Abstract
Enterovirus 71 (EV71) caused hand, foot and mouth disease (HFMD) is a serious threat to the health of young children. Although type I interferon (IFN-I) has been proven to control EV71 replication, the key downstream IFN-stimulated gene (ISG) remains to be clarified and investigated. Recently, we found that 2′-5′-oligoadenylate synthetases 3 (OAS3), as one of ISG of IFN-β1b, was antagonized by EV71 3C protein. Here, we confirm that OAS3 is the major determinant of IFN-β1b-mediated EV71 inhibition, which depends on the downstream constitutive RNase L activation. 2′-5′-oligoadenylate (2-5A) synthesis activity deficient mutations of OAS3 D816A, D818A, D888A, and K950A lost resistance to EV71 because they could not activate downstream RNase L. Further investigation proved that EV71 infection induced OAS3 but not RNase L expression by IFN pathway. Mechanically, EV71 or IFN-β1b-induced phosphorylation of STAT1, but not STAT3, initiated the transcription of OAS3 by directly binding to the OAS3 promoter. Our works elucidate the immune regulatory mechanism of the host OAS3/RNase L system against EV71 replication.
Keywords: Enterovirus 71 (EV71), 2′-5′-oligoadenylate synthetases 3 (OAS3), RNase L, IFN-β1b, JAK/STAT
Highlights
-
•
OAS3 contributes important inhibition effect for IFN-β1b against EV71.
-
•
OAS3 resistance to EV71 replication depends on RNase L activation.
-
•
STAT1 initiates the transcription of OAS3 by directly binding to the OAS3 promoter.
1. Introduction
As an important subtype of infectious enterovirus (EV), the EV71 epidemic continues to cause outbreaks of HFMD worldwide (Rasti et al., 2019). The lack of understanding on the molecular mechanisms of the virus-host interactions limited the development of specific anti-EV71 strategies (Swain et al., 2022).
Critically important to understanding antiviral innate immunity is determining which host proteins are responsible for inhibiting different types of viruses. Previous study proved that IFN-I has shown the ability to control the replication of EVs (Lu et al., 2015). For instance, IFN-I could prevent hepatonecrosis and viral dissemination in coxsackievirus B3 (CVB3)-infected mice (Koestner et al., 2018). And IFN-β effectively reduced viral production in CVB3-infected myocardial fibroblasts (Kandolf et al., 1985). Also, IFN-I had been proved to increase survival rates with decreased viral loads in IFN-I treated mice (Yi et al., 2011; Rasti et al., 2019). However, there remain huge gaps about functions and regulatory mechanisms of the EV71 infection-associated key ISG. The OAS-RNase L system possesses potent Antiviral function induced by IFN (Samuel, 2001; Kristiansen et al., 2011). OASs are common dsRNA-associated pattern-recognition receptors for many types of RNA viruses. Four OAS members are identified in humans, of which OAS1, OAS2, and OAS3 catalyze 2-5A synthesis from ATP upon binding dsRNA. When RNase L was activated by 2-5A, viral and cellular RNAs were endonucleolytic cleaved for blocking viral replication and causing cell death. Amino acids D816, D818, D888, and K950 of OAS3 have been proved to be crucial for its 2-5A synthesis activity by sequence alignment and activity determination (Ibsen et al., 2014). OASL could not catalyze the synthesis of 2-5A but activates RIG-I signaling to respond viral infection (Choi et al., 2015). Although the antiviral effect of OAS/RNase L system has been illuminated on some viruses, the role of OAS/RNase L system in EV71 replication is still unknown.
In our study, we verified that OAS3 could inhibit EV71 replication depending on RNase L activation. OAS3 but not RNase L could be upregulated by EV71 infection. And the induction of OAS3 depended on IFN pathway. As we known, IFN-I produced by the immune response interacts with the IFN-I receptor (IFNAR1) to induce the phosphorylation of STAT1 and STAT2 through the JAK-STAT pathway (Schoggins and Rice, 2011; Stark and Darnell, 2012). Phosphorylated STAT1 and STAT2 form a heterodimer and translocate to the nucleus, where they bind to the IFN-stimulated response element (ISRE) and subsequently promote ISGs expression (Ivashkiv and Donlin, 2014). Although the OAS family members are proven to be IFN-induced proteins, the molecular mechanism by which IFN induces OAS3 remains unclear. Here, we unveiled that IFN-β1b but not other IFNs significantly inhibited EV71 replication and viral cytopathic effect, and proved that OAS3 contributed important inhibition effect for IFN-β1b against EV71. Further studies revealed the complete IFN-β1b/JAK/STAT1/OAS3 innate immune response pathway for the early stage of EV71 infection. Compared with previous studies, our studies provide a reliable molecular mechanism for the clinical use of IFN-β1b to treat EV71 infection.
2. Materials and methods
2.1. Cells, viruses, and agents
Human HEK293T cells (ATCC catalog no. CRL-11268) were purchased and preserved in our lab. Cell culture was performed as previously described (Zhou et al., 2022). EV71 CC063 was isolated and preserved by our lab (Xu et al., 2020). Viruses were amplified using RD cells. Viral titers in cultural supernatants were determined by plaque assay.
Recombinant human IFN-β1b, IFN-γ, and IL-29 were purchased from BBI LIFE SCIENCES CORPORATION, Shanghai, China (C600038, C600039, C600117). The used concentration of IFNs was 250 U/mL for IFN-β1b, 250 U/mL for IFN-γ, and 250 U/mL for IL-29).
2.2. Plasmid construction
OAS3 overexpression and silencing plasmids were constructed as previously described (Zhou et al., 2022). OAS3 mutants D816A, D818A, D888A, and D950A were generated by using standard site-directed mutagenesis. For RNase L knockout, RNase L-specific sgRNA was cloned into the CRISPR-control (K010) plasmid (Applied Biological Materials, Richmond, BC, Canada). Plasmids of STAT1, STAT1-knockdown (STAT1-KD), STAT3-knockdown (STAT3-KD), IFNR1-knockdown (IFNR1-KD) were kind gifts from Prof. Guangyun Tan (Tan et al., 2018). sgRNA sequences are shown in Supplementary Table S1.
2.3. Stable silencing and CRISPR/Cas9 knockdown cell lines
OAS3 stable silencing cell line was constructed and screened as previously described (Zhou et al., 2022), For RNase L, STAT1, STAT3, and IFNR1 knockdown, plasmids expressing Cas9, sgRNA, and a puromycin selection marker were co-transfected into HEK293T cells. After 36 h of culture, puromycin (2 μg/mL) were added to select cells. After 48–72 h, 10 × dilution of living cells was perform to obtain single clone per well on a 96-well plate. The efficiency of gene KD was measured by Western blot.
