Abstract
Polo-like-kinase (PLK) 1 activity is associated with maintaining the functional and physical properties of the centrosome’s pericentriolar matrix (PCM). In this study, we use a multimodal approach of human cells (HeLa), zebrafish embryos, and phylogenic analysis to test the role of a PLK1 binding protein, cenexin, in regulating the PCM. Our studies identify that cenexin is required for tempering microtubule nucleation by maintaining PCM cohesion in a PLK1-dependent manner. PCM architecture in cenexin-depleted zebrafish embryos was rescued with wild-type human cenexin, but not with a C-terminal cenexin mutant (S796A) deficient in PLK1 binding. We propose a model where cenexin’s C terminus acts in a conserved manner in eukaryotes, excluding nematodes and arthropods, to sequester PLK1 that limits PCM substrate phosphorylation events required for PCM cohesion.
INTRODUCTION
The centrioles and surrounding pericentriolar matrix (PCM) define the centrosome as one of the most complex nonmembranous organelles in the cell (Mennella et al., 2014; Vertii et al., 2016). Despite the centrosome’s structural and molecular complexity, the most characterized function of the centrosome is to nucleate and organize polarized microtubule (MT) arrays that generate cell polarity and form the structural framework for the mitotic spindle (Vertii et al., 2016). One way this function is regulated is by the centrosome acting as a scaffold to regulatory molecules, such as the mitotic kinase, polo-like kinase 1 (PLK1). PLK1 is a major regulator of bipolar spindle formation through PLK1-scaffold interactions at mitotic centrosomes/spindle poles. Once PLK1 is recruited to the centrosome, it modulates the phosphorylation and assembly of centrosome components, such as pericentrin and Cep215, which are needed for γ-tubulin and γ-TuRC recruitment resulting in PCM expansion (Wueseke et al., 2016; Colicino and Hehnly, 2018; Ohta et al., 2021). This expansion, termed centrosome maturation, plays a crucial role in mitotic centrosome function during division (Lee and Rhee, 2011; Ohta et al., 2021). Following maturation, continued PLK1 activity has been shown to be essential for PCM cohesion (Cabral et al., 2019; Mittasch et al., 2020; Rathbun et al., 2020).
Centrosome localized PLK1 scaffolds include Sas-4 (Novak et al., 2016), Ana1(Alvarez-Rodrigo et al., 2020), Cep192 (Gomez-Ferreria et al., 2007), and Odf2 isoform 9, called cenexin (Soung et al., 2006, 2009; Chang et al., 2013). Sas-4 and Ana1 are reported to recruit PLK1 to the centrioles (Novak et al., 2016; Alvarez-Rodrigo et al., 2020), whereas Cep192 is reported to recruit an initial population of PLK1 to the PCM during bipolar spindle formation in mammalian, Drosophila, and Caenorhabditis elegans dividing cells (Kemp et al., 2004; Conduit et al., 2014; Joukov et al., 2014; Meng et al., 2015). However, much less is known about cenexin’s role during this time and the studies that have been performed were done primarily in murine or human cell models (Soung et al., 2006, 2009; Chang et al., 2013) with little known about cenexin’s conservation across phyla.
Previous studies identified a cenexin-dependent enrichment of PLK1 at the oldest mitotic centrosome in human cells (Colicino et al., 2019). Owing to the nature of centriole duplication, the two mitotic centrosomes that make up a spindle are inherently asymmetric from one another. The older (mother) mitotic centrosome is enriched with the centriole appendage protein cenexin, compared with the younger mitotic centrosome (daughter; Ishikawa et al., 2005; Tateishi et al., 2013; Hung et al., 2016; Colicino et al., 2019; Hall and Hehnly, 2021). This enrichment has been linked to higher kinetochore-MT stability and preferential misalignment of chromosomes toward the cenexin-rich (older) mitotic centrosome when mitotic errors occur (Gasic et al., 2015; Colicino et al., 2019). Cenexin is unique from other Odf2 isoforms in that it bears a C-terminal extension where the PLK1 binding site is situated (Chang et al., 2013). Cenexin binds to PLK1 following cyclin-dependent kinase (CDK)1 phosphorylation of serine at position 796 (S796; Soung et al., 2009). However, the relationship between PLK1 and cenexin on PCM organization is unknown. We found that cenexin’s C terminus acts in a conserved manner in eukaryotes, excluding nematodes and arthropods, to anchor PLK1 moderating its potential to phosphorylate PCM substrates required for PCM integrity and function.
RESULTS AND DISCUSSION
Cenexin is needed for PCM cohesion
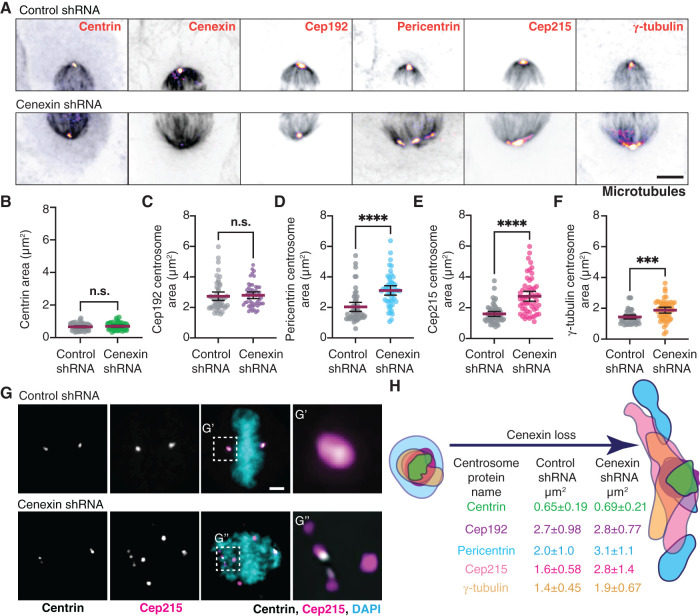
To examine the role of cenexin in PCM organization during metaphase, cells were depleted of cenexin using short hairpin RNA (shRNA; cell lines in Hung et al., 2016; Figure 1A, and Supplemental Figure S1, A and B). PCM components Cep192, pericentrin, Cep215, and γ-tubulin, along with a centriole marker centrin are presented as maximum projections from metaphase cells with the MT-based spindle labeled (Figure 1A, and Supplemental Figure S1A). To identify centrosome organization defects, the two-dimensional area of centrosome proteins was measured in cenexin-depleted cells compared with control cells (Figure 1, B–F). The centriole marker, centrin, and the PCM protein, Cep192, had no significant changes in centrosome area between cenexin shRNA and control cells (Figure 1, A–C). However, significant increases in pericentrin, Cep215, and γ-tubulin area occurred in cenexin shRNA cells compared with controls (Figure 1, A and D–F, and Supplemental Figure S1A). This significant increase in PCM area was associated with significant increases in PCM fragmentation that were characterized as splayed or scattered (Supplemental Figure S1C, and example of scattered in Figure 1G). With scattered phenotypes we found Cep215 puncta throughout the spindle, but centrin-decorated centrioles were still nicely positioned at the polar ends of the spindle (Figure 1G). No significant defects in centriole number were noted (Supplemental Figure S1D). Pericentrin and γ-tubulin fragments were consistently found to colocalize with one another (Supplemental Figure S1E) and their fragmentation corresponded with defects in chromosome alignment (Figure 1G), a phenotype identified in Colicino et al. (2019). These studies suggest that cenexin loss is potentially disrupting pericentrin, Cep215, and γ-tubulin cohesion within the PCM, but not Cep192 organization or centriole number (Figure 1H). This is interesting in regard to the studies by Soung et al. that reported abnormal/multipolar spindle morphologies with cenexin loss (Soung et al., 2006, 2009). Our findings suggest that the multipolar spindle phenotype with cenexin loss may be a result of PCM fragmentation and not due to a gain in number of centrioles/centrosomes.
