Abstract
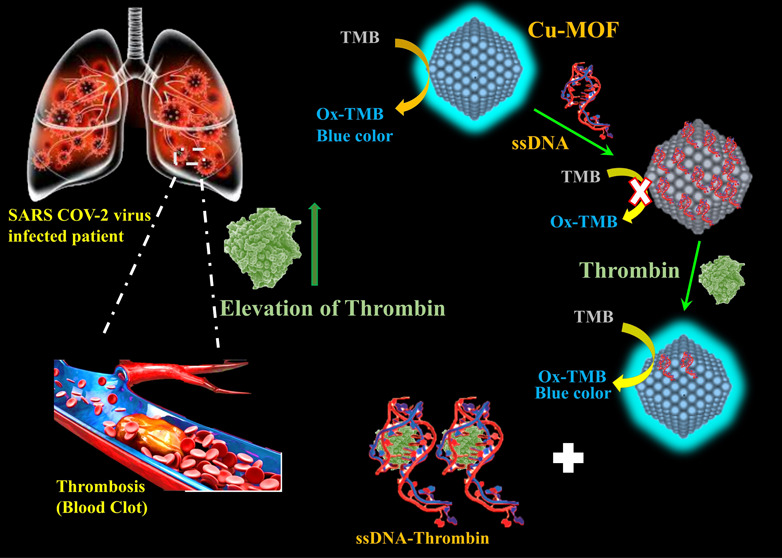
An essential tool in the management and control of the COVID-19 pandemic is the development of a fast, selective, sensitive, and inexpensive COVID-19 biomarkers detection method. Herein, an ultrasensitive and label-free biosensing strategy was described for the colorimetric and fluorimetric detection of thrombin. A dual-mode aptasensing method based on integrating engineered ssDNA with a stimulated fluorescent enzyme-mimetic copper-based metal–organic framework (Cu-MOF) as a molecular recognition element for thrombin was investigated. Cu-MOFs displayed stimulated fluorescence and enzyme-mimetic peroxidase activities that oxidize the chromogenic colorless substance TMB to blue-colored oxTMB. The thrombin-based aptamer (ssDNA) can be immobilized on the Cu-MOF surface to form a functionalized composite, ssDNA/MOF, and quench the stimulated fluorescence emission and the enzymatic activity of the Cu-MOF. Later, addition of thrombin recovers the fluorescence and enzymatic activity of the MOF. Thus, a turn-on colorimetry/fluorimetry aptasensing probe was designed for the detection of thrombin. Based on colorimetric assay, 350 pM was recorded as the lower limit of detection (LOD), while based on the fluorescence mode, 110 fM was recorded as the LOD (when S/N = 3). The label-free aptasensing probe was used successfully for the detection of thrombin in COVID-19 patients with satisfactory recoveries, 95–98%. Since the detection time of our aptasensor is relatively rapid (45 min) and due to the low-cost precursors and easy-to-operate characteristics, we believe that it has great potential to be used in point-of-care testing (POCT).
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an infectious and deadly coronavirus that has killed millions around the world; it was first reported in China in 2019; then, COVID-19 soon became a pandemic illness, and it is now a severe threat to global health.1−4 The most prognostic parameters of severe COVID-19 are lymphopenia, thrombocytopenia, leukocytosis, CRP, PCT, LDH, AST, ALT, and D-dimer.5,6 Excessive inflammation, hypoxia, immobility, and microvascular damage may all be reasons for thrombosis.7−11 In COVID-19 patients, a high thrombin generation capacity that remains within normal values despite heparin therapy and hypofibrinolysis are observed.12,13 The coagulopathy linked to coronavirus disease 2019 (COVID-19) is characterized by a prothrombotic condition.14 Patients with COVID-19 showed high levels of thrombin generation (TG), when they were diagnosed. Intermediate subtherapeutic thromboprophylaxis has reduced TG more successfully.15
In the literature, there are many methods and techniques to detect thrombin, such as fluorescence, colorimetric, and electrochemical. Techniques such as immunoassay-based and radioassay-based methods are in use as a benchmark.16,17 However, the aforementioned methods are time-consuming, high-cost, and affected by various pHs and temperatures.18 For instance, immunoassay uses natural antibodies, which are temperature-dependent and suffer from denaturation and a short shelf-life.16,19,20 Thus, one should think about stable, robust, selective, and sensitive alternatives with low-cost precursors.