2.4. RNA extraction and RT-qPCR
Total RNA extraction and RT-qPCR were performed as previously described (Xu et al., 2020). The primers are listed in Supplementary Table S2. The mRNA levels of target genes were normalized to GAPDH levels, and relative mRNA levels were compared by the 2−ΔΔCt method.
2.5. Western blot analysis and antibodies
Preparation of cell lysate, SDS-PAGE, and Western blot was performed as previously described (Zhou et al., 2022). The following antibodies and reagents were used: human OAS3 (#21915-1-AP, Proteintech, Wuhan, China), RNase L (#22577-1-AP, Proteintech, Wuhan, China), IFNAR1 (#MA5-35051, ThermoFisher, Shanghai, China), STAT1 (#D120084, BBI, Shanghai, China), p-STAT1 (#D155017, BBI, Shanghai, China), STAT3 (#D220083, BBI, Shanghai, China), p-STAT3 (#D155018, BBI, Shanghai, China), β-tubulin (#RM2003, Ray Antibody Biotech, Beijing, China). Polyclonal antibodies (pAbs) against EV71 were obtained from rabbits immunized with EV71 whole viruses in our laboratory (Xu et al., 2020). Goat anti-rabbit peroxidase-conjugated IgG antibody (Millipore; AP132P) and goat anti-mouse peroxidase-conjugated IgG Antibody (Millipore; AP124P) were used as secondary antibodies.
2.6. Cell viability
Residual cell viability was measured by CCK8 kit (#FC101, TransGen) according to the manufacturer's instructions.
2.7. OAS3 promoter reporter and dual-luciferase reporter assay
As the promoter region of genes is usually the sequence upstream of the initiation codon of genes, 2000 bp upstream sequence of OAS3 was selected and cloned into a pGL4.11[luc2P] (#E6661, Promega) as promoter region. HEK293T or STAT1 KD cells were co-transfected with the respective OAS3 promoter reporter and pGL4.74[hRluc/TK] (#E6921, Promega) reporter. After 24 h, cells were treated with IFN-β1b or EV71 for 24 h. The Dual-Luciferase Reporter Assay System (#E1910, Promega, Madison, WI, USA) was used to measure the activity of firefly luciferase and Renilla luciferase in the lysates.
2.8. Chromatin immunoprecipitation (ChIP)
Briefly, the treated cells were cross-linked with 1% formaldehyde, sheared to an average size of ∼ 500 bp, and then immunoprecipitated with IgG or anti-bodies against STAT1 or STAT3. The proximal promoter regions containing putative four STAT1-binding sites within the OAS3 promoter were amplified by quantitative RT-qPCR.
2.9. Statistical analyses
Statistical results were analyzed using SPSS 17.0. Data are reported as means ± standard deviation (SD) from three independent experiments. Statistical significance was evaluated by Student's t-test. Significant differences are indicated as follows: ∗P < 0.05, ∗∗P < 0.01. # stands for no significance.
3. Results
3.1. OAS3 resistance to EV71 replication depends on RNase L activation
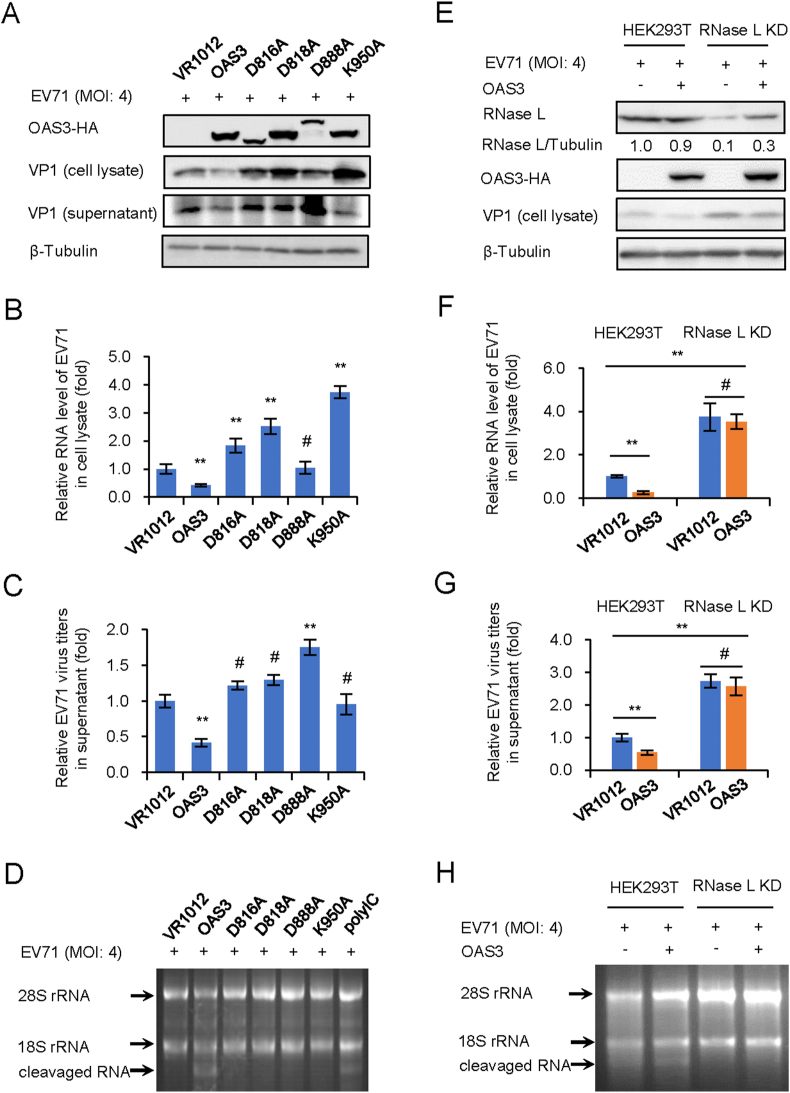
Previous reports have proved that OAS3 has a higher 2-5A synthesis activity than OAS1 and OAS2 (Li et al., 2016), and our recent study found that OAS3 has a stronger anti-EV71 ability than OAS1 and OAS2 (unpublished data). We hypothesized that OAS3 resistance to EV71 depends on 2-5A synthase activity, although EV71 genome is ssRNA rather than dsRNA. To verify the hypothesis, HEK293T shOAS3 cells were constructed and transfected with OAS3 wild type (WT) or inactivated mutants (D816A, D818A, D888A, and K950A) for 12 h prior to EV71 infection at an MOI of 4 for 48 h. Western blot showed that OAS3 significantly reduced EV71 VP1 expression (Fig. 1A). RT-qPCR analysis indicated that OAS3 overexpression reduced intracellular viral RNA level (Fig. 1B). And extracellular viral titers were also downregulated by OAS3 (Fig. 1C). On the contrary, four inactivated mutants did not show any inhibitory effect on EV71 replication. Agarose electrophoresis of total rRNA of cells indicated that WT OAS3 expression obviously induced intracellular RNase L activation and RNA cleavage, but inactivated mutants did not (Fig. 1D). In order to confirm the role of RNase L in OAS3-mediated EV71 inhibition, negative control pLKO.1 and RNase L KD HEK293T cells were transfected with VR1012 or OAS3 plasmid for 12 h prior to EV71 infection at an MOI of 4. After 48 h, we found that RNase L KD facilitated EV71 replication, and the inhibitory effect of OAS3 on EV71 replication was significantly attenuated in RNase L KD cells (Fig. 1E–G). This was attributed to the impairment of RNase L-dependent intracellular RNA degradation (Fig. 1H). These data demonstrated that OAS3 suppressed EV71 replication in an RNase L-dependent manner.