FIGURE 1:
Cenexin is needed for PCM cohesion. (A) Metaphase HeLa cells mitotic centrosomes labeled for centrosome markers: centrin, cenexin, Cep192, pericentrin, Cep215, γ-tubulin (Fire LUT) and MT marker, and α-tubulin (gray). Control shRNA (top) and cenexin shRNA (bottom) -treated cells shown. Scale bar, 5 μm. (B–F) Representative scatter plots depicting two-dimensional areas (μm2) of centrin (B), Cep192 (C), pericentrin (D), Cep215 (E), and γ-tubulin (F) in control and cenexin shRNA-treated metaphase cells. Mean with 95% confidence intervals shown. Unpaired, two-tailed Student’s t tests; n.s., not significant; ***, p < 0.001; ****, p < 0.0001. (G) Control shRNA (top) and cenexin shRNA (bottom) metaphase cell projection. Centrin (gray), Cep215 (magenta), and DNA (DAPI; cyan) shown. Insets magnified 3× from G′ and G″. Scale bar, 5 μm. (H) Model depicting representative centrosome protein outline from a single representative mitotic centrosome reflecting changes resulting from cenexin loss. Mean two-dimensional areas (μm2) ± SD are provided. For graphs: detailed statistical analysis in Supplemental Table S1. See Supplemental Figure S1.
To identify if there was any change in the amount of centrin, Cep192, pericentrin, Cep215, and γ-tubulin recruited to the centrosome, a ratio of mean centrosome intensity of cenexin shRNA to control shRNA-treated cells was calculated. If a ratio of 1 is obtained (gray dashed line, Supplemental Figure S1F), it would indicate that there is no significant change in protein levels at the centrosome. No significant deviation from 1 occurred for centrin, Cep192, pericentrin, Cep215, and γ-tubulin (Supplemental Figure S1F), suggesting the expanded area that pericentrin, Cep215, and γ-tubulin occupy is unlikely due to increased protein abundance at the centrosome. We also found no overt differences in Cep215, pericentrin, Cep192, and γ-tubulin protein abundance when performing a Western blot on whole cell lysates (Supplemental Figure S1G), along with no overt defects in interphase centrosome morphology (Supplemental Figure S1, H and I). Based on these findings, we propose that there isn’t a defect in the ability to recruit PCM proteins to the centrosome, but that a defect in maintaining PCM cohesion occurs during metaphase with loss of cenexin. This seems to be the case when we image PCM live using the PCM marker RFP-pericentrin AKAP450 C-terminal centrosome targeting domain (PACT) in control and cenexin-depleted cells. PCM is assembled nicely in cenexin-depleted cells, but then starts to splay with time in metaphase (Supplemental Figure S1J). We propose a model where the packing of Cep215, pericentrin, and γ-tubulin complexes are compromised in cenexin-depleted cells resulting in a loss of PCM cohesion that presents as fragmentation (Figure 1H).
Cenexin tempers MT nucleation by mediating pericentrin-associated acentrosomal nucleation sites
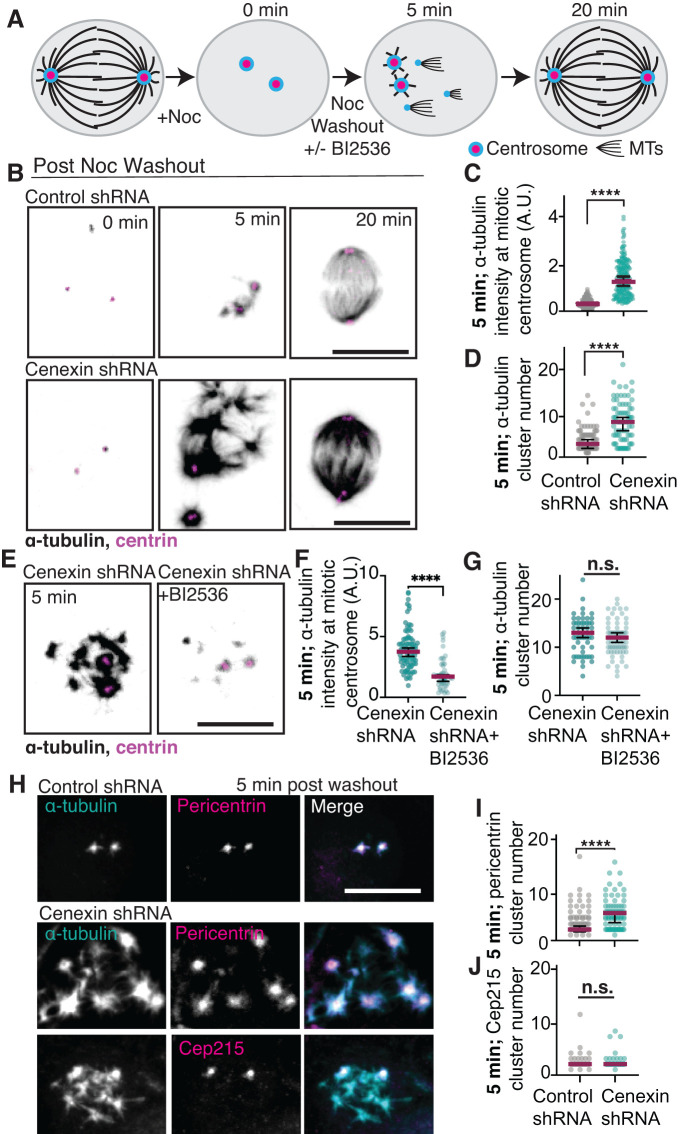
To examine whether centrosome MT nucleation is dependent on cenexin and its identified binding partner, PLK1 (Soung et al., 2006, 2009), we performed a centrosome-mediated MT-renucleation assay (Figure 2A). MT-based spindles were disassembled in control and cenexin-depleted cells with the MT-disrupting drug nocodazole. After nocodazole treatment, nocodazole was washed out with medium that contained or lacked the PLK1 inhibitor BI2536. Centrin-labeled mitotic centrosomes were examined at different time points post nocodazole washout for MT renucleation (Figure 2, A–G, and Supplemental Figure S2, A–C). At 0 min washout there was little to no detectable α-tubulin at mitotic centrosomes in both control and cenexin-depleted cells (Figure 2B), suggesting that the nocodazole treatment was successful in MT disassembly. After 5 min of MT regrowth, mitotic centrosomes in cenexin-depleted cells demonstrated an increased ability to nucleate MTs compared with control cells (Figure 2, B and C, and Supplemental Figure S2, A and B). We also noted that an increased number of acentrosomal MT nucleation clusters occurred in cenexin-depleted cells (8.0 ± 0.502 sites) compared with control (4.03 ± 0.293 sites; Figure 2, B and D, and Supplemental Figure S2C). By 20 min both control and cenexin-depleted cells formed a bipolar spindle (Figure 2B, and Supplemental Figure S2A). This suggests that the acentrosomal MT clusters can transport to the main mitotic centrosomes to assist in the formation of a bipolar mitotic spindle (Rusan et al., 2002; Tulu et al., 2003).