Aptamers are oligonucleotides, single-stranded DNA (ssDNA) or RNA, that were created using the systematic evolution of ligands by exponential enrichment (SELEX) in vitro selection and polymerase chain reaction technique.21−25 They have high affinity and specificity for their targets. As a result, aptamers can be utilized as artificial antibodies.26,27 They have unique properties that make them highly promising for biosensing applications, such as high specificity, a wide range of targets, easy synthesis, nonimmunogenicity, stability under various pHs and temperatures, and low cost. Different types of targets can be bound to aptamers, such as organic dyes, bacteria, biomarkers, and proteins.28 This binding is due to changing spatial conformation, stacking aromatic ring interactions, electrostatic forces, and/or hydrogen bonding.29,30
Metal–organic frameworks (MOFs) are crystalline nanomaterials made up of metal ions and organic ligands.31−33 MOFs are widely used in a variety of fields, including separation,34 gas adsorption,35 energy storage,36 and catalysis,37 due to their huge specific areas, simple synthesis procedures, plentiful functional groups, enzyme-mimetic behavior, and chemical stability.38,39 Because of these benefits, MOFs are particularly well-suited for the construction of biosensors with a wide range of applications, particularly in the biomedical domains.40−46 Recently, researchers are interested in integrating nucleic acids with MOFs for biosensing.47−49 Moreover, the enzyme-mimetic activity or fluorescence emission of some MOFs allows for using this entity in colorimetric or fluorescence assay.50−54 To this end, rational selection of MOFs with enzyme-mimetic behavior and fluorescence emission gives the best route toward functionalization with a suitable aptamer.
Aptamer-functionalized MOFs for the detection of thrombin have been reported in the literature. Wang et al.55 have prepared and used Fe-MIL-88A for colorimetric detection of thrombin with 10 nM as a LOD. Due to the peroxidase activity of Fe-MIL-88A, they just used this colorimetric property for the detection strategy. Others such as Zhang et al.56 used an electrochemical method for aptamer-functionalized Zr-based MOFs for the detection of thrombin. They obtained 0.37 pg·mL–1 as a LOD. To this end, we think of a dual-mode sensing strategy combined with aptamers for the detection of thrombin.
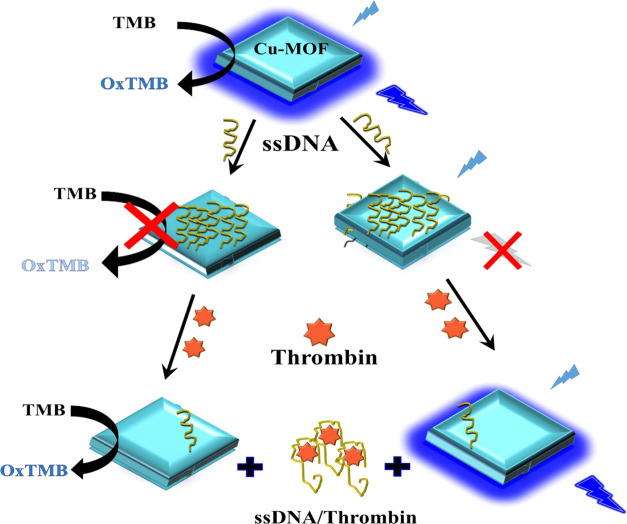
In the present work, Cu-MOFs have been prepared that have both stimulated fluorescence and mimetic peroxidase-like enzymatic activity. An engineered thrombin-binding aptamer (ssDNA) has been integrated with Cu-MOFs to combine the mimetic behavior and fluorescence of the MOFs with high affinity of the ssDNA toward thrombin detection (Scheme 1). Combining an aptamer platform with MOFs will enhance the affinity and sensitivity of thrombin detection. Femtomolar detection of thrombin was achieved using our low-cost, robust, and highly stable aptasensor.
Scheme 1. Preparation and Detection Mechanisms of Cu-MOF Integrated with the ssDNA Biosensor for Thrombin Detection.
2. Experimental Section
2.1. Chemicals
A thrombin amine aptamer (ssDNA) was synthesized and purified by Bioneer Co., Ltd. (Daejeon, south Korea) with sequences (5-(NH2)-(CH2)6-CCA-TCT-CCA-CTT-GGT-TGG-TGT-GGT-TGG-3). Dimethylformamide (DMF), 3, 3′ 5, 5′-tetramethylbenzidine (TMB), terephthalic acid (TA), and thrombin protein extracted from human plasma were all purchased from Merck-Sigma-Aldrich (Baden-Württemberg, Germany) and used as received. Cu(NO3)2·3H2O and hydrogen peroxide (H2O2) 30% were purchased from Biochem chemopharma Co., Ltd. (Biochem, ZA Cosne sur Loire, France). TE buffer (Tris-EDTA, pH 8.0) was purchased from EMB corporation (EMB Co.). Phosphate buffer saline (pH 7) was ordered from (CDH Co., Ltd. Delhi, India).
2.2. Instruments
Field emission scanning electron microscopy (FE-SEM) was used for taking the image of the MOF particles (Germany ZEISS Gemini SEM). UV–vis absorption spectra were recorded on a Cary 60 Spectrophotometer (Agilent technologies). Fluorescence measurements (PL) were recorded on a Cary Eclipsed fluorescence Spectrophotometer (Agilent Technologies).
2.3. MOF Synthesis
A Cu-metal–organic framework (Cu-MOF) was prepared based on reports in the literature with minor modifications.57 In brief, 0.97 g of Cu(NO3)2·3H2O and 0.67 g of TA (terephthalic acid) were first dissolved in 20 mL of DMF. The resulting solution was then transferred to a 50 mL Teflon-lined stainless-steel autoclave for reaction at 150 °C for 3 h. Blue crystals were collected by centrifugation and rinsed with adequate ethanol and deionized water alternately. To prepare a stock solution (100 μg·mL–1) of Cu MOF in water, 10 mg of the Cu-MOF was dispersed in 100 mL of deionized water.
