Abstract
Microglia are the primary resident innate immune cells of the CNS. They possess branched, motile cell processes that are important for their cellular functions. To study the pathways that control microglial morphology and motility under physiological and disease conditions, it is necessary to quantify microglial morphology and motility precisely and reliably. Several image analysis approaches are available for the quantification of microglial morphology and motility. However, they are either not automated, not freely accessible, and/or limited in the number of morphology and motility parameters that can be assessed. Thus, we have developed MotiQ, an open-source, freely accessible software for automated quantification of microglial motility and morphology. MotiQ allows quantification of a diverse set of cellular motility and morphology parameters, including the parameters that have become the gold standard in the microglia field. We demonstrate that MotiQ can be applied to in vivo, ex vivo, and in vitro data from confocal, epifluorescence, or two-photon microscopy, and we compare its results to other analysis approaches. We suggest MotiQ as a versatile and customizable tool to study microglia.
INTRODUCTION
Microglia are the parenchymal tissue resident macrophages of the CNS. In the intact adult brain, microglia are highly ramified and have a complex three-dimensional (3D) branch structure. Intravital two-photon microscopy in mice with microglia-specific expression of enhanced green fluorescent protein (EGFP) has shown that microglial cell branches are highly motile and continuously undergo extension and retraction (Davalos et al., 2005; Nimmerjahn et al., 2005). This motility enables microglia to perform the surveillance of their environment to then execute their functions, including limiting synaptic excitability (Badimon et al., 2020), partial phagocytosis of synaptic structures (Weinhard et al., 2018), synaptic pruning during development and in disease (Hong et al., 2016), and formation and maintenance of synapses (Miyamoto et al., 2016). It has been estimated that the cortical parenchyma is completely scanned by microglia every few hours (Nimmerjahn et al., 2005).
Microglial morphology and motility change during systemic inflammation (Gyoneva et al., 2014), in neurodegenerative and neuropsychiatric diseases (Bolmont et al., 2008; De Picker et al., 2021; Franco-Bocanegra et al., 2021), upon aging (Conde and Streit, 2006; Hefendehl et al., 2014), and in response to specific deletions of microglial receptors (Pagani et al., 2015; Sipe et al., 2016; Madry et al., 2018; Merlini et al., 2021). To study the pathways that underlie these changes and to determine their functional impact, it is necessary to precisely and reliably analyze microglial cell motility. A set of parameters for microglial morphology has become standard, such as the surface area-to-volume ratio (e.g., Orr et al., 2009; Madry et al., 2018; Plescher et al., 2018), the total length of the process tree as well as the numbers of branches and tips (e.g., Nimmerjahn et al., 2005; Hefendehl et al., 2014; Erny et al., 2015), or the convex hull around the cell (e.g., Fontainhas et al., 2011; Baron et al., 2014; Fernández-Arjona et al., 2017). Current analysis approaches 1) are only partly automated (Fernández-Arjona et al., 2017; Young and Morrison, 2018) and provide low throughput, 2) rely on manual tracing of cells (Cătălin et al., 2017; Paris et al., 2018; Basilico et al., 2019; Sun et al., 2019), 3) are limited to a two-dimensional (2D) analysis (Cătălin et al., 2017; Fernández-Arjona et al., 2017; Kluge et al., 2017; Madry et al., 2018; Young and Morrison, 2018; Kyriazis et al., 2019), and/or 4) use custom-written, lab-specific software with which a limited number of microglial parameters can be determined (Abdolhoseini et al., 2016; Kluge et al., 2017; Madry et al., 2018; Paris et al., 2018; Kyriazis et al., 2019). Commercial image analysis software such as Imaris (Bitplane) has been used in previous studies, but these commercial solutions are not available to every lab due to cost considerations and often use proprietary algorithms.
Here, we introduce MotiQ, a versatile and open-source software for the automatized and comprehensive quantification of microglial motility and morphology in 2D and 3D microscopic image series. In addition to data from two-photon microscopy, MotiQ is applicable to a variety of other microscopic data sets and allows for the quantification of a broad set of more than 60 parameters of microglial morphology, motility, and signaling. MotiQ is a freely accessible resource for microglia research and represents an open-source platform for researchers to establish standards for microglial quantification.
RESULTS
MotiQ workflow
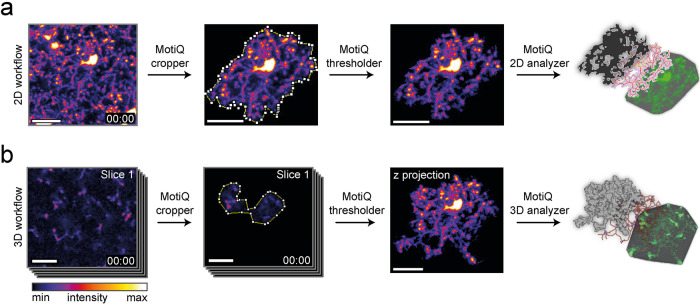
We developed MotiQ primarily to quantify microglial morphology and motility in 3D time-lapse images acquired by in vivo two-photon microscopy. For open access and high applicability MotiQ is composed of modular ImageJ plug-ins. The source code, all plug-ins, and a manual are freely accessible online (https://github.com/hansenjn/MotiQ). MotiQ allows two types of analyses: a more rapid 2D analysis of maximum-intensity projections (Figure 1a) and a more comprehensive analysis of original 3D z stacks (Figure 1b). Both MotiQ quantification approaches are performed with a three-step workflow using the plug-ins 1) MotiQ cropper for single-cell image generation, 2) MotiQ thresholder for image segmentation, and 3) MotiQ 2D analyzer or MotiQ 3D analyzer for quantification. This three-step plug-in format allows one to replace or adapt individual steps, to include additional plug-ins in the workflow, or to use individual MotiQ plug-ins for applications other than microglial cell analysis.
FIGURE 1:
MotiQ workflow. MotiQ offers two main approaches for microglial cell analysis: (a) 2D time-lapse image analysis and (b) analysis of 3D time-lapse image stacks. Both workflows are composed of three steps: single-cell selection with MotiQ cropper, image segmentation with MotiQ thresholder, and cell quantification using (a) MotiQ 2D analyzer or (b) MotiQ 3D analyzer. Cell volume, cell surface, cell skeleton, and the convex hull of the cell are reconstructed and analyzed and serve as a basis for the calculation of more than 60 parameters of microglial morphology, dynamics, and fluorescence kinetics. MotiQ automatically generates 3D image visualizations by implementing Volume Viewer (ImageJ plug-in by Kai Uwe Barthel, Internationale Medieninformatik, HTW Berlin, Berlin, Germany). Scale bars, 20 µm.
The first plug-in, MotiQ cropper, aids in the selection of single microglia from recordings and generates images comprising one individual microglial cell (for technical details see Supplemental Note 1 and the manual provided on the GitHub page). We implemented this user-dependent, single-cell selection approach to realize the analysis of microglia under pathological conditions, for example, Alzheimer’s disease (AD), where microglia and their cell processes are densely packed and closely interlaced at the cell border (Condello et al., 2015), which renders automated single-cell detection imprecise. This challenge is commonly addressed by manual tracing approaches, in which a trained investigator traces the individual processes of a single cell, which can be time consuming (Gyoneva and Traynelis, 2013; Baron et al., 2014; Cătălin et al., 2017; Paris et al., 2018; Basilico et al., 2019; Sun et al., 2019). In contrast, in MotiQ cropper a region of interest (ROI) that contains an individual cell is selected. The cell detection and process tracing itself is then fully automated based on the other MotiQ plug-ins. The single-cell selection with MotiQ is therefore faster than careful manual tracing, because the user does not need to precisely track individual processes but only needs to roughly encircle a ROI. Additionally, by guiding the user through z stacks and time frames, by transferring selections from image to image, and by automatically saving the resulting single-cell image and metadata pertaining to the selected ROIs, MotiQ cropper renders the whole process more time efficient. The output image, together with corresponding metadata, can be used to retrospectively validate the selection process.
The next step in the MotiQ workflow is image segmentation, which is accomplished using MotiQ thresholder (Supplemental Note 2). In other image analysis methods for microglia, such as multistep protocols for ImageJ (e.g., Fontainhas et al., 2011; Fernández-Arjona et al., 2017; Young and Morrison, 2018), users adjust settings and threshold values to segment cells from background, image by image. Such manual multistep workflows are time consuming and error prone unless fully automated macros are developed. MotiQ thresholder standardizes and automatizes image preprocessing and segmentation, even for users who are not able to develop computer code. MotiQ thresholder’s pipeline, including image preprocessing steps, local or global auto-threshold methods, and image segmentation, is easily customizable by the user through the plug-in’s setting dialogues.
