Abstract
Neurodevelopmental research has traditionally focused on development of individual structures, yet multiple lines of evidence indicate parallel development of large-scale systems, including canonical neural networks (i.e., default mode, frontoparietal). However, the relationship between region- vs. network-level development remains poorly understood. The current study tests the ability of a recently developed multi-task coactivation matrix approach to predict canonical resting state network engagement at baseline and at two-year follow-up in a large and cohort of young adolescents. Pre-processed tabulated neuroimaging data were obtained from the Adolescent Brain and Cognitive Development (ABCD) study, assessing youth at baseline (N = 6073, age = 10.0 ± 0.6 years, 3056 female) and at two-year follow-up (N = 3539, age = 11.9 ± 0.6 years, 1726 female). Individual multi-task co-activation matrices were constructed from the beta weights of task contrasts from the stop signal task, the monetary incentive delay task, and emotional N-back task. Activation-based predictive modeling, a cross-validated machine learning approach, was adopted to predict resting-state canonical network engagement from multi-task co-activation matrices at baseline. Note that the tabulated data used different parcellations of the task fMRI data (“ASEG” and Desikan) and the resting-state fMRI data (Gordon). Despite this, the model successfully predicted connectivity within the default mode network (DMN, rho = 0.179 ± 0.002, p < 0.001) across participants and identified a subset of co-activations within parietal and occipital macroscale brain regions as key contributors to model performance, suggesting an underlying common brain functional architecture across cognitive domains. Notably, predictive features for resting-state connectivity within the DMN identified at baseline also predicted DMN connectivity at two-year follow-up (rho = 0.258). These results indicate that multi-task co-activation matrices are functionally meaningful and can be used to predict resting-state connectivity. Interestingly, given that predictive features within the co-activation matrices identified at baseline can be extended to predictions at a future time point, our results suggest that task-based neural features and models are valid predictors of resting state network level connectivity across the course of development. Future work is encouraged to verify these findings with more consistent parcellations between task-based and resting-state fMRI, and with longer developmental trajectories.
Abbreviations: ABCD, Adolescent Brain and Cognitive Development; APM, Activation-Based Predictive Modeling; CPM, Connectome-Based Predictive Modeling; DMN, Default Mode Network; FP, Fronto-parietal Network; MID, Monetary Incentive Delay Task; PC, Principal Components; ROI, Regions of Interest; SAL, Salience Network; SST, Stop Signal Task
Keywords: Co-activation, Adolescence, Development, Predictive modeling, Resting-state connectivity, Cognition
Highlights
-
•
Multi-task co-activation matrices were computed for > 3000 youth at ages 10 and 12.
-
•
Matrices were entered into cross-validated activation-based predictive models (APM).
-
•
At age 10, APM robustly predicted resting state connectivity and cognitive function.
-
•
Models developed at age 10 had higher predictive accuracy when used on age-12 data.
1. Introduction
Adolescence is a time of significant neural change (Casey et al., 2019, Kilford et al., 2016, Larsen and Luna, 2018, Meruelo et al., 2017). While research has traditionally focused on development of individual structures, significant research over recent years indicates parallel development of large-scale systems (Morgan et al., 2018, Power et al., 2010), such as the default mode network (Fan et al., 2021, Sherman et al., 2014, Supekar et al., 2010), the frontoparietal network and the salience network (Fair et al., 2007, Wendelken et al., 2016). These three networks all exhibit significant change during childhood and adolescence and together subserve critical processes including development of executive control and affective/salience encoding processes (Fair et al., 2008, Marek and Dosenbach, 2018, Snyder et al., 2021, Wendelken et al., 2016). In addition, while many studies have sought to characterize developmental trajectories of task-based activation patterns (Lamm et al., 2014, Padmanabhan et al., 2011) and of resting state networks (Grayson and Fair, 2017, Scheinost et al., 2015) separately, very little is known about the relationship between these two developmental processes. The current study explores the relationship between canonical network engagement and task-based activation patterns using a recently developed multi-task, predictive modeling approach (Li et al., 2021), with a special focus on young adolescents when significant neural change takes place. Specifically, we test the ability of individual-level task-based coactivation patterns to predict individual-level resting-state network connectivity at two time points using data from a large cohort of young adolescents (N ∼ 6000) from the Adolescent Brain and Cognitive Development (ABCD) Study (Bjork et al., 2017, Volkow et al., 2018).
Multi-task co-activation matrices were chosen for our analysis as a tool to summarize whole-brain functional activity across performance of different cognitive tasks targeting different cognitive domains (Laird et al., 2013, Li et al., 2021). Constructed from task effect estimates across different cognitive domains, multi-task co-activations identify consistently correlated or anti-correlated activity between pairwise brain regions across tasks. Thus, unlike functional connectivity matrices – which are traditionally computed using nodal time-courses without consideration of specific events of interest – functional co-activation matrices provide a simple method for summarizing individual participant whole-brain patterns of functional engagement across diverse cognitive events. Further, previous meta-analyses have suggested that, at the group level, multi-task co-activations capture brain activity common to different cognitive domains and represent fundamental components of brain’s functional architecture, and studies have demonstrated that the structure of group-level multi-task co-activations matches the organization of many resting-state networks (RSNs) (Laird et al., 2013, Smith et al., 2009). This work can therefore also identify such fundamental components and investigate whether such components can predict resting-state network connectivity.
The current study highlights a few innovations and improvements upon existing work. First, the study is the first to investigate the relationship between task-based co-activations and resting-state network connectivity and their co-development at an individual level. To achieve this goal, we extended a previously validated connectome-based predictive modeling (CPM) approach (Finn et al., 2015, Shen et al., 2017) to conduct activation-based predictive modeling (APM). As with CPM, APM uses cross-validation to identify predictive features within the co-activation matrices, lowering the chance of overfitting. The distribution of predictive features can be easily visualized, enabling high interpretability. We report APM performances on predictions of resting-state connectivity and cognitive measures at baseline, as well as the performance of the baseline model at two-year follow-up. Given parallel development trajectories of individual brain regions and large-scale networks, we anticipated that the relationship between whole-brain co-activations and resting-state connectivity would remain relatively stable with neural development, even if considerable changes would occur within the co-activations and resting-state connectivity themselves. Additionally, a practical advantage of our approach is that it may be easily implemented using publicly available curated data, without requiring individual labs to compute connectivity matrices from raw data (e.g., ABCD’s curated data release includes task beta weights for ROIs but does not include timeseries data). Thus, we anticipate that this approach will be more easily accessible to smaller labs without significant computational resources.
