Graphical abstract

Keywords: Arsenic, Microbial genomics, Machine Learning, Human microbiome
Abstract
Arsenic is a ubiquitous toxic element, the global cycle of which is highly affected by microbial redox reactions and assimilation into organoarsenic compounds through sequential methylation reactions. While microbial biotransformation of arsenic has been studied for decades, the past years have seen the discovery of multiple new genes related to arsenic metabolism. Still, most studies focus on a small set of key genes or a small set of cultured microorganisms. Here, we leveraged the recently greatly expanded availability of microbial genomes of diverse organisms from lineages lacking cultivated representatives, including those reconstructed from metagenomes, to investigate genetic repertoires of taxonomic and environmental controls on arsenic metabolic capacities. Based on the collection of arsenic-related genes, we identified thirteen distinct metabolic guilds, four of which combine the aio and ars operons. We found that the best studied phyla have very different combinations of capacities than less well-studied phyla, including phyla lacking isolated representatives. We identified a distinct arsenic gene signature in the microbiomes of humans exposed or likely exposed to drinking water contaminated by arsenic and that arsenic methylation is important in soil and in human microbiomes. Thus, the microbiomes of humans exposed to arsenic have the potential to exacerbate arsenic toxicity. Finally, we show that machine learning can predict bacterial arsenic metabolism capacities based on their taxonomy and the environment from which they were sampled.
1. Introduction
Arsenic is ubiquitous in nature. It is commonly found in one of two inorganic forms; the pentavalent arsenate (As(V)) and the trivalent arsenite (As(III)), both of which are extremely toxic [1]. As(V) is taken up via phosphate transporters and As(III) via aqua-glyceroporins. Because arsenic uptake is incidental, it is assumed that all organisms have some coping mechanisms to deal with the toxicity [2]. Both arsenite oxidation and arsenate reduction can also support respiration, and these pathways are believed to have evolved before the split between the archaeal and bacterial domains [3]. Microorganisms play a major role in the global arsenic cycle, driving both the mineral precipitation and dissolution. Microbes are also capable of assimilating arsenic into more complex compounds or forming volatilized methylated forms of arsenic [4]. Taken together, microbes have a huge potential effect on human exposure to arsenic. While Bangladesh is a prime example of the detrimental effects of groundwater contamination, other hotspots are known around the world, exposing millions of people to arsenic-contaminated drinking water [5].
The last decade has seen an exponential growth in the availability of sequenced genomes, and with it the discovery and expansion of the known arsenic transforming genes in microorganisms [6]. Currently, there are four types of operons, one dedicated to detoxification of arsenic (ars operon) and three respiratory operons using arsenic as the electron acceptor (dissimilatory arsenate reduction by the arr operon) or electron donor (respiratory arsenite oxidation by aio/arx operons) [7]. The ars operon can be split into an inorganic path (the canonical reduction of arsenate and excretion of arsenite), and an organic pathway. The organic pathway starts with the methylation of arsenite by the ArsM enzyme, forming monomethylarsonous acid (MMAs(III)). MMAs(III) can be further methylated by ArsM and volatilized, excreted from the cells via the ArsP efflux system, or oxidized into less toxic compounds via the ArsH enzyme. An alternative efflux system (ArsJ) can excrete arsenate by forming an unstable intermediate with glyceraldehyde 3-phosphate.
To date, studies have either focused on specific arsenic metabolizing microbes or surveyed the presence of a small number of key genes in larger datasets [8], [9], [10], [11], [12]. A few studies that investigated larger numbers of genes did not look at co-localization of these genes, which is necessary to improve confidence in pathway identification. In this study, we utilized a relatively comprehensive set of bacterial and archaeal genomes, thus a very large set of genes involved in arsenic transformation, to analyze how arsenic-related genes are combined in microorganisms. We also relied on gene co-localization to help overcome low homology or low levels of differentiation of auxiliary and regulatory genes. Our research resulted in a database of genes related to arsenic biotransformation, within which we identified the core arsenic microbial guilds. We show that the combination of microbial taxonomy and environmental information predict the resident arsenic microbial guilds. Further, we show that arsenic exposure shapes the inventories of arsenic-relevant capacities in human microbiomes, thus could increase arsenic toxicity.
2. Results
2.1. Genomic sampling of microbial diversity across environments
We constructed a database of genomes (GAsDb) that sampled the broadest possible taxonomic and environmental diversity, drawing upon both the RefSeq genomes and ∼3500 metagenomes that our laboratories and collaborators have assembled, primarily from terrestrial environments. We categorized each metagenome (and, for RefSeq, genome) in terms of its ecosystem of origin: terrestrial near-surface, marine, deep subsurface, engineered and host-associated. Each metagenome was also assigned an environment type (101 categories, including soil, freshwater, human) and a geographic location (432 locations, divided into country/province/state), and climate (22 types - derived from the dominant climate of a given location). To assign taxonomy to 91,685 genomes, we used GTDB-Tk [13], for an initial assignment, but in some cases corrected the taxonomy to reflect more appropriate standard nomenclature. The genomes in the database are from both the Bacterial and Archaeal domains, with representatives from 92 phyla, 309 classes, 785 orders, 1,610 families, and 3,411 genera.
2.2. Distribution of arsenic genes and arsenic loci in genomes
For the purpose of this work, we designed 29 profile hidden Markov models (HMMs) targeting five genes of the aio operon, five genes of the arx operon, five genes of the arr operon (one gene was targeted with two HMMs), and 14 genes of the ars operon. Sequences were assigned a single annotation. When a sequence was found by multiple HMMs, assignments were made based on HMM scores. Following maximum likelihood tree constructions, we identified four The four new clades that were all monophyletic with one of the 29 clades, and closely enough related that we assumed that they have related functions (see methods and Supplementary information 1). The HMM for ArsR did not yield robust results so it is not included in this work.
In our analyses, we also considered the co-localization of genes to support functional assignments based on HMMs. Sets of co-localized genes involved in arsenic transformations were used to define arsenic loci. Loci with no discernible functionality (e.g. all the genes were auxiliary), or loci with a high ratio of hits below their HMM score threshold were removed from the dataset. The GAsDb contains 949 unique arsenic loci (see methods and Supplementary Tables 1-3). Genomes carry between 1 and 12 arsenic loci, with a median of 2 loci. 41% of genomes only have a single locus and 40% of loci appear in a single genome. Only ten arsenic loci appear in > 5% of the genomes, all of them containing genes from the ars operon involved in arsenic resistance. Five of the ten have an arsenic efflux system (four loci for export of arsenite and one for export of methylated arsenic). Another, arsM, is a single gene locus. Two genes, arsC and arsH, appear in multiple loci.
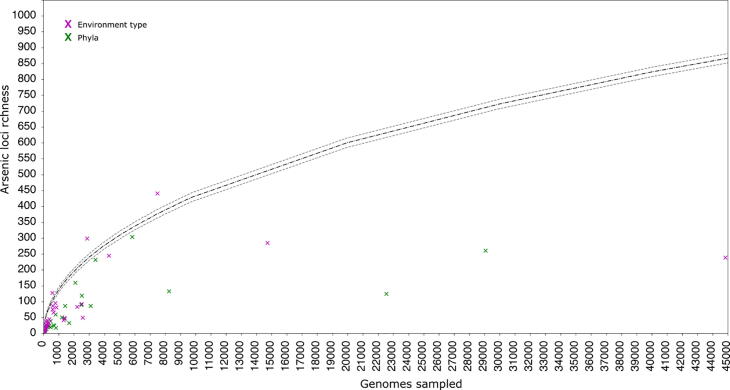
The richness of microbes, grouped by taxonomy or environment, was compared to random sampling of the whole dataset (Fig. 1). Groups with significantly higher richness compared to random sampling (marks above standard deviation) are enriched for arsenic metabolism. No phylum was significantly enriched and only three types of environments had significantly high arsenic loci richness: groundwater, hotsprings, and sites associated with mining.
Fig. 1.
