Abstract
Objective
To investigate the bacterial distribution and antimicrobial resistance profile of clinical isolates from Gram-negative bacteria bloodstream infections (GNBSI) in China.
Methods
The clinical bacterial strains isolated from blood culture were collected during April 2019 to December 2021 in 21 member hospitals of China Bloodstream Gram-negative Pathogens Antimicrobial Resistance and Virulence Surveillance Network (CARVIS-NET). Antibiotic susceptibility test was conducted by broth microdilution method recommended by Clinical and Laboratory Standards Institute (CLSI, United States). WHONET 2021 and SPSS 22.0 were used to analyze data.
Results
During the study period, 1939 Gram-negative bacteria were collected from 21 hospitals, among which 1,724 (88.9%) were Enterobacteriaceae, 207 (10.7%) were non-fermenting Gram-negative bacteria and 8 (0.4%) were others. The top five bacterial species were Escherichia coli (46.2%), Klebsiella pneumoniae (31.6%), Pseudomonas aeruginosa (4.9%), Acinetobacter baumannii (4.2%) and Enterobacter cloacae (3.0%). For K. pneumoniae, antibiotic resistance was mainly prevalent in hospital-associated bloodstream infections, while for A. baumannii, antibiotic resistance was mainly prevalent in community-associated bloodstream infections. It is worth mentioning that 94.1% of the 1939 Gram-negative isolates were susceptible to polymyxin B. The sensitivity of the strains involved in our investigation to polymyxin B is highly correlated with their sensitivity to colistin.
Conclusion
The surveillance results in CARVIS-NET-2021 showed that the main pathogens of GNBSI in China were Enterobacteriaceae, while E. coli was the most common pathogen. The resistance rates of K. pneumonia, P. aeruginosa, A. baumannii, and E. cloacae to multiple antibiotics kept on a high level. In many cases, polymyxin B and colistin has become the last-resort agents to combat bloodstream infections caused by multidrug-resistant (MDR) Gram-negative bacteria.
Keywords: Gram-negative bacteria, drug resistance, bloodstream infection, resistance surveillance, polymyxin B
Introduction
Good surveillance programs are the key to combat antibiotic resistance, including preservation and transportation of strains, determination of antibiotic susceptibility rates and detection of the expression of resistance genes that spread through different populations(Critchley and Karlowsky, 2004). Many current surveillance programs have problems to be solved such as obvious duplicate clones, lacking of clinical information or incorrect identification of colonizing and infecting strains. Therefore, in order to ensure the quality of monitoring, it is necessary to designate a center laboratory for quality confirmation, and to analyze the collected data after making sure the collected pathogens are reliable (Critchley and Karlowsky, 2004; Johnson, 2015).
Gram-negative bacteria bloodstream infection (GNBSI) has been a global public health problem (Huh et al., 2020; Nutman et al., 2021). Drug resistance in Gram-negative bacteria, including those that cause bloodstream infections, has increased dramatically over the past 20 years (Ergonul et al., 2016; Huh et al., 2020). Currently, the pathogenic spectrum and the resistance characteristics of bloodstream infection in China are changing, and they are not consistent in different regions. Therefore, this topic deserves further study (Liu et al., 2018; Liang et al., 2021).
Gram-negative pathogens that produce extended-spectrum β-lactamases (ESBLs) are now common and are highly associated with nonstandard drug administration and high mortality. In addition, carbapenem resistance is also becoming increasingly severe (Rodriguez-Bano et al., 2018; Tamma et al., 2021). These objective conditions have limited clinical medication decision, so a better understanding of local epidemiology might be helpful to optimize the therapeutic regimen (Critchley and Karlowsky, 2004). It is a pity that the development of antimicrobials has not kept pace with the increasing spread of resistance genes, especially for Gram-negative bacteria. Therefore, it is critical to make better use of existing antibiotics and strict infection control scheme to prevent these life-threatening infections (Collignon and McEwen, 2019).
Polymyxins are a group of polycationic antimicrobial lipopeptides biosynthesized by bacteria belonging to the Genus Bacillus. Polymyxin B and polymyxin E (also known as colistin) are the major representatives of polymyxins which have been used in clinical practice since the 1950s (El-Sayed Ahmed et al., 2020). Subsequently, the emergence of significant side-effects such as neurotoxicity, nephrotoxicity and anaphylactoid reaction limited their use (Zhan et al., 2019). Nevertheless, the increase of infections caused by MDR bacteria has led to a renewed clinical emphasis on polymyxins. Polymyxins have strong antibacterial activity and are considered to be the last therapeutic agent for infections caused by a variety of multidrug-resistant Gram-negative bacteria (Nang et al., 2021).
In view of the above, we designed this study to investigate the bacterial distribution and drug resistance of clinical isolates from GNBSI in China by implementing a well-designed surveillance plan, with a view to providing factual evidence for more rational use of antibiotics in clinic.
Materials and methods
Bacterial strains
Gram-negative bacteria were consecutively and non-repetitively collected from patients with clinical- and laboratory-confirmed bloodstream infection between 2019 and 2021 in 21 centers located in 20 Chinese cities. All organisms were considered clinically significant by local hospital criteria and were isolated from high-quality specimens of each patient’s first positive blood culture. In this study, community-associated bloodstream infection was defined as bloodstream infection that occurred in the community (e.g., outpatient, emergency-patient blood culture-positive cases) or within 48 h of hospitalization (positive blood culture reported within 48 h of hospitalization). While nosocomial-associated bloodstream infection was defined as bloodstream infection after 48 h of hospitalization (positive blood cultures sent after 48 h of hospitalization). All isolates were sent to the central clinical microbiology laboratory of Peking Union Medical College Hospital (PUMCH) for identification confirmation by MALDI-TOF MS (Vitek MS, biomérieux, France). The Human Research Ethics Committee of PUMCH approved this study (Ethics Approval Number: HS2755).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was conducted by broth microdilution method as per the CLSI recommendations. MICs were interpreted following the CLSI M100-S32 guidelines (CLSI, 2022) or European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2022) Breakpoint tables for interpretation of MICs and zone diameters Version 12.0. The breakpoint for E. coli, K. pneumoniae and E. cloacae of aztreonam/avibactam and sitafloxacin refers to the breakpoint of aztreonam and moxifloxacin, respectively. For P. aeruginosa, the breakpoint of aztreonam/avibactam, sitafloxacin and cefoperazone/sulbactam refers to the breakpoint of aztreonam, levofloxacin and cefoperazone, respectively. For A. baumannii, the breakpoint of ceftazidime/avibactam and sitafloxacin refers to the breakpoint of ceftazidime and levofloxacin, respectively. E. coli ATCC 25922, P. aeruginosa ATCC 27853 and K. pneumoniae ATCC 700603 were used as quality controls.
Statistical analyses
Statistical analysis was performed with WHONET 2021 and SPSS 22.0. MIC50 and MIC90 refer to the minimum inhibitory concentration required to inhibit the growth of 50 and 90% of the tested bacteria. The 95% confidence intervals were calculated using the adjusted Wald method. Comparison of rates was assessed using a chi-squared test. Binary logistic analysis was used for independent risk factor analysis. A value of p < 0.05 was considered statistically significant, and p < 0.001 was extremely significant.
Results
Demographic information and pathogen distribution of patients with Gram-negative bacteria bloodstream infection
A total of 1939 Gram-negative bacteria isolated from 1939 patients with clinical- and laboratory-confirmed bloodstream infection were involved in this study. 1,102 (56.8%) were males and 837 (43.2%) were females. Enterobacteriaceae was the main family of pathogens isolated (1724, 88.9%), with E. coli (896, 46.2%) and K. pneumoniae (612, 31.6%) being the top two most commonly isolated species. Non-fermenting Gram-negative bacteria comprised 10.7% of the observed pathogens, of which P. aeruginosa (94, 4.9%) and A. baumannii (82, 4.2%) were the most common (Supplementary Table S1). When the infected population was divided into three groups with the boundaries of 18 and 65 years old, it was found that E. coli and K. pneumoniae were the top two Gram-negative bacteria in all the three age groups. Meanwhile, the proportion of E. coli appeared to increase with age (Supplementary Table S1).