Second, the current study constructed co-activation matrices by examining interactions between all possible pairs of brain regions. As a comparison, existing multi-task co-activation research use a seed-based approach to construct the matrices (Laird et al., 2013, Li et al., 2021), which only captures the interactions between one specific brain region (the “seed”) with the rest of the brain and does not represent all possible interactions between brain regions. The current study’s approach includes much more information and can therefore draw conclusions with much more confidence.
Third, while previous analysis suggested multi-task co-activations as a representation of the “core” functional architecture of the brain and discovered structural similarity between task-based co-activations and resting-state networks (Laird et al., 2013, Smith et al., 2009), it was conducted on group-level data only. Here, we extend this work to focus on co-activation patterns at the individual level (unlike prior meta-analytic work that has focused on co-activation patterns across individuals). We hypothesized that individual whole-brain co-activation matrices would successfully predict canonical network engagement at rest and that predictive performance would be strongest for models built using data from all three available ABCD tasks (reward, inhibition and affective working memory tasks). For comparison, we also examined the performance of whole-brain multi-task co-activation matrices for predicting a measure of general cognitive ability. Within this context, we hypothesized that, if multi-task co-activations indeed capture general brain functions, they should be able to reliably predict general cognitive ability.
2. Materials and methods
2.1. The adolescent brain cognitive development study
Data used in this study were obtained from the Adolescent Brain Cognitive Development (ABCD) study release 3.0 (https://nda.nih.gov/abcd, DAR ID #7566, see Supplementary materials Table S1 for access dates). The ABCD study is an ongoing longitudinal cohort study including 21 unique data collection sites across the United States and comprises 11,878 children aged 9–10 at baseline (Bjork et al., 2017, Volkow et al., 2018). The study will track biological and behavioral development through adolescence into young adulthood, and the current release includes data from the full cohort from the baseline, six-month follow-up, and one-year follow-up, and data from part of the full cohort from the 18-month follow-up and the two-year follow-up.
2.2. Neuroimaging data
The ABCD study includes three fMRI tasks (Casey et al., 2018): the monetary incentive delay task (MID) (Knutson et al., 2000), the stop signal task (SST) (Logan, 1994), and the emotional N-back task (EnBack) (Cohen et al., 2016). The MID measures domains of reward processing, including the anticipation and receipt of rewards and losses. The SST engages domains of impulsivity and impulse control. The EnBack task engages working memory using emotional stimuli.
The curated ABCD study release 3.0 (which includes the corrected data from Philips scanners) includes mean beta weights for 98 brain regions of interest (ROIs) covering the cortex, subcortex and cerebellum for each task across different task contrasts (e.g., correct stop versus correct go for the SST, reward anticipation versus neutral for the MID, emotional faces versus neutral faces for the EnBack). These mean beta weights were used to construct multi-task co-activation matrices for each participant, as described below and summarized in Fig. 1A.
Fig. 1.
Data processing and modeling pipeline. (A) Multi-task co-activation matrices were constructed by calculating the Pearson correlation between brain regions across task contrasts. (B) Adapted from connectome-based predictive modeling (Shen et al., 2017), activation-based predictive modeling consists of four steps: select predictive features, sum edge weights, fit predictive model, and apply model to novel data. Note: All pre-processing and computation of task beta-weights was conducted by the ABCD Consortium, details in Supplemental Materials.
All image pre-processing and first-level analytic steps (e.g., the calculation of beta weights for a given task) were conducted by the ABCD Consortium and are briefly described in the Supplemental Materials and Methods. For more details on image collection, preprocessing, and analysis, please refer to the official ABCD publication (Hagler et al., 2019).
2.3. Construction of multi-task co-activation matrices
Quality control was conducted using the ABCD imaging inclusion recommendations (package name: abcd_imgincl01) for 11,788 participants at baseline and 5700 participants at two-year follow-up with data at the time of access. Details on inclusion criteria can be found in ABCD 3.0 Release Notes (https://nda.nih.gov/study.html?id=901) - MRI Quality Control Recommended Inclusion. For functional MRIs, this includes a series of automated and manual quality control steps such as checking the average framewise displacement as well as the number of seconds with framewise displacements less than 0.2, 0.3, or 0.4 mm, according to recommendations by Power et al. (2012). At each timepoint, only participants whose fMRI data on the SST, MID, and EnBack tasks followed the ABCD imaging inclusion recommendations were considered for the construction of the multi-task co-activation matrices, which resulted in 6075 participants at baseline, and 3539 participants at two-year follow-up.
Fig. 1 A provides a schematic overview of the construction of co-activation matrices. Beta weights were downloaded for task-based fMRI in 30 subcortical (“ASEG”) and 68 cortical (“APARC”) ROIs (Desikan et al., 2006, Fischl et al., 2002) averaged across all runs (package names: mrisst02, nback_bwroi02, midaparc03, and midaparcp203). Eighty-six ROIs remained after exclusion of cerebral white matter, cerebellum white matter, ventricles, brain stem, and cerebrospinal fluid (Supplementary materials Table S2). Twelve task contrasts were selected (Table 1), and the corresponding beta weights were extracted for each ROI. Using these data, an 86 by 86 multi-task co-activation matrix was constructed for each participant by calculating the Pearson correlation between regional task contrasts (Fig. 1A). Two participants had missing contrast data at baseline and were excluded. A total of 6073 matrices at baseline (10.0 ± 0.6 years, female: 3056) and 3539 matrices at two-year follow-up (11.9 ± 0.6 years, female: 1726) were generated. The number of participants remaining after each step of preprocessing is also summarized in Supplementary materials Table S3.
Table 1.
List of contrasts used to construct multi-task co-activation matrices.
| Task | Contrast |
|---|---|
| SST | correct stop versus correct go |
| incorrect stop versus correct go | |
| correct stop versus incorrect stop | |
| incorrect go versus correct go | |
| MID | all anticipation of loss versus neutral |
| all anticipation of reward versus neutral | |
| all loss positive versus negative feedback contrast | |
| all reward positive versus negative feedback | |
| EnBack | 2 back versus 0 back |
| face versus place | |
| negative face versus neutral face | |
| positive face versus neutral face |
2.4. Activation-based predictive modeling (APM)
Predictive models were run using an adapted version of connectome-based predictive modeling (CPM). CPM is a cross-validated machine-learning approach that takes whole-brain connectomes as input and identifies positive and negative features in the connectomes that are predictive of target variables, such as behavioral measurements (Shen et al., 2017). Although traditionally run using functional connectivity matrices as input, CPM may also take other types of imaging data as input (Shen et al., 2017). We therefore extended this approach to include multi-task functional co-activation matrices as input for the first time (Fig. 1B), hereafter referred to as activation-based predictive modeling (APM). For both CPM and APM, participant-level differences in the input matrices and the target variables drive the training and evaluation of the predictive models.
