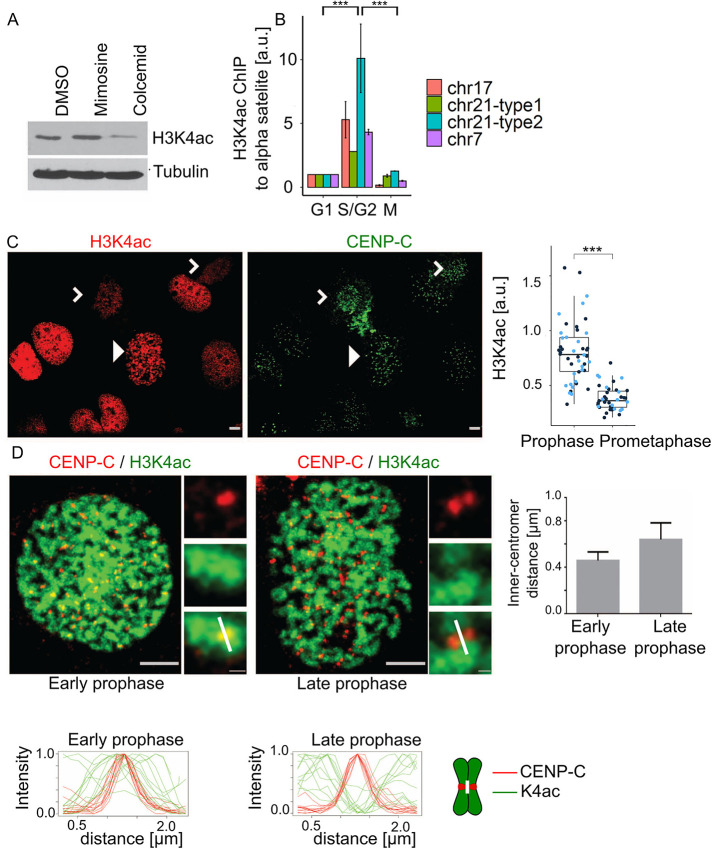
We next investigated the occurrence of H3K4ac across the cell cycle. We confirmed that the H3K4ac antibody has much higher reactivity against an H3K4ac peptide than a set of other modified histone tail peptides (Supplemental Figure S2A). The antibody does not recognize a peptide that is both T3ph and K4ac, which suggests that the positive signal from K4ac staining comes from histone H3 tails that are K4 acetylated and not T3 phosphorylated. We used this antibody to probe lysates from U2OS cells arrested in G1/S (mimosine arrest) or mitosis (colcemid arrest). These analyses found that H3K4ac was decreased in mitotic cells (Figure 2A). We next analyzed the temporal control of centromeric H3K4ac by chromatin immunoprecipitation (ChIP). We precipitated chromatin using the H3K4ac antibody from cells arrested in G1, S/G2, or M phase and probed for centromeric chromatin using four different α-satellite specific primers by quantitative PCR. We found that centromeres are specifically acetylated on H3K4 in S/G2 and the signal is reduced during mitosis and G1 (Figure 2B). The reduction of H3K4ac during mitosis was confirmed by immunofluorescence. The nuclei of most interphase cells had robust H3K4ac staining. The staining was also high in prophase cells but much lower in prometaphase cells (Figure 2C; arrowhead indicates a prophase cell; white carets indicate two prometaphase cells). In Supplemental Figure 2B we show a field with two prophase cells, and one is high in H3K4ac, while the other is low. This suggests that the mark is removed during prophase, which is the period when the CPC accumulates at inner centromeres (Wheatley et al., 2001; Li et al., 2006). We asked whether inner centromeres were deacetylated before other chromosome regions during prophase by costaining cells with a centromere marker (anti-CENP-C) and with the H3K4ac antibody. Prophase cells were divided into either early or late prophase based on the average distance between sister CENP-C foci, which are separated by chromosome condensation during prophase (Figure 2D; Supplemental Figure S2B). We quantified the CENP-C and H3K4ac signals in inner centromeres by quantifying along 2 µm lines between two sister kinetochores but running perpendicular to the axis defined by the two sister kinetochores (Figure 2D; note white line in inset merged images and on the chromosome cartoon). Line graphs of multiple centromeres were normalized and aligned to the peak of the CENP-C signal. H3K4ac peaked near the inner centromeres of all early prophase cells. In contrast, the inner centromere was the lowest point of H3K4ac in late prophase cells (Figure 2D). We conclude that H3K4ac inhibits CPC binding to histone tails in vitro (Figure 1) and the levels of this mark in the inner centromere change during midprophase (Figure 2). Thus the H3K4ac mark anticorrelates both spatially and temporally to the localization of the CPC to inner centromeres.
FIGURE 2:
Histone H3K4 is acetylated from S/G2 until midprophase during cell cycle progression. (A) Histone H3 K4ac level in lysates of U2OS either not synchronized (DMSO), arrested in G1/S with mimosine, or in mitosis with colcemid. (B) ChIP analysis where chromatin was immunoprecipitated with histone H3K4ac antibodies and probed by real-time PCR with primers that recognize four unique α-satellite sequences on the specified chromosomes. Note the increase in K4 acetylation signal in S/G2 (G2) relative to G1 and M phase. Experiment was performed twice. For statistical analysis an Anova test was applied; p value = 1.1⋅10–5; Tukey test was performed for pairwise comparisons; ***P < 0.001. (C) Immunofluorescence analysis of histone H3K4ac shows high level of H3K4ac in prophase cells (white arrowhead) n = (25, 14) but reduced H3K4ac signal during prometaphase (white carets) n = (25, 14); p value1 = (4.488⋅10–7, 2.564⋅10–5). For statistical analysis Welch’s t test was applied; ***P < 0.001. Red cells stained with anti-H3K4ac; green cells stained with anti-CENP-C; scale bar, 3 µm. The two blue-colored dots in the quantification represent the two experimental replicates. (D) The histone H3K4ac mark is removed from centromeres in midprophase. Images of an example of early and late prophase cells that were defined by the distance of CENP-C foci (plot on the right); the white mark in the merged inset shows a typical line portrayed in the intensity profiles in graphs below; scale bar, 3 µm; inset bar, 0.3 µm. Line graphs below images represent the fluorescent intensities of a 2.5-µm line drawn perpendicular to the axis of sister kinetochores are tiled for cells in either early or late prophase. Line graphs were normalized between 0 and 1, and the CENP-C peaks were centered; CENP-C (red, n = 12) and histone H3K4 (green, n = 12). A white line on the chromosome scheme next to line graphs highlights the region taken for analysis. There was enough CENP-C in inner centromere regions in late prophase chromosomes to define a central peak.