Age characteristics of patients with Gram-negative bacteria bloodstream infection
Analysis of the source of bloodstream infection showed that in the juveniles’ population, Gram-negative bacteria infection from respiratory tract contributed the most to bloodstream infection, accounting for 24.7% of all juveniles with bloodstream infection, followed by gastrointestinal infection (10.1%) and urinary tract infections (4.5%). For adults aged 19 to 65, respiratory tract infection (17.2%), urinary tract infections (12.9%) and abdominal infection of organs except for liver and gallbladder (9.5%) were the top three sources of bloodstream infections, respectively. Yet for the elderly over 65 years old, they are urinary tract infection (16.8%), respiratory tract infection (16.3%) and biliary tract infection (15.2%) in order (Figure 1). With increasing age, the proportion of primary and bloodstream Gram-negative bacteria infections that came from the community rather than the hospital increased, and so did the mortality rate (p < 0.001; Supplementary Table S2).
Figure 1.
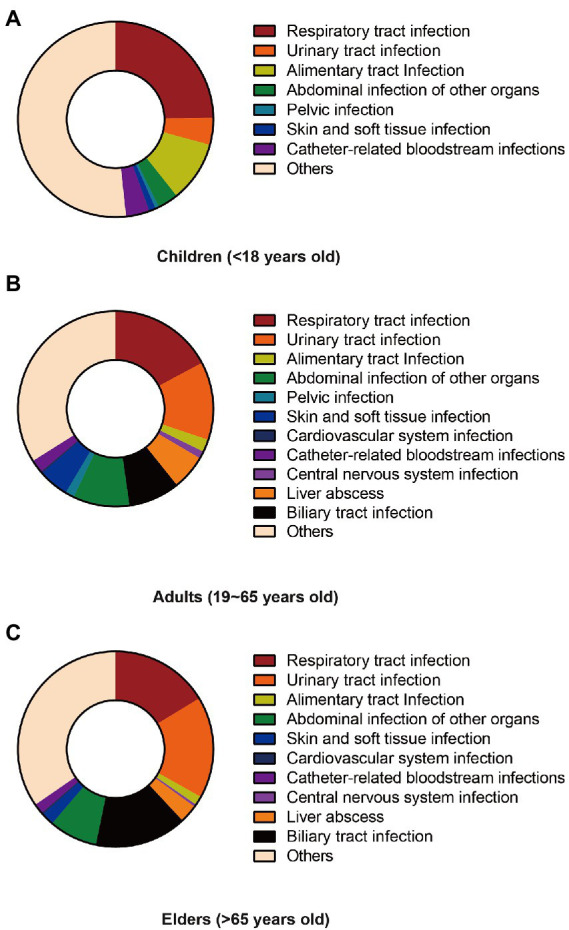
Difference infection sources of different age groups. (A) Composition of sources of infection among children under 18. (B) Composition of sources of infection among adults aged 19–65. (C) The composition of the source of infection in the elderly over 65 years old.
Susceptibility of Gram-negative bacteria to antibiotics and their clinical features
Escherichia coli
As is shown in Supplementary Table S3, majority of the 896 strains of E. coli had high susceptibility to polymyxin B, colistin, ceftazidime/avibactam, aztreonam/avibactam, amikacin, meropenem, imipenem, piperacillin/tazobactam, ertapenem, cefoxitin and cefoperazone/sulbactam. Nevertheless, ceftazidime, cefepime, sitafloxacin, levofloxacin, ceftriaxone and trimethoprim/sulfamethoxazole showed poor performance in anti-E. coli in bloodstream infection, with resistance rates of 23.2, 32.5, 50.9, 52.1, 57.8 and 65.3%, respectively.
Further analysis indicated that E. coli isolated from patients over 65 years old were more susceptible to trimethoprim/sulfamethoxazole (p = 0.003). Patients whose primary or bloodstream infection associated with medical institutions had lower frequency of susceptibility to trimethoprim/sulfamethoxazole (p < 0.01), while patients whose bloodstream infection occurred after 48 h of hospitalization had lower frequency of susceptibility to ceftazidime as well (p = 0.046). Among patients admitted to intensive care unit (ICU), the frequency of drug resistance to ertapenem, ceftriaxone, cefepime and ceftazidime was significantly higher than that of other patients. Curiously, patients who were more susceptible to trimethoprim/sulfamethoxazole appeared to have a higher mortality rate (p = 0.047; Table 1). Further binary logistic analysis found that age greater than 65 years, ICU admission, and agranulosis were independent risk factors for death in patients with E. coli bloodstream infection (Figure 2A).
Table 1.
Comparison of the antimicrobial susceptibility rates of Escherichia coli isolates in clinical features.
| Antibiotic | %S | MIC50 | MIC90 | Gender (%) | Age (%) | Primary infection(%) | Bloodstream infection(%) | ICU admission(%) | Clinical outcome(%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female N = 442 |
Male N = 454 |
p-Value | ≤ 65 N = 506 | > 65 N = 385 | p-Value | CO N = 497 | HA N = 348 | p-Value | CO N = 504 | HA N = 390 | p-Value | Yes N = 127 | No N = 729 | p-Value | Death N = 36 |
Others N = 834 |
p-Value | ||||
| Polymyxin B | 99.0 | 0.25 | 0.5 | 98.6 | 99.3 | 0.335 | 98.8 | 99.2 | 0.739 | 99.4 | 98.6 | 0.285 | 99.2 | 98.7 | 0.698 | 98.8 | 99.0 | 1.000 | 97.2 | 99.0 | 0.318 |
| Colistin | 98.5 | 0.25 | 0.5 | 98.2 | 98.9 | 0.375 | 98.0 | 99.2 | 0.140 | 98.8 | 98.3 | 0.742 | 98.8 | 98.2 | 0.454 | 98.2 | 98.6 | 0.956 | 97.2 | 98.7 | 0.400 |
| Ceftazidime/avibactam | 99.8 | 0.064 | 0.064 | 99.8 | 99.8 | 1.000 | 99.8 | 99.7 | 1.000 | 99.8 | 99.7 | 1.000 | 100.0 | 99.5 | 0.190 | 100.0 | 99.7 | 1.000 | 100.0 | 99.8 | 1.000 |
| Aztreonam/Avibactam | 99.7 | 0.064 | 0.064 | 99.5 | 99.8 | 0.620 | 99.4 | 100.0 | 0.263 | 99.8 | 99.4 | 0.572 | 100.0 | 99.2 | 0.083 | 98.8 | 99.9 | 0.091 | 100.0 | 99.6 | 1.000 |
| Amikacin | 98.0 | 2 | 4 | 98.0 | 98.0 | 0.954 | 97.6 | 98.4 | 0.393 | 98.4 | 98.0 | 0.663 | 98.0 | 97.9 | 0.943 | 96.4 | 98.4 | 0.190 | 100.0 | 98.1 | 1.000 |
| Meropenem | 96.9 | 0.064 | 0.064 | 97.1 | 96.7 | 0.755 | 96.4 | 97.7 | 0.293 | 97.0 | 96.8 | 0.906 | 97.4 | 96.2 | 0.281 | 97.0 | 96.8 | 0.914 | 100.0 | 96.9 | 0.565 |
| Imipenem | 96.5 | 0.25 | 0.5 | 96.6 | 96.5 | 0.915 | 96.4 | 97.1 | 0.560 | 97.0 | 96.0 | 0.430 | 97.4 | 95.4 | 0.099 | 97.0 | 96.4 | 0.715 | 100.0 | 96.5 | 0.507 |
| Piperacillin/Tazobactam | 94.8 | 8 | 8 | 95.7 | 93.8 | 0.210 | 93.9 | 96.1 | 0.136 | 95.2 | 94.3 | 0.554 | 95.6 | 93.6 | 0.174 | 95.8 | 94.5 | 0.498 | 100.0 | 94.6 | 0.295 |
| Ertapenem | 92.7 | 0.064 | 0.25 | 93.9 | 91.6 | 0.192 | 92.1 | 93.8 | 0.338 | 92.8 | 93.1 | 0.847 | 92.9 | 92.6 | 0.867 | 88.0 | 93.8 | 0.009* | 100.0 | 92.4 | 0.166 |
| Cefoxitin | 86.6 | 2 | 16 | 88.0 | 85.2 | 0.224 | 86.2 | 87.5 | 0.551 | 87.7 | 85.6 | 0.375 | 87.9 | 84.9 | 0.188 | 89.2 | 86.0 | 0.271 | 91.7 | 86.2 | 0.492 |
| Cefoperazone/Sulbactam | 85.0 | 8 | 32 | 86.2 | 83.9 | 0.339 | 83.8 | 86.8 | 0.220 | 85.7 | 83.9 | 0.470 | 85.9 | 83.8 | 0.391 | 80.8 | 86.0 | 0.091 | 86.1 | 85.3 | 0.887 |
| Ceftazidime | 69.0 | 1 | 64 | 71.3 | 66.7 | 0.143 | 67.4 | 71.2 | 0.227 | 71.8 | 66.1 | 0.075 | 71.6 | 65.4 | 0.046* | 59.3 | 71.2 | 0.003* | 80.6 | 68.3 | 0.121 |
| Cefepime | 53.5 | 2 | 128 | 54.1 | 52.9 | 0.717 | 52.0 | 55.6 | 0.285 | 55.3 | 52.3 | 0.384 | 55.0 | 51.3 | 0.274 | 44.3 | 55.6 | 0.009* | 58.3 | 53.2 | 0.548 |
| Sitafloxacin | 49.1 | 0.5 | 2 | 49.3 | 48.9 | 0.899 | 46.8 | 52.2 | 0.112 | 50.3 | 49.7 | 0.866 | 48.8 | 49.5 | 0.841 | 45.5 | 49.9 | 0.302 | 38.9 | 49.5 | 0.212 |
| Levofloxacin | 42.6 | 2 | 16 | 45.0 | 40.3 | 0.154 | 40.1 | 45.7 | 0.094 | 43.9 | 44.0 | 0.976 | 42.1 | 43.3 | 0.703 | 41.3 | 42.9 | 0.703 | 36.1 | 42.8 | 0.426 |
| Ceftriaxone | 42.2 | 64 | 256 | 42.8 | 41.6 | 0.732 | 40.3 | 44.9 | 0.167 | 44.1 | 42.2 | 0.599 | 43.8 | 40.0 | 0.248 | 35.3 | 43.8 | 0.047* | 44.4 | 42.0 | 0.768 |
| Trimethoprim/sulfamethoxazole | 34.7 | 8 | 8 | 34.4 | 35.0 | 0.842 | 30.4 | 40.0 | 0.003* | 38.8 | 29.0 | 0.003* | 39.3 | 28.7 | 0.001* | 34.1 | 34.8 | 0.862 | 50.0 | 33.9 | 0.047* |
p < 0.05.