We implemented image preprocessing steps in MotiQ thresholder that allow the production of an image in which foreground (microglia) and background intensities are more distinguished: 1) downscaling of image resolution, 2) rescaling the intensity histogram for better coverage of the intensity range, 3) including multiple time frames or z stacks into the histogram, and 4) converging the relationship of fore- and background pixel numbers toward equilibrium, for example, by using maximum-intensity projections of multiple time step or z stack images. The user can select one or a combination of those methods. For a detailed description and illustration of the rationale behind these methods, see Supplemental Note 2 and Supplemental Figures 1 and 2. For subsequent threshold calculation, MotiQ thresholder uses one of the intensity thresholding algorithms available in ImageJ. The threshold algorithm to be used is defined by the user, based on the best performance in separating fore- and background in the user’s images. Importantly, MotiQ thresholder performs image preprocessing and threshold calculation in an automatically generated copy of the image, while the intensity threshold is finally applied to the unsegmented, original input image. Thereby, the preprocessing methods do not result in a loss of resolution or dimension and MotiQ thresholder not only generates a binary image but can also produce a background-removed version of the original image, containing fully preserved fluorescence intensity information. MotiQ thresholder also allows for separate threshold calculation and application in individual time frames, which is important for time-lapse data with instable fluorescence intensity levels, for example, due to photobleaching. Because the user may need to optimize the MotiQ thresholder settings for individual data sets, we provide default settings for the analysis of different types of data in Supplemental Table 1, which may serve as a starting point for further optimizations.
In the last step of the MotiQ workflow, cell analysis and visualization are performed using MotiQ 2D analyzer or MotiQ 3D analyzer, depending on the respective image type (Supplemental Note 3). First, connected pixels are detected and bundled to individual objects via a Flood-Filler algorithm. Objects smaller than a predefined size are removed, which allows the removal of noise pixels, falsely detected as foreground. The remaining larger objects are considered to belong to the selected microglial cell and are quantified. In this step, the microglial cell, its cell skeleton, and its convex hull are automatically reconstructed from these objects, and more than 60 cellular parameters that represent microglial morphology (Supplemental Table 2), motility (Supplemental Table 3), and signaling (Supplemental Table 4) are automatically determined. We collected the major part of this list of parameters based on the parameters that have been applied to microglia in the literature and that are considered standard parameters (details below). We aimed to make the application of MotiQ as versatile as possible and allow the best comparability of output data to previous literature reports. Furthermore, MotiQ 3D analyzer contains a tool that illustrates the analysis results in 3D visualizations (Figure 1).
Analysis of in vivo and ex vivo two-photon microscopy data sets
To demonstrate which microglial morphology and dynamic parameters can be determined with MotiQ, we subjected recordings from intravital time-lapse two-photon microscopy in CX3CR1-EGFP/wt mice (Jung et al., 2000) to the MotiQ 2D and 3D analysis workflow (Supplemental Videos 1 and 2).
Movie S1.
2D analysis of in vivo two‐photon microscopy data
Movie S2.
3D analysis of in vivo two‐photon microscopy data
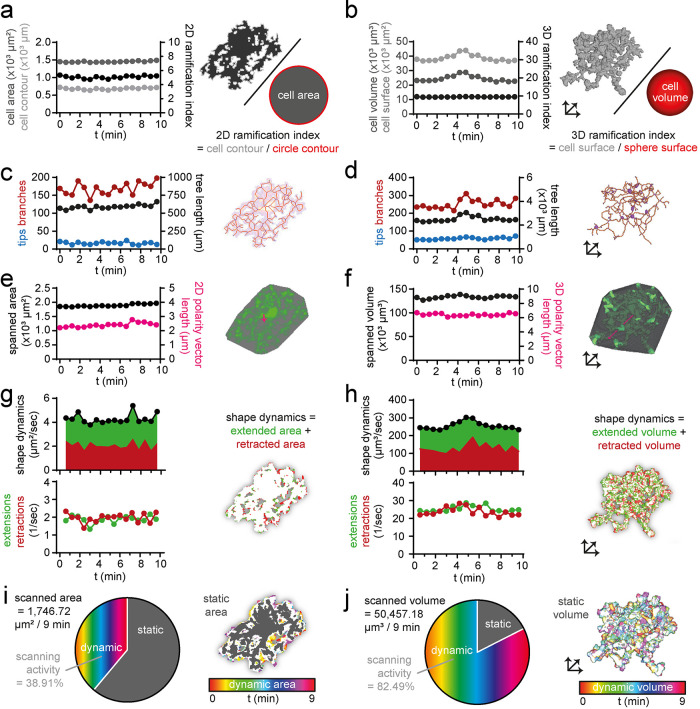
The morphology of a microglial cell is predominantly characterized by its ramification, that is, the complexity of its branch structure. Microglial ramification is measured by two main approaches: 1) by determining the relationship of cell surface to cell volume (ramification index) (e.g., Orr et al., 2009; Madry et al., 2018; Plescher et al., 2018) and 2) by analyzing the microglial cell skeleton (e.g., Nimmerjahn et al., 2005; Hefendehl et al., 2014; Erny et al., 2015). The ramification index describes the overall complexity of the microglial cell, and the cell skeleton parameters provide additional information about subcomponents of the microglial process tree.
More specifically, the 2D ramification index is defined as the ratio of the cell contour to the circumference of a circle with the same cell area as the respective cell (Figure 2a) and the 3D ramification index is defined as the ratio of the cell surface area to the surface area of a sphere with the same volume as the respective cell (Figure 2b). For skeletonization and analysis of the resulting cell skeleton, we implemented the ImageJ plug-ins Skeletonize3D and AnalyzeSkeleton (Arganda-Carreras et al., 2010) in MotiQ, allowing us to quantify a variety of cell skeleton characteristics (Supplemental Table 2), including number of tips, number of branches, and total tree length (Figure 2, c and d). Other morphological parameters that are included in MotiQ are the brain area (2D) or brain volume (3D) spanned by a microglial cell, that is, spanned area (Figure 2e) or spanned volume (Figure 2f), which are defined as the size of the cell’s convex hull and represent the territory of an individual cell. Thus, these parameters reveal the potential influence of the microglial cell on the surrounding brain parenchyma. As a measure for cell polarity, that is, the asymmetric distribution of processes from the soma, the vector from the cell’s center of mass to the center of the convex hull is included in MotiQ (Figure 2, e and f).
FIGURE 2:
Analysis of in vivo two-photon microscopy data. Selected morphological and dynamic parameters of a representative cortical microglial cell in CX3CR1GFP/wt mice imaged with in vivo two-photon microscopy and analyzed with MotiQ: (a) 2D ramification index, (b) 3D ramification index, (c, d) number of tips, number of branches, and process tree length, (e, f) spanned area or spanned volume and cell polarity vector length (5× magnified for better visualization), (g, h) shape dynamics and number of extensions and retractions as parameter for cell shape alteration, and (i, j) scanned area or scanned volume represent the brain area or brain volume that has been occupied by a microglial cell over a selected time span (here 9 min).
In addition to morphological parameters, we included parameters in MotiQ that represent dynamic changes of the microglial cell and its process tree. For example, MotiQ allows determining the number and area (2D) or volume (3D) of individual microglial process extensions and retractions in consecutive time steps (Figure 2, g and h). Individual extensions or retractions are defined as objects consisting of a group of connected pixels that newly emerge or disappear in an image compared with the previous time step. The parameter shape dynamics is the sum of the total extended and retracted area (2D) or volume (3D) and serves as a measure for the overall change of the microglial cell shape. Furthermore, as a readout for the interaction of a microglial cell with its environment, MotiQ allows the determination of the scanned area (2D; Figure 2i) or scanned volume (3D; Figure 2j), which are defined as the brain area or the brain volume that is occupied by a microglial cell and its processes over a longer time span. A similar parameter, called “volume fill,” has been applied in 2D before (Nimmerjahn et al., 2005) for a whole stack of images containing many cells, demonstrating that microglial motility allows microglia to sample the brain volume over time. We introduce this parameter for single cells as a measure for microglial scanning activity and add that the scanned area or volume is subdivided into the static area or volume, in which the cell is present in all analyzed time frames (usually the soma and larger primary cell branches), and the dynamic area or volume, in which the cell compartment is only transiently present (Figure 2, i and j). The scanning activity of the cell is determined as the percentage of the scanned area or volume that is dynamic.
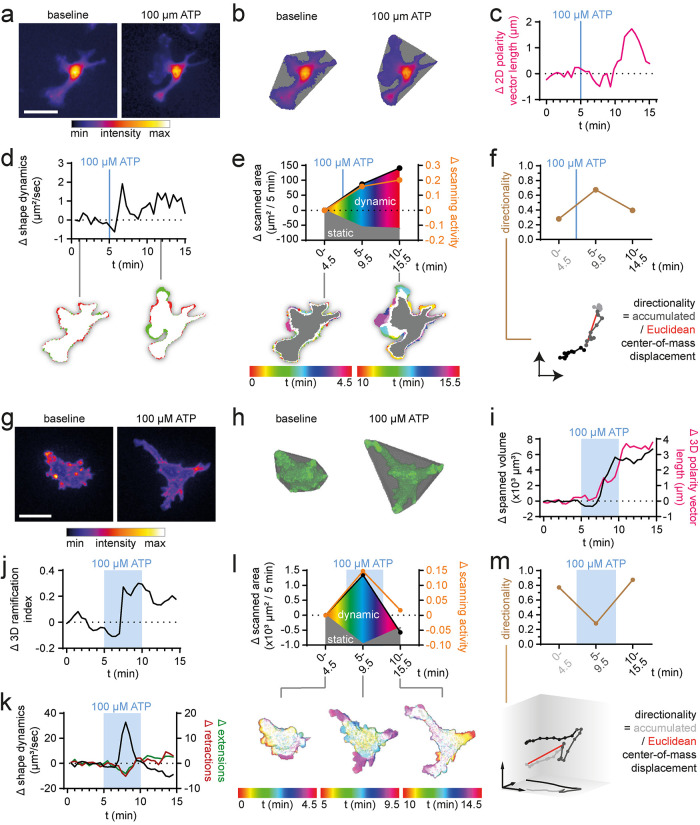
To test whether MotiQ sensitively detects changes in morphology and motility, we next analyzed microglia before and after stimulation in ex vivo time-lapse, two-photon microcopy recordings from acute cerebral slices with MotiQ. Acute slice preparations have been used to study microglial responses to extracellular stimuli such as extracellular adenosine triphosphate (ATP) (Fontainhas et al., 2011; Dissing-Olesen et al., 2014; Matyash et al., 2017). We perfused cerebral slices with ATP to induce a change in microglial morphology and motility. We then analyzed the microscopic data with MotiQ 2D and 3D (Figure 3). We sensitively and reliably detected ATP-induced changes in morphology and dynamic parameters, such as an increase in the ramification index (Figure 3, c and d), number of branches, tree length (Figure 3, e and f), spanned area (2D; Figure 3g) or volume (3D; Figure 3h), shape dynamics (Figure 3, i and j), and microglial scanning parameters (Figure 3, i–l).
