Abstract
Porcine epidemic diarrhea (PED) caused by porcine epidemic diarrhea virus (PEDV) is one of the most devastating diseases in the global pig industry due to its high mortality rate in piglets. Maternal vaccines can effectively enhance the gut-mammary gland-secretory IgA axis to boost lactogenic immunity and passive protection of nursing piglets against PEDV challenge. From 2017 to 2021, we collected 882 diarrhea samples from 303 farms in China to investigate the epidemiology of PEDV. The result showed that about 52.15% (158/303) of the farms were positive for PEDV with an overall detection rate of 63.95% (564/882) of the samples. The S1 fragments of S gene from 104 strains were sequenced for the phylogenetic analysis. A total of 71 PEDV strains (68.27%) sequenced in this study were clustered into the predominant G2c subgroup, while the newly-defined G2d strains (9.62%) were identified in three provinces of China. The NH-TA2020 strain of G2c subgroup was isolated and cultured, and its infection to piglets caused watery diarrhea within 24 h, indicating its strong pathogenicity. Oral administration of NH-TA2020 strain to pregnant gilts stimulated high levels of IgA antibody in colostrum. The piglets fed by the gilts above were challenged with NH-TA2020 strain or CH–HeB-RY-2020 strain from G2d subgroup, and the clinical symptoms and virus shedding were significantly reduced compared to the mock group. Our findings suggest that G2c subgroup is the predominant branch circulating in China from 2017 to 2021. Oral administration of NH-TA2020 enhances maternal IgA and lactogenic immune responses, which confer protection against the homologous and emerging G2d PEDV strains challenges in neonates.
Keywords: Porcine epidemic diarrhea virus (PEDV), Molecular epidemiological investigation, Virus isolation, Lactogenic immune, IgA
Highlights
-
•
From 2017 to 2021, PEDV positive rate of Chinese farms and samples tested in this study was 52.15% and 63.95%, respectively.
-
•
A total of 71 sequenced PEDV strains (68.27%) were clustered into the predominant G2c subgroup.
-
•
The newly-defined G2d strains (9.62%) were identified in three provinces of China.
-
•
NH-TA2020 strain belonging to the G2c subgroup was isolated and its strong pathogenicity was confirmed.
-
•
The milk containing high levels of IgA antibody induced by NH-TA2020 strain could protect piglets against PEDV challenge.
1. Introduction
Porcine epidemic diarrhea (PED) is an acute and highly contagious intestinal infectious disease caused by porcine epidemic diarrhea virus (PEDV) infection. It results in elevated body temperature, anorexia, vomiting, severe diarrhea, and dehydration. The disease is transmitted through fecal-oral transmission and causes very high mortality in suckling pigs. PEDV S glycoprotein is responsible for the induction of neutralizing antibodies and exhibits major genetic variations among different strains (Chen et al., 2013; Sun et al., 2015). The deletion and insertion mutations of the S gene are potentially associated with the pathogenicity and tissue tropism of PEDV (Lin et al., 2016). The S1 region of the S protein contains sialic acid-binding activity and is undergoing more rapid mutation than the S2 region (Jarvis et al., 2016; Li et al., 2016). Most importantly, the S1 protein is responsible for receptor binding and viral entering cells to initiate infection (Li et al., 2017; Thavorasak et al., 2022). Thus, patterns of S1 region mutations are an important target for a better understanding of the PEDV prevalence.
PED was initially reported in England and Belgium in the 1970s and reported in China as early as the 1980s (Li et al., 2012; Pensaert and de Bouck, 1978). During the period until 2010, PEDV strains from the classical subgroup (G1a) caused a widespread epidemic in East Asia, but only sporadic and regional PED cases were documented in China (Wang et al., 2016a). With the emergence of higher pathogenicity strains of PEDV since October 2010, the subgroup G2a PEDV strains have caused large-scale PED outbreaks with considerable economic losses to the Asian swine industry (Huang et al., 2013; Lara-Romero et al., 2018). Since 2013, there has been an emergence of a highly virulent PEDV that is classified into G2b subgroup in North America, and soon afterward, it rapidly swept across several countries worldwide (Stevenson et al., 2013). Following the outbreaks of G2b strains, novel variants emerged in the United States with deletions and insertions in the S gene, which are classified into S-INDEL subgroup (Wang et al., 2014). Moreover, a more detailed classification called G2c subgroup was defined, which consists of a large number of circulating strains identified since 2010 in China (Tian et al., 2021). It was reported that antigenic variations between classical and emerging PEDV strains may result in the failure of traditional attenuated vaccines (Song et al., 2015). Continued monitoring and precise classification of global PEDV strains, therefore, is important for the development of strategies to prevent and control PED (Lin et al., 2016).
Vaccination is an important method to control PED. Since 1994, the inactivated and live attenuated PEDV CV777 strain vaccines have been developed (Sun et al., 2015). The inactivated and the attenuated bivalent Transmissible Gastroenteritis Virus (TGEV) and PEDV vaccines have been used extensively in the Chinese pig population, which played an important role in the control of PEDV infections. In addition to the traditional vaccines stemming from classical strains, the PEDV inactivated vaccine and attenuated vaccine based on AJ1102 strain, the first G2b strain used in PEDV vaccines, were successfully marketed in December 2017 and widely used, resulting in more effective control of the disease (Bi et al., 2012; Wang et al., 2016a). However, epidemiological investigation reports in recent years demonstrated that PED remains one of the main diseases afflicting the swine industry, which puts forward urgent requirements for developing more effective control measures (Gao et al., 2021; Su et al., 2020).
In this study, we conducted a long-term investigation of PEDV in China from 2017 to 2021. We found a series of PEDV strains that significantly differed from traditional strains appeared and were prevalent in many regions of China, including the predominant G2c strains and the recombinant G2d strains. A newly emerged PEDV variant named NH-TA2020 of G2c subgroup was isolated and its high pathogenicity to piglets was confirmed. We demonstrated that oral challenge by NH-TA2020 strain induced high level of PEDV IgA antibody in the colostrum of pregnant gilts. Moreover, piglets can gain passive protection against the homologous and heterologous G2d PEDV strain challenges from the inoculated gilts, suggesting that PEDV NH-TA2020 strain may be an effective vaccine candidate to prevent and control the newly emerged strains of the devastating disease.