2.4. Colorimetric Assay
For the colorimetric assay protocol, 150 μL of 100 μg·mL–1 Cu-MOF solution was mixed with 100 μL of ssDNA solution (1.6 μM) and then incubated for 5 h at 35 °C. The ssDNA with the presence of different amounts, 0.0–7.0 nM, of thrombin was stirred for 15 min in PBS buffer, followed by addition of 150 μL (100 μg·mL–1) of the Cu-MOF to it with stirring for 5 h. Peroxidase enzymatic activity was measured by taking 100 μL from every solution mixed with 100 μL of 5 mM TMB as a substrate with 100 μL of 30 mM H2O2 solution; then, the volume was calibrated to 1.0 mL using a buffer, pH 4. The color change was observed and recorded after 45 min.
2.5. Fluorimetric Assay
For the fluorescence-based assay, 150 μL of 100 μg·mL–1 Cu-MOF solution was mixed with 100 μL of ssDNA (1.6 μM) and then stirred for 5 h at 35 °C. Recovery response was obtained with addition of different amounts, 0.0–7.0 nM, of thrombin and stirring for 15 min in PBS buffer, with 100 μL of 30 mM H2O2 solution added and then incubated for 8 h in room temperature. The fluorescence spectra were recorded at 320 nm excitation wavelength.
2.6. Stirring Time Effect of Cu-MOF/ssDNA
This study has been done by mixing 15 ng·mL–1 MOF with 160 nM aptamer in PBS; then, 100 μL has been taken from the mixture solution in different periods of time, 0.5, 1, 3, 5, and 24 h; then, 0.5 mM TMB substrate was added and100 μL of 30 mM H2O2 solution was added; the volume was calibrated to 1 mL using a buffer (pH 4); then, the color change was observed and recorded after 45 min.
3. Results and Discussion
3.1. Characterizations
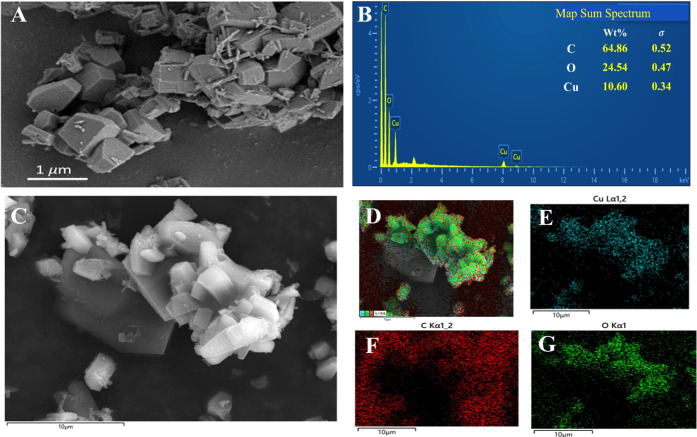
Based on FE-SEM, the Cu-MOF product forms polydisperse decahedral crystals in the micrometer-to-sub-micrometer range (Figure 1A). EDS elemental mapping and spectra were characterized and are shown in Figure 1B–G. As one can notice, the crystals are made of C, O, and Cu elements, confirming the formation of the Cu-MOF.
Figure 1.
(A) FE-SEM of the Cu-MOF; (B) EDX mapping spectra, and (C–G) EDX distribution of the Cu-MOF at the 10 μm scale bar.