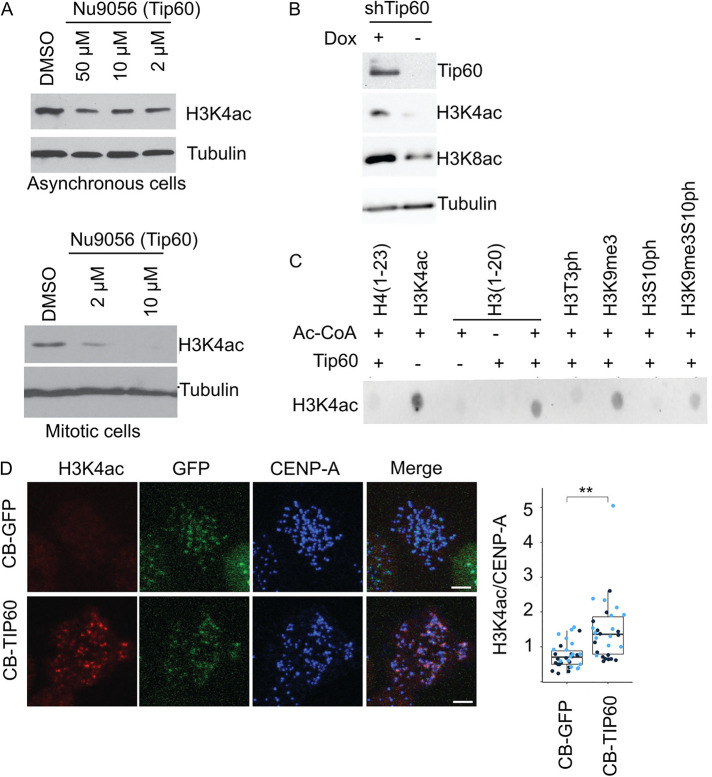
We next identified the H3K4 acetyltransferase that regulates this centromeric mark. We found inhibitors of Tip60 (Nu9056) but not GCN5 (CTPH2) or p300/CBP (Inhibitor VI) resulted in decreased H3K4ac levels in U2OS cells (Figure 3A; Supplemental Figure S3A). The small amount of H3K4 that remained in mitotic cells was also sensitive to the Tip60 inhibitor (Figure 3A). Cells depleted of Tip60 by shRNA also reduced H3K4ac (Figure 3B; Supplemental Figure S3D). Tip60 has been implicated as an S/G2 H3K4 acetyltransferase in fission yeast (Xhemalce and Kouzarides, 2010), and it also acetylates histone H4 on K118, which is adjacent to the site phosphorylated by Bub1 to control Sgo1 binding (Lee et al., 2018) and indirectly controls CPC levels. Interestingly, Tip60 also contains a chromodomain that binds the H3K9me3 mark, which can target it to heterochromatin (Xhemalce and Kouzarides, 2010; Rajagopalan et al., 2018) and Aurora B phosphorylates H3S10 to control binding of this mark by HP1 (Fischle et al., 2005). We confirmed that Tip60 acetylates H3K4 in vitro and found that this activity was retained on H3K9me3 peptides but the signal was missing when Tip60 was incubated with H3T3ph or H3S10ph peptides (Figure 3C; Supplemental Figure S3E). The H3K4ac signal was partially recovered on peptides that had both H3K9me3 and H3S10ph (Supplemental Figure S3E). Thus these data suggest Haspin and Aurora kinases may inhibit Tip60, which may underlie a switchlike response. To test if Tip60 could generate H3K4ac in vivo we generated cells that expressed a fusion of CENP-B-GFP and Tip60 (Figure 3D). CENP-B binds directly to α-satellite DNA and the fusion protein was exogenously targeted to centromere. Fusion of CENP-B to Tip60 was sufficient to generate H3K4ac at mitotic centromeres, where it is normally low. A control where we targeted GFP alone did not generate H3K4ac signal (Figure 3D). We conclude that Tip60 acetylates H3K4 in centromeric heterochromatin in S/G2 cells.
FIGURE 3:
KAT5/Tip60 (Tip60) is the major histone acetyltransferase (HAT) for H3K4 in U2OS cells. (A) Histone H3 K4 acetylation detected by anti-H3K4ac immunoblot analysis from asynchronous and mitotic U2OS cells treated with Nu9056 inhibitor of Tip60’s acetyltransferase activity. Bottom cells arrested in nocodazole were treated with Nu9056 for 1 h. (B) Tip60 shRNA depleted cells show reduced level of histone H3K4ac. Cells were engineered to express an shRNA against Tip60 after addition of doxycyclin (Dox). (C) In vitro Tip60 HAT activity on histone H3 peptides was detected using histone H3K4ac immunoblot; 1 µg of each peptide was incubated with the specified reagents and then transferred to a membrane for immunodetection. (D) Targeting of Tip60 to centromeres increases histone H3K4ac to that region of mitotic chromosomes. Cells were engineered to express either CENP-B:GFP (CB-GFP) or CENP-B:GFP:Tip60 (CB-Tip60) after the addition of doxycyclin. Cells in doxycycline were stained for the indicated antibodies and mitotic cells were imaged by immunofluorescence. Dark and light blue dots in the plot represent independent experimental replicates. CB-GFP n = (11, 23); CB-Tip60 n = (16, 14) p value = (0.004; 0.005). For statistical analysis Welch’s t test was applied; **P < 0.01.