FIGURE 2:
Cenexin tempers MT nucleation by mediating pericentrin-associated acentrosomal nucleation sites. (A) Model of MT-renucleation assay. (B) Control and cenexin shRNA-treated HeLa cells are incubated with nocodazole, then washed with fresh media (washout). At 0 min, 5 min, and 20 min post washout, cells are fixed and immunostained for α-tubulin (inverted gray) and centrin (magenta). Scale bar, 10 μm. (C, D) Scatter plots of α-tubulin intensity at metaphase centrosomes (C) or α-tubulin cluster number (D) 5 min post nocodazole washout in control shRNA (gray) and cenexin shRNA (cyan) -treated cells. Mean (magenta) with 95% confidence intervals displayed. Unpaired, two-tailed Student’s t tests; ****, p < 0.0001. (E) Cenexin shRNA HeLa cells treated with or without the PLK1 inhibitor BI2536 at 5 min post nocodazole washout immunostained for α-tubulin (inverted gray) and centrin (magenta). Scale bar, 10 μm. (F, G) Scatter plots of α-tubulin intensity at metaphase centrosomes (F) or α-tubulin cluster number (G) 5 min post nocodazole washout. Mean (magenta) with 95% confidence intervals are displayed. Unpaired, two-tailed Student’s t tests; ****, p < 0.0001. (H) α-Tubulin (gray; cyan in merge), pericentrin (gray; magenta in merge), and Cep215 (gray; magenta in merge) metaphase cell projections 5 min post nocodazole washout. Scale bar, 10 μm. (I, J) Scatter plot depicting the number of pericentrin clusters (I) and Cep215 clusters (J) 5 min post nocodazole washout in control shRNA and cenexin shRNA cells. Mean (magenta) with 95% confidence intervals displayed. Unpaired, two-tailed Student’s t tests; ****, p < 0.0001; n.s., not significant. For graphs: statistical analysis in Supplemental Table S1. See Supplemental Figure S2.
We focused on the 5 min nocodazole washout time point to examine the role of PLK1 in the elevated MT nucleation events identified in metaphase cells depleted of cenexin. The significant increase in intensity of α-tubulin signal at centrin-marked mitotic centrosomes in cenexin-depleted cells was reduced when treated with the PLK1 inhibitor, BI2536 (Figure 2, E and F). These studies suggest that cenexin is needed to temper MT nucleation at mitotic centrosomes and that the increase in MT nucleation caused by cenexin loss is driven by PLK1. However, PLK1 inhibition did not diminish the α-tubulin acentrosomal clusters found with cenexin depletion (Figure 2G), suggesting that these may be PCM fragments from the metaphase cells prenocodazole treatment that could be associating with acentrosomal sites during the MT regrowth assay following nocodazole washout.
To test whether PCM associates with acentrosomal MT nucleation sites that form 5 min post nocodazole washout, cells were immunostained for two PCM components that are known to form a complex, Cep215 and pericentrin (Lee and Rhee, 2011; Chen et al., 2014), and acentrosomal MT clusters. Colocalization of Cep215 and pericentrin with α-tubulin was calculated using a Mander’s overlap coefficient to measure the degree of colocalization (0 to 1, with 1 being perfectly colocalized) from metaphase cells 5 min post nocodazole washout. In control cells, we found that Cep215 and pericentrin colocalized with α-tubulin (above 0.5 coefficient; Supplemental Figure S2, D and E). However, with cenexin loss pericentrin maintained its localization with α-tubulin, whereas Cep215 did not colocalize to the same extent (0.41 coefficient; Supplemental Figure S2, D and E). Along these same lines, we find that pericentrin associates with α-tubulin clusters/acentrosomal sites in cenexin-depleted cells, whereas Cep215 is not present at these sites (Figure 2H). Consistent with this, cenexin-depleted cells on average have more pericentrin clusters compared with Cep215 clusters (Figure 2, I and J). This finding suggested that Cep215 and pericentrin can no longer maintain a stable association with cenexin loss resulting in pericentrin becoming more associated with acentrosomal MT sites.
Cenexin is required for PCM cohesion and PLK1 for PCM dispersion
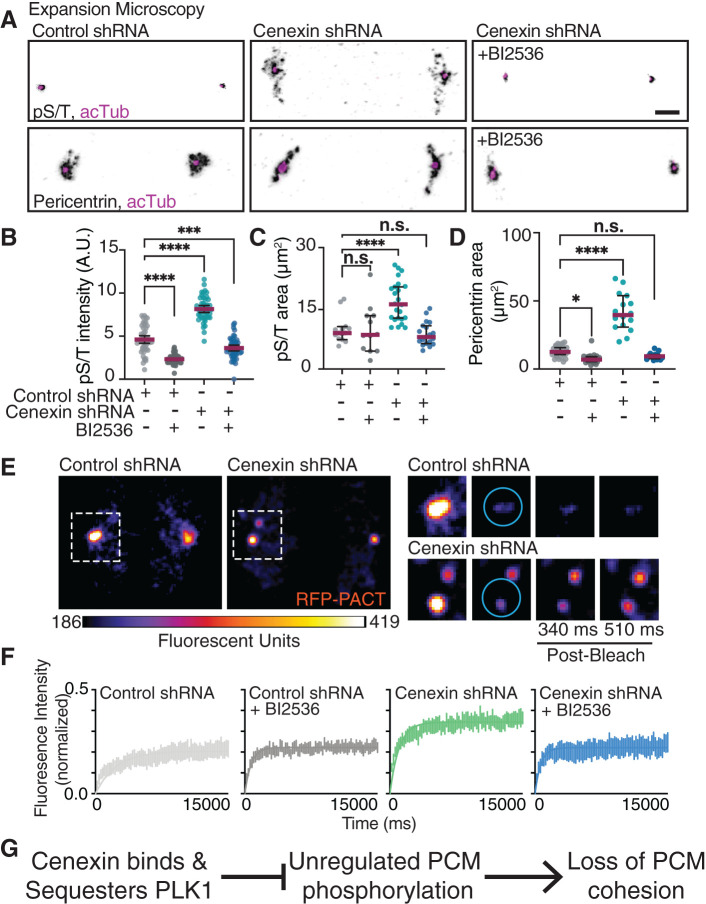
We examined whether PCM organization is modulated by cenexin through its identified role as a PLK1 binding partner (Soung et al., 2006, 2009). Specifically, we wanted to examine if cenexin loss causes changes in active PLK1 at centrosomes and/or PCM substrate phosphorylation that associates with PCM fragmentation. To test for PCM substrate phosphorylation we immunostained for phosphorylated serine/threonine (pS/T) modified proteins in relation to the PCM using confocal (Supplemental Figure S3A) or expansion microscopy (ExM; Figure 3A). ExM is an imaging protocol that provides subdiffraction limited details of centrosome organized components (Chozinski et al., 2016; Asano et al., 2018; Sahabandu et al., 2019). Using both ExM and quantitative confocal microscopy, cenexin-depleted mitotic cells demonstrated elevated PCM substrate phosphorylation (pS/T) compared with control conditions (Figure 3, A and B, and Supplemental Figure S3A). The elevated phosphorylation state found in cenexin-depleted cells was alleviated when cells were treated with the PLK1 small molecule inhibitor, BI2536 (Figure 3, A and B, and Supplemental Figure S3A), suggesting that elevated pS/T was specific to PLK1. Using ExM, we found that pS/T modified centrosome substrates and pericentrin encompassed a significantly larger area in cenexin-depleted cells (16.68 ± 4.99 μm2 for pS/T, and 41.46 ± 13.64 μm2 for pericentrin) compared with control cells (9.72 ± 3.35 μm2 for pS/T, and 12.92 ± 4.17 μm2 for pericentrin; Figure 3, A, C, and D). Centrioles were noted in expanded cells by immunostaining for acetylated tubulin (Figure 3A, and Supplemental Video 1). Notably, we identified that cenexin-depleted cells treated with BI2536, had normal PCM area measured by either Cep215 in non-ExM cells (Supplemental Figure S3, B and G) or pericentrin in ExM cells (Figure 3, A and D, and Supplemental Video 1). These studies suggest that cenexin-depleted mitotic cells have fragmented PCM potentially due to elevated PLK1-dependent phosphorylation events on PCM substrates.