3.2. Enzymatic Activity and Stimulated Fluorescence of the Cu-MOF
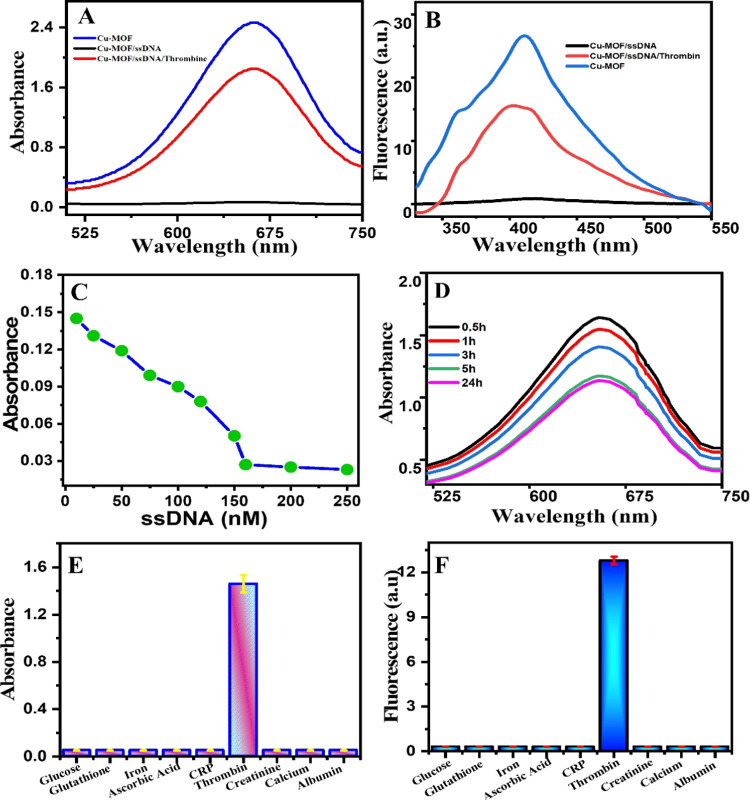
The Cu-MOF showed a peroxidase-mimetic enzyme activity with the appearance of a clear blue color via oxidation of the colorless TMB substrate to blue oxidized TMB in the presence of H2O2. When amine-terminated ssDNA was added, it adsorbed on the surface of Cu-MOFs via the π–π interaction, and the enzymatic activity of the Cu-MOF was inhibited.58 However, when thrombin protein was added, the enzymatic activity of the Cu-MOF was shown again and the blue color recovered due to the selective binding of the ssDNA with the thrombin protein (Figure 2A). The high affinity of NH2-ssDNA binding with thrombin led to a sensitive colorimetric probe for the detection of the thrombin protein. Interestingly, due to the stimulated fluorescence of Cu-MOFs with H2O2 (λemission 410 when λexcitation at 320 nm), the emission of Cu-MOFs was quenched after adding thrombin ssDNA and recovered after addition of the thrombin protein. Thus, we designed a fluorescence-based probe in parallel with colorimetric assay exploiting the fluorescence and enzymatic behaviors of the Cu-MOFs (Figure 2B).
Figure 2.
Aptamer-based assay responses on the Cu-MOF after addition of thrombin. (A) Colorimetry-based assay. (B) Fluorescence-based assay. (C) Optimum ssDNA concentration. (D) Stirring time effect on the adsorption capacity of the ssDNA on the Cu-MOF (15 ng·mL–1), ssDNA (160 nM). (E, F) Selectivity study of the proposed biosensor, Cu-MOF (15 ng·mL–1), ssDNA (160 nM), and thrombin (5 nM), and the concentrations of other interferences are all 500 nM. (E) Colorimetric assay and (F) fluorescence assay response.
3.3. Optimizations
To obtain the best catalytic activity, variable experimental parameters were optimized, such as stirring time and ssDNA concentrations. Figure 2C shows the effect of stirring time of the Cu-MOF with ssDNA on the enzymatic activity. With increasing time of stirring, the enzymatic activity of the Cu-MOF decreases gradually until 5 h, due to adsorption of ssDNA on the MOF surface. Beyond 5 h and until 24 h, there were very slight changes in the enzymatic activity, which indicates that 5 h is the optimum time for stirring. The concentration of ssDNA was also optimized as shown in Figure 2D. At 160 nM, the whole catalytic activity of the Cu-MOF was totally inhibited, suggesting that 160 nM is the best concentration of ssDNA to start with for an off–on experiment.
3.4. Selectivity Study
To evaluate the selectivity of our aptamer-based sensor, common ions, biomolecules, and proteins that usually coexist with thrombin in the serum matrix were tested. Interferences such as glucose, glutathione, ascorbic acid, iron, C-reactive protein (CRP), creatinine, albumin, and calcium were selected for this study. The concentration of all of them in selective experiments was 100 times that of thrombin. As shown in Figure 2E,F, the corresponding absorbance and fluorescence intensity of thrombin were much greater than those of the other proteins and metals. These results indicate that the Cu-MOF/ssDNA sensor has high selectivity for the detection of thrombin, which makes the Cu-MOF/ssDNA sensor a good probe for the real serum fluid sample.