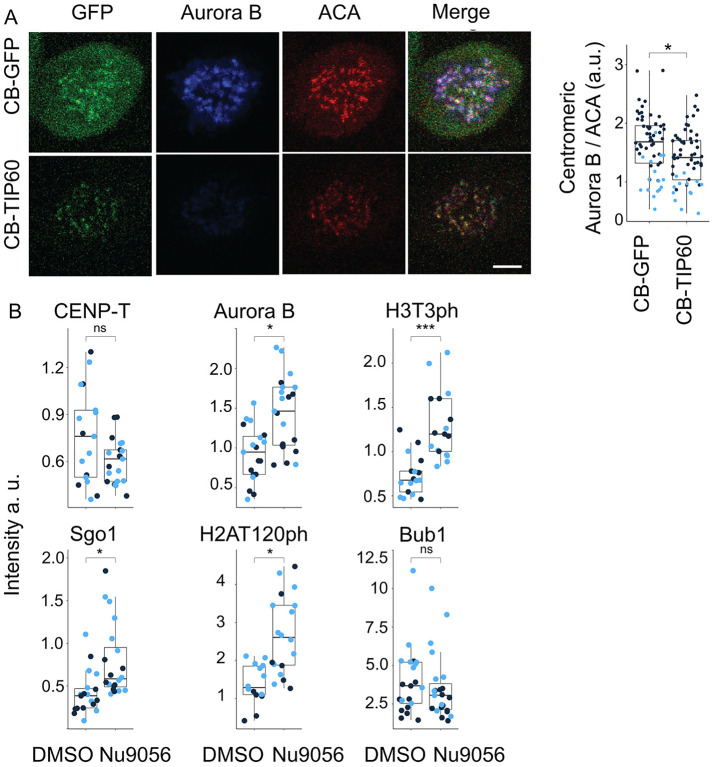
The CENP-B-GFP-Tip60 fusion construct also enabled a simple test for whether Tip60 targeting could displace the CPC from inner centromeres in mitosis. Cells expressing this construct had dramatically reduced levels of Aurora B at prometaphase and metaphase centromeres relative to controls (Figure 4A). These experiments were performed in nocodazole to limit regulation associated with mitotic progression (Figure 4A). We utilized this system to measure the ability of Tip60 to control most of the identified steps of CPC recruitment. Specifically, we compared the levels of regulators with those after the addition of a Tip60 inhibitor and found that inhibition of Tip60 stimulated the levels of Aurora B and most of its regulated steps (Figure 4B). The observed changes to Aurora B levels correlated with changes in H3T3ph and H2AT120 levels, which is consistent with Tip60 acetylation of both H3K4 and H2AK118 regulating CPC recruitment (Figure 4B). These data are consistent with the acetylation of histone H3K4 by Tip60 controlling both the kinetochore- and the cohesin-based branches of CPC recruitment in addition to preventing the binding of Survivin (Lee et al., 2018). We conclude that Tip60 is a master negative regulator of CPC localization.
FIGURE 4:
Targeting of Tip60 to centromeres dysregulates the localization of the Aurora B and other proteins of the Centromere Signaling Network (CSN). (A) Aurora B at centromeres is reduced when Tip60 is targeted to centromeres. Cells were arrested in mitosis with nocodazole and stained for the indicated antibody. CB-GFP n = (44, 16); CB-Tip60 n = (43, 13); p value = (0.0015; 0.033); scale bar, 5 µm. (B) Tip60 targeting affects many kinetochore/inner centromere events in an acetyltransferase dependent manner. Cells expressing CB-Tip60 were treated with either DMSO or the Tip60 acetyltransferase inhibitor Nu9056, and the localization of the indicated protein or PTM were imaged by immunofluorescence. Light and dark blue dots indicate two independent experimental replicates. Quantification of fluorescence signal of Aurora B nDMSO = (10, 9), nNU9056 = (11, 10), p value = (0.01, 0.0034); Sgo1 nDMSO = (10, 10), nNU9056 = (9, 11), p value = (0.049, 0.042); CENP-T nDMSO = (6, 11), nNU9056 = (10, 11), p value = (0.486, 0.051); Bub1 nDMSO = (11, 10), nNU9056 = (12, 11); p value = (0.4184, 0.6519); histone H3T3ph nDMSO = (8, 9), nNU9056 = (7, 9); p value (0.0018, 0.0025) and histone H2AT120ph nDMSO = (5, 9), nNU9056 = (6, 11) p value = (0.029, 0.0017) at centromeres in cells with Tip60 targeted to centromeres by CENP-B fusion and treated with Nu9016. For statistical analysis Welch’s t test was applied. ***P < 0.001, **P < 0.01, *P < 0.05, and ns P > 0.05.
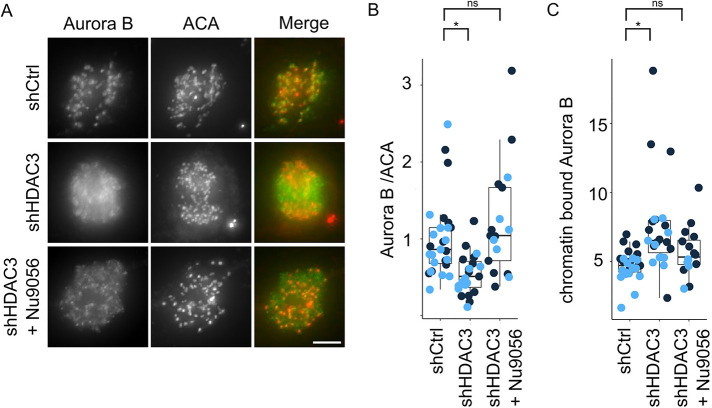
The histone deacetylase 3 (HDAC3) was previously shown to remove H3K4ac during mitosis (Eot-Houllier et al., 2008). We therefore asked if HDAC3 acts in opposition to Tip60 to localize the CPC. Depletion of HDAC3 by shRNA reduced the levels of Aurora B in the inner centromere (Supplemental Figure S3, B–D), suggesting that HDAC3 must remove H3K4ac to enable CPC localization. Interestingly, inhibition of Tip60 during S/G2 in cells depleted of HDAC3 restores CPC levels (Figure 5). The simplest explanation of these data is that HDAC3 removes the inhibitory mark on H3K4 put down by Tip60 to control CPC localization.