Fig. 1.
OAS3 inhibits EV71 replication depending on RNase L activation. 2-5A synthesis activity of OAS3 determines the inhibitory effect on EV71 replication.
A WT and inactivated mutants of OAS3 transfected HEK293T cells were infected with EV71 at an MOI of 4. The expression levels of exogenous OAS3 and VP1 were examined by Western blotting. B Viral RNA in cell lysate was examined by RT-qPCR. The RNA level of VR1012 was set as “1”. C Viral titers in the supernatants were determined by the plaque assay. D RNase L activation was examined by agarose electrophoresis of total rRNA of cells. E The negative control pLKO.1 and RNase L knockdown (RNase L KD) HEK293T cells were transfected with OAS3-expressing or control plasmids. Cells were infected with EV71 at an MOI of 4. The expression levels of endogenous RNase L, exogenous OAS3, and VP1 were examined by Western blotting. F Viral RNA in cell lysate was examined by RT-qPCR. The RNA level of VR1012 was set as “1”. G Viral titers in the supernatants were determined by the plaque assay. H RNase L activation was examined by agarose electrophoresis of total rRNA of cells. The results represent the means ± standard deviation from three independent experiments. Statistical significance was analyzed using Student's t-test (# no significance, ∗∗P < 0.01).
3.2. EV71 infection induces OAS3 but not RNase L expression by IFN pathway
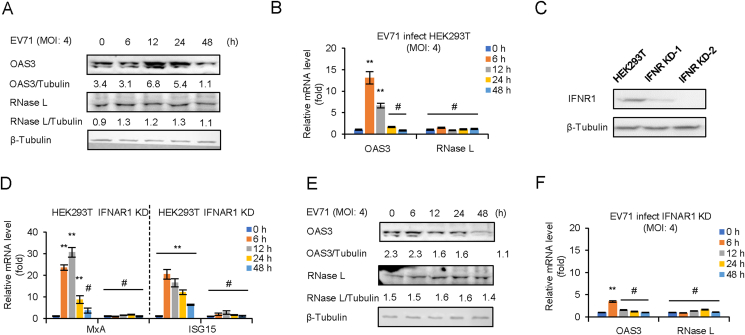
Negative control pLKO.1 and IFNAR1 KD HEK293T cells were infected with EV71 at an MOI of 4 for 0–48 h mRNA level of OAS3 and RNase L was determined by RT-qPCR and Western blot. The results indicated that the protein level of OAS3 was increased at 12–24 h post-infection (h.p.i.), and the transcription of OAS3 was upregulated at 6–12 h.p.i. in pLKO.1 HEK293T cells. However, neither mRNA nor protein levels of RNase L were affected by viral infection (Fig. 2A and B). Since IFN response is the early event of viral infection, IFNAR1 KD HEK293T cells were constructed and screened (Fig. 2C). Then, the mRNA levels of MxA and ISG15, two IFN-β1b inducible ISGs, were detected to confirm the IFNAR1 knockdown efficiency (Fig. 2D). RT-qPCR indicated that IFNAR1 knockdown completely blocked IFN-β1b-induced expression of MxA and ISG15. Both mRNA and protein levels of OAS3 were no longer induced by viral infection in IFNAR1 KD HEK293T cells (Fig. 2E and F). Our findings verified that OAS3 but not RNase L was upregulated in the early stage of EV71 infection. And the induction of OAS3 depended on IFN pathway.
Fig. 2.
EV71 infection induces OAS3 but not RNase L expression by IFN pathway. EV71 infection upregulates the expression of OAS3 but not RNase L. HEK293T cells were infected with EV71 at an MOI of 4, and the expression levels of OAS3 and RNase L were examined by Western blotting (A). The mRNA levels of OAS3 and RNase L were examined by RT-qPCR (B). The RNA level of “0 h” was set as “1”. C Construction of IFNAR1 CRISPR/Cas9 knockdown cell line. The efficiency of knockdown was examined by Western blotting. D The negative control pLKO.1 and IFNAR1 knockdown (IFNAR1 KD) HEK293T cells were treated with 250 U/mL IFN-β1b for 48 h, and the expression levels of MxA and ISG15 were examined by RT-qPCR at different time points. The RNA level of “0 h” was set as “1”. E IFNAR1 KD HEK293T cells were infected with EV71 at an MOI of 4, and the expression levels of OAS3 and RNase L were examined by Western blotting. F The mRNA levels of OAS3 and RNase L were examined by RT-qPCR. The RNA level of “0 h” was set as “1”. The results represent the means ± standard deviation from three independent experiments. Statistical significance was analyzed using Student's t-test (# no significance, ∗∗P < 0.01).