4. Dual-Mode Colorimetric and Fluorometric Assay
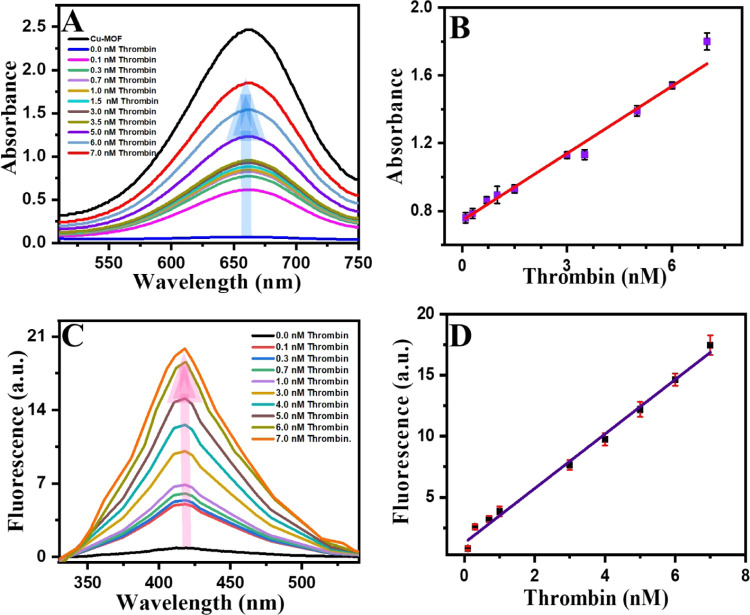
In the colorimetric assay, the intensity of blue color increased as the amount of thrombin increased (Figure 3A) due to removal of the immobilized ssDNA on the surface of the Cu-MOF. Hence, the enzymatic activity will be restored. A linear relationship was constructed from 0.7 to 7.0 nM (Figure 3B) with a lower limit of detection (LOD) as small as 360 pM being calculated. Regarding the fluorescence-based assay, different concentrations of thrombin were added to the Cu-MOF/ssDNA solutions and the fluorescence recovery was observed, as shown in Figure 3C. A dynamic linear relationship was obtained as the thrombin concentration increased (R2 = 0.998), and a LOD as small as 110 fM was calculated (Figure 3D). The LOD and LOQ were determined according to S/N =3 and S/N = 10 criteria, respectively (S was estimated to SD (n = 10) obtained from the current intensity of the lowest concentration of thrombin in the calibration 0.7 nM). Such a small LOD is attributed to the synergistic effect of the luminescence of the Cu-MOF with high affinity of ssDNA toward thrombin. Both LODs are much lower than the normal level of thrombin in normal adults and in patients with COVID-19. Our proposed biochemical sensor showed excellent LODs and a dynamic range in both assays, colorimetric and fluorimetry modes. Table 1 shows a comparison between common MOF-based methods for the detection of thrombin. As one can notice, our results showed very low LODs compared with those of others.
Figure 3.
Thrombin concentration-dependent response profiles; (A, C) Colorimetric and fluorescence spectra, respectively. (B, D) Calibration curves of thrombin using colorimetry and fluorescent aptamer-based assays, respectively. Error bars are for n = 3.
Table 1. MOF-Based Aptasensors Using MOFs as Signal Probes.
In general, fluorimetric assays are more sensitive and selective than colorimetric assays. Dynamic ranges are broader and LODs are lower in fluorescence-based assays. Thus, fluorescence-based methods are more favorable for trace analysis. However, colorimetric-based assays are simple requiring low-cost instruments for the analysis.
5. Applications
Plasma samples of patients with COVID-19 were taken for thrombin analysis from Shahid-Hemn Hospital (Slemani City, Kurdistan Region, Iraq). The plasma sample collection was done following the guidelines of the ethical committee at the hospital, and the patients were informed and before collecting blood, permission was acquired. The samples were collected from the emergency and intensive care units of the hospital. Plasma samples were 1000-fold diluted with double-deionized water (to convert the thrombin range to our biosensor’s linearity range) and then spiked with 1.0 and 3.5 nM standard thrombin solution. Table 2 shows various colorimetric and fluorescence results using our proposed biosensor. Spike recoveries were very satisfactory, ranging from 86 to 94% for colorimetric assay and from 88 to 98% for fluorescence assay. Thus, a quantitative thrombin COVID-19 biomarker in real serum was achieved. In one word, Cu-MOF/ssDNA already demonstrated its utility in clinical diagnosis as a novel biochemical assay. For the intra-assay, the precision (repeatability) reached a RSD of 4% (n = 3) for the same day, and the interassay precession (reproducibility) reached 6% (n = 3) for different days.
Table 2. Various Colorimetric and Fluorescence Recoveries with RSDs, Number of Replications, n = 3.
| sample | unspiked (nM) | added (nM) | spiked (nM) | recovery % | RSD % |
|---|---|---|---|---|---|
| Colorimetric Assay | |||||
| 1 | 1.15 ± 0.02 | 1 | 2.01 ± 0.01 | 86.00 ± 1.52 | 1.71 |
| 1.64 ± 0.021 | 3.5 | 4.94 ± 0.02 | 94.35 ± 1.03 | 1.33 | |
| 2 | 3.03 ± 0.01 | 1 | 3.91 ± 0.03 | 88.00 ± 2.07 | 0.51 |
| 5.35 ± 0.20 | 3.5 | 8.64 ± 0.02 | 94.08 ± 1.95 | 0.34 | |
| Fluorometric Assay | |||||
| 1 | 1.17 + 0.02 | 1 | 2.10 ± 0.01 | 93.00 ± 2.16 | 0.72 |
| 1.40 + 0.02 | 3.5 | 4.50 ± 0.03 | 88.00 ± 1.38 | 0.33 | |
| 2 | 2.95 + 0.02 | 1 | 3.89 ± 0.02 | 94.20 ± 2.46 | 1.53 |
| 3.21 + 0.01 | 3.5 | 6.65 ± 0.07 | 98.34 ± 2.63 | 0.22 | |
6. Conclusions
A dual-mode and label-free aptasensor based on integration of ssDNA with a stimulated fluorescent peroxidase Cu-MOF was developed for the detection of thrombin. The biosensor enabled femtomolar detection of thrombin based on the fluorescence mode and picomolar detection based on the colorimetry mode. The detection limits by both modes, colorimetry and fluorimetry, were much lower than the physiological thrombin level in human serum. The colorimetric and fluorometric assays were successfully evaluated, and satisfactory results were obtained for the detection of thrombin in the serum of COVID-19 patients. This label-free aptasensor will pave the way toward designing more functionality-sensitive fluorescent nanozymes.
