Abstract
Arrays of actin filaments (F-actin) near the apical surface of epithelial cells (medioapical arrays) contribute to apical constriction and morphogenesis throughout phylogeny. Here, superresolution approaches (grazing incidence structured illumination, GI-SIM, and lattice light sheet, LLSM) microscopy resolve individual, fluorescently labeled F-actin and bipolar myosin filaments that drive amnioserosa cell shape changes during dorsal closure in Drosophila. In expanded cells, F-actin and myosin form loose, apically domed meshworks at the plasma membrane. The arrays condense as cells contract, drawing the domes into the plane of the junctional belts. As condensation continues, individual filaments are no longer uniformly apparent. As cells expand, arrays of actomyosin are again resolved—some F-actin turnover likely occurs, but a large fraction of existing filaments rearrange. In morphologically isotropic cells, actin filaments are randomly oriented and during contraction are drawn together but remain essentially randomly oriented. In anisotropic cells, largely parallel actin filaments are drawn closer to one another. Our images offer unparalleled resolution of F-actin in embryonic tissue, show that medioapical arrays are tightly apposed to the plasma membrane and are continuous with meshworks of lamellar F-actin. Medioapical arrays thereby constitute modified cell cortex. In concert with other tagged array components, superresolution imaging of live specimens will offer new understanding of cortical architecture and function.
INTRODUCTION
Apical constriction, driven by contractility in medioapical arrays and junctional belts, is a mechanism utilized by epithelial cells across phylogeny to reduce apical area and promote cell and tissue reorganization (reviewed in Martin and Goldstein, 2014; Gorfinkiel, 2016; Coravos et al., 2017; Hunter and Fernandez-Gonzalez, 2017; Harris, 2018; Sutherland and Lesko, 2020). Sustained contraction causes bends in cell sheets and can result in cell ingression, extrusion, or subduction. Alternatively, cells can undergo cycles of contraction and expansion—they pulsate or oscillate. In some cases such pulsations lead to a sustained contraction through proposed clutch- and ratchet-like functions (Franke et al., 2005; Fernandez et al., 2007; Martin et al., 2008, 2010; Ma et al., 2009; Solon et al., 2009; Blanchard et al., 2010; David et al., 2010; Azevedo et al., 2011; Gorfinkiel and Blanchard, 2011; Roh-Johnson et al., 2012; Sokolow et al., 2012; Hara et al., 2016; Jurado et al., 2016; Marston et al., 2016; Mason et al., 2016; Lardennois et al., 2019; Miao and Blankenship, 2020).
In dorsal closure, a morphogenic process occurring halfway through embryogenesis of Drosophila, two sheets of lateral epidermis close over a dorsal hole (reviewed in Hayes and Solon, 2017; Kiehart et al., 2017; Aristotelous et al., 2018). The amnioserosa cells filling this hole pulsate and then ingress from the tissue plane and apoptose. They are integrated into a continuous epidermal cell sheet and pulsate asynchronously, so that both cell autonomous and cell nonautonomous forces (produced by neighboring cells) play a role in their shape changes (Franke et al., 2005; Jayasinghe et al., 2013; Hunter et al., 2014; Machado et al., 2015). Nonmuscle myosin II (hereafter myosin) in concert with actin filaments forms the medioapical arrays that along with junctional belts contribute to apical constriction of amnioserosa cells (Franke et al., 2005; Fernandez et al., 2007; Ma et al., 2009; Solon et al., 2009; Blanchard et al., 2010; David et al., 2010; Azevedo et al., 2011; Gorfinkiel and Blanchard, 2011; Sokolow et al., 2012; Hara et al., 2016; Jurado et al., 2016; Sumi et al., 2018; Dehapiot et al., 2020; Miao and Blankenship, 2020).
Contraction correlates temporally with increased levels of actin and myosin, seen by confocal microscopy as mobile, fluorescent patches that fluctuate in intensity during both transient, pulsatile constrictions and ingression (Franke et al., 2005; Ma et al., 2009; Blanchard et al., 2010; David et al., 2010; Azevedo et al., 2011; Saravanan et al., 2013; Fischer et al., 2014; Munjal et al., 2015; Duque and Gorfinkiel, 2016; Pasakarnis et al., 2016; Goodwin et al., 2017; Dehapiot et al., 2020). Manipulation of forces, through laser surgery, or manipulation of actin or myosin, with drugs or through genetic or molecular genetic strategies that include mutations in myosin and cytoskeletal regulators, abolishes or alters the kinetics and duration of contractions (Kiehart et al., 2000; Franke et al., 2005; Fernandez et al., 2007; Peralta et al., 2008; Solon et al., 2009; Blanchard et al., 2010; David et al., 2010; Azevedo et al., 2011; Fischer et al., 2014; Machado et al., 2015; Duque and Gorfinkiel, 2016; Sumi et al., 2018). Nevertheless, limitations in resolution for live imaging have prevented a detailed structural investigation of the properties of the medioapical arrays and their relationship to the cell’s membrane, cortex, and junctional belts.
To understand better how actin and myosin are arranged to cause medioapical array function and the apical pulsations that they drive, we image genetically encoded, fluorescently tagged cytoskeletal or junctional proteins to observe real-time cytoskeletal and junctional kinematics using superresolution approaches on living Drosophila embryos undergoing dorsal closure. Grazing incidence structured illumination microscopy (GI-SIM; Guo et al., 2018) provides increased spatial resolution (below 100 nm), allowing us to track individual actin and myosin filaments during and across cycles of cellular contraction and expansion. Lattice light sheet microscopy (LLSM; Chen et al., 2014) provides a three-dimensional (3D) view of dorsal closure and shows that the medioapical arrays lie juxtaposed to the domed apical surface of the amnioserosa cells and thereby establishes that medioapical arrays are modified actin cortex and not an additional, actomyosin-rich structure. Interestingly, in morphologically isotropic cells, actin filaments are arranged with random orientations. When such isotropic cells contract the filaments condense—when such cells expand the filamentous networks relax and F-actin and bipolar myosin filaments remain randomly distributed. In contrast, in morphologically anisotropic cells actin filaments are predominantly oriented along the long axis of the cell. When such cells contract the actin filaments move closer to one another and maintain their preferential orientation. Together, GI-SIM and LLSM provide unprecedented, direct visualization of actomyosin dynamics during apical constriction in three dimensions. The detailed kinematics of filament orientation and localization provides new information to guide models of apical constriction. Moreover, the use of these imaging techniques with additional fluorescent proteins will illuminate molecular mechanisms driving apical constriction in dorsal closure and other biological processes.
RESULTS AND DISCUSSION
GI-SIM images individual actin filaments in medioapical arrays
GI-SIM images actin filaments in the medioapical arrays of kinematically active amnioserosa cells with unparalleled spatial and temporal resolution (Figure 1 and Supplemental Movies 1 and 2; Guo et al., 2018; see Materials and Methods for a brief overview of GI-SIM). Below, we provide evidence that the filaments that we resolve are individual actin filaments. We recommend that the reader watch Supplemental Movie 1, read the descriptions of cell shape and actin filament (F-actin) kinematics below, and refer to stills captured in Figure 1 (i.e., take advantage of the psychophysics of motion vision; Burr and Ross, 1986; Nishida et al., 2018, and references therein). The movie shows all or part of 13 aminoserosal cells that pulsate asynchronously. As expected, at various times, the pulsations appear to propagate across all or part of the field of view in a wave-like manner (see references above). Figure 1 is part of the field of view imaged in Supplemental Movie 1.
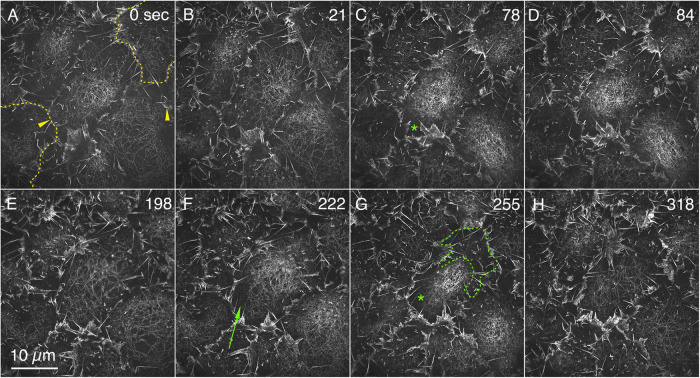
FIGURE 1:
Actin filaments are resolved by GI-SIM in condensing and relaxing medioapical arrays. F-actin is labeled with GFP-MoeABD. (A–H) Stills are a ROI from a time-lapsed sequence of amnioserosa cells in various states of apical contraction (Supplemental Movie 1, select portions magnified in Supplemental Movies 5 and 8). Comparable patterns of F-actin are seen labeled with Lifeact-mEGFP (Supplemental Movie 2). The dotted yellow lines in A show our estimate of the cell outlines of two cells at the periphery of the image (the relationship between the lamellipodia and filopodia and cell junctions is shown in Supplemental Figure 1 and Supplemental Movie 3). Yellow arrowheads in A show where filaments extend toward a bright band of lamellipodia and filopodia that project parallel to the optic axis. The green arrow in F points to filaments in a dark region of the cell that would appear as a low level of background fluorescence (a dark patch) by standard confocal methods. The green outline in G shows the upper half of a dark halo that surrounds the concentration of filaments in this cell. The green asterisks in C and G also label the dark halos in that central cell. Time is in seconds from the start of imaging.
Movie S1.
GI-SIM of amnioserosa cells ubiquitously expressing GFP-MoeABD to label F-actin. The field of view of the movie is larger than the region of interest shown in Figure 1. The movie is of a time-lapsed sequence sped up 30 times. Stills shown in Figure 1 and at high magnification in Figures 2 and 4.
Movie S2.
GI-SIM of amnioserosa cells expressing Lifeact-mEGFP in epidermal cells. Single frames from this movie were used for Figure 3B and Supplemental Figure 2. A magnified view of the cell to the far right is the region of interest shown in Supplemental Movie 7. The movie is from a time-lapsed sequence sped up 60 times.