FIGURE 3:
Cenexin is required for PCM cohesion and PLK1 for PCM dispersion. (A) Expansion microscopy of control and cenexin shRNA metaphase cell centrosomes with or without BI2536. Centrosomes immunolabeled for pS/T (inverted gray, top) or pericentrin (inverted gray, bottom), centrioles decorated with acetylated tubulin (acTub; magenta). Scale bar, 5 μm. (B–D) Representative scatter plots depicting pS/T mean intensity at metaphase centrosomes (B), and pS/T (C) and pericentrin (D) expanded two-dimensional areas (μm2) in control shRNA and cenexin shRNA metaphase cells with or without BI2536. Mean (magenta) with 95% confidence intervals displayed. One-way ANOVA with multiple comparisons to control cells; n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. (E) FRAP examples of control and cenexin shRNA metaphase cell centrosomes marked with PCM marker RFP-PACT. Magnified insets of boxed centrosomes depict examples of prebleach, bleach, and postbleach time points. Cyan circles represent the region in which a 405-nm laser was applied for photobleaching. Scale bar, 5 μm. (F) FRAP curves for metaphase control shRNA (gray), control shRNA plus BI2536 (black), cenexin shRNA (green), and cenexin shRNA plus BI2536 (blue) cells. SEM for n > 6 cells. (G) A proposed model that cenexin binds and sequesters PLK1 inhibiting PLK1’s potential to have unregulated phosphorylation on PCM substrates that results in PCM cohesion loss. For graphs: statistical analysis in Supplemental Table S1. See Supplemental Figure S3.
Movie S1.
Volumetric rendering of Pericentrin (grey) and centrioles (acetylated tubulin, magenta) at mitotic centrosomes. Control and Cenexin shRNA treated cells plus/minus BI2536 shown. Inset shows Pericentrin (green) and DAPI (blue).
We tested whether the elevated amounts of pS/T in cenexin-depleted cells resulted from either too much PLK1 at the centrosome or an overabundance of active PLK1 at the centrosome by immunostaining for phospho-PLK1(T210) (Supplemental Figure S3C) and PLK1 (Supplemental Figure S3D) in the presence or absence of the PLK1 small molecule inhibitor (BI2536). BI2536 treatment resulted in a robust and significant decrease in pPLK1(T210) (Supplemental Figure S3, C and E) in both control and cenexin-depleted cells, demonstrating that BI2536 was specifically decreasing PLK1 activity. With cenexin depletion, no significant change in PLK1 at mitotic centrosomes was identified (Supplemental Figure S3, D and F). We were surprised to find no increase in active pPLK1(T210) between control and cenexin-depleted conditions (Supplemental Figure S3, C and E) due to the elevated substrate phosphorylation levels identified in cenexin-depleted cells (Figure 3B). One potential interpretation from our findings is that under cenexin-depleted conditions, the normally cenexin-bound population of pPLK1(T210) is free to act on PCM substrates resulting in a loss of PCM cohesion (Figure 3G).
We addressed whether the biophysical state of the PCM was altered with loss of cenexin. A PCM marker, RFP-PACT, was expressed in control and cenexin-depleted cells treated with and without the PLK1 inhibitor BI2536 (Figure 3, E and F). Using fluorescence recovery after photobleaching (FRAP), we examined the mobility of RFP-PACT at metaphase centrosomes, where we found significantly different dynamics between cenexin-depleted cells and control (Figure 3F, and Supplemental Figure S3H). Control cells mean mobile fraction was significantly less than the mobile fraction of cenexin-depleted cells (Figure 3F, and Supplemental Figure S3H). PLK1 inhibition caused no significant effect in PCM mobility in control cells, but the increased PCM mobility found in cenexin-depleted cells was partially rescued with BI2536 (Figure 3F, and Supplemental Figure S3H). Together these studies suggest that the increased substrate phosphorylation through PLK1-cenexin disruption is likely causing increased PCM mobility. This increase in PCM mobility is suggestive of a model where the packing of PCM is compromised in cenexin-depleted cells, ultimately resulting in a loss of PCM cohesion (Figure 3G).
Cenexin phosphorylation at its conserved C-terminal PLK1 binding site is required for maintenance of PCM in vivo
Human cenexin (Odf2 isoform 9) is unique from other human Odf2 isoforms in that it bears an N- and C-terminal extension (1–42 amino acid stretch for N terminus, 701–805 amino acid stretch for C terminus), with the PLK1 binding site being situated within the C terminus (Figure 4, A and C; Chang et al., 2013). Previous studies identified that human cenexin binds to PLK1 following CDK1 phosphorylation of S796 (Soung et al., 2009). Here, we examined whether human cenexin, and its N- and C-terminal extensions, are present across phyla by performing BLASTp searches of the human sequences against 67 proteomes through National Center for Biotechnology Information’s (NCBI’s) webserver (Figure 4B). Because sequence hits for several species were not detected on the web server due to insufficient similarity, we performed BLASTp searches using a local BLAST installation to identify possible orthologous sequences (Supplemental Figure S4A). With these studies, we found that cenexin is present in all vertebrates (chordates) along with two other centrosome proteins tested—the PLK1 scaffold CEP192 (Joukov et al., 2014), and the centriole protein centrin (Levy et al., 1996; Salisbury et al., 2002). Cenexin is also present in hemichordates, mollusks, annelids, rotifers, platyhelminths, and cnidarias, but absent in arthropods and nematodes, along with ctenophores, poriferas, and placozoa. The presence of cenexin in each phylum requires the presence of centrioles (e.g., chordates); however, the presence of centrioles does not necessarily guarantee that cenexin is present (e.g., nematodes; Figure 4B, and Supplemental Figure S4A). When performing BLASTp searches through the webserver, the C-terminal part of cenexin is conserved in all vertebrates (chordates, excluding genus pongo; Figure 4B), hemichordates, mollusks, annelids, rotifers, platyhelminths, and cnidarias, whereas the N terminus is only present in chordates (excluding genus pongo, danios, and petromyzon) and hemichordates (Figure 4B). However, using a local BLAST installation, pongo and danio were identified with high confidence to include both the N and the C terminus, but oryctolagus along with petromyzon excludes the N terminus (Supplemental Figure S4A). These findings suggest that the C terminus containing the PLK1 binding site evolved first and is highly conserved in its function and potential regulation of the centrosome across phyla. Interestingly, our analyses reveal that the PLK1 scaffold Cep192 (Joukov et al., 2014; Wong et al., 2021) is detected in arthropods where the cenexin C-terminal extension is lacking. Additionally, a Cep192-“like” centrosome protein, spd-2, that acts as a PLK1 scaffold, was identified in C. elegans that was not identified by BLASTp analysis (Kemp et al., 2004). This suggests that Cep192 may have some redundancy in PLK1 scaffolding functions allowing cenexin to be potentially dispensable in arthropods and C. elegans (Figure 4B).
