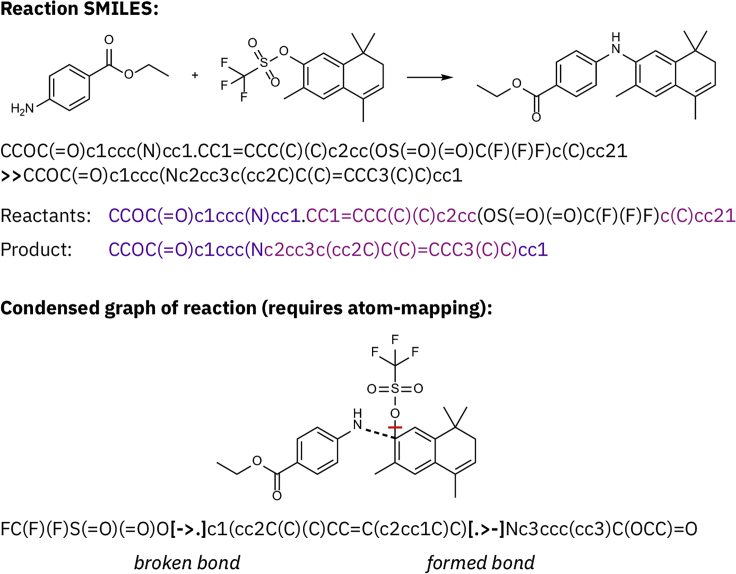
Summary
Artificial intelligence (AI) and machine learning (ML) are expanding in popularity for broad applications to challenging tasks in chemistry and materials science. Examples include the prediction of properties, the discovery of new reaction pathways, or the design of new molecules. The machine needs to read and write fluently in a chemical language for each of these tasks. Strings are a common tool to represent molecular graphs, and the most popular molecular string representation, Smiles, has powered cheminformatics since the late 1980s. However, in the context of AI and ML in chemistry, Smiles has several shortcomings—most pertinently, most combinations of symbols lead to invalid results with no valid chemical interpretation. To overcome this issue, a new language for molecules was introduced in 2020 that guarantees 100% robustness: SELF-referencing embedded string (Selfies). Selfies has since simplified and enabled numerous new applications in chemistry. In this perspective, we look to the future and discuss molecular string representations, along with their respective opportunities and challenges. We propose 16 concrete future projects for robust molecular representations. These involve the extension toward new chemical domains, exciting questions at the interface of AI and robust languages, and interpretability for both humans and machines. We hope that these proposals will inspire several follow-up works exploiting the full potential of molecular string representations for the future of AI in chemistry and materials science.
The bigger picture
Artificial intelligence for the discovery of new functional molecules can bring enormous societal and technological progress. Here, one crucial question is how to write molecules such that computers can easily process them. In this perspective, we analyze Selfies, a relatively young method for representing molecules in a computer. Since its invention 2 years ago, Selfieshas since simplified and enabled numerous workflows for artificial intelligence (AI) in chemistry and material science.
We take an in-depth look into the future of Selfies and molecular string representations. We detail 16 new future research directions, ranging from new AI applications in chemistry, to the development of robust languages for large chemical domains, to questions about the readability of different chemical languages for humans and machines. Thereby, we hope to open a myriad of exciting doors with consequences in materials science and beyond.
This community paper discusses Selfies, a relatively new representation for molecules at the computer. Selfieswas developed to overcome critical issues concerning the robustness of previously state-of-the-art representations in artificial intelligence applications. We overview the history of molecular string representations and the applications of Selfieswithin the last 2 years. We point out 16 concrete future research directions that will hopefully inspire the community and push ideas of robust representations in the realm of artificial intelligence and machine learning.
Introduction
The discovery of new materials and molecules with exceptional properties could lead to enormous scientific, technological, and ultimately societal impact. In the last few years, digital discoveries—that is, in silico discoveries using computers—have been significantly reinforced through machine-learning (ML) applications and other artificial intelligence (AI) tools for chemistry. Specifically, recent advances in AI and ML have sparked numerous new applications in quantum chemistry,1, 2, 3, 4, 5, 6, 7 molecular dynamics simulations,8, 9, 10 prediction of molecular properties11, 12, 13 and reactivity,14, 15, 16, 17 artificial molecular design,18, 19, 20, 21, 22 and the formulation of design heuristics.23,24 One germane question in all these applications is which language should be used to symbolically represent molecules and materials?
Since the 1980s, simplified molecular-input line-entry system (Smiles) strings have been a very prominent graph representation in computational chemistry. However, questions have arisen as to whether Smiles is an ideal language for computer applications that are tasked to discover new structures. For example, Smiles are not robust on their own, which means that generative models are likely to create strings that do not represent valid molecular graphs. A large body of work has been devoted to resolving this issue in recent years. Many of the advances came from model-dependent solutions, fixing the problem inside ML algorithms.25,26
In 2020, some of us introduced SELF-referencing embedded string (Selfies; Selfies can be installed via pip install selfies at https://github.com/aspuru-guzik-group/selfies).27 This new string-based representation circumvents the issue of robustness by defining a formal grammar that always leads to a valid molecular graph. This new molecular graph representation has simplified numerous applications in cheminformatics and even enabled new ones. Given this exciting potential, the authors assembled (in a virtual mini-workshop in August 2021 organized by IOP and the Acceleration Consortium, on the topic of this paper) to jointly discuss the future of Selfies in terms of generalizations and new applications. Here, we present an overview of the progress as well as outstanding questions, formulating 16 concrete projects and challenging ideas for the next years.
The perspective is structured as follows: we first summarize briefly the 250-year-long history of molecular representations. Then, we look at modern representations and discuss their strengths and weaknesses. This motivates a look into the future, where many open questions remain. In our journey, we also visit stochastic macromolecules and crystals. We will go further down the rabbit hole of inorganic chemistry and look at the potential for modeling and predicting chemical reactions. Then, we analyze the performance of string-based and non-string-based representations in terms of ML, and finally, we also investigate questions about the general interpretability of chemical languages—for both human and artificial scientists. During our journey through different fields of chemistry and AI research, we propose 16 independent stand-alone research projects that could define the future of molecular representations for AI in chemistry. Some of the proposed projects are well-defined and can (so we hope) directly be implemented, while other tasks indicate important problems in molecular representations that still need new conceptual insights to achieve a solution.
Our perspective mainly focuses on the new opportunities of Selfies. For more detailed reviews of general molecular representations, we refer the interested reader to Warr28 and Wigh et al.29 We want to be clear: Smiles has had a tremendous impact on cheminformatics since the 1980s and will certainly continue to be an impactful tool. Canonical Smiles together with structure normalization enables the definition of uniqueness, which is the current working pharmaceutical industry standard.30 For industrial applications, we note that Smiles was originally developed as a commercial tool, while Selfies is entirely open source and freely available, which is an important opportunity for Selfies for commercial products.
Historical review
Shaping the future of molecular representation is only sensible if we comprehend its history. Here, we briefly describe the 250-year evolution of chemical notations and the advent of modern string representations for molecules. Detailed accounts of the history can be found in other papers.31, 32, 33, 34, 35, 36
1787: The origin of chemical nomenclature is rooted in the seminal work Méthode de nomenclature chimique, with contributions from Lavoisier and others.37 This work ushered in the modern, post-alchemy era of chemical nomenclature.
1808: Dalton developed his atomic theory and used symbols to represent elements and compounds.38 These symbols resembled those used in the prior, alchemical era. For example, the elements hydrogen and sulfur were represented by and , respectively, while the compound water was represented as . However, such highly specialized symbols had two major drawbacks. Firstly, they were non-intuitive and therefore cumbersome for others to learn and apply. Secondly, they were incompatible with contemporaneous printing methods, resulting in limited circulation of Dalton’s work.
1813: Berzelius sought to address this by proposing a terminology where the first letters of the Latin names of a substance were used instead of symbols.39 This new notation represented chemical ratios rather than molecular structures.
1889–1911: International committees were formed to standardize the chemical nomenclature. The International Chemistry Committee published the Geneva Rules for Organic Chemistry in 1889. This was the first attempt to standardize chemical nomenclature.35 Nomenclature reforms continued with the International Association of Chemical Societies, which convened in 1911 in Paris. However, the proceedings were interrupted by the outbreak of World War I.40
1919–1930: The International Union of Pure and Applied Chemistry (IUPAC) was formed following the conclusion of World War I. In 1921, the Union continued to advance chemical nomenclature, culminating in 1930 with the so-called Liège Rules.36
1944–1947: While the outbreak of World War II interrupted the work of IUPAC, Dyson independently published a seminal work entitled A Notation for Organic Compounds in 1944.41 A revised version, A New Notation and Enumeration System for Organic Compounds, was subsequently accepted by IUPAC in 1947.33,42 The latter received criticism for not adding to the problem of chemical nomenclature, and those better explanations would be found in the original lecture in 1944. The claims in Dyson’s work were taken with reservations, especially the affirmation that there was only one possible cipher for any one chemical compound when there was not enough evidence and little scrutiny by the chemistry community.43 There was a feeling that he was prescribing a sledgehammer to crush a nut.
1949–1951: With the advent of computers, there was a new necessity to adapt chemical formulas to line notation using ASCII, thereby eliminating, among other features, the use of subscript and Greek letters.44 In 1949, the IUPAC Commission on Codification, Ciphering, and Punched Card Techniques opened a call for proposals regarding an international notation system. The criteria for the proposed annotation system included simplicity of use and ease of printing and typewriting. In 1951, the commission reviewed line notations with contributions from seven different proposals.45 From those, Dyson’s ciphering remained the standard, though many alternatives were used in practice. Among these, the Wiswesser Line Notation (WLN)31 is the most noteworthy. It provided a “compact way of uniquely and unambiguously representing the complete topology of a chemical molecule” and was preferred by scientists for many decades thereafter.34
1961–1969: During this era, the WLN method became the de facto standard in computer and punched card approaches to storing large datasets of chemical compounds.46 Subsequent efforts focused on automated hardware specially designed to codify molecules, like the Army Chemical Typewriter (Figure 1), or, alternatively, on improving machine readability and storage capacity, for example, the Hayward Notation (1961)47 and the Skolnik Notation (1969).48 In the former, the aim was to establish a basis for a one-to-one relationship between structure, cipher, and nomenclature, while for the latter it was to have the notations conform to the accepted chemical structures and invoke relatively few rules.
Figure 1.
Special-purpose type writer for chemistry
(A) Typical tape obtained with the Army Chemical Typewriter (ACT) built by members of the Walter Reed Army Institute of Research.
(B) The ACT, a mechanical typewriter for the encoding of chemical structures.
(C) Typed characters from the ACT. Image from Feldman et al.49
Modern molecular string representations
The development of molecular string representations has continued in the direction laid out by IUPAC in 1949. However, advances in computer power and cheminformatics applications have accelerated development far beyond the use cases originally envisioned. In the following section, we discuss four molecular string representations that are widely used today, with a focus on their applications in AI for chemistry and material science.
Smiles
Weininger published Smiles in 1988 with the goal to serve the needs of “modern chemical information processing.”50,51 The development of Smiles focused on the implementation of molecular graph theory, to allow for rigorous structure specification with a grammar that is both minimal and natural. Smiles has since become the de facto standard representation in cheminformatics.
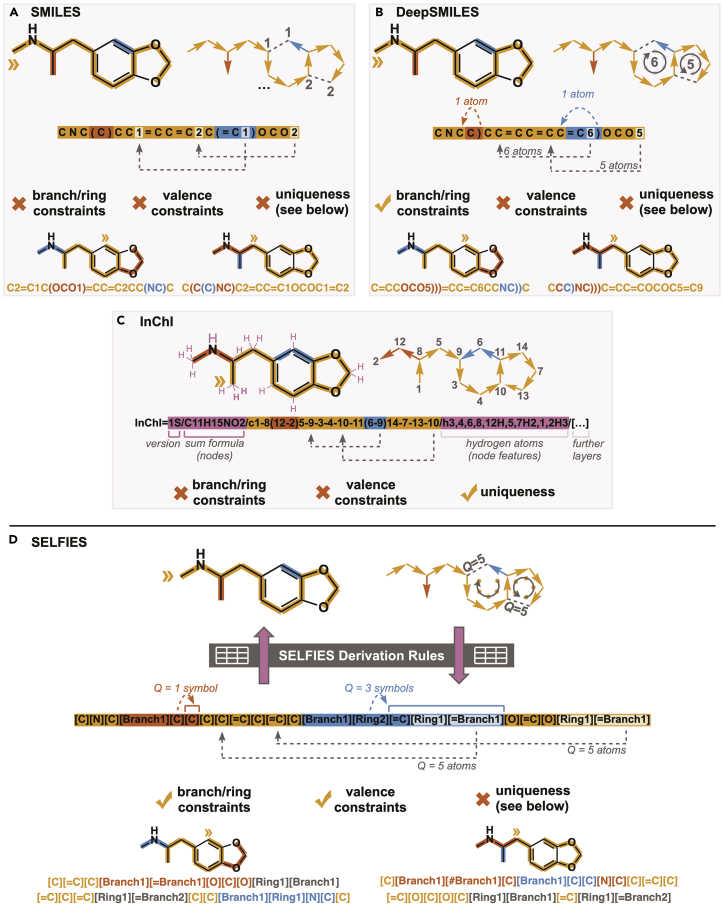
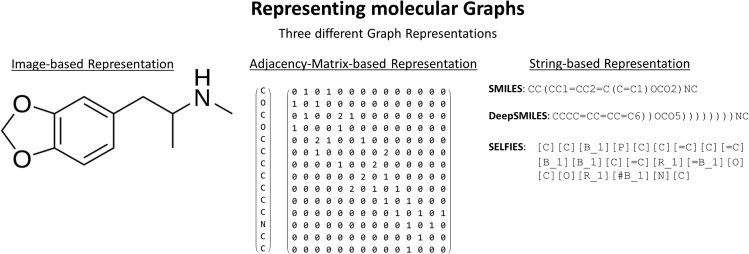
An example of the Smiles representation is shown in Figure 2. In Smiles, molecules are defined as a chain of atoms, which are written as letters in a string. Branches in the molecule are defined within parentheses, while ring closures are indicated by two matching numbers. The Smiles grammar, though simple, allows for the description of complex structures as well as properties such as stereochemistry, aromatic bonds, chirality, ions, and isotopes.
Figure 2.
Molecular string representations
(A–C) Derivation of established string representations (A) Smiles, (B) DeepSmiles, and (C) InChI from molecular structures using 3,4-methylenedioxymethamphetamine (MDMA) as an example. Branches and ring closures are represented by specific syntax based on the main path (orange).
(D) Derivation of a Selfies string from the molecular structure, building on the corresponding derivation rules.
While Smiles has been a workhorse for cheminformatics over the last three decades, in recent years, new applications in cheminformatics have exposed several weaknesses, which motivated the introduction of new molecular string representations. Firstly, multiple different Smiles strings can represent the same molecule (e.g., see Figure 2A). This weakness has been addressed by a different representation called the International Chemical Identifier (InChI), which we will explain below, and can be enforced by post-processing canonicalization via tools such as RDKit.52
Another weakness is that Smiles has no mechanism to ensure that molecular strings are valid with respect to syntax and physical principles. An example of the former is CC(CCCC, a string with an unpaired open parenthesis. This string has no valid interpretation as a molecular graph. Semantic errors involve strings that form valid graphs but do not reflect valid chemical structures. For example, the string CO=CC represents a molecular graph with an oxygen atom that has three bonds—a violation of the maximum number of bonds that neutral oxygen can form.
The lack of syntactic and semantic robustness has a significant impact with respect to the validity of computer-designed molecules based on evolutionary or deep-learning methods.18,53,54 One solution has been the design of special ML models that attempt to enforce robustness.25,55,56 A more fundamental solution is the modification of the molecular representation itself. O’Boyle and Dalke pioneered this approach by developing DeepSmiles, a modification of Smiles that obviates most syntactic errors, though semantic mistakes were still possible.57 Finally, 2020 witnessed the release of Selfies—a molecular string representation27 that is 100% robust to both syntactic and semantic errors.