APM was used to generate predictive models of average resting-state connectivity within the default mode network (DMN), salience network (SAL), and fronto-parietal network (FP) (obtained from the curated ABCD study release, package name: abcd_betnet02). The canonical networks are based on the Gordon parcellation, a functionally defined parcellation based on resting-state functional connectivity patterns (Gordon et al., 2016). Details on the preprocessing of resting-state imaging data, as well as the calculation of average connectivity within networks, can be found in Supplemental materials and Methods and the official ABCD publication (Hagler et al., 2019). Not all participants with co-activation matrices also had resting-state connectivity data. Overall, 5605 participants had both co-activation matrices and resting-state connectivity data at baseline that met the ABCD image inclusion criteria (package name: abcd_imgincl01), and the co-activation matrices of these participants were entered into APMs to predict resting state network connectivity (10.0 ± 0.6 years, female: 2888).
To identify co-activation patterns associated with cognitive measures, a common target for CPM, we also ran APMs on the three general neurocognitive components identified by Thompson et al. (2019) using data from the ABCD neurocognitive battery (Luciana et al., 2018). The components were determined by running Bayesian principal component analysis on nine neurocognitive measures, including measures from seven NIH Toolbox tests (e.g., the Picture Vocabularty Task, the List Sorting Working Memory Test) as well as the Rey Auditory Verbal Learning Test and the little man task, as reported previously (Thompson et al., 2019). The three principal components (PCs) were shown to encompass general cognitive ability (PC1), executive function (PC2), and learning/memory (PC3). These principal components were obtained through the ABCD release 3.0 RDS file (https://nda.nih.gov/study.html?id=1042). Overall, 5689 participants had both co-activation matrices and cognitive PCs data at baseline and these participants were entered into APM (10.0 ± 0.6 years, female: 2852).
All APM analyses were conducted using custom scripts in Python, based on Shen et al. (2017). The scripts and the code used to generate task-based coactivation matrices are available at our GitHub repository https://github.com/fye92/abcd_fy. A 10-fold cross-validation APM was adopted for all analyses. For each fold, 90 % of the participants was assigned as training data and the remaining 10 % was assigned as testing data. During training, Pearson correlation coefficient (r) was calculated across participants between edge weights in the input co-activation matrices and a target measure, e.g., DMN connectivity. Edges that positively correlated with the target measure with p < 0.05 were identified as the positive features (“positive edges”), whereas edges that negatively correlated with the target measure with p < 0.05 were identified as the negative features (“negative edges”). A summary statistic was then calculated for each individual by subtracting the sum of negative edge weights from the sum of positive edge weights, and a linear regression model was trained on this statistic to predict the target measure. The predictive features identified in the training data were then extracted from the co-activation matrices from the testing data, and the trained models were then applied to the summary statistic of the testing data to generate predictions. Model performance was quantified as the Spearman correlation (rho) between the testing-data predictions and actual values across the whole sample. To further improve the reliably of our results, 10-fold APM was repeated 100 times to generate 100 Spearman rho values, where subject order is randomly shuffled for each iteration. To prevent over-fitting to a given iteration, the final predictive network retains only positive and negative features that were shared across every fold and every iteration.
2.5. Statistical analysis
Permutation testing was adopted to measure the significance of the observed Spearman rho values. For 1000 iterations, the target measures (resting-state connectivity or cognitive PCs) were permuted and then fed into 10-fold APM. The resulting 1000 Spearman rho values formed a null distribution and a one-tailed p-value was calculated by contrasting the actual Spearman rho against the null distribution:
Note that one-tailed p-values were chosen over two-tailed p-value because was expected to be positive (see Table 2 in the Results section), and only null-model rhos more positive than were considered significant.
Table 2.
APM model performance across 100 iterations.
| #Iter of null model | #Iter of true model | Spearman rho | One-tailed p-value | |
|---|---|---|---|---|
| Average resting-state connectivity - DMN | 1000 | 100 | 0.1792 (0.0022) | < 0.001 |
| Average resting-state connectivity – FP | 1000 | 100 | 0.1207 (0.0065) | < 0.001 |
| Average resting-state connectivity - SAL | 1000 | 100 | 0.0443 (0.0017) | 0.001 |
| Cognitive PC1 (general ability) | 1000 | 100 | 0.1404 (0.0060) | < 0.001 |
| Cognitive PC2 (executive function) | 1000 | 100 | 0.0592 (0.0016) | < 0.001 |
| Cognitive PC3 (learning/memory) | 1000 | 100 | 0.0803 (0.0018) | < 0.001 |
Results reported here use both positive and negative features (i.e., combined model performance). Model performance is quantified as the Spearman correlation (rho) between the test-set predictions and actual values across the whole sample. Standard deviations are shown in parentheses. Bold font indicates the best performing models for each domain.
2.6. Model stability from age 10 to age 12
To determine the stability of identified predictive models, the predictive ability of successful models derived at baseline were tested at two-year follow-up. As the cognitive PCs are not yet available for two-year follow-up, these analyses were constrained to models that successfully predicted average connectivity within resting-state networks. For these analyses, predictive features identified in baseline APMs were extracted from two-year follow-up co-activation matrices. For each participant, the summary statistic of the predictive features was calculated, and a prediction of age-12 average resting-state connectivity was made by entering the summary statistic into a linear regression model using the mean slope and intercept obtained during training at age 10. Spearman correlation was then calculated across participants between the prediction and the actual age-12 average resting-state connectivity.
2.7. Post-hoc testing
To gauge the contribution of each individual task to the models’ predictive performance, co-activation matrices were re-computed removing one task at a time. Excluding participants with missing data, removing the SST and removing the EnBack both resulted in 6074 matrices at baseline, and removing the MID resulted in 6073 matrices at baseline. APMs were run using task-removed co-activation matrices to predict baseline average resting-state connectivity and cognitive PCs. Overall, 5605 participants had both MID-removed/EnBack-removed co-activation matrices and resting-state connectivity data at baseline (10.0 ± 0.6 years, female: 2888), and 5606 participants had both SST-removed co-activation matrices and resting-state connectivity data at baseline (10.0 ± 0.6 years, female: 2888). For prediction of general cognitive measures, 5689 participants had both MID-removed co-activation matrices and cognitive PCs at baseline (10.0 ± 0.6 years, female: 2852), and 5690 participants had both SST-removed/EnBack-removed co-activation matrices and cognitive PCs at baseline (10.0 ± 0.6 years, female: 2852). Identical to previous analyses, 10-fold APM was run with p-value threshold 0.05 and was repeated 100 times to generate 100 Spearman rho values. To test for between-model differences in accuracy, Steiger’s test for dependent correlations was used to quantify the significance of the differences between rhos.