3.3. IFN-β1b significantly inhibits EV71 replication and viral cytopathic effect
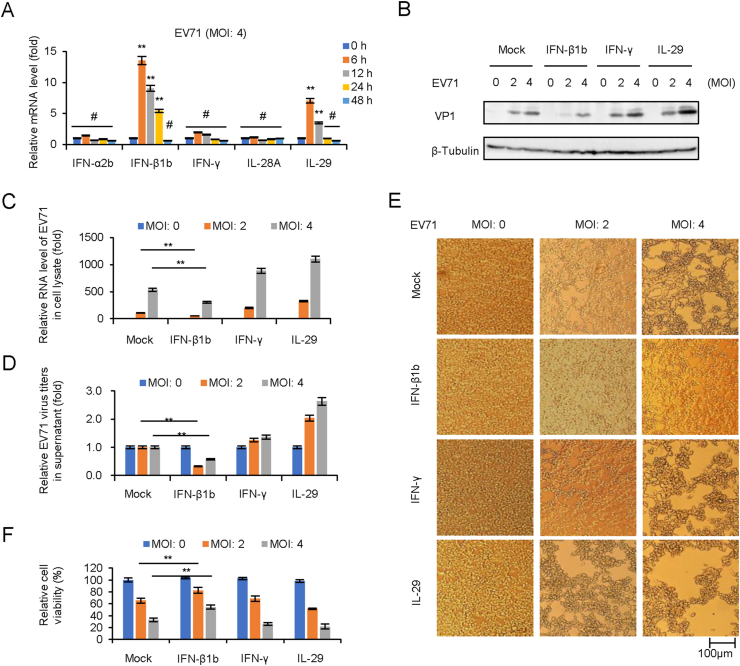
Since the upregulation of OAS expression in Fig. 2A might attribute to IFN-I, this prompted us to screen and confirm IFNs expression in pLKO.1 cells in case of EV71 infection in Fig. 2A by RT-qPCR. The results indicated that IFN-β1b and IL-29 (IFN-λ1) were upregulated from 6 to 12 h. p.i (Fig. 3A). In order to evaluate the antiviral effect of different type IFNs, HEK293T cells were pretreated with IFNs (250 U/mL IFN-β1b, 250 U/mL IFN-γ, and 250 U/mL IL-29) for 24 h prior to EV71 infection at MOIs of 2 and 4. Cell culture and supernatants were collected at 48 h.p.i. for viral protein expression, total viral RNA, and infectious viral titration detection. Western blot detection showed that viral VP1 expression in cell lysate was inhibited by IFN-β1b treatment (Fig. 3B). RT-qPCR indicated that IFN-β1b reduced intracellular viral RNA transcription (Fig. 3C). Titer determination indicated that IFN-β1b, but not IFN-γ and IL-29, could reduce extracellular viral titers (Fig. 3D). Furthermore, apparent cytopathic effect (CPE) and cell viability determination showed that IFN-β1b exhibited better cytopathic inhibition effect than IFNγ and IL-29 (Fig. 3E and F).
Fig. 3.
IFN-β1b significantly inhibits EV71 replication and viral cytopathic effect. A EV71 infection induces the expression of IFN-β1b in HEK293T cells. HEK293T cells were infected with EV71 at an MOI of 4, and the expression levels of IFNs were examined by RT-qPCR at different time points. The RNA level of “0 h” was set as “1”. B IFN-β1b plays an antiviral effect in EV71 infected HEK293T cells. After 250 U/mL IFN-β1b, 250 U/mL IFN-γ, and 250 U/mL IL29 treated HEK293T cells for 24 h, cells were infected EV71 at different MOI. The expression levels of VP1 were examined by Western blotting. C The expression levels of EV71 viral RNA were examined by RT-qPCR. The RNA level at an MOI of 0 was set as “1”. D Viral titers in the supernatants were determined by the plaque assay. E Apparent CPE was observed at 48 h.p.i. F Cell viability in the experiment of Fig. 3E was measured by CCK8. The cell viability at an MOI of 0 was set as “1”. The results represent the means ± standard deviation from three independent experiments. Statistical significance was analyzed using Student's t-test (# no significance, ∗∗P < 0.01).
3.4. OAS3 contributes important inhibition effect for IFN-β1b against EV71
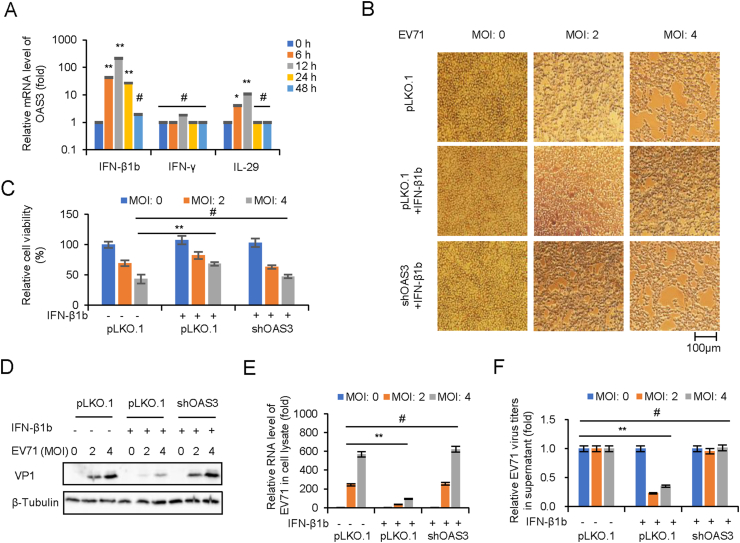
Importantly, mRNA level of OAS3 was found to be strongly induced by IFN-β rather than IFN-γ and IL-29 in the experiment of Fig. 4A. In order to explore the role of OAS3 in IFN-β-mediated inhibition of EV71 replication, HEK293T shRNA control (pLKO.1) and shOAS3 cells were pretreated with or without IFN-β1b for 24 h prior to EV71 infection at MOIs of 2 and 4. Apparent CPE and quantitative cell viability assay showed that compared with IFN-β1b treated pLKO.1 group, OAS3 silencing remarkably diminished the protective effect of IFN-β1b (Fig. 4B and C). RT-qPCR and Western blot showed that OAS3 silencing enhanced RNA transcription and protein expression of EV71 in presence of IFN-β1b (Fig. 4D and E). Also, extracellular viral production were upregulated by OAS3 silencing (Fig. 4F). Taken together, these findings showed that EV71 infection stimulated the innate immune response of IFN-β1b/OAS3, and OAS3 contributes important inhibition effect for IFN-β1b against EV71.
Fig. 4.