Figure 2.
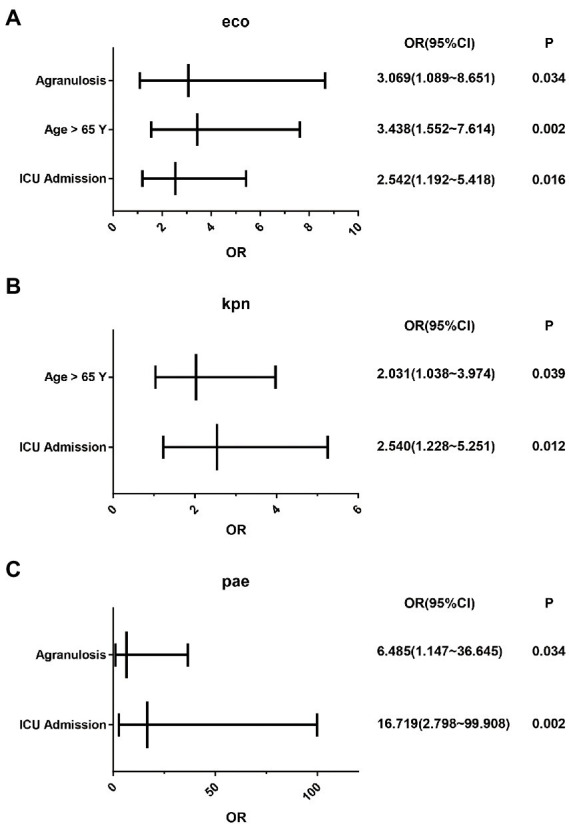
Analysis of independent risk factors for death in patients with Gram-negative bacteria bloodstream infection. (A) Independent risk factors for death in patients with bloodstream infection with E. coli. (B) Independent risk factors for death in patients with bloodstream infection with K. pneumoniae. (C) Independent risk factors for death in patients with bloodstream infection with P. aeruginosa.
Klebsiella pneumoniae
In the cases of K. pneumoniae, only polymyxin B, colistin, aztreonam/avibactam and ceftazidime/avibactam had sensitivity rates greater than 80%. Conversely, amikacin, meropenem, imipenem, ertapenem, piperacillin/tazobactam, cefoperazone/sulbactam, cefoxitin, sitafloxacin, ceftazidime, levofloxacin, cefepime, ceftriaxone and trimethoprim/sulfamethoxazole showed poor performance in anti-K. pneumoniae bloodstream infection (Supplementary Table S4).
Further analysis indicated that K. pneumoniae isolated from patients over 65 years old were more susceptible to meropenem (p = 0.039) and imipenem (p = 0.022). For antibiotics except for aztreonam/avibactam, ceftazidime/avibactam, polymyxin B and colistin, isolates derived from patients whose infection occurred after 48 h of hospitalization had lower frequency of susceptibility to the antibiotics with poor performance (p < 0.05). Among patients admitted to ICU, the frequency of drug resistance to sitafloxacin, amikacin, ertapenem, meropenem, piperacillin/tazobactam, cefepime, cefoperazone/sulbactam, ceftriaxone, ceftazidime, cefoxitin, imipenem and levofloxacin was significantly higher than that of other patients (p < 0.05). The mortality of these patients was higher than others as well (p < 0.001; Table 2). Binary logistic analysis found that age greater than 65 years and ICU admission were independent risk factors for death in patients with K. pneumoniae bloodstream infection (Figure 2B).
Table 2.
Comparison of the antimicrobial susceptibility rates of K. pneumoniae isolates in clinical features.
| Antibiotic | %S | MIC50 | MIC90 | Gender (%) | Age (%) | Primary infection(%) | Bloodstream infection(%) | ICU admission(%) | Clinical outcome(%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female N = 219 |
Male N = 393 |
p-value | ≤ 65 N = 401 | > 65 N = 200 | p-Value | CO N = 308 | HA N = 280 | p-Value | CO N = 292 | HA N = 317 | p-Value | Yes N = 196 |
No N = 416 |
p-Value | Death N = 55 |
Others N = 528 |
p-Value | ||||
| Polymyxin B | 97.1 | 0.5 | 0.5 | 96.3 | 97.5 | 0.437 | 97.3 | 96.4 | 0.575 | 97.7 | 97.9 | 0.915 | 96.9 | 97.2 | 0.860 | 96.9 | 97.1 | 0.904 | 94.5 | 97.2 | 0.236 |
| Colistin | 97.1 | 0.25 | 0.5 | 96.3 | 97.5 | 0.437 | 97.3 | 96.4 | 0.575 | 97.7 | 97.9 | 0.915 | 96.9 | 97.2 | 0.860 | 96.9 | 97.1 | 0.904 | 94.5 | 97.2 | 0.236 |
| Aztreonam/Avibactam | 99.8 | 0.064 | 0.064 | 100.0 | 99.7 | 1.000 | 100.0 | 99.5 | 0.722 | 100.0 | 99.6 | 0.962 | 100.0 | 99.7 | 1.000 | 99.5 | 100.0 | 0.700 | 98.2 | 100.0 | 0.165 |
| Ceftazidime/avibactam | 99.5 | 0.064 | 0.064 | 100.0 | 99.2 | 0.489 | 99.5 | 99.5 | 1.000 | 100.0 | 98.9 | 0.214 | 100.0 | 99.1 | 0.277 | 100.0 | 99.3 | 0.568 | 100.0 | 99.4 | 1.000 |
| Amikacin | 78.3 | 1 | 128 | 78.1 | 78.4 | 0.934 | 77.5 | 80.2 | 0.446 | 86.0 | 71.1 | 0.000* | 87.7 | 69.4 | 0.000* | 66.3 | 83.9 | 0.000* | 47.3 | 81.1 | 0.000* |
| Meropenem | 74.8 | 0.064 | 256 | 78.1 | 73.3 | 0.189 | 73.0 | 80.7 | 0.039* | 86.0 | 64.3 | 0.000* | 87.7 | 62.8 | 0.000* | 60.7 | 81.5 | 0.000* | 49.1 | 76.7 | 0.000* |
| Imipenem | 73.7 | 0.25 | 128 | 76.7 | 72.0 | 0.205 | 71.5 | 80.2 | 0.022* | 84.1 | 63.9 | 0.000* | 85.6 | 62.5 | 0.000* | 61.2 | 79.6 | 0.000* | 49.1 | 75.6 | 0.000* |
| Ertapenem | 71.4 | 0.064 | 256 | 74.0 | 70.0 | 0.294 | 70.0 | 76.1 | 0.118 | 82.1 | 61.1 | 0.000* | 83.9 | 59.6 | 0.000* | 57.7 | 77.9 | 0.000* | 45.5 | 73.3 | 0.000* |
| Piperacillin/Tazobactam | 71.4 | 8 | > 256 | 74.9 | 69.5 | 0.155 | 70.0 | 76.6 | 0.090 | 80.8 | 62.5 | 0.000* | 83.6 | 59.9 | 0.000* | 57.7 | 77.9 | 0.000* | 47.3 | 73.5 | 0.000* |
| Cefoperazone/Sulbactam | 67.2 | 8 | 256 | 65.3 | 68.2 | 0.465 | 66.6 | 70.1 | 0.394 | 77.9 | 56.8 | 0.000* | 80.1 | 54.9 | 0.000* | 51.5 | 74.5 | 0.000* | 36.4 | 69.3 | 0.000* |
| Cefoxitin | 64.7 | 2 | 128 | 64.8 | 64.6 | 0.959 | 63.6 | 69.0 | 0.190 | 75.6 | 55.4 | 0.000* | 76.7 | 53.9 | 0.000* | 53.1 | 70.2 | 0.000* | 34.5 | 67.2 | 0.000* |