InChI
Smiles are not unique representations of molecular graphs, i.e., a structure can be represented by multiple strings and custom identifiers. This makes it difficult to construct large-scale databases where each structure has to map to a unique label, and vice versa. InChI was created in 2013 by IUPAC as an open-source software to encode molecular structures in order to standardize searching across databases and the internet.58 InChI strings are composed of six main layers and multiple sublayers, where each layer represents a specific category of information about the molecule (sublayers include chemical formula, atomic connections, charges, and stereochemistry). There are several advantages introduced by the InChI syntax. The first is that molecules have a canonical representation, which allows straightforward linking in databases. O’Boyle created a method based on this feature of InChI that generates universal Smiles strings to standardize the output from different cheminformatics toolkits.59 Another benefit of InChI is that the layered structure encodes hierarchical information, and so two molecules that are derivatives of each other will have the same parent structure. Finally, InChI is more expressive than Smiles and can encode more information. For example, InChI can specify which hydrogen atoms are mobile and which are immobile.58 This allows for tautomers of the same molecule to be represented by the same InChI string, while with the Smiles framework, each tautomer is represented by a different string. Also, Smiles requires explicit notation of double-bond locations, while InChI infers them. Consequently, resonance structures are represented by a single InChI string but potentially multiple Smiles strings. There are also a number of disadvantages with the use of InChI strings. The first is that the hierarchical structure and syntax make the notation difficult to read by humans (although this is a point of contention, as the readability improves with usage; we come back to this aspect in comparing strings, adjacency matrices, and images as molecular graph representations for ML). The complicated syntax also makes it more difficult to employ InChI in generative modelling, as there are a number of arithmetic and grammatical rules that are difficult to enforce when sampling a new molecule from deep-learning models. Moreover, the current standard InChI consistently disconnects bonds to metal atoms, which leads to the loss of important stereochemical and bonding information. However, this behavior might change in future versions.60 In practice, it has been found that InChI performs worse than Smiles in ML-based applications, likely due to the above-mentioned reasons.54
DeepSmiles
Deep neural networks are increasingly used to create generative models for the design of new molecules.18 Many models were trained using molecules encoded as Smiles strings. These models are subsequently queried to generate Smiles strings representing molecules with specific target properties. However, the resulting Smiles may have unmatched parentheses or ring closure symbols, rendering the molecule invalid. To resolve these issues, O’Boyle and Dalke created DeepSmiles, which encodes into a syntax more suitable for automated inverse design such as deep generative models.57 The DeepSmiles grammar only uses one symbol to represent ring closures (instead of two). This symbol is a number that indicates how far back in the string the ring is connected. Branching is represented by one or more closing parentheses, where the number indicates branch length. Thereby, DeepSmiles resolves most cases of syntactical mistakes. This advance leads to greater robustness compared with Smiles with respect to random mutations and deep generative models.27 However, DeepSmiles strings still allow for semantically incorrect strings, i.e., molecules that violate basic physical constraints. This factor points to a need for an even more robust molecular grammar.
Selfies
Introduced in 2020, Selfies is a 100% robust molecular string representation.27 That is, Selfies cannot produce an invalid molecule, as every combination of symbols in the Selfies alphabet maps to a chemically valid graph. Let us imagine the same for a natural language, such as English. In the overwhelming majority of cases, an arbitrary combination of letters from the Latin alphabet (a–z) will not lead to a valid word. In this sense, English is not robust, while Selfies is robust with respect to chemistry.
Selfies is a formal grammar (or automaton) with derivation rules. This can be understood as a small computer program with minimal memory to achieve 100% robust derivation. The Selfies grammar is designed with the explicit aim of eliminating syntactically and semantically invalid molecules, for example in generative tasks.
In Smiles, syntactic invalidity consists of unbalanced parentheses or ring identifiers. For instance, a generative model using Smiles may generate a string that includes an open parenthesis with no corresponding closing parenthesis. The resulting string would represent an invalid graph. The problem stems from the non-local definition of rings and branches, which has already been addressed through the introduction of DeepSmiles.57 To resolve these issues, Selfies follows a different approach. Here, rings and branches are both defined at one single location. Special symbols (such as [Branch1] or [Ring1]) start a branch or ring. Instead of using an end symbol, the subsequent token in the string defines the length of the branch or ring. To achieve that, the next symbol is overloaded (similar to function overloading in programming languages allowing the creation of multiple functions with identical names but different implementations) by a number (see the concrete overloading list of Selfies v.2.0 in Table 1). We show one concrete example. The Selfies expression [C][Branch1][Ring2][C][C][C][C][C][C] has a branch symbol at the second position. Thereby, the subsequent symbol ([Ring2]) is overloaded and now defines the size of the branch (corresponding to the Q value in Figure 2). We see in Table 1 that [Ring2] stands for the number Q = 2. The length of the branch in Selfies is defined as (Q + 1), therefore the corresponding Smiles string is C(CCC)CCC’. Analogously, the sizes of rings can be described. In Selfies, we use base 16 to describe numbers (see Table 1). If we want to define branches or rings longer than 16 symbols, we can use [RingN]. Here, N stands for the number of subsequent symbols that are overloaded and combined (as a hex number) to describe long branches and rings. With these ideas, all syntactic mistakes are resolved.
Table 1.
List of Selfies symbols that are overloaded with numeric values if they appear after a ring or branch token
| Index | Symbol | Index | Symbol |
|---|---|---|---|
| 0 | [C] | 8 | [#Branch2] |
| 1 | [Ring1] | 9 | [O] |
| 2 | [Ring2] | 10 | [N] |
| 3 | [Branch1] | 11 | [=N] |
| 4 | [=Branch1] | 12 | [=C] |
| 5 | [#Branch1] | 13 | [#C] |
| 6 | [Branch2] | 14 | [S] |
| 7 | [=Branch2] | 15 | [P] |
| All other symbols are assigned index 0 | |||
It is a hexadecimal system, and larger numbers can be represented by overloading the next n symbols.
Semantic mistakes lead to molecular graphs that violate physical constraints. They are avoided by applying another concept from theoretical computer science—formal grammar or formal automata.61 The formal automaton derives the molecules, and every derivation step can change the state of the automaton. As the state defines the rules for the next derivation step, it can be used as a minimal memory that encodes physical constraints and ensures that only meaningful molecules are derived. Selfies can be seen as a very simple programming language for chemistry, and a Selfies string is a program that creates a valid molecular graph upon execution. This leads to interesting consequences and possibilities, which we will discuss in strings as programming languages.
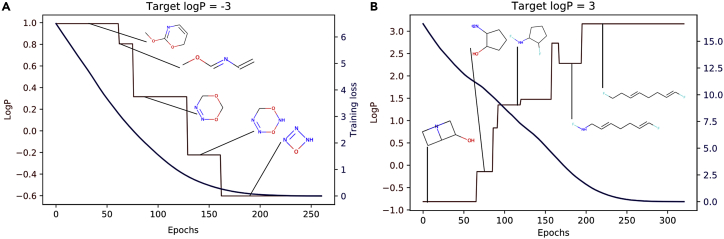
Robustness can be demonstrated by inspecting the internal latent space of a deep-learning model that is trained once with Smiles and once with Selfies (Figure 3). Without changing anything inside the ML model, every Selfies output is physically valid. Not surprisingly, Selfies has already been shown to improve, simplify, or even enable new AI-driven applications in cheminformatics. These include genetic algorithms,62 curiosity-based exploration,63 efficient combinatorial methods,64 and many other topics to be discussed later.
Figure 3.
Decoding points from the internal representation (latent space) of a variational autoencoder (VAE)
Green stands for valid and blue for invalid molecules. The left image is trained using Smiles strings, most of its latent space representing invalid molecular strings. The right image shows the latent space of a VAE trained with Selfies. Every point stands for a physically meaningful molecule. Figure from Krenn et al.27
The library contains two core functions that facilitate the translation between Smiles and Selfies representations, alongside other peripheral functions for manipulating Selfies strings. The following depicts a simple use case of Selfies:
import selfies as sf
benzene = “c1ccccc1”
# SMILES to SELFIES
benzene_sf = sf.encoder(benzene)
# [C][=C][C][=C][C][=C][Ring1][=Branch1]
# SELFIES to SMILES
benzene_smi = sf.decoder(benzene_sf)
# C1=CC=CC=C1
In this example, benzene is first translated to Selfies and then back to Smiles. The initial Smiles string is dearomatized to encode the molecule robustly in Selfies.
Current capabilities of Selfies
Currently, Selfies can represent ordinary organic molecules, including isotopes, and charged and radical species. Furthermore, it can represent chirality and stereochemistry by using an analogous approach to that of Smiles.
Selfies can not yet fully represent macromolecules, crystals, and molecules with complicated bonds. We will explain the context, the challenges, and potential ways to generalize Selfies to tackle these current shortcomings and to develop an even more general, 100% robust string representation for ML in chemistry.
Additionally, while the robustness can be guaranteed, it is not necessary that all molecules generated by a Selfies string can also be synthesized or are interesting or useful for specific tasks.
General mappings
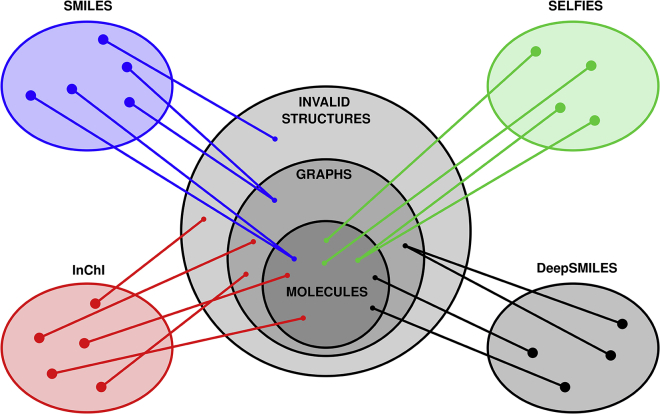
Selfies, Smiles, InChI, and DeepSmiles are representations of a molecular graph. They all aim to map a string of tokens to a molecular graph, as illustrated in Figure 4. Smiles is a surjective representation from strings to structures that include molecular graphs but also non-molecular (semantically invalid) graphs and other structures that cannot be interpreted as graphs (syntactically invalid). InChI has the same codomain, but its mapping is bijective, meaning each string corresponds to only one structure, and vice versa. DeepSmiles makes the first important advance in terms of validity and can be seen as a surjective mapping from strings to general (not necessarily molecular) graphs. Finally, Selfies is a surjective mapping from strings to molecular graphs. Both Smiles and Selfies can be made bijective through post-processing. For example, canonicalization (as provided by a number of tools such as RDKit) leads to a restricted domain, where each element maps to exactly one structure. It remains open whether a bijective mapping from strings to molecular graphs will be possible without post-selection. In the remaining text, we will discuss generalizations of Selfies and other molecular string representations along with important open questions. We will raise a number of concrete future projects, which can be seen as stand-alone projects that aim to further the development of molecular string representation and their applications in ML for cheminformatics.
Figure 4.
Graphical representation of the mapping from strings to their corresponding structures
Smiles maps to general structures that include molecules but also non-molecular graphs or invalid (non-graph) structures. InChI maps to the same space, although in a unique, bijective way. DeepSmiles maps strings to general graphs, not all of which stand for molecular graphs. Finally, Selfies is the only representation that maps in a surjective way only to molecular graphs.
Future project 1: metaSelfies—100% domain-agnostic robustness directly from data
So far, the discussion has focused on Selfies as a robust representation for molecular graphs. However, Selfies can also be thought of as a domain-independent robust representation for any graph in which vertices and edges have different semantic constraints. Selfies presently uses domain-dependent constraints, which limit the maximum number of bonds that can be used by an atom. Mathematically, this constraint can be formulated in terms of the maximum vertex degree in a molecular graph. Interestingly, the domain-dependent rules could be obtained directly from large datasets in a deterministic way, without using ML. A technical description of such an algorithm is presented in the supplemental information of Krenn et al.27
The derivation rules of Selfies are defined to satisfy the number of bonds a certain atom can form. In the language of graph theory, it constrains the vertex degree for each vertex type. Given a large enough dataset of example graphs, one can directly approximate the maximum allowed vertex degree for every vertex type. Thus, Selfies obtains its defining feature of robust derivation rules.
It is important to realize that vertex degree constraints can not only be formulated for molecules in chemistry but also for many other graph-based databases in the natural sciences. Examples include quantum optical experiments, where each individual optical element has a well-defined vertex degree constraint.65 In quantum circuits for quantum computers, individual gates have well-defined vertex degree constraints. RNA origamis66 in biology also have vertex degree constraints (in addition to other constraints) that can be extracted from large databases.
Therefore, the robust generation of graphs can be seen as the basis of Selfies (metaSelfies), while the vertex degree constraints define the scientific domain. The opportunity of extracting the full Selfies language from data only and the understanding that this language can be applied in diverse domains open up exciting opportunities. Given a particular dataset, it would immediately, without training, be able to generate 100% robust samples in the new domain, without anybody ever having to craft the language by hand. Additionally, a model could learn to solve design tasks in multiple domains. Given highly diverse training datasets, the opportunity for the generation of creative new solutions exists. For instance, one could us metaSelfies directly as the input of a variational autoencoder (VAE) or a generative adversarial network (GAN). The quality of this approach will significantly depend on the size and diversity of the dataset.
One can envision that domain-specific derivation rules could be shared in a standardized form in a Selfies registry, facilitating reuse by the community.
Future project 2: The effect of token overloading in generative models
One innovation in Selfies is the encoding of the sizes of branches and rings in a robust way. This is referred to as overloading and is done by enumerating the subsequent symbol(s) after the defining branch or ring token. Thereby, a token is interpreted as a hex number according to a table. A drawback of this way to ensure robustness is that it makes some Selfies more difficult to read. One important question is to understand how overloading impacts ML models and whether the index alphabet—which is currently heuristically composed—can be improved to enhance performance in ML models. It might be interesting, using attention mechanisms, to study how these models understand overloading and comtrast that with the way humans think about it.
Macromolecules
A challenging task in computational chemistry and biology is the simulation of macromolecules, which include biomolecules (nucleic acids, proteins, carbohydrates, and lipids) and synthetic polymers (e.g., plastics and synthetic fibers). Some macromolecules, such as polymers, are largely stochastic in nature and often feature a wide distribution over multiple chemical structures. In contrast, Smiles representations were created to describe deterministic structures such as small molecules, indicating that a new way of representing stochastic systems is needed.
One of the earliest macromolecule syntaxes developed was CurlySmiles,67 which provides a method for encoding repetitive units such as monomers. This method encodes monomers as well-defined structures. Thus, it is unable to capture any stochasticity or complex connectivity between monomers. To address this issue, Lin et al. developed BigSmiles,68 a polymer extension of Smiles that provides principles to represent the stochastic nature of polymers. A few syntax rules were added regarding the type of monomers and connectivity in the polymer. A schematic BigSmiles representation from Lin et al. is shown in Figure 5. BigSmiles therefore provides a list of building blocks that can be assembled stochastically at run time. Since BigSmiles inherited the basic syntax of Smiles and introduced new symbols that require matching, it also suffers from the invalidity of some representations.
Figure 5.
Schematic of BigSmiles representations from Lin et al.68
Polymers are represented as monomers (repeating units) enclosed within curly brackets; the curly brackets indicate that the molecule is a stochastic object. The monomers are represented as Smiles strings, with additional information expressing the connectivity between monomeric units.
Zhang et al. proposed Helm69 as a hierarchical way to represent large biomolecules. Unlike BigSmiles, which emphasizes the stochastic nature of synthetic polymers, Helm represents the full structure of a biomolecule with monomers replaced by their unique identifiers. That means that Helm does not represent individual atoms, but larger substructures are represented by symbols with the potential of repetitions. This idea allows the representation of much larger structures in a concise way. Helm, however, has the same drawback as Smiles with respect to reliance on matching parentheses, leading to reduced robustness for its usage in generative models.
Next, we describe two interesting stand-alone projects that could advance molecular string representations and their application in AI for macromolecules.
Future project 3: BigSELFIES—stochastically assembling building blocks for 100% robust polymers
Selfies can naturally be extended to biomolecule representations by combining the best of BigSmiles (stochastic repeating patterns) and Helm (amino acids). A sequence of amino acids can be encoded with standardized symbols (for example, V = valine), and every possible amino acid sequence is a valid representation. For the development of Helm-Selfies, one will need to identify grammatical rules for the entry and exit points of the amino acid sequence monomers or other macro-components. A challenge is that those rules likely go beyond individual bonding constraints, but this could be solved by adding more complex derivation states (i.e., memory during the derivation).
From these rules, BigSelfies, an extension of Selfies to stochastic derivation using predefined lists of monomers, will follow directly. This is because Helm-Selfies will need to work for every combination of monomers. During derivation, it will not matter whether the structure is built deterministically or stochastically. We anticipate that BigSelfies and Helm-Selfies will first be developed as stand-alone projects and, afterward, incorporated into the main Selfies language.
Such a new representation will allow for the application of generative models to large molecules and polymers, with minimal hand-crafted features in the model. The ML algorithm can directly work on the string representation, and all outputs are valid and interpretable structures. This approach will allow for the applications of both simple and fast algorithms that have been proven successful for organic molecule design.64 Furthermore, many deep generative models can directly be applied to design questions without any in-model conditioning or post-selection.