FIGURE 4:
Cenexin phosphorylation at its conserved C-terminal PLK1 binding site is required for maintenance of PCM in vivo. (A) Schematic representation of human hODF2 and cenexin (Cnxn; ODF2 isoform 9). The blue and magenta boxes highlight the N- and C-terminal extensions unique to cenexin. (B) Phylogenetic tree of cenexin and its N terminus and C terminus in relation to Cep192 and centrin across animal phyla. Cyan and orange boxes indicate detection of cenexin and centrosome components within representative species of each phylum. (C) Amino acid alignment between human and zebrafish cenexin C-terminal PLK1 binding motif. The serine highlighted in magenta represents the known human cenexin-PLK1 site (S796) and potential zebrafish binding site (S829). Letters in the middle between two sequences represent identical amino acids, and the + sign represents an amino acid of functional identity. Representative confocal maximum projection of cenexin from expanded (ExM) human cells and from a zebrafish embryo cell shown. Scale bar, 0.05 μm. (D–F) Representative cells from 512-cell zebrafish embryos under cenexin depletion conditions (cenexin MO; D), or rescue conditions (cenexin MO plus mCh-cenexin or mCh-cenexin-S796A, magenta in E and F) fixed and immunostained for γ-tubulin (inverted gray; D and E) or pST (inverted gray; F). Insets (D′, E′, E″, F′, and F″) at 5× magnification; corresponding areas outlined (μm2). Scale bar, 5 μm. (G) Scatter plot depicting γ-tubulin area (μm2) at mitotic centrosomes. Mean (magenta) with 95% confidence intervals shown. One-way ANOVA with multiple comparisons to control cells; n.s., not significant; **, p < 0.01; ****, p < 0.0001. (H) Scatter plot depicting pS/T area (μm2) at mitotic centrosomes. Unpaired, two-tailed Student’s t tests; ****, p < 0.0001. For graphs: statistical analysis in Supplemental Table S1. See also Supplemental Figure S4.
To determine whether the role cenexin plays in PCM organization is conserved between human cells and an in vivo chordate model identified to have the C-terminal extension, we aligned the amino acid sequence of human cenexin to zebrafish (Danio rerio) cenexin (Figure 4, B and C). The two sequences shared 50.36% identity with a 70% positivity rate that accounts for amino acids with functional similarity, suggesting high conservation between human and zebrafish cenexin. Human cenexin amino acids spanning the S796 site (781–798) were aligned to zebrafish cenexin where S829 corresponded with human S796 (Figure 4C). This provides further support that the C-terminal tail containing the PLK1 binding site has a conserved function across species. To test this, we depleted cenexin from zebrafish embryos using previously published translational morpholinos (MOs) to block the translation of cenexin mRNAs (Novorol et al., 2013) and rescued with human wild-type cenexin (mCherry-cenexin) or cenexin deficient in PLK1 binding (mCherry-cenexin-S796A). mCherry-cenexin displayed a compact ring-like centriole organization at zebrafish 512-cell stage centrioles comparable to immunostaining for endogenous cenexin using ExM in human cells (Figure 4C, and Supplemental Figure S4B). Both mCherry-cenexin and -cenexin-S796A had concentrated organization toward the centrosome center (Figure 4E) with no significant difference in cenexin area occurring between the two (Supplemental Figure S4D). When depleting endogenous zebrafish cenexin, γ-tubulin became more fragmented and occupied a significantly larger area (5.32 μm2 in Figure 4, D and G) compared with control conditions (Figure 4G, and Supplemental Figure S4C) or wild-type cenexin (mCherry-cenexin) rescue conditions (3.68 μm2 in Figure 4, E and G). However, mCherry-cenexin-S796A was unable to rescue centrosome area (7.19 μm2 in Figure 4, E and G), suggesting that the ability of cenexin to bind to PLK1 at S796 is required for its function to modulate PCM organization.
These results prompted us to examine whether PCM substrate phosphorylation relies on cenexin in vivo. To do this, we fixed and immunostained cenexin-depleted embryos rescued with mCherry-cenexin or mCherry-cenexin-S796A for pS/T modified proteins in relation to the PCM (Figure 4, F and H). With mCherry-cenexin rescue conditions, pS/T signal was tightly organized (0.88 ± 0.30 μm2; Figure 4, F and H). Whereas, with mCherry-cenexin-S796A rescues, pS/T signal was misorganized and dispersed (1.5 ± 0.71 μm2 compared with controls at 0.88 ± 0.30 μm2; Figure 4, F and H). These results agree with what we found in vitro (Figure 3G), where inhibition of a cenexin-PLK1 interaction results in expanded pS/T distribution (Figure 4, F and H). Altogether, our experiments indicate that the conserved PLK1 binding motif in the C-terminal tail of cenexin is required for PCM integrity and function.
In summary, we speculate that cenexin sequesters a population of PLK1 preventing it from overphosphorylating PCM proteins (Figure 3G). If cenexin is absent, the cenexin-bound population of PLK1 is now free and available to phosphorylate PCM proteins. This may be why we don’t see losses of PLK1 at the centrosome (Supplemental Figure S3F) or significant increases in pPLK1(T210), but we do find a significant increase in pS/T substrates with cenexin loss that is PLK1 dependent (Figure 3, B and C). We predict that this hyperphosphorylation of PCM proteins disrupts the cohesion of the PCM making the PCM more mobile as evidenced by our FRAP experiment (Figure 3F, and Supplemental Figure S3H) and potentially allowing the PCM to be more competent for microtubule nucleation events (Figure 2) but does not necessarily affect the recruitment of PCM proteins to the centrosome (Supplemental Figure S1F). We conclude that when cenexin is present, the cenexin-bound population of PLK1 is unavailable to PCM proteins resulting in normal PCM cohesion and microtubule nucleation.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Resource availability
Lead contact.
For further information or to request resources/reagents, contact the Lead Contact, Heidi Hehnly (hhehnly@syr.edu).
Materials availability.
No new materials were generated for this study.
Data and code availability.
All data sets analyzed for this study are displayed. A supplemental CSV file is provided with the species used in the phylogenetic analysis with a FTP link to download their proteomes (Supplemental File SF1A, Metazoan_taxon_list_FTP_links.csv), the tree file is provided in Newick format (File SF1B, nuclear_genes_tree.nwk), the R script file for phylogeny analysis is available as a supplemental file SF1 (File SF1C, phylo_analysis.html), and the BLAST results are provided in a supplemental table formatted in Supplemental File SF1 (File SF1D, NCBI_BLAST_results.zip; File SF1E, Homebrew_BLAST_results.zip).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Zebrafish
All zebrafish lines were maintained using standard procedures approved by the Syracuse University IACUC committee (protocol #18-006). For details, see Rathbun et al. (2020) and Aljiboury et al. (2021). See Key Resources Table for a list of zebrafish transgenic lines used.