2. Materials and methods
2.1. Sample collection, cDNA synthesis and PEDV RNA detection
From 2017 to 2021, clinical samples including diarrhea feces and small intestines of dead piglets were collected from 303 pig farms experiencing severe diarrhea throughout China. Samples of feces or homogenized intestines were diluted with 2 vol of phosphate-buffered saline (PBS), and then clarified by centrifugation for 10 min at 1800×g. The total RNA of each sample (200 μL) was extracted by Virus DNA/RNA Extraction Kit II (Geneaid, Taiwan) and reverse transcribed into cDNA by EasyScript First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing) as described in the handbooks without modification. To detect PEDV nucleic acid, the cDNA was used as the template in the SYBR green quantitative real-time PCR (qPCR) assay (Applied Biosystems, Foster City, CA, USA), with the PEDV-N-F and PEDV-N-R primers shown in Supplementary Table S1.
2.2. Gene amplification, sequencing and analysis
The positive samples whose CT values are less than 30 were selected for S1 region sequencing. To obtain the S1 region sequences, PCR was conducted with the specific primer pair of PEDV-S1-F and PEDV-S1-R listed in Supplementary Table S1. To amplify the S gene of the NH-TA2020 strain, four pairs of primers were used for PCR, including PEDV-S-1-F&PEDV-S-1-R, PEDV-S-2-F&PEDV-S-2-R, PEDV-S-3-F&PEDV-S-3-R, PEDV-S-4-F&PEDV-S-4-R (Supplementary Table S1). PCR products were analyzed by electrophoresis in 1.0% agarose gels. The fragments were cloned into pEASY-blunt vectors (TransGen Biotech, Beijing), and the positive clones were sequenced at Sangon Biotech (Qingdao, China). The phylogenetic tree was constructed using the neighbor-joining method implemented in MEGA v.7.1.0 (44) software with the default settings (1000 bootstrap replicates) based on the PEDV S gene sequences sequenced in this study and reference sequences collected in GenBank. Initial tree was drawn to scale, with branch lengths measured in the number of substitutions per site. The resulting tree was visualized using iTOL v.6 (Interactive Tree of Life, http://itol.embl.de/).
2.3. Recombination analysis
To identify potential recombination between the PEDV strains, seven recombination detection algorithms (RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, and 3Seq) were implemented in the Recombination Detection Program version.4.101 (RDP v.4.101) with default settings and a threshold P-value of 0.01 were utilized. The window size of RDP, Bootscan, SiScan, MaxChi and Chimaera was 30, 200, 200, 214 and 60, respectively. The step size of Bootscan and SiScan was 20 and 20, respectively. Putative recombination events were only considered credible when supported by at least five of the aforementioned methods.
2.4. Virus isolation and propagation
All animal experiments were approved by the Technology Ethics Committee of the Dezhou University. All animals were humanely euthanized by penetrating captive bolt method and confirmed insensible, according to the American Veterinary Medical Association Euthanasia Guidelines (Underwood and Anthony, 2020). The PEDV-positive intestinal sample for virus isolation was detected by PCR for the possible causative agents including TGEV, PDCoV, PoRV, and PEDV, using the specific primers (TGEV-F&TGEV-R, PDCoV-F&PDCoV-R, PoRV-F&PoRV-R, and PEDV-F&PEDV-R, respectively) listed in Supplementary Table S1. The pure PEDV positive sample was diluted by serum-free Dulbecco's Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA), then centrifuged at 1800×g for 10 min at 4 °C. The supernatants were collected and filtered through the syringe filter with a 0.22 μm pore size. Then 200 μL supernatant was mixed with 200 μL DMEM including 5 μg/mL trypsin (Sigma-Aldrich, St. Louis, MO, USA) and inoculated onto the monolayers of Vero cells (American Type Culture Collection, ATCC), which were rinsed twice with PBS in advance. After incubation at 37 °C for 1 h with constant shaking, 1 mL maintenance media with 10 μg/mL trypsin were added and the plates were placed at 37 °C in an incubator with 5% CO2 for 3–5 days. Then the plates were treated with a freeze-thaw cycle twice to obtain the first passage (F1) viruses. After centrifuging at 1800×g for 1 min at 4 °C, the supernatant was inoculated onto the monolayers of Vero cells grown in T25 cell culture flask for further propagation until the cells developed a visible cytopathic effect (CPE).
2.5. Immunofluorescence assay (IFA)
Vero cells were infected with PEDV NH-TA2020 strain (GenBank NO. ON155919) or HN1303 strain (GenBank NO. KR080551.1) for 24 h and the cell cultures on coverslips were fixed in 80% acetone at 4 °C for 30 min. The cells were then stained with a PEDV monoclonal antibody (Pulike Biological Engineering Inc., Henan) at 37 °C for 1 h. After incubation, cells were washed with PBS three times and stained with FITC-conjugated rabbit anti-mouse IgG (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 1 h. The cells were examined under a fluorescence microscope after washing three times with PBS.
2.6. Pathogenicity analysis
Ten 17-day-old piglets with no previous herd history of PED outbreak or PEDV vaccination were purchased from Xiajin New Hope Liuhe Agriculture and Animal Husbandry Co., Ltd. (Dezhou, Shandong, China). These piglets were confirmed as negative for PEDV, TGEV, PRoV, and PDCoV by PCR (data not shown). The piglets were assigned randomly into two groups. Piglets in the experimental group were infected with NH-TA2020 viruses (1.0 × 107 TCID50 orally for each) and piglets in the mock group were treated with the same volume of DMEM. During the experiment, the clinical signs including diarrhea and vomiting were recorded daily. The rectal swabs were collected for virus shedding detection by qPCR. Intestinal segments including duodenum, jejunum, ileum, and colon were collected from one pig of each group when the piglets had obvious clinical symptoms for immunohistochemical (IHC) examinations and Hematoxylin and Eosin (HE) analysis.
2.7. Detection of the PEDV IgA levels
The colostrum samples were collected within 12 h after the parturition. The PEDV IgA levels were detected by the IDEXX PEDV IgA Test Kit (06-55550-01) according to the instructions. Measure and record absorbance values at 650 nm using a Microplate Reader (Multiskan FC, Thermo Fisher Scientific, Waltham, MA, USA). S/P value = (Sample-NC)/(PC-NC). NC: negative control; PC: positive control.
2.8. Passive protection assessment
A total of 12 commercial crossbred gilts with the same parity and expected farrowing date were purchased from Xiajin New Hope Liuhe Agriculture and Animal Husbandry Co., Ltd. (Dezhou, Shandong, China). Six gilts were infected with 2 × 107.0 TCID50 NH-TA2020 viruses orally 60 days before delivery. Another six gilts were fed with the same volume of DMEM. One week after the challenge, the piggery environment was disinfected thoroughly with Virkon S (1:200) and 1% Disinfectant Foaming Matrix, which makes the disinfectant visible. The feces samples of gilts were collected by cotton swabs and ground samples were collected by gauze. Disinfection was performed every day and PEDV RNA was detected every week by qPCR until no PEDV nucleic acid was detected three consecutive times (Supplementary Table S2). The pregnant gilts were divided randomly into four groups and each group included three NH-TA2020 viruses challenged gilts or DMEM-treated gilts. Anti-PEDV IgA in colostrum samples of each pig was detected using ELISA. On the third day after birth, piglets were infected with 106.5 TCID50 NH-TA2020 viruses orally or CH–HeB-RY-2020 tissues (a G2d strain, 1/10 homogenized small intestines per piglet). All piglets were monitored daily for clinical symptoms including diarrhea, vomiting, and dehydration. Fecal swabs were collected every two days and analyzed by qPCR to detect virus shedding.