To quantify how each macroscale brain region contributed to the predictive power of APM models, “virtual lesioning” (Lichenstein et al., 2021, Yip et al., 2020) was carried out by removing one macroscale brain region at a time from the multi-task co-activation matrices and re-running APM. The 86 ROIs were assigned to nine macroscale brain regions, based on FreeSurfer’s classification (Klein and Tourville, 2012): cerebellum, cingulate, frontal, insula, limbic, occipital, parietal, striatum, and temporal networks (Supplementary Table S2). For these analyses, the cohort remained the same as in the original APM analysis (N = 5605 for resting-state connectivity and N = 5689 for cognitive PCs). As done in previous analyses, 10-fold APM was run with a p-value threshold of 0.05 and was repeated 100 times. For each “lesioned” macroscale brain region, performance of the “lesioned” co-activation matrices was quantified by the 100 Spearman rho values. Steiger’s test for dependent correlations was used to quantify the significance of the differences between rhos (i.e., to test for between-model differences in accuracy).
2.8. Data and code availability
The data that support the findings of this study are openly available in the ABCD study release 3.0 (https://nda.nih.gov/abcd, see Supplementary materials Table S1 for package names and access dates). The code for APM is available at our GitHub repository https://github.com/fye92/abcd_fy. The list of participant IDs used in the analysis will be available upon direct request to the corresponding author and review by all authors.
3. Results
3.1. Activation-based modeling using multi-task co-activation matrices
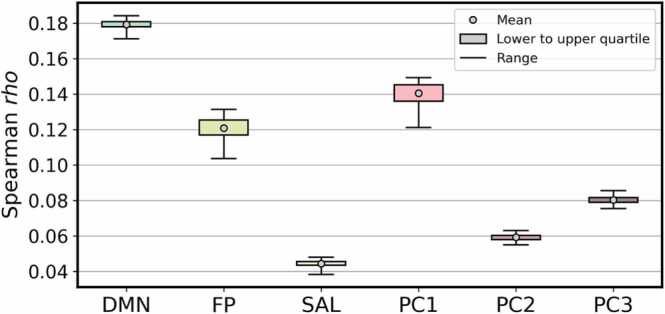
Model performance of APMs run using multi-task co-activation matrices are summarized in Fig. 2 and Table 2. Multi-task co-activation matrices successfully predicted average DMN connectivity across participants (rho = 0.1792 ± 0.0022, p-value < 0.001). While prediction of other canonical networks was also statistically significant, effect sizes (rho) were smaller (FP: rho = 0.1207 ± 0.0065, p < 0.001; SAL: rho = 0.0443 ± 0.0017, p = 0.001).
Fig. 2.
APM model performance across 100 iterations. From left to right: for prediction of DMN connectivity, FP connectivity, SAL connectivity, cognitive PC1 (general ability), PC2 (executive function), and PC3 (learning/memory). Gray circles represent mean, boxes represent lower to upper quartile, and whiskers represent range.
Among the three principal components identified by Thompson et al. (2019), the co-activation matrices were best at predicting PC1 “general ability” (rho = 0.1404 ± 0.0060, p < 0.001). While prediction of the other two PCs was also statistically significant, effect sizes (rho) were smaller (PC2 “executive function”: rho = 0.0592 ± 0.0016, p < 0.001; PC3 “learning/memory” rho = 0.0803 ± 0.0018, p < 0.001).
3.2. Distribution of significant features in macroscale brain regions
Here, we focus on the best performing models for resting-state connectivity (DMN) and cognition (PC1), respectively. Fig. 3 summarizes the distribution of positive and negative features identified from predictions of DMN connectivity and cognitive PC1 in and across macroscale brain regions, based on FreeSurfer’s classification (Klein and Tourville, 2012).
Fig. 3.
Distribution of positive and negative predictive features in and across macroscale brain regions. Top panel: (A) Absolute number of edges and (B) ratio of edges, from prediction of average resting-state connectivity within the DMN. Bottom panel: (C) Absolute number of edges and (D) ratio of edges, from prediction of cognitive PC1.
For prediction of average DMN connectivity (Fig. 3A), more positive features (i.e., edges in the co-activation matrices that were significantly positively correlated with DMN connectivity) were identified than negative features (i.e., edges in the co-activation matrices that were significantly negatively correlated with DMN connectivity). Positive features are those for which increased co-activation between brain regions is a positive predictor of DMN strength and negative features are those for which decreased co-activation between brain regions is a positive predictor of DMN strength. Positive features were more frequently distributed between the frontal and temporal networks (116 features), the frontal and limbic networks (101 features), and between the temporal and limbic networks (61 features). Negative features were primarily between the frontal and parietal networks (26 features) and within the parietal network (23 features). Fig. 3B further summarizes predictive features as the ratio of total edges within and between macroscale brain regions. The occipital network contributed the most to positive features, with 64 % of its connections being statistically significant. The parietal network contributed the most to negative features, with 51 % of its connections being statistically significant. The cerebellum and insula only contain two ROIs and therefore the ratio of significant features within these regions are either 0 or 1 and thus uninformative. The importance of each macroscale brain region to the prediction of DMN connectivity was further investigated in Section 3.5.
For the prediction of cognitive PC1 (“general ability”, Fig. 3C), positive features are those for which increased co-activation between brain regions is a positive predictor of general cognitive ability and negative features are those for which decreased co-activation between brain regions is a positive predictor of general cognitive ability.. Positive features were primarily identified between the frontal network and cerebellum (19 features), the frontal and parietal networks (19 features), the parietal network and cerebellum (14 features), the frontal and occipital networks (13 features), and the parietal network and cingulate (11 features). Negative features were distributed primarily within the frontal network (21 features), between the frontal and temporal networks (17 features), the frontal and limbic networks (12 features), the frontal and parietal networks (12 features), within the parietal network (12 features), and between the parietal and limbic networks (10 features). Fig. 3D further summarizes predictive features as the ratio of total edges within and between macroscale brain regions. Connections between the parietal network and cerebellum (70 %) and between the frontal network and cerebellum (43 %) contributed the most to positive features, whereas the parietal network contributed the most to negative features (27 %). The importance of each macroscale brain region to the prediction of general cognitive ability was further investigated in Section 3.5.