FIGURE 5:
The histone deacetylase HDAC3 activity opposes Tip60 activity to relocalize Aurora B from chromatin to inner centromeres. Cells treated to reduce HDAC3 by shRNA had reduced amounts of Aurora B but this was rescued by concurrent inhibition of Tip60 acetyltransferase activity. (A) Immunofluorescence images showing Aurora B inner centromeric localization in prometaphase cells treated with shHDAC3 knockdown with and without Tip60 inhibition. Note that most of the Aurora B remains on noninner centromeric (chromosome arm) regions after HDAC3 depletion. However, if Tip60 was inhibited in the previous S/G2 phase then HDAC3 is not required to localize Aurora B to inner centromeres; scale bar, 5 µm. (B) Quantification of Aurora B signal at centromeres after the indicated treatment: shCtrl n = (11, 13); shHDAC3 n = (13, 10); shHDAC3+Nu9056 n = (11, 5); p valueshCtrl-shHDAC3 = (0.0211, 0.0046); p valueshCtrl-shHDAC3+Nu9056 (0.49, 0.63). (C) Quantification of chromatin bound Aurora B signal after the centromeric signals have been subtracted to measure the amount of Aurora B on chromosome arms after the indicated treatments. shCtrl n = (11, 13); shHDAC3 n = (13, 10); shHDAC3+Nu9056 n = (11, 5); p valueshCtrl-shHDAC3 = (0.02678, 0.00065); p valueshCtrl-shHDAC3+Nu9056 (0.25, 0.41). For statistical analysis Welch’s t test was applied; ns > 0.05, *P < 0.05. Light and dark blue dots indicate two independent experimental replicates.
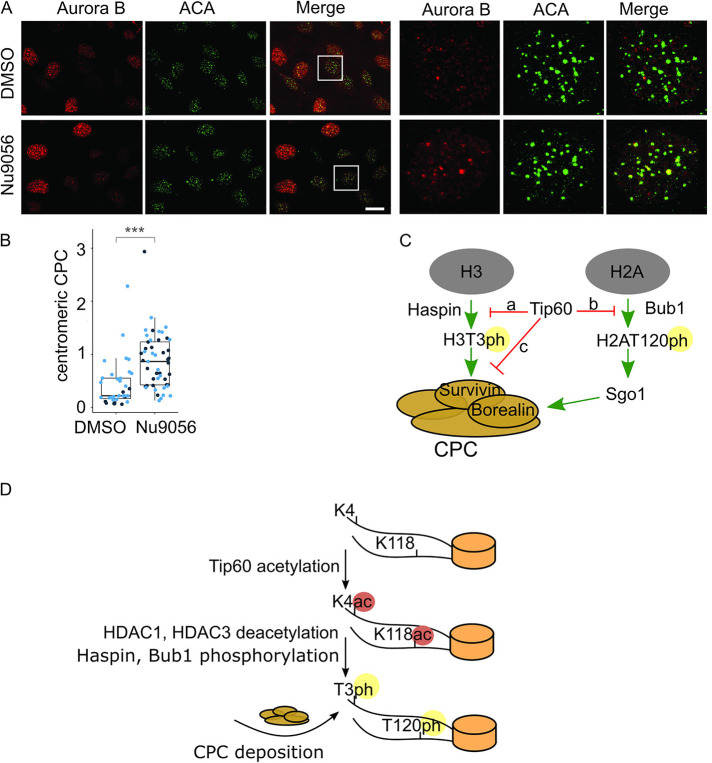
It was previously shown that CPC levels are higher in G2 nuclei than G1 nuclei, although the levels are lower than in mitotic centromeres (Monier et al., 2007). Our data suggest that Tip60 acetylates H3K4 in S/G2 cells on centromeric sequences. This predicts that we should find higher levels of CPC proteins on centromeres of interphase cells after inhibition of Tip60. In fact, Aurora B rarely colocalizes with the ACA centromere marker in interphase cells. However, both the levels of Aurora B and the number of Aurora B positive centromeres increased in cells where Tip60 had been inhibited (Figure 6, A and B). We conclude that Tip60 prevents premature localization of the CPC to centromeres.
FIGURE 6:
Inhibition of Tip60 promotes premature centromeric targeting of Aurora B in interphase cells. (A) Inhibition of Tip60 by Nu9056 leads to Aurora B localization at centromeres in interphase (S/G2) cells as shown by immunofluorescence; scale bar, 20 µm. (B) Quantification of S/G2 centromeric fluorescence signals for Aurora B in control cells (n = 9, 26) and in Tip60-inhibited cells (n = 26, 23) p value (2.833⋅10–7, 8.152⋅10–5). Aurora B signal that overlaps with an ACA mask was quantified in S/G2 cells as defined by Aurora B signal. G1 cells have low Aurora B signal since Aurora B is an APC substrate. For statistical analysis Welch’s t test was applied. ***P < 0.001. (C) A proposed model for Aurora B localization regulated by acetylation of H3K4 of H3 by Tip60. Our data suggest that histone H3K4ac by Tip60 during S/G2 prevents Haspin phosphorylation of H3T3 (arrow a) and Survivin binding to chromatin to inhibit CPC localization to centromeres (arrow c). During the middle of prophase HDAC3 removes H3K4ac mark to enable CPC accumulation at inner centromeres. Previous studies have shown that Tip60 inhibits Bub1 binding to H2AT120 (arrow b) (Lee et al., 2018) and together with our work it suggests that Tip60 is a master regulator of CPC localization. (D) A model to control the timing of CPC localization to inner centromeres through changes in chromatin PTMs. Tip60 acetylates histone H3 on K4 and histone H2A on K118 in S/G2 to prevent CPC localization. In midprophase HDAC1 and HDAC3 deacetylate these histone marks to enable Bub1 phosphorylation and subsequent Sgo1 recruitment, Haspin phosphorylation of histone H3T3 and subsequent Survivin binding of H3T3ph. Together these events drive CPC recruitment to inner centromeres in midprophase.
DISCUSSION
We demonstrate that Tip60 acetylates H3K4 to temporally control the localization of the CPC. Tip60 acetylates H3K4 during S/G2 and prevents Haspin phosphorylation of H3T3 and Survivin binding to chromatin to inhibit CPC localization to centromeres. During the middle of prophase HDAC3 removes this mark to enable CPC accumulation at inner centromeres (Figure 6C). Tip60 also inhibits Histone H2A phosphorylation, which is a second histone mark that controls CPC localization. These data complement previous studies that demonstrated that Tip60 acetylates H2AK118 and can prevent Haspin phosphorylation of H3T3 in vitro. Together, these data suggest that Tip60 is a master negative regulator of CPC recruitment to centromeres (Figure 6D). We also show that HDAC3, which has previously been shown to deacetylate H3K4 in mitosis, acts in opposition to Tip60 to temporally control CPC localization. These data complement the finding that RSF1/HDAC1 is localized to mitotic centromeres to deacetylate H2AK118 (Lee et al., 2018). Thus two independent deacetylases (HDAC1 and HDAC3) temporally control the initiation of CPC localization in prophase (Figure 6D). We note that many CPC studies were performed in HeLa cells where this regulation would be missing because the E6 protein targets Tip60 for degradation in cells transformed by oncogenic papilloma viruses (Jha et al., 2010).