| Sitafloxacin | 63.7 | 0.125 | 16 | 65.3 | 62.8 | 0.546 | 63.6 | 65.0 | 0.744 | 72.1 | 56.1 | 0.000* | 73.6 | 54.3 | 0.000* | 51.5 | 69.5 | 0.000* | 32.7 | 66.1 | 0.000* |
| Ceftazidime | 63.1 | 0.5 | 256 | 64.8 | 62.1 | 0.499 | 61.4 | 68.0 | 0.113 | 73.4 | 53.9 | 0.000* | 76.0 | 51.4 | 0.000* | 53.6 | 67.5 | 0.001* | 32.7 | 65.5 | 0.000* |
| Levofloxacin | 58.7 | 0.5 | 64 | 58.4 | 58.8 | 0.936 | 58.4 | 59.9 | 0.729 | 66.6 | 51.8 | 0.000* | 67.8 | 49.8 | 0.000* | 45.4 | 64.9 | 0.000* | 25.5 | 61.2 | 0.000* |
| Cefepime | 57.0 | 0.125 | 256 | 53.4 | 59.0 | 0.179 | 55.9 | 60.4 | 0.299 | 69.2 | 45.0 | 0.000* | 71.6 | 43.2 | 0.000* | 45.9 | 62.3 | 0.000* | 32.7 | 58.5 | 0.000* |
| Ceftriaxone | 52.9 | 0.25 | 256 | 48.4 | 55.5 | 0.093 | 52.0 | 55.8 | 0.374 | 66.6 | 39.6 | 0.000* | 68.8 | 38.5 | 0.000* | 43.9 | 57.2 | 0.002* | 25.5 | 54.7 | 0.000* |
| Trimethoprim/sulfamethoxazole | 52.0 | 2 | 8 | 48.4 | 53.9 | 0.188 | 52.2 | 51.3 | 0.825 | 60.1 | 42.5 | 0.000* | 62.0 | 42.3 | 0.000* | 49.0 | 53.4 | 0.311 | 40.0 | 52.3 | 0.083 |
p < 0.05.
Pseudomonas aeruginosa
Toward P. aeruginosa, polymyxin B, colistin, aztreonam/avibactam, ceftazidime/avibactam, sitafloxacin and amikacin performed well. All of the other antibiotics involved in this investigation had poor sensitivities, among which there were naturally resistant ones (Supplementary Table S5). Patients primary infected in community had significantly higher frequency of susceptibility to Sitafloxacin (p = 0.022). Among patients admitted to ICU, the frequency of drug resistance to Levofloxacin was significantly higher than that of other patients (p = 0.024; Table 3). Binary logistic analysis found that agranulosis and ICU admission were independent risk factors for death in patients with P. aeruginosa bloodstream infection (Figure 2C).
Table 3.
Comparison of the antimicrobial susceptibility rates of P. aeruginosa isolates in clinical features.
| Antibiotic | %S | MIC50 | MIC90 | Gender (%) | Age (%) | Primary infection(%) | Bloodstream infection(%) | ICU admission(%) | Clinical outcome(%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female N = 31 |
Male N = 64 |
p-Value | ≤ 65 N = 66 | > 65 N = 28 | p-Value | CO N = 34 | HA N = 56 | p-Value | CO N = 33 | HA N = 62 | p-Value | Yes N = 32 | No N = 63 | p-Value | Death N = 13 |
Others N = 82 |
p-Value | ||||
| Polymyxin B | 98.9 | 1 | 1 | 100.0 | 98.4 | 1.000 | 98.5 | 100.0 | 1.000 | 100.0 | 98.2 | 1.000 | 100.0 | 98.4 | 1.000 | 100.0 | 98.4 | 1.000 | 100.0 | 98.8 | 1.000 |
| Colistin | 98.9 | 1 | 1 | 100.0 | 98.4 | 1.000 | 98.5 | 100.0 | 1.000 | 100.0 | 98.2 | 1.000 | 100.0 | 98.4 | 1.000 | 100.0 | 98.4 | 1.000 | 100.0 | 98.8 | 1.000 |
| Ceftazidime/avibactam | 96.8 | 1 | 4 | 100.0 | 95.3 | 0.548 | 95.5 | 100.0 | 0.552 | 97.1 | 98.2 | 1.000 | 97.0 | 96.8 | 1.000 | 100.0 | 95.2 | 0.526 | 100.0 | 96.3 | 1.000 |
| Amikacin | 94.7 | 2 | 8 | 96.8 | 93.8 | 0.897 | 95.5 | 92.9 | 0.991 | 100.0 | 92.9 | 0.286 | 100.0 | 91.9 | 0.233 | 93.8 | 95.2 | 1.000 | 92.3 | 95.1 | 0.529 |
| Sitafloxacin | 90.5 | 0.125 | 1 | 93.5 | 89.1 | 0.744 | 92.4 | 85.7 | 0.530 | 100.0 | 85.7 | 0.022* | 100.0 | 85.5 | 0.053 | 87.5 | 92.1 | 0.728 | 92.3 | 90.2 | 1.000 |
| Aztreonam/Avibactam | 85.3 | 4 | 16 | 83.9 | 85.9 | 1.000 | 84.8 | 85.7 | 1.000 | 94.1 | 83.9 | 0.272 | 90.9 | 82.3 | 0.407 | 81.3 | 87.3 | 0.631 | 76.9 | 86.6 | 0.623 |
| Cefepime | 81.1 | 2 | 32 | 77.4 | 82.8 | 0.529 | 80.3 | 82.1 | 0.836 | 79.4 | 83.9 | 0.587 | 75.8 | 83.9 | 0.337 | 84.4 | 79.4 | 0.556 | 92.3 | 79.3 | 0.463 |
| Ceftazidime | 81.1 | 2 | 64 | 77.4 | 82.8 | 0.529 | 80.3 | 82.1 | 0.836 | 79.4 | 82.1 | 0.748 | 78.8 | 82.3 | 0.681 | 81.3 | 81.0 | 0.972 | 100.0 | 78.0 | 0.135 |
| Piperacillin/Tazobactam | 76.8 | 8 | 128 | 77.4 | 76.6 | 0.926 | 80.3 | 67.9 | 0.192 | 76.5 | 76.8 | 0.973 | 78.8 | 75.8 | 0.743 | 78.1 | 76.2 | 0.833 | 92.3 | 74.4 | 0.285 |
| Meropenem | 74.7 | 0.5 | 64 | 77.4 | 73.4 | 0.675 | 75.8 | 71.4 | 0.660 | 88.2 | 71.4 | 0.063 | 84.8 | 69.4 | 0.098 | 75.0 | 74.6 | 0.966 | 69.2 | 75.6 | 0.882 |
| Levofloxacin | 73.7 | 0.5 | 8 | 71.0 | 75.0 | 0.676 | 78.8 | 64.3 | 0.140 | 82.4 | 71.4 | 0.242 | 78.8 | 71.0 | 0.410 | 59.4 | 81.0 | 0.024* | 61.5 | 75.6 | 0.465 |
| Imipenem | 67.4 | 2 | 32 | 71.0 | 65.6 | 0.603 | 68.2 | 64.3 | 0.713 | 73.5 | 67.9 | 0.569 | 69.7 | 66.1 | 0.724 | 65.6 | 68.3 | 0.796 | 69.2 | 67.1 | 1.000 |
p < 0.05.