At t = 0 the center cell in Figure 1, A and B, is in the process of expansion, and filaments are randomly oriented and differentially distributed across the apical end of the cell. The filamentous meshwork is most condensed near the center right of the cell with a relaxed part of the meshwork observed near the top of the cell. In optimal images, filaments extend to the cell margin (e.g., in the upper left of the center cell, filaments are seen to extend toward the bright band of lamellipodia and filopodia that project parallel to the optic axis; arrowheads in Figures 1A and 2). In some images, a dark halo surrounds much, if not all, of the medioapical array, apparently separating it from the cell margin (e.g., asterisks in Figure 1, C and G). The 3D geometry of the medioapical arrays, so apparent when viewed with LLSM, suggests that this gap is partially a consequence of the domed, medioapical array leaving the imaging plane and is partially a reflection of the spatial heterogeneity of the arrays (see below).
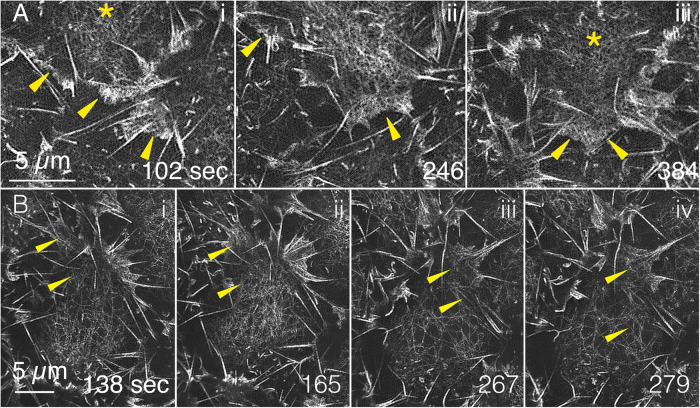
FIGURE 2:
GI-SIM shows that medioapical arrays are continuous with lamellipodia. F-actin labeled with GFP-MoeABD. (A) Three panels are magnified ROIs from Supplemental Movie 1 that show medioapical arrays (yellow asterisks) where they merge with the lamellipodia (yellow arrowheads). (B) Four additional panels (from a different embryo) also demonstrate continuity between medioapical arrays and lamellipodia. The top panel in Supplemental Movie 5 includes each of the time points shown in the top row of Figure 2; the bottom panel in Supplemental Movie 5 shows time points 120–330 s.
With time (21 s; Figure 1B), the distributions of filaments in the cells change dramatically. The most condensed portion of the actin meshwork in the central cell has shifted toward the top right and the most relaxed portion occupies the bottom left of the cell, that is, the condensed region traverses the cell in a wave-like manner as described previously, at lower resolution (Ma et al., 2009; Blanchard et al., 2010; David et al., 2010; Azevedo et al., 2011; Saravanan et al., 2013; Fischer et al., 2014; Munjal et al., 2015; Duque and Gorfinkiel, 2016; Pasakarnis et al., 2016; Goodwin et al., 2017). Connections between the medioapical array and the cell margin are most apparent along the bottom right side and bottom of the cell. At 78 s, the center cell is in its most contracted state (Figure 1C)—some individual filaments can still be resolved, but a dense meshwork is the most conspicuous feature of the image. A clear gap between the meshwork and the bottommost margin of the cell is also observed (asterisk in Figure 1C). At 84 s (Figure 1D), while the cell as a whole has begun to expand, the most concentrated part of the meshwork shifts leftward. Subsequently, further expansion ensues until at 198 s (Figure 1E), the meshwork is most relaxed. Condensation in the top regions of the cell follows (Figure 1F)—a wave of contraction proceeds toward the bottom of the micrograph, with the cell maximally contracted at ∼255 s (Figure 1G). Again, relaxation ensues, with maximal relaxation in this cycle at ∼318 s (Figure 1H). Cell contractions and expansions follow, with the maximally condensed meshwork seen at ∼402, 546, and 723 s and the maximally relaxed state observed at ∼492, 645, and 786 s, respectively. In this sequence, cells and meshworks continued to oscillate for the interval over which they were followed. However, with time, it was more difficult to resolve individual actin filaments, presumably due to photobleaching and/or photodamage. The kinematics of F-actin can also be observed in embryos expressing Lifeact-mEGFP (Supplemental Movie 2), GFP-Utrophin, and GFP-Actin (unpublished data; Lifeact-mEGFP and GFP-MoeABD provided superior images). The behaviors of actin filaments described here are representative of more than 80 cells from 18 F-actin or actin-labeled embryos.
The position of the margins of individual amnioserosa cells can be estimated by the location of the kinematically active, F-actin–rich lamellipodia and filopodia that project from cell junctions into the perivitelline space (yellow dashed lines in Figure 1A estimate the position of cell margins as examples; see also Supplemental Figure 1 and Supplemental Movies 1–3; refer also to Figure 6 and Supplemental Movie S17 from Chen et al., 2014). Such projections are also seen at amnioserosa cell junctions in embryos that ectopically and ubiquitously express GFP-Synaptogamin (GFP-Syt; Zhang et al., 2002), a transmembrane protein (Supplemental Movie 4), indicating that these protrusions are not artifacts of genetically encoded labels for actin or F-actin. Cadherin-rich cell junctions typically lie too deep in the tissue to be imaged reliably with GI-SIM but can be imaged with LLSM for reference (Supplemental Figure 1 and Supplemental Movie 3). Together, high-resolution GI-SIM and LLSM images indicate that the cadherin-rich cell junctions, with associated actin nucleators (e.g., Arp2/3; Homem and Peifer, 2008, 2009; Dominguez and Holmes, 2011; Brieher and Yap, 2013; Nowotarski et al., 2014; Pollard, 2016), are responsible for promoting the assembly of diverse arrays of F-actin–rich structures. These include actomyosin-rich junctional belts, the cores of filopodia and dendritic networks, or meshworks of filaments that drive lamellipodia extension and integrity.
Movie S3.
LLSM surface view of amnioserosa cells from an embryo double labeled with GFP-MoeABD (green) and dECadherin-mTomato knockIn (magenta) to label F-actin and cell junctions, respectively. The movie shows the protrusion of lamellipodia and filopodia at or very near cell junctions and waves of F-actin meshwork condensation and relaxation commensurate with apical contraction and expansion. The movie is from a time-lapsed sequence sped up 97 times. Stills are shown in Supplemental Figure 1.
Movie S4.
GI-SIM of Syt-GFP labeled plasma membrane shows a comparable proliferation of lamellipodia and filopodia as that seen in embryos expressing constructs that label F-actin (GFP-MoeABD and Lifeact-mEGFP, Supplemental Movie 1 and 2, respectively). The movie is from a time-lapsed sequence sped up 60 times.
Our images show that the dendritic actin meshworks of lamelli are continuous with the condensing and relaxing medioapical arrays (Figure 2 and Supplemental Movie 5). Previous imaging by standard confocal microscopy yields fluorescent patches of actin that do not readily distinguish between these diverse actin- and actomyosin-rich structures, each of which has very different roles in cellular function. Indeed, a recent high-resolution analysis of the Caenorhabditis elegans cortex showed that a previously reported relationship between putative concentrations of PIP2 in the plasma membrane and potential signaling “hubs” responsible for promoting actin nucleation and cell polarization was in fact due to membrane folds associated with filopodia (Hirani et al., 2019). Overall, the unparalleled views of the cytoskeleton in embryos, afforded by a combination of GI-SIM and LLSM, set the stage for superior understanding of cellular function at a molecular level and an important overview for even higher-resolution studies that will require cryoelectron microscopy and tomography.
Movie S5.
Medioapical arrays extending into the lamelli and actin filament severing. GI-SIM regions of interest from Supplemental Movie 1 and Figure 1 (embryo labeled for F-actin with GFP-MoeABD). Medioapical arrays extend into cell junctions and the lamellipodia and filopodia that emanate from them. The movie is from a time-lapsed sequence sped up 20 times. Stills are shown in Figure 2.
In sum, apical cell areas pulsate through cycles of contraction and expansion while the meshworks rhythmically condense and relax on a comparable timescale. In expanded cells, medioapical arrays are seen as a meshwork of F-actin (Figure 1) that can be followed for a substantial portion of the pulsatile cycle. In a given cell, array condensation and relaxation are neither temporally or spatially uniform (Supplemental Movies 1 and 2).
Individual actin filaments are resolved with GI-SIM
The majority of the filaments that we resolve in the relaxed F-actin arrays are most likely single actin filaments. The fluorescence intensity along the filament length is remarkably uniform (especially when viewed in movies that include sparse distributions of filaments in relaxed networks). In such regions, the psychophysics of motion integrates out specular features of still images that are generated by processing for structured illumination microscopy. The specular nature of the still, GI-SIM images has thus far precluded informative, quantitative analysis of intensity distributions along the length of the imaged filaments. Nevertheless, the distribution of intensities along the length of a filament appears to be consistent with a uniformly labeled, single actin filament. Only where two or more filaments of the meshwork cross one another or lie adjacent to one another over some distance do filaments that are sparsely distributed have substantially increased intensity. Moreover, the intensities of what we interpret as individual actin filaments in relaxed networks are characterized by fluorescence intensities that are much lower than that observed in the bundled arrays of actin in the tips of even the smallest filopodia (Supplemental Figure 2). In addition, the overall morphology of the filaments compares favorably with the morphology of actin filaments imaged by total internal reflection fluorescence (TIRF) microscopy in vitro (e.g., Fujiwara et al., 2002; reviewed in Wioland et al., 2022). We do not know of any reports of bundles of F-actin with uniform numbers of filaments (e.g., doublets or triplets). Moreover, we observe uniform labeling with four genetically encoded fluorescent F-actin tags (compare Supplemental Movies 1 and 2; also unpublished data), suggesting that this uniform fluorescence is not due to a filament-labeling artifact.
GI-SIM images that resolve individual filaments during medioapical condensation and resolve medioapical array structure provides a structural basis for understanding the heterogeneity in the distribution of fluorescence seen in confocal views of the arrays. For example, dark patches in confocal images contain no information about filament distribution, whereas the GI-SIM images show filaments extending into such dark regions (Figure 1F, arrow). This observation suggests that GI-SIM should be a particularly useful tool for the analysis of the distribution of F-actin during the formation and repopulation of cellular blebs that form when the plasma membrane rips free of its underlying cortex (Charras et al., 2008).