FIGURE 3:
Analysis of ex vivo two-photon microscopy data. 2D and 3D analysis of microglial morphology and dynamics in ex vivo two-photon microscopy data of a representative microglial cell in acute cerebral slices of CX3CR1EGFP/wt mice before and after perfusion with 100 µM ATP (blue boxes). (a, b) Spanned area (2D) or volume (3D) before (t = 5 min) and during (t = 45 min) ATP perfusion. (c, d) Ramification index, (e, f) tree length, number of tips, and number of branches, (g, h) spanned area (2D) or volume (3D), cell polarity vector length, (i, j) shape dynamics, number of extensions and retractions, and (k, l) scanned area (2D) or volume (3D) during ATP perfusion. Scale bar, 20 µm.
Analysis of histological data sets
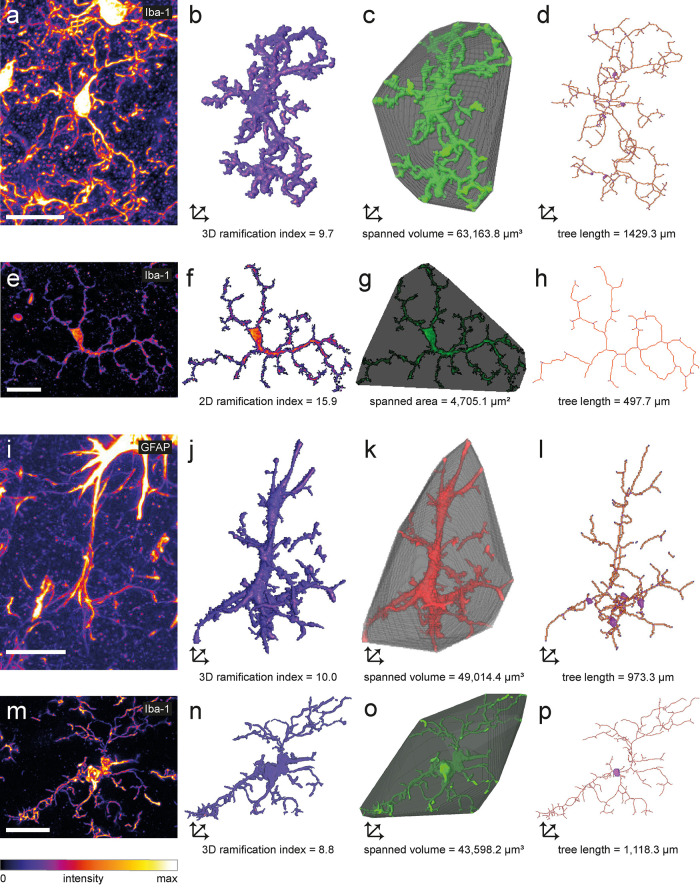
Because morphometric endpoint analysis of microglia in fixed tissue is one of the most commonly used techniques in microglia research, we next analyzed confocal images of immunofluorescently labeled Iba1-positive murine microglia in fixed cerebral (Figure 4, a–d) and retinal (Figure 4, e–h) tissue sections. For both data sets, we used MotiQ to reconstruct microglia and analyze their morphological characteristics. To further illustrate the range of applications of MotiQ, we additionally analyzed confocal images of immunofluorescently GFAP-positive astrocytes (Figure 4, i–l) and Iba1-positive human microglia (Figure 4, m–p). Here we show that MotiQ could also be used to analyze a broad range of other cell types.
FIGURE 4:
Analysis of cellular morphology in tissue sections. Maximum-intensity projections of confocal z stacks of an (a) Iba1-labeled murine cortical microglial cell, (e) Iba1-labeled murine retinal microglial cell, (i) hippocampal GFAP-labeled murine astrocyte, and (m) Iba1-labeled human microglial cell. 2D or 3D images of (b, f, j, n) reconstructed cell, (c, g, k, o) spanned volume (3D) or spanned area (2D), and (d, h, l, p) cell skeleton illustrating selected morphology parameters of a, e, i, and m analyzed with MotiQ. Scale bars, 20 µm.
Fully automated analysis of histological data sets
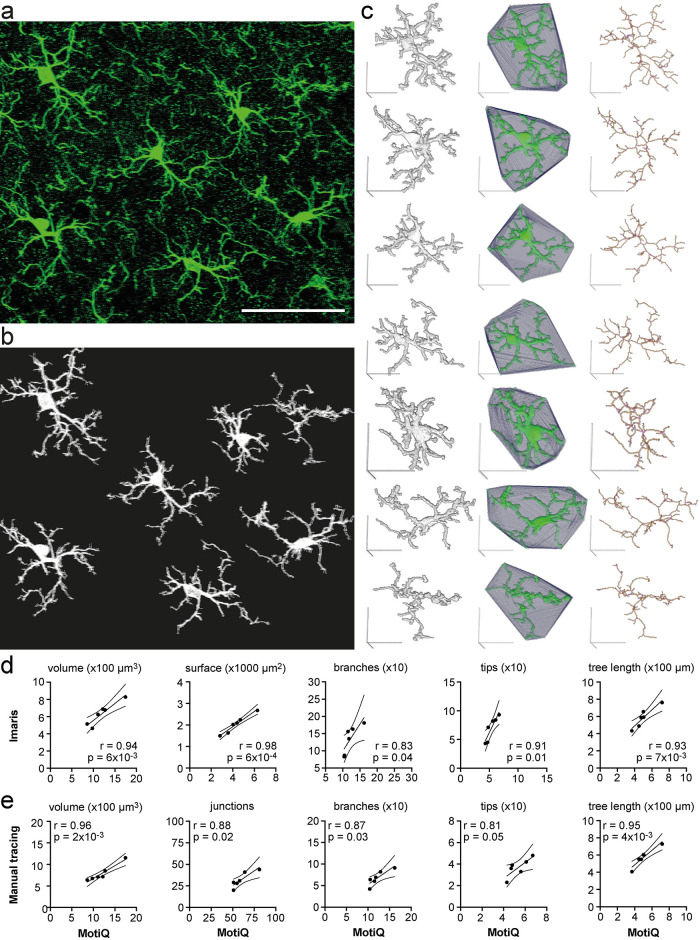
MotiQ 3D analyzer can automatically separate microglial cells without user-dependent cell selection by MotiQ cropper. This is, however, reliably applicable only to high-resolution 3D microscopy data with a high stack depth, as in these data sets cells that are next to each other can be clearly separated and many cells are completely inside the stack so that their entire process tree can be continuously reconstructed. To demonstrate the high-throughput MotiQ analysis, we subjected a whole confocal image of immunofluorescently labeled Iba1-positive murine microglia in fixed cerebral tissue sections to the MotiQ workflow without MotiQ cropper (Figure 5a). In this workflow, MotiQ 3D analyzer was set to track all particles by overlap in the image and analyze each particle separately (see Supplemental Note 3). Indeed, MotiQ could user independently separate and reconstruct individual cells in the stack (Figure 5b) and revealed their morphological characteristics (Figure 5c). To scrutinize the validity and accuracy of this approach, we also subjected the same data set to two other established methods for quantifying microglial morphology: 1) An automated analysis approach based on the commercial software Imaris (Figure 5d) (Gyoneva and Traynelis, 2013; Gyoneva et al., 2014; Hefendehl et al., 2014) and 2) a manual tracing of the individual cells (Baron et al., 2014; Cătălin et al., 2017; Basilico et al., 2019; Sun et al., 2019) based on the Simple Neurite Tracer plug-in for Fiji (Longair et al., 2011; Arshadi et al., 2021) (Figure 5e). Of note, approach 2 required a trained observer about 30 min of user interaction per cell. Importantly, the resulting output parameters significantly correlated with the respective MotiQ parameters (Figure 5, d and e), showing that MotiQ output parameters are qualitatively comparable to these other approaches.