Acinetobacter baumannii
For A. baumannii, only polymyxin B and colistin had susceptibility greater than 90%, whose susceptibility were both 96.3%. Ceftazidime/avibactam and sitafloxacin performed fairly well with susceptibility of 84.1 and 85.4%, respectively. Except for antibiotics which had no breakpoints to be referred yet, the drug resistance rate of all other antibiotics reached 60.0% and above (Supplementary Table S6). Different from E. coli and K. pneumoniae, patients infected with A. baumannii in the community had higher frequency of resistance to cefepime, imipenem and levofloxacin. For ICU patients, the frequency of drug resistance to amikacin, trimethoprim/sulfamethoxazole, meropenem, piperacillin/tazobactam, cefepime, ceftriaxone and levofloxacin was significantly higher than that of other patients (p < 0.05). Patients infected with strains resistant to trimethoprim/sulfamethoxazole and amikacin had higher mortality (p < 0.05; Table 4).
Table 4.
Comparison of the antimicrobial susceptibility rates of A. baumannii isolates in clinical features.
| Antibiotic | %S | MIC50 | MIC90 | Gender (%) | Age (%) | Primary infection(%) | Bloodstream infection(%) | ICU admission(%) | Clinical outcome(%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female N = 42 |
Male N = 40 |
p-Value | ≤ 65 N = 59 | > 65 N = 23 | p-Value | CO N = 29 | HA N = 53 | p-Value | CO N = 20 | HA N = 62 | p-Value | Yes N = 45 | No N = 37 | p-Value | Death N = 19 | Others N = 62 | p-Value | ||||
| Polymyxin B | 96.3 | 0.5 | 1 | 100.0 | 92.5 | 0.112 | 96.6 | 95.7 | 1.000 | 96.6 | 96.2 | 1.000 | 100.0 | 95.2 | 1.000 | 95.6 | 97.3 | 1.000 | 94.7 | 96.8 | 0.557 |
| Colistin | 96.3 | 0.5 | 1 | 100.0 | 92.5 | 0.112 | 96.6 | 95.7 | 1.000 | 96.6 | 96.2 | 1.000 | 100.0 | 95.2 | 1.000 | 95.6 | 97.3 | 1.000 | 94.7 | 96.8 | 0.557 |
| Sitafloxacin | 85.4 | 1 | 4 | 85.7 | 85.0 | 0.927 | 84.7 | 87.0 | 1.000 | 75.9 | 90.6 | 0.140 | 80.0 | 87.1 | 0.677 | 82.2 | 89.2 | 0.374 | 94.7 | 82.3 | 0.332 |
| Ceftazidime/avibactam | 84.1 | 4 | 16 | 81.0 | 87.5 | 0.417 | 83.1 | 87.0 | 0.922 | 79.3 | 86.8 | 0.568 | 75.0 | 87.1 | 0.349 | 86.7 | 81.1 | 0.491 | 89.5 | 82.3 | 0.695 |
| Amikacin | 35.4 | 128 | 128 | 31.0 | 40.0 | 0.392 | 37.3 | 30.4 | 0.560 | 27.6 | 39.6 | 0.276 | 30.0 | 37.1 | 0.564 | 24.4 | 48.6 | 0.023* | 15.8 | 40.3 | 0.049* |
| Trimethoprim/sulfamethoxazole | 28.0 | 8 | 8 | 26.2 | 30.0 | 0.701 | 32.2 | 17.4 | 0.180 | 13.8 | 35.8 | 0.034 | 15.0 | 32.3 | 0.135 | 17.8 | 40.5 | 0.022* | 5.3 | 35.5 | 0.011* |
| Imipenem | 28.0 | 64 | 128 | 28.6 | 27.5 | 0.914 | 32.2 | 17.4 | 0.180 | 13.8 | 35.8 | 0.034* | 15.0 | 32.3 | 0.135 | 20.0 | 37.8 | 0.074 | 15.8 | 32.3 | 0.164 |
| Ceftazidime | 25.6 | 64 | 256 | 21.4 | 30.0 | 0.374 | 28.8 | 17.4 | 0.287 | 13.8 | 32.1 | 0.070 | 20.0 | 27.4 | 0.509 | 17.8 | 35.1 | 0.073 | 15.8 | 29.0 | 0.394 |
| Meropenem | 24.4 | 64 | 128 | 23.8 | 25.0 | 0.900 | 27.1 | 17.4 | 0.357 | 13.8 | 30.2 | 0.098 | 15.0 | 27.4 | 0.409 | 13.3 | 37.8 | 0.010* | 5.3 | 30.6 | 0.052 |
| Piperacillin/Tazobactam | 23.2 | 256 | >256 | 23.8 | 22.5 | 0.888 | 27.1 | 13.0 | 0.175 | 13.8 | 28.3 | 0.137 | 15.0 | 25.8 | 0.489 | 13.3 | 35.1 | 0.020* | 5.3 | 29.0 | 0.067 |
| Levofloxacin | 23.2 | 8 | 32 | 21.4 | 25.0 | 0.702 | 25.4 | 17.4 | 0.439 | 10.3 | 30.2 | 0.042* | 15.0 | 25.8 | 0.489 | 11.1 | 37.8 | 0.004* | 10.5 | 27.4 | 0.226 |
| Cefepime | 19.5 | 64 | 256 | 20.0 | 19.0 | 0.913 | 22.0 | 13.0 | 0.540 | 6.9 | 26.4 | 0.033* | 10.0 | 22.6 | 0.363 | 8.9 | 32.4 | 0.007* | 5.3 | 24.2 | 0.138 |
| Ceftriaxone | 13.4 | 256 | 256 | 15.0 | 11.9 | 0.681 | 13.6 | 13.0 | 1.000 | 3.4 | 18.9 | 0.105 | 5.0 | 16.1 | 0.372 | 4.4 | 24.3 | 0.021* | 5.3 | 16.1 | 0.408 |
| Ertapenem | 1.2 | 256 | 256 | 0.0 | 2.5 | 0.488 | 0.0 | 4.3 | 0.280 | 0.0 | 1.9 | 1.000 | 0.0 | 1.6 | 1.000 | 0.0 | 2.7 | 0.451 | 0.0 | 1.6 | 1.000 |
p < 0.05.
Enterobacter cloacae
Enterobacter cloacae isolated in this study were highly susceptible to ceftazidime/avibactam, aztreonam/avibactam, amikacin and meropenem, while their susceptibility to ertapenem, piperacillin/tazobactam and cefoperazone/sulbactam also exceeded 80%. However, for other antibiotics involved in this study, the drug resistance of E. cloacae in bloodstream infection could not be underestimated (Supplementary Table S7). It was found that patients over 65 years old were more susceptible to trimethoprim/sulfamethoxazole (p = 0.035) and cefepime (p = 0.036; Table 5). Notably, patients who were resistant to cefoperazone/sulbactam had a higher mortality rate (p = 0.027; Table 5).
Table 5.
Comparison of the antimicrobial susceptibility rates of E. cloacae isolates in clinical features.