The use of both an inhibitory histone mark and an activating mark to regulate Survivin histone reading not only adds extra steps of regulation but also greatly increases the affinity change for this histone code reader. Survivin binding to H3 is only two- to fivefold stimulated by phosphorylation (Wang et al., 2010; Niedzialkowska et al., 2012), so it was unclear how this could generate a switch from exclusion to the recruitment of the CPC. We suggest that acetylation of H3K4 actively inhibits recruitment of CPC thereby considerably lowering the affinity of Survivin binding in G2. A positive feedback loop between the CPC and Haspin ensures rapid recruitment of the CPC (Wang et al., 2011). Our data suggest that future studies should test whether HDAC3 is a target of this regulation. The fact that these steps are both inhibited until midprophase provides a need for rapid switchlike activation of steps that localize the CPC.
Our studies suggest a number of important areas for future studies. First, it is critical to understand the mitotic regulation of HDAC3 and HDAC1. We detect specific deacetylation of H3K4 at inner centromeres in prophase that precedes the bulk deacetylation that happens by prometaphase. We suggest that there must be mechanisms to recruit HDAC3 to the inner centromere in prophase and independent steps to recruit HDAC1. It is also interesting that Tip60 binds to H3 that is methylated on K9 in response to double-strand breaks (Sun et al., 2005, 2009; Ayrapetov et al., 2014). This mark might ensure that this regulation is confined to heterochromatin. Second, it is also important to determine if the recognition of Tip60 is controlled by histone H3 S10 phosphorylation which could inhibit Tip60 binding and tip the equilibrium to HDAC3 to initiate the cascade. Third, a new pool of the CPC has been identified at the inner kinetochore, and it is unclear if Tip60 also regulates this pool, which became difficult to observe after HDAC3 depletion (Broad et al., 2020; Hadders et al., 2020; Liang et al., 2020).
Tip60 is an important tumor suppressor, and, like the CPC, it has been implicated in chromosome instability (Grezy et al., 2016). Interestingly oncogenic papillomaviruses degrade both Tip60 and p53 and generate strong chromosome instability (Scheffner et al., 1990, 1993; Jha et al., 2010). We suggest that reduced Tip60 misregulates CPC to lower the fidelity of chromosome segregation, while the simultaneous loss of p53 would allow these cells to remain proliferative to drive tumorigenesis.
MATERIALS AND METHODS
Cell lines
All experiments were performed in U2OS cells unless specified. Since the papilloma virus expressed E6 targets Tip60 for degradation, it was critical to avoid HeLa cells in these experiments which express high levels of E6 (Jha et al., 2010). U2OS cells were grown in McCoy’s 5A modified media (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies). For plasmid transfection cells were grown to 80–90% confluency. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. The human lentiviral shRNAmir pGIPZ constructs were obtained from Open Biosystems and grown and purified according to their protocol. The targeting sequences of the shRNAs used in this study are HDAC3 (AGAAGTCCACTACCTGGTT) and control (nonsilencing, TCGCTTGGGCGAGAGTAAG). To package virus, 1.5 × 107 Lenti-X 293T cells were cotransfected with 18 μg pGIPZ plasmid, 6 μg pMD2G plasmid, and 12 μg psPAX2 plasmid. Medium were replenished 24 h after transfection and supernatants containing virus were collected and filtered through 0.2-μm filters 48 h after transfection. U2OS cells were incubated with virus for 24 h at 50% confluency; 48 h after infection with virus, cells were trypsinized, transferred to a 10-cm dish, and grown for a week in the presence of 3 μg/ml puromycin. For Aurora B localization experiments (Figure 5), U2OS cells were incubated with 0.5 μg/ml puromycin and 1 μg/ml doxocycline for 72 h to induce expression of shRNA; 10 μM of Nu9056 (Sigma) were added 1 h before fixation. A stable U2OS cell line expressing CENP-B fused to Tip60 (in pcDNA5.0/FRT/TO-CB-Tip60) was generated by cotransfecting these constructs with pOG44 (Invitrogen) into Flp-In U2OS T-REx cells and selection with hygromycin (300 µg/ml, Invitrogen) for 2 wk. For CB-Tip60 targeting experiments (Figure 4), cells were incubated with 1 μg/ml doxocycline for 24 h and then with 3.3 μM nocodazole for a total 2 h; 10 μM Nu9056 were added for 1 h after 1 h of incubation with nocodazole, and cells were fixed.
Immunoblotting and immunofluorescence
These antibodies were purchased from the following sources and used for immunoblotting at a dilution as indicated: Aurora-B (1:500 Bethyl, A300-431A), CENP-T (gift from Dan Foltz, 1:1000), tubulin (Sigma, T4026, 1:5000), histone H3 T3 phosphorylation (Cell Signaling, 1:1:1000), histone H3 K4 acetylation (H3K4ac Millipore, 1:1000), Tip60 (Santa Cruz 1:2000), Aurora-B pT232 (Rockland, 1:200) ACA (anti-centromere antigen, Antibodies Inc., 15-234-0001, 1:500), Sgo1 (ThermoFisher, 1:250), CENP-C (Millipore 1:1000), CENP-A (Abcam, 1:1000), and Bub1 (Genetex, 1:250). U2OS cells were synchronized to mitosis with 100 ng/ml colcemid for 16 h or arrested in 2 mM thymidine blocked to arrest cells in S phase.