Richness of arsenic loci in phyla (green) and environment types (magenta), compared to the mean richness from random sampling (1000 permutations) of the dataset (black line). Grey dashed lines indicate one standard deviation from the mean. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. Arsenic biotransformation guilds and core arsenic loci
To examine if microbes can be classified into arsenic guilds with a similar set of genes, we first grouped the genomes to unique arsenic profiles (AsPRO). This was based on the gene content and gene count over the whole genome, resulting in 7,178 unique arsenic profiles. We further reduced the data to 7,141 AsPRO by eliminating profiles containing a single gene type. While the efflux systems for arsenite (arsB, ACR3), the efflux system for arsenate (arsJ), and methylation to volatilization (arsM) can be considered standalone detoxification pathways, we preferred to remove such genomes for several reasons. First, our dataset consists of metagenome-assembled genomes (MAGs) of unknown completeness, due to the large size of the database. That meant that we could not easily distinguish between true single gene genomes and noise from low-quality MAGs (this is not a concern for other AsPRO since the loci analysis shows the genes are co-localized in genomes). Second, the single gene profiles added too much noise when analyzing the metabolism of the clusters and their removal provided improved metabolic resolution, with little effect on the overall clustering structure (see methods).
The chosen AsPROs were then clustered based on their genes (Fig. 2). Clustering of AsPROs was conducted twice, once with a focus on global structure and once with a focus on local structure (see methods). In global structure, higher weight is given to the most abundant genes, while in local structure, smaller gene differences are weighted more. The former enabled a more refined analysis of metabolism in some of the larger groups. The representative metabolism for each cluster was based on the proportion of genomes harboring the gene (Table 1). Genes were considered representative if they were present in at least 50% of the genomes of a cluster. An exception was made for the cluster containing ArxAB as this was the only one containing these genes.
Fig. 2.
Clustering of AsBT-Guilds and AsBT-SubGuilds. A two-dimensional UMAP projection of unique arsenic profiles (AsPRO). Black oval indicates global structure clustering while colors indicate sub clustering based on local structure. Small black points indicate AsPROs that were not clustered (considered noise).
Table 1.
AsBT-Guilds/AsBT-SubGuilds (a) size and (b) representative arsenic metabolism.
| Guild | Subguild | Size (# genomes) |
|---|---|---|
| AsBT-Guild_1 | AsBT-SubGuild_09 | 166 |
| AsBT-Guild_2 | AsBT-SubGuild_03 | 404 |
| AsBT-Guild_3 | AsBT-SubGuild_13 | 687 |
| AsBT-SubGuild_14 | 3680 | |
| AsBT-Guild_4 | AsBT-SubGuild_06 | 975 |
| AsBT-Guild_5 | AsBT-SubGuild_21 | 98 |
| AsBT-SubGuild_18 | 172 | |
| AsBT-SubGuild_25 | 382 | |
| AsBT-SubGuild_20 | 457 | |
| AsBT-SubGuild_24 | 1996 | |
| AsBT-SubGuild_15 | 3674 | |
| AsBT-SubGuild_19 | 6472 | |
| AsBT-SubGuild_23 | 13464 | |
| AsBT-SubGuild_22 | 13594 | |
| AsBT-Guild_6 | AsBT-SubGuild_01 | 6167 |
| AsBT-Guild_7 | AsBT-SubGuild_16 | 607 |
| AsBT-SubGuild_08 | 1143 | |
| AsBT-SubGuild_17 | 1593 | |
| AsBT-SubGuild_12 | 1691 | |
| AsBT-Guild_8 | AsBT-SubGuild_05 | 2574 |
| AsBT-Guild_9 | AsBT-SubGuild_04 | 2113 |
| AsBT-Guild_10 | AsBT-SubGuild_00 | 738 |
| AsBT-Guild_11 | AsBT-SubGuild_10 | 118 |
| AsBT-SubGuild_11 | 235 | |
| AsBT-Guild_12 | AsBT-SubGuild_07 | 365 |
| AsBT-Guild_13 | AsBT-SubGuild_02 | 205 |
| AsBT-Guild_1 | AsBT-Guild_2 | AsBT-Guild_3 | AsBT-Guild_4 | AsBT-Guild_5 | AsBT-Guild_6 | AsBT-Guild_7 | AsBT-Guild_8 | AsBT-Guild_9 | AsBT-Guild_10 | AsBT-Guild_11 | AsBT-Guild_12 | AsBT-Guild_13 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AsBT_SubGuild_9 | AsBT_SubGuild_3 | AsBT_SubGuild_13 | AsBT_SubGuild_14 | AsBT_SubGuild_6 | AsBT_SubGuild_21 | AsBT_SubGuild_18 | AsBT_SubGuild_25 | AsBT_SubGuild_20 | AsBT_SubGuild_24 | AsBT_SubGuild_15 | AsBT_SubGuild_19 | AsBT_SubGuild_23 | AsBT_SubGuild_22 | AsBT_SubGuild_1 | AsBT_SubGuild_16 | AsBT_SubGuild_8 | AsBT_SubGuild_17 | AsBT_SubGuild_12 | AsBT_SubGuild_5 | AsBT_SubGuild_4 | AsBT_SubGuild_0 | AsBT_SubGuild_10 | AsBT_SubGuild_11 | AsBT_SubGuild_7 | AsBT_SubGuild_2 | |
| aioA | 20.48 | 21.71 | 0 | 0 | 0 | 0 | 2.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0.06 | 0 | 0.12 | 0.09 | 0.14 | 100 | 99.57 | 5.48 | 8.29 |
| aioB | 18.07 | 21.71 | 0 | 0 | 0 | 0 | 2.33 | 0 | 0.22 | 0 | 0.03 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0.06 | 0 | 0 | 0.09 | 0 | 100 | 100 | 5.48 | 8.78 |
| aioR | 73.49 | 49.61 | 100 | 95.52 | 25.54 | 0 | 11.63 | 1.31 | 0.88 | 2.4 | 2.59 | 53.58 | 18.14 | 0.05 | 0.08 | 0.33 | 33.07 | 0.38 | 2.72 | 3.7 | 6.58 | 5.28 | 23.73 | 90.64 | 73.15 | 47.8 |
| aioS | 75.9 | 28.68 | 100 | 99.97 | 13.23 | 0 | 0.58 | 0 | 0.22 | 0 | 1.25 | 0 | 0 | 0.06 | 0.03 | 0.16 | 0 | 0.06 | 0 | 0.66 | 7.1 | 6.37 | 21.19 | 94.89 | 57.81 | 40.98 |
| aioX | 81.33 | 9.3 | 0 | 0 | 4.31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0.08 | 0 | 0 | 5.08 | 100 | 50.14 | 40 |
| aioX_like | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.55 | 4.39 |
| arxA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 21.95 |
| arxB1 | 0 | 0.78 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 38.54 |
| arxB1_like | 0 | 39.53 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.05 | 0 | 0 | 0.43 | 25.21 | 11.71 |
| arxB2 | 0 | 0.78 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.38 | 27.8 |
| arxC | 0 | 40.31 | 0 | 0 | 2.15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.23 | 0 | 0 | 0 | 0 | 5.21 | 35.61 |
| arxD | 0 | 4.65 | 0 | 0 | 5.85 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 27.4 | 28.29 |
| arrA | 0 | 0 | 0.44 | 0 | 5.64 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0.31 | 0.05 | 0 | 0 | 0 | 74.52 | 19.51 |
| arrB | 0 | 0.78 | 0 | 0 | 5.95 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 66.03 | 8.78 |
| arrC | 0 | 1.55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 47.32 |
| arrD | 0 | 0 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.09 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 73.7 | 10.24 |