3.3. Model performances at two-year follow-up
For the prediction of resting-state connectivity within the DMN, the predictive network derived at baseline was also successful at predicting the DMN connectivity at two-year follow-up (N = 3383, age = 12.0 ± 0.6 years, 1676 female). The Spearman correlation between prediction of year-two DMN connectivity and actual year-two DMN connectivity was rho = 0.258 (Fig. 4). The prediction was generated by applying baseline predictive network and linear regression model to year-two co-activation matrices. Predictions of the FP and SAL connectivity at two-year follow-up are shown in Supplementary Fig. S1.
Fig. 4.
Using age-ten predictive network to predict age-twelve average resting-state connectivity within the DMN. The predictive network was identified by running APM between baseline co-activation matrices and baseline average resting-state connectivity within the DMN (N = 5605, age = 10.0 ± 0.6 years, 2888 female). This prediction was generated by applying baseline predictive network and linear regression model identified at baseline to year-two co-activation matrices: mean slope (m = 8.57e-5) and intercept (b = 0.22). Data shown using hexagonal binning plots for which color scale indicates the number of individuals represented by each hexagon.
3.4. Model performances after task removal
Tasks used to construct the co-activation matrices affected the performances of APM models (Fig. 5). Removing the MID significantly lowered the model performances for prediction of resting-state DMN connectivity (rho = −0.0304, Steiger’s p = 1.08e-5), and removing the EnBack significantly lowered the model performances for cognitive PC1 (rho = −0.0583, Steiger’s p = 3.69e-6). Removing the MID also lowered model performances for the prediction of cognitive PC1 and removing the EnBack also lowered model performances for the prediction of DMN connectivity, though the differences were not statistically significant.
Fig. 5.
Model performances after task removal. Y axis marks the relative differences in the mean Spearman correlation coefficient across 100 iterations between the task-removed models and the original models. The bar plots describe the relative model performance with MID removed (yellow), with EnBack removed (light green), and with SST removed (dark green). The analysis only included subjects present in the original analysis and all three task-removed analyses. Left: Model performances for predicting resting-state connectivity within the DMN (N = 5605, age = 10.0 ± 0.6 years, 2888 female). Right: Model performance for predicting cognitive PC1 (N = 5689, age = 10.0 ± 0.6 years, 2852 female). The asterisks right above each bar mark the statistical significance of the difference between the task-removed rho and the original rho. The asterisks above the brackets mark the statistical significance of the difference between the task-removed rhos. *: 0.01 < p < 0.05, **: 0.001 < p < = 0.01, ***: p < = 0.001.
In contrast, removing the SST significantly improved model performances for the prediction of cognitive PC1 (rho = 0.0308, Steiger’s p = 0.0015), while removing the SST task mildly lowered model performances for the prediction of DMN connectivity. The difference between the SST-removed and the original models was not statistically significant.
Model performances after task removal for predictions of rs-fMRI SAL, rs-fMRI FP, cognitive PC2, and cognitive PC3 can be found in Supplementary materials Fig. S2. Model performances using only positive or negative features are provided in Supplementary materials Fig. S3. In general, removing the MID or EnBack hindered model performance, whereas removing the SST usually kept model performance relatively unchanged, or even improved model performance.
3.5. Model performances after removal of macroscale brain regions
For the prediction of resting-state DMN connectivity, the “virtual lesioning” analyses revealed that removing the parietal network from the co-activation matrices decreased the performance of APM the most (rho = 0.1551 ± 0.0013, Steiger’s p = 3.33E-15). Removing the occipital network also decreased the performance of APM considerably (rho = 0.1644 ± 0.0015, Steiger’s p = 9.39E-8). For the prediction of cognitive PC1, removing the parietal network from the co-activation matrices decreased the performance of APM the most (rho = 0.1232 ± 0.0047, Steiger’s p = 0.0033).
The “virtual lesioning” results for all macroscale brain regions and the corresponding p-values can be found in Supplementary materials Fig. S4, and Table S4-S7. Reversed “virtual lesioning” analyses were also conducted by keeping one macroscale brain region at a time in the co-activation matrices, and results can be found in Supplementary materials Fig. S5, and Table S8-S11.
4. Discussion
This study tested the hypothesis that multi-task co-activation matrices could be used to predict canonical resting-state network connectivity and cognitive function in large sample of youth. To that end, we adapted an existing pattern learning algorithm – connectome-based predictive modeling—to create activation-based predictive modeling (APM). This novel approach was successful in generating accurate predictive models of individual participant resting-state patterns of DMN connectivity and general cognitive ability in young adolescents (∼10 years). Moreover, predictive features for resting-state connectivity identified at baseline also predicted connectivity at two-year follow-up (∼12 years), suggesting the validity of the predictive features and models over the course of development.
Our approach builds on prior work. Meta-analytic functional brain co-activations across multiple tasks spanning different cognitive domains were first introduced in a PET study by Koski and Paus (2000) and were subsequently applied to task-based fMRI (Laird et al., 2013). While earlier research focused on specific brain regions of interest (Koski and Paus, 2000, Postuma and Dagher, 2006), Toro et al. extended the concept of multi-task co-activation to the whole brain (Toro et al., 2008) and utilized a large amount of experiments available in the BrainMap database (Fox and Lancaster, 2002, Laird et al., 2005; http://brainmap.org). Despite differences in the exact approach used to construct co-activation maps, these meta-analyses generally suggest that—at the group level—multi-task co-activations capture brain activity common to different cognitive domains and represent fundamental components of brain’s functional architecture (Laird et al., 2013). Moreover, studies have demonstrated that the structure of group-level multi-task co-activations matches the organization of many resting-state networks (RSNs) (Laird et al., 2013, Smith et al., 2009). Such convergence between task-based activations (whether single-task or multi-task) and resting-state connectivity implies that there may be a “core” network active in the brain’s functional architecture, regardless of whether the subjects are in an internally induced mind-wandering state or an externally stimulated state such as during task-based fMRI (Laird et al., 2013). Our findings are consistent with this theory and for the first time demonstrate this relationship between multi-task co-activations and resting-state connectivity at an individual level and in a developmental cohort using a robust, cross-validated approach.
Consistent with recent prior work in ABCD, overall effect sizes were somewhat modest. Studies have shown that such values are expected in a large study with thousands of participants and that these effect sizes are more reliable than the inflated effect sizes reported in much smaller samples (e.g., N < 100) (Chaarani et al., 2021, Marek et al., 2022, Owens et al., 2021, Rapuano et al., 2020). Here we follow the recently proposed guidelines indicating that effects from in the ABCD study corresponding to r ≥ 0.10 are in fact “above average” effect sizes (Owens et al., 2021). Thus, findings from the highest performing models meeting or exceeding this threshold within each domain (i.e., models predicting DMN connectivity and cognitive PC1) are focused on below.