Isotropic and anisotropic condensations and contractions
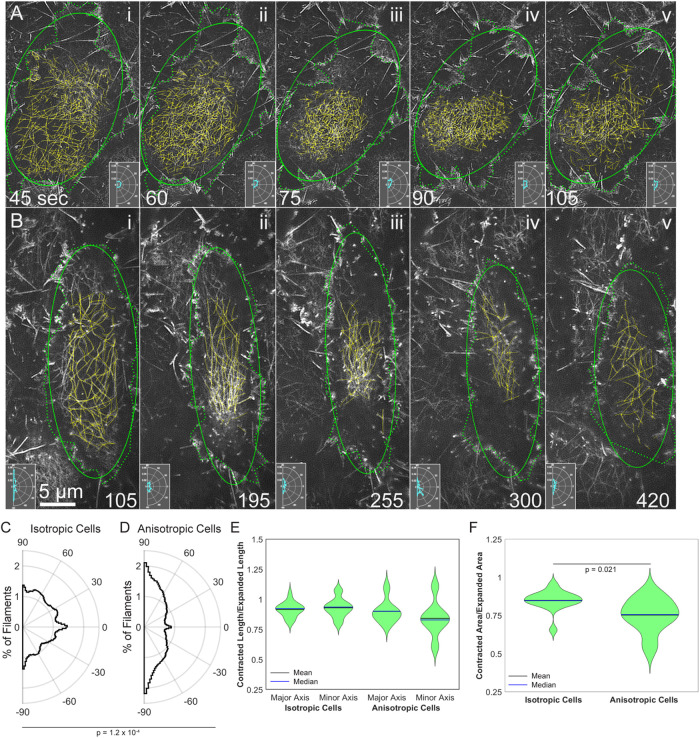
We see instances of both isotropic and anisotropic cell contractions and medioapical array condensations. To better understand how cell shape changes are related to changes in filament distribution, we traced the outlines of cells and their actin filaments in morphologically isotropic (i.e., isodiametric; Figure 3A) and anisotropic (i.e., elongated; Figure 3B) amnioserosa cells during the course of closure. The outlined cell boundaries were used to estimate the apical area of the cell and then were fitted with an ellipse. The ellipse provided us with a measure of the orientation of the cell and allowed us to compare cell contraction along the major and minor axes. Anisotropic cells remained anisotropic, and the major axis of the ellipse varied by an average of 3.5 ± 2.9° (n = 7 contractions from six anisotropic cells). The isotropic cells also remained isotropic, though the major axis of the ellipse varied more from expanded to contracted state (27.1 ± 40.9°). Because the major and minor axis are similar in length in the isotropic cells, they often switch (i.e., major becomes minor and vice versa); this accounts for the increased variation in orientation that we observe. Filaments were manually traced, and a polar histogram inset was generated to track the orientation of filaments in each panel (Figure 3 and Supplemental Movies 6 and 7).
FIGURE 3:
Actin filament array condensation and relaxation parallels amnioserosa cell contraction and expansion in (A) an isotropically and (B) an anisotropically contracting cell. F-actin labeled with (A) GFP-MoeABD from Supplemental Movie 1 or (B) Lifeact-mEGFP from Supplemental Movie 2. Dotted green lines—outline of cells based on the position of lamellipodia and filopodia. Solid green ellipse—depicts an ellipse fitted to the cell outline. Yellow lines—manual traces of individual actin filaments. Polar histogram insets depict the orientation of traced actin filaments. (C) Average distribution of filament orientation in six isotropic cells from four embryos. (D) Average distribution of filament orientation in six anisotropic cells from four embryos. The distributions shown in C and D are significantly different with a p = 0.00012. (E) Ratio of contracted axis length to expanded axis length in isotropic and anisotropic cells; n = 15 contractions from the six isotropic cells traced and n = 7 contractions from the six anisotropic cells traced. (F) Ratio of contracted area to expanded area in isotropic and anisotropic cells; n = 15 contractions from the six isotropic cells traced and n = 7 contractions from the six anisotropic cells traced. Anisotropic cells showed a greater reduction in area with p = 0.021. Time is in seconds from the start of imaging. Animated versions of F-actin dynamics are shown in Supplemental Movies 6 and 7 for an isotropic and anisotropic cell, respectively.
Movie S6.
Filament dynamics in an isotropically contracting and expanding amnioserosa cell. Four movie panels relate pulsations in cell area (area was approximated from the location of actin-rich protrusions emanating from cell junctions), filament orientation, the overall orientation of the cell and polar histograms of the orientation of the filaments. (Left) - GI-SIM images of F-actin in an amnioserosa cell from a GFP-MoeABD labeled embryo (approximate boundary shown with a dashed yellow outline). (Middle) Traces of the actin filaments from the left panel. (Right upper) Cell area (black), major (red) and minor (blue) axes of ellipses fit to the cell area shown as a function of time in the cell outlines from the left panel. (Right lower) Polar histograms of filament orientation as a function of time (see Materials and Methods). The movie in the left panel is from a time-lapsed sequence sped up 30 times. Animations of filament traces, graphs of area, major and minor ellipse axes and polar histograms correspond in time to the movie. Stills are shown in Figure 3.
Movie S7.
Filament dynamics in an anisotropically contracting and expanding amnioserosa cell. 4 movie panels relate pulsations in cell area (area approximated by the location of actin-rich protrusions emanating from cell junctions), filament orientation, the overall orientation of the cell and polar histograms of the orientation of the filaments. (Left) GI-SIM images of F-actin in an amnioserosa cell from a Lifeact-mEGFP labeled embryo (approximate boundary shown with a dashed yellow outline). (Middle) Traces of the actin filaments from the left panel. (Right upper) Cell area (black), major (red) and minor (blue) axes of ellipses fit to the cell area shown as a function of time in the cell outlines from left panel. (Right lower) Polar histograms of filament orientation as a function of time. The movie in Left panel is from a time-lapsed sequence sped up 30 times. Animations of filament traces, graphs of area, major and minor ellipse axes and polar histograms correspond in time to the movie. Stills are shown in Figure 3.
In morphologically isotropic, expanded cells, actin filaments appear to be more or less randomly oriented—during isotropic contraction, they remain roughly so (Figure 3A, i–v). During the course of contraction, cell area drops by as much as 34% due to changes in the length of both the major and minor axis of the ellipse—that is, the contraction is more or less isotropic and filaments remain relatively randomly oriented throughout the contraction and the beginning of cell expansion (Supplemental Movie 6).
In contrast, in expanded anisotropic cells filaments are substantially aligned along the major axis. During contraction, the area drops by as much as 45% while the orientation of the long axis of the ellipse and the filaments remains almost constant—filament spacing is reduced as they become even more preferentially aligned along the major axis. While both the major and minor axes contract, they do so to different extents (Figure 3E). This suggests that contraction perpendicular to the major axis and perpendicular to the long axes of the actin filaments is the major cause of area change in anisotropic cells (Figure 3B, i–v, and 3EE and Supplemental Movie 7). In contrast, during anisotropic cell expansion and filament relaxation, filament orientation remains predominantly anisotropic, but less so.
Unfortunately, in the data sets that we recovered, we did not see cells that changed between isotropic (which predominate early in closure) to anisotropic (which are more frequently seen later in closure, especially near the canthi and the junction between the amnioserosa and the dorsal-most epidermal cells of the lateral epidermis, e.g., see Sokolow et al., 2012). This may reflect that the length of time that we could image embryos at high resolution was limited, in part due to photobleaching and/or photodamage. In addition, the traces overlaid on the images in Figure 3 are consistent with differences in filament length observed in isotropic versus anisotropic cells. Nevertheless, by comparing images without the traces overlaid (Supplemental Figure 3) it is clear that there are long filaments present in both isotropic and anisotropic cells. More detailed analysis will be required to determine whether or not systematic differences in filament length characterize isotropic versus anisotropic cells (see Materials and Methods).
Because transgenic mosaic studies show that myosin produces force for cell contraction in the amnioserosa, we surmise that this is the main source of contractility in both isotropic and anisotropic cells (Franke et al., 2005). Our working hypothesis is that while actin filaments slide past one another to effect isotropic contraction, they are predominanty reoriented and drawn closer together to effect anisotropic contraction. The anisotropic distribution of filaments leads to greater variability in the contraction along the axis of the cell perpendicular to filament alignment (Figure 3E), resulting in a greater decrease in apical area in anisotropic cells (compare 34 to 45% for the cells shown and Figure 3F).
Previous imaging techniques identified dynamic changes in isotropic and anisotropic cell shapes but failed to capture the differences in filament position and orientation that are responsible for such changes. We see no overt evidence for obvious changes in filament length that we can unambiguously attribute to filament growth or shortening as opposed to movement of filament ends into or out of the GI-SIM’s optical section. This observation might lead us to conclude that the new arrangements of filaments seen during relaxation are likely due to the displacement of preexisting, individual actin filaments driven by contractile forces exerted by myosin and/or by changes in the function and distribution of actin cross-linkers. However, when we measure mean fluorescence intensity as a proxy for the number of filaments, we see substantial changes that seem inconsistent with our failure to observe growth or shrinkage of individual filaments (see Supplemental Figure 4). Indeed, such overall changes in fluorescence are consistent with compelling evidence that there are two populations of actin filaments in amnioserosa cells during closure, one that is persistent and one that pulsates during cycles of cell contraction and explansion (Dehapiot et al., 2020). The limited depth of imaging in GI-SIM and the three dimensionality of the amnioserosa makes this system highly susceptible to filaments leaving the image plane, which likely also contribute to the fluorescence fluctuations that we observe. More detailed analysis of filament kinematics using GI-SIM and F-actin tagged with GFP-Actin, GFP-MoeABD, Lifeact-mEGFP or GFP-Utrophin (which the Dehapiot et al., 2020, study used) and appropriate experiments designed to alter actin dynamics in the amnioserosa cells will be required to evaluate quantitatively the extent to which the arrays we see are stable, pulsating, or a combination of both.