FIGURE 5:
Fully automated analysis of cellular morphology in tissue sections. (a) Maximum-intensity projection of a confocal z stack of Iba1-labeled murine cortical microglia. Scale bar, 50 µm. (b) Maximum-intensity projection of automatically detected cells after analyzing (a) with MotiQ 3D analyzer (using a high size threshold [of 10,000 voxels] to reconstruct cells that are moreover complete in the stack). (c) MotiQ visualizations of each cell: reconstructed cell (left), spanned volume (middle), and cell skeleton (right). Scale bars, 20 µm. (d, e) Direct comparison of the results obtained with MotiQ (c) to results obtained using (d) an alternative automated approach based on the commercial software Imaris or (e) a manual approach using the Fiji plug-in Simple Neurite Tracer (Arshadi et al., 2021). Each data point represents one cell. MotiQ results are indicated on the x-axis. Imaris (d) or manual tracing (e) results are indicated on the y-axis. Lines show mean and 95% confidence interval of a linear regression. r and p values for a Pearson correlation are indicated. Note: Imaris and the manual approach did not allow detection of the top right cell in the image (a), as the cell body is not completely within the stack. Thus, this cell is missing in the correlations.
Analysis of in vitro live-cell data sets
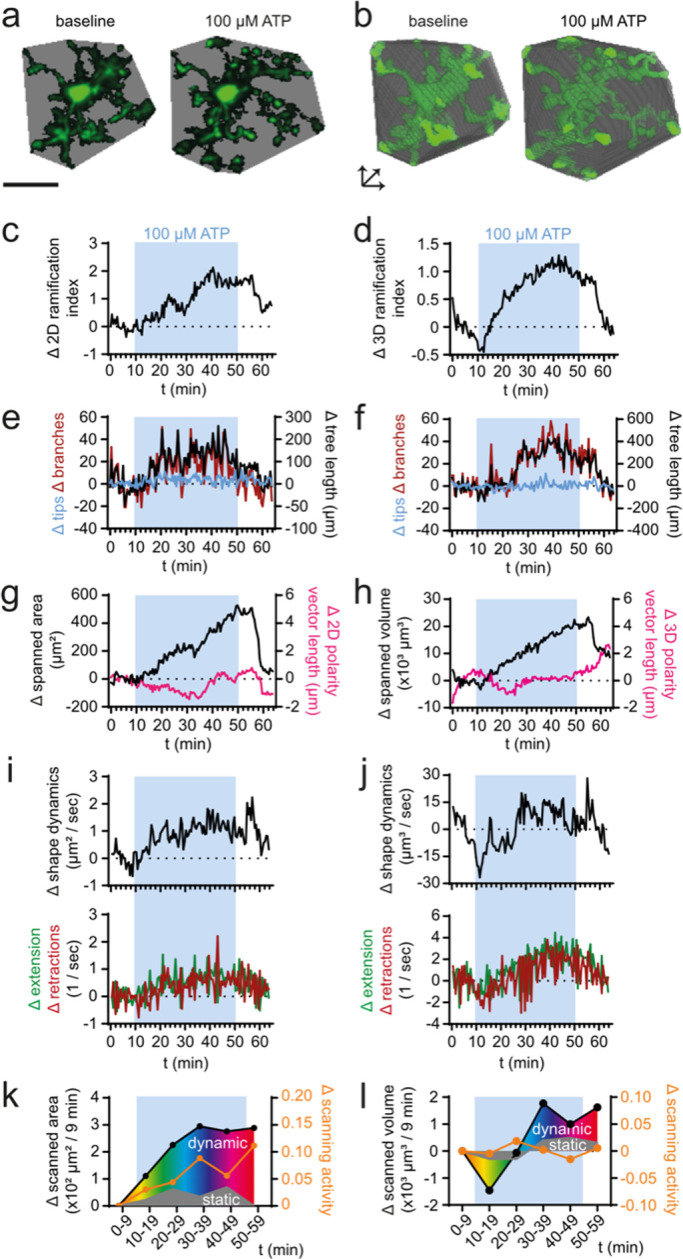
Next, we tested whether MotiQ can be applied to data sets from cultured microglia. The morphology and motility of cultured murine or human microglia can be imaged in a 2D culture system on a plastic or glass surface (Honda et al., 2001; Koizumi et al., 2007) or using 3D culture systems in which microglia are cultured in a 3D matrix, such as Matrigel (Orr et al., 2009) or collagen (Haw et al., 2014), in coculture with other CNS cells (Takata et al., 2017), or in cerebral organoids (Ormel et al., 2018). Here, we acquired time-lapse images of GFP-positive murine primary microglia in a 2D culture system using epifluorescence microscopy and in a 3D Matrigel culture system using spinning-disk confocal microscopy. We added ATP during the imaging experiments to induce changes in microglial morphology and motility.
We subjected the data set of each experimental design to the MotiQ 2D or 3D analysis workflow (Figure 6; Supplemental Videos 3 and 4). Here, we selected slightly adapted MotiQ thresholder settings for optimized image segmentation (Supplemental Table 1). Again, with MotiQ we sensitively detected experimentally induced changes in microglial morphology and motility, that is, in cell polarity (Figure 6, f and m), shape dynamics (Figure 6, d and k), and scanning activity (Figure 6, e and l). Of note, directionality parameters (Figure 6, f and m) are also implemented in MotiQ and can be used when studying chemokinesis or chemotaxis.
FIGURE 6:
Analysis of 2D- and 3D-cultured microglia. (a–f) MotiQ 2D analysis of epifluorescence time-lapse images of a representative primary microglial cell from CX3CR1eGFP/+ mice, cultured in a 2D culture system. (a) False-colored fluorescence intensity images and (b) spanned area before (t = 0 min) and after (t = 11 min 30 s) addition of ATP (100 μM). (c) Cell polarity, (d) shape dynamics, (e) scanned area, scanning activity, and (f) directionality (=accumulated/Euclidean center of mass displacement) of a selected microglial cell after addition of ATP. (g–m) MotiQ 3D analysis of spinning-disk confocal time-lapse images of a representative primary murine microglial cell from Lifeact–GFP+/− mice, cultured in a 3D Matrigel culture system. (g) Maximum-intensity projection images during baseline recording (t = 0 min) and ATP (t = 9 min 30 s) perfusion. (h, i) Spanned volume, (i) polarity vector, (j) 3D ramification index, (k) shape dynamics, (l) scanned area and scanning activity, and (m) directionality. Insets visualize selected measurements. Scale bar, 20 µm.
Movie S3.
Analysis of 2D‐cultured primary microglia
Movie S4.
Analysis of 3D‐cultured primary microglia
Bcause the image segmentation step in MotiQ fully preserves fluorescence intensity information of the cell, MotiQ can also be used to quantify cellular fluorescence intensity levels, kinetics, and distribution. This allows for a direct correlation of microglial morphology and dynamics with cellular fluorescence parameters. As more chemical and genetically encoded fluorescent indicators become available for signaling molecules such as protons, calcium, or second messengers, MotiQ can be used for studying signaling pathways that underlie cellular morphology and motility. To demonstrate such correlative measurements of morphology and motility and fluorescence kinetics, we generated time-lapse images of primary murine macrophages expressing the genetically encoded calcium sensor GCaMP3 during high-dose ATP treatment (5 mM). We then analyzed an exemplary data set using MotiQ (Supplemental Video 5), which revealed a temporal correlation of a transient intracellular/cytosolic calcium increase (Figure 7a) with changes in macrophage ramification (Figure 7b) and scanning activity (Figure 7c) during ATP perfusion.
FIGURE 7:
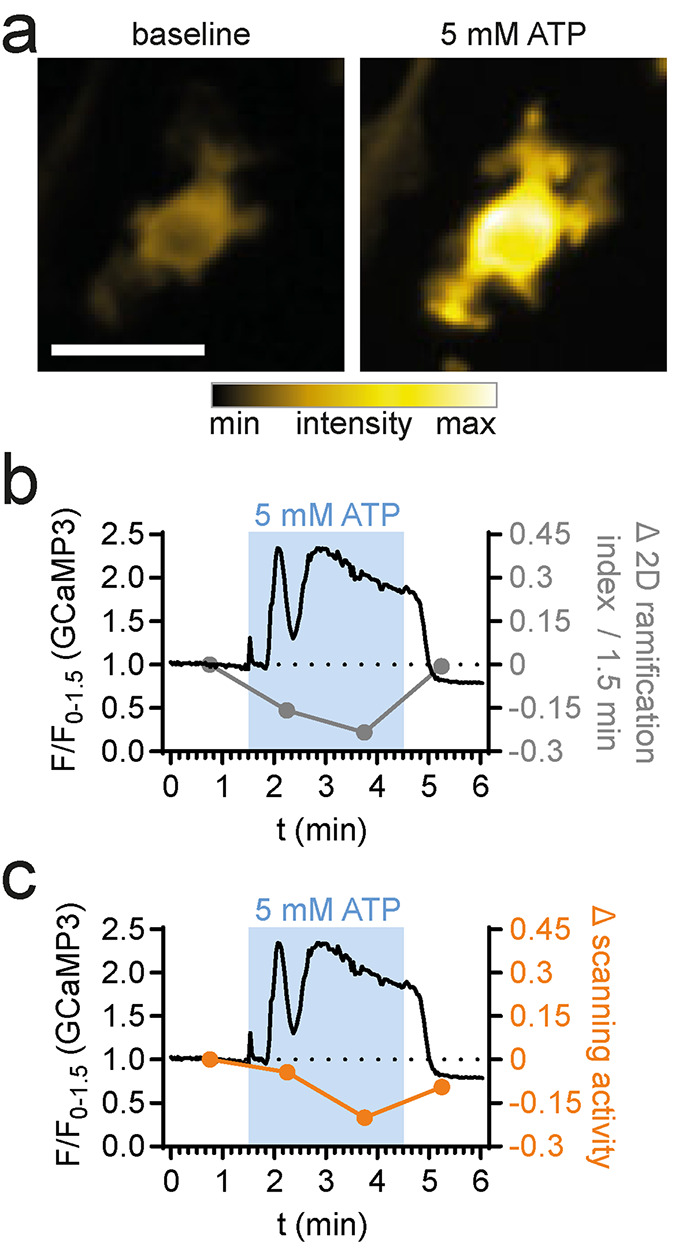
Correlative analysis of morphology, dynamics, and fluorescence kinetics in cultured macrophages. MotiQ 2D analysis of epifluorescent time-lapse images of a 2D-cultured bone marrow–derived macrophage, expressing the genetically encoded calcium sensor GCaMP3. (a) False-colored fluorescence intensity images before and during perfusion with ATP (5 mM). Calcium signals were recorded for 90 s before perfusion with ATP (180 s) and washout. (b, c) Upon ATP perfusion, a calcium signal was detected and temporally correlated to changes in the cell’s (b) 2D ramification index and (c) scanned area. Scale bar, 20 µm.