Crystals
A crystal is a periodic arrangement of atoms or molecules, commonly described by a set of lattice parameters, atomic coordinates, and symbols denoting symmetries other than translations. This description was standardized decades ago in the form of the crystallographic information file (CIF), which is widely accepted by the crystallography community.70,71 The connectivity between atoms/building blocks is often a useful abstraction for thinking about chemical structures and materials that can be represented as a graph. The introduction of molecular graphs can be traced back to the 1870s,72 but it was not until the late 1970s that periodic graphs were introduced to describe crystals.73,74 Such abstractions led to various applications in solid-state chemistry. Prominent examples include the “chemical diagrams” used in the Cambridge Structural Database (CSD) for structure search,75 connected coordination polyhedra to classify oxysalts,76 and net topologies in reticular chemistry.77
One can envisage an augmented version of Selfies that can be used to represent connectivity between atoms (the bond topology) in crystal structures robustly. String representations that have been explored for bond topology, such as the extended point symbols used in TOPOS78 for periodic graphs and the layered assemblies notation (LAN)79 for two-dimensional (2D) materials, are either non-invertible (the graph cannot be constructed from strings without a lookup table) or based on a structural prototype. Selfies, however, provides a mapping that loses no information when converting between sequence and connectivity and an explicit description of the connectivity. This allows for generative learning across the chemical space and supervised learning on sequences instead of crystal structures or graphs. String-based graph representations are ubiquitous in chemistry and biophysics because strings are easy to use, process, and store, and there is a vibrant ecosystem of tools like RDKit and deep-learning models for sequences that interface directly with strings. A robust string-based graph representation of crystals could inherit these advantages and transform materials informatics.
Net and quotient graph
What is the “crystal graph” that can be represented by a string? To answer this question, first, the basic terminology used in this section is introduced. For more formal definitions, see Delgado-Friedrichs and O’Keeffe.80 A crystal structure can be abstracted to a periodic graph, called a net, whose vertices represent the atoms (not atomic coordinates) and whose edges represent bonds between atoms. In practice, it might not be obvious which net best describes a crystal. The definition of edges can be ambiguous due to non-directional bonding or complicated coordination environments. For the latter, readers are referred to a recent benchmark of coordination number determination.81
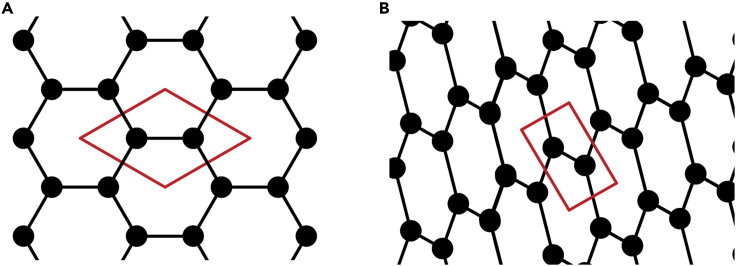
A net is an infinite, connected, undirected, simple (i.e., no loops and no multiple edges between a pair of vertices) graph. A net is n periodic (1 ≤ n ≤ 3) if it permits translations in n independent directions. Assigning coordinates to vertices constructs an embedding of a net. An embedding is faithful if edges do not intersect each other and only contain their respective end vertices. Two faithful embeddings of the same net are shown in Figure 6. Note how they share the same net even though they differ in their coordinates and cell parameters. Thus, to represent the connectivity in a crystal as a string requires representing a net that has a faithful embedding corresponding to the crystal’s real space structure.
Figure 6.
Nets for representing crystals
(A) Crystal structure of graphene (2D honeycomb lattice).
(B) 2D carbon structure of an orthorhombic lattice. The structures are two different faithful 2D embeddings of the same underlying net. This shows that a net, unlike its real space realization, does not bear spatial information (e.g., bond lengths, coordinates). Inspired by the success of Selfies in representing finite molecular graphs, in the section "Net and quotient graph," we discuss how Selfies can be extended to represent crystal nets.
Generally, a graph with an infinite number of edges cannot be described by a string of finite length. Fortunately, a net can be represented by a finite graph, known as its quotient graph.82 There are two variants of quotient graphs, one with directed, labeled edges, and one with undirected, unlabeled edges. Here, the focus will be on the former, which seems more suitable for developing crystal-Selfies (vide infra), since only the first uniquely determines a net.
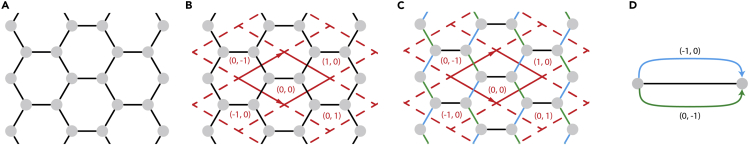
The procedure to generate quotient graphs is depicted in Figure 7 using graphene as an example:
-
1.
Start from an embedding E of the net N.
-
2.
For embedding E, define a coordinate system C including an origin and a set of basis vectors (2 vectors for 2D, 3 vectors for 3D) representing the periodicity of E. Index all cells by their positions with respect to the origin. For instance, the cell containing the origin is the (0, 0) cell.
-
3.
Group translationally invariant edges into edge classes (black, green, and blue in Figure 7).
-
4.
For each edge class, select one edge connecting a vertex in the (0, 0) cell and a vertex in the cell. Direct the edge starting from the vertex in the (0, 0) cell and label this edge as , where are restricted to .
Figure 7.
Construction of the labeled quotient graph (LQG) for the underlying net of graphene
(A) Embed the net corresponding to graphene.
(B) Define a coordinate system with two basis vectors (solid arrows) and an origin in the (0, 0) cell encompassed by solid lines. Index cells by their positions relative to the cell containing the origin.
(C) Group bonds into three bond classes (black, blue, green) by translational invariance.
(D) The result is the LQG. The label of (0, 0) bonds is dropped by convention.
The finite graph generated from this procedure is called the labeled quotient graph (LQG) of the embedding of the net N with coordinate system C. On the one hand, LQGs uniquely determine crystallographic nets up to isomorphism. An LQG can be converted to a net by choosing an arbitrary coordinate system or to a crystallographic net through its automorphism group.83 On the other hand, LQGs with two different labelings can represent a pair of isomorphic nets. Such labelings are called equivalent. Methods to check for equivalent LQGs can be found in a study by Chung et al.82
An unlabeled QG (UQG) can be obtained by removing edge labels and edge directions from an LQG. UQGs are more similar to molecular graphs and preserve the neighborhoods of vertices. Unfortunately, the same (up to isomorphic) UQG could be derived from two nets that are not isomorphic, and vice versa.84 Thus, UQG alone cannot be used to describe a net. However, it is possible to enumerate LQGs from a UQG by enumerating edge labels.85
Future project 4: LQGs in Selfies
From the above definitions, it appears that LQGs are most suited for string representation since they (1) are finite and (2) uniquely determine a net. LQGs have already been used in previous studies to represent crystals. A numerical encoding of LQG, the Systre key,86 was implemented to identify nets. More recently, the LQG implementation was employed in crystal structure generation using a VAE.87 While the current Selfies scheme is able to represent molecules with localized bonds robustly, to represent an LQG, several improvements are needed:
-
1.
Edges in a quotient graph (LQG or UQG) can be self-loops or parallel edges; these are not allowed in the current Selfies. The solution may be to treat them as size 1 and size 2 rings, respectively.
-
2.
There should be symbols for edge directions and edge labels such that the edge properties of an LQG can be represented.
-
3.
The choices for edge direction and edge label are finite, and not all labelings are allowed; for example, parallel edges cannot have the same labeling vector . There should be additional grammar that respects such (often local) restrictions.
-
4.
While an LQG uniquely determines a net, two non-isomorphic LQGs can represent the same net. This can happen in many cases, such as constructing an LQG from a supercell or from the aforementioned label equivalence. Thus, a canonicalization process is desired such that every net can have a canonical crystal-Selfies.
Future project 5: Crystal-Selfies in generative models
The search space for theoretical materials is practically infinite. While high-throughput virtual screening methods are now common in materials informatics and valuable for exploring new regions of materials space, generative models could provide a more systematic direction for targeted materials design. Generative models also aim to reduce systematic bias in the exploration of chemical space, allowing for a higher chance of discovery. By solving the missing pieces in the previous future project, Selfies could be augmented to crystal-Selfies, a lightweight and robust string representation of crystal (bond) topology that could improve crystal structure generation.
Currently, a few different approaches are followed to construct generative models for crystal structures. The first approach, employed mainly in the field of metal-organic frameworks (MOFs),88,89 starts from a net that is usually selected from established datasets. Appropriate building blocks are then chosen as nodes and their connections as edges of the net. The generation resembles the isoreticular expansion of MOFs. Such a method relies on predefined nets in addition to a set of available building blocks.
Another approach is to focus solely on embeddings. The embedding can be represented by a set of parameters based on a structural prototype,90,91 which may not be generalizable. Alternatively, embedding representations can be learned92,93 from datasets. Such representations are often continuous and thus suitable for inverse design. However, since bond topology information is not explicitly included, it is unclear whether this approach can generate topologically diverse structures.
Alternatively, it is possible to start with generating LQGs: in 2004, Thimm demonstrated that structures can be generated with minimal specifications (number of atoms in a unit cell and vertex degree for each atom) by (1) generating a UQG based on the specifications, (2) enumerating LQGs from the UQG, (3) unfolding the LQGs to nets, and (4) obtaining faithful embeddings from the nets.85 This method allows us to control the formation of types of nets over generated structures and does not rely on predefined nets. In addition, as discussed earlier, both LQGs and UQGs can be represented by crystal-Selfies. Thus, following Thimm’s approach, structure generation using crystal-Selfies can be, for example, a mapping: chemical composition UQG (crystal-Selfies) LQG (crystal-Selfies) net embedding.
A shortcoming of net-based representations is the obscure connections between the net of a crystal and the physical/chemical properties of that crystal. From a Smiles string or a molecular graph, properties (e.g., 2D descriptors) like logP can be readily estimated without embedding the graph (i.e., molecular conformations). However, for crystals, currently, both physical and chemical properties are calculated from embeddings. Thus, a calculator connecting net and crystal properties would greatly benefit the development of this field. It has been demonstrated that the dimensionality of a crystal structure can be derived from its LQG.94 More information regarding relations between a net and its embeddings can be found in a study by Blatov and Proserpio.95
Finally, for crystal generative models using Selfies, some general considerations are listed here:
-
1.
The alphabet of Selfies can be extended to include building units and linkers used in reticular or inorganic chemistry. This also helps to minimize the space of LQGs by reducing the number of vertices. An alternative would be to use contraction operations.
-
2.
It has been demonstrated that the symmetry and topological features of an LQG are related to that of the corresponding net.96,97 Thus, the model can be conditioned on these features.
-
3.
While a UQG does not determine a net, it does preserve neighborhoods. This means that it is possible to generate nets with specific local structures by making the neighbors of a vertex immutable.
-
4.
Some nets cannot be (faithfully) embedded in 3D. Crystal generative models should be conditioned such that these “pathological nets” are excluded from generations. Some properties used to identify such nets are introduced by Thimm.85
Beyond organic chemistry: Complicated bonds
In this section, we discuss the challenges and prospects of extending Selfies beyond organic chemistry. In contrast to organic molecules,60 transition metal, lanthanide, actinide, and main-group metal compounds are difficult to handle with current digital molecular representations28 due to special bonding situations and intricate 3D structures, combined with technical limitations that have evolved for historical reasons. Most problems trace back to (1) the assumption that bonding is localized and thus can be described with valence bond (VB) theory, (2) the non-explicit representation of terminal hydrogen atoms, which are added to the heavy (non-H) atoms based on rules derived from VB models in an approach called “implicit hydrogens,” and (3) the inability to describe stereochemistry that goes beyond the usual restrictions of organic chemistry, i.e., stereogenic carbon centers plus some cases of cis/trans isomerism in C=C double bonds and cumulenes. While organic chemistry has plenty of examples of more advanced stereochemistry such as planar and helical chirality,98, 99, 100 current digital molecular representations are generally not equipped to handle those.
Therefore, any approach toward a general digital molecular representation covering all elements of the periodic table will fail if it is unable to handle the issues mentioned above. Here, we will illustrate a number of prominent examples that highlight the urgent need to improve the situation, as otherwise, a major part of chemical space will remain inaccessible to modern cheminformatics and AI approaches.2
Complex, “fuzzy” bonding situations versus VB theory
One reason for including connectivity information in a molecular string representation is that it allows chemists to describe structures in a simple way, for example by decomposing them into substructures. Furthermore, from an ML perspective, connectivity information might also be thought of as an additional inductive bias that can help a model to generalize.101
However, bonding information turns into a significant technical problem if there is no algorithmically unambiguous way to define it102 and when there is a wide array of possible interactions of different strength and origin. This ambiguity in defining bonds has led some chemists to call them “convenient fiction,”103 which is also reflected in the widespread use of the bond type “any” for substructure queries in databases such as the CSD to ensure no entries are missed. In some domains of chemistry, VB theory provides a convenient and intuitive way to think about chemical bonding that is easy to encode in widely used data structures. In standard organic chemistry, for instance, most bonding situations can be described as two-center two-electron (2c-2e) bonds, a scenario that translates well into molecular string representations where atoms are nodes and covalent bonds between two atoms sharing two electrons are edges of a molecule graph. However, as the OpenSmiles standard notes, “This simple mental model has little resemblance to the underlying quantum mechanical reality of electrons, protons, and neutrons …”104
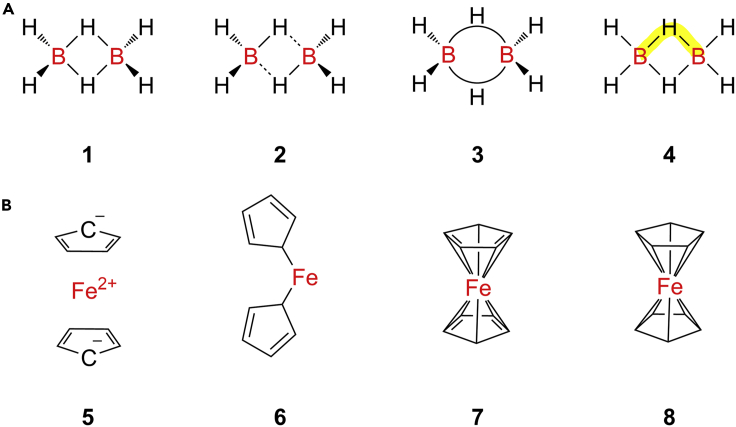
Two prominent examples from main-group element and transition-metal compounds, respectively, will be discussed here to outline the corresponding major issues. Figure 8A shows four different molecular structural models for diborane (B2H6), an important reducing agent and key reactant for hydroboration reactions. Most (inorganic) chemists, when asked to sketch the molecule, will likely draw structure 1, which properly captures the two bridging -hydrido ligands but results in an incorrect valence electron (VE) count of 16 VEs instead of the proper 12 VEs, when each line connecting two element symbols is assumed to represent two electrons. In order to preserve the electron-counting function of the lines representing 2c-2e bonds, sometimes structure 2 is used, wherein additional interactions between the two BH3 subunits are indicated by dashed lines, which are assumed not to contribute to the electron counting and thus have been termed “zero-order bonds” by Clark.105 However, this structure 2 incorrectly implies the symmetry of the molecule to be C2h, while X-ray structure analysis has demonstrated that diborane belongs to the D2h point group. All four terminal B–H bonds are equivalent at approximately 1.09 Å, and the four B–H distances in the B2H6 “diamond-shaped core” are also essentially equivalent at about 1.24 Å. Notably, the observed differences of 0.03 Å in these formally equivalent B–H bond distances are possibly caused by packing effects.106 Therefore, some chemistry textbooks use structure 3 with two bent “banana bonds,” with the two arched lines each representing two VEs. Such a representation, although it gives the correct VE count, cannot be used in standard molecular graphs, which assume that each edge connects two—and only two—nodes (atoms). A better description of the structure of diborane makes use of 3c-2e bonds, where two electrons are fully delocalized over the B–H–B unit, as highlighted in yellow in structure 4.
Figure 8.
Examples of molecules with complicated bonds
(A) Different structural representations for diborane (B2H6), where 1 properly accounts for the symmetrical B2H6 “diamond core” but gives an incorrect valence electron (VE) count; 2 uses zero-order bonds, indicated as dashed lines, to preserve the VE count but features a molecular symmetry that is too low; 3 attempts to capture the actual three-center two-electron (3c-2e) bonding by use of arced “banana bonds” but cannot be used in molecular graph approaches, which only allow for each edge to connect two nodes (atoms); and 4 shows the full delocalization of an electron pair over the B–H–B unit.
(B) Lewis structures of ferrocene (C10H10Fe), where 5 is unfortunately used by PubChem but is wrong, as the compound is not ionic. 6 and 7 cannot account for the 1H and 13C NMR spectra, both of which feature only one singlet, indicative of ten chemically equivalent CH units. Only 8 is fully in line with crystallographic and spectroscopic data but at the expense of making electron counting impossible.