For immunofluorescence U2OS cells were seeded onto coverslips coated with poly-l-Lysine (Sigma 1 d before staining. The cells were cofixed with 4% paraformaldehyde, PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, and 4 mM MgCl2, pH 6.9) and 0.5% Triton-X 100 for 20 min at room temperature (RT). After washing with TBS-T three times, cells were blocked with 3% bovine serum albumin (BSA) for 60 min. Immunostaining was performed with the following primary antibodies at the indicated dilution for 1 h at RT. After washing three times with TBST-T, cells were incubated with fluorescent secondary antibodies (Jackson ImmunoResearch) for 30 min at RT. After washing two times with PBS, the cells were counterstained with 0.5 µg/ml DAPI for 5 min. After two more washes with water, the coverslips were mounted onto slides using ProlongGold Antifade (Invitrogen) and sealed with nail polish. Image acquisition was performed using at 63× on Zeiss Observer Z1 widefield microscope. Images were processed and analyzed using FIJI. Z stacks were projected using maximum intensity Z projection. To quantify fluorescence levels at centromeres, a thresholding algorithm was used to mark centromeres on the basis of ACA mask in projected images. Background was subtracted using the in-built rolling ball algorithm in FIJI. Indicated statistical analysis was performed using R (v.3.6.3). All box and whisker graphs represent the median (central line), 25th–75th percentile (bounds of the box), and 5th–95th percentile (whiskers). Data for line graphs of prophase U2OS cells were generated using Volocity software (PerkinElmer V6.3). To distinguish early and late prophase cells, lines were drawn between the two paired CENP-C spots and the inner centromere distance was calculated for each of the pairs. To visualize the depletion of H3K4 between early and late prometaphase, lines were drawn between the paired CENP-C foci and the lines were perpendicular to the plane linking the paired CENP-C centromere foci. Line graphs were normalized so that they had identical upper and lower intensities and aligned with each of the lines so that the peak of the CENP-C intensity was centered. The R script is available on request.
H3K4 ChIP experiments
U2OS cells were arrested using double thymidine block and released for 6 h (S/G2), nocodazole block (M), or nocodazole block and release (G1). ChIP was performed as described (Kuscu et al., 2019) using a set of previously described PCR primers against unique α-satellite repeats at the centromeres of each of the specified chromosomes (Warburton et al., 1991).
Protein purification, crystallization, and data collection
Protein purification was carried out as described previously (Niedzialkowska et al., 2012). hSurvivin (UniProt ID: O15392) was expressed from p8HIS vector (DNA sequence confirmed by sequencing) in BL21(DE3)RIPL cells in the presence of kanamycin 100 µg/ml and chloramphenicol 34 µg/ml. Protein expression was induced when the culture reached OD600 of 1.0 with the final concentration of 0.150 mM of IPTG; 60 µg/l of ZnCl2 were added to media to facilitate hSurvivin expression. Cells were incubated overnight at 18°C. The cell pellet was resuspended in a lysis buffer composed of 50 mM Tris, pH = 7.8, 2 mM imidazole, 120 mM NaCl, and 10 mM β-mercaptoethanol, and cells were lysed using Avestin EmulsiFlex C3 homogenizer. Cell lysate was clarified by ultracentrifugation for 40 min at 4°C in Beckman Coulter Type 45 Ti fixed angle rotor at 30 000 rpm. Lysate was applied on NiNTA resin, and beads were washed with a wash buffer composed of 50 mM Tris, pH = 7.8, 10 mM imidazole, 500 mM NaCl, and 10 mM β-mercaptoethanol. Protein was eluted in the elution buffer, and 8His tag was removed by overnight thrombin digestion during dialysis in a buffer composed of 50 mM Tris, pH = 7.8, 250 mM imidazole, 500 mM NaCl, and 10 mM β-mercaptoethanol. Digested protein was applied on NiNTA resin equilibrated with digestion buffer composed of 50 mM Tris, pH = 8.0, 150 mM NaCl, and 10 mM β-mercaptoethanol. Flow-through containing hSurvivin was applied on Superdex 200 column attached to AKTA system with a flow 1 ml/min in a buffer composed of 5 mM HEPES 7.5 and 10 mM β-mercaptoethanol. Fractions containing purified Survivin were pooled together and concentrated to 12 mg/ml. For cocrystallization and soaking experiments, lyophilized peptides (GenScript, Piscataway, NJ) were suspended in bacteriostatic water to 50 mM concentration. Crystals of hSurvivin were grown in hanging drops at 16°C using EasyXtal Tools crystallization plates. The crystallization drop was composed of 1 μl of protein in 5 mM HEPES, pH = 7.5, 10 mM β-mercaptoethanol, and 1 μl of mother liquor solution; 33% ethylene glycol in crystal mother liquor was used as a cryoprotectant; in the case of hSurvivin in complex with H3T3phK4me2 LV oil was used. The list of crystallization solutions is presented in Supplemental Table S1. The complexes of hSurvivin with H3T3phK4me1(1-12), H3T3phK4me3(1-12), and H3T3phK4ac(1-12) were obtained from cocrystallization experiments, while the complex of hSurvivin with H3T3phK4me2(1-12) peptide was obtained from soaking procedure.
Data were collected at 100 K on sector 19 and sector 21 at Argonne National Laboratory (Argonne, IL). Structures of wild-type hSurvivin with H3T3phK4me1(1-12), H3T3phK4me2(1-12), H3T3phK4me3(1-12), and H3T3phK4ac(1-12) were solved by molecular replacement using 3UEC (Niedzialkowska et al., 2012) structure as a model. Data processing and model building were done with HKL-3000 (Minor et al., 2006). Molecular replacement was performed using HKL-3000 and MOLREP (Murshudov et al., 1997). The resulting model was further refined with REFMAC5 (Murshudov et al., 1997) and COOT (Emsley et al., 2010). The structure was validated using MOLPROBITY (Chen et al., 2010) and ADIT (Yang et al., 2004) tools. Figures were prepared using PyMOL (www.pymol.org). The diffraction images are available on the Integrated Resource for Reproducibility in Macromolecular Crystallography Web site (https://proteindiffraction.org/) (Grabowski et al., 2016). The ligands in the active site and the electron density maps, including omit maps, can be inspected interactively using Molstack (Porebski et al., 2018) at https://molstack.bioreproducibility.org/c/YNYK/.