| arrS | 0 | 51.94 | 0.15 | 0 | 90.67 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10.17 | 0 | 8.77 | 5.85 |
| ACR3 | 59.64 | 75.19 | 19.51 | 19.62 | 76.1 | 95.92 | 81.98 | 76.7 | 56.67 | 0.35 | 71.77 | 32.39 | 1.1 | 55.23 | 81.24 | 17.63 | 1.14 | 91.27 | 0.71 | 71.13 | 17.98 | 64.91 | 43.22 | 69.36 | 78.36 | 69.76 |
| arsA | 15.66 | 31.78 | 1.46 | 1.55 | 2.46 | 13.27 | 1.74 | 100 | 0 | 1.2 | 0.3 | 0.8 | 0 | 0.1 | 2.12 | 99.18 | 98.34 | 99.06 | 100 | 1.83 | 0.19 | 5.96 | 9.32 | 11.06 | 19.45 | 13.17 |
| arsB | 2.41 | 19.38 | 0.44 | 0.65 | 1.03 | 1.02 | 2.33 | 17.8 | 26.7 | 100 | 8.98 | 62.48 | 98.37 | 0.02 | 25.33 | 79.08 | 99.48 | 12.12 | 100 | 2.88 | 0.76 | 4.61 | 7.63 | 16.17 | 0 | 2.44 |
| arsC | 19.88 | 13.18 | 3.49 | 7.09 | 65.03 | 80.61 | 5.81 | 37.96 | 74.84 | 41.93 | 32.34 | 61.91 | 50.78 | 59.38 | 47.72 | 6.43 | 100 | 59.89 | 99.65 | 93.93 | 8.76 | 50.95 | 44.07 | 44.68 | 8.22 | 7.32 |
| arsC_LMWP | 52.41 | 71.32 | 39.45 | 36.06 | 73.44 | 83.67 | 72.09 | 45.55 | 66.74 | 61.72 | 48.58 | 31.98 | 24.61 | 40.07 | 46.47 | 90.77 | 4.37 | 78.97 | 2.31 | 16.81 | 41.17 | 68.02 | 51.69 | 69.36 | 58.63 | 56.59 |
| arsD | 13.86 | 20.93 | 0.58 | 1.22 | 2.15 | 0 | 0.58 | 0 | 0 | 0 | 0.05 | 0.02 | 0 | 0.1 | 0.92 | 100 | 100 | 100 | 100 | 0.93 | 0.05 | 3.66 | 6.78 | 8.94 | 13.15 | 8.78 |
| arsT | 0 | 0 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0.01 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0.09 | 100 | 0 | 2.13 | 0.55 | 0 |
| arsO | 1.81 | 23.26 | 0 | 0.38 | 0.82 | 100 | 0.58 | 6.02 | 100 | 0 | 0.3 | 1.25 | 0 | 0.08 | 3.26 | 0 | 0 | 3.2 | 0 | 6.58 | 1.99 | 47.02 | 8.47 | 15.32 | 0 | 0 |
| arsO_like | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| arsH | 21.69 | 46.51 | 8.88 | 6.09 | 65.74 | 22.45 | 5.81 | 34.03 | 51.2 | 88.78 | 54.46 | 52.9 | 80.58 | 39.94 | 78.32 | 14.83 | 83.73 | 12.99 | 97.93 | 14.05 | 14.72 | 62.2 | 27.12 | 62.55 | 9.86 | 8.29 |
| arsI | 6.63 | 59.69 | 10.92 | 8.13 | 41.13 | 57.14 | 0.58 | 12.57 | 0.88 | 0.05 | 99.97 | 0.79 | 0 | 0.15 | 74.62 | 0 | 0 | 3.08 | 0 | 1.75 | 21.44 | 7.59 | 47.46 | 28.94 | 3.56 | 3.41 |
| arsJ | 3.01 | 13.18 | 0.29 | 0 | 0.21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.15 | 0 | 0 | 0 | 0 | 100 | 0.14 | 0.41 | 0.85 | 1.7 | 1.37 | 2.44 |
| arsM | 45.18 | 64.34 | 68.12 | 60.6 | 22.87 | 7.14 | 50.58 | 15.18 | 8.75 | 0.5 | 15.24 | 16.87 | 0.79 | 38.03 | 0.18 | 8.24 | 0.09 | 14.81 | 0.06 | 6.81 | 77.61 | 23.44 | 36.44 | 15.32 | 63.01 | 59.02 |
| arsP | 9.64 | 47.29 | 5.97 | 4.1 | 5.64 | 1.02 | 100 | 0 | 1.97 | 0 | 0.16 | 100 | 0 | 0.01 | 0.28 | 99.84 | 33.6 | 0.13 | 2.84 | 6.23 | 0.38 | 5.56 | 20.34 | 28.51 | 36.44 | 29.76 |
| arsP_like | 1.2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50.85 | 0.66 | 0 | 0 | 0 | 0.19 | 100 | 0.41 | 0.85 | 0 | 0 | 0 |
Clustering resulted in 13 arsenic biotransformation guilds (AsBT-Guild), 12 of which originated from the global structure and one unique to the local structure (the AsPRO were dispersed among several guilds or considered noise with global structuring). In addition, several AsBT-Guilds could be further separated into subclusters (AsBT-SubGuild). AsBT-Guild 11 and 13 contain two subclusters, AsBT-Guild 7 contains 4 subclusters, and AsBT-Guild 5 (the largest group) is divided into 8 subclusters (Fig. 2). All but two AsBT-Guilds contained the inorganic pathway of the ars operon, and all but one AsBT-Guild had at least one gene of the organic pathway of the ars operon as part of their representative detoxification system. Multiple AsBT-SubGuilds had an additive proportion exceeding 100% of the two arsC variants (TRX-like protein family and LMWP protein family). The arsenite pump variants (arsB and ACR3) on the other hand, rarely overlapped within genomes. Each of the respiration operons arr, arx, and aio were represented in a separate AsBT-Guild. Of these, the AsBT-Guild enriched for arx did not have it as representative metabolism (even though it was unique to it). The AsBT-Guild represented by the aio operon was further divided into two AsBT-SubGuilds, one containing the short version of the operon (aioBA) and the other the long version (aioBAXSR).
We discovered four new AsBT-Guilds in our work. The two larger AsBT-Guilds (comprising 4,297 and 2,070 genomes) did not have the inorganic ars path but rather relied on arsM as the sole detoxification enzyme. The first group (AsBT-Guild 3) paired the aio two-component system (aioSR) with arsM, and the second group (AsBT-Guild 9) had arsM and the sibling clade of arsP (arsP-like), the MMAs efflux system. Members of the two AsBT-Guilds (3 and 9) are predominantly found in soil environments, where they account for just over half of the soil derived genomes (2,918 and 1,698 genomes respectively of a total of 9,181 soil genomes). Genomes of AsBT-Guild 3 are also found in groundwater and sediment environments in large numbers (650 and 541 genomes respectively). The other AsBT-Guilds were smaller (comprising 404 and 166 genomes). These AsBT-Guilds paired aio regulation genes with ars detoxification. Genomes in the larger group (AsBT-Guild 2) contain multiple aio and arx regulatory and auxiliary genes combined with a short ars operon and arsM. >75% of the genomes have five or more unique arsenic genes (not accounting for gene counts). Genomes in the smaller group (AsBT-Guild 1) have the full set of regulatory aio genes (aioXSR) paired with a short ars operon.
Next, we wanted to examine whether we could identify a core set of arsenic loci. The core arsenic loci is the minimal set of loci found (in various combinations) in more than half of the genomes of each phylum. We filtered GAsDB based on the per phylum frequency of the different loci and identified that the core arsenic loci consists of 83 loc (Supplementary Table 2). Over 99% of all genomes are represented by the core arsenic loci (i.e. all these genomes have at least one of the loci in the core set), with a mean phylum representation of 97.6% and 46 of 92 phyla fully recovered. One phylum (Candidatus Sumerlaeota) retained 50% of its genomes and the phylum with the second-lowest retention (Aquificae) was at 81.5%. All the studied genes were accounted for within the core loci, except for arsenite oxidation via arxA.