Acknowledgments
GKA thanks the Ministry of Health and Directorate of Health in Sulaymaniyah Governorate for the study leave.
The authors declare no competing financial interest.
References
- Krammer F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Wu D.; Wu T.; Liu Q.; Yang Z. The SARS-CoV-2 outbreak: what we know. Int. J. Infect. Dis. 2020, 94, 44–48. 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasöksüz M.; Kiliç S.; Saraç F. Coronaviruses and sars-cov-2. Turk. J. Med. Sci. 2020, 50, 549–556. 10.3906/sag-2004-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.; Chang L.; Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): Current status, challenges, and countermeasures. Rev. Med. Virol. 2020, 30, e2106 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-j.; Ni Z. y.; Hu Y.; et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippi G.; Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin. Chem. Lab. Med. 2020, 58, 1063–1069. 10.1515/cclm-2020-0240. [DOI] [PubMed] [Google Scholar]
- Nougier C.; Benoit R.; Simon M.; et al. Hypofibrinolytic state and high thrombin generation may play a major role in SARS-COV2 associated thrombosis. J. Thromb. Haemostasis 2020, 18, 2215–2219. 10.1111/jth.15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W.; Varley J.; Johnston A.; et al. Thrombotic complications of patients admitted to intensive care with COVID-19 at a teaching hospital in the United Kingdom. Thromb. Res. 2020, 191, 76. 10.1016/j.thromres.2020.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ani F.; Chehade S.; Lazo-Langner A. Thrombosis risk associated with COVID-19 infection. A scoping review. Thromb. Res. 2020, 192, 152–160. 10.1016/j.thromres.2020.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provazníková D.; et al. Seventeen novel SERPINC1 variants causing hereditary antithrombin deficiency in a Czech population. Thromb. Res. 2020, 189, 39–41. 10.1016/j.thromres.2020.02.025. [DOI] [PubMed] [Google Scholar]
- Ranucci M.; Ballotta A.; Di Dedda U.; et al. The procoagulant pattern of patients with COVID-19 acute respiratory distress syndrome. J. Thromb. Haemostasis 2020, 18, 1747–1751. 10.1111/jth.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings M. J.; Baldwin M. R.; Abrams D.; et al. Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. Lancet 2020, 395, 1763–1770. 10.1016/S0140-6736(20)31189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medcalf R. L.; Keragala C. B.; Myles P. S. Fibrinolysis and COVID-19: a plasmin paradox. J. Thromb. Haemostasis 2020, 18, 2118–2122. 10.1111/jth.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranucci M.; Sitzia C.; Baryshnikova E.; et al. Covid-19-associated coagulopathy: biomarkers of thrombin generation and fibrinolysis leading the outcome. J. Clin. Med. 2020, 9, 3487. 10.3390/jcm9113487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campello E.; Bulato C.; Spiezia L.; et al. Thrombin generation in patients with COVID-19 with and without thromboprophylaxis. Clin. Chem. Lab. Med. 2021, 59, 1323–1330. 10.1515/cclm-2021-0108. [DOI] [PubMed] [Google Scholar]
- Wang C.; Sun Y.; Zhao Q. A sensitive thrombin-linked sandwich immunoassay for protein targets using high affinity phosphorodithioate modified aptamer for thrombin labeling. Talanta 2020, 207, 120280 10.1016/j.talanta.2019.120280. [DOI] [PubMed] [Google Scholar]
- Kurantsin-Mills J.; Ofosu F. A.; Safa T. K.; Siegel R. S.; Lessin L. S. Plasma factor VII and thrombin–antithrombin III levels indicate increased tissue factor activity in sickle cell patients. Br. J. Haematol. 1992, 81, 539–544. 10.1111/j.1365-2141.1992.tb02989.x. [DOI] [PubMed] [Google Scholar]
- Van Veen J. J.; Gatt A.; Makris M. Thrombin generation testing in routine clinical practice: are we there yet?. Br. J. Haematol. 2008, 142, 889–903. 10.1111/j.1365-2141.2008.07267.x. [DOI] [PubMed] [Google Scholar]