Another complex bonding situation arises in organometallic “sandwich” complexes such as ferrocene (C10H10Fe), which are common building blocks in organic chemistry and have important industrial applications, for example in Ziegler-Natta catalysis.107 Some databases such as PubChem108 utilize ionic structure 5, as shown in Figure 8B, assuming a “naked” Fe(II) cation without any coordinated ligands, combined with two separate cyclopentadienyl anions. This structure, however, is utterly wrong, as ferrocene is a compound without separate charged ions that can be purified by vacuum sublimation and is insoluble in polar solvents such as water but dissolves well in non-polar organic solvents such as n-hexane and toluene. The uncharged structure 6 would be in line with these properties but does not account for the 1H and 13C nuclear magnetic resonance (NMR) spectra, which both exhibit only one single peak, indicating that all ten CH units are chemically equivalent, while the NMR spectra of representation 6 would feature three different peaks for each nucleus. Furthermore, two-coordinate iron centers are exceedingly rare and require very bulky ligands to be stabilize.109 Alternatively, structure 7 has the Fe(II) center sandwichedbetween the two cyclopentadienyl rings but still cannot account for the NMR spectra due to the combination of two localized C=C double bonds and one carbanionic center per ring. Only structure 8 correctly captures both the NMR properties and the X-ray data, which indicate ten equivalent Fe–C and C–H bonds and an identical length for all ten C–C bonds.110 This, however, goes at the expense of any kind of VE counting, as the actual bonding requires a molecular orbital (MO) treatment that at least considers both the cyclopentadienyl system and the iron d orbitals. The situation becomes even more complicated when one attempts to capture not only covalent bonds but also weaker agostic interactions, in which the two electrons of a C–H bond interact with empty metal d orbitals in another example of 3c-2e bonding. The same applies to other weak interactions such as hydrogen bonds, raising important questions as to which interactions should actually be captured in a digital molecular representation as a “bond” (and which should not) and how to automatically detect them from a set of atomic coordinates, ultimately leading to a rather arbitrary distinction between bonded and non-bonded. To quote Democritus: “Nothing exists except atoms and empty space; everything else is only opinion.”
No “standard” valences
Current molecular string representations make use of models based in VB theory, as it allows the definition of standard valences for the different elements. Missing hydrogen atoms are inferred and inserted implicitly, which allows for a more compact representation. These standard valences are usually fixed to satisfy the octet rule, which is not generally applicable. Even many main-group elements do not follow that rule. For the d and f elements, such a rule is largely irrelevant due to strongly delocalized bonding with significant mixing between metal and ligand orbitals that require an MO theory treatment, something that cannot be captured by structural representations exclusively based on 2c-2e bonds.
For example, while the noble gas elements have to be formally assigned a standard valence of zero, many stable compounds with them, such as XeOF4, are known and readily prepared. Even carbon does not necessarily obey the octet rule, as the catalytic center of nitrogenase, the enzyme that is central for biological nitrogen fixation, contains an FeMo cofactor with the composition [Fe7MoS9C] that is built around a carbide center with a formal charge of −IV and six equivalent Fe–C bonds, as demonstrated by X-ray crystallography.111 Beyond such surprising structural motifs created by nature itself, inorganic chemists in particular constantly look for new oxidation states112 and bond orders.113,114 Furthermore, there needs to be a critical discussion of the term valence itself, as in inorganic chemistry, it is normally used to describe the physical oxidation state (related to the spectroscopically accessible d-electron count) of a metal center (e.g., trivalent iron is Fe(III), which is usually six coordinate), while in the context of InChI and Smiles, it refers to the number of bonds to neighboring atoms. Therefore, any approach to generally applicable digital molecular representations should not make use of standard valences and needs to treat all hydrogen atoms explicitly.
Stereochemistry beyond the tetrahedron
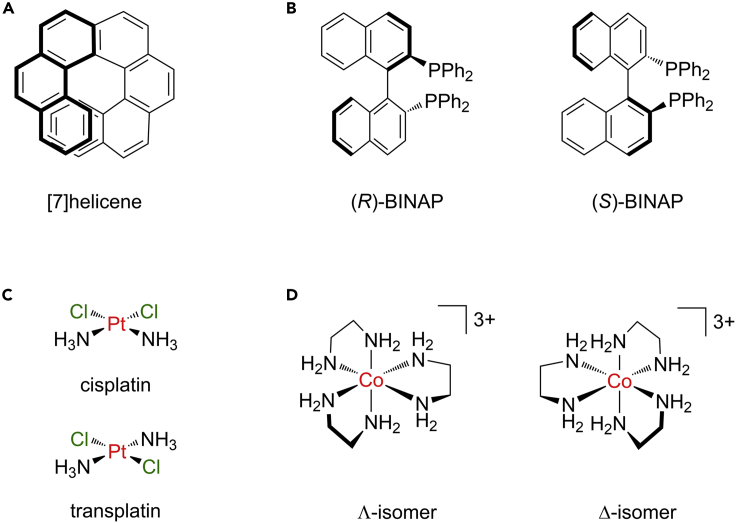
Most organic molecules feature either linear sp, planar sp2, or tetrahedral sp3 carbon centers, and, thus, their stereochemistry is usually restricted to point chirality from stereogenic centres, cis/trans isomerism of C=C double bonds in alkenes, or axial chirality in allenes/cumulenes. However, in more complex structures, even within organic chemistry, planar or axial chiral elements can additionally come into play. Prominent examples of the latter include ortho-condensed polycyclic aromatic compounds from the class of the [n]helicenes (Figure 9A). Such systems are far from academic curiosities, as axial chirality is important to enantioselective catalysis. This is apparent in the BINAP class of ligands, for which Noyori was awarded the 2001 Nobel Prize in Chemistry (Figure 9B).
Figure 9.
More examples of molecules with complicated bonds
(A and B) Examples of (A) helical and (B) axial chirality in organic compounds
(C) Diastereomeric coordination compounds: cisplatin is an approved anticancer drug, while its isomer transplatin is inactive.
(D) Helical chirality in metal complexes.
Furthermore, metal complexes are characterized by a wide range of coordination geometries with coordination numbers in the range of 2–16. The structural motif assumed is often dictated by electronic ligand field (LF) effects rather than steric repulsion, as in the widely used VSEPR model applicable to main group chemistry. For example, a metal center with four ligands, in addition to a tetrahedral structure, could also assume a square-planar coordination environment, where the central metal atom and the ligands are in one plane, with L-M-L angles of 90° and 180°, respectively. In MA2B2-type compounds, this gives rise to two stereoisomers, with cis- and trans-[PtCl2(NH3)2] as some of the most important examples (Figure 9C). The compound cisplatin is an approved anticancer drug with wide applications in chemotherapy and annual multibillion-dollar sales, while transplatin shows no biological activity. Unfortunately, PubChem considers both compounds simply as “synonyms” and thus provides an incorrect record for them.115 The reason for this is rooted in the erroneous application of the concept of standard valences. Since the Pt(II) center is assigned a valence of two, the compound is incorrectly represented as a mixture of a bent(!) PtCl2 unit and two separate NH3 molecules to also preserve the standard valence of three for nitrogen. However, the two ammine ligands are bonded to the metal in a fashion that is comparable to covalent bonds in organic chemistry, and in aqueous solution, it is actually the chlorido ligands that are exchangeable to water, not the ammine ligands. When moving from four to six coordination, the range of accessible structures becomes even broader, and one has to additionally consider new stereocenters generated by fixation of ligand atoms to the metal, which can lead to helical structures, as discovered by Alfred Werner more than 100 years ago116 (Figure 9D). To complicate matters even further, coordination numbers of 12 and higher have been reported. One example is [Ph4P][Hf(BH4)5], in which each borohydride unit [BH4]− act as either bi- or tridentate ligands to the Hf(IV) metal center, which leads to a maximum possible coordination number of 5 × 3 = 15.117
Alternative approaches
Many alternative molecular representations that have been put forward try to be more faithful in representing chemical concepts such as multicenter bonds or stereochemistry.
Separation of - and -electron systems
In conventional molecular string representations (e.g., Smiles and Selfies), atoms are considered to be nodes and bonds to be edges of a molecular graph. These are then assigned numerical values such as atomic number, number of unshared electrons, and bond order, which are considered invariants of the graph, as they do not depend on the labeling scheme of the nodes (atoms).118 Most approaches allow all edges to connect just two nodes, in line with the standard 2c-2e bonds that dominate most of organic chemistry.
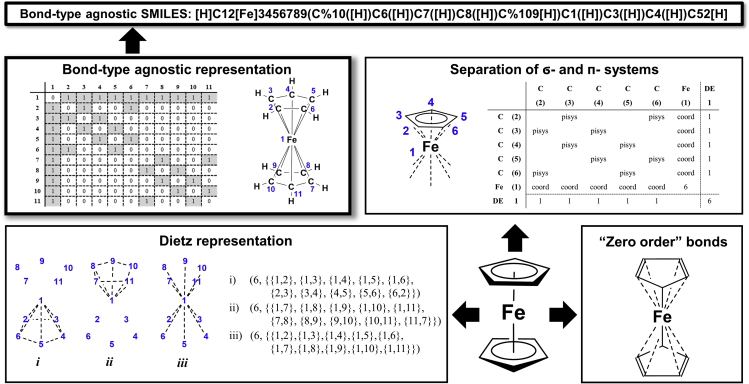
In the symbolically extended BE (sXBE) matrices,119, 120, 121, 122 however, delocalized electron systems are encoded using special bond types such as pisys (e.g., benzene) or edsys (for electron-deficient systems such as boranes). Therefore, these representations allow for a better representation of the true multicenter bonding nature of some systems such as diborane or ferrocene (Figures 10A and 10B).
Figure 10.
Current possibilities to represent molecules with complicated bonds (here ferrocene)
Top left: bond-agnostic edges neglect some physical constraints and can be written as Smiles or a graph. Top right: separation of - and -electron systems. Bottom left: Dietz representation. Bottom right: zero-order bonds.
Dietz representation
As an alternative, Dietz suggested a hypergraph concept123 where edges are allowed to contain more than two nodes, accounting for multicenter bonding (Figure 10C). However, the approach of Dietz, Ugi, and Stein is based on groups of nodes and edges, which are additionally characterized by the number of unshared VEs and delocalized electrons.118 This approach tries to exactly capture the electronic structure but leads to complicated nested sets of brackets that may be hard to comprehend. Furthermore, a clear assignment of VEs is often not possible in transition-metal chemistry due to extensive delocalization. Consequently, as the resulting representation and terminology is difficult to tackle, to our knowledge, they have not been used in any digital structure representation to date. Furthermore, as noted by Bauerschmidt and Gasteiger, the Dietz system (and all others described so far) cannot easily distinguish between different spin states of the electrons.124 This is relevant for carbenes, where the singlet and triplet states have a vastly different reactivity, and also applies to molecules as simple as dioxygen. Hence, together with its complexity, this representation has not found widespread use.
Zero-order bonds
To address the issue of multicenter bonding, non-specified bond orders, and the related problems with implicit hydrogens, in 2011, Clark proposed two backward-compatible modifications to connection table (CT)-based molecular representations.105 In that work, it was suggested to allow for a bond order of zero for all interactions or bonds that do not fit the conventional scheme and to add a property that explicitly describes the number of connected hydrogen (Figure 10D). Interestingly, the zero-bond order reflects the fact that, due to the ambiguity of bond orders, many chemists perform database substructure searches with “any” as the bond type. However, as discussed in the previous section (Figure 8A, structure 2), this can lead to an incorrect decrease in molecular symmetry. There are also cases where ambiguities appear regarding which bonds should be denoted as zero order and which ones should not. A common resort to be expected in that context is that many users will then simply label all bonds as zero order.
Thus, it should be stressed again that in d- and f-block chemistry, as well as main-group organometallic compounds, it is often impossible to assign any particular bond orders without high-level quantum chemical calculations, due to the highly delocalized nature of the bonding, where electrons are often spread out over a significant number of atoms, including the metal center itself, the immediately coordinated atoms, and additional ligand groups. In summary, despite more than 25 years of research into the issue, little progress has been made toward a generally applicable and domain-independent digital molecular representation, as some of the concepts that representations are built upon (standard valences, 2c-2e bonding, and the possibility to assign bonds and bond orders unambiguously) are ill defined for many compounds outside of classic organic chemistry.
Tooling and the value of simplicity
In this section, a number of essentials characterizing molecular assemblies of atoms and what is needed to create a digital representation thereof are outlined. The high variability of metal complexes, in particular in terms of electronic structure and coordination geometry, calls for a flexible and extensible “layer approach,” in which the essentials strictly required to describe a molecular structure are included in a base layer, while all domain-specific information is covered by additional and user-definable property layers, which can be used or ignored depending on the users’ goals.
-
1.
Base layer (domain independent): The nodes (atoms) “carry” the atomic number and (non-standard) isotope distribution. Edges (bonds) indicate strong pairwise attractive interactions, although it remains to be defined which interactions should be captured and which ones not.
-
2.
Property layer #1 (domain dependent): Nodes carry information about local stereochemistry and charge; edges carry bond order and type information (such as single, double, triple, aromatic bonds).
-
3.
Higher-level property layer #2 (domain dependent): Information from ML models, handcrafted information, experimental data such as NMR chemical shifts, “strategic bonds” for either retrosynthesis or reactivity prediction.
An interesting aspect of the additional property layers is that, beyond certain values assigned based on user interaction or software-encoded domain-specific models, these might also be generated from ML approaches, which could allow for a more nuanced picture than simple binary assignments often governing current models.125 To conclude, the need to describe all of chemical space is at odds with imposing strong rules on the allowed valence or connectivity, and more elaborate derivation rules need to be developed.
Future project 6: Generalization of Selfies and automatic compilation of complex rules from data
Many of the properties described above could directly be implemented into string-based representations, following IUPAC recommendations. For example, to represent non-tetrahedral metal complexes, the coordination environment can be specified by adding the “polyhedral symbol”126 to the Selfies string. A general approach for the representation itself is outlined in the previous section (tooling and the value of simplicity).
These thoughts are applicable to general string-based representations. We now focus on the possibility of defining a robust generalization of Selfies that incorporates molecules beyond VBs. The following idea is one possibility to achieve this goal—however, it is clear that it requires more clever ideas or a modified way to practically achieve a robust representation of molecules with complex bonds.
Most chemists may possibly agree on which structures are “correct” (and which are not) by visual inspection of structural formulas. As this ability is based on knowledge obtained by inspection of other compounds and the underlying trends that govern their bonding, it should be possible to train an ML model to deduce these rules (i.e., the necessary extended Selfies grammatical rules) for general Selfies from an appropriate dataset. This project is a further extension of the topic described in future project 1 (metaSelfies). One of the most extensive and curated structure collections is the CSD. However, one has to keep in mind that there will be biases in such a dataset that need to be accounted for. For example, the CSD only contains compounds that could be crystallized and were deemed to be of sufficient interest for X-ray structure analysis. This could potentially be corrected by supplementing the model with data from other databases and by the addition of manually selected structures. Furthermore, state-of-the-art quantum chemical calculations are nowadays able to provide optimized geometries that often approach the accuracy of experimentally obtained structures and might thus also be of interest to feed to such models. One potential means of progression is to create a neural network that learns to classify compounds into “correct” or “incorrect” categories. After training, symbolic regression127 could be used to extract symbolic rules that can be used directly by Selfies.
Reactions
So far, we have discussed only representations of molecules. However, a significant part of chemistry consists of the modifications of molecules via reactions. In this section, the applications of ML in reactions are discussed and what role molecular representations play.
A chemical reaction can be divided into four distinct parts: reactants, agents, products, and overall conditions. Products are the outcome of the reaction or the molecule(s) obtained once the reaction is done. Reactants are the building blocks of the product(s): the initial compounds containing atoms that will be incorporated into the product. Agents can be anything from catalysts to solvents that are added to the reaction mixture but will not be part of the product molecule(s). (This is a simplification, as sometimes it is not possible to identify which molecule contributes to the product, such as in reactions involving protic catalysts.) Conditions are, for example, the temperature and pressure at which the reaction is run or other more complex variables such as heating profiles, the order of addition of reactants and agents, and so on. The agents and conditions describe the environment in which the reaction happens. Depending on the available dataset, conditions and agents may not always be fully described.