All crystals diffracted to similar resolution (around 2.6 Å). Most of the complexes crystallized in the C2 space group except for H3T3phK4me2, which crystallized in the I222 space group. The former complex was more difficult to crystallize and handle; the few obtained crystals diffracted poorly and the resulting data and refinement were worse than for other complexes. In all cases, similarly to previous studies, the electron density for only the first four amino acids was interpretable. In the case of the structure of hSurvivin in the complex with H3T3phK4me2 peptide, it was difficult to obtain crystals of the complex from cocrystallization or soaking experiments and the crystals were very sensitive to any cryoprotectant used and the process of flash cooling in liquid nitrogen. The data obtained from diffraction experiments of the crystal of hSurvivin in complex with H3T3phK4me2 were of poor quality and refinement statistics. Data collection and refinement statistics are presented in Supplemental Table S2.
Isothermal titration calorimetry
Before each ITC experiment, the protein was dialyzed overnight against the proper buffer. Lyophilized peptides (GenScript, Piscataway, NJ) with sequences corresponding to the first 12 amino acids of histone H3 were dissolved in bacteriostatic water to obtain 50 mM stock concentration. Before each experiment the peptides were diluted with a dialysis buffer to a desired working concentration. ITC measurements were performed using an ITC200 isothermal titration calorimeter (GE Healthcare). The binding experiments were carried out in 50 mM citric acid, HEPES, and CHES buffer system at pH equaled 7.2. Citric acid, HEPES, and CHES buffers were prepared as described (Newman, 2004). Protein concentration equaled 0.1 mM and the peptide concentration equaled 1 mM. The first titration was carried out using 0.5 µl of peptide solution, followed by 35 1.5-µl injections applied 180 s apart. Proper mixing of titrated peptide with protein in a chamber was ensured by constant stirring applied at the speed of 700 rpms during the experiment. Titration experiments were conducted at 25°C. Binding isothermal fit was done with Origin software using single binding site model with stoichiometry, ΔH, association constant (Ka) as variable.
Histone acetylation and antibody dot blot assays
Acetylation reaction was done for 1 h at 30°C. Reaction was done in a buffer composed of 50 mM Tris, pH 8.0, 0.1 mM EDTA, and 5% glycerol and contained 0.01 µg/µl Tip60 (SignalChem, K314-380G-05), 1 mM AcCoA (Sigma), 1 mM dithiothreitol, and 10 ηg/µl of BSA. As a substrate 1000, 500, 250, 125, and 62.5 ηg of histone H3 peptide were used. Following the HAT assay, dot blot analysis was performed as described in that work (Perez-Burgos et al., 2004) with minor modifications. Briefly, 2 µl of the TIP60 acetylation reactions were spotted on polyvinylidene difluoride membranes. The membranes were then blocked for 1 h at RT in TBST (0.1% Tween) and 3% BSA followed by overnight incubation at 4°C with the H3K4ac antibody (Millipore) at 1:5000 in the same buffer and immunoblot detection.
Accession numbers
PDB ID: 7LBO – hSurvivin with H3T3phK4me1 peptide
PDB ID: 7LBQ – hSurvivin with H3T3phK4me2 peptide
PDB ID: 7LBK – hSurvivin with H3T3phK4me3 peptide
PDB ID: 7LBP – hSurvivin with H3T3phK4ac peptide
Supplementary Material
Acknowledgments
The work described here was supported by NIH Grants No. GM118798 and No. GM124042 to P.T.S., No. GM126900 to B.D.S., No. GM053163 to W.M.; E.N. was supported by a grant from the Schiff foundation. The structural data shown in this report used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the SBC 19-ID sector was supported by the Structural Biology Center (SBC). Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant No. 085P1000817).
Abbreviations used:
- ChIP
chromatin immunoprecipitation
- CPC
chromosome passenger complex
- CSN
centromere signaling network
- Dox
doxycyclin
- H3K4ac
histone H3 lysine 4
- H3T3ph
histone H3 threonine 3
- HAT
histone acetyltransferase
- HDAC3
histone deacetylase 3
- INCENP
inner centromere protein
- ITC
isothermal titration calorimetry
- PP2A
protein phosphatase 2
- PTMs
posttranslational modifications
- RT
room temperature.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-06-0283) on June 2, 2022.
REFERENCES
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD (2014). DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci USA 111, 9169–9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broad AJ, DeLuca KF, DeLuca JG (2020). Aurora B kinase is recruited to multiple discrete kinetochore and centromere regions in human cells. J Cell Biol 219, e201905144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Pinson X, Platani M, Salloum Z, Xu Z, Clark A, Macisaac F, Ogawa H, Eggert U, Glover DM, et al. (2012). The chromosomal passenger complex activates Polo kinase at centromeres. PLoS Biol 10, e1001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Heck MM, Earnshaw WC (1987). The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol 105, 2053–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Higgins JM (2005). Haspin: a mitotic histone kinase required for metaphase chromosome alignment. Cell Cycle 4, 665–668. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eot-Houllier G, Fulcrand G, Watanabe Y, Magnaghi-Jaulin L, Jaulin C (2008). Histone deacetylase 3 is required for centromeric H3K4 deacetylation and sister chromatid cohesion. Genes Dev 22, 2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD (2005). Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122. [DOI] [PubMed] [Google Scholar]
- Goto Y, Yamagishi Y, Shintomi-Kawamura M, Abe M, Tanno Y, Watanabe Y (2017). Pds5 Regulates Sister-Chromatid Cohesion and Chromosome Bi-orientation through a Conserved Protein Interaction Module. Curr Biol 27, 1005–1012. [DOI] [PubMed] [Google Scholar]
- Grabowski M, Langner KM, Cymborowski M, Porebski PJ, Sroka P, Zheng H, Cooper DR, Zimmerman MD, Elsliger MA, Burley SK, Minor W (2016). A public database of macromolecular diffraction experiments. Acta Crystallogr D Struct Biol 72, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grezy A, Chevillard-Briet M, Trouche D, Escaffit F (2016). Control of genetic stability by a new heterochromatin compaction pathway involving the Tip60 histone acetyltransferase. Mol Biol Cell 27, 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadders MA, Hindriksen S, Truong MA, Mhaskar AN, Wopken JP, Vromans MJM, Lens SMA (2020). Untangling the contribution of Haspin and Bub1 to Aurora B function during mitosis. J Cell Biol 219, e201907087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Lee KH, Hyun S, Lee NJ, Lee SJ, Hwang H, Yu J (2011). Methylation-mediated control of aurora kinase B and Haspin with epigenetically modified histone H3 N-terminal peptides. Bioorg Med Chem 19, 2373–2377. [DOI] [PubMed] [Google Scholar]