Our findings demonstrated that individual participants' task-based fingerprints could be used to predict the same participant’s resting-state DMN connectivity at age ten, and we speculate that the predictive network identified by APM captures the aforementioned “core” network of the brain. This speculation is further supported by our finding that the co-activations are better at predicting general cognitive ability (PC1) than domain-specific behaviors (PC2 and PC3). The model did not work as well in predicting resting-state FP and SAL connectivity at the same age. One possible explanation is that the DMN, a network traditionally considered active “at rest” during processes such as remembering and envisioning the future (Buckner and DiNicola, 2019), may be most consistent with what the multi-task co-activations were designed to capture – the backbone of essential brain functions active regardless of brain states. The FP network, which is critical for cognitive control (Marek and Dosenbach, 2018), and the SAL network, which detects and integrates emotional and sensory stimuli (Seeley, 2019), may only be activated during specific brain states. However, further work is needed to test this specific theory within the context of the ABCD dataset.
Post-hoc, virtual lesioning analyses revealed that removing parietal (including bilateral inferior parietal regions, precuneus, postcentral regions, superior parietal regions, and supramarginal regions) and occipital (including bilateral cuneus, lateral occipital regions, lingual regions, and pericalcarine) regions had the greatest impacts on predictive model performance. This is consistent with the distribution of predictive edges identified by APM (Fig. 2), where occipital regions contributed the most to positive features (64 % of its edges) and parietal regions contributed the most to negative features (51 % of its edges). While some regions in the parietal and occipital networks overlap with the DMN (parts of inferior and superior parietal regions, precuneus, supramarginal regions and lateral occipital regions), many do not. Additionally, removing frontal regions from co-activation matrices did not affect APM performance, even though the frontal network also extensively overlaps with the DMN. This observation echoes the hypothesis that, although there is a convergence between the structure of multi-task co-activations and the organization of many of the resting-state networks, the “core” network identified by APM is different from both. Notably, parietal regions were essential for the prediction of both DMN activity and general cognitive ability, indicating the network may be an important component of the “core” network.
Task-removal analyses indicated that the SST was not as important as the MID and EnBack for the prediction of DMN connectivity and general cognitive ability. From a developmental perspective, this may be explained by previous findings suggesting that inhibitory control develops at a later stage than reward processing or working memory capacity (Casey, 2014, Yip and Potenza, 2018). Given that APM models were trained at age ten, it is likely that inhibitory control has not yet fully developed to become relevant at such an early stage and/or that individual differences in inhibitory control processes are not yet sufficiently varied to yield robust predictions. While it is also possible that previously described design issues of the SST task in ABCD may have led to diminished predictive utility (Bissett et al., 2021), this possibility requires direct testing.
Our approach has several advantages relative to prior work. First, while most of the existing literature focuses on seed-based methods (see, for example, (Bolton et al., 2020; De Marco et al., 2018; Laird et al., 2013; Pelland et al., 2017)), the current study examined the co-activations between all possible pairs of brain regions. This pair-wise co-activation method is ideal for data-driven methods such as APM. Second, thanks to the recent coordinated efforts of establishing multi-site large-cohort studies, task contrasts data in the current study were obtained from a “single” dataset (ABCD), in the sense that it is a more unified effort than the previous meta-analyses where thousands of papers with very different experiment designs and research interests were pooled. This not only lowers undesired variability in the data due to different study protocols, but also enabled us to extract multi-task co-activations for each individual, instead of relying on activation maps summarized from each pooled experiment (see, for example, (Smith et al., 2009)). Furthermore, the ABCD dataset provides longitudinal data for the same cohort of participants, allowing the investigation of the relationship between multi-task co-activations and resting-state connectivity across development. Practically, our approach is also relatively easy to implement as it may be done using publicly available curated data (e.g., summary metrics for different ROIs, as in the ABCD data release) without requiring individual labs to compute connectivity matrices from raw data (e.g., as would be needed for connectome-based modeling of ABCD data). Thus, we anticipate that this approach will be more easily accessible to smaller labs without significant computational resources and may therefore advance open science approaches to this unparalleled dataset. Finally, APM produces reliable results through 100 iterations of 10-fold cross-validation, where subject order is randomly shuffled for each iteration. The method warrants representative sampling of subjects during training and is effective in avoiding overfitting. Given that the distribution of site sizes in our sample is relatively even, it is unlikely that the successful predictions were merely driven by a specific site.
Future work should further investigate the concept of the “core” network identified in this paper, especially in the developmental context. For example, do the identified predictive features and their associated developmental trajectories change with age? As age increases, does APM become better at predicting PC2, PC3, FP network strength, and SAL network strength? It would also be interesting to study whether the identified predictive features and their predictive performance differ between healthy and clinical populations. Additionally, future work should investigate if the SST task becomes more relevant at an older age, given previous findings that inhibitory control develops at a later stage than reward processing or working memory capacity (Casey, 2014, Yip and Potenza, 2018). While it is out of the scope of the current paper, it would also be interesting to see whether a model trained using age-10 data predicts change in network strengths between age 10 and age 12, or even further into the developmental trajectory. We also recognize that one inherent limitation with using the tabulated data from ABCD is the discrepancy in the parcellations of the task fMRI data (“ASEG” and Desikan) and the resting-state fMRI data (Gordon). Future work with the right resources should verify whether similar results can be obtained with more consistent parcellations between task-based and resting-state fMRI.
5. Conclusions
Evidence presented here suggests that, using activation-based predictive modeling (APM), multi-task co-activation patterns (based on the “ASEG” and Desikan parcellation) can predict resting-state DMN connectivity (Gordon parcellation) and general cognitive ability at an individual level in a large cohort of young adolescents. Interestingly, predictive models identified at age ten can be applied to age-twelve co-activation patterns to predict age-twelve resting-state DMN connectivity, suggesting the validity of the predictive features and models even over the course of development. The identified predictive features and their associated developmental trajectories may advance our understanding of brain states and provide a new perspective on studying brain functions and their development in healthy and clinical populations.
Funding
This work was supported by National Institutes of Health grants R01AA027553, K08DA051667, and T32DA007238.