Movie S5.
Analysis of calcium signal and cellular motility
Analysis of microglia in a mouse model of AD
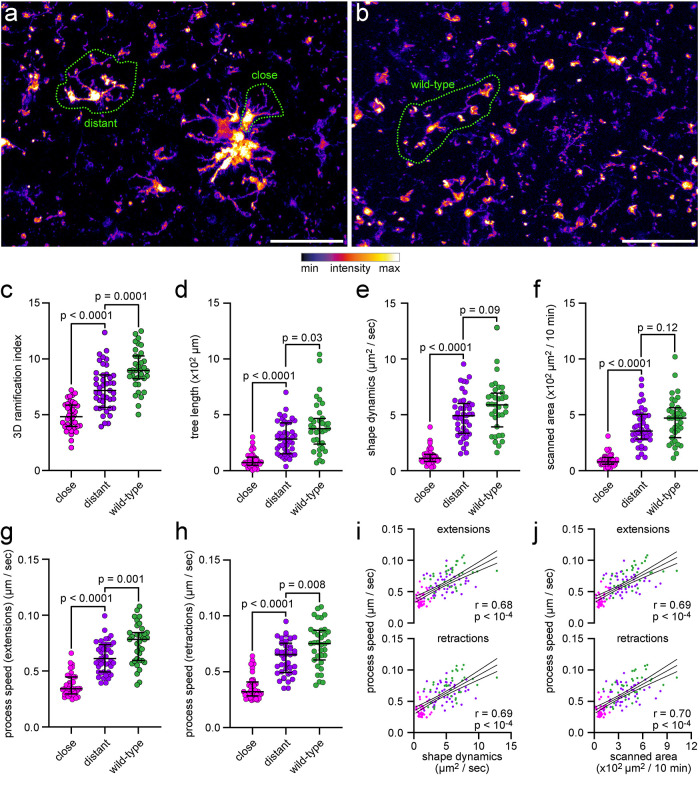
Finally, we assessed the applicability and accuracy of MotiQ on a benchmark data set. We chose to analyze the morphology and motility of microglia in the APP/PS1 mouse model of AD (Jankowsky et al., 2003). It has previously been shown that microglia in AD mouse models show reduced ramification and motility (Bolmont et al., 2008; Koenigsknecht-Talboo et al., 2008; Krabbe et al., 2013; Plescher et al., 2018) in particular when in vicinity to Abeta plaques (Bolmont et al., 2008; Plescher et al., 2018). Thus, we aimed to test whether MotiQ can reliably reveal these pathological changes as well. We obtained acute cerebral slices from APP/PS1 CX3CR1-EGFP/wt mice and “wild-type” CX3CR1-EGFP/wt mice, acquired images with a two-photon microscope (Figure 8, a and b), and subjected them to MotiQ analysis. MotiQ 3D analysis revealed a significant reduction of ramification (Figure 8c) and tree length (Figure 8d) in plaque-associated and plaque-distant microglia, while plaque-associated microglia were mostly affected, in line with previous reports in another AD mouse model (Plescher et al., 2018). Similarly, MotiQ analysis revealed impaired microglial motility in APP/PS1 CX3CR1-EGFP/wt mice, especially at the plaque, reported by the MotiQ parameters shape dynamics (Figure 8e) and scanned area (Figure 8f). We next sought to compare these results to a previously described manual approach for quantifying microglial motility (Nimmerjahn et al., 2005; Bolmont et al., 2008): A trained observer tracked the position of individual microglial process tips manually for all cells. This showed that process extensions and retractions were slower in the AD mouse model, especially at the plaque (Figure 8, g and h). Importantly, the results from manual tracking correlated with the motility parameters determined with MotiQ analysis (Figure 8, i and j).
FIGURE 8:
Analysis of microglial morphology and dynamics in a cerebral amyloidosis mouse model of AD. Analysis of microglial morphology and dynamics in ex vivo two-photon microscopy data of microglia in acute cerebral slices of APP/PS1 CX3CR1EGFP/wt mice or wild-type CX3CR1EGFP/wt. (a, b) Maximum-intensity projection of images from (a) APP/PS1 CX3CR1EGFP/wt mice or (b) wild-type CX3CR1EGFP/wt mice. Scale bar, 50 µm. (c) Ramification index and (d) tree length, determined by MotiQ 3D analysis. (e) Shape dynamics and (f) scanned area, determined by MotiQ 2D analysis. (g–j) To verify the parameter results for shape dynamics and scanned area, for each analyzed cell, all visible processes were also manually tracked in 2D. Based on this tracking, for each cell, the average process speed of (g) process extensions or (h) retractions was determined. (i, j) Correlation of the manually determined process speeds with the MotiQ results shape dynamics (i, shape dynamics plotted alone in e) and scanned area (j, scanned area plotted alone in f). Bar plots (c–h) show the median ± interquartile range; individual data points represent individual cells (n = 3 animals per group; close: 9–14 cells per animal, distant: 14–17 cells per animal, wild type: 10–13 cells per animal); p values for a two-sided Mann–Whitney test are indicated. Data points in (i) and (j) also represent individual cells and are colored as in (c–h). Magenta: plaque-close cells. Purple: plaque-distant cells. Dark green: wild-type cells. Lines show mean and 95% confidence interval of a linear regression. r and p values for a Pearson correlation are indicated.
DISCUSSION
Owing to the paucity of freely accessible and easy-to-use standard tools for image analysis of microglia, approaches for the quantification of microglial morphology and dynamics have largely varied between studies, making comparison of data difficult. To fill this gap, we developed the open-source ImageJ-based software MotiQ that enables the quantification of more than 60 parameters of microglial morphology, motility, and fluorescence kinetics. We furnished MotiQ with a modular structure so that each individual MotiQ plug-in can be potentially used for different approaches. For example, MotiQ thresholder is a flexible tool for image preprocessing and segmentation that offers a variety of thresholding approaches. MotiQ thresholder automatizes image preprocessing and segmentation workflows, fosters reproducibility by logging the individual image preprocessing and segmentation steps, and is readily applicable to other data and workflows.
In MotiQ we integrated several parameters that have become standard for the quantification of microglial morphology and motility to allow the best comparability of output data to those of previous studies. For example, the parameter ramification index, which describes the overall complexity of a microglial cell, has previously been used by us (Fülle et al., 2018; Plescher et al., 2018; Schmöle et al., 2018) and others and is similar to the described parameters surface area-to-volume ratio (Orr et al., 2009; Gyoneva and Traynelis, 2013) or cell circularity (Fernández-Arjona et al., 2017). Parameters that characterize the microglial process tree have been previously used in several studies to quantify microglial morphology changes (Fontainhas et al., 2011; Baron et al., 2014; Hefendehl et al., 2014; Erny et al., 2015; Catalin et al., 2017; Kluge et al., 2017; Fulle et al., 2018; Plescher et al., 2018). The parameters spanned area (2D) or spanned volume (3D), which focus on the interaction of the microglial cell with its environment, have been used in custom-written analysis approaches (Fontainhas et al., 2011; Baron et al., 2014; Fernández-Arjona et al., 2017). In previous studies the number of process extensions and retractions (Koenigsknecht-Talboo et al., 2008) or the speed of single processes (Davalos et al., 2005; Nimmerjahn et al., 2005; Bolmont et al., 2008; Gyoneva et al., 2014; Hefendehl et al., 2014; Paris et al., 2018) have been—mostly manually—quantified as an important parameter of the dynamic behavior of microglia. With MotiQ the number and extent of individual process extensions and retractions are automatically determined, similar to previous custom-written analysis approaches for microglial motility (Kluge et al., 2017; Madry et al., 2018), which, however, were limited to 2D analysis, in contrast to MotiQ, which provides a 3D implementation. Of note, we show that MotiQ’s parameters for microglial morphology and motility directly correlate to results from manual process tracking, highlighting that MotiQ can automatize the manual tracking of processes while revealing functionally similar results.