3. Results
3.1. Detection of PEDV in China from 2017 to 2021
A total of 882 feces samples and small intestines of dead piglets were collected from 303 farms in 16 provinces of China from 2017 to 2021. According to the geographical distribution, the 16 provinces are divided into five regions, including eastern China (Shandong/Jiangsu), southwestern China (Sichuan), northern China (Hebei/Liaoning/Beijing), southern China (Hubei/Hunan/Anhui/Jiangxi/Guangxi), and midwestern China (Gansu/Shanxi/Shaanxi/Henan/Inner Mongolia). RT-qPCR analysis showed that 52.15% (158/303) of the detected farms were positive for PEDV nucleic acid in the five years. The positive rate at the farm level differed from 46.94% to 55.00% in the five regions. A total of 564 of 882 samples were found to be positive for PEDV. The positive rate at the sample level differed from 53.85% to 68.51%, with an overall positive rate of 63.95% (Table 1).
Table 1.
The prevalence and genotypes of PEDVs in China during 2017–2021.
| Prevalence of PEDV infection |
Genotypes of PEDV strains (n = 104) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Province | Year | Tested sample | Positive sample | Positive rate at sample-level | Tested farm | Positive farm | Positive rate at farm-level | S-INDEL | G2a | G2b | G2c | G2d |
| Eastern China (Shandong/Jiangsu) | 2017 | 64 | 44 | 68.75% | 25 | 14 | 56.00% | 5 | 0 | 0 | 6 | 0 |
| 2018 | 41 | 23 | 56.10% | 15 | 8 | 53.33% | 3 | 0 | 0 | 4 | 0 | |
| 2019 | 23 | 15 | 65.22% | 7 | 3 | 42.86% | 0 | 0 | 0 | 1 | 0 | |
| 2020 | 69 | 51 | 73.91% | 29 | 16 | 55.17% | 1 | 0 | 1 | 11 | 0 | |
| 2021 | 38 | 28 | 73.68% | 15 | 7 | 46.67% | 0 | 0 | 0 | 3 | 0 | |
| Total | 235 | 161 | 68.51% | 91 | 48 | 52.75% | 9 | 0 | 1 | 25 | 0 | |
| 25.71% | 0.00% | 2.86% | 71.43% | 0.00% | ||||||||
| Southwestern China (Sichuan) | 2017 | 15 | 9 | 60.00% | 5 | 3 | 60.00% | 0 | 1 | 0 | 0 | 0 |
| 2018 | 69 | 50 | 72.46% | 18 | 12 | 66.67% | 2 | 0 | 2 | 2 | 2 | |
| 2019 | 53 | 37 | 69.81% | 19 | 10 | 52.63% | 0 | 1 | 3 | 0 | 3 | |
| 2020 | 48 | 28 | 58.33% | 15 | 7 | 46.67% | 0 | 0 | 1 | 2 | 1 | |
| 2021 | 10 | 7 | 70.00% | 3 | 1 | 33.33% | 0 | 0 | 0 | 0 | 1 | |
| Total | 195 | 131 | 67.18% | 60 | 33 | 55.00% | 2 | 2 | 6 | 4 | 7 | |
| 9.52% | 9.52% | 28.57% | 19.05% | 33.33% | ||||||||
| Northern China (Hebei/Liaoning/Beijing) | 2017 | 13 | 8 | 61.54% | 2 | 1 | 50.00% | 0 | 0 | 0 | 1 | 0 |
| 2018 | 35 | 25 | 71.43% | 5 | 2 | 40.00% | 0 | 0 | 0 | 2 | 0 | |
| 2019 | 8 | 5 | 62.50% | 5 | 2 | 40.00% | 0 | 0 | 0 | 0 | 0 | |
| 2020 | 48 | 27 | 56.25% | 16 | 10 | 62.50% | 0 | 0 | 2 | 4 | 2 | |
| 2021 | 67 | 42 | 62.69% | 25 | 13 | 52.00% | 0 | 0 | 0 | 8 | 0 | |
| Total | 171 | 107 | 62.57% | 53 | 28 | 52.83% | 0 | 0 | 2 | 15 | 2 | |
| 0.00% | 0.00% | 10.53% | 78.95% | 10.53% | ||||||||
| Southern China (Hubei/Hunan/Anhui/Jiangxi/Guangxi) | 2017 | 15 | 8 | 53.33% | 5 | 3 | 60.00% | 0 | 0 | 0 | 0 | 0 |
| 2018 | 5 | 3 | 60.00% | 2 | 1 | 50.00% | 0 | 0 | 0 | 0 | 0 | |
| 2019 | 8 | 5 | 62.50% | 5 | 2 | 40.00% | 0 | 0 | 0 | 0 | 0 | |
| 2020 | 37 | 19 | 51.35% | 9 | 4 | 44.44% | 0 | 0 | 0 | 2 | 0 | |
| 2021 | 91 | 49 | 53.85% | 29 | 16 | 55.17% | 0 | 0 | 1 | 13 | 0 | |
| Total | 156 | 84 | 53.85% | 50 | 26 | 52.00% | 0 | 0 | 1 | 15 | 0 | |
| 0.00% | 0.00% | 6.25% | 93.75% | 0.00% | ||||||||
| Midwestern China (Gansu/Shaanxi/Shanxi/Henan/Inner Mongolia) | 2017 | 5 | 3 | 60.00% | 2 | 1 | 50.00% | 0 | 0 | 0 | 0 | 0 |
| 2018 | 7 | 5 | 71.43% | 5 | 2 | 40.00% | 0 | 0 | 0 | 0 | 0 | |
| 2019 | 6 | 3 | 50.00% | 3 | 1 | 33.33% | 0 | 0 | 0 | 0 | 0 | |
| 2020 | 38 | 25 | 65.79% | 12 | 6 | 50.00% | 0 | 0 | 0 | 3 | 0 | |
| 2021 | 69 | 45 | 65.22% | 27 | 13 | 48.15% | 0 | 0 | 0 | 9 | 1 | |
| Total | 125 | 81 | 64.80% | 49 | 23 | 46.94% | 0 | 0 | 0 | 12 | 1 | |
| 0.00% | 0.00% | 0.00% | 92.31% | 7.69% | ||||||||
| Total | 2017 | 112 | 72 | 64.29% | 39 | 22 | 56.41% | 5 | 1 | 0 | 7 | 0 |
| 2018 | 157 | 106 | 67.52% | 45 | 25 | 55.56% | 5 | 0 | 2 | 8 | 2 | |
| 2019 | 98 | 65 | 66.33% | 39 | 18 | 46.15% | 0 | 1 | 3 | 1 | 3 | |
| 2020 | 240 | 150 | 62.50% | 81 | 43 | 53.09% | 1 | 0 | 4 | 22 | 3 | |
| 2021 | 275 | 171 | 62.18% | 99 | 50 | 50.51% | 0 | 0 | 1 | 33 | 2 | |
| Total | 882 | 564 | 63.95% | 303 | 158 | 52.15% | 11 | 2 | 10 | 71 | 10 | |
| 10.58% | 1.92% | 9.62% | 68.27% | 9.62% | ||||||||
3.2. Phylogenetic analysis based on S1 region