When we observed the proportions of the core loci in different phyla (Fig. 3a) we discover that there exists a stark difference in the loci content of the five most represented phyla in GAsDB (Gammaproteobacteria, Betaproteobacteria, Alphaproteobacteria, Firmicutes, and Actinobacteria), compared to all other phyla. To see if the difference in loci is also translated into a difference in functionality we reverted back to examining the proportions of the different genes in the genomes. Within GAsDB, the most common genes are for arsenate reduction (ArsC variants) and arsenite excretion (ArsB/ACR3), followed by MMAs oxidation (ArsH) and arsenite methylation (ArsM) (Fig. 3b). But, when the proportion is calculated per phylum a very different picture appears (Fig. 3c). The median proportion of arsenate reduction (ArsC variants) in phyla is 0.4, while > 60% of genomes in the database have the genes. MMAs oxidation (ArsH) is similar with a median proportion < 0.1 in phyla but found in nearly half of the genomes. The difference between phyla proportion for arsenite excretion (ArsB, ACR3) and its overall presence is smaller but follows a similar trend. Arsenite methylation shows the opposite trend, with a median proportions of ∼ 0.55 across phyla, even though it is present in 20% of genomes. The answer to the difference in proportion lies with the five largest phyla in GAsDB. In these instances (and in other genes) the largest phyla are outliers in the distribution of proportions. These five phyla account for nearly 80% of the genomes in the dataset so they can highly distort the overall view of arsenic biotransformation. Moreover, the distortion also occurs in the literature, since these phyla are also the most studied phyla in microbiology [14].
Fig. 3.
Core loci and gene functionality proportions in genomes. (a) the proportion of the 20 most frequent loci across all phyla. Lighter color in heatmap indicates higher proportions within the phyla. Loci are sorted from the top in descending order by their overall frequency. Phyla are sorted left to right in descending order based on their loci richness. (b) the proportion of functions in all of the genomes. (c) the distribution of function proportion by phyla. Each phylum is represented on the boxplot as a green-shaded dot. The darkness of the dot reflects the order of magnitude size of the phylum. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.4. Taxonomic and environmental drivers of arsenic biotransformation
The above analysis has mostly been independent of taxonomic and environmental data (referred to below as metadata features) associated with the genomes. Furthermore, the classification of AsBT-Guilds was based on AsPRO and not on individual genomes. Still, the arsenic loci accumulation hinted at significant enrichment in some environments, while the core arsenic loci analysis revealed a stark difference between the well-studied phyla compared to all other phyla. In this section we will examine which metadata features are enriched in the AsBT-Guilds, and how well the metadata features predict the AsBT-Guilds.
To examine the level of metadata feature specificity to AsPROs (Supplementary Table 4) we examined two parameters. First we checked what is the variation within the features (Fig. 4a). Feature variables are the instances of a metadata feature (e.g Actinobacteria is a feature variable of the Phyla feature). Second we examined if one feature variable accounted for the majority of the genomes (i.e. the dominant feature variant) in a given AsPRO (Fig. 4b). These parameters are important for downstream analysis that test how predictive metadata features are of metabolism. The higher the within variation of a feature, and the more evenly the feature variables are distributed within AsPROs, the less predictive they would be.
Fig. 4.
Specificity of metadata in AsPROs. (a) mean variable counts for metadata features. Error bars indicate confidence intervals at 95%. (b) distribution of the proportion of the dominant variable in the features. White stars indicate the mean and black lines indicate median (when median is not observed it is equal to 1).
In both cases, AsPROs that account for a single genome were excluded from the analysis to prevent bias. In addition, we compared the statistics for AsPROs comprising ten or more genomes (n = 468) to AsPROs of less than ten genomes (n = 1,357). The environmental features have a lower variable count (except for location). The high variation in the location feature was an important indication that the data does not suffer from sampling bias. The taxonomic features show an increase in variability with higher taxonomic levels. Large AsPROs had higher variable counts than small AsPROs for all metadata features but Domain. The difference between the means was much more substantial compared to differences in median values or even the third quartile, indicating the data are skewed by a few highly variable large AsPROs. The average proportion of the dominant variable ranged from 0.65 (climate) to 0.99 (Domain) with most features above 0.75. Large AsPROs had only slightly lower mean and median values compared to the smaller AsPROs. This means that although their variable counts are higher there still exists a single variable for each feature that accounts for most of the genomes associated with AsPROs. Moving forward, we chose parameters that had low variability at the AsPRO level: ecosystem and environment types for environmental metadata, and Domain, Phyla, and Class as the taxonomic metadata.
To test if certain metadata was enriched in the AsBT-Guilds (and AsBT-SubGuilds) we used the Fisher’s Exact test (Supplementary Tables 5–6). We consider only metadata with odds ratio higher than 1 and bonferroni corrected p-value < 0.05 to be significantly enriched in a given AsBT-Guild. We also note that enrichment does not necessarily equate to the most common metadata feature in an AsBT-Guild. The enrichment results varied greatly between AsBT-Guilds from very specific (a single environment type, phyla, and class) to 70 different variables across four features. At least one variable of Phyla, Class, and environment type was enriched in each AsBT-Guilds and AsBT-SubGuilds. An ecosystem variable was enriched in all but a single AsBT-SubGuild. A median of one ecosystem, three environment types, three phyla, and five classes were enriched in each guild/subguild. Of the four new AsBT-Guilds, 1 and 2 (pairing aio/arx regulation with the inorganic path of the ars operon) were enriched in samples from mining sites. AsBT-Guilds 3 and 9 (AioSR paired with ArsM) was enriched in multiple phyla from terrestrial ecosystems, with AsBT-Guild 9 highly specific to soil environments. AsBT-Guild 8 was noteworthy, because while it was enriched in several environment types, most of them marine (seawater, shrimp, mollusca, zooplankton, fish, and cephalopods). Oxygen levels (deduced from the environment type) were an important factor for the respiration operons. AsBT-Guild 12 (arr operon) was enriched in anaerobic environments, while AsBT-Guild 11 (aio operon) was enriched in aerobic environments. The latter was also more associated with contaminated sites. An interesting split occurred within the subguilds of AsBT-Guild 7 (long ars operon), based on the reductase and efflux pump variants. Human associated Gammaproteobacteria have the TRX-like ArsC variant and ArsB pump, while human associated Firmicutes have the LMWP ArsC variant and ArsB pump. A third subguild with the LMWP ArsC variant and ACR3 pump is enriched within both Gammaproteobacteria and Firmicutes from food products.
The results so far showed that the metadata are specific to AsPROs and that similar AsPROs had similar metadata. Now we turned the analysis around to test if metadata can be used to predict the AsBT-Guild of genomes. We chose to use Phyla, Class, Ecosystem, Environment type, and Climate in order to predict AsBT-Guild membership by genome. Phyla and Class can robustly be identified with gene markers and research groups generating data from their own samples know where the samples are from so can assign environmental information. A subset of the data that contained information for all features (n = 46170, split 8:2 for training and validation sets) was used to create a Random Forest Classification model (Supplementary information 2). A grid search was used to refine the model parameters and the best overall model was chosen based on its accuracy. The selected model had the following performance indices for the validation set: accuracy of 0.773, MSE of 0.203, logloss of 0.993, and r2 of 0.977. While the overall accuracy was relatively high, the mean per-calls error was 0.576. From the confusion matrix it is clear that the model accuracy stems from high precision in predicting four AsBT-Guilds. AsBT-Guild 5 was detected at 0.9 accuracy while accounting for 62% of the genomes in the validation set. AsBT-Guild 3 (n = 814, accuracy = 0.86), AsBT-Guild 6 (n = 709, accuracy = 0.76), and AsBT-Guild 9 (n = 374, accuracy = 0.78) accounted for an additional 21% of the genomes in the validation set. It is worth mentioning that the two AsBT-Guilds (3 and 9) that lack the short inorganic ars operon were accurately predicted.
The most important metadata features in the model (Fig. 5a) are phyla (∼50%) and environment type (∼30%), followed by climate (∼10%), class (∼8%) and ecosystem (∼2%). When the predicted AsBT-Guild is compared to the observed AsBT-Guild by phyla or environment type the accuracy of the results are much better (Fig. 5b-c). For a given environment type, the lowest true positive (TP) % is 50% and for 36/46 environments the TP is equal or>75%. Two important environments with < 75% TP are sediment (74%) and soil (71%). These are prone to annotation errors in reported metadata. Other noteworthy environments that require improved annotation are food products, groundwater, and freshwater.