Cell culture
HeLa cells treated with either control or cenexin shRNA (Hung et al., 2016) were used throughout this study. Cells were selected in puromycin (3 μg/ml). See Key Resources Table for a list of cell lines used.
METHOD DETAILS
Immunofluorescence
Cells were plated on #1.5 coverslips until they reached 90% confluence and fixed using ice-cold methanol for 10 min. Standard immunostaining procedures were performed (described in Colicino et al., 2018). Coverslips were rinsed with diH2O and mounted on glass slides using ProLong Gold mounting media (Thermo Fisher Scientific; P36934). Zebrafish embryos were fixed using 4% paraformaldehyde (PFA) at 4°C overnight, for immunostaining (see Rathbun et al., 2020; Aljiboury et al., 2021). See Key Resources Table for a list of antibodies used in this study.
Chemical inhibitors
Chemical inhibitors include nocodazole used on cells at 100 nM or 10 μm and BI2536 used on cells at 100 nM. ProTAME (Fisher; I44001M) was used at 10 μM to obtain mitotic cells. See Key Resources Table for more information on inhibitors used in this study.
Morpholino injections
Anti-cenexin vivo translational morpholinos (Gene Tools; Novorol et al., 2013) or vivo standard control morpholinos (Gene Tools) were constituted as 1 mM stock in water and injected into zebrafish yolks at one cell stage in a final concentration of 2 ng/nl. Injection protocols are detailed in Aljiboury et al. (2021).
Plasmid constructs and mRNA
Gibson cloning methods were used to generate mCherry-cenexin-WT and mCherry-cenexin-S796A plasmids (NEBuilder HiFi DNA assembly kit), then purified using a DNA maxi-prep kit (Bio Basic; 9K-006-0023). mRNA was generated from plasmids using a mMESSAGE mMACHINE SP6 transcription kit (Thermo Fisher Scientific; AM1340).
FRAP
FRAP experiments were performed using a Leica DMi8 STP800 spinning-disk confocal microscope using a 40×/1.10 NA water immersion objective. A region of interest (ROI) was placed over one of the spindle poles, where a 405-nm laser was applied. RFP-PACT fluorescence was bleached within the ROI after administration of the laser. Fluorescence recovery time as well as signal intensity were measured every 100 ms to determine the mobile fraction.
MT-renucleation assay
HeLa cells were treated with 1 μM nocodazole in media for 30 min. Cells were washed 20 times with 1X phosphate-buffered saline (PBS) and placed in media at 37°C for 0 s, 30 s, 1 min, 2 min, 5 min, or 20 min. Cells were fixed using methanol overnight at –20°C. Cells were immunostained and then imaged for analysis.
Expansion microscopy
Protocol used was modified from previously published expansion protocols (Chozinski et al., 2016; Sahabandu et al., 2019). A modified protocol is described below. See Key Resources Table for chemicals used.
Cell preparation.
HeLa cells were grown on glass coverslips and synchronized with 10 μM proTAME for 1 h, then fixed with ice-cold methanol for 10 min or 4% PFA in 1X PBS for 1 h at 22–25°C.
Immunostaining and acrylamide incubation.
Cells were blocked in PBSΔT for 1 h at 22–25°C if methanol fixed or in 0.1% Triton X-100 with 5% donkey serum in 1X PBS for 15 min at 22–25°C if PFA fixed. Cells were then incubated with primary antibodies in blocking buffer overnight at 4°C, then with secondary antibodies for 4 h at 22–25°C. Cells were incubated in 30% acrylamide solution in 1X PBS overnight at 40°C.
Gelation, cell punching, and digestion.
Following three 10-min 1X PBS washes (PFA fixed) or 10X PBSΔT washes (if methanol fixed), the slides (cell side up) and the parafilm-covered Petri dish were placed on an ice bath. Gelation reagents were placed on the coverslips in chilled 1X PBS: 20% acrylamide, 7% sodium acrylate, and 0.04% bisacrylamide, with ammonium presulfate (APS) and tetramethylethylenediamine) (TEMED) added just before application. Gelation solution was added to each coverslip, incubated for 20 min on ice, and then incubated at 30°C for 1.5 h. A 4-mm biopsy punch was utilized to excise punches from gelled samples. Punches were incubated with digestion buffer (Triton X + EDTA + Tris pH 8 + NaCl in water) overnight at 22–25°C in the dark.
Postexpansion staining to enhance fluorescence.
Cell punches were washed and blocked with appropriate buffer, then incubated with primary antibodies followed by secondary antibodies in blocking buffer for 4 h at 22–25°C.
Expansion and mounting.
After digestion, samples were expanded in dH2O for 2 h at 22–25°C with dH2O exchange every 20 min. Samples were left in dH2O and 1,4-Diazabicyclo [2.2.2] octane (DABCO) overnight to expand and protect immunofluorescent signal. Samples were mounted in a MatTek plate the following day for imaging.
Imaging
Tissue culture cells and zebrafish embryos were imaged using a Leica DMi8 STP800 (Leica, Bannockburn, IL) equipped with an X-light V2 confocal unit spinning disk and an 89 North-LDI laser launch with a Photometrics Prime-95B camera or a Leica SP8 laser scanning confocal microscope (LSCM; Leica, Bannockburn, IL). Optics used on Leica DMi8 were HC PL APO 63×/1.40 NA oil CS2 or HC PL APO 40×/1.10 NA CORR WCS2 water. The optics used on the SP8 LSCM is HC PL APO 40×/1.10 NA CORR CS2 0.65 water objective or HC PL APO 63×/1.30 NA Glyc CORR CS2 glycerol objective. Visiview software (Lecia DMi8) or LasX (SP8 LSCM) were used to acquire images.
For live zebrafish imaging, fluorescent transgenic or mRNA-injected embryos (injection protocols in Aljiboury et al., 2021) were mounted in 2% low melting agarose gel (Thermo Fisher Scientific; 16520100) at 512-cell stage and imaged using a SP8 LSCM or spinning-disk confocal microscope.
Image and statistical analysis
Images were processed using FIJI/ImageJ software. All analyses was performed on maximum projections unless otherwise stated. The spindle pole area was measured using the freehand selection tool to draw a boundary of the poles and calculating the area within the shape (Aljiboury et al., 2021). Signal intensities were measured by placing a ROI over the site of interest (poles or clusters). Signal intensity was calculated by subtracting the minimum intensity from the mean intensity of the measured region, unless otherwise noted. All figures were created in Adobe Illustrator, and graphs were created using Graphpad Prism software. Statistical analyses (unpaired Student’s t tests and analysis of variance ANOVA) were performed using Graphpad Prism. **, p < 0.01; **, p < 0.001; and ***, p < 0.0001.