To reveal the characteristics of these PEDV strains, samples of the 158 positive farms were selected for S1 sequencing, 104 of which succeeded (Supplementary Material 1). The sequences were deposited in GenBank with accession No. ON168750–ON168853, respectively (Supplementary Table S3). The phylogenetic tree was constructed using S genes from 34 reference PEDV strains available in GenBank, and the 104 S1 sequences obtained in this study as described previously (Huang et al., 2013). As shown in Fig. 1A, all PEDV strains fell clearly into two genetic subgroups, designated as traditional G1 and variant G2. No classic G1a or G1b strain was detected in this study, while a total of 11 S-INDEL (also known as G1c) strains were detected, nine of which were from Shandong Province and the other two were from Sichuan Province. The only two classic G2a strains were found in Sichuan Province. AJ1102, a widely used attenuated vaccine strain in China, was clustered into G2b subgroup, together with 10 strains from four provinces. It is reported that a mass of circulating strains identified since 2010 in China was divided into a distinct subgroup named G2c (Tian et al., 2021; Wen et al., 2021). Obviously, 71 PEDV strains (68.27%) sequenced in this study were grouped into the G2c subgroup, together with the representative strains (CH-SCMY-1-2014, CH/SCZY103/2017, CH-HNLH-2015, CH-JXJA-2017) of this group. Interestingly, we found a new subgroup of G2 group, here we named G2d, which is quite different from G2a, G2b and G2c subgroups. The 10 G2d strains (9.62%) were identified in three provinces, including Sichuan (7 strains), Hebei (2 strains), and Henan (1 strain).
Fig. 1.
Detection of PEDV in China from 2017 to 2021. A Phylogenetic trees of sequenced strains in this study and the representative strains of PEDV were constructed based on the S1 gene sequences. The tree was performed by the neighbor-joining method using MEGA v7.0.26 software with 1000 bootstrap replicates. The name, GenBank accession number, year, and region of each sequence are shown in order. B The regional distribution of PEDV subgroups identified in this study at the farm-level. C The yearly distribution of PEDV subgroups identified in this study at the farm-level.
The yearly detected quantities of each genetic subgroup in the five regions were listed in Table 1, and the proportion of each genetic subgroup was shown in Fig. 1B. G2c was the dominant subgroup of the eastern China, northern China, southern China and mid-western China with the proportion of 68.27%–93.75%. However, G2d subgroup (33.33%) and G2b subgroup (28.57%) had a greater share of prevalence than G2c subgroup (19.05%) in southwestern China. The yearly proportion of each genetic subgroup in all six regions was shown in Fig. 1C. G2c was the predominant subgroup throughout the five years besides 2019, when only eight farms were sequenced successfully.
3.3. Recombination analysis of G2c and G2d representative strains
The S1 fragments of S gene sequenced in this study were compared with the representative strains of each subgroup to further investigate the genetic characteristics of G2c and G2d strains (Fig. 2A). As a result, the G2c strains shared higher nucleotide homology with G2 strains and S-INDEL strains, rather than G1 strains. The similarity comparisons between NH-TA2020 strain (G2c) and other subgroup strains indicated that the N-terminal sequence of NH-TA2020 strain was more like G2 strains, while the middle and C-terminal sequence was more like OH851 strain (S-INDEL) (Fig. 2B). Furthermore, the position 974–4122 of the S gene of NH-TA2020 strain (G2c) was predicted as a recombinant between AJ1102 (major parent, G2b) and OH851 (minor parent, S-INDEL) (Fig. 2D), which was supported by seven detection methods.
Fig. 2.
Recombination analysis of G2c and G2d representative strains. A PEDV S1 gene homology analysis of the sequenced strains and the representative strains. Homology of sequences was analyzed by the Clustal W methods using DNASTAR Lasergene MegAlign V.7.1.0 (44). The ranges of homology between the strains were shown. B The homology analysis of the S gene of NH-TA2020 strain and the representative strains using SimPlot v3.5.1 with the default settings. The data was visualized using Microsoft Excel. C The homology analysis of S gene of CH-SCMY-2018 strain and the representative strains. D Detection of possible recombination events in the NH-TA2020, AJ1102 and OH851 strains. The recombination software (RDP v4.101) was used to recognize recombination events. Seven recombination detection algorithms (RDP, GENECONV, BootScan, MaxChi, Chimaera, SiScan, and 3Seq) were implemented with default settings and a threshold P-value of 0.01 was utilized. The window size of RDP, Bootscan, SisScan, MaxChi and Chimaera was 30, 200, 200, 214 and 60, respectively. The step size of Bootscan and SisScan was 20 and 20, respectively. E Detection of possible recombination events in the CH-SCMY-2018, AH2012 and OH851 strains.
Similarly, the nucleotide sequence of G2d strains had a higher identity to the G2 strains and S-INDEL strain (Fig. 2A) than G1 strains. It is interesting to note that the position around 700–1200 of the S gene of CH-SCMY-2018 strain (G2d) shared higher nucleotide homology with AH2012 strain (G2a) than OH851 strain (S-INDEL), while the rest sequences were more like OH851 strain than AH2012 strain (Fig. 2C). What's more, seven detection methods by RDP4 software indicated the position 684–1226 of the S gene of the CH-SCMY-2018 was likely produced by intragroup recombination between OH851 (major parent) and AH2012 (minor parent) with a high degree of reliability (Fig. 2E).