Fig. 5.
Random Forest Classifier model for arsenic metabolism. a) Variable importance in the Random Forest Classifier model. The bar plot shows the scaled importance of each of the metadata features. The insert table shows the relative importance and percent contribution to prediction. b) Percent of true positive predictions for validation and test datasets based on phyla. c) Percent of true positive predictions for validation and test datasets based on environment type.
For phyla, the results are similar with 32 of 46 having 75% TP or higher. Important phyla with low TP were Gammaproteobacteria, Betaproteobacteria and Deltaproteobacteria. Incorrect predictions for the Proteobacteria were assigning them to AsBT-Guild 5 and for Deltaproteobacteria assigning them to AsBT-Guild 3.
Since climate only accounted for 10% of the prediction, a second set containing all other genomes that did not have climate information, but had the other features (n = 16682) was also tested with the model. The accuracy for the second test set was 0.767 (MSE = 0.209, logloss = 1.18), but the per class error was very high (0.81) and only two AsBT-Guild (6 and 5) had low error rates. These two account for most of the samples in the test data (81.2%).
The breakdown of environments again had soil and sediment at low TP (Fig. 5c). Mining related sites also showed low TP (∼70%) and were the largest group in the low TP group. Wastewater, salt water (grouping seawater, brackish water, and hypersaline water), and mollusca had ≤ 45% TP and were relatively large (group sizes > 80 genomes).
For phyla (Fig. 5b), most of the error stemmed from the Alphaproteobacteria, Betaproteobacteria and Gammaproteobacteria. Large groups that performed well in both data sets were the Actinobacteria, Firmicutes, and Bacteroidetes (Epsilonproteobacteria was also good but not that large). Zetaproteobacteria prediction improved significantly in the test data from 46% in the validation (n = 26) to 94% in the test (n = 53). Although the method in which climate was assigned to genomes was very generalized (to the most common climate region in a given location), and its importance in the model is low, it does improve the model predictions. Climate is easy to include if the geographic location of sampling is known.
2.5. Effect of arsenic exposure on the human microbiome
Human exposure to arsenic through groundwater and food is a worldwide concern, most prominently in Bangladesh [15]. A recent paper analyzed the probability of groundwater arsenic contamination and identified additional hotspots [5]. In the following analysis we sought to investigate if human microbiomes from areas of higher arsenic exposure risk cluster together, based on their arsenic metabolism. This would indicate that arsenic exposure affects the composition of the human microbiome. Within our data, we had one group of gut samples from humans from Laksam Upazila (Bangladesh) in which arsenicosis from arsenic exposure was confirmed [16]. We used this group as an indicator to locate the cluster of interest and called it the “arsenic exposed” cluster (the quotes are meant to convey we cannot prove arsenic exposure). Human associated microbial genomes were grouped based on their refined location and bodily source (see methods), and clustered by the counts of AsBT-SubGuilds in those groups (Fig. 6a). The “arsenic exposed” cluster was found to be Cluster 16).
Fig. 6.
Arsenic metabolism in human-associated microbial genomes. a) Clustering of genomes grouped by their geographic location and bodily source of sampling. b) Geographic location of the “arsenic-exposed” cluster. Locations in orange are part of the cluster. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We next evaluated the other locations that were clustered with Laksam Upazila. We found that many (but not all) locations in the cluster have been shown to have heightened arsenic contamination in groundwater and soil (Fig. 6b). Asian locations included West Bengal, Pakistan, Maharashtra (India), several provinces in China (Liaoning, Shandong, Zhejiang), Taiwan, southeast asian countries (Thailand, Viet Nam), Japan, and Saudi Arabia. Locations in the Americas included Mexico, Argentina (Buenos Aires), Columbia (Antioquia), Ecuador, several US states (Nebraska, California, Michigan, Montana, Florida), and several Canadian provinces (British Columbia and Manitoba). African locations included sub-saharan nations (Tanzania, Kenya, Mali, and Zambia) while European locations included Denmark, France, Italy, and the United Kingdom. The main locations in the cluster that match previous reporting [5] are West Bengal, Pakistan, Liaoning, Mexico and Buenos Aires. Other areas with increase risk of exposure (04.-0.6 probability As > 10 ppb) were Maharashtra, Thailand (central area), Viet Nam (southern area), Saudi Arabia, California, Tanzania, Kenya, Mali, and Zambia.
The AsBT-SubGuilds enriched in the “arsenic exposed” cluster were AsBT-SubGuilds 19 and 18. A unique feature of both subtypes is that all of their genomes contain the arsP efflux system (excreting MMAs from the cells). In AsBT-SubGuild 18 just over 50% of the genomes also contained arsM. The high prevalence of the arsP efflux system indicates that the microbiome of humans exposed to arsenic further exacerbates the toxic effect since MMAs are more toxic to humans than arsenate and arsenite [17]. The arsenic exposed human samples have a much lower occurrence of AsBT-SubGuild 1, which are human-associated actinobacteria. AsBT-SubGuilds 18 is found in very low numbers in the entire set of human associated samples (n = 5) compared to AsBT-SubGuild 19, but it is only found in the “arsenic cluster” and adjacent cluster 11, that has additional genomes from Dhaka, Bangladesh and are known cholera patients. The cholera-patient samples have some overlap of locations with the arsenic-exposed cluster and the samples are enriched with AsBT-SubGuilds 5 (all members have the arsJ efflux pump for As(V)). The human source of the cholera-patient sample are feces (or clinical samples) while the arsenic-exposed samples also have multiple samples from the urinary system.
3. Discussion
In this paper we curated a large genomic database of arsenic related genes from Bacteria and Archaea, using 29 custom designed HMMs. Utilizing co-localization of genes, we enhanced the yield of identified genes to include more distantly related sequences, as well as new putative genes. Overall we defined nearly 949 unique arsenic loci (based on gene presence in a given loci), far higher than the few dozen known to date [8], [18]. Our examination of the core arsenic loci (83 loci) and gene proportions in different phyla revealed that some of the most studied phyla in microbiology (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria, and Firmicutes) [14] have a very different arsenic metabolism from all other phyla. While they may be dominant in some environments, it is clear that they are not representative of the microbial arsenic metabolism and more focus should be given to other phyla.
Clustering of the genomes based on their arsenic related genes generated 13 types of representative arsenic metabolic/transformation capabilities (AsBT-Guilds), some further divided into more refined groups, totaling at 26 AsBT-SubGuilds. Of these AsBT-Guilds, four represent newly defined metabolic profiles. Interestingly, three of the four AsBT-Guilds (guilds 1, 2, and 3) paired the aio regulation system (aioR, aioS, and to a lesser extent aioX) [19] with the ars operon. While in two of those AsBT-Guilds a subset of genomes also have the catalytic aio units (aioBA, these are found at much lower proportions (10–20%) compared to the corresponding arsenate reductases of the ars operon (50–80%). Even though most of the genomes used in this work are considered to be draft genomes, we would argue that the results indicating the presence of aioSR without the aioX or the catalytic unit (aioBA) is strong. When identifying loci in the genomes we allowed for gaps between genes (up to five genes apart), but the majority of loci were of sequential genes (81.5% no gaps, 8.25% a single gene gap). The loci containing aioSR alone was one of the most commonly found loci (identified in 5870 genomes), while the loci aioXSR are found in 132 genomes. Other loci had the genes associated with different genes, even further adding to the results. One interesting, but unverified, hypothesis is the possibility that aioSR may also activate other genes (including ars operon genes) in response to arsenite.