Openly available datasets are derived from either patents128,129 or chemical journals130 and, more rarely, experimental procedures directly.131 These datasets are distributed using Smiles as a representation for the reaction itself and usually include extra information in various formats. There is no standard format that allows for conveying information about reactions and their details simultaneously. Initially intended for organic chemists, these datasets also attracted the attention of computational chemists, as they enabled the development of new methods and algorithms. The Open Reaction Database provides a centralized platform to collect and access reaction datasets.132
Chemical reactions are commonly investigated in ML for chemistry regarding two broad categories: reaction completion and property prediction. Usually, the full reaction is provided when running property predictions. A typical variable to predict could be the yield of the reaction or the energy profile. Reaction completion consists of completing a reaction scheme, where some of the molecules or conditions are missing. Two subcategories of interest are reaction prediction, where the goal is to predict a product based on a given set of reactants, and retrosynthesis, where the goal is to predict a set of reactants given a particular product. Likewise, prediction of reaction conditions and/or agents represents a major current challenge.
-
1.
Reaction completion
-
(a)
Reaction prediction
-
(b)
Retrosynthesis
-
(c)
Condition and agent prediction
-
2.
Property prediction
Reaction completion is the category of tasks where the representation matters most, as algorithms not only take molecules as input but also need to output molecules. Therefore, the main discussion here will be about possible algorithms and representations of reactions with respect to reaction completion.
There are three broad categories of methods designed for reaction completion:
-
1.
Template-based methods
-
2.
Graph-based methods
-
3.
Text-based methods.
Template-based methods use a set of reaction templates that encode the possible changes effected during a reaction. These templates are either written by domain experts133 or are directly extracted from data using atom mapping.134,135 Atom mapping links the product atoms with the corresponding reactant atoms and, hence, specifies the reaction center. In template-based reaction completion methods, it is common to see the outcome of these templates ranked by a neural network134,135 to define which reaction is the most likely to happen. Graph-based methods134,136 typically use graph neural networks (GNNs). Generally, this kind of method splits the project into two subtasks: the first step localizes where the changes in the graph should happen by selecting atoms, and in a later step, the changes are performed. Similar to template-based methods, the bond changes used for training of the graph-based methods are extracted from atom mapping. Therefore, their performance depends on the quality of the underlying atom mapping.137
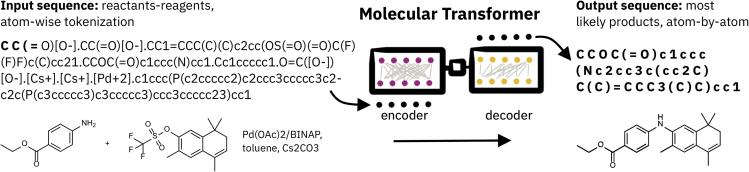
Text-based methods use textual representations of molecules to take advantage of models initially developed for neural machine translation, such as the transformer model138 (see Figure 11). Such sequence-2-sequence methods for forward prediction, retrosynthesis, and agent completion can be atom-mapping independent, as the reactant and product atoms do not have to be linked in the training reactions.139, 140, 141
Figure 11.
An example of a molecular transformer, which uses Smiles to represent and transform reactant and agent molecules into the product of the reaction, as used by Schwaller et al.139
The tokenization of the Smiles is shown by the bold characters separated with spaces.
All these reaction completion methods could benefit from improving the underlying representation of the reactions they are using. The following paragraphs will focus on the most promising improvements, and we will discuss how the three methods presented will benefit from it.
The reactions present in the current datasets are rarely balanced, meaning not every atom from the left-hand side of the chemical equation can theoretically be mapped to an atom to the right-hand side. Indeed, in the literature, parts of a reaction are often omitted when they are either considered irrelevant or are unknown (for instance, not mentioning the side products) or so obvious it does not need to be mentioned (for instance, disregarding counterions or necessary byproducts such as CO2). While this makes sense when a human reads a reaction, since it improves clarity, it would be beneficial if the reactions were complete for an algorithm to learn from them. For graph-based methods, this would reduce the number of graph edits that need to be predicted as there would be less variation on both sides of the reaction. For text-based methods, this would allow a user to enforce an atom count at inference, which would most likely improve the performance. Finally, template-based methods would also benefit, as the templates extracted from the data would be more consistent.
A way to enforce the atom count of a reaction would be to describe only one side of the reaction, for instance the reactants, and then describe only the changes happening during the reaction.142 This would not only enforce balanced reactions but also remove the unnecessary redundancy of the current representation, as illustrated in Figure 12. Bort et al.143 proposed the use of such a text-based condensed graph of reaction (CGR) representation to perform property prediction. Extra symbols were added to the reactants to describe the reaction. This representation is well-suited for template-based methods, as it turns every reaction into a ready-to-use template. This would also be convenient for graph-based methods, as there is no need to extract the graph edits. Further work is required to make this kind of representation useful for text-based methods. The application of such methods is difficult if there is no separation between the changes and the initial molecules, which to some extent also applies to graph-based methods.
Figure 12.
In most cases, the changes happening during the reaction affect only a small fraction of the molecule, and everything else is left unchanged
However, current representations, like reaction Smiles, do not capture that, and major parts of the molecules are actually repeated. In contrast, condensed graphs of representation (CGRs) represent the bond changes in the reactions. To generate a CGR from a reaction Smiles, the atom mapping has to be determined first. Agents and conditions are not shown in the figure.
However, the atom mapping that enables extracting reaction templates or graph edits and building CGRs is typically not directly available for experimentally observed reactions. Moreover, human labeling is prohibitively time consuming for large databases. Traditionally, automated atom mapping was performed using extended-connectivity-, maximum common substructure-, and optimization-based approaches.144 Schwaller et al.137 recently showed that accurate atom mapping could be learned from reactions represented as Smiles without existing atom mapping through unsupervised training.
So far, we have discussed methods to improve the representation but have not considered extending Selfies to represent reactions. We will consider two cases: a representation that is syntactically robust, and one that is semantically robust. A syntactically robust representation would ensure the validity of the graph edits proposed. However, this would not guarantee that the results make sense chemically. This is the goal of the semantically correct representation. In the following project, we will discuss the benefits and the feasibility of such a representation.
Future project 7: Graph edit rules and metaSelfies for reactions
A syntactically robust reaction representation would most likely improve the performance of predictive models, as it is no longer possible to predict an invalid representation or an invalid graph edit sequence. To achieve this representation, the rule set that defines Selfies has to be extended significantly. Although they are significantly more comprehensive, it should still be possible to write down the set of rules corresponding to the possible graph edits.
The first important semantic constraint that should be implemented in a 100% robust representation of reactions are physical conservation laws. For example, a representation should allow only reactions that conserve the number of atoms of different elements and the total charge of the involved compounds.
More advanced semantic constraints in reaction representations will be harder to achieve. The number of rules needed is probably extremely high. Our best estimate of the number of rules needed is from the work of Szymkuć,133 with over 50,000 rules. Applying a similar approach to reaction Selfies will be quite an endeavor and will not be scalable, as the number of rules is too high. A more suitable approach would be to extract the rules from the data directly. Such rules could either be extracted using hand-crafted algorithms (similar to the project on metaSelfies for organic molecules) or could be learned with ML. The latter case requires the extraction of rules from the ML model, which could be achieved with symbolic regression of a trained neural network. This project is conceptually related with the project for molecules with complicated bonds.
Strings as programming languages
String representations such as Smiles or Selfies are often considered less expressive and powerful than true “graph-based” representations, for instance those used in GNNs. However, fundamentally, quite the opposite is true for two very appealing reasons:
-
•
Strings and matrices can represent graphs: Often, graph-based representations are understood implicitly as adjacency matrices. However, graphs are abstract objects and can indeed be represented in diverse ways, for example by adjacency matrices but also by strings (or other ways such as images). In that sense, both strings and matrices can be representations for graphs.
-
•
Strings can store Turing-complete programming languages: In the most general case, one can store the source code of computer programs as strings. For example, a Python file is a simple string, which is executed by the Python interpreter. Python is, of course, a Turing-complete language, which means that strings can encode the most powerful computational algorithms. Coming back to graph representations, one can imagine that Smiles or Selfies are programming languages that are executed by an interpreter (for instance, by RDKit). The output of the program is a graph.
Arguably, Smiles and Selfies are rather simple programming languages, but this way of thinking indicates that one can develop much more powerful string-based molecular graph representations. These new molecular programming languages can be Turing complete and thus can encode arbitrary properties of a molecule that can be encoded in a computer. What follows now are a number of interesting future research questions that study the consequences of these ideas.
Future project 8: A molecular programming languages
Besides the performance of current string-based representations, the question remains how to extend string representations or Selfies to incorporate more prior information without losing desirable properties such as robustness. In the following, we propose two possible extensions to Selfies:
-
•
Including 3D information such as bond angles and dihedral angles: By incorporating 3D information, a Selfies could directly map to a specific molecular conformer, which could be beneficial in structure generation and embedding methods.145 In practice, extensive conformer searches could be circumvented if a specific configuration is already defined in a Selfies. A possible implementation of such 3D-Selfies could be envisioned through the use of pointer variables that locate positions in memory. The positions cannot directly be encoded using coordinates, as they do not necessarily correspond to valid structures. Rather, a more implicit encoding (such as those of rings and branches) could be envisioned by overloading symbols. Clearly, more conceptual ideas are necessary for implementing this idea.
-
•
Including meta-characters for loops and logic: Another important extension would include basic expressions of programming languages that can be used to enable different types of logic such as for loops to repeat substructures or characters for symmetric branches. Such characters could be of immense value to generate Selfies for larger and more complicated molecules (such as polymers or crystals, as discussed in previous sections). The general idea of meta-characters goes hand in hand with the creation of a general purpose and domain-independent representation (i.e., metaSelfies), as discussed in future project 1.
Future project 9: A 100% robust programming language
The discussion in the previous project motivates another leap: the possibility of a Turing-complete programming language that is 100% robust, i.e., every combination of elements in the instruction set gives a valid computer program. This question goes beyond chemistry but follows directly from the previous discussion. As such, we chose to add it as one exciting future project that might be impactful for AI research in general.
The question of deep generative models for code generation has just recently seen impressive progress in OpenAI’s Codex, a GPT language model clone that was trained on all Python codes on GitHub.146 It would be exciting to explore possibilities for generative ML models that have access to a scripting language that produces valid code in every instance. Interestingly, the question of robust programming languages has been discussed in the field of artificial life since the pioneering 1993 work of Tierra.147,148 Extensions of these ideas have since been applied to studies on artificial evolution.149,150 We hope inspiration can be taken from that field of study.
Comparing strings, adjacency matrices, and images as molecular graph representations for ML
Strings may be graph representations in the same way as adjacency matrix representations or image-based representations (cf. Figure 13). Since strings are directly related to programming languages, they are in general the most expressive of all graph representations. A very important question is how these different graph representations differ in actual ML applications.
Figure 13.
Graphs can be represented in numerous ways, for example using images, adjacency matrices, or strings
All of them are graph representations. By relating string-based representations to programming languages, we show that they are in general the most expressive representations. For Selfies, B1 and R1 are abbreviations for Branch1 and Ring1, respectively.
To answer this, it is interesting to note that different representations are suitable for different, specialized neural network architectures. Image-based representations can benefit from convolutional neural networks (CNNs), adjacency matrix-based representations are the foundations for GNNs, and string-based representations work well for language models such as recurrent neural networks (RNNs) and transformers.
The question of how these representations and their related ML models compete in the same task is so far underexplored. One very recent study has shown that chemical language models (using Selfies) and RNNs are powerful enough to generate very complex molecular distributions, including the largest molecules from PubChem.151 So far, GNN-based generative models struggle with this task and do not yet scale to these large sizes.
The comparison between the representations (and their corresponding models) leads to a number of interesting questions:
-
•
Memory footprint: As vehicles for storing molecular data, both strings and matrices should provide characteristic descriptions of the data. A fundamental principle for data description in ML is minimal description length (MDL). That is, the best description of the data is given by the model that compresses it best. One example of MDL is Kolmogorov complexity,152 which is defined as the length of the shortest computer program that produces the sequence of data. Even though Kolmogorov complexity itself is not computable, practical approximations of Kolmogorov complexity can be used to quantify the memory footprint of the molecular representation. This is especially important when using the strings or matrices as input to downstream algorithms for molecular property prediction or molecular generation. The level of physical memory burden incurred from using different representations can have significant impact on the execution speed, processor utilization, and energy cost of the program.
-
•
Optimization difficulty: Even if representations have the same memory footprint, their impact on the outcome of the ML algorithms may still vary. One reason is the difficulty of non-convex optimization. The resulting deep-learning model may not be able to fully exploit the information in the data. The choice of input representation may also have an effect on the loss landscape of the neural network optimization problem, which would certainly influence training dynamics. Different molecular representations could lead to distinct local optima, producing models that differ in terms of generalization performance and sensitivity to input perturbation.
-
•
Computational efficiency: From a computational perspective, string versus graph representation can also have different complexities due to the differences in numerical algorithms. For example, for strings of different lengths, one can either use sequential processing models such as RNNs or transformers with padding, which can be easily parallelized. However, the padded strings would have different sparsity structures (the patterns of zeros) than the matrix representations. These sparsity structures can be utilized to a varying degree in order to accelerate numerical operations including addition, multiplication, or eigenvalue decomposition. The efficiency of the entire program, thus, can be easily affected.
To shed light onto these different properties, we suggest the following project.
Future project 10: Comparisons in various data regimes in a regression task
While string-based representations tend to be more expressive and easier to generate, adjacency matrices in conjunction with GNNs have important advantages, such as permutation invariance. Images of the molecular graphs (which can be understood as another graph representation) could take advantage of extremely efficient, pretrained CNNs. A suitable experiment could be a discriminative task in the various data regimes. This of course depends on the target property to be learned. For example, for learning coordinate-dependent properties, it is still unknown how much prior information is actually necessary and whether string-based representations will outperform graph-based representations in the high data regime for specific tasks.
We suggest the development of a benchmark to compare image, adjacency matrix, and string representations for graphs in various data regimes for discriminative tasks. The PCQM4M-LSC dataset may be useful for these comparisons: with approximately 3.8 million molecules and their associated highest occupied molecular orbital-lowest unoccupied molecular orbital (HOMO-LUMO) energy gaps (as estimated by density functional theory [DFT] simulation), it poses a formidable chemical regression task.153,154
The comparison should measure all three models in (at least) the prediction quality over the following characteristics:
-
•
The number of training epochs.
-
•
The number of model parameters.
-
•
Various numbers of examples in the training data.
-
•
Various sizes (measured in edges) of the largest molecules in the training dataset.
These experiments will give insightful answers about the characteristics of different data modalities in ML tasks and will give experimental evidence about which models should be used in which situations in future practical applications.
Future project 11: Comparisons in generative tasks
A main motivation of Selfies is its application in generative, inverse-design tasks. We therefore suggest the development of new generative model benchmarks. For that, a number of important precautions need to be considered. First, when Selfies is used, a comparison among models based on their ability to generate valid molecules is no longer a useful design objective.155 Interestingly, previously used benchmarks155,156 have also placed great importance on distributional learning metrics. However, this approach is reported to have multiple flaws in the form of edge cases.157 For instance, simple algorithms that place carbon atoms at random positions within molecules have been shown to perform well on distribution matching objectives. Additionally, the recent proposal of the STONED algorithm,64 which makes use of random Selfies mutations, has demonstrated ease in matching the structural distribution of molecules. FastFlows158 uses normalizing flows to model distributions of molecules represented as Selfies and achieve fast sampling speeds. Another class of methods used for comparing molecular generative models can be classified as goal-directed benchmarks. In these, generative models compete among one another to optimize one or more molecular property functions. It can also be important to generate dense local chemical spaces, for example to create counterfactuals to explain black-box models.159 Many of these tasks are provided within GuacaMol;156 however, given the current rise of more sophisticated models, these benchmarks have become outdated. Recently, many generative models have been able to achieve perfect results on many of the GuacaMol tasks,160, 161, 162 making it difficult to establish comparisons between models. Therefore, to compare deep generative models, one needs more sophisticated objectives that reflect the complexity of real-world molecular design. We anticipate that the next generations of benchmarks will estimate more complex and physically relevant properties within catalysis, drug discovery, and materials science using semi-empirical quantum chemistry and DFT.
Interpretability and usability of string-based representations
For humans
Historically, representations have been developed with humans in mind for reading and writing molecules. String-based representations are more difficult to interpret than images of molecules, and an important question is their understandability for humans. On the one hand, human chemists might want to write molecules quickly as text instead of drawing them, might be able to get a quick understanding of the structure without inserting it into a plotting tool, or might be interested in identifying substructures. On the other hand, readability for humans might not always be necessary. For example, InChI strings are broadly used despite the fact that the human readability was considered to be of low importance when InChI was designed.163 It is also worth pointing out that while human readability is one of the often-cited advantages of Smiles, figuring out what a Smiles actually stands for can require significant intellectual effort. We just have to look at the Smiles for a simple steroid such as testosterone to see that this is the case:
O=C1CC[C@]2(C)[C@@]3([H])CC[C@]4(C)[C@@H](O)CC[C@@]4([H])[C@]3([H])CCC2=C1.