Phylogenetic analysis of cenexin
To analyze the pattern of evolutionary conservation of cenexin (Cnxn; Odf2 isoform 9) among metazoan phyla, a metazoan phylogeny was built using a single protein isoform (isoform 1) of three nuclear genes (SMCA1A, SMC1B, and MCM5) for 67 species that represent the major metazoan phyla and for which annotated proteomes are available on NCBI (Supplemental Figure SF1, A and B). Saccharomyces cerevisiae and Neurospora crassa were used as outgroups. The three protein sequences were first extracted from the Homo sapiens proteome and orthologues in the remaining species were identified using BLASTp (Altschul et al., 1990). A multiple sequence alignment of the three proteins was constructed using Muscle (Edgar, 2004), and individual alignments were subsequently concatenated. A maximum likelihood phylogenetic tree was built with PhyML using default parameters (Guindon et al., 2010). To identify orthologues of cenexin and its N and C termini, we first performed BLASTp searches of the human sequences against the 67 proteomes on the NCBI web server. Sequence hits for several species were not detected on the web server due to insufficient similarity, therefore we performed BLASTp searches using a local BLAST installation to identify possible orthologous sequences. Because we obtained a BLASTp hit for each species, we classified hit confidence as potential orthologues based on hit length, percent identity, and NCBI annotation. Specifically, we considered a hit a “high confidence” orthologue if the protein was annotated by NCBI as an Odf2 orthologue or had a matched alignment length >100 a.a. (Cnxn), >80 a.a. (C terminus), or >30 a.a. (N terminus) and a percent identity >60% (Cnxn) or >40% (C terminus and N terminus). Alternatively, we considered a hit a “medium confidence” orthologue if the matched alignment length was between 30 a.a. and 100 a.a. (Cnxn), 30 a.a. and 80 a.a. (C terminus), or 40 a.a. and 30 a.a. and a percent identity >40%. Finally, alignment matches that were shorter than 30 a.a. (Cnxn and C terminus) or 15 a.a. (N terminus) and percent identity <40% were considered “low confidence” hits. Any hit that did not match our criteria was considered an “unlikely orthologue.” All phylogenetic analyses were performed on R primarily using the ggtree package (Yu et al., 2017). The phylogenetic analysis script and input files are available in Supplemental Files SF1, C–E.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-Tubulin-AF555 (mouse) | EMD Millipore | Cat# 05-829-AF555 |
| CDK5RAP2/Cep215 (rabbit) | Bethyl laboratories | Cat# IHC-00063, RRID: AB_2076863 |
| Cep192 (rabbit) | Bethyl laboratories | Cat# A302-324A, |
| Centrin (mouse) | EMD Millipore | Cat# 04-1624, RRID: AB_10563501 |
| DAPI | SigmaAldrich | Cat# D9542-10MG |
| Donkey anti-mouse AF647 | Fisher Scientific | Cat# A31571, AB_162542 |
| Donkey anti-rabbit AF 488 | Life Technologies | Cat# A21206, RRID: AB_2535792 |
| Donkey anti-rabbit AF 568 | Life Technologies | Cat# A10042, AB_2534017 |
| Donkey anti-rabbit AF 647 | Life Technologies | Cat# A31573, RRID: AB_2536183 |
| Gamma-tubulin (mouse) | Abcam Biochemicals | Cat# 11316, RRID: AB_297920 |
| Gamma-tubulin (rabbit) | Sigma Aldrich | Cat# T5192, RRID: AB_261690 |
| Pericentrin (rabbit) | Abcam Biochemicals | Cat# ab4448 AB_304461 |
| PLK1 (rabbit) | Cell Signaling Technology | Cat# 4513S, RRID: AB_2167409 |
| Phospho-PLK1 (Thr210) (rabbit) | Cell Signaling Technology | Cat# 5472, RRID: AB_10698594 |
| Phospho-(Ser/Thr) (rabbit) | Cell Signaling Technology | Cat# 9631, RRID: AB_330308 |
| ODF2/cenexin (rabbit) | Thomas Scientific | Cat# 12058-1-AP |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Acrylamide solution, 40% | Sigma Aldrich | Cat# A4058-100ML |
| Acrylamide-bisacrylamide solution | EMD Millipore | Cat# 1300-500ML |
| Agarose | Thermo Fisher | Cat# 16520100 |
| APS (ammonium presulfate) | Fisher Bioreagents | Cat# BP179-100 |
| BI2536 | Selleck Chemicals | Cat# S1109 |
| Bovine serum albumin | Fisher Scientific | Cat# BP1600-100 |
| Dimethylsulfoxide | Fisher Scientific | Cat# BP231100 |
| DABCO (1,4-diazabicyclo [2.2.2] octane, 97%) | Fisher Scientific | Cat# AC112470250 |
| EDTA | Fisher Scientific | Cat# BP120-500 |
| NaCl | Fisher Bioreagents | Cat# BP358-212 |
| Nocodazole | Fisher Scientific | Cat# AC358240500 |
| Paraformaldehyde | Fisher Scientific | Cat# AA433689M |
| Phosphate-buffered saline | Fisher Scientific | Cat# 10010023 |
| Prolong gold | Invitrogen | Cat# P36934 |
| Sodium acrylate, 97% | Sigma Aldrich | Cat# 408220-25G |
| ProTAME | R&D Systems | Cat# I-440-01M |
| TEMED | Fisher Scientific | Cat# BP150-100 |
| Tris base | Fisher Bioreagents | Cat# BP152-5 |
| Triton X-100 | Fisher Scientific | Cat# BP151500 |
| Tween 20 | Thermo Fisher | Cat# BP337500 |
| Experimental Models: Organisms/Strains | ||
| Human HeLa cells control shRNA | Hung et al., 2016 | N/A |
| Human HeLa cells cenexin shRNA | Hung et al., 2016 | N/A |
| Zebrafish | Zebrafish International Resource Center (ZIRC) | TAB (wild type) |
| Zebrafish | Gift from Solnica-Krezel Lab, generated by Harris Lab | Tg(-5actb2:cetn4-GFP) |
| Software and Algorithms | ||
| Adobe Illustrator | ||
| ImageJ/FIJI | Schindelin et al., 2012 | https://imagej.net/Fiji |
| Prism8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| LAS-X software | Leica microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| R version 4.1.2; package ggtree | The R project for statistical computing | https://www.r-project.org |
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants no. R01GM-127621 (H.H.) and no. R01GM-130874 (H.H.). This work was supported by the U.S Army Medical Research Acquisition Activity through the FY16 Prostate Cancer Research Programs under Award no. W81XWH-20-1-0585 (H.H.). Opinions, interpretations, conclusions, and recommendations are those of the authors and not necessarily endorsed by the Department of Defense.
Abbreviations used:
- CDK1
cyclin-dependent kinase 1
- Cep192
centrosome protein 192
- Cep215
centrosome protein 215
- Cnxn
cenexin
- FRAP
fluorescence recovery after photobleaching
- MTs
microtubules
- ODF2
outer dense fiber 2
- PACT
pericentrin/AKAP450 centrosomal targeting domain
- PCM
pericentriolar matrix
- PLK1
polo-like kinase 1
- pS/T
phospho-serine/threonine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-01-0015) on May 24, 2022.