Even more noteworthy are the two large AsBT-Guilds (3 and 9) that are lacking in the inorganic pathway of the ars operon. Instead these groups have ArsM that sequentially methylates As(III). Not only are AsBT-Guilds 3 and 9 predominant in soil environments, they represent some of the more dominant phyla in soil [20], [21], [22], [23]. AsBT-Guild 3 contains two thirds of the Acidobacteria (1,649), the majority of Deltaproteobacteria (8 4 1), Rokubacteria (6 3 9), Gemmatimonadetes (6 0 6), and a large portion of Nitrospirae (126, 23.6% of the phylum) in the GAsDb. AsBT-Guild 9 contains most of the Dormibacteria (4 5 8) and close to half of Verrucomicrobia (190 accounting for 47.5%). It also contains a large number of Actinobacteria (9 1 6) which account for nearly 40% of non-human microbiome Actinobacteria. This indicates that arsenic methylation in soil may be much more prominent than in other environments [10], [12]. Arsenic methylation can be used by bacteria as an allelopathic agent [24]. To counteract the toxicity of MMAs, bacteria can demethylate it and excrete the As(III). Nearly 60% of genomes in AsBT-Guild 5, the other large AsBT-Guild in soil, have the arsH gene, which encodes the oxidation of trivalent MMAs to a less toxic form of pentavalent MMAs [25]. AsBT-SubGuild 15 (within AsBT-Guild 5) is common in soil environments (1247) and nearly all genomes contain the arsI gene that encodes the demethylation of MMAs. This group contains Firmicutes, Betaproteobacteria, and Alphaproteobacteria. Here we can show a competitive relationship between different phyla based on their arsenic metabolism (Fig. 7).
Fig. 7.
Schematic diagram of interaction between different groups of soil microbes based on their arsenic metabolism. AsBT-Guilds 3 and 9 methylate arsenite which is transported out of the cells. The methylated arsenic can be oxidized by members of AsBT-Guild 5 or demethylated back to arsenite by AsBT-SubGuild 15 (part of AsBT-Guild 5), excreting it back into the environment. Dashed lines indicate expected transport by an unknown mechanism.
Many other AsBT-Guilds were enriched with a particular combination of taxa and environment types. AsBT-Guild 2 (and more specifically AsBT-SubGuild 3) was enriched in Chloroflexi and Betaproteobacteria from leachate reactors of mining operations. The genomes in this sub guild have a wide arsenic metabolic capacity. Genomes in this group had a mean of 8.6 unique genes and 35% had 12 or more unique genes. Overall, 11 of 32 genes were present in at least 40% of genomes. Originating from a highly contaminated environment the bacteria need multiple pathways to protect them from arsenic exposure.
AsBT-Guild 8 was enriched in Gammaproteobacteria and Zetaproteobacteria from seawater and marine organisms. The possession of both the inorganic ars pathway and the arsJ efflux system that excretes As(V) is compatible with As(V) being the dominant arsenic species in the marine ecosystem [4]. AsBT-Guild 11 (aio operon) and AsBT-Guild 12 (arr operon) are enriched in environments that match their oxidation reduction potential needs. Genomes of AsBT-Guild 11 are enriched in aerobic environments while genomes of AsBT-Guild 12 are enriched in anaerobic environments. Of the sub-guilds of AsBT-Guild 7 (long ars operon), three are enriched in human microbiomes (AsBT-SubGuilds 8, 12, and 16) while the fourth (AsBT-SubGuild 17) is enriched in food products.
Building on the strong relationship found between the metadata and arsenic metabolism we created a random forest classifier model that is able to predict the AsBT-Guild a bacterium belongs to, based on its metadata (Phyla and Class for taxonomy. Ecosystem, climate, and environment type for environmental information). The model has an overall accuracy > 75% which is also consistent for most phyla and environment types, the two most important parameters contributing to the model. Even though the model's accuracy is high, the data are still very noisy and improvements of metadata curation would further improve the model. The taxonomic assignment was relatively robust, as it was based on a well established method [13]. Still, the genomes themselves were not filtered for genome completeness so we were not able to assign taxonomy to the entire set of genomes we started with. Assignment of environmental information was even more subjective. While the curation of metagenomic samples from sites sampled by our group and collaborators is robust, NCBI-derived genomes are both lacking in information and the information present is at times misleading or incorrect. That said, we believe the predictive model would be very valuable to groups and organizations that are unable to conduct in-depth genomic analyses. Taxonomy can easily be derived from 16S rRNA sequencing, while the environmental information would be known to the people generating the data.
An important focus of this work was the analysis of arsenic exposure in human microbiomes. Our analysis shows that arsenic exposure changes the human microbiome, enriching for specific metabolic types. Grouping genomes by their location and clustering the locations by AsBT-SubGuild abundance, an “arsenic exposed” cluster was identified. The cluster contained genomes sampled from humans known to be exposed to arsenic in Laksam Upazila [16] that were used as an indicator of the cluster of interest. Examination of the other location groups in the cluster showed several known hotspots for arsenic contamination, and more from increased exposure (04.-0.6 probability of As > 100 ppb) [5]. Most of the genomes in the “arsenic-exposed” cluster are from the gut microbiome but another prominent source was the urinary system. Previous research on arsenic exposed humans in Mexico showed elevated MMAs in their urine [26]. The AsBT-SubGuild 19, characterized by the presence of the arsP efflux pump, was most prominent in the “arsenic-exposed” cluster. Another unique aspect of this sub guild is that it is the only one within AsBT-Guild 5 that has aioR at high proportions (>50% compared to a mean of 4.6% in the other sub guilds). The arsP efflux pump excretes trivalent MMAs from microbial cells [6], which are more toxic to humans than As(III) [17]. The microbial driven increase in methylated arsenic was experimentally shown in previous research [27], [28], while other work has shown increased methylated arsenic in urine of exposed individuals [29]. By increasing the host exposure to trivalent MMAs, the bacteria further exacerbate the detrimental effect of arsenic exposure to their hosts. AsBT-Guild 6 of human associated Actinobacteria was the most reduced in the “arsenic-exposed” cluster. This group contains both arsH that can oxidize trivalent MMAs into lesser toxic pentavalent MMAs [25], as well as arsI that can demethylate MMAs.
Our work shows the importance of analyzing the full spectrum of genes related to arsenic metabolism as well as utilizing co-localization data to support gene annotation. While sequence based analysis cannot provide proof of functionality, the consistent patterns across thousands of genomes lends support to the potential functions. We were able to identify microbial guilds with unique metabolic profiles and linked the aio two component regulation system to the ars operon. Arsenic methylation was revealed to be significant both in soil environments as well as in the human microbiome. Our predictive model can be used to further identify with high accuracy the metabolic potential of bacteria in most environments and could support decision making and improve monitoring of the potential for arsenic exposure around the world.
4. Abbreviations
GAsDb - The Genomic database for arsenic biotransformation. This includes all the genes found by the HMMs, as well as information (taxonomic and environmental) about the genomes from which they were found.
AsPRO - Unique arsenic gene profile in genomes. Each AsPRO represents all the genomes that contain the same genes (including gene frequency).
AsBT-Guild - Arsenic biotransformation guild. A cluster of AsPRO that shares a representative set of genes that can be translated into a function.
AsBT-SubGuild - a subset of AsBT-Guild derived from local clustering of the AsPRO.
Methods
4.1. Creating the genomic database
Sequence data was downloaded from NCBI Reference Sequence Database (89,253 genome assemblies) and the Banfield lab database (3,512 binning projects) were downloaded in 2018 . In both cases, open reading frames (ORFs) were predicted by Prodigal [30] with amino acid sequences as the output. For the Banfield lab database an additional filtering step to remove unbinned scaffolds was needed before gene prediction. Due to the size and complexity of the data, it was not possible to access the quality of the metagenome-assembled genomes (MAGs). To compensate for that, we applied strict filtering parameters (see following sections) and removed data we were less confident about.
Genome taxonomy was assigned using GTDB-Tk [13], followed by name and taxonomic hierarchy corrections to comply with the generally accepted knowledge.