3.4. Isolation and culture of PEDV NH-TA2020 strain
To further study the characteristics of the predominant strains in China, we next attempted to isolate the emerging strains grouped in the G2c branch. The small intestines sample named NH-TA2020 was taken from a piglet that exhibited severe watery diarrhea, vomiting, and dehydration in Tai'an City, Shandong Province. The sample was confirmed as positive for PEDV and negative for TGEV, PDCoV, and PoRV (Supplementary Fig. S1A). The complete sequence of PEDV S gene was divided into four fragments and amplified by four pairs of primers (Supplementary Fig. S1B). The PCR products were cloned and sequenced, respectively. The results indicate that there were 4161 nucleotides in the S gene of NH-TA2020 sample (GenBank No. ON168803).
The supernatant of intestinal contents of NH-TA2020 sample, as well as the positive control named PEDV HN1303 strain, were inoculated onto the monolayers of Vero cells, respectively. Both the NH-TA2020 sample and HN1303 strain caused obvious CPE at 24 h post-inoculation (Fig. 3). Immunofluorescence (IFA) assay using a PEDV monoclonal antibody indicated that PEDV was successfully propagated in Vero cells. The newly isolated virus was named as NH-TA2020 strain and the whole genome sequence (GenBank accession NO. ON155919, Supplementary materials 1) was obtained by Sanger sequencing. The NH-TA2020 strain was steadily propagated and the titer of third generation virus was determined as 106.5 TCID50/mL (data not shown).
Fig. 3.
Cytopathic effect observation and IFA assay of NH-TA2020 strain. The monolayers of Vero cells were inoculated with 0.1 MOI PEDV HN1301 or NH-TA2020 strain for 24 h. Then the cytopathic effect was observed using a microscope and the cells were harvested for IFA assay using a PEDV monoclonal antibody.
3.5. The pathogenicity of NH-TA2020 in piglets
To test the pathogenicity of NH-TA2020, five 17-day-old piglets were orally challenged with the viruses (1.0 × 107 TCID50). Three of five of the piglets showed vomiting and watery diarrhea in 24 h after infection, while all the five piglets showed typical symptoms in 48 h and lasted for 3–5 days (Table 2). The PEDV RNA test results of the challenged piglets were positive, while the control piglets treated with DMEM were negative (data not shown) and showed no clinical symptoms from beginning to end. The small intestines of two piglets from two groups were collected for pathological slice observation. The hematoxylin-eosin (HE) staining analysis showed that there was no obvious pathological lesion in the duodenum and colon. The jejunum shows mucosal epithelial degeneration with vacuolation and necrosis. Mucosal epithelial cells degeneration could be found in the ileum (Fig. 4A). The IHC analysis using a PEDV monoclonal antibody demonstrated that the duodenum and colon of the NH-TA2020 challenged piglets were negative for PEDV, while the jejunum and ileum were positive (Fig. 4B). These results suggested the NH-TA2020 strain is infectious and highly pathogenic to piglets.
Table 2.
Pathogenicity analysis of NH-TA2020 strain.
| Challenged with | Days post challenge |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Number of piglets with clinical symptom | NH-TA2020 | 3/5 | 5/5 | 4/4a | 3/4 | 3/4 | 2/4 | 0/4 |
| DMEM | 0/5 | 0/5 | 0/4a | 0/4 | 0/4 | 0/4 | 0/4 | |
Note: Five 17-day-old piglets were orally challenged with 1.0 × 107 TCID50 NH-TA2020 strain. Piglets in the control group were treated with the same volume of DMEM. Numbers of piglets with clinical symptom were recorded daily including vomiting, mushy and watery diarrhea. a: two piglets were dissected for IHC and HE analysis.
Fig. 4.
NH-TA2020 infection caused typical pathological lesions in small intestines. The 17-day-old piglet was orally infected with 1.0 × 107 TCID50 NH-TA2020 viruses and the piglets in the control group were treated with the same volume of DMEM. Intestinal segments including duodenum, jejunum, ileum, and colon were collected when the first piglet had obvious clinical symptoms. A The pathological slice observation results of Hematoxylin and Eosin analysis. B The pathological slice observation results of immunohistochemical examinations using a PEDV monoclonal antibody.
3.6. The lactogenic immune protection by oral administration of NH-TA2020
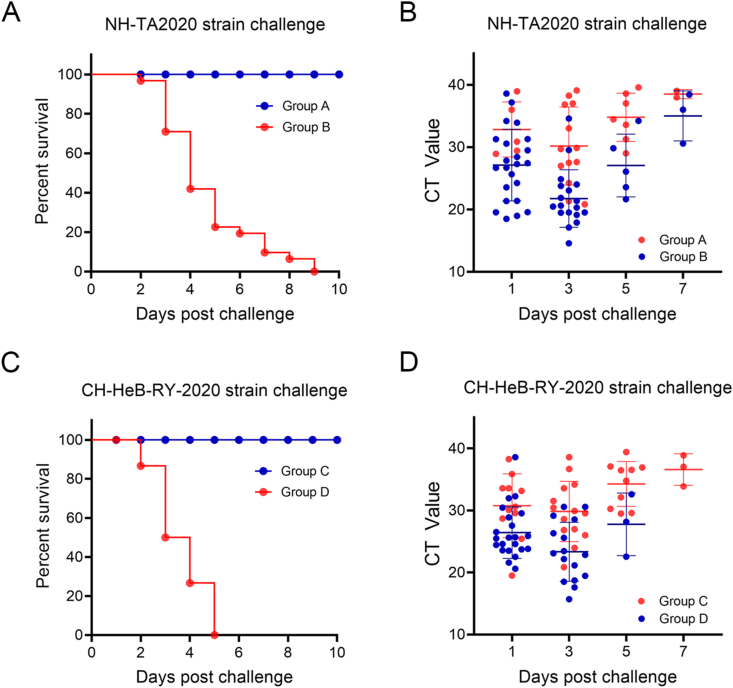
Pregnant gilts were orally infected with NH-TA2020 strain 60 days before delivery. The clinical symptoms including depression, diarrhea and anorexia were found in all six gilts after immunization (Supplementary Fig. S2). The piglets were challenged with NH-TA2020 strain three days after the birth. Compared with the piglets of control gilts (group B), the piglets of the immunized gilts (group A) had less diarrhea proportion and symptoms after the challenge (Table 3). Diarrhea in the group A piglets stopped within 7 days without dehydration or death. Meanwhile, all the piglets of group B had diarrhea within 2 days after the challenge, which dehydrated and died within 8 days (Fig. 5A). The fecal swabs of the piglets were collected to detect the PEDV nucleic acid contents. As is shown in Fig. 5B, with the development of the disease, the virus shedding of both group A and group B first increased and then decreased. The mean virus shedding of the group A piglets is less than the group B piglets in the whole process.