As novel dynamic parameters, we implemented scanning activity (2D/3D), scanned area (2D), and scanned volume (3D) in MotiQ to quantify the brain volume that is sampled by an individual microglial cell. Moreover, MotiQ offers parameters that describe the directionality of cellular movement, which are important when measuring the chemotactic response of microglia toward stimuli, especially in in vitro experiments, but potentially also in vivo. Importantly, MotiQ enables correlative studies on morphology, dynamics, and fluorescence kinetics in a cell to address the role of cellular signaling pathways using fluorescent indicators such as calcium- or pH-sensitive dyes.
MotiQ can be applied to a variety of data sets, including in vivo and ex vivo two-photon microscopy of microglia, confocal endpoint analysis of microglial morphology in fixed tissue sections, and epifluorescence and confocal live-cell microscopy of cultured microglia. Of note, in high-resolution images, where adjacent microglial process can be clearly separated, MotiQ could fully automatically reconstruct microglial cells without any user-dependent cell selection, while revealing similar results compared with those of a time-consuming manual analysis. Thus, under these conditions, MotiQ allows analyzing microglial morphology with high throughput and, thereby, performing detailed morphological studies on microglia with high statistical power.
Furthermore, MotiQ is not restricted to study murine or human microglia but could also be used to characterize other glial cells such as astrocytes or the dynamics of subcellular structures such as neuronal growth cones or primary cilia.
The only comparable widely used alternative for studying microglial morphology and dynamics is the commercial software Imaris. However, Imaris is not affordable for every lab, and the underlying code and algorithms are not freely available. In contrast, the source code of MotiQ is freely accessible (https://github.com/hansenjn/MotiQ), making the analysis procedure transparent and customizable.
MotiQ offers a precise and comprehensive analysis of microglial morphology and motility. We envision that MotiQ will become a tool in microglial research used by many labs, fostering comparability of data and facilitating investigations on pathways controlling microglial morphology and the interaction of microglia with their environment.
MATERIALS AND METHODS
Animals
Lifeact–GFP (Clausen et al., 1999; Riedl et al., 2010), LysMcre (Clausen et al., 1999), Ai38GCaMP (Zariwala et al., 2012), APP/PS1 (Jankowsky et al., 2003), and CX3CR1EGFP/wt (Jung et al., 2000) mice have been described previously. C57BL/6J mice were purchased from Janvier Labs. CX3CR1EGFP/wt and Ai38-GCaMP mice were purchased from Jackson Laboratories. Lifeact–GFP mice were provided by Frank Bradke (DZNE, Bonn, Germany) with permission of Michael Sixt (IST, Austria). Mice were housed in groups on a 12-h light/dark cycle with food and water available ad libitum. Research and animal care procedures were approved by the Landesamt für Natur, Umwelt und Verbraucherschutz of North Rhine-Westphalia (Germany).
Human brain tissue
Postmortem tissue samples from a nonneurological, nonneuropathological 62-yr-old control case patient were obtained from the University Hospital Southampton NHS Foundation Trust as part of the UK Brain Archive Information Network (BRAIN UK), which is funded by the Medical Research Council, and Brain Tumour Research. The use of human tissue was covered by the ethical approval provided by BRAIN UK (Research Ethics Committee South Central Hampshire B, reference 14/SC/0098).
In vivo two-photon imaging
CX3CR1EGFP/wt mice were anesthetized with isoflurane (1.5–3% in 100% oxygen at 0.7–1 l min–1). Body temperature was kept at 37°C. Hair, skin, and periosteum were removed, and the skull was fixed in a custom-made metal frame using a cyano veneer fast/cyano veneer powder mixture (Hager & Werken, Duisburg, Germany). The metal frame was fixed in a custom-made stereotactic device. A cranial window (3 mm diameter) was created using a dental drill, filled with agarose (37°C; 3% in phosphate-buffered saline) and sealed with a cover glass (Hugo Sachs). Two-photon imaging of anesthetized CX3CR1EGFP/wt mice was performed with an upright two-photon microscope (Trim Scope; LaVision Biotec, Bielefeld, Germany) equipped with a 20× objective (1.0 NA; Zeiss). EGFP was excited at 870 nm with a Ti:Sapphire laser (Chameleon Ultra II, pumped by an 18 W laser; Coherent) and scanned at 516–556 nm. Laser power below the objective was 10–20 mW. The image frame rate was 1.6 s at a resolution of 993 × 993 pixels. Image stacks were recorded starting at ∼40 µm below the pial surface of the cortex (36 s intervals, z stack size 60 µm with z slice intervals of 3 µm).
Ex vivo two-photon imaging of acute cerebral slices
CX3CR1EGFP/wt mice were killed and decapitated, and the brains were immediately transferred to ice-cold aCSF (NaCl 134 mM; KCl 2.5 mM; MgCl2 1.3 mM; CaCl2 2 mM; K2HPO4 1.26 mM; NaHCO3 26 mM, d-glucose 10 mM; pH 7.4; saturated with carbogene). Serial coronal cerebral sections (300 µm) were prepared using a Leica VT1200S vibratome (speed: 0.1 mm/s; amplitude: 1.5 mm). Acute slices were fixed in a custom-built perfusion chamber using a harp slice grip (ALA Scientific Instruments) and perfused at a speed of 1.5 ml/min with aerated aCSF. Imaging was performed using the same two-photon system as described above. Image stacks were recorded starting 20 µm below the coronal surface of the acute cerebral slice (30.8 s stack intervals, z stack size 60 µm with z slice intervals of 3 µm). After recording for 10 min, 100 µM ATP (Sigma-Aldrich) dissolved in aCSF was washed in for 40 min. Finally, ATP was washed out for 10 min.
Preparation, staining, and imaging of fixed tissue sections
For visualization and analysis of cortical microglia, the brains of 8-mo-old mice (C57BL6J/C3HHeJ) were immersion-fixed in 4% paraformaldehyde (PFA) and microglia were stained in 100-µm-thick coronal cryosections with anti-Iba1 (Wako; 019-19741; 1 µg/ml) and goat anti-rabbit Alexa Fluor 488 (Invitrogen; 3 µg/ml). Confocal imaging was performed with a Nikon Ti-E Confocal Microscope (Nikon, Tokyo, Japan; z stack size 40 µm with z slice intervals of 1 µm; Figure 4a) or with a Zeiss LSM 700 Confocal Microscope (Carl Zeiss, Germany; z stack size 20 µm with z slice intervals of 0.5 µm; Figure 5). Astrocytes were visualized in 40-µm-thick sagittal vibratome sections from 8–12-wk-old mice (C57BL/6J-RCCHsd). Astrocytes were stained in free-floating sections with anti-GFAP (Abcam; ab7260; 1:500) and goat anti-rabbit Alexa Fluor 594 (Invitrogen; 4 µg/ml) and imaged with a Zeiss LSM 780 Confocal Microscope (Carl Zeiss AG, Oberkochen, Germany; z stack size 30–40 µm with z slice intervals of 1 µm). For analysis of retinal microglia, the eyes of 3-mo-old C57BL/6J mice were enucleated and fixed and stained as described previously (Dannhausen et al., 2015). Retinal whole mounts were imaged using an AxioImager.M2plus ApoTome2 microscope (Carl Zeiss, Germany).
For visualization of human microglia, 50-µm-thick Formalin-fixed, paraffin-embedded sections from a postmortem sample of human temporal cortex were incubated with anti-Iba1 (Wako; 019-19741; 1:500) for 24 h after heat retrieval and subsequently incubated with goat anti-rabbit Alexa Fluor 488 (Invitrogen; 1:200; 2 h), followed by 70% Sudan Black incubation to reduce autofluorescence. Counterstaining was obtained with SYTOX Orange (Invitrogen; 25 nM). Confocal images were generated in the gray matter using a Leica TCS SP8 laser scanning confocal microscope system equipped with a 63× objective, using a 3.0× digital zoom. Z stacks were acquired with a z slice interval of 0.5 µm.
Primary microglia
Primary mixed glial cultures were generated to obtain cultured microglia as described (Krabbe et al., 2013). Briefly, newborn CX3CR1EGFP/wt or Lifeact–GFP+/− mice were decapitated. The brains were removed and stripped of their meninges. The brains were mechanically dissociated after incubation with trypsin-EDTA (0.05%) and cultivated in poly-l-lysine (PLL)-coated culture flasks in DMEM (Life Technologies, Darmstadt, Germany), supplemented with 10% fetal bovine serum (FBS) (Biochrom, Berlin, Germany) and 1% penicillin/streptomycin (Life Technologies, Darmstadt, Germany). After 5–7 d, proliferation of microglia was stimulated by adding supernatant of the murine fibroblastic cell line L929.
Epifluorescence imaging of primary microglia in a 2D culture system
Microglia were obtained from the culture supernatant of primary mixed glial cultures after vigorous shaking and seeded on PLL-coated eight-well plates (Ibidi, Munich, Germany) in phenol red–free and FBS-free DMEM (Life Technologies, Darmstadt, Germany; supplemented with 1% glutamate, 1% pyruvate, 1% penicillin/streptomycin). Cells were imaged at 37°C and 10% CO2 using an epifluorescence microscope (Nikon Ti-E; Nikon, Tokyo, Japan) equipped with a 20× objective (Nikon, Tokyo, Japan; NA 0.7) and an EGFP filter set (AHF Analysetechnik, Tübingen, Germany).