- Shuman M. A.; Majerus P. W. The measurement of thrombin in clotting blood by radioimmunoassay. J. Clin. Invest. 1976, 58, 1249–1258. 10.1172/JCI108579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G.; Zhang S.; Hu X. Signal enhancement in a lateral flow immunoassay based on dual gold nanoparticle conjugates. Clin. Biochem. 2013, 46, 1734–1738. 10.1016/j.clinbiochem.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Irvine D.; Tuerk C.; Gold L. Selexion: Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J. Mol. Biol. 1991, 222, 739–761. 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- Ellington A. D.; Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Cuenoud B.; Szostak J. W. A DNA metalloenzyme with DNA ligase activity. Nature 1995, 375, 611–614. 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]
- Szostak J. W. In vitro genetics. Trends Biochem. Sci. 1992, 17, 89–93. 10.1016/0968-0004(92)90242-2. [DOI] [PubMed] [Google Scholar]
- Avino A.; Fabrega C.; Tintore M.; Eritja R. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. 10.2174/138161212799958387. [DOI] [PubMed] [Google Scholar]
- Adachi T.; Nakamura Y. Aptamers: A review of their chemical properties and modifications for therapeutic application. Molecules 2019, 24, 4229 10.3390/molecules24234229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.; Suess B. Identification of RNA aptamers with riboswitching properties. Methods 2016, 97, 44–50. 10.1016/j.ymeth.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Ali G. K.; Omer K. M. Molecular imprinted polymer combined with aptamer (MIP-aptamer) as a hybrid dual recognition element for bio(chemical) sensing applications. Review. Talanta 2022, 236, 122878 10.1016/j.talanta.2021.122878. [DOI] [PubMed] [Google Scholar]
- Jarczewska M.; Górski Ł.; Malinowska E. Electrochemical aptamer-based biosensors as potential tools for clinical diagnostics. Anal. Methods 2016, 8, 3861–3877. 10.1039/C6AY00499G. [DOI] [Google Scholar]
- Hołubowski R.; Jarczewska K. The combination of multi-step differential transformation method and finite element method in vibration analysis of non-prismatic beam. Int. J. Appl. Mech. 2017, 9, 1750010 10.1142/S1758825117500107. [DOI] [Google Scholar]
- Yaghi O. M.; Li G.; Li H. Selective binding and removal of guests in a microporous metal–organic framework. Nature 1995, 378, 703–706. 10.1038/378703A0. [DOI] [Google Scholar]
- Shatery O. B. A.; Omer K. M. Selectivity Enhancement for Uric Acid Detection via In Situ Preparation of Blue Emissive Carbon Dots Entrapped in Chromium Metal–Organic Frameworks. ACS omega 2022, 7, 16576–16583. 10.1021/acsomega.2c00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Xu Q.; Jiang H.-L. Metal–organic frameworks meet metal nanoparticles: synergistic effect for enhanced catalysis. Chem. Soc. Rev. 2017, 46, 4774–4808. 10.1039/C6CS00724D. [DOI] [PubMed] [Google Scholar]
- Altintas C.; Avci G.; Daglar H.; et al. Database for CO2 separation performances of MOFs based on computational materials screening. ACS Appl. Mater. Interfaces 2018, 10, 17257–17268. 10.1021/acsami.8b04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M. J.; Lee R. G.; Brandt T. A.; et al. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N. Engl. J. Med. 2017, 377, 222–232. 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- McFarland J., National Center for Education Statistics ; et al. The Condition of Education 2017. NCES 2017-144. 2017.
- Chughtai A.; Byrne M.; Flood B. Linking ethical leadership to employee well-being: The role of trust in supervisor. J. Bus. Ethics 2015, 128, 653–663. 10.1007/s10551-014-2126-7. [DOI] [Google Scholar]
- Gao T.; Han X.; Zhu H.. et al. FewRel 2.0: Towards more challenging few-shot relation classification, arXiv Prepr. arXiv1910.07124, 2019 10.48550/arXiv.1910.07124. [DOI]
- Liu X.Syntheses, Structures and Properties of Metal-Organic Frameworks, 2015.