| Antibiotic | %S | MIC50 | MIC90 | Gender (%) | Age (%) | Primary infection(%) | Bloodstream infection(%) | ICU admission(%) | Clinical outcome(%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Female N = 20 |
Male N = 38 |
p-Value | ≤ 65 N = 37 | > 65 N = 21 | p-Value | CO N = 18 | HA N = 38 | p-Value | CO N = 16 | HA N = 42 | p-Value | Yes N = 36 | No N = 22 | p-Value | Death N = 5 |
Others N = 50 |
p-Value | ||||
| Polymyxin B | 81.0 | 0.5 | 128 | 70.0 | 86.8 | 0.229 | 81.1 | 81.0 | 1.000 | 72.2 | 86.8 | 0.337 | 75.0 | 83.3 | 0.727 | 77.3 | 83.3 | 0.821 | 80.0 | 80.0 | 1.000 |
| Colistin | 79.3 | 0.25 | 128 | 70.0 | 84.2 | 0.353 | 81.1 | 76.2 | 0.917 | 70.2 | 84.2 | 0.487 | 75.0 | 81.0 | 0.891 | 72.7 | 83.3 | 0.526 | 60.0 | 80.0 | 0.298 |
| Ceftazidime/avibactam | 100.0 | 0.064 | 0.064 | 100.0 | 100.0 | - | 100.0 | 100.0 | - | 100.0 | 100.0 | - | 100.0 | 100.0 | - | 100.0 | 100.0 | - | 100.0 | 100.0 | - |
| Aztreonam/Avibactam | 98.3 | 0.064 | 0.125 | 100.0 | 97.4 | 1.000 | 97.3 | 100.0 | 1.000 | 100.0 | 100.0 | - | 100.0 | 97.6 | 1.000 | 100.0 | 97.2 | 1.000 | 100.0 | 98.0 | 1.000 |
| Amikacin | 96.6 | 1 | 4 | 95.0 | 97.4 | 1.000 | 94.6 | 100.0 | 0.530 | 94.4 | 100.0 | 0.321 | 93.8 | 97.6 | 0.479 | 100.0 | 94.4 | 0.521 | 100.0 | 96.0 | 1.000 |
| Meropenem | 91.4 | 0.064 | 0.25 | 85.0 | 94.7 | 0.328 | 86.5 | 100.0 | 0.148 | 88.9 | 94.7 | 0.587 | 87.5 | 92.9 | 0.609 | 90.9 | 91.7 | 1.000 | 100.0 | 90.0 | 1.000 |
| Ertapenem | 84.5 | 0.125 | 2 | 80.0 | 86.8 | 0.762 | 78.4 | 95.2 | 0.184 | 83.3 | 86.8 | 1.000 | 81.3 | 85.7 | 0.989 | 86.4 | 83.3 | 1.000 | 100.0 | 82.0 | 0.578 |
| Piperacillin/Tazobactam | 82.8 | 8 | 32 | 80.0 | 84.2 | 0.970 | 78.4 | 90.5 | 0.418 | 77.8 | 86.8 | 0.636 | 75.0 | 85.7 | 0.564 | 81.8 | 83.3 | 1.000 | 60.0 | 86.0 | 0.184 |
| Cefoperazone/Sulbactam | 82.8 | 8 | 32 | 80.0 | 84.2 | 0.970 | 78.4 | 90.5 | 0.418 | 77.8 | 84.2 | 0.831 | 75.0 | 85.7 | 0.564 | 72.7 | 88.9 | 0.221 | 40.0 | 88.0 | 0.027* |
| Imipenem | 79.3 | 0.5 | 4 | 65.0 | 86.8 | 0.107 | 75.7 | 85.7 | 0.569 | 72.2 | 84.2 | 0.487 | 75.0 | 81.0 | 0.891 | 81.8 | 77.8 | 0.972 | 100.0 | 78.0 | 0.571 |
| Sitafloxacin | 77.6 | 0.012 | 2 | 80.0 | 76.3 | 1.000 | 78.4 | 76.2 | 1.000 | 72.2 | 81.6 | 0.654 | 75.0 | 78.6 | 1.000 | 77.3 | 77.8 | 1.000 | 80.0 | 76.0 | 1.000 |
| Cefepime | 77.6 | 0.064 | 128 | 70.0 | 81.6 | 0.500 | 67.6 | 95.2 | 0.036* | 72.2 | 81.6 | 0.654 | 68.8 | 81.0 | 0.520 | 77.3 | 77.8 | 1.000 | 100.0 | 74.0 | 0.324 |
| Levofloxacin | 72.4 | 0.064 | 8 | 70.0 | 73.7 | 0.765 | 70.3 | 76.2 | 0.628 | 61.1 | 78.9 | 0.278 | 68.8 | 73.8 | 0.955 | 77.3 | 69.4 | 0.517 | 80.0 | 70.0 | 1.000 |
| Ceftazidime | 65.5 | 0.5 | 128 | 60.0 | 68.4 | 0.521 | 59.5 | 76.2 | 0.198 | 61.1 | 68.4 | 0.589 | 56.3 | 69.0 | 0.359 | 59.1 | 69.4 | 0.421 | 40.0 | 68.0 | 0.327 |
| Ceftriaxone | 51.7 | 1 | 256 | 55.0 | 50.0 | 0.717 | 43.2 | 66.7 | 0.086 | 55.6 | 50.0 | 0.698 | 50.0 | 52.4 | 0.871 | 40.9 | 58.3 | 0.198 | 20.0 | 56.0 | 0.178 |
| Trimethoprim/sulfamethoxazole | 48.3 | 4 | 8 | 45.0 | 50.0 | 0.717 | 37.8 | 66.7 | 0.035* | 55.6 | 44.7 | 0.449 | 56.3 | 45.2 | 0.453 | 59.1 | 41.7 | 0.198 | 60.0 | 46.0 | 0.659 |
| Cefoxitin | 10.3 | 128 | 128 | 15.0 | 7.9 | 0.405 | 10.8 | 9.5 | 1.000 | 5.6 | 13.2 | 0.652 | 0.0 | 14.3 | 0.173 | 4.5 | 13.9 | 0.392 | 0.0 | 12.0 | 1.000 |
p < 0.05.
Correlation between susceptibility of polymyxin B/colistin and clinical background of patients
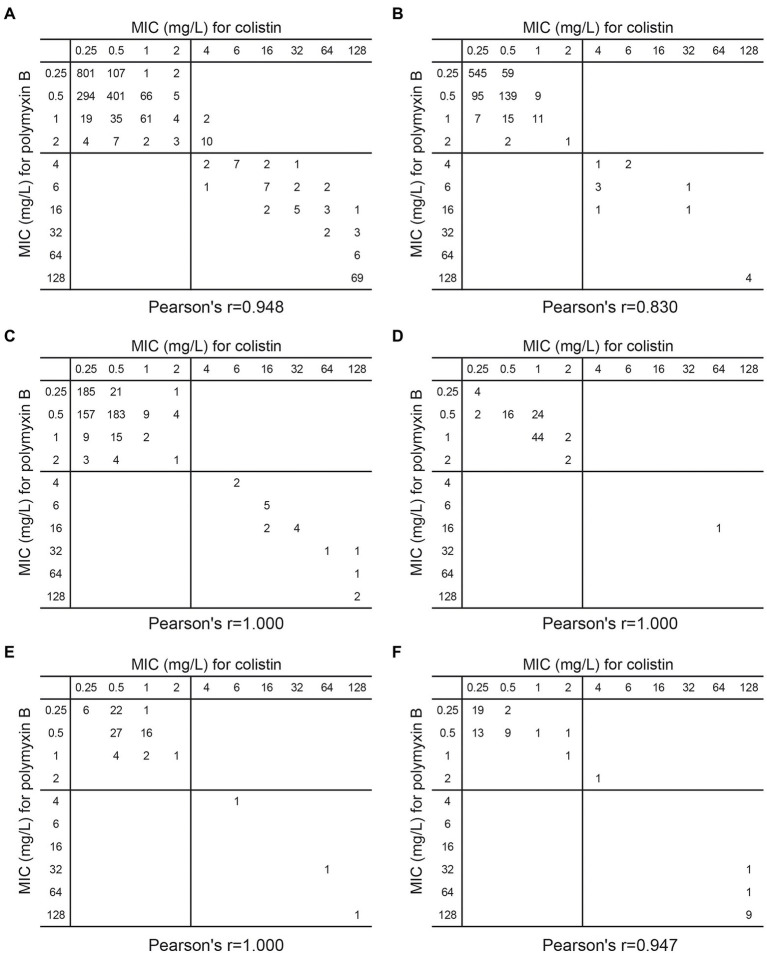
Since both polymyxin B and colistin belonged to polypeptide antibiotics, it was wondered whether there was a correlation between the susceptibility of different pathogens towards polymyxin B and colistin. Scatterplot analyses and Fisher’s exact test revealed that when polymyxin B and colistin were used on Gram-negative bacteria causing bloodstream infection, their minimum inhibitory concentrations(mics) were significantly correlated (Pearson’s r = 0.948, p < 0.001). This conclusion was also valid when E. coli (Pearson’s r = 0.830; p < 0.001), K. (Pearson’s r = 1.000, p < 0.001), P. aeruginosa (Pearson’s r = 1.000; p < 0.001), A. baumannii (Pearson’s r = 1.000; p < 0.001) and E. cloacae (Pearson’s r = 0.947; p < 0.001) were analyzed separately (Figure 3).
Figure 3.
Scatterplot of MIC (mg/l) of colistin versus polymyxin B against (A) 1,939 Gram-negative bacteria (B) 896 E. coli (C) 612 K. pneumoniae (D) 95 P. aeruginosa (E) 82 A. baumannii (F) 58 E. cloacae which caused bloodstream infection in China.