Environmental metadata information for the Refseq genomes was parsed from the genomes genbank file, and for the Banfield lab genomes information was known for each binning project. For parameters were chosen: Ecosystem, Environment type, geography location, Climate. The information was curated manually to the best of our capability, recognising that user input may be fuzzy or mislabeled. Ecosystem of origin included: terrestrial near-surface, marine, deep subsurface, engineered and host-associated. Environment type was a more refined parameter including 101 categories. The environmental and engineered ecosystems shared most of the environment type categories (e.g. sediment, freshwater) with a few unique categories per ecosystem (e.g. hydrothermal vent in the marine ecosystem or food product in the engineered ecosystem). The environment type categories for the host-associated ecosystem were based on the host identity (e.g human, sponge). Geographic locations (432 locations) were either country or state/province for the largest countries (e.g. USA, China, Russia), as well as countries with a long north–south axis (e.g. Chile, Argentina). For climate we used the Köppen climate classification (retrieved from https://en.climate-data.org/) for terrestrial locations and a more general climate description (i.e. polar, temperate, tropical) for marine locations. When a location contained multiple climate regions the most common climate was chosen.
4.2. Designing Hidden Markov Model profiles for arsenic related genes
The initial seed sequences for the HMMs were either taken from TIGRFAM (arsA, arsB, ACR3, arsC Trx type, arsC low molecular weight type, arsH, aioA, aioB) or from literature describing confirmed enzyme function (et al. 2017 [6] and references within). To each gene, additional putative seed sequences were added based on pBLAST searches [31] targeting varying phyla to increase the sequence diversity of the seed sequences.
To verify the monophyletic clustering of the seed sequences a reference set of sequences was used. Using the NCBI Conserved Domain Database [32], sequences of each gene family were downloaded. Protein sequences were align using MAFFT [33], followed by tree construction with FastTree [34] and visualized in iTOL [35].
After clustering verification, the seed sequences were used to build an HMM with HMMER [36]. Threshold scores were assigned by searching the HMMs back against the reference sequence set and locating the highest score of a none-seed sequence. The threshold scores were reevaluated again by randomly subsetting 2000 hits from the Refseq HMM search outputs four times and checking scores against the seeds.
We were unsuccessful in creating a robust HMM for arsR so this gene was not included in the study.
4.3. Filtering hits and identifying loci
Initial data filtering was done in R using the Rstudio integrated development environment [37], [38]. Output tables from the HMM searches were loaded into Rstudio and merged into a single data table. Sequences with multiple HMMs hits were located and the best match was chosen based on the HMM score. Sequences were then filtered by their size, allowing a range between 50% and 200% of the mean length of the seed sequences. Thresholds were applied to indicate if a sequence was reliable or unreliable (but sequences were not filtered out).
Monophyletic clustering of sequence verification was conducted on the sequences as a final filtering step. To reduce computational requirements, the sequences were first clustered to representative sequences using MMseqs2 [39]. The representatives were then aligned with the seed and reference sequences and built a maximum-likelihood tree using IQ-Tree [40]. Representative clustering with the seeds were used to subset the hit database to include only verified hits.
As loci were identified based on co-localization of genes on scaffolds. This was done by parsing the scaffold feature numbers output and locating sequential ORFs. While initially a gap of up to five features was allowed, the mean distance between ORFs was 1.17 (n = 386334, sd = 0.44). Following the identification of loci, a primary putative function was assigned to each of them, based on gene presence. An hierarchical assignment of primary function priorities oxidoreduction transformations (arsC types, aioA, arrA, arxA), followed by methyl transformations (arsM, arsI, arsH), transport (arsB, ACR3, arsJ, arsP), regulation (aioR, aioS, aioX, arrS), and auxiliary genes (all else). Function based filtering included removing all loci with auxiliary primary function and keeping regulatory loci that contained > 50% reliable hits.
4.4. Modeling genomes with unsupervised and supervised methods
Unsupervised clustering was done using Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) [41] combined with Hierarchical Density-Based Spatial Clustering of Applications with Noise (HDBSCAN) [42], [43]. These methods were applied to cluster the arsenic biotransformation guilds and subguilds (AsBT-Guild and AsBT-SubGuild respectively) as well as the human microbiome by location clustering. For AsBT-Guild/AsBT-SubGuild clustering the input table consisted of gene counts for each unique genomic profile (AsPRO). The human associated microbiome clustering input was AsBT-Guild counts for each group, based on location and bodily source. Data was Z-transformed prior to fit transforming with UMAP. For global structure clustering UMAP was run with n_neighbors = 120, and HDBSCAN was run with min_cluster_size = 100, and no value set for min_samples. For local structure clustering UMAP was run with n_neighbors = 30, and HDBSCAN was run with min_cluster_size = 100 and min_samples = 10. The first iteration of AsBT-Guild/AsBT-SubGuild clustering included all AsPROs. The largest resulting cluster did not have any defined metabolism and further investigation showed that the main driver defining the cluster was a low number of gene types in the AsPROs, primarily AsPROs with a single gene. To achieve better cluster metabolic resolution, the single genes AsPROs were removed and the clustering was redone using the same parameters. The removal of the single gene AsPROs did not affect the global structure and within the subclusters of the local structure split the large AsBT-Guild into subclusters with resolved metabolism.
Supervised modeling of the relation between genome metadata and their assigned AsBT-Guild was done usingDistributed Random Forest with the H2O.ai machine learning software [44]. The software was selected since it enables the use of categorical predictor variables without the need for one-hot encoding, thus keeping the number of predictor variables low. Train and validation sets were split at 8:2 ratio. Fixed model parameters included balancing class distribution (balance_classes = True), and including all predictor columns at each level (mtries = -2). Additional parameters, selected by grid search were the method of histogram aggregation ('AUTO', 'Random', 'UniformAdaptive'), maximum tree depth (20,40,80), minimum number of observations for a leaf in order to split (1,10, 50), the number of bins to be included in the histogram (5, 50, 100, 500, 1000), and the number of trees to build in the model (50, 200). The best model from the grid search was selected by the mean per class accuracy index (mean_per_class_accuracy = 0.424). The resulting model had the following parameter: histogram_type = ‘UniformAdaptive’, max_depth = 20, min_rows = 1, nbins_cats = 100, ntrees = 200. Feature importance was calculated as follows: “H2O-3 looks at the squared error before and after the split using a particular variable. The difference is the improvement·H2O uses the improvement in squared error for each feature that was split on (rather than the accuracy). Each feature’s improvement is then summed up at the end to get its total feature importance (and then scaled between 0 and 1)” (https://docs.h2o.ai/h2o/latest-stable/h2o-docs/variable-importance.html).
5. Supplementary information
Supplementary information can be found in the below link: https://figshare.com/projects/Arsenic_genomics/117447.
CRediT authorship contribution statement
Ray Keren: Conceptualization, Methodology, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Raphaël Méheust: Methodology, Formal analysis, Data curation, Writing – review & editing. Joanne M. Santini: Resources, Data curation, Writing – review & editing. Alex Thomas: Resources, Writing – review & editing. Jacob West-Roberts: Methodology, Writing – review & editing. Jillian F. Banfield: Resources, Writing – original draft, Writing – review & editing, Supervision. Lisa Alvarez-Cohen: Resources, Writing – original draft, Writing – review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was funded by NIEHS Superfund Basic Research Program, R42 ES004705-19. The authors would like to thank all research groups that generate valuable sequence data with proper metadata information.