- Hindriksen S, Lens SMA, Hadders MA (2017). The Ins and Outs of Aurora B Inner Centromere Localization. Front Cell Dev Biol 5, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A (2010). Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell 38, 700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H (2010). Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 330, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Mammadov R, Czikora A, Unlu H, Tufan T, Fischer NL, Arslan S, Bekiranov S, Kanemaki M, Adli M (2019). Temporal and Spatial Epigenome Editing Allows Precise Gene Regulation in Mammalian Cells. J Mol Biol 431, 111–121. [DOI] [PubMed] [Google Scholar]
- Lee HS, Lin Z, Chae S, Yoo YS, Kim BG, Lee Y, Johnson JL, Kim YS, Cantley LC, Lee CW, et al. (2018). The chromatin remodeler RSF1 controls centromeric histone modifications to coordinate chromosome segregation. Nat Commun 9, 3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kao GD, Garcia BA, Shabanowitz J, Hunt DF, Qin J, Phelan C, Lazar MA (2006). A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev 20, 2566–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Zhang Z, Chen Q, Yan H, Zhang M, Zhou L, Xu J, Lu W, Wang F (2020). Centromere-localized Aurora B kinase is required for the fidelity of chromosome segregation. J Cell Biol 219, e20190709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Jia L, Yu H (2013). Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Curr Biol 23, 1927–1933. [DOI] [PubMed] [Google Scholar]
- Mahen R, Koch B, Wachsmuth M, Politi AZ, Perez-Gonzalez A, Mergenthaler J, Cai Y, Ellenberg J (2014). Comparative assessment of fluorescent transgene methods for quantitative imaging in human cells. Mol Biol Cell 25, 3610–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006). HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62, 859–866. [DOI] [PubMed] [Google Scholar]
- Mo F, Zhuang X, Liu X, Yao PY, Qin B, Su Z, Zang J, Wang Z, Zhang J, Dou Z, et al. (2016). Acetylation of Aurora B by TIP60 ensures accurate chromosomal segregation. Nat Chem Biol 12, 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier K, Mouradian S, Sullivan KF (2007). DNA methylation promotes Aurora-B-driven phosphorylation of histone H3 in chromosomal subdomains. J Cell Sci 120, 101–114. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997). Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Newman J (2004). Novel buffer systems for macromolecular crystallization. Acta Crystallogr D Biol Crystallogr 60, 610–612. [DOI] [PubMed] [Google Scholar]
- Niedzialkowska E, Wang F, Porebski PJ, Minor W, Higgins JM, Stukenberg PT (2012). Molecular basis for phosphospecific recognition of histone H3 tails by Survivin paralogues at inner centromeres. Mol Biol Cell 23, 1457–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Burgos L, Peters AH, Opravil S, Kauer M, Mechtler K, Jenuwein T (2004). Generation and characterization of methyl-lysine histone antibodies. Methods Enzymol 376, 234–254. [DOI] [PubMed] [Google Scholar]
- Pfister K, Pipka JL, Chiang C, Liu Y, Clark RA, Keller R, Skoglund P, Guertin MJ, Hall IM, Stukenberg PT (2018). Identification of drivers of aneuploidy in breast tumors. Cell Rep 23, 2758–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski PJ, Sroka P, Zheng H, Cooper DR, Minor W (2018). Molstack-Interactive visualization tool for presentation, interpretation, and validation of macromolecules and electron density maps. Protein Sci 27, 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan D, Tirado-Magallanes R, Bhatia SS, Teo WS, Sian S, Hora S, Lee KK, Zhang Y, Jadhav SP, Wu Y, et al. (2018). TIP60 represses activation of endogenous retroviral elements. Nucleic Acids Res 46, 9456–9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert JG, Samejima K, Platani M, Molina O, Kimura H, Jeyaprakash AA, Ohta S, Earnshaw WC (2018). HP1alpha targets the chromosomal passenger complex for activation at heterochromatin before mitotic entry. EMBO J 37, e97677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002). Active genes are tri-methylated at K4 of histone H3. Nature 419, 407–411. [DOI] [PubMed] [Google Scholar]
- Sapountzi V, Logan IR, Robson CN (2006). Cellular functions of TIP60. Int J Biochem Cell Biol 38, 1496–1509. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Huibregtse JM, Vierstra RD, Howley PM (1993). The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75, 495–505. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM (1990). The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63, 1129–1136. [DOI] [PubMed] [Google Scholar]
- Storchova Z, Becker JS, Talarek N, Kogelsberger S, Pellman D (2011). Bub1, Sgo1, and Mps1 mediate a distinct pathway for chromosome biorientation in budding yeast. Mol Biol Cell 22, 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005). A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 102, 13182–13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD (2009). Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 11, 1376–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P, Stukenberg PT (2016). A centromere-signaling network underlies the coordination among mitotic events. Trends Biochem Sci 41, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JM (2010). Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 330, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Ulyanova NP, van der Waal MS, Patnaik D, Lens SM, Higgins JM (2011). A positive feedback loop involving Haspin and Aurora B promotes CPC accumulation at centromeres in mitosis. Curr Biol 21, 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Greig GM, Haaf T, Willard HF (1991). PCR amplification of chromosome-specific alpha satellite DNA: definition of centromeric STS markers and polymorphic analysis. Genomics 11, 324–333. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC (2001). INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol 11, 886–890. [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Kouzarides T (2010). A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev 24, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y, Honda T, Tanno Y, Watanabe Y (2010). Two histone marks establish the inner centromere and chromosome bi-orientation. Science 330, 239–243. [DOI] [PubMed] [Google Scholar]
- Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD (2004). Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr D Biol Crystallogr 60, 1833–1839. [DOI] [PubMed] [Google Scholar]
- Zhiteneva A, Bonfiglio JJ, Makarov A, Colby T, Vagnarelli P, Schirmer EC, Matic I, Earnshaw WC (2017). Mitotic post-translational modifications of histones promote chromatin compaction in vitro. Open Biol 7, 170076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.