Comparing the clinical background of different isolates, it was found that the susceptibility of these Gram-negative bacteria to polymyxin B and colistin did not vary significantly depending on whether the patients were suffering from pulmonary diseases, hepatobiliary diseases, digestive system diseases, cardiovascular system diseases, nervous system diseases, urinary system diseases, diabetes, agranulosis or any form of tumors. The smoking or splenectomy history did not affect the response of Gram-negative pathogens to polymyxin B or colistin either. Strangely, among strains involved in this study, which were isolated from patients with hypoproteinemia seemed to be more susceptible to polymyxin B, while those isolated from patients with history of alcohol intake were more susceptible to colistin (Supplementary Table S8). These associations needed to be further analyzed and confirmed. When it comes to recent medical operation, it was revealed that most common medical procedures occurred in recent 3 months did not seem to affect the efficacy of polymyxin B and colistin, including medical institution admission, surgery, hormone usage, immunosuppressor usage, indwelling catheter usage and antibiotics usage history. Similarly, whether the patient’s fever exceeds 39°C at this visit has no effect yet (Supplementary Table S8).
When it comes to the issue of the difference in susceptibility of Gram-negative bacteria from different infection sources to polymyxin B and colistin, it was implied that the bloodstream infection isolates from catheter-related bloodstream infections had the lowest susceptibility to polymyxin B (90.4%; Supplementary Table S9). And isolates from abdominal infection of other organs except for liver and biliary tract infection had the lowest susceptibility to colistin (90.0%; Supplementary Table S9). For alimentary tract infection and cardiovascular system infection derived Gram-negative bacterial bloodstream infection, the susceptibility of polymyxin B reached 100.0% (Supplementary Table S9).
The general geographical differences in polymyxin B and colistin susceptibilities were presented in Supplementary Table S10; Supplementary Figure S1. It was shown that Gram-negative strains from East China had the highest resistance rate to polymyxin B and colistin, which were 9.3 and 10.1% respectively, while in South China, their resistance rates were both 3.1%. To our relief, Gram-negative bacteria prevalent in most regions of China were still highly susceptible to polymyxin B (3.1% ~ 9.3%) and colistin (3.1% ~ 10.1%). The results of deep analysis showed that for E. coli, K. pneumoniae, P. aeruginosa, and A. baumannii, the susceptibility of polymyxin B in different regions of China were between 92.3 and 100.0%. While for E. cloacae, its sensitivities in North, East, Central, South and Northeast China were 87.5, 82.4, 71.4, 33.3 and 75.0%, separately (Supplementary Table S11).
Discussion
According to the data collected by CARVIS-NET, in China, the pathogenic spectrum and infection source distribution of Gram-negative bacteria bloodstream infection patients of different age groups had different characteristics. And different age groups responded differently to different antibiotics. Levofloxacin has been used in China as early as 1990s, which might be the reason why the susceptibility rate of adult and elderly patients to it and its similar antibiotic amikacin was lower. Similarly, for several newer antibiotics, the susceptibility rate in the minor patient group was relatively low. Perhaps due to the poor immune function of the elderly, the proportion of patients with primary and bloodstream infections from the community and the mortality rate gradually increased with age. Whether there is a deeper reason for this remains to be further investigated.
From the drug resistance monitoring data, it could be seen that drug resistance of Gram-negative bacteria to antibiotics is still a serious problem in China. For the 2nd generation cephalosporins represented by cefoxitin, only E. coli had a relative low resistance rate of 9.1%, and all the other species had a resistance rate higher than 30%, or MIC50 reached 128 μg/ml. For the 3rd generation cephalosporins represented by ceftriaxone and ceftazidime, among the five common gram-negative bacteria involved in in this study, except for P. aeruginosa, whose resistance rate to ceftazidime was 13.7%, the resistance rates of the other four bacteria were all higher than 20%. Similarly, for cefepime, a 4th generation cephalosporin, the resistance rates of E. coli, K. pneumoniae and A. baumannii all exceeded 30%. It should be emphasized that the drug resistance rates of A. baumannii to almost all the cephalosporins involved were more than 74.0%. Based on these facts, it could be inferred that the effective rates of monotherapy with cephalosporins to treat gram-negative bacterial bloodstream infection was not satisfactory. Further investigation on susceptibility of ceftazidime combined with avibactam, a novel β-lactamase inhibitor, pointed that this combination could significantly improve the efficiency of ceftazidime. Resistance to ceftazidime for 99.1% of E. coli, 98.5% of K. pneumoniae, 100.0% of A. baumannii, and 100.0% of E. cloacae were offset by the use of Avibactam. Yet for 16.9% of ceftazidime-resistant P. aeruginosa strains, drug resistance was still not improved. Therefore, the presence of β-lactamase was still a great challenge for the usage of β-lactam antibiotics.
Early studies had found that many bacteria could not directly use the folic acid from environment, which was necessary for the synthesis of purine and pyrimidine(Gleckman et al., 1981). Trimethoprim/sulfamethoxazole(SXT) was designed to target on the first two steps of folic acid anabolism, respectively, so that the synthesis of folic acid could be fundamentally blocked(Smilack, 1999). At first, the effect of SXT was satisfactory. However, with its rapid promotion clinically, Gram-negative bacteria had gradually optimized their metabolic pathways under the strong evolutionary pressure. Therefore, the current situation of resistance to sulfonamides was also not optimistic.
For Gram-negative bacteria, quinolones mainly target bacterial DNA gyrase(Hooper, 1998; Hooper and Jacoby, 2015). The application of quinolones began in the 1980s. With the extensive application in the past 40 years, the response rate of quinolones in Gram-negative bacteria was getting lower and lower, and the problem of drug resistance could not be ignored either.
In addition to the antibiotics with common drug resistance problem above, different bacteria have their own characteristics in response to other antibiotics. For E. coli, glycylcyclones/halotetracyclines, carbapenems, penicillins or cephalosporins combined with β-lactamase inhibitors performed well. For K. pneumoniae, whose problem of drug resistance was getting increasingly serious, β-lactams or the third-generation cephalosporins acted only when β-lactamase inhibitor avibactam was combined. P. aeruginosa was naturally resistant to ertapenem and ceftriaxone involved in this investigation. When ceftazidime was combined with avibactam, its inhibitory effect was also remarkable. When it comes to E. cloacae, the sensitivity rate when ceftazidime was used for single drug treatment was only 65.5%, yet when combined with avibactam, the sensitivity rate was greatly improved to 100%. The underlying mechanism was that in recent years, the prevalence of ESBL among Gram-negative bacteria has shown an upward trend, and many bacteria have acquired β-lactamases, which could lead β-lactam antibiotics (including ceftazidime) to be hydrolyzed, thus losing the inhibitory effect on the strain. Avibactam belongs to β-lactamase inhibitor. When ceftazidime and avibactam are applied simultaneously, the β-lactamases will be inhibited so that they no longer have the ability to degrade ceftazidime, thus they are still sensitive to ceftazidime (Castanheira et al., 2021; Yahav et al., 2021). Besides, amikacin, an aminoglycoside, performed well against P. aeruginosa and E. cloacae in bloodstream infections as well.
Polymyxins were important antibiotics for the treatment of Gram-negative bacteria, especially MDR Gram-negative bacteria(Nang et al., 2021). In the past, colistin have been widely used in animal husbandry. The long-term irregular application has led to the gradual development of resistance of gram-negative bacteria from animals to colistin. In 2015, MCR-1, the first plasmid mediated colistin resistance gene in enterobacteriaceae in china, was detected. Some researchers found that the detection rates of MCR-1 in samples of raw meat, animals and infected inpatients were 15, 21 and 1%, respectively. The difference was because that colistin have large side effects on humans, especially for kidney, so their use in humans was limited. Recently, due to the increase of infections caused by multidrug-resistant Gram-negative bacteria, colistin and polymyxin have been reapplied globally as “the last antibiotic,” but it is far less than its early application in animal husbandry in terms of frequency and dosage. Therefore, although the resistance rate of bacteria from animals is high at present, it still maintains high sensitivity rate in humans (Pitt et al., 2003; Landman et al., 2005; Keen 3rd et al., 2010; Gales et al., 2011; Quan et al., 2017). However, its high drug resistance in animal husbandry was a potentially hidden danger thence monitoring the susceptibility of them also had important academic and clinical value (Rhouma et al., 2016). It is worth emphasizing that in this research, polymyxins represented by polymyxin B and colistin had outstanding performance against Gram-negative bacteria represented by E. coli, K. pneumoniae, P. aeruginosa and A. baumannii. When used to resist the bloodstream infection of these four bacteria, the susceptibility rates reached 95% unexceptionally. In this cohort, polymyxin B showed relatively poor performance in anti-E. cloacae. Considering the limited sample size, large sample studies were still necessary.