Table 3.
The efficiency of lactogenic immune protection by oral administration of NH-TA2020.
| Group | Pregnant gilts challenged with | Mean S/P value of IgA | Piglets challenged with | Days post challenge |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||||
| A | NH-TA2020 | 8.849 | NH-TA2020 | 6/36 | 14/36 | 17/36 | 16/36 | 9/36 | 8/36 | 3/36 | 0/36 | 0/36 | 0/36 |
| B | DMEM | 0.134 | NH-TA2020 | 25/29 | 28/28 | 20/20 | 11/11 | 5/5 | 4/4 | 3/3 | 1/1 | – | – |
| C | NH-TA2020 | 7.872 | CH–HeB-RY-2020 | 12/35 | 17/35 | 18/35 | 15/35 | 13/35 | 6/35 | 3/35 | 2/35 | 0/35 | 0/35 |
| D | DMEM | 0.238 | CH–HeB-RY-2020 | 27/33 | 29/29 | 18/18 | 11/11 | 3/3 | – | – | – | – | – |
Note: Six pregnant gilts were infected with 2 × 107.0 TCID50 NH-TA2020 viruses orally 60 days before delivery. Another pregnant six gilts were fed with the same dose of DMEM. All the gilts were divided randomly into four groups and each group included three NH-TA2020 viruses challenged gilts (group A and C) or DMEM challenged gilts (group B and D). Anti-PEDV IgA in colostrum samples of each pig was detected using ELISA and the mean S/P values of each group were shown. On the third day after birth, piglets were infected with 106.5 TCID50 NH-TA2020 viruses (group A and B) or CH–HeB-RY-2020 strain tissues (1/10 homogenized intestines per piglet) (group C and D) orally. Numbers of piglets with clinical symptoms were recorded daily including vomiting, diarrhea, and dehydration.
Fig. 5.
The efficiency of lactogenic immune protection by oral administration of NH-TA2020. Piglets' survival curves of group A and group B (A), as well as group C and group D (C) were shown respectively, corresponding to the grouping of Table 3. Fecal swabs of all piglets for passive protection analysis corresponding to Table 3 were collected every two days and analyzed by qPCR to detect PEDV shedding. The PEDV shedding of piglets challenged with NH-TA2020 strain (B) or CH–HeB-RY-2020 strain (D) was shown (mean with SD), respectively.
Meanwhile, each of the other piglets of the immunized and control gilts was fed with one-tenth of the homogenized small intestine from CH–HeB-RY-2020 positive piglets, a G2d strain whose S1 region homology with NH-TA2020 strain was 94.0%. As shown in Table 3, about one-third (12/35) of the piglets from the immunized gilts (group C) had diarrhea on the first day and about half of the piglets (18/35) had diarrhea on the third day. Then the number of piglets with diarrhea gradually decreased. On the 9th day after the challenge, all diarrhea stopped without dehydration or death. In the meantime, four piglets in group D died while all the alive piglets had diarrhea on the second day after the challenge (Fig. 5C). Another 11 piglets in the group D died and all the alive piglets were dehydrated on the third day. All the piglets died within 5 days after the challenge. What's more, the mean virus shedding of the piglets from group C was less than the piglets from group D (Fig. 5D). These results demonstrated that oral administration of NH-TA2020 by gilts can partly protect piglets against PEDV NH-TA2020 strain and CH–HeB-RY-2020 strain challenges by breastfeeding.
4. Discussion
Since the variant strains emerged in late 2010, PEDV has become the most important intestinal pathogen of swine in China, and the infections have been reported in most provinces (Wang et al., 2016b). In this study, a high prevalence of infection with PEDV, 63.95% (564/882) and 52.15% (158/303), was detected in diarrhea samples and pig farms, respectively. The circulating strains were genetically classified into at least five subgroups by the S1 fragment sequences analysis, including S-INDEL, G2a, G2b, G2c, and G2d. Only two classic G2a strain was detected, suggesting that the classic G2a, as well as G1a and G1b strains, were rarely circulating recently in China. Although there was an outbreak in 2011 in China (Li et al., 2012), it was reported that S-INDEL strains exhibited a low detection rate compared with the G2 variants (Su et al., 2020). Consistently, we also identified S-INDEL strains circulating in only 2 of 16 provinces with relatively low prevalence (10.58%) from 2017 to 2021 (Table 1). The predominant strains identified in this study were classified into G2c (68.27%), a subgroup that clustered into the classic G2a group in the previous studies. However, the representative strains, such as LYG, CH/HNLH/2015 and CH-JXJA-2017, were listed in an independent branch compared with other classic G2a strains in these studies (Wang et al., 2016a; Zhang et al., 2019). Therefore, a more detailed classification named G2c subgroup consisting of a large number of circulating strains identified since 2010 was performed (Tian et al., 2021).
Previous studies have reported on the occurrence of recombination in different PEDV strains (Boniotti et al., 2016; Chen et al., 2017; Li et al., 2018). It was reported that CH-JXJA-2017, which was clustered into G2c subgroup in this study, shared the highest nucleotide identity (99.2%) with CH/ZMDZY/11, a recombinant S-INDEL-like strain from China (Li et al., 2018; Wang et al., 2016a). In this study, the S-gene scale similarity comparisons and recombinant analyses confirmed the chimeric nature of the G2c subgroup: positions 1 to 974 from G2b group, and positions 975 to the end from S-INDEL group. Collectively, the phylogenies suggest that the G2c subgroup evolved from a recombination event, with the 5′ part of the S gene acquired from G2b group and the 3′ part genomic regions acquired from S-INDEL. Interestingly, we also identified a new subgroup here called G2d, which was distinctly different from the other subgroups (Fig. 1A). The recombinant analyses indicated that G2d strains were possibly generated via recombination between S-INDEL strains (major parent) and G2a strains (minor parent). However, the recombinant pattern of G2d was significantly different with G2c: positions 684–1226 of the S gene of G2d were from G2a strains, while the rest sequences of the S gene of G2d were from S-INDEL. G2d was the predominant strain in Southwestern China (Fig. 1B), and was prevalent in Northern China (10.53%) and Midwestern China (7.69%), which therefore needs close attention to the possible pandemic.