Spinning-disk confocal imaging of primary microglia in a 3D culture system
Phenol red–free Corning Matrigel Matrix (VWR, Radnor, PA) was thawed and placed on top of 5 mm coverslips (gel thickness ∼100 µm). After solidification of the gel at 37°C (20–30 min), microglia (in DMEM, 10% FBS, 1% penicillin/streptomycin) obtained from primary mixed glial cultures of Lifeact–GFP+/− mice were put on top of the gel in order to enter the gel. During imaging, the prepared coverslips were continuously perfused with Hank’s balanced salt solution (HBSS) buffer. Imaging was performed with a spinning-disk confocal microscope (Olympus, Tokyo, Japan; 60× objective; time interval 30 s; z stacks 25–35 µm with z slice intervals of 1 µm). After 5 min of baseline recording, perfusion buffer was switched to HBSS containing 100 µM ATP (Sigma-Aldrich) for 5 min and then switched again to pure HBSS for 5 min.
Primary bone marrow–derived macrophage cultures
The tibia and femur of heterozygous Ai38-GCaMP:LysMcre mice were prepared, and the bone marrow was washed out and collected in RPMI medium. After lysis of blood cells, the cell pellet was resuspended in 5 ml of RPMI medium, passed through a 40 µm cell strainer, and cultivated in RPMI medium containing 10% FBS and 1% penicillin/streptomycin at 37°C and 5% CO2. Cells were used for experiments after day in vitro 10.
Epifluorescence imaging of primary macrophages
Ai38-GCaMP–positive bone marrow–derived macrophages were seeded on glass coverslips 1 d before experiments. Imaging was performed at room temperature, and a gravity-driven perfusion system was used for continuous perfusion with a flow rate between 0.5 and 1 ml/min. Images were acquired every 2 s with a 20× objective (Nikon, Tokyo, Japan; NA 0.7) using an epifluorescence microscope (Nikon Ti-E; Nikon, Tokyo, Japan) equipped with an EGFP filter set (AHF Analysetechnik, Tübingen, Germany) at room temperature. A focus system (Nikon PFS, Nikon, Tokyo, Japan) was used to correct z drift.
Image preprocessing
Epifluorescence microscopy images of microglia in a 2D culture were preprocessed by Bleach Correction using the ImageJ plug-in CorrectBleach by Kota Miura (correction method: histogram matching). Time-lapse images were registered using the ImageJ plug-in MultiStackReg (Thevenaz et al., 1998). 3D images were intensity normalized in the z direction using the ImageJ function Enhance contrast (0.5%; normalized). All images were preprocessed using the ImageJ function Smooth with two exceptions: 1) The confocal image of human microglia was preprocessed with Gaussian Blur (2 pixel), as it harbored a higher resolution than other images requiring a higher smoothing than performed by the Smooth function; 2) the confocal image stack of Iba1-labeled microglia that was used for full-automated cell separation of individual microglia was preprocessed using the ImageJ function Subtract Background (50 pixel) to reduce regional intensity differences.
MotiQ plug-ins and microglial parameters
The MotiQ analysis workflow is described in the manual provided via the GitHub repository https://github.com/hansenjn/MotiQ. For a detailed description of all MotiQ plug-ins, see Supplemental Notes 1–4. MotiQ thresholder settings for image processing of data shown here are listed in Supplemental Table 1. Microglial morphological, dynamic, and signaling parameters are described in detail in Supplemental Tables 2–4.
Analysis based on Imaris
In Imaris, the surface module was used to select the area covered by microglia (Iba1 channel; smoothing 0.5 µm). To eliminate noise, a manual intensity threshold (18.9338) was applied and a second threshold was adjusted manually to filter for complete cells (>∼30,000 voxels). A mask was created based on the surface and used as input for filament remodeling (7 µm was set as soma size for starting points, 0.5 µm as process diameter for seed points). The thresholds for starting points (28.984) and seed points (110.201) were adjusted manually. Seed points that were within 14 µm around the starting points and disconnected segments were removed and smoothing (0.1 µm) was applied. To obtain continuous processes, an automated maximal gap length (7 µm) was used. The parameters dendrite volume, dendrite area, filament length, number of dendrite branch points, number of dendrite terminal points, and number of dendrite segments were chosen for comparison to the respective parameter in MotiQ (3D volume, surface, tree length, number of junctions, number of tips, and number of branches).
Manual tracing of the process tree in Fiji using Simple Neurite Tracer
Manual microglial process tracing was performed using the Simple Neurite Tracer plug-in in Fiji (Arshadi et al., 2021). All primary branches of a reconstructed cell were combined in a shared root. The skeleton parameters cable length, number of branch points, number of tips, and number of branches were used to compare the manual tracing approach parameters to the respective parameters in MotiQ (process tree length, number of junctions, number of tips, and number of branches). To determine the cellular volume, the “fill” option (threshold set at 0.010614749425259) was used.
Manual tracking of process tips
Manual tracking of the microglial process tips was performed in ImageJ. For each clearly visible process, a trained observer set a point-ROI to the center of the process tip in each time step and noted whether the process extended or retracted in that time step. From the point-ROI the coordinates were extracted using ImageJ. The speed of the extension or retraction was then determined as the displacement of the process tip between two consecutive time steps.
Statistics
Statistical analysis was performed in GraphPad Prism, Version 9.3.1. All correlations (Figures 5, d and e, and 8, i and j) have been tested for linearity (requirement for a Pearson correlation): A simple linear regression including a runs test was applied. No significant departure from linearity was detected for any correlation tested, and thus, a Pearson correlation was used for all correlations. Column data sets (Figure 8, c–h) were tested for normal distribution using a D’Agostino and Pearson normality test. Not all conditions were normal distributed. Thus, a Mann–Whitney test was selected for statistical comparison.
Software and code availability
MotiQ source code and software, along with a manual, are available online (https://github.com/hansenjn/MotiQ, license GPL-3.0).
Supplementary Material
Acknowledgments
We thank Yvonne Biederbick and Antje Baumgartner for excellent technical assistance. We also thank Luis Alvarez for helping with the visualization of the center of mass displacement over time in 3D. This work was supported by grants from the German Science Foundation (EXC 1023 ImmunoSensation), Helmholtz Impuls- und Vernetzungsfonds and CoEN 5008. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC2151-390873048.
Abbreviations used:
- ATP
adenosine triphosphate
- 2D
two dimensional
- 3D
three dimensional.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0585) on June 22, 2022.