In conclusion, the surveillance results in CARVIS-NET showed that the main pathogens of Gram-negative bacteria bloodstream infection in China were Enterobacteriaceae, while E. coli accounted for the vast majority. The resistance rate of K. pneumoniae, P. aeruginosa, A. baumannii and E. cloacae to multiple antibiotics kept on a high level. In many cases, polymyxin B and colistin has become the last-resort agents to combat bloodstream infections caused by MDR Gram-negative bacteria.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
QY conceived and designed the study. JX analyzed the data and wrote the manuscript. PJ, YZ, WY, QY, and YX revised the manuscript. JX, JZ, HG, WK, GZ, JL, and TW performed the experiments. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82072318) and National Key Research and Development Program of China (2021YFC2301002). This work was also supported by Wuhan Healcare Pharmaceuticals Co., Ltd. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We are grateful to all the investigators of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to all the investigators of this study.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1017488/full#supplementary-material
References
- Castanheira M., Doyle T. B., Deshpande L. M., Mendes R. E., Sader H. S. (2021). Activity of ceftazidime/avibactam, meropenem/vaborbactam and imipenem/relebactam against carbapenemase-negative carbapenem-resistant Enterobacterales isolates from US hospitals. Int. J. Antimicrob. Agents 58:106439. doi: 10.1016/j.ijantimicag.2021.106439, PMID: [DOI] [PubMed] [Google Scholar]
- CLSI (2022). Performance Standards for Antimicrobial Susceptibility Testing, Document M100, (32nd Edn.) Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Collignon P. J., McEwen S. A. (2019). One health-its importance in helping to better control antimicrobial resistance. Trop. Med. Infect. Dis. 4:22. doi: 10.3390/tropicalmed4010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley I. A., Karlowsky J. A. (2004). Optimal use of antibiotic resistance surveillance systems. Clin. Microbiol. Infect. 10, 502–511. doi: 10.1111/j.1469-0691.2004.00911.x [DOI] [PubMed] [Google Scholar]
- El-Sayed Ahmed M. A. E., Zhong L. L., Shen C., Yang Y., Doi Y., Tian G. B. (2020). Colistin and its role in the era of antibiotic resistance: an extended review (2000-2019). Emerg. Microbes Infect. 9, 868–885. doi: 10.1080/22221751.2020.1754133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergonul O., Aydin M., Azap A., Basaran S., Tekin S., Kaya S., et al. (2016). Turkish Society of Clinical M and Infectious Diseases H-RISG. Healthcare-associated gram-negative bloodstream infections: antibiotic resistance and predictors of mortality. J. Hosp. Infect. 94, 381–385. doi: 10.1016/j.jhin.2016.08.012, PMID: [DOI] [PubMed] [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) . (2022). Clinical breakpoints and dosing of antibiotics. Available at https://www.eucast.org/clinical_breakpoints/. Accessed November 11, 2022.
- Gales A. C., Jones R. N., Sader H. S. (2011). Contemporary activity of colistin and polymyxin B against a worldwide collection of gram-negative pathogens: results from the SENTRY antimicrobial surveillance program (2006-09). J. Antimicrob. Chemother. 66, 2070–2074. doi: 10.1093/jac/dkr239, PMID: [DOI] [PubMed] [Google Scholar]
- Gleckman R., Blagg N., Joubert D. W. (1981). Trimethoprim: mechanisms of action, antimicrobial activity, bacterial resistance, pharmacokinetics, adverse reactions, and therapeutic indications. Pharmacotherapy 1, 14–19. doi: 10.1002/j.1875-9114.1981.tb03548.x, PMID: [DOI] [PubMed] [Google Scholar]
- Hooper D. C. (1998). Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin. Infect. Dis. 27, S54–S63. doi: 10.1086/514923, PMID: [DOI] [PubMed] [Google Scholar]
- Hooper D. C., Jacoby G. A. (2015). Mechanisms of drug resistance: quinolone resistance. Ann. N. Y. Acad. Sci. 1354, 12–31. doi: 10.1111/nyas.12830, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh K., Chung D. R., Ha Y. E., Ko J. H., Kim S. H., Kim M. J., et al. (2020). Song JH and Korean antimicrobial resistance surveillance network I. impact of difficult-to-treat resistance in gram-negative bacteremia on mortality: retrospective analysis of Nationwide surveillance data. Clin. Infect. Dis. 71, e487–e496. doi: 10.1093/cid/ciaa084, PMID: [DOI] [PubMed] [Google Scholar]
- Johnson A. P. (2015). Surveillance of antibiotic resistance. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 370:20140080. doi: 10.1098/rstb.2014.0080, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen E. F., 3rd, Robinson B. J., Hospenthal D. R., Aldous W. K., Wolf S. E., Chung K. K., et al. (2010). Prevalence of multidrug-resistant organisms recovered at a military burn center. Burns 36, 819–825. doi: 10.1016/j.burns.2009.10.013, PMID: [DOI] [PubMed] [Google Scholar]
- Landman D., Bratu S., Alam M., Quale J. (2005). Citywide emergence of Pseudomonas aeruginosa strains with reduced susceptibility to polymyxin B. J. Antimicrob. Chemother. 55, 954–957. doi: 10.1093/jac/dki153, PMID: [DOI] [PubMed] [Google Scholar]
- Liang T., Xu C., Cheng Q., Tang Y., Zeng H., Li X. (2021). Epidemiology, risk factors, and clinical outcomes of bloodstream infection due to extended-Spectrum Beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in hematologic malignancy: a retrospective study from central South China. Microb. Drug Resist. 27, 800–808. doi: 10.1089/mdr.2020.0033, PMID: [DOI] [PubMed] [Google Scholar]
- Liu S., Wang M., Zheng L., Guan W. (2018). Antimicrobial resistance profiles of nosocomial pathogens in regional China: a brief report from two tertiary hospitals in China. Med. Sci. Monit. 24, 8602–8607. doi: 10.12659/MSM.911229, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nang S. C., Azad M. A. K., Velkov T., Zhou Q. T., Li J. (2021). Rescuing the last-line Polymyxins: achievements and challenges. Pharmacol. Rev. 73, 679–728. doi: 10.1124/pharmrev.120.000020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutman A., Tellapragada C., Giske C. G., Yahav D. (2021). New evidence for managing gram-negative bloodstream infections. Curr. Opin. Infect. Dis. 34, 599–610. doi: 10.1097/QCO.0000000000000784, PMID: [DOI] [PubMed] [Google Scholar]
- Pitt T. L., Sparrow M., Warner M., Stefanidou M. (2003). Survey of resistance of Pseudomonas aeruginosa from UK patients with cystic fibrosis to six commonly prescribed antimicrobial agents. Thorax 58, 794–796. doi: 10.1136/thorax.58.9.794, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J., Li X., Chen Y., Jiang Y., Zhou Z., Zhang H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/S1473-3099(16)30528-X, PMID: [DOI] [PubMed] [Google Scholar]
- Rhouma M., Beaudry F., Letellier A. (2016). Resistance to colistin: what is the fate for this antibiotic in pig production? Int. J. Antimicrob. Agents 48, 119–126. doi: 10.1016/j.ijantimicag.2016.04.008, PMID: [DOI] [PubMed] [Google Scholar]
- Rodriguez-Bano J., Gutierrez-Gutierrez B., Machuca I., Pascual A. (2018). Treatment of infections caused by extended-Spectrum-Beta-lactamase-, AmpC-, and Carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev. 31, 13–23. doi: 10.1128/CMR.00079-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilack J. D. (1999). Trimethoprim-sulfamethoxazole. Mayo Clin. Proc. 74, 730–734. doi: 10.4065/74.7.730 [DOI] [PubMed] [Google Scholar]
- Tamma P. D., Aitken S. L., Bonomo R. A., Mathers A. J., van Duin D., Clancy C. J. (2021). Infectious Diseases Society of America guidance on the treatment of extended-Spectrum beta-lactamase producing Enterobacterales (ESBL-E), Carbapenem-resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 72, 1109–1116. doi: 10.1093/cid/ciab295, PMID: [DOI] [PubMed] [Google Scholar]
- Yahav D., Giske C. G., Gramatniece A., Abodakpi H., Tam V. H., Leibovici L. (2020). New beta-lactam-beta-lactamase inhibitor combinations. Clin. Microbiol. Rev. 34, e00115–20. doi: 10.1128/CMR.00115-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Ma N., Liu R., Wang N., Zhang T., He L. (2019). Polymyxin B and polymyxin E induce anaphylactoid response through mediation of mas-related G protein-coupled receptor X2. Chem. Biol. Interact. 308, 304–311. doi: 10.1016/j.cbi.2019.05.014, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.