To prevent and control PED, initiating and maintaining herd immunity is commonly performed through feedback or vaccination. On one hand, feedback is generally considered to lack of widely recognized standard protocol of high efficacy. As a consequence, feedback procedures utilized in the field always result in inconsistent immune responses (Niederwerder and Hesse, 2018). What's more, how to get rid of the risks of the other potential pathogens in the feedback materials remains challenging. On the other hand, several attenuated vaccines against PEDV have been developed due to the high prevalence of severe PED outbreaks, which are widely used in Asian countries for the high immunogenicity and long-lasting immune responses (Li et al., 2020; Sato et al., 2011; Song et al., 2007). However, most of the attenuated vaccines were produced from G1a strains and have become less protective against highly pathogenic strains that have appeared since 2013. Two live attenuated vaccines based on the AJ1102 strain and KNU–141113S-DEL5/ORF3 strain of the G2b subgroup were used in China and Korea in recent years (Bi et al., 2012; Wang et al., 2016a; Won et al., 2020). About 10 of 104 strains (9.62%) detected in this study were clustered into G2b subgroup, suggesting that the commercial attenuated vaccine based on G2b strains perhaps is effective to control diarrhea in these farms. Besides, the gilts of the two farms where we collected NH-TA2020 sample and CH–HeB-RY-2020 sample were immunized with commercial inactivated vaccines based on CV777 strain three times per year, followed by a booster injection 3 weeks later, respectively. However, diarrhea occurs almost every year in these farms, suggesting that it is necessary to develop vaccines from emerging strains, such as G2c and G2d strains, based on the latest epidemiological investigation. Though no increased abortions, stillbirths or pre-weaning mortality was observed in this study, the negative impact of NH-TA2020 strain on the performance of pregnant gilts was still unclear because of the limited number of experimental animals. Close attentions in the future research and development of vaccines based on this strain are needed.
Stimulating gastrointestinal immunity in sows and replacement gilts through controlled exposure to infectious tissues increases the beneficial lactogenic immunity through IgA in colostrum or milk consumed by suckling piglets (Goede et al., 2015; Langel et al., 2019; Niederwerder and Hesse, 2018). The neutralizing antibody level in milk was mainly related to specific IgA during lactation (Song et al., 2016). In this study, the NH-TA2020 strain of the G2c subgroup was isolated and cultured as a substitute for the infectious tissues. Compared with the feedback materials, PEDV culture has higher purity and can be accurately quantified. The high immunogenicity in the pregnant gilts was verified by lactic IgA testing. The protective efficacy against the homologous and G2d PEDV strains challenge was confirmed with lower fecal virus shedding, less pathogenic signs and less mortality in the piglets. It is noteworthy that a part of the suckling piglets fed with high IgA antibody levels of milk also exhibited various degrees of diarrhea, though without dehydration and death. This may partly be due to the high virulence and dose of the challenging strains. Practically, exposure to highly virulent strains commonly happens to replacement gilts, and there should be no such high-dose viruses exposed to the piglets under production conditions. What's more, PEDV is an enveloped virus and can be effectively inactivated by a wide range of disinfectants (Holtkamp et al., 2017; Huss et al., 2017). Thus, the risk of the highly virulent strains exposure to the replacement gilts is therefore controllable for the farms, which may be a considerable method to establish the immune in piglets.
5. Conclusions
Altogether, we performed a broad epidemiological investigation of clinical diarrhea samples for PEDV in China from 2017 to 2021 and identified the predominant G2c subgroup and emerging G2d subgroup. Both the G2c and G2d subgroups evolved from the recombination events, but the recombinant patterns were different. To investigate the characteristic of G2c strains, a novel PEDV virus named NH-TA2020 was isolated and cultured. The oral immunization of NH-TA2020 induced a high level of maternal IgA in the colostrum and provided certain protections against the homologous strain and a G2d strain challenge. This study demonstrated that the NH-TA2020 strain had high immunogenicity and was potential as a live vaccine candidate for lactogenic immune protection in nursing piglets.
Data availability
All the data generated during the current study are included in the manuscript. The data of qPCR assay in this study are available from the corresponding author, Dr. Zhichun Yan (Jasonynh@126.com), upon reasonable request.
Ethics statement
All animal experiments were approved by the Technology Ethics Committee of the Dezhou University (DZU/IACUC_2021-3-23-1).
Author contributions
Xiaowen Li: conceptualization, funding acquisition, resources, writing – review & editing. Yang Li: data curation, project administration, visualization, writing – original draft. Jiapei Huang: data curation, methodology, validation. Yali Yao: data curation, methodology. Wenying Zhao: data curation, methodology. Yunjing Zhang: data curation, methodology. Jie Qing: formal analysis. Jing Ren: funding acquisition, software. Zhong Yan: formal analysis. Zewei Wang: investigation. Xiaofang Hu: investigation. Duli Kang: resources. Hongqiang Liu: resources. Zhichun Yan: resources, supervision, writing – review & editing.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported by the National Key R&D Program of China (2021ZD0113803), the "Pioneer" and "Leading Goose" R&D Program of Zhejiang (2022C02031), and the National Natural Science Foundation of China (No. 31701424).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.08.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bi J., Zeng S., Xiao S., Chen H., Fang L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012;86:10910–10911. doi: 10.1128/JVI.01919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniotti M.B., Papetti A., Lavazza A., Alborali G., Sozzi E., Chiapponi C., Faccini S., Bonilauri P., Cordioli P., Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus. Italy. Emerg Infect Dis. 2016;22:83–87. doi: 10.3201/eid2201.150544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu X., Lang H., Wang Z., Shi D., Shi H., Zhang X., Feng L. Genetic variation of nucleocapsid genes of porcine epidemic diarrhea virus field strains in China. Arch. Virol. 2013;158:1397–1401. doi: 10.1007/s00705-013-1608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Li S., Zhou R., Zhu M., He S., Ye M., Huang Y., Li S., Zhu C., Xia P., Zhu J. Two novel porcine epidemic diarrhea virus (PEDV) recombinants from a natural recombinant and distinct subtypes of PEDV variants. Virus Res. 2017;242:90–95. doi: 10.1016/j.virusres.2017.09.013. [DOI] [PubMed] [Google Scholar]