We expect that GI-SIM, in concert with two-color imaging of fluorescent actin filaments decorated with proteins such as fluorescent capping proteins, Enabled or formins, will provide additional information about the behavior of filament ends. Because our current experiments cannot rule out non–cell autonomous contributions to cell shape changes, further analysis of filament distributions and rearrangements in mosaic embryos that position contractile cells adjacent to noncontractile cells will be required (Franke et al., 2005). Ultimately, how such GI-SIM data on filament behavior in vivo map onto the kinetics of actin assembly in vitro may provide interesting insight into the activities of a variety of actin binding proteins, including monomer binding proteins, filament binding proteins, nucleators, and barbed and pointed end capping proteins and how they function synergistically (reviewed in Pollard, 2016, 2017; Carlier and Shekhar, 2017).
Discerning individual actin filament dynamics in vivo
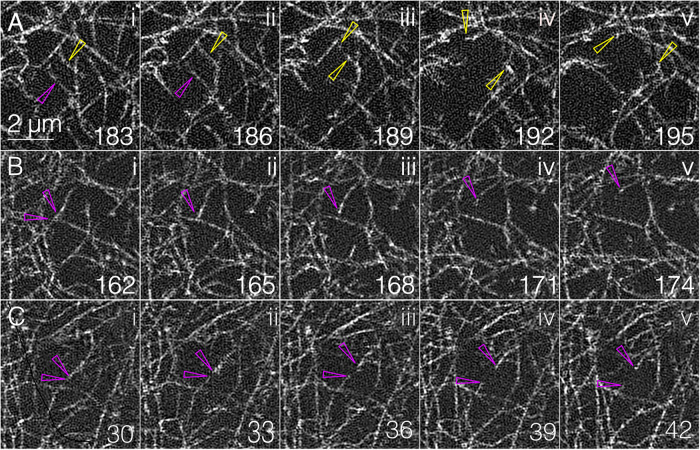
The ability to see individual filaments suggests that GI-SIM might also be used to examine key characteristics of filament dynamics in the medioapical arrays such as severing, elongation, and shortening, cross-linking, compression, and the formation of branches. Indeed, through frame-by-frame inspection of the GI-SIM movie sequence, we can consistently track severing events and to date have seen them only in expanding cells (Figure 4). Curved (slack) actin filaments straighten (presumably under tension) and then snap apart (this is most apparent in Supplemental Movie 8), presumably as a consequence of applied tension. Such events are readily observed in expanding cells, while filamentous meshworks are relaxing and when the domes of medioapical arrays remain relatively fixed in the GI-SIM imaging plane. Inspection of 11 cells in three different embryos revealed 30 unambiguous severing events. In any given cell, the frequency with which filaments were severed ranged from simultaneous severing of two or three filaments in close proximity to one another to an average duration between observed severing events of 41.2 s.
FIGURE 4:
GI-SIM shows single-filament severing. (A) Five panels from Supplemental Movie 1 show filament severing. Hollow yellow and magenta arrowheads in i show a top and bottom filament that become stretched in ii. Both filaments are severed between ii and iii, and the bottom filament leaves the image plane. Hollow yellow arrowheads in iii–v point to the filament ends that spring apart under tension. (B, C) Two additional severing events; magenta arrowheads show the filaments before (panels i) and after (panels ii–v) severing. Filament breakage is also shown in Supplemental Movie 8.
Movie S8.
Animation of Figure 4 showing actin filament severing. The movie shows each time point from Figure 4 six times before transitioning to the subsequent time point. The 5 panel sequence is repeated 5 times by the end of the movie. The yellow asterisk appears in the frame in which severing of the two filaments is apparent (e.g., severing occurred in the 3 sec between Figure 4Aii, 186 sec, and 4Aiii, 189 sec). The times shown are from Supplemental Movie 1, from which these severing sequences are taken). Similar animation was used for the bottom two panels, showing different severing events.
Thus far, we have been unable to document unambiguously other events, including filament growth, shortening, compression, and branching. GI-SIM produces a projected image, that is, all captured filaments appear in a single plane—the images provide no information about depth. Thus, a branch may be a branch or may appear as such because a filament end lies above or below another filament. Similarly, we are unable to distinguish between filament ends and filaments that extend up or down, out of the image plane. Some through focus analysis or 3D SIM may help us conclusively determine at what frequency such events occur in medioapical arrays, but the increased number of frames required to generate high-resolution, 3D-SIM images leads to significant photobleaching and difficulty in analyzing multiple cycles of cell oscillation—new, more stable fluorophores may facilitate more extensive 3D-SIM analysis.
GI-SIM images of bipolar myosin filaments in medioapical arrays
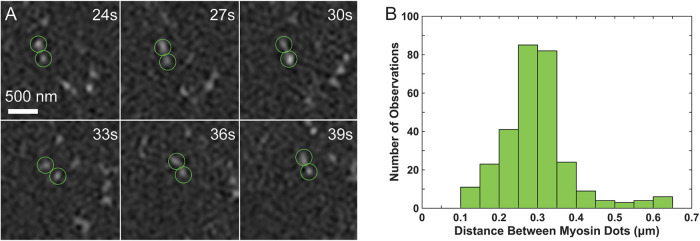
GI-SIM imaging also showed small, bipolar myosin filaments in the medioapical arrays undergoing cycles of condensation and relaxation (Figures 5 and 6 and Supplemental Movie 9). We imaged embryos in which every myosin head is endogenously labeled with GFP. Thus bipolar filaments appear as two bright, paired dots that comigrate (we defined comigration as dots that migrated together over the course of at least 9 s). A histogram of the distance between these paired dots shows a peak at ∼0.3 µm (Figure 6)—this distance is consistent with GFP tagging the myosin head and the size of fly nonmuscle myosin II bipolar filaments measured by platinum shadowed electron microscopy of purified fly nonmuscle myosin (see Figure 5 in Kiehart and Feghali, 1986). We interpret the range of separations between co-migrating dots as differences in bipolar filament orientation.
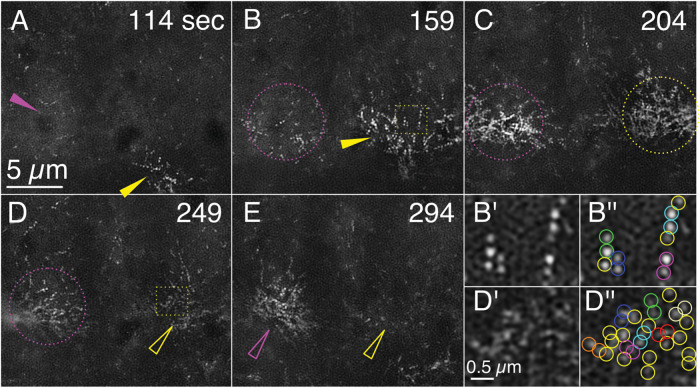
FIGURE 5:
Myosin filaments are resolved as paired dots by GI-SIM in condensing and relaxing medioapical arrays. Panels A–E are still images from Supplemental Movie 9 taken at the timepoints shown. Maximum condensations of myosin arrays in two adjacent cells are depicted and tracked by magenta and yellow arrowheads, hollow arrowheads, or dotted circles. Solid arrows track the direction of movement of the condensation. Dotted circles indicate an array that is condensing but whose centroid is roughly fixed. Hollow arrowheads point to an array that is relaxing. Dotted yellow boxes in B and D are magnified in B′ and B′′ and D′ and D′′), respectively. B′′ and D′′ are identical to B′ and D′, respectively, except that dots that contribute to bipolar filaments identified by comigration are shown circled in blue, cyan, green, orange, magenta, and red, whereas dots that could not be unambiguously identified as a member of a pair are circled in yellow. Dynamics of myosin filament distributions are shown in Supplemental Movie 9.
FIGURE 6:
Myosin filaments can be tracked through time and have lengths (0.30 ± 0.01 µm) consistent with previous reports. (A) Putative myosin bipolar filaments can be tracked for multiple frames. Time points are from Supplemental Movie 9. (B) The distance between the heads of putative pairs is ∼300 nm; n = 292 measuements from 50 pairs.
Movie S9.
GI-SIM of myosin bipolar filaments during medioapical array condensation and relaxation. Amnioserosa cells from an embryo homozygous for GFP knocked into coding sequence of the zipper/Myosin II locus that encodes the myosin head. Myosin bipolar filaments tracked during condensation and relaxation. Approximately 1/3 of this movie is shown in Figure 5. The movie is from a time-lapsed sequence sped up 30 times. Stills are shown in Figures 5 and 6.
Six distinct foci of myosin filaments form as a consequence of condensation during the course of the sequence shown in Figure 5 (a cropped version of the field shown in Supplemental Movie 9). Again, through the psychophysics of motion vision, viewing Supplemental Movie 9 will greatly facilitate interpretation of the still panels depicted in Figure 5. Owing to the area that surrounds each of the medioapical arrays, and by comparison to the condensed arrays of F-actin seen in amnioserosal cell contraction and expansion (see above), we surmise that each focus is in an individual, contracted cell (myosin does not label the filopodia and lamellipodia that serve to outline the amnioserosa cells in embryos expressing a fluorescent marker for F-actin). The two cells that are most instructive are expanding at the beginning of the sequence. Pairs of dots are easily distinguished at 114 s (Figure 5A). These pairs of dots can be tracked for up to 12 s while the meshwork is relaxed and open. At 159 s pairs of dots become more organized in the right cell—up to six dots are organized into a single line (Figure 5, B and B′, pairs color coded in B′′). We interpret these strings of pairs as individual filaments arrayed along an actin filament. As the meshwork continues to condense, the pairs of dots are not as easily distinguished, that is, it is hard to discern which dots are components of a single bipolar filament (Figure 5, C–E). Nevertheless, even when the cell is maximally contracted, some dot pairs move in concert with one another and can be identified in regions where myosin filaments are sparse (Figure 5, D′ and D′′).