CRediT authorship contribution statement
Fengdan Ye: Conceptualization, Methodology, Software, Validation, Formal Analysis, Investigation, Data Curation, Writing – Original Draft, Writing – Review & Editing, Visualization Robert Kohler: Conceptualization, Data Curation, Writing – Review & Editing Bianca Serio: Data Curation, Writing – Review & Editing Sarah Lichenstein: Conceptualization, Data Curation, Writing – Review & Editing Sarah W Yip: Conceptualization, Methodology, Software, Validation, Formal Analysis, Investigation, Data Curation, Resources, Writing – Original Draft, Writing – Review & Editing, Supervision, Project administration.
Data statement
The data that support the findings of this study are openly available in the ABCD study release 3.0 (https://nda.nih.gov/abcd, see Supplementary materials Table S1 for package names and access dates). The code for APM is available at our GitHub repository https://github.com/fye92/abcd_fy. The list of participant IDs used in the analysis will be available upon direct request to the corresponding author and review by all authors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dcn.2022.101160.
Appendix A. Supplementary material
Supplementary material
.
Data availability
Data will be made available on request.
References
- Bissett P.G., Hagen M.P., Jones H.M., Poldrack R.A. Design issues and solutions for stop-signal data from the adolescent brain cognitive development (ABCD) study. eLife. 2021;10 doi: 10.7554/eLife.60185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork J.M., Straub L.K., Provost R.G., Neale M.C. The ABCD study of neurodevelopment: identifying neurocircuit targets for prevention and treatment of adolescent substance abuse. Curr. Treat. Options Psychiatry. 2017;4:196–209. doi: 10.1007/s40501-017-0108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton T.A.W., Tuleasca C., Wotruba D., et al. TbCAPs: a toolbox for co-activation pattern analysis. Neuroimage. 2020;211 doi: 10.1016/j.neuroimage.2020.116621. [DOI] [PubMed] [Google Scholar]
- Buckner R.L., DiNicola L.M. The brain's default network: updated anatomy, physiology and evolving insights. Nat. Rev. Neurosci. 2019;20:593–608. doi: 10.1038/s41583-019-0212-7. [DOI] [PubMed] [Google Scholar]
- Casey B.J. Beyond simple models of self-control to circuit-based accounts of adolescent behavior. Annu Rev. Psychol. 2014;66:295–315. doi: 10.1146/annurev-psych-010814-015156. [DOI] [PubMed] [Google Scholar]
- Casey B.J., Cannonier T., Conley M.I., et al. The adolescent brain cognitive development (ABCD) study: imaging acquisition across 21 sites. Dev. Cogn. Neurosci. 2018;32:43–54. doi: 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey B.J., Heller A.S., Gee D.G., Cohen A.O. Development of the emotional brain. Neurosci. Lett. 2019;693:29–34. doi: 10.1016/j.neulet.2017.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaarani B., Hahn S., Allgaier N., et al. Baseline brain function in the preadolescents of the ABCD study. Nat. Neurosci. 2021;24:1176–1186. doi: 10.1038/s41593-021-00867-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A.O., Conley, M.I., Dellarco, D.V., Casey, B.J., 2016. The impact of emotional cues on short-term and long-term memory during adolescence. Program No. 90.25 Neuroscience Meeting Planner, San Diego, CA: Society for Neuroscience.
- De Marco M., Meneghello F., Pilosio C., Rigon J., Venneri A. Up-regulation of DMN connectivity in mild cognitive impairment via network-based cognitive training. Curr. Alzheimer Res. 2018;15:578–589. doi: 10.2174/1567205015666171212103323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R.S., Segonne F., Fischl B., et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Fair D.A., Dosenbach N.U., Church J.A., et al. Development of distinct control networks through segregation and integration. Proc. Natl. Acad. Sci. USA. 2007;104:13507–13512. doi: 10.1073/pnas.0705843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair D.A., Cohen A.L., Dosenbach N.U., et al. The maturing architecture of the brain's default network. Proc. Natl. Acad. Sci. USA. 2008;105:4028–4032. doi: 10.1073/pnas.0800376105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F., Liao X., Lei T., et al. Development of the default-mode network during childhood and adolescence: a longitudinal resting-state fMRI study. Neuroimage. 2021;226 doi: 10.1016/j.neuroimage.2020.117581. [DOI] [PubMed] [Google Scholar]
- Finn E.S., Shen X., Scheinost D., et al. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci. 2015;18:1664–1671. doi: 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Salat D.H., Busa E., et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fox P.T., Lancaster J.L. Opinion: Mapping context and content: the BrainMap model. Nat. Rev. Neurosci. 2002;3:319–321. doi: 10.1038/nrn789. [DOI] [PubMed] [Google Scholar]
- Gordon E.M., Laumann T.O., Adeyemo B., Huckins J.F., Kelley W.M., Petersen S.E. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex. 2016;26:288–303. doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson D.S., Fair D.A. Development of large-scale functional networks from birth to adulthood: a guide to the neuroimaging literature. Neuroimage. 2017;160:15–31. doi: 10.1016/j.neuroimage.2017.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler D.J., Jr., Hatton S., Cornejo M.D., et al. Image processing and analysis methods for the adolescent brain cognitive development study. Neuroimage. 2019;202 doi: 10.1016/j.neuroimage.2019.116091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilford E.J., Garrett E., Blakemore S.J. The development of social cognition in adolescence: an integrated perspective. Neurosci. Biobehav Rev. 2016;70:106–120. doi: 10.1016/j.neubiorev.2016.08.016. [DOI] [PubMed] [Google Scholar]
- Klein A., Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front. Neurosci. 2012;6:171. doi: 10.3389/fnins.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson B., Westdorp A., Kaiser E., Hommer D. FMRI visualization of brain activity during a monetary incentive delay task. Neuroimage. 2000;12:20–27. doi: 10.1006/nimg.2000.0593. [DOI] [PubMed] [Google Scholar]
- Koski L., Paus T. Functional connectivity of the anterior cingulate cortex within the human frontal lobe: a brain-mapping meta-analysis. Exp. Brain Res. 2000;133:55–65. doi: 10.1007/s002210000400. [DOI] [PubMed] [Google Scholar]
- Laird A.R., Lancaster J.L., Fox P.T. BrainMap: the social evolution of a human brain mapping database. Neuroinformatics. 2005;3:65–78. doi: 10.1385/ni:3:1:065. [DOI] [PubMed] [Google Scholar]