- Wang M.; Cao R.; Zhang L.; et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayani K. F.; Omer K. M. A red luminescent europium metal organic framework (Eu-MOF) integrated with a paper strip using smartphone visual detection for determination of folic acid in pharmaceutical formulations. New J. Chem. 2022, 46, 8152–8161. 10.1039/D2NJ00601D. [DOI] [Google Scholar]
- Xu Z.; Zhang Z.; She Z.; et al. Aptamer-functionalized metal-organic framework-coated nanofibers with multi-affinity sites for highly sensitive, selective recognition of ultra-trace microcystin-LR. Talanta 2022, 236, 122880 10.1016/j.talanta.2021.122880. [DOI] [PubMed] [Google Scholar]
- Wang N.; Liu Z.; Wen L.; et al. Aptamer-binding zirconium-based metal-organic framework composites prepared by two conjunction approaches with enhanced bio-sensing for detecting isocarbophos. Talanta 2022, 236, 122822 10.1016/j.talanta.2021.122822. [DOI] [PubMed] [Google Scholar]
- Pirot S. M.; Omer K. M. Surface imprinted polymer on dual emitting MOF functionalized with blue copper nanoclusters and yellow carbon dots as a highly specific ratiometric fluorescence probe for ascorbic acid. Microchem. J. 2022, 182, 107921 10.1016/j.microc.2022.107921. [DOI] [Google Scholar]
- Sh Mohammed Ameen S.; Sher Mohammed N. M.; Omer K. M. Visual monitoring of silver ions and cysteine using bi-ligand Eu-based metal organic framework as a reference signal: Color tonality. Microchem. J. 2022, 181, 107721 10.1016/j.microc.2022.107721. [DOI] [Google Scholar]
- Pirot S. M.; Omer K. M. Designing of robust and sensitive assay via encapsulation of highly emissive and stable blue copper nanocluster into zeolitic imidazole framework (ZIF-8) with quantitative detection of tetracycline. J. Anal. Sci. Technol. 2022, 13, 22 10.1186/s40543-022-00333-6. [DOI] [Google Scholar]
- Lin S.; et al. Aptamer-functionalized stir bar sorptive extraction coupled with gas chromatography–mass spectrometry for selective enrichment and determination of polychlorinated biphenyls in fish samples. Talanta 2016, 149, 266–274. 10.1016/j.talanta.2015.11.062. [DOI] [PubMed] [Google Scholar]
- Huang W.; Xu Y.; Wang Z.; et al. Dual nanozyme based on ultrathin 2D conductive MOF nanosheets intergraded with gold nanoparticles for electrochemical biosensing of H2O2 in cancer cells. Talanta 2022, 249, 123612 10.1016/j.talanta.2022.123612. [DOI] [PubMed] [Google Scholar]
- Guo J.; et al. Label-free fluorescence detection of hydrogen peroxide and glucose based on the Ni-MOF nanozyme–induced self-ligand emission. Microchim. Acta 2022, 189, 219 10.1007/s00604-022-05313-6. [DOI] [PubMed] [Google Scholar]
- Mishra N.; Rohaninejad M.; Chen X.; Abbeel P.. A simple neural attentive meta-learner, arXiv Prepr. arXiv1707.03141, 2017.
- Wang Q.; Hong G.; Liu Y.; Hao J.; Liu S. Dual enzyme-like activity of iridium nanoparticles and their applications for the detection of glucose and glutathione. RSC Adv. 2020, 10, 25209–25213. 10.1039/D0RA05342B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y.-p.; Liu S.; Gao Z.; Ning B.; Wang Y. State-of-the-art progress of switch fluorescence biosensors based on metal-organic frameworks and nucleic acids. Microchim. Acta 2021, 188, 168 10.1007/s00604-021-04827-9. [DOI] [PubMed] [Google Scholar]
- Liu S.; Bai J.; Huo Y.; et al. A zirconium-porphyrin MOF-based ratiometric fluorescent biosensor for rapid and ultrasensitive detection of chloramphenicol. Biosens. Bioelectron. 2020, 149, 111801 10.1016/j.bios.2019.111801. [DOI] [PubMed] [Google Scholar]
- Isho R. D.; Sher Mohammad N. M.; Omer K. M. Enhancing enzymatic activity of Mn@Co3O4 nanosheets as mimetic nanozyme for colorimetric assay of ascorbic acid. Anal. Biochem. 2022, 654, 114818 10.1016/j.ab.2022.114818. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhu Y.; Binyam A.; et al. Discovering the enzyme mimetic activity of metal-organic framework (MOF) for label-free and colorimetric sensing of biomolecules. Biosens. Bioelectron. 2016, 86, 432–438. 10.1016/j.bios.2016.06.036. [DOI] [PubMed] [Google Scholar]
- Zhang Z.-H.; et al. Aptamer-embedded zirconium-based metal–organic framework composites prepared by de novo bio-inspired approach with enhanced biosensing for detecting trace analytes. ACS Sens. 2017, 2, 982–989. [DOI] [PubMed] [Google Scholar]
- Li X.; et al. Three hidden talents in one framework: a terephthalic acid-coordinated cupric metal–organic framework with cascade cysteine oxidase-and peroxidase-mimicking activities and stimulus-responsive fluorescence for cysteine sensing. J. Mater. Chem. B 2018, 6, 6207–6211. 10.1039/C8TB02167H. [DOI] [PubMed] [Google Scholar]
- Wibowo A.; Marsudi M. A.; Pramono E.; et al. Recent improvement strategies on metal-organic frameworks as adsorbent, catalyst, and membrane for wastewater treatment. Molecules 2021, 26, 5261 10.3390/molecules26175261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.; Li G.; Hu Y. Aptamer-involved fluorescence amplification strategy facilitated by directional enzymatic hydrolysis for bioassays based on a metal-organic framework platform: highly selective and sensitive determination of thrombin and oxytetracycline. Microchim. Acta 2017, 184, 2365–2373. 10.1007/s00604-017-2263-7. [DOI] [Google Scholar]
- Ren Q.; Xu X.; Cao G.; et al. Electrochemical thrombin aptasensor based on using magnetic nanoparticles and porous carbon prepared by carbonization of a zinc (II)-2-methylimidazole metal-organic framework. Microchim. Acta 2019, 186, 659 10.1007/s00604-019-3781-2. [DOI] [PubMed] [Google Scholar]