This suggests a trade-off in the necessity of readability and concrete computational applications. However, there is certainly a natural question of how well humans can interpret molecular string representations, which has not been investigated experimentally to the best of our knowledge. Therefore, we suggest the following project.
Future project 12: Experiment on readability of molecular string representations
We suggest an experiment that tests the human readability of Smiles-, DeepSmiles-, Selfies-, and adjacency matrix-based representations of molecules. We envision a study with 50 or more participants from different countries. None of the participants may be previously familiar with these representations, to guarantee a fair comparison. The participants will get instructions for understanding each of the representations, with which they should familiarize themselves before the experiments start.
At the evaluation phase, the participants are asked to solve a number of tasks, such as substructure identification and translating the representation from and to molecular graphs. The participants will also be asked to solve some tasks in which they need to actively choose their preferred representation(s). The results might help us to understand which representations are easiest to read by analyzing the accuracy, speed, and participant’s preference of representations. Post-hoc interviews could then elaborate on the challenges of different representations and might help to design a potential Esperanto for Chemistry—an easy-to-understand language for molecules.
For many chemistry applications, readability is not necessary, as the human operator can readily translate molecular strings to 2D graph of the molecule. However, we argue that beyond human readability, such an experiment might allow us to compare and contrast which properties of representations are challenging for humans compared with computers. These results could potentially lead to interesting findings on the differences between humans and machines, thus showing where we should place our trust in our intuitions around ML for chemistry.
For machines
An interesting question is how ML models interpret different representations. Specifically, if Selfies is used in a generative model, all generated molecules are correct. In this case, how can one be sure that the model’s output is meaningful concerning some metrics such as usefulness and not just a collection of random strings, which, by construction, lead to valid molecules? Furthermore, how can the machine interpretability of different representations be compared, specifically between Smiles and Selfies? In other words, which one is “easier” to learn for machines?
In deep generative models using VAEs, the latent space using Smiles consists of numerous, scattered, valid regions that exist within invalid valleys (see Figure 3). In contrast, the entire latent space corresponds to valid molecular structures if Selfies is employed instead. This fact allows for the application of continuous gradient descent optimization in the latent space, where the optimizer will always provide meaningful structures. The robustness, however, does not necessarily correspond to a smooth encoding in the latent space, per se, where small changes in the latent space lead to small modifications in the molecule. Therefore, it remains to be seen whether generative models can actually learn structure-property relations using Selfies.
Deep molecular dreaming
One experiment that tackles the problem of interpretability and smoothness to a certain extent employs the technique of DeepDreaming.164 The generative model denoted as Pasithea consists of a single neural network that is used for the generation of molecules in two steps. In the first of these, the network learns to predict a chemical property given a one-hot encoding of a Selfies. In the second step, the neural network weights are frozen, and a target value of the property is fixed. Gradient descent is then used with respect to the one-hot encoding, meaning that the input molecule is continuously modified. The results of two design processes are shown in Figure 14. While the model continuously decreases the loss, the one-hot encoding of the molecule is changed within the discrete space. It is apparent that the target property increases/decreases for positive/negative target values of logP in a nearly monotonous way. This indicates that the model has indeed understood an essence of logP and its relation to the structure of the molecule and is not exploiting only the robustness of Selfies. A complementary approach is to use directly invertible neural networks for generative models, such as presented in Hu.165
Figure 14.
Pasithea, the DeepDreaming generative model
While the model continuously decreases the loss, the molecule changes in discrete steps. The target property was logP of the molecule. The network is able to increase or decrease the molecular property almost steadily, which indicates a certain “understanding” of the representation. Image from Shen et al.164
DECIMER
Optical chemical structure recognition (OCSR) tools have been developed to extract chemical structures and convert them into a computer-readable format. The best-performing OCSR tools are mostly rule-based algorithms. To address the OCSR problem by using the latest computational intelligence techniques and provide an automated open-source software solution, deep learning for chemical image recognition (DECIMER) was launched (Figure 15).166 One of the biggest challenges in developing DECIMER was to use the string representation of chemical structures in a meaningful way. The issue encountered initially with Smiles was splitting them into meaningful tokens during training and evaluation, when the predicted Smiles were syntactically and semantically incorrect, reducing the accuracy of the tool. As a result of using Selfies, this issue was resolved, leading to better training of models. Additionally, it demonstrates how efficiently neural networks can be trained to read and write Selfies strings.
Figure 15.
DECIMER and STOUT
A framework for translating images or strings to Smiles. Experiments show that the application of Selfies as an intermediate representation improves the results, which indicates that ML models find it easier to read and write Selfies compared with Smiles. These indications are surprising because it is not clear how the model exploits Selfies’s robustness to improve results. Image from Rajan et al.166,167
STOUT
A conceptually related tool is Smiles-to-IUPAC name translator (STOUT). It was developed to translate between the IUPAC names and string representations of molecules. IUPAC developed a naming scheme for chemistry based on a set of rules. Due to the complexity of this rule set, assigning a chemical name is challenging for humans, and there are a limited number of rule-based cheminformatics applications available to assist with this process, all of which are commercial. STOUT is an open-source, deep-learning-based neural machine translation approach developed to generate the IUPAC name for a given molecule from its Smiles string and carry out the reverse translation.167 One key observation was that STOUT works better when using Selfies as an internal representation than with Smiles. Therefore, the Smiles strings are internally converted into Selfies before the input is processed by the model. Likewise, the predicted Selfies are decoded back into Smiles during reverse translation. This is another indication that Selfies is understood better than Smiles for some complex deep-learning tasks. The precise reason for the advantage is not well understood, therefore it will be very interesting to understand the behavior of more complex grammars in deep neural networks (future projects 2 and 14). This will then hopefully indicate other tasks that could benefit from Selfies or other advanced representations.
Selfies in a language model
It was shown recently that an RNN language model trained on Selfies is more robust to overfitting than with Smiles.151 This is understood from the larger novelty of the generated molecules at similar quality of the learned distribution.
There are numerous future experiments that could shed light into the “understandability” of different representations. We summarize a few of them here.
Future project 13: Translation between different representations
It would be interesting to train a neural network that can translate between different representations of molecular graphs, including (current or future) string-based representations, adjacency matrix representations, or images of molecular graphs. This would be exciting for two reasons. Firstly, if the neural network learns to work with three entirely different representations, it might build up an interesting and robust internal representation, which could subsequently be analyzed. Secondly, it gives the opportunity to combine three of the most powerful ML methods at the same time, namely GNNs for the adjacency matrix representation, transformers for strings, and CNNs for the images of molecular graphs. A concrete use case could look like this: the goal is to predict a molecular property from a molecule that is encoded as a Selfies. The neural network translates the Selfies to an adjacency matrix and an image, producing a latent meta-representation of the molecule in one of its hidden layers in the process. All or some of these four representations are provided to downstream models with appropriate architectures (e.g., GNN for an adjacency matrix or transformer for a string), which are then ensembled to produce better predictions and overcome deficiencies in each individual chemical representation. Note that some important progress has already been achieved in translation tasks. Examples are image-to-string representation translations166,168 and string-to-IUPAC translations.167,169
Future project 14: Which string-based representation allows for simpler models and faster training?
Several experiments could be performed to determine how the use of different representations for training ML models on the same set of regression tasks impacts learning and final quality metrics, such as accuracy. Initially, these projects should comprise the usual benchmark endpoints for ML prediction, such as boiling points, logP, and pKa. In addition, tasks known to be influenced by the 3D structure of the compounds, such as predicting HOMO or LUMO energies or activity toward a biological target, could also be explored.
In a first experiment, models with the same end goal could be trained to determine how different representations impact the final accuracy and how they impact the model’s ability to achieve better performance with less training time. In another experiment, the numbers of neurons and layers of neural networks would be decreased, and the number of episodes necessary to reach a certain quality would be recorded. This project would allow us to verify the ability of models trained on Selfies to generalize better, provided the performance after these model simplifications does not decrease as fast as for models trained on different representations.
One of the reasons why this future project might be important is the following: there are studies that investigate DeepSmiles in deep neural networks and indicate that the advanced grammar has a detrimental effect on the learning capability in some specific tasks.170 The overloading of symbols certainly is a complex operation (related to task 2), thus it will be interesting to investigate the learning capability of Selfies.
Future project 15: Smoothness of latent space in deep generative models
Another interesting experiment would be to investigate the smoothness of latent spaces of VAEs trained with Smiles, DeepSmiles, and Selfies. If one wants to use gradient-based optimizers in the latent space, it would be desirable if the properties of the generated molecules changed to a small extent when sampling from closely related points in the latent space. We suggest measuring a set of properties for each generated molecule while continuously wandering in the latent space. Notably, the design of such an ML experiment needs to take the invalid regions of the latent space into account.
Future project 16: Learning what the machine has learned in the latent space
The latent space represents the intrinsic representation that has been learned by the model to solve a given task. It will be exciting to understand what this representation stands for. If one understands how a VAE encodes and decodes molecules to and from the latent space, some of the questions presented above can likely be answered even without performing further experiments. To that end, t-stochastic neighbor embedding (t-SNE)171 and other dimensionality reduction tools are expected to be challenging to interpret, thus one direction could be the applications of latent spaces with only two or three dimensions, which can be displayed without projections. Related projects have rediscovered interesting physical concepts such as the heliocentric coordinates,172 the arrow of time,173 or interpretation in quantum optics,174,175 and we expect similar exciting possibilities in materials science and chemistry.
Conclusion
The resolution of the 16 proposed challenges could significantly advance the applicability of AI in diverse fields of chemistry and beyond. Furthermore, questions about the interpretability of languages for machines could help us understand how a machine solves complex tasks in chemistry—what principles or concepts it uses. This could be a path for human scientists to learn ideas from AI in chemistry. We hope that our journey of possibilities will inspire researchers in the cheminformatics and applied AI community and lead to exciting new results and advances in molecular string representations.
About the authors
The authors assembled in a publicly announced, open online mini-workshop organized by IOP Publishing and the Acceleration Consortium Toronto, on the topic of Selfies and the future of molecular string representation. All participants were invited to jointly write a perspective paper that extended the discussions and ideas developed in the workshop. Writing of the paper was organized through Discord. The participants range from undergraduate students to university professors and professionals in related industries, with a background in chemistry, physics, engineering, and computer science. The 31 authors come from 14 different countries on four continents.
Acknowledgments
The authors thank Greg Landrum, Daniel Flam-Shepherd, Suliman Sharif, and Bettina Lier for valuable comments on the manuscript. The authors also thank Sara Bebbington of IOP Publishing and Zamyla Chan and Erin Warner of the University of Toronto Acceleration Consortium for helping to organize the Selfies workshop. M.K. acknowledges support from the FWF (Austrian Science Fund) via the Erwin Schrödinger fellowship no. J4309. R.F.L. received a PhD Scholarship from the São Paulo Research Foundation (FAPESP) – grant #2021/01633-3. This study was financed in part by CAPES – Finance Code 001. R.P. acknowledges funding through a Postdoc.Mobility fellowship by the Swiss National Science Foundation (SNSF; project no. 191127). A.W. would like to thank the Natural Sciences and Engineering Council of Canada (NSERC) for financial support via a CGS-M scholarship. G.T. acknowledges financial support from NSERC via the PGS-D scholarship. R.Y. acknowledges support from the US Department of Energy, Office of Science, AWS Machine Learning Research Award, and NSF grant #2037745. D.L. and G.F.v.R. were supported by the von Lilienfeld lab at the University of Vienna. A.D.W. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM137966. K.M.J. and B.S. acknowledge funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 666983, MaGic). J.M.N.-D. acknowledges support by the National Council for Science and Technology (CONACYT) under award number CVU 105568. P.S. acknowledges support from the NCCR Catalysis (grant number 180544), a National Centre of Competence in Research funded by the Swiss National Science Foundation. S.M.M. was supported by the Swiss National Science Foundation (SNSF) under grant P2ELP2_195155. U.S. acknowledges support from the Deutsche Forschungsgemeinschaft (DFG) within NFDI4Chem (grant no. NFDI4-1). Q.A. acknowledges support from the National Science Foundation (grant no. DMR-1928882). A.A.G. acknowledges support from the Canada 150 Research Chairs Program, the Google Focused Award, and Dr. Anders G. Frøseth.
Contributor Information
Mario Krenn, Email: mario.krenn@mpl.mpg.de.
Alán Aspuru-Guzik, Email: alan@aspuru.com.