REFERENCES
- Aljiboury AA, Mujcic A, Cammerino T, Rathbun LI, Hehnly H (2021). Imaging the early zebrafish embryo centrosomes following injection of small-molecule inhibitors to understand spindle formation. STAR Protoc 2, 100293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990). Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Alvarez-Rodrigo I, Wainman A, Raff JW (2020). Ana1 recruits PLK1 to mother centrioles to promote mitotic PCM assembly and centriole elongation. BioRxiv 2020.08.11.244194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano SM, Gao R, Wassie AT, Tillberg PW, Chen F, Boyden ES (2018). Expansion microscopy: Protocols for imaging proteins and RNA in cells and tissues. Curr Protoc Cell Biol 80, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G, Laos T, Dumont J, Dammermann A (2019). Differential requirements for centrioles in mitotic centrosome growth and maintenance. Dev Cell 50, 355–366.e6. [DOI] [PubMed] [Google Scholar]
- Chang J, Seo SG, Lee KH, Nagashima K, Bang JK, Kim BY, Erikson RL, Lee KW, Lee HJ, Park JE, et al. (2013). Essential role of cenexin1, but not Odf2, in ciliogenesis. Cell Cycle 12, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CT, Hehnly H, Yu Q, Farkas D, Zheng G, Redick SD, Hung HF, Samtani R, Jurczyk A, Akbarian S, et al. (2014). A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr Biol 24, 2327–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chozinski TJ, Halpern AR, Okawa H, Kim H-J, Tremel GJ, Wong ROL, Vaughan JC (2016). Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods 136, 485–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicino EG, Garrastegui AM, Freshour J, Santra P, Post DE, Kotula L, Hehnly H (2018). Gravin regulates centrosome function through PLK1. Mol Biol Cell 29, 532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicino EG, Hehnly H (2018). Regulating a key mitotic regulator, polo-like kinase 1 (PLK1). Cytoskeleton 75, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colicino EG, Stevens K, Curtis E, Rathbun L, Bates M, Manikas J, Amack J, Freshour J, Hehnly H (2019). Chromosome misalignment is associated with PLK1 activity at cenexin-positive mitotic centrosomes. Mol Biol Cell 30, 1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Richens JH, Wainman A, Holder J, Vicente CC, Pratt MB, Dix CI, Novak ZA, Dobbie IM, Schermelleh L, et al. (2014). A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife 3, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasic I, Nerurkar P, Meraldi P (2015). Centrosome age regulates kinetochore–microtubule stability and biases chromosome mis-segregation. Elife 4, e07909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, Sharp DJ (2007). Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol 17, 1960–1966. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Hall NA, Hehnly H (2021). A centriole’s subdistal appendages: contributions to cell division, ciliogenesis and differentiation. Open Biol 11, 200399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung HF, Hehnly H, Doxsey S (2016). The mother centriole appendage protein cenexin modulates lumen formation through spindle orientation. Curr Biol 26, 793–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S, Tsukita S (2005). Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7, 517–524. [DOI] [PubMed] [Google Scholar]
- Joukov V, Walter JC, De Nicolo A (2014). The Cep192-organized aurora A-Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol Cell 55, 578–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF (2004). Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell 6, 511–523. [DOI] [PubMed] [Google Scholar]
- Lee K, Rhee K (2011). PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J Cell Biol 195, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Lai EY, Remillard SP, Heintzelman MB, Fulton C (1996). Centrin is a conserved protein that forms diverse associations with centrioles and MTOCs in naegleria and other organisms. Cell Motil Cytoskeleton 33, 298–323. [DOI] [PubMed] [Google Scholar]
- Meng L, Park J-E, Kim T-S, Lee EH, Park S-Y, Zhou M, Bang JK, Lee KS (2015). Bimodal interaction of mammalian polo-like kinase 1 and a centrosomal scaffold, Cep192, in the regulation of bipolar spindle formation. Mol Cell Biol 35, 2626–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Agard DA, Huang B, Pelletier L (2014). Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol 24, 188–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittasch M, Tran VM, Rios MU, Fritsch AW, Enos SJ, Gomes BF, Bond A, Kreysing M, Woodruff JB (2020). Regulated changes in material properties underlie centrosome disassembly during mitotic exit. J Cell Biol 219, e201912036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak ZAA, Wainman A, Gartenmann L, Raff JWW (2016). Cdk1 phosphorylates Drosophila Sas-4 to recruit polo to daughter centrioles and convert them to centrosomes. Dev Cell 37, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novorol C, Burkhardt J, Wood KJ, Iqbal A, Roque C, Coutts N, Almeida AD, He J, Wilkinson CJ, Harris WA (2013). Microcephaly models in the developing zebrafish retinal neuroepithelium point to an underlying defect in metaphase progression. Open Biol 3, 130065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Zhao Z, Wu D, Wang S, Harrison JL, Gómez-Cavazos JS, Desai A, Oegema KF (2021). Polo-like kinase 1 independently controls microtubule-nucleating capacity and size of the centrosome. J Cell Biol 220, e202009083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbun LI, Aljiboury AA, Bai X, Hall NA, Manikas J, Amack JD, Bembenek JN, Hehnly H (2020). PLK1- and PLK4-mediated asymmetric mitotic centrosome size and positioning in the early Zebrafish embryo. Curr Biol 30, 4519–4527.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Serdar Tulu U, Fagerstrom C, Wadsworth P (2002). Reorganization of the microtubule array in prophase/prometaphase requires cytoplasmic dynein-dependent microtubule transport. J Cell Biol 158, 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahabandu N, Kong D, Magidson V, Nanjundappa R, Sullenberger C, Mahjoub MR, Loncarek J (2019). Expansion microscopy for the analysis of centrioles and cilia. J Microsc 276, 145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL, Suino KM, Busby R, Springett M (2002). Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol 12, 1287–1292. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung N-K, Kang YH, Kim K, Kamijo K, Yoon H, Seong Y-S, Kuo Y-L, Miki T, Kim SR, Kuriyama R, et al. (2006). Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes. Mol Cell Biol 26, 8316–8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung NK, Park JE, Yu LR, Lee KH, Lee JM, Bang JK, Veenstra TD, Rhee K, Lee KS (2009). Plk1-dependent and -independent roles of an ODF2 splice variant, hCenexin1, at the centrosome of somatic cells. Dev Cell 16, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S (2013). Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J Cell Biol 203, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu US, Rusan NM, Wadsworth P (2003). Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr Biol 13, 1894–1899. [DOI] [PubMed] [Google Scholar]
- Vertii A, Hehnly H, Doxsey S (2016). The centrosome, a multitalented renaissance organelle. Cold Spring Harb Perspect Biol 8, a025049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S-S, Wilmott ZM, Saurya S, Zhou FY, Chau K-Y, Goriely A, Raff JW (2021). Mother centrioles generate a local pulse of Polo/PLK1 activity to initiate mitotic centrosome assembly. BioRxiv 2021.10.26.465695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wueseke O, Zwicker D, Schwager A, Wong YL, Oegema K, Jülicher F, Hyman AA, Woodruff JB (2016). Polo-like kinase phosphorylation determines Caenorhabditis elegans centrosome size and density by biasing SPD-5 toward an assembly-competent conformation. Biol Open 5, 1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Smith DK, Zhu H, Guan Y, Lam TTY (2017). ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8, 28–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data sets analyzed for this study are displayed. A supplemental CSV file is provided with the species used in the phylogenetic analysis with a FTP link to download their proteomes (Supplemental File SF1A, Metazoan_taxon_list_FTP_links.csv), the tree file is provided in Newick format (File SF1B, nuclear_genes_tree.nwk), the R script file for phylogeny analysis is available as a supplemental file SF1 (File SF1C, phylo_analysis.html), and the BLAST results are provided in a supplemental table formatted in Supplemental File SF1 (File SF1D, NCBI_BLAST_results.zip; File SF1E, Homebrew_BLAST_results.zip).