Similar to the actin filaments, we observed the arrays of myosin bipolar filaments (i.e., comigrating dots) rhythmically condensing and relaxing on a timescale comparable to that observed for the actin filaments. In addition we observe that the focus of maximal condensation (arrowheads) of myosin dots migrating across the image similar to the condensation of actin filaments moving across the cell described above (Figure 5). The behaviors of bipolar myosin filaments described here are representative of more than 50 cells from eight myosin-labeled embryos. We further interpret these foci in three dimensions below.
While instructive, the myosin GI-SIM images are not as compelling as those of F-actin. First, because GFP-Myosin is not in filopodia and lamellipodia, the margins of the cells are not visible for reference and much of the imaging field appears relatively empty. Second, whereas the F-actin is imaged as continuous filaments up to 10–13 µm in length, the bipolar myosin filaments are paired dots and thus fail to have the same visual impact. In particular, the continuity of the actin filaments is easy to interpret as a section through a mat of filaments—the paired dots are less obviously so. Finally, the tags we employ for F-actin (detailed in Materials and Methods) are expressed from genomically inserted, recombinant transgenes (and therefore by definition are overexpressed), which makes imaging of F-actin less susceptible to photobleaching. Thus we captured more cycles of condensation and relaxation than with myosin (imaged with a GFP-Myosin knockin). Whether or not bipolar myosin filaments are stable throughout cycles of myosin filament condensation and relaxation remains an open question.
Robust, two-color labeling of F-actin and myosin filaments (and other protein components of the medioapical arrays) in the same embryos/cells would greatly facilitate unambiguous interpretation of medioapical array structure and function. We attempted two-color imaging of F-actin and myosin filaments in the same embryos/cells; however, we were unable to obtain a satisfactory data set because the red fluorophores bleached too rapidly. These difficulties could potentially be overcome by excitation at a different wavelength or by generating new genetic constructs with more photostable fluorophores. Importantly, the advent of straightforward CRISPR methods to introduce fluorophores at endogenous loci (Port et al., 2014; Gratz et al., 2015) and/or the use of HaloTag or SNAP-tag approaches (e.g., Erdmann et al., 2019; Thirukkumaran et al., 2019) that use brighter, more stable, small-molecule fluorphores will allow for more flexibility in observing myosin in vivo. In the future, we expect GI-SIM imaging coupled with systematic through focus studies using 3D SIM and novel approaches to live embryo specimen preparation to greatly expand the usefulness of GI-SIM. Further, quantitative analysis of actin arrays decorated with the various proteins with which they interact should be instructive. Paired with experimental manipulations that alter actin and myosin filament assembly (e.g., constitutively active myosin light chain kinases, actin nucleators, F-actin capping proteins or other cytoskeletal regulators), the extent to which medioapical array behavior depends on the redistribution of existing filaments, the assembly or disassembly of filaments, or some combination of the two may well be addressed.
LLSM images medioapical arrays in three dimensions
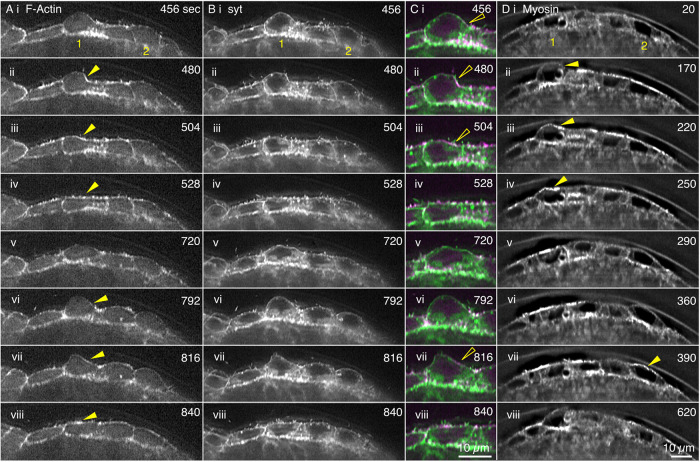
We use LLSM microscopy to better understand the morphology of the medioapical arrays and to better interpret the GI-SIM images of both F-actin and myosin in three dimensions. In LLSM images, fluorescence can be viewed in apical, basal, volume rendered, and cross-sectional views (Chen et al., 2014). LLSM cross-sections show that F-actin lies tightly apposed to the plasma membrane as confirmed by labeling both F-actin and plasma membranes (Figure 7 and Supplemental Movie 10). They also show that the apical surfaces of individual amnioserosa cells are conspicuously domed and can transiently bulge far into the perivitelline space on the same timescale as oscillations. There is a low level of F-actin and myosin associated, nonuniformly with the apical membrane as the bulges protrude, and both F-actin and myosin are recruited to the membrane before and concomitant with bulge retraction. (Note that the red fluorescent protein–tagged F-actin bleaches with time and during later stages of a time-lapsed sequence, low levels of F-actin become visible only when images are appropriately adjusted for brightness and constrast.) Supplemental Figure 5 shows a total of 12 cycles of cell bulging and flattening in four different cells wherein the length of the apical margin and the mean fluorescence intensity of myosin measured along that apical margin are plotted as a function of time. Cell bulging correlates well with loss of apical myosin, and cell flattening reflects additional myosin recruitment to the apical margin of the cell. In some instances the correlation is not as tight as might be expected. We attribute this to our one dimensional analysis of a 3D bulge—concentrations of F-actin and myosin in the apical regions of the cell tend to be heterogeneous, displaying patches of high concentration when the apical regions of the aminoserosa are viewed enface (see figures herein and in Ma et al., 2009; Blanchard et al., 2010; David et al., 2010; Azevedo et al., 2011; Saravanan et al., 2013; Fischer et al., 2014; Munjal et al., 2015; Duque and Gorfinkiel, 2016; Pasakarnis et al., 2016; Goodwin et al., 2017). Thus, in such instances, we surmise that the yz planes that we use to analyze cell bulging lie in a section of a bulge that is outside of a major patch of myosin (and F-actin) accumulation.
FIGURE 7:
Cross-sections from LLSM data sets show that amnioserosa cells bulge into the perivitelline space, actin and myosin accumulate to “rescue” bulges, and the actin cortex colocalizes with the plasma membrane. Images from time-lapse sequences of the amnioserosa in two different embryos. The (anti-)correlation between bulge retraction and the accumulation of myosin is shown in Supplemental Figure 5. (A–C) Amnioserosa cells in an embryo double labeled for F-actin with RFP-MoeABD and the plasma membrane with Syt-GFP shows two cells (1 and 2) that bulge into the perivitelline space. (A, B) Black-and-white images of approximately six cells in the amnioserosa. (C) Magnified view of cell 1 double labeled with F-actin in magenta and plasma membrane in green. (D) GFP-Myosin knockin-labeled amnioserosa. Cells 1 and 2 bulge into the perivitelline space. Yellow arrowheads in A and D show the accumulation of actin and myosin, respectively, on a bulge and then its rescue. Hollow yellow arrowheads show colocalization of medioapical array (F-actin) and plasma membrane (Syt-GFP). Scale bar in Cviii is for all panels in C; scale bar in Dviii is for all panels in A, B, and D. Time is in seconds from the start of imaging. Dynamics of actin, syt, and myosin are shown in Supplemental Movie 10.
Movie S10.
Cross sections of amnioserosa cells show protrusion into the perivitelline space and "rescue" via the recruitment of actin and myosin. Datasets from two LLSM experimental runs. Corresponds to Figure 7. (A - C) Embryo labeled for F-actin with RFP-Moe-ABD (A&C, magenta in C) and for plasma membrane with Syt-GFP (B&C, green in C). The movie is from a time-lapsed sequence sped up 170 times. (D) Embryo labeled with GFP-Myosin knockIn. The movie is from a time-lapsed sequence sped up 140 times. Stills are shown in Figure 7.
Medioapical arrays are modified cortex
To mediate cell and amnioserosa cell sheet contraction, the medioapical arrays must be mechanically coupled to the junctional belts and cells must be tightly adherent to one another. A robust cortex would be expected to contribute to such functions. Nevertheless, the medioapical arrays of F-actin are not as uniformly ordered and compact as has been observed in other cell cortices (Charras et al., 2006; Morone et al., 2006; Ecke et al., 2020). Indeed, while medioapical arrays have sometimes been assumed to be cell cortex, we believe that our study provides the most compelling confirmation of that assumption. First, in optimal optical sections, the medioapical arrays appear to be continuous with the lamellipodial and filopodial projections at the apical surface (Figures 2 and 7 and Supplemental Movies 5 and 10). Second, we were unable to image cortex distinct from medioapical arrays—in LLSM cross-sections F-actin colocalizes with a fluorescent plasma membrane marker (Supplemental Movie 10). In GI-SIM apical views show that this colocalized F-actin behaves as the medioapical arrays, condensing and relaxing as the cells oscillate (Supplemental Movies 1 and 2). Third, the range of mesh sizes of the medioapical arrays varies considerably but during cell contraction is comparable to the mesh size in the lamellipodia (unpublished data). Fourth, previous studies report substantial diversity in cortical mesh sizes (approximately fourfold) that depend on cell type and function (50–200 nm; see Charras et al., 2006; Morone et al., 2006; Chugh et al., 2017). Our measured mesh sizes are ∼120 nm larger. We conclude that the medioapical arrays constitute a modified and dynamic cell cortex—to what extent such a remarkably heterogeneous cortical array is required or enables amnioserosal cell function for producing forces for closure, as a target for signaling, or for other unknown tasks is an open research question. The large mesh size that we observe in expanded cells (with relaxed filament networks; Supplemental Figure 6) may be necessary for the rapid and extensive expansions of the apical membrane into the perivitelline space—the purpose of such expansions remains a mystery.
Experiments that overexpressed constitutively active diaphanous, a fly formin that is expected to increase the number of filaments in amnioserosa cells, resulted in decreased area fluctuations and decreased recoil velocity after laser ablation while nevertheless maintaining a kinematically active medioapical array population (Fischer et al., 2014). This suggests that these cells have viscoelastic properties that are distinct from those of wild-type amnioserosa cells, perhaps because they have more filaments in their medioapical arrays—future inspection of such cells with GI-SIM should be very revealing.