OAS3 plays an important role in IFN-β1b-mediated inhibition of EV71. A IFN-β treatment induces the expression of OAS in HEK293T cells. HEK293T cells were treated with 250 U/mL IFN-β1b, 250 U/mL IFN-γ, and 250 U/mL IL-29 for 48 h, and the expression levels of OAS3 were examined by RT-qPCR at different time points. The RNA level of “0 h” was set as “1”. B HEK293T shRNA control (pLKO.1) and shOAS3 cells were pretreated with or without 250 U/mL IFN-β for 24 h prior to EV71 infection at MOIs of 2 and 4. Apparent CPE was observed at 48 h.p.i. C Cell viability in the experiment of Fig. 4B was measured by CCK8. D The expression levels of VP1 were examined by Western blotting. E The expression levels of EV71 viral RNA were examined by RT-qPCR. The RNA level at an MOI of 0 was set as “1”. F Viral titers in the supernatants were determined by the plaque assay. The results represent the means ± standard deviation from three independent experiments. Statistical significance was analyzed using Student's t-test (# no significance, ∗∗P < 0.01).
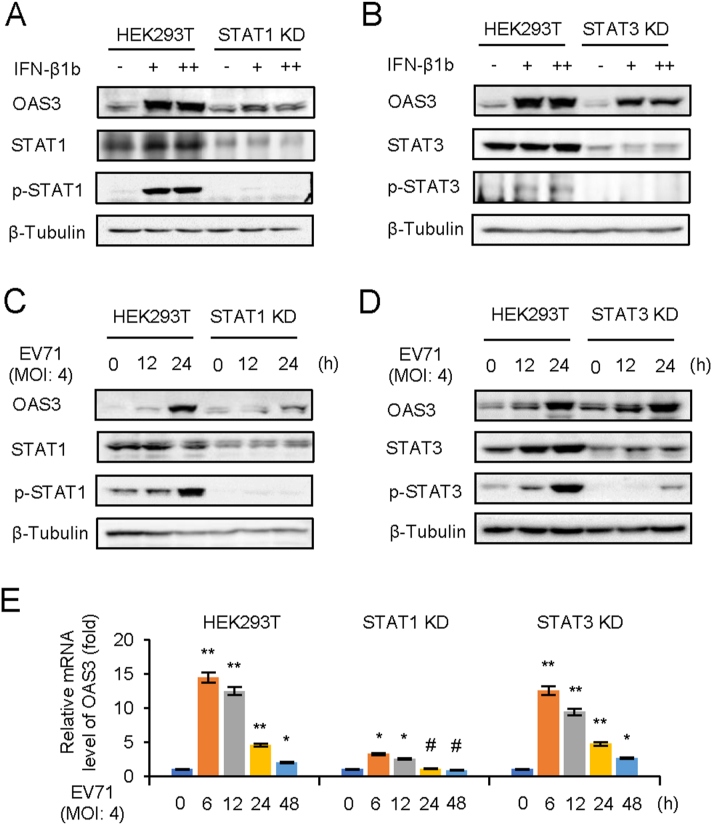
3.5. IFN-β1b-mediated OAS3 induction depends on STAT1 phosphorylation
The classical IFN-I pathway is that IFN-I induces phosphorylation of STAT1 and STAT2 through the IFNR1/JAK/STAT pathway (Stark and Darnell, 2012). STAT3 is also a multifunctional factor that regulated by JAK/STAT pathway (Gough et al., 2009; Wan et al., 2010). And the phosphorylation of STAT3 induced by EV71 infection has been reported (Wang et al., 2019). Nevertheless, the expression of STAT3-induced downstream ISG during viral infection remains unclear. In order to unveil the mechanisms through which IFN-β1b induces OAS3, STAT1 or STAT3 KD HEK293T cells were constructed. Negative control pLKO.1 and STAT KD HEK293T cells were treated with different doses of IFN-β1b for 24 h. Endogenous STAT1, p-STAT1, STAT3, p-STAT3, and OAS3 were detected by Western blot. Interestingly, IFN-β1b could not induce OAS3 in STAT1 KD cells but not in STAT3 KD cells (Fig. 5A and B). Also, EV71 infection at a MOI of 4 for 0–24 h induced IFN-β1b expression. STAT1, but not STAT3, are important for OAS3 upregulation by EV71 infection (Fig. 5C and D). The transcription level of OAS3 determined by RT-qPCR was consistent with the above results (Fig. 5E). Taken together, we speculate that JAK/STAT1 pathway is essential for IFN-β1b-induced OAS3 expression.
Fig. 5.
IFN-β1b-mediated OAS3 induction depends on STAT1 phosphorylation. A The negative control pLKO.1 and STAT1 knockdown (KD) HEK293T cells were treated with 125 U/mL (+) and 250 U/mL (++) IFN-β1b for 24 h as indicated, and the expression of endogenous OAS3, STAT1, and p-STAT1 were examined by Western blotting. B The negative control pLKO.1 and STAT3 KD HEK293T cells were treated with 125 U/mL (+) and 250 U/mL (++) IFN-β1b for 24 h as indicated, and the expression of endogenous OAS3, STAT1, and p-STAT1 were examined by Western blotting. C The negative control pLKO.1 and STAT1 KD HEK293T cells were infected with EV71 at an MOI of 4, and the expression of endogenous OAS3, STAT1, and p-STAT1 were examined by Western blotting at different time points. D The negative control pLKO.1 and STAT3 KD HEK293T cells were infected with EV71 at an MOI of 4, and the expression of endogenous OAS3, STAT1, and p-STAT1 were examined by Western blotting at different time points. E The negative control pLKO.1, STAT1 KD, and STAT3 KD HEK293T cells were infected with EV71 at an MOI of 4, and the expression levels of OAS3 were examined by RT-qPCR at different time points. The RNA level of “0 h” was set as “1”. The results represent the means ± standard deviation from three independent experiments. Statistical significance was analyzed using Student's t-test (# no significance, ∗P < 0.05, ∗∗P < 0.01).