REFERENCES
- Abdolhoseini M, Walker F, Johnson S (2016). Automated tracing of microglia using multilevel thresholding and minimum spanning trees. Annu Int Conf IEEE Eng Med Biol Soc 2016, 1208–1211. [DOI] [PubMed] [Google Scholar]
- Arganda-Carreras I, Fernández-González R, Muñoz-Barrutia A, Ortiz-De-Solorzano C (2010). 3D reconstruction of histological sections: application to mammary gland tissue. Microsc Res Tech 73, 1019–1029. [DOI] [PubMed] [Google Scholar]
- Arshadi C, Günther U, Eddison M, Harrington KIS, Ferreira TA (2021). SNT: a unifying toolbox for quantification of neuronal anatomy. Nat Methods 18, 374–377. [DOI] [PubMed] [Google Scholar]
- Badimon A, Strasburger HJ, Ayata P, Chen X, Nair A, Ikegami A, Hwang P, Chan AT, Graves SM, Uweru JO (2020). Negative feedback control of neuronal activity by microglia. Nature 586, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R, Babcock AA, Nemirovsky A, Finsen B, Monsonego A (2014). Accelerated microglial pathology is associated with Aβ plaques in mouse models of Alzheimer’s disease. Aging Cell 13, 584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilico B, Cortese B, Ratano P, Angelantonio SD, Ragozzino D (2019). Time-lapse whole-field fluorescence imaging of microglia processes motility in acute mouse hippocampal slices and analysis. Bio Protoc 9, e3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolmont T, Haiss F, Eicke D, Radde R, Mathis CA, Klunk WE, Kohsaka S, Jucker M, Calhoun ME (2008). Dynamics of the microglial/amyloid interaction indicate a role in plaque maintenance. J Neurosci 28, 4283–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cătălin B, Stopper L, Bălşeanu T-A, Scheller A (2017). The in situ morphology of microglia is highly sensitive to the mode of tissue fixation. J Chem Neuroanat 86, 59–66. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8, 265–277. [DOI] [PubMed] [Google Scholar]
- Conde JR, Streit WJ (2006). Microglia in the aging brain. J Neuropathol Exp Neurol 65, 199–203. [DOI] [PubMed] [Google Scholar]
- Condello C, Yuan P, Schain A, Grutzendler J (2015). Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat Commun 6, 6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannhausen K, Karlstetter M, Caramoy A, Volz C, Jägle H, Liebisch G, Utermöhlen O, Langmann T (2015). Acid sphingomyelinase (aSMase) deficiency leads to abnormal microglia behavior and disturbed retinal function. Biochem Biophys Res Commun 464, 434–440. [DOI] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, Gan W-B (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nat Neurosci 8, 752–758. [DOI] [PubMed] [Google Scholar]
- De Picker LJ, Victoriano GM, Richards R, Gorvett AJ, Lyons S, Buckland GR, Tofani T, Norman JL, Chatelet DS, Nicoll JAR, Bochec D (2021). Immune environment of the brain in schizophrenia and during the psychotic episode: a human post-mortem study. Brain Behav Immun 97, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissing-Olesen L, LeDue JM, Rungta RL, Hefendehl JK, Choi HB, MacVicar BA (2014). Activation of neuronal NMDA receptors triggers transient ATP-mediated microglial process outgrowth. J Neurosci 34, 10511–10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D, Hrabě de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, Keren-Shaul H, Mahlakoiv T, Jakobshagen K, Buch T, et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 18, 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Arjona MDM, Grondona JM, Granados-Durán P, Fernández-Llebrez P, López-Ávalos MD (2017). Microglia morphological categorization in a rat model of neuroinflammation by hierarchical cluster and principal components analysis. Front Cell Neurosci 11, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontainhas AM, Wang M, Liang KJ, Chen S, Mettu P, Damani M, Fariss RN, Li W, Wong WT (2011). Microglial morphology and dynamic behavior is regulated by ionotropic glutamatergic and GABAergic neurotransmission. PLoS One 6, e15973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Bocanegra DK, Gourari Y, McAuley C, Chatelet DS, Johnston DA, Nicoll JAR, Boche D (2021). Microglial morphology in Alzheimer’s disease and after Aβ immunotherapy. Sci Rep 11, 15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fülle L, Offermann N, Hansen JN, Breithausen B, Erazo AB, Schanz O, Radau L, Gondorf F, Knöpper K, Alferink J, et al. (2018). CCL17 exerts a neuroimmune modulatory function and is expressed in hippocampal neurons. Glia 66, 2246–2261. [DOI] [PubMed] [Google Scholar]
- Gyoneva S, Davalos D, Biswas D, Swanger SA, Garnier-Amblard E, Loth F, Akassoglou K, Traynelis SF (2014). Systemic inflammation regulates microglial responses to tissue damage in vivo. Glia 62, 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyoneva S, Traynelis SF (2013). Norepinephrine modulates the motility of resting and activated microglia via different adrenergic receptors. J Biol Chem 288, 15291–15302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haw RTY, Tong CK, Yew A, Lee HC, Phillips JB, Vidyadaran S (2014). A three-dimensional collagen construct to model lipopolysaccharide-induced activation of BV2 microglia. J Neuroinflammation 11, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefendehl JK, Neher JJ, Sühs RB, Kohsaka S, Skodras A, Jucker M (2014). Homeostatic and injury-induced microglia behavior in the aging brain. Aging Cell 13, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Sasaki Y, Ohsawa K, Imai Y, Nakamura Y, Inoue K, Kohsaka S (2001). Extracellular ATP or ADP induce chemotaxis of cultured microglia through Gi/o-coupled P2Y receptors. J Neurosci 21, 1975–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Dissing-Olesen L, Stevens B (2016). New insights on the role of microglia in synaptic pruning in health and disease. Curr Opin Neurobiol 36, 128–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Xu G, Fromholt D, Gonzales V, Borchelt DR (2003). Environmental enrichment exacerbates amyloid plaque formation in a transgenic mouse model of Alzheimer disease. J Neuropathol Exp Neurol 62, 1220–1227. [DOI] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR (2000). Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20, 4106–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge MG, Kracht L, Abdolhoseini M, Ong LK, Johnson SJ, Nilsson M, Walker FR (2017). Impaired microglia process dynamics post-stroke are specific to sites of secondary neurodegeneration. Glia 65, 1885–1899. [DOI] [PubMed] [Google Scholar]
- Koenigsknecht-Talboo J, Meyer-Luehmann M, Parsadanian M, Garcia-Alloza M, Finn MB, Hyman BT, Bacskai BJ, Holtzman DM (2008). Rapid microglial response around amyloid pathology after systemic anti-Abeta antibody administration in PDAPP mice. J Neurosci 28, 14156–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, Shinozaki Y, Ohsawa K, Tsuda M, Joshi BV, Jacobson KA, Kohsaka S, Inoue K (2007). UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature 446, 1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krabbe G, Halle A, Matyash V, Rinnenthal JL, Eom GD, Bernhardt U, Miller KR, Prokop S, Kettenmann H, Heppner FL (2013). Functional impairment of microglia coincides with beta-amyloid deposition in mice with Alzheimer-like pathology. PLoS One 8, e60921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriazis AD, Noroozizadeh S, Refaee A, Choi W, Chu L-T, Bashir A, Cheng WH, Zhao R, Namjoshi DR, Salcudean SE, et al. (2019). An end-to-end system for automatic characterization of Iba1 immunopositive microglia in whole slide imaging. Neuroinformatics 17, 373–389. [DOI] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD (2011). Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453–2454. [DOI] [PubMed] [Google Scholar]
- Madry C, Kyrargyri V, Arancibia-Cárcamo IL, Jolivet R, Kohsaka S, Bryan RM, Attwell D (2018). Microglial ramification, surveillance, and interleukin-1β release are regulated by the two-pore domain K+ channel THIK-1. Neuron 97, 299–312.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash M, Zabiegalov O, Wendt S, Matyash V, Kettenmann H (2017). The adenosine generating enzymes CD39/CD73 control microglial processes ramification in the mouse brain. PLoS One 12, e0175012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini M, Rafalski VA, Ma K, Kim KY, Bushong EA, Rios Coronado PE, Yan Z, Mendiola AS, Sozmen EG, Ryu JK, et al. (2021). Microglial Gi-dependent dynamics regulate brain network hyperexcitability. Nat Neurosci 24, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto A, Wake H, Ishikawa AW, Eto K, Shibata K, Murakoshi H, Koizumi S, Moorhouse AJ, Yoshimura Y, Nabekura J (2016). Microglia contact induces synapse formation in developing somatosensory cortex. Nat Commun 7, 12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318. [DOI] [PubMed] [Google Scholar]
- Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, Johansen LE, van Dijk RE, Scheefhals N, van Berlekom AB, et al. (2018). Microglia innately develop within cerebral organoids. Nat Commun 9, 4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr AG, Orr AL, Li X-J, Gross RE, Traynelis SF (2009). Adenosine A(2A) receptor mediates microglial process retraction. Nat Neurosci 12, 872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Paolicelli RC, Murana E, Cortese B, Angelantonio SD, Zurolo E, Guiducci E, Ferreira TA, Garofalo S, Catalano M, et al. (2015). Defective microglial development in the hippocampus of Cx3cr1 deficient mice. Front Cell Neurosci 9, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris I, Savage JC, Escobar L, Abiega O, Gagnon S, Hui C-W, Tremblay M-È, Sierra A, Valero J (2018). ProMoIJ: a new tool for automatic three-dimensional analysis of microglial process motility. Glia 66, 828–845. [DOI] [PubMed] [Google Scholar]
- Plescher M, Seifert G, Hansen JN, Bedner P, Steinhäuser C, Halle A (2018). Plaque-dependent morphological and electrophysiological heterogeneity of microglia in an Alzheimer’s disease mouse model. Glia 66, 1464–1480. [DOI] [PubMed] [Google Scholar]
- Riedl J, Flynn KC, Raducanu A, Gärtner F, Beck G, Bösl M, Bradke F, Massberg S, Aszodi A, Sixt M, Wedlich-Söldner R (2010). Lifeact mice for studying F-actin dynamics. Nat Methods 7, 168–169. [DOI] [PubMed] [Google Scholar]
- Schmöle A-C, Lundt R, Toporowski G, Hansen JN, Beins E, Halle A, Zimmer A (2018). Cannabinoid receptor 2-deficiency ameliorates disease symptoms in a mouse model with Alzheimer’s disease-like pathology. J Alzheimers Dis 64, 379–392. [DOI] [PubMed] [Google Scholar]
- Sipe GO, Lowery RL, Tremblay MÈ, Kelly EA, Lamantia CE, Majewska AK (2016). Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat Commun 7, 10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Suzuki K, Toptunov D, Stoyanov S, Yuzaki M, Khiroug L, Dityatev A (2019). In vivo two-photon imaging of anesthesia-specific alterations in microglial surveillance and photodamage-directed motility in mouse cortex. Front Neurosci 13, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K, Kozaki T, Lee CZW, Thion MS, Otsuka M, Lim S, Utami KH, Fidan K, Park DS, Malleret B, et al. (2017). Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity 47, 183–198.e6. [DOI] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, Unser M (1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7, 27–41. [DOI] [PubMed] [Google Scholar]
- Weinhard L, di Bartolomei G, Bolasco G, Machado P, Schieber NL, Neniskyte U, Exiga M, Vadisiute A, Raggioli A, Schertel A, et al. (2018). Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat Commun 9, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K, Morrison H (2018). Quantifying microglia morphology from photomicrographs of immunohistochemistry prepared tissue using ImageJ. J Vis Exp 2018, 57648. [DOI] [PMC free article] [PubMed]
- Zariwala HA, Borghuis BG, Hoogland TM, Madisen L, Tian L, De Zeeuw CI, Zeng H, Looger LL, Svoboda K, Chen T-W (2012). A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J Neurosci 32, 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.