Concluding remarks
Superresolution images from GI-SIM and LLSM provide unparalleled views of the structure and function of actin and myosin in the medioapical arrays found in embryonic tissue at the level of individual actin and bipolar myosin filaments. The arrays are tightly apposed to the plasma membrane and constitute the apical cortex in the amnioserosa—the arrays are continuous with the lamellipodia and filopodia emanating from junctional belts and with the junctional belts themselves and move on a timescale of seconds. These arrays undergo cycles of condensation and relaxation that are consistent with cell area contraction and expansion—moreover, they can occur isotropically or anisotropically on a timescale of minutes (i.e., ∼2 orders of magnitude slower). On a similar timescale, amnioserosa cells bulge into the perivitelline space, as the medioapical arrays condense and pull the meshwork basally toward the plane of the junctional belts. Together, these kinematically rapid, rich, and stunning movements of actin and myosin in medioapical arrays, lamellipodia, and filopodia are integrated with contractions of circumferential actomyosin in junctional belts to drive the steady and slow emergent movements that define dorsal closure with a timescale of hours. Thus, a fascinating feature of this morphological process is that while actin- and actomyosin-based motilities occur on subsecond and second timescales, and cycles of cell contraction and expansion occur on a minute timescale, cell closure occurs on an hour timescale fully three to four orders of magnitude slower than the actin- and actomyosin-based movements on which closure is based. The remarkable resolution of GI-SIM and LLSM images of dorsal closure should allow investigators to assess the realities of ratchets and clutches. Furthermore, how the vigorous motility of filopodia and lamellipodia contribute to amnioserosa function is also of interest. Finally, the superresolution images that we provide should contribute new levels of detail for mechanistic models of closure.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Fly stocks
GAL4 Drivers: e22c driver stock is w; E22c-GAL4 (Lawrence et al., 1995); daughterless driver stock is w*; P[GAL4-da.g32]UH1 (Wodarz et al., 1995).
F-actin tags: spaghetti squash driven GFP-MoeABD (aka sGMCA; Kiehart et al., 2000); UAS-RFP-MoeABD (Singh, 2012); UAS-Lifeact-mEGFP (Spracklen et al., 2014).
Myosin tag: GFP-Myosin knockin is encoded by w*; P[PTT-GC]zipCC01626 (Buszczak et al., 2007).
Cell junction tag: DE-Cadherin-mTomato knockin stock is y1 w*; TI[TI] shg mTomato (Huang et al., 2009).
Plasma membrane tag: UAS-Synaptotagmin-1-GFP (Syt-GFP, Zhang et al., 2002).
It is important to note that the F-actin and plasma membrane tags were encoded by transgenes whose transcription was driven by a selected, exogenous promoter/enhancer cassette (e.g., sGMCA, which is expressed ubiquitously via genomic sequnces that drive expression of the nonmuscle myosin regulatory light chain; Kiehart et al., 2000) or by the bipartite GAL4-UAS system (Brand and Perrimon, 1993), whereas the fluorescent myosin and cadherin constructs were “knockins” and embryos imaged were homozyogous for those knockins, so that all copies of each protein were labeled and expression was from endogenous regulatory elements.
Expression of GFP-tagged proteins, via heterologous promoter/enhancer cassettes or via knockins that utilize endogenous regulators of transcription, may cause unintentional artifacts (Edwards et al., 1997; Waterman-Storer et al., 1998; Wu and Pollard, 2005; Belin et al., 2013; Spracklen et al., 2014; Flores et al., 2019). It is highly unlikely that such artifacts, if they exist, contribute in any major adverse or misleading way to the distributions of F-actin and myosin that we report here. First, we note that changes in the distribution of F-actin and myosin that we see at high resolution are consistent with the changes seen as fluorescent patches when imaged at lower resolution (however, improved resolution alters our view of dark patches in cells imaged at high resolution). Second, flies homozygous for GFP-MoeABD (also seen in the literature as sGMCA, e.g., Kiehart et al., 2000) and for myosin and cadherin knockins are viable as homozygous stocks (with as many as four copies of GFP-MoeABD per fly genome; unpublished data). Third, embryos from such stocks have developmental times that are comparable to those of controls. Fourth, previous studies suggest that GFP-MoeABD binds to all known actin-rich structures in all Drosophila tissues that have been investigated, including the actin-rich cortex, lamellipodia, and filopodia, actin-rich chordotonal organs, ring canals, cytokinetic furrows, and muscle, etc. (e.g., Edwards et al., 1997). Finally, and perhaps most importantly, we note that the overall structure and behaviors of lamellipodia and filopodia in the presence of two different actin tags (GFP-MoeABD and Lifeact-mEGFP) and a tag for membranes (Syt-GFP) are indistinguishable. Although female flies that express Lifeact-mEGFP at high levels are sterile (Spracklen et al., 2014), embryos that are expressing Lifeact-mEGFP with the driver we use (e22C-GAL4) complete embryogenesis and hatch in a timeframe comparable to that of controls. A further, more detailed quantitation of the behavior of these GFP-tagged proteins could reveal some artifactual effects on the distribution of these proteins. However, for the reasons outlined above, it is highly unlikely that such effects would substantially alter the behavior of membranes and cytoskeletal proteins that we describe here.
Embryo collection and preparation for imaging
Embryo handling.
Embryos were prepared for imaging as previously described (Kiehart et al., 1994). Adult flies were allowed to lay eggs for 3–4 h at 25°C. Embryos were aged at 16–18°C for 24 h or selected from overnight egg lays and then dechorionated for 1.25 min in 50% bleach and washed extensively with deionized water. Embryos early in dorsal closure were lined up on an agar pad, picked up with an embryo glue–coated coverslip, covered with halocarbon oil, and imaged with modifications to standard protocals (Kiehart et al., 1994; Kiehart et al., 2006) as described below.
Preparations for imaging by GI-SIM.
Embryos were sometimes desiccated on the coverslip in a tightly sealed jar containing Drierite (W. A. Hammond Drierite Company, Ltd.), typically for 6 min. Desiccation helps flatten the embryos against the coverslip and places the surface of the amnioserosa ∼1.9–3.5 μm from the top of the coverslip. The desiccated embryos typically hatch, and when rescued to a food source, develop normally.
Preparing for imaging by LLS.
Embryos were mounted on 5 mm round coverslips prepared as previously described (Planchon et al., 2011; Chen et al., 2014), with the ventral side of the embryo glued to the coverslip (for excitation and imaging of the dorsal side).
Microscopes and imaging
GI-SIM overview.
(See Guo et al., 2018, for details.) SIM is a preferred form of superresolution microscopy because of its low excitation intensity, its use of conventional fluorescent labels, and its ability to reconstruct a superresolution image from just nine raw, 2D images. Combined with TIRF illumination strategies, SIM eliminates out-of-focus noise by exciting fluorochromes that lie within the penetration distance of TIRF’s evanescent wave. In GI-SIM, the illumination is adjusted so that it enters the back aperture of the objective just inside the critical angle for TIRF, thereby creating an illumination field that extends into the specimen a distance comparable to the depth of focus of the objective. Thus the illumination field, generated at the coverslip/specimen interface, leaves the coverslip to penetrate the vitelline envelope, the perivitelline space, the plasma membrane, and a thin swath of cytoplasm just inside the apical margins of cells, in our case, the amnioserosa cells found on the dorsal side of the embryo. GI-SIM provides ∼97 nm resolution in the x,y plane.
GI-SIM imaging.
Medioapical arrays and junctional belts lie too deep in the specimen for standard TIRF microscopy. Nevertheless, with GI-SIM we were able to image the medioapical arrays near the apical surface of the amnioserosa. Embryos were prepared as described above and live imaged using the protocol and lasers described in Guo et al. (2018). For GI-SIM we used a Zeiss 1.49NA TIRF objective.
LLSM overview.
(See Chen et al., 2014, for details.) LLSM addresses the twin, mutually opposed goals of light microscopists interested in imaging live, biological specimens—the desire to image fine structural details at ever higher spatial resolution and to do so with higher and higher temporal resolution. A laser light sheet (in this case a sheet of Bessel beams) illuminates the specimen with a thin, penetrating sheet of light, situated perpendicular to the optic axis of the imaging objective and 45° from the surface of a small coverslip, on top of which sits the specimen (in this case a Drosophila embryo, dorsal side up). A high numerical aperture dipping objective collects fluorescent light emanating from the illuminated plane—images of a significant part of the embryo are collected by passing the embryo through the sheet while collecting images. This strategy minimizes phototoxicity by ensuring that only the specimen plane that is being imaged is irradiated. Images collected during a single pass are assembled as a 3D data set. By repeating the process (with a large specimen like the Drosophila embryo, it takes ∼10 s to collect the ∼300 images that are assembled into a single data set), a 4D data set can be assembled. Outstanding resolution in z allows imaging analysis of sections perpendicular to the optic axis of a typical microscope (Figure 7). LLSM provides 230 nm resolution in the x,y and 370 nm resolution in z (Chen et al., 2014).
LLSM imaging.
Embryos were prepared as described above and were imaged as described in Chen et al. (2014). To ensure oxygenation, air- or oxygen-saturated phosphate-buffered saline or Schneider’s media was perfused over the embryos.
Spinning-disk confocal imaging.
Some micrographs (panels G, J, and K of Supplemental Figure 3) were also taken using a Yokogawa W1 Spinning Disk head attached to a Zeiss Axiovert 200M using a 63 × 1.4 NA oil immersion objective with a 1.25× optivar using standard protocols (Kiehart et al., 1994, 2006).
Quantitative analysis
Actin filament alignment.