References
- 1.Oremland R.S., Stolz J.F. The ecology of arsenic. Science. 2003;300(5621):939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- 2.Slyemi D., Bonnefoy V. How prokaryotes deal with arsenic. Environ Microbiol Rep. 2012;4:571–586. doi: 10.1111/j.1758-2229.2011.00300.x. [DOI] [PubMed] [Google Scholar]
- 3.van Lis R., Nitschke W., Duval S., Schoepp-Cothenet B. Arsenics as bioenergetic substrates. Biochim Biophys Acta-Bioenerg. 2013;1827:176–188. doi: 10.1016/j.bbabio2012.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Neff J.M. Ecotoxicology of arsenic in the marine environment. Environ Toxicol Chem. 1997;16(5):917–927. doi: 10.1002/etc.5620160511. [DOI] [Google Scholar]
- 5.Podgorski J., Berg M. Global threat of arsenic in groundwater. Science. 2020;368(6493):845–850. doi: 10.1126/science.aba1510. [DOI] [PubMed] [Google Scholar]
- 6.Zhu Y.-G., Xue X.-M., Kappler A., Rosen B.P., Meharg A.A. Linking Genes to Microbial Biogeochemical Cycling: Lessons from Arsenic. Environ Sci Technol. 2017;51(13):7326–7339. doi: 10.1021/acs.est.7b00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan G.e., Chen X., Du S., Deng Z., Wang L., Chen S. Genetic mechanisms of arsenic detoxification and metabolism in bacteria. Curr Genet. 2019;65(2):329–338. doi: 10.1007/s00294-018-0894-9. [DOI] [PubMed] [Google Scholar]
- 8.Andres J., Bertin P.N., Danchin A. The microbial genomics of arsenic. Fems Microbiol Rev. 2016;40(2):299–322. doi: 10.1093/femsre/fuv050. [DOI] [PubMed] [Google Scholar]
- 9.Saunders J.K., Fuchsman C.A., McKay C., Rocap G. Complete arsenic-based respiratory cycle in the marine microbial communities of pelagic oxygen-deficient zones. Proc Natl Acad Sci. 2019;116(20):9925–9930. doi: 10.1073/pnas.1818349116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunivin T.K., Yeh S.Y., Shade A. A global survey of arsenic-related genes in soil microbiomes. BMC Biol. 2019;17:45. doi: 10.1186/s12915-019-0661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben Fekih I., Zhang C., Li Y.P., Zhao Y.i., Alwathnani H.A., Saquib Q., et al. Distribution of Arsenic Resistance Genes in Prokaryotes. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y.i., Su J.-Q., Ye J., Rensing C., Tardif S., Zhu Y.-G., et al. AsChip: A High-Throughput qPCR Chip for Comprehensive Profiling of Genes Linked to Microbial Cycling of Arsenic. Environ Sci Technol. 2019;53(2):798–807. doi: 10.1021/acs.est.8b0379810.1021/acs.est.8b03798.s00110.1021/acs.est.8b03798.s002. [DOI] [PubMed] [Google Scholar]
- 13.Chaumeil P.-A., Mussig A.J., Hugenholtz P., Parks D.H. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schloss P.D., Girard R.A., Martin T., Edwards J., Thrash J.C., Delong E.F., et al. Status of the Archaeal and Bacterial Census: an Update. MBio. 2016;7(3) doi: 10.1128/mBio.00201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yunus F., Khan S., Chowdhury P., Milton A., Hussain S., Rahman M. A Review of Groundwater Arsenic Contamination in Bangladesh: The Millennium Development Goal Era and Beyond. Int J Environ Res Public Health. 2016;13(2):215. doi: 10.3390/ijerph13020215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devoto A.E., Santini J.M., Olm M.R., Anantharaman K., Munk P., Tung J., et al. Megaphages infect Prevotella and variants are widespread in gut microbiomes. Nat Microbiol. 2019;4(4):693–700. doi: 10.1038/s41564-018-0338-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petrick J.S., Ayala-Fierro F., Cullen W.R., Carter D.E., Vasken Aposhian H. Monomethylarsonous Acid (MMAIII) Is More Toxic Than Arsenite in Chang Human Hepatocytes. Toxicol Appl Pharmacol. 2000;163(2):203–207. doi: 10.1006/taap.1999.8872. [DOI] [PubMed] [Google Scholar]
- 18.Santini J.M., Ward S.A., Ward S.A. Microbial arsenic response and metabolism in the genomics era. Metab Arsenite. 2018 doi: 10.1201/b12350-13. [DOI] [Google Scholar]
- 19.Sardiwal S., Santini J.M., Osborne T.H., Djordjevic S. Characterization of a two-component signal transduction system that controls arsenite oxidation in the chemolithoautotroph NT-26. FEMS Microbiol Lett. 2010;313:20–28. doi: 10.1111/j.1574-6968.2010.02121.x. [DOI] [PubMed] [Google Scholar]
- 20.Crits-Christoph A., Olm M.R., Diamond S., Bouma-Gregson K., Banfield J.F. Soil bacterial populations are shaped by recombination and gene-specific selection across a grassland meadow. ISME J. 2020;14(7):1834–1846. doi: 10.1038/s41396-020-0655-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharrar A.M., Crits-Christoph A., Méheust R., Diamond S., Starr E.P., Banfield J.F., et al. Bacterial Secondary Metabolite Biosynthetic Potential in Soil Varies with Phylum, Depth, and Vegetation Type. mBio. 2020;11(3) doi: 10.1128/mBio.00416-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017;15(10):579–590. doi: 10.1038/nrmicro.2017.87. [DOI] [PubMed] [Google Scholar]
- 23.Janssen P.H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S rRNA and 16S rRNA Genes. Appl Environ Microbiol. 2006;72(3):1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J., Rosen B.P. The Arsenic Methylation Cycle: How Microbial Communities Adapted Methylarsenicals for Use as Weapons in the Continuing War for Dominance. Front. Environ Sci. 2020;8 doi: 10.3389/fenvs.2020.00043. [DOI] [Google Scholar]
- 25.Chen J., Bhattacharjee H., Rosen B.P. ArsH is an organoarsenical oxidase that confers resistance to trivalent forms of the herbicide monosodium methylarsenate and the poultry growth promoter roxarsone. Mol Microbiol. 2015;96(5):1042–1052. doi: 10.1111/mmi.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urinary Trivalent Methylated Arsenic Species in a Population Chronically Exposed to Inorganic Arsenic | Environmental Health Perspectives | Vol. 113, No. 3 n.d. https://ehp.niehs.nih.gov/doi/full/10.1289/ehp.7519 (accessed May 23, 2021). [DOI] [PMC free article] [PubMed]
- 27.Van de Wiele T., Gallawa C.M., Kubachk K.M., Creed J.T., Basta N., Dayton E.A., et al. Arsenic Metabolism by Human Gut Microbiota upon in Vitro Digestion of Contaminated Soils. Environ Health Perspect. 2010;118(7):1004–1009. doi: 10.1289/ehp.0901794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu H., Wu B., Zhang X.-X., Liu S.u., Yu J., Cheng S., et al. Arsenic Metabolism and Toxicity Influenced by Ferric Iron in Simulated Gastrointestinal Tract and the Roles of Gut Microbiota. Environ Sci Technol. 2016;50(13):7189–7197. doi: 10.1021/acs.est.6b0153310.1021/acs.est.6b01533.s001. [DOI] [PubMed] [Google Scholar]
- 29.Navarro Serrano I., Llorente Ballesteros M.T., Sánchez Fernández Pacheco S., Izquierdo Álvarez S., López Colón J.L. Total and speciated urinary arsenic levels in the Spanish population. Sci Total Environ. 2016;571:164–171. doi: 10.1016/j.scitotenv.2016.07.134. [DOI] [PubMed] [Google Scholar]
- 30.Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K., Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 34.Price M.N., Dehal P.S., Arkin A.P., Poon A.F.Y. FastTree 2 – Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43(W1):W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team Rs . RStudio, Inc.; Boston, MA: 2015. RStudio: Integrated Development for R. [Google Scholar]
- 38.R Development Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2012.
- 39.Mirdita M., Steinegger M., Söding J., Hancock J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics. 2019;35(16):2856–2858. doi: 10.1093/bioinformatics/bty1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. McInnes J. Healy N. Saul L. Großberger UMAP: Uniform Manifold Approximation and Projection JOSS 3 29 861 10.21105/joss 10.21105/joss.00861
- 42.Campello RJGB, Moulavi D, Sander J. Density-Based Clustering Based on Hierarchical Density Estimates. In: Pei J, Tseng VS, Cao L, Motoda H, Xu G, editors. Adv. Knowl. Discov. Data Min., Berlin, Heidelberg: Springer; 2013, p. 160–72. https://doi.org/10.1007/978-3-642-37456-2_14.
- 43.McInnes L, Healy J. Accelerated Hierarchical Density Clustering 2017. https://doi.org/10.1109/ICDMW.2017.12.
- 44.H2O.ai. Python Interface for H2O, Python module version 3.10.0.8. 2016.