References
- 1.Zubatiuk T., Isayev O. Development of multimodal machine learning potentials: toward a physics-aware artificial intelligence. Acc. Chem. Res. 2021;54:1575–1585. doi: 10.1021/acs.accounts.0c00868. [DOI] [PubMed] [Google Scholar]
- 2.Huang B., von Lilienfeld O.A. Ab initio machine learning in chemical compound space. Chem. Rev. 2021;121:10001–10036. doi: 10.1021/acs.chemrev.0c01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behler J. Four generations of high-dimensional neural network potentials. Chem. Rev. 2021;121:10037–10072. doi: 10.1021/acs.chemrev.0c00868. [DOI] [PubMed] [Google Scholar]
- 4.Westermayr J., Marquetand P. Machine learning for electronically excited states of molecules. Chem. Rev. 2021;121:9873–9926. doi: 10.1021/acs.chemrev.0c00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keith J.A., Vassilev-Galindo V., Cheng B., Chmiela S., Gastegger M., Müller K.R., Tkatchenko A. Combining machine learning and computational chemistry for predictive insights into chemical systems. Chem. Rev. 2021;121:9816–9872. doi: 10.1021/acs.chemrev.1c00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dral P.O., Barbatti M. Molecular excited states through a machine learning lens. Nat. Rev. Chem. 2021;5:388–405. doi: 10.1038/s41570-021-00278-1. [DOI] [PubMed] [Google Scholar]
- 7.von Lilienfeld O.A., Müller K.R., Tkatchenko A. Exploring chemical compound space with quantum-based machine learning. Nat. Rev. Chem. 2020;4:347–358. doi: 10.1038/s41570-020-0189-9. [DOI] [PubMed] [Google Scholar]
- 8.Glielmo A., Husic B.E., Rodriguez A., Clementi C., Noé F., Laio A. Unsupervised learning methods for molecular simulation data. Chem. Rev. 2021;121:9722–9758. doi: 10.1021/acs.chemrev.0c01195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unke O.T., Chmiela S., Sauceda H.E., Gastegger M., Poltavsky I., Schütt K.T., Tkatchenko A., Müller K.R. Machine learning force fields. Chem. Rev. 2021;121:10142–10186. doi: 10.1021/acs.chemrev.0c01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friederich P., Häse F., Proppe J., Aspuru-Guzik A. Machine-learned potentials for next-generation matter simulations. Nat. Mater. 2021;20:750–761. doi: 10.1038/s41563-020-0777-6. [DOI] [PubMed] [Google Scholar]
- 11.Walters W.P., Barzilay R. Applications of deep learning in molecule generation and molecular property prediction. Acc. Chem. Res. 2021;54:263–270. doi: 10.1021/acs.accounts.0c00699. [DOI] [PubMed] [Google Scholar]
- 12.Deringer V.L., Bartók A.P., Bernstein N., Wilkins D.M., Ceriotti M., Csányi G. Gaussian process regression for materials and molecules. Chem. Rev. 2021;121:10073–10141. doi: 10.1021/acs.chemrev.1c00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nandy A., Duan C., Taylor M.G., Liu F., Steeves A.H., Kulik H.J. Computational discovery of transition-metal complexes: from high-throughput screening to machine learning. Chem. Rev. 2021;121:9927–10000. doi: 10.1021/acs.chemrev.1c00347. [DOI] [PubMed] [Google Scholar]
- 14.Gallegos L.C., Luchini G., St John P.C., Kim S., Paton R.S. Importance of engineered and learned molecular representations in predicting organic reactivity, selectivity, and chemical properties. Acc. Chem. Res. 2021;54:827–836. doi: 10.1021/acs.accounts.0c00745. [DOI] [PubMed] [Google Scholar]
- 15.Żurański A.M., Martinez Alvarado J.I., Shields B.J., Doyle A.G. Predicting reaction yields via supervised learning. Acc. Chem. Res. 2021;54:1856–1865. doi: 10.1021/acs.accounts.0c00770. [DOI] [PubMed] [Google Scholar]
- 16.Meuwly M. Machine learning for chemical reactions. Chem. Rev. 2021;121:10218–10239. doi: 10.1021/acs.chemrev.1c00033. [DOI] [PubMed] [Google Scholar]
- 17.Jorner K., Tomberg A., Bauer C., Sköld C., Norrby P.O. Organic reactivity from mechanism to machine learning. Nat. Rev. Chem. 2021;5:240–255. doi: 10.1038/s41570-021-00260-x. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Lengeling B., Aspuru-Guzik A. Inverse molecular design using machine learning: generative models for matter engineering. Science. 2018;361:360–365. doi: 10.1126/science.aat2663. [DOI] [PubMed] [Google Scholar]
- 19.Terayama K., Sumita M., Tamura R., Tsuda K. Black-box optimization for automated discovery. Acc. Chem. Res. 2021;54:1334–1346. doi: 10.1021/acs.accounts.0c00713. [DOI] [PubMed] [Google Scholar]
- 20.Janet J.P., Duan C., Nandy A., Liu F., Kulik H.J. Navigating transition-metal chemical space: artificial intelligence for first-principles design. Acc. Chem. Res. 2021;54:532–545. doi: 10.1021/acs.accounts.0c00686. [DOI] [PubMed] [Google Scholar]
- 21.Pollice R., Dos Passos Gomes G., Aldeghi M., Hickman R.J., Krenn M., Lavigne C., Lindner-D’Addario M., Nigam A.K., Ser C.T., Yao Z., Aspuru-Guzik A. Data-driven strategies for accelerated materials design. Acc. Chem. Res. 2021;54:849–860. doi: 10.1021/acs.accounts.0c00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White A.D. Deep learning for molecules and materials. Liv. J. Comput. Mol. Sci. 2021;3:1499. doi: 10.33011/livecoms.3.1.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford J.M., Kingston C., Toste F.D., Sigman M.S. Data science meets physical organic chemistry. Acc. Chem. Res. 2021;54:3136–3148. doi: 10.1021/acs.accounts.1c00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jablonka K.M., Ongari D., Moosavi S.M., Smit B. Big-data science in porous materials: materials genomics and machine learning. Chem. Rev. 2020;120:8066–8129. doi: 10.1021/acs.chemrev.0c00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin W., Barzilay R., Jaakkola T. ICML; 2018. Junction Tree Variational Autoencoder for Molecular Graph Generation. [Google Scholar]
- 26.Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4:eaap7885. doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krenn M., Häse F., Nigam A.K., Friederich P., Aspuru-Guzik A. Self-referencing embedded strings (SELFIES): a 100% robust molecular string representation. Mach. Learn Sci. Technol. 2020;1:045024. [Google Scholar]
- 28.Warr W.A. Representation of chemical structures. WIREs. Comput. Mol. Sci. 2011;1:557–579. [Google Scholar]
- 29.Wigh D.S., Goodman J.M., Lapkin A.A. A review of molecular representation in the age of machine learning. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2022:e1603. [Google Scholar]
- 30.Hähnke Volker D., Kim S., Bolton E.E. Pubchem chemical structure standardization. J. Cheminf. 2018;10:1–40. doi: 10.1186/s13321-018-0293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiswesser W.J. The Wiswesser line formula notation. Chem. Eng. News Archive. 1952;30:3523–3526. [Google Scholar]
- 32.Donald Lyle Dorward . University of Illinois at Urbana-Champaign; 1965. Words about Words: Nonconventional Methods of Handling Chemical Information,” Occasional Papers; p. 76. [Google Scholar]
- 33.Fletcher J.H., Dermer O.C., Fox R.B. American Chemical Society; 1974. Nomenclature of Organic Compounds. [Google Scholar]
- 34.Warr W.A. Diverse uses and future prospects for Wiswesser line-formula notation. J. Chem. Inf. Comput. Sci. 1982;22:98–101. [Google Scholar]
- 35.Hepler-Smith E. ‘Just as the structural formula does’: names, diagrams, and the structure of organic chemistry at the 1892 Geneva nomenclature congress. Ambix. 2015;62:1–28. doi: 10.1179/1745823414y.0000000006. [DOI] [PubMed] [Google Scholar]
- 36.Fauque D. 1919-1939: the first life of the union. Chem. Int. 2019;41:2–6. [Google Scholar]
- 37.de Morveau Louis-Bernard Guyton, Lavoisier A.L., Berthollet C.-L., de Fourcroy Antoine-Francois, Hassenfratz J.-H., Adet P.-A. Cuchet; 1787. Methode de nomenclature chimique. [Google Scholar]
- 38.Dalton J. Deansgate for R Bickerstaff; 1808. A New System of Chemical Philosophy, Part 1. Printed by S Russell, 125. [Google Scholar]
- 39.Berzelius J.J. Essay on the cause of chemical proportions, and on some circumstances relating to them; together with a short and easy method of expressing them. Ann. Philos. 1813;2:443–454. [Google Scholar]
- 40.International Association of Chemical Societies Nature. 1912;89:245–246. [Google Scholar]
- 41.Dyson G.M. A notation for organic compounds. Nature. 1944;154:114. [Google Scholar]
- 42.Dyson G. Longmans, Green and Co.; 1947. A New Notation and Enumeration System for Organic Compounds. [Google Scholar]
- 43.Brightman R. Names into cipher. Nature. 1947;160:175. doi: 10.1038/160615a0. [DOI] [PubMed] [Google Scholar]
- 44.Raos N., Miličević A. Methods of writing constitutional formulas. Kemija u industriji/J. Chem. Chem. Eng. 2012;61:435–449. [Google Scholar]
- 45.Wiswesser William., J. Notational systems for structural formulas. Chem. Eng. News Archive. 1952;30:407–410. [Google Scholar]
- 46.Wiswesser W.J. How the WLN began in 1949 and how it might be in 1999. J. Chem. Inf. Comput. Sci. 1982;22:88–93. [Google Scholar]
- 47.Hayward H.W. Office of Research and Development, Patent Office; 1961. A New Sequential Enumeration and Line Formula Notation System for Organic Compounds. [Google Scholar]
- 48.Skolnik H., Clow A. A notation system for indexing pesticides. J. Chem. Doc. 1964;4:221–227. [Google Scholar]
- 49.Feldman A., Holland D.B., Jacobus D.P. The automatic encoding of chemical structures. J. Chem. Doc. 1963;3:187–189. [Google Scholar]
- 50.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988;28:31–36. [Google Scholar]
- 51.Weininger D., Weininger A., Weininger J.L. SMILES. 2. Algorithm for generation of unique SMILES notation. J. Chem. Inf. Comput. Sci. 1989;29:97–101. [Google Scholar]
- 52.Landrum G. RDKit; 2013. RDKit: A Software Suite for Cheminformatics, Computational Chemistry, and Predictive Modeling. [Google Scholar]
- 53.Schneider G., Fechner U. Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov. 2005;4:649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 54.Gómez-Bombarelli R., Wei J.N., Duvenaud D., Hernández-Lobato J.M., Sánchez-Lengeling B., Sheberla D., Aguilera-Iparraguirre J., Hirzel T.D., Adams R.P., Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018;4:268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma T., Chen J., Xiao C. Constrained generation of semantically valid graphs via regularizing variational autoencoders. arXiv. 2018 doi: 10.48550/arXiv.1809.02630. Preprint at. [DOI] [Google Scholar]
- 56.Qi L., Allamanis M., Brockschmidt M., Gaunt A.L. Constrained graph variational autoencoders for molecule design. arXiv. 2018 doi: 10.48550/arXiv.1805.09076. Preprint at. [DOI] [Google Scholar]
- 57.Noel O’Boyle, Dalke A. DeepSMILES: an adaptation of SMILES for use in machine-learning of chemical structures. ChemRxiv. 2018 doi: 10.26434/chemrxiv.7097960.v1. Preprint at. [DOI] [Google Scholar]
- 58.Heller S., McNaught A., Stein S., Tchekhovskoi D., Pletnev I. InChI - the worldwide chemical structure identifier standard. J. Cheminf. 2013;5:7–9. doi: 10.1186/1758-2946-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Boyle N.M. Towards a universal SMILES representation - a standard method to generate canonical SMILES based on the InChI. J. Cheminf. 2012;4:1–14. doi: 10.1186/1758-2946-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodman J.M., Pletnev I., Thiessen P., Bolton E., Heller S.R. InChI version 1.06: now more than 99.99% reliable. J. Cheminf. 2021;13:40–48. doi: 10.1186/s13321-021-00517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hopcroft J.E., Motwani R., Ullman J.D. Introduction to automata theory, languages, and computation. SIGACT News. 2001;32:60–65. [Google Scholar]
- 62.Nigam A.K., Friederich P., Krenn M., Aspuru-Guzik A. International Conference on Learning Representations. 2020. Augmenting genetic algorithms with deep neural networks for exploring the chemical space. [Google Scholar]
- 63.Thiede L.A., Krenn M., Nigam A.K., Aspuru-Guzik A. Curiosity in exploring chemical space: intrinsic rewards for deep molecular reinforcement learning. arXiv. 2020 doi: 10.48550/arXiv.2012.11293. Preprint at. [DOI] [Google Scholar]
- 64.Nigam A.K., Pollice R., Krenn M., Gomes G.D.P., Aspuru-Guzik A. Beyond generative models: superfast traversal, optimization, novelty, exploration and discovery (STONED) algorithm for molecules using SELFIES. Chem. Sci. 2021;12:7079–7090. doi: 10.1039/d1sc00231g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krenn M., Malik M., Fickler R., Lapkiewicz R., Zeilinger A. Automated search for new quantum experiments. Phys. Rev. Lett. 2016;116:090405. doi: 10.1103/PhysRevLett.116.090405. [DOI] [PubMed] [Google Scholar]
- 66.Han D., Qi X., Myhrvold C., Wang B., Dai M., Jiang S., Bates M., Liu Y., An B., Zhang F., et al. Single-stranded DNA and RNA origami. Science. 2017;358:eaao2648. doi: 10.1126/science.aao2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drefahl A. CurlySMILES: a chemical language to customize and annotate encodings of molecular and nanodevice structures. J. Cheminf. 2011;3:1–7. doi: 10.1186/1758-2946-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin T.-S., Coley C.W., Mochigase H., Beech H.K., Wang W., Wang Z., Woods E., Craig S.L., Johnson J.A., Kalow J.A., et al. BigSMILES: a structurally-based line notation for describing macromolecules. ACS Cent. Sci. 2019;5:1523–1531. doi: 10.1021/acscentsci.9b00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang T., Li H., Xi H., Stanton R.V., Rotstein S.H. A hierarchical notation language for complex biomolecule structure representation. J. Chem. Inf. Model. 2012;52:2796–2806. doi: 10.1021/ci3001925. [DOI] [PubMed] [Google Scholar]
- 70.Hall S.R., Allen F.H., Brown I.D. The crystallographic information file (CIF): a new standard archive file for crystallography. Acta Crystallogr. A. 1991;47:655–685. [Google Scholar]
- 71.Brown I.D., McMahon B. CIF: the computer language of crystallography. Acta Crystallogr. B. 2002;58:317–324. doi: 10.1107/s0108768102003464. [DOI] [PubMed] [Google Scholar]
- 72.Cayley P. LVII. On the mathematical theory of isomers. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1874;47:444–447. [Google Scholar]
- 73.O’Keefe M., Hyde B.G. Plane nets in crystal chemistry. Philos. Trans. Royal Soc. A. 1980;295:553–618. [Google Scholar]
- 74.Wells A.F. Wiley; 1977. Three Dimensional Nets and Polyhedra. [Google Scholar]
- 75.Groom C.R., Bruno I.J., Lightfoot M.P., Ward S.C. The Cambridge structural database. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 2016;72:171–179. doi: 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krivovichev Sergey V. Vol. 22. Oxford University Press; 2009. (Structural Crystallography of Inorganic Oxysalts). [Google Scholar]
- 77.O’Keeffe M., Peskov M.A., Ramsden S.J., Yaghi O.M. The reticular chemistry structure resource (RCSR) database of, and symbols for, crystal nets. Acc. Chem. Res. 2008;41:1782–1789. doi: 10.1021/ar800124u. [DOI] [PubMed] [Google Scholar]
- 78.Blatov V.A., Shevchenko A.P., Proserpio D.M. Applied topological analysis of crystal structures with the program package ToposPro. Cryst. Growth Des. 2014;14:3576–3586. [Google Scholar]
- 79.Tritsaris G.A., Xie Y., Rush A.M., Carr S., Mattheakis M., Kaxiras E. LAN: a materials notation for two-dimensional layered assemblies. J. Chem. Inf. Model. 2020;60:3457–3462. doi: 10.1021/acs.jcim.0c00630. [DOI] [PubMed] [Google Scholar]
- 80.Delgado-Friedrichs O., O’Keeffe M. Crystal nets as graphs: terminology and definitions. J. Solid State Chem. 2005;178:2480–2485. [Google Scholar]
- 81.Pan H., Ganose A.M., Horton M., Aykol M., Persson K.A., Zimmermann N.E.R., Jain A. Benchmarking coordination number prediction algorithms on inorganic crystal structures. Inorg. Chem. 2021;60:1590–1603. doi: 10.1021/acs.inorgchem.0c02996. [DOI] [PubMed] [Google Scholar]
- 82.Chung S.J., Hahn T., Klee W.E. Nomenclature and generation of three-periodic nets: the vector method. Acta Crystallogr. A. 1984;40:42–50. [Google Scholar]
- 83.Klee W.E. Crystallographic nets and their quotient graphs. Cryst. Res. Technol. 2004;39:959–968. [Google Scholar]
- 84.Bader M., Klee W.E., Thimm G. The 3-regular nets with four and six vertices per unit cell. Z. für Kristallogr. - Cryst. Mater. 1997;212:553–558. [Google Scholar]
- 85.Thimm G. Crystal structures and their enumeration via quotient graphs. Z. Kristallog. - Crystal. Mater. 2004;219:528–536. [Google Scholar]
- 86.Delgado-Friedrichs O., Hyde S.T., O’Keeffe M., Yaghi O.M. Crystal structures as periodic graphs: the topological genome and graph databases. Struct. Chem. 2017;28:39–44. [Google Scholar]
- 87.Tian X., Fu X., Ganea O.-E., Barzilay R., Jaakkola T. Crystal diffusion variational autoencoder for periodic material generation. arXiv. 2021 doi: 10.48550/arXiv.2110.06197. Preprint at. [DOI] [Google Scholar]
- 88.Yao Z., Sánchez-Lengeling B., Bobbitt N.S., Bucior B.J., Kumar S.G.H., Collins S.P., Burns T., Woo T.K., Farha O.K., Snurr R.Q., Aspuru-Guzik A. Inverse design of nanoporous crystalline reticular materials with deep generative models. Nat. Mach. Intell. 2021;3:76–86. [Google Scholar]
- 89.Colón Y.J., Gómez-Gualdrón D.A., Snurr R.Q. Topologically guided, automated construction of metal–organic frameworks and their evaluation for energy-related applications. Cryst. Growth Des. 2017;17:5801–5810. [Google Scholar]