- Gao Q., Zheng Z., Wang H., Yi S., Zhang G., Gong L. The new porcine epidemic diarrhea virus outbreak may mean that existing commercial vaccines are not enough to fully protect against the epidemic strains. Front. Vet. Sci. 2021;8:697839. doi: 10.3389/fvets.2021.697839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goede D., Murtaugh M.P., Nerem J., Yeske P., Rossow K., Morrison R. Previous infection of sows with a "mild" strain of porcine epidemic diarrhea virus confers protection against infection with a "severe" strain. Vet. Microbiol. 2015;176:161–164. doi: 10.1016/j.vetmic.2014.12.019. [DOI] [PubMed] [Google Scholar]
- Holtkamp D.J., Myers J., Thomas P.R., Karriker L.A., Ramirez A., Zhang J., Wang C. Efficacy of an accelerated hydrogen peroxide disinfectant to inactivate porcine epidemic diarrhea virus in swine feces on metal surfaces. Can. J. Vet. Res. 2017;81:100–107. [PMC free article] [PubMed] [Google Scholar]
- Huang Y.W., Dickerman A.W., Pineyro P., Li L., Fang L., Kiehne R., Opriessnig T., Meng X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio. 2013;4:e00737. doi: 10.1128/mBio.00737-13. 00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss A.R., Schumacher L.L., Cochrane R.A., Poulsen E., Bai J., Woodworth J.C., Dritz S.S., Stark C.R., Jones C.K. Elimination of porcine epidemic diarrhea virus in an animal feed manufacturing facility. PLoS One. 2017;12:e0169612. doi: 10.1371/journal.pone.0169612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis M.C., Lam H.C., Zhang Y., Wang L., Hesse R.A., Hause B.M., Vlasova A., Wang Q., Zhang J., Nelson M.I., Murtaugh M.P., Marthaler D. Genomic and evolutionary inferences between American and global strains of porcine epidemic diarrhea virus. Prev. Vet. Med. 2016;123:175–184. doi: 10.1016/j.prevetmed.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langel S.N., Paim F.C., Alhamo M.A., Lager K.M., Vlasova A.N., Saif L.J. Oral vitamin A supplementation of porcine epidemic diarrhea virus infected gilts enhances IgA and lactogenic immune protection of nursing piglets. Vet. Res. 2019;50:101. doi: 10.1186/s13567-019-0719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Romero R., Gomez-Nunez L., Cerriteno-Sanchez J.L., Marquez-Valdelamar L., Mendoza-Elvira S., Ramirez-Mendoza H., Rivera-Benitez J.F. Molecular characterization of the spike gene of the porcine epidemic diarrhea virus in Mexico, 2013-2016. Virus Gene. 2018;54:215–224. doi: 10.1007/s11262-017-1528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li W., Lucio de Esesarte E., Guo H., van den Elzen P., Aarts E., van den Born E., Rottier P.J.M., Bosch B.J. Cell attachment domains of the porcine epidemic diarrhea virus spike protein are Key targets of neutralizing antibodies. J. Virol. 2017;91:e00273. doi: 10.1128/JVI.00273-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Song D., Zhang F., Gong W., Guo N., Li A., Zhou X., Huang D., Ye Y., Tang Y. Complete genome sequence of a recombinant porcine epidemic diarrhea virus strain, CH/JXJA/2017, isolated in jiangxi, China, in 2017. Genome Announc. 2018;6:e01590. doi: 10.1128/genomeA.01590-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Ma Z., Li Y., Gao S., Xiao S. Porcine epidemic diarrhea virus: molecular mechanisms of attenuation and vaccines. Microb. Pathog. 2020;149:104553. doi: 10.1016/j.micpath.2020.104553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Saif L.J., Marthaler D., Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016;226:20–39. doi: 10.1016/j.virusres.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederwerder M.C., Hesse R.A. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound Emerg Dis. 2018;65:660–675. doi: 10.1111/tbed.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Takeyama N., Katsumata A., Tuchiya K., Kodama T., Kusanagi K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Gene. 2011;43:72–78. doi: 10.1007/s11262-011-0617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin Exp Vaccine Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D.S., Oh J.S., Kang B.K., Yang J.S., Moon H.J., Yoo H.S., Jang Y.S., Park B.K. Oral efficacy of Vero cell attenuated porcine epidemic diarrhea virus DR13 strain. Res. Vet. Sci. 2007;82:134–140. doi: 10.1016/j.rvsc.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., Stone S., Drebes D., Greiner L.L., Dvorak C.M.T., Murtaugh M.P. Characterization of anti-porcine epidemic diarrhea virus neutralizing activity in mammary secretions. Virus Res. 2016;226:85–92. doi: 10.1016/j.virusres.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Su M., Li C., Qi S., Yang D., Jiang N., Yin B., Guo D., Kong F., Yuan D., Feng L., Sun D. A molecular epidemiological investigation of PEDV in China: characterization of co-infection and genetic diversity of S1-based genes. Transbound Emerg Dis. 2020;67:1129–1140. doi: 10.1111/tbed.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Ma J., Wang Y., Wang M., Song W., Zhang W., Lu C., Yao H. Genomic and epidemiological characteristics provide new insights into the phylogeographical and spatiotemporal spread of porcine epidemic diarrhea virus in Asia. J. Clin. Microbiol. 2015;53:1484–1492. doi: 10.1128/JCM.02898-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavorasak T., Chulanetra M., Glab-Ampai K., Teeranitayatarn K., Songserm T., Yodsheewan R., Sae-Lim N., Lekcharoensuk P., Sookrung N., Chaicumpa W. Novel neutralizing epitope of PEDV S1 protein identified by IgM monoclonal antibody. Viruses. 2022;14:125. doi: 10.3390/v14010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Yang X., Li H., Ma B., Guan R., Yang J., Chen D., Han X., Zhou L., Song Z., Xie X., Wang H. Molecular characterization of porcine epidemic diarrhea virus associated with outbreaks in southwest China during 2014-2018. Transbound Emerg Dis. 2021;68:3482–3497. doi: 10.1111/tbed.13953. [DOI] [PubMed] [Google Scholar]
- Underwood W., Anthony R. 2020. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition. Retrieved on March 2013, 2020–2021. [Google Scholar]
- Wang D., Fang L., Xiao S. Porcine epidemic diarrhea in China. Virus Res. 2016;226:7–13. doi: 10.1016/j.virusres.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Guo D., Li C., Wei S., Wang Z., Liu Q., Zhang B., Kong F., Feng L., Sun D. Molecular characterization of the ORF3 and S1 genes of porcine epidemic diarrhea virus non S-indel strains in seven regions of China. PLoS One. 2016;11:e0160561. doi: 10.1371/journal.pone.0160561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F., Yang J., Li A., Gong Z., Yang L., Cheng Q., Wang C., Zhao M., Yuan S., Chen Y., El-Ashram S., Li Y., Yu H., Guo J., Huang S. Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019. PLoS One. 2021;16:e0253622. doi: 10.1371/journal.pone.0253622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H., Lim J., Noh Y.H., Yoon I., Yoo H.S. Efficacy of porcine epidemic diarrhea vaccines: a systematic review and meta-analysis. Vaccines (Basel) 2020;8:642. doi: 10.3390/vaccines8040642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Luo S., Gu J., Li Z., Li K., Yuan W., Ye Y., Li H., Ding Z., Song D., Tang Y. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019;15:470. doi: 10.1186/s12917-019-2212-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript. The data of qPCR assay in this study are available from the corresponding author, Dr. Zhichun Yan (Jasonynh@126.com), upon reasonable request.