- Laird A.R., Eickhoff S.B., Rottschy C., Bzdok D., Ray K.L., Fox P.T. Networks of task co-activations. Neuroimage. 2013;80:505–514. doi: 10.1016/j.neuroimage.2013.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm C., Benson B.E., Guyer A.E., et al. Longitudinal study of striatal activation to reward and loss anticipation from mid-adolescence into late adolescence/early adulthood. Brain Cogn. 2014;89:51–60. doi: 10.1016/j.bandc.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen B., Luna B. Adolescence as a neurobiological critical period for the development of higher-order cognition. Neurosci. Biobehav Rev. 2018;94:179–195. doi: 10.1016/j.neubiorev.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Dahmani L., Wang D., et al. Co-activation patterns across multiple tasks reveal robust anti-correlated functional networks. Neuroimage. 2021;227 doi: 10.1016/j.neuroimage.2020.117680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichenstein S.D., Scheinost D., Potenza M.N., Carroll K.M., Yip S.W. Dissociable neural substrates of opioid and cocaine use identified via connectome-based modelling. Mol. Psychiatry. 2021 doi: 10.1038/s41380-019-0586-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan G.D. Academic Press; San Diego, CA, US: 1994. On the Ability to Inhibit Thought and Action: A Users’ Guide to the Stop Signal Paradigm Inhibitory Processes in Attention, Memory, and Language; pp. 189–239. [Google Scholar]
- Luciana M., Bjork J.M., Nagel B.J., et al. Adolescent neurocognitive development and impacts of substance use: overview of the adolescent brain cognitive development (ABCD) baseline neurocognition battery. Dev. Cogn. Neurosci. 2018;32:67–79. doi: 10.1016/j.dcn.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Dosenbach N.U.F. The frontoparietal network: function, electrophysiology, and importance of individual precision mapping. Dialog-. Clin. Neurosci. 2018;20:133–140. doi: 10.31887/DCNS.2018.20.2/smarek. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S., Tervo-Clemmens B., Calabro F.J., et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022;603:654–660. doi: 10.1038/s41586-022-04492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meruelo A.D., Castro N., Cota C.I., Tapert S.F. Cannabis and alcohol use, and the developing brain. Behav. Brain Res. 2017;325:44–50. doi: 10.1016/j.bbr.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan S.E., White S.R., Bullmore E.T., Vertes P.E. A network neuroscience approach to typical and atypical brain development. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. 2018;3:754–766. doi: 10.1016/j.bpsc.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens M.M., Potter A., Hyatt C.S., et al. Recalibrating expectations about effect size: a multi-method survey of effect sizes in the ABCD study. PLOS One. 2021;16 doi: 10.1371/journal.pone.0257535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A., Geier C.F., Ordaz S.J., Teslovich T., Luna B. Developmental changes in brain function underlying the influence of reward processing on inhibitory control. Dev. Cogn. Neurosci. 2011;1:517–529. doi: 10.1016/j.dcn.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelland M., Orban P., Dansereau C., Lepore F., Bellec P., Collignon O. State-dependent modulation of functional connectivity in early blind individuals. Neuroimage. 2017;147:532–541. doi: 10.1016/j.neuroimage.2016.12.053. [DOI] [PubMed] [Google Scholar]
- Postuma R.B., Dagher A. Basal ganglia functional connectivity based on a meta-analysis of 126 positron emission tomography and functional magnetic resonance imaging publications. Cereb. Cortex. 2006;16:1508–1521. doi: 10.1093/cercor/bhj088. [DOI] [PubMed] [Google Scholar]
- Power J.D., Fair D.A., Schlaggar B.L., Petersen S.E. The development of human functional brain networks. Neuron. 2010;67:735–748. doi: 10.1016/j.neuron.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Barnes K.A., Snyder A.Z., Schlaggar B.L., Petersen S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapuano K.M., Rosenberg M.D., Maza M.T., et al. Behavioral and brain signatures of substance use vulnerability in childhood. Dev. Cogn. Neurosci. 2020;46 doi: 10.1016/j.dcn.2020.100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinost D., Finn E.S., Tokoglu F., et al. Sex differences in normal age trajectories of functional brain networks. Hum. Brain Mapp. 2015;36:1524–1535. doi: 10.1002/hbm.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley W.W. The salience network: a neural system for perceiving and responding to homeostatic demands. J. Neurosci. 2019;39:9878–9882. doi: 10.1523/JNEUROSCI.1138-17.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Finn E.S., Scheinost D., et al. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc. 2017;12:506–518. doi: 10.1038/nprot.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman L.E., Rudie J.D., Pfeifer J.H., Masten C.L., McNealy K., Dapretto M. Development of the default mode and central executive networks across early adolescence: a longitudinal study. Dev. Cogn. Neurosci. 2014;10:148–159. doi: 10.1016/j.dcn.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Fox P.T., Miller K.L., et al. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl. Acad. Sci. USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder W., Uddin L.Q., Nomi J.S. Dynamic functional connectivity profile of the salience network across the life span. Hum. Brain Mapp. 2021;42:4740–4749. doi: 10.1002/hbm.25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K., Uddin L.Q., Prater K., Amin H., Greicius M.D., Menon V. Development of functional and structural connectivity within the default mode network in young children. Neuroimage. 2010;52:290–301. doi: 10.1016/j.neuroimage.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson W.K., Barch D.M., Bjork J.M., et al. The structure of cognition in 9 and 10 year-old children and associations with problem behaviors: Findings from the ABCD study's baseline neurocognitive battery. Dev. Cogn. Neurosci. 2019;36 doi: 10.1016/j.dcn.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R., Fox P.T., Paus T. Functional coactivation map of the human brain. Cereb. Cortex. 2008;18:2553–2559. doi: 10.1093/cercor/bhn014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow N.D., Koob G.F., Croyle R.T., et al. The conception of the ABCD study: from substance use to a broad NIH collaboration. Dev. Cogn. Neurosci. 2018;32:4–7. doi: 10.1016/j.dcn.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendelken C., Ferrer E., Whitaker K.J., Bunge S.A. Fronto-parietal network reconfiguration supports the development of reasoning ability. Cereb. Cortex. 2016;26:2178–2190. doi: 10.1093/cercor/bhv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S.W., Potenza M.N. Application of research domain criteria to childhood and adolescent impulsive and addictive disorders: implications for treatment. Clin. Psychol. Rev. 2018;64:41–56. doi: 10.1016/j.cpr.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S.W., Kiluk B., Scheinost D. Toward addiction prediction: an overview of cross-validated predictive modeling findings and considerations for future neuroimaging research. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. 2020;5:748–758. doi: 10.1016/j.bpsc.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The data that support the findings of this study are openly available in the ABCD study release 3.0 (https://nda.nih.gov/abcd, see Supplementary materials Table S1 for package names and access dates). The code for APM is available at our GitHub repository https://github.com/fye92/abcd_fy. The list of participant IDs used in the analysis will be available upon direct request to the corresponding author and review by all authors.
Data will be made available on request.