- 90.Fung V., Zhang J., Hu G., Ganesh P., Sumpter B.G. Inverse design of two-dimensional materials with invertible neural networks. arXiv. 2021 doi: 10.48550/arXiv.2106.03013. Preprint at. [DOI] [Google Scholar]
- 91.Nouira A., Sokolovska N., Crivello J.-C. CrystalGAN: learning to discover crystallographic structures with generative adversarial networks. arXiv. 2018 doi: 10.48550/arXiv.1810.11203. Preprint at. [DOI] [Google Scholar]
- 92.Court C.J., Yildirim B., Jain A., Cole J.M. 3-D inorganic crystal structure generation and property prediction via representation learning. J. Chem. Inf. Model. 2020;60:4518–4535. doi: 10.1021/acs.jcim.0c00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Noh J., Kim J., Stein H.S., Sanchez-Lengeling B., Gregoire J.M., Aspuru-Guzik A., Jung Y. Inverse design of solid-state materials via a continuous representation. Matter. 2019;1:1370–1384. [Google Scholar]
- 94.Gao H., Wang J., Guo Z., Sun J. Determining dimensionalities and multiplicities of crystal nets. NPJ Comput. Mater. 2020;6:143. [Google Scholar]
- 95.Blatov V.A., Proserpio Davide M. In: Modern Methods of Crystal Structure Prediction. Oganov A.R., editor. Wiley-VCH; 2010. Periodic-graph approaches in crystal structure prediction; pp. 1–28. Chapter 1. [Google Scholar]
- 96.Thimm G. Crystal topologies – the achievable and inevitable symmetries. Acta Crystallogr. A. 2009;65:213–226. doi: 10.1107/S0108767309003638. [DOI] [PubMed] [Google Scholar]
- 97.Eon J.-G. Topological features in crystal structures: a quotient graph assisted analysis of underlying nets and their embeddings. Acta Crystallogr. A Found. Adv. 2016;72:268–293. doi: 10.1107/S2053273315022950. [DOI] [PubMed] [Google Scholar]
- 98.Pfaltz A., Drury W.J. Design of chiral ligands for asymmetric catalysis: from C2-symmetric P, P- and N, N-ligands to sterically and electronically nonsymmetrical P, N-ligands. Proc. Natl. Acad. Sci. USA. 2004;101:5723–5726. doi: 10.1073/pnas.0307152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Narcis M.J., Takenaka N. Helical-chiral small molecules in asymmetric catalysis. Eur. J. Org. Chem. 2014;2014:21–34. [Google Scholar]
- 100.López R., Palomo C. Planar chirality: a mine for catalysis and structure discovery. Angew. Chem. Int. Ed. 2022;61 doi: 10.1002/anie.202113504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wilson A.G., Izmailov P. Bayesian deep learning and a probabilistic perspective of generalization. arXiv. 2020 doi: 10.48550/arXiv.2002.08791. [DOI] [Google Scholar]
- 102.Gonthier J.F., Steinmann S.N., Wodrich M.D., Corminboeuf C. Quantification of “fuzzy” chemical concepts: a computational perspective. Chem. Soc. Rev. 2012;41:4671–4687. doi: 10.1039/c2cs35037h. [DOI] [PubMed] [Google Scholar]
- 103.Ball P. Beyond the bond. Nature. 2011;469:26–28. doi: 10.1038/469026a. [DOI] [PubMed] [Google Scholar]
- 104.James C.A. 2015. OpenSMILES Specification. [Google Scholar]
- 105.Clark A.M. Accurate specification of molecular structures: the case for zero-order bonds and explicit hydrogen counting. J. Chem. Inf. Model. 2011;51:3149–3157. doi: 10.1021/ci200488k. [DOI] [PubMed] [Google Scholar]
- 106.Warren Smith H., Lipscomb W.N. Single-crystal X-ray diffraction study of β-diborane. J. Chem. Phys. 1965;43:1060–1064. [Google Scholar]
- 107.Halterman R.L., Togni A., editors. Metallocenes: Synthesis, Reactivity, Applications. Wiley-VCH; 1998. [Google Scholar]
- 108.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sharpe H.R., Geer A.M., Taylor L.J., Gridley B.M., Blundell T.J., Blake A.J., Davies E.S., Lewis W., McMaster J., Robinson D., Kays D.L. Selective reduction and homologation of carbon monoxide by organometallic iron complexes. Nat. Commun. 2018;9:3757–3758. doi: 10.1038/s41467-018-06242-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dunitz J.D., Orgel L.E., Rich A. The crystal structure of ferrocene. Acta Crystallogr. 1956;9:373–375. [Google Scholar]
- 111.Einsle O., Rees D.C. Structural enzymology of nitrogenase enzymes. Chem. Rev. 2020;120:4969–5004. doi: 10.1021/acs.chemrev.0c00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu H.S., Truhlar D.G. Oxidation state 10 exists. Angew. Chem. 2016;128:9150–9152. doi: 10.1002/anie.201604670. [DOI] [PubMed] [Google Scholar]
- 113.La Macchia G., Aquilante F., Veryazov V., Roos B.O., Gagliardi L. Bond length and bond order in one of the shortest Cr–Cr bonds. Inorg. Chem. 2008;47:11455–11457. doi: 10.1021/ic801537w. [DOI] [PubMed] [Google Scholar]
- 114.Nguyen T., Sutton A.D., Brynda M., Fettinger J.C., Long G.J., Power P.P. Synthesis of a stable compound with fivefold bonding between two chromium(I) centers. Science. 2005;310:844–847. doi: 10.1126/science.1116789. [DOI] [PubMed] [Google Scholar]
- 115.National Center for Biotechnology Information PubChem compound summary for CID 5702198, cisplatin. 2021. https://pubchem.ncbi.nlm.nih.gov/compound/Cisplatin
- 116.Werner A. Nobel Lecture; 1913. On the Constitution and Configuration of Higher-Order Compounds. [Google Scholar]
- 117.Makhaev V.D., Borisov A.P., Boiko G.N., Tarasov B.P. Anionic zirconium and hafnium borohydride complexes. Russ. Chem. Bull. 1990;39:1081–1087. [Google Scholar]
- 118.Krotko D.G. Atomic ring invariant and modified CANON extended connectivity algorithm for symmetry perception in molecular graphs and rigorous canonicalization of SMILES. J. Cheminf. 2020;12:1–11. doi: 10.1186/s13321-020-00453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ugi I., Gillespie P. Beschreibung chemischer Systeme und ihrer Umwandlungen durch be-Matrizen und ihre Transformations-Eigenschaften. Angew. Chem. 1971;83:980–981. [Google Scholar]
- 120.Ugi I., Stein N., Knauer M., Gruber B., Bley K., Weidinger R. New elements in the representation of the logical structure of chemistry by qualitative mathematical models and corresponding data structures. in ‘computer chemistry. Top. Curr. Chem. 1993;166:199–233. [Google Scholar]
- 121.Stein N. New perspectives in computer-assisted formal synthesis design-treatment of delocalized electrons. J. Chem. Inf. Comput. Sci. 1995;35:305–309. [Google Scholar]
- 122.Stein N. Technical University Munich; 1993. Das sXBE- und sXR-Modell der konstitutionellen Chemie. Ph.D. thesis. [Google Scholar]
- 123.Dietz A. Yet another representation of molecular structure. J. Chem. Inf. Comput. Sci. 1995;35:787–802. [Google Scholar]
- 124.Bauerschmidt S., Gasteiger J. Overcoming the limitations of a connection table description: a universal representation of chemical species. J. Chem. Inf. Comput. Sci. 1997;37:705–714. [Google Scholar]
- 125.Jablonka K.M., Ongari D., Moosavi S.M., Smit B. Using collective knowledge to assign oxidation states of metal cations in metal–organic frameworks. Nat. Chem. 2021;13:771–777. doi: 10.1038/s41557-021-00717-y. [DOI] [PubMed] [Google Scholar]
- 126.Damhus T., Hartshorn R.M., Hutton A.T. Nomenclature of Inorganic Chemistry: Iupac Recommendations 2005. Chem. Int. 2005 [Google Scholar]
- 127.Cranmer M., Sanchez-Gonzalez A., Battaglia P., Xu R., Cranmer K., Spergel D., Ho S. NeurIPS; 2020. Discovering Symbolic Models from Deep Learning with Inductive Biases. [Google Scholar]
- 128.Lowe D.M. University of Cambridge; 2012. Extraction of Chemical Structures and Reactions from the Literature. Ph.D. thesis. [Google Scholar]
- 129.Lowe D. Chemical reactions from US patents (1976-Sep2016) 2017. https://figshare.com/articles/dataset/Chemical_reactions_from_US_patents_1976-Sep2016_/5104873
- 130.Jiang S., Zhang Z., Zhao H., Li J., Yang Y., Lu B.-L., Xia N. When SMILES smiles, practicality judgment and yield prediction of chemical reaction via deep chemical language processing. IEEE Access. 2021;9:85071–85083. [Google Scholar]
- 131.Buitrago Santanilla A., Regalado E.L., Pereira T., Shevlin M., Bateman K., Campeau L.-C., Schneeweis J., Berritt S., Shi Z.-C., Nantermet P., et al. Nanomole-scale high-throughput chemistry for the synthesis of complex molecules. Science. 2015;347:49–53. doi: 10.1126/science.1259203. [DOI] [PubMed] [Google Scholar]
- 132.Kearnes S.M., Maser M.R., Wleklinski M., Kast A., Doyle A.G., Dreher S.D., Hawkins J.M., Jensen K.F., Coley C.W. The open reaction database. J. Am. Chem. Soc. 2021;143:18820–18826. doi: 10.1021/jacs.1c09820. [DOI] [PubMed] [Google Scholar]
- 133.Szymkuć S., Gajewska E.P., Klucznik T., Molga K., Dittwald P., Startek M., Bajczyk M., Grzybowski B.A. Computer-assisted synthetic planning: the end of the beginning. Angew Chem. Int. Ed. Engl. 2016;55:5904–5937. doi: 10.1002/anie.201506101. [DOI] [PubMed] [Google Scholar]
- 134.Coley C.W., Jin W., Rogers L., Jamison T.F., Jaakkola T.S., Green W.H., Barzilay R., Jensen K.F. A graph-convolutional neural network model for the prediction of chemical reactivity. Chem. Sci. 2019;10:370–377. doi: 10.1039/c8sc04228d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Segler M.H.S., Preuss M., Waller M.P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- 136.Jin W., Coley C., Barzilay R., Jaakkola T. NeurIPS; 2017. Predicting Organic Reaction Outcomes with Weisfeiler-Lehman Network. [Google Scholar]
- 137.Schwaller P., Hoover B., Reymond J.-L., Strobelt H., Laino T. Extraction of organic chemistry grammar from unsupervised learning of chemical reactions. Sci. Adv. 2021;7:eabe4166. doi: 10.1126/sciadv.abe4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I. NeurIPS. 2017. Attention is all you need. [Google Scholar]
- 139.Schwaller P., Laino T., Gaudin T., Bolgar P., Hunter C.A., Bekas C., Lee A.A. Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Cent. Sci. 2019;5:1572–1583. doi: 10.1021/acscentsci.9b00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schwaller P., Petraglia R., Zullo V., Nair V.H., Haeuselmann R.A., Pisoni R., Bekas C., Iuliano A., Laino T. Predicting retrosynthetic pathways using transformer-based models and a hyper-graph exploration strategy. Chem. Sci. 2020;11:3316–3325. doi: 10.1039/c9sc05704h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Vaucher A.C., Schwaller P., Laino T. Completion of partial reaction equations. ChemRxiv. 2020 doi: 10.26434/chemrxiv.13273310.v1. Preprint at. [DOI] [Google Scholar]
- 142.Frank H., Lachiche N., Varnek A., Wagner A. Condensed graph of reaction: considering a chemical reaction as one single pseudo molecule. Int. J. Artif. Intell. Tool. 2011;20:253–270. [Google Scholar]
- 143.Bort W., Baskin I.I., Gimadiev T., Mukanov A., Nugmanov R., Sidorov P., Marcou G., Horvath D., Klimchuk O., Madzhidov T., Varnek A. Discovery of novel chemical reactions by deep generative recurrent neural network. Sci. Rep. 2021;11:3178–3215. doi: 10.1038/s41598-021-81889-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen W.L., Chen D.Z., Taylor K.T. Automatic reaction mapping and reaction center detection. WIREs. Comput. Mol. Sci. 2013;3:560–593. [Google Scholar]
- 145.Lemm D., von Rudorff G.F., von Lilienfeld O.A. Machine learning based energy-free structure predictions of molecules, transition states, and solids. Nat. Commun. 2021;12:4468–4510. doi: 10.1038/s41467-021-24525-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen M., Tworek J., Jun H., Yuan Q., de Oliveira Pinto H.P., Kaplan J., Edwards H., Burda Y., Joseph N., Brockman G., et al. Evaluating large language models trained on code. arXiv. 2021 doi: 10.48550/arXiv.2107.03374. Preprint at. [DOI] [Google Scholar]
- 147.Ray T.S. An evolutionary approach to synthetic biology: zen and the art of creating life. Artif. Life. 1993;1:179–209. [Google Scholar]
- 148.Adami C. Springer Science & Business Media; 1998. Introduction to Artificial Life. [Google Scholar]
- 149.Lenski R.E., Ofria C., Pennock R.T., Adami C. The evolutionary origin of complex features. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- 150.Wilke C.O., Wang J.L., Ofria C., Lenski R.E., Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412:331–333. doi: 10.1038/35085569. [DOI] [PubMed] [Google Scholar]
- 151.Flam-Shepherd D., Zhu K., Aspuru-Guzik A. Keeping it simple: language models can learn complex molecular distributions. arXiv. 2021 doi: 10.48550/arXiv.2112.03041. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kolmogorov A.N. On tables of random numbers. Sankhya: Indian J. Stat., Series A. 1963;25:369–376. [Google Scholar]
- 153.Nakata M., Shimazaki T. PubChemQC project: a large-scale first-principles electronic structure database for data-driven chemistry. J. Chem. Inf. Model. 2017;57:1300–1308. doi: 10.1021/acs.jcim.7b00083. [DOI] [PubMed] [Google Scholar]
- 154.Wu Z., Ramsundar B., Feinberg E.N., Gomes J., Geniesse C., Pappu A.S., Leswing K., Pande V. Moleculenet: a benchmark for molecular machine learning. Chem. Sci. 2018;9:513–530. doi: 10.1039/c7sc02664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Polykovskiy D., Zhebrak A., Sanchez-Lengeling B., Golovanov S., Tatanov O., Belyaev S., Kurbanov R., Artamonov A., Aladinskiy V., Veselov M., et al. Molecular sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 2020;11:1931. doi: 10.3389/fphar.2020.565644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brown N., Fiscato M., Segler M.H.S., Vaucher A.C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 2019;59:1096–1108. doi: 10.1021/acs.jcim.8b00839. [DOI] [PubMed] [Google Scholar]
- 157.Renz P., Van Rompaey D., Wegner J.K., Hochreiter S., Klambauer G. On failure modes in molecule generation and optimization. Drug Discov. Today Technol. 2019;32:55–63. doi: 10.1016/j.ddtec.2020.09.003. [DOI] [PubMed] [Google Scholar]
- 158.Frey N.C., Gadepally V., Ramsundar B. FastFlows: flow-based models for molecular graph generation. arXiv. 2022 doi: 10.48550/arXiv.2201.12419. Preprint at. [DOI] [Google Scholar]
- 159.Wellawatte G.P., Seshadri A., White A.D. Model agnostic generation of counterfactual explanations for molecules. Chem. Sci. 2022;13:3697–3705. doi: 10.1039/d1sc05259d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nigam A.K., Pollice R., Aspuru-Guzik A. Janus: parallel tempered genetic algorithm guided by deep neural networks for inverse molecular design. arXiv. 2021 doi: 10.48550/arXiv.2106.04011. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ahn S., Kim J., Lee H., Shin J. Guiding deep molecular optimization with genetic exploration. arXiv. 2020 doi: 10.48550/arXiv.2007.04897. Preprint at. [DOI] [Google Scholar]
- 162.Winter R., Montanari F., Steffen A., Briem H., Noé F., Clevert D.A. Efficient multi-objective molecular optimization in a continuous latent space. Chem. Sci. 2019;10:8016–8024. doi: 10.1039/c9sc01928f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Heller S.R., McNaught A., Pletnev I., Stein S., Tchekhovskoi D. InChI, the IUPAC international chemical identifier. J. Cheminf. 2015;7:23–34. doi: 10.1186/s13321-015-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shen C., Krenn M., Eppel S., Aspuru-Guzik A. Deep molecular dreaming: inverse machine learning for de-novo molecular design and interpretability with surjective representations. Mach. Learn, Sci. Technol. 2021;2:03LT02. [Google Scholar]
- 165.Hu W. Inverse molecule design with invertible neural networks as generative models. J. Biomed. Sci. Eng. 2021;14:305–315. [Google Scholar]
- 166.Rajan K., Zielesny A., Steinbeck C. DECIMER: towards deep learning for chemical image recognition. J. Cheminf. 2020;12:65–69. doi: 10.1186/s13321-020-00469-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rajan K., Zielesny A., Steinbeck C. STOUT: SMILES to IUPAC names using neural machine translation. J. Cheminf. 2021;13:1–14. doi: 10.1186/s13321-021-00512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Clevert D.A., Le T., Winter R., Montanari F. Img2Mol – accurate SMILES recognition from molecular graphical depictions. Chem. Sci. 2021;12:14174–14181. doi: 10.1039/d1sc01839f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Winter R., Montanari F., Noé F., Clevert D.A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem. Sci. 2019;10:1692–1701. doi: 10.1039/c8sc04175j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Arús-Pous J., Johansson S.V., Prykhodko O., Bjerrum E.J., Tyrchan C., Reymond J.-L., Chen H., Engkvist O. Randomized SMILES strings improve the quality of molecular generative models. J. Cheminf. 2019;11:71. doi: 10.1186/s13321-019-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van der Maaten L., Hinton G. Visualizing data using t-sne. J. Mach. Learn. Res. 2008;9 [Google Scholar]
- 172.Iten R., Metger T., Wilming H., Del Rio L., Renner R. Discovering physical concepts with neural networks. Phys. Rev. Lett. 2020;124:010508. doi: 10.1103/PhysRevLett.124.010508. [DOI] [PubMed] [Google Scholar]
- 173.Seif A., Hafezi M., Jarzynski C. Machine learning the thermodynamic arrow of time. Nat. Phys. 2021;17:105–113. [Google Scholar]
- 174.Krenn M., Erhard M., Zeilinger A. Computer-inspired quantum experiments. Nat. Rev. Phys. 2020;2:649–661. doi: 10.1103/PhysRevLett.125.050501. [DOI] [PubMed] [Google Scholar]
- 175.Flam-Shepherd D., Wu T., Gu X., Cervera-Lierta A., Krenn M., Aspuru-Guzik A. Learning interpretable representations of entanglement in quantum optics experiments using deep generative models. arXiv. 2021 doi: 10.48550/arXiv.2109.02490. Preprint at. [DOI] [Google Scholar]