3.6. STAT1 promotes transcription of OAS3 by binding to the OAS3 promoter
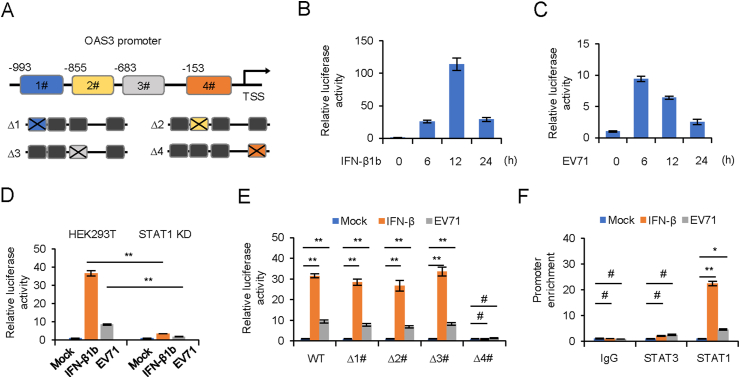
In order to identify the potential binding sites of transcription factor STAT1 in the promoter of OAS3, we predicted four potential binding regions in OAS3 promoter by JASPAR and TRANSFAC (Fig. 6A) (Matys et al., 2003; Mathelier et al., 2016). Full length and four binding sites individual deletion forms of OAS3 promoter were cloned into a luciferase reporter vector. Dual-luciferase reporter assay was performed to detect the transcription regulation of OAS3. Our results showed that both IFN-β1b treatment and EV71 infection significantly induced OAS3 transcription (Fig. 6B and C), whereas this induction was attenuated by STAT1 KD (Fig. 6D). Furthermore, STAT1 binding site #4 was seemingly crucial for OAS3 transcription upon IFN-β1b treatment or EV71 infection (Fig. 6E). Finally, the direct interaction of STAT1 with the OAS3 promoter was identified by the ChIP assay. As shown in Fig. 6F, compared with IgG and STAT3, STAT1 could bind more strongly to the OAS3 promoter in the presence of IFN-β1b or EV71. This demonstrated that IFN-β1b treatment or EV71 infection significantly promoted recruitment of STAT1, but not STAT3, to OAS3 promoter Site #4 region. These results suggested the crucial role of STAT1 in promoting OAS3 transcription upon IFN-β1b treatment.
Fig. 6.
STAT1 binds to the OAS3 promoter. A Schematic illustration showing four potential STAT1 binding sites in the OAS3 promoter region. B, C HEK293T cells were transfected with firely luciferase reporter harboring wild type (WT) promoter regions of OAS3 together with Renilla luciferase reporter for 12 h. Cells were treated with 250 U/mL IFN-β1b or infected with EV71 at an MOI of 4 for 24 h, and dual-luciferase reporter assay was measured at different time points. Relative luciferase activity of “0 h” was set as “1”. D The negative control pLKO.1 and STAT1 knockdown (KD) HEK293T cells were transfected with firefly luciferase reporter harboring WT promoter regions of OAS3 together with Renilla luciferase reporter for 12 h. Cells were treated with 250 U/mL IFN-β1b or infected with EV71 at an MOI of 4 and dual-luciferase reporter assay was measured at 24 h. Relative luciferase activity of “Mock” was set as “1”. E HEK293T cells were transfected with firefly luciferase reporter harboring WT or mutant promoter regions of OAS3 together with Renilla luciferase reporter for 12 h. Cells were treated with 250 U/mL IFN-β1b or infected with EV71 at an MOI of 4 and dual-luciferase reporter assay was measured at 24 h. Relative luciferase activity of “Mock” was set as “1”. F HEK293T cells were treated with 250 U/mL IFN-β1b or infected with EV71 at an MOI of 4 for 24 h. A chromatin immunoprecipitation assay was performed to confirm the binding of STAT1 on OAS3 promoter. The RNA level of “Mock” was set as “1”. The results represent the means ± standard deviation from three independent experiments. Statistical significance was analyzed using Student's t-test (# no significance, ∗P < 0.05, ∗∗P < 0.01).
4. Discussion
Although the association of OAS/RNase L system with virus replication have been studied extensively, the Picornaviridae shows the best relevance between activation of the OAS/RNase L pathway and inhibition of virus replication. For example, Chinese hamster ovary cells constitutively expressing the 40-kDa OAS homologue are resistant to Mengo virus but not VSV or herpesviruses (Chebath et al., 1987). Similar results are reported within human OAS1-overexpressing human and mouse cells. Human T98G cells that express OAS1 and mouse NIH 3T3 cells that express murine OAS display a resistance to encephalomyelitis virus (EMC virus) replication but not to VSV replication (Rysiecki et al., 1989; Coccia et al., 1990). The resistance to EMC virus replication correlates with the OAS enzyme activity. Likewise, mouse cells stably expressing human OAS2 show a partial antiviral response to EMC virus (Marié et al., 1999). Constitutive expression of OAS2 in human HT1080 cells likewise causes inhibition of EMC virus replication but not of VSV, Sendai Virus (SeV), or reovirus replication (Ghosh et al., 2000). Li et al. reported that the activation of RNase L was dependent on OAS3 expression during infection with diverse human viruses. OAS3 displayed a higher affinity for dsRNA than either OAS1 or OAS2, consistent with its dominant role in RNase L activation (Li et al., 2016). Our recent work investigated the anti-EV71 effect of OAS1, OAS2 and OAS3, and found that OAS3 had a stronger antiviral effect than OAS1 and OAS2 (unpublished data). Here, we further confirm that OAS3 inhibition of EV71 is depended on 2-5A synthetic activity and RNase L activation (Fig. 1). The viral RNA level in HEK293T cells was significantly lower than that in RNase L knockdown cells (Fig. 1F). Meanwhile, the endogenous rRNA in HEK293T cells but not RNase L knockdown cells was degraded with EV71 infection (Fig. 1H). We suggested that intracellular constitutive RNase L could be activated by EV71 infection to degraded viral RNA. Previous studies have shown that RNase L is constitutively present in most types of cells (Floyd-Smith and Denton, 1988), and treatment with IFN-α/β enhances RNase L activity in some types of cells (Jacobsen et al., 1983; Floyd-Smith, 1988). We next demonstrated that EV71 infection could induce OAS3 but not RNase L expression (Fig. 2), while the induced OAS3 further activated latent RNase L to play an anti-EV71 role. The dependence on RNase L activation seems to demonstrate that OAS3 does not detract from mechanisms other than OAS/RNase L in its anti-EV71 activity. Our conclusion is mutually corroborated with the results reported by Birdwell et al. that activation of RNase L does not require murine coronavirus-induced IFN but rather correlates with adequate levels of basal Oas gene expression, maintained by basal IFN signaling (Birdwell et al., 2016).