To document how the actin filaments were changing over time, we measured the angle filaments made with respect to the laboratory frame of reference during contraction and expansion cycles in amnioserosa cells. Filaments were traced by hand, and the traces were used in conjunction with the Directionality plug-in in Fiji/ImageJ (Schindelin et al., 2012; Gomez et al., 2016). Owing to the mottled character of GI-SIM images, making high-fidelity traces of individual filaments is not straightforward, and doing so with high confidence is further compromised when the density of filaments in the meshwork is high. The 3D geometry of the amnioserosa cells paired with the projected image we obtain from GI-SIM means that an observed filament “end” may be a true filament end or instead may be due to a filament that leaves the imaging plane. In some cells, the traces appeared shorter than others (compare Figure 3, A and B). This may be the case—better tools to define ends of filaments or through focus analysis will be needed to determine this unambiguously. Supplemental Figure 3 provides stills from all of the cells traced without the traces overlaid for comparison. In these images it is clear that there are long filaments in both isotropic and anisotropic cells. We traced filaments and approximated apical area by outlining the cell and using the lamellipodia and filopodia protrusions as the cell boundary, as shown by the dotted green lines in Figure 3, A and B. We analyzed six isotropic cells from four different embryos and five anisotropic cells from four different embryos. The output from the Directionality plug-in describes the percentage of the filaments at each angle ranging from –90° to 90°. These percentages were averaged for all time points for all isotropic cells and reported in Figure 3C and for all time points for all anisotropic cells and reported in Figure 3D. We analyzed the significance of the difference between these two distributions using the nonparametric, two-sample Kolmorgorov–Smirnov test. We traced filaments shown in isotropic and anisotropic cells expressing GFP-MoeABD or Lifeact-mEGFP; still images from each data set traced are shown in Supplemental Figure 3 for comparison. We did not note significant differences in the intensity or behavior of the arrays when comparing these genetic backgrounds to each other, suggesting that the data from the filament traces are not due to an artifact of our labeling actin filaments.
Fluorescence intensity of medioapical arrays.
We measured fluorescence intensity changes of the medioapical arrays using two different actin markers, GFP-MoeABD and Lifeact-mEGFP. In FIJI/ImageJ, we outlined the medioapical arrays as separate regions of interest (ROIs) for 31–51 time points covering a duration of 615 s. We then measured the mean pixel intensity (total fluorescence intensity/number of pixels) for each ROI. The mean fluorescence intensity was normalized by dividing the measured fluorescence intensity at each time point by the maximum fluorescence intensity measured for that cell. Calculations were done and graphs were generated in Origin (OriginLab). The data are reported in Supplemental Figure 4.
Tracking myosin minifilaments.
To track myosin in GI-SIM images, we used the Spots tool in Imaris (Bitplane). This resolved numerous fluorescent dots due to GFP, which was “knocked in” to the 5′ end of the zipper locus that encodes the the sole fly nonmuscle myosin II heavy chain locus (Buszczak et al., 2007). Some of the dots comigrate as pairs and are consistent with putative myosin bipolar filaments (see Figure 5). Pairs of dots that moved together for a minimum of four frames (9 s) were selected as putative bipolar filaments. The distance between pairs of dots was measured using the X and Y coordinates of the centroid of each dot. The histogram in Figure 6 contains measurements from 50 tracked minifilaments over time for a total of 292 measurements. All measurements were taken from an ∼45 × 45 μm area with ∼10 cells.
Fitting ellipses to cell area.
The ROI described above was used to make a binary image of the approximated, hand-drawn cell outline, which was then fitted with an ellipse in Fiji/ImageJ (Schindelin et al., 2012). The lengths of the major and minor axes of this ellipse were measured in order to determine which axis of the cell was contributing most to changes in shape.
Measuring actin mesh size of medioapical arrays.
The mesh size was measured using a semiautomatic segmentation with the Surface tool in Imaris. A surface was generated for the actin filaments and used to make a mask. A new surface was then made from this mask, which detected the gaps between the actin filaments. Errors in the segmentation were manually corrected using the Cut tool. We report mesh size as the square root of the segmented area, as previously reported (Charras et al., 2006; Morone et al., 2006).
Correlation between myosin accumulation and the retraction of cell bulges.
Time-lapsed LLSM image sequences were loaded into Imaris (Bitplane, Oxford Instruments) and then rotated to display a plane (or “orthoslice” in yz), perpendicular to both the embryo’s surface and its anterior posterior axis (which we define as the x axis)—that is, a cross-section of the apical end of the embryo with the dorsal side up. The orthoslice was translated incrementally (by 2 µm) along the anterior posterior, x-axis. Each orthoslice was animated to generate a stack of images in time. Stacks were analyzed in FIJI/ImageJ as 16-bit images, and the apical margin of a target cell was traced using a 5-pixel-wide line to measure the average fluorescence intensity along the line pathlength. The data were normalized for each cell by dividing the apical pathlength at a given time point by the maximum length observed for that cell over the time interval examined and by dividing the mean myosin intensity by the maximum mean myosin intensity observed for that cell. Normalization, plots of apical pathlength, and mean myosin intensity versus time were generated using Origin software.
Supplementary Material
Acknowledgments
We thank Brenton Hoffman, Amy Bejsovec, and Richard Mortensen and members of the Kiehart and Christoph Schmidt labs for useful discussions regarding experimental design and data analysis. We thank Jennifer Lippincott-Schwartz at the HHMI Janelia Research Campus and Xiaolei Wang of the Duke Advanced Light Imaging and Spectroscopy Facility for access to their LLSMs. R.P.M., A.H.C., and S.M.F. were funded by National Institutes of Health (NIH) Grant T32-GM007184. W.R.L. acknowledges support from the Searle Scholars Program, the Beckman Young Investigator Program, an NIH New Innovator Award (DP2GM136653), and the Packard Fellows Program. S.M.F. was funded by National Science Foundation-Division of Graduate Education (DGE) award 1644868, and this work was supported by NIH R01GM033830 and Maximizing Investigators’ Research Award R35GM127059 awarded to D.P.K.
Abbreviations used:
- F-actin
actin filaments or filamentous actin
- GI-SIM
grazing incidence structured illumination microscopy
- LLSM
lattice light sheet microscopy
- TIRF
total internal reflection fluorescence.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-11-0537) on May 11, 2022.
REFERENCES
- Aristotelous AC, Crawford JM, Edwards GS, Kiehart DP, Venakides S (2018). Mathematical models of dorsal closure. Prog Biophys Mol Biol 137, 111–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo D, Antunes M, Prag S, Ma X, Hacker U, Brodland GW, Hutson MS, Solon J, Jacinto A (2011). DRhoGEF2 regulates cellular tension and cell pulsations in the amnioserosa during Drosophila dorsal closure. PloS One 6, e23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD (2013). Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell 24, 982–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard GB, Murugesu S, Adams RJ, Martinez-Arias A, Gorfinkiel N (2010). Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development 137, 2743–2752. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Brieher WM, Yap AS (2013). Cadherin junctions and their cytoskeleton(s). Curr Opin Cell Biol 25, 39–46. [DOI] [PubMed] [Google Scholar]
- Burr D, Ross J (1986). Visual processing of motion. Trends Neurosci 9, 304–307. [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, et al. (2007). The Carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175, 1505–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Shekhar S (2017). Global treadmilling coordinates actin turnover and controls the size of actin networks. Nat Rev Mol Cell Biol 18, 389–401. [DOI] [PubMed] [Google Scholar]
- Charras GT, Coughlin M, Mitchison TJ, Mahadevan L (2008). Life and times of a cellular bleb. Biophys J 94, 1836–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras GT, Hu CK, Coughlin M, Mitchison TJ (2006). Reassembly of contractile actin cortex in cell blebs. J Cell Biol 175, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA 3rd, Liu Z, et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P, Clark AG, Smith MB, Cassani DAD, Dierkes K, Ragab A, Roux PP, Charras G, Salbreux G, Paluch EK (2017). Actin cortex architecture regulates cell surface tension. Nat Cell Biol 19, 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coravos JS, Mason FM, Martin AC (2017). Actomyosin pulsing in tissue integrity maintenance during morphogenesis. Trends Cell Biol 27, 276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David DJ, Tishkina A, Harris TJ (2010). The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development 137, 1645–1655. [DOI] [PubMed] [Google Scholar]
- Dehapiot B, Clement R, Alegot H, Gazso-Gerhat G, Philippe JM, Lecuit T (2020). Assembly of a persistent apical actin network by the formin Frl/Fmnl tunes epithelial cell deformability. Nat Cell Biol 22, 791–802. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC (2011). Actin structure and function. Annu Rev Biophys 40, 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque J, Gorfinkiel N (2016). Integration of actomyosin contractility with cell-cell adhesion during dorsal closure. Development 143, 4676–4686. [DOI] [PubMed] [Google Scholar]
- Ecke M, Prassler J, Tanribil P, Muller-Taubenberger A, Korber S, Faix J, Gerisch G (2020). Formins specify membrane patterns generated by propagating actin waves. Mol Biol Cell 31, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KA, Demsky M, Montague RA, Weymouth N, Kiehart DP (1997). GFP-moesin illuminates actin cytoskeleton dynamics in living tissue and demonstrates cell shape changes during morphogenesis in Drosophila. Dev Biol 191, 103–117. [DOI] [PubMed] [Google Scholar]
- Erdmann RS, Baguley SW, Richens JH, Wissner RF, Xi Z, Allgeyer ES, Zhong S, Thompson AD, Lowe N, Butler R, et al. (2019). Labeling Strategies matter for super-resolution microscopy: a comparison between HaloTags and SNAP-tags. Cell Chem Biol 26, 584–592.e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez BG, Arias AM, Jacinto A (2007). Dpp signalling orchestrates dorsal closure by regulating cell shape changes both in the amnioserosa and in the epidermis. Mech Dev 124, 884–897. [DOI] [PubMed] [Google Scholar]