In EVs patients, IFN treatment remains an important clinical therapeutic option. However, due to the associated adverse effects and low efficiency, IFN treatment is not widely used in clinical therapeutics. Understanding mechanism of IFN-mediated inhibition of EVs replication is relative imperative. Our results suggested that EV71 infection up-regulated IFN-β1b and IL-29. Among of them, only IFN-β1b significantly inhibited EV71 replication and viral cytopathic effect (Fig. 3). Huang et al. also observed that IFN-β expression was upregulated in EV71-infected neural cells via pattern recognition receptors (PRRs) sensing of virus RNA (Huang et al., 2019). However, Su et al. revealed type III IFNs, rather than type I IFNs, as the dominant production through TLR3/IRF1 signaling upon multiple human enterovirus infection including EV71 (Su et al., 2020). And IFN-λ subsequently induced antiviral activity against EV replication in vitro and in vivo. Our previous studies have shown that EV71 can infect multiple organs in mice by intracranial challenge (Xu et al., 2020). And the virus has also been found in multiple organs of EV71-infected people, causing different symptoms. We suggest that these seemingly contradictory results may be caused by differences in infection systems.
IFN-I response is the first line of defense against viral infection through producing hundreds of ISGs to suppress viral replication. IFN-I induce the ISGs through the JAK/STAT signal pathway (Stark and Darnell, 2012). First, IFN-I bind to heterodimeric receptors consisting of IFNAR1 and 2, causing transphosphorylation and activation of the “Janus” tyrosine kinases Tyk2 and JAK1. Then the STATs are recruited to the receptor-bound JAKs and phosphorylated at tyrosine. Phosphorylated STAT1/2 subsequently associates with IFN-regulatory factor 9 (IRF9) to activate transcription of ISGs by binding the ISREs (Ivashkiv and Donlin, 2014). STAT3 exhibits dual regulatory roles during viral infection (Kuchipudi, 2015; Roca Roca Suarez et al., 2018). Some of the viral infections were facilitated by STAT3 since specific inhibitors or siRNAs of STAT3 repressed viral replication (Ho and Ivashkiv, 2006; Wang et al., 2011; Tsai and Lee, 2018). However, with respect to vaccinia virus and influenza A virus, STAT3 knockdown increased viral replication with decreased expression levels of ISGs (Mahony et al., 2017). Previous reports suggested that STAT3 could regulate EV71-mediated OAS1 and MxA expression by interfering with the nuclear entry of STAT1 (Wang et al., 2019). In our work, we identified OAS3, as a STAT1-dependent ISG, is a key downstream anti-EV71 factor (Fig. 4, Fig. 5). Further investigation indicated that STAT1 directly binds to the OAS3 promoter, which is responsible for IFN-β1b or EV71-induced OAS3 induction (Fig. 6).
5. Conclusions
In conclusion, our studies shed new light on the complete IFN-β1b/JAK/STAT1/OAS3 innate immune response pathway for the early stage of EV71 infection. It provides new data for further understanding the relationship between OAS/RNase L system and enterovirus infection.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author contributions
Baisong Zheng: conceptualization, methodology, software, writing-original draft, writing-reviewing and editing. Xiaolei Zhou: investigation, data curation, methodology. Li Tian: visualization, investigation. Jian Wang: investigation. Wenyan Zhang: conceptualization, supervision, validation, funding acquisition, resources.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by funding from the National Key R&D Program of China (2021YFC2301900 and 2301904), National Natural Science Foundation of China (81930062), Science and Technology Department of Jilin Province (YDZJ202201ZYTS590), Health Commission of Jilin Province (2020J059), the Key Laboratory of Molecular Virology, Jilin Province (20102209).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.07.013.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Birdwell L.D., Zalinger Z.B., Li Y., Wright P.W., Elliott R., Rose K.M., Silverman R.H., Weiss S.R. Activation of rnase l by murine coronavirus in myeloid cells is dependent on basal oas gene expression and independent of virus-induced interferon. J. Virol. 2016;90:3160–3172. doi: 10.1128/JVI.03036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebath J., Benech P., Revel M., Vigneron M. Constitutive expression of (2'-5') oligo a synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. doi: 10.1038/330587a0. [DOI] [PubMed] [Google Scholar]
- Choi U.Y., Kang J.S., Hwang Y.S., Kim Y.J. Oligoadenylate synthase-like (oasl) proteins: dual functions and associations with diseases. Exp. Mol. Med. 2015;47:e144. doi: 10.1038/emm.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia E.M., Romeo G., Nissim A., Marziali G., Albertini R., Affabris E., Battistini A., Fiorucci G., Orsatti R., Rossi G.B., et al. A full-length murine 2-5a synthetase cdna transfected in nih-3t3 cells impairs emcv but not vsv replication. Virology. 1990;179:228–233. doi: 10.1016/0042-6822(90)90292-y. [DOI] [PubMed] [Google Scholar]
- Floyd-Smith G. (2'-5')an-dependent endoribonuclease: enzyme levels are regulated by ifn beta, ifn gamma, and cell culture conditions. J. Cell. Biochem. 1988;38:13–21. doi: 10.1002/jcb.240380103. [DOI] [PubMed] [Google Scholar]
- Floyd-Smith G., Denton J.S. A (2'-5')an-dependent endonuclease: tissue distribution in balb/c mice and the effects of ifn-beta treatment and anti-ifn-alpha/beta immunoglobulin on the levels of the enzyme. J. Interferon Res. 1988;8:517–525. doi: 10.1089/jir.1988.8.517. [DOI] [PubMed] [Google Scholar]
- Ghosh A., Sarkar S.N., Sen G.C. Cell growth regulatory and antiviral effects of the p69 isozyme of 2-5 (a) synthetase. Virology. 2000;266:319–328. doi: 10.1006/viro.1999.0085. [DOI] [PubMed] [Google Scholar]
- Gough D.J., Corlett A., Schlessinger K., Wegrzyn J., Larner A.C., Levy D.E. Mitochondrial stat3 supports ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H.H., Ivashkiv L.B. Role of stat3 in type i interferon responses. Negative regulation of stat1-dependent inflammatory gene activation. J. Biol. Chem. 2006;281:14111–14118. doi: 10.1074/jbc.M511797200. [DOI] [PubMed] [Google Scholar]