- Fischer SC, Blanchard GB, Duque J, Adams RJ, Arias AM, Guest SD, Gorfinkiel N (2014). Contractile and mechanical properties of epithelia with perturbed actomyosin dynamics. PLoS One 9, e95695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores LR, Keeling MC, Zhang X, Sliogeryte K, Gavara N (2019). Lifeact-GFP alters F-actin organization, cellular morphology and biophysical behaviour. Sci Rep 9, 3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke JD, Montague RA, Kiehart DP (2005). Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol 15, 2208–2221. [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Takahashi S, Tadakuma H, Funatsu T, Ishiwata S (2002). Microscopic analysis of polymerization dynamics with individual actin filaments. Nat Cell Biol 4, 666–673. [DOI] [PubMed] [Google Scholar]
- Gomez JM, Chumakova L, Bulgakova NA, Brown NH (2016). Microtubule organization is determined by the shape of epithelial cells. Nat Commun 7, 13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin K, Lostchuck EE, Cramb KM, Zulueta-Coarasa T, Fernandez-Gonzalez R, Tanentzapf G (2017). Cell-cell and cell-ECM adhesions cooperate to organize actomyosin networks and maintain force transmission during dorsal closure. Mol Biol Cell 28, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N (2016). From actomyosin oscillations to tissue-level deformations. Dev Dyn 245, 268–275. [DOI] [PubMed] [Google Scholar]
- Gorfinkiel N, Blanchard GB (2011). Dynamics of actomyosin contractile activity during epithelial morphogenesis. Curr Opin Cell Biol 23, 531–539. [DOI] [PubMed] [Google Scholar]
- Gratz SJ, Rubinstein CD, Harrison MM, Wildonger J, O’Connor-Giles KM (2015). CRISPR-Cas9 genome editing in Drosophila. Curr Protoc Mol Biol 111, 31.2.1–2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li D, Zhang S, Yang Y, Liu JJ, Wang X, Liu C, Milkie DE, Price R, Tulu US, et al. (2018). Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond time scales. Cell 175, 1430–1442.e17. [DOI] [PubMed] [Google Scholar]
- Hara Y, Shagirov M, Toyama Y (2016). Cell boundary elongation by non-autonomous contractility in cell oscillation. Curr Biol 26, 2388–2396. [DOI] [PubMed] [Google Scholar]
- Harris TJC (2018). Sculpting epithelia with planar polarized actomyosin networks: principles from Drosophila. Semin Cell Dev Biol 81, 54–61. [DOI] [PubMed] [Google Scholar]
- Hayes P, Solon J (2017). Drosophila dorsal closure: an orchestra of forces to zip shut the embryo. Mech Dev 144, 2–10. [DOI] [PubMed] [Google Scholar]
- Hirani N, Illukkumbura R, Bland T, Mathonnet G, Suhner D, Reymann AC, Goehring NW (2019). Anterior-enriched filopodia create the appearance of asymmetric membrane microdomains in polarizing C. elegans zygotes. J Cell Sci 132, jcs230714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem CC, Peifer M (2008). Diaphanous regulates myosin and adherens junctions to control cell contractility and protrusive behavior during morphogenesis. Development 135, 1005–1018. [DOI] [PubMed] [Google Scholar]
- Homem CC, Peifer M (2009). Exploring the roles of diaphanous and enabled activity in shaping the balance between filopodia and lamellipodia. Mol Biol Cell 20, 5138–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhou W, Dong W, Watson AM, Hong Y (2009). Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci USA 106, 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GL, Crawford JM, Genkins JZ, Kiehart DP (2014). Ion channels contribute to the regulation of cell sheet forces during Drosophila dorsal closure. Development 141, 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MV, Fernandez-Gonzalez R (2017). Coordinating cell movements in vivo: junctional and cytoskeletal dynamics lead the way. Curr Opin Cell Biol 48, 54–62. [DOI] [PubMed] [Google Scholar]
- Jayasinghe AK, Crews SM, Mashburn DN, Hutson MS (2013). Apical oscillations in amnioserosa cells: basolateral coupling and mechanical autonomy. Biophys J 105, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurado J, de Navascues J, Gorfinkiel N (2016). alpha-Catenin stabilises cadherin-catenin complexes and modulates actomyosin dynamics to allow pulsatile apical contraction. J Cell Sci 129, 4496–4508. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Crawford JM, Aristotelous A, Venakides S, Edwards GS (2017). Cell sheet morphogenesis: dorsal closure in Drosophila melanogaster as a model system. Annu Rev Cell Dev Biol 33, 169–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R (1986). Cytoplasmic myosin from Drosophila melanogaster. J Cell Biol 103, 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA (2000). Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J Cell Biol 149, 471–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart DP, Montague RA, Rickoll WL, Foard D, Thomas GH (1994). High-resolution microscopic methods for the analysis of cellular movements in Drosophila embryos. Methods Cell Biol 44, 507–532. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Tokutake Y, Chang M-S, Hutson MS, Wiemann J, Peralta XG, Toyama Y, Wells AR, Rodriguez A, Edwards GS (2006). Ultraviolet laser microbeam for dissection of Drosophila embryos. In: Cell Biology: A Laboratory Handbook, 3rd ed., vol. 3, ed. Celis JE, San Diego, CA: Elsevier Academic Press, 87–103. [Google Scholar]
- Lardennois A, Pasti G, Ferraro T, Llense F, Mahou P, Pontabry J, Rodriguez D, Kim S, Ono S, Beaurepaire E, et al. (2019). An actin-based viscoplastic lock ensures progressive body-axis elongation. Nature 573, 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PA, Bodmer R, Vincent JP (1995). Segmental patterning of heart precursors in Drosophila. Development 121, 4303–4308. [DOI] [PubMed] [Google Scholar]
- Ma X, Lynch HE, Scully PC, Hutson MS (2009). Probing embryonic tissue mechanics with laser hole drilling. Phys Biol 6, 036004. [DOI] [PubMed] [Google Scholar]
- Machado PF, Duque J, Etienne J, Martinez-Arias A, Blanchard GB, Gorfinkiel N (2015). Emergent material properties of developing epithelial tissues. BMC Biol 13, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston DJ, Higgins CD, Peters KA, Cupp TD, Dickinson DJ, Pani AM, Moore RP, Cox AH, Kiehart DP, Goldstein B (2016). MRCK-1 drives apical constriction in C. elegans by linking developmental patterning to force generation. Curr Biol 26, 2079–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Gelbart M, Fernandez-Gonzalez R, Kaschube M, Wieschaus EF (2010). Integration of contractile forces during tissue invagination. J Cell Biol 188, 735–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Goldstein B (2014). Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development 141, 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, Wieschaus EF (2008). Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason FM, Xie S, Vasquez CG, Tworoger M, Martin AC (2016). RhoA GTPase inhibition organizes contraction during epithelial morphogenesis. J Cell Biol 214, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Blankenship JT (2020). A nudge or a shove: altering actomyosin pulse profiles in vivo. Dev Cell 52, 135–136. [DOI] [PubMed] [Google Scholar]
- Morone N, Fujiwara T, Murase K, Kasai RS, Ike H, Yuasa S, Usukura J, Kusumi A (2006). Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J Cell Biol 174, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munjal A, Philippe JM, Munro E, Lecuit T (2015). A self-organized biomechanical network drives shape changes during tissue morphogenesis. Nature 524, 351–355. [DOI] [PubMed] [Google Scholar]
- Nishida S, Kawabe T, Sawayama M, Fukiage T (2018). Motion Perception: From Detection to Interpretation. Annu Rev Vis Sci 4, 501–523. [DOI] [PubMed] [Google Scholar]
- Nowotarski SH, McKeon N, Moser RJ, Peifer M (2014). The actin regulators Enabled and Diaphanous direct distinct protrusive behaviors in different tissues during Drosophila development. Mol Biol Cell 25, 3147–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasakarnis L, Frei E, Caussinus E, Affolter M, Brunner D (2016). Amnioserosa cell constriction but not epidermal actin cable tension autonomously drives dorsal closure. Nat Cell Biol 18, 1161–1172. [DOI] [PubMed] [Google Scholar]
- Peralta XG, Toyama Y, Kiehart DP, Edwards GS (2008). Emergent properties during dorsal closure in Drosophila morphogenesis. Phys Biol 5, 015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E (2011). Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods 8, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (2016). Actin and actin-binding proteins. Cold Spring Harb Perspect Biol 8, a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (2017). What we know and do not know about actin. Handb Exp Pharmacol 235, 331–347. [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, Bullock SL (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci USA 111, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M, Shemer G, Higgins CD, McClellan JH, Werts AD, Tulu US, Gao L, Betzig E, Kiehart DP, Goldstein B (2012). Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science 335, 1232–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan S, Meghana C, Narasimha M (2013). Local, cell-nonautonomous feedback regulation of myosin dynamics patterns transitions in cell behavior: a role for tension and geometry? Mol Biol Cell 24, 2350–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V (2012). Analysis of crinkled Function in Drosophila melanogaster Hair and Bristle Morphogenesis. PhD Dissertation. Durham, NC: Department of Biology, Duke University. [Google Scholar]
- Sokolow A, Toyama Y, Kiehart DP, Edwards GS (2012). Cell ingression and apical shape oscillations during dorsal closure in Drosophila. Biophys J 102, 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J, Kaya-Copur A, Colombelli J, Brunner D (2009). Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell 137, 1331–1342. [DOI] [PubMed] [Google Scholar]
- Spracklen AJ, Fagan TN, Lovander KE, Tootle TL (2014). The pros and cons of common actin labeling tools for visualizing actin dynamics during Drosophila oogenesis. Dev Biol 393, 209–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi A, Hayes P, D’Angelo A, Colombelli J, Salbreux G, Dierkes K, Solon J (2018). Adherens junction length during tissue contraction is controlled by the mechanosensitive activity of actomyosin and junctional recycling. Dev Cell 47, 453–463.e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland A, Lesko A (2020). Pulsed actomyosin contractions in morphogenesis. F1000Res 9, F1000 Faculty Res-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirukkumaran OM, Wang C, Asouzu NJ, Fron E, Rocha S, Hofkens J, Lavis LD, Mizuno H (2019). Improved HaloTag ligand enables BRET imaging with NanoLuc. Front Chem 7, 938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Bulinski JC, Salmon ED (1998). Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr Biol 8, 1227–1230. [DOI] [PubMed] [Google Scholar]
- Wioland H, Jegou A, Romet-Lemonne G (2022). Celebrating 20 years of live single-actin-filament studies with five golden rules. Proc Natl Acad Sci USA 119, e2109506119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E (1995). Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia of Drosophila. Cell 82, 67–76. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD (2005). Counting cytokinesis proteins globally and locally in fission yeast. Science 310, 310–314. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Rodesch CK, Broadie K (2002). Living synaptic vesicle marker: synaptotagmin-GFP. Genesis 34, 142–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.