- Huang H.I., Lin J.Y., Chen S.H. Ev71 infection induces ifnβ expression in neural cells. Viruses. 2019;11:1121. doi: 10.3390/v11121121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibsen M.S., Gad H.H., Thavachelvam K., Boesen T., Desprès P., Hartmann R. The 2'-5'-oligoadenylate synthetase 3 enzyme potently synthesizes the 2'-5'-oligoadenylates required for rnase l activation. J. Virol. 2014;88:14222–14231. doi: 10.1128/JVI.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type i interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen H., Czarniecki C.W., Krause D., Friedman R.M., Silverman R.H. Interferon-induced synthesis of 2-5a-dependent rnase in mouse jls-v9r cells. Virology. 1983;125:496–501. doi: 10.1016/0042-6822(83)90222-2. [DOI] [PubMed] [Google Scholar]
- Kandolf R., Canu A., Hofschneider P.H. Coxsackie b3 virus can replicate in cultured human foetal heart cells and is inhibited by interferon. J. Mol. Cell. Cardiol. 1985;17:167–181. doi: 10.1016/s0022-2828(85)80019-5. [DOI] [PubMed] [Google Scholar]
- Koestner W., Spanier J., Klause T., Tegtmeyer P.K., Becker J., Herder V., Borst K., Todt D., Lienenklaus S., Gerhauser I., Detje C.N., Geffers R., Langereis M.A., Vondran F.W.R., Yuan Q., van Kuppeveld F.J.M., Ott M., Staeheli P., Steinmann E., Baumgärtner W., Wacker F., Kalinke U. Interferon-beta expression and type i interferon receptor signaling of hepatocytes prevent hepatic necrosis and virus dissemination in coxsackievirus b3-infected mice. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen H., Gad H.H., Eskildsen-Larsen S., Despres P., Hartmann R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J. Interferon Cytokine Res. 2011;31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- Kuchipudi S.V. The complex role of stat3 in viral infections. J Immunol Res. 2015;2015 doi: 10.1155/2015/272359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Banerjee S., Wang Y., Goldstein S.A., Dong B., Gaughan C., Silverman R.H., Weiss S.R. Activation of rnase l is dependent on oas3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2241–2246. doi: 10.1073/pnas.1519657113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Yi L., Ke C., Zhang Y., Liu R., Chen J., Kung H.F., He M.L. The interaction between human enteroviruses and type i ifn signaling pathway. Crit. Rev. Microbiol. 2015;41:201–207. doi: 10.3109/1040841X.2013.813903. [DOI] [PubMed] [Google Scholar]
- Mahony R., Gargan S., Roberts K.L., Bourke N., Keating S.E., Bowie A.G., O'Farrelly C., Stevenson N.J. A novel anti-viral role for stat3 in ifn-α signalling responses. Cell. Mol. Life Sci. 2017;74:1755–1764. doi: 10.1007/s00018-016-2435-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marié I., Rebouillat D., Hovanessian A.G. The expression of both domains of the 69/71 kda 2',5' oligoadenylate synthetase generates a catalytically active enzyme and mediates an anti-viral response. Eur. J. Biochem. 1999;262:155–165. doi: 10.1046/j.1432-1327.1999.00361.x. [DOI] [PubMed] [Google Scholar]
- Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R., Zhang A.W., Parcy F., Lenhard B., Sandelin A., Wasserman W.W. Jaspar 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Kloos D.U., Land S., Lewicki-Potapov B., Michael H., Münch R., Reuter I., Rotert S., Saxel H., Scheer M., Thiele S., Wingender E. Transfac: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasti M., Khanbabaei H., Teimoori A. An update on enterovirus 71 infection and interferon type i response. Rev. Med. Virol. 2019;29 doi: 10.1002/rmv.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca Suarez A.A., Van Renne N., Baumert T.F., Lupberger J. Viral manipulation of stat3: evade, exploit, and injure. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rysiecki G., Gewert D.R., Williams B.R. Constitutive expression of a 2',5'-oligoadenylate synthetase cdna results in increased antiviral activity and growth suppression. J. Interferon Res. 1989;9:649–657. doi: 10.1089/jir.1989.9.649. [DOI] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Rice C.M. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol. 2011;1:519–525. doi: 10.1016/j.coviro.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G.R., Darnell J.E., Jr. The jak-stat pathway at twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R., Shereen M.A., Zeng X., Liang Y., Li W., Ruan Z., Li Y., Liu W., Liu Y., Wu K., Luo Z., Wu J. The tlr3/irf1/type iii ifn axis facilitates antiviral responses against enterovirus infections in the intestine. mBio. 2020;11 doi: 10.1128/mBio.02540-20. e02540-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain S.K., Gadnayak A., Mohanty J.N., Sarangi R., Das J. Does enterovirus 71 urge for effective vaccine control strategies? Challenges and current opinion. Rev. Med. Virol. 2022:e2322. doi: 10.1002/rmv.2322. [DOI] [PubMed] [Google Scholar]
- Tan G., Xu F., Song H., Yuan Y., Xiao Q., Ma F., Qin F.X., Cheng G. Identification of trim14 as a type i ifn-stimulated gene controlling hepatitis b virus replication by targeting hbx. Front. Immunol. 2018;9:1872. doi: 10.3389/fimmu.2018.01872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.H., Lee C.K. Stat3 cooperates with phospholipid scramblase 2 to suppress type i interferon response. Front. Immunol. 2018;9:1886. doi: 10.3389/fimmu.2018.01886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J., Fu A.K.Y., Ip F.C.F., Ng H.-K., Hugon J., Page G., Wang J.H., Lai K.-O., Wu Z., Ip N.Y. Tyk2/stat3 signaling mediates β-amyloid-induced neuronal cell death: implications in alzheimer's disease. J. Neurosci. 2010;30:6873–6881. doi: 10.1523/JNEUROSCI.0519-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yuan M., Wang S., Zhang L., Zhang R., Zou X., Wang X., Chen D., Wu Z. Stat3 regulates the type i ifn-mediated antiviral response by interfering with the nuclear entry of stat1. Int. J. Mol. Sci. 2019;20:4870. doi: 10.3390/ijms20194870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.B., Levy D.E., Lee C.K. Stat3 negatively regulates type i ifn-mediated antiviral response. J. Immunol. 2011;187:2578–2585. doi: 10.4049/jimmunol.1004128. [DOI] [PubMed] [Google Scholar]
- Xu N., Yang J., Zheng B., Zhang Y., Cao Y., Huan C., Wang S., Chang J., Zhang W. The pyrimidine analog fnc potently inhibits the replication of multiple enteroviruses. J. Virol. 2020;94:e00204–e00220. doi: 10.1128/JVI.00204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., He Y., Chen Y., Kung H.F., He M.L. Potent inhibition of human enterovirus 71 replication by type i interferon subtypes. Antivir. Ther. 2011;16:51–58. doi: 10.3851/IMP1720. [DOI] [PubMed] [Google Scholar]
- Zhou X., Tian L., Wang J., Zheng B., Zhang W. Ev71 3c protease cleaves host anti-viral factor oas3 and enhances virus replication. Virol. Sin. 2022;37:418–426. doi: 10.1016/